Abstract
The acute respiratory distress syndrome (ARDS) is a common complication of severe COVID-19 (coronavirus disease 2019) caused by SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) infection. Knowledge of molecular mechanisms driving host responses to SARS-CoV-2 is limited by the lack of reliable preclinical models of COVID-19 that recapitulate human illness. Further, existing COVID-19 animal models are not characterized as models of experimental acute lung injury (ALI) or ARDS. Acknowledging differences in experimental lung injury in animal models and human ARDS, here we systematically evaluate a model of experimental acute lung injury as a result of SARS-CoV-2 infection in Syrian golden hamsters. Following intranasal inoculation, hamsters demonstrate acute SARS-CoV-2 infection, viral pneumonia, and systemic illness but survive infection with clearance of virus. Hamsters exposed to SARS-CoV-2 exhibited key features of experimental ALI, including histologic evidence of lung injury, increased pulmonary permeability, acute inflammation, and hypoxemia. RNA sequencing of lungs indicated upregulation of inflammatory mediators that persisted after infection clearance. Lipidomic analysis demonstrated significant differences in hamster phospholipidome with SARS-CoV-2 infection. Lungs infected with SARS-CoV-2 showed increased apoptosis and ferroptosis. Thus, SARS-CoV-2 infected hamsters exhibit key features of experimental lung injury supporting their use as a preclinical model of COVID-19 ARDS.
Abbreviations: ARDS, acute respiratory distress syndrome; ALI, acute lung injury; COVID-19, coronavirus disease 2019; SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus-2; ACE2, angiotensin converting enzyme 2; BAL, bronchoalveolar lavage; IFIT, INF-induced protein with tetratricopeptide repeats; IFN, interferon; IHC, immunohistochemistry; IL, interleukin; MPO, myeloperoxidase; NGS, next generation sequencing; OPLS-DA, orthogonal projection of latent structures - discriminate analysis; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PFU, plaque forming unit; PLA2, phospholipase A2; TFRC, transferrin receptor protein 1; VIP, variable importance in projection; VOC, variant of concern
AT A GLANCE COMMENTARY.
Bednash JS, et al.
Background
The acute respiratory distress syndrome (ARDS) is a common complication of severe COVID-19. Animal models of SARS-CoV-2 ARDS are crucial to understanding molecular mechanisms driving disease.
Translational Significance
We systematically evaluated Syrian golden hamsters as an experimental model of SARS-CoV-2 ARDS, based on biophysical, transcriptomic, and lipidomic data. Hamsters demonstrate key features of ARDS, including hypoxemia. Detailed molecular analysis demonstrates upregulation of interferon signaling, recapitulating findings of human illness. We also present the first and most robust lipidomic analysis in an animal model of SARS-CoV-2 infection. These findings demonstrate utility of hamsters as an experimental model of COVID-19 ARDS.
Alt-text: Unlabelled box
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of a global pandemic of coronavirus disease 2019 (COVID-19).1 Respiratory failure and the acute respiratory distress syndrome (ARDS) are primary drivers of mortality in COVID-19.2, 3, 4 In humans, ARDS is characterized by diffuse, bilateral, radiographic, non–cardiogenic pulmonary edema, and hypoxemic respiratory failure.5 Reliable animal models that closely mimic human host response in COVID-19, including key features of respiratory failure and ARDS,6 , 7 are essential to investigate pathobiology and develop therapeutics. Such preclinical models of SARS-CoV-2 ARDS are lacking, a major obstacle to the development of targeted countermeasures to COVID-19, and its complications.
SARS-CoV-2 is a member of the betacoronavirus (β-CoV) genera, which are enveloped, positive-strand RNA viruses8 that primarily infect bats but can also infect humans, and rodents.9 Other well-known betacoronaviruses include severe acute respiratory syndrome (SARS)-CoV that emerged in the early 2000s and Middle East Respiratory (MERS-)CoV. These viruses all gain entry to mammalian host cells through interactions of viral spike glycoproteins with the host angiotensin-converting enzyme 2 (ACE2).10 Structural modeling of ACE2 identified 25 essential amino acids for interaction of SARS-CoV-2 spike protein with human (h)ACE211, 12, 13 and suggested that differences in ACE2 determine the range of hosts susceptible to infection.14 , 15
Differences in ACE2 structure and expression contribute to phenotypic differences in animal models of COVID-19. Compared to human (h)ACE2, mouse (Mus musculus) ACE2 contains only 16/25 amino acids essential for SARS-CoV-2 binding.14 Wild type mice inoculated with SARS-CoV-2 do not demonstrate viral infection.16 While other animals demonstrate SARS-CoV-2 infection, including ferrets,17 cats,18 cynomolgus macaques19 and Rhesus macaques,20, 21, 22, 23 their utility as an experimental system is limited by practical issues related to size of the animals, availability of animals and facilities, and expense. To facilitate COVID-19 research in mouse models, hACE2 transgenic mice were developed.24 In these transgenic models, the severity of disease ranges from mild to lethal, depending on the design of the promotor system governing expression of hACE2.24, 25, 26 These mice display variable patterns of disease and often succumb to lethal encephalitis,27 an uncommon finding in humans, raising concerns about applicability of this model to human host response. Another strategy that has been explored is a mouse-adapted SARS-CoV-2. This approach allows for use of currently available reagents including an opportunity to exploit mouse genetics (transgenic mice) with the caveat that mice develop only mild disease.28 Another limitation is that adaptation of SARS-CoV-2 to mice limits the ability to test novel circulating strains as they arise.
Comparison of vertebrate ACE2 sequences identified Syrian golden hamsters (Mesocricetus auratus, “hamster” from here forward) as a potential small animal model of COVID-19. Among the 25 amino acids necessary for binding of SARS-CoV-2 spike protein, hamster, and human ACE2 share 22/25 identical residues.14 Experimental studies have shown hamsters to be an attractive model for the study of coronaviridae29 and COVID-19, as wild type hamster ACE2 allows uptake of strains SARS-CoV-2 circulating among humans.15 Indeed, in experimental models, hamsters display viral infection in the lung with histologic lung injury,15 , 30, 31, 32 increased transcription of inflammatory markers in the lung,32 and the animals display transmission of SARS-CoV-2 among cage-mates.33 However, hamsters have not been adequately characterized as models of SARS-CoV-2 ARDS. Specifically, the characteristic physiologic derangements seen in ARDS (decreased lung compliance and tissue oxygenation) and the key molecular signatures have not been examined.
Here we evaluated hamsters as a model of SARS-CoV-2 ARDS. Although animal models do not fully replicate all features of human ARDS,7 , 34 they do exhibit some features consistent with experimental acute lung injury (ALI).35 , 36 Consistent with human clinical disease and criteria of experimental ALI,7 SARS-CoV-2 infected hamsters exhibit histologic lung injury, increased alveolar capillary permeability, acute lung inflammation, and physiologic impairment with hypoxemia. To reveal the molecular mechanisms that contribute to ALI, we performed both transcriptomic and lipidomic analysis of hamster lung tissue and BAL. RNAseq of hamster lungs demonstrated upregulation of inflammatory mediators and interferon signaling that persist post infection. Mass Spectrometric (MS) lipidomic analysis demonstrated upregulation of several lipid molecular species with SARS-CoV-2 infection, consistent with pro-oxidant conditions, and accumulation of species identified as ferroptotic cell death signals. We conclude that with SARS-CoV-2 infection, hamsters exhibit key features of experimental ALI. Molecular profiling recapitulates many mechanisms reported in human observational studies, validating utility of hamsters as a preclinical model of COVID-19 ARDS.
MATERIALS AND METHODS
Virus and biosafety
All procedures and experiments involving live virus followed approved operating procedures of the Ohio State University Biosafety Level-3 facility (IBC Protocol # 2020R00000046). In accordance with BSL-3 protocols, all potentially infectious materials underwent treatment for virus inactivation prior to removal from the BSL-3 facility for downstream analysis of biological materials. SARS-CoV-2, isolate USA-WA1/2020, was obtained from Biodefense and Emerging Infections Research Resources Repository (BEI Resources, Batch # 70034262). The viral stock was amplified by passage in VeroE6 cells (ATCC). Briefly, a viral batch isolate was thawed and diluted 1:10,000 in incomplete DMEM (Gibco #11995-065, supplemented with 4.5 g/L D-glucose, 110 mg/L sodium pyruvate), before addition to cultured confluent Vero cells. Cells were incubated with virus for 1h (37°C, 5% CO2). Media was replaced with complete DMEM (DMEM as above plus 4% heat-inactivated fetal bovine serum), and cells were incubated for 3d (37°C, 5% CO2) to allow viral propagation. After 3d, visual inspection under light microscopy demonstrated near complete death of Vero cells. Supernatant was carefully collected, centrifuged at low speed to remove cell debris, aliquoted, frozen, and stored at -80°C. The live virus titer in frozen aliquots was determined to be ∼107 plaque forming unit (PFU) per mL using a modified version of a plaque assay developed by the Diamond laboratory37 described below.
