Abstract
Transforming growth factor β (TGF-β) plays important roles in the regulation of proliferation, differentiation, apoptosis, and carcinogenesis. To identify genes responsible for maintaining the phenotype induced by TGF-β, we performed a retrovirus-mediated gene trap screening designed to isolate TGF-β-responsive genes in human lung carcinoma cell line A549. After screening 249 trap lines, 21 were found to express the reporter β-galactosidase gene in a TGF-β-responsive manner. Interestingly, in large proportions of these trap lines, the reporter gene was responsive also to phorbol ester and was suppressed by gamma interferon. Fragments of all these trapped genes were recovered by 5′- and 3′-rapid amplification of cDNA ends (RACE), and in 15 out of 21 cases (71%), the TGF-β responsiveness of the endogenous genes was confirmed by RNA blot hybridization. In at least five cases, the TGF-β-induced upregulation was found to be cycloheximide resistant, suggesting the roles of the genes in the TGF-β-induced primary responses. Sequence analyses revealed that 43% (9 of 21) of the trapped genes were novel and that the remainder included genes previously reported to be upregulated by TGF-β, such as epidermal growth factor receptor and β1 integrin, documenting the validity of this approach. Other known genes include the ones encoding the proteins associated with cell proliferation (ribosomal proteins S15a, hNRP/NAP-1, and lipocortin II), focal adhesions (paxillin), and transcriptional regulation (thyroid hormone receptor activator molecule 1 [TRAM-1]).
Transforming growth factor β (TGF-β) represents a large family of secreted polypeptides playing important roles in the regulation of various cellular responses (34). The prototype member, TGF-β1, is of particular importance in cancer research, since it inhibits the growth of certain types of cells in vitro (32, 38), and since mutational inactivation of its signaling molecules is found in human tumors (17, 34). Recent studies have revealed the nature of a TGF-β signaling pathway where SMAD family proteins play central roles in transmitting a signal from the receptor serine/threonine kinase at the cell surface directly to the nuclear transcription machinery (21, 34, 52). The TGF-β signal induces the expression of several inhibitors for cyclin-dependent kinases, such as p21Cip1, p15Ink4B, and p27Kip1, which have previously been proposed as mediators for TGF-β-induced growth inhibition (7, 18, 37). It is therefore likely that other important factors for TGF-β functions can also be found among the genes transcriptionally regulated by the TGF-β signal.
The study of differentially expressed genes has been one of the most promising approaches to identifying genes important for growth, differentiation, and development. Recent technical advances and improvements are accelerating the analysis of gene expression profiles at the transcript level. These methods include suppression subtractive hybridization (9), differential display (23, 30), comparative expressed sequence tag analysis (29, 50), serial analysis of gene expression (51), and DNA microarrays (42). One drawback to these methods, however, is that since they use mRNA as a starting material, they tend to preferentially detect abundantly expressed genes.
In this study, we have employed a supplementary approach to screen and identify genes responsive to various external stimuli in cultured cells. Our method is based on a “gene trap” initially designed for random insertional mutagenesis in mouse embryonic stem cells and uses a retroviral vector to introduce a promoterless reporter gene into the host chromosomes (12, 13, 16). Retroviral DNA is known to integrate into numerous sites in the host chromosomes with no obvious sequence specificity, although transcriptionally active regions may be preferred targets for integration (43). The reporter gene of this vector can only be expressed if it inserts within an intron, an exon, or a promoter of a transcriptionally active gene in the right orientation. This allows one to select for the cells (trap line) in which the insertion has occurred and to obtain the sequences of the trapped genes useful for gene identification as well as subsequent cloning. Furthermore, the responses of the trapped genes to various factors can be readily examined by staining the trap lines with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) after exposure to these factors. Such examination may provide important information not only on the behavior of the trapped genes themselves but also on the interactions between the signals evoked by these factors.
Several groups have previously used the gene trap to identify genes whose expression was either induced or suppressed during cellular differentiation (11, 26), oncogenic transformation (2), and apoptosis (39). By modifying the reporter/selection marker system, we adapted this method so as to be suitable for rapid screening of differentially expressed genes in cultured cells. Our initial attempt to apply this method to the screening of genes upregulated by TGF-β led to the identification of a series of interesting genes, including several novel genes, and has demonstrated the feasibility and efficacy of this approach.
MATERIALS AND METHODS
Cell culture.
The A549 human lung adenocarcinoma cell line (15) (obtained from the American Type Culture Collection) was maintained in Dulbecco modified Eagle medium supplemented with 5% fetal calf serum, penicillin (100 U/ml), and streptomycin (100 μg/ml) (growth medium [GM]). Additional medium factors and their concentrations were as follows: 5 ng of recombinant human epidermal growth factor (EGF; Toyobo)/ml, 5 ng of recombinant human platelet-derived growth factor (PDGF) AB heterodimer (R & D Systems)/ml, 5 ng of recombinant human insulin-like growth factor (IGF-1; Roche Diagnostics)/ml, 1 to 2.5 ng of recombinant human transforming growth factor β1 (TGF-β1; R & D Systems)/ml, 200 IU of recombinant human gamma interferon (IFN-γ); Roche Diagnostics)/ml, 10 μM T3 (Sigma), 10 ng of phorbol 12-myristate 13-acetate (PMA; Sigma)/ml, and 0.1 mM 6N,2′-O-dibutyryladenosine 3′:5′-cyclic monophosphate (dbcAMP; Sigma).
Vector construction and virus preparation.
pROSA-nGBT plasmid was constructed by modifying pROSAβ-geo (12). First, the 5′ HindIII-BamHI fragment corresponding to the β-galactosidase (β-Gal) portion of β-geo was replaced with the nuclear localization signal (NLS) of simian virus 40 large T antigen (3) using synthetic oligonucleotides. Second, the BamHI-XbaI fragment corresponding to the neo coding sequences of β-geo was replaced with a herpes simplex virus (HSV) thymidine kinase (TK) gene fragment (1). Third, a DNA fragment encoding blasticidin-S (BLA-S) deaminase (24) was amplified by PCR using pUCSV-BSD plasmid (Kaken) as a template and inserted into the BamHI site located between the β-Gal and HSV TK gene sequences. The ends of each fragment were designed so that the final construct, designated nGBT, encoded a contiguous fusion protein. Finally, the nGBT gene was inserted between the HindIII and XbaI sites of the original pROSAβ-geo vector, resulting in the replacement of β-geo with nGBT. pROSA-nGBT DNA was transfected into an amphotropic packaging host (GP+envAm12) (33), and freshly harvested culture supernatant was used as the ROSA-nGBT virus preparation.
