Graphical abstract
Keywords: STORM, STochastic Optical Reconstruction Microscopy; STED, STimulated Emission Depletion; TIRF, Total Internal Reflection Fluorescence; ACE2, Angiotensin-Converting Enzyme 2; TMPRSS2, TransMembrane PRoteaSe Serine 2; SMLM, Single Molecule Localization Microscopy; ISM, Image Scanning Microscopy
Keywords: SARS-CoV-2 spike, B.1.1.7 variant of concern, Late entry, Clathrin, STED, dSTORM
Abstract
We exploited a multi-scale microscopy imaging toolbox to address some major issues related to SARS-CoV-2 interactions with host cells. Our approach harnesses both conventional and super-resolution fluorescence microscopy and easily matches the spatial scale of single-virus/cell checkpoints. After its validation through the characterization of infected cells and virus morphology, we leveraged this toolbox to reveal subtle issues related to the entry phase of SARS-CoV-2 variants in Vero E6 cells. Our results show that in Vero E6 cells the B.1.1.7 strain (aka Alpha Variant of Concern) is associated with much faster kinetics of endocytic uptake compared to its ancestor B.1.177. Given the cell-entry scenario dominated by the endosomal “late pathway”, the faster internalization of B.1.1.7 could be directly related to the N501Y mutation in the S protein, which is known to strengthen the binding of Spike receptor binding domain with ACE2. Remarkably, we also directly observed the central role of clathrin as a mediator of endocytosis in the late pathway of entry. In keeping with the clathrin-mediated endocytosis, we highlighted the non-raft membrane localization of ACE2. Overall, we believe that our fluorescence microscopy-based approach represents a fertile strategy to investigate the molecular features of SARS-CoV-2 interactions with cells.
1. Introduction
Since late 2019, SARS-CoV-2 has rapidly spread worldwide generating a pandemic with devastating social consequences. The development of a handful of novel and effective vaccines [1] represented a brilliant scientific achievement and it holds promise for a rapid end of the pandemic. Nonetheless, the way out of pandemic could be slowed by the emergence of novel SARS-CoV-2 lineages endowed with better ability to spread and infect humans while featuring lower in vitro susceptibility to neutralizing monoclonal and serum antibodies [2]. In this context, elucidation of structure–property relationships that modulate virus-cell host checkpoints, such as entry, replication, and egress, is crucial to assess the role of genome mutation on virus infectivity.
SARS-CoV-2 contains four structural proteins, namely spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins (Scheme 1a). S is a ∼180 kDa glycoprotein anchored in the viral membrane and protruding as homotrimers from the viral surface (the “corona”) [3]. S plays the most important roles in viral attachment, fusion and entry [4], [5]. The N-terminal S1 subunit contains the receptor binding domain (RBD) that mediates SARS-CoV-2 binding to the cell membrane receptor ACE2 [6]. Yet, this interaction could not be unique in accounting for virus interaction at cell membrane, as the host role of heparan sulfates [7], [8], [9] and neuropilin-1 [10] were recently highlighted. The C-terminal S2 subunit (Scheme 1b) is responsible for the fusion of the viral envelope with cellular membranes to deliver the viral RNA [11]. The S-mediated membrane fusion follows two proteolytic events: i) the “priming” cleavage that occurs at the S1/S2 interface, which yields S1 and S2 non-covalently bound in a pre-fusion conformation, and ii) the “activation” cleavage that occurs within the S2 subunit (S2′) to trigger the fusion process [12]. For several CoVs, including SARS-CoV-2, fusion can occur either at the plasma or endosomal membrane, according to the “early” and the “late” pathways of entry [13]. Availability of the transmembrane-bound protease TMPRSS2 favors virus entry through the early pathway [14]. Conversely, in TMPRSS2-negative cell lines, CoVs internalize by the late pathway and fusion is triggered by the Cathepsin B/L proteases [13].
Scheme 1.
Structure of SARS-CoV-2 and differences between B.1.177, B.1.1.7, and B.1Δ in the Spike protein. (a) SARS-CoV-2 structure. (b) Scheme of S protein. S is composed of the S1 and S2 subunits, which are further subdivided into SP: signal peptide, NTD: N-terminal domain, RBD: receptor binding domain, RBM: receptor Binding Motif, FP: fusion peptide, HR1-2: repetitive heptapeptides, TM: transmembrane domain, CP: cytoplasmic peptide. In the present study, three viral strains B.1.1.7 (aka: Alpha Variant of Concern), B.1.177, and B.1Δ were considered. All strains shared the D614G mutations, which became dominant worldwide in 2020 on account of promoting enhanced virus transmissibility. Compared to the original Wuhan strain (GenBank accession number NC_045512), the three strains contained the following mutations in the Spike protein: B.1Δ (D614G, R685del, S686del), B.1.177 (A222V, D614G, Q675H), B.1.1.7 (H69del, V70del, Y144del, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H). In the scheme, the two most relevant mutations of B.1.1.7 with respect to B.1.177 and B.1Δ are reported: N501Y in RBM and P681H at the S1/S2 cleavage site (indicated by the dashed line). On bottom, genome (NC_045512 used as reference genome) and primary sequence map of the S1/S2 boundary for virus isolates belonging to B.1.177, B.1.1.7, and B.1Δ strains. The proteolytic cleavage site 1 (furin) and 2 (Cathepsin L) are enclosed into dashed lines [19], and the relevant proteolyzed linkages are highlighted by blue and red wedges, respectively.
Quite remarkably, SARS-CoV-2 bears a polybasic amino acid insert (PRRA) at the S1/S2 junction (Scheme 1b). This site is potentially cleavable by furin, a protease commonly found in the secretory pathway of most cell lines [15]. Accordingly, a few studies suggest that SARS-CoV-2 may be primed at S1/S2 by furin (or related proteases) during maturation, thereby harboring a cleaved S protein during egress and secondary infection [16], [17]. The cleaved S protein seems to be advantageous for activating the early pathway in TMPRSS2-expressing cells [18].
The mechanistic knowledge of virus entry in cells is relevant for developing drugs tailored to prevent infection [20], [21]. For instance, therapeutic strategies aimed at inhibiting TMPRSS2 protease activity are currently under evaluation [22], [23]. The proposed, yet unclear and non-exclusive involvement of clathrin and caveolin-1 as mediators of endocytosis in the late pathway may afford further molecular targets to stop pathogenesis [24]. Nonetheless, mutations in the S protein may be crucial for the first step of viral transmission of novel SARS-CoV-2 variants, with significant epidemiological consequences. In particular, D614G became a dominant mutation in SARS-CoV-2 lineages that have been circulating worldwide since spring 2020. Mutation D614G seems associated with a selective infectivity advantage [25], possibly due to a more favorable entry phase [26] compared to wild-type SARS-CoV-2. A further evolutionary step of the virus occurred in late 2020 in the UK, where the B.1.1.7 SARS-CoV-2 variant (recently denominated as “Alpha” Variant of Concern by WHO) was firstly detected [25]. B.1.1.7 rapidly outcompeted older D614G strains in many countries [27], [28], due to its 40–70% higher transmissibility that could be associated with ∼ 30% higher mortality rates [29]. Although B.1.1.7 retains D614G, the enhanced transmissibility of this lineage appears to be related to two additional mutations in the S protein: 1) N501Y, which resides in the Receptor Binding Motif of S, and 2) P681H, which is next to the furin cleavage S1/S2 site (Scheme 1b). Yet, the exact mechanism that confers dominance to B.1.1.7 over the older D614G strains is still unclear, although it has been proposed that N501Y may increase the affinity for ACE2 receptor [30], [31] and/or P681H may modulate the amount of cleaved S protein harbored by infecting viruses thereby influencing their entry mechanism [32], [33].
Recent advances in fluorescence microscopy opened the way to individual virus imaging as a tool to understand viral life cycle. The dynamic and heterogeneous nature of virus-cell interactions is the perfect framework for highly sensitive imaging systems such as confocal fluorescence microscopy and Total Internal Reflection Fluorescence (TIRF) microscopy. Of note, TIRF enables imaging of a 100–150 nm layer above the coverslip where 2D cell cultures are adhered, it is thus ideally tailored to follow dynamic processes occurring at the cell membrane like viral entry. Yet, viruses such as CoVs have a size around 100 nm, i.e. well below the optical resolution of confocal and TIRF microscope on the focal plane (200–300 nm), and details of single viral particles interacting with subcellular structures may be only partially revealed with these techniques. Optical super-resolution methods that break the light-diffraction barrier either by leveraging on the photophysical properties of the fluorescent probe or by structuring the excitation light, may easily reach the 20–150 nm spatial scale [34]. Indeed, STimulated Emission Depletion (STED) and Single Molecule Localization Microscopy (SMLM) have been recently applied to image single viruses of different families at < 100 nm, also in the cellular context [35], [36], [37]. To our knowledge, however, no super-resolution imaging of full (or pseudotyped) SARS-CoV-2 interacting with cells was yet described in the literature, albeit some studies relying on conventional diffraction-limited microscopy have appeared [38].
In this study, we deploy for the first time a multi-scale fluorescence microscopy toolbox to investigate entry checkpoints of SARS-CoV-2 with two general goals: 1) demonstrate that imaging SARS-CoV-2 at single virus level does help answering biological questions that can only be partially addressed by in vitro techniques, and 2) highlight the ability of super-resolution techniques to afford morphology details of virus structure and molecular interactions with the cell. Our multi-scale toolbox was organized according to the resolution capability of each technique: confocal and TIRF microscopy (200–300 nm) were applied to visualize interactions at cell level; super-resolution microscopy techniques (image scanning microscopy, ISM [39], in airyscan mode [40], [41]: 120–180 nm, STED: 70–100 nm, SMLM: 25–40 nm) were applied (and validated) to reveal single-virus morphology and interactions with cell substructures. By our approach we shed light on the different endocytic uptake kinetics of variant B.1.1.7 compared to B.1.177, an older D614G lineage with large diffusion in Europe in late 2020 [42], as well as on the role of clathrin and caveolin in mediating the endocytic uptake of the virus in the late pathway in Vero E6 cells. Beside their own relevance, we believe that our results are representative of a new and fertile approach for the study of SARS-CoV-2 interactions with cells.
