Abstract
Background
Metastasis occurs in the majority of pancreatic ductal adenocarcinoma (PDAC) patients at diagnosis or following resection. Patients with liver metastasis and those with lung metastasis have significantly different prognosis. Here, we sought to understand how cancer-associated fibroblasts (CAFs) play roles in the development of organ-specific metastasis.
Methods
PDAC tumor cell lines established from the primary tumors with liver and lung metastasis potentials, respectively, in Kras/p53 mutation conditional knock-in (KPC) mice were co-cultured with matched CAFs or mouse mesenchymal stem cells. CAFs were isolated from metastases and subjected to DNA methylation and whole transcriptomic RNA sequencing analysis.
Results
The ability of mouse PDAC tumor cell lines in developing liver or lung-specific metastases was demonstrated in orthotopic models. Tumor cells associated with liver metastasis potential, but not those associated with lung metastasis potential, induced the methylation of metabolism genes including NQO1 and ALDH1a3 and subsequent downregulated mRNA expression of a broader group of metabolism genes in CAFs. DNA methylation and downregulation of metabolism genes in CAFs in liver metastasis, but not those in lung metastasis, appeared to be regulated by DNA methyltransferase. Tumor cells associated with liver metastasis potential, but not those associated with lung metastasis potential, induce inflammatory CAF (iCAF) signatures. CAFs from liver metastasis demonstrated a more homogenous iCAF phenotype, whereas CAFs from lung metastasis maintained the heterogeneity.
Conclusions
PDAC with organ-specific metastatic potentials has different capacities in inducing methylation of metabolism genes in CAFs, modulating CAF phenotypes, and resulting in different levels of heterogeneity of CAFs in different metastatic niches.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13045-021-01203-1.
Keywords: DNA methylation, Cancer-associated fibroblast, Heterogeneity, Organ-specific metastasis, Pancreatic cancer
To the editor
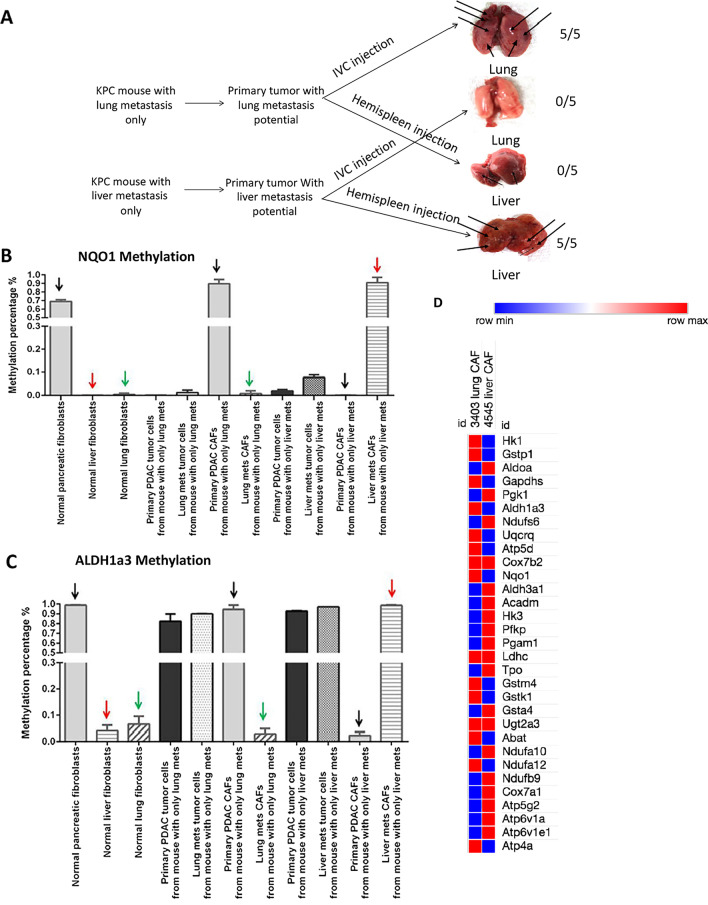
Our clinical observations revealed disease heterogeneity among pancreatic ductal adenocarcinoma (PDAC) patients with different distant metastatic sites, resulting in distinct clinical outcomes [1]. We sought to investigate the mechanism of organ-specific metastasis in the transgenic KPC mouse model [2] and established two cell lines from primary tumors of KPC mouse with liver metastasis only and KPC mouse with lung metastasis only (Fig. 1a; Additional file 1: Fig. S1A). Previously, we demonstrated that PDAC tumor cells induce DNA methylation globally in CAFs [3, 4] and in macrophages [5] including a wide range of metabolism genes resulting in their downregulation. We chose to examine methylation of ALDH1a3 in the glucose metabolism pathway and NQO-1 in the oxidative phosphorylation (OXIPHOS) pathway (Additional file 1: Fig. S1B) and mRNA expression of their related genes (Additional file 1: Table S1). CAFs from liver metastases and lung metastases demonstrated different levels of methylation and mRNA expression in these genes (Fig. 1b–d).
