Abstract
A typical DYT1 dystonia is caused by a heterozygous GAG deletion (c.907–909) in the TOR1A gene (ΔE, p.Glu303del) and the pathogenesis is not clear. In this study, human induced pluripotent stem cell (hiPSC) lines carrying the heterozygous or homozygous GAG deletion in TOR1A gene were generated by genetic modification of a healthy hiPSC line (WTC11, UCSFi001-A). These hiPSC lines showed the normal stem cell morphology and karyotype, expressed the same pluripotency markers as their parental line, and had the capacity to differentiate into three germ layers, providing a valuable resource in determining the pathogenesis of human DYT1 dystonia.
1. Resource Table
| Unique stem cell lines identifier | ULLi002-A-49 |
| ULLi002-A-51 | |
| Institution | University of Louisiana at Lafayette, LA USA |
| Contact information of the reported cell line distributor | Baojin Ding (Baojin.Ding@Louisiana.edu) |
| Type of cell lines | iPSC |
| Origin | Human |
| Additional origin info (applicable for human ESC or iPSC) | Age: 30 YR Sex: Male Ethnicity: Asian |
| Cell Source | Skin fibroblasts. |
| Method of reprogramming | N/A |
| Clonality | Clonal |
| Evidence of the reprogramming transgene loss (including genomic copy if applicable) | RT/q-PCR |
| Cell culture system used | Serum-free and feeder-free medium |
| Type of Genetic Modification | Induced mutation |
| Associated disease | DYT1 dystonia |
| Gene/locus | TOR1A c.907_909delGAG (p.Glu303del)/9q34.11 |
| Multiline rationale | Isogenic clones carrying heterozygous or homozygous of the same gene mutation. |
| Method of modification/site-specific nuclease used | CRISPR/Cas9 |
| Site-specific nuclease (SSN) delivery method | Electroporated with a 4D-Nucleofector (Lonza) using CA-137 program. |
| All genetic material introduced into the cells | gRNA vector MLM3636 (Addgene #43860) Cas9 vector p3s-Cas9HC (Addgene #43945) |
| Unique stem cell lines identifier | ULLi002-A-49 ULLi002-A-51 |
| Analysis of the nuclease-targeted allele status | Sequencing of the targeted allele |
| Method of the off-target nuclease activity surveillance | Targeted PCR/sequencing |
| Name of transgene | N/A |
| Eukaryotic selective agent resistance (including inducible/gene expressing cell-specific) | N/A |
| Inducible/constitutive system details | N/A |
| Date archived/stock date | 4/29/2021 |
| Cell line repository/bank |
https://hpscreg.eu/cell-line/ULLi002-A-49
https://hpscreg.eu/cell-line/ULLi002-A-51 |
| Ethical/GMO work approvals | Genetic modification was performed at Genome Engineering and iPSC Center (GEiC) at Washington University in St. Louis. |
| Addgene/public access repository recombinant DNA sources’ disclaimers (if applicable) | N/A |
2. Resource utility
Although rodent models of DYT1 dystonia provide insights into disease mechanisms, significant species-dependent differences exist because animals with the identical heterozygous mutation fail to show pathology. These hiPSC lines will provide a valuable resource to develop human cellular systems in modeling DYT1 dystonia. Table 1
Table 1.
Characterization and validation.