SARS-CoV-2 plaque assay
At least 18 hours prior to the assay, 12-well plates were seeded with a sufficient number of Vero cells so that each well was confluent by the assay start; plates were incubated overnight at 37°C. On the day of the assay, serial dilutions of virus-containing material were prepared in cDMEM (1:101, 1:102, 1:103, 1:104) and warmed to 37°C. Media from each well of the 12-well plate was gently removed via pipette and replaced with 500 uL of each virus serial dilution, the volume pipetted down the side of the well so as not to disturb the Vero cell monolayer. The plate was incubated for 1 hour at 37°C, 5% CO2. During that infection incubation period, a solution comprising a 1:0.7 mixture of cDMEM and 2% methylcellulose (viscosity: 4000 cP) was freshly made and warmed to 37°C in a water bath. After the 1-hour infection incubation period, the supernatant was removed from each well, and replaced with 1 mL of the warmed cDMEM/methylcellulose mixture. The culture plate was then returned to the incubator and left undisturbed for 3 day. On the final day, cDMEM/methylcellulose mixture was removed from each well, cells were fixed with 4% para-formaldehyde in PBS (20 minutes, room temperature), washed with PBS, and stained with 0.05% crystal violet (in 20% methanol). After rinsing plates with distilled water, plates were dried, and plaques were counted under a light microscope at 20x magnification.
Hamster infection with SARS-CoV-2
All studies were reviewed and approved by the local Institutional Animal Care and Use Committee (IACUC) prior to their onset. 10–12-week-old, male Golden Syrian hamsters were purchased from Envigo (North America) and socially housed with littermate controls in a USDA- and AALAC-accredited Animal Biosafety Level 3 (ABSL3) facility. At the time of infection, each animal was anesthetized with isoflurane, weighed, and held at a semi-supine position while 100 μL of virus-containing media at 5 × 105 PFU was given via intranasal (i.n.) instillation. Control hamsters were given the same volume of sterile media, using the same anesthesia and an i.n. instillation protocol. After i.n. instillation, each hamster was returned and housed in its home cage and monitored daily for changes in weight or body condition.
Lung physiology measurements
7 days post-inoculation, hamsters were removed from cages individually and placed into a chamber delivering vaporized isoflurane to facilitate handling. Once sedate, hamsters were weighed and anesthetized with ketamine (Vedco, St. Joseph, Missouri) at 150mg/kg and xylazine (Akorn, Lake Forest, Illinois) at 15 mg/kg. Using the MouseOx system (Starr Life Sciences, Oakmont, Pennsylvania), pulse oximetry measurements were obtained. Tracings were analyzed for quality, and values obtained during a steady state reading of at least 30 sec duration. Next, a 15-gauge ½ inch metal tracheostomy tube was placed, and hamsters were mechanically ventilated using the FlexiVent FX system (SCIREQ, Montreal, Canada). Measurements of lung elastance and airway resistance were obtained from the constant phase model38 using the integrated flexiWare software. Standardized recruitment maneuvers were performed prior to each physiologic measurement to prevent atelectasis and standardize volume history.39
Bronchoalveolar lavage
At the end of the mechanical ventilation protocol, animals were euthanized by anesthetic overdose. Bronchoalveolar lavage (BAL) was performed by instilling 2 mL of sterile PBS twice via the tracheostomy tube. The BAL fluid was divided into 2 aliquots and then centrifuged for 6 minutes at 500 g at 4°C. Pelleted cells from one aliquot underwent viral inactivation by resuspension in 4% paraformaldehyde, were counted by an automated cell counter (Bio-Rad), and cytospins were performed with Diff-Quick staining (Fisher Scientific) to obtain differential cell counts. Supernatant from one aliquot was treated with Triton X-100 (1% final concentration) to inactivate virus and underwent Lowry protein assay (Bio-Rad). Remaining BAL aliquots were prepared for lipid analysis as described below. Virus inactivation in lipid samples was achieved by storage in at least 60% ethanol.
RNA extraction
Following BAL, hamsters underwent organ harvest. Lung tissue was divided to facilitate multiple parallel analyses. One small lung lobe was preserved in Trizol (Invitrogen), which inactivates virus, prior to homogenization. Total cellular RNA was collected from lung tissue using the Direct-Zol Minimprep Kit (Zymogen), followed by in-tube/off-column DNAse I treatment (New England Biolabs) and RNA cleanup with Monarch RNA cleanup Kit (50 μg) (New England Biolabs), following the manufacturer's protocol. 1 μg cellular RNA was then used to create cDNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer's protocol. qPCR was performed on a C1000 Touch Thermocycler (Bio-Rad) using SYBR Select Master Mix (Applied Biosystems) according to the manufacturer's protocol with 20 ng cDNA as a template and primer concentration of 200 nM. Each biological replicate was performed in at least technical duplicate; data were analyzed using the ΔΔCq method. qPCR primer sequences are available in Supplementary Table 1.
SARS-CoV-2 genomic sequencing
Previously extracted RNA underwent first and second strand cDNA synthesis (NEBNext 133 Ultra# II Non–Directional RNA Second Strand Synthesis Module, NEB, Ipswich, MA) followed by sequencing using amplicon-based next-generation sequencing (NGS) methods.40 Library preparations were performed using the CovidSeq kit (Illumina, San Diego, CA) per the manufacturer's protocol and sequenced on the NextSeq 550 sequencer (Illumina). The analysis pipeline included BaseSpace and the Dragen COVID lineage plug-in (version 3.5.1, Illumina), as well as a custom-developed pipeline using GATK tools, for low-level quantitation of variant. SARS-CoV-2 strain typing employed the Pangolin COVID-19 Lineage assigner (https://pangolin.cog-uk.io). Variant quantitation for the 2 different assays was cross-validated using a set of 20 SARS-CoV-2 positive samples.
Transcriptome next generation sequencing
Total RNA was extracted from hamster lungs as per above. Total RNA was quantified using the Invitrogen Qubit RNA HS Assay kit (Invitrogen, Carlsbad, California) and RNA quality was assessed by RNA integrity scoring using Agilent 2100 Bioanalyzer and/or 2200 TapeStation (Agilent, Santa Clara California). RNA seq libraries were prepared using kits from New England Biolabs (Ipswich, Massachusetts) with 100 ng total RNA by targeted depletion of rRNA (NEB E#E6310x). Fragmentation and amplification were done using NEBNext Ultra II Directional (stranded) RNA Library Prep Kit (NEB#E7760L) and NEBNext (E64490S/L). Multiplex Oligos for Illumina Unique Dual Index Primer Pairs. Samples were sequenced to a depth of 40 million 2 × 150 bp clusters on the Illumina NovaSeq platform (Illumina, Inc, San Diego, California).
Sequence alignment, gene count generation, and differential expression analysis
RNA sequencing samples reads were aligned to the Syrian golden hamster genome (GCF_000349665.1_MesAur1.0) and gene counts were obtained for each sample using STAR 2.7.7a.41 These counts were used for Differential Expression Analysis using the DESeq2 package.42 For visualization and ranking of genes, we performed effect size shrinkage of the data using the apeglm method43 and variance stabilizing transformation on the raw count data to generate heatmaps. Genes with human orthologs demonstrating >2.5 Log2 fold change increase and P > 0.05 were subjected to STRING network analysis (v11.0, string-db.org).44
Lung histology and staining
Hamster lungs were inflated with 10% formalin at a pressure of 20 cm H2O and fixed for >24 hours and embedded in paraffin. 4 µm sections were stained with hematoxylin and eosin for histopathological examinations (by HistoWiz). Tissue slides to be stained for immunohistochemistry were deparaffinized and rehydrated before heat induced epitope retrieval with 0.01 M citrate buffer (pH 6.0). Slides were incubated in 3% H2O2 (Fisher, H325) in PBS for 5 minutes to block endogenous peroxidases and blocked in 1% normal swine serum (Vector Laboratories, S-4000) in PBS for 15 minutes. Primary antibodies were diluted in 1% normal swine serum in PBS, and sections were treated with primary antibody solutions and incubated at 4°C overnight. See table 2 for antibodies and concentrations. After washing with PBS, sections were treated for 1 h at room temperature with secondary antibodies conjugated with biotin (1:1500) (Vector Laboratories, SA-5004). Slides were washed and incubated in Streptavidin-HRP [1:200] (Sigma-Aldrich, AP132) for 45 minutes. Slides were washed again followed by color development using 3,3′-diaminobenzidine solution in PBS with H2O2 and counterstaining in Hematoxylin Stain, Mayer Modified (Newcomer Supply, 1202B) for 1 minutes. Slides were sealed with DPX mounting media (Sigma, 06522) and imaged on an EVOS M7000 digital microscope (Invitrogen). Images were collected with the M7000 software (Invitrogen). Negative controls without primary antibody were run in parallel.
Table 2.
Top 15 most differentially expressed transcripts from hamsters infected with SARS-CoV-2 vs controls at 7 dpi
| Gene | log2FoldChange | Adjusted P |
|---|---|---|
| Spp1 | 8.01 | 3.7E-58 |
| Gbx2 | 7.13 | 6.0E-13 |
| Akr1b10 | 6.97 | 2.2E-26 |
| Gdf3 | 6.74 | 1.1E-13 |
| Aicda | 6.49 | 3.2E-22 |
| Clec4f | 6.35 | 3.1E-07 |
| Gzmk | 6.29 | 1.2E-28 |
| Ms4a7 | 5.50 | 3.1E-22 |
| Cd207 | 5.40 | 2.6E-11 |
| Slamf9 | 5.07 | 6.8E-27 |
| Cxcl9 | 4.75 | 4.2E-35 |
| Siglec1 | 4.70 | 2.1E-41 |
| Arhgap40 | 4.70 | 1.0E-11 |
| Spata19 | 4.62 | 1.6E-06 |
| Ccna1 | 4.48 | 5.1E-10 |
Lung injury scoring
H&E-stained lung tissue sections were scored as described previously32 by 2 independent scorers to assess the severity of lung damage. At least 10 areas were scored for gross examination at 4X (low magnification), and at least 20 areas were scored for 20X (higher magnification) scoring.
