ABSTRACT
Background
Human milk is a rich source of human milk oligosaccharides (HMOs) and bacteria. It is unclear how these components interact within the breast microenvironment.
Objectives
The objectives were first, to investigate the association between maternal characteristics and HMOs, and second, to assess the association between HMOs and microbial community composition and predicted function in milk from women with high rates of gestational glucose intolerance.
Methods
This was an exploratory analysis of a previously completed prospective cohort study (NCT01405547) where milk samples (n = 107) were collected at 3 mo postpartum. Milk microbiota composition was analyzed by V4-16S ribosomal RNA gene sequencing and HMOs by rapid high-throughput HPLC. Data were stratified and analyzed by maternal secretor status phenotype and associations between HMOs and microbiota were determined using linear regression models (ɑ-diversity), Adonis (B-diversity), Poisson regression models (differential abundance), and general linear models (predicted microbial function).
Results
Prepregnancy BMI, race, and frequency of direct breastfeeding, but not gestational glucose intolerance, were found to be significantly associated with a number of HMOs among secretors and non-secretors. Fucosyllacto-N-hexaose was negatively associated with microbial richness (Chao1) among secretors [B-estimate (SE): −9.3 × 102 (3.4 × 102); P = 0.0082] and difucosyllacto-N-hexaose was negatively associated with microbiota diversity (Shannon index) [−1.7 (0.78); P = 0.029] among secretors. Lacto-N-neotetraose (LNnT) was associated with both microbial B-diversity (weighted UniFrac R2 = 0.040, P = 0.036) and KEGG ortholog B-diversity (Bray-Curtis R2 = 0.039, P = 0.043) in secretors. Additionally, difucosyllactose in secretors and disialyllacto-N-hexaose and LNnT in non-secretors were associated with enrichment of predicted microbial genes encoding for metabolism- and infection-related pathways (P-false discovery rate < 0.1).
Conclusions
HMOs are associated with the microbial composition and predicted microbial functions in human milk at 3 mo postpartum. Further research is needed to investigate the role these relations play in maternal and infant health.
Keywords: human milk microbiota, human milk oligosaccharides, gestational diabetes, gestational glucose intolerance, microbiome, maternal BMI, secretor status, breastfeeding, lactation, human milk
Introduction
Human milk is home to a myriad of microorganisms, including bacteria. The bacteria in human milk, termed the human milk microbiota, are believed to play a role in the colonization of the infant's gastrointestinal (GI) microbiota and may contribute to both short- and long-term health outcomes (1, 2). In addition to these microbes, human milk is also a rich source of biologically active components, including growth factors, antibodies, oligosaccharides, hormones, cytokines, chemokines, and antimicrobial compounds (3). Previous studies have focused on potential external modifiers of the milk microbiota, such as maternal diet, antibiotic exposure, and mode of delivery (4–8); however, very few reports have examined how components within human milk may act as potential internal modifiers by interacting with the milk microbiota and vice versa.
One of the bioactive components of interest are human milk oligosaccharides (HMOs). HMOs are unconjugated, structurally diverse carbohydrates, which are the third most abundant solid component in human milk after lactose and lipids (9). HMO composition is dictated in large part by the expression of a single gene, fucosyltransferase-2 (FUT2), and mothers with an active allele of this gene are known as “secretors,” whereas those lacking an active copy (homozygous for the inactive allele) are “non-secretors” (9). Emerging literature has reported that maternal secretor status is associated with a number of parameters of human health, from maternal risk of infection and chronic disease to her child's GI microbiota composition (10, 11). Maternal characteristics including secretor status, parity, lactation period, race, breastfeeding exclusivity, geographical region, maternal BMI, and mode of delivery have been shown to be associated with specific HMOs (12–14). Only 1 group has examined the association between gestational glucose metabolism in metabolically healthy women and specific HMOs in milk, but none have investigated the association between gestational diabetes mellitus (GDM) and individual HMOs, along with the interplay with the milk microbiota (15–17). GDM has been found to be associated with altered activities of cellular glycosyltransferases, but its impact on individual HMOs is unknown (16). Due to the risk of maternal glucose intolerance impacting the HMOs present in milk, further investigation into this association is necessary (16).
When consumed by an infant, HMOs can act as prebiotics, or fermentable fibers, and act as a carbon source for specific bacteria in the infant GI tract (9). Within the infant GI tract, HMOs have been shown to be utilized by Bifidobacterium and Bacteroides species, while in vitro, they have also been shown to interact with other bacteria, such as Staphylococcus (18–21). While it has been hypothesized that bacteria in human milk may interact with HMOs within the mammary gland in a similar manner as to how they interact within the GI tract, this has not been systematically investigated. Only 4 studies to date have examined the association between HMOs and milk microbiota in mature human milk (22–25). These 4 studies have shown varying associations between specific HMOs and particular taxa within the milk microbiota; however, 3 of these 4 studies were limited by small sample sizes (n = 16, n = 25, n = 30, n = 393) and homogeneous cohorts in terms of race and metabolically healthy women. Additionally, none of these 4 studies examined the associations between HMOs and predicted microbial functions of the milk microbiota (22–25).
To address these gaps in the literature, the objective of this current study was to determine the association between HMOs and the human milk microbiota (microbial community structure and predicted functions) in a racially diverse prospective cohort of women with high rates of gestational glucose intolerance. Additionally, we aimed to determine how maternal demographic, metabolic, and obstetrical clinical parameters were associated with HMO composition. To the best of our knowledge, this is the first study to assess the relation between the milk microbiota, HMOs, and gestational impaired glucose tolerance (IGT).
Methods
Study participants and design
This was an exploratory analysis of bio-banked human milk samples and maternal demographic, metabolic, and obstetrical health data from a previously completed prospective cohort study (ClinicalTrials.gov: NCT01405547) (26, 27). The study protocol and findings of associations between these maternal clinical variables and the microbial composition of human milk have been previously published (7, 8, 26). Pregnant participants (n = 216) were recruited from Sinai Health (formerly, Mount Sinai Hospital) in Toronto, Canada, and in late pregnancy completed a 3-h 100-g oral-glucose-tolerance test (OGTT) between March 2009 and July 2010. For this present analysis, women were included if sufficient volumes of their mature milk sample from 3 mo postpartum was available for both HMO and milk microbiota analyses. The study protocol was approved by the Sinai Health Human Research Ethics Board and consent was obtained from all participants (26). A total of 117 women with term-born infants provided a mature milk sample at 3 mo postpartum, with 111 samples available for the present study (Supplemental Figure 1).
Maternal demographic, metabolic, and obstetrical data collection
A standardized self-report questionnaire was given during the first study visit (occurring at 30 wk of gestation; 95% CI: 25, 33 wk) to gather demographic, anthropometric, and obstetrical data. Prepregnancy BMI (kg/m2) was calculated and categorized as normal weight (18.5–24.9), overweight (25.0–29.9), and obese (≥30). Maternal race was categorized as White, Asian, or other (pooled Black, South Asian, other); the latter was combined due to low frequency of these races in the cohort. Mode of delivery was categorized as vaginal, scheduled cesarean delivery, unscheduled cesarean delivery). Glucose tolerance status was categorized as GDM, gestational IGT, or normoglycemia, and was diagnosed based on the results of the OGTT (26).
Collection of human milk samples
Mothers were asked to pump a complete breast expression of milk using a double electric breast pump (Medela, Inc.) with a sterile pumping kit at their 3-mo postpartum visit. No pumping or breastfeeding was to take place 2 h prior to the study visit. The collected mature whole-milk samples were placed in aliquots and stored at –80°C. Information regarding frequency of direct breastfeeding per day and human milk exclusivity (yes/no) was also collected at this visit.