Fig. 1.
Distinct DNA methylation patterns in metabolism genes were observed in CAFs isolated from liver metastasis versus lung metastasis. a Establishment of primary mouse PDAC tumor cell lines with organ-specific metastasis potentials. Different KPC tumor cell lines were established from primary pancreatic tumor of KPC mice that developed liver metastasis only and lung metastasis only, respectively. These KPC cell lines were tested for their capacities to generate metastasis in mice through the hemispleen injection model of liver metastasis and the inferior vena cava injection model of lung metastases as described previously [6, 7]. Five mice per group were used to test each KPC cell line using both mouse models, respectively. Numbers of mice that developed large metastatic lesions were indicated. Longer arrows indicate large metastatic lesions; shorter arrows indicate small metastatic lesions. Note that the KPC cell line established from primary tumor with liver metastases consistently gave rise to liver metastasis macroscopically but not lung metastasis (5/5), whereas the KPC cell line established from primary tumor with lung metastases consistently gave rise to lung metastasis (5/5). The lung met tumor cell line was only able to develop a small number of macroscopically visualized metastatic foci when the cell line was injected by the hemispleen technique (Additional file 1: Fig. S1A). Liver and lung were examined microscopically and confirmed that the liver met tumor cell line only developed micro-metastases in lung (Additional file 1: Fig. S1A). b and c Percentages of methylation in the ALDH1a3 and NQO-1 gene were measured in tumor cells and CAFs isolated from primary pancreatic tumor and liver or lung metastases from KPC mouse with only liver or lung metastasis, respectively. Four different CAFs including: (1) CAFs isolated from primary KPC tumors of a mouse that developed liver metastasis only; (2) CAFs isolated from primary KPC tumors of a mouse that developed lung metastasis only; (3) CAFs isolated from liver metastases of a KPC mouse that developed liver metastasis only (liver mets CAFs); (4) CAFs isolated from lung metastases of a KPC mouse that developed lung metastasis only (lung mets CAFs) were all compared to normal pancreas (indicated with black arrow), liver (indicated with red arrow) and lung (indicated with green arrow) fibroblasts. Note that, for both NQO-1 and ALDH1a3 genes, DNA methylation levels were found to be elevated in liver mets CAFs from KPC mouse that developed liver metastasis compared to normal liver fibroblasts. NQO-1 and ALDH1a3 did not show an elevated DNA methylation in lung mets CAFs from KPC mouse that developed lung metastasis compared to normal lung fibroblasts, even though they demonstrated a high-level DNA methylation in CAFs from primary tumors that developed lung metastasis only. The methylation of NQO-1 and ALDH1a3 remained the same in tumor cells from both primary tumors and liver/lung metastases although the methylation level of ALDH1a3 was higher than that of NOQ-1 in tumor cells. Methylation percentage was quantified by MethySYBR real-time PCR (MSP). Black, red, green arrows indicate different comparison groups. Triplicate experimental results are presented as mean ± SEM. *Unpaired t test, p < 0.05. Independent experiments were conducted twice. d Heatmap was generated from the