| Classification | Test | Result | Data |
|---|---|---|---|
| Morphology | Photography | Typical primed pluripotent human stem cell morphology. | Fig. 1 Panel A |
| Pluripotency status evidence for the described cell line | Qualitative analysis | Immunocytochemistry showed expression of pluripotency markers: OCT4, SOX2, NANOG, SSEA4. | Fig. 1 Panel D |
| Quantitative analysis | Compared to DAPI, % of positive cell (ULLi002-A-49, ULLi002-A-51) OCT4: 98%, 98%; SOX2: 99%, 98%; NANOG: 97%, 95%; SSEA-4: 97%, 98%. RT-PCR showed highly express OCT4, SOX2, NANOG, KLF4. | Fig. 1 Panel D and F | |
| Karyotype | Karyotype (G-banding) and resolution | 46, XY, Resolution 400 | Fig. 1 Panel C |
| Genotyping for the desired genomic alteration/allelic status of the gene of interest | PCR across the edited site and deep sequencing analysis | Heterozygous or homozygous GAG deletion (c.907–909) in TOR1A gene. | Fig. 1 Panel B and Supplementary Fig. S1A and B. |
| Transgene-specific PCR | N/A | N/A | |
| Verification of the absence of random plasmid integration events | PCR/Southern | Off Target Analysis of gRNA showed 100% minus a weighted sum of off-target hit-scores in the target genome. | N/A |
| Parental and modified cell line genetic identity evidence | STR analysis, microsatellite PCR (mPCR) or specific (mutant) allele seq | STR analysis of 18 loci, all matched. | Supplementary Fig. S1C |
| Mutagenesis/genetic modification outcome analysis | Sequencing (genomic DNA PCR or RT-PCR product) | The sequencing results of genomic DNA all matched with the parental line. | Fig. 1 Panel B and Supplementary Fig. S1A and B |
| PCR-based analyses | The sequencing results PCR products all matched with the parental line. | Fig. 1 Panel B and Supplementary Fig. S1A and B | |
| Southern Blot or WGS; western blotting (for knock-outs, KOs) | N/A | N/A | |
| Off-target nuclease analysis | PCR across top 5/10 predicted top likely off-target sites, whole genome/exome sequencing | N/A | N/A |
| Specific pathogen-free status | Mycoplasma | Tested by MycoAlert PLUS kit: Negative | Fig. S1D |
| Multilineage differentiation potential | Embryoid body formation, RT-PCR | Upregulation of trilineage markers PAX6, OTX1 (ectoderm), DCN, IGF2, GATA2 (mesoderm), and SOX7, SOX17 (endoderm). | Fig. 1 Panel G |
| Donor screening (OPTIONAL) | HIV 1 + 2 Hepatitis B, Hepatitis C | N/A | N/A |
| Genotype additional info (OPTIONAL) | Blood group genotyping | N/A | N/A |
| HLA tissue typing | N/A | N/A |
3. Resource details
Dystonia is a common movement disorder characterized by sustained or intermittent muscle contractions causing abnormal movements and/or postures. Because the clinical characteristics and underlying causes of dystonia are very heterogeneous, the pathological mechanisms of dystonia remain largely unknown (Ding, 2022). The diagnosis and etiological definition of this disorder remain challenging. The childhood-onset DYT1 dystonia represents the most frequent and severe form of dystonia, providing an excellent example to understand the pathogenesis of this disease. The typical DYT1 dystonia is caused by a heterozygous GAG deletion (c.907–909) in the TOR1A gene (ΔE, p. Glu303del) (Ozelius et al., 1997). Interestingly, mice with the identical Tor1a mutation as a heterozygote failed to show any pathological phenotypes (Goodchild et al., 2005), suggesting that the significant species-dependent differences exist between DYT1 mouse models and human patients. However, the limited access to patient neurons and the lack of in vitro human neuronal systems greatly impede the progress of research in dystonia. Generation of hiPSC lines containing disease-causing mutations and their isogenic controls has been shown to be a powerful tool in modeling human neurological diseases (Sepehrimanesh and Ding, 2020; Ding et al., 2021). In this study, we genetically modified a well-characterized hiPSC line (WTC11, UCSFi001-A) with CRISPR/Cas9 method and obtained two iPSC clones carrying the heterozygous (ULLi002-A-49, or USCFi001-A-49) and homozygous (ULLi002-A-51, or USCFi001-A-51) GAG deletion in the TOR1A gene, respectively. These hiPSC lines and their isogenic controls provide a valuable resource in modeling DYT1 dystonia.