Redox lipidomics
Lipids from BAL, cells and lungs were extracted using the Folch procedure as previously described.45 Phospholipids were analyzed by LC/MS using a Dionex Ultimate 3000 HPLC system coupled on-line to Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific) using a normal phase column (Luna 3 μm Silica (2) 100 Å, 150 × 2.0 mm, (Phenomenex)). The column was maintained at 35°C. Gradient solvents A: propanol/hexane/water (285:215:5, v/v/v) and B: propanol/hexane/water (285:215:40, v/v/v) containing 10 mM ammonium acetate were used. The column was eluted for 0–23 minutes with a linear gradient from 10%–32% B; 23–32 minutes from 32%–65% B; 32–35 minutes from 65%–100% B; 35–62 minutes held at 100% B; 62–64 minutes from 100%–10% B followed by equilibration from 64–80 minutes at 10% B. MS and MS2 analysis was performed in negative ion mode at a resolution of 140,000 for the full MS scan and 17,500 for the MS2 scan in a data-dependent mode. Analysis of data was performed using the software package Compound Discoverer (Thermo Fisher Scientific) with an in-house–generated analysis workflow and phospholipid database. Phospholipids were filtered by retention time and confirmed by fragmentation analysis. SIMCA 16.1 software was used to explore the major effects that potentially drive the differences in lipid profiles in SARS-CoV-2 infected hamsters.
Immunoblotting
After harvest, lung tissue was submerged in cold RIPA buffer, homogenized, and clarified by centrifugation. Lysates were diluted in SDS protein sample buffer. Proteins were separated by electrophoresis and transferred to a nitrocellulose membrane. Blots were blocked in 5% milk, followed by probing overnight with antibodies (see Supplementary Table 2). Following addition of secondary antibodies (goat anti–mouse, Biorad, 170–6516, and goat anti–rabbit, Biorad, 170–6515; 1:2000 dilution), membranes were developed using a Western Bright Sirius immunoblotting detection kit (Advansta) and imaged using Biorad imaging software. Single band intensity was quantified using ImageJ software.
Study approval
All procedures and experiments involving live virus followed approved operating procedures of the Ohio State University Biosafety Level-3 facility (IBC Protocol #2020R00000046). Animal experiments followed approved protocols of the Ohio State University Office of Responsible Research Practices (IACUC Protocol #2020A00000044).
RESULTS
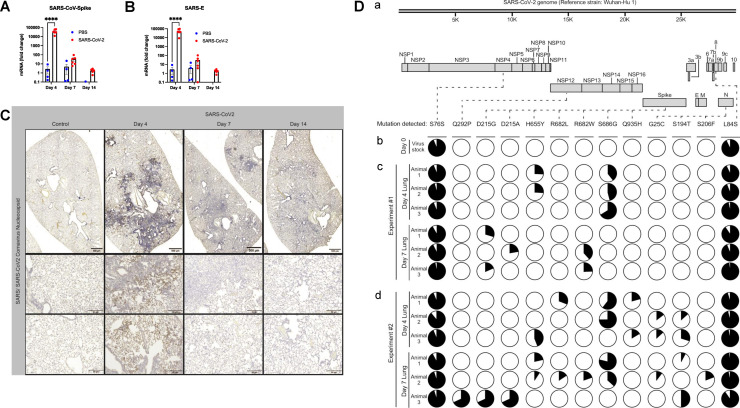
Hamsters demonstrate SARS-CoV-2 viral infection and SARS-CoV-2 intra-host sequence diversity. We first evaluated characteristics of viral infection in the hamster model. Male hamsters around 16 weeks of age were inoculated with 5 × 105 PFU SARS-CoV-2 or PBS control via i.n. instillation. Hamsters were euthanized 4, 7, and 14 days post inoculation (dpi). At each experimental endpoint, we performed numerous procedures including (1) assessment of lung mechanics; (2) pulse oximetry; (3) bronchoalveolar lavage; and (4) organ harvest. To confirm SARS-CoV-2 infection, we measured SARS-CoV-2-spike and SARS-E RNA from lung tissue showing significant increases in viral RNA on 4 dpi with a significant reduction in viral load by 7 dpi (Figs. 1 , A and B). Similarly, SARS-CoV-2 spike protein was present by immunohistochemistry in lung tissue sections from infected hamsters compared to PBS controls with most intense staining on 4 dpi and resolution by 7 and 14 dpi (Fig. 1C). Variants of SARS-CoV-2 are currently in circulation among humans, and SARS-CoV-2 exhibits intra-host sequence diversity which may contribute to immune escape variants.46 To determine if this feature of SARS-CoV-2 pathogenesis also occurs in the lungs of experimentally infected hamsters, we extracted viral RNA from the lungs of infected hamsters and sequenced via next-generation sequencing (NGS) using a validated clinical assay.40 The virus stock used to infect hamsters at Day 0 (Isolate USA-WA1/2020) was sequenced in the same manner, so as to identify mutations that arise in individual hamsters by either 4 or 7 dpi. Consistent with the sequencing results of the USA-WA1/2020 isolate provider, our virus stock had mutations in 2 genes relative to reference strain Wuhan-Hu-1: NSP4 (S76S) and ORF8 (L84S) (Fig. 1D). These mutations were stable through the course of infection, as the proportion of viral reads containing NSP4 (S76S) and ORF8 (L84S) did not substantially differ between the virus stock used at Day 0 and all lung samples collected on 4 or 7 dpi. However, eleven sequence variations developed in virus sequenced from individual animals that were not present in our virus stock: one in the polymerase gene, NSP12 (Q292P), 7 in the surface glycoprotein gene, SPIKE (D215G, D215A, H655Y, R682L, R682W, S686G, Q935H), and 3 in the nucleocapsid gene, N (G25C, S194T, S206F). Notably, SPIKE mutations H655Y and S686G have previously been observed in domestic cats and ferrets, respectively, and are predicted to be immune-evasive.47 , 48 A SPIKE mutation (D215G) is present in the Variant of Concern (VOC) B.1.351, which displays resistance to antibody-mediated neutralization.49 , 50 Collectively, these results demonstrate an effective acute viral infection in hamsters with SARS-COV-2 inoculation with accumulation of mutations in at least 3 viral genes (NSP12, SPIKE, and N) during the course of infection, including mutations associated with clinically relevant VOCs.
Fig 1.
SARS-CoV-2 inoculation leads to infection in Syrian golden hamsters. (A,B) Total RNA was harvested from hamster lung tissue, followed by analysis by qPCR. SARS-CoV-2 infected hamsters demonstrated significantly increased SARS-CoV-2-spike and SARS-E mRNA at 4 dpi but no significant differences at 7 or 14 dpi. n >= 4 hamsters and/or group. Bar graph represents mean fold change ± SEM. Dots represent individual animals. ****P < 0.0001 as determined by ANOVA with post-hoc Sidak's multiple comparisons test. (C) Representative images from hamster lung tissue demonstrate increased staining for SARS-CoV-2 spike protein by immunohistochemistry (IHC) at 4 dpi but no significant increased viral protein at 7 or 14 dpi. (D) SARS-CoV-2 intra-host sequence analysis. (Da) Depiction and annotation of the SARS-CoV-2 genome, as modified from Gordon et al.40 to show the locations of thirteen mutations detected in (Db) viral RNA extracted from the SARS-CoV-2 stock used to infect all the hamsters in this study, (Dc) viral RNA extracted from individual hamster lungs at post-infection Day 4 or post-infection Day 7 (n = 3 hamsters and/or time point), as collected during one experiment (Experiment #1), and (Dd) viral RNA extracted from individual hamster lungs at the same time points, as collected during a second independent experiment (Experiment #2). From left to right, these mutations were in the following genes and nucleotide positions (protein changes are indicated in parentheses): NSP4.228C→T (S76S), NSP12.875A→C (Q292P), Spike.644A→G (D215G), Spike.644A→C (D215A), Spike.1963C→T (H655Y), Spike.2044C→T(R682W), Spike.2045G→T(R682L), Spike.2056A→G(S686G), Spike.2805A→T(Q935H), N.73G→T(G25C), N.580T→A(S194T), N.617C→T(S206F), and ORF8.251T→C(L84S). The pie charts under each mutation indicate the proportion of reads in a given sample that either contained the mutation (the black portion of the pie) or wild type nucleotide (the white portion of the pie).
