Abstract
Objectives:
This study aimed to determine the effect of Enterococcus faecalis on the cell growth and hyphal formation of Candida albicans and to understand the exact mechanism of candidal inhibition by the existence of E. faecalis by metabolomic analysis.
Materials and Methods:
Single- and dual-species biofilms of E. faecalis and C. albicans were formed in a microtiter plate, and the metabolomic profiles of both biofilms was determined by gas chromatography-mass spectrometry. The hyphal cell growth of C. albicans after treatment with both the supernatant and biofilm cells of E. faecalis was examined microscopically. The expression levels of Efg1 and the images of C. albicans cell wall in single- and dual-species biofilms were determined by real-time quantitative polymerase chain reaction and transmission electron microscopy, respectively. The violacein levels produced by Chromobacterium violaceum were measured to determine the quorum sensing (QS) inhibitory activity of single- and dual-species biofilms.
Results:
The biofilm cell growth, Efg1 expression, and hyphal development of C. albicans were inhibited by E. faecalis. Compared to single-species biofilms, alterations in carbohydrate, amino acid, and polyamine metabolites were observed in the dual-species biofilm for both microorganisms. Putrescine and pipecolic acid were detected at high levels in dual-species biofilm. A thicker β-glucan chitin and a denser and narrower fibrillar mannan layer of C. albicans cell wall were observed in dual-species biofilm. QS inhibitory activity was higher in dual-species biofilm suspensions of E. faecalis and C. albicans than in their single-species biofilms.
Conclusion:
E. faecalis inhibited the hyphal development and biofilm formation of C. albicans. Biofilm suspensions of C. albicans and E. faecalis showed an anti-QS activity, which increased even further in the environment where the two species coexisted. Investigation of putrescine and pipecolic acid can be an important step to understand the inhibition of C. albicans by bacteria.
Keywords: Dual-biofilm, Candida albicans, Enterococcus faecalis, fungal inhibition, metabolomic
INTRODUCTION
Biofilms formed in non-sterile mucosal sites are polymicrobial, and interspecies interactions in biofilms vary. They can interact either in a synergistic or antagonistic manner.1,2,3,4 Candida albicans and Enterococcus faecalis are frequently found together in biofilm-related infections.5,6,7 They have common features such as strong biofilm-forming capability that complicates the treatment of chronic infections, especially infections associated with foreign bodies.8,9
Microbial metabolomics has attracted great attention in microbiology in recent years.10,11 To better understand the biofilm structure of microorganisms, metabolic differences between planktonic and biofilm forms of the same microorganism have been investigated, but the results of the polymicrobial biofilm environment containing multiple species have not been reported in the literature yet.6
Studies concerning the details of the relationship between C. albicans and E. faecalis are limited.4 Thus, this study aimed to investigate the interactions at the metabolic level in the dual-species biofilm model formed by E. faecalis and C. albicans. The metabolic profile that both cells exhibit alone and in a common biofilm environment were compared by gas chromatography-mass spectrometry (GC-MS)-based metabolic analysis. Besides metabolomics analysis, the effects of each other were also investigated by several analyses including microscopy, quorum sensing (QS), and mRNA expression.
MATERIALS AND METHODS
Microbial strains
E. faecalis ATCC 47077/OG1RF and C. albicans ATCC MYA-2876 were cultured in brain heart infusion broth (BHI) (Oxoid, Basingstoke, UK) overnight at 37°C. Chromobacterium violaceum ATCC 12472 was grown in Luria Bertani broth (Merck, Darmstadt, Germany).
Effect of E. facealis on C. albicans hyphal morphogenesis
C. albicans were cultured in Yeast extract-peptone-dextrose broth (Merck, Darmstadt, Germany) at 30°C for 24 h. The inoculum suspension of the cell pellet was prepared in Roswell Park Memorial Institute Medium (RPMI) as 105 cfu/mL. After the addition of 1 mL of the inoculum to the wells of cell culture slides, which were coated with 20% fetal bovine serum (FBS), they were incubated for 90 min at 30°C. After the incubation period, the wells were rinsed with phosphate-buffered saline (PBS); then, RPMI containing 20% FBS and E. faecalis supernatant at a ratio of 1:1 (v/v) were transferred into wells.
