Abstract
Background
MKI67 plays a vital role in the tumour microenvironment (TME) and congenital immunity. The present work focuses on exploring the prognosis prediction performance of MKI67 and its associations with T cell activity and immune infiltration within numerous cancers, especially hepatocellular liver carcinoma (LIHC).
Methods
Oncomine, GEPIA2, and HPA were adopted to analyse MKI67 levels in different types of cancers. The prognostic prediction performance of MKI67 was evaluated through the TCGA portal, GEPIA2, LOGpc, and Kaplan–Meier Plotter databases. The associations of MKI67 with related gene marker sets and immune infiltration were inspected through TISIDB, GEPIA2, and TIMER. We chose MKI67 to analyse biological processes (BPs) and KEGG pathways related to the coexpressed genes. Furthermore, the gene–miRNA interaction network for MKI67 in liver cancer was also examined based on the miRWalk database.
Results
MKI67 expression decreased in many cancers related to the dismal prognostic outcome of LIHC. We found that MKI67 significantly affected the prognosis of LIHC in terms of histology and grade. Increased MKI67 levels were directly proportional to the increased immune infiltration degrees of numerous immune cells and functional T cells, such as exhausted T cells. In addition, several critical genes related to exhausted T cells, including TIM-3, TIGIT, PD-1, LAG3, and CXCL13, were strongly related to MKI67. Further analyses showed that MKI67 was associated with adaptive immunity, cell adhesion molecules (CAMs), and chemokine/immune response signal transduction pathways.
Conclusion
MKI67 acts as a prognostic prediction biomarker in several cancers, particularly LIHC. Upregulation of MKI67 elevates the degree of immune infiltration of many immune cell subtypes, including functional T cells, CD4+ T cells, and CD8+ T cells. Furthermore, MKI67 shows a close correlation with T cell exhaustion, which plays a vital role in promoting T cell exhaustion within LIHC. Detection of the MKI67 level contributes to prognosis prediction and MKI67 modulation within exhausted T cells, thus providing a new method to optimize the efficacy of anti-LIHC immunotherapy.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12876-021-01984-2.
Keywords: Immunotherapy, Immune infiltration, T cells exhaustion, Prognosis, MKI67
Introduction
Liver hepatocellular carcinoma (LIHC), a frequently occurring liver cancer, affects 6/100,000 people every year and is a primary cause of cancer-associated mortality [1, 2]. Current targeted drug therapy cannot provide a satisfactory therapeutic effect because of several diverse factors, including LIHC drug resistance, biological behaviour, and clinical factors [3]. The uncertainty of the molecular mechanisms related to cancer genesis and development further complicates efficient anti-LIHC treatment [4]. In addition, the lack of disease stage- or tumour type-specific markers has dramatically hindered the prevention and management of LIHC. In this regard, it is necessary to elucidate the tumour-immune interaction phenotypes and identify new immune-associated therapeutic targets for liver cancers.
MKi67 expression is found within proliferating cells alone under general conditions [5]. Under normal conditions, MKi67 shows cortical nucleolar localization during interphase and is recruited into condensed chromosomes in mitosis [6, 7]. The MKi67 gene located on chromosome 10q25-ter mainly functions to encode 2 MKi67 isoforms (345 and 395 kDa, respectively) [8–10]. Additionally, the MKi67 level is elevated from the G1 phase to mitosis and shows a rapid decrease later. MKi67 protein expression can be evaluated within cell nuclei at the G1/S/G2 phase and mitosis rather than within quiescent cell nuclei at the G0 phase [11, 12]. As a result, MKi67 expression represents cell proliferation status. Ki67 shows high expression within cancer cells and can be regarded as a prognostic prediction factor for cancer [13, 14]. Furthermore, MKi67 has been studied extensively in retrospective articles as a candidate prognostic prediction factor for cancer proliferation [15, 16]. Plenty of evidence supports the role of MKi67 in diagnosing cancer [17–20]. Cancer cells show high MK167 protein expression, and the positive MKI67 rate (referred to as the labelling index) is related to the clinicopathological characteristics and survival of diverse cancers, such as LIHC [21]. In an article enrolling LIHC cases receiving surgery, high MK167 expression was identified in cancer tissues, which predicted greater tumour grade and early cancer relapse [22, 23]. Furthermore, p53 (encoded via a tumour suppressor gene) and MK167 staining have been extensively adopted to predict LIHC survival postoperatively or even after liver transplantation [24, 25]. The above results indicate that MKI67 plays a vital role in cancer migration, invasion, and progression.
The present work focuses on detecting MKI67 levels and mutations in LIHC cases derived from publicly accessible databases such as The Cancer Genome Atlas (TCGA). MKI67-related functional networks and genomic alterations within LIHC were evaluated based on multidimensional analysis, where the function of MKI67 in tumour immunity was also explored. The findings of this work can help identify novel diagnostic and therapeutic targets for LIHC.
Materials and methods
Analysis based on oncomine database
Oncomine is an integrative database covering 86,733 samples and 715 gene expression profiling datasets developed to facilitate data mining [26]. It is utilized in the present work to assess MKI67 levels and patient prognosis in different types of cancers. (https://www.oncomine.org/resource/login.html).
Analysis based on TIMER database
Tumour Immune Estimation Resource (TIMER, cistrome. shinyapps.io/timer), a kind of easy-to-use web interface, has provided a computational approach for oncology investigators to comprehensively and dynamically analyse and monitor cancer genomic and immunologic data [27]. It contains gene expression profiling data of 10,897 samples covering 32 different kinds of TCGA-derived cancers to estimate six tumour-infiltrating immune cells (TIICs), including CD4+ T cells, CD8+ T cells, dendritic cells (DCs), B cells, neutrophils, and macrophages. This study adopted constrained least-squares fitting for specific gene levels, which negatively correlated with the tumour purity of all cancers [28], for predicting the 6 TIIC subpopulation abundances. Furthermore, “Gene module” and “Diff Exp module” were utilized to analyse the MKI67 level within diverse cancers, as well as the associations of MKI67 level with 6 TIIC subpopulation abundances. The Wilcoxon test assessed the significant difference in MKI67 expression levels. We adopted statistical significance and purity-adjusted partial Spearman’s correlation to evaluate the association between MKI67 levels and immune infiltration. Tumour infiltration degrees for MKI67 across different cancers showing distinct somatic copy number alterations (SCNA) were compared using the “SCNA module” defined by GISTIC 2.0. Typically, the module consists of high amplification (2), arm-level gain (1), diploid/normal (0), arm-level deletion (− 1), and deep deletion (− 2) [29]. Furthermore, we utilized the “Correlation module” to explore the associations of MKI67 levels with TIIC gene markers carefully chosen based on previously published articles, including markers for T cells, B cells, effector T cells, CD8+ T cells, central memory T cells, effector memory T cells, exhausted T cells, resident memory T cells, effector Treg cells, resting Treg cells, neutrophils, T-helper 1 (Th1), dendritic cells (DCs), macrophages, mast cells, and natural killer cells (NK cells) [30–33]. We designed scatterplots of MKI67 gene expression in specific cancers using this module based on statistical significance and Spearman’s correlation analysis. We also displayed gene expression data in the form of a log2 RSEM (RNA-Seq by Expectation–Maximization).
Analysis based on OnCoLnc database
OncoLnc covers the survival data for 21 TCGA-derived cancers along with the corresponding MiTranscriptome, mRNA, and miRNA data http://www.oncolnc.org/. The cases can be classified into different groups based on gene expression to obtain results using this database. OncoLnc contributes to viewing the Kaplan–Meier plot results for at least one cancer simultaneously. It offers Cox regression data and allows the extraction of sufficient data for analysis. In addition, users can also examine the prognostic significance of the tested genes within 21 cancers simultaneously, facilitating the investigation of the vital functions of specific genes in cancer survival.
Analysis based on GEPIA2 database
The Gene Expression Profiling Interactive Analysis 2 database (GEPIA2, http://gepia2.cancer-pku.cn/) represents a web-based approach to investigate gene expression and interactions in cancer tissues and noncarcinoma tissues based on Genotype-Tissue Expression (GTEx) and TCGA-derived data, which can further provide customizable functions, such as profiling plotting, differential expression analysis, patient survival analysis, dimensionality reduction analysis, correlation analysis, and similar gene identification. [34] This study adopted “survival analysis” to examine the association of MKI67 levels with the survival of diverse TCGA-derived cancers. In addition, we used Spearman’s correlation analysis to examine the association of MKI67 with the TIIC gene. Both cancer and noncarcinoma sample datasets were used in subsequent analyses.
Analysis based on Kaplan–Meier plotter database
The Kaplan–Meier Plotter database was developed as an online approach for rapidly assessing the influence of gene expression on 21 cancer survivors, as well as the four significant datasets, namely, breast cancer (BC, n = 6234), lung cancer (LC, n = 3452), ovarian cancer (n = 2190), and gastric cancer (GC, n = 1440) [35]. It was adopted to evaluate the associations of MKI67 levels with the survival of these four cancers. A pan-cancer dataset was used to study MKI67 levels within diverse LIHC subtypes. Meanwhile, we determined HRs (95% CIs) and log-rank P values and plotted the survival curves (http://kmplot.com/).
