Abstract
Werner's syndrome (WS) is a human disease with manifestations resembling premature aging. The gene defective in WS, WRN, encodes a DNA helicase. Here, we describe the generation of mice bearing a mutation that eliminates expression of the C terminus of the helicase domain of the WRN protein. Mutant mice are born at the expected Mendelian frequency and do not show any overt histological signs of accelerated senescence. These mice are capable of living beyond 2 years of age. Cells from these animals do not show elevated susceptibility to the genotoxins camptothecin or 4-NQO. However, mutant fibroblasts senesce approximately one passage earlier than controls. Importantly, WRN−/−;p53−/− mice show an increased mortality rate relative to WRN+/−;p53−/− animals. We consider possible models for the synergy between p53 and WRN mutations for the determination of life span.
Werner's Syndrome (WS) is a recessive genetic disease which shows premature onset of many pathologies normally associated with old age (18). Patients with WS appear normal during the first decade of life. The first manifestation of this disease is typically growth failure during adolescence. Subsequently, these patients suffer prematurely from a variety of age-related disorders: skin changes, osteoporosis, diabetes, accelerated atherosclerosis, and cancer, particularly sarcomas. Fibroblasts derived from individuals with WS divide many fewer times prior to senescence than do fibroblasts from age-matched control individuals (13). Genomic instability has been observed in WS cells, as chromosomal rearrangements (5, 19, 21) and as mutations within the hypoxanthine phosphoribosyltransferase gene (HPRT); in vivo, an increased frequency of HPRT mutant cells has been observed in patients with WS (2, 3, 14). The gene defective in WS, WRN, encodes a protein of 1,432 amino acids with similarity to the RecQ subfamily of DNA helicases (26). Although mutations throughout the WRN gene have been observed in the homozygous state, homozygosity for a mutation very near the 3′ end of the WRN open reading frame is sufficient to lead to the disease (15).
A mouse knockout (KO) of the WRN gene has been described (10). Lebel and Leder deleted exons III and IV in the catalytic helicase domain of the WRN locus, a mutation predicted to eliminate catalytic function. Cells containing this mutation express an internally deleted, nearly full-length WRN protein. Homozygous mutant mice are viable, indicating that this particular mutation is not lethal. However Lebel and Leder showed a decreased embryonic survival of their mutant: on a C57BL/6-129/SvEv outbred background and on a 129/SvEv inbred background, the ratios of +/+:+/−:−/− mice born are 1:2.0:0.8 and 1:1.9:0.6, respectively. Mutant embryonic stem (ES) cells have an approximately sixfold increased mutation rate at the HPRT locus. They are also 10-fold more sensitive to camptothecin, a topoisomerase I inhibitor, and are two- to threefold more sensitive to etoposide, a topoisomerase II inhibitor. Late-passage mutant embryonic fibroblasts also show decreased saturation density in culture, although this was not evident in early-passage cells. The mice themselves, however, are healthy and fertile, showing no signs of premature organismic aging or increased rates of tumor formation. Thus, this KO does not recapitulate many of the phenotypes of human WS.
Here, the generation and characterization of a WRN-null mouse mutant is described. Most phenotypes in the mutant are remarkably similar to the wild type. Cells from these animals are not hypersensitive to camptothecin, unlike those of Lebel and Leder. Most interestingly, the WRN−/− homozygous animal displays a shorter life span in the p53−/− background. We discuss this shortening with respect to a possible aging phenotype.
MATERIALS AND METHODS
Cloning of WRN.
A size-selected murine cDNA plasmid library was screened by standard methods (20) by using an 820-bp probe derived from the 3′ end of the human WRN-coding sequence. This probe was generated by PCR from human cDNA with the following oligonucleotides: 5′ AGG TCC AGA TTG GAT CAT TGC 3′ and 5′ GGC CAA CAT GGC AGC TTT GCC 3′. Hybridizations were performed at 55°C. Twenty-two clones were isolated, and preliminary restriction mapping and 5′ sequencing suggested that they were all products of the same gene. The largest clone was sequenced on both strands.