HMO analysis
HPLC was used to characterize HMOs in human milk as previously described (28). Briefly, oligosaccharides were extracted by high-throughput solid-phase extraction over C18 and Carbograph microcolumns and fluorescently labeled with 2-aminobenzamide. Labelled oligosaccharides were analyzed by HPLC on an amide-80 column (Tosoh Bioscience) with online fluorescence detection. Absolute concentrations were calculated based on standard response curves for each of the annotated HMOs. Total concentration of HMOs was calculated as the sum of the annotated oligosaccharides (milligrams per milliliter). HMO-bound fucose and HMO-bound sialic acid were calculated on a molar basis (nanomoles per milliliter). HMO Simpson's diversity index was calculated based on relative abundances of all annotated HMOs. Maternal secretor status was determined by the high abundance (secretor) or near absence (non-secretor) of the HMO 2′-fucosyllactose (2′FL) in the respective milk samples. The 19 HMOs examined in these analyses included the following: 2′FL, 3-fucosyllactose (3FL), 3′-sialyllactose (3′SL), 6′-sialyllactose (6′SL), difucosyllactose (DFLac), difucosyllacto-N-hexaose (DFLNH), difucosyllacto-N-tetrose (DFLNT), disialyllacto-N-hexaose (DSLNH), disialyllacto-N-tetraose (DSLNT), fucodisialyllacto-N-hexaose (FDSLNH), fucosyllacto-N-hexaose (FLNH), lacto-N-fucopentaose-I/II/III (LNFP I/II/III), lacto-N-hexaose (LNH), lacto-N-neotetraose (LNnT), lacto-N-tetrose (LNT), sialyl-lacto-N-tetraose-b/c (LSTb/c).
DNA extraction, amplification, and sequencing of human milk microbiota
DNA from the human milk samples that had never been previously thawed was extracted using the NucleoSpin Food DNA isolation kit (Macherey-Nagel), as described previously (7, 8, 29). Briefly, the 515F and 806R primers were used during PCR (28 cycles) to amplify the V4 hypervariable region (30, 31). To confirm amplification quality, a negative control (sterile water), positive control (Pseudomonas aeruginosa), and mock community were also run alongside the DNA samples. An Illumina MiSeq instrument (MiSeq-V2-300 cycle chemistry) was used to sequence the quantified library to produce 150-bp paired end reads.
Bioinformatic and data analyses of the milk microbiota
Raw paired end sequences have been deposited in the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under the BioProject accession number PRJNA516669. The UPARSE pipeline in USEARCH processed the sequencing reads. The Ribosomal Database Project (RDP) 16S gold database (USEARCH) was used to detect and discard chimeric sequences (32). Sequences were grouped together into operational taxonomic units (OTUs) at 97% similarity using de novo clustering and the RDP 16S gold database was used to assign taxonomy to these OTUs. A FastTree QIIME python script was run to produce a phylogenetic tree (33).
A specialized package in R (version 3.6.1; R Foundation for Statistical Computing) intended for microbiome analysis, Phyloseq (1.25.2), was used to analyze microbiota composition (34). Singleton and doubleton OTUs were removed, as were OTUs that mapped to cyanobacteria/chloroplasts. Samples with low read counts were removed (n = 4) and data were rarefied to 20,000 reads per sample. Phyloseq was subsequently used to calculate and visualize relative abundances of predominant taxa (phylum and genus levels), ɑ-diversity (Chao1 and Shannon indices), and B-diversity (weighted UniFrac distance, Bray-Curtis dissimilarity) of the milk microbiota.
Statistical analyses
Associations between maternal characteristics, secretor status, and HMO composition
Chi-square tests (R version 3.6.1) were used to examine associations between categorical maternal clinical data and secretor status (secretor vs. non-secretor), whereas Wilcoxon rank-sum tests were run to determine associations between continuous maternal clinical data and secretor status. The categorical maternal data of interest included prepregnancy BMI (healthy, overweight, obese), glucose tolerance status (normoglycemic, IGT, GDM), mode of delivery (vaginal, scheduled cesarean delivery, unscheduled cesarean delivery), race (White, Asian, other), and human milk exclusivity [exclusive vs. nonexclusive (mixed formula feeding)]. The continuous maternal data included frequency of direct breastfeeding per day (n/day) and BMI (kg/m2). Wilcoxon rank-sum tests were also run to determine the relation between secretor status and total HMO concentration (milligrams per milliliter), HMO-bound sialic acid concentration (nanomoles per milliliter), and HMO-bound fucose concentration (nanomoles per milliliter). The significance level for these tests was set at P < 0.05.
After assessing relations between secretor status and HMO composition patterns, data were stratified by secretor status (secretor, n = 78; non-secretor, n = 29) and univariable linear regression models (SAS version 9.4; SAS Institute) were run to determine associations between maternal clinical data and each of the 19 HMOs within each secretor status stratum. Clinical data included the following: categorical prepregnancy BMI (healthy, overweight, obese), prepregnancy BMI as a continuous variable (kg/m2), glucose tolerance status (normoglycemic, IGT, GDM), mode of delivery (vaginal, scheduled cesarean delivery, unscheduled cesarean delivery), race (White, Asian, other), human milk exclusivity (vs. nonexclusive), and frequency of direct breastfeeding (n/day). To avoid skewing the data in the analyses, FUT2-dependent HMOs (2′FL, LNFPI, DFLac, DFLNH), known to be almost exclusively found in secretor milk, were not examined within the non-secretor strata for the present, and all subsequent, analyses.
HMO composition and milk microbiota
Using the total cohort for this present study (n = 107), associations between secretor status and microbial ɑ-diversities (Chao1 and Shannon indices) were determined by multivariable linear regression models (PROC MIXED) in SAS version 9.4, while adjusting for microbial DNA extraction batch and PCR sequencing batch. The data were then stratified by secretor status and models were built to assess the association between the top 19 HMOs (milligrams per milliliter), HMO diversity (Simpson's index), total HMO concentration (milligrams per milliliter), HMO-bound sialic acid concentration (nanomoles per milliliter), and HMO-bound fucose concentration (nanomoles per milliliter) and ɑ-diversity (Chao1 and Shannon indices). All models were adjusted by microbial DNA extraction and PCR batch effects. Due to sample size constraints from stratifying on the basis of secretor status, we were not able to adjust for any additional maternal covariates of interest. The significance level for these analyses was set at P < 0.05.
B-Diversities (weighted UniFrac distance, Bray-Curtis dissimilarity) of the human milk microbiota were calculated and visualized using principal coordinate analysis. All HMO concentrations were made categorical (Bisect 1 or 2) and run grouped as Bisect 1 (concentrations below the median) and Bisect 2 (concentrations above the median) for B-diversity analyses. Associations within the overall cohort (n = 107) between secretor status and microbial B-diversities (weighted UniFrac distance, Bray-Curtis dissimilarity) were determined by Adonis (vegan package version 2.5-3 in R) (30) while adjusting for microbial DNA extraction batch and PCR sequencing batch. Data were then stratified by secretor status and associations between HMOs and B-diversity measures were ascertained using Adonis (vegan package version 2.5-3 in R), as previously described (7, 8, 30). All HMO models were first run unadjusted in the Adonis model and then run adjusted for microbial DNA extraction and PCR batch effects. The significance level was set at P < 0.05.
Relative abundances of the top 5 phyla and top 8 genera of milk microbiota were explored as these were the predominant taxa consistent in both secretor and non-secretor milk samples. Using the overall cohort (n = 107), Poisson regression models (PROC GENMOD) were built in SAS (version 9.4) to evaluate the association between secretor status as a covariate and the relative abundance of predominant microbial taxa as outcomes, while adjusting for microbial DNA extraction and PCR batch effects. Data were then stratified by secretor status and the associations between HMOs and predominant microbial taxa were assessed. All models were adjusted for microbial DNA extraction and PCR batch effects. To address overdispersion, the SEs of these models were adjusted using Pearson scaling. To account for multiple comparisons, the Benjamini-Yekutieli cut-point approach was used (35); based on 5 phyla (5 tests), significance was set at P ≤ 0.022 for the phylum models, and for 8 genera (8 tests), significance was set at P ≤ 0.018 for the genus models.
HMO composition and predicted microbial functions
Microbial functions were inferred using Piphillin (https://piphillin.secondgenome.com/) (36). The Kyoto Encyclopedia of Genes and Genomes (KEGG; https://www.genome.jp/kegg/) was used as a reference database to create a gene feature table from the 16S ribosomal RNA (rRNA) sequence data. A random subset of samples was used to accommodate the Piphillin analysis (n <100 samples required), resulting in 93 samples available for statistical analyses. KEGG pathways not expressed in bacteria were subsequently removed. All KEGG analyses were run stratified by secretor status (secretor, n = 67; non-secretor, n = 26). Statistically significant associations between HMOs and KEGG pathways were assessed in 2 ways: first, examining the association between HMOs and B-diversity of predicted KEGG orthologs (Bray-Curtis dissimilarity) using Adonis models in R adjusted for microbial DNA extraction and PCR sequencing batch effects (P < 0.05); second, individual HMOs associated with differentially expressed predicted functional KEGG pathways were assessed using MaAsLin2 in R [false discovery rate (FDR) adjusted, P < 0.1], while adjusting for microbial DNA extraction and PCR sequencing batch effects.