Gene trap screening.
In the first experiment, A549 cells (106 cells in 10 60-mm-diameter dishes) were infected with ROSA-nGBT virus and after 48 h were treated with TGF-β1 (2.5 ng/ml) for 14 h and then with TGF-β1 plus BLA-S (8 μg/ml) for 7 days. The BLA-S-resistant colonies were trypsinized, pooled, and plated (splitting ratio, 1:10) into new dishes with GM. After a 5-day incubation, the cells were treated with ganciclovir (GCV) (0.2 μM; Roche) for 5 days. In the second experiment, a total of 2.6 × 106 A549 cells (2 × 105 cells per 100-mm-diameter dish) were infected with ROSA-nGBT virus. TGF-β1 was added 3 days later, and BLA-S (2 μg/ml) was added on the following day. The selection medium was renewed once every 3 to 4 days. Colonies that survived these treatments were isolated and expanded, and their aliquots were plated in duplicate onto 24-well plates (one well without TGF-β and the other with TGF-β). After 48 h of incubation, the cells were stained with X-Gal (see below). Trap lines that stained deeper in the presence of TGF-β1 than in its absence were subjected to further analyses.
cDNA isolation.
5′-Rapid amplification of cDNA ends (5′-RACE) and 3′-RACE were performed following the protocols of Parry and Alphey (36) with slight modifications. Briefly, for 5′-RACE, total RNA was isolated using MagExtractor (Toyobo) from a trap line grown to subconfluence on a 60-mm-diameter dish. The negative-strand cDNA that contained the vector sequence was selectively reverse transcribed from the RNA sample using Revertra-Ace (Toyobo) and a β-Gal-specific primer (Gal-rev: 5′-CAA CCA CCG CAC GAT AGA GAT TC-3′). The reaction products were treated with RNase H and purified using a QiaQuick PCR purification kit (Qiagen). A deoxyribosyladenine tail was added using terminal deoxynucleotidyl transferase, and the reaction products were amplified by nested PCR. The first PCR (30 cycles) was performed in the presence of 10% dimethyl sulfoxide (DMSO) with the forward tail-anchored primer UNIZAP-dT (5′-GAG AGA GAG AGA GAG AGA GAA CTA GTC TCG AGT TTT TTT TTT TTT TTT TT-3′) and the NLS-based reverse primer NLS-3S (5′-GAT CAA CCT TCC TCT TCT TCT TAG-3′). The second PCR (30 cycles) was performed in the absence of DMSO with the same forward primer, UNIZAP-dT, and the splice acceptor-based reverse primer SA-REV (5′-TCG ATC CCC ACT GGA AAG ACC GCG A-3′). For 3′-RACE, poly(A)+ RNA was isolated from total RNA of untreated A549 cells using an oligo(dT) cellulose column and reverse transcribed with semirandom primers (WALK-A [5′-GTA ATA CGA CTC ACT ATA GGG CAC GCG TGG NNN GTS AC-3′] or WALK-B [5′-GTA ATA CGA CTC ACT ATA GGG CAC GCG TGG NNN GAW TC-3′]). The first PCR (30 cycles) was performed using a forward primer specific to each gene (gene-specific primer 1) and arbitrary reverse primer AP-1 (5′-GTA ATA CGA CTC ACT ATA GGG C-3′). The second PCR (30 cycles) was performed using another forward primer corresponding to a downstream sequence of the gene (gene-specific primer 2) and reverse primer AP-2S (5′-ACT ATA GGG CAC GCG TGG T-3′). Amplified fragments of both 3′- and 5′-RACE were subcloned into pBluescript II SK(−) and sequenced using DyeTerminator Bigdye (ABI). Standard protocols (40) were used for general techniques in DNA manipulation and nucleic acid analyses.
RNA blot hybridization.
A549 cells were cultured in GM, GM containing 2.5 ng of TGF-β/ml, or GM containing 2.5 ng of TGF-β and 10 μg of cycloheximide (CHX)/ml for 48 h, and their total RNA was extracted by the guanidinium-cesium chloride method as described previously (40). The RNA (20 μg) was subjected to electrophoresis in a 1% agarose gel containing formaldehyde and transferred to a Hybond-N (Amersham) nylon filter using 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). The resulting filter was hybridized with each of the following probes: 5′-RACE products for A4 (800 bp), A21 (450 bp), A82 (100 bp), and A143 (140 bp); 3′-RACE products for A26 (1.7 kb), A70 (1.4 kb), A161 (420 bp), A162 (370 bp), and A164 (210 bp); reverse transcription-PCR products containing coding regions for A80 (1.3 kb), A96 (280 bp), A89 (700 bp), A100 (1 kb), A114 (1.2 kb), A116 (1.4 kb), A126 (290 bp), A152 (1.2 kb), and A153 (340 bp); an 850-bp fragment containing the full coding region of A83; a 1.4-kb fragment containing the pyruvate kinase coding region (35) for A14; and a 2.4-kb fragment containing the 5′ coding region of thyroid hormone receptor activator molecule 1 (TRAM-1) cDNA (46) for A88. All the probes were labeled with [α-32P]dCTP using Ready-To-Go DNA-labeling beads (Amersham), followed by purification using MicroSpin columns (Amersham).
β-Gal assay and X-Gal staining.
Cells (105/well) were seeded onto six-well plates in duplicate, and on the following day TGF-β (1 ng/ml) was added to one of a pair. The cells were further cultured for 48 h, trypsinized, counted, and lysed with 1× reporter lysis buffer (Promega) to yield the lysates with the same cell concentration (106 cells/ml). The lysates (100 μl) were subjected to the colorimetric enzyme assay in triplicate using the β-Gal enzyme assay system (Promega). The experiments were repeated at least three times. To examine the response of trapped genes to various other agents (Fig. 3, Table 1), cells (104/well) were seeded onto a 24-well-plate, and on the following day, appropriate growth factors and/or chemicals were added. After 48 h of incubation, the cells were fixed and treated with an X-Gal staining solution (5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 1 mM MgCl2, 0.04% X-Gal) at 37°C for 24 h (41). The experiments were repeated twice with similar results.
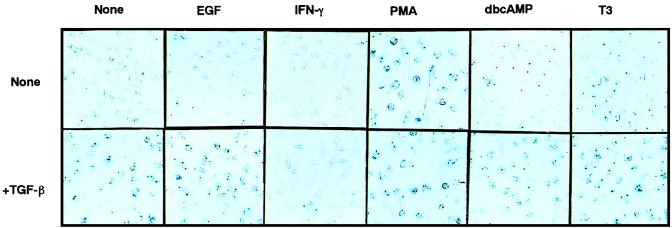
FIG. 3.