TISIDB database analysis
The TISIDB database covers 988 immune-associated anticancer genes reported in data from previous studies, noncarcinoma multiomics data, molecular profiling data, high-throughput screening (HTS) technologies, and different immunological data resources collected based on seven publicly accessible databases [36]. It analyses the associations between the screened genes and chemokines, lymphocytes, and immunomodulators. The present work adopted TISIDB to assess the associations of Annexin levels with LIHC clinical stages and investigate the relationships of MKI67 levels with immunomodulators and lymphocytes (http://cis.hku.hk/TISIDB).
Analysis based on human protein atlas database
The HPA (https://www.proteinatlas.org/) database covers all pathological and gene expression data collected from numerous studies conducted using diverse cell lines and tissue types [37]. This database was implemented in the present work to examine MKI67 levels within diverse tissues along with MKI67 localization in cells. The direct links to these images according to the human protein atlas are as follows.
MEXPRESS database analysis
MEXPRESS (https://mexpress.be/) represents a way to visualize data related to DNA methylation status, TCGA expression, clinical information, and the underlying associations [38]. Here, we utilized MEXPRESS to investigate the SPC25 gene methylation status and the association of SPC25 mRNA levels with various clinical features among BC cases.
Analysis based on LinkedOmics database
LinkedOmics signifies an openly accessible database covering multiomics data of 32 TCGA-derived cancers [39]. We conducted Pearson’s test for statistical analyses of MKI67 coexpression by LinkedOmics of “LinkFinder.” The data are presented as heat map/volcano map/scatter plots. In addition, we utilized LinkedOmics of the “LinkInterpreter” module for Gene Ontology (GO, Biological Process (BP)) annotation, Kyoto Encyclopedia of Genes and Genomes (KEGG) for pathway analysis, and Gene Set Enrichment Analysis (GSEA) for transcription factor-target/miRNA-target/kinase-target enrichments using the threshold of false discovery rate (FDR) < 0.05 for 1000 iterations (http://www.linkedomics.org).
Analysis based on miRWalk database
Target genes. MiRNAs screened through the miRWalk approaches were enrolled as possible MKI67-regulating miRNAs [40]. (http://mirwalk.umm.uni-heidelberg.de/).
Statistical methods
The Oncomine database-derived data was presented as ranking, fold-change (FC), and P-values. Survival curves were plotted from TCGA portal, GEPIA2, KM Plotter, LoGPC, and TIMER, (Cox) P-value and HR by the log-rank test. Wilcoxon rank-sum test (two-sided) was used for comparing the infiltration degree of every SCNA category with normal tissue. Spearman correlation was also adopted to assess the association between MKI67 level and other gene or immune infiltration levels in specific cancer types. P ≤ 0.05 indicated statistical significance, shown in the figures.
Results
MKI67 mRNA expression in diverse human cancers
We measured MKI67 mRNA expression in tumour and noncarcinoma samples from distinct cancers based on the Oncomine database to determine the differential MKI67 expression between cancer and noncarcinoma samples. According to our results, MKI67 expression was increased in bladder cancer, CNS and brain cancers, breast cancer (BC), head and neck cancer (HNC), colorectal cancer (CRC), oesophageal cancer (EC), cervical cancer, gastric cancer (GC), liver cancer, lung cancer (LC), lymphoma, ovarian cancer, pancreatic cancer, and sarcoma compared with noncarcinoma samples (Fig. 1a). In addition, other datasets also showed decreased levels of MKI67 mRNA expression in CNS and brain cancers and BC, kidney, and leukaemia cancers. Additional file 1: Table S1 provides more data about MKI67 levels in diverse cancers. To better evaluate MKI67 levels in human cancers, we analysed MKI67 levels in various TCGA-derived cancers based on RNA-seq data. Figure 1b shows MKI67 levels within cancer and noncarcinoma samples in TCGA-derived cancers. MKI67 levels decreased significantly in skin cutaneous melanoma (SKCM) relative to matched noncarcinoma samples. However, MKI67 levels increased within BRCA (invasive breast carcinoma), BLCA (urothelial bladder carcinoma), COAD (colon adenocarcinoma), CHOL (cholangiocarcinoma), HNSC (head and neck cancer), ESCA (oesophageal carcinoma), KIRP (kidney renal papillary cell carcinoma), KIRC (kidney renal clear cell carcinoma), KICH (kidney chromophobe), LIHC (hepatocellular liver carcinoma), PRAD (prostate adenocarcinoma), LUSC (lung squamous cell carcinoma), LUAD (lung adenocarcinoma), STAD (stomach adenocarcinoma), READ (rectum adenocarcinoma), UCEC (uterine corpus endometrial carcinoma) and THCA (thyroid carcinoma) relative to noncarcinoma samples.
Fig. 1.
MKI67 expression in diverse human cancer types. a MKI67 up-regulation or down-regulation in diverse cancer datasets relative to non-carcinoma samples obtained from Oncomine database. b MKI67 expression in diverse TCGA-derived cancers measured using TIMER
Prognosis prediction performance of MKI67 in cancers
To assess the prognosis prediction performance of LIHC in cancer, the associations between the LIHC level and cancer survival were determined from three large-scale cancer databases containing a variety of samples. Typically, this study first determined the impact of LIHC expression on cancer prognostic outcome based on the OncoLnc database. Conspicuously, the LIHC level was markedly associated with the prognosis of eight cancer types: KIRC, BRCA, LGG, KIRP, LUAD, LIHC, SKCM, and PAAD (Table 1).
Table 1.
Relation between MKI67 expression and patient prognosis of different cancer in OncoLnc database
| Cancer | Cox | P value | FDR | Rank | Median | Mean |
|---|---|---|---|---|---|---|
| BLCA | 0.017 | 8.30e−01 | 9.25e−01 | 14,656 | 2546.83 | 2892.19 |
| BRCA | 0.196 | 3.00e−02 | 3.18e−01 | 1558 | 1701.38 | 2293.89 |
| CESC | 0.007 | 9.60e−01 | 9.86e−01 | 15,876 | 4153.29 | 4477.01 |
| COAD | − 0.036 | 7.20e−01 | 9.11e−01 | 12,906 | 3605.44 | 3927.73 |
| ESCA | − 0.154 | 2.20e−01 | 9.72e−01 | 3737 | 5406.79 | 6246.63 |
| GBM | 0.051 | 5.70e−01 | 9.34e−01 | 10,197 | 809.93 | 1006.12 |
| HNSC | 0.037 | 6.10e−01 | 8.42e−01 | 12,004 | 3773.88 | 4310.74 |
| KIRC | 0.211 | 1.30e−02 | 3.84e−02 | 5613 | 388.75 | 544.72 |
| KIRP | 0.996 | 7.80e−09 | 2.07e−06 | 62 | 140.49 | 283.54 |
| LAML | − 0.011 | 9.20e−01 | 9.76e−01 | 14,335 | 4542.33 | 5680.33 |
| LGG | 0.249 | 1.10e−02 | 2.62e−02 | 7023 | 289.2 | 551.11 |
| LIHC | 0.382 | 4.50e−05 | 4.76e−03 | 148 | 624.13 | 964.54 |
| LUAD | 0.281 | 2.30e−04 | 1.34e−02 | 288 | 1309.51 | 1749.87 |
| LUSC | − 0.027 | 7.00e−01 | 9.33e−01 | 12,679 | 2607.62 | 2905.06 |
| OV | 0.069 | 3.40e−01 | 8.37e−01 | 6799 | 2414.05 | 2886.55 |
| PAAD | 0.358 | 1.30e−03 | 2.86e−02 | 772 | 1145.59 | 1363.72 |
| READ | − 0.073 | 7.70e−01 | 9.86e−01 | 12,829 | 3665.46 | 3858.49 |
| SARC | 0.157 | 1.10e−01 | 3.52e−01 | 4966 | 2138.02 | 2519.99 |
| SKCM | 0.172 | 1.10e−02 | 6.66e−02 | 2623 | 1768.63 | 2055.23 |
| STAD | − 0.151 | 7.40e−02 | 3.95e−01 | 3156 | 5150.04 | 5609.92 |
| UCEC | 0.06 | 6.10e−01 | 9.95e−01 | 10,064 | 2149.23 | 2492.38 |
Significantly different results are displayed in this table (cox P < 0.05)
We conducted comprehensive analysis on the associations of MKI67 level with patient survival from 3 large-scale cancer databases consisting of sufficient samples to assess the prognosis prediction performance of MKI67 for cancer. Figure 2a illustrates the influence of MKI67 level on survival in cancer. The poor overall survival (OS) and disease-free survival (DFS) of LIHC was significantly related to higher MKI67 levels (P = 4.5e−4, HR = 1.9; P = 4.2e−5, HR = 1.9, respectively) (Fig. 2b). MKI67 upregulation predicted the survival of KIRP (OS: P = 0.00059, HR = 3); PFS: P = 6.9e−06, HR = 4); ACC (OS: (P = 3.6e−08, HR = 12); PFS: P = 0.00048, HR = 3.3); LGG (OS: P = 0.00015, HR = 2); PFS: P = 0.035, HR = 1.4) (Fig. 2c–e). Poor prognosis was also correlated with higher MKI67 expression in PAAD (OS: P = 6.7e−05, HR = 2.28); PFS: P = 0.014, HR = 2.85); SARC (OS: P = 0.0028, HR = 1.82); PFS: P = 0.0025, HR = 1.82); and STAD (OS: P = 0.01, HR = 0.65); PFS: P = 0.0042, HR = 0.39) (Additional file 1: Figure S1F–H). Nonetheless, poor prognosis in ESCA was associated with lower MKI67 expression (OS: P = 0.011, HR = 0.18; PFS: P = 0.045, HR = 0.37) (Additional file 1: Figure S1F). Furthermore, the level of MKI67 expression significantly differed in disease-free survival and overall survival with PADD (pancreatic adenocarcinoma), SARC (sarcoma), and UVM (uveal melanoma) (Additional file 1: Figure S2A–C). Thereafter, we examined the association of MKI67 level with patient survival from 4 large-scale cancer datasets (including BC, LC, liver cancer and ovarian cancer) based on Kaplan–Meier Plotter. Likewise, MKI67 upregulation predicted the dismal prognostic outcome of LIHC (OS: P = 0.00011, HR = 1.96 [1.38–2.77]; PFS: P = 1.1e−05, HR = 2.14 [1.51–3.02]), BRCA (OS: P = 1e−06, HR = 1.73 [1.39–2.16]; PFS: P = 1.7e−10, HR = 1.43 [1.28–1.59]), OV (OS: P = 0.0079, HR = 1.23 [1.05–1.43]; PFS: P = 0.028, HR = 1.17 [1.02–1.35]), and LUAD (OS: P < 1e−16, HR = 1.95 [1.69–2.25]; PFS: P = 5e−11, HR = 2.32 [1.79–3]) (Fig. 2f–i).
