Abstract
Remdesivir is one of a few antiviral drugs approved for treating severe cases of coronavirus 2 (SARS-CoV-2) infection in hospitalized patients. The prodrug is a nucleoside analog that interferes with viral replication by inhibiting viral RNA-dependent RNA polymerase. The drug has also been shown to be a weak inhibitor of human mitochondrial RNA polymerase, leaving open the possibility of mitochondrial off-targets and toxicity. The investigation was designed to explore whether remdesivir causes mitochondrial toxicity, using both genomic and functional parameters in the assessment. Human-derived HepG2 liver cells were exposed for up to 48 h in culture to increasing concentrations of remdesivir. At sub-cytotoxic concentrations (<1 μM), the drug failed to alter either the number of copies or the expression of the mitochondrial genome. mtDNA copy number was unaffected as was the relative rates of expression of mtDNA-encoded and nuclear encoded subunits of complexes I and IV of the mitochondrial respiratory chain. Consistent with this is the observation that remdesivir was without effect on mitochondrial respiration, including basal respiration, proton leak, maximum uncoupled respiration, spare respiratory capacity or coupling efficiency. We conclude that although remdesivir has weak inhibitory activity towards mitochondrial RNA polymerase, mitochondria are not primary off-targets for the mechanism of cytotoxicity of the drug.
Keywords: Mitochondria, Off-Target, COVID, Remdesivir
1. Introduction
Remdesivir (Veklury®, Gilead Sciences) is a phosphoramidate substituted nucleotide analog that is incorporated by RNA-dependent RNA polymerase into and truncates replication of viral RNA. The drug possesses potent and broad spectrum antiviral activity, including severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Agostini et al., 2018). Based on recent favorable clinical trials (Beigel et al., 2020; Goldman et al., 2020; Spinner et al., 2020), the drug was approved by the U.S. FDA in 2020 for the treatment of hospitalized patients 12 years of age and older and weighing at least 40 kg with severe COVID-19 (Grein et al., 2020).
Remdesivir (RDV) is a prodrug and when administered to mammals is hydrolyzed to the monophosphate metabolite, which is subsequently phosphorylated to the pharmacologically active adenosine triphosphate analog (RDV-TP). This phosphorylated nucleoside analog is recognized as a substrate for RNA-dependent RNA polymerase and is incorporated into and inhibits replication of the viral RNA (Tchesnokov et al., 2019). As a result, RDV-TP interrupts by truncating the synthesis of the viral genome, including that of SARS-CoV-2 (Gordon et al., 2020). The critical factor that endows RDV its specificity as an antiviral agent is that the phosphorylated nucleoside analog is a favorable substrate for viral but not human mitochondrial RNA polymerase (Tchesnokov et al., 2019; Warren et al., 2016). The dissociation constants (Km) for RDV inhibiting Ebola virus, respiratory syncytial virus and human mitochondrial RNA polymerase are 5.7 μM, 0.5 μM, and 21 μM, respectively (Tchesnokov et al., 2019). Preclinical safety studies with a broad panel of 87 cellular and biochemical assays demonstrated that clinically relevant and non-cytotoxic concentrations of RDV and its monophosphorylated analog have minimal off-target effects in numerous primary and transformed human cells lines (Xu et al., 2021).
Being a nucleoside analog, however, raises concerns regarding the well-established clinical history of mitochondrial toxicity associated with nucleoside analog antiviral drugs (Kakuda, 2000). This concern is exemplified by the nucleoside analog reverse transcriptase inhibitors (NARTIs) effective in limiting replication of the AIDS virus. In addition to truncating the viral genome, these NARTIs also inhibit mitochondrial gene replication leading to mitochondrial depletion and metabolic dysfunction in affected patients (Fosslien, 2001; Simpson et al., 1989). Being an analog of ATP, the phosphorylated metabolite of RDV may also interfere with adenine nucleotide-dependent metabolic regulation, including mitochondrial oxidative phosphorylation (Feng, 2018; Lund and Wallace, 2004a, Lund and Wallace, 2004b). In light of the growing concerns for mitochondrial off-targets of drug action (Wallace and Starkov, 2000), we investigated the potential hazard of RDV causing depletion of the mitochondrial genome or transcriptome and/or deficits in mitochondrial function that could possibly compromise the clinical efficacy of this drug in treating critical COVID-19 positive patients.
2. Methods
2.1. Cell culture
The human hepatoma cell line HepG2/C3A was purchased from American Tissue Culture Collection (ATCC; Manassas, VA) and grown in 75 cm2 flasks with Eagle's Minimum Essential Media (EMEM) supplemented with 10% fetal bovine serum (FBS) at 37 °C in a humidified atmosphere of 5%CO2. Each replicate was a culture started from a single vial of frozen cells, with a passage number between 9 and 10 from original stock. For SRB, mtDNA copy number, and RT-qPCR assays, cells were passaged to 24-well tissue culture plates at 80% confluence and allowed to attach overnight. For mitochondrial function assay the cells were passaged to 96-well Seahorse Bioscience® XF96 cell culture plates at a density of 40 K cells per well.
