Abstract
Heterozygous gain-of-function (GOF) mutations in STAT1 in patients with chronic mucocutaneous candidiasis (CMC) and hypothyroidism were discovered in 2011. CMC is the recurrent or persistent mucocutaneous infection by Candida fungi, and hypothyroidism results from autoimmune thyroiditis. Patients with these diseases develop other infectious diseases, including viral, bacterial, and fungal diseases, and other autoimmune manifestations, including enterocolitis, immune cytopenia, endocrinopathies, and systemic lupus erythematosus. STAT1-GOF mutations are highly penetrant with a median age at onset of 1 year and often underlie an autosomal dominant trait. As many as 105 mutations at 72 residues, including 65 recurrent mutations, have already been reported in more than 400 patients worldwide. The GOF mechanism involves impaired dephosphorylation of STAT1 in the nucleus. Patient cells show enhanced STAT1-dependent responses to type I and II interferons (IFNs) and IL-27. This impairs Th17 cell development, which accounts for CMC. The pathogenesis of autoimmunity likely involves enhanced type I IFN responses, as in other type I interferonopathies. The pathogenesis of other infections, especially those caused by intramacrophagic bacteria and fungi, which are otherwise seen in patients with diminished type II IFN immunity, has remained mysterious. The cumulative survival rates of patients with and without severe disease (invasive infection, cancer, and/or symptomatic aneurysm) at 60 years of age are 31% and 87%, respectively. Severe autoimmunity also worsens the prognosis. The treatment of patients with STAT1-GOF mutations who suffer from severe infectious and autoimmune manifestations relies on hematopoietic stem cell transplantation and/or oral JAK inhibitors.
Keywords: STAT1, chronic mucocutaneous candidiasis, Mendelian susceptibility to mycobacterial diseases, gain-of-function, virus
Introduction
Human signal transducer and activator of transcription 1 (STAT1), one of seven members of the STAT family, is a latent cytoplasmic transcription factor that mediates cell signaling in response to multiple stimuli, including type I, II, and III interferons (IFNs) and interleukin (IL)-27 (1–8). When these ligands bind the two chains of their receptors, Janus kinases (JAKs), JAK1 and JAK2, and/or TYK2 that are constitutively associated with the receptor chains are brought together and activated. These tyrosine-protein kinases phosphorylate the receptors, which recruit STAT1, which is then phosphorylated at tyrosine 701 (Y701) by JAKs. Phosphorylated STAT1 can form a homodimer known as gamma-interferon activation factor (GAF), after stimulation by type II IFN and IL-27 and, to a lesser extent, type I and III IFNs. GAF translocates to the nucleus and binds gamma-interferon-activated sites (GAS) to up- or downregulate IFN-stimulated and IFN-regulated genes (ISGs and IRGs, respectively) (9). Phosphorylated STAT1 also forms a heterotrimer known as interferon-stimulated gene factor 3 (ISGF3) with STAT2 and IRF9 after stimulation by type I or III IFNs. ISGF3 then binds the interferon-stimulated response element (ISRE) and induces or regulates gene transcription (9).
In humans, STAT1 plays a nonredundant role in type I, II, and III IFN and IL-27 signaling (3, 7, 8, 10, 11). Type II IFN-induced GAF-mediated signaling is essential to eliminate intramacrophagic pathogens such as mycobacteria, whereas type I IFN-induced ISRE-mediated signaling plays a pivotal role in host immunity against viruses (12). Inborn errors of human STAT1 with loss-of-function (LOF) mutations cause three types of primary immunodeficiencies (PIDs): i) autosomal recessive (AR) complete STAT1 deficiency (13–16), ii) AR partial STAT1 deficiency (17–19), and iii) autosomal dominant (AD) STAT1 deficiency (20–27). Patients with AR complete STAT1 deficiency have biallelic complete LOF mutations of STAT1. These patients develop life-threatening viral infections, especially herpesvirus and mycobacterial infections, reflecting the lack of STAT1-mediated type I, II, and III IFN and IL-27 signaling (13–17). Hematopoietic stem cell transplantation can resolve these life-threatening conditions (12, 16). AR partial STAT1 deficiency, caused by biallelic hypomorphic STAT1 mutations, is a milder form of disease, as patients with this disorder suffer from mild viral and mycobacterial disease (17–19). The clinical penetrance of AR STAT1 deficiency in patients is complete (14–19). In contrast, patients with AD STAT1 deficiency develop Mendelian susceptibility to mycobacterial diseases (MSMD), a PID characterized by selective predisposition to mycobacteria in the absence of other prominent immunodeficiencies (20–28). These patients carry heterozygous STAT1-LOF mutations for both the type I and II IFN pathways; however, paradoxically, heterozygosity selectively impairs type II IFN-induced GAF-mediated signaling without disturbing type I IFN-induced ISGF3-mediated signaling (20). The clinical penetrance of STAT1 mutations in AD STAT1 deficiency is incomplete (21–23, 27).
STAT1 gain-of-function mutations as the genetic etiology of syndromic CMC
Chronic mucocutaneous candidiasis (CMC) is characterized by recurrent or persistent infection of the nails, skin, and oral and genital mucosa by Candida species. CMC is among the broad infectious manifestations of severe T cell immunodeficiencies (29–32). Most patients with AD hyper-IgE syndrome (mutations in STAT3 but not IL6ST) (33–36) and most patients with AR RORγT (37) or ZNF341 (38, 39) deficiency present CMC with fewer other infections than those presented by patients with severe T cell immunodeficiencies. All of these conditions exhibit a scarcity of IL-17-producing T cells and are categorized as syndromic CMC (SCMC). AR AIRE deficiency also causes SCMC by the production of neutralizing autoantibodies against IL-17A and/or IL-17F (40, 41).
The first genetic etiologies of the isolated form of CMC (ICMC), in which CMC is the major clinical manifestation in otherwise healthy individuals, were deciphered with the identification of AR IL-17RA and AD IL-17F deficiencies (42). This discovery was compatible with previous mouse studies showing the essential roles of IL-17 cytokines in the host defense against mucosal immunity to C. albicans (43–46). The apparent lack of other infections, in addition to mucocutaneous bacterial infections in patients with AR IL-17RA deficiency, was surprising and indicated the high redundancy of this pathway in humans (47, 48). The identification of these monogenic diseases clearly indicated that inborn errors of human IL-17 immunity underlie ICMC. However, ICMC due to inborn errors of IL-17 cytokines or receptors is relatively rare, with 5 (IL17F), 23 (IL17RA), and 3 (IL17RC) patients reported to date (42, 48–50). Mutations in TRAF3IP2, which encodes ACT1, downstream of the IL-17 receptors have been found in only 7 patients (31, 51–53). In addition, very recently, a heterozygous loss-of-expression and LOF mutation in MAPK8, which encodes JNK1, was identified in a multiplex family with CMC and connective tissue disorder (54). This discovery indicates that the human JNK1-dependent MAPK signaling pathway is essential for IL-17A- and IL-17F-dependent mucocutaneous immunity to Candida.
In 2011, whole-exome sequencing led to the identification of germline STAT1 mutations as a genetic etiology of CMC in patients with hypothyroidism (55, 56). These STAT1 mutations were shown to be GOF mutations due to hyperphosphorylation of STAT1 at Y701 in response to stimulation with type I and II IFNs and IL-27 (55). Follow-up studies found that STAT1-GOF mutations can account for more than half of cases of inherited CMC, which in most cases is syndromic (57–60). Indeed, STAT1-GOF mutations were shown to cause broader than expected infectious and noninfectious phenotypes in addition to CMC (57, 58). Therefore, STAT1-GOF mutations are currently thought to cause SCMC.
Genetics of STAT1-GOF mutant alleles
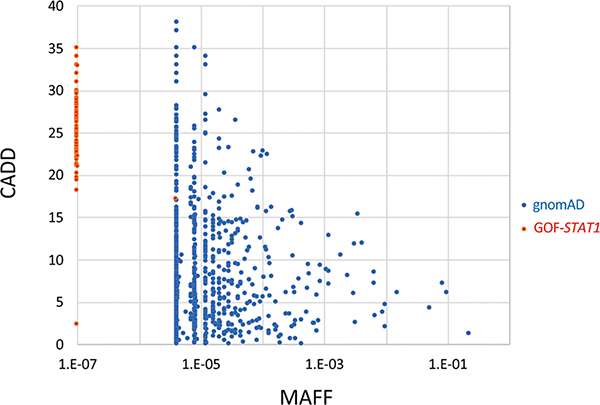
As many as 105 STAT1-GOF mutations have been reported (Fig. 1A, Table 1) (55–137). In contrast, fewer STAT1-LOF mutations that cause AR complete STAT1 deficiency (4 mutations) (14–16), AR partial STAT1 deficiency (4 mutations) (17–19), and AD STAT1 deficiency (10 mutations) (20–26) have been reported thus far (Fig. 1B, Table 2–4). STAT1-GOF mutations were originally identified in the coiled-coil domain (CCD) of STAT1 (55, 56). They were also later found in other domains of STAT1 (58, 103, 105). The majority (87.6%) of these GOF mutations are in the CCD and DNA-binding domain (DBD) of STAT1; GOF mutations in the CCD alone account for half (52.6%) of the cases (58). Four mutations, A267V, R274Q, R274W and T385M, are thought to be recurrent mutations due to hotspot events, but not founder events (Table 1). Moreover, different mutations at the same residue, such as R274Q, R274W, and R274G, have been found for as many as 24 residues. Among 105 mutations, 83 mutations were demonstrated to be GOF mutations by transient gene expression experiments, such as reporter assays (Table 1). The pathogenesis of disease due to the other 22 mutations, excluding four mutations (R70P, T133A, E284K and L354V) (67, 137), has been proven by detecting increased STAT1 phosphorylation or impaired dephosphorylation using patient cells. None of these STAT1-GOF mutations, excluding the T133A mutation which was inherited from an asymptomatic mother (137), can be found in the Genome Aggregation Database (gnomAD), which contains whole-exome sequencing data from 123,136 individuals. The combined annotation-dependent depletion (CADD) score and the minor allele frequency (MAF: based on gnomAD) of all the heterozygous variants of STAT1 found in gnomAD and STAT1-GOF mutations were compared as previously described (138) (Fig. 2). Compared with common variants found in gnomAD, STAT1-GOF mutations showed lower MAFs and relatively higher CADD scores.
Figure 1.
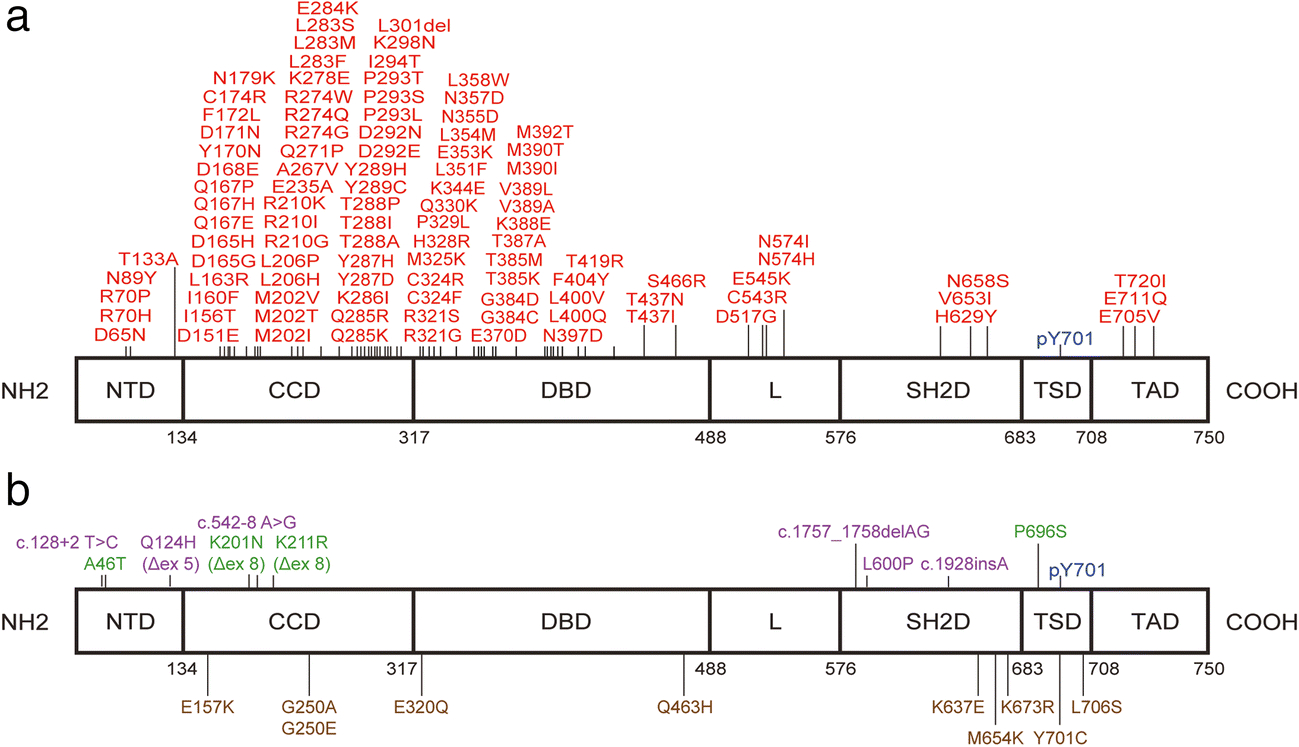
Known GOF and LOF mutations of STAT1
GOF mutations are shown in red, while LOF mutations that cause AD-STAT1 deficiency, AR-STAT1 complete deficiency and AR-STAT1 partial deficiency are shown in brown, purple and green, respectively. N-terminal domain (NTD), coiled-coil domain (CCD), DNA-binding domain (DBD), linker (L) domain, SH2 domain (SH2D), tail segment domain (TSD), transactivation domain (TAD).