RNA sequencing analysis of mouse CAFs isolated from a KPC mouse that developed lung metastasis (3404LungCAF) only and from a KPC mouse that developed liver metastasis only (4545LiverCAF) to compare expression of selected metabolic genes. Mouse homologs of genes in the ALDH1-associated metabolism pathway and NQO-1-associated oxidative phosphorylation pathway previously found to have a significantly increased methylation level and also a significantly decreased mRNA expression level in CAFs following co-culture with human PDAC tumor cells [3] were selected. Heatmap was generated using transcripts per million (TPM) scores
We hypothesized that different methylation patterns in CAFs from different metastatic sites were induced by PDAC tumor cells with different metastasis potentials. We thus studied tumor-induced DNA methylation in the mouse mesenchymal stem cell line (moMSC), the precursor of CAFs, by co-culturing with three different primary PDAC tumor cell lines derived from a KPC mouse without metastasis (PancPrimaryTumorCell), a KPC mouse with liver metastasis only (LiverMetTumorCell), and a KPC mouse with lung metastasis only (LungMetTumorCell). Remarkably, different tumor cell lines showed differential capacities in inducing DNA methylation on NQO-1 and ALDH1a3 in moMSCs (Additional file 1: Fig. S2A and B). mRNA expression of NQO-1 as well as other metabolism genes were also downregulated in moMSC co-cultured with LiverMetTumorCells (Additional file 1: Fig. S2C–E). These results suggested that different PDAC tumor cells show different capacities in possibly shaping metabolic states of CAFs in different metastatic sites. We also developed CAF lines from primary KPC tumor with liver metastasis only (4545PancCAF) or that with lung metastasis only (3403PancCAF) and co-cultured them with matched tumor cells. Induced DNA methylation changes in NQO1 and ALDH1a3 were observed at higher degrees in 4545PancCAF from tumor co-culture compared to its mono-culture than those in 3403PancCAF (Additional file 1: Fig. S2F). Consistently, NQO-1 expression was downregulated in 4545PancCAF from tumor co-culture (Additional file 1: Fig. S2G). ALDH1a3 expression was not downregulated in 4545PancCAF from tumor co-culture, suggesting that ALDH1a3 expression is regulated by a more complex mechanism (Additional file 1: Fig. S2H).
We next sought to understand if tumor-induced gene methylation occurred in the metastatic niches where tumor cells metastasized to. We orthotopically implanted KPC cells with both liver and lung metastasis potentials (Fig. 2a). The transplanted mice developed liver and/or lung metastases. NQO-1 methylation was elevated in CAFs isolated from liver metastases compared to normal liver fibroblasts (Fig. 2b), but remained at baseline levels in CAFs from lung metastases (Fig. 2c). Supporting this notion, tumor-induced methylation of NQO-1 and ALDH1a3 and downregulation of their mRNA expression or expression of their related metabolism genes in CAFs from liver metastasis could be reversed by treatment of DNA demethylating agent, decitabine, whereas methylation in CAFs from lung metastasis was not affected (Additional file 1: Fig. S3). Thus, although the primary tumors harbor an organ-specific metastatic potential, metastatic organs possibly harbor contributive factors for an organ-specific tumor-induced metabolism gene methylation.