To evaluate the direct effect of E. faecalis cells on hyphal cells, 50 µL of E. faecalis suspension was transferred to C. albicans, which had previously adhered to slides via incubation for 90 min. Finally, 950 µL of Spider medium containing 20% FBS was transferred onto slides and incubated at 37°C for 24 h.4
To assess the effect of E. faecalis supernatant on the development of C. albicans hyphal cells, the supernatant of E. faecalis was used instead of its cell suspension in the same method above. Slides containing biofilms were rinsed with PBS, and microscopic images were acquired using an inverted microscope (Thermo Scientific, MA, USA).
Development of single- and dual-species biofilm models
Inoculum suspensions with final concentrations of ~106 cfu/mL for E. faecalis and 105 cfu/mL for C. albicans were made in BHI. Mature biofilms were formed as described previously.12 Our experimental conditions include the biofilm formation of E. facealis and C. albicans alone and culturing both microorganisms together.
For the quantification of biofilm cells, plates containing biofilms were sonicated after 5 min of vortexing, thereby allowing biofilm cells to break out of the wells.12 Tryptic soy agar [(TSA); Merck, Darmstadt, Germany] and sabouraud dextrose agar [(SDA); Merck] were used for the enumeration of single-species E. faecalis and C. albicans biofilm cells, respectively. For the enumeration of E. faecalis and C. albicans cells in dual-species biofilms, TSA media with amphotericin B (0.025 mg/mL) and SDA media with vancomycin (0.100 mg/mL) were used.
Quantitative real-time polymerase chain reaction (PCR)
C. albicans biofilms (single and dual) were harvested as described above. The mRNA expression changes of Efg1 in C. albicans biofilms were evaluated using qPCR method adopted from a study.12 The sequence of each primer was compared in C. albicans database using Basic Local Alignment Search Tool to assess its specifity.13,14
Quantification of violacein in single- and dual-species biofilms
The production of purple-colored violacein, which is regulated by the QS system in C. violaceum, is an easily observable and measurable marker and is widely used in QS research.15 In the present study, after obtaining E. faecalis and C. albicans cells and supernatants in single- and dual-species biofilms as described above, QS activities were evaluated by slightly modified violacein measurement analysis according to methods by Sankar Ganesh and Ravishankar Rai15 The amounts of violacein produced by C. violaceum after separate treatment with both cell and supernatant solutions of single- and dual-species biofilms were compared with each other.
Metabolomic analysis
As mentioned above, the biofilms (single and dual) were formed in 96-well micro plates with minor revisions. Shortly, C. albicans (106 cfu/mL) was attached for 4 h individually. After transferring E. faecalis (106 cfu/mL) to the culture medium after 4 h, the coculture was incubated at 37°C for 24 h.16
The preparation of samples and GC-MS-dependent conditions was conducted following methods reported by previous a study.16
Freeze-substitution transmission electron microscopy (TEM) analysis
TEM analysis was applied as described previously.17 Briefly, C. albicans biofilm cells were harvested by sonication and centrifugation as described above. Briefly, the cell pellets were mixed in 1% agarose and moved to the sample carriers. After freeze-substitution of the cells in liquid nitrogen, the samples were embedded in epoxy resin. Ultra-thin sections were obtained (100 nm thickness). Samples were visualized with a Hitachi HT7800 TEM.
Statistical analysis
SPSS version 23 (SPSS, Chicago, IL, USA) was used for the statistical analyses. Groups were compared by Student’s t-test. P values <0.05 were significant, and each test was performed at least three times.
Ethics committee approval
The authors declared that an ethics committee approval was not needed for this study.