2. Results
2.1. Viral strains and imaging conditons
Three different SARS-CoV-2 strains, B.1Δ, B.1.177 and B.1.1.7, were used for all experiments (see Scheme 1 for Spike sequences). All strains were isolated on Vero E6 cells from nasopharyngeal swabs from COVID-19 patients at Laboratory of Microbiology and Virology, Vita-Salute San Raffaele University. The genomes of B.1.177 (hCoV-19/Italy/LOM-UniSR10/2021, GISAID Accession ID: EPI_ISL_2544194) and B.1.1.7 (hCoV-19/Italy/LOM-UniSR7/2021, GISAID Accession ID: EPI_ISL_1924880) were checked after the amplification passages to check the integrity of the spike “furin” polybasic cleavage site (CS) 681–686 (Scheme 1b). Western Blot analysis showed that in actual B.1.1.7 viruses the S protein was largely uncleaved, whereas 31% of S1/S2 cleavage was observed in B.1.177 virions (Supplementary Information, Fig. S1)
B.1Δ originated from a B.1 strain (hCoV-19/Italy/LOM-UniSR1/2020, GISAID Accession ID: EPI_ISL_413489) whose passaging in VeroE6 yielded a CS devoid of 685RS686 sequence (Scheme 1). For this reason, B.1Δ cannot be proteolytically cleaved by trypsin/furin-like proteases during virus maturation. In our study, B.1Δ has two benchmark roles: 1) it retains a mutation pattern of S protein like B.1.177, whose B.1 is direct ancestor, 2) it is a good structural benchmark of S protein of our B.1.1.7 strain, whose S protein in not S1/S2 cleaved as witnessed by Western Blot analysis (Supplementary Information, Fig. S1).
Adherent Vero E6 cells infected by B.1.177, B.1.1.7 or B.1Δ were methanol-fixed and immunostained by orthogonal anti-S or anti-N antibodies followed by fluorescently labeled secondary antibodies. The concomitant use of Alexa488 and Alexa647 dyes was suitable for both confocal/ISM (airyscan) and direct STORM (dSTORM). dSTORM exploits the intrinsic cycling of these fluorophores between bright (on) and dark (off) states to image and localize sparse single molecules at different times across a large field of view and reconstruct a pointillist super-resolved map of the labeled specimen [43]. Albeit Alexa488 is less efficient than Alexa647 in providing fluorescent photocycling, these two common dyes may be used together when two-color dSTORM is carried out by adopting tailored buffer solutions [44]. Conversely, STED nanoscopy requires stable and non-blinking fluorophores because the resolution improvement is performed by the targeted detection of non-depleted fluorophores and high photon flux is necessary [45]. Thus, we selected Atto594 and Atto647 for two-color STED imaging. Two-color STED images were acquired according to the separation of photons by lifetime tuning modality [46] (hereafter abbreviated as τ-STED). In some cases, we acquired two-color STED/Confocal images on samples prepared for dSTORM, by using Alexa488 as reporter for STED and Alexa647 as reporter for confocal imaging.
2.2. Imaging of infected Vero E6 cells
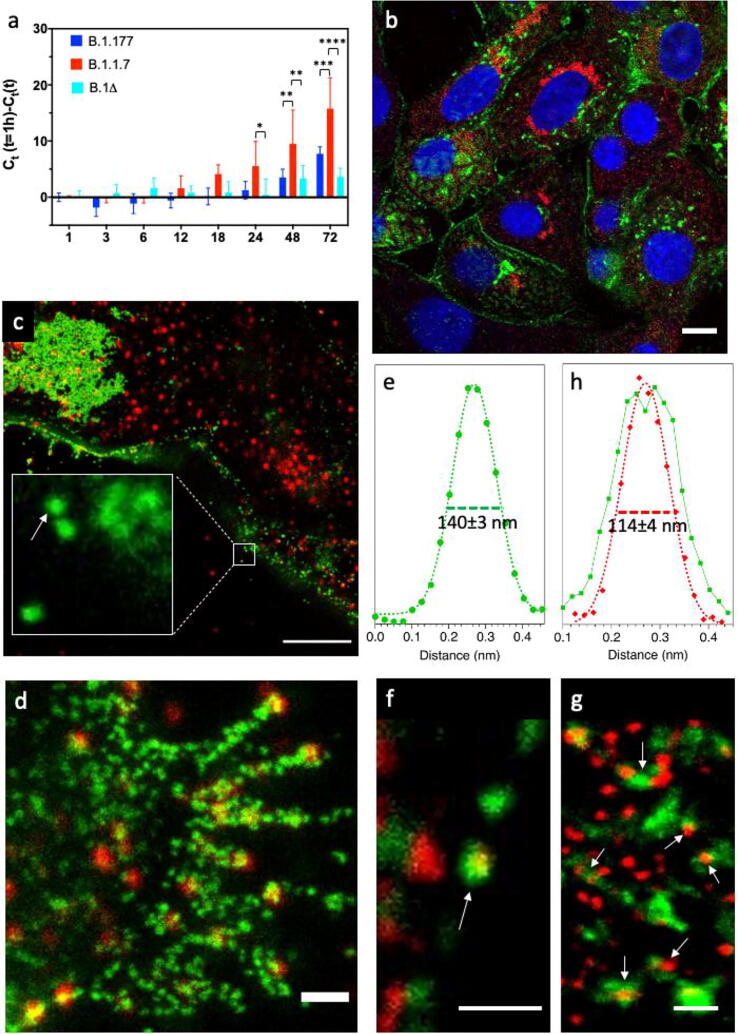
At first, we assessed the relative infection capabilities of Vero E6 by our three strains. Cells were inoculated for 1 h at 37 °C at 0.001 multiplicity of infection (MOI) and the amount of virus in the external medium was checked from 6 to 72 h post infection (hpi) by reverse transcriptase-polymerase chain reaction (RT-PCR). Significant virus production started at 24–48 h, depending on the strain (Fig. 1a). Of note, from 48 hpi onward the cells infected by B.1.1.7 resulted the most productive. Instead, B.1.177 and B.1Δ were characterized by statistically undistinguishable virus production.
Fig. 1.
Imaging of infected Vero E6 cells at 48 hpi. (a) Growth curves showing the release of viral genome into the medium of Vero E6 cells incubated for 1 h with viruses at 0.001 MOI.; Ct: threshold cycle; Ct(1 h): 44.9 (B.1.1.7), 33.0 (B.1.177), 34 (B.1Δ). (b) Confocal image of cells infected by B.1Δ. (c, d) STED (green)/Confocal (red) images of cell membrane regions at different zoom at hpi by B.1.177; inset of (c): single viral particles and small viral clusters attached to the cell membrane. (e) Horizontal intensity profile of the of the green fluorescence associated with the virus particle indicated by the arrow in inset of panel (c). (f, g) Dual color τ-STED images of cell regions at different zoom by B.1.177; arrows indicate a virion labeled on both the corona and core (f) and N-packed viral genome particles that are coating with the envelope and the spike corona (g). (h) Horizontal intensity profiles of the green and red fluorescence associated with the virus particle indicated by the arrow in (f). For all images, Blue: Hoechst 33342, Green: S protein, Red: N protein. Scale bar: 10 μm (b), 5 μm (c), 1 μm (d), 500 nm (f,g). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Infected cells at 48 hpi were directly investigated by dual-color confocal and STED microscopy upon orthogonal immunolabeling of S and N proteins. Coherently with a sustained viral maturation and egressing process, confocal imaging highlighted a significant fluorescent signal of S protein in the cytoplasm and along the plasma membrane of Vero E6 cells (Fig. 1b, green). Large patches of virions accumulated close to the plasma membrane (Fig. 1c, green), in structures that may be identified with virus morphogenesis matrix vesicae (VMMV) by comparison with electron microscopy images [47]. Leveraging on its nanoscale resolution (70 nm), STED demonstrated that S-fluorescent particles along membrane were composed of single viruses and very small clusters of about 2–3 virions (Fig. 1c, e). Indeed, a single S-labeled particle detected by STED was characterized by 140 mm FWHM (Fig. 1e): considering the STED resolution (FWHM) of 70 nm, we empirically determined the particle diameter d by the equation d = [(FWHMobj)2- (FWHMSTED)2]0.5 = 120 nm, a value in good agreement with the actual virion diameter including the spike corona (vide infra).
The N protein was found to accumulate in perinuclear regions (Fig. 1b, red) and in small clusters near the plasma membrane (Fig. 1c, red). These regions may be identified with sites of N-production or of N/RNA complexes in liquid-phase separated assemblies [48], [49]. Very interestingly, S and N protein colocalized only in a subset of egressing virions along the membrane (Fig. 1d).
In these spots, a smaller N-particle was embedded into a larger S-particle (Fig. 1g, h), in agreement with the core–shell structure of SARS-CoV-2. Dual-color τ-STED clearly highlighted in the cytoplasm several N-particles surrounded by a banana-shaped S-enriched region (Fig. 1h). These systems likely correspond to ERGIC regions where the envelope coating of the N-packed virus genome is taking place [47]. Of note, both S-coated and bare N-particles were characterized by FWHM 110 nm (Fig. 1h), yielding d = 85 nm, which is again in good agreement with the actual diameter of the virus core (vide infra).
2.3. Nanoscale determination of viral particle size
Given the higher resolution of dSTORM (average localization precision: 30 nm) and the strong z-sectioning of TIRF imaging mode (100–120 nm), we set out to investigate the size of single viral particles detected by dSTORM-TIRF on the basal plasma membrane during the entry (vide infra) and egress phases (Fig. 2).
Fig. 2.