Table 1.
Known STAT1-GOF mutations
| Functional domains | GOF mutations | Cellular assay* | Functional test** | References |
|---|---|---|---|---|
| N-terminal domain (NTD) | D65N | Proved | n/a | (118–123) |
| R70H | n/a | GOF | (58) | |
| R70P | n/a | n/a | (67) | |
| N89Y | n/a | GOF | (58) | |
| T133A | n/a | n/a | (137) | |
| Coiled coil domain (CCD) | D151E | n/a | GOF | (58) |
| I156T | n/a | GOF | (58, 96, 116) | |
| I160F | n/a | GOF | (58) | |
| L163R | n/a | GOF | (58, 91) | |
| D165G | Proved | GOF | (55, 58, 60, 86) | |
| D165H | Proved | GOF | (55, 58, 116, 128) | |
| Q167E | n/a | GOF | (58) | |
| Q167H | n/a | GOF | (58) | |
| Q167P | n/a | GOF | (58) | |
| D168E | Proved | GOF | (58, 134) | |
| Y170N | n/a | GOF | (55, 58, 135) | |
| D171N | n/a | GOF | (58) | |
| F172L | Proved | GOF | (57, 83, 98) | |
| C174R | n/a | GOF | (55, 58) | |
| N179K | n/a | GOF | (58, 60) | |
| M202I | Proved | GOF | (55, 58, 109) | |
| M202T | n/a | GOF | (58, 111) | |
| M202V | Proved | GOF | (55, 57–59, 110) | |
| L206H | n/a | GOF | (58) | |
| L206P | Proved | GOF | (65) | |
| R210G | n/a | GOF | (58) | |
| R210I | Proved | GOF | (58, 108) | |
| R210K | n/a | GOF | (58, 69) | |
| E235A | Proved | n/a | (96, 116) | |
| A267V | Proved | GOF | (55, 56, 58–60, 66, 68, 70, 79, 84, 93, 98, 104, 120, 124, 128, 132) | |
| Q271P | n/a | GOF | (55, 58) | |
| R274G | Proved | GOF | (94, 98) | |
| R274Q | Proved | GOF | (55, 57–60, 78, 80, 86, 92, 111, 113, 115, 118–123, 126, 128, 132, 133) | |
| R274W | Proved | GOF | (55–58, 60, 69, 81, 86, 92, 101, 106, 107, 109, 117, 128) | |
| K278E | Proved | n/a | (114) | |
| L283F | Proved | n/a | (118–123) | |
| L283M | Proved | GOF | (70, 104) | |
| L283S | Proved | n/a | (102, 132) | |
| E284K | n/a | n/a | (67) | |
| Q285K | Proved | n/a | (109) | |
| Q285R | n/a | GOF | (58, 60) | |
| K286I | n/a | GOF | (55, 58) | |
| Y287D | Proved | GOF | (57, 58) | |
| Y287H | n/a | GOF | (58) | |
| T288A | Proved | GOF | (55, 58, 118–123, 127) | |
| T288I | Proved | GOF | (58, 124) | |
| T288P | n/a | GOF | (58) | |
| Y289C | Proved | GOF | (58, 109) | |
| Y289H | n/a | GOF | (58) | |
| D292E | Proved | GOF | (58, 86) | |
| D292N | n/a | GOF | (58) | |
| P293L | n/a | GOF | (58, 62) | |
| P293S | Proved | GOF | (57, 58) | |
| P293T | n/a | GOF | (58) | |
| I294T | Proved | GOF | (58, 86, 100) | |
| K298N | Proved | n/a | (88) | |
| L301del | n/a | GOF | (58) | |
| DNA binding domain (DBD) | R321G | n/a | GOF | (58) |
| R321S | Proved | GOF | (58, 128) | |
| C324F | Proved | n/a | (64, 82) | |
| C324R | n/a | GOF | (58, 87, 99, 100) | |
| M325K | Proved | n/a | (118) | |
| H328R | Proved | GOF | (86, 109, 116, 128) | |
| P329L | Proved | GOF | (59, 109, 111) | |
| Q330K | Proved | n/a | (118–123) | |
| K344E | n/a | GOF | (58) | |
| L351F | Proved | GOF | (58, 70, 104, 118–123) | |
| E353K | Proved | GOF | (58, 96, 98, 109, 116, 117, 128) | |
| L354M | Proved | GOF | (58, 59) | |
| L354V | n/a | n/a | (137) | |
| N355D | n/a | GOF | (58) | |
| N357D | n/a | GOF | (58, 111) | |
| L358F | Proved | n/a | (124, 129) | |
| L358W | Proved | GOF | (58, 108, 116) | |
| E370D | Proved | GOF | (58, 85) | |
| G384C | n/a | GOF | (58) | |
| G384D | Proved | GOF | (58, 114) | |
| T385K | Proved | n/a | (57) | |
| T385M | Proved | GOF | (57, 58, 60, 64, 70–73, 75, 82, 86, 95, 98, 104, 105, 108, 109, 115, 116, 118, 121, 122, 128) | |
| T387A | Proved | GOF | (58, 70, 74, 77, 104) | |
| K388E | Proved | GOF | (57, 58, 73, 111) | |
| V389A | n/a | GOF | (58, 132) | |
| V389L | Proved | GOF | (76) | |
| M390I | Proved | GOF | (58, 124) | |
| M390T | Proved | GOF | (58, 59, 86, 132) | |
| M392T | n/a | GOF | (58) | |
| N397D | Proved | GOF | (57, 58, 63, 86) | |
| L400Q | Proved | GOF | (58, 115) | |
| L400V | Proved | GOF | (58, 70, 104) | |
| F404Y | Proved | GOF | (57, 58, 61) | |
| T419R | n/a | GOF | (58) | |
| T437I | Proved | n/a | (111) | |
| T437N | Proved | GOF | (131) | |
| S466R | Proved | GOF | (57, 58, 86, 125) | |
| Linker domain (L) | D517G | n/a | GOF | (58) |
| C543R | n/a | GOF | (176) | |
| E545K | Proved | n/a | (112) | |
| N574H | Proved | n/a | (136) | |
| N574I | n/a | GOF | (58) | |
| SH2 domain (SH2D) | H629Y | Proved | n/a | (94, 103, 116, 128) |
| V653I | Proved | n/a | (89, 90) | |
| N658S | n/a | GOF | (58) | |
| Transactivation domain (TAD) | E705V | Proved | GOF | (97) |
| S708F | Proved | n/a | (118–123) | |
| E711Q | n/a | GOF | (58, 123) | |
| T720I | n/a | GOF | (58, 67) |
Increased STAT1 phosphorylation or impaired dephosphorylation was demonstrated with patient cells.
GOF was confirmed by gene expression assays, including reporter assays.
n/a: data not available
Table 2.
Known LOF mutations that cause AR complete STAT1 deficiency.
Table 4.
Known LOF mutations that cause AD STAT1 deficiency.
Figure 2.
Population genetics for STAT1. CADD score (y-axis) vs. minor allele frequency (MAF, x-axis) for all the heterozygous variants of STAT1 found in gnomAD (blue) and STAT1-GOF mutations (red).
Cellular features
Enhanced STAT1 phosphorylation in response to stimulation with type I and II IFNs and IL-27 was first characterized by immunoblot analysis of Epstein-Barr virus (EBV)-transformed lymphoblastoid cell lines (EBV-LCLs) derived from patients (55). This finding has been repeatedly confirmed by further studies in which whole peripheral blood mononuclear cells (PBMCs), CD3+ or CD4+ T cells, NK cells, and monocytes were analyzed upon stimulation with type I and II IFNs or IL-27 (57, 59, 64–66, 74, 77, 80, 82–84, 88–90, 94, 96, 104, 109, 112, 116, 117, 131, 132, 139–141). Fibroblast cell lines from a patient also showed increased STAT1 phosphorylation upon type II IFN stimulation (76). An enhanced response to type III IFN is also expected but has not yet been proven. PBMCs (59, 80) and EBV-LCLs (60, 95, 98) from patients showed persistent STAT1 phosphorylation in the presence of the protein kinase inhibitor staurosporine. On one hand, increased STAT1 protein levels in primary cells from patients with STAT1-GOF mutations have been reported in some studies (104, 118, 128, 141).
Molecular and biological features
In patients with AR complete or partial STAT1 deficiency, STAT1 mutations result in the complete or partial loss of STAT1 protein expression, leading to the impairment of STAT1-mediated type I, II, and III IFN and IL-27 signaling (14–19). MSMD-related STAT1 mutations do not disturb STAT1 protein expression but exert a dominant-negative effect on WT STAT1-mediated type II IFN signaling (20–27). In contrast, CMC-related STAT1 mutations are GOF mutations that lead to hyperphosphorylation of STAT1 in response to stimulation with type I and II IFNs and IL-27 (55). The underlying mechanism occurs through enhanced STAT1 phosphorylation due at least in part to impairment of the nuclear dephosphorylation of activated STAT1 (55). Liu et al. first reported that a STAT1-null fibrosarcoma cell line (U3C cells) with transient expression of mutant STAT1 showed persistent STAT1 phosphorylation in the presence of the protein kinase inhibitor staurosporine (55). This observation suggests that the impairment of nuclear dephosphorylation is the molecular mechanism underlying STAT1 hyperphosphorylation. Impaired or delayed dephosphorylation was repeatedly confirmed in several subsequent studies through transient gene expression experiments (97, 114, 130) and the use of patient cells (59, 60, 95, 98, 118).
STAT1 forms two types of homodimers depending on its phosphorylation status, parallel dimers and antiparallel dimers (142, 143). Phosphorylated STAT1 forms parallel dimers, whereas unphosphorylated STAT1 preferentially forms antiparallel dimers with the reciprocal binding of CCD and DBD (CCD/DBD) (144). Kagawa et al. hypothesized that CCD/DBD play an important role in controlling STAT1 activity (23). They mutagenized 342 individual wild-type amino acids in the CCD and DBD to alanine and functionally investigated the effects of those alanine substitutions by measuring type II IFN-induced GAS transcriptional activity. This assay, called systemic alanine scanning, correctly predicted 100% of previously reported LOF mutations and 78.1% of known GOF mutations in the CCD/DBD of STAT1. The majority of the GOF alanine substituents were located at the interface of the antiparallel STAT1 dimer, suggesting that GOF mutations disrupt dimerization for the formation of antiparallel STAT1 structures. Formation of the antiparallel STAT1 dimer facilitates phosphatase access by presenting phosphorylated Y701 at both ends of the antiparallel dimer for its ready dephosphorylation (142, 144). Therefore, impairment of the dimerization of STAT1 to form antiparallel dimers may lead to resistance against dephosphorylation by phosphatase enzymes. Known and candidate phosphatases that dephosphorylate STAT1 include TCPTP (PTPN2) and SHP-2 (PTPN11) (145). Whether their activities against STAT1-GOF mutant proteins are decreased remains to be tested.