2.2. Chemical exposure
Remdesivir was purchased form Cayman Chemicals (Item no. 30354) as the monophosphoramidate prodrug and 1000× stock solutions were prepared in anhydrous DMSO. Remdesivir stock was diluted in growth media and serial dilutions made such that the concentration of DMSO was 0.1% in all dilutions. On the day of exposure, the media was removed from each well and replaced with treated media and the cells incubated for 24 to 72 h.
2.3. Cytotoxicity
To establish an appropriate does range for RDV, cell killing was measured by sulphorhodamine B (SRB) binding assay (Skehan P. et al. 1990). Cells were seeded at 50% confluence on 24-well culture plates. After exposure, the cells were washed twice with PBS then fixed with ice cold 1% acetic acid in methanol for 30 min. The plate was then rinsed with 1% acetic acid and stained with 0.5% SRB in 1% acetic acid for 30 min. The SRB stain was discarded and the plate was rinsed with 1% acetic acid until the wash was clear. After air drying, the bound SRB was solubilized in 10 mM Tris Base and the absorbance measured at 540 nm on a SpectraMax® M3 microplate reader (Moledular Devices, Inc.). The amount of dye bound by the cells is directly proportional to the number of attached cells and thus a reliable indicator of cell proliferation and killing.
2.4. Mitochondrial function (Seahorse)
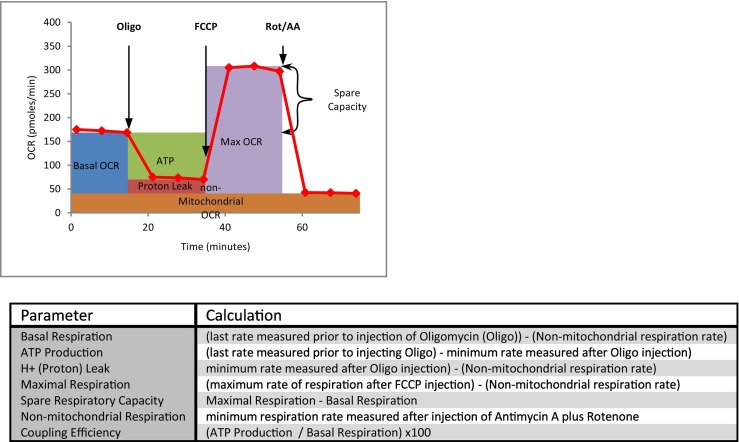
HepG2/C3A cells plated at 40 K and 20 K per well on Agilent Seahorse 96-well culture plates were exposed to RDV in EMEM growth media or 24 and 48 h respectively. On the day of the assay, the media was replaced with Seahorse XF DMEM Medium, pH 7.4 (Agilent, 103575–100) containing 5 mM HEPES, 10 mM glucose, 1 mM sodium pyruvate, and 2 mM l-glutamine and equilibrated in a non-CO2, 37 °C incubator for 30 min prior to starting the assay. The Seahorse Cell Mito Stress Test assay consisted of sequential additions of optimized concentrations of oligomycin (2 μM), FCCP (2 μM) and rotenone/antimycin A (0.5 μM) recording the rate of oxygen consumption between intervals of addition. Parameter calculations were made using the Seahorse XF Cell Mito Stress Test Report Generator application (Fig. 4; Agilent).
Fig. 4.
Illustration and Definition of Respiratory Parameters. The upper panel illustrates the sequence of additions for the Seahorse XF Extracellular Flux Analyzer Mito Stress Test and the areas reflecting the corresponding respiratory parameters as listed and defined in the lower panel.
2.5. RT-qPCR
Gene expression was measured by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). After exposure, the cells were washed with phosphate buffered saline (PBS) and total RNA was extracted using the RNeasy Plus Mini Kit (Qiagen). 350 μl lysis buffer was added directly to each well and the plate mixed by vortex. The cell lysate was homogenized with 5 passes through a 27-gauge needle fitted to a tuberculin syringe and transferred to the gDNA eliminator spin column provided in the kit. Washing and centrifugation steps were carried out according to the manufacturer's protocol. RNA was eluted in RNAse free water and quantified spectrophotometrically at 260 nm on a NanoDrop ND1000 (NanoDrop Technologies Inc. Wilmington, DE). cDNA libraries were prepared by reverse transcription using the Omniscript RT Kit (Qiagen, 205113) with 250 ng of total RNA. Gene expression was measured by quantitative PCR (qPCR) using the FastStart Essential DNA Green Master mix (Roche, 06924204001) on a LightCycler 96 system (Roche Molecular Systems Inc.). Gene specific primers (Table 1 ) were designed using the National Center for Biotechnology Information (NCBI) Primer-Blast application (Ye, 2012). Quantification was made against a five-point, ten-fold serial dilution of gene target specific PCR product. 2′-C-methyladeonsoine (2-C-MA; Santa Cruz Biotechnology) was included as a positive control for inhibiting mitochondrial RNA Polymerase.
Table 1.