Size of virions by dSTORM-TIRF. (a) TIRF image of a Vero E6 cell at 1 hpi (B.1.1.7 infection). (b) TIRF image of a Vero E6 cell at 72 hpi (B.1.177 infection). (c) Cluster analysis (DBSCAN) of the region of interest (ROI) enclosed in the white squares of (a); clusters are colored according to the relative number of localizations on 0–255 pseudocolor scale; pixel size: 10 nm, average localization precision (±SD): 32 ± 8 nm. (d) Zoom of the region enclosed in the white square of panel (c). (e, f) Distributions of viral particle radius (r) as obtained by cluster analysis: each dataset was fitted by Gaussian curve yielding: <r> = 50.3 ± 1.6 nm, SD: 13.4 ± 2.3 nm (B.1.1.7, S protein), <r> = 51.4 ± 1.1 nm, SD: 19.4 ± 2.6 nm (B.1.177, S protein), <r> = 43.9.4 ± 0.9 nm, SD: 15.2 ± 1.2 nm (B.1.177, N protein). Green: S protein (a), N protein (b). Scale bar: 5 μm (a,b), 1 μm (c), 500 nm (d). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To retrieve the particle size, single-molecule localization data were analyzed by density-based spatial clustering of applications with noise (DBSCAN), a clustering algorithm based on localization maps that can discover clusters of arbitrary shapes [50]. Remarkably, in the localization maps DBSCAN identified several clusters (Fig. 2c,d) characterized by high labeling density (20,000–600,000 localizations/μm2). Size-distributions of the clusters yielded the average particle radius for both B.1.177 and B.1.1.7 under different labeling conditions (Fig. 2e,f). We found out <r> = 50.3 ± 1.6 nm and <r> = 51.4 ± 1.1 nm for B.1.1.7 and B.1.177 when the S protein was labeled, and <r> = 43.9 ± 0.9 nm for B.1.177 when the N protein was labeled. According to our previous measurements (vide supra), we identified the S-positive particles on the membrane with fully enveloped viruses. As expected, our findings demonstrate that full B.1.1.7 and B.1.177 virions do not differ significantly in size. Notably, the smaller size of N-positive particles supports their identification with the N-packed virus genome, which is found also devoid of the Spike corona in the pre-coating phase (Fig. 1g).
To check whether our experimental results are in keeping with the published morphology data, we simulated the TIRF excitation of a virion labeled by a primary/secondary antibody couple on its corona and how the localization density of the S proteins resulted on the image plane (Supplementary Information, Fig. S2).
Given the cylindrical symmetry of the illumination system, we calculated the localization density as a function of the distance (ρ) from the center of a viral particle located at different distances from the basal plane (Supplementary Information, Fig. S2), to mimic different experimental conditions. Our simulation showed that the localization density grows up from ρ = 0 nm to ρ = 48–58 nm, depending on the labeling site on the S protein, to decrease slowly farther off (Supplementary Information, Fig. S2). This implies that the maximum fluorescent intensity of an S-labeled virus must be expected slightly above its envelope radius, in good agreement with our cluster analysis results.
2.4. Clathrin-mediated “Late pathway” of virus entry in Vero E6
Next, we set out to investigate the biological mechanism of virus access to the Vero E6 cells by our spatial multiscale imaging platform. Recent data pointed out how the entry pathway of SARS-CoV-2 is critically dependent on the presence of TMPRSS2 protease on the cell membrane [51]. Accordingly, we first checked for the presence of TMPRSS2 on our Vero E6 cell line. In full agreement with literature data [52], we found out that Vero E6 express almost no TMPRSS2 (Fig. 3a). In this experiment, HepG2 and Caco-2 cells were used as positive control of low and high TMPRSS2 expression.
Fig. 3.
Size of virions by dSTORM-TIRF. (a) RNA expression of TMPRSS2 in Vero E6, HepG2, and Caco-2 cells by quantitative real-time PCR. (b) Difference between released viral genome at 48 hpi following treatment with Cathepsin L inhibitor E64d and without the same inhibitor in Vero E6 cells and the cell lineage Vero E6TMPRSS2(+) permanently overexpressing TMPRSS2. Ct: threshold cycle.
In absence of TMPRSS2, SARS-CoV-2 is thought to enter cells by the late pathway, i.e. by the endosomal route [17], [51], whose milestone is the cleavage of S protein by endosomal cathepsins, particularly Cathepsin L [4]. For that reason, we set out to test the effect of cathepsin inhibitor E64d [33] on the viral infection in normal Vero E6 and a cell lineage permanently overexpressing the protease TMPRSS2 (Vero E6TMPRSS2(+)). At 48 hpi, infection by B.1.177 and B.1.1.7 was largely inhibited by E64d in Vero E6 (Fig. 3b). Conversely, E64d exerted a negligible effect when applied to Vero E6TMPRSS2(+). Taken together, these findings clearly demonstrate that the late pathway represents the predominant route of virus entry in Vero E6 cells.
The early endocytic events of the “late pathway” were investigated by our multiscale imaging platform adopting an infection scheme that enabled synchronization of virus entry [53]. Cells were pre-incubated with our strains for 1–3 h at 4 °C, allowing membrane attachment of the virus but preventing its endocytosis. After the chilling step, the non-attached virions were removed, and cells were incubated at 37 °C to promote viral entry until fixation at 1–3 hpi. This time span ensured the absence of almost any virus production and egress [51], [54].
As early as 1 hpi the cells inoculated with B.1.1.7 were significantly populated by virions likely undergoing their endocytic process (Fig. 4a). Conversely, cells infected by B.1.177 and B.1Δ displayed only a few scattered spots attributable to full viral particles (Fig. 4b, c). The signal due to N protein witnessed the ongoing release of virus genome in B.1.1.7-infected cells, but not in the other models (Fig. 4d, e). Nonetheless, more cells bearing membrane-adhered viral particles became visible at 2–3 hpi also for B.1.177 and B.1Δ (Fig. 5). These findings are in keeping with previous electron-microscopy studies that showed membrane-attached single and pairs of Wuhan SARS-CoV-2 viruses up to 6 hpi at onset of their internalization process [47], [55].
Fig. 4.
Imaging of the early phase (1 hpi) of virus entry. Cells were exposed to B.1.177 for 1 h at 4 °C, washed, kept for 1 h at 37 °C, and then fixed and immunolabeled. (a–c) STED (green)/Confocal (red) images of cells infected by B.1.1.7 (a), B.1.177 (b), B.1Δ (c) at 1hpi; the dotted lines depict the profile of the cells. (d, e) Zoomed perinuclear region of a cell infected by B.1.177 (d) or B.1.1.7 (e). In all panels the arrows indicate single virions with 130–140 nm size. For all images, Blue: Hoechst 33342, Green: S protein, Red: N protein. Scale bar: 5 μm (a-c), 1 μm (c,d). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 5.

Imaging of the early phase (2–3 hpi) of virus entry. Cells were exposed to B.1.177 for 1 h at 4 °C, washed, kept for 2 h at 37 °C, and then fixed and immunolabeled. Several viruses were located onto the cell plasma membrane, as witnessed by a 3D stack in confocal mode; a medial XY plane of the cell is visible in the main panel; on the right and below are reported the YZ and XZ sections corresponding to the dotted line, respectively. Blue: Hoechst 33342, Green: S protein, scale bar: 10 µm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Clathrin-mediated and caveolar endocytosis represent the most common initial step of virus endocytosis [56]. Accordingly, we investigated the endocytosis pathway of B.1.177 by quantitative colocalization imaging [57], by concomitantly labeling the S protein and clathrin or caveolin-1. Remarkably, dual-color ISM images alleged a significant colocalization between viral particles and clathrin, but not caveolin-1 (Fig. 6). This pattern was quantitatively confirmed by Pearson's coefficient R, which measures the stoichiometric correlation between the two fluorescent partners as a proxy of their functional association (Table 1).
Fig. 6.
During the early entry phase virions on the membrane colocalize with clathrin but not with Caveolin-1. (a) Confocal images of B.1.177 at 3 hpi (in red) with Caveolin-1 (in green); Green: Caveolin-1, red: S protein, scale bar: 10 µm. (b) Same as in (a) but Caveolin-1 is replaced by clathrin. (c-f) ISM images of regions in (a) or (b) enclosed in cyan and orange squares. Scale bar: 10 µm (a,b), 2 µm (c-f). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 1.
Pearson's coefficients for molecular partners imaged on cell membrane.
| ACE2 | RBD | Clathrin | Caveolin-1 | CD71 | |
|---|---|---|---|---|---|
| SARS-CoV-2 (S) | – | – | 0.37 ± 0.06 | 0.06 ± 0.04 | – |
| ACE2 | 0.69 ± 0.01 | 0.66 ± 0.07 | – | 0.01 ± 0.05 | 0.15 ± 0.02 |
Perfect stoichiometric correlation (R = 1) can never be achieved, owing to incomplete labeling, fluorescence background, and slight spatial mismatch of colors due to residual chromatic aberration. Accordingly, a positive control made of green/far-red doubly immunostained ACE2 receptor set the maximum achievable R to 0.69 ± 0.01. With this reference, we found a medium/strong functional association of S with clathrin, but a poor or negligible association with caveolin-1 (Table 1).
Most notably, nanoscale imaging by dSTORM-TIRF demonstrated that single virions fully overlap with clathrin clusters at the basal membrane (Fig. 7). This supports clathrin-mediated endocytosis of the full virus, which was questioned by recent electron microscopy results in Vero E6 showing some clathrin pits at 50–100 nm from membrane-attached virions likely to endocytose released viral material [47].
Fig. 7.
Virions are embedded into clathrin-coated pits. (a) TIRF image of Vero E6 cell after 3 h post-incubation at 4C (B.1.177 infection). (b) Two-color map of dSTORM-TIRF localization density of clathrin (green) and S (red): colocalized particles appear yellow; pixel size: 10 nm, average localization precision (±SD): 32 ± 10 nm (both channels). Green: clathrin, red: S protein. Scale bar: 10 µm (a), 1 µm (b). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
A further support to clathrin-mediated late entry of SARS-CoV-2 was provided by τ-STED measurements, which addressed the apical submembrane level where most colocalized signal was visible (Fig. 8). τ-STED images clearly showed single virions embedded into larger clathrin vesicles (170 ± 90 nm) that, albeit not resolved into the structural triskelion, can safely be attributed to clathrin pits.
Fig. 8.