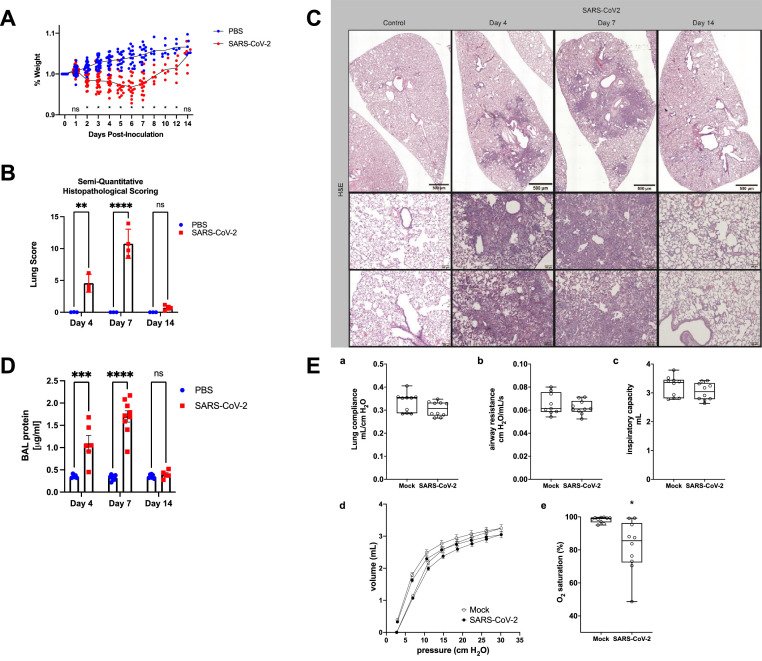
SARS-CoV-2 infected hamsters demonstrate key features of experimental ALI. Following demonstration of viral infection with peak of 4 dpi (Fig. 1), we next assessed hamster host response to SARS-CoV-2. Suggestive of systemic illness, hamsters infected with SARS-CoV-2 demonstrated weight loss compared to controls with nadir at day 6 of infection (Fig. 2 A). Next, we specifically evaluated hamsters for experimental ALI. The main features of experimental ALI include: (1) histologic evidence of tissue injury; (2) disruption of the alveolar capillary membrane; (3) acute inflammatory response; and (4) physiologic impairment of lung function.7 To assess lung tissue damage, we performed hematoxylin and eosin staining of hamster lung tissue with semi-quantitative histopathological scoring.32 SARS-CoV-2 infection resulted in significantly increased injury scores at 4, 7, and 14 dpi compared to control with peak of injury at day 7 (Fig. 2B). Representative low magnification images show a dense patchy infiltrate present on 4 and 7 dpi (Fig. 2C, top). At high magnification, we observed inflammatory cell infiltration and obliteration of the airspaces at 4 and 7 dpi. At 14 dpi, we see improvement in the infiltrate with persistent thickening the alveolar membranes (Fig. 2C, bottom), suggesting persistence of tissue injury and damage. We also performed Trichrome staining of lung sections and did not observe significant fibrotic changes at any time point (Sup. Fig. 1). To assess for disruption of the alveolar capillary membrane, we measured BAL protein, which demonstrated significant increases at 4 and 7 dpi with resolution by 14 dpi (Fig. 2D). Following an automated perturbation protocol using a FlexiVent rodent ventilator, we assessed lung compliance (Fig. 2Ea), airway resistance (Fig. 2Eb), inspiratory capacity (Fig. 2Ec) and pressure-volume loops (Fig. 2Ed) at 7 dpi and interestingly observed no significant differences between infected and control hamsters. We chose 7 dpi as our pilot experiments suggested 7 dpi as the peak of lung injury. We did observe significant deficits in oxygenation, as measured by oximetry in SARS-CoV-2 infected hamsters compared to controls (Fig. 2Ee), indicative of impaired gas-exchange with SARS-CoV-2 infection in hamsters.
Fig 2.
SARS-CoV-2-infected hamsters show key features of experimental ARDS. (A) Hamsters inoculated with SARS-CoV-2 or PBS mock were weighed daily and demonstrate significant weight loss with nadir around 6 dpi. Trend lines demonstrate average daily weights for SARS-CoV-2 group and PBS control. n >= 6 hamsters and/or group at each timepoint. Data points represent individual animals. *P < 0.05 as determined by ANOVA with post-hoc Sidak's multiple comparisons test. (B) Lung H & E tissue sections were scored by 2 independent investigators using a scoring rubric. Hamsters demonstrate increased lung tissue injury at 4, 7, and 14 dpi with most severe injury at 7 dpi. n = 4 hamsters and/or group. Graph represents mean injury score ± SEM. Data points represent individual animals. *P < 0.05 as determined by ANOVA with post-hoc Sidak's multiple comparisons test. (C) Representative images from H & E staining of hamster lung tissue demonstrates increased cell infiltration with a patchy distribution in SARS-CoV-2 inoculated hamsters compared to PBS controls. (D) Hamsters underwent tracheostomy and BAL. Protein concentrations were elevated in SARS-CoV-2-infected hamsters compared to control. n >= 6 hamsters and/or group for all panels. Graph represents mean protein concentration ± SEM. Data points represent individual animals. *P < 0.05 as determined by ANOVA with post-hoc Sidak's multiple comparisons test. (E) (a-c) Lung compliance, airway resistance, and inspiratory capacity were measured using the Flexivent system 7 dpi. d) Pressure volume loops were obtained from SARS-CoV-2 and mock infected mice at Day 7. e) Oxygen saturations were measured by pulse oximetry 7 dpi. Data not normally distributed, *P < 0.05 by Mann-Whitney test. n=10 hamsters and/or group for all panels. Box blots show median +/- interquartile range and whiskers define minutes and max values. Data points represent individual animals.
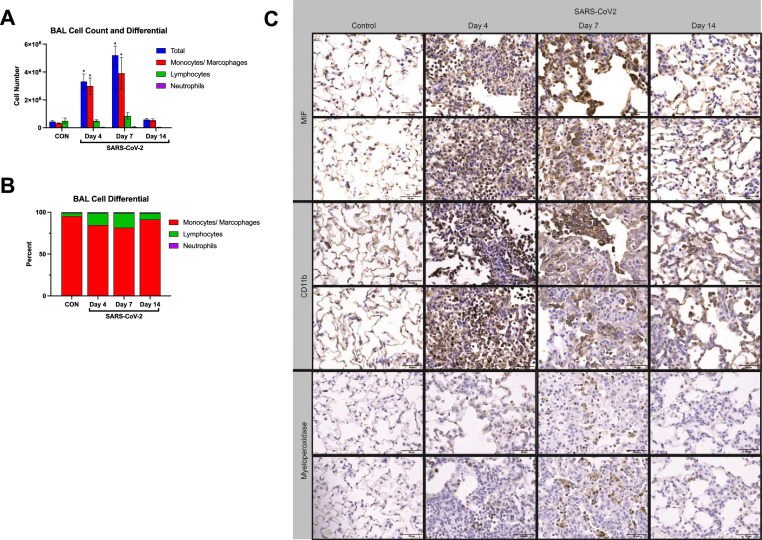
Monocytes and macrophages dominate cellular response to SARS-CoV-2 infection in hamsters. We next characterized inflammatory responses in COVID-19 hamsters. BAL cell counts, performed at each endpoint, showed increases in BAL cell counts at 4 and 7 dpi, again with return to baseline by 14 dpi (Fig. 3 A). Monocytes and macrophages were the predominant cell type at all time points and accounted for the increases in total BAL cell counts (Fig. 3B). To further support this observation, we assessed inflammatory cell infiltration in lung tissue sections. Consistent with H & E staining, we observed significant inflammatory cell infiltration and debris in the alveolar spaces by immunohistochemical staining. Notably, the predominant cell types were monocytes and macrophages, as evidenced by significant increases in MIF (macrophage-inhibitory factor) and CD11b staining (Fig. 3C), consistent with findings by BAL analysis. These results indicate that inflammatory cells, predominantly of myeloid lineage, infiltrate the lung early in the course of SARS-CoV-2 infection with a peak that lags behind improvement in viral infection. Further, we observed evidence of persistent increases in monocytes and macrophages in the alveolar space at 14 dpi (Fig. 3C). These data demonstrate infiltration of the lung tissue and alveolar space with inflammatory cells with SARS-CoV-2 infection in hamsters.
Fig 3.
Infection with SARS-CoV-2 causes significant lung inflammation in hamsters. (A) BAL total cell count and differential shows increased inflammatory cell infiltration with SARS-CoV-2 infection in hamsters. (B) BAL cell differential is dominated by monocytes and macrophages across all timepoints. (C) Representative images from hamster lung tissue demonstrate increased staining for MIF, Cd11b, and myeloperoxidase by immunohistochemistry (IHC) at 4, 7, and 14 dpi with the majority of cellular infiltrate staining positive for MIF and Cd11b at all 3 time points.
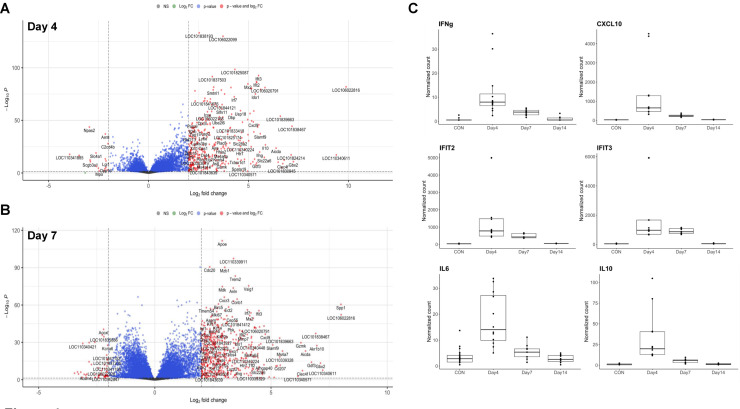
SARS-CoV-2 infection induces significant changes in the lung tissue transcriptome of hamsters. To explore molecular mechanisms promoting development of ALI, we harvested total cellular RNA from hamster lung tissue, prepared total transcriptome RNA libraries following ribosomal RNA depletion, and performed next generation RNA sequencing. Reads from RNA sequencing mapped to 22,405 distinct transcripts that were used for differential expression analysis. Notably, not all mapped hamster transcripts have human or mouse orthologs. We detected significant differences in the transcriptome of hamsters following SARS-CoV-2 infection compared to control at 4 and 7 dpi (Figs. 4 A, 4B). With few exceptions, transcriptomic differences mostly resolved by 14 dpi in this model (Sup. Fig. 2A). There was significant overlap in genes upregulated at both 4 and 7 dpi (Sup. Fig. 2B). Top differentially expressed genes at 4 dpi included interferon (IFN)-γ, IFN responsive genes such as IFIT2 and IFIT3 (INF-induced protein with tetratricopeptide repeats 2 and 3), and chemokines, CXCL9 and CXCL10, as shown in Table 1 . We also observed increased interleukin (IL)-6 and IL-10 transcripts at 4 dpi. Top differentially expressed genes at 7 dpi are shown in Table 2 .