Recently, other possible mechanisms to explain the increased STAT1 phosphorylation observed in patients with STAT1-GOF mutations have been suggested (141). Bernasconi et al. identified increased STAT1 protein levels in patient cells (128). Based on this discovery, they reported that increased STAT1 phosphorylation in patients is the consequence of not only impaired dephosphorylation but also increased amounts of total STAT1. Moreover, Zimmerman et al. reported that STAT1-GOF mutations cause increased STAT1 protein levels, leading to high levels of STAT1 phosphorylation with normal levels of STAT1 dephosphorylation. Peterson et al. also reported that STAT1-GOF mutations cause the premature nuclear import of phosphorylated STAT1 without altering the phosphorylation or dephosphorylation rate (133). The mechanism underlying this increase in the amount of STAT1 protein remains to be characterized, but increased expression of STAT1 mRNA was found in patients with STAT1-GOF mutations (104, 141). In addition, increased STAT1 protein levels with high STAT1 mRNA levels were confirmed in a knock-in mouse model with a mutation equivalent to the R274Q mutation in humans (146). Further evidence is required to conclude the molecular mechanisms underlying the hyperactivation of STAT1 in patients with STAT1-GOF mutations.
Immunological features
A study of a large cohort of patients with STAT1-GOF mutations (n= 274) revealed the reduced frequency of CD4+ T cells (in 28% of the patients studied), CD8+ T cells (in 16% of the patients studied), CD19+ B cells (in 19% of the patients studied), CD19+CD27+ memory B cells (in 49% of the patients studied), and CD16+CD56+ NK cells (in 25% of the patients studied) (58). Impairment of the terminal maturation of NK cells, as shown by the decreased proportions of CD56dim NK cell subsets, with decreased NK cell cytotoxic function was noted (104, 109). However, these findings are not diagnostically relevant; therefore, it is difficult to speculate on this congenital disorder based on the results of general immunological tests. Some patients display dysgammaglobulinemia involving high serum levels of total IgG (in 20% of the patients tested) or low serum levels of total IgG (in 3% of the patients tested), IgG2 (in 38% of the patients tested) or IgG4 (in 50% of the patients tested). The impairment of antigen-specific antibodies against tetanus, diphtheria toxoid, or poliovirus was found in 23% of the patients tested.
A lower proportion of Th17 cells in the peripheral blood, which can explain at least in part the cause of CMC, is frequently, but not always, observed in patients with STAT1-GOF mutations (55). However, the molecular mechanisms accounting for the decrease in Th17 cells remain to be elucidated (31). Patient cells showed enhanced STAT1-dependent responses to type I and II IFNs and IL-27 (55). Type I and II IFNs and IL-27, which predominantly signal via STAT1, are known to inhibit IL-17 T cell development in mice and humans (147). Therefore, the enhancement of type I and II IFN- and/or IL-27-induced STAT1 signaling might explain the inhibition of Th17 cells in those patients. On one hand, IL-6, IL-21 and IL-23 promote Th17 development mainly via the activation of STAT3-mediated signaling (12, 55). Patients with impaired STAT3 signaling due to a dominant negative mutation in STAT3 (STAT3-DN) develop CMC with a decreased frequency of Th17 cells (148). Zhang et al. investigated the mechanism underlying this decrease in Th17 cells by focusing on the clinical and cellular similarities between STAT1-GOF and STAT3-DN patients (116). They found that naïve T cells upregulate PD-L1 after IL-27 stimulation, leading to the inhibition of Th17 differentiation, in both disorders. The upregulation of PD-L1 was also observed at the basal level in naïve CD4+ T cells from patients with STAT1-GOF mutations and is thought to be correlated with the inhibition of Th17 differentiation (96).
Ma et al. intensively investigated CD4+ helper T cells and identified that patients with STAT1-GOF mutations show increased Th1 cell numbers, decreased Th17 cell numbers, and almost normal Th2 and Tfh cell numbers (149). They also found altered proportions of circulating follicular helper (cTfh) cells, which regulate the development of antigen-specific B cell immunity, in patients with STAT1-GOF mutations, similar to patients with STAT3-DN mutations. Both types of patients presented significant reductions in the number of CCR6+CXCR3− cTfh cells, the most proficient B-helper cTfh cell population. They also showed that cTfh cells from these patients produced high levels of IFN-γ, resulting in the inhibition of Tfh-induced B-cell differentiation (149). The strong similarities in the effects of STAT1-GOF and STAT3-DN mutations on CD4+ T cell differentiation may indicate a putative inhibitory effect of hypermorphic STAT1 mutations on activity of STAT3 (139, 150, 151). The altered naive CD4+ T cell differentiation for STAT1-GOF suggest the humoral immune defects are CD4+ T cell intrinsic. However, intrinsic B cell defects also contributes to poor antibody responses in STAT1-GOF (150). IL-21 plays an important role in isotype switching, affinity maturation, antibody production, and differentiation of B cells (152–154). But, the naïve B cells from patient with STAT1-GOF show poor response to IL-21, resulting in reduced secretion of IgM, IgG, and IgA (150). The increased B-cell apopotosis is also pointed out in patients with STAT1-GOF (96).
Inborn error of immunity due to upregulated type I IFN signaling, called type I interferonopathy, causes severe inflammatory phenotypes and autoimmunity (155–157). Type I interferonopathies, first proposed in 2011, are defined as Mendelian disorders associated with upregulated type I IFN signaling due to inappropriate stimulation of the type I IFN response pathway or defective negative regulation of the type I IFN system (156). In addition, PBMCs from patients with STAT1-GOF mutations also show an enhanced response to type I IFNs associated with the hyperphosphorylation of STAT1 (125, 158). Whether diseases caused by STAT1-GOF mutations should be included among type I interferonopathies is under discussion. As of 2016, diseases caused by STAT1-GOF mutations were not included in this disease category because of insufficient evidence of a functional relationship between ISG production and clinical phenotype (155).
Inappropriate exposure to type I IFN is detrimental to mammals (159). Indeed, patients treated with recombinant type I IFNs sometimes present autoimmune conditions, including SLE, autoimmune thyroid disease, type 1 diabetes mellitus, Sjögren’s syndrome, hemolytic anemia, thrombocytopenia, hypothyroidism, inflammatory myositis, Raynaud’s disease and vitiligo (160, 161). Patients with type I interferonopathies, monogenic diseases in which type I IFN production is constitutively upregulated, develop various developmental manifestations, as well as signs of immunodeficiency and autoinflammation (162). They also develop several autoimmune manifestations with neurological and dermatological phenotypes, particularly SLE (155, 157, 159). In general, patients with STAT1-GOF mutations do not present the typical neurological and/or dermatological phenotypes associated with type I interferonopathies. However, SLE is a representative complication in patients with STAT1-GOF mutations. In addition, autoimmunity is a common feature of patients with STAT1-GOF mutations. Indeed, this autoimmunity can be early and severe and occasionally resembles the series of symptoms in immune dysregulation, polyendocrinopathy, enteropathy, X-linked (IPEX) syndrome (108). Autoimmunity in patients with IPEX syndrome results from defects in regulatory T cells (Tregs) due to mutations in FOXP3, a master transcriptional factor in Tregs (163). In contrast, Treg numbers and function were found to be intact in STAT1-GOF patients with severe autoimmunity that resembled IPEX syndrome (these patients were diagnosed with an IPEX-like syndrome) (108). Kaleviste et al. focused on this clinical observation and investigated type I IFN signaling in STAT1-GOF patients (132). These STAT1-GOF patients showed the clear expression of genes with an interferon signature, whereas circulating levels of IFN-α were not persistently elevated. The authors also examined enrichment in trimethylation of the lysine 4 residue of histone 3 (H3K4me3), an epigenetic modification associated with the activation of transcription, in areas associated with ISGs in STAT1-GOF patients. Together with calcification of the blood vessels and SLE-like disease, which are characteristics shared between type I interferonopathies and STAT1-GOF, this suggests that STAT1-GOF patients are predisposed to IFN-related autoimmunity. Currently, STAT1-GOF are not included among type I interferonopathies, but we see no reason why they should not be, given that STAT1-GOF mutations have been shown to result in increased type I IFN responses in various cell types (55, 57, 83, 84, 90, 131, 132, 139, 155).
Clinical manifestations
The largest systematic study to investigate the clinical manifestations of patients with STAT1-GOF mutations was reported in 2016 (58). This study investigated 274 patients proven to have STAT1-GOF mutations from 167 families originating from 40 countries (58). Among these 274 cases, 167 (61%) were familial cases (60 families). The penetrance of STAT1-GOF mutations was complete, with 98% of patients who developed CMC showing a median age at onset of one year. None of the remaining 2% of cases (6 patients) were totally asymptomatic. Indeed, five had an invasive bacterial infection, one had an invasive fungal infection, 4 had hypothyroidism, and 1 had cerebral aneurysm. The global penetrance of STAT-GOF is therefore considered to be almost complete. The male/female ratio among the patients was 1.03, suggesting that this disorder is distributed equally among sexes. This large cohort study revealed detailed infectious and noninfectious manifestations of STAT1-GOF mutations in patients.
A). Infectious manifestations
The majority of the patients developed CMC (98%: n=268), with a median age at onset of one year (58). Mucocutaneous fungal infections, mostly caused by Candida albicans, affected the oral mucosa (93%: n=254), skin (57%: n=155), esophageal/genital mucosa (56%: n=153), nails (56%: n=56), and/or scalp (20%: n=55). Superficial dermatophytic infection of the scalp, skin or nails was suspected in 16% of patients with microbiological confirmation (Trichophyton spp. or Microsporon spp. isolated in 52% of those patients). Twenty-eight patients (10%) developed an invasive fungal infection, including invasive candidiasis (4%: n=10), fungal pneumonia (6%: n=17), or cryptococcal meningitis (1%: n=3). The causative agents of these invasive fungal infections were Candida spp. (29% of the isolated fungi), Pneumocystis jirovecii (18%), Cryptococcus spp. (18%), Aspergillus spp. (15%), and others (20%).
Many of the patients in this cohort study (74%: n=202) also developed bacterial infections, including lower respiratory infections (47%: n=129), ENT infections (44%: n=121), and/or skin infections (28%: n=77) (58). Recurrent or chronic sinusitis or otitis media account for most of the ear, nose, and throat infections, and folliculitis was the main cause of the skin infections. Bacteria were isolated from 99 patients (36%), and the causative agents were found to be Staphylococcus aureus (36% of the isolated bacteria), Streptococcus spp. (20% of the isolated bacteria), Pseudomonas aeruginosa (13% of the isolated bacteria), Haemophilus influenzae (9% of the isolated bacteria)), and other bacteria (21%). Mycobacterial infections were found in 6% of patients with STAT1-GOF mutations, and these infections were mostly caused by Mycobacterium tuberculosis (35% of the mycobacteria isolated), Bacille Calmette-Guérin (BCG) (30% of the mycobacteria isolated), or other mycobacteria (35% of the mycobacteria isolated), suggesting that patients with STAT1-GOF mutations are at risk for mycobacterial infection.
The molecular mechanism underlying mycobacterial disease in patients with STAT1-GOF mutations has remained elusive. IFN-γ-induced GAF-mediated signaling is essential to eliminate mycobacteria. IFN-γ production in STAT1-GOF patients has been investigated in several studies. Some studies showed that IFN-γ production in response to cytokine or polyclonal stimulation was normal upon the stimulation of PBMCs (73, 78, 89, 100, 114) or CD4+ T cells (65, 88, 96, 98, 105, 114, 150) with PMA and ionomycin, whereas other studies found decreased IFN-γ production by PBMCs (56, 93, 100, 119–123, 129) or CD4+ T cells (66, 101, 118–123, 149). In addition, some studies detected increased type II IFN production by PBMCs (94), CD4+ T cells (64, 74, 82, 112), or CD8+ T cells (82, 112). These results suggest that IFN-γ production in response to cytokine or polyclonal stimulation is highly variable among patients with STAT1-GOF mutations. IFN-γ production by PBMCs (60, 89, 114) and CD4+ T cells (114) in response to Candida antigen was found to be normal, but IFN-γ was barely detected upon PBMC stimulation in other studies (73, 100, 129). Weak IFN-γ production by patient PBMCs upon stimulation with heat-killed S. aureus (73), β-glucan (78), and Penicillium marneffei (129) was also reported. In our experience, the purified protein derivatives of tuberculin (PPD) skin test in a patient with mycobacterial infections was negative at 3 and 6 years of age, indicating a defective T cell response (95). Kataoka et al. identified a patient with an R274G STAT1-GOF mutation and disseminated M. tuberculosis infection despite the negative results of QuantiFERON-TB Gold Plus and PPD skin tests. They hypothesized that the pathogenesis of the infection was the exhaustion of specific immune subsets sensitive to the aberrant activation of STAT1 (80). In addition, an impaired response to type II IFN restimulation is thought to be responsible for mycobacterial susceptibility (98). Additional cases and experimental verification are required to explain the host susceptibility to mycobacteria observed in STAT1-GOF patients.