Gene specific primer sequences and mRNA/DNA source sequence identification.
| Gene Symbol | mRNA accession | Forward primer | Reverse primer |
|---|---|---|---|
| B2M | NG_012920.2 | TGT TCC TGC TGG GTA GCT CT | CCT CCA TGA TGC TGC TTA CA |
| COX5B | NM_001862.3 | TTG CTG CTA GTC GCG GAC GC | TCC CTC TCC AAC CCA GTC GCC |
| DDIT3 | NM_001195053.1 | AGT CAT TGC CTT TCT CCT TCG GGA | AAG CAG GGT CAA GAG TGG TGA AGA |
| MT-CO2 (COX2) | NC_012920.1 | CGA GTA CAC CGA CTA CGG CGG | AGT CGC AGG TCG CCT GGT TCT |
| mtDNA 262–388 | NC_012920.1 | CAC TTT CCA CAC AGA CAT CA | TGG TTA GGC TGG TGT TAG GG |
| MT-ND2 | NC_012920.1 | CAC CCC TCT GAC ATC CGG CCT | TCC ACC TCA ACT GCC TGC TAT GAT |
| NDUFB3 v1 | NM_002491.3 | TTC CGG TTG GCT CCG GTT GCA | CGG CCC CAT GGA TCC CTT AGC |
2.6. Mitochondrial DNA copy number
Mitochondrial (mtDNA) and nuclear (nDNA) DNA were measured by quantitative PCR (qPCR) using the FastStart Essential DNA Green Master mix (Roche, 06924204001) on a LightCycler 96 system (Roche Molecular Systems Inc.). Gene specific primers (Table 1.) for the nuclear encoded B2M and the 262–388 bp range of mitochondrial DNA were purchased from Integrated DNA Technologies (IDT, Corralville, IA) using previously published sequences (Malik et al., 2011). Quantification of gene target was made against a five-point, ten-fold serial dilutions of gene specific DNA standard the values are reported as the ratio of mtDNA copies per nDNA copies. 40 nM (ddC; Sigma-Aldrich) served as a noncytotoxic positive control for mtDNA depletion.
2.7. Statistical analyses
Data were analyzed by Single Factor Analysis of Variance (ANOVA) followed by Dunnett's multiple comparison procedure (Dunnett, 1955) with alpha of 0.05 using JMP pro statistical analysis software version 12.0.1 (SAS, Cary, NC.).
3. Results
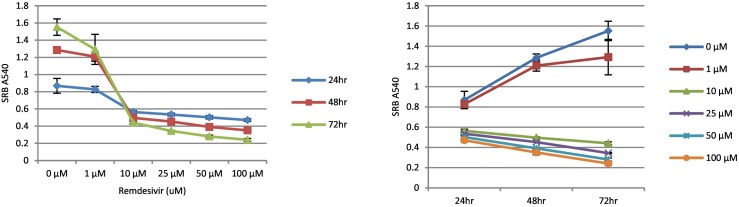
The goal of the investigation was to test the hypothesis that RDV interferes with the molecular regulation of bioenergetic properties of human-derived cells at sub-cytotoxic and clinically relevant drug concentrations. Accordingly, the first objective was to establish the time- and dose-dependent cytotoxicity of RDV, which are illustrated in Fig. 1 . Panel A shows the dose-dependent decrease in cell viability at each of three time points (1, 2, and 3 days), as reflected by the concentration of SRB-reactive protein. In the absence of RDV, SRB absorbance increased as a function of time in culture consistent with cell proliferation under control conditions. In contrast, concentrations of 10 μM RDV and higher caused near complete loss of cells at all three time points. 1 μM Remdesivir caused a loss of viable cells, the degree to which increased with increasing exposure time. The concentration-dependent decrease in cell viability is further illustrated in panel 1B, which shows that at 10 μM remdesivir and above SRB-reactive protein decreased over time whereas SRB absorbance increased with time for control cells as well as cells exposed to 1 μM drug. Based on these results, we established the standard sub-cytotoxic exposure for all subsequent experiments as 1 μM RDV for 48 h.
Fig. 1.
Remdesivir Time- and Dose-dependent Cytotoxicity – HepG2/C3A cells were exposed to remdesivir for up to 72 h in 24-well plates containing increasing μM concentrations of drug. At the end of the exposure the cells were fixed with methanol and stained with sulphorhodamine B (SRB). The bound SRB was solubilized in 10 mM Tris base and the absorbance measured 540 nm. Error bars reflect the standard deviation of duplicate wells (n = 1) for each concentration of remdesivir.
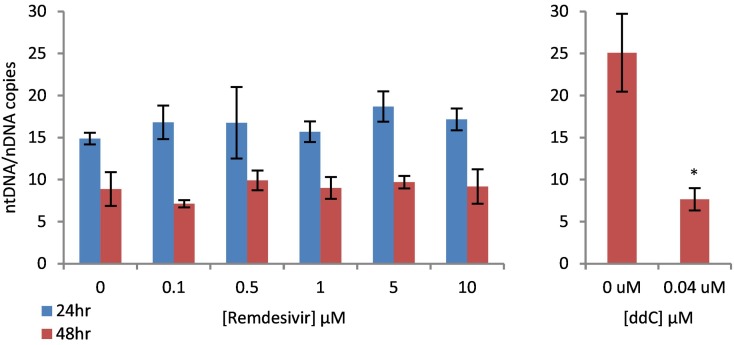
Fig. 2 illustrates the effect of increasing concentrations of RDV on mitochondrial DNA (mtDNA) copy number at both 24 and 48 h time points. mtDNA copy number is measured as the ratio of mitochondrial versus nuclear encoded gene targets recommended by Malik et al. (Malik et al., 2011). As demonstrated, mtDNA copy number for HepG2 cells was between 5 and 25 for all concentrations of RDV at both 24 and 48 h; there was no statistically significant treatment effect. It is noteworthy that mtDNA copy number was consistently higher at 24 h compared to 48 h for all treatments, which we attribute to possibly dilution bias regarding mtDNA as described by Malik et al. (Malik et al., 2011). Regardless, the results demonstrate that RDV, under these exposure conditions, does not interfere with mitochondrial DNA replication in HepG2 cells in culture, even at the cytostatic or cytotoxic concentration of 10 μM. Conversely, the positive control 2′, 3′ –dideoxycytidine (ddC) caused significant mtDNA depletion at the noncytotoxic concentration of 40 nM (panel 2B).