RESULTS
Effects of E. faecalis supernatant and biofilm cells on C. albicans hyphal morphogenesis and biofilm development
When grown in the common medium, E. faecalis biofilm cells prevented the growth of C. albicans cells. However, no significant change was seen in the growth of E. faecalis (Figure 1). Although it was not statistically significant, C. albicans biofilm cell counts treated with biofilm culture supernatant of E. faecalis decreased (Figure 1).
Figure 1.
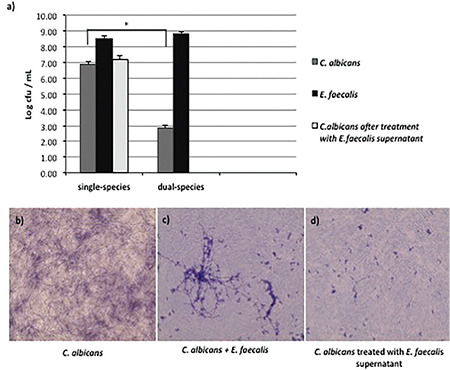
Enterococcus faecalis biofilm cells prevent the proliferation and hyphal development of Candida albicans. a) Proliferation of cells in single- and dual-species biofilms (cfu/mL). Compared with C. albicans in single-species biofilm, E. faecalis prevented the proliferation of C. albicans cells in dual-species biofilm (*p<0.05). Optical microscope images of b) C. albicans biofilm cells formed in six-well cell culture plate. c) C. albicans biofilms with E. faecalis cells and d) C. albicans biofilms exposed to the supernatant of biofilm culture of E. faecalis
To analyze the influence of both E. faecalis cells and factors released by E. faecalis on C. albicans hyphal cell formation, C. albicans biofilms were formed on the slides. At 48 h of mature C. albicans single-species biofilm formation, a significant number of hyphal cells were observed (Figure 1b). However, the hyphal formation of C. albicans cells was inhibited by both E. faecalis biofilm cells and its supernatant when they were incubated together (Figure 1c, d).
EFG1 gene expression profile in C. albicans
To research the inhibitory activity of biofilm cells and supernatant of E. faecalis on C. albicans hyphae formation, Efg1 expression in C. albicans was determined by RT-qPCR. The expression of Efg1 gene in C. albicans was significantly downregulated for both treatment (p<0.05) (Figure 2).
Figure 2.
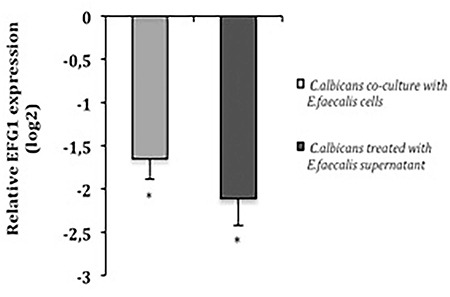
Expression of Efg1 gene in Candida albicans. It was significantly downregulated both in the presence of Enterococcus faecalis cells and in treatment with biofilm culture supernatant of E. faecalis. Statistical significance (*p<0.05) was relative to untreated C. albicans single-species biofilm
Changed metabolite levels in the single- and dual-species biofilms
In this study, GC-MS-based metabolomic analyses were performed to understand how the presence of one microbial species in the dual-species biofilm environment developed by E. faecalis and C. albicans affects the other at the metabolic level. A total of 172 different metabolites were determined, and 112 of them were identified by the index library. Partial least squares discriminant analysis methods were used for both multivariate statistical analysis of GC-MS metabolomic results and the determination of the differences in metabolomic profiles between single- and dual-species biofilms (Figure 3). First, statistical analysis of the models was performed using R2 and Q2 values. All biofilms with values >0.7 show that the method was valid and the models were stable.
Figure 3.