Approximately 38% of STAT1-GOF patients showed susceptibility to viral infections, developing recurrent mucocutaneous viral infections (32%: n=88) or at least one systemic or atypical viral infection (8%: n=18) (58). The main causes of mucocutaneous viral infections were herpes simplex virus (HSV) (27% of the viruses isolated) and varicella-zoster virus (VZV) (31%), which causes severe chickenpox or shingles in childhood. Recurrent molluscum contagiosum or warts were also frequent and found in 12% (n=32) of patients with STAT1-GOF mutations. Systemic viral infection was mainly associated with cytomegalovirus (CMV) or Epstein-Barr virus (EBV) infection. Uncontrollable CMV infection requiring antiviral treatment occurred in 8 patients (3%). In contrast, 10 patients (4%) developed a chronic active EBV infection that was not severe and did not require specific treatment. Although the frequency was low, systemic or atypical viral infections associated with human herpesvirus 6 (HHV6), parvovirus, BK virus, and hepatitis C virus (HCV) were also reported. Furthermore, two patients developed live vaccine (smallpox and measles)-induced severe disease.
In addition to the infections reported in this large cohort study (58), several individual case reports or small studies on a series of patients with rare infections have been reported. In terms of viral infections, progressive multifocal leukoencephalopathy caused by the polyomavirus JC virus (n=4) (115) and Orf infection, a zoonotic infection caused by a dermatotropic parapoxvirus (n=1) (81), have been described. In terms of fungal infections, cutaneous infection caused by Fusarium solani (fusariosis) (110) or Demodex spp. (demodicidosis) (99, 126) and disseminated infection caused by Coccidioides immitis (coccidioidomycosis) (58, 98), Histoplasma capsulatum (histoplasmosis) (98), Penicillium marneffei (124, 129), or Apophysomyces trapeziformis (mucormycosis) (58, 85) have been reported. Both coccidioidomycosis and histoplasmosis have been reported in patients with autosomal dominant partial IFN-γR1 deficiency (164, 165). In addition, Penicillium marneffei infections, coccidioidomycosis, and histoplasmosis have been reported in patients with anti-type II IFN antibody (166, 167). These observations, together with their susceptibility to mycobacteria, suggest that patients with STAT1-GOF mutations may disturb type II IFN induced host immune response.
B). Autoimmunity
More than one third of patients with STAT1-GOF mutations presented with autoimmune manifestations (37%: n=60) in the large cohort study (58). The male/female ratio was 0.79, suggesting that affected women have higher risk of autoimmunity. Endocrine organs were main target of autoimmunity, including hypothyroidism (22%: n=60), type 1 diabetes mellitus (4%: n=11), and hyperthyroidism (n=1). Some of the patients developed cutaneous diseases (10%: n=28), including vitiligo, alopecia, or psoriasis. Five female patients developed SLE and one patient had scleroderma. Some of the patients developed autoimmune hepatitis (2%: n=6), hematological autoimmunity (4%: n=11) which cause hemolytic anemia or autoimmune thrombocytopenia, and autoantibody positive pernicious anemia (n=1) or celiac disease (n=4). Most patients with autoimmune manifestations were positive for autoantibodies (65% of the patients tested for the presence of autoantibodies). Inflammatory bowel disease was found in 6 patients, including Crohn’s disease (n=2), ulcerative colitis (n=2), and enteropathy with lymphocytic infiltration (n=2).
In addition to this study, Uzel et al. reported 5 children who presented severe autoimmune symptoms, resembling those found in patients with IPEX syndrome (108). These patients presented severe autoimmunity from infancy (one from toddler), such as protein losing enteropathy (n=2), villous blunting or atrophy (n=3), hematological autoimmunity (n=2), and type 1 diabetes mellitus (n=3). These patients also presented CMC (n=4) together with bacterial (n=5) and/or viral (n=3) infections. Leiding et al. also described 5 patients with IPEX-like syndrome, who were treated with hematopoietic stem cell transplantation (86). Interestingly, 5 of these 10 unrelated patients with IPEX-like syndrome carried the T385M STAT1 mutation, suggesting that specific GOF mutations may be related to the development of severe autoimmunity
C). Others
Patients with STAT1-GOF mutations present a broad clinical spectrum of variable severity. Some of the patients suffer from life-threatening infections (64, 86, 100). Persistent or recurrent lower respiratory tract infections result in bronchiectasis (14–21%) (57, 58, 66, 108). Persistent or recurrent mucocutaneous fungal infections may lead to the development of squamous cell carcinoma. Indeed, squamous cell carcinomas which affect cutaneous, gastrointestinal, or laryngeal regions, were found in 4% of the patients with STAT1-GOF mutations (58). As for the other rare symptoms, a case with enamel defect and delayed dental shedding (n=1), and a case with psoriasiform hyperkeratosis (n=1) have also been reported (73, 93).
Aneurysm occurs at a higher rate in patients with CMC than in healthy individuals (168–170). The large cohort study revealed that 6% (n=17) of patients with STAT1-GOF mutations had aneurysms (58). Most aneurysms were located in the cerebral vascular system and were found to be multiple aneurysms (55, 56, 58, 66, 106). In contrast, extracerebral aneurysms were less common (68). Indeed, only one genetically confirmed patient who developed recurrent abdominal and thoracic aortic aneurysm with signs of vasculitis has been reported (106). In terms of extracerebral vascular disease, a patient with systemic, inflammatory large-vessel vasculitis (87) and a patient with aortic calcification (102) have been reported. The cause of aneurysm in these patients remains unclear. In some cases, Candida hyphae were identified in the aneurysm tissue by histology, suggesting that they were mycotic aneurysms (60, 86, 168, 169). On the other hand, aneurysm coexisted with autoimmune symptoms such as atopy or thyroid dysfunction in three-quarters of the patients studied (12 of 16 patients with aneurysm) (58). In addition, the coexistence of severe autoimmunity and aneurysm, or the coexistence of vasculitis and aneurysms, was found in several patients with STAT1-GOF mutations (57, 106, 108). Aneurysm was reported in patients with STAT3-LOF mutations (171) but not in other patients with SCMC. These clinical observations may reflect the link between aneurysm and autoimmunity in patients with STAT1-GOF mutations.
Outcome and treatment
Approximately 12% of patients with STAT1-GOF mutations died at a median age of 30 years with severe infection (38%), cancer (24%) and/or cerebral hemorrhage associated with aneurysm (15%) (58). Therefore, invasive infection, cancer and/or symptomatic aneurysm are predictors of a poor outcome. Indeed, the cumulative survival rate at 60 years of age in STAT1-GOF patients with these predictors was 31%, whereas that of STAT1-GOF patients without these predictors was 87% (58). Failure to thrive was found in 12% of patients. Approximately 21% of patients develop bronchiectasis and cystic pulmonary lesions associated with recurrent pneumonia or bronchitis. Recurrent esophageal candidiasis resulted in secondary gastrointestinal complications, such as dysphagia (6.9%) or esophageal stenosis (4.4%).
Most patients with STAT1-GOF mutations required long-term topical and/or systemic antifungal treatment (57, 58). Triazoles are frequently used for topical treatment, and nystatin is a good alternative (57). A large cohort study provided an overview of the treatments used for STAT1-GOF patients (58). Fluconazole is the main first-line oral therapy, followed by itraconazole and/or posaconazole. Approximately 39% of patients treated with long-term antifungal therapy showed clinical resistance to at least one antifungal agent. These patients required second- or third-line treatments, including voriconazole, echinocandins, terbinafine or liposomal amphotericin B. Antibacterial prophylaxis, mainly co-trimoxazole, against recurrent lower respiratory infection was used in 24% of patients. Polyvalent immunoglobulins were used in 13% of patients who suffered from recurrent pneumonia. In terms of noninfectious manifestations, the patients occasionally developed severe autoimmune disorder and were treated with immunosuppressive agents (58, 87, 108).
Hematopoietic stem cell transplantation (HSCT) can be a curable treatment in patients with PIDs and has also been applied in patients who suffer from severe infectious and/or autoimmune manifestations (63, 75, 82, 86). Leiding et al. investigated 15 patients with STAT1-GOF mutations treated with HSCT (86). All patients in this cohort suffered from severe infection, and five suffered from severe autoimmunity with a diagnosis of IPEX-like syndrome. The symptoms associated with STAT1-GOF mutations disappeared after HSCT, suggesting that HSCT can be a curative treatment for this congenital disorder. However, HSCT simultaneously significantly increased the risk of secondary graft failure, which was found in 50% of patients with primary engraftment, and transplant-related mortality. Indeed, the three-year overall survival after transplantation was only 40%. This study also revealed that GOF mutations in the STAT1 DBD might be associated with more severe clinical manifestations. Indeed, the GOF mutations in 10 of the 15 patients were in the DBD (86), whereas approximately two-thirds of the mutations were identified in the CCD (58). Among the 15 patients treated with HSCT, 5 patients carried the T385M GOF mutation, which is frequently found in patients with IPEX-like syndrome (86, 108) and/or combined immunodeficiency (64, 100). In addition, some studies have suggested that severe clinical manifestations are correlated with specific STAT1-GOF mutations, such as C284R, I294T, C324R, C324F, and T385M (64, 87, 100). These observations suggest a genotype-phenotype correlation, at least for some of the STAT1-GOF mutations identified.
Several patients were administered the Janus kinase (JAK) inhibitor ruxolitinib for the treatment of severe clinical manifestations (65, 77, 109, 112, 117, 172, 173). Ruxolitinib seemed to improve CMC and autoimmune manifestations in the majority of the patients treated (65, 77, 109, 112, 172, 173). Forbes et al. investigated 11 patients administered ruxolitinib for the treatment of autoimmunity or immune dysregulation not controlled with other therapies (172). Ten of these patients showed significant clinical improvement due to oral ruxolitinib, suggesting ruxolitinib as a possible therapeutic choice in most patients. In contrast, the therapeutic failure of ruxolitinib with worsening fungal infections, such as CMC and coccidioidomycosis, or herpes zoster infection have also been reported (117, 172). Thus, acyclovir prophylaxis has been used to prevent herpes virus infection during ruxolitinib treatment (172). Ruxolitinib suppresses the hyperresponsiveness of STAT1 to ligand stimulation, leading to the normalization of Th1 and follicular T helper cell responses (112), and the partial rescue of NK cell differentiation and function (109). The effect of ruxolitinib on Th17 cell differentiation and/or IL-17 cytokine production is controversial. Some reports describe a positive effect (112, 173), whereas the frequency of circulating Th17 cells in some patients did not change following treatment (65, 117). Another JAK inhibitor, baricitinib, was used in one patient (90). The patient showed remarkable clinical improvement in response to oral baricitinib with partial restoration of IL-17A production by PBMCs. A few patients were treated with GM-CSF or G-CSF; however, the effectiveness of these treatments is controversial (113, 174, 175).
Conclusion
STAT1-GOF mutations are the genetic etiology of a unique disorder that combines the manifestations of SCMC and autoimmunity. CMC is caused by impaired IL-17-mediated immunity, but its mechanisms have remained elusive. Thyroiditis is a type I interferonopathy, although the specific nature of this manifestation is also elusive. Most other infectious and noninfectious manifestations are unexplained. Determining the molecular mechanism underlying mycobacterial disease is particularly challenging, as these infections are typically seen in patients with impaired type II IFN immunity. Remarkably, the discovery of STAT1-GOF mutations has paved the way for the treatment of patients with JAK inhibitors, illustrating the therapeutic impact of the genetic and immunological dissection of human infectious diseases. Immunosuppression with these inhibitors not only improves autoimmunity but also paradoxically improves host defense. The potential risks and benefits of HSCT should be considered in this context. Further studies are required to better understand the pathogenesis of the diverse manifestations seen in patients with STAT1-GOF mutations as a prerequisite to better manage these patients.
Table 3.
Known LOF mutations that cause AR partial STAT1 deficiency.
| Functional domains | LOF mutations | References |
|---|---|---|
| N-terminal domain (NTD) | A46T* | (19) |
| Coiled coil domain (CCD) | K201N (Δexon 8) | (18) |
| K211R* (Δexon 8) | (19) | |
| Tail segment domain (TSD) | P696S | (17) |
A46T and K211R were identified as compound heterozygous mutations.