Results
Participant description
Human milk samples were expressed and collected at 3 ± 1 mo postpartum (mean ± SD). Maternal clinical data, including prepregnancy BMI, gestational glucose tolerance, mode of delivery, race, frequency of direct breastfeeding, and human milk exclusivity (vs. nonexclusive) are described in Table 1 according to secretor status. Mode of delivery was significantly associated with secretor status following chi-square analyses (P = 0.0032; Table 1); no other statistically significant differences were identified between maternal clinical data and secretor status.
TABLE 1.
Summary of maternal demographic, metabolic, and obstetric characteristics stratified by secretor status phenotype1
| Baseline characteristics | Cohort | Secretors | Non-secretors |
|---|---|---|---|
| Race | |||
| White | 60 (56) | 47 (60) | 13 (45) |
| Asian | 26 (24) | 16 (21) | 10 (34) |
| Other | 21 (20) | 15 (19) | 6 (21) |
| Prepregnancy BMI, kg/m2 | |||
| Mean ± SD | 24.4 ± 4.6 | 24.1 ± 4.5 | 25.4 ± 5.0 |
| Healthy (18.5–24.9) | 67 (63) | 51 (65) | 16 (55) |
| Overweight (25–29.9) | 29 (27) | 21 (27) | 8 (28) |
| Obese (>30) | 11 (10) | 6 (8) | 5 (17) |
| Gestational glucose tolerance status | |||
| Normoglycemia | 65 (61) | 50 (64) | 15 (52) |
| Impaired glucose tolerance | 18 (17) | 13 (17) | 5 (17) |
| Gestational diabetes | 24 (22) | 15 (19) | 9 (31) |
| Mode of delivery2 | |||
| Unscheduled cesarean delivery | 27 (25) | 13 (17) | 14 (48) |
| Scheduled cesarean delivery | 21 (20) | 18 (23) | 3 (10) |
| Vaginal | 59 (55) | 47 (60) | 12 (41) |
| Human milk exclusivity | |||
| Exclusive | 54 (50.5) | 39 (50) | 15 (52) |
| Nonexclusive | 53 (49.5) | 39 (50) | 14 (48) |
| Frequency of direct breastfeeding,3n/d | 7.0 (5.5, 8.5) | 7.0 (5.5, 8.0) | 7.0 (4.9, 8.1) |
Values are n (%) unless otherwise specified. Maternal race was categorized as white, Asian, or other (pooled Black, South Asian, other); the latter was combined due to low frequency of these races. Cohort n = 107; secretors, n = 78; non-secretors, n = 29.
A statistically significant association was observed between mode of delivery and secretor status phenotype following a chi-square test (P = 0.0032) of categorical data (vaginal vs. scheduled cesarean delivery vs. unscheduled cesarean delivery) (P < 0.05).
Frequency of direct breastfeeding is expressed as median (IQR).
Secretor status is a predictor of HMO composition at 3 mo postpartum
Among secretor mothers, the top 3 most abundant HMOs by concentration (milligrams per milliter) included 2’FL, DFLNT, and LNFPII (Figure 1; Supplemental Table 1). Among non-secretor mothers, the most abundant HMOs by concentration (milligrams per milliter) included LNFPII, FDSLNH, and LNT. Secretor mothers expressed significantly higher concentrations (milligrams per milliter) of total HMOs in their milk compared with non-secretor mothers [median (IQR): 11.4 (10.9, 12.0) vs. 8.13 (7.91, 8.22) mg/mL; P < 0.0001], mainly due to the higher quantities of 2’FL. Secretor mothers also expressed over double the concentration (nanomoles per milliliter) of HMO-bound fucose in their milk compared with non-secretors [1.5 × 104 (1.4 × 104, 1.6 × 104) vs. 5.7 × 103 (4.8 × 103, 6.5 × 103); P < 0.0001] (Figure 1; Supplemental Table 1).
FIGURE 1.
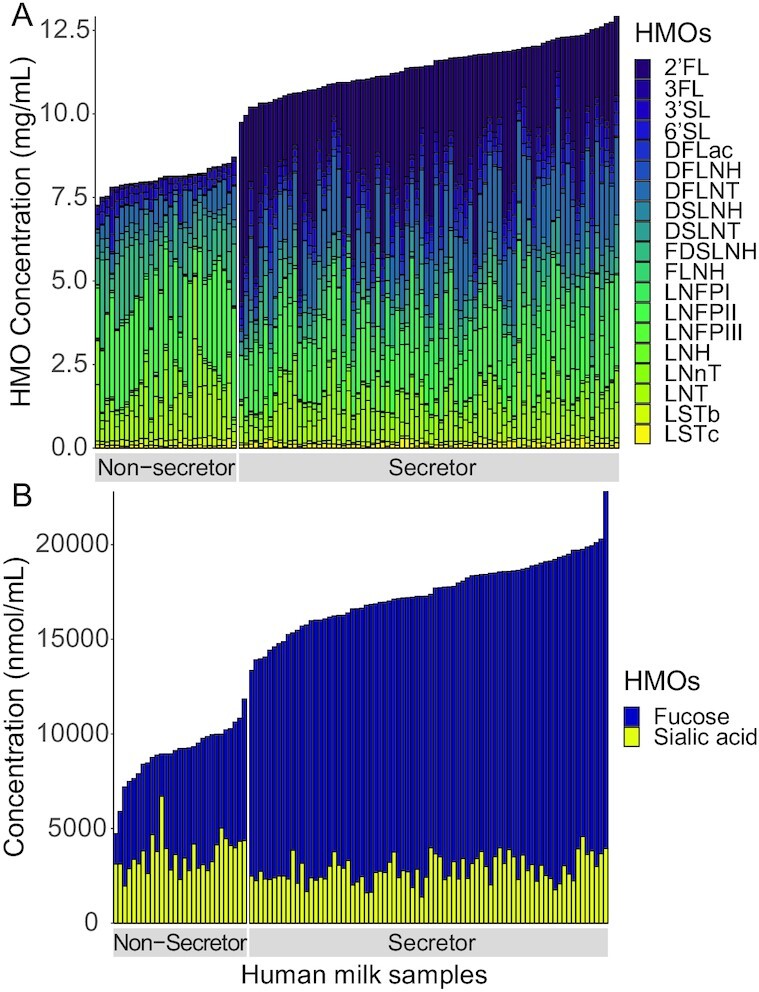
(A) HMO concentrations (mg/mL) and (B) HMO-bound sialic acid and fucose concentrations (nmol/mL) in mature human milk by secretor status phenotype (secretor, n = 78; non-secretor, n = 29). HMOs measured were 2′FL, 3FL, 3′SL, 6′SL, DFLac, DFLNH, DFLNT, DSLNH, DSLNT, FDSLNH, FLNH, LNFP I/II/III, LNH, LNnT, LNT, and LSTb/c. DFLac, difucosyllactose; DFLNH, difucosyllacto-N-hexaose; DFLNT, difucosyllacto-N-tetrose; DSLNH, disialyllacto-N-hexaose; DSLNT, disialyllacto-N-tetraose; FDSLNH, fucodisialyllacto-N-hexaose; FLNH, fucosyllacto-N-hexaose; HMO, human milk oligosaccharide; LNFP I/II/III, lacto-N-fucopentaose-I/II/III; LNH, lacto-N-hexaose; LNnT, lacto-N-neotetraose; LNT, lacto-N-tetrose; LSTb/c, sialyl-lacto-N-tetraose-b/c; 2′FL, 2′-fucosyllactose; 3FL, 3-fucosyllactose; 3′SL, 3′-sialyllactose; 6′SL, 6′-sialyllactose.
Maternal prepregnancy BMI, race, and frequency of direct breastfeeding are associated with the HMO profiles of human milk
Relations between maternal clinical data and HMO composition were examined separately for secretors and non-secretors. Within the secretor strata, prepregnancy BMI (kg/m2) was found to be positively associated with DFLac (P = 0.011), while higher concentrations of DSLNT were observed in the milk of Asian mothers compared with other races (P = 0.045) (Table 2). Negative dose-dependent relations were also observed for secretor mothers with frequency of direct breastfeeding (n/day) for LNFPI (P = 0.0034), LSTb (P = 0.043), DSLNT (P = 0.0088), and total HMO (P = 0.021) concentrations.