Fig. 2.
Dose-dependent Effect of Remdesivir on Mitochondrial DNA Copy Number in HepG2 Cells – A) HepG2 cells were exposed to increasing μM concentrations of remdesivir for 24 or 48 h. At the end of the exposure, DNA was extracted and quantified by qPCR using the nuclear (nDNA) and mitochondrial (mtDNA) gene targets recommended by Malik at el. (2011). B) HepG2 cells were exposed to sub-cytotoxic concentration of 2′, 3′ –dideoxycytidine (40 nM ddC) for 48 h as a positive control for mtDNA depletion. The values are expressed as mtDNA/nDNA copies and expressed as the mean ± S.D. for 3 independent measures. Asterisks indicate statistically significant difference from vehicle control (Student's t-test P < 0.05).
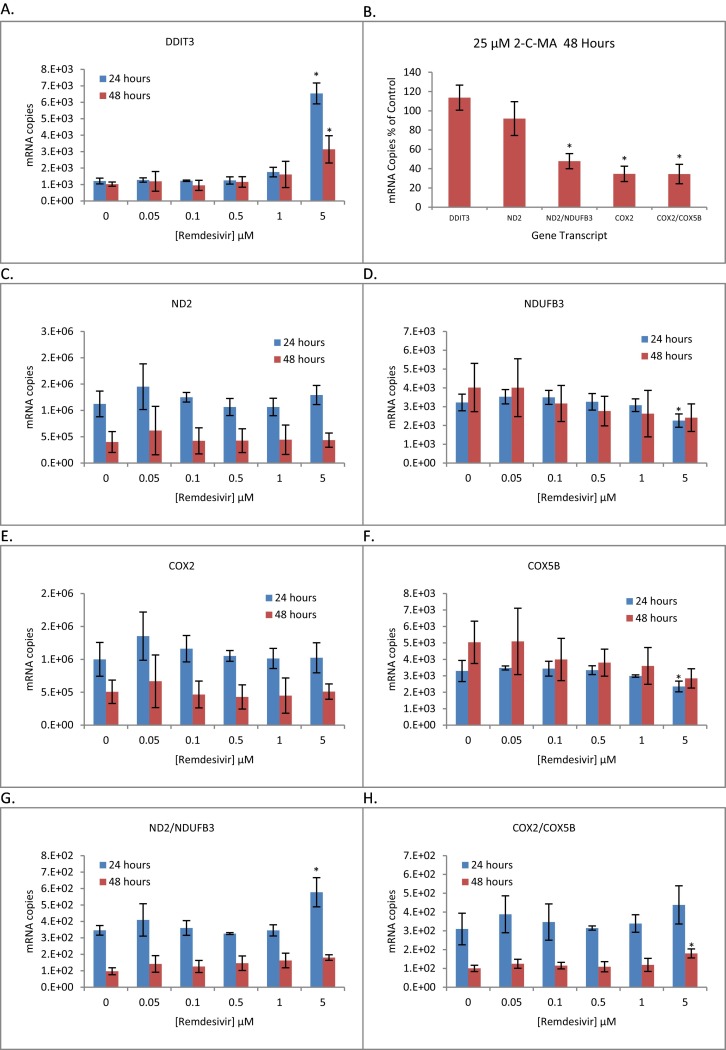
Similarly, we found no evidence that RDV interferes with mitochondrial gene transcription under these conditions (Fig. 3 ). This contrasts with the results for the positive control 2-C-MA, which at 25 μM inhibited mitochondria-specific gene expression at noncytotoxic concentrations (Ddit3 gene expression). The criteria we employed as a measure of mitochondrial transcription was a treatment-related effect on the ratio of RT-qPCR quantified transcripts for genes encoded by the mitochondrial versus nuclear genomes using two paired gene sets ND2/NDUFB3 and COX2/COX5B; ND2 and COX2 being mitochondrial encoded genes of respiratory complexes I and IV, respectfully, and NDUFB3 and COX5B being encoded by the nuclear genome. For this set of experiments, we conducted a second preliminary study to exclude cytotoxicity as a potential contributing factor by quantitative PCR of the DNA damage inducible transcript 3 (DDIT3) gene transcript, which is part of the unfolded protein response to ER stress. Ddit3 is induced under conditions of severe cellular stress (Los et al., 1999; Oyadomari and Mori, 2004) and we have used this transcript previously as a sensitive and specific quantifiable measure of early cell injury (Bjork and Wallace, 2009). The results for the time and dose dependent cytotoxicity measured with the DDIT3 transcript (Fig. 3A) is consistent with that observed using the SRB assay (Fig. 1); 5 μM RDV caused significant cytotoxicity and was not considered when evaluating the direct effects of the drug. Ddit3 gene expression with 1 μM RDV was increased slightly, but was not statistically significant with an n = 3 samples.