Generation of antibodies against WRN.
A polyclonal antiserum was raised in chickens (Covance) against a His6-tagged protein fragment corresponding to amino acid residues 1191 to 1390 of the WRN protein. Immunoglobulin Y was isolated from eggs by using a commercially available kit (EGGstract; Promega) and was further purified over a diaminopropylamine column (Pierce) containing 5 mg of bound immunizing antigen.
Tissue Western blotting.
Fragments of various mouse tissues were placed in Laemmli buffer, macerated with a polytron, and boiled. Equal amounts of protein were loaded into each lane and assayed by Coomassie blue staining of a duplicate gel (20). Horseradish peroxidase-conjugated antichicken antibodies were used to detect bound anti-WRN. ECL reagent (Amersham) was used to develop the bound secondary antibody.
Targeting the WRN locus.
Several genomic clones in lambda phage encoding portions of the WRN locus were recovered by screening a genomic library in EMBL 3A with a full-length WRN-coding region probe by standard methods (20). Two clones encoding portions of the catalytic helicase domain were subcloned into pBR322 and were extensively mapped with restriction enzymes. To construct the 5′ homology arm of the targeting vector, a 3.0-kb SalI/HindIII fragment from the larger pBR322 clone was subcloned into the SalI/HindIII sites of pSL1190 (Pharmacia). For the 3′ homology arm, a 4.9-kb BamHI/ScaI fragment was ligated into the BamHI/SmaI sites of the pSL1190 vector. A KpnI/NotI fragment containing the β-geo cassette (β-galactosidase/neomycinr fusion gene) was then inserted into the KpnI/NotI sites of the pSL1190 construct. An internal SalI site in the β-geo cassette was obliterated by partial digestion followed by blunting and religation, and then the completed cassette was excised from the vector via SalI digestion. All cell culture and mouse embryo manipulations were as previously described except that no negative selection step was employed (11). Chimeric founders were crossed with BALB/c animals, and progeny from these matings were intercrossed to obtain homozygotes.
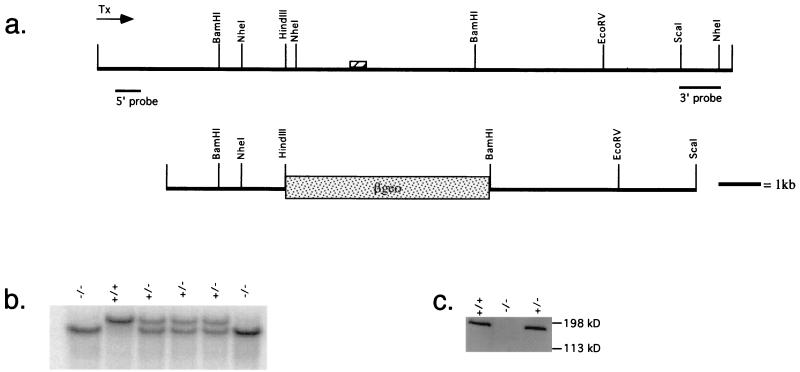
The genotypes of neomycin-resistant ES cell clones were checked with two external flanking probes. The 3′ probe consisted of a 1.2-kb ScaI/NheI restriction fragment (see Fig. 2a) which yielded a 9-kb wild-type band and a 6-kb mutant band on genomic DNA digested with NheI. The 5′ probe consisted of an NdeI/KpnI fragment which was isolated and further digested with RsaI, and the uppermost ∼800-bp fragment was used as the probe. On EcoRV-restricted genomic DNA, this probe detected a large (>13 kb) band representing the wild-type allele and an approximately 10-kb band representing the mutant allele.
FIG. 2.