Fig. 2.
Prognosis prediction performance of MKI in diverse cancers measured using GEPIA2 (a–e) and Kaplan–Meier Plot (f–i). a Survival heatmap for MKI in 33 TCGA-derived cancers. Heatmap showing log10 HRs for MKI67. Blue and red blocks represent decreased and increased risks, separately. Rectangles with frames indicate the significance upon prognosis analysis. DFS and OS curves for b LIHC (n = 362), c KIRP (n = 281), d ACC (n = 76), e LGG (n = 514). OS and PFS survival curves in f liver cancer (n = 364, n = 316), g BC (n = 1402, n = 3951), h ovarian cancer (n = 1656, n = 1435), and i lung cancer (n = 1925, n = 982). OS overall survival, RFS relapse-free survival, DFS disease-free survival, PFS progression-free survival
Effect of MKI67 on regulating immune molecules
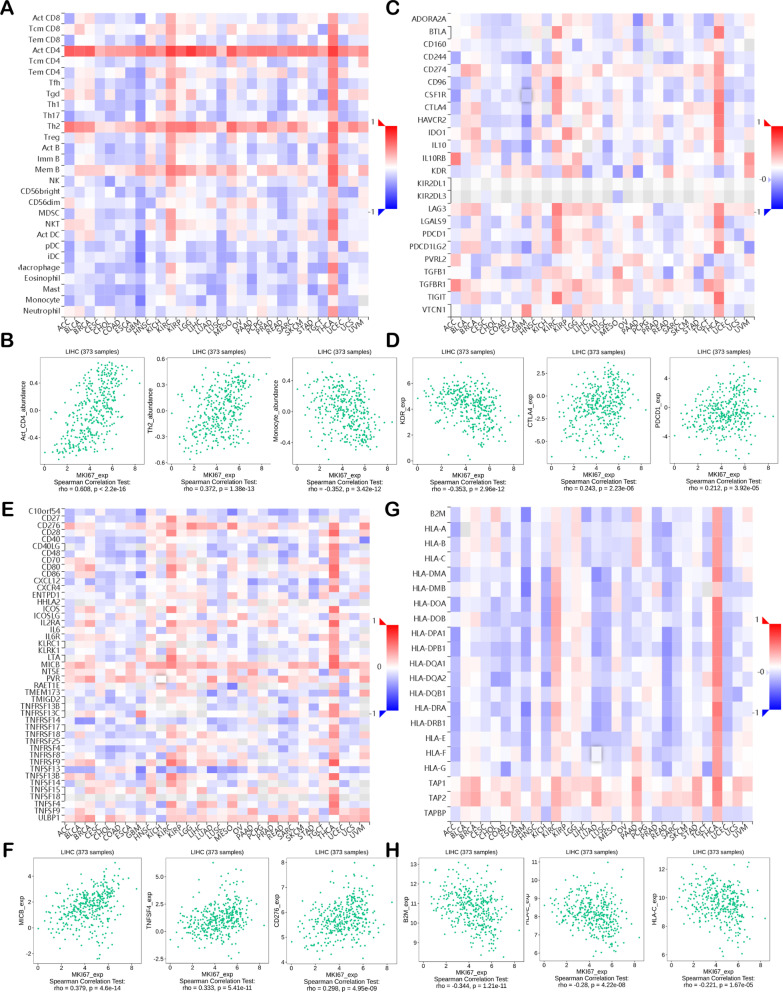
Here, we examined Spearman’s correlations of MKI67 levels with immunomodulators based on the TISIDB database (Fig. 3). Figure 3a displays the association of MKI67 levels with TILs. The most significantly correlated lymphocytes were activated CD4 T cells (Act-cd4; Spearman: ρ = 0.608, P < 2.2e−16), type 2 helper cells (Th2; Spearman: ρ = 0.372, P = 1.38e−13), and monocytes (monocytes; Spearman: ρ = -0.352, P = 3.42e−12) (Fig. 3b). Immunomodulators are divided into major histocompatibility complex (MHC) molecules, immune inhibitors, and immunostimulators. Figure 3c reveals the association of MKI67 expression with immunoinhibitors, among which the most significantly correlated ones were KDR (Spearman: ρ = -0.353, P = 2.96e−12), PDCD1 (Spearman: ρ = 0.212, P = 3.92e−05) and CTLA4 (Spearman: ρ = 0.243, P = 2.23e−06) (Fig. 3d). Figure 3e displays the association of MKI67 levels with immunostimulators, where the most significantly correlated ones were MICB (Spearman: ρ = 0.379, P = 4.6e−14), CD276 (Spearman: ρ = 0.298, P = 4.95e−09), and TNFSF4 (Spearman: ρ = 0.333, P = 5.41e−11) (Fig. 3f). Figure 3g represents the association of MKI67 levels with MHC molecules, among which the most significantly correlated are B2M (Spearman: ρ = 0.344, P = 1.21e−11), HLA-C (Spearman: ρ = 0.221, P = 1.67e−05), and HLA-E (Spearman: ρ = -0.28, P = 4.22e−08) (Fig. 3h). Therefore, MKI67 is perhaps related to the regulation of these immune molecules.
Fig. 3.
Spearman’s correlation between MKI67 and immunomodulators and lymphocytes (TISIDB). a Association of TILs abundances with MKI67 level; b the three most significant TILs with the highest Spearman’s correlation with MKI67 levels; c association of increasing immune inhibitors with MKI67 level; d the three most significant immune inhibitors with the maximum Spearman’s correlation with MKI67 level; e association of immunostimulators abundances with MKI67 level; f the four most significant immunostimulators with the maximum Spearman’s correlation with MKI67 levels; g association of MHC molecules with MKI67 level; h the three most significant MHC molecules with the highest Spearman’s correlation with MKI67 level. Blue and red cells stand for negative and positive correlations, separately. Color intensity is directly proportional to correlation strength. MHC major histocompatibility complex, TILs tumor-infiltrating lymphocytes
MKI67 is related to immune infiltration degrees within LIHC
We can estimate lymph node metastasis (LNM) and the survival of cancer cases separately based on lymphocyte infiltration degrees within a tumour [41–43]. Therefore, we further analysed the association of MKI67 levels with immune cell infiltration degrees within 39 cancers based on the TIMER database (Additional file 1: Figure S2). Consequently, MKI67 levels were significantly related to tumour purity within 14 cancers and B cell infiltration degrees within 23 cancers. MKI67 was also related to CD4+ T cell infiltration degrees in 22 cancers, CD8+ T cell infiltration degrees in 19 cancers, DC infiltration degrees in 25 cancers, neutrophil infiltration degrees in 24 cancers, and macrophage infiltration degrees in 17 cancers. However, MKI67 expression was not related to the infiltration degrees of CD4+ T cells, CD8+ T cells, B cells, macrophages, DCs, or neutrophils within cholangiocarcinoma (CHOL) (Additional file 1: Figure S2H). In LIHC, MKI67 expression indicated an increase in the infiltration degrees of CD4+ T cells (R = 0.381, P = 2.58e−13), CD8+ T cells (R = 0.331, P = 3.26e−10), DCs (R = 0.471, P = 3.55e−20), neutrophils (R = 0.41, P = 2.10e−15), and macrophages (R = 0.475, P = 1.30e−20) (Fig. 4a). Furthermore, in some types of cancers, including THCA, BRCA, SKCM, KIRC, and GBM, the immune infiltration degrees were markedly associated with MKI67 (Additional file 1: Figure S2). This study also presented Kaplan–Meier plots based on the TIMER database to explore the association of immune cell infiltration degrees with MKI67 expression in LIHC. Hence, MKI67 levels were not significantly related to the immune cell infiltration degree or survival of LIHC (Fig. 4b). We also determined the infiltration degrees in LIHC showing diverse SCNA for MKI67 (Fig. 4c). According to the results obtained, MKI67 exerts a vital role in modulating the infiltration degrees of immune cells in LIHC, especially for neutrophils and DCs (Fig. 4c). This study revealed that MKI67 had a stimulating effect on the degree of immune infiltration, particularly for CD4+ T cells, neutrophils, B cells, and dendritic cells, in LIHC.
Fig. 4.