Fig. 3.
Dose-dependent Effect of Remdesivir on Mitochondrial and Nuclear Gene Expression. Panels A and C–H) HepG2 cells were exposed in culture to increasing concentrations of remdesivir for 24 and 48 h. RNA was extracted and RT-qPCR performed to quantify gene transcripts. Values are mRNA copies normalized to 18 s ribosomal RNA. B) HepG2 cells were exposed to a sub-toxic concentration of 2′-C-methyladeonsoine (25 μM 2-C-MA) as a positive control for mitochondrial RNA polymerase inhibition. Values are normalized to 18 s RNA and expressed as a percent of control. The plots represent the mean ± S.D. for 3 independent replications and asterisks indicate a statistically significant difference compared to the 0 μM dose group (Dunnett's p < 0.05).
Viewing the individual gene transcripts (Fig. 3, panels B–E), RDV failed to alter the quantitative expression of the mitochondrial encoded genes at any concentration and effected the nuclear encoded genes only at a 5 μM concentration, which is cytotoxic to the cells (Fig. 3A). Likewise, the ratio of mitochondrial to nuclear encoded gene transcripts (Fig. 3F and G) was not altered by concentrations of RDV that were not also cytotoxic to the cells.
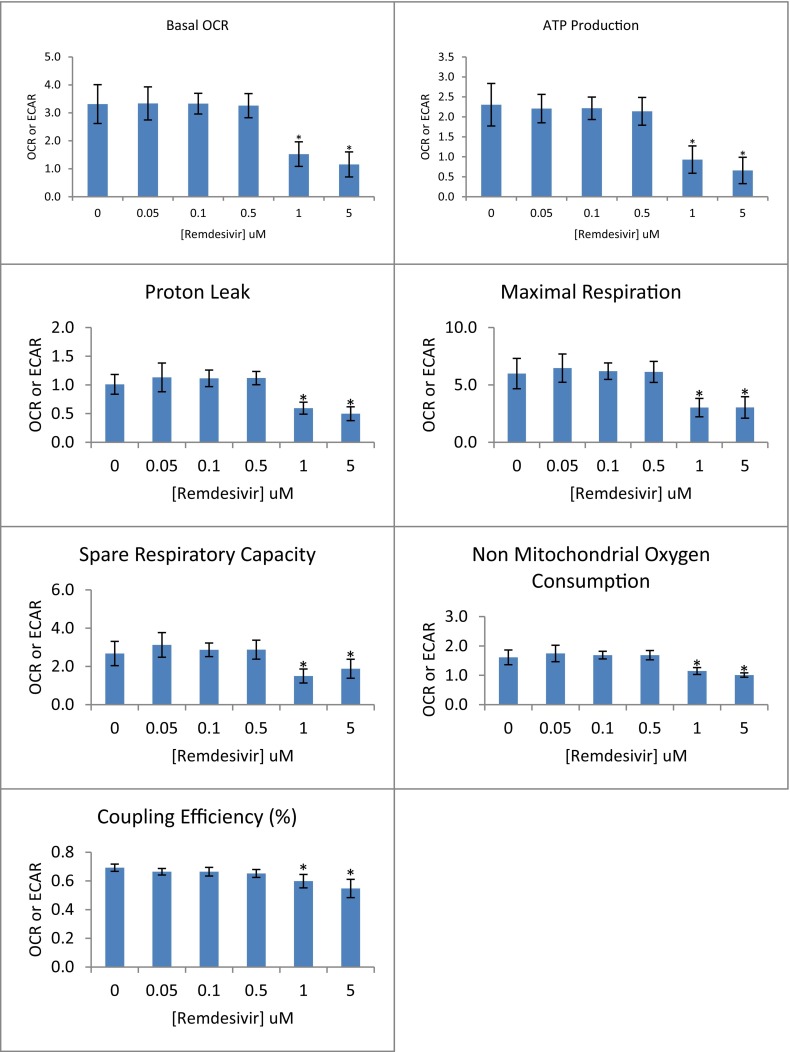
Our final objective, despite having no effect on mitochondrial gene replication or expression, was to examine if RDV at these same concentrations alters mitochondrial function, employing the Seahorse® XF96 (Beeson et al., 2010; Nadanaciva et al., 2012) Extracellular Flux Analyzer technology for measuring whole cell oxygen consumption rate under various conditions. Fig. 4 illustrates the conduct of this assay along with the method of calculation for each reported parameter. This figure serves as reference for interpreting the results reported in Fig. 5 for the dose-dependent effect of RDV on cellular respiration at 48 h of exposure. What was observed is that RDV failed to significantly alter any of the respiratory parameters at concentrations of 0.5 μM or less. It was only at concentrations that approached or caused significant cytotoxicity (1 μM and 5 μM; Fig. 1) that RDV had a significant effect on suppressing mitochondrial respiration. Similar results were observed after 24 h of exposure to 5 μM RDV; However, 1 μM RDV did not alter mitochondrial respiration at this earlier time point (data not shown). The fact that all measured parameters were suppressed at high concentrations of RDV suggests a generalized decrement of mitochondrial respiration that was likely secondary to cell morbidity and not specific to the molecular mechanism of RDV toxicity.