Response of the A116 trap line to various agents. A114 cells were incubated in medium containing the indicated growth factors and/or chemicals for 48 h and were stained with X-Gal.
TABLE 1.
Responses of the reporter genes in the trap lines to various agents
| Trap line | Relative intensity of staininga with indicated agentb in:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Absence of TGF-β
|
Presence of TGF-β
|
||||||||||||||
| EGF | IFN-γ | PMA | PDGF | IGF-1 | dbcAMP | T3 | No agent | EGF | IFN-γ | PMA | PDGF | IGF-1 | dbcAMP | T3 | |
| A14 | − | −c | + | − | − | − | − | + | + | − | ++ | + | + | + | + |
| A80 | − | −c | + | − | − | − | − | + | + | − | ++ | + | + | ++ | + |
| A82 | − | − | + | − | − | − | − | + | + | + | ++ | + | + | + | + |
| A96 | − | − | + | − | − | − | − | + | + | − | ++ | + | + | + | + |
| A100 | − | −c | + | − | − | − | − | + | + | + | ++ | + | + | + | + |
| A114 | − | − | + | − | − | − | − | + | + | − | + | + | + | + | + |
| A116 | − | −c | + | − | − | − | − | + | + | − | + | + | + | + | + |
| A126 | − | −c | − | − | − | − | − | + | + | − | + | + | + | + | + |
| A143 | − | −c | − | − | − | − | − | + | + | − | + | + | + | + | + |
| A152 | − | −c | + | − | − | − | − | + | + | − | + | + | + | + | + |
| A153 | − | −c | + | − | − | − | − | + | + | − | + | + | + | + | + |
| A161 | − | −c | + | − | − | − | − | + | + | − | + | + | + | + | + |
| A162 | − | −c | + | − | − | − | − | + | + | − | ++ | + | + | + | + |
| A164 | − | −c | − | − | − | − | − | + | + | − | + | + | + | + | + |
Relative intensity of staining is indicted qualitatively as follows: −, same as background; +, thicker than background; ++, thicker than TGF-β-treated sample.
Concentrations: EGF, PDGF, IGF-1, 5 ng/ml; IFN-γ, 200 U/ml; PMA, 10 ng/ml; dbcAMP, 0.1 mM; T3, 1 mM; TGF-β1, 1 ng/ml.
Reduction in background staining was observed.
RESULTS
Construction of gene trap vector.
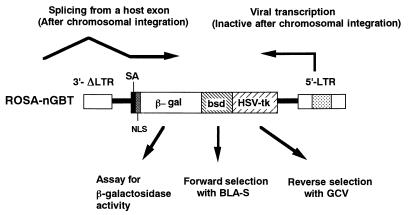
To facilitate screening for differentially expressed genes in cultured cells, we constructed a gene trap vector carrying a newly developed reporter gene, nGBT, next to the splice acceptor site in the self-inactivating retroviral vector pROSAβ-geo (12). The nGBT gene encodes a fusion protein consisting of four parts: NLS, β-Gal, BLA-S deaminase, and HSV TK (Fig. 1). When ROSA-nGBT virus infects the cells and the vector integrates into an active transcriptional unit of the host genome in the appropriate orientation, a chimeric transcript containing a 5′ segment of the host gene (trapped gene) spliced into the splice acceptor sequence located at 5′ end of the nGBT reporter gene will be expressed (Fig. 1). Such cell clones (trap lines) will become resistant to BLA-S and sensitive to GCV. The expression profile of the trapped gene can be monitored using β-Gal activity.
FIG. 1.
Gene trap strategy with ROSA-nGBT vector. In the cells infected with this virus, the integrated proviral DNA became transcriptionally silent. The nGBT reporter gene lacks its own promoter but is preceded by a strong splice acceptor sequence. When the viral DNA is integrated in an active transcriptional unit in the host genome, the nGBT gene can be transcribed as a part of the chimeric mRNA whose 5′ portion is derived from the host transcript. SA, splice acceptor.
Isolation of gene trap cell lines responsive to TGF-β.
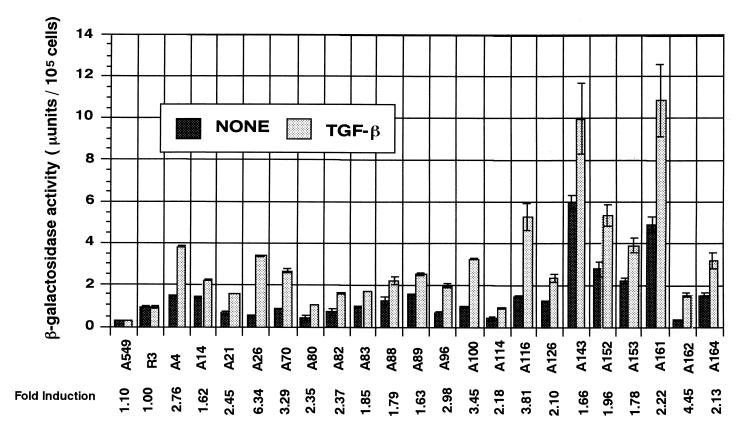
To detect the genes persistently upregulated in response to TGF-β, we infected human lung carcinoma cell line A549 with ROSA-nGBT virus and selected the cells with BLA-S in the presence of TGF-β for a week. TGF-β is known to suppress the anchorage-independent growth of A549 cells (38). We observed that in the presence of 1 ng of TGF-β1/ml the rate of their anchorage-dependent growth was also decreased to one-third that of their untreated counterparts and that their morphology became flatter (data not shown). In the first experiment, to eliminate the trap lines constitutively expressing a high level of reporter gene, counter-selection with GCV in the absence of TGF-β was performed. A total of 117 trap lines were obtained after these selections, and the induction of the reporter expression was examined by X-Gal staining in the presence or absence of TGF-β. Thirteen trap lines (lines A4 to A114) were stained deeper in the presence of TGF-β and isolated for further analysis. We found that the majority of trap lines expressed the reporter gene at certain levels without TGF-β treatment (Fig. 2; see below) and that GCV damaged the cells at the concentration we used (0.2 mM). Therefore in the second experiment, the GCV selection was omitted, and out of 132 BLA-S-resistant clones, eight additional trap lines (A116, A126, A143, A152, A153, A161, A162, and A164) showing TGF-β responsiveness were isolated for further analysis. To estimate the extent of reporter gene induction, we performed a quantitative β-Gal assay with each trap line incubated in the presence and absence of TGF-β. The induction index (β-Gal activity for TGF-β-treated cells divided by β-Gal activity for untreated cells) was found to range from 1.62 to 6.34 (Fig. 2).