Association of MKI67 with immune infiltration degrees within LIHC. a Association of MKI67 level with immune infiltration degrees of CD8+ T cells, CD4+ T cells, B cells, neutrophils, macrophages, DCs, and tumor purity; b Kaplan–Meier plots concerning immune infiltration and MKI67 expression within LIHC; c tumor-infiltration degrees of LIHC displaying diverse SCNAs defined using GISTIC 2.0, which include high amplification (2), arm-level gain (1), diploid/normal (0), arm-level deletion (− 1), and deep deletion (− 2)
Association of MKI67 with TIIC gene maker
We analysed the association of MKI67 levels with TIIC gene markers in LIHC based on GEPIA2 and TIMER to reveal the underlying relationship of MKI67 expression with tumour immune infiltration degree. Based on our previous study, we adopted commonly used TIIC gene markers and diverse functional T cells, including B cells, T cells, CD8+ T cells, monocytes, M1/M2 macrophages, TAMs, natural killer (NK) cells, neutrophils, mast cells, DCs, Tfhs, effector T cells, Th1/Th2/Th17 cells, central memory T cells, effector memory T cells, exhausted T cells, resident memory T cells, effector Treg T cells and resting Treg T cells. Table 2 reveals the tumour purity-adjusted results of the correlation analysis for LIHC. MKI67 was markedly related to gene markers of monocytes, B cells, M1/M2 macrophages, TAMs, CD8+ T cells, neutrophils, T cells, DCs, mast cells, NK cells, and many functional T cells, including effector memory T cells, effector T cells, resident memory T cells, central memory T cells, resting Treg T cells, exhausted T cells, Th1/Th2/Th17/Tfh cells, and effector Treg T cells. Interestingly, such observations verified that MKI67 was related to T cells, B cells, and functional T cells, as reported previously. It also illustrated the new relationship of MKI67 with exhausted T cells; however, there is little relevant research confirming this relationship.
Table 2.
Association of MKI67 with immune cell gene markers in LIHC detected through TIMER
| Immune cell | Gene markers | None | Purity | ||
|---|---|---|---|---|---|
| Cor | P value | Cor | P value | ||
| CD8+ T cell | CD8A | 0.218 | *** | 0.211 | *** |
| CD8B | 0.180 | ** | 0.182 | ** | |
| T cell | CD6 | 0.214 | *** | 0.215 | *** |
| CD3D | 0.279 | *** | 0.278 | *** | |
| CD3E | 0.217 | *** | 0.214 | *** | |
| SH2D1A | 0.195 | ** | 0.194 | ** | |
| TRAT1 | 0.183 | ** | 0.196 | ** | |
| CD3G | 0.279 | *** | 0.274 | *** | |
| CD2 | 0.224 | *** | 0.227 | *** | |
| B cell | CD19 | 0.247 | *** | 0.242 | *** |
| FCRL2 | 0.202 | *** | 0.214 | *** | |
| KIAA0125 | 0.146 | * | 0.146 | * | |
| TNFRSF17 | 0.133 | 0.01 | 0.142 | * | |
| SPIB | 0.376 | *** | 0.357 | *** | |
| PNOC | 0.197 | ** | 0.211 | *** | |
| CD79A | 0.176 | ** | 0.172 | * | |
| Monocyte | CD86 | 0.316 | *** | 0.314 | *** |
| CD115(CSF1R) | 0.184 | ** | 0.190 | ** | |
| TAM | CD68 | 0.272 | *** | 0.263 | *** |
| IL10 | 0.249 | *** | 0.240 | *** | |
| M1 Macrophage | IRF5 | 0.412 | *** | 0.411 | *** |
| COX2(PTGS2) | 0.138 | * | 0.151 | * | |
| M2 Macrophage | CD163 | 0.106 | 0.041 | 0.109 | 0.042 |
| MS4A4A | 0.119 | 0.022 | 0.128 | 0.017 | |
| Neutrophils | FPR1 | 0.239 | *** | 0.244 | *** |
| SIGLEC5 | 0.296 | *** | 0.304 | *** | |
| CSF3R | 0.307 | *** | 0.302 | *** | |
| FCGR3B | 0.158 | * | 0.153 | * | |
| CEACAM3 | 0.120 | 0.021 | 0.136 | 0.017 | |
| CD116(ITGAM) | 0.296 | *** | 0.312 | *** | |
| Natural killer cell | XCL2 | 0.210 | *** | 0.226 | *** |
| KIR2DL1 | − 0.003 | 0.955 | − 0.023 | 0.668 | |
| KIR2DL3 | 0.177 | ** | 0.189 | ** | |
| KIR2DL4 | 0.177 | ** | 0.167 | * | |
| Dendritic cell | CCL13 | 0.181 | ** | 0.162 | * |
| CD209 | 0.171 | ** | 0.180 | ** | |
| HSD11B1 | − 0.352 | *** | − 0.344 | *** | |
| HLA-DPB1 | 0.185 | ** | 0.185 | ** | |
| HLA-DQB1 | 0.148 | * | 0.148 | * | |
| HLA-DRA | 0.196 | ** | 0.199 | ** | |
| HLA-DPA1 | 0.176 | ** | 0.186 | ** | |
| BCDA-1(CD1C) | 0.136 | * | 0.141 | * | |
| BDCA-4(NRP1) | 0.291 | *** | 0.296 | *** | |
| CD11c(ITGAX) | 0.363 | *** | 0.354 | *** | |
| Mast cell | TPSB2 | − 0.028 | 0.595 | − 0.044 | 0.413 |
| HDC | − 0.166 | * | − 0.175 | * | |
| Th1 | IFN-γ(IFNG) | 0.269 | *** | 0.282 | *** |
| TNF-α(TNF) | 0.288 | *** | 0.299 | *** | |
| STAT4 | 0.259 | *** | 0.263 | *** | |
| STAT1 | 0.379 | *** | 0.379 | *** | |
| Th2 | GATA3 | 0.250 | *** | 0.258 | *** |
| STAT6 | 0.162 | * | 0.157 | * | |
| STAT5A | 0.329 | *** | 0.329 | *** | |
| Tfh | BCL6 | 0.172 | ** | 0.179 | ** |
| IL21 | 0.172 | ** | 0.178 | ** | |
| Th17 | STAT3 | 0.200 | ** | 0.198 | ** |
| IL17A | 0.100 | 0.054 | 0.099 | 0.065 | |
| Effector T cell | CX3CR1 | 0.199 | ** | 0.209 | *** |
| FGFBP2 | − 0.140 | * | − 0.120 | 0.026 | |
| FCGR3A | 0.305 | *** | 0.309 | *** | |
| Effector memory T cell | PD-1 (PDCD1) | 0.344 | *** | 0.332 | *** |
| DUSP4 | 0.324 | *** | 0.330 | *** | |
| Central memory T cell | CCR7 | 0.131 | 0.011 | 0.136 | 0.011 |
| SELL | 0.175 | ** | 0.188 | ** | |
| IL7R | 0.176 | ** | 0.185 | ** | |
| Resident memory T cell | CD69 | 0.176 | ** | 0.174 | * |
| ITGAE | 0.306 | *** | 0.297 | *** | |
| CXCR6 | 0.201 | *** | 0.203 | ** | |
| MYADM | 0.391 | *** | 0.375 | *** | |
| Exhausted T cell | TIM-3 (HAVCR2) | 0.344 | *** | 0.345 | *** |
| TIGIT | 0.325 | *** | 0.333 | *** | |
| LAG3 | 0.306 | *** | 0.295 | *** | |
| CXCL13 | 0.273 | *** | 0.295 | *** | |
| LAYN | 0.227 | *** | 0.243 | *** | |
| Resting Treg T cell | FOXP3 | 0.194 | ** | 0.223 | *** |
| IL2RA | 0.308 | *** | 0.309 | *** | |
| Effector Treg T cell | CTLA4 | 0.354 | *** | 0.353 | *** |
| CCR8 | 0.444 | *** | 0.456 | *** | |
| TNFRSF9 | 0.371 | *** | 0.386 | *** | |
TAM, tumor-associated macrophage; LIHC, Liver hepatocellular carcinoma; Treg, regulatory T cells; Tfh, Follicular helper T cells; Th, T helper cells; Purity, tumor purity-adjusted correlation. None, unadjusted correlation; Cor, ρ-value upon Spearman correlation. Significance levels: * P < 0.05; ** P < 0.01; *** P < 0.001
This study also examined correlations of MKI67 levels with TIIC gene markers in LIHC and noncarcinoma tissues using GEPIA2 (Table 3) to confirm the above observations. MKI67 correlated positively with TAMs (Cor = 0.27, P < 0.0001), monocytes (Cor = 0.28, P < 0.0001), CD8+ T cells (Cor = 0.2, P < 0.001), neutrophils (Cor = 0.37, P < 0.0001), M1 macrophages (Cor = 0.31, P < 0.0001), and several functional T cells, particularly effector memory T cells (Cor = 0.4, P < 0.0001) and effector T cells (Cor = 0.26, P < 0.0001). Here, MKI67 was significantly related to several critical genes involved in T cell exhaustion, including TIM-3 (Cor = 0.14, P < 0.01), PD-1 (Cor = 0.15, P < 0.01), CXCL13 (Cor = 0.085, P < 0.1), and TIGHT (Cor = 0.21, P < 0.0001). These genes have an essential effect on existing antitumour immunotherapies. However, for the remaining immune cell types, such as NK cells, DCs, and neutrophils, we found statistically significant differences but a weak correlation strength compared with the above cell types. In comparison, MKI67 expression did not have a significant link with TIIC gene markers in noncarcinoma tissues.