TABLE 2.
Significant associations between maternal characteristics and individual HMOs stratified by secretor status1
| HMOs and maternal characteristics | B-Estimate (SE) | P |
|---|---|---|
| Secretors | ||
| DFLac, mg/mL | ||
| Prepregnancy BMI (kg/m2) | 0.0099 (0.0038) | 0.011 |
| DSLNT, mg/mL | ||
| Race | 0.0312 | |
| White vs. other | −0.0076 (0.060) | 0.903 |
| Asian vs. other | 0.15 (0.073) | 0.0453 |
| LNFPI, mg/mL | ||
| Frequency of direct breastfeeding (n/d) | −0.072 (0.024) | 0.0034 |
| LSTb, mg/mL | ||
| Frequency of direct breastfeeding (n/d) | −0.0051 (0.0025) | 0.043 |
| DSLNT, mg/mL | ||
| Frequency of direct breastfeeding (n/d) | −0.025 (0.0091) | 0.0088 |
| Total HMOs, mg/mL | ||
| Frequency of direct breastfeeding (n/d) | −0.075 (0.032) | 0.021 |
| Non-secretors | ||
| FLNH, mg/mL | ||
| Prepregnancy BMI (kg/m2) | −0.0030 (0.0012) | 0.024 |
| LNT, mg/mL | ||
| Race | 0.0292 | |
| White vs. other | −0.67 (0.27) | 0.0213 |
| Asian vs. other | −0.65 (0.29) | 0.0323 |
| LNFPII, mg/mL | ||
| Race | 0.00222 | |
| White vs. other | 1.04 (0.28) | 0.00093 |
| Asian vs. other | 0.77 (0.29) | 0.0133 |
| DFLNT, mg/mL | ||
| Race | 0.00472 | |
| White vs. other | 0.43 (0.13) | 0.00253 |
| Asian vs. other | 0.36 (0.13) | 0.0133 |
| LNnT, mg/mL | ||
| Race | 0.00242 | |
| White vs. other | −0.46 (0.15) | 0.00443 |
| Asian vs. other | −0.52 (0.15) | 0.00243 |
| 6’SL, mg/mL | ||
| Race | 0.0412 | |
| White vs. other | −0.16 (0.072) | 0.0353 |
| Asian vs. other | −0.17 (0.075) | 0.0363 |
| DSLNT, mg/mL | ||
| Race | 0.0122 | |
| White vs. other | −0.35 (0.12) | 0.00793 |
| Asian vs. other | −0.32 (0.13) | 0.0203 |
| HMO-bound fucose, nmol/mL | ||
| Race | 0.00022 | |
| White vs. other | 2.4 × 103(5.4 × 102) | 0.00013 |
| Asian vs. other | 2.1 × 103 (5.7 × 102) | 0.00083 |
| FDSLNH, mg/mL | ||
| Frequency of direct breastfeeding (n/d) | 0.10 (0.040) | 0.015 |
| 3FL, mg/mL | ||
| Frequency of direct breastfeeding (n/d) | −0.014 (0.0067) | 0.049 |
| 3’SL, mg/mL | ||
| Frequency of direct breastfeeding (n/d) | −0.020 (0.0091) | 0.042 |
Secretors n = 78; non-secretors, n = 29. P values less than 0.05 were significant. DFLac, difucosyllactose; DFLNH, difucosyllacto-N-hexaose; DFLNT, difucosyllacto-N-tetrose; DSLNT, disialyllacto-N-tetraose; FDSLNH, fucodisialyllacto-N-hexaose; FLNH, fucosyllacto-N-hexaose; HMO, human milk oligosaccharide; LNFP I/II, lacto-N-fucopentaose-I/II; LNH, lacto-N-hexaose; LNnT, lacto-N-neotetraose; LNT, lacto-N-tetrose; LSTb, sialyl-lacto-N-tetraose-b; 3FL, 3-fucosyllactose; 3′SL, 3′-sialyllactose; 6′SL, 6′-sialyllactose.
Values are overall group-effect P value. Associations between maternal clinical data and HMOs stratified by secretor status were assessed via univariable linear regression models (PROC MIXED; SAS Institute). Maternal race was categorized as White, Asian, or other (pooled Black, South Asian, other); the latter was combined due to low frequency of these races.
Values are the pairwise comparison P-value.
Among non-secretors, race was a strong predictor of individual HMO concentrations (Table 2). Lower concentrations of LNT were observed among both White (P = 0.021) and Asian (P = 0.032) mothers; in addition, reduced concentrations of 6’SL, LNnT, and DSLNT were observed in White and Asian mothers compared with other races. Conversely, a higher concentration of LNFPII was observed among White (vs. other) mothers (P = 0.0009) and Asian (vs. other) mothers (P = 0.013), along with a greater concentration of HMO-bound fucose compared with other races (Table 2). Additionally, dose-dependent relations were again observed with frequency of direct breastfeeding among the non-secretor strata; lower concentrations of 3FL (P = 0.049) and increased concentrations of FDSLNH (P = 0.015) were observed with each additional time an infant was fed directly at the breast.
Individual HMOs are associated with microbial diversity in human milk
No statistically significant associations were found between secretor status as a variable and microbial ɑ-diversity as an outcome measure when examining the overall cohort (n = 107) [Chao1 index, B-estimate (SE): −12.7 (35.8), P = 0.72; Shannon index: 0.07 (0.12), P = 0.55]. However, within the secretor strata, 3’SL and FLNH were negatively associated with milk microbiota richness (Chao1 index; P = 0.049 and P = 0.0082, respectively) and DFLNH was negatively associated with milk microbiota diversity (Shannon index; P = 0.029) (Supplemental Figure 2; Supplemental Table 2). Among non-secretor samples, LNT was also negatively associated with milk microbiota richness (P = 0.025).
No statistically significant associations were found between secretor status as a variable and microbial B-diversity as an outcome measure when examining the overall cohort (n = 107; weighted UniFrac distance: R2 = 0.018, P = 0.11; Bray-Cutis dissimilarity: R2 = 0.013, P = 0.19). Among secretors, LNnT was associated with milk microbiota B-diversity (adjusted Weighted UniFrac distance: R2 = 0.040, P = 0.036) (Supplemental Figure 3, Supplemental Table 3); however, among the non-secretor strata, no individual HMOs were associated with milk microbiota B-diversity (Supplemental Table 4).
Individual HMOs are associated with microbial taxa in human milk
Milk microbiota composition stratified by secretor strata is outlined in Supplemental Table 5 and visualized in Supplemental Figure 4. Milk microbiota composition was found to be relatively consistent with minor statistically significant differences between the 2 secretor strata. When looking at the overall cohort, secretor status (secretor vs. non-secretor) was associated with a reduced incidence of Actinobacteria (P = 0.016) (Supplemental Table 6); however, no associations were observed between secretor status and predominant genera within the overall cohort.
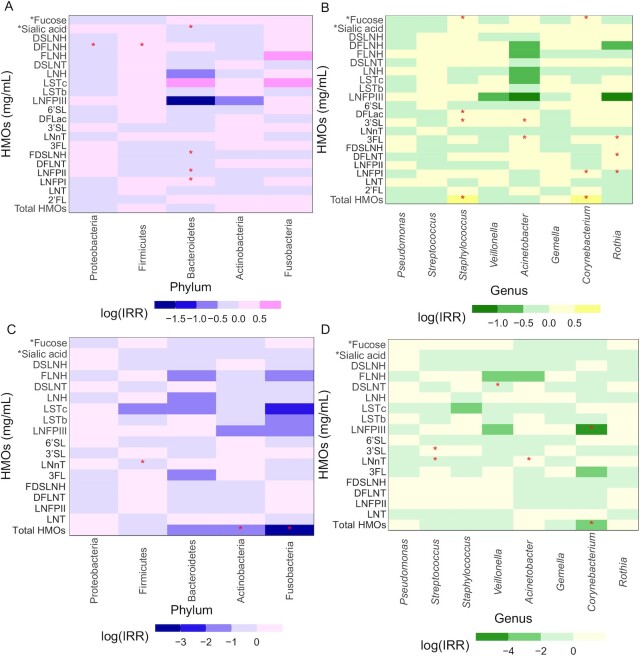
Within the secretor strata, DFLNH concentration was associated with a significantly reduced incidence of Proteobacteria (P = 0.013) and an increased incidence of Firmicutes (P = 0.017), while LNFPII (P = 0.001), FDSLNH (P = 0.0064), and HMO-bound sialic acid concentration (nanomoles per milliliter) (P = 0.019) were all associated with a significantly reduced incidence of Bacteroidetes (Figure 2, Supplemental Table 6). Total HMO concentration was associated with a significantly increased incidence of Staphylococcus (P = 0.0054) and Corynebacterium (P = 0.012). LNFPI was associated with a significantly reduced incidence of Rothia (P = 0.0057) (Figure 2, Supplemental Table 6).