Fig. 5.
Cellular Oxygen Consumptions Parameters Associated with Remdesivir Exposure. Oxygen consumption rates (OCR) were measured for HepG2 cells exposed to increasing concentrations of remdesivir for 48 h. Each panel represents a parameter calculated according to the formula listed in Fig. 4 and each bar reflects the mean ± S.D. for 4 independent experimental observations. Asterisks (*) indicate a statistically significant difference compared to the zero remdesivir control (Dunnett's, p ≤ 0.05).
4. Discussion
Originally developed as a treatment for Ebola virus, remdesivir also shows potent inhibition of SARS-CoV-2 corona virus and was approved in 2020 as an effective antiviral agent for treating hospitalized patients 12 years and older and weighing at least 40 kg infected with the COVID-19 virus (Grein et al., 2020). The phosphorylated nucleoside metabolite of RDV is mistakenly recognized by viral RNA-dependent RNA polymerase (Gordon et al., 2020), which is essential for replicating the viral genome. As a result, the elongating RNA strand is truncated and viral replication halted. The drug has undergone clinical testing including phase III clinical trials with few reported minor adverse events, most frequently an elevation in serum aminotransferase enzymes (Beigel et al., 2020; Goldman et al., 2020; Spinner et al., 2020), which some have attributed to the viral infection itself.
The concern that RDV might produce adverse clinical events due to a lack of specificity towards RNA-dependent RNA polymerase has been addressed in-part previously. The drug has been demonstrated to inhibit purified human mitochondrial RNA polymerase, but only at concentrations more than 100-fold in excess of that required to inhibit Ebola or RSV RNA-dependent RNA polymerase (Tchesnokov et al., 2019). Xu and coworkers (Xu et al., 2021) conducted an extensive in vitro screen of potential off-target assays, some of which were mitochondrial, in characterizing the preclinical toxicity of RDV. Their format was a panel of 87 high throughput cellular and biochemical assays across 8 different human cell types. What they found was that the concentration of cellular ATP decreased after 5 days of continuous exposure to RDV, the 50% effective concentration (EC50) ranging between 2 and 20 μM RDV for the individual cell types. Associated with this was a decrease in spare respiratory capacity, as measured by Seahorse XF Flux analyzer, at concentrations exceeding 1 μM after 3 days RDV exposure, which the authors attribute not to a primary mitochondrial effect, but rather to a generalized cytotoxicity. This is further supported by the lack of any observed effect of RDV on mtDNA copy number, measured by qPCR, or protein synthesis measured by western blot. The authors conclude that, overall, there was a lack of a specific effect of RDV on cellular respiration among the four cell systems examined (Xu et al., 2021).
The results presented in this investigation are targeted specifically to effects of RDV on mitochondria and are consistent with and substantively extend the findings published by Xu et al. (Xu et al., 2021). First, using both the SRB assay as well as the more sensitive and specific measure of Ddit3 gene expression, we find that the EC50 for cytotoxicity for HepG2/3CA cells (1–10 μM RDV for 2–3 days) is in the same range as that reported by Xu et al. using ATP luminescence as a measure of viability for this same cell line (3.7 μM at 5 days). We also confirm the observation that, using qPCR but with different gene targets, the nucleoside analog does not alter mtDNA copy number in HepG2 cells.
Xu and coworkers demonstrate that the EC50 of RDV for affecting the concentration of mtDNA-encoded cytochrome oxidase subunit I (COX—I) protein is not different from that for the nuclear-encoded SDH-A, which they interpret as the drug not preferentially inhibiting mitochondrial protein synthesis, i.e., gene translation (Xu et al., 2021). In the present investigation we extend this observation by demonstrating that at sub-cytotoxic concentrations RDV also fails to inhibit mitochondrial gene transcription. This is reflected by the relative expression of mitochondrial genes encoding subunits for either of two complexes of the electron transport chain, complex I (NDH) and complex IV (COX). This lack of effect of RDV on mtDNA transcription is a more direct measure than is mitochondrial protein synthesis for mtRNA polymerase activity.
Of perhaps greatest significance is the observation that RDV, under the conditions employed, does not alter mitochondrial respiratory function. Xu and coworkers using the Seahorse Extracellular Flux Analyzer technology reported that RDV does not alter spare respiratory capacity in HepG2 cells (Xu et al., 2021). We expanded this observation to demonstrate that RDV, at concentrations that did not alter cell viability, did not alter any of the parameters associated with cell respiration in this human hepatic cell line. This included not only spare respiratory capacity, but also basal respiration, ATP production, proton leak, maximum respiration, non-mitochondrial respiration and coupling efficiency. An exception is 1 μM RDV, which caused an increase in Ddit3 gene expression that was not statiscally significant with 3 sample replicates. Although this might be interpreted that RDV interferes with mitochondrial respiration at non-cytotoxic concentrations, it is also possible that at 1 μM RDV the cells are stressed but not to the point of statistical significance. At 24 h 1 μM RDV did not interfere with cell respiration (data not shown). The lack of an effect of RDV on hepatic cell respiration is consistent with the absence of an effect of the drug on the mitochondrial genome (gene dose or expression). From this we conclude that despite being a nucleoside analog that is a potent inhibitor of RNA polymerase, this activity is specific in sparing the mitochondria as demonstrated by Tchesnokov et al. (Tchesnokov et al., 2019). Xu et al. demonstrated that RDV has greater cytotoxic and mitochondrial potency in HepG2 cells than the parent nucleoside or nucleoside triphosphate (Xu et al., 2021), indicating that the lack of activity observed with RDV is not due to a lack metabolic activation of the prodrug. Thus, we conclude that remdesivir does not cause mitochondrial toxicity and that the cytotoxicity that is observed occurs by a mechanism that does not involve mitochondrial genomic or functional features as primary molecular off-targets.