Targeting the murine WRN locus. (a) Restriction map of a WRN lambda genomic clone (top map) or the mouse WRN-targeting construct (bottom map). The position of an exon encoding the 3′-most region of the helicase domain is indicated by a hatched box. The direction of transcription, as revealed by genomic sequencing, as well as the position of 5′ and 3′ probes used to genotype ES clones and mice are indicated. (b) Genotyping of a litter derived from a heterozygous cross. Hybridization was performed with the 5′ probe as indicated in Fig. 2a. (c) Cells derived from mutant animals do not express any detectable WRN protein. Extracts from ear fibroblasts from mice of the indicated genotypes were probed with chicken affinity-purified anti-WRN antibody. Similar results were obtained probing extracts derived from ES cells of various genotypes (data not shown).
A PCR genotyping assay was also developed based on the results of genomic sequencing. The oligonucleotides used were as follows: pSL3093, 5′ GCC TGC AGC TGG CGC CAT C 3′; COMMON.2, 5′ CAA TAA CCA ATG GAA TTC TAA GC 3′; and WT.1, 5′ TAC ATT TGC CAT TTT AAG GTG GC 3′. The PCR conditions were 95°C for 3 min, followed by 30 cycles of 94°C, 30 s, 57°C, 30 s, and 72°C, 30 s, followed by a final 5-min incubation at 72°C. This combination of oligonucleotides produces an approximately 250-bp band in the presence of the mutant allele and an approximately 150-bp band in the presence of the wild-type allele.
Splenocyte culture.
Spleens were isolated from mice of the indicated genotypes, erythrocytes were lysed, and the splenocytes were resuspended at a concentration of 2 × 106/ml in plating media (10% fetal bovine serum–Gln–HEPES–6.0 × 10−5 M β-mercaptoethanol in RPMI medium [Gibco]). To determine response to mitogenic stimulation, 0.5 × 105 lymphocytes/well were plated in triplicate in 96-well plates. Anti-CD3 was added at the indicated dilution, and cells were cultured for 72 h. During the last 24 h, the cultures were pulsed with 1 μCi of [3H]thymidine per well. The wells were then harvested, and proliferation was quantified on a scintillation counter. The results shown are representative of two separate experiments.
Embryonic fibroblasts.
Murine embryonic fibroblasts were generated from day-13.5 embryos as previously described (6). Fibroblasts were cultured in media consisting of 10% fetal bovine serum in Dulbecco modified Eagle medium (Gibco). To measure genotoxin sensitivity, murine embryonic fibroblasts were plated at 25,000 cells/well in a 96-well plate. The next day, the indicated concentrations of toxins were added. The cells were then cultured for 3 days, and cell proliferation was subsequently quantitated by using the Boehringer-Mannheim Cell Proliferation Kit II, following the manufacturer's instructions. In each case, two independent cell lines of each genotype were treated in two wells each, and the results were averaged.
Nucleotide sequence accession number.
The 6,476-nucleotide cDNA sequence encoding the murine WRN protein has been submitted to GenBank under accession no. AF241636.
RESULTS
Cloning and protein expression studies of the mouse WRN homolog.
A size-selected murine cDNA library derived from activated lymph node and spleen was screened at reduced stringency with a hybridization probe derived from the 3′ end of the human WRN-coding sequence. The largest clone was sequenced in its entirety on both strands. This 6,476-nucleotide cDNA encodes a putative protein of 1,401 amino acids which is 72% identical to the human WRN protein at the amino acid level. Others have independently cloned the mouse WRN homolog (7). The inferred protein sequence reported by Imamura et al. is identical to that reported here at all but three residues: the Imamura et al. sequence contains a Q rather than a K at position 800, an A rather than a T at position 1145, and a V rather than an L at position 1181. These differences may represent polymorphisms between the strains of mice used to generate the libraries from which these cDNAs were derived or errors in reverse transcription. Outside the coding region, the Imamura et al. sequence shows several nucleotide differences from that described here. The WRN nucleotide sequence reported here also contains two exons in the 5′ and 3′ untranslated regions absent in the Imamura et al. sequence as well as 1,347 nucleotides of 3′-UTR sequence not reported by Imamura et al.