FIG. 2.
Quantitative analysis of the reporter gene activities in trap lines. Each trap line was cultured in the presence (TGF-β) or absence (None) of TGF-β1 for 48 h, and β-Gal activities in the cell lysates were determined. Each bar represents the average activity and the standard error from triplicate cultures. The experiments were repeated at least three times, and the data from a typical experiment are presented. The induction index (activity of TGF-β-treated cells divided by activity of untreated cells) is shown at the bottom. Parental A549 cells and R3, a trap line whose X-Gal staining pattern was found to be unaffected by TGF-β, were included as controls. The P value for each pair of data (Student's t test) was at most 2 × 10−4 (i.e., significant difference).
Response of reporter gene in the trap lines to other agents.
Southern blot analysis indicated that each of the trap lines isolated under these conditions contained a single copy of integrated vector DNA (data not shown). The behavior of the reporter gene in a trap line must therefore reflect the nature of a trapped promoter, which can be readily monitored by X-Gal assay. Taking advantage of this readiness, we attempted to classify these trap lines based on the spectra of their responses to various stimuli applied alone or in conjunction with TGF-β. An example of such experiments is shown in Fig. 3. The results with some of the trap lines (Table 1) revealed several interesting patterns. First, a majority of the trap lines tested (11 of 14) also responded to a phorbol ester, PMA. Second, IFN-γ suppressed the TGF-β-induced reporter expression in many trap lines (12 of 14). Third, neither induction nor suppression of the reporter gene was detected after treatment with EGF, IGF-1, PDGF, a cyclic AMP analogue (dbcAMP), or thyroid hormone (tri-iodo-l-thyronine; T3) in these trap lines. A slight induction of the reporter gene was observed in A80 when treated with dbcAMP in the presence of TGF-β.
Response of endogenous genes to TGF-β in A549 cells.
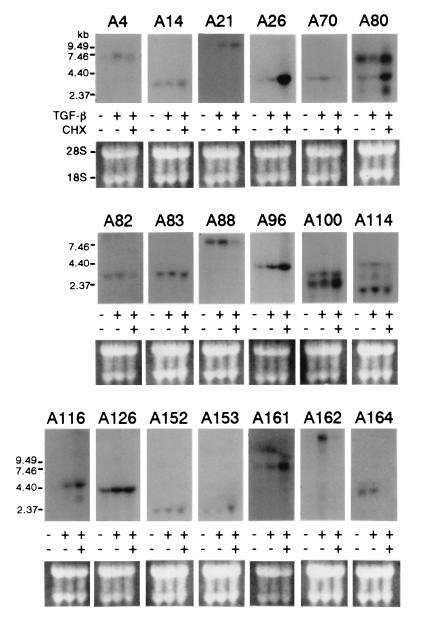
Since the β-Gal assay and X-Gal staining evaluate the expression levels of the reporter protein in the context of the integrated proviral DNA, they may not exactly reflect the behavior of the corresponding endogenous genes. We therefore analyzed the response of the endogenous mRNA to TGF-β in the parental A549 cells by RNA blot hybridization using the cDNA fragment of each trapped gene (see Materials and Methods) as a probe (Fig. 4). Strong upregulation by TGF-β was detected in five cases (A4, A21, A26, A116, and A162), and weak but significant upregulation was detected in seven cases (A70, A96, A100, A114, A126, A152, and A161). More than one band was detected with the A100 and A114 probes, and the upper band and all three bands, respectively, seem to be responsive to TGF-β. A slight but reproducible upregulation was also detected with the A83 probe. On the other hand, no clear upregulation was observed in four cases (A14, A80, A152, and A164), and no hybridization bands were detected in two cases (A89 and A143). Taking all the data together, upregulation by TGF-β was confirmed by RNA blot hybridization in 15 out of 21 cases (71%).
FIG. 4.
Northern blot analysis of the endogenous transcripts. Endogenous transcripts corresponding to the trapped genes were detected by Northern blot analysis with total RNA from parental A549 cells cultured in either GM, 2.5-ng/ml TGF-β, or 2.5-ng/ml TGF-β plus 10-μg/ml CHX for 48 h. Probes used are described in Materials and Methods. For the estimation of the quality and quantity of RNA samples, ethidium bromide-stained RNA on the blot is shown in the lower portion of each panel.
To examine whether these genes are direct targets for TGF-β or not, we compared the response of these genes to TGF-β in the presence or absence of CHX, an inhibitor of de novo protein synthesis (Fig. 4). In most cases (A14, A21, A26, A80, A96, A100, A116, A153, and A161), their transcripts further increased with CHX treatment, and in some cases (A70, A82, A88, A114, A162, and A164) their transcripts decreased. The levels of expression of the rest (A4, A83, A126, and A152) were not altered by CHX treatment (summarized in Table 2).
TABLE 2.
Summary of the TGF-β-responsive genes identified in this study
| Trap line | Identity of protein (accession no.) or homology | Reported function (reference[s]) | Induction by TGF-β
|
||
|---|---|---|---|---|---|
| β-Gala | RNAb | CHXc | |||
| Known genes | |||||
| A21 | EGF receptor (U48722) | Cell growth (31, 47) | 2.45 | ++ | ↑ |
| A82 | Ribosomal protein S15a (AA364270) | Protein synthesis (4) | 2.37 | ± | ↓ |
| A88 | TRAM-1 (AF016031) | T3 receptor coactivator (46) | 1.79 | + | ⇓ |
| A96 | Paxillin (U14588) | Integrin signal (48) | 2.98 | + | ↑ |
| A100 | hnRNP-K/tump (X72727) | Cell cycle progression (8) | 3.45 | ±* | ↑ |
| A114 | hNRP/NAP-1 (D28430) | Nucleosome assembly (44) | 2.18 | +* | ↓ |
| A116 | ZNF 185 LIM-domain protein (Y09538) | (20) | 3.81 | ++ | ↑ |
| A152 | Lipocortin II (D00017) | EGF receptor substrate (6, 22) | 1.96 | + | → |
| A126 | β1-Integrin (X07979) | ECMd protein receptor (19) | 2.10 | + | → |
| Unknown genes | |||||
| A4 | Zinc finger motif | 2.76 | ++ | → | |
| A26 | 6.34 | ++ | ⇑ | ||
| A70 | 3.29 | + | ⇓ | ||
| A83 | Thioredoxin domain | 1.85 | ± | → | |
| A161 | 2.22 | + | ↑ | ||
| A162 | 4.45 | ++ | ⇓ | ||
| Unconfirmed genesf | |||||
| A14 | Pyrunvate kinase (M23725) | Cytosolic T3-binding protein (25) | 1.62 | − | ↑ |
| A80 | Paxillin-related LIM protein (AF061258) | 2.35 | −* | ↑ | |
| A89 | FRA7H linked | 1.63 | N/D | ||
| A143 | BACe clone | 1.66 | N/D | ||
| A153 | cDNA clone | 1.78 | − | ↑ | |
| A164 | U19H snoRNA (AJ224167) | Ribosome biogenesis (5) | 2.13 | − | ⇓ |
Induction index of β-Gal activity in trap lines (Fig. 2).