Fig 4.
Differential gene expression in hamsters with SARS-CoV-2 infection. Volcano plots of overall differential expression analysis show differences in transcriptome of control and SARS-CoV-2 infected hamsters at 4 dpi (A) and 7 dpi (B). The x-axis corresponds to the log (base 2) of the fold change between infected and control hamsters. The y-axis represents the negative log (base 10) of p-values. Cutoffs for effect size and significance are set at >|2| Log2 fold change and P < 0.05. (C) Box plots of several of the most differentially expressed genes depicting normalized counts for each experimental endpoint. n >= 6 hamsters and/or group for all panels.
Table 1.
Top 15 most differentially expressed transcripts from hamsters infected with SARS-CoV-2 vs controls at 4 dpi
| Gene | log2FoldChange | Adjusted P |
|---|---|---|
| Gbx2 | 7.25 | 4.3E-10 |
| Clec4f | 6.72 | 2.6E-08 |
| Aicda | 6.38 | 2.4E-22 |
| Il10 | 5.85 | 2.4E-25 |
| Slc22a6 | 5.79 | 7.0E-14 |
| Rsad2 | 5.62 | 5.4E-77 |
| Slamf9 | 5.59 | 9.4E-36 |
| Ifng | 5.56 | 9.4E-19 |
| Ifit3 | 5.51 | 1.4E-89 |
| Cxcl10 | 5.41 | 2.6E-83 |
| Gdf3 | 5.41 | 7.1E-09 |
| Ifit2 | 5.40 | 8.5E-84 |
| Ido1 | 5.35 | 9.9E-73 |
| Il21 | 5.30 | 2.4E-12 |
| Cxcl9 | 5.24 | 1.1E-46 |
To identify primary networks contributing to transcriptomic changes with SARS-CoV-2 infection, we focused on the most differentially expressed genes at 4, and 7 dpi. We identified genes with robust differential expression, defined as >|3| Log2 fold change increase that met significance criteria and performed STRING network analysis [44]. We excluded transcripts without human orthologs from this analysis. STRING analysis did not identify relevant datasets from hamsters, but human orthologs suggest network upregulation of IL-10 signaling, exocytosis of tertiary granule lumen proteins, catalysis, and expression of IFN-induced genes at 4 dpi (Sup. Fig. 2C). At 7 dpi, STRING network analysis reveals upregulation of expression of IFN-induced genes, IFNα and IFNβ signaling and exocytosis of tertiary granule lumen proteins based on human ortholog datasets (Sup. Fig. 2D). Taken together, these data demonstrate persistent inflammatory signaling at 4, and 7 dpi with upregulation of interferon-induced genes (Fig. 4C).
SARS-CoV-2 infection induces changes in the BAL and lung tissue phospholipidome of hamsters. To assess differences in the lipidome between control and SARS-CoV-2 infected hamsters, we performed global redox phospholipidomics analysis of BAL fluid, cells obtained from BAL, and lung tissue samples at 4, 7, and 14 dpi. In BAL we were able to detect 430 phospholipid (PL) molecular species including 100 oxygenated phospholipid (PLox) species and 46 lyso-phospholipids (lyso-PL). In cells, mainly represented by monocytes and macrophages, the phospholipidome consisted of 403 molecular species. Among those, 97 were represented by PLox species, and 32 species were lyso-PL. Analysis of lung samples revealed the presence of 512 PL molecular species, including 72 PLox and 47 lyso-PL species. In addition, in all samples we were able to detect lipid mediators, including pro-inflammatory mediators such as LTB4, HXA3, and anti–inflammatory, LXA4. However, we did not observe any significant difference in these lipid mediators (Sup. Fig. 3).
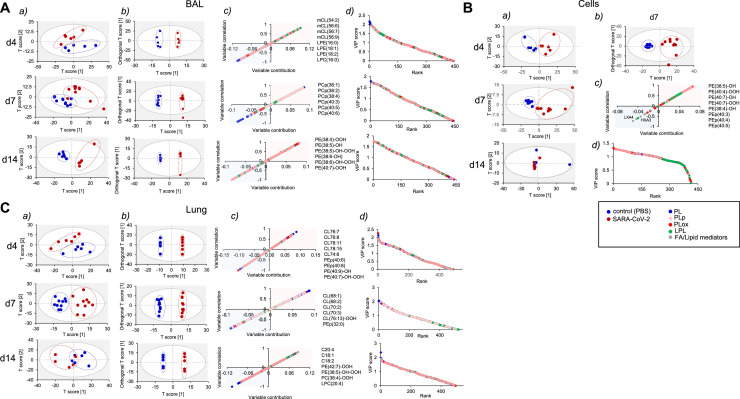
To better characterize the differences in the hamster lipidome induced by SARS-CoV-2, we used multivariate analysis. First, all identified lipids were subjected to principal component analysis (PCA) to find the differences in the lipidome between healthy and SARS-CoV-2 infected animals. The PCA analysis revealed that BAL samples (Fig. 5 Aa), cell samples (except samples obtained on 14 dpi) (Fig. 5Ba) and lung samples (Fig. 5Ca) in control groups clustered tightly, while samples in the groups of infected animals were distributed diffusely, demonstrating heterogeneity in the lipids associated with the viral infection. No differences in cell samples obtained on day 14 after infection with the virus were detected (Fig. 5Ba). To uncover the driving forces among the variables detected in the lipidome we used orthogonal projection of latent structures - discriminant analysis (OPLS-DA). The score plots in the OPLS-DA showed better clustering tendency in BAL (4, 7, and 14 dpi) (Fig. 5Ab), lung samples (4, 7, and 14 dpi) (Fig. 5Cb) and cell samples (d 7) (Fig. 5Bb). Data on lipidome obtained from cell samples at 4 and 14 dpi were unable to be fitted into the OPLS-DA model; thus we were not able to make score plots to visualize the differences. This indicates that the cell lipidome of infected hamsters does not have distinct changes from control groups. Next, we used S-plots to assess the variable correlation vs variable contribution for the OPLS-DA model (Fig. 5Ac, Bc, and Cc) as well as the variable importance in projection (VIP) score plots that reflected the significance of those variables for the OPLS-DA model (Fig. 5Ad, Bd, Cd). We established that at different time points, different phospholipids were contributing to the lipidome differences between control groups and groups of hamsters infected with SARS-CoV-2. Thus, lyso-PLs, phosphatidylcholine plasmalogen species and oxygenated phosphatidylethanolamine (PE) species were the major contributors in BAL samples on 4, 7 and 14 dpi, respectively (Fig. 5Ac, Ad). Cardiolipins were the predominant phospholipid species accumulated in lung of infected animals on 4 and 7 dpi (Fig. 5Cc, Cd). The significant differences in lipidome of cells were detected only at 7 dpi. These differences were mainly due to oxygenated PE species and PE plasmalogens. (Fig. 5Bc, Bd). Oxygenated PE species, including hydroperoxy-PE species that were previously identified as ferroptotic cell death signals, were among the top 10 phospholipid metabolites that are mainly responsible for the group differentiating in cell samples on day 7 as well as in BAL and in lung samples on 14 dpi (Fig. 5Ac, Bc, Cc).
Fig 5.
SARS-CoV-2 induced changes in lipidome of BAL (A), cells (B), and lung (C) of infected hamsters. (A, B, C) Score plots of PCA (a) OPLS-DA (b) analysis show the differences in lipidomes of control and SARS-CoV-2 treated hamsters. (c) S-plot of OPLS-DA analysis shows variable correlation vs variable contribution for the OPLS-DA model. Positive correlation implies accumulation in infected group. (d) The variable importance in projection (VIP) score plots reflect the significance of variables for the OPLS-DA models. The VIP >1 was considered to be a statistically significant difference between the 2 groups. PL molecular species that have highest positive VIP score are listed. n >= 6 hamsters and/or group for all panels.