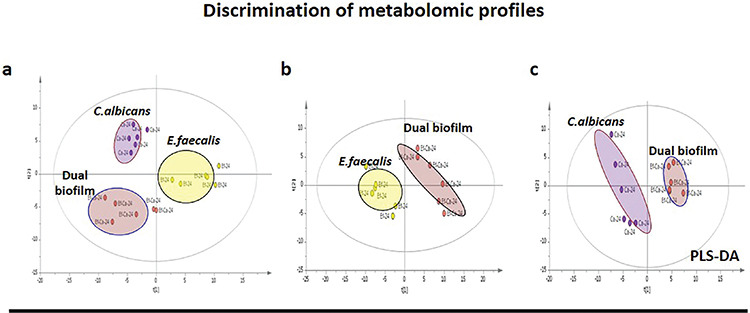
a) PLS-DA score graphs of single- and dual-species biofilm of Candida albicans for metabolomic profile comparison. b) PLS-DA score plots show clear separation between Enterococcus faecalis and its dual-species biofilm. c) PLS-DA score plots demonstrate apparent distinction with C. albicans and its dual-species biofilm. Each circle represents the sharp metabolomic distinction in the biofilms
PLS-DA: Partial least squares discriminant analysis
The changed metabolite levels determined in the biofilms (single and dual) are shown separately in Table 1. No significant difference was found in the amounts of the remaining tricarboxylic acid (TCA) cycle intermediates, except for succinate and citric acid in both biofilms of E. faecalis (Table 1). This result is not surprising considering that E. faecalis lacks the TCA cycle. C. albicans has lower concentrations of TCA intermediates in the dual-species biofilm than in the single-species biofilm (Table 1). Levels of maltose, glucose, and leucrose were high in C. albicans biofilm alone. The existence of E. faecalis in the same environment caused a significant decline in the amounts of these metabolites.
Table 1. Relative metabolite amounts in the biofilms of Enterococcus faecalis or Candida albicans.
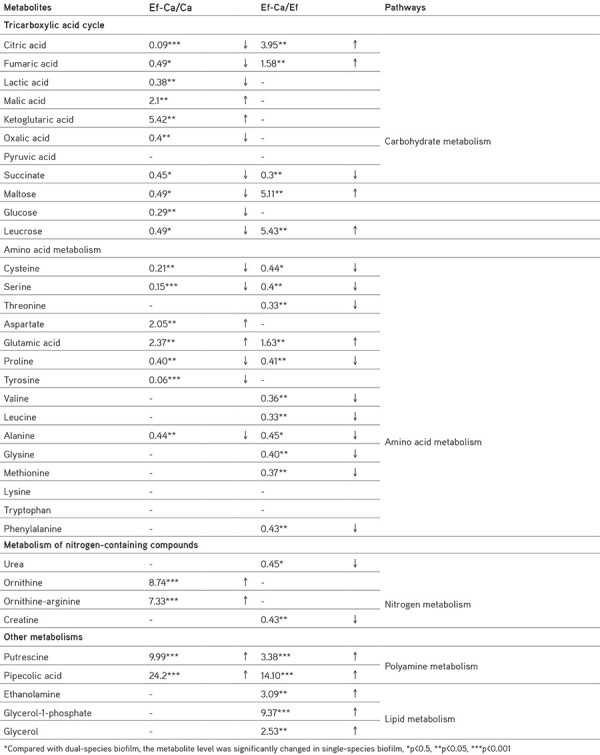
When comparing both biofilms (single and dual), the concentrations of valine, leucine, glycine, methionine, threonine, and phenylalanine were significantly reduced, specifically for E. faecalis, and a decrease in the level of tyrosine was also notable for C. albicans. Putrescine and pipecolic acid concentrations in the dual-species biofilm remained significant, which are the most promising results of this study.
Changes in Candida cell wall architecture in single- and dual-species biofilms
The cell wall biomass was significantly different in dual-species biofilm including the thicker β-glucan-chitin layer and the more dense and narrower fibrillar layer of mannan, when compared with the cells in biofilm alone (Figure 4).
Figure 4.