FIGURE 2.
Differential abundance of the top 5 most abundant phyla and top 8 most abundant genera in human milk microbiota according to HMO composition by secretor strata. (A) Phylum secretor strata, (B) genus secretor strata, (C) phylum non-secretor strata, (D) genus non-secretor strata. Secretor, n = 78; non-secretor, n = 29. *Sialic acid and *Fucose denote that these HMO-bound molecules were run in the Poisson regression model as nmol/mL; all other HMOs were run as mg/mL, as indicated in the y-axis label. Heatmaps represent the results from multivariable Poisson regression models (PROC GENMOD; SAS Institute), adjusted for microbial DNA extraction and PCR sequencing batches with the top 5 phyla and top 8 genera as the outcome variables; the colored boxes refer to the incidence rate ratio on a log scale, while asterisks refer to statistically significant results. *P ≤ 0.022 for phylum, P ≤ 0.018 for genus. To account for multiple comparisons, the Benjamini-Yekutieli cut-point approach was used. DFLac, difucosyllactose; DFLNH, difucosyllacto-N-hexaose; DFLNT, difucosyllacto-N-tetrose; DSLNH, disialyllacto-N-hexaose; DSLNT, disialyllacto-N-tetraose; FDSLNH, fucodisialyllacto-N-hexaose; FLNH, fucosyllacto-N-hexaose; HMO, human milk oligosaccharide; LNFP I/II/III, lacto-N-fucopentaose-I/II/III; LNH, lacto-N-hexaose; LNnT, lacto-N-neotetraose; LNT, lacto-N-tetrose; LSTb/c, sialyl-lacto-N-tetraose-b/c; 2′FL, 2′-fucosyllactose; 3FL, 3-fucosyllactose; 3′SL, 3′-sialyllactose; 6′SL, 6′-sialyllactose.
Among the non-secretor strata, total HMO concentration was associated with a significantly reduced incidence of Actinobacteria (P = 0.0029), Fusobacteria (P = 0.002), and Corynebacterium (P < 0.0001) (Figure 2, Supplemental Table 6). 3’SL was associated with a significantly increased incidence of Streptococcus (P = 0.006), while LNnT was associated with a reduced incidence of Streptococcus (P = 0.010). LNnT was associated with a significantly increased incidence of Acinetobacter (P = 0.017), while DSLNT and LNFPIII were associated with reduced incidences of Veillonella (P = 0.010) and Corynebacterium (P = 0.014), respectively.
Individual HMOs are associated with the predicted functional capabilities of microbiota in human milk
A number of changes were observed in the abundance of predicted KEGG pathways in relation to individual HMOs. Among the secretor strata, DFLac was significantly associated with an enrichment of predicted pathways encoding for metabolism and infection, including lipoic acid metabolism, bacterial invasion of epithelial cells, and Staphylococcus aureus infection (P-FDR < 0.1) (Table 3). When examining the non-secretor strata, LNnT and DSLNH were associated with an enrichment in microbial pathways involved in the degradation of aromatic hydrocarbons and biosynthesis of antibiotics (P-FDR < 0.1).
TABLE 3.
Significant relations between individual HMOs and predicted microbial KEGG pathways in human milk stratified by secretor status1
| HMO and KEGG pathway | Coefficient (SD)2 | P 3 |
|---|---|---|
| Secretors | ||
| DFLac | ||
| Bacterial invasion of epithelial cells | 1.97 × 10^−5 (5.06 × 10^−6) | 0.056 |
| Staphylococcus aureus infection | 0.00041 (0.00012) | 0.095 |
| Lipoic acid metabolism | 4.66 × 10^−5 (1.18 × 10^−5) | 0.056 |
| Pyruvate metabolism | 0.00018 (4.71 × 10^−5) | 0.059 |
| Secondary bile acid biosynthesis | 1.31 × 10^−5 (3.55 × 10^−6) | 0.065 |
| Tuberculosis | −3.31 × 10^−5 (8.32 × 10^−6) | 0.056 |
| Biosynthesis of secondary metabolites | 0.00056 (0.00016) | 0.077 |
| Biosynthesis of vancomycin group antibiotics | −1.34 × 10^−5 (3.83 × 10^−6) | 0.077 |
| Carbon fixation pathways in prokaryotes | 0.00014 (3.89 × 10^−5) | 0.077 |
| Vibrio cholerae infection | 4.52 × 10^−6 (1.31 × 10^−6) | 0.077 |
| Carbon metabolism | 0.00025 (7.73 × 10^−5) | 0.095 |
| d-Arginine and d-ornithine metabolism | 1.43 × 10^−5 (4.29 × 10^−6) | 0.095 |
| Non-secretors | ||
| LNnT | ||
| Toluene degradation | 0.00017 (3.51 × 10^−5) | 0.015 |
| Chlorocyclohexane and chlorobenzene degradation | 0.00012 (3.06 × 10^−5) | 0.094 |
| DSLNH | ||
| Biosynthesis of enediyne antibiotics | 9.38 × 10^−7 (1.93 × 10^−7) | 0.019 |
Associations between individual HMOs and secretor status stratified differentially expressed predicted functional KEGG pathways were assessed using MaAsLin2. All models were adjusted for microbial DNA extraction batch and PCR sequencing batch. Secretors, n = 67; non-secretors, n = 26. DFLac, difucosyllactose; DSLNH, disialyllacto-N-hexaose, HMO, human milk oligosaccharide; KEGG, Kyoto Encyclopedia of Genes and Genomes; LNnT, lacto-N-neotetraose.
Values are shown as coefficient (SD) from MaAsLin2 models that were adjusted for microbial DNA extraction and PCR batch effects.
P values are false discovery rate (FDR) adjusted. A cutoff of PFDR < 0.1 was deemed statistically significant.
In addition to KEGG pathways, associations between HMOs and KEGG ortholog (KO) B-diversity (Bray-Curtis dissimilarity) were also explored. Similar to our findings with microbial B-diversity, LNnT was significantly associated with KO B-diversity among secretors (R2 = 0.039, P = 0.043) (Supplemental Table 7). Within the non-secretor strata, DFLNT was also found to be significantly associated with KO B-diversity in adjusted models (R2 = 0.14, P = 0.007) (Supplemental Table 8).
Discussion
Maternal glucose intolerance during pregnancy increases a mother's risk of developing type 2 diabetes post-pregnancy as well as increases the risk of morbidities in her neonate, such as hypoglycemia (37–40). Little is known about the role of perturbed glucose metabolism on human milk components; however, our results are the first to show that 1) not GDM but prepregnancy BMI, race, frequency of direct breastfeeding (n/day), and secretor status are associated with specific HMOs in mature human milk and 2) that HMOs are associated with microbial diversity, abundance of predominant taxa, and predicted microbial functions, regardless of glucose tolerance status.