Acknowledgments
SO acknowledges Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (16H05355 and 19H03620), the Promotion of Joint International Research from the Japan Society for the Promotion of Science (18KK0228), and the Practical Research Project for Rare/Intractable Diseases from the Japan Agency for Medical Research and Development, AMED. KM acknowledges a fellowship grant from the Japan Foundation for Pediatric Research and the EURO CMC grant (ANR-14-RARE-0005-02). JLC acknowledge National Institutes of Health grants (grant no. R01AI127564). AP acknowledge grants from the French National Research Agency (ANR) under the “ Lymphocyte T helper (Th) cell differentiation in patients with inborn errors of immunity to Mycobacterium and/or Candida species” program (ANR-FNS LTh-MSMD-CMCD, ANR-18-CE93-0008-01). We thank Peng Zhang and Shohei Eto for their assistance.
Footnotes
Conflicts of interest
The authors declare that they have no relevant conflicts of interest.
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References
- 1.Dale TC, Imam AM, Kerr IM, Stark GR. Rapid activation by interferon alpha of a latent DNA-binding protein present in the cytoplasm of untreated cells. Proc Natl Acad Sci U S A. 1989;86(4):1203–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levy DE, Kessler DS, Pine R, Darnell JE Jr. Cytoplasmic activation of ISGF3, the positive regulator of interferon-alpha-stimulated transcription, reconstituted in vitro. Genes Dev. 1989;3(9):1362–71. [DOI] [PubMed] [Google Scholar]
- 3.Darnell JE Jr., Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264(5164):1415–21. [DOI] [PubMed] [Google Scholar]
- 4.Leonard WJ, O’Shea JJ. Jaks and STATs: biological implications. Annu Rev Immunol. 1998;16:293–322. [DOI] [PubMed] [Google Scholar]
- 5.Subramaniam PS, Torres BA, Johnson HM. So many ligands, so few transcription factors: a new paradigm for signaling through the STAT transcription factors. Cytokine. 2001;15(4):175–87. [DOI] [PubMed] [Google Scholar]
- 6.Casanova JL, Holland SM, Notarangelo LD. Inborn errors of human JAKs and STATs. Immunity. 2012;36(4):515–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4(1):69–77. [DOI] [PubMed] [Google Scholar]
- 8.Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol. 2003;4(1):63–8. [DOI] [PubMed] [Google Scholar]
- 9.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annual review of biochemistry. 1998;67:227–64. [DOI] [PubMed] [Google Scholar]
- 10.Levy DE, Darnell JE Jr. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3(9):651–62. [DOI] [PubMed] [Google Scholar]
- 11.Schindler C, Levy DE, Decker T. JAK-STAT signaling: from interferons to cytokines. J Biol Chem. 2007;282(28):20059–63. [DOI] [PubMed] [Google Scholar]
- 12.Boisson-Dupuis S, Kong XF, Okada S, Cypowyj S, Puel A, Abel L, et al. Inborn errors of human STAT1: allelic heterogeneity governs the diversity of immunological and infectious phenotypes. Curr Opin Immunol. 2012;24(4):364–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sancho-Shimizu V, Perez de Diego R, Jouanguy E, Zhang SY, Casanova JL. Inborn errors of anti-viral interferon immunity in humans. Curr Opin Virol. 2011;1(6):487–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dupuis S, Jouanguy E, Al-Hajjar S, Fieschi C, Al-Mohsen IZ, Al-Jumaah S, et al. Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat Genet. 2003;33(3):388–91. [DOI] [PubMed] [Google Scholar]
- 15.Chapgier A, Wynn RF, Jouanguy E, Filipe-Santos O, Zhang S, Feinberg J, et al. Human complete Stat-1 deficiency is associated with defective type I and II IFN responses in vitro but immunity to some low virulence viruses in vivo. J Immunol. 2006;176(8):5078–83. [DOI] [PubMed] [Google Scholar]
- 16.Vairo D, Tassone L, Tabellini G, Tamassia N, Gasperini S, Bazzoni F, et al. Severe impairment of IFN-gamma and IFN-alpha responses in cells of a patient with a novel STAT1 splicing mutation. Blood. 2011;118(7):1806–17. [DOI] [PubMed] [Google Scholar]
- 17.Chapgier A, Kong XF, Boisson-Dupuis S, Jouanguy E, Averbuch D, Feinberg J, et al. A partial form of recessive STAT1 deficiency in humans. J Clin Invest. 2009;119(6):1502–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kong XF, Ciancanelli M, Al-Hajjar S, Alsina L, Zumwalt T, Bustamante J, et al. A novel form of human STAT1 deficiency impairing early but not late responses to interferons. Blood. 2010;116(26):5895–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kristensen IA, Veirum JE, Moller BK, Christiansen M. Novel STAT1 alleles in a patient with impaired resistance to mycobacteria. J Clin Immunol. 2011;31(2):265–71. [DOI] [PubMed] [Google Scholar]
- 20.Dupuis S, Dargemont C, Fieschi C, Thomassin N, Rosenzweig S, Harris J, et al. Impairment of mycobacterial but not viral immunity by a germline human STAT1 mutation. Science. 2001;293(5528):300–3. [DOI] [PubMed] [Google Scholar]
- 21.Chapgier A, Boisson-Dupuis S, Jouanguy E, Vogt G, Feinberg J, Prochnicka-Chalufour A, et al. Novel STAT1 alleles in otherwise healthy patients with mycobacterial disease. PLoS Genet. 2006;2(8):e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsumura M, Okada S, Sakai H, Yasunaga S, Ohtsubo M, Murata T, et al. Dominant-negative STAT1 SH2 domain mutations in unrelated patients with Mendelian susceptibility to mycobacterial disease. Hum Mutat. 2012;33(9):1377–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kagawa R, Fujiki R, Tsumura M, Sakata S, Nishimura S, Itan Y, et al. Alanine-scanning mutagenesis of human signal transducer and activator of transcription 1 to estimate loss- or gain-of-function variants. J Allergy Clin Immunol. 2017;140(1):232–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirata O, Okada S, Tsumura M, Kagawa R, Miki M, Kawaguchi H, et al. Heterozygosity for the Y701C STAT1 mutation in a multiplex kindred with multifocal osteomyelitis. Haematologica. 2013;98(10):1641–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sampaio EP, Bax HI, Hsu AP, Kristosturyan E, Pechacek J, Chandrasekaran P, et al. A novel STAT1 mutation associated with disseminated mycobacterial disease. J Clin Immunol. 2012;32(4):681–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ueki M, Yamada M, Ito K, Tozawa Y, Morino S, Horikoshi Y, et al. A heterozygous dominant-negative mutation in the coiled-coil domain of STAT1 is the cause of autosomal-dominant Mendelian susceptibility to mycobacterial diseases. Clin Immunol. 2017;174:24–31. [DOI] [PubMed] [Google Scholar]
- 27.Bustamante J, Boisson-Dupuis S, Abel L, Casanova JL. Mendelian susceptibility to mycobacterial disease: genetic, immunological, and clinical features of inborn errors of IFN-gamma immunity. Semin Immunol. 2014;26(6):454–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosain J, Kong XF, Martinez-Barricarte R, Oleaga-Quintas C, Ramirez-Alejo N, Markle J, et al. Mendelian susceptibility to mycobacterial disease: 2014–2018 update. Immunol Cell Biol. 2019;97(4):360–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Puel A, Cypowyj S, Marodi L, Abel L, Picard C, Casanova JL. Inborn errors of human IL-17 immunity underlie chronic mucocutaneous candidiasis. Curr Opin Allergy Clin Immunol. 2012;12(6):616–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lanternier F, Cypowyj S, Picard C, Bustamante J, Lortholary O, Casanova JL, et al. Primary immunodeficiencies underlying fungal infections. Current opinion in pediatrics. 2013;25(6):736–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okada S, Puel A, Casanova JL, Kobayashi M. Chronic mucocutaneous candidiasis disease associated with inborn errors of IL-17 immunity. Clin Transl Immunology. 2016;5(12):e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puel A Human inborn errors of immunity underlying superficial or invasive candidiasis. Hum Genet. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minegishi Y, Saito M, Tsuchiya S, Tsuge I, Takada H, Hara T, et al. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature. 2007;448(7157):1058–62. [DOI] [PubMed] [Google Scholar]
- 34.Holland SM, DeLeo FR, Elloumi HZ, Hsu AP, Uzel G, Brodsky N, et al. STAT3 mutations in the hyper-IgE syndrome. N Engl J Med. 2007;357(16):1608–19. [DOI] [PubMed] [Google Scholar]
- 35.Spencer S, Kostel Bal S, Egner W, Lango Allen H, Raza SI, Ma CA, et al. Loss of the interleukin-6 receptor causes immunodeficiency, atopy, and abnormal inflammatory responses. J Exp Med. 2019;216(9):1986–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beziat V, Tavernier SJ, Chen YH, Ma CS, Materna M, Laurence A, et al. Dominant-negative mutations in human IL6ST underlie hyper-IgE syndrome. J Exp Med. 2020;217(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okada S, Markle JG, Deenick EK, Mele F, Averbuch D, Lagos M, et al. IMMUNODEFICIENCIES. Impairment of immunity to Candida and Mycobacterium in humans with bi-allelic RORC mutations. Science. 2015;349(6248):606–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beziat V, Li J, Lin JX, Ma CS, Li P, Bousfiha A, et al. A recessive form of hyper-IgE syndrome by disruption of ZNF341-dependent STAT3 transcription and activity. Sci Immunol. 2018;3(24). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frey-Jakobs S, Hartberger JM, Fliegauf M, Bossen C, Wehmeyer ML, Neubauer JC, et al. ZNF341 controls STAT3 expression and thereby immunocompetence. Sci Immunol. 2018;3(24). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kisand K, Boe Wolff AS, Podkrajsek KT, Tserel L, Link M, Kisand KV, et al. Chronic mucocutaneous candidiasis in APECED or thymoma patients correlates with autoimmunity to Th17-associated cytokines. J Exp Med. 2010;207(2):299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Puel A, Doffinger R, Natividad A, Chrabieh M, Barcenas-Morales G, Picard C, et al. Autoantibodies against IL-17A, IL-17F, and IL-22 in patients with chronic mucocutaneous candidiasis and autoimmune polyendocrine syndrome type I. J Exp Med. 2010;207(2):291–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puel A, Cypowyj S, Bustamante J, Wright JF, Liu L, Lim HK, et al. Chronic mucocutaneous candidiasis in humans with inborn errors of interleukin-17 immunity. Science. 2011;332(6025):65–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Conti HR, Shen F, Nayyar N, Stocum E, Sun JN, Lindemann MJ, et al. Th17 cells and IL-17 receptor signaling are essential for mucosal host defense against oral candidiasis. J Exp Med. 2009;206(2):299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kagami S, Rizzo HL, Kurtz SE, Miller LS, Blauvelt A. IL-23 and IL-17A, but not IL-12 and IL-22, are required for optimal skin host defense against Candida albicans. J Immunol. 2010;185(9):5453–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cypowyj S, Picard C, Marodi L, Casanova JL, Puel A. Immunity to infection in IL-17-deficient mice and humans. Eur J Immunol. 2012;42(9):2246–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sparber F, LeibundGut-Landmann S. Interleukin-17 in Antifungal Immunity. Pathogens. 2019;8(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Casanova JL, Abel L. Human genetics of infectious diseases: Unique insights into immunological redundancy. Semin Immunol. 2018;36:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levy R, Okada S, Beziat V, Moriya K, Liu C, Chai LY, et al. Genetic, immunological, and clinical features of patients with bacterial and fungal infections due to inherited IL-17RA deficiency. Proc Natl Acad Sci U S A. 2016;113(51):E8277–E85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ling Y, Cypowyj S, Aytekin C, Galicchio M, Camcioglu Y, Nepesov S, et al. Inherited IL-17RC deficiency in patients with chronic mucocutaneous candidiasis. J Exp Med. 2015;212(5):619–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fellmann F, Angelini F, Wassenberg J, Perreau M, Arenas Ramirez N, Simon G, et al. IL-17 receptor A and adenosine deaminase 2 deficiency in siblings with recurrent infections and chronic inflammation. J Allergy Clin Immunol. 2016;137(4):1189–96 e2. [DOI] [PubMed] [Google Scholar]