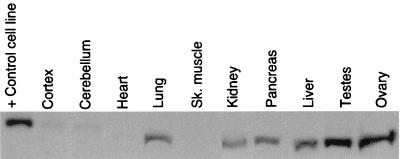
In order to examine the tissue distribution of the WRN protein, polyclonal antiserum to a C-terminal fragment of the WRN protein corresponding to amino residues 1191 to 1390 was raised in chickens. This region was chosen because it lies outside the catalytic helicase domain and was therefore unlikely to contain epitopes cross-reactive with other helicases. Affinity-purified antiserum was used to probe a Western blot containing lysates of various murine tissues (Fig. 1). The band corresponding to the murine WRN protein migrates at roughly 170 kDa. Murine WRN protein is expressed in lung, kidney, pancreas, liver, testes, and ovary but is present only at very low levels in cortex, cerebellum, heart, and skeletal muscle.
FIG. 1.
Expression profile of murine WRN protein. Tissue Western blot of WRN protein expression. The blot was probed with an antibody directed against the C terminus of the mouse WRN protein. The lane marked + Control cell line contains a lysate of X3, an epithelial cell line that expresses high levels of the WRN protein, a gift of B. Panning. The apparent molecular weight discrepancy between WRN derived from X3 and from the murine tissues is an electrophoresis artifact.
Targeting the WRN locus.
Mice bearing a targeted mutation in the murine WRN gene were generated. The full-length murine WRN cDNA was used as a hybridization probe to recover several clones from a 129/SvJ genomic library (library courtesy of the Housman laboratory). In turn, these clones were used to generate a targeting construct in which the 3′-most exon encoding a portion of the catalytic helicase domain is replaced by a β-geo cassette (Fig. 2a). If there should be splicing around this cassette, this mutation is also predicted to introduce a frameshift mutation. Homozygous mutations in the helicase domain or near the 3′ end of the WRN open reading frame are sufficient to confer the WS phenotype in humans (15). This construct was electroporated into ES cells; of 88 neomycin-resistant clones selected, 11 were heterozygous for the WRN mutation, yielding a targeting frequency of 12.5%. Two correctly targeted clones were used to generate chimeric founders. These mice, representing two independent ES cell clones, were used to generate heterozygotes, and these heterozygotes were subsequently intercrossed to obtain homozygotes. Mice of different genotypes are distinguished by Southern blotting (Fig. 2b) or by PCR assay. Western blot analysis of whole-cell extracts of ear fibroblasts from mice of different genotypes using an antibody directed against the C terminus of the murine WRN protein demonstrates that there is no detectable WRN expression in KO cells (Fig. 2c). Probing of these extracts with antiserum directed against the N terminus of the WRN protein did not reveal a truncated WRN protein in mutant animals (data not shown).
WRN KO animals are viable.
Heterozygous crosses have produced offspring in the ratio of 108 +/+ to 173 +/− to 105 −/−. Crosses between heterozygous mice and mutant mice have yielded mice in the ratio of 98 +/− to 93 −/−. Since these are close to the ratios predicted by simple Mendelian segregation, it seems unlikely that the WRN mutation described in this work confers any prenatal lethality, in contrast to that described elsewhere (10). Mutant animals are grossly normal, and all KO animals tested are fertile. The oldest homozygote obtained is over 2 years old and is still healthy. Histological examination of several aged KO animals ranging in age between 3 and 17 months failed to uncover any unusual lesions, with the exception of bone marrow hyperplasia in one.
Mutant splenocytes proliferate normally.