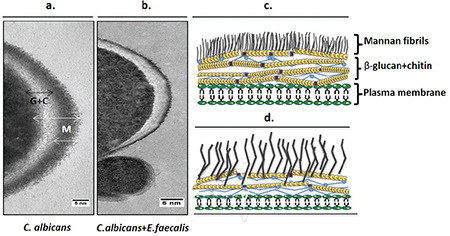
Visualizaton of Candida albicans cell walls grown in single-species (a) and dual-species biofilms (b). (Figures consisted of ≈100 cell images); bar, 5 nm. G + C, β-glucan and chitin; M, mannan. Drawings representing the possible structural changes are shown in c (for the cell wall of C. albicans in dual-species biofilm) and d (for the cell wall of C. albicans in single-species biofilm)
Measurement of violacein in single- and dual-species biofilms
The amount of violacein produced by C. violaceum was determined in single- and dual-species biofilms formed by E. faecalis and/or C. albicans (Figure 5). Compared with untreated media containing only C. violaceum (control), C. violaceum produced less violacein after separate treatment of E. faecalis and C. albicans single- and dual-species biofilms with both supernatant and cell culture suspensions. When single- and dual-species biofilms of both microorganisms were compared, C. violaceum, which was treated with both cell and supernatant suspensions of the dual-species biofilms, produced less violacein for all test conditions, except for the supernatant of E. faecalis.
Figure 5.
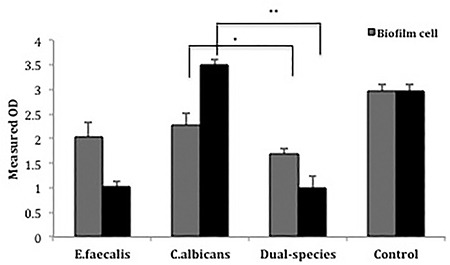
Quantitative measurement of violacein in both single- and dual-species biofilms. Asterisks indicate significance (p<0.05). A significant decrease was shown for all test conditions compared with the control
DISCUSSION
Infections are often considered and treated as a condition caused by a single microorganism; however, in several microorganisms, coexistence of many human microbiome members is observed. These microorganisms live together harmoniously under physiological conditions. Many environmental factors may disrupt this balance; consequently, single or several species become dominant in the environment.18
In this study, the effect of the interaction between E. faecalis and C. albicans on biofilm formation was investigated based on microscopy and metabolomics. The results revealed that in dual-species biofilms, the proliferation of E. faecalis is not affected by the presence of C. albicans; however, the existence of these species in the same environment has an antagonistic effect on the growth of C. albicans (Figure 1). Compared with controls, the reduction of the production of violacein, which provides QS signal communication in C. violaceum treated by single-biofilm cells of C. albicans, also indicates the presence of a molecule that provides C. albicans-induced anti-QS activity in the environment.
In this study, compared with the untreated C. albicans cells, the number of C. albicans hyphal cells decreased when treated with cell suspension or supernatant of E. faecalis biofilm (Figure 2). Therefore, both E. faecalis cells and factors released into the medium have been found to inhibit the hyphal development of Candida. Similar to this finding, in recent studies, bacterial-fungal cooccurrence has been reported to have an antagonizing effect on Candida cell growth. A study that investigated the interference between C. albicans and Lactobacillus species showed that C. albicans did not grow on the surface of the vaginal mucosa because of the lactic acid produced by the Lactobacillus species.19 The coexistence of Staphylococcus aureus and C. albicans in the biofilm environment leads to a substantial increase in the attachment and colonization abilitity of S. aureus. Thus, S. aureus can use C. albicans hyphal cells as a scaffold to the development of a biofilm.20
In this study, the coexistence of E. faecalis and C. albicans in the biofilm model developed may have supported the formation of an anaerobic environment because of increased oxygen consumption. Under this condition, Candida relies on the glycolytic pathway to produce energy. No significant difference was found in the single- and dual-species biofilms of E. faecalis in glucose consumption. The elevated levels of maltose and leucrose in the dual-species biofilm are thought to be caused by the existence of C. albicans. The bacteria within the biofilm are exposed to various environmental conditions, causing the population to be highly heterogeneous in terms of oxygen content.21 Fox et al.22 showed that the hypoxic nature of C. albicans biofilms supports the growth of anaerobic bacteria that share the same environment.