Both lines showed a typical pluripotent stem cell morphology with a high nucleus/cytoplasm ratio (Fig. 1A). Sanger DNA sequencing of polymerase chain reaction (PCR) products (Fig. 1B) and deep sequencing analysis (Supp. Fig. S1A and S1B) confirmed that these iPSC lines contain heterozygous (ULLi002-A-49) and homozygous (ULLi002-A-51) GAG deletion (c.907–909) in the TOR1A gene. Chromosomes from iPSCs were harvested and analyzed using the GTW banding method. Of 20 metaphase cells examined of each clone, all cells are characteristic of a chromosomally normal male karyotype, 46, XY (Fig. 1C). Short tandem repeat (STR) analysis at 18 loci indicated that ULLi002-A-49 and ULLi002-A-51 clones completely matched the parental hiPSC (UCSFi001-A) identity (Supp. Fig. S1C). Immunostaining indicated that these iPSCs highly expressed pluripotency markers of OCT4, NANOG, SOX2, and SSEA4 (Fig. 1D). Quantitative RT-PCR analysis demonstrated that the pluripotency markers of OCT4, SOX2, NANOG, and KLF4 were expressed at similar levels as their parental line (UCSFi001A) (Fig. 1F). Following spontaneous differentiation, embryoid bodies (EBs) (Fig. 1E) derived from ULLi002-A-49 and ULLi002-A-51 displayed dramatic upregulation of markers of the ectoderm (PAX6, OTX1), mesoderm (DCN, IGF2, GATA2) and endoderm (SOX7, SOX17) lineages. The expression levels of these trilineage markers were consistent with their parental line UCSFi001-A and much higher than undifferentiated iPSCs (Fig. 1G). PCR screening demonstrated that ULLi002-A-49 and ULLi002-A-51 were negative for mycoplasma (Fig. S1 D).