In addition to general description of lipidomic changes with SARS-CoV-2 infection, our lipidomic analysis focused on characterization of lipid species associated with major inflammatory pathways. As we observed transcriptional upregulation of interferon signaling in our RNAseq dataset, we hypothesized that changes in lipidome may reflect activation of phospholipase A2 (PLA2), an important inflammatory mediator, activated in response to IFN signaling.51 , 52 In BAL and cells at 4 and 7 dpi, SARS-CoV-2 samples contained elevated levels of lysophospholipids, lysophosphatidylethanolamine (LPE) and lysophosphatidylcholine (LPC), with saturated and mono-unsaturated fatty acids in sn-1 position. These lysophospholipids are formed from phosphatidylethanolamine (PE) and phosphatidylcholine (PC) via canonical PLA2-driven reactions (BAL 4 dpi and 7 dpi, Sup. Fig. 4A; cells 4 dpi and 7 dpi, Sup. Fig. 4B). Although we observed predominantly monocytes and macrophages in hamster BAL and lung tissue, lipidome data showed accumulation of species associated with myeloperoxidase (MPO) activation. Samples from hamsters with SARS-CoV-2 contained increased contents of LPE containing polyunsaturated fatty acids in sn-2 position (BAL at 4 dpi, Sup. Fig 5A; cells at 4 and 7 dpi, Sup. Fig. 5B). These products are usually generated in pro-oxidant conditions due to activation of MPO (ClO-), expressed abundantly in neutrophil granulocytes, or iNOS/NO•/O2•- (ONOO-).
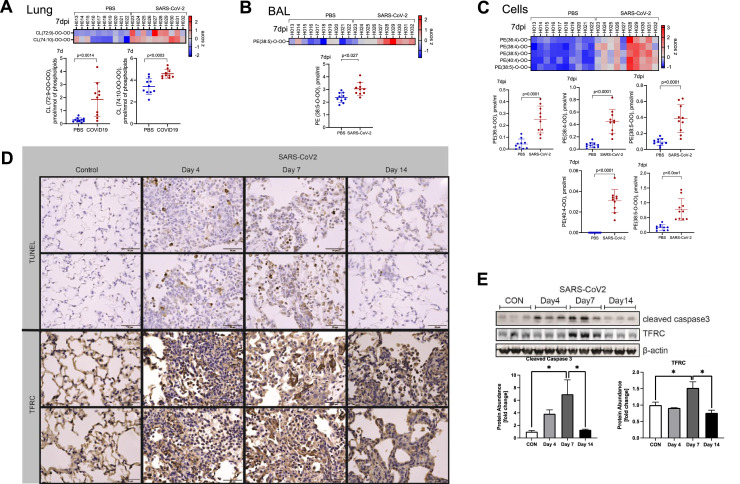
SARS-CoV-2 infection leads to apoptosis and ferroptosis in hamster lung tissue. Acute lung injury and ARDS are often characterized by a neutrophilic infiltrate and necrotic cell death. In COVID-19, apoptotic cell death in the lung is known to occur in both humans53 and the hamster model.15 Consistent with these reports, our lipidomic analysis of lung tissue showed that SARS-CoV-2 infection caused accumulation of oxidized cardiolipin species that are apoptotic cell death signals (Fig. 6 A). We also identified accumulation of oxidized PE species that are ferroptotic death signals with SARS-CoV-2 infection in BAL at 7 dpi (Fig. 6B) and cells at 14 dpi (Fig. 6C) with persistence of these signals at 14 dpi (Sup. Fig. 5C, D). To further support these findings, we stained hamster lung tissue sections and observe positive TUNEL staining, consistent with apoptosis, with peak at 7 dpi and increased TFRC (Transferrin Receptor Protein 1), a specific marker of ferroptosis,54 , 55 staining by IHC, again most evident at 7 dpi (Fig. 6D). We performed immunoblotting to further support these findings and observed significant increases in protein abundance of cleaved caspase-3, a marker of apoptosis,56 , 57 and TFRC (Fig. 6E). Both proteins peaked at 7 dpi with return to baseline abundance by 14 dpi. These data suggest that both apoptosis and ferroptosis contribute significantly to cell death and lung injury in hamsters infected with SARS-CoV-2.
Fig 6.
Hamster lungs demonstrate increased apoptosis and ferroptosis with SARS-CoV-2 infection. Heatmaps show relative differences and plots depict concentrations of (A) oxidized cardiolipins in lung tissue at 7 dpi and oxidized PE species in (B) BAL and (C) cells at 7 dpi. Data points represent individual animals. (D) Representative images from hamster lung tissue demonstrate increased TUNEL staining and increased staining for TFRC protein by immunohistochemistry (IHC) at 4, 7 and 14 dpi. n = 3 hamsters and/or group. (E) Immunoreactive cleaved caspase 3 increased at 4 and 7 dpi, and immunoreactive TFRC increased at 7 dpi in lung tissue lysates from hamsters infected with SARS-CoV-2. Below, densitometric analysis of cleaved caspase 3 and TFRC signal vs time, normalized to β-actin. n = 3 hamsters and/or group. Data shown as mean ± SEM. *P < 0.05 as determined by ANOVA with post-hoc Sidak's multiple comparisons test.
DISCUSSION
The new contributions in this study are a focused evaluation of the utility of Syrian golden hamsters as a model of experimental ALI with SARS-CoV-2 infection, based on biophysical, transcriptomic, and lipidomic data. Although prior studies have demonstrated infection of hamsters with SARS-CoV-2 virus,15 , 31 we present granular data on the pro-inflammatory lipid mediators, molecular networks, and lung mechanical properties within SARS-CoV-2 infected hamsters that provides a pathophysiologic basis for our observations of acute lung injury seen in this experimental model of ARDS. Importantly, we identified potential molecular drivers of SARS-CoV-2 acute lung injury, with increases in interferon signaling and upregulation of interferon inducible genes by RNA-seq transcriptomic analysis, and increases in both oxygenated PE, associated with ferroptosis, and PE plasmalogens by lipidomic profiling. The hamster model, however, has limitations in the absence histologic evidence of hyaline membranes, neutrophil-predominant lung inflammation, and severely reduced compliance and increased stiffness typically seen in humans with ARDS. This might be attributed to a lower viral burden, a unique pathobiological response in hamsters, or simply that we may have missed these changes given the kinetics of our analysis. Notably, established ALI animal models are known to demonstrate differences from human ARDS.7 , 34, 35, 36 Another key drawback to the use of hamsters as an experimental model of SARS-CoV-2 ARDS is the relative lack of validated reagents for detailed molecular analysis.
This study is the first to systematically evaluate hamsters infected with SARS-CoV-2 as a model of ARDS. A notoriously heterogeneous syndrome, human ARDS is inflammatory lung injury characterized by increased alveolar capillary permeability, non–cardiogenic edema, and hypoxemic respiratory failure.58 ARDS caused by SARS-CoV-2 in humans satisfies these criteria.59 In 2011, the American Thoracic Society (ATS) published a workshop report describing key features of experimental ALI in animals. Further, this report detailed relevant measurements to evaluate each criterion.7 Using this guideline, we evaluated each key feature. We demonstrate (1) histologic lung tissue injury by accumulation of debris in the alveolar space, thickening of the alveolar walls (Fig. 2C), and enhanced injury as determined by histology scoring (Fig. 2B); (2) disruption of the alveolar capillary barrier by an increase in BAL protein concentration (Fig. 2D); (3) an inflammatory response evidenced by infiltration of the lung tissue with inflammatory cells (Fig. 3A – C) with an increase in proinflammatory cytokines in the lung tissue (Fig. 4C); and physiologic dysfunction by hypoxemia (Fig. 2E). Despite observing histologic lung injury, hyaline membranes were notably absent in this model. Hyaline membranes, a characteristic finding in human ARDS, are uncommon in rodent models of experimental ALI,6 , 7 , 34 highlighting the limitations to preclinical modeling of human ARDS. A notable difference from other preclinical models of ARDS was the predominance of monocytes and macrophages throughout the course of lung injury, as opposed to neutrophilic infiltrate (Fig. 3A – C). However, other reports have noted the higher incidence of inflammatory monocytes in COVID-1960 , 61 and other viral illnesses.62 A recent report noted neutrophilia in only 31% of patients with severe COVID-19.63 We did observe evidence of MPO activation in our lipidome data (Sup. Fig. 5A, B), suggesting that neutrophilic infiltration may have lessened by our first endpoint of 4 dpi. Given the degree of injury based on tissue and BAL findings, we expected to observe impairments in lung compliance and resistance but we only observed significant hypoxemia (Fig. 2E). This finding may be due to the patchy nature of the infiltrates as demonstrated on tissue sections, leading to ventilation – perfusion (V/Q) mismatching. The normal or preserved lung in between patches may have been sufficient to mask or overcome any impairment in lung compliance and airway resistance as detected by the rodent ventilator. Hypoxemia in the absence of dyspnea and minimal changes in lung compliance has been described in humans as silent (or “happy”) hypoxemia.64, 65, 66, 67 Our model may mirror this particular clinical course.
One current limitation of hamsters as an animal model is the lack of validated molecular reagents, such as antibodies, ELISA kits, and primers of PCR. The hamster genome has been previously mapped (Ensembl MesAur1.0, GCA_000349665.1), facilitating RNA-seq transcriptomic analysis of hamster lung tissue. Many of the differentially expressed genes did not have human orthologs (Fig. 4), limiting pathways analysis. Of note, we performed bulk RNA-seq from lung tissue, which allowed for inactivation of virus upon tissue harvest but did limit the resolution of analysis compared to single cell sequencing. Our analysis revealed upregulation of IFN signaling, specifically IFN-γ, and IFN inducible genes, such as IFIT2 and IFIT3, at 4 and 7 dpi. Our data demonstrating monocyte predominance in the BAL and increased IFN-γ and CXCL10 signaling recapitulates findings from BAL fluid from patients with respiratory failure secondary to COVID-1963 Both human and mouse studies have established the vital role of Type I IFN in modulating response to COVID-19.68, 69, 70 Early in the course of disease, IFN signaling is vital to host defense and numerous studies suggest that dysregulation or deficiency of Type I IFNs leads to severe, life-threatening disease.71, 72, 73, 74 Hamsters also demonstrated significant increases in pro-inflammatory cytokines associated with severe COVID-19 and poor prognosis in humans, specifically CXCL10,75 and IL-6.76 These findings demonstrate the utility of hamsters as a model to study the host response to SARS-CoV-2 complicated by ARDS.