Compared with C. albicans alone, reduced amounts of citric acid, fumaric acid, and oxalic acid in the dual-species biofilm indicate that C. albicans need more energy in the presence of E. faecalis. A study reported that α-ketoglutarate dehydrogenase, a TCA cycle enzyme, is suppressed by Efg1, which is a crucial factor for the hyphal development of C. albicans.23 The downregulation of Efg1 in C. albicans obtained in our study may have led to the suppression of a-ketoglutarate dehydrogenase, which may lead to the transition of C. albicans into the glyoxylate cycle. Thus, it can be a reason for the accumulation of large amounts of ketoglutaric acid and malic acid in the dual-species biofilm.
Glycerol metabolism is an important pathway for the synthesis of lipids and (lipo) teichoic acids in E. faecalis. Lipids, one of the main membrane components, are needed for energy accumulation.24 E. faecalis has increased lipid-related metabolite synthesis when grown with C. albicans. This increase indicates the greater need for lipid-related cell membrane products such as phospholipids and/or lipoteichoic acids in E. feacalis.
Putrescine, an important polyamine in cellular survival, does not support cell proliferation in low amounts; by contrast, the overabundant quantity of internal cells led to the inhibition of cell proliferation.25,26 In this study, one of the most important differences was the concentration of putrescine. Compared with C. albicans biofilm alone, it enhanced approximately by 10- and 3.4-fold in dual-species biofilm and E. faecalis, respectively. In our previous study, the high level of putrescine detected in the dual-species biofilms formed by C. albicans and Proteus mirabilis supports our current data.16
Another interesting result of our study was that the pipecolic acid level increased by 24- and 14-fold for C. albicans and E. faecalis in dual-species biofilm environment when compared with both C. albicans and E. faecalis single biofilms, respectively. The naturally occurring alkyl derivatives of pipecolic acid (piperidine-2-carboxylic acid) are structural components of many biologically active compounds.27 Detailed studies have also shown that the organic compound pipecolic acid is an osmoprotectant and plays a role in protecting macromolecules from denaturation. In the osmoregulation stages, which are generally the same in all living organisms, the first stage is the accumulation of potassium and glutamate, followed by the accumulation of small organic compounds by intracellular synthesis or uptake by external media.28 In our study, higher levels of sugars such as maltose and leucrose in the dual-species biofilm than E. faecalis biofilm alone may have been a threat for E. faecalis because of increased osmolarity. E. faecalis may have synthesized pipecolic acid known to be an osmoprotectant to deal with this threat. The synthesis of bacterial pipecolic acid is a byproduct during the catalysis of the proline amino acid, which may explain the low level of proline in the dual-species biofilm obtained from our study.
The alterations in the yeast cell wall as an adaptation to osmotic stress have been highlighted in the literature.29 We detected the more dense and shorter mannan layer and thicker β-glucan-chitin layer in Candida cell wall grown in dual-species biofilm than in the single-species biofilm. In both cases, alterations in the cell wall of C. albicans are similar to those in cells with and without salt-induced osmotic stress in the study of Ene et al.29 This strengthens the possibility of increased osmotic stress in the dual-species biofilm environment.
Compared with the single-species biofilm of both microorganisms, significant decrease was observed in many amino acid levels in the dual-species biofilm. This reduction in amino acid levels in the dual-species biofilm shows that anabolic reactions are dominant for both species to grow, develop, and multiply. Clearly, amino acid synthesis is required for C. albicans biofilm development.
CONCLUSION
The metabolite diversity of both microorganisms, which was affected by each other by increasing the cellular stress due to high carbohydrate consumption, more energy needs, etc., was demonstrated in our results. The high levels of putrescine and pipecolic acid synthesized as osmoprotectant by both species may have suppressed the growth of Candida. This study provided preliminary data for a detailed investigation of the possible role of putrescine and pipecolic acid in the prevention of C. albicans via bacterial species.