Author credit
This research was conceived by KBW and executed by JAB. Both authors participated equally in the design, analysis and interpretation of the investigation.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgement
This work was supported in-part by an unrestricted research grant from the 3M Company. 3M Company did not contribute to the study concept or participate in any part of the experimental design, data analysis and interpretation, or in preparation of this manuscript.
Editor: Lawrence Lash
References
- Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X., Smith E.C., Case J.B., Feng J.Y., Jordan R., Ray A.S., Cihlar T., Siegel D., Mackman R.L., Clarke M.O., Baric R.S., Denison M.R. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio. 2018;9 doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeson C.C., Beeson G.C., Schnellmann R.G. A high-throughput respirometric assay for mitochondrial biogenesis and toxicity. Anal. Biochem. 2010;404:75–81. doi: 10.1016/j.ab.2010.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., Lopez de Castilla D., Finberg R.W., Dierberg K., Tapson V., Hsieh L., Patterson T.F., Paredes R., Sweeney D.A., Short W.R., Touloumi G., Lye D.C., Ohmagari N., Oh M.D., Ruiz-Palacios G.M., Benfield T., Fatkenheuer G., Kortepeter M.G., Atmar R.L., Creech C.B., Lundgren J., Babiker A.G., Pett S., Neaton J.D., Burgess T.H., Bonnett T., Green M., Makowski M., Osinusi A., Nayak S., Lane H.C., Members A.-S.G. Remdesivir for the treatment of Covid-19 - final report. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjork J.A., Wallace K.B. Structure-activity relationships and human relevance for perfluoroalkyl acid-induced transcriptional activation of peroxisome proliferation in liver cell cultures. Toxicol. Sci. 2009;111:89–99. doi: 10.1093/toxsci/kfp093. [DOI] [PubMed] [Google Scholar]
- Dunnett C. A multiple comparison procedure for comparing several treatments with a control. J. Amer. Stat. Assn. 1955;50:1096–1121. [Google Scholar]
- Feng J.Y. Addressing the selectivity and toxicity of antiviral nucleosides. Antivir. Chem. Chemother. 2018;26 doi: 10.1177/2040206618758524. 2040206618758524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fosslien E. Mitochondrial medicine--molecular pathology of defective oxidative phosphorylation. Ann. Clin. Lab. Sci. 2001;31:25–67. [PubMed] [Google Scholar]
- Goldman J.D., Lye D.C.B., Hui D.S., Marks K.M., Bruno R., Montejano R., Spinner C.D., Galli M., Ahn M.Y., Nahass R.G., Chen Y.S., SenGupta D., Hyland R.H., Osinusi A.O., Cao H., Blair C., Wei X., Gaggar A., Brainard D.M., Towner W.J., Munoz J., Mullane K.M., Marty F.M., Tashima K.T., Diaz G., Subramanian A., Investigators G-U Remdesivir for 5 or 10 days in patients with severe Covid-19. N. Engl. J. Med. 2020;383:1827–1837. doi: 10.1056/NEJMoa2015301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon C.J., Tchesnokov E.P., Woolner E., Perry J.K., Feng J.Y., Porter D.P., Gotte M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020;295:6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grein J., Ohmagari N., Shin D., Diaz G., Asperges E., Castagna A., Feldt T., Green G., Green M.L., Lescure F.X., Nicastri E., Oda R., Yo K., Quiros-Roldan E., Studemeister A., Redinski J., Ahmed S., Bernett J., Chelliah D., Chen D., Chihara S., Cohen S.H., Cunningham J., D’Arminio Monforte A., Ismail S., Kato H., Lapadula G., L’Her E., Maeno T., Majumder S., Massari M., Mora-Rillo M., Mutoh Y., Nguyen D., Verweij E., Zoufaly A., Osinusi A.O., DeZure A., Zhao Y., Zhong L., Chokkalingam A., Elboudwarej E., Telep L., Timbs L., Henne I., Sellers S., Cao H., Tan S.K., Winterbourne L., Desai P., Mera R., Gaggar A., Myers R.P., Brainard D.M., Childs R., Flanigan T. Compassionate use of remdesivir for patients with severe Covid-19. N. Engl. J. Med. 2020;382:2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakuda T.N. Pharmacology of nucleoside and nucleotide reverse transcriptase inhibitor-induced mitochondrial toxicity. Clin. Ther. 2000;22:685–708. doi: 10.1016/S0149-2918(00)90004-3. [DOI] [PubMed] [Google Scholar]