Table 3.
The correlations of MKI67 with immune cell gene markers for LIHC by GEPIA2
| Immune cell | Gene markers | Tumor | Tumor-sum | Normal | Tumor-sum | ||||
|---|---|---|---|---|---|---|---|---|---|
| Cor | P value | Cor | P value | Cor | P value | Cor | P value | ||
| CD8+ T cell | CD8A | 0.2 | ** | 0.2 | ** | 0.39 | * | 0.37 | * |
| CD8B | 0.23 | *** | 0.31 | 0.026 | |||||
| T cell | CD6 | 0.12 | 0.024 | 0.2 | ** | 0.24 | 0.093 | 0.41 | * |
| CD3D | 0.23 | *** | 0.42 | * | |||||
| CD3E | 0.13 | * | 0.34 | 0.017 | |||||
| SH2D1A | 0.13 | 0.014 | 0.39 | * | |||||
| TRAT1 | 0.049 | 0.35 | 0.36 | * | |||||
| CD3G | 0.24 | *** | 0.35 | 0.013 | |||||
| CD2 | 0.15 | * | 0.3 | 0.035 | |||||
| B cell | CD19 | 0.096 | 0.066 | 0.17 | * | 0.28 | 0.051 | 0.44 | * |
| FCRL2 | 0.068 | 0.19 | 0.5 | ** | |||||
| KIAA0125 | 0.052 | 0.32 | 0.52 | ** | |||||
| TNFRSF17 | 0.06 | 0.25 | 0.43 | * | |||||
| SPIB | 0.13 | 0.01 | 0.22 | 0.12 | |||||
| PNOC | 0.071 | 0.18 | 0.48 | ** | |||||
| CD79A | 0.062 | 0.24 | 0.38 | * | |||||
| Monocyte | CD86 | 0.32 | *** | 0.28 | *** | 0.21 | 0.15 | 0.28 | 0.052 |
| CD115(CSF1R) | 0.25 | *** | 0.27 | 0.056 | |||||
| TAM | CD68 | 0.26 | *** | 0.27 | *** | 0.2 | 0.15 | 0.21 | 0.14 |
| IL10 | 0.21 | *** | 0.15 | 0.29 | |||||
| M1 Macrophage | IRF5 | 0.31 | *** | 0.31 | *** | 0.13 | 0.37 | 0.07 | 0.63 |
| COX2(PTGS2) | 0.078 | 0.14 | − 0.032 | 0.82 | |||||
| M2 Macrophage | CD163 | 0.15 | * | 0.12 | 0.018 | 0.18 | 0.22 | 0.25 | 0.078 |
| MS4A4A | 0.16 | * | 0.22 | 0.12 | |||||
| Neutrophils | FPR1 | 0.25 | *** | 0.37 | *** | − 0.035 | 0.81 | 0.15 | 0.3 |
| SIGLEC5 | 0.31 | *** | − 0.011 | 0.94 | |||||
| CSF3R | 0.22 | *** | 0.11 | 0.43 | |||||
| FCGR3B | 0.05 | 0.34 | 0.17 | 0.24 | |||||
| CEACAM3 | 0.19 | ** | 0.064 | 0.66 | |||||
| CD116(ITGAM) | 0.33 | *** | 0.36 | 0.011 | |||||
| CD8+ T cell | CD8A | 0.2 | ** | 0.2 | ** | 0.39 | * | 0.37 | * |
| CD8B | 0.23 | *** | 0.31 | 0.026 | |||||
| T cell | CD6 | 0.12 | 0.024 | 0.2 | ** | 0.24 | 0.093 | 0.41 | * |
| CD3D | 0.23 | *** | 0.42 | * | |||||
| CD3E | 0.13 | * | 0.34 | 0.017 | |||||
| SH2D1A | 0.13 | 0.014 | 0.39 | * | |||||
| TRAT1 | 0.049 | 0.35 | 0.36 | * | |||||
| CD3G | 0.24 | *** | 0.35 | 0.013 | |||||
| CD2 | 0.15 | * | 0.3 | 0.035 | |||||
| B cell | CD19 | 0.096 | 0.066 | 0.17 | * | 0.28 | 0.051 | 0.44 | * |
| FCRL2 | 0.068 | 0.19 | 0.5 | ** | |||||
| KIAA0125 | 0.052 | 0.32 | 0.52 | ** | |||||
| TNFRSF17 | 0.06 | 0.25 | 0.43 | * | |||||
| SPIB | 0.13 | 0.01 | 0.22 | 0.12 | |||||
| PNOC | 0.071 | 0.18 | 0.48 | ** | |||||
| CD79A | 0.062 | 0.24 | 0.38 | * | |||||
| Monocyte | CD86 | 0.32 | *** | 0.28 | *** | 0.21 | 0.15 | 0.28 | 0.052 |
| CD115(CSF1R) | 0.25 | *** | 0.27 | 0.056 | |||||
| TAM | CD68 | 0.26 | *** | 0.27 | *** | 0.2 | 0.15 | 0.21 | 0.14 |
| IL10 | 0.21 | *** | 0.15 | 0.29 | |||||
| M1 Macrophage | IRF5 | 0.31 | *** | 0.31 | *** | 0.13 | 0.37 | 0.07 | 0.63 |
| COX2(PTGS2) | 0.078 | 0.14 | − 0.032 | 0.82 | |||||
| M2 Macrophage | CD163 | 0.15 | * | 0.12 | 0.018 | 0.18 | 0.22 | 0.25 | 0.078 |
| MS4A4A | 0.16 | * | 0.22 | 0.12 | |||||
| Neutrophils | FPR1 | 0.25 | *** | 0.37 | *** | − 0.035 | 0.81 | 0.15 | 0.3 |
| SIGLEC5 | 0.31 | *** | − 0.011 | 0.94 | |||||
| CSF3R | 0.22 | *** | 0.11 | 0.43 | |||||
| FCGR3B | 0.05 | 0.34 | 0.17 | 0.24 | |||||
| CEACAM3 | 0.19 | ** | 0.064 | 0.66 | |||||
| CD116(ITGAM) | 0.33 | *** | 0.36 | 0.011 | |||||
| Central memory T cell | CCR7 | 0.063 | 0.23 | 0.12 | 0.023 | 0.37 | * | 0.38 | * |
| SELL | 0.1 | 0.055 | 0.24 | 0.092 | |||||
| IL7R | 0.042 | 0.43 | 0.31 | 0.031 | |||||
| Resident memory T cell | CD69 | 0.063 | 0.23 | 0.28 | *** | 0.3 | 0.037 | 0.25 | 0.075 |
| ITGAE | 0.21 | *** | − 0.046 | 0.75 | |||||
| CXCR6 | 0.12 | 0.02 | 0.25 | 0.085 | |||||
| MYADM | 0.27 | *** | − 0.092 | 0.52 | |||||
| Exhausted T cell | TIM-3(HAVCR2) | 0.14 | * | 0.3 | *** | 0.15 | 0.31 | 0.37 | * |
| PD-1 (PDCD1) | 0.15 | * | 0.46 | ** | |||||
| TIGIT | 0.21 | *** | 0.34 | 0.015 | |||||
| LAG3 | 0.16 | * | 0.15 | 0.31 | |||||
| CXCL13 | 0.085 | 0.1 | 0.28 | 0.049 | |||||
| LAYN | 0.12 | 0.024 | 0.26 | 0.073 | |||||
| Resting Treg T cell | FOXP3 | 0.011 | 0.84 | 0.18 | ** | 0.29 | 0.038 | 0.47 | ** |
| IL2RA | 0.2 | *** | 0.18 | 0.21 | |||||
| Effector Treg T cell | CTLA4 | 0.21 | *** | 0.27 | *** | 0.36 | * | 0.4 | * |
| CCR8 | 0.23 | *** | 0.37 | * | |||||
| TNFRSF9 | 0.018 | 0.74 | 0.36 | * | |||||
Cor, ρ value of Spearman's correlation. Tumor, single gene marker correlation analysis in LIHC tissue. Normal, single gene marker correlation analysis in normal tissue; Cor, ρ-value upon Spearman correlation. Significance levels: *P < 0.05; **P < 0.01; ***P < 0.001
Expression levels of MKI67 in LIHC
This study also examines MKI67 levels in diverse LIHC immune subtypes in the TISIDB. As a result, we detected MKI67 expression in 4 subtypes, namely, C1 (wound healing), C2 [interferon γ (IFN-γ) dominance], C3 (inflammation), C4 (lymphocyte depletion), C5 (immunological quiet), and C6 (TGF-b dominance). The greatest MKI67 expression was detected in the C1 type, whereas the lowest was measured in the C3 type (Fig. 5a). We also measured MKI67 levels in diverse LIHC molecular subtypes (iCluster:1, iCluster:2, and iCluster:3) in TISIDB [44]. According to our results, the greatest and lowest MKI67 levels were found in the iCluster: 1 and iCluster: 2 subtypes, respectively (Fig. 5b). Furthermore, MKI67 was closely associated with the tumour immune microenvironment (TIME). Shmulevich’s work revealed that six immune subtypes were clustered for cancer [45]. According to the GEPIA database, in comparison with LIHC at diverse stages, the upregulated level was detected at stage III, while the downregulated level was detected at stages I and IV (Fig. 5c). As revealed by HPA-based analysis, more intense MKI67 staining was detected within LIHC samples than in noncarcinoma samples (Fig. 5d). In addition, according to MEXPRESS-based analysis, MKI67 levels were related to sample type, simplified tumour stage, and fibrosis risk score (Fig. 5e).