Virions are mostly embedded into submembrane clathrin-coated pits. (a) τ-STED image of clathrin and B.1.177 viruses attached to cell membranes; Green: S protein, red: clathrin. (b) Zoom of the region enclosed in the white square in (a). (c) Histogram of diameters (D) of clathrin (red) and viral (blue) particles found in (b): <D>=170 + -90 nm (clathrin), <D>=120 + -36 nm (virions). Scale bar: 2 µm (a), 200 nm (b). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The clathrin-mediated endocytosis of SARS-CoV-2 is at odds with the controversial hypothesis that ACE2 resides in caveolin-enriched raft regions of the cell membrane in several cell lines, including Vero E6 [58], [59]. Accordingly, we set out to investigate the localization and functionality of ACE2 in the Vero E6 membrane by our microscopy toolbox. Confocal and TIRF imaging confirmed that ACE2 shows a prevalent membrane localization (Fig. 9a), with some minor cytoplasmic staining.
Fig. 9.
ACE2 and its molecular partners onto the membrane of Vero E6. (a) Membrane distribution of ACE2. (b) ISM colocalization image of ACE2 and SARS-CoV-2 RBD. (c) ISM colocalization image of ACE2 and CD71. (d) ISM colocalization image of ACE2 (red) and Caveolin-1 (green). Red: ACE2, green: RBD, Caveolin-1, CD71. Scale bar: 10 µm (a), 2 µm (b–d). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The functional receptor activity of membrane ACE2 towards SARS-CoV-2 was corroborated by the large colocalization with recombinant RBD of the S protein (Fig. 9b, Table 1). Also, we found a significant degree of colocalization of ACE2 with CD71, the transferrin receptor (Fig. 9b, Table 1). CD71 is known as a marker of the non-raft regions of the cell membrane [60] and its clathrin-mediated endocytosis upon stimulation is well documented [61]. Conversely, ISM images highlighted negligible ACE2 colocalization with caveolin-1 (Fig. 9c, Table 1).
We can conclude that ACE2 localizes poorly in caveolin-1-enriched membrane regions in Vero E6 cells, in agreement with the absence of caveolar endocytosis of SARS-CoV-2. Additionally, the observed colocalization with CD71 supports the presence of ACE2 in non-raft regions of the cell membrane, wherefrom it may activate clathrin-mediated endocytosis upon contact with the S protein of SARS-CoV-2.
3. Discussion
The ongoing COVID-19 pandemic makes imperative the full understanding of virus-host interactions. In this context, the pivotal role of the surface Spike (S) protein was early recognized since it mediates both the docking with the host cell receptor (ACE2) and the fusion process. The subtle interplay of S with the ACE2 receptor, its ability to hijack the cell endocytic machinery, and its intrinsic tunable fusogenic properties are directly related to the viral tropism. The SARS-CoV-2 S is the antigen encoded by available vaccines [1]. Also, the S glycoprotein represents the main target of therapeutic approaches aimed at neutralizing virus infectivity. S appears also key to viral adaptation to humans under selective pressure, and its sequence variability has already enabled the emergence of dominant viral variants harboring mutations such as the D614G present in lineage B.1.177 and the Alpha Variants of Concern (B.1.1.7 lineage).
In spite of the accumulated knowledge, the thorough elucidation of unclear checkpoints of S-mediated entry requires a research approach focused more on single-virus interaction with the host cell [36]. In this perspective, for the first time in virus-cell studies we leveraged the combination of conventional microscopy (confocal, TIRF) with super-resolution microscopy techniques (ISM, STED, dSTORM) whose common ability to image biological samples at < 200 nm scale is properly tailored to the size of SARS-CoV-2. This toolbox enables the imaging of single-virus interactions with cells at different spatial scales, affording both functional and structural details. Of note, we applied our spatial multiscale imaging toolbox to the real virus, because virus models may not fully recapitulate the complex arrangement of S protein on virus envelope and its interaction with the target cells [62].
In the first part of this work, we validated our multiscale imaging platform by addressing infected cells during the replication/assembly/release phases of virus cycle. Indeed, several studies based on electron microscopy have detailed the rich morphology of intracellular structures and complexes that sustain virus packing, budding, and egress. This knowledge can be exploited to benchmark collected optical data. By our approach, we were able to distinguish single integer viruses, virus assemblies such as virus morphogenesis matrix vesicae (VMMV), and the coating phase of N-packed virus genome particles attached to ERGIC. Additionally, leveraging on the nanoresolution of STED and dSTORM, we properly identified virus morphology by the characteristic diameters of its N-packed RNA core and of the outer S-corona.
In the second part of the work, we applied our multiscale imaging toolbox to investigate the early phase of virus entry in cells. The B.1.177 has been one of the dominant clades in several European countries from summer 2020 to early 2021, to be afterwards rapidly outcompeted by B.1.1.7, aka the Alpha variant of SARS-CoV-2, suggesting a transmission advantage likely conferred by its genome changes. Several effects can contribute to improved transmissibility, including more rapid viral replication, faster entry, and the ability to escape the innate immunity of the host [63]. Although present data are somewhat conflicting [64], [65], a recent report hinted at the ability of P681H mutation of B.1.1.7 to influence cleavability at the furin S1/S2 site (Scheme 1) in TMPRSS2 expressing cells, thereby modulating the cell entry of the virus [66]. Yet, the (verified) absence of TMPRSS2 on the surface of Vero E6 cells avoids the kinetic interference of protease-mediated priming and activation of S at the cell plasma membrane. Indeed, we verified that both strains gain access to Vero E6 cells through the “late pathway” i.e. via endosomal internalization and cathepsin-mediated cleavage and activation of S protein.
The “late pathway” of SARS-CoV-2 entry can be split in two different phases: first the activation of endocytic internalization of the viral particle (the “penetration” phase), then the endosome maturation which ends in the cathepsin-mediated activation of the Spike protein and the release of viral genome into the cytoplasm (the “uncoating” phase [67]). While recent studies shed light on many mechanistic details of the second phase, including its crucial dependence on structural characteristics of S such as the S1/S2 cleavage ratio, the first phase is still rather obscure. By direct imaging, we highlighted that B.1.1.7 is much faster in activating the endocytic internalization than B.1.177 and B.1Δ. This property does not seem to depend on the S1/S2 cleavage ratio, as B.1.1.7 and B.1Δ retains mostly full-length S protein, whereas B.1.177 shows 31% cleavage (Supplementary Information, Fig. S1). We are tempted to attribute the faster phenotype of B.1.1.7 to the N501Y replacement, which is thought to boost the binding interaction with the ACE2 receptor on the cell membrane [30], [31] and could thereby trigger more rapid activation of the endosomal machinery. Together with other factors (related to the mutation pattern of the viral genome), the quicker endosomal uptake may significantly contribute to the faster infection kinetics of B.1.1.7 upon 1 h exposure that we indeed observed.
The second question we addressed is the actual roles of clathrin and caveolin-1 in the late pathway of SARS-CoV-2. To our knowledge, no direct visualization of the protein mediator of endosomal entry followed by SARS-CoV-2 has been reported yet. Structural similarities with other CoVs such as NL63 [68] and SARS-CoV [69] allegedly point out clathrin as the likeliest endocytic mediator of SARS-CoV-2 entry. Indeed, clathrin-mediated internalization of the naked S protein and S-pseudotyped lentivirus has been recently demonstrated in HEK293T stably expressing ACE2, [70] whose lack of TMPRSS2 activates solely the late entry pathway. Surprisingly, however, other authors suggested for the same cell line an endocytic mechanism of S-pseudotyped lentivirus mediated by neither clathrin nor caveolin-1 [71]. These conflicting results question the use of pseudotyped virus models to study virus entry in cells. Discrepancies in cell entry mechanisms of pseudotyped virus have been encountered also in other contexts, and they could be related to differences in Spike protein trimer assembly and presentation in pseudotype models as obtained from different preparations [62]. The absence of other constitutive SARS-CoV-2 proteins from pseudoviruses could be another factor that limits the use of these models in studies of virus entry. At any rate, our results highlight the role of clathrin vesicles as major carriers of the integer SARS-CoV-2 virus from the surface to the early endosome in TMPRSS2-lacking Vero E6 cells. Conversely, caveolin-1 seems not to participate significantly in virus entry. This finding is in excellent agreement with the large exclusion of the ACE2 receptor from the caveolin-enriched raft domains of the cell membrane that we observed. The latter results are particularly interesting, as some authors alleged the role of lipid rafts and caveolin-1 in SARS-CoV entry [59], [72]. Yet, exclusion of ACE2 from these membrane regions was reported by other authors [73], [74] and a major role of caveolin-1 in ACE2 distribution and virus entry is not easy to reconcile with the popular hypothesis of a clathrin-mediated first step of late entry in CoVs.
4. Conclusions
A fluorescence microscopy imaging toolbox, which harnesses both conventional and super-resolution fluorescence microscopy and easily matches the spatial scale of single virus-cell checkpoints, was shown to tackle some relevant issues related to the entry of SARS-CoV-2 in Vero E6. This cell line constitutes a common model to study infection in non-airway cellular epithelia lacking the membrane TMPRSS2 protease, and therefore invaded by SARS-CoV-2 according to the “late pathway” of cell entry. The late pathway involves endocytic uptake (the “penetration” phase) of the virus followed by cathepsin-mediated activation of the S protein (the “uncoating” phase), which occurs at cleavage sites downstream of the classical “furin” polybasic cleavage site (CS) 681–686 [75]. Our attention focused on three strains of SARS-CoV-2 all bearing the D614G mutation: B.1.1.7 (aka: Alpha Variant of Concern), its ancestor B.1.177 (which was outcompeted in Europe by B.1.1.7 at the end of 2020), and B.1Δ, i.e. a parent B.1 strain whose CS does not contain the 685RS686 sequence and therefore cannot be proteolytically cleaved by trypsin/furin-like proteases during virus maturation. Notably, the S1/S2 uncleavable B.1Δ was selected as it represents a reference for B.1.177 mutations on S, while being a benchmark of S structure (S1/S2 cleavage) of our B.1.1.7 strain.
At first, we validate our multiscale imaging toolbox by revealing morphological features of infected cells and viruses that were previously described by electron microscopy (EM) experiments. Of note, nanoscopy imaging (STED or dSTORM) can provide enough information to identify different typologies of viral particles in the cell context, thereby adding functional information to the morphological inspection of EM.