Of the maternal characteristics we examined, race (White, Asian, other) appeared to be the most consistent determinant of HMO composition in our cohort, and this association has also been reported by others (13). Although there does appear to be a relation between race and FUT2 status, associations appear to differ geographically, perhaps reflecting different single nucleotide polymorphisms in the FUT2 gene (41–43). We also found maternal prepregnancy BMI to be positively associated with DFLac, an HMO that has previously been shown to be positively correlated with increased weight gain, weight velocity, and length gain in infants at 5 mo postpartum (44). Despite associations with prepregnancy BMI, it is interesting that no associations were observed between glucose tolerance status and HMO composition, given the potential for altered glucose tolerance to modify glycosylation patterns of human milk glycoproteins (16, 45). Interestingly, the 1 study to date (n = 24, milk collected 2 wk postpartum, United States) to examine associations between GDM and total HMO abundances also observed no differences in total HMO concentrations in milk from mothers with or without GDM; however, the authors did not examine individual HMOs (16). Recently, third-trimester glucose homeostasis in normoglycemic women (n = 136, United States) was found to be associated with secretor status, with non-secretors displaying significantly greater fasting insulin and HOMA-IR values than secretors; however, these values were still within a healthy range for pregnant women in their third trimester (17). The non-secretor group was also shown to have a significantly higher BMI than secretors. It is unknown why these different associations were observed compared with our study findings, but it is important to emphasize that the women in the study by Saben et al. (17) were metabolically healthy and did not meet the diagnostic criteria for either gestational IGT or GDM. Finally, Azad et al. (13) also found associations between frequency of direct breastfeeding and HMO composition, hypothesizing that the regularity of milk removal and the volume of milk production both have the potential to alter the quantities and types of HMOs in human milk (13, 46, 47). This hypothesis continues to hold true for our study, as we also found a number of associations with this variable, regardless of secretor strata, indicating that the frequency of direct breastfeeding is a probable modulator of HMO composition.
Minor variations in microbiota composition were observed based on secretor status, a finding that is in line with previous investigations (48, 49); however, we did identify differences in milk microbiota diversity according to specific HMOs. FLNH was associated with milk microbiota richness as previously reported by Moossavi et al. (22), although our directionality was opposite. LNnT was also found to be associated with human milk microbiota B-diversity, although the magnitude was quite small; however, this finding may suggest that the microbial communities as a whole may differ depending on the HMO concentrations that a mother expresses in her milk. Differences in the B-diversity of the overall composition of predicted KEGG orthologs were also observed with LNnT. LNnT has been previously studied for its role in infant nutrition, especially as a supplemental addition to infant formula, as well as its inverse association with weight and height in children at 5 y of age (50, 51). For this reason, this observed association between LNnT and human milk microbiota B-diversity at both the taxonomic and predicted functional levels requires further investigation due to its potential implications in infant and child health.
Overall, the associations between HMOs and differentially abundant taxa in the milk microbiota were modest. Previous research has found total HMO content of human milk to be positively associated with Staphylococcus relative abundance (21). This supports our findings showing a positive association between overall HMO concentration and Staphylococcus genus relative abundance among secretor mothers. Intriguingly, a previous in vitro analysis reported that HMOs can stimulate the growth of Staphylococcus in a variety of media (21). Future directions validating this mechanism in vitro are warranted to determine whether HMOs are able to stimulate the growth of Staphylococcus.
No other study to our knowledge has investigated beyond taxa associations with HMOs and evaluated how predicted microbial functions differ depending on HMO composition. DFLac was found within the secretor strata to be associated with a number of predicted pathways encoding for bacterial invasion of epithelial cells and S. aureus infection. Fascinatingly, previous work examining secretor status and the risk of infection has shown a differing susceptibility for risk of a number of infections (i.e., mumps, measles, and norovirus), with either an increased or decreased risk based on secretor genotype and the illness in question (10, 52, 53). S. aureus is one of the causative agents of mastitis, and interestingly, previous work has shown an increased abundance of infection-related genes in the milk microbiota of women with a confirmed diagnosis of mastitis (54, 55). Although we were unable to assess if secretor status was associated with the risk of mastitis in our cohort, assessing this relation in future studies may be warranted given that the HMO glycosylation pattern may directly impact the development of mastitis.
The present study had many strengths, including a metabolically heterogenous population, along with secretor phenotype stratification to help us better understand HMO composition. Our cohort was also enriched with mothers diagnosed with altered gestational glucose tolerance, allowing us to investigate the role (or lack thereof) this metabolic variant plays in HMO composition. We recognize there are also limitations to the present study. First, we were unable to assess for the FUT3 genotype in our cohort, which would have provided further resolution into the genetic components of HMO composition by classifying mothers based on Lewis blood group, which is not as straightforward to determine based on HMO composition alone. Future research integrating the genetic heterozygosity and classifications of all 4 potential milk subgroups of FUT2/FUT3 would provide a deeper understanding of how maternal demographic, metabolic, and obstetrical factors are associated with HMO composition, when accounting for known genetic factors. Second, although a large proportion of our cohort had gestational IGT (∼40%), there is the potential that this sample size of women, especially when stratified by secretor status, was inadequate to detect some differences for some comparisons. Third, our use of 16S rRNA gene sequencing data to infer microbial functions using Piphillin may not reflect the true microbiota function and requires confirmation with shotgun metagenomic sequencing. In addition, our milk samples were stored at –80°C for approximately 6 y before 16S rRNA gene sequencing was conducted; although the literature suggests this does not result in a substantial change to the microbiota or HMO compositions, this is a potential limitation of our study (56, 57). Last, our study was cross-sectional, meaning we were not able to investigate longitudinal changes in HMOs over time, or how their relation with maternal clinical variables or microbiota may change over the postpartum period.
Our study is the first to analyze the interplay between HMOs and the human milk microbiota composition in a cohort of women with varying degrees of gestational glucose intolerance, a metabolic profile that deserves greater attention given its potential association with altered glycosylation patterns of HMOs. Our microbial predicted function results may point to the need to investigate risk of infections, such as mastitis, during lactation, and to determine whether secretor status may confer protection from risk of additional postpartum morbidities in both the mother and infant.
Supplementary Material
Acknowledgments
The authors’ responsibilities were as follows—SHL, AJH, and DLO: designed the original study; LL-N, CY, SHL, JKC, and PWW: conducted the research; LL-N, CY, and JKC: analyzed data; MRA and AK: were involved in performing statistical analyses; LL-N: wrote the initial draft of the manuscript; DLO: had primary responsibility for final content; and all authors including LB, SU, JB, and AS: participated in writing and critical revision of the manuscript for important intellectual content and all authors read and approved the final manuscript.
Notes
Supported by the Canadian Institutes of Health Research (MOP#125997) and a Canadian Diabetes Association Operating Grant (#OG-3-09-2393). SHL was supported by grant P20GM109036 from the National Institute of General Medical Sciences of the National Institutes of Health. LL-N is a recipient of a Canadian Institutes of Health Research Doctoral Award (The Frederick Banting and Charles Best Canada Graduate Scholarship Doctoral Award).
Author disclosures: AS is a co-founder of MedBiome, a clinical microbiomics company. DLO is a member of the Journal of Nutrition’s Editorial Board. The authors report no conflicts of interest. The sources of support had no role in the research study design or conduct, sample preparation, data or statistical analysis, interpretation, or writing of the manuscript.
Supplemental Figures 1–4 and Supplemental Tables 1–8 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents available on https://academic.oup.com/jn.
Abbreviations used: DFLac, difucosyllactose; DFLNH, difucosyllacto-N-hexaose; DFLNT, difucosyllacto-N-tetrose; DSLNH, disialyllacto-N-hexaose; DSLNT, disialyllacto-N-tetraose; FDR, false discovery rate; FDSLNH, fucodisialyllacto-N-hexaose; FLNH, fucosyllacto-N-hexaose; FUT2, fucosyltransferase-2; FUT3, fucosyltransferase-3; GDM, gestational diabetes mellitus; GI, gastrointestinal; HMO, human milk oligosaccharide; IGT, impaired glucose tolerance; KEGG, Kyoto Encyclopedia of Genes and Genomes; KO, KEGG ortholog; LNFP, lacto-N-fucopentaose; LNnT, lacto-N-neotetraose; LNT, lacto-N-tetrose; LSTb/c, sialyl-lacto-N-tetraose-b/c; OGTT, oral-glucose-tolerance test; OTU, operational taxonomic unit; rRNA, ribosomal RNA; 2′FL, 2′-fucosyllactose; 3FL, 3-fucosyllactose; 3′SL, 3′-sialyllactose; 6′SL, 6′-sialyllactose.
Contributor Information
Lauren LeMay-Nedjelski, Department of Nutritional Sciences, University of Toronto, Toronto, Ontario, Canada; Translational Medicine Program, The Hospital for Sick Children, Toronto, Ontario, Canada.
Chloe Yonemitsu, Department of Pediatrics and Mother-Milk-Infant Center of Research Excellence (MOMI CORE), University of California, San Diego, La Jolla, CA, USA.
Michelle R Asbury, Department of Nutritional Sciences, University of Toronto, Toronto, Ontario, Canada; Translational Medicine Program, The Hospital for Sick Children, Toronto, Ontario, Canada.