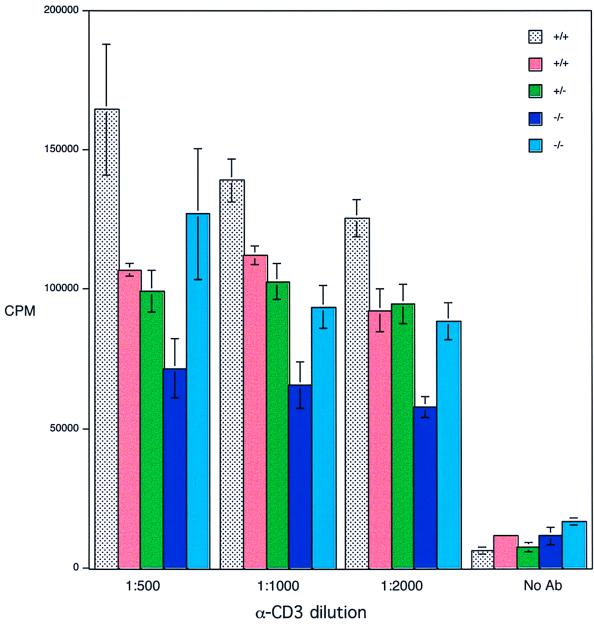
The proliferation of cells derived from mutant animals was examined in culture. Splenocytes were derived from two mutant animals, two wild-type animals, and one heterozygote and were treated with various dilutions of anti-CD3, a mitogenic stimulus; the response was measured by [3H]thymidine uptake 3 days later (Fig. 3). No significant differences were noted between mutant and control animals.
FIG. 3.
Proliferation of WRN−/− or control splenocytes in response to anti-CD3. Splenocytes were isolated from animals of the indicated genotypes (five animals total) and were induced to proliferate by using the indicated dilutions of anti-CD3 supernatant (hamster clone 145-2C11). Three days later, the cultures were pulsed with tritiated thymidine, and the proliferation was quantified by scintillation counting.
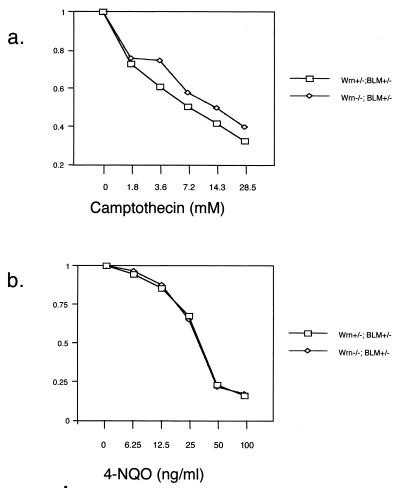
No heightened susceptibility to camptothecin or 4-NQO in mutant embryonic fibroblasts.
WS patient cells show sensitivity to the DNA-damaging agent 4-NQO (4, 16), and WRN KO ES cells described by Lebel and Leder show sensitivity to the topoisomerase I poison camptothecin (10). In order to determine whether WRN mutation would confer sensitivity to these agents, WRN+/− or WRN−/− embryonic fibroblasts were cultured in the presence of these agents, and the number of viable cells was quantitated by using an assay to detect viable cells via their mitochondrial respiration (Cell Proliferation Kit II; Boehringer-Mannheim) 3 days later. These cell lines were also heterozygous for a mutation in the BLM gene (G. Luo and A. Bradley, unpublished data). Neither camptothecin (Fig. 4a) nor 4-NQO (Fig. 4b) affected WRN mutants differentially.
FIG. 4.
No elevated sensitivity to camptothecin or 4-NQO in WRN−/−;BLM+/− embryonic fibroblasts. (a) Camptothecin treatment. (b) 4-NQO treatment. y axis represents the number of cells in the treated well divided by the number of cells in the control well.
Modestly accelerated senescence in WRN−/− embryonic fibroblasts.