We set out to investigate the endocytic uptake of the virus. Direct imaging of virions at 1 h after inoculation clearly indicate that B.1.1.7 outcompetes B.1.177 and B.1Δ in terms of a much faster endosomal uptake in Vero E6 cells. Given the structural similarity of S in B.1.1.7 and B.1Δ, the faster cell uptake of B.1.1.7 could be related to the stronger interactions of its S protein with the ACE2 receptor due to the N501Y substitution, which is not present in B.1.177 and B.1Δ
A second result of our approach was the clear and direct demonstration of clathrin as mediator of endocytosis in the late pathway, a mechanism that had been previously suggested only by analogy with other CoVs and from experiments with pseudotyped virus models. Notably, our data also reconciled clathrin-mediated endocytosis with ACE2 localization on membranes, showing that the receptor does not colocalize with caveolar-enriched regions, as previously reported.
In conclusion, we believe that our fluorescence microscopy imaging toolbox represents a fertile strategy to address urgent questions on virus-cell checkpoints at the single virus level, while avoiding the conflicting results sometimes obtained using models unable to recapitulate the desired viral phenotypes.
5. Materials and methods
For safety reasons, all the procedures of infection were performed in a Biohazard Safety Level 3 facility.
5.1. Cell lines and culture
African green monkey kidney cells (Vero E6) were obtained from ATCC (CRL-1586). Vero E6 were cultured in DMEM high glucose medium supplemented with heat-inactivated 10% fetal bovine serum (FBS) (Sigma-Aldrich, Milan, Italy), 2 mM L-glutamine, 10 U/ml penicillin, and 10 mg/ml streptomycin (Sigma-Aldrich, Milan, Italy), at 37 °C in the presence of 5% CO2.
TMPRSS2-expressing Vero E6 cells (Vero E6TMRPSS2(+)) were kindly provided by the National Institute for Biological Standards and Control (NIBSC) and were supplemented with 10% FBS and 1 mg/mL geneticin (G418).
5.2. Virus isolation and amplification
Clinical isolates of SARS-CoV-2, B.1.177 and B.1.1.7 (hCoV-19/Italy/LOM-UniSR10/2021, GISAID Accession ID: EPI_ISL_2544194 and hCoV-19/Italy/LOM-UniSR7/2021, GISAID Accession ID: EPI_ISL_1924880) were isolated on Vero E6 cells from nasopharyngeal swabs from COVID-19 patients in BSL-3 facility at Laboratory of Microbiology and Virology, Vita-Salute San Raffaele University.
An aliquot (0.8 mL) of the transport medium of the nasopharyngeal swab (COPAN's kit UTM® universal viral transport medium—COPAN) was mixed with an equal volume of DMEM without FBS and supplemented with a double concentration of P/S and Amphotericin B. The mixture was added to 80% confluent Vero E6 cells monolayer seeded into a 25 cm2 tissue culture flask. After 6 h adsorption at 37 °C, 1 mL of DMEM supplemented with 2% FBS and Amphotericin B were added. Twenty-four hours post-infection (hpi), 3 mL of DMEM supplemented with 2% FBS and Amphotericin B were added after a PBS wash. Live images were acquired (Olympus CKX41 inverted phase-contrast microscopy) daily for evidence of cytopathic effects (CPE), and aliquots were collected for viral RNA extraction and In-house one-step real-time RT-PCR assay. Five days post-infection (dpi) cells and supernatant were collected, aliquoted, and stored at − 80 °C (P1). For secondary (P2) virus stock, Vero E6 cells seeded into 25 cm2 tissue culture flasks were infected with 0.5 mL of P1 stored aliquot, and infected cells and supernatant were collected 48 hpi and stored at − 80 °C. For tertiary (P3) virus stock, Vero E6 cells seeded into 25 cm2 tissue culture flasks were infected with 0.2 mL of P2 stored aliquot and prepared as above described.
5.3. Virus titration
P3 virus stocks were titrated using both Plaque Reduction Assay (PRA, PFU/mL) and Endpoint Dilutions Assay (EDA, TCID50/mL). For PRA, confluent monolayers of Vero E6 cells seeded in 24-well plates were infected with 10-fold-dilutions of virus stock. After 1 h of adsorption at 37 °C, the cell-free virus was removed, and cells washed with PBS. Cells were then incubated for 46 h in DMEM containing 2% FBS and 0.5% agarose. After fixing and staining the cell monolayers, viral plaques were counted. For EDA, Vero E6 cells (3∙105 cells/mL) were seeded into 96-well plates and infected with base 10 dilutions of virus stock. After 1 h of adsorption at 37 °C, the cell-free virus was removed, and complete medium was added to cells after a PBS wash. After 72 h, cells were observed to evaluate CPE. TCID50/mL was calculated according to the Reed–Muench method.
5.4. Kinetic study of virus growth in cells
Vero E6 cells (3∙105 cells/mL) were seeded into 96-well plates and cultured for 1 day at 37 °C. Then, the cells were washed three times with PBS after 1 h of virus adsorption (0.001 MOI) at 37 °C, and cells were incubated for 1, 3, 6, 12, 18, 24, 48, and 72 hpi. The cell supernatants were collected at the different time points as well. Viral genome from the supernatants of both experimental settings was extracted and analyzed by real-time RT-PCR.
5.5. Inhibition of SARS-CoV2 infection by E64d
Vero E6 cells (3∙105 cells/mL) or Vero E6TMPRSS2(+) were seeded into 96-well plates and cultured for 1 day at 37 °C. Subsequently, cells were incubated 1 h before infection with 10 µM E64d, a Cathepsin L inhibitor, (Sigma Aldrich, Milan, Italy). Then, cells were infected for 1 h at 37 °C with B.1.177 or B.1.1.7 (0.001 MOI) virus. After virus adsorption, the cells were washed three times with PBS and incubated for 48 h with 10 µM E64d. Cells infected without E64d treatment were added as well as experimental control. The experiment was performed in triplicate, and the cell supernatants were collected for viral genome extraction and subsequent analysis by RT-PCR.
5.6. Virion purification and western blotting
Vero E6 cells (3∙105 cells/mL) were seeded into a 6-well plate and cultured for 1 day at 37 °C. Then, cells were infected for 1 h at 37 °C with B.1.177 or B.1.1.7 (0.01 MOI) virus. After virus adsorption, the cells were washed three times with PBS and incubated for 72 h. Cell supernatant was collected, heat inactivated (56 °C for 30 min) and clarified by low-speed spin. Virions in the medium were pelleted by ultracentrifugation through a 20% sucrose cushion at 26,000 rpm for 3 h at 4 °C in a BeckmanSW28 rotor. For western blot analysis, sample reducing agent NuPAGE 10x (Thermo Fisher Scientific) and LDS NuPAGE 4x (Thermo Fisher Scientific) were added to samples, which were heated at 85 °C for 3 min. Samples were loaded in equal volumes (5 and 15 μL) into a NuPAGE 4–12% Bis-Tris Gel (Thermo Fisher Scientific) and electrophoresed by SDS-Page (200 V for 50 min) in MOPS 1x buffer (Thermo Fisher Scientific). Protein was then transferred to a polyvinylidene difluoride (PVDF) membrane at 4 °C for 90 min in ice cold Western Transfer Buffer (25 mM Tris, 192 mM Glycine, MeOH 20% (v/v)). Membranes were then blotted with a pool of SARS-CoV Spike (S) specific antibodies (ab97387, Abcam; GTX632604, GeneTex), followed by probing with horseradish peroxidase (HRP)-conjugated anti-human antibody (A0293, Merck) and anti-Mouse (A4416, Merck) as secondary. Signal was developed by treating membranes with SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific) imaging on a ChemiDoc MP System (Bio-Rad #12003154). Densitometry was performed using ImageLab 6.0.1 (Bio-Rad #12012931).
5.7. Real-time RT-PCR of SARS-CoV-2
SARS-CoV-2 RNA relative amounts detected for each experimental condition as a cycle threshold (Ct) value were compared, with a mean Ct value determined for the positive infection control. The viral RNA was purified from 100 μL of all cell-free culture supernatant, using the QIAamp Viral RNA Mini Kit (Qiagen). The purified RNA was then used to perform the synthesis of first-strand complementary DNA, using the SuperScript First-Strand Synthesis System for RT-PCR (Thermo Fisher Scientific).
Real-time PCR, using the SYBR Green dye-based PCR amplification and detection method, was performed to detect the complementary DNA. We used the SYBR Green PCR Master Mix (Thermo Fisher Scientific), with the forward primer N2F (TTA CAA ACA TTG GCC GCA AA), the reverse primer N2R (GCG CGA CAT TCC GAA GAA). The PCR conditions were 95 °C for 2 min, 45 cycles of 95 °C for 20 s, annealing at 55 °C for 20 s and elongation at 72 °C for 30 s, followed by a final elongation at 72 °C for 10 min. RT-PCR was performed using the ABI-PRISM 7900HT Fast Real-Time instrument (Applied Biosystems) and optical-grade 96-well plates. Samples were run in duplicate, with a total volume of 20 μL.
5.8. Analysis of TMPRSS2 expression by real-time RT-PCR
Vero E6 cells were seeded in 6-well plates at 2∙105 cells/well and cultured for 1 day at 37 °C. Three wells were washed once with PBS, after that 1 mL TRIZOL (Thermo Fisher Scientific) was added. Samples were then transferred in 4 mL tubes and processed for total RNA extraction with RNA micro kit (ZYMO REAGENT) according to company protocol. RNA quantity and integrity were assessed with Qubit 4.0 fluorometer (Thermo Fisher Scientific) using Qubit RNA BR kit (Thermo Fisher) and Qubit RNA IQAssay kit respectively. 500 ng of each sample were reverse transcribed with iScript gDNA clear cDNA Synthesis kit (BioRad) according to kit protocol and 10 ng of cDNA were analyzed for TMPRSS2 expression by real-time RT-PCR on a CFX Connect Real Time System using SsoAdvancedSybrGreen Supermix (BioRad). Forward primer: ACTTCATCCTTCAGGTGTA, reverse primer: TTGTGTTCAGTTTCATAAAG. The amplification protocol was 2 min at 95 °C, 40 two-step cycles of 10 s at 95 °C and 30 s at 60 °C, final ramping from 65 °C to 95 °C with 0.5 °C increments at 5 sec/step, for amplicon melting profile. The cDNA of Caco-2 and HepG2 cells was used as a positive control of TMPRSS2 amplification. All values were normalized by the housekeeping gene RPL13A. All samples were run in duplicate.