There are several reports documenting lipid profiles of plasma in COVID-19 patients emphasizing marked changes in phospholipids, neutral lipids (particularly, triacylglycerols, cholesterol esters) and non–esterified fatty acids related to the severity and transition from the pro-inflammatory to the resolution phases of the disease, as well as in cured patients.77, 78, 79, 80 In few cases, lipidomics studies were conducted in COVID-19 BAL samples, thus reflecting lipidomics changes in the lungs of intubated patients. The results revealed increased levels of fatty acids and inflammatory lipid mediators with both pro-inflammatory (thromboxanes, prostaglandins, leukotrienes) as well as anti–inflammatory (lipoxin A4 and the D-series resolvins) capacities.81 In other words, the lipid mediator storm occurring in severe COVID-19 engages mechanisms operating with both pro- and anti–inflammatory lipid mediators.
Lipidomics studies using animal models of COVID-19 are sparse. In one of them, an untargeted LC-MS based lipidomic analysis of the nasal samples was performed during viral shedding and post-shedding in an asymptomatic SARS-CoV-2 ferret model.82 Statistically significant changes were found only in 3 lipids (out of a total of 325 identified) belonging to the class of phosphatidylethanolamines (PEs) - PE(20:4/22:6), PE(20:5/22:5), and PE(22:6/20:4) - which were all downregulated in the post-shedding group. In the current study, we employed redox lipidomics approach and identified not only individual molecular species in all 6 major classes of phospholipids (PE, PC, PG, PS, PI, CL) but also their oxidative and hydrolytic (lyso-phospholipids) metabolites. Importantly, multivariate analysis showed that both non–oxidized phospholipids as well as their peroxidized and hydrolyzed metabolites contributed to the separation of changes between the control group of hamsters from the groups of infected with SARS-CoV-2. Thus, our study represents the first and most robust lipidomic analysis in an animal model of SARS-CoV-2.
Understanding mechanisms that propagate acute lung injury in SARS-CoV-2 addresses a profound unmet need to develop specific therapeutics to mitigate COVID-19 disease and promote greater understanding of specific biologic processes that lead to ARDS. A fundamentally heterogeneous syndrome, study of specific ARDS endotypes has utility beyond each cause. Hamsters demonstrate utility as an experimental model of ALI in COVID-19 and with modern sequencing and MS techniques, detailed molecular analysis is possible. Further characterization of this model may provide benefits in development of novel therapeutics to SARS-CoV-2 and establish a platform to assess emerging highly virulent variants of novel pandemic strains of coronaviridae.
AUTHOR CONTRIBUTIONS
Research studies were designed by JSB, JAE, JDL, RKM, and RTR. Animal experiments were performed by JSB, JAE, DF, HL, ORM, MR-F, JDL, and RTR. Assays and analysis of materials obtained from the animal model were conducted by JSB, VEK, DF, YYT, VAT, SNS, LF, AE, FJ, DJ, JDL, and RTR. Data analysis was performed by JSB, VEK, JAE, YYT, VAT, SNS, LF, LJ, AM, DJ, JDL, RKM, and RTR. The manuscript was prepared by JSB, VEK, YYT, JDL, RKM, and RTR.
ACKNOWLEDGMENTS
This work was supported by NIH grants UH3HL123502, R01HL096376, R01HL097376, R01HL098174, R01HL081784, and P01HL114453 to R.K.M., R01HL142767 to J.A.E., and R01AI121212 to R.T.R. The project was also supported by Award Number Grant UL1TR002733 and KL2TR002734 from the National Center for Advancing Translational Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Advancing Translational Sciences or the National Institutes of Health. RNA library preparation and sequencing was performed by The Ohio State University Genomics Shared Resource (supported in part by P30CA016058) with special thanks to Pearlly Yan, PhD and Estela Puchulu-Campanella for their contributions. The authors acknowledge the journal's policy on disclosure of potential conflicts of interest, and hereby declare that they have no financial or personal conflicts of interest to disclose. The authors also acknowledge and agree to the journal's authorship statement.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.trsl.2021.10.007.
Appendix. Supplementary materials
REFERENCES
- 1.Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richardson S, Hirsch JS, Narasimhan M, et al. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with covid-19 in the New York city area. JAMA. 2020;323:2052–2059. doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hariri L, Hardin CC. Covid-19, angiogenesis, and ARDS endotypes. N Engl J Med. 2020;383:182–183. doi: 10.1056/NEJMe2018629. [DOI] [PubMed] [Google Scholar]
- 5.Thompson BT, Chambers RC, Liu KD. Acute respiratory distress syndrome. N Engl J Med. 2017;377:562–572. doi: 10.1056/NEJMra1608077. [DOI] [PubMed] [Google Scholar]
- 6.Aeffner F, Bolon B, Davis IC. Mouse models of acute respiratory distress syndrome: a review of analytical approaches, pathologic features, and common measurements. Toxicol Pathol. 2015;43:1074–1092. doi: 10.1177/0192623315598399. [DOI] [PubMed] [Google Scholar]
- 7.Matute-Bello G, Downey G, Moore BB, et al. An official American Thoracic Society workshop report: features and measurements of experimental acute lung injury in animals. Am J Respir Cell Mol Biol. 2011;44:725–738. doi: 10.1165/rcmb.2009-0210ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan JF, Kok KH, Zhu Z, et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan JF, To KK, Tse H, Jin DY, Yuen KY. Interspecies transmission and emergence of novel viruses: lessons from bats and birds. Trends Microbiol. 2013;21:544–555. doi: 10.1016/j.tim.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271-80 e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 13.Sun J, He WT, Wang L, et al. COVID-19: epidemiology, evolution, and cross-disciplinary perspectives. Trends Mol Med. 2020;26:483–495. doi: 10.1016/j.molmed.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Damas J, Hughes GM, Keough KC, et al. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc Natl Acad Sci U S A. 2020;117:22311–22322. doi: 10.1073/pnas.2010146117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan JF, Zhang AJ, Yuan S, et al. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in a golden syrian hamster model: implications for disease pathogenesis and transmissibility. Clin Infect Dis. 2020;71:2428–2446. doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94(7):e00127-20. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim YI, Kim SG, Kim SM, et al. Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host Microbe. 2020;27:704-9 e2. doi: 10.1016/j.chom.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi J, Wen Z, Zhong G, et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rockx B, Kuiken T, Herfst S, et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 2020;368:1012–1015. doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chandrashekar A, Liu J, Martinot AJ, et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. 2020;369:812–817. doi: 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu J, Tostanoski LH, Peter L, et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369:806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williamson BN, Feldmann F, Schwarz B, et al. Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2. Nature. 2020;585:273–276. doi: 10.1038/s41586-020-2423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Doremalen N, Lambe T, Spencer A, et al. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature. 2020;586:578–582. doi: 10.1038/s41586-020-2608-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bao L, Deng W, Huang B, et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 25.McCray PB, Jr., Pewe L, Wohlford-Lenane C, et al. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J Virol. 2007;81:813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tseng CT, Huang C, Newman P, et al. Severe acute respiratory syndrome coronavirus infection of mice transgenic for the human Angiotensin-converting enzyme 2 virus receptor. J Virol. 2007;81:1162–1173. doi: 10.1128/JVI.01702-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rathnasinghe R, Strohmeier S, Amanat F, et al. Comparison of transgenic and adenovirus hACE2 mouse models for SARS-CoV-2 infection. Emerg Microbes Infect. 2020;9:2433–2445. doi: 10.1080/22221751.2020.1838955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leist SR, Dinnon KH, Schafer A, 3rd, et al. A mouse-adapted SARS-CoV-2 induces acute lung injury and mortality in standard laboratory mice. Cell. 2020;183 doi: 10.1016/j.cell.2020.09.050. 1070-85 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gretebeck LM, Subbarao K. Animal models for SARS and MERS coronaviruses. Curr Opin Virol. 2015;13:123–129. doi: 10.1016/j.coviro.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osterrieder N, Bertzbach LD, Dietert K, et al. Age-dependent progression of SARS-CoV-2 infection in syrian hamsters. Viruses. 2020;12(7):779. doi: 10.3390/v12070779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Imai M, Iwatsuki-Horimoto K, Hatta M, et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc Natl Acad Sci U S A. 2020;117:16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee AC, Zhang AJ, Chan JF, et al. Oral SARS-CoV-2 Inoculation Establishes Subclinical Respiratory Infection with Virus Shedding in Golden Syrian Hamsters. Cell Rep Med. 2020;1 doi: 10.1016/j.xcrm.2020.100121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sia SF, Yan LM, Chin AWH, et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583:834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matute-Bello G, Frevert CW, Martin TR. Animal models of acute lung injury. Am J Physiol Lung Cell Mol Physiol. 2008;295:L379–L399. doi: 10.1152/ajplung.00010.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.FR D'Alessio. Mouse Models of Acute Lung Injury and ARDS. Methods Mol Biol. 2018;1809:341–350. doi: 10.1007/978-1-4939-8570-8_22. [DOI] [PubMed] [Google Scholar]