- Los G., Benbatoul K., Gately D.P., Barton R., Christen R., Robbins K.T., Vicario D., Kirmani S., Orloff L.A., Weisman R., Howell S.B. Quantitation of the change in GADD153 messenger RNA level as a molecular marker of tumor response in head and neck cancer. Clin. Cancer Res. 1999;5:1610–1618. [PubMed] [Google Scholar]
- Lund K.C., Wallace K.B. Direct effects of nucleoside reverse transcriptase inhibitors on rat cardiac mitochondrial bioenergetics. Mitochondrion. 2004;4:193–202. doi: 10.1016/j.mito.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Lund K.C., Wallace K.B. Direct, DNA pol-gamma-independent effects of nucleoside reverse transcriptase inhibitors on mitochondrial bioenergetics. Cardiovasc. Toxicol. 2004;4:217–228. doi: 10.1385/ct:4:3:217. [DOI] [PubMed] [Google Scholar]
- Malik A.N., Shahni R., Rodriguez-de-Ledesma A., Laftah A., Cunningham P. Mitochondrial DNA as a non-invasive biomarker: accurate quantification using real time quantitative PCR without co-amplification of pseudogenes and dilution bias. Biochem. Biophys. Res. Commun. 2011;412:1–7. doi: 10.1016/j.bbrc.2011.06.067. [DOI] [PubMed] [Google Scholar]
- Nadanaciva S., Rana P., Beeson G.C., Chen D., Ferrick D.A., Beeson C.C., Will Y. Assessment of drug-induced mitochondrial dysfunction via altered cellular respiration and acidification measured in a 96-well platform. J. Bioenerg. Biomembr. 2012;44:421–437. doi: 10.1007/s10863-012-9446-z. [DOI] [PubMed] [Google Scholar]
- Oyadomari S., Mori M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 2004;11:381–389. doi: 10.1038/sj.cdd.4401373. [DOI] [PubMed] [Google Scholar]
- Simpson M.V., Chin C.D., Keilbaugh S.A., Lin T.S., Prusoff W.H. Studies on the inhibition of mitochondrial DNA replication by 3′-azido-3′-deoxythymidine and other dideoxynucleoside analogs which inhibit HIV-1 replication. Biochem. Pharmacol. 1989;38:1033–1036. doi: 10.1016/0006-2952(89)90245-1. [DOI] [PubMed] [Google Scholar]
- Spinner C.D., Gottlieb R.L., Criner G.J., Arribas Lopez J.R., Cattelan A.M., Soriano Viladomiu A., Ogbuagu O., Malhotra P., Mullane K.M., Castagna A., Chai L.Y.A., Roestenberg M., Tsang O.T.Y., Bernasconi E., Le Turnier P., Chang S.C., SenGupta D., Hyland R.H., Osinusi A.O., Cao H., Blair C., Wang H., Gaggar A., Brainard D.M., McPhail M.J., Bhagani S., Ahn M.Y., Sanyal A.J., Huhn G., Marty F.M., Investigators G-U Effect of remdesivir vs standard care on clinical status at 11 days in patients with moderate COVID-19: a randomized clinical trial. JAMA. 2020;324:1048–1057. doi: 10.1001/jama.2020.16349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchesnokov E.P., Feng J.Y., Porter D.P., Gotte M. Mechanism of inhibition of ebola virus RNA-dependent RNA polymerase by Remdesivir. Viruses. 2019;11 doi: 10.3390/v11040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace K.B., Starkov A.A. Mitochondrial targets of drug toxicity. Annu. Rev. Pharmacol. Toxicol. 2000;40:353–388. doi: 10.1146/annurev.pharmtox.40.1.353. [DOI] [PubMed] [Google Scholar]
- Warren T.K., Jordan R., Lo M.K., Ray A.S., Mackman R.L., Soloveva V., Siegel D., Perron M., Bannister R., Hui H.C., Larson N., Strickley R., Wells J., Stuthman K.S., Van Tongeren S.A., Garza N.L., Donnelly G., Shurtleff A.C., Retterer C.J., Gharaibeh D., Zamani R., Kenny T., Eaton B.P., Grimes E., Welch L.S., Gomba L., Wilhelmsen C.L., Nichols D.K., Nuss J.E., Nagle E.R., Kugelman J.R., Palacios G., Doerffler E., Neville S., Carra E., Clarke M.O., Zhang L., Lew W., Ross B., Wang Q., Chun K., Wolfe L., Babusis D., Park Y., Stray K.M., Trancheva I., Feng J.Y., Barauskas O., Xu Y., Wong P., Braun M.R., Flint M., McMullan L.K., Chen S.S., Fearns R., Swaminathan S., Mayers D.L., Spiropoulou C.F., Lee W.A., Nichol S.T., Cihlar T., Bavari S. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Barauskas O., Kim C., Babusis D., Murakami E., Kornyeyev D., Lee G., Stepan G., Perron M., Bannister R., Schultz B.E., Sakowicz R., Porter D., Cihlar T., Feng J.Y. Off-target in vitro profiling demonstrates that remdesivir is a highly selective antiviral agent. Antimicrob. Agents Chemother. 2021;65 doi: 10.1128/AAC.02237-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., et al. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinf. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]