Fig. 1.
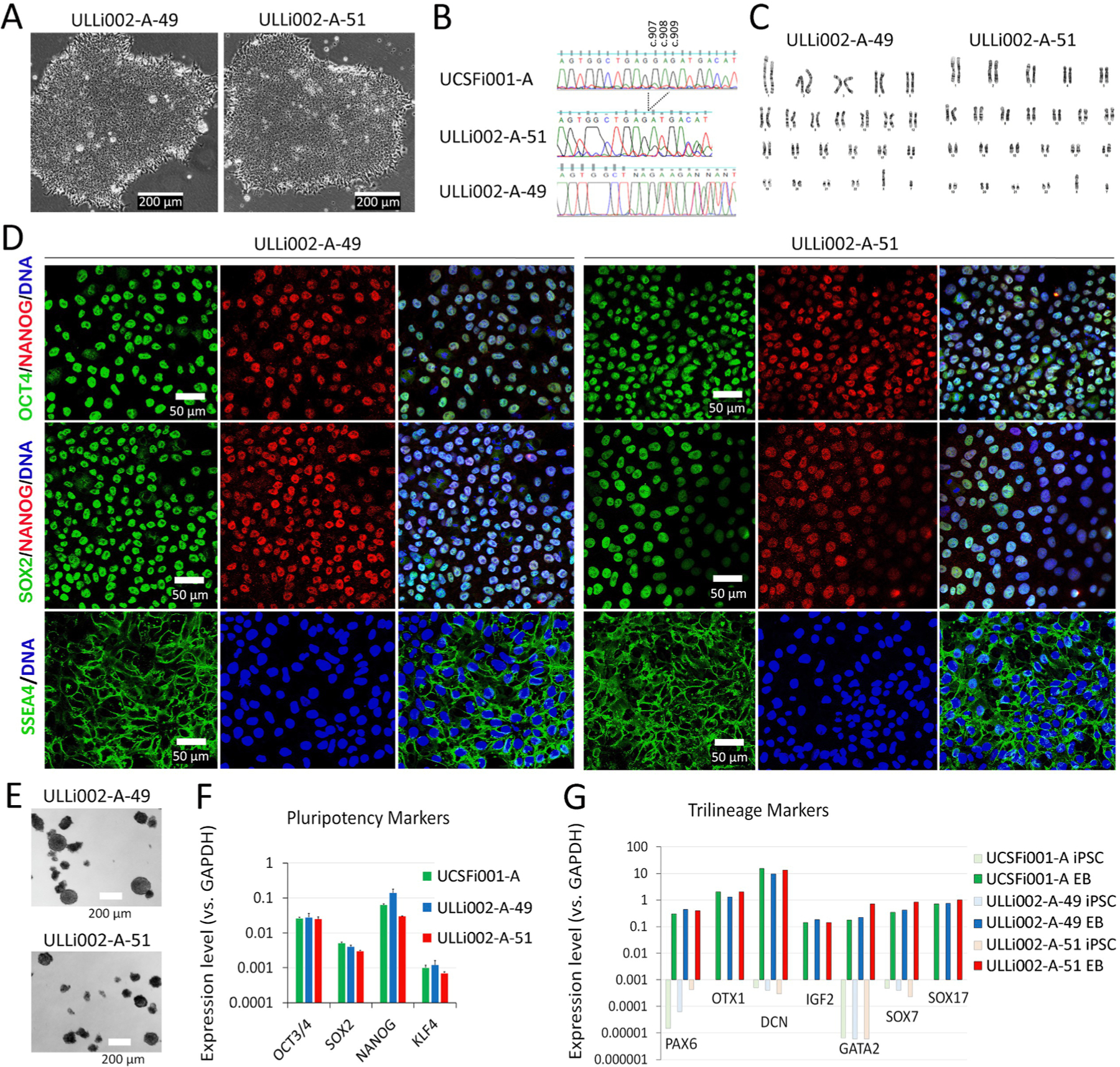
Characterization of ULLi002-A49 and ULLi002-A51 iPSC lines.
4. Materials and methods
4.1. Generate and culture TOR1A mutant iPSCs
Human TOR1A mutant clones were generated from the WTC11 iPSC line (Gladstone institute, UCSFi001-A) by the Genome Engineering and iPSC center (GEiC) at Washington University in St. Louis. Briefly, approximately 1×106 single cells were resuspended in P3 primary buffer (Lonza) with gRNA/Cas9 ribonucleoprotein (RNP) complex (200 pmol synthetic gRNA and 80 pmol SpCas9 protein) and hTOR1A mutant single-stranded oligo donor (ssODN) (Table 2). Subsequently, cells were electroporated with a 4D-Nucleofector (Lonza) using CA-137 program. Following nucleofection, the editing efficiency was confirmed by targeted deep sequencing using primer sets specific to target regions, and then the pool was single-cell sorted. Single cell clones were screened with targeted deep sequencing analysis. All iPSCs were cultured with mTeSR Plus (STEMCELL Technology) on matrigel-coated plates at 37 °C in a humidified, 5% CO2 incubator and passage at a 1:6 ratio using gentle cell dissociation reagent (Versene, Gibco).
Table 2.
Reagents details.