Fig. 5.
MKI67 expression levels in liver hepatocellular carcinoma. a MKI67 expression in TISIDB-derived LIHC having diverse molecular subtypes; b MKI67 expression in TISIDB-derived LIHC having diverse immune subtypes; c MKI67 expression in GEPIA-derived LIHC of diverse stages (MKI67 gene levels were calculated relative to the log counts per million mapped reads (log2CPM) in (a, b) and log2(TPM + 1) in (c); d immunohistochemistry (IHC) for MKI67 based on the HPA database; e MKI67 level related to PR/ER status and sample type. T: Protein levels of MKI67 in tumor tissue (staining: high; intensity: strong, quantity: 75–25%) https://www.proteinatlas.org/ENSG00000148773-MKI67/pathology/liver+cancer#img; N: Protein levels of MKI67 in normal tissue (staining: Not detected; intensity: weak, quantity: < 25%) https://www.proteinatlas.org/ENSG00000148773-MKI67/tissue/liver#img
MKI67 co-expression networks within LIHC
To further understand MKI67’s biological significance in LIHC, LinkedOmics of the “LinkFinder” module was adopted to check the MKI67 coexpression pattern. Figure 6a reveals that 13,073 genes (red dots) showed a positive correlation with MKI67, whereas 6848 (green dots) showed a negative correlation (P < 0.05). Figure 6b, c display the heatmaps for the 50 most significant MKI67-related genes (both positive and negative). According to the GSEA-annotated GO terms, MKI67 coexpression genes were mainly associated with DNA recombination, chromosome segregation, and mitotic cell cycle phase transition in contrast to cellular amino acid metabolic process, translational elongation, steroid metabolic process, protein maturation, lipid catabolic process, cofactor biosynthetic process, and mitochondrial respiratory chain complex assembly (Fig. 6d). As revealed by KEGG analysis, these genes were primarily associated with the cell cycle, microRNAs in cancer, pyrimidine metabolism, spliceosomes, etc. (Fig. 6e). Notably, the 50 genes with the most significant positive correlation became high-risk markers for LIHC, among which 49 showed great HRs (HR, P < 0.05) (Fig. 6f). In contrast, 21 of the 50 genes with a significant negative correlation showed low HRs (P < 0.05) (Fig. 6g).
Fig. 6.
MKI67 co-expression genes in LIHC (LinkedOmics). a MKI67-related genes in LIHC discovered by Pearson’s test. Green and red dots indicate genes with significant negative and positive correlations with MKI67, separately; b, c Heatmaps displaying 50 most significant MKI67-related genes in LIHC (both positive and negative); d, e Significantly associated GO: BP annotations together with KEGG pathway analysis for MKI67 in LIHC; f, g survival heatmaps displaying the 50 most significant MKI67-related genes in LIHC (both positive and negative). Survival heatmaps displaying the log10 HRs of diverse genes. Blue and red blocks represent decreased and increased risks distinctly. Rectangles with frames represent significantly positive and negative outcomes upon prognosis analysis (P < 0.05). GO Gene Ontology, KEGG Kyoto Encyclopedia of Genes and Genomes, FDR false discovery rate, LIHC Liver hepatocellular carcinoma; h MKI67 and its predicted miRNAs (MKI67 presented in yellow circles and targeted miRNAs presented in blue circles. The interaction between the MKI67 and related miRNAs is represented in the form of lines)
Among these pathways, the hsa04151:PI3K-Akt signalling pathway, hsa04115: p53 signalling pathway, hsa04010: MAPK signalling pathway, hsa04310:Wnt signalling pathway, hsa04350:TGF-beta signalling pathway, and hsa04110: cell cycle pathway were involved in the tumorigenesis and pathogenesis of liver hepatocellular carcinoma (Fig. 7).
Fig. 7.
Hepatocellular carcinoma pathway regulated by the MKI67 alteration in LIHC
miRNA screening of regulatory MKI67
We applied miRWalk to screen the targeted miRNAs of MKI67. Then, we drew the miRNA-gene network using miRWalk. As illustrated in 6H, the interaction network consisted of MKI67 and 1003 miRNAs. Moreover, the contribution level of the miRNAs to MKI67 is represented as the number of lines. Additionally, the top 20 miRNAs targeting MKI67 are presented in Fig. 6h.
Discussion
Risk factors for HCC include viral hepatitis infection, alcoholism, autoimmune diseases, and cholestasis [46–49]. The combination of FFAs and cholesterol induces severe inflammation and steatohepatitis and, at the cellular level, impairs mitochondrial function and biogenesis [50]. Recently, medical experts believe that obesity-related nonalcoholic steatohepatitis (NASH) or nonalcoholic fatty liver disease (NAFLD) is the main cause of HCC [51, 52]. Some scholars have shown that sartan significantly improved the recurrence time of liver cancer patients after rf ablation but did not improve the five-year survival rate of LIHC. Stiffness declines significantly after in hepatitis virus patients undergoing antiviral therapy [53, 54]. Plenty of evidence supports the role of MKi67 in diagnosing cancer [17–20]. MKi67, located on chromosome 10q25-ter, can encode 2 isoforms (345 and 395 kDa, respectively) [8–10]. The positive MKI67 rate (referred to as the labelling index) is suggested to be related to the clinicopathological characteristics or the survival of cancers, such as LIHC [21]. MK167 upregulation in cancer tissues is related to early cancer relapse and advanced tumour grade, according to an article enrolling LIHC cases receiving surgery [22, 23]. In addition, p53 and MK167 staining are used extensively for predicting LIHC prognosis postoperatively or even after liver transplantation [24, 25]. Such results indicate the vital role of MKI67 in tumour invasion, migration, and development. Nonetheless, the association of MKI67 levels with T cell activity, prognosis, and immune infiltration in diverse cancers remains unclear. We obtained cancer samples from some large-scale databases for analysis. According to our results, MKI67 expression is related to the survival of diverse cancers, such as LIHC. Furthermore, MKI67 coexpression genes were suggested to play a significant role in the prognosis prediction of cancer. More investigations suggest that MKI67 levels are related to the degree of immune infiltration within LIHC. Therefore, MKI67 was identified as a candidate prognostic biomarker for LIHC, which offers a new direction for understanding the association of MKI67 with T cell activity and immune infiltration.
The present study analysed MKI67 expression with prognosis expression profiles in different types of cancers based on individual datasets from Oncomine and 33 TCGA-derived cancers in GEPIA2. MKI67 expression levels in cancer and noncarcinoma samples were studied. Based on the Oncomine database, MKI67 showed high expression in bladder cancer, CNS and brain, breast cancer (BC), colorectal cancer (CRC), cervical cancer, oesophageal cancer (EC), head and neck cancer (HNC), gastric cancer (GC), liver cancer, ovarian cancer, lung cancer (LC), lymphoma, sarcoma, and pancreatic cancer compared with noncarcinoma tissues. However, according to particular datasets, MKI67 was expressed at low levels in CNS and brain cancers, BC, leukaemia, and kidney cancer (Fig. 1a). However, TCGA-based data analysis revealed high expression of MKI67 in BLCA, KIRC, BRCA, CHOL, COAD, ESCA, HNC, KICH, KIRP, LUSC, LUAD, LIHC, READ, PRAD, STAD, UCEC, and THCA and low expression within SKCM relative to noncarcinoma samples (Fig. 1b). Human Protein Atlas data further verified MKI67 upregulation in liver cancer through immunohistochemistry (Fig. 5d). Different MKI67 levels measured in diverse cancers from distinct databases can provide a novel data extraction method and help illustrate the mechanisms associated with diverse biological characteristics. However, as discovered from the above databases, MKI67 expression is related to the prognosis of LIHC, KIRP, ACC, BRCA, and LUAD. According to TCGA-based analysis, MKI67 upregulation is associated with many cancers’ dismal prognostic outcomes (PAAD, SARC, KIRP, and UVM). However, in STAD and ESCA, MKI67 downregulation predicted favourable patient survival. Concerning the GEPIA2 datasets, MKI67 upregulation might serve as a factor to independently predict the dismal survival of LIHC and ACC (Fig. 2b, d). According to the Kaplan–Meier Plotter database, MKI67 upregulation related to high HRs predicted OS and PFS for LIHC, BRCA, and LUAD (Fig. 2f, g, i). Collectively, the above results suggest the feasibility of MKI67 as a prognostic biomarker for LIHC. The present work evaluated the association of MKI67 with immunity based on the TISIDB database. According to our results, MKI67L is closely related to lymphocytes (including monocytes, type 2 helper cells, and activated CD4 T cells), immunostimulators (such as MICB, CD276, and TNFSF4), immunoinhibitors (including KDR and PDCD1 CTLA4), and MHC molecules (including B2M and HLA-C HLA-E). Therefore, MKI67 can act as a novel target to investigate immune escape in LIHC cells and a therapeutic target for anti-LIHC immunotherapy.