James Butcher, Ottawa Institute of Systems Biology and Department of Biochemistry, Microbiology, and Immunology, University of Ottawa, Ottawa, Ontario, Canada.
Sylvia H Ley, Department of Epidemiology, Tulane University School of Public Health and Tropical Medicine, New Orleans, LA, USA.
Anthony J Hanley, Department of Nutritional Sciences, University of Toronto, Toronto, Ontario, Canada.
Alex Kiss, Department of Research Design and Biostatistics, Sunnybrook Research Institute, Toronto, Ontario, Canada.
Sharon Unger, Department of Nutritional Sciences, University of Toronto, Toronto, Ontario, Canada; Translational Medicine Program, The Hospital for Sick Children, Toronto, Ontario, Canada; Department of Pediatrics, University of Toronto, Toronto, Ontario, Canada; Department of Pediatrics, Sinai Health, Toronto, Ontario, Canada; Lunenfeld-Tanenbaum Research Institute, Sinai Health, Toronto, Ontario, Canada.
Julia K Copeland, Centre for the Analysis of Genome Evolution and Function, University of Toronto, Ontario, Canada.
Pauline W Wang, Centre for the Analysis of Genome Evolution and Function, University of Toronto, Ontario, Canada.
Alain Stintzi, Ottawa Institute of Systems Biology and Department of Biochemistry, Microbiology, and Immunology, University of Ottawa, Ottawa, Ontario, Canada.
Lars Bode, Department of Pediatrics and Mother-Milk-Infant Center of Research Excellence (MOMI CORE), University of California, San Diego, La Jolla, CA, USA.
Deborah L O'Connor, Department of Nutritional Sciences, University of Toronto, Toronto, Ontario, Canada; Translational Medicine Program, The Hospital for Sick Children, Toronto, Ontario, Canada.
References
- 1. Togo A, Dufour J-C, Lagier J-C, Dubourg G, Raoult D, Million M. Repertoire of human breast and milk microbiota: a systematic review. Future Microbiol. 2019;14:623–41. [DOI] [PubMed] [Google Scholar]
- 2. Ruiz L, García-Carral C, Rodriguez JM. Unfolding the human milk microbiome landscape in the omics era. Front Microbiol. 2019;10:1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Garwolińska D, Namieśnik J, Kot-Wasik A, Hewelt-Belka W. Chemistry of human breast milk-a comprehensive review of the composition and role of milk metabolites in child development. J Agric Food Chem. 2018;66:11881–96. [DOI] [PubMed] [Google Scholar]
- 4. Moossavi S, Sepehri S, Robertson B, Bode L, Goruk S, Field CJ, Lix LM, de Souza RJ, Becker AB, Mandhane PJet al. . Composition and variation of the human milk microbiota are influenced by maternal and early-life factors. Cell Host Microbe. 2019;25:324–35, e4. [DOI] [PubMed] [Google Scholar]
- 5. Padilha M, Danneskiold-Samsøe NB, Brejnrod A, Hoffmann C, Cabral VP, Iaucci J de M, Sales CH, Fisberg RM, Cortez RV, Brix Set al. . The human milk microbiota is modulated by maternal diet. Microorganisms. 2019;7(11):502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Williams JE, Carrothers JM, Lackey KA, Beatty NF, York MA, Brooker SL, Shafii B, Price WJ, Settles ML, McGuire MAet al. . Human milk microbial community structure is relatively stable and related to variations in macronutrient and micronutrient intakes in healthy lactating women. J Nutr. 2017;147:1739–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. LeMay-Nedjelski L, Butcher J, Ley SH, Asbury MR, Hanley AJ, Kiss A, Unger S, Copeland JK, Wang PW, Zinman Bet al. . Examining the relationship between maternal body size, gestational glucose tolerance status, mode of delivery and ethnicity on human milk microbiota at three months post-partum. BMC Microbiol. 2020;20:219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. LeMay-Nedjelski L, Asbury MR, Butcher J, Ley SH, Hanley AJ, Kiss A, Unger S, Copeland JK, Wang PW, Stintzi Aet al. . Maternal diet and infant feeding practices are associated with variation in the human milk microbiota at 3 months postpartum in a cohort of women with high rates of gestational glucose intolerance. J Nutr. 2021;151(2):320–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bode L. The functional biology of human milk oligosaccharides. Early Hum Dev. 2015;91:619–22. [DOI] [PubMed] [Google Scholar]
- 10. Azad MB, Wade KH, Timpson NJ. FUT2 secretor genotype and susceptibility to infections and chronic conditions in the ALSPAC cohort. Wellcome Open Res. 2018;3:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Smith-Brown P, Morrison M, Krause L, Davies PSW. Mothers secretor status affects development of childrens microbiota composition and function: a pilot study. PLoS One. 2016;11:e0161211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McGuire MK, Meehan CL, McGuire MA, Williams JE, Foster J, Sellen DW, Kamau-Mbuthia EW, Kamundia EW, Mbugua S, Moore SEet al. . What's normal? Oligosaccharide concentrations and profiles in milk produced by healthy women vary geographically. Am J Clin Nutr. 2017;105:1086–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Azad MB, Robertson B, Atakora F, Becker AB, Subbarao P, Moraes TJ, Mandhane PJ, Turvey SE, Lefebvre DL, Sears MRet al. . Human milk oligosaccharide concentrations are associated with multiple fixed and modifiable maternal characteristics, environmental factors, and feeding practices. J Nutr. 2018;148:1733–42. [DOI] [PubMed] [Google Scholar]
- 14. Samuel TM, Binia A, de Castro CA, Thakkar SK, Billeaud C, Agosti M, Al-Jashi I, Costeira MJ, Marchini G, Martínez-Costa Cet al. . Impact of maternal characteristics on human milk oligosaccharide composition over the first 4 months of lactation in a cohort of healthy European mothers. Sci Rep. 2019;9:11767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jantscher-Krenn E, Treichler C, Brandl W, Schönbacher L, Köfeler H, van Poppel MNM. The association of human milk oligosaccharides with glucose metabolism in overweight and obese pregnant women. Am J Clin Nutr. 2019;110:1335–43. [DOI] [PubMed] [Google Scholar]
- 16. Smilowitz JT, Totten SM, Huang J, Grapov D, Durham HA, Lammi-Keefe CJ, Lebrilla C, German JB. Human milk secretory immunoglobulin a and lactoferrin N-glycans are altered in women with gestational diabetes mellitus. J Nutr. 2013;143:1906–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Saben JL, Abraham A, Bode L, Sims CR, Andres A. Third-trimester glucose homeostasis in healthy women is differentially associated with human milk oligosaccharide composition at 2 months postpartum by secretor phenotype. Nutrients. 2020;12(8):2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kirmiz N, Robinson RC, Shah IM, Barile D, Mills DA. Milk glycans and their interaction with the infant-gut microbiota. Annu Rev Food Sci Technol. 2018;9:429–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Korpela K, Salonen A, Hickman B, Kunz C, Sprenger N, Kukkonen K, Savilahti E, Kuitunen M, Vos WM de. Fucosylated oligosaccharides in mother's milk alleviate the effects of caesarean birth on infant gut microbiota. Sci Rep. 2018;8:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Asakuma S, Hatakeyama E, Urashima T, Yoshida E, Katayama T, Yamamoto K, Kumagai H, Ashida H, Hirose J, Kitaoka M. Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J Biol Chem. 2011;286:34583–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hunt KM, Preuss J, Nissan C, Davlin CA, Williams JE, Shafii B, Richardson AD, McGuire MK, Bode L, McGuire MA. Human milk oligosaccharides promote the growth of staphylococci. Appl Environ Microbiol. 2012;78:4763–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moossavi S, Atakora F, Miliku K, Sepehri S, Robertson B, Duan QL, Becker AB, Mandhane PJ, Turvey SE, Moraes TJet al. . Integrated analysis of human milk microbiota with oligosaccharides and fatty acids in the CHILD cohort. Front Nutr. 2019;6:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cabrera-Rubio R, Kunz C, Rudloff S, García-Mantrana I, Crehuá-Gaudiza E, Martínez-Costa C, Collado MC. Association of maternal secretor status and human milk oligosaccharides with milk microbiota: an observational pilot study. J Pediatr Gastroenterol Nutr. 2019;68:256–63. [DOI] [PubMed] [Google Scholar]