Effects of TGF-β on the levels of endogenous mRNA in A549 cells as assessed by RNA blot hybridization (Fig. 4), indicated qualitatively as follows: ++, clear induction; +, slight but significant induction; ±, possible induction; −, no apparent induction; N/D, mRNA not detectable. *, multiple bands were detected.
Effects of CHX on the levels of endogenous mRNA in A549 cells incubated in the presence of TGF-β as assessed by RNA blot hybridization (Fig. 4), indicated qualitatively as follows: →, no apparent effects; ↑, slight upregulation; ⇑, strong upregulation; ↓, slight downregulation; ⇓, strong downregulation.
ECM, extracellular matrix.
BAC, bacterial artificial chromosome.
Genes whose induction could not be confirmed by RNA blot hybridization.
Sequence analysis of the trapped genes.
To identify the trapped genes, the cellular sequences contained in the chimeric transcripts expressed in the trap lines were analyzed by 5′-RACE. The 5′-RACE products, ranging from 50 to 500 bp in length (average, 100 bp), were sequenced, and the data were compared to GenBank and SwissProt databases using the BLASTN or BLASTX algorithm, respectively, to search for sequence similarities. In 12 cases, identical sequences were detected in the databases at this stage (Table 2; genes with accession numbers). For the remaining cases, 3′-RACE with RNA from TGF-β-treated A549 cells was performed to analyze sequences of the trapped genes further downstream. The lengths of the 3′-RACE products ranged from 200 to 2,000 bp (average, 400 bp). No additional identical sequences were detected in the databases at this stage. The known genes whose upregulation was confirmed by RNA blot hybridization include two previously described TGF-β-responsive genes, EGF receptor (47) and β1-integrin (19). TGF-β regulation has not been described for the other genes. The known TGF-β-responsive genes can be roughly classified into three categories based on the properties of their products: (i) growth-associated proteins (EGF receptor, ribosomal protein S15a, hnRNP-K/tump, hNRP/NAP-1, lipocortin II), (ii) focal adhesion-associated proteins (paxillin, β1-integrin), and (iii) thyroid hormone receptor coactivator (TRAM-1) (Table 2). The insertional events in 10 out of 12 known genes have occurred in the 5′-noncoding region and in the other two cases near the 5′ end within the coding region (data not shown).
DISCUSSION
In this study, we have attempted to apply a retrovirus-mediated gene trap strategy to identify genes responsive to TGF-β. In this first trial, we could identify 15 genes whose transcripts were upregulated after TGF-β treatment. Two of these genes, the EGF receptor and β1-integrin genes, have previously been reported to be upregulated by TGF-β (19, 47), seven others are known genes not previously noticed to be responsive to TGF-β, and the remainder (six genes) are either novel or cloned but poorly characterized. Although the initial screening by X-Gal staining yielded 6 additional trap lines whose reporter activities were responsive to TGF-β, we failed to detect the upregulation of the corresponding endogenous genes in parental A549 cells after TGF-β treatment by RNA blot hybridization (6 out of 21 lines; 29%). Even among the genes whose differential expression was confirmed by RNA blot hybridization, the induction of β-Gal activity in trap lines and the induction of mRNA in A549 cells were not always proportional (compare Fig. 2 and 4; qualitatively summarized in Table 2). Since most trap lines have retroviral insertions at or near the 5′ ends of target genes, the reporter gene expression should reflect the upstream promoter activity but not the effects of internal regulatory elements or mRNA stability of the target genes. This may be a reason for the discrepancy between β-Gal data and RNA blot data. Nevertheless, the 71% (15 of 21) success rate would justify further application of the present approach as a first screening for differentially expressed genes in cultured cells. Furthermore, we could isolate novel genes at a very high frequency (9 of 21; 43%), suggesting that our approach can detect a gene population rarely recognized by the other common methods to isolate cDNA. Since this method targets chromosomal DNA, the influence of mRNA abundance on the repertoire of the genes identified is expected to be minimum. In addition, the selection drug employed in this study (BLA-S) has several advantages over other commonly used drugs such as G418. BLA-S can rapidly kill many types of cells at relatively low concentrations (less than 1 μg/ml) even at high cell densities. Thus low-level expression of the marker gene (bsd) can confer clear-cut resistance to the cells. We could therefore minimize the bias due to the enrichment of the cells expressing relatively high levels of marker genes during the initial drug selection step.
One important advantage of this approach is that we obtain trap lines before isolating trapped genes. Information on the response of the reporter gene to various stimuli helps to classify the trap lines and to decide which lines should be further characterized. Trap lines themselves may also be useful in finding interactions between different signaling pathways. For instance, a majority of the trap lines tested in this study (11 of 14) responded to a phorbol ester, PMA (Table 1). This seems consistent with the recent finding by Zhang et al. (53) that a Smad3–Smad4 complex can activate a tetradecanoyl phorbol acetate (another phorbol ester similar to PMA)-responsive element (TRE) directly or in conjunction with c-Fos and c-Jun. Thus, it will be interesting to see whether the associated genes have TRE in their promoters and to examine whether these genes are indeed regulated directly by these signals. As another example, IFN-γ was found to suppress the TGF-β-induced reporter expression in many trap lines (12 of 14). This is also consistent with the recent finding by Ulloa et al. (49) that IFN-γ induces the expression of an antagonistic SMAD (Smad7) thereby inhibiting receptor-mediated Smad3 phosphorylation, a triggering event in SMAD-mediated signal transduction. Thus, studies with a panel of trap lines and various reagents or stimuli may help in finding novel interactions between apparently independent signaling pathways.