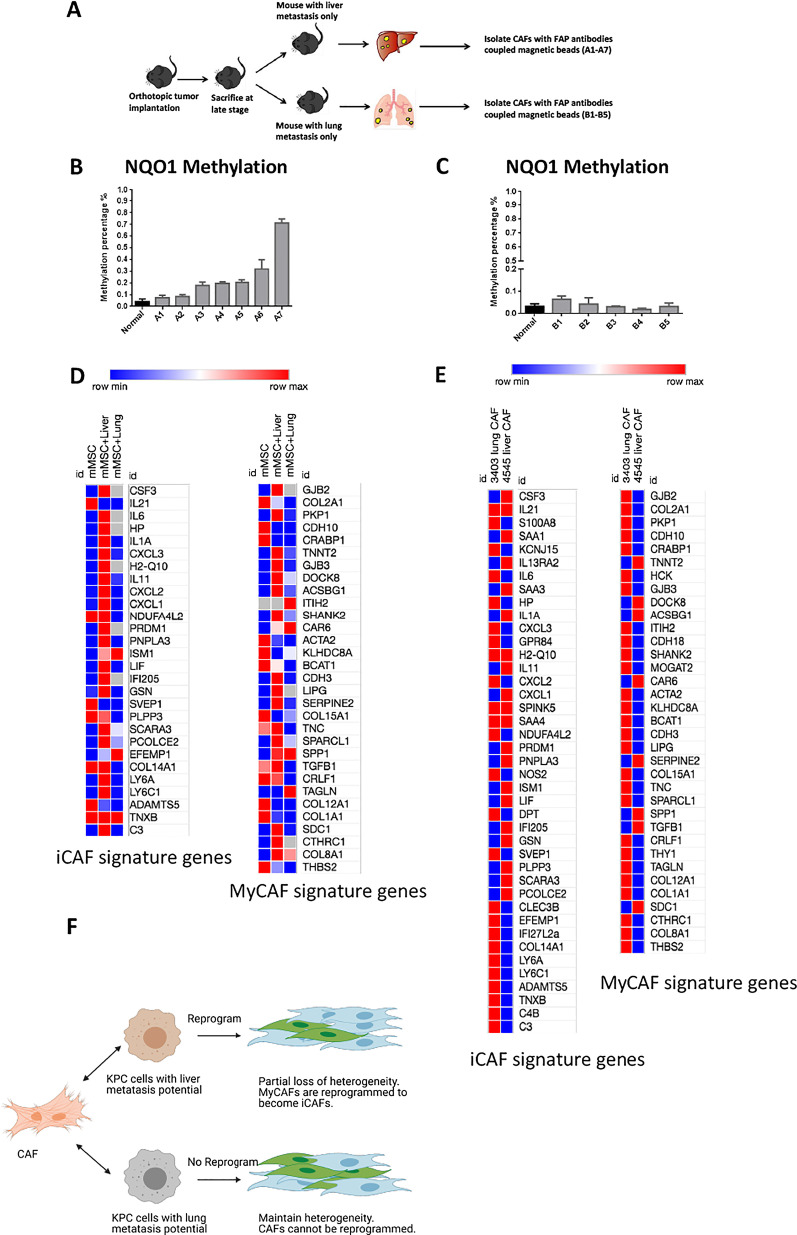
Fig. 2.
PDAC tumor cells with different organ-specific metastatic potentials showed different capacities in modulating the metabolism gene methylation in CAFs and also the heterogeneity of CAFs in metastatic sites. a The schema shows that wild-type C56Bl/6 mice orthotopically implanted with primary pancreatic KPC tumors spontaneously developed liver or lung metastases, which were dissected for CAF isolation by using magnetic beads coupled with anti-FAP antibodies. Two KPC cell lines from mice that developed both liver and lung metastases were orthotopically implanted in the pancreas of 40 mice (20 mice per cell line). As previously shown [6, 7], the majority of mice died from primary tumor growth without metastases. Of 40 mice, 7 of them developed liver metastasis only and 5 developed lung metastasis only. CAFs were isolated from dissected liver and lung metastases. b Percentage of methylation in the NQO-1 gene in CAFs from multiple liver metastases was measured by MSP. Methylation percentage of NQO-1 in normal liver fibroblasts was used as a baseline methylation level for comparison. A1-7: 7 mice developed liver metastasis spontaneously. Note that methylation levels of the NQO-1 gene were elevated in CAFs isolated from liver metastases compared to normal liver fibroblasts. c Percentage of methylation in the NQO1 gene in CAFs from multiple lung metastases was measured by MSP. Methylation percentage of NQO-1 in normal lung fibroblasts was used as a baseline methylation level for comparison. B1-5: 5 mice developed lung metastasis spontaneously. Note that the gene methylation levels of NQO-1 remained at baseline levels in CAFs isolated from lung metastases compared to normal lung fibroblasts. d Heatmap was generated using TPM scores based on the RNA sequencing analysis of mouse moMSC (mMSC) after co-cultured with Liver Met tumor cells (mMSC + liver) and Lung Met tumor cells (mMSC + lung) to compare the expression of iCAF and myCAF signature genes. Red color indicates upregulation in co-culture compared to mono-culture, blue color indicates downregulation in co-culture compared to mono-culture. e Heatmap was generated using TPM based on the RNA sequencing analysis of mouse CAFs isolated from liver metastasis (4545 liver CAF) and from lung metastasis only (3403 lung CAF) to compare the expression of iCAF and myCAF signature genes. f Diagram illustration shows that CAFs in liver metastasis are reprogrammed by PDAC cells with liver metastasis potential and subsequently lose part of their heterogeneity. Note that CAFs in lung metastasis are not reprogrammed by KPC cells with lung metastasis potential and thus maintain their heterogeneity. Image created using Biorender