- 51.Boisson B, Wang C, Pedergnana V, Wu L, Cypowyj S, Rybojad M, et al. An ACT1 mutation selectively abolishes interleukin-17 responses in humans with chronic mucocutaneous candidiasis. Immunity. 2013;39(4):676–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhattad S, Dinakar C, Pinnamaraju H, Ganapathy A, Mannan A. Chronic Mucocutaneous Candidiasis in an Adolescent Boy Due to a Novel Mutation in TRAF3IP2. J Clin Immunol. 2019;39(6):596–9. [DOI] [PubMed] [Google Scholar]
- 53.Nemer G, El-Hachem N, Eid E, Hamie L, Bardawil T, Khalil S, et al. A novel TRAF3IP2 variant causing familial scarring alopecia with mixed features of discoid lupus erythematosus and folliculitis decalvans. Clin Genet. 2020. [DOI] [PubMed] [Google Scholar]
- 54.Li J, Ritelli M, Ma CS, Rao G, Habib T, Corvilain E, et al. Chronic mucocutaneous candidiasis and connective tissue disorder in humans with impaired JNK1-dependent responses to IL-17A/F and TGF-beta. Sci Immunol. 2019;4(41). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu L, Okada S, Kong XF, Kreins AY, Cypowyj S, Abhyankar A, et al. Gain-of-function human STAT1 mutations impair IL-17 immunity and underlie chronic mucocutaneous candidiasis. J Exp Med. 2011;208(8):1635–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van de Veerdonk FL, Plantinga TS, Hoischen A, Smeekens SP, Joosten LA, Gilissen C, et al. STAT1 mutations in autosomal dominant chronic mucocutaneous candidiasis. N Engl J Med. 2011;365(1):54–61. [DOI] [PubMed] [Google Scholar]
- 57.Depner M, Fuchs S, Raabe J, Frede N, Glocker C, Doffinger R, et al. The Extended Clinical Phenotype of 26 Patients with Chronic Mucocutaneous Candidiasis due to Gain-of-Function Mutations in STAT1. J Clin Immunol. 2016;36(1):73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Toubiana J, Okada S, Hiller J, Oleastro M, Lagos Gomez M, Aldave Becerra JC, et al. Heterozygous STAT1 gain-of-function mutations underlie an unexpectedly broad clinical phenotype. Blood. 2016;127(25):3154–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mizoguchi Y, Tsumura M, Okada S, Hirata O, Minegishi S, Imai K, et al. Simple diagnosis of STAT1 gain-of-function alleles in patients with chronic mucocutaneous candidiasis. J Leukoc Biol. 2014;95(4):667–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Soltesz B, Toth B, Shabashova N, Bondarenko A, Okada S, Cypowyj S, et al. New and recurrent gain-of-function STAT1 mutations in patients with chronic mucocutaneous candidiasis from Eastern and Central Europe. J Med Genet. 2013;50(9):567–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Al Rushood M, McCusker C, Mazer B, Alizadehfar R, Grimbacher B, Depner M, et al. Autosomal dominant cases of chronic mucocutaneous candidiasis segregates with mutations of signal transducer and activator of transcription 1, but not of Toll-like receptor 3. J Pediatr. 2013;163(1):277–9. [DOI] [PubMed] [Google Scholar]
- 62.Aldave Becerra JC, Cachay Rojas E. A 3-Year-Old Girl with Recurrent Infections and Autoimmunity due to a STAT1 Gain-of-Function Mutation: The Expanding Clinical Presentation of Primary Immunodeficiencies. Front Pediatr. 2017;5:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aldave JC, Cachay E, Nunez L, Chunga A, Murillo S, Cypowyj S, et al. A 1-year-old girl with a gain-of-function STAT1 mutation treated with hematopoietic stem cell transplantation. J Clin Immunol. 2013;33(8):1273–5. [DOI] [PubMed] [Google Scholar]
- 64.Baris S, Alroqi F, Kiykim A, Karakoc-Aydiner E, Ogulur I, Ozen A, et al. Severe Early-Onset Combined Immunodeficiency due to Heterozygous Gain-of-Function Mutations in STAT1. J Clin Immunol. 2016;36(7):641–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bloomfield M, Kanderova V, Parackova Z, Vrabcova P, Svaton M, Fronkova E, et al. Utility of Ruxolitinib in a Child with Chronic Mucocutaneous Candidiasis Caused by a Novel STAT1 Gain-of-Function Mutation. J Clin Immunol. 2018;38(5):589–601. [DOI] [PubMed] [Google Scholar]
- 66.Breuer O, Daum H, Cohen-Cymberknoh M, Unger S, Shoseyov D, Stepensky P, et al. Autosomal dominant gain of function STAT1 mutation and severe bronchiectasis. Respir Med. 2017;126:39–45. [DOI] [PubMed] [Google Scholar]
- 67.Carey B, Lambourne J, Porter S, Hodgson T. Chronic mucocutaneous candidiasis due to gain-of-function mutation in STAT1. Oral Dis. 2019;25(3):684–92. [DOI] [PubMed] [Google Scholar]
- 68.Dadak M, Jacobs R, Skuljec J, Jirmo AC, Yildiz O, Donnerstag F, et al. Gain-of-function STAT1 mutations are associated with intracranial aneurysms. Clin Immunol. 2017;178:79–85. [DOI] [PubMed] [Google Scholar]
- 69.Dhalla F, Fox H, Davenport EE, Sadler R, Anzilotti C, van Schouwenburg PA, et al. Chronic mucocutaneous candidiasis: characterization of a family with STAT-1 gain-of-function and development of an ex-vivo assay for Th17 deficiency of diagnostic utility. Clin Exp Immunol. 2016;184(2):216–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dotta L, Scomodon O, Padoan R, Timpano S, Plebani A, Soresina A, et al. Clinical heterogeneity of dominant chronic mucocutaneous candidiasis disease: presenting as treatment-resistant candidiasis and chronic lung disease. Clin Immunol. 2016;164:1–9. [DOI] [PubMed] [Google Scholar]
- 71.Eren Akarcan S, Ulusoy Severcan E, Edeer Karaca N, Isik E, Aksu G, Migaud M, et al. Gain-of-Function Mutations in STAT1: A Recently Defined Cause for Chronic Mucocutaneous Candidiasis Disease Mimicking Combined Immunodeficiencies. Case Reports Immunol. 2017;2017:2846928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eslami N, Tavakol M, Mesdaghi M, Gharegozlou M, Casanova JL, Puel A, et al. A gain-of-function mutation of STAT1: A novel genetic factor contributing to chronic mucocutaneous candidiasis. Acta Microbiol Immunol Hung. 2017;64(2):191–201. [DOI] [PubMed] [Google Scholar]
- 73.Frans G, Moens L, Schaballie H, Van Eyck L, Borgers H, Wuyts M, et al. Gain-of-function mutations in signal transducer and activator of transcription 1 (STAT1): chronic mucocutaneous candidiasis accompanied by enamel defects and delayed dental shedding. J Allergy Clin Immunol. 2014;134(5):1209–13 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Giardino G, Somma D, Cirillo E, Ruggiero G, Terrazzano G, Rubino V, et al. Novel STAT1 gain-of-function mutation and suppurative infections. Pediatr Allergy Immunol. 2016;27(2):220–3. [DOI] [PubMed] [Google Scholar]
- 75.Grunebaum E, Kim VH, Somers GR, Shammas A, Roifman CM. Bone marrow transplantation for monoallelic signal transducer and activator of transcription 1 deficiency. J Allergy Clin Immunol. 2016;138(2):612–5 e1. [DOI] [PubMed] [Google Scholar]
- 76.Hartono SP, Vargas-Hernandez A, Ponsford MJ, Chinn IK, Jolles S, Wilson K, et al. Novel STAT1 Gain-of-Function Mutation Presenting as Combined Immunodeficiency. J Clin Immunol. 2018;38(7):753–6. [DOI] [PubMed] [Google Scholar]
- 77.Higgins E, Al Shehri T, McAleer MA, Conlon N, Feighery C, Lilic D, et al. Use of ruxolitinib to successfully treat chronic mucocutaneous candidiasis caused by gain-of-function signal transducer and activator of transcription 1 (STAT1) mutation. J Allergy Clin Immunol. 2015;135(2):551–3. [DOI] [PubMed] [Google Scholar]
- 78.Hori T, Ohnishi H, Teramoto T, Tsubouchi K, Naiki T, Hirose Y, et al. Autosomal-dominant chronic mucocutaneous candidiasis with STAT1-mutation can be complicated with chronic active hepatitis and hypothyroidism. J Clin Immunol. 2012;32(6):1213–20. [DOI] [PubMed] [Google Scholar]
- 79.Huh HJ, Jhun BW, Choi SR, Kim YJ, Yun SA, Nham E, et al. Bronchiectasis and Recurrent Respiratory Infections with a De Novo STAT1 Gain-of-Function Variant: First Case in Korea. Yonsei Med J. 2018;59(8):1004–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kataoka S, Muramatsu H, Okuno Y, Hayashi Y, Mizoguchi Y, Tsumura M, et al. Extrapulmonary tuberculosis mimicking Mendelian susceptibility to mycobacterial disease in a patient with signal transducer and activator of transcription 1 (STAT1) gain-of-function mutation. J Allergy Clin Immunol. 2016;137(2):619–22 e1. [DOI] [PubMed] [Google Scholar]
- 81.Kilic SS, Puel A, Casanova JL. Orf Infection in a Patient with Stat1 Gain-of-Function. J Clin Immunol. 2015;35(1):80–3. [DOI] [PubMed] [Google Scholar]
- 82.Kiykim A, Charbonnier LM, Akcay A, Karakoc-Aydiner E, Ozen A, Ozturk G, et al. Hematopoietic Stem Cell Transplantation in Patients with Heterozygous STAT1 Gain-of-Function Mutation. J Clin Immunol. 2019;39(1):37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kobbe R, Kolster M, Fuchs S, Schulze-Sturm U, Jenderny J, Kochhan L, et al. Common variable immunodeficiency, impaired neurological development and reduced numbers of T regulatory cells in a 10-year-old boy with a STAT1 gain-of-function mutation. Gene. 2016;586(2):234–8. [DOI] [PubMed] [Google Scholar]
- 84.Koo S, Kejariwal D, Al-Shehri T, Dhar A, Lilic D. Oesophageal candidiasis and squamous cell cancer in patients with gain-of-function STAT1 gene mutation. United European Gastroenterol J. 2017;5(5):625–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kumar N, Hanks ME, Chandrasekaran P, Davis BC, Hsu AP, Van Wagoner NJ, et al. Gain-of-function signal transducer and activator of transcription 1 (STAT1) mutation-related primary immunodeficiency is associated with disseminated mucormycosis. J Allergy Clin Immunol. 2014;134(1):236–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Leiding JW, Okada S, Hagin D, Abinun M, Shcherbina A, Balashov DN, et al. Hematopoietic stem cell transplantation in patients with gain-of-function signal transducer and activator of transcription 1 mutations. J Allergy Clin Immunol. 2018;141(2):704–17 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Maeshima K, Ishii K, Shibata H. An Adult Fatal Case with a STAT1 Gain-of-function Mutation Associated with Multiple Autoimmune Diseases. J Rheumatol. 2019;46(3):325–7. [DOI] [PubMed] [Google Scholar]
- 88.Martinez-Martinez L, Martinez-Saavedra MT, Fuentes-Prior P, Barnadas M, Rubiales MV, Noda J, et al. A novel gain-of-function STAT1 mutation resulting in basal phosphorylation of STAT1 and increased distal IFN-gamma-mediated responses in chronic mucocutaneous candidiasis. Mol Immunol. 2015;68(2 Pt C):597–605. [DOI] [PubMed] [Google Scholar]