5.9. Cell infection (synchronized) for immunofluorescence study of virus endocytosis
105 Vero E6 cells were seeded in 35 mm glass bottom dishes (Willco, Amsterdam) with 2 mL of culture medium and cultured for 1 day at 37 °C. Cells were then pre-chilled by incubation at 4 °C for 30 min. Subsequently, the medium was removed, and cells were infected at MOI 0.1 for 3 h keeping the temperature at 4 °C. At the end of incubation, the viral inoculum was removed, cells were gently washed with ice-cold PBS, and cell culture medium was added. Cells were incubated at 37 °C and 5% CO2 for 1,3,6, and 48 h. Next, the medium was removed, cells were washed 3 times with 500 µL of PBS, and finally fixed and permeabilized with ice-cold 100% methanol for 15 min. After methanol was removed, cells were rinsed again three times in PBS.
5.10. Primary antibodies for immunofluorescence studies
-
●
anti-S IgG rabbit monoclonal antibody (40592-V05H, Sino Biological), dilution: 1:200
-
●
anti-N IgG rabbit monoclonal antibody (40143-R019, Sino Biological), dilution: 1:200
-
●
anti-N IgG mouse monoclonal antibody (#33717, Cell Signaling), dilution: 1:6400
-
●
anti-ACE2 IgG rabbit monoclonal antibody (ab15348, AbCam), dilution: 1:200
-
●
anti-clathrin IgG mouse monoclonal antibody (sc-12734, SantaCruz), dilution: 1:200
-
●
anti-caveolin-1 IgG mouse monoclonal antibody (sc-70516, SantaCruz), dilution: 1:200
-
●
anti-CD71 IgG mouse monoclonal antibody (sc-65882, SantaCruz), dilution: 1:100
-
●
anti-α-tubulin IgG mouse monoclonal antibody (T5168, Merck), dilution: 1:1000
5.11. Secondary antibodies for immunofluorescence studies and combinations
-
●
donkey anti-rabbit IgG Alexa488-labeled monoclonal antibody (a21206, ThermoFisher), dilution: 1:500 (confocal, ISM, and dSTORM-TIRF experiments)
-
●
donkey anti-rabbit IgG Alexa647-labeled monoclonal antibody (a31573, ThermoFisher), dilution: 1:500 (confocal, ISM, and dSTORM-TIRF experiments)
-
●
donkey anti-mouse IgG Alexa488-labeled monoclonal antibody (a21202, ThermoFisher), dilution: 1:500 (confocal, ISM, and dSTORM-TIRF experiments)
-
●
donkey anti-mouse IgG Alexa647-labeled monoclonal antibody (a31571, ThermoFisher), dilution: 1:500 (confocal, ISM, and dSTORM-TIRF experiments)
-
●
goat anti-rabbit IgG Atto647N-labeled monoclonal antibody (40839, Merck), dilution: 1:200 (τ-STED experiments)
-
●
goat anti-mouse IgG Atto594-labeled monoclonal antibody (76085, Merck), dilution: 1:200 (τ-STED experiments)
5.12. Immunostaining of infected cells
Methanol-fixed infected cells and a methanol-fixed negative control were incubated overnight at 4C° with 150 μL of a solution of anti-S IgG or anti-N IgG in PBS + 3% BSA (Sigma-Aldrich, Milan, Italy). For colocalization experiments, the incubation solution was supplemented with anti-clathrin IgG or anti-caveolin-1 IgG. In one experiment on cells at 72 hpi, the incubation solution was supplemented with anti-tubulin IgG. After rinsing four times with PBS + 0.5% BSA (PBB), infected cells and negative controls were incubated for 1 h with a solution of 1–2 fluorescently-labeled secondary antibody/ies in PBB (see secondary antibody section in Materials and Methods) and then rinsed four times with PBB and three times with PBS. When required, cell nuclei were stained by exposure for 5 min to 1 mg/100 mL Hoechst 33,342 (ThermoFisher) in water.
5.13. Immunostaining of non-infected cells
105 Vero-E6 cells were seeded in 35 mm glass bottom dishes (Willco) with 2 mL of culture medium and cultured for 1 day at 37 °C. Then, cells were fixed with PFA 2% in PBS for 15 min, rinsed three times with PBS, permeabilized for 15 min with Triton-X 100 (Sigma) 0.1% in PBS for 15 min, and rinsed four times with PBS + 0.5% BSA (PBB). Fixed cells were incubated overnight at 4°C with 200 μL of a solution of anti-ACE2 IgG, and either Spike RBD-mFC Recombinant Protein (40592-V05H-100, SinoBiological), or anti-caveolin-1 IgG, or anti-CD71 IgG. After rinsing four times with PBB, immunolabeled cells and negative controls were incubated for 1 h with a solution of 2 fluorescent-labeled secondary antibodies in PBB (see secondary antibody section in Materials and Methods) and then rinsed four times with PBB and three times with PBS. When required, cell nuclei were stained by exposure for 5 min to 1 mg/100 mL Hoechst 33,342 (Thermo Fisher Scientific) in water.
5.14. Confocal and ISM microscopy
Fluorescence was measured by a confocal Zeiss LSM 880 with Airyscan (Carl Zeiss, Jena, Germany), supplied with GaAsP detectors (Gallium:Arsenide:Phosphide). Samples were viewed with a 63x Apochromat NA = 1.4 oil-immersion objective. We adopted 0.9x zoom for imaging multiple cells (1 pixel = 220 nm), and 2-6x zoom for imaging single cells; ISM imaging was carried out at zoom > 3. The pinhole size was set to 44 μm, which corresponds to 1 airy unit (AU) for the green acquisition channel. Pixel dwell time was adjusted to 1.52 us and 512x512 pixel or 1024x1024 images were collected. In confocal mode, we carried out concomitant acquisition for all channels line by line with line-average set to 4. In airyscan mode, we carried out a sequential acquisition for all channels with frame-average set to 4. The acquisition channels were set as follows:
-
●
Blue (Hoechst 33342): λex = 405 λem = 420–500 nm
-
●
Green (Alexa488): λex = 488, λem = 500–560 nm
-
●
Far-red (Alexa647): λex = 640, λem = 650–700 nm
Images were visualized and processed by the open-source software Fiji (NIH, Bethesda). Colocalization of the green and far-red images was quantified by Pearson's coefficient R according to the method by Costes et al. [76] by the colocalization threshold and colocalization test routines of Fiji.
5.15. Single molecule localization by dSTORM-TIRF
A commercial N-STORM TIRF microscope (Nikon Instruments), equipped with an oil immersion objective (CFI Apo TIRF 100×, NA 1.49, oil; Nikon) was used to acquire 40,000 frames at a 33 Hz frame rate using TIRF illumination. Excitation intensities were as follows: ∼0.5–1 KW/cm2 for the 647 nm readout (200 mW laser; MPB Communications), ∼0.1–0.2 KW/cm2 for the 488 nm readout (50 mW laser; Oxxius), and ∼35 W/cm2 for the 404 activation (100 mW laser; Coherent). For single color measurements, we set a repeating cycle of 1 activation frame at 404 nm / 3 readout frames at 647 nm or 488 nm. For double color measurements, we set a repeating cycle of 1 activation frame at 405 nm / 3 readout frames at 488 nm / 1 activation frame at 405 nm/3 readout frames at 647. Image detection was performed with an EMCCD camera (Andor iXon DU-897; Andor Technologies) with EM gain activated and set to 300. We set full TIRF excitation of the sample by changing the objective back-aperture illumination through the acquisition software of the Microscope (NIS Elements AR 5.20.01, Nikon). The Perfect Focus System (Nikon) was used during the entire recording process. Fluorescence-emitted signal was spectrally selected by the four-color dichroic mirrors (ZET405/488/561/647; Chroma) and filtered by a quadribandpass filter (ZT405/488/561/647; Chroma).
For imaging conditions, STORM imaging buffer was used containing a glucose oxidase solution as an oxygen scavenging system. Imaging buffer was prepared as follows. 690 μL of 50 mM Tris buffer (pH 8.0), containing 10 mM of NaCl and 10% w/v of glucose, were mixed with 25 μL of DL lactate (60% w/w syrup in water, Sigma Aldrich, L1375-100 ML) and 3.5 μL of COT (200 mM in DMSO). The solution was stored at 4C and filtered (220 nm) before use. Immediately prior to the use, the solution was mixed with 3.5 μL of GLOX solution, 3.5 μL of 2-mercaptoethanol, 25 μL of Cysteamine (1 M in H2O, Sigma Aldrich, 30070-10G), and 45 μL of Oxyrase. The resulting solution was added to the petri dish, which was sealed with aluminum tape. GLOX solution was composed of glucose oxidase (14 mg) and Catalase (50 μL, 17 mg/mL) dissolved in buffer A (200 μL). GLOX solution was stored at 4C for a maximum of 14 days.
5.16. Single molecule localization analysis
Acquired dSTORM stacks were processed by Thunderstorm, a Fiji plugin for PALM and STORM data analysis [77]. At first, we set the properties of acquisition by the “Camera setup” menu: pixel size = 158.7 nm, Photoelectrons per A/D count: 2.5, Base level: 100 counts, EM gain: 300. Then, we carried out the localization algorithm (“Run analysis”), setting the following parameters: a) pre-filter: difference of averaging filters with 3 and 6 pixels as first and second kernel size, respectively; b) approximate localization of molecules by local maximum method with threshold 200 and 8-neighbourhood connectivity; c) sub-pixel localization by the Integrated Gaussian method, performing least-squares multi-fitting (threshold p = 1E-6) with initial sigma 1.6 pixels and fitting radius 3 pixels, maximum 5 molecules for fitting region with limit intensity range 1–1000 photons. Eventually, we cleaned the obtained results from drift and those localizations not strictly lying on the focal plane by the following post-filtering algorithm: a) removal of first 500 frames; b) drift correction by correlation; c) merging reactivated molecules (max distance: 20 nm, max off frames: 1, limited frames per molecule); d) removal of localizations with: (intensity = 1000 AND sigma > 180 nm AND uncertainty > 130 nm).