Liver cancer is not a single disorder but is further classified into numerous molecular subtypes. According to the TISIDB database analysis, the MKI67 gene displayed the greatest expression within the iCluster: 1 subtype, while iCluster: 3 type ranked second, whereas MKI67 was expressed at low levels in the iCluster: 2 type. Differential MKI67 expression in LIHC of diverse immune subtypes was detected. The results suggested that the C1 type displayed the most significant expression relative to those in the remaining three subtypes. Comprehensive analysis of MKI67 levels within diverse LIHC subtypes from diverse databases suggests the vital role of MKI67 in microenvironment characteristics.
Given that MKI67 has an important effect on the immune system and predicts the prognosis of LIHC, this study examined the associations of MKI67 with immune infiltration degrees within LIHC (Fig. 4a). MKI67 upregulation is closely related to the degree of immune infiltration of many immune cell subpopulations, such as B cells, neutrophils, macrophages, DCs, CD4+ T cells and CD8+ T cells (Fig. 4a). The varied SCNA for MKI67 did not significantly affect the immune infiltration degrees of CD4+ T cells or B cells within LIHC, and considerable attention was devoted to the close links between MKI67 and immune cells (Fig. 4c). According to subsequent analyses of the relationships of MKI67 with immune cell gene markers, MKI67 interacted with many immune cells and diverse functional T cells, including central memory T cells, effector T cells, and exhausted T cells (Tables 2 and 3). T cell exhaustion accounts for a leading reason for weak anticancer immunity [, and the measures for preventing T cell exhaustion represent a key role in anticancer immunotherapy [55–57]. As revealed by our results, MKI67 upregulation showed a positive correlation with several critical genes related to exhausted T cells, such as TIM-3, PD-1, LAG3, and TIGIT. These act as therapeutic targets for immunotherapy [58, 59].
Interestingly, this study suggested that the dual role of MKI67 is specific. It also predicted that MKI67 upregulation has prognostic outcomes for several cancers, including LIHC, whereas inducing T cell exhaustion shows ineffective anticancer immunity. The above two inverse trends were not contradictory. Recently, some articles have shed more light on the exact related mechanisms. Consequently, MKI67 plays a critical and distinct role in normal immunity development and regulating the TME, which is essential to identify a specific stage.
Conclusion
The results in this work indicate the potential of MKI67 as a prognostic biomarker for several cancers, particularly LIHC. MKI67 upregulation is associated with more significant immune infiltration degrees of B cells, CD4+ T cells, CD8+ T cells, neutrophils, DCs, and many functional T cells. MKI67 exerts a stimulating effect on immunity, and it also shows a high correlation with exhausted T cells, which may serve as an essential factor to promote T cell exhaustion within LIHC. The detection of MKI67 levels possibly adds to prognosis prediction and modulation of MKI67 levels in exhausted T cells, which offers novel management for enhancing the efficacy of anti-LIHC immunotherapy.
Supplementary Information
Additional file 1: Supplementary Figure 1. Correlation of MKI67 expression with prognostic values in diverse type cancers. Supplementary Figure 2. Correlation of MKI67 expression with immune infiltration level in diverse type cancers via TIMER database. Supplementary Table 1. Mki67 expression in cancers verus normal tissue in oncomine database.
Acknowledgements
We acknowledge TCGA, TIMER, and so on databases for providing their platforms and contributors for uploading their meaningful datasets. The authors sincerely thank all participants involved in this study.
Abbreviations
- MKI67
Marker of proliferation Ki-67
- LIHC
Liver hepatocellular carcinoma
- CAM
Cell adhesion molecule
- TIN
Tumor-infiltrating neutrophil
- TAM
Tumor-associated macrophages
- KM
Kaplan–Meier
- DC
Dendritic cell
- SCNA
Somatic copy number alteration
- FDR
False discovery rate
- FC
Fold-change
- LNM
Lymph node metastasis
- TME
Tumor microenvironment
Authors' contributions
SYW and PL conceived the project and wrote the manuscript. SYW, PL, LYY, QYZ, and ZYX participated in data analysis, discussion, and language editing. JD and HTS reviewed the manuscript. All authors read and approved the final manuscript.
Funding
Not applicable.
Availability of data and materials
The datasets used for the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
All methods of this study were carried out in accordance with relevant guidelines and regulations. The data of this study are from TCGA, TIMER, Oncomine, GEPIA2 database, and do not involve animal experiments and human specimens, no ethics-related issues.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Shi-yi Wu and Pan Liao have contributed equally to this work
Contributor Information
Jie Dong, Email: dongjie202107@163.com.
Hong-tao Sun, Email: sunhongtao2019@163.com.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag HB, Kanwal F. Epidemiology of hepatocellular carcinoma in the United States: Where are we? Where do we go? Hepatology. 2014;60:1767–1775. doi: 10.1002/hep.27222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lohitesh K, Chowdhury R, Mukherjee S. Resistance a major hindrance to chemotherapy in hepatocellular carcinoma: an insight. Cancer Cell Int. 2018;18:44. doi: 10.1186/s12935-018-0538-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DiStefano JK, Davis B. Diagnostic and prognostic potential of AKR1B10 in human hepatocellular carcinoma. Cancers (Basel) 2019 doi: 10.3390/cancers11040486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scholzen T, Gerdes J. The Ki-67 protein: from the known and the unknown. J Cell Physiol. 2000;182:311–322. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 6.Isola J, Helin H, Kallioniemi OP. Immunoelectron-microscopic localization of a proliferation-associated antigen Ki-67 in MCF-7 cells. Histochem J. 1990;22:498–506. doi: 10.1007/BF01007235. [DOI] [PubMed] [Google Scholar]
- 7.Verheijen R, Kuijpers HJ, Schlingemann RO, Boehmer AL, van Driel R, Brakenhoff GJ, Ramaekers FC. Ki-67 detects a nuclear matrix-associated proliferation-related antigen. I. Intracellular localization during interphase. J Cell Sci. 1989;92(Pt 1):123–130. doi: 10.1242/jcs.92.1.123. [DOI] [PubMed] [Google Scholar]
- 8.Schlüter C, Duchrow M, Wohlenberg C, Becker MH, Key G, Flad HD, Gerdes J. The cell proliferation-associated antigen of antibody Ki-67: A very large, ubiquitous nuclear protein with numerous repeated elements, representing a new kind of cell cycle-maintaining proteins. J Cell Biol. 1993;123:513–522. doi: 10.1083/jcb.123.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fonatsch C, Duchrow M, Rieder H, Schlüter C, Gerdes J. Assignment of the human Ki-67 gene (MK167) to 10q25-qter. Genomics. 1991;11:476–477. doi: 10.1016/0888-7543(91)90163-9. [DOI] [PubMed] [Google Scholar]
- 10.Duchrow M, Schlüter C, Wohlenberg C, Flad HD, Gerdes J. Molecular characterization of the gene locus of the human cell proliferation-associated nuclear protein defined by monoclonal antibody Ki-67. Cell Prolif. 1996;29:1–12. doi: 10.1111/j.1365-2184.1996.tb00090.x. [DOI] [PubMed] [Google Scholar]
- 11.Du Manoir S, Guillaud P, Camus E, Seigneurin D, Brugal G. Ki-67 labeling in postmitotic cells defines different Ki-67 pathways within the 2c compartment. Cytometry. 1991;12:455–463. doi: 10.1002/cyto.990120511. [DOI] [PubMed] [Google Scholar]
- 12.Gerdes J, Lemke H, Baisch H, Wacker HH, Schwab U, Stein H. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J Immunol. 1984;133:1710–1715. [PubMed] [Google Scholar]
- 13.Rioux-Leclercq N, Turlin B, Bansard J, Patard J, Manunta A, Moulinoux JP, et al. Value of immunohistochemical Ki-67 and p53 determinations as predictive factors of outcome in renal cell carcinoma. Urology. 2000;55:501–505. doi: 10.1016/s0090-4295(99)00550-6. [DOI] [PubMed] [Google Scholar]
- 14.Visapää H, Bui M, Huang Y, Seligson D, Tsai H, Pantuck A, et al. Correlation of Ki-67 and gelsolin expression to clinical outcome in renal clear cell carcinoma. Urology. 2003;61:845–850. doi: 10.1016/s0090-4295(02)02404-4. [DOI] [PubMed] [Google Scholar]
- 15.Karamitopoulou E, Perentes E, Tolnay M, Probst A. Prognostic significance of MIB-1, p53, and bcl-2 immunoreactivity in meningiomas. Hum Pathol. 1998;29:140–145. doi: 10.1016/s0046-8177(98)90224-6. [DOI] [PubMed] [Google Scholar]
- 16.Geyer FC, Rodrigues DN, Weigelt B, Reis-Filho JS. Molecular classification of estrogen receptor-positive/luminal breast cancers. Adv Anat Pathol. 2012;19:39–53. doi: 10.1097/PAP.0b013e31823fafa0. [DOI] [PubMed] [Google Scholar]
- 17.Zini L, Porpiglia F, Fassnacht M. Contemporary management of adrenocortical carcinoma. Eur Urol. 2011;60:1055–1065. doi: 10.1016/j.eururo.2011.07.062. [DOI] [PubMed] [Google Scholar]