Another important advantage of this approach is its reliability in identifying trapped genes. In this experiment, we could obtain 5′-RACE products from all the trap lines isolated and could eventually isolate full coding sequences of some of the novel genes by 3′-RACE (Akiyama et al., unpublished data); 3′-RACE was effective enough to clone a cDNA of up to 2 kb, which was sufficient for recovering the coding regions of relatively small genes. For confirmation of their sequences, we also screened some commercially available phage cDNA libraries (human placenta and testis) with radiolabeled 3′-RACE products (more than 1.4 kb in length) as probes. In most cases, the frequencies of hybridization-positive clones were less than 10−5, indicating low expression of these genes in placenta and testis. Importantly, the cDNA fragments initially isolated by 5′-RACE usually correspond to the 5′-terminal regions of the trapped genes. This is in contrast to the other methods designed to detect differential gene expression, which often target the 3′ ends of mRNAs, a region suitable for gene identification because of the sequence diversity even among members of the same gene family. Although 5′-noncoding sequences should serve the same purpose, one drawback at the moment is that the sequence information for this region is often lacking in the database because of a technical reason: isolation of this region has been relatively difficult by the classical procedures for cDNA synthesis. The gene trap approach is expected to enrich such information and also facilitate the analysis of the promoter regions of the trapped genes.
The regulation of trapped genes was first screened by X-Gal staining, then examined quantitatively by β-Gal assay, and finally confirmed by RNA blot hybridization. The β-Gal assay was sensitive enough to detect a difference of expression as small as 1.6-fold (Fig. 2), although the biological significance of such small changes remains to be clarified. Since the present format of the gene trap approach relies on the initial drug selection of trap lines, which usually takes several hours to days, the genes upregulated only transiently after application of a signal probably escape the screening. The genes isolated by this approach would therefore represent genes whose expression is persistently altered, directly or indirectly, by the signal. We found that in some cases analysis of the effects of CHX by RNA blot hybridization may clarify this point. For instance, CHX had little influence on the TGF-β responsiveness of some of the genes identified in this study (e.g., A4, A83, A126, and A152), suggesting that the response of these genes did not require de novo protein synthesis and that therefore the genes may represent the primary targets for the TGF-β signal. In contrast, CHX seems to cancel the upregulating effects of TGF-β on some other genes, such as A82 and A114, suggesting that they may not be the primary targets. In some other cases (A70, A88, A162, and A164), however, CHX reduced the amounts of mRNA to levels well below those in the naive A549 cells (no addition of TGF-β), suggesting that these genes require some CHX-sensitive component(s) to maintain their basal activity or mRNA stability. Yet in some other cases (A26, A96, and A161), CHX increased the amounts of mRNA to levels significantly higher than those in the TGF-β-treated cells, suggesting the involvement of either CHX-sensitive negative regulators (which may or may not be specific to TGF-β signaling) or CHX-induced positive regulators. For the last two groups of genes, the effects of TGF-β may be masked by the effects of CHX, and therefore it seems difficult to judge through this method whether these genes represent the primary targets for the TGF-β signal or not.
We initially expected that counter-selection with GCV may facilitate the elimination of trap lines with constitutively expressed genes. Inclusion of counter-selection, however, did not significantly improve the efficiency of trap line isolation: 13 of 117 trap lines (11%) after counter-selection versus 8 of 132 trap lines (6.1%) without counter-selection. A quantitative comparison of levels of reporter gene expression (Fig. 2) suggests that the counter-selection might have served to select for trap lines with low background expression but that, on the other hand, it might have eliminated trap lines with strongly expressed genes. Moreover, high concentrations of GCV nonspecifically damage the cells, and this may interfere with the subsequent X-Gal assay because of an endogenous β-Gal-like activity (10, 27) induced in damaged cells. Thus, we could not take full advantage of counter-selection in this study. The counter-selection may, however, be useful in cases where genes are regulated in a more stringent fashion.
Among the nine genes confirmed to respond to TGF-β in this study, five genes (the EGF receptor, ribosomal protein S15a, hnRNP-K/tump, hNRP/NAP-1, and lipocortin II genes) can be classified as growth associated. In fact, these genes are known to be upregulated in growing and/or malignant cells (4, 6, 8, 22, 31, 44). It has been known for some time that TGF-β induces EGF receptor expression (47). Our finding indicates that induction of growth-associated genes by TGF-β is not a rare event. Importantly, other growth factors, such as EGF, PDGF, and IGF-1, failed to induce these growth-associated genes. This may explain why TGF-β exhibits a paradoxical growth-promoting activity when present with other growth factors in certain types of cells (38). On the other hand, preliminary experiments indicated that the overexpression of the A83 gene suppressed the growth of A549 cells (Matsuo et al., unpublished data). Thus, these novel genes may play some roles in TGF-β-induced growth inhibition. The two focal adhesion-associated proteins detected in this study (β1-integrin and paxillin) may also contribute to the growth-inhibitory effects (14, 48). Although the extent of induction was small, TRAM-1 was found to respond to TGF-β. This might suggest a possible molecular basis for the cooperative effects between thyroid hormone and TGF-β observed in certain systems (28, 45).
In conclusion, we could detect, by using retrovirus-mediated gene trap screening, a series of known and novel genes persistently upregulated by TGF-β. We could also examine the patterns of their responses to various stimuli using the trap lines. Based on these results, we conclude that this approach has several unique advantages over other methods and should provide a useful tool to detect and isolate genes that are responsive to various external stimuli in cultured cells. Further characterization of the novel genes isolated in this study may yield important insights into the mechanism of action of TGF-β.
ACKNOWLEDGMENTS
We thank P. Soriano for pROSAβ-geo, A. Takeshita for TRAM-1 cDNA, T. Noguchi for pyruvate kinase cDNA, Toyobo Co. Ltd. for providing some materials, Mariko Sasa, Yasuko Ono, and Takashi Kawai for technical assistance, and Aki Miyazaki for secretarial assistance.
This work was supported by research grants from the Ministry of Education, Science, Sports, and Culture of Japan and from Sankyo Co., Ltd.