5.17. Single molecule localization density maps and cluster analysis
Single molecule localization maps and cluster analysis were collected by the LocAlization Microscopy Analyzer software (LAMA), available for download at http://share.smb.uni-frankfurt.de/index.php/software-menue/lama. Before LAMA analysis, the localization list exported by Thunderstorm was converted to the Molecular Accuracy Localization Keep (MALK) format used by the LAMA by the localization converter routine of the LAMA software.
Single molecule localization density maps were obtained by the Visualization routine of LAMA and consisted of 2D histograms of the localization list obtained by using a pixel size of 10x10 nm and codified into a 0–255 color map. For dual color images, before visualization, the localization lists of both colors were spatially registered by the register cabinet of the LAMA by using localization lists of multicolor beads. This procedure is extensively described in the documentation file accompanying the LAMA software, which can be downloaded at https://share.smb.uni-frankfurt.de/index.php/component/jdownloads/download/4-lama-tutorial/8-lama-documentation.
Hierarchical Cluster Analysis (HCA) was performed by the Density-Based Algorithm for Discovering Clusters in Large Spatial Databases with Noise (DBSCAN) preceded by Ordering Points To Identify the Clustering Structure (OPTICS) algorithm [50]. OPTICS-DBSCAN was performed an 8x8 um Region of Interest (ROI) (3–5 for each cell) by setting the minimum cluster size to 5 localizations and the noise level to 10%.
5.18. Stimulated emission depletion microscopy (τ-STED)
Lifetime-tuning STED (τ-STED) measurements were performed using a Leica STELLARIS 8 Falcon τ-STED (Leica Microsystems, Mannheim, Germany) inverted confocal/STED microscope. Excitation was provided by a White Light Laser and selecting the following wavelengths by the acoustic-optical tunable filter (AOTF): 488 nm, 560 nm, and 638 nm. Detection has been performed by the embedded tunable spectrometer in the 500–550 nm, 570–630 nm, 660–750 nm ranges respectively, and three Power HyD detectors. The pinhole was set to 0.6–1 Airy size. Line scanning speed ranged from 10 to 1400 Hz in standard acquisition mode. In τ-STED mode, the 775 nm pulsed laser beam is superimposed at a typical power of 100–250 mW before the objective. Two-colors τ-STED has been performed sequentially by line for the red and far-red fluorophores. Green fluorophores are not affected by the depletion beam at 775 nm.
5.19. Graphics and statistics
Graphs were prepared using Prism 7 (GraphPad) and IgorPro8 (Wavemetrics) software. Data are shown as the mean +/- SEM. Statistical analysis was performed by Prism 7 (GraphPad). For comparisons amongst Cycle thresholds (Ct) the 2way ANOVA multiple comparisons with corrections were performed by using Prism 9 (GraphPad).
Author contributions
BS, FB, AD, MP, RB designed the study; BS, PQ, NC, PB, GS, RB planned and performed research; CDP, NC, NM, EC, PGS, VC, GL, EP performed research; BS, PQ, CDP, NC, NM, EC, VC, EP, MC, PB, GS, RB analyzed data; BS, PQ, CDP, NC, GF, ML, MC, FB, AD, MP, RZ, PB, GS, RB wrote and revised the paper.
Funding
This research was supported by MIUR, Progetto di Ricerca di Interesse Nazionale, (bando PRIN 2017, Project n. 2017KM79NN), Regione Lombardia-Fondazione CARIPLO, bando POR-FERS 2014-2020 (grant PAN-ANTICOVID-19). All funding sources had no involvement in study design, collection, analysis, interpretation of data, writing the report; and decision to submit the article for publication.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Dr. Pasqualantonio Pingue (NEST, Scuola Normale Superiore) and Dr. Michele Oneto (IIT Nanophysics) are gratefully acknowledged for technical assistance and support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.10.038.
Contributor Information
Paolo Bianchini, Email: paolo.bianchini@iit.it.
Giovanni Signore, Email: g.signore@fpscience.it.
Ranieri Bizzarri, Email: ranieri.bizzarri@unipi.it.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Li D.D., Li Q.H. SARS-CoV-2: vaccines in the pandemic era. Military Med Res. 2021;8 doi: 10.1186/s40779-020-00296-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen R.E., et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat Med. 2021 doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yao H., Song Y., Chen Y., Wu N., Xu J., Sun C., et al. Molecular architecture of the SARS-CoV-2 Virus. Cell. 2020;183(3):730–738.e13. doi: 10.1016/j.cell.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Comm. 2020;11(1) doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saponaro F., Rutigliano G., Sestito S., Bandini L., Storti B., Bizzarri R., et al. ACE2 in the Era of SARS-CoV-2: controversies and novel perspectives. Front Mol Biosci. 2020;7:588618. doi: 10.3389/fmolb.2020.588618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clausen T.M., et al. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell. 2020;183:1043. doi: 10.1016/j.cell.2020.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hudak A., Letoha A., Szilak L., Letoha T. Contribution of syndecans to the cellular entry of SARS-CoV-2. Int J Mol Sci. 2021;22:5336. doi: 10.3390/ijms22105336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qiao Y.Y., et al. Targeting transcriptional regulation of SARS-CoV-2 entry factors ACE2 and TMPRSS2. Proc Natl Acad Sci USA. 2021;118 doi: 10.1073/pnas.2021450118. Artn Bsr20210290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daly J.L., et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020;370:861. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., et al. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci U S A. 2020;117(21):11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benton D.J., et al. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature. 2020;588 doi: 10.1038/s41586-020-2772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chambers J.P., Yu J., Valdes J.J., Arulanandam B.P., Franciosa G. SARS-CoV-2, early entry events. J Pathog. 2020;2020:1–11. doi: 10.1155/2020/9238696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaimes J.A., Millet J.K., Whittaker G.R. Proteolytic Cleavage of the SARS-CoV-2 Spike Protein and the Role of the Novel S1/S2 Site. iScience. 2020;23 doi: 10.1016/j.isci.2020.101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang T., Bidon M., Jaimes J.A., Whittaker G.R., Daniel S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antivir Res. 2020;178:104792. doi: 10.1016/j.antiviral.2020.104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang T., Jaimes J.A., Bidon M.K., Straus M.R., Daniel S., Whittaker G.R. Proteolytic Activation of SARS-CoV-2 Spike at the S1/S2 Boundary: Potential Role of Proteases beyond Furin. ACS Infect Dis. 2021;7(2):264–272. doi: 10.1021/acsinfecdis.0c00701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffmann M., Kleine-Weber H., Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol Cell. 2020;78(4):779–784.e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furinlike cleavage site absent in CoV of the same clade. Antivir Res. 2020;176 doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seyedpour S., Khodaei B., Loghman A.H., Seyedpour N., Kisomi M.F., Balibegloo M., et al. Targeted therapy strategies against SARS-CoV-2 cell entry mechanisms: a systematic review of in vitro and in vivo studies. J Cell Physiol. 2021;236(4):2364–2392. doi: 10.1002/jcp.30032. [DOI] [PubMed] [Google Scholar]
- 21.Xia S., et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J., Zhan P., Liu X.Y. Targeting the entry step of SARS-CoV-2: a promising therapeutic approach. Signal Transduct Tar. 2020;5 doi: 10.1038/s41392-020-0195-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qiao Y.Y., et al. Targeting transcriptional regulation of SARS-CoV-2 entry factors ACE2 and TMPRSS2. Proc Natl Acad Sci USA. 2021;118 doi: 10.1073/pnas.2021450118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glebov O.O. Understanding SARS-CoV-2 endocytosis for COVID-19 drug repurposing. Febs J. 2020 doi: 10.1111/febs.15369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Volz E., et al. Evaluating the Effects of SARS-CoV-2 Spike Mutation D614G on Transmissibility and Pathogenicity. Cell. 2021;184:64. doi: 10.1016/j.cell.2020.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ozono S., Zhang Y., Ode H., Sano K., Tan T.S., Imai K., et al. SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nat Commun. 2021;12(1) doi: 10.1038/s41467-021-21118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cyranoski D. Alarming covid variants show key role of genomic surveillance. Nature. 2021;589:337–338. doi: 10.1038/d41586-021-00065-4. [DOI] [PubMed] [Google Scholar]
- 28.Washington N.L., et al. Emergence and rapid transmission of SARS-CoV-2 B.1.1.7 in the United States. Cell. 2021;184:2587. doi: 10.1016/j.cell.2021.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iacobucci G. Covid-19: New UK variant may be linked to increased death rate, early data indicate. Bmj-Brit Med J. 2021;372 doi: 10.1136/bmj.n230. [DOI] [PubMed] [Google Scholar]
- 30.Laffeber C, de Koning K, Kanaar R, Lebbink JH. Experimental evidence for enhanced receptor binding by rapidly spreading SARS-CoV-2 variants. bioRxiv. 2021 doi: 10.1101/2021.02.22.432357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ali F., Kasry A., Amin M. The new SARS-CoV-2 strain shows a stronger binding affinity to ACE2 due to N501Y mutant. Med Drug Discov. 2021;10:100086. doi: 10.1016/j.medidd.2021.100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davies N.G., et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372:149. doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peacock T.P., Goldhill D.H., Zhou J., Baillon L., Frise R., Swann O.C., et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat Microbiol. 2021;6(7):899–909. doi: 10.1038/s41564-021-00908-w. [DOI] [PubMed] [Google Scholar]