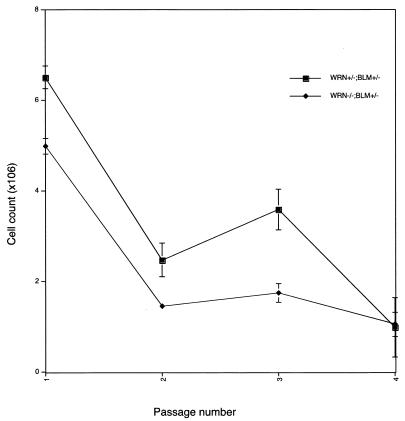
In humans, a cardinal feature of WS is accelerated senescence in patient skin fibroblasts. Experiments were undertaken in order to determine whether this phenotype might be recapitulated in the WRN KO mouse. WRN+/−;BLM+/− and WRN−/−;BLM+/− fibroblasts were serially passaged in culture; 106 cells were plated at each passage, and the number of cells present at confluence was determined several days later (Fig. 5). The number of cells at confluence has been used as a measure of replicative potential in previous studies (25). WRN KO cultures cease growing approximately one passage earlier than controls.
FIG. 5.
Premature replicative senescence in WRN−/−;BLM+/− embryonic fibroblasts. Cells were generated from two independent embryos of each genotype. At each passage, 106 cells were plated; the cells were harvested between 3 and 5 days later when all the cultures were visually judged to be confluent. The cells were then trypsinized and counted, and 106 cells were subsequently replated. The cell counts at the end of each passage are recorded.
Homozygous WRN mutations accelerate mortality in p53−/− animals.
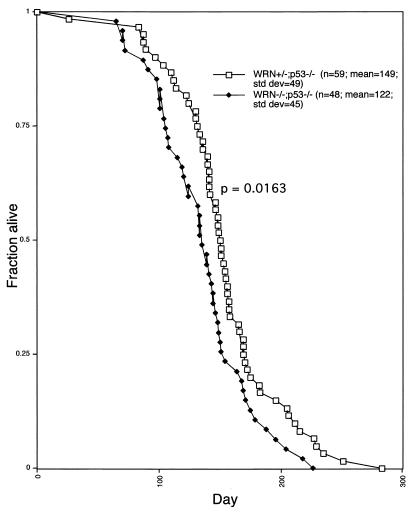
In humans, WS is associated with a heightened susceptibility to tumors. In order to accentuate any predisposition to tumors in WRN KO mice, WRN mutants were bred to p53 mutants (8). Animals with the genotypes WRN−/−;p53−/− or WRN+/−;p53−/− were monitored over time (Fig. 6). Whereas WRN+/−;p53−/− animals had an average life span of 149 days, WRN−/−;p53−/− animals lived for an average 122 days. The survival curves are statistically different from one another by the Wilcox ranked sum test (P = 0.0163). The possible implications of this result are discussed below.
FIG. 6.
Mortality of WRN+/−;p53−/− and WRN−/−;p53−/− mice. The health status of the mice was monitored several times per week. Mice were sacrificed when obviously moribund or, in some cases, died on their own. The difference between the curves was judged significant (P = 0.0163) by the Wilcox ranked sum test.
DISCUSSION
Here, the cloning of a highly conserved murine homolog of the WRN protein is described. Despite the high degree of sequence identity between these two proteins, the human and mouse WRN homologs do not show similar immunolocalization patterns (12). Mice bearing a targeted mutation in the catalytic helicase domain of WRN are viable and fertile, they do not show any histological signs of premature aging, and they are capable of surviving until at least 2 years of age. Splenocytes from these animals proliferate normally in response to a mitogenic stimulus; however, cells from these animals senesce prematurely in cell culture.