REFERENCES
- 1.Akiyama N, Alexander D, Aoki Y, Noda M. Characterization of mutations induced by 300 and 320 nm UV radiation in a rat fibroblast cell line. Mutat Res. 1996;372:119–131. doi: 10.1016/S0027-5107(96)00179-0. [DOI] [PubMed] [Google Scholar]
- 2.Andreu T, Beckers T, Thoenes E, Hilgard P, von Melchner H. Gene trapping identifies inhibitors of oncogenic transformation. J Biol Chem. 1998;273:13848–13854. doi: 10.1074/jbc.273.22.13848. [DOI] [PubMed] [Google Scholar]
- 3.Benditt J O, Mayer C, Fasold H, Barnard F C, Riedel N. Interaction of a nuclear location signal with isolated nuclear envelopes and identification of signal-binding proteins by photoaffinity labeling. Proc Natl Acad Sci USA. 1989;86:9327–9331. doi: 10.1073/pnas.86.23.9327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonham-Smith P C, Oancia T L, Moloney M M. Cytoplasmic ribosomal protein S15a from Brassica napus: molecular cloning and developmental expression in mitotically active tissues. Plant Mol Biol. 1992;18:909–919. doi: 10.1007/BF00019205. [DOI] [PubMed] [Google Scholar]
- 5.Bortolin M L, Kiss T. Human U19 intron-encoded snoRNA is processed from a long primary transcript that possesses little potential for protein coding. RNA. 1998;4:445–454. [PMC free article] [PubMed] [Google Scholar]
- 6.Cole S P, Pinkoski M J, Bhardwaj G, Deeley R G. Elevated expression of annexin II (lipocortin II, p36) in a multidrug resistant small cell lung cancer cell line. Br J Cancer. 1992;65:498–502. doi: 10.1038/bjc.1992.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Datto M B, Li Y, Panus J F, Howe D J, Xiong Y, Wang X F. Transforming growth factor beta induces the cyclin-dependent kinase inhibitor p21 through a p53-independent mechanism. Proc Natl Acad Sci USA. 1995;92:5545–5549. doi: 10.1073/pnas.92.12.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dejgaard K, Leffers H, Rasmussen H H, Madsen P, Kruse T A, Gesser B, Nielsen H, Celis J E. Identification, molecular cloning, expression and chromosome mapping of a family of transformation upregulated hnRNP-K proteins derived by alternative splicing. J Mol Biol. 1994;236:33–48. doi: 10.1006/jmbi.1994.1116. [DOI] [PubMed] [Google Scholar]
- 9.Diatchenko L, Lau Y-F C, Campbell A P, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov E D, Siebert P D. Suppression subtractive hybridization: a method for generating differentially regulated or tissue specific cDNA probes and libraries. Proc Natl Acad Sci USA. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dimri G P, Lee X, Basile G M, Acosta M, Scott G, Roskelley C, Medrano E E, Linskens M, Rubel I, Pereira-Smith O, Peacocke M, Campisi J. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forrester L M, Nagy A, Sam M, Watt A, Stevenson L, Bernstein A, Joyner A L, Wurst W. An induction gene trap screen in embryonic stem cells: identification of genes that respond to retinoic acid in vitro. Proc Natl Acad Sci USA. 1996;93:1677–1682. doi: 10.1073/pnas.93.4.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedrich G, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 13.Fredrich G, Soriano P. Insertional mutagenesis by retroviruses and promoter traps in embryonic stem cells. Methods Enzymol. 1993;225:681–701. doi: 10.1016/0076-6879(93)25044-3. [DOI] [PubMed] [Google Scholar]
- 14.Giancotti F G, Ruoslahti E. Elevated levels of the α5β1 fibronectin receptor suppress the transformed phenotype of Chinese hamster ovary cells. Cell. 1990;60:849–859. doi: 10.1016/0092-8674(90)90098-y. [DOI] [PubMed] [Google Scholar]
- 15.Giard D J, Aaronson S A, Todaro G J, Arnstein P, Kersey J H, Dosik H, Parks W P. In vitro cultivation of human tumors: establishment of cell lines derived from a series of solid tumors. J Natl Cancer Inst. 1973;51:1417–1423. doi: 10.1093/jnci/51.5.1417. [DOI] [PubMed] [Google Scholar]
- 16.Gossler A, Zachgo J. Gene and enhancer trap screens in ES cell chimeras. In: Joyner A L, editor. Gene targeting: a practical approach. New York, N.Y: Oxford University Press; 1993. pp. 181–277. [Google Scholar]
- 17.Hahn S A, Schutte M, Hoque A T, Moskaluk C A, de Costa L T, Rosenblum E, Weinstein C L, Fischer A, Xeo C J, Hruban R H, Kern S E. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 18.Hannon G J, Beach D. P15INK4B is a potential effecter of TGF-β-induced cell cycle arrest. Nature. 1995;371:257–261. doi: 10.1038/371257a0. [DOI] [PubMed] [Google Scholar]
- 19.Heino J, Ignotz R A, Hemler M E, Crouse C, Massague J. Regulation of cell adhesion receptors by transforming growth factor-β. Concomitant regulation of integrins that share a common beta 1 subunit. J Biol Chem. 1989;264:380–388. [PubMed] [Google Scholar]
- 20.Heiss N S, Gloeckner G, Bächner D, Kioschis P, Klauck S M, Hinzmann B, Rosenthal A, Herman G E, Pouska A. Genomic structure of a novel LIM domain gene (ZNF185) in Xq28 and comparisons with the orthologous murine transcript. Genomics. 1997;43:329–338. doi: 10.1006/geno.1997.4810. [DOI] [PubMed] [Google Scholar]
- 21.Heldin C H, Miyazono K, ten Dijke P. TGF-β signaling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 22.Huang K S, Wallner B P, Mattaliano R J, Tizard R, Burne C, Frey A, Hession C, McGray P, Sinclair L K, Chow E P. Two human 35 kd inhibitors of phospholipase A2 are related to substrates of pp60v-src and of the epidermal growth factor receptor/kinase. Cell. 1986;46:191–199. doi: 10.1016/0092-8674(86)90736-1. [DOI] [PubMed] [Google Scholar]
- 23.Hubank M, Schatz S G. Identifying differences in mRNA expression by representational difference analysis of cDNA. Nucleic Acids Res. 1994;22:5640–5648. doi: 10.1093/nar/22.25.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamakura T, Kobayashi K, Tanaka T, Yamaguchi I, Toyoshige E. Cloning and expression of a new structural gene for blasticidin S deaminase, a nucleoside aminohydrolase. Agric Biol Chem. 1987;51:3165–3168. [Google Scholar]