| Antibodies used for immunocytochemistry | |||
|---|---|---|---|
| Antibody | Dilution | Company Cat # and RRID | |
| Pluripotency Markers | Mouse anti-OCT4 | 1:200 | Santa Cruz Cat# sc-5279, RRID: AB_628051 |
| Mouse anti-SOX2 | 1:200 | Santa Cruz Biotechnology Cat# sc-365823, RRID:AB_10842165 | |
| Mouse anti-SSEA4 | 1:200 | Abcam Cat# ab16287, RRID:AB_778073 | |
| Rabbit anti-Nanog | 1:100 | Abcam Cat# ab21624, RRID:AB_446437 | |
| Secondary antibodies | Donkey anti-Mouse IgG (H + L), Alexa Fluor 488 | 1:500 | Jackson ImmunoResearch Labs Cat# 715-545-150, RRID:AB_2340846 |
| Donkey Anti-Rabbit IgG (H + L), Alexa Fluor 594 | 1:500 | Jackson ImmunoResearch Labs Cat# 711-585-152, RRID:AB_2340621 | |
| Nuclear stain | Hoechst33342 | 1 μg/mL | Invitrogen Cat # H3570. RRID: NOT FOUND |
| Site-specific nuclease | |||
| Nuclease information | Cas9 | Cas9 vector p3s-Cas9HC (Addgene #43945) | |
| Delivery method | electroporation | 4D-Nucleofector (Lonza, Cat # AAF-1002B) | |
| Selection/enrichment strategy | sorted into 96-well plates with one cell per well | Single cell clones were screened and expanded | |
| Primers and Oligonucleotides used in this study | |||
| Target | Forward/Reverse primer (5’−3’) | ||
| Pluripotency marker | OCT 3/4 | CGAGAGGATTTTGAGGCTGC/CGAGGAGTACAGTGCAGTGA | |
| Pluripotency marker | SOX 2 | AGGATAAGTACACGCTGCCC/TTCATGTGCGCGTAACTGTC | |
| Pluripotency marker | NANOG | TGTCTTCTGCTGAGATGCCT/CAGAAGTGGGTTGTTTGCCT | |
| Pluripotency marker | KLF4 | TCTCCAATTCGCTGACCCAT/CGGATCGGATAGGTGAAGCT | |
| Differentiation marker | PAX6 | GGGCGGAGTTATGATACCTACA/ATATCAGGTTCACTTCCGGGAA | |
| Differentiation marker | OTX1 | TACGCCCTCCTCTTCCTACT/GCATGTGGGTGGTGATGATG | |
| Differentiation marker | DCN | CTGAAGAACCTTCACGCATTGA/GGCAATTCCTTCAGCTGATTCT | |
| Differentiation marker | IGF2 | CAATATGACACCTGGAAGCAGT/GTAGAGCAATCAGGGGACGG | |
| Differentiation marker | GATA2 | ACCTGTTGTGCAAATTGTCAGA/ATCCCTTCCTTCTTCATGGTCA | |
| Differentiation marker | SOX7 | ACTCCACTCCAACCTCCAAG/TTCATTGCGATCCATGTCCC | |
| Differentiation marker | SOX17 | ATCGGGGACATGAAGGTGAA/TCCTTAGCCCACACCATGAA | |
| House-Keeping Genes | GAPDH | CAAATTCCATGGCACCGTCA/GGACTCCACGACGTACTCAG | |
| Genotyping-PCR | TOR1A | ACAGCAGCTTAATTGACCGGA/ATCATGAGCCCTGCGATGAG | |
| Sequencing | TOR1A | GTGTATCCGAGTGGAAATGC | |
| hTOR1A gRNA (IDT) | TOR1A | TGAAGACATTGTAAGCAGAG | |
| hTOR1A ssODN (IDT) | TOR1A | AATGTGTATCCGAGTGGAAATGCAGTCCCGAGGCTATGAAATTGATGAAGACATTGTAAGTAGAGTGGCTGAGATGACATTTTTCCCCAAAGAGGAGAGAGTTTTCTCAGATAAAGGCTGCA | |
4.2. Embryoid bodies (EB) formation
hiPSCs were dissociated with Versene and transferred to low attachment 10-cm petri dishes in KOSR medium (DMEM/F12 medium containing 20% KnockOut Serum Replacement, 1% GlutaMax, 1% non-essential amino acids, 50 μM β-mercaptoethanol, and 1% penicillin–streptomycin) supplemented with 10 μM Y-27632. The medium was changed every other day and EBs gradually formed. After 7 days of suspension culture, EBs were digested with 0.25% Trypsin and cultured on gelatin coated plates with KOSR medium for another 7 days. The total RNAs were extracted for RT-PCR analysis.