- 89.Meesilpavikkai K, Dik WA, Schrijver B, Nagtzaam NM, van Rijswijk A, Driessen GJ, et al. A Novel Heterozygous Mutation in the STAT1 SH2 Domain Causes Chronic Mucocutaneous Candidiasis, Atypically Diverse Infections, Autoimmunity, and Impaired Cytokine Regulation. Frontiers in immunology. 2017;8:274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Meesilpavikkai K, Dik WA, Schrijver B, Nagtzaam NMA, Posthumus-van Sluijs SJ, van Hagen PM, et al. Baricitinib treatment in a patient with a gain-of-function mutation in signal transducer and activator of transcription 1 (STAT1). J Allergy Clin Immunol. 2018;142(1):328–30 e2. [DOI] [PubMed] [Google Scholar]
- 91.Mekki N, Ben-Mustapha I, Liu L, Boussofara L, Okada S, Cypowyj S, et al. IL-17 T cells’ defective differentiation in vitro despite normal range ex vivo in chronic mucocutaneous candidiasis due to STAT1 mutation. J Invest Dermatol. 2014;134(4):1155–7. [DOI] [PubMed] [Google Scholar]
- 92.Niehues H, Rosler B, van der Krieken DA, van Vlijmen-Willems I, Rodijk-Olthuis D, Peppelman M, et al. STAT1 gain-of-function compromises skin host defense in the context of IFN-gamma signaling. J Allergy Clin Immunol. 2019;143(4):1626–9 e5. [DOI] [PubMed] [Google Scholar]
- 93.Nielsen J, Kofod-Olsen E, Spaun E, Larsen CS, Christiansen M, Mogensen TH. A STAT1-gain-of-function mutation causing Th17 deficiency with chronic mucocutaneous candidiasis, psoriasiform hyperkeratosis and dermatophytosis. BMJ Case Rep. 2015;2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ovadia A, Sharfe N, Hawkins C, Laughlin S, Roifman CM. Two different STAT1 gain-of-function mutations lead to diverse IFN-gamma-mediated gene expression. NPJ Genom Med. 2018;3:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pedraza-Sanchez S, Lezana-Fernandez JL, Gonzalez Y, Martinez-Robles L, Ventura-Ayala ML, Sadowinski-Pine S, et al. Disseminated Tuberculosis and Chronic Mucocutaneous Candidiasis in a Patient with a Gain-of-Function Mutation in Signal Transduction and Activator of Transcription 1. Frontiers in immunology. 2017;8:1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Romberg N, Morbach H, Lawrence MG, Kim S, Kang I, Holland SM, et al. Gain-of-function STAT1 mutations are associated with PD-L1 overexpression and a defect in B-cell survival. J Allergy Clin Immunol. 2013;131(6):1691–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sampaio EP, Ding L, Rose SR, Cruz P, Hsu AP, Kashyap A, et al. Novel signal transducer and activator of transcription 1 mutation disrupts small ubiquitin-related modifier conjugation causing gain of function. J Allergy Clin Immunol. 2018;141(5):1844–53 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sampaio EP, Hsu AP, Pechacek J, Bax HI, Dias DL, Paulson ML, et al. Signal transducer and activator of transcription 1 (STAT1) gain-of-function mutations and disseminated coccidioidomycosis and histoplasmosis. J Allergy Clin Immunol. 2013;131(6):1624–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Second J, Korganow AS, Jannier S, Puel A, Lipsker D. Rosacea and demodicidosis associated with gain-of-function mutation in STAT1. J Eur Acad Dermatol Venereol. 2017;31(12):e542–e4. [DOI] [PubMed] [Google Scholar]
- 100.Sharfe N, Nahum A, Newell A, Dadi H, Ngan B, Pereira SL, et al. Fatal combined immunodeficiency associated with heterozygous mutation in STAT1. J Allergy Clin Immunol. 2014;133(3):807–17. [DOI] [PubMed] [Google Scholar]
- 101.Smeekens SP, Plantinga TS, van de Veerdonk FL, Heinhuis B, Hoischen A, Joosten LA, et al. STAT1 hyperphosphorylation and defective IL12R/IL23R signaling underlie defective immunity in autosomal dominant chronic mucocutaneous candidiasis. PLoS One. 2011;6(12):e29248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Smyth AE, Kaleviste E, Snow A, Kisand K, McMahon CJ, Cant AJ, et al. Aortic Calcification in a Patient with a Gain-of-Function STAT1 Mutation. J Clin Immunol. 2018;38(4):468–70. [DOI] [PubMed] [Google Scholar]
- 103.Sobh A, Chou J, Schneider L, Geha RS, Massaad MJ. Chronic mucocutaneous candidiasis associated with an SH2 domain gain-of-function mutation that enhances STAT1 phosphorylation. J Allergy Clin Immunol. 2016;138(1):297–9. [DOI] [PubMed] [Google Scholar]
- 104.Tabellini G, Vairo D, Scomodon O, Tamassia N, Ferraro RM, Patrizi O, et al. Impaired natural killer cell functions in patients with signal transducer and activator of transcription 1 (STAT1) gain-of-function mutations. J Allergy Clin Immunol. 2017;140(2):553–64 e4. [DOI] [PubMed] [Google Scholar]
- 105.Takezaki S, Yamada M, Kato M, Park MJ, Maruyama K, Yamazaki Y, et al. Chronic mucocutaneous candidiasis caused by a gain-of-function mutation in the STAT1 DNA-binding domain. J Immunol. 2012;189(3):1521–6. [DOI] [PubMed] [Google Scholar]
- 106.Tanimura M, Dohi K, Hirayama M, Sato Y, Sugiura E, Nakajima H, et al. Recurrent inflammatory aortic aneurysms in chronic mucocutaneous candidiasis with a gain-of-function STAT1 mutation. Int J Cardiol. 2015;196:88–90. [DOI] [PubMed] [Google Scholar]
- 107.Toth B, Mehes L, Tasko S, Szalai Z, Tulassay Z, Cypowyj S, et al. Herpes in STAT1 gain-of-function mutation [corrected]. Lancet. 2012;379(9835):2500. [DOI] [PubMed] [Google Scholar]
- 108.Uzel G, Sampaio EP, Lawrence MG, Hsu AP, Hackett M, Dorsey MJ, et al. Dominant gain-of-function STAT1 mutations in FOXP3 wild-type immune dysregulation-polyendocrinopathy-enteropathy-X-linked-like syndrome. J Allergy Clin Immunol. 2013;131(6):1611–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vargas-Hernandez A, Mace EM, Zimmerman O, Zerbe CS, Freeman AF, Rosenzweig S, et al. Ruxolitinib partially reverses functional natural killer cell deficiency in patients with signal transducer and activator of transcription 1 (STAT1) gain-of-function mutations. J Allergy Clin Immunol. 2018;141(6):2142–55 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang X, Lin Z, Gao L, Wang A, Wan Z, Chen W, et al. Exome sequencing reveals a signal transducer and activator of transcription 1 (STAT1) mutation in a child with recalcitrant cutaneous fusariosis. J Allergy Clin Immunol. 2013;131(4):1242–3. [DOI] [PubMed] [Google Scholar]
- 111.Wang X, Zhang R, Wu W, Wang A, Wan Z, van de Veerdonk FL, et al. New and recurrent STAT1 mutations in seven Chinese patients with chronic mucocutaneous candidiasis. Int J Dermatol. 2017;56(2):e30–e3. [DOI] [PubMed] [Google Scholar]
- 112.Weinacht KG, Charbonnier LM, Alroqi F, Plant A, Qiao Q, Wu H, et al. Ruxolitinib reverses dysregulated T helper cell responses and controls autoimmunity caused by a novel signal transducer and activator of transcription 1 (STAT1) gain-of-function mutation. J Allergy Clin Immunol. 2017;139(5):1629–40 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wildbaum G, Shahar E, Katz R, Karin N, Etzioni A, Pollack S. Continuous G-CSF therapy for isolated chronic mucocutaneous candidiasis: complete clinical remission with restoration of IL-17 secretion. J Allergy Clin Immunol. 2013;132(3):761–4. [DOI] [PubMed] [Google Scholar]
- 114.Yamazaki Y, Yamada M, Kawai T, Morio T, Onodera M, Ueki M, et al. Two novel gain-of-function mutations of STAT1 responsible for chronic mucocutaneous candidiasis disease: impaired production of IL-17A and IL-22, and the presence of anti-IL-17F autoantibody. J Immunol. 2014;193(10):4880–7. [DOI] [PubMed] [Google Scholar]
- 115.Zerbe CS, Marciano BE, Katial RK, Santos CB, Adamo N, Hsu AP, et al. Progressive Multifocal Leukoencephalopathy in Primary Immune Deficiencies: Stat1 Gain of Function and Review of the Literature. Clin Infect Dis. 2016;62(8):986–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang Y, Ma CA, Lawrence MG, Break TJ, O’Connell MP, Lyons JJ, et al. PD-L1 up-regulation restrains Th17 cell differentiation in STAT3 loss- and STAT1 gain-of-function patients. J Exp Med. 2017;214(9):2523–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zimmerman O, Rosler B, Zerbe CS, Rosen LB, Hsu AP, Uzel G, et al. Risks of Ruxolitinib in STAT1 Gain-of-Function-Associated Severe Fungal Disease. Open Forum Infect Dis. 2017;4(4):ofx202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chen X, Xu Q, Li X, Wang L, Yang L, Chen Z, et al. Molecular and Phenotypic Characterization of Nine Patients with STAT1 GOF Mutations in China. J Clin Immunol. 2020;40(1):82–95. [DOI] [PubMed] [Google Scholar]
- 119.Haake K, Wustefeld T, Merkert S, Luttge D, Gohring G, Auber B, et al. Human STAT1 gain-of-function iPSC line from a patient suffering from chronic mucocutaneous candidiasis. Stem Cell Res. 2020;43:101713. [DOI] [PubMed] [Google Scholar]
- 120.Kwon WK, Choi S, Kim HJ, Huh HJ, Kang JM, Kim YJ, et al. Flow Cytometry for the Diagnosis of Primary Immunodeficiency Diseases: A Single Center Experience. Allergy Asthma Immunol Res. 2020;12(2):292–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lee JS, An Y, Yoon CJ, Kim JY, Kim KH, Freeman AF, et al. Germline gain-of-function mutation of STAT1 rescued by somatic mosaicism in immune dysregulation-polyendocrinopathy-enteropathy-X-linked-like disorder. J Allergy Clin Immunol. 2020;145(3):1017–21. [DOI] [PubMed] [Google Scholar]
- 122.Moriya K, Suzuki T, Uchida N, Nakano T, Katayama S, Irie M, et al. Ruxolitinib treatment of a patient with steroid-dependent severe autoimmunity due to STAT1 gain-of-function mutation. Int J Hematol. 2020. [DOI] [PubMed] [Google Scholar]
- 123.Saez-de-Ocariz M, Suarez-Gutierrez M, Migaud M, P OF-R, Casanova JL, Segura-Mendez NH, et al. Rosacea as a striking feature in family members with a STAT1 gain-of-function mutation. J Eur Acad Dermatol Venereol. 2020. [DOI] [PubMed] [Google Scholar]
- 124.Lee PP, Lao-Araya M, Yang J, Chan KW, Ma H, Pei LC, et al. Application of Flow Cytometry in the Diagnostics Pipeline of Primary Immunodeficiencies Underlying Disseminated Talaromyces marneffei Infection in HIV-Negative Children. Frontiers in immunology. 2019;10:2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Stellacci E, Moneta GM, Bruselles A, Barresi S, Pizzi S, Torre G, et al. The activating p.Ser466Arg change in STAT1 causes a peculiar phenotype with features of interferonopathies. Clin Genet. 2019;96(6):585–9. [DOI] [PubMed] [Google Scholar]
- 126.Molho-Pessach V, Meltser A, Kamshov A, Ramot Y, Zlotogorski A. STAT1 gain-of-function and chronic demodicosis. Pediatr Dermatol. 2020;37(1):153–5. [DOI] [PubMed] [Google Scholar]
- 127.Tirosh I, Spielman S, Barel O, Ram R, Stauber T, Paret G, et al. Whole exome sequencing in childhood-onset lupus frequently detects single gene etiologies. Pediatr Rheumatol Online J. 2019;17(1):52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zimmerman O, Olbrich P, Freeman AF, Rosen LB, Uzel G, Zerbe CS, et al. STAT1 Gain-of-Function Mutations Cause High Total STAT1 Levels With Normal Dephosphorylation. Frontiers in immunology. 2019;10:1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lee PP, Mao H, Yang W, Chan KW, Ho MH, Lee TL, et al. Penicillium marneffei infection and impaired IFN-gamma immunity in humans with autosomal-dominant gain-of-phosphorylation STAT1 mutations. J Allergy Clin Immunol. 2014;133(3):894–6 e5. [DOI] [PubMed] [Google Scholar]