Nevertheless, it was unknown whether CAFs from different metastatic niches are functionally different. The functions of the subpopulations of CAFs were previously characterized and showed different transcriptomic signatures [8–10]. In our RNA sequencing analysis, mono-cultured moMSCs showed low expression of the majority of inflammatory CAF (iCAF) signature genes (Additional file 1: Table S2) and high expression of some of myofibroblastic CAF (myCAF) signature genes (Fig. 2d). However, upon co-cultured with LiverMetTumorCells, moMSC was programmed to express essentially all iCAF signature genes and the majority of myCAF signature genes. However, expression of the majority of iCAF and myCAF genes in moMSC co-cultured with LungMetTumorCell remained low, suggesting that LungMetTumorCell was not able to program moMSC. As expected, CAFs from lung metastasis expresses both iCAF genes and myCAF genes (Fig. 2e). By contrast, CAFs from liver metastasis did not demonstrate a myCAF signature, but expressed some iCAF genes that were not upregulated in CAFs from lung metastasis. Taken together, CAFs in liver metastasis are more homogeneous likely as a result of reprogramming of myCAF into iCAF by tumor cells that metastasize to liver, whereas CAFs in lung metastasis remain to be heterogenous (Fig. 2f). A change in the heterogeneity of CAFs to a more homogeneous iCAF phenotype may be responsible for the aggressive feature of liver metastasis and is potentially targetable by anti-IL-1β antibody [11].
Supplementary Information
Additional file 1. Representative images of lung and liver metastases from KPC mice implanted with KPC tumor cells with lung and liver metatasis potentials and schema of how represntative metabolism genes were selected for study.
Acknowledgements
Not applicable.
Abbreviations
- PDAC
Pancreatic ductal adenocarcinoma
- CAFs
Cancer-associated fibroblasts
- KPC
Kras/p53 mutation conditional knock-in
- moMSC
Mouse mesenchymal stem cells
- RNAseq
RNA sequencing
- iCAF
Inflammatory CAF
- myCAFs
Myofibroblastic CAFs
- apCAFs
Antigen-presenting CAFs
- TME
Tumor microenvironment
Authors' contributions
LZ and JZ conceived the concept; XP, JZ, and LZ designed the research studies. XP, JZ, QX, MZ, and KF conducted experiments, acquired and analyzed the data. XP, JZ, and LZ wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by an NIH grant R01 CA169702 (LZ), an NIH Grant R01 CA197296 (LZ), a National Cancer Institute Specialized Programs of Research Excellence in Gastrointestinal Cancers Grant P50 CA062924 (LZ), and a Sidney Kimmel Comprehensive Cancer Center Grant P30 CA006973.