- 34.Diaspro A., Bianchini P. Optical Nanoscopy. Riv Nuovo Cimento. 2020;43(8):385–455. doi: 10.1007/s40766-020-00008-1. [DOI] [Google Scholar]
- 35.Chojnacki J., Eggeling C. Super-resolution fluorescence microscopy studies of human immunodeficiency virus. Retrovirology. 2018:15. doi: 10.1186/s12977-018-0424-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Castelletto S, Boretti A. Viral particle imaging by super-resolution fluorescence microscopy. Chemical Physics Impact. 2021:2. [Google Scholar]
- 37.Banerjee S., Maurya S., Roy R. Single-molecule fluorescence imaging: Generating insights into molecular interactions in virology. J Biosciences. 2018;43(3):519–540. doi: 10.1007/s12038-018-9769-y. [DOI] [PubMed] [Google Scholar]
- 38.Putlyaeva LV, Lukyanov KA. Studying SARS-CoV-2 with Fluorescence Microscopy. Int J Mol Sci 2021; 22. ARTN 6558 10.3390/ijms22126558. [DOI] [PMC free article] [PubMed]
- 39.Castello M., Tortarolo G., Buttafava M., Deguchi T., Villa F., Koho S., et al. A robust and versatile platform for image scanning microscopy enabling super-resolution FLIM. Nat Methods. 2019;16(2):175–178. doi: 10.1038/s41592-018-0291-9. [DOI] [PubMed] [Google Scholar]
- 40.Huff J. The Airyscan detector from ZEISS: confocal imaging with improved signal-to-noise ratio and super-resolution. Nat Methods. 2015;12(12):i–ii. doi: 10.1038/nmeth.f.388. [DOI] [Google Scholar]
- 41.Storti B., Civita S., Faraci P., Maroni G., Krishnan I., Levantini E., et al. Fluorescence imaging of biochemical relationship between ubiquitinated histone 2A and Polycomb complex protein BMI1. Biophys Chem. 2019;253:106225. doi: 10.1016/j.bpc.2019.106225. [DOI] [PubMed] [Google Scholar]
- 42.Hodcroft E.B., et al. Spread of a SARS-CoV-2 variant through Europe in the summer of 2020. Nature. 2021;595:707-+. doi: 10.1038/s41586-021-03677-y. [DOI] [PubMed] [Google Scholar]
- 43.Grove J. Super-resolution microscopy: a virus’ eye view of the cell. Viruses. 2014;6:1365–1378. doi: 10.3390/v6031365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nahidiazar L, Agronskaia AV, Broertjes J, van den Broek B, Jalink K. Optimizing Imaging Conditions for Demanding Multi-Color Super Resolution Localization Microscopy. PLoS ONE 2016; 11. ARTN e0158884 10.1371/journal.pone.0158884. [DOI] [PMC free article] [PubMed]
- 45.Vicidomini G., Bianchini P., Diaspro A. STED super-resolved microscopy. Nat Methods. 2018;15(3):173–182. doi: 10.1038/nmeth.4593. [DOI] [PubMed] [Google Scholar]
- 46.Lanzano L, Hernandez IC, Castello M, Gratton E, Diaspro A, Vicidomini G. Encoding and decoding spatio-temporal information for super-resolution microscopy. Nat Comm. 2015;6 doi: 10.1038/ncomms7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brahim Belhaouari D., Fontanini A., Baudoin J.-P., Haddad G., Le Bideau M., Bou Khalil J.Y., et al. The strengths of scanning electron microscopy in deciphering SARS-CoV-2 infectious Cycle. Front Microbiol. 2020;11 doi: 10.3389/fmicb.2020.02014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jack A, et al. SARS-CoV-2 nucleocapsid protein forms condensates with viral genomic RNA. Plos Biol 2021; 19. ARTN e3001425 10.1371/journal.pbio.3001425. [DOI] [PMC free article] [PubMed]
- 49.Zhao M, et al. GCG inhibits SARS-CoV-2 replication by disrupting the liquid phase condensation of its nucleocapsid protein. Nat Comm 2021; 12. ARTN 2114 10.1038/s41467-021-22297-8. [DOI] [PMC free article] [PubMed]
- 50.Khater I.M., Nabi I.R., Hamarneh G. A review of super-resolution single-molecule localization microscopy cluster analysis and quantification methods. Patterns (N Y) 2020;1(3):100038. doi: 10.1016/j.patter.2020.100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koch J, Uckeley ZM, Doldan P, Stanifer M, Boulant S, Lozach PY. TMPRSS2 expression dictates the entry route used by SARS-CoV-2 to infect host cells. Embo J 2021; 40. ARTN e107821 10.15252/embj.2021107821. [DOI] [PMC free article] [PubMed]
- 52.Matsuyama S., Nao N., Shirato K., Kawase M., Saito S., Takayama I., et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci U S A. 2020;117(13):7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tai C.J., Li C.L., Tai C.J., Wang C.K., Lin L.T. Early viral entry assays for the identification and evaluation of antiviral compounds. Jove-J Vis Exp. 2015 doi: 10.3791/53124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ogando NS, et al. SARS-coronavirus-2 replication in Vero E6 cells: replication kinetics, rapid adaptation and cytopathology. J Gen Virol. 2020;101:925–940. doi: 10.1099/jgv.0.001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Caldas L.A., Carneiro F.A., Higa L.M., Monteiro F.L., da Silva G.P., da Costa L.J., et al. Ultrastructural analysis of SARS-CoV-2 interactions with the host cell via high resolution scanning electron microscopy. Sci Rep. 2020;10(1) doi: 10.1038/s41598-020-73162-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mercer J., Schelhaas M., Helenius A. Virus Entry by Endocytosis. Annu Rev Biochem. 2010;79(1):803–833. doi: 10.1146/annurev-biochem-060208-104626. [DOI] [PubMed] [Google Scholar]
- 57.Zinchuk V., Zinchuk O., Okada T. Quantitative colocalization analysis of multicolor confocal immunofluorescence microscopy images: Pushing pixels to explore biological phenomena. Acta Histochem Cytochem. 2007;40(4):101–111. doi: 10.1267/ahc.07002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang H, Yuan Z, Pavel MA, Hansen SB. The role of high cholesterol in age-related COVID19 lethality. bioRxiv. 2020 doi: 10.1101/2020.05.09.086249. [DOI] [Google Scholar]
- 59.Lu Y., Liu D.X., Tam J.P. Lipid rafts are involved in SARS-CoV entry into Vero E6 cells. Biochem Bioph Res Co. 2008;369(2):344–349. doi: 10.1016/j.bbrc.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harder T., Scheiffele P., Verkade P., Simons K. Lipid domain structure of the plasma membrane revealed by patching of membrane components. J Cell Biol. 1998;141:929–942. doi: 10.1083/jcb.141.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rappoport J.Z., Simon S.M. Real-time analysis of clathrin-mediated endocytosis during cell migration. J Cell Sci. 2003;116:847–855. doi: 10.1242/jcs.00289. [DOI] [PubMed] [Google Scholar]
- 62.Daniloski Z, et al. The Spike D614G mutation increases SARS-CoV-2 infection of multiple human cell types. Elife. 2021 doi: 10.7554/eLife.65365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thorne LG, et al. Evolution of enhanced innate immune evasion by the SARS-CoV-2 B.1.1.7 UK variant. bioRxiv. 2021 doi: 10.1101/2021.06.06.446826. [DOI] [Google Scholar]
- 64.Walker AS, et al. Increased infections, but not viral burden, with a new SARS-CoV-2 variant. medRxiv. 2021 doi: 10.1101/2021.01.13.21249721. [DOI] [Google Scholar]
- 65.Golubchik T, et al. Early analysis of a potential link between viral load and the N501Y mutation in the SARS-COV-2 spike protein. medRxiv. 2021 doi: 10.1101/2021.01.12.20249080. [DOI] [Google Scholar]
- 66.Brown JC, et al. Increased transmission of SARS-CoV-2 lineage B.1.1.7 (VOC 2020212/01) is not accounted for by a replicative advantage in primary airway cells or antibody escape. bioRxiv. 2021 doi: 10.1101/2021.02.24.432576. [DOI] [Google Scholar]
- 67.Ryu W.-S. Academic Press; 2017. In: Molecular Virology of Human Pathogenic Viruses) [Google Scholar]
- 68.Milewska A, et al. Entry of Human Coronavirus NL63 into the Cell. J Virol. 2018:92. doi: 10.1128/JVI.01933-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Inoue Y., Tanaka N., Tanaka Y., Inoue S., Morita K., Zhuang M., et al. Clathrin-dependent entry of severe acute respiratory syndrome coronavirus into target cells expressing ACE2 with the cytoplasmic tail deleted. J Virol. 2007;81(16):8722–8729. doi: 10.1128/JVI.00253-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bayati A., Kumar R., Francis V., McPherson P.S. SARS-CoV-2 infects cells following viral entry via clathrin-mediated endocytosis. J Biol Chem. 2021;100306 doi: 10.1016/j.jbc.2021.100306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li X., Zhu W., Fan M., Zhang J., Peng Y., Huang F., et al. Dependence of SARS-CoV-2 infection on cholesterol-rich lipid raft and endosomal acidification. Comput Struct Biotech J. 2021;19:1933–1943. doi: 10.1016/j.csbj.2021.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Glende J., Schwegmann-Wessels C., Al-Falah M., Pfefferle S., Qu X., Deng H., et al. Importance of cholesterol-rich membrane microdomains in the interaction of the S protein of SARS-coronavirus with the cellular receptor angiotensin-converting enzyme 2. Virology. 2008;381(2):215–221. doi: 10.1016/j.virol.2008.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li G.-M., Li Y.-G., Yamate M., Li S.-M., Ikuta K. Lipid rafts play an important role in the early stage of severe acute respiratory syndrome-coronavirus life cycle. Microbes Infect. 2007;9(1):96–102. doi: 10.1016/j.micinf.2006.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Warner F.J., Lew R.A., Smith A.I., Lambert D.W., Hooper N.M., Turner A.J. Angiotensin-converting enzyme 2 (ACE2), but not ACE, is preferentially localized to the apical surface of polarized kidney cells. J Biol Chem. 2005;280(47):39353–39362. doi: 10.1074/jbc.M508914200. [DOI] [PubMed] [Google Scholar]
- 75.Bollavaram K., et al. Multiple sites on SARS-CoV-2 spike protein are susceptible to proteolysis by cathepsins B, K, L, S, and V. Protein Sci. 2021;30:1131–1143. doi: 10.1002/pro.4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Costes S.V., Daelemans D., Cho E.H., Dobbin Z., Pavlakis G., Lockett S. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys J. 2004;86(6):3993–4003. doi: 10.1529/biophysj.103.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ovesny M., Krizek P., Borkovec J., Svindrych Z.K., Hagen G.M. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics. 2014;30:2389–2390. doi: 10.1093/bioinformatics/btu202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.