- 24. Williams JE, Price WJ, Shafii B, Yahvah KM, Bode L, McGuire MA, McGuire MK. Relationships among microbial communities, maternal cells, oligosaccharides, and macronutrients in human milk. J Hum Lact. 2017;33:540–51. [DOI] [PubMed] [Google Scholar]
- 25. Ayoub Moubareck C, Lootah M, Tahlak M, Venema K. Profiles of human milk oligosaccharides and their relations to the milk microbiota of breastfeeding mothers in Dubai. Nutrients. 2021;13(4):1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ley SH, O'Connor DL, Retnakaran R, Hamilton JK, Sermer M, Zinman B, Hanley AJ. Impact of maternal metabolic abnormalities in pregnancy on human milk and subsequent infant metabolic development: methodology and design. BMC Public Health. 2010;10:590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ley SH, Hanley AJ, Sermer M, Zinman B, O'Connor DL. Associations of prenatal metabolic abnormalities with insulin and adiponectin concentrations in human milk. Am J Clin Nutr. 2012;95:867–74. [DOI] [PubMed] [Google Scholar]
- 28. Autran CA, Kellman BP, Kim JH, Asztalos E, Blood AB, Spence ECH, Patel AL, Hou J, Lewis NE, Bode L. Human milk oligosaccharide composition predicts risk of necrotising enterocolitis in preterm infants. Gut. 2018;67:1064–70. [DOI] [PubMed] [Google Scholar]
- 29. LeMay-Nedjelski L, Copeland J, Wang PW, Butcher J, Unger S, Stintzi A, O'Connor DL. Methods and strategies to examine the human breastmilk microbiome. Methods Mol Biol Clifton NJ. 2018;1849:63–86. [DOI] [PubMed] [Google Scholar]
- 30. Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O'Hara RB, Simpson GL, Solymos Pet al. . vegan: Community Ecology Package [Internet]. 2019. [Accessed 2021 Feb 20]. Available from: https://CRAN.R-project.org/package=vegan. [Google Scholar]
- 31. Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer Met al. . Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang Q, Garrity GM, Tiedje JM, Cole JR. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Price LB, Liu CM, Melendez JH, Frankel YM, Engelthaler D, Aziz M, Bowers J, Rattray R, Ravel J, Kingsley Cet al. . Community analysis of chronic wound bacteria using 16S rRNA gene-based pyrosequencing: impact of diabetes and antibiotics on chronic wound microbiota. PLoS One. 2009;4(7):e6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McMurdie PJ, Holmes S. Phyloseq: a bioconductor package for handling and analysis of high-throughput phylogenetic sequence data. Pac Symp Biocomput. 2012;235–46. [PMC free article] [PubMed] [Google Scholar]
- 35. Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001;29:1165–88. [Google Scholar]
- 36. Iwai S, Weinmaier T, Schmidt BL, Albertson DG, Poloso NJ, Dabbagh K, DeSantis TZ. Piphillin: improved prediction of metagenomic content by direct inference from human microbiomes. PLoS One. 2016;11:e0166104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Plows JF, Stanley JL, Baker PN, Reynolds CM, Vickers MH. The pathophysiology of gestational diabetes mellitus. Int J Mol Sci. 2018;19(11):3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Spaight C, Gross J, Horsch A, Puder JJ. Gestational diabetes mellitus. In: Stettler C, Christ E, Diem P editors. Endocrine development [Internet]. S. Karger AG; 2016. p. 163–78. Available from: https://www.karger.com/Article/FullText/439413. [DOI] [PubMed] [Google Scholar]
- 39. Farrar D, Simmonds M, Bryant M, Sheldon TA, Tuffnell D, Golder S, Lawlor DA. Treatments for gestational diabetes: a systematic review and meta-analysis. BMJ Open. 2017;7(6):e015557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yamamoto JM, Kellett JE, Balsells M, García-Patterson A, Hadar E, Solà I, Gich I, van der Beek EM, Castañeda-Gutiérrez E, Heinonen Set al. . Gestational diabetes mellitus and diet: a systematic review and meta-analysis of randomized controlled trials examining the impact of modified dietary interventions on maternal glucose control and neonatal birth weight. Diabetes Care. 2018;41:1346–61. [DOI] [PubMed] [Google Scholar]
- 41. Payne DC, Currier RL, Staat MA, Sahni LC, Selvarangan R, Halasa NB, Englund JA, Weinberg GA, Boom JA, Szilagyi PGet al. . Epidemiologic association between FUT2 secretor status and severe rotavirus gastroenteritis in children in the United States. JAMA Pediatr. 2015;169:1040–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ferrer-Admetlla A, Sikora M, Laayouni H, Esteve A, Roubinet F, Blancher A, Calafell F, Bertranpetit J, Casals F. A natural history of FUT2 polymorphism in humans. Mol Biol Evol. 2009;26:1993–2003. [DOI] [PubMed] [Google Scholar]
- 43. Soejima M, Koda Y. Distinct single nucleotide polymorphism pattern at the FUT2 promoter among human populations. Ann Hematol. 2008;87:19–25. [DOI] [PubMed] [Google Scholar]
- 44. Larsson MW, Lind MV, Laursen RP, Yonemitsu C, Larnkjær A, Mølgaard C, Michaelsen KF, Bode L. Human milk oligosaccharide composition is associated with excessive weight gain during exclusive breastfeeding—an explorative study. Front Pediatr Frontiers; 2019;7:297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rudman N, Gornik O, Lauc G. Altered N-glycosylation profiles as potential biomarkers and drug targets in diabetes. FEBS Lett. 2019;593(13):1–615. [DOI] [PubMed] [Google Scholar]
- 46. Ballard O, Morrow AL. Human milk composition: nutrients and bioactive factors. Pediatr Clin North Am. 2013;60:49–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stelwagen K, Singh K. The role of tight junctions in mammary gland function. J Mammary Gland Biol Neoplasia. 2014;19:131–8. [DOI] [PubMed] [Google Scholar]
- 48. Moossavi S, Azad MB. Origins of human milk microbiota: new evidence and arising questions. Gut Microbes. 2019;0:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cabrera-Rubio R, Collado MC, Laitinen K, Salminen S, Isolauri E, Mira A. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am J Clin Nutr. 2012;96:544–51. [DOI] [PubMed] [Google Scholar]
- 50. Hegar B, Wibowo Y, Basrowi RW, Ranuh RG, Sudarmo SM, Munasir Z, Atthiyah AF, Widodo AD, Supriatmo, Kadim Met al. . The role of two human milk oligosaccharides, 2′-fucosyllactose and lacto-n-neotetraose, in infant nutrition. Pediatr Gastroenterol Hepatol Nutr. 2019;22:330–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lagström H, Rautava S, Ollila H, Kaljonen A, Turta O, Mäkelä J, Yonemitsu C, Gupta J, Bode L. Associations between human milk oligosaccharides and growth in infancy and early childhood. Am J Clin Nutr. 2020;11(4):769–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nordgren J, Svensson L. Genetic susceptibility to human norovirus infection: an update. Viruses. 2019;11:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sharma S, Hagbom M, Carlsson B, Nederby Öhd J, Insulander M, Eriksson R, Simonsson M, Widerström M, Nordgren J. Secretor status is associated with susceptibility to disease in a large GII.6 norovirus foodborne outbreak. Food Environ Virol. 2020;12:28–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Patel SH, Vaidya YH, Patel RJ, Pandit RJ, Joshi CG, Kunjadiya AP. Culture independent assessment of human milk microbial community in lactational mastitis. Sci Rep. 2017;7:7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jiménez E, de Andrés J, Manrique M, Pareja-Tobes P, Tobes R, Martínez-Blanch JF, Codoñer FM, Ramón D, Fernández L, Rodríguez JM. Metagenomic analysis of milk of healthy and mastitis-suffering women. J Hum Lact. 2015;31:406–15. [DOI] [PubMed] [Google Scholar]
- 56. Tap J, Cools-Portier S, Pavan S, Druesne A, Öhman L, Törnblom H, Simren M, Derrien M. Effects of the long-term storage of human fecal microbiota samples collected in RNAlater. Sci Rep. 2019;9:601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lyons KE, Fouhy F, Shea C-AO, Ryan CA, Dempsey EM, Ross RP, Stanton C. Effect of storage, temperature, and extraction kit on the phylogenetic composition detected in the human milk microbiota. MicrobiologyOpen. 2021;10:e1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.