4.3. Immunostaining and confocal microscopy
Briefly, cultured iPSCs were fixed with 4% paraformaldehyde (PFA) in PBS for 15 min at room temperature followed by incubation in blocking buffer (3% bovine serum albumin in PBS) with (for nuclear markers) or without (for cell surface marker SSEA4) 0.2% Triton X-100 for 1 h. Cells were then incubated with primary antibodies (Table 2) in blocking buffer at 4 °C overnight and then followed by washing and incubation with fluorophore-conjugated corresponding secondary antibodies at room temperature for 2 hrs. Nuclei were stained with Hoechst 33342 (Invitrogen). Images were obtained with a Leica SP5 confocal microscope.
4.4. Quantitative PCR analysis
iPSCs and EBs were collected from cultured plates and lysed in TRIzol (Invitrogen). Total RNAs were isolated by the phenol/chloroform extracting method, and then were reverse-transcribed into cDNAs with the SuperScript™ III Reverse Transcriptase (Invitrogen). Quantitative PCR analysis was performed using SYBR Green PCR Master Mix (Applied Biosystems) and run on StepOne qPCR machine (Applied Biosystems). The gene expression data were analyzed using the ΔΔCT method and the values were normalized to the expression of the GAPDH housekeeping gene (Fig. 1F and G). Primers used in this study were listed in Table 2.
4.5. Karyotyping
Chromosomes from iPSC clones were analyzed using the GTW banding method at GEiC at Washington University in St. Louis.
4.6. STR analysis
Short tandem repeat (STR) analysis of 18 loci (Fig. S1C) were performed at GEiC at Washington University in St. Louis.
4.7. Mycoplasma test
Mycoplasma test (Fig. S1D) was performed by MycoAlert PLUS kit (Lonza) at GEiC at Washington University in St. Louis.
Supplementary Material
Acknowledgements
We thank Genome Engineering & iPSC Center (GEiC) at Washington University in St. Louis for their excellent services, and members of the Ding laboratory (Dr. Masood Sepehrimanesh, Mr. Jacob Stagray, Mr. Casey Coutee, and Mr. Md Abir Hosain) for help and discussion. This work was supported by National Institute of Neurological Diseases and Stroke (No. NIH/NINDS NS112910 to B.D.) and Department of Defense (DoD) Peer Reviewed Medical Research Program (PRMRP) Discovery Award (No. W81XWH2010186 to B.D.).
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scr.2021.102536.
References
- Ding B, 2022. Novel insights into the pathogenesis of DYT1 dystonia from induced patient-derived neurons. Neural Regen Res 17 (3), 561. 10.4103/1673-5374.320978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B, Tang Y.u., Ma S, Akter M, Liu M-L, Zang T, Zhang C-L, 2021. Disease Modeling with Human Neurons Reveals LMNB1 Dysregulation Underlying DYT1 Dystonia. J Neurosci 41 (9), 2024–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodchild RE, Kim CE, Dauer WT, 2005. Loss of the dystonia-associated protein torsinA selectively disrupts the neuronal nuclear envelope. Neuron 48 (6), 923–932. [DOI] [PubMed] [Google Scholar]
- Ozelius LJ, Hewett JW, Page CE, Bressman SB, Kramer PL, Shalish C, de Leon D, Brin MF, Raymond D, Corey DP, Fahn S, Risch NJ, Buckler AJ, Gusella JF, Breakefield XO, 1997. The early-onset torsion dystonia gene (DYT1) encodes an ATP-binding protein. Nat Genet 17 (1), 40–48. [DOI] [PubMed] [Google Scholar]
- Sepehrimanesh M, Ding B, 2020. Generation and Optimization of Highly Pure Motor Neurons from Human Induced Pluripotent Stem Cells via Lentiviral Delivery of Transcription Factors. Am J Physiol Cell Physiol 319 (4), C771–C780. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