- 25.Kato H, Fukuda T, Parkinson C, McPhie P, Cheng S Y. Cytosolic thyroid hormone-binding protein is a monomer of pyruvate kinase. Proc Natl Acad Sci USA. 1989;86:7861–7865. doi: 10.1073/pnas.86.20.7861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kerr W G, Heller M, Herzenberg L A. Analysis of lipopolysaccharide-response genes in B-lineage cells demonstrates that they can have differentiation stage-restricted expression and contain SH2 domains. Proc Natl Acad Sci USA. 1996;93:3947–3952. doi: 10.1073/pnas.93.9.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuo C H, Wells W W. β-Galactosidase from rat mammary gland. Its purification, properties, and role in the biosynthesis of 6β-O-d-galactopyranosyl myo-inositol. J Biol Chem. 1978;253:3550–3556. [PubMed] [Google Scholar]
- 28.LaKatos P, Stern P H. Effects of cyclosporins and transforming growth factor beta 1 on thyroid hormone action in cultured fetal rat limb bones. Calcif Tissue Int. 1992;50:123–128. doi: 10.1007/BF00298788. [DOI] [PubMed] [Google Scholar]
- 29.Lee N H, Weinstock K G, Kirkness E F, Earle-Hughes J A, Fuldner R A, Marmaros S, Glodek A, Gocayne J D, Adams M D, Kerlavage A R, Fraser C M, Venter J C. Comparative expressed-sequence-tag analysis of differential gene expression profiles in PC-12 cells before and after nerve growth factor treatment. Proc Natl Acad Sci USA. 1995;92:8303–8307. doi: 10.1073/pnas.92.18.8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang P, Pardee A B. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 31.Libermann T A, Nusbaum H R, Razon N, Kris R, Lax I, Soreq H, Whittle N, Waterfield M D, Ullrick A, Schlessinger J. Amplification, enhanced expression and possible rearrangement of EGF receptor gene in primary human brain tumors of glial origin. Nature. 1985;313:144–147. doi: 10.1038/313144a0. [DOI] [PubMed] [Google Scholar]
- 32.Markowitz S D, Roberts A B. Tumor suppressor activity of the TGF-β pathway in human cancers. Cytokine Growth Factor Rev. 1996;7:93–102. doi: 10.1016/1359-6101(96)00001-9. [DOI] [PubMed] [Google Scholar]
- 33.Markowitz D, Goff S, Bank A. Construction and use of a safe and efficient amphotropic packaging call line. Virology. 1988;167:400–406. [PubMed] [Google Scholar]
- 34.Massague J. TGF-β signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 35.Noguchi T, Inoue H, Tanaka T. The M1- and M2-type isozymes of rat pyruvate kinase are produced from the same gene by alternative RNA splicing. J Biol Chem. 1986;261:13807–13812. [PubMed] [Google Scholar]
- 36.Parry H D, Alphey L. The utilization of cloned DNAs to study gene organization and expression. In: Glover D M, Hames B D, editors. DNA cloning 1, core techniques. 2nd ed. Oxford, United Kingdom: IRL Press; 1987. pp. 177–178. [Google Scholar]
- 37.Reynisdottir I, Polyak K, Iavarone A, Massague J. Kip/Cip and Ink4 Cdk inhibitors cooperate to induce cell cycle arrest in response to TGF-β. Genes Dev. 1995;9:1831–1845. doi: 10.1101/gad.9.15.1831. [DOI] [PubMed] [Google Scholar]
- 38.Roberts A B, Anzano M A, Wakefield L M, Roche N S, Stern F D, Sporn M B. Type β transforming growth factor: a bifunctional regulator of cellular growth. Proc Natl Acad Sci USA. 1985;82:119–123. doi: 10.1073/pnas.82.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Russ A P, Friedel C, Ballas K, Kalina U, Zahn D, Strebhardt K, von Melchner H. Identification of genes induced by factor deprivation in hematopoietic cells undergoing apoptosis using gene-trap mutagenesis and site-specific recombination. Proc Natl Acad Sci USA. 1996;93:15279–15284. doi: 10.1073/pnas.93.26.15279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1987. [Google Scholar]
- 41.Sanes J R, Rubenstein J L, Nicolas J F. Use of a recombinant retrovirus to study post-implantation cell lineage in mouse embryos. EMBO J. 1986;5:3133–3142. doi: 10.1002/j.1460-2075.1986.tb04620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schena M, Shalon K, Davis R W, Brown P O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 43.Scheridin U, Rhodes K, Breindl M. Transcriptionally active genome regions are preferred targets for retrovirus integration. J Virol. 1990;64:907–912. doi: 10.1128/jvi.64.2.907-912.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simon H U, Mills G B, Kozlowski M, Hogg D, Branch D, Ishimi Y, Siminovitch K A. Molecular characterization of hNRP, a cDNA encoding a human nucleosome-assembly-protein-1-related gene product involved in the induction of cell proliferation. Biochem J. 1994;297:389–397. doi: 10.1042/bj2970389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sumitani S, Kasayama S, Sato B. Tyroid hormone inhibits androgen-enhanced DNA synthesis in Shionogi carcinoma 115 cells without affecting autocrine growth factor mRNA expression. J Steroid Biochem Mol Biol. 1994;50:5–11. doi: 10.1016/0960-0760(94)90166-x. [DOI] [PubMed] [Google Scholar]
- 46.Takeshita A, Cardona G R, Koibuchi N, Suen C S, Chin W W. TRAM-1 a novel 160 kDa thyroid hormone receptor activator molecule, exhibits distinct properties from steroid receptor coactivator-1. J Biol Chem. 1997;272:27629–27634. doi: 10.1074/jbc.272.44.27629. [DOI] [PubMed] [Google Scholar]
- 47.Thompson K L, Assoian R, Rosner M R, Marshall C J. Transforming growth factor-β increases transcription of the genes encoding the epidermal growth factor receptor and fibronectin in normal rat kidney fibroblasts. J Biol Chem. 1988;263:19519–19524. [PubMed] [Google Scholar]
- 48.Tong X, Howley P M. The bovine papillomavirus E6 oncoprotein interacts with paxillin and disrupts the actin cytoskeleton. Proc Natl Acad Sci USA. 1997;94:4412–4417. doi: 10.1073/pnas.94.9.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ulloa L, Doody J, Massague J. Inhibition of transforming growth factor-β/SMAD signalling by interferon-γ/STAT pathway. Nature. 1999;397:710–713. doi: 10.1038/17826. [DOI] [PubMed] [Google Scholar]
- 50.Vasmatzis G, Essand M, Brinkmann U, Lee B, Pastan I. Discovery of three genes specifically expressed in human prostate by expressed sequence tag database analysis. Proc Natl Acad Sci USA. 1998;95:300–304. doi: 10.1073/pnas.95.1.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Velculescu V E, Zhang L, Vogelstein B, Kizler K W. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Y, Derynck R. Regulation of Smad signaling by protein associations and signaling crosstalk. Trends Cell Biol. 1999;9:274–279. doi: 10.1016/s0962-8924(99)01579-2. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Y, Feng X-H, Derynck R. Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-β-induced transcription. Nature. 1998;394:909–913. doi: 10.1038/29814. [DOI] [PubMed] [Google Scholar]