- 18.Zizi-Sermpetzoglou A, Moustou E, Petrakopoulou N, Arkoumani E, Tepelenis N, Savvaidou V. Atypical polypoid adenomyoma of the uterus. A case report and a review of the literature. Eur J Gynaecol Oncol. 2012;33:118–121. [PubMed] [Google Scholar]
- 19.Viale G. Pathological work up of the primary tumor: getting the proper information out of it. Breast. 2011;20(Suppl 3):S82–S86. doi: 10.1016/S0960-9776(11)70300-9. [DOI] [PubMed] [Google Scholar]
- 20.Bertolini M, Sobue T, Thompson A, Dongari-Bagtzoglou A. Chemotherapy induces oral mucositis in mice without additional noxious stimuli. Transl Oncol. 2017;10:612–620. doi: 10.1016/j.tranon.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo Y, Ren F, Liu Y, Shi Z, Tan Z, Xiong H, et al. Clinicopathological and prognostic significance of high Ki-67 labeling index in hepatocellular carcinoma patients: a meta-analysis. Int J Clin Exp Med. 2015;8:10235–10247. [PMC free article] [PubMed] [Google Scholar]
- 22.D'Errico A, Grigioni WF, Fiorentino M, Baccarini P, Grazi GL, Mancini AM. Overexpression of p53 protein and Ki67 proliferative index in hepatocellular carcinoma: an immunohistochemical study on 109 Italian patients. Pathol Int. 1994;44:682–687. doi: 10.1111/j.1440-1827.1994.tb02947.x. [DOI] [PubMed] [Google Scholar]
- 23.Nakanishi K, Sakamoto M, Yamasaki S, Todo S, Hirohashi S. Akt phosphorylation is a risk factor for early disease recurrence and poor prognosis in hepatocellular carcinoma. Cancer. 2005;103:307–312. doi: 10.1002/cncr.20774. [DOI] [PubMed] [Google Scholar]
- 24.Guzman G, Alagiozian-Angelova V, Layden-Almer JE, Layden TJ, Testa G, Benedetti E, et al. p53, Ki-67, and serum alpha feto-protein as predictors of hepatocellular carcinoma recurrence in liver transplant patients. Mod Pathol. 2005;18:1498–1503. doi: 10.1038/modpathol.3800458. [DOI] [PubMed] [Google Scholar]
- 25.Stroescu C, Dragnea A, Ivanov B, Pechianu C, Herlea V, Sgarbura O, et al. Expression of p53, Bcl-2, VEGF, Ki67 and PCNA and prognostic significance in hepatocellular carcinoma. J Gastrointest Liver Dis. 2008;17:411–417. [PubMed] [Google Scholar]
- 26.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li B, Li JZ. A general framework for analyzing tumor subclonality using SNP array and DNA sequencing data. Genome Biol. 2014;15:473. doi: 10.1186/s13059-014-0473-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Danaher P, Warren S, Dennis L, D'Amico L, White A, Disis ML, et al. Gene expression markers of Tumor Infiltrating Leukocytes. J Immunother Cancer. 2017;5:18. doi: 10.1186/s40425-017-0215-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhattacharya S, Dunn P, Thomas CG, Smith B, Schaefer H, Chen J, et al. ImmPort, toward repurposing of open access immunological assay data for translational and clinical research. Sci Data. 2018;5:180015. doi: 10.1038/sdata.2018.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nirmal AJ, Regan T, Shih BB, Hume DA, Sims AH, Freeman TC. Immune cell gene signatures for profiling the microenvironment of solid tumors. Cancer Immunol Res. 2018;6:1388–1400. doi: 10.1158/2326-6066.CIR-18-0342. [DOI] [PubMed] [Google Scholar]
- 33.Guo X, Zhang Y, Zheng L, Zheng C, Song J, Zhang Q, et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat Med. 2018;24:978–985. doi: 10.1038/s41591-018-0045-3. [DOI] [PubMed] [Google Scholar]
- 34.Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagy Á, Lánczky A, Menyhárt O, Győrffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8:9227. doi: 10.1038/s41598-018-27521-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ru B, Wong CN, Tong Y, Zhong JY, Zhong SSW, Wu WC, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35:4200–4202. doi: 10.1093/bioinformatics/btz210. [DOI] [PubMed] [Google Scholar]
- 37.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 38.Koch A, de Meyer T, Jeschke J, van Criekinge W. MEXPRESS: Visualizing expression, DNA methylation and clinical TCGA data. BMC Genomics. 2015;16:636. doi: 10.1186/s12864-015-1847-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: Analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46:D956–D963. doi: 10.1093/nar/gkx1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sticht C, de La Torre C, Parveen A, Gretz N. miRWalk: An online resource for prediction of microRNA binding sites. PLoS ONE. 2018;13:e0206239. doi: 10.1371/journal.pone.0206239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ravelli A, Roviello G, Cretella D, Cavazzoni A, Biondi A, Cappelletti MR, et al. Tumor-infiltrating lymphocytes and breast cancer: beyond the prognostic and predictive utility. Tumour Biol. 2017;39:1010428317695023. doi: 10.1177/1010428317695023. [DOI] [PubMed] [Google Scholar]
- 42.Azimi F, Scolyer RA, Rumcheva P, Moncrieff M, Murali R, McCarthy SW, et al. Tumor-infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma. J Clin Oncol. 2012;30:2678–2683. doi: 10.1200/JCO.2011.37.8539. [DOI] [PubMed] [Google Scholar]
- 43.Ohtani H. Focus on TILs: prognostic significance of tumor infiltrating lymphocytes in human colorectal cancer. Cancer Immun. 2007;7:4. [PMC free article] [PubMed] [Google Scholar]
- 44.Konecny GE, Wang C, Hamidi H, Winterhoff B, Kalli KR, Dering J, et al. Prognostic and therapeutic relevance of molecular subtypes in high-grade serous ovarian cancer. J Natl Cancer Inst. 2014 doi: 10.1093/jnci/dju249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang T-H, et al. The immune landscape of cancer. Immunity. 2018;48:812–830.e14. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schaper M, Rodriguez-Frias F, Jardi R, Tabernero D, Homs M, Ruiz G, et al. Quantitative longitudinal evaluations of hepatitis delta virus RNA and hepatitis B virus DNA shows a dynamic, complex replicative profile in chronic hepatitis B and D. J Hepatol. 2010;52:658–664. doi: 10.1016/j.jhep.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 47.de Martel C, Maucort-Boulch D, Plummer M, Franceschi S. World-wide relative contribution of hepatitis B and C viruses in hepatocellular carcinoma. Hepatology. 2015;62:1190–1200. doi: 10.1002/hep.27969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fan J-H, Wang J-B, Jiang Y, Xiang W, Liang H, Wei W-Q, et al. Attributable causes of liver cancer mortality and incidence in china. Asian Pac J Cancer Prev. 2013;14:7251–7256. doi: 10.7314/apjcp.2013.14.12.7251. [DOI] [PubMed] [Google Scholar]
- 49.Bosetti C, Turati F, La Vecchia C. Hepatocellular carcinoma epidemiology. Best Pract Res Clin Gastroenterol. 2014;28:753–770. doi: 10.1016/j.bpg.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 50.Bellanti F, Villani R, Tamborra R, Blonda M, Iannelli G, Di Bello G, et al. Synergistic interaction of fatty acids and oxysterols impairs mitochondrial function and limits liver adaptation during nafld progression. Redox Biol. 2018;15:86–96. doi: 10.1016/j.redox.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Petrick JL, Braunlin M, Laversanne M, Valery PC, Bray F, McGlynn KA. International trends in liver cancer incidence, overall and by histologic subtype, 1978–2007. Int J Cancer. 2016;139:1534–1545. doi: 10.1002/ijc.30211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Facciorusso A. The influence of diabetes in the pathogenesis and the clinical course of hepatocellular carcinoma: recent findings and new perspectives. Curr Diabetes Rev. 2013;9:382–386. doi: 10.2174/15733998113099990068. [DOI] [PubMed] [Google Scholar]
- 53.Facciorusso A, Del Prete V, Crucinio N, Muscatiello N, Carr BI, Di Leo A, Barone M. Angiotensin receptor blockers improve survival outcomes after radiofrequency ablation in hepatocarcinoma patients. J Gastroenterol Hepatol. 2015;30:1643–1650. doi: 10.1111/jgh.12988. [DOI] [PubMed] [Google Scholar]
- 54.Facciorusso A, Del Prete V, Turco A, Buccino RV, Nacchiero MC, Muscatiello N. Long-term liver stiffness assessment in hepatitis C virus patients undergoing antiviral therapy: results from a 5-year cohort study. J Gastroenterol Hepatol. 2018;33:942–949. doi: 10.1111/jgh.14008. [DOI] [PubMed] [Google Scholar]
- 55.Zarour HM. Reversing T-cell dysfunction and exhaustion in cancer. Clin Cancer Res. 2016;22:1856–1864. doi: 10.1158/1078-0432.CCR-15-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol. 2015;15:486–499. doi: 10.1038/nri3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wherry EJ. T cell exhaustion. Nat Immunol. 2011;12:492–499. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- 58.Anderson AC, Joller N, Kuchroo VK. Lag-3, Tim-3, and TIGIT: co-inhibitory receptors with specialized functions in immune regulation. Immunity. 2016;44:989–1004. doi: 10.1016/j.immuni.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science. 2018;359:1350–1355. doi: 10.1126/science.aar4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Figure 1. Correlation of MKI67 expression with prognostic values in diverse type cancers. Supplementary Figure 2. Correlation of MKI67 expression with immune infiltration level in diverse type cancers via TIMER database. Supplementary Table 1. Mki67 expression in cancers verus normal tissue in oncomine database.
Data Availability Statement
The datasets used for the current study are available from the corresponding author on reasonable request.