Footnotes
Conflict of interest: No conflict of interest was declared by the authors. The authors are solely responsible for the content and writing of this paper.
Funding sources: This work was supported by funding from TUBITAK (grant no. 115S550).
References
- 1.Burmolle M, Ren DW, Bjarnsholt T, Sorensen SJ. Interactions in multispecies biofilms: do they actually matter? Trends Microbiol. 2014;22:84–91. doi: 10.1016/j.tim.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 2.Morales DK, Grahl N, Okegbe C, Dietrich LEP, Jacobs NJ, Hogan DA. Control of Candida albicans metabolism and biofilm formation by Pseudomonas aeruginosa phenazines. mBio. 2013;4:e00526–12. doi: 10.1128/mBio.00526-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tampakakis E, Peleg AY, Mylonakis E. Interaction of Candida albicans with an intestinal pathogen, Salmonella enterica Serovar Typhimurium. Eukaryot Cell. 2009;8:732–737. doi: 10.1128/EC.00016-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cruz MR, Graham CE, Gagliano BC, Lorenz MC, Garsin DA. Enterococcus faecalis inhibits hyphal morphogenesis and virulence of Candida albicans. Infect Immun. 2013;81:189–200. doi: 10.1128/IAI.00914-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ten Oever J, Netea MG. The bacteriome-mycobiome interaction and antifungal host defense. Eur J Immunol. 2014;44:3182–3191. doi: 10.1002/eji.201344405. [DOI] [PubMed] [Google Scholar]
- 6.Pfaller MA, Diekema DJ. Epidemiology of invasive mycoses in North America. Crit Rev Microbiol. 2010;36:1–53. doi: 10.3109/10408410903241444. [DOI] [PubMed] [Google Scholar]
- 7.Wenner JJ, Rettger LF. A systematic study of the Proteus group of bacteria. J Bacteriol. 1919;4:331–353. doi: 10.1128/jb.4.4.331-353.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nobile CJ, Johnson AD. Candida albicans biofilms and human disease. Annu Rev Microbiol. 2015;69:71–92. doi: 10.1146/annurev-micro-091014-104330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayer FL, Wilson D, Hube B. Candida albicans pathogenicity mechanisms. Virulence. 2013;4:119–128. doi: 10.4161/viru.22913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reaves ML, Rabinowitz JD. Metabolomics in systems microbiology. Curr Opin Biotechnol. 2011;22:17–25. doi: 10.1016/j.copbio.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu YJ, Wang CS, Ho WE, Ong CN. Recent developments and applications of metabolomics in microbiological investigations. Trac-Trend Anal Chem. 2014;56:37–48. [Google Scholar]
- 12.Kart D, Tavernier S, Van Acker H, Nelis HJ, Coenye T. Activity of disinfectants against multispecies biofilms formed by Staphylococcus aureus, Candida albicans and Pseudomonas aeruginosa. Biofouling. 2014;30:377–383. doi: 10.1080/08927014.2013.878333. [DOI] [PubMed] [Google Scholar]
- 13.Bandara HM, Cheung BP, Watt RM, Jin LJ, Samaranayake LP. Secretory products of Escherichia coli biofilm modulate Candida biofilm formation and hyphal development. J Investig Clin Dent. 2013;4:186–199. doi: 10.1111/jicd.12048. [DOI] [PubMed] [Google Scholar]
- 14.Altschul SF, Madden TL, Schaffer AA, Zhang J, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sankar Ganesh P, Ravishankar Rai V. Attenuation of quorum-sensing-dependent virulence factors and biofilm formation by medicinal plants against antibiotic resistant Pseudomonas aeruginosa. J Tradit Complement Med. 2018;8:170–177. doi: 10.1016/j.jtcme.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kart D, Yabanoglu Ciftci S, Nemutlu E. Altered metabolomic profile of dual-species biofilm: interactions between Proteus mirabilis and Candida albicans. Microbiol Res. 2020;230:126346. doi: 10.1016/j.micres.2019.126346. [DOI] [PubMed] [Google Scholar]