- 130.Fujiki R, Hijikata A, Shirai T, Okada S, Kobayashi M, Ohara O. Molecular mechanism and structural basis of gain-of-function of STAT1 caused by pathogenic R274Q mutation. J Biol Chem. 2017;292(15):6240–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Henrickson SE, Dolan JG, Forbes LR, Vargas-Hernandez A, Nishimura S, Okada S, et al. Gain-of-Function STAT1 Mutation With Familial Lymphadenopathy and Hodgkin Lymphoma. Front Pediatr. 2019;7:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kaleviste E, Saare M, Leahy TR, Bondet V, Duffy D, Mogensen TH, et al. Interferon signature in patients with STAT1 gain-of-function mutation is epigenetically determined. Eur J Immunol. 2019;49(5):790–800. [DOI] [PubMed] [Google Scholar]
- 133.Petersen J, Staab J, Bader O, Buhl T, Ivetic A, Meyer T. Identification of a distinct subset of disease-associated gain-of-function missense mutations in the STAT1 coiled-coil domain as system mutants. Mol Immunol. 2019;114:30–40. [DOI] [PubMed] [Google Scholar]
- 134.van Zelm MC, Bosco JJ, Aui PM, De Jong S, Hore-Lacy F, O’Hehir RE, et al. Impaired STAT3-Dependent Upregulation of IL2Ralpha in B Cells of a Patient With a STAT1 Gain-of-Function Mutation. Frontiers in immunology. 2019;10:768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Baghad B, Benhsaien I, El Fatoiki FZ, Migaud M, Puel A, Chiheb S, et al. [Chronic mucocutaneous candidiasis with STAT1 gain-of-function mutation associated with herpes virus and mycobacterial infections]. Ann Dermatol Venereol. 2020;147(1):41–5. [DOI] [PubMed] [Google Scholar]
- 136.Al Dhanhani H, Al Shehri T, Lilic D, Buddles M, Kisand K, Maccari ME, et al. Double Trouble? CMC with a Mutation in both AIRE and STAT1. J Clin Immunol. 2018;38(6):635–7. [DOI] [PubMed] [Google Scholar]
- 137.Rudilla F, Franco-Jarava C, Martinez-Gallo M, Garcia-Prat M, Martin-Nalda A, Riviere J, et al. Expanding the Clinical and Genetic Spectra of Primary Immunodeficiency-Related Disorders With Clinical Exome Sequencing: Expected and Unexpected Findings. Frontiers in immunology. 2019;10:2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhang P, Bigio B, Rapaport F, Zhang SY, Casanova JL, Abel L, et al. PopViz: a webserver for visualizing minor allele frequencies and damage prediction scores of human genetic variations. Bioinformatics. 2018;34(24):4307–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zheng J, van de Veerdonk FL, Crossland KL, Smeekens SP, Chan CM, Al Shehri T, et al. Gain-of-function STAT1 mutations impair STAT3 activity in patients with chronic mucocutaneous candidiasis (CMC). Eur J Immunol. 2015;45(10):2834–46. [DOI] [PubMed] [Google Scholar]
- 140.Zimmerman O, Rosen LB, Swamydas M, Ferre EMN, Natarajan M, van de Veerdonk F, et al. Autoimmune Regulator Deficiency Results in a Decrease in STAT1 Levels in Human Monocytes. Frontiers in immunology. 2017;8:820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bernasconi AR, Yancoski J, Villa M, Oleastro MM, Galicchio M, Rossi JG. Increased STAT1 Amounts Correlate with the Phospho-STAT1 Level in STAT1 Gain-of-function Defects. J Clin Immunol. 2018;38(7):745–7. [DOI] [PubMed] [Google Scholar]
- 142.Mao X, Ren Z, Parker GN, Sondermann H, Pastorello MA, Wang W, et al. Structural bases of unphosphorylated STAT1 association and receptor binding. Mol Cell. 2005;17(6):761–71. [DOI] [PubMed] [Google Scholar]
- 143.Zhong M, Henriksen MA, Takeuchi K, Schaefer O, Liu B, ten Hoeve J, et al. Implications of an antiparallel dimeric structure of nonphosphorylated STAT1 for the activation-inactivation cycle. Proc Natl Acad Sci U S A. 2005;102(11):3966–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mertens C, Zhong M, Krishnaraj R, Zou W, Chen X, Darnell JE Jr. Dephosphorylation of phosphotyrosine on STAT1 dimers requires extensive spatial reorientation of the monomers facilitated by the N-terminal domain. Genes Dev. 2006;20(24):3372–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bohmer FD, Friedrich K. Protein tyrosine phosphatases as wardens of STAT signaling. JAKSTAT. 2014;3(1):e28087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tamaura M, Satoh-Takayama N, Tsumura M, Sasaki T, Goda S, Kageyama T, et al. Human gain-of-function STAT1 mutation disturbs IL-17 immunity in mice. Int Immunol. 2020;32(4):259–72. [DOI] [PubMed] [Google Scholar]
- 147.Liu H, Rohowsky-Kochan C. Interleukin-27-mediated suppression of human Th17 cells is associated with activation of STAT1 and suppressor of cytokine signaling protein 1. J Interferon Cytokine Res. 2011;31(5):459–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ma CS, Chew GY, Simpson N, Priyadarshi A, Wong M, Grimbacher B, et al. Deficiency of Th17 cells in hyper IgE syndrome due to mutations in STAT3. J Exp Med. 2008;205(7):1551–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ma CS, Wong N, Rao G, Avery DT, Torpy J, Hambridge T, et al. Monogenic mutations differentially affect the quantity and quality of T follicular helper cells in patients with human primary immunodeficiencies. J Allergy Clin Immunol. 2015;136(4):993–1006 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ma CS, Wong N, Rao G, Nguyen A, Avery DT, Payne K, et al. Unique and shared signaling pathways cooperate to regulate the differentiation of human CD4+ T cells into distinct effector subsets. J Exp Med. 2016;213(8):1589–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hiller J, Hagl B, Effner R, Puel A, Schaller M, Mascher B, et al. STAT1 Gain-of-Function and Dominant Negative STAT3 Mutations Impair IL-17 and IL-22 Immunity Associated with CMC. J Invest Dermatol. 2018;138(3):711–4. [DOI] [PubMed] [Google Scholar]
- 152.Bryant VL, Ma CS, Avery DT, Li Y, Good KL, Corcoran LM, et al. Cytokine-mediated regulation of human B cell differentiation into Ig-secreting cells: predominant role of IL-21 produced by CXCR5+ T follicular helper cells. J Immunol. 2007;179(12):8180–90. [DOI] [PubMed] [Google Scholar]
- 153.Avery DT, Deenick EK, Ma CS, Suryani S, Simpson N, Chew GY, et al. B cell-intrinsic signaling through IL-21 receptor and STAT3 is required for establishing long-lived antibody responses in humans. J Exp Med. 2010;207(1):155–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wu Y, van Besouw NM, Shi Y, Hoogduijn MJ, Wang L, Baan CC. The Biological Effects of IL-21 Signaling on B-Cell-Mediated Responses in Organ Transplantation. Frontiers in immunology. 2016;7:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Rodero MP, Crow YJ. Type I interferon-mediated monogenic autoinflammation: The type I interferonopathies, a conceptual overview. J Exp Med. 2016;213(12):2527–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Crow YJ. Type I interferonopathies: a novel set of inborn errors of immunity. Ann N Y Acad Sci. 2011;1238:91–8. [DOI] [PubMed] [Google Scholar]
- 157.Crow YJ, Lebon P, Casanova JL, Gresser I. A Brief Historical Perspective on the Pathological Consequences of Excessive Type I Interferon Exposure In vivo. J Clin Immunol. 2018;38(6):694–8. [DOI] [PubMed] [Google Scholar]
- 158.Maródi L Mucocutaneous Candidiasis. Stiehm’s Immune Deficiencies 2014. p. 775–802. [Google Scholar]
- 159.Crow YJ, Manel N. Aicardi-Goutieres syndrome and the type I interferonopathies. Nat Rev Immunol. 2015;15(7):429–40. [DOI] [PubMed] [Google Scholar]
- 160.Selmi C, Lleo A, Zuin M, Podda M, Rossaro L, Gershwin ME. Interferon alpha and its contribution to autoimmunity. Curr Opin Investig Drugs. 2006;7(5):451–6. [PubMed] [Google Scholar]
- 161.Briggs TA, Rice GI, Daly S, Urquhart J, Gornall H, Bader-Meunier B, et al. Tartrate-resistant acid phosphatase deficiency causes a bone dysplasia with autoimmunity and a type I interferon expression signature. Nat Genet. 2011;43(2):127–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Meyts I, Casanova JL. A human inborn error connects the alpha’s. Nat Immunol. 2016;17(5):472–4. [DOI] [PubMed] [Google Scholar]
- 163.Bennett CL, Christie J, Ramsdell F, Brunkow ME, Ferguson PJ, Whitesell L, et al. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet. 2001;27(1):20–1. [DOI] [PubMed] [Google Scholar]
- 164.Vinh DC, Masannat F, Dzioba RB, Galgiani JN, Holland SM. Refractory disseminated coccidioidomycosis and mycobacteriosis in interferon-gamma receptor 1 deficiency. Clin Infect Dis. 2009;49(6):e62–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zerbe CS, Holland SM. Disseminated histoplasmosis in persons with interferon-gamma receptor 1 deficiency. Clin Infect Dis. 2005;41(4):e38–41. [DOI] [PubMed] [Google Scholar]
- 166.Pruetpongpun N, Khawcharoenporn T, Damronglerd P, Suthiwartnarueput W, Apisarnthanarak A, Rujanavej S, et al. Disseminated Talaromyces marneffei and Mycobacterium abscessus in a Patient With Anti-Interferon-gamma Autoantibodies. Open Forum Infect Dis. 2016;3(2):ofw093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Browne SK, Burbelo PD, Chetchotisakd P, Suputtamongkol Y, Kiertiburanakul S, Shaw PA, et al. Adult-onset immunodeficiency in Thailand and Taiwan. N Engl J Med. 2012;367(8):725–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Leroy D, Dompmartin A, Houtteville JP, Theron J. Aneurysm associated with chronic mucocutaneous candidiasis during long-term therapy with ketoconazole. Dermatologica. 1989;178(1):43–6. [DOI] [PubMed] [Google Scholar]
- 169.Marazzi MG, Bondi E, Giannattasio A, Strozzi M, Savioli C. Intracranial aneurysm associated with chronic mucocutaneous candidiasis. European journal of pediatrics. 2008;167(4):461–3. [DOI] [PubMed] [Google Scholar]
- 170.Kirkpatrick CH. Chronic mucocutaneous candidiasis. Pediatr Infect Dis J. 2001;20(2):197–206. [DOI] [PubMed] [Google Scholar]
- 171.Chandesris MO, Azarine A, Ong KT, Taleb S, Boutouyrie P, Mousseaux E, et al. Frequent and widespread vascular abnormalities in human signal transducer and activator of transcription 3 deficiency. Circ Cardiovasc Genet. 2012;5(1):25–34. [DOI] [PubMed] [Google Scholar]
- 172.Forbes LR, Vogel TP, Cooper MA, Castro-Wagner J, Schussler E, Weinacht KG, et al. Jakinibs for the treatment of immune dysregulation in patients with gain-of-function signal transducer and activator of transcription 1 (STAT1) or STAT3 mutations. J Allergy Clin Immunol. 2018;142(5):1665–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Mossner R, Diering N, Bader O, Forkel S, Overbeck T, Gross U, et al. Ruxolitinib Induces Interleukin 17 and Ameliorates Chronic Mucocutaneous Candidiasis Caused by STAT1 Gain-of-Function Mutation. Clin Infect Dis. 2016;62(7):951–3. [DOI] [PubMed] [Google Scholar]
- 174.van de Veerdonk FL, Koenen HJ, van der Velden WJ, van der Meer JW, Netea MG. Immunotherapy with G-CSF in patients with chronic mucocutaneous candidiasis. Immunol Lett. 2015;167(1):54–6. [DOI] [PubMed] [Google Scholar]
- 175.van de Veerdonk FL, Netea MG. Treatment options for chronic mucocutaneous candidiasis. J Infect. 2016;72 Suppl:S56–60. [DOI] [PubMed] [Google Scholar]
- 176.Acker KP, Borlack R, Iuga A, Remotti HE, Soderquist C, Okada S, et al. Ruxolitinib Response in an Infant with Very-Early-Onset Inflammatory Bowel Disease and Gain-of-Function STAT1 Mutation. J Pediatr Gastroenterol Nutr. 2020. [DOI] [PubMed] [Google Scholar]
- 177.Sakata S, Tsumura M, Matsubayashi T, Karakawa S, Kimura S, Tamaura M, et al. Autosomal recessive complete STAT1 deficiency caused by compound heterozygous intronic mutations. Int Immunol. 2020. [DOI] [PubMed] [Google Scholar]