Availability of data and materials
RNA sequencing data are provided as supplement materials.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
LZ receives grant support from Bristol-Meyer Squibb, Merck, iTeos, Amgen, NovaRock, Inxmed, and Halozyme, and received the royalty for licensing GVAX to Aduro Biotech. LZ is a paid consultant/Advisory Board Member at Biosion, Alphamab, NovaRock, Akrevia/Xilio, Ambrx, Novagenesis, Datarevive, Snow Lake Capitals, and Mingruzhiyao. LZ holds shares at Alphamab and Mingruzhiyao.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xingyi Pan and Jiaojiao Zhou have contributed equally to this work and should be considered as first authors.
Contributor Information
Jiaojiao Zhou, Email: zhoujj@zju.edu.cn.
Lei Zheng, Email: lzheng6@jhmi.edu.
References
- 1.Wangjam T, Zhang Z, Zhou XC, Lyer L, Faisal F, Soares KC, et al. Resected pancreatic ductal adenocarcinomas with recurrence limited in lung have a significantly better prognosis than those with other recurrence patterns. Oncotarget. 2015;6(34):36903–36910. doi: 10.18632/oncotarget.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7(5):469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 3.Xiao Q, Zhou D, Rucki AA, Williams J, Zhou J, Mo G, et al. Cancer-Associated fibroblasts in pancreatic cancer are reprogrammed by tumor-induced alterations in genomic DNA methylation. Cancer Res. 2016;76(18):5395–5404. doi: 10.1158/0008-5472.CAN-15-3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan X, Zheng L. Epigenetics in modulating immune functions of stromal and immune cells in the tumor microenvironment. Cell Mol Immunol. 2020;17(9):940–953. doi: 10.1038/s41423-020-0505-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang M.W. PXY, et al. Pancreatic cancer cells render tumor-associated macrophages metabolically reprogrammed through a GARP-dependent and DNA methylation-mediated mechanism. Signal Transduct Target Ther. 2021;In Press. [DOI] [PMC free article] [PubMed]
- 6.Foley K, Rucki AA, Xiao Q, Zhou D, Leubner A, Mo G, et al. Semaphorin 3D autocrine signaling mediates the metastatic role of annexin A2 in pancreatic cancer. Sci Signal. 2015;8(388):ra77. doi: 10.1126/scisignal.aaa5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rucki AA, Foley K, Zhang P, Xiao Q, Kleponis J, Wu AA, et al. Heterogeneous stromal signaling within the tumor microenvironment controls the metastasis of pancreatic cancer. Cancer Res. 2017;77(1):41–52. doi: 10.1158/0008-5472.Can-16-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biffi G, Oni TE, Spielman B, Hao Y, Elyada E, Park Y, et al. IL1-induced JAK/STAT signaling is antagonized by TGFβ to shape CAF heterogeneity in pancreatic ductal adenocarcinoma. Cancer Discov. 2019;9(2):282–301. doi: 10.1158/2159-8290.Cd-18-0710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elyada E, Bolisetty M, Laise P, Flynn WF, Courtois ET, Burkhart RA, et al. Cross-species single-cell analysis of pancreatic ductal adenocarcinoma reveals antigen-presenting cancer-associated fibroblasts. Cancer Discov. 2019;9(8):1102. doi: 10.1158/2159-8290.CD-19-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Öhlund D, Handly-Santana A, Biffi G, Elyada E, Almeida AS, Ponz-Sarvise M, et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med. 2017;214(3):579–596. doi: 10.1084/jem.20162024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das S, Shapiro B, Vucic EA, Vogt S, Bar-Sagi D. Tumor cell-derived IL1β promotes desmoplasia and immune suppression in pancreatic cancer. Cancer Res. 2020;80(5):1088–1101. doi: 10.1158/0008-5472.Can-19-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Representative images of lung and liver metastases from KPC mice implanted with KPC tumor cells with lung and liver metatasis potentials and schema of how represntative metabolism genes were selected for study.
Data Availability Statement
RNA sequencing data are provided as supplement materials.