- 17.Netea MG, Gow NA, Munro CA, Bates S, Collins C, Ferwerda G, Hobson RP, Bertram G, Hughes HB, Jansen T, Jacobs L, Buurman ET, Gijzen K, Williams DL, Torensma R, McKinnon A, MacCallum DM, Odds FC, Van der Meer JW, Brown AJ, Kullberg BJ. Immune sensing of Candida albicans requires cooperative recognition of mannans and glucans by lectin and Toll-like receptors. J Clin Invest. 2006;116:1642–1650. doi: 10.1172/JCI27114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gulati M, Nobile CJ. Candida albicans biofilms: development, regulation, and molecular mechanisms. Microbes Infect. 2016;18:310–321. doi: 10.1016/j.micinf.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strus M, Kucharska A, Kukla G, Brzychczy-Wloch M, Maresz K, Heczko PB. The in vitro activity of vaginal Lactobacillus with probiotic properties against Candida. Infect Dis Obstet Gynecol. 2005;13:69–75. doi: 10.1080/10647440400028136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kean R, Rajendran R, Haggarty J, Townsend EM, Short B, Burgess KE, Lang S, Millington O, Mackay WG, Williams C, Ramage G. Candida albicans mycofilms support Staphylococcus aureus colonization and enhances miconazole resistance in dual-species interactions. Front Microbiol. 2017;8:258. doi: 10.3389/fmicb.2017.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blank LM, Sauer U. TCA cycle activity in Saccharomyces cerevisiae is a function of the environmentally determined specific growth and glucose uptake rates. Microbiology (Reading). 2004;150:1085–1093. doi: 10.1099/mic.0.26845-0. [DOI] [PubMed] [Google Scholar]
- 22.Fox EP, Cowley ES, Nobile CJ, Hartooni N, Newman DK, Johnson AD. Anaerobic bacteria grow within Candida albicans biofilms and induce biofilm formation in suspension cultures. Curr Biol. 2014;24:2411–2416. doi: 10.1016/j.cub.2014.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan J, Hu C, Yu JH. Lipid biosynthesis as an antifungal target. J Fungi (Basel). 2018;4:50. doi: 10.3390/jof4020050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doedt T, Krishnamurthy S, Bockmühl DP, Tebarth B, Stempel C, Russell CL, Brown AJ, Ernst JF. APSES proteins regulate morphogenesis and metabolism in Candida albicans. Mol Biol Cell. 2004;15:3167–3180. doi: 10.1091/10.1091/mbc.E03-11-0782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Porat Z, Wender N, Erez O, Kahana C. Mechanism of polyamine tolerance in yeast: novel regulators and insights. Cell Mol Life Sci. 2005;62:3106–3116. doi: 10.1007/s00018-005-5341-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valdes-Santiago L, Ruiz-Herrera J. Stress and polyamine metabolism in fungi. Front Chem. 2014;1:42. doi: 10.3389/fchem.2013.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hibi M, Mori R, Miyake R, Kawabata H, Kozono S, Takahashi S, Ogawa J. Novel enzyme family found in filamentous fungi catalyzing trans-4-hydroxylation of l-pipecolic acid. Appl Environ Microbiol. 2016;82:2070–2077. doi: 10.1128/AEM.03764-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gouesbet G, Jebbar M, Talibart R, Bernard T, Blanco C. Pipecolic acid is an osmoprotectant for Escherichia coli taken up by the general osmoporters ProU and ProP. Microbiology (Reading). 1994;140:2415–2422. doi: 10.1099/13500872-140-9-2415. [DOI] [PubMed] [Google Scholar]
- 29.Ene IV, Walker LA, Schiavone M, Lee KK, Martin-Yken H, Dague E, Gow NA, Munro CA, Brown AJ. Cell wall remodeling enzymes modulate fungal cell wall elasticity and osmotic stress resistance. mBio. 2015;6:e00986. doi: 10.1128/mBio.00986-15. [DOI] [PMC free article] [PubMed] [Google Scholar]