- 36.Chimenti L, Morales-Quinteros L, Puig F, et al. Comparison of direct and indirect models of early induced acute lung injury. Intensive Care Med Exp. 2020;8:62. doi: 10.1186/s40635-020-00350-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Case JB, Bailey AL, Kim AS, Chen RE, Diamond MS. Growth, detection, quantification, and inactivation of SARS-CoV-2. Virology. 2020;548:39–48. doi: 10.1016/j.virol.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Irvin CG, Bates JH. Measuring the lung function in the mouse: the challenge of size. Respir Res. 2003;4:4. doi: 10.1186/rr199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ingenito EP, Mora R, Cullivan M, Marzan Y, Haley K, Mark L, et al. Decreased surfactant protein-B expression and surfactant dysfunction in a murine model of acute lung injury. Am J Respir Cell Mol Biol. 2001;25:35–44. doi: 10.1165/ajrcmb.25.1.4021. [DOI] [PubMed] [Google Scholar]
- 40.Wang H, Jean S, Eltringham R, et al. Mutation-specific SARS-CoV-2 PCR screen: rapid and accurate detection of variants of concern and the identification of a newly emerging variant with spike L452R mutation. J Clin Microbiol. 2021;59(8):e0092621. doi: 10.1128/JCM.00926-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu A, Ibrahim JG, Love MI. Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics. 2019;35:2084–2092. doi: 10.1093/bioinformatics/bty895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–DD13. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kagan VE, Mao G, Qu F, et al. Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nat Chem Biol. 2017;13:81–90. doi: 10.1038/nchembio.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lythgoe KA, Hall M, Ferretti L, et al. SARS-CoV-2 within-host diversity and transmission. Science. 2021;372(6539):eabg0821. doi: 10.1126/science.abg0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Braun KM, Moreno GK, Halfmann PJ, et al. Transmission of SARS-CoV-2 in domestic cats imposes a narrow bottleneck. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sawatzki K, Hill NJ, Puryear WB, Foss AD, Stone JJ, Runstadler JA. Host barriers to SARS-CoV-2 demonstrated by ferrets in a high-exposure domestic setting. Proc Natl Acad Sci U S A. 2021;118(18) doi: 10.1073/pnas.2025601118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Madhi SA, Baillie V, Cutland CL, et al. Efficacy of the ChAdOx1 nCoV-19 Covid-19 vaccine against the B.1.351 Variant. N Engl J Med. 2021;384:1885–1898. doi: 10.1056/NEJMoa2102214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wibmer CK, Ayres F, Hermanus T, et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat Med. 2021;27:622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- 51.Ponzoni M, Cornaglia-Ferraris P. Interferon-gamma-stimulated and GTP-binding-proteins-mediated phospholipase A2 activation in human neuroblasts. Biochem J. 1993;294(Pt 3):893–898. doi: 10.1042/bj2940893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pruzanski W, Vadas P. Phospholipase A2–a mediator between proximal and distal effectors of inflammation. Immunol Today. 1991;12:143–146. doi: 10.1016/S0167-5699(05)80042-8. [DOI] [PubMed] [Google Scholar]
- 53.Liu Y, Garron TM, Chang Q, et al. Cell-Type Apoptosis in Lung during SARS-CoV-2 Infection. Pathogens. 2021;10(5):509. doi: 10.3390/pathogens10050509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feng H, Schorpp K, Jin J, et al. Transferrin Receptor Is a Specific Ferroptosis Marker. Cell Rep. 2020;30:3411-23 e7. doi: 10.1016/j.celrep.2020.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu Y, Yang Q, Su Y, et al. MYCN mediates TFRC-dependent ferroptosis and reveals vulnerabilities in neuroblastoma. Cell Death Dis. 2021;12:511. doi: 10.1038/s41419-021-03790-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fernandes-Alnemri T, Litwack G, Alnemri ES. CPP32, a novel human apoptotic protein with homology to Caenorhabditis elegans cell death protein Ced-3 and mammalian interleukin-1 beta-converting enzyme. J Biol Chem. 1994;269:30761–30764. [PubMed] [Google Scholar]
- 57.Nicholson DW, Ali A, Thornberry NA, et al. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature. 1995;376:37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- 58.Force ADT, Ranieri VM, Rubenfeld GD, et al. Acute respiratory distress syndrome: the Berlin definition. JAMA. 2012;307:2526–2533. doi: 10.1001/jama.2012.5669. [DOI] [PubMed] [Google Scholar]
- 59.Grasselli G, Zangrillo A, Zanella A, et al. Baseline characteristics and outcomes of 1591 patients infected with SARS-CoV-2 admitted to ICUs of the lombardy region. Italy. JAMA. 2020;323:1574–1581. doi: 10.1001/jama.2020.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu X, Han M, Li T, et al. Effective treatment of severe COVID-19 patients with tocilizumab. Proc Natl Acad Sci U S A. 2020;117:10970–10975. doi: 10.1073/pnas.2005615117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cillo AR, Somasundaram A, Shan F, et al. Bifurcated monocyte states are predictive of mortality in severe COVID-19. bioRxiv. 2021 2021.02.10.430499. [Google Scholar]
- 62.Kwissa M, Nakaya HI, Onlamoon N, et al. Dengue virus infection induces expansion of a CD14(+)CD16(+) monocyte population that stimulates plasmablast differentiation. Cell Host Microbe. 2014;16:115–127. doi: 10.1016/j.chom.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grant RA, Morales-Nebreda L, Markov NS, et al. Circuits between infected macrophages and T cells in SARS-CoV-2 pneumonia. Nature. 2021;590:635–641. doi: 10.1038/s41586-020-03148-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haryalchi K, Heidarzadeh A, Abedinzade M, Olangian-Tehrani S, Ghazanfar Tehran S. The importance of happy hypoxemia in COVID-19. Anesth Pain Med. 2021;11 doi: 10.5812/aapm.111872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lemyze M, Mallat J, Nigeon O, et al. Rescue therapy by switching to total face mask after failure of face mask-delivered noninvasive ventilation in do-not-intubate patients in acute respiratory failure. Crit Care Med. 2013;41:481–488. doi: 10.1097/CCM.0b013e31826ab4af. [DOI] [PubMed] [Google Scholar]
- 66.Tobin MJ. Basing respiratory management of COVID-19 on physiological principles. Am J Respir Crit Care Med. 2020;201:1319–1320. doi: 10.1164/rccm.202004-1076ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kashani KB. Hypoxia in COVID-19: sign of severity or cause for poor outcomes. Mayo Clin Proc. 2020;95:1094–1096. doi: 10.1016/j.mayocp.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Israelow B, Song E, Mao T, et al. Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. J Exp Med. 2020;217(12):e20201241. doi: 10.1084/jem.20201241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee JS, Park S, Jeong HW, et al. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci Immunol. 2020;5(49) doi: 10.1126/sciimmunol.abd1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Major J, Crotta S, Llorian M, et al. Type I and III interferons disrupt lung epithelial repair during recovery from viral infection. Science. 2020;369:712–717. doi: 10.1126/science.abc2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bastard P, Rosen LB, Zhang Q, et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020:370. doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bastard P, Zhang Q, Cobat A, et al. Insufficient type I IFN immunity underlies life-threatening COVID-19 pneumonia. C R Biol. 2021;344:19–25. doi: 10.5802/crbiol.36. [DOI] [PubMed] [Google Scholar]
- 73.Lopez L, Sang PC, Tian Y, Sang Y. Dysregulated interferon response underlying severe COVID-19. Viruses. 2020;12(12):1433. doi: 10.3390/v12121433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Q, Bastard P, Liu Z, et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370(6515):eabd4570. doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Laing AG, Lorenc A, Del Molino Del Barrio I, et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat Med. 2020;26:1623–1635. doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- 76.Del Valle DM, Kim-Schulze S, Huang HH, et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat Med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nguyen M, Bourredjem A, Piroth L, et al. High plasma concentration of non-esterified polyunsaturated fatty acids is a specific feature of severe COVID-19 pneumonia. Sci Rep. 2021;11:10824. doi: 10.1038/s41598-021-90362-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lam SM, Zhang C, Wang Z, et al. A multi-omics investigation of the composition and function of extracellular vesicles along the temporal trajectory of COVID-19. Nat Metab. 2021;3:909–922. doi: 10.1038/s42255-021-00425-4. [DOI] [PubMed] [Google Scholar]
- 79.Bai Y, Huang W, Li Y, et al. Lipidomic alteration of plasma in cured COVID-19 patients using ultra high-performance liquid chromatography with high-resolution mass spectrometry. Biosci Rep. 2021;41(3) doi: 10.1042/BSR20204305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu J, Zhou M, Luo P, et al. Plasma metabolomic profiling of patients recovered from COVID-19 with pulmonary sequelae 3 months after discharge. Clin Infect Dis. 2021 doi: 10.1093/cid/ciab147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Archambault AS, Zaid Y, Rakotoarivelo V, et al. High levels of eicosanoids and docosanoids in the lungs of intubated COVID-19 patients. FASEB J. 2021;35:e21666. doi: 10.1096/fj.202100540R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Beale DJ, Shah R, Karpe AV, et al. Metabolic profiling from an asymptomatic ferret model of SARS-CoV-2 infection. Metabolites. 2021;11(5):327. doi: 10.3390/metabo11050327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.