Homozygous WRN mutations accelerate mortality in p53−/− animals. There are two general possible explanations for this synthetic interaction between the WRN and p53 genes. First, homozygous mutations in WRN may exacerbate the cancer-prone phenotype of p53−/− animals. The genome instability reported in cases of WS could be the molecular basis of this interaction. WRN and p53 have recently been shown to interact physically, further suggesting that these proteins may cooperate to maintain genome stability (1, 23). A second possibility is that the homozygous WRN mutant animals do have a slightly accelerated aging phenotype. This phenotype might be first evident in the p53−/− background because of its short life span. In this view, the cancer phenotype itself would be under the control of the aging program of mice. Thus, speeding up this program would advance all of the regulated phenotypes, including cancer in a wild-type or p53−/− cancer-prone strain. This model predicts that the WRN−/− animals will also display a slightly shortened life span in the p53 wild-type background. Although some WRN−/− animals are now over 2 years old, it is still too early to know whether their life span will be shortened compared to that of the wild type.
Lebel and Leder have described a WRN KO bearing a helicase domain mutation which shows several phenotypes (10). Mutant ES cells are highly sensitive to camptothecin and show an elevated mutation rate, and late-passage embryonic fibroblasts possess a shortened in vitro life span compared with that of wild-type cells. In addition, mutant animals are born at less than the expected frequency, suggesting that this mutation confers some prenatal lethality. By contrast, mutant embryonic fibroblasts described herein are not hypersensitive to camptothecin. The former difference may stem from biological differences between embryonic fibroblasts and ES cells. WRN−/− embryonic fibroblasts generated in this work do possess a modestly shortened in vitro proliferative capacity, in accord with the results of Lebel and Leder; however, we find that WRN mutant mice are born at the expected frequency. Several possible explanations exist for these discrepancies. Modifying loci in ES cells and/or mouse strains may alter the phenotypic consequences of WRN mutations. The nature of the WRN alleles generated represents another potential reason for these discrepant results. The allele described herein deletes an exon in the catalytic helicase domain and introduces a frameshift mutation, resulting in no detectable protein expression, as assayed by immunofluorescence (12) and Western blotting using an anti-C-terminal antibody. As the nuclear localization signal of the human WRN protein lies at the distal C terminus of the protein, it seems likely that this mutation should represent a functional null. By contrast, the mutation described by Lebel and Leder results in the expression of an internally deleted fragment that still has the potential to localize to the nucleus, where it might exert unpredictable effects. Thus, the effects noted by Lebel and Leder might not represent those of a true null allele in the WRN gene.
Several possible explanations exist as to why murine WRN mutants do not recapitulate the full spectrum of effects seen in human WS patients. Mice may possess more than one WRN homolog; disruption of the putative second WRN gene or both genes in the same animal might be required to recapitulate the human phenotype. Several observations argue against this hypothesis. In this study, 22 clones, all derived from the same gene, were isolated via reduced-stringency hybridization of a splenic library. This same gene has been isolated by using degenerate reverse transcriptase PCR (7, 10). Hence, if there is a second WRN gene in mice, it must be expressed at much lower levels and/or be significantly diverged in sequence from the one that has been described. The WRN gene lies in a chromosomal region in the mouse which is syntenic to human chromosome 8p, the location of the human WRN gene (10, 26). Screening of Northern blots at reduced stringency does not reveal any transcripts which might correspond to a second WRN gene (D. B. Lombard, unpublished data). Finally, antibodies derived against the WRN protein and antibodies against the human WRN protein only recognize the known WRN protein in the mouse (D. B. Lombard and R. Marciniak, unpublished results). Thus, it is unlikely, though still formally possible, that more than one WRN gene exists in the mouse.
Another possible explanation for the failure to produce a strong WS-like phenotype in the mouse is simply divergence between mice and humans in WRN function and/or, more generally, in DNA repair functions. In humans, the WRN protein is concentrated in the nucleolus, whereas the murine WRN protein is spread diffusely throughout the nucleoplasm (12). This suggests that some divergence in WRN function may have occurred between mice and humans. It is also possible that murine WRN is functionally redundant with another helicase, either a RecQ family member or perhaps a member of a different helicase family altogether. In addition, mice may show milder effects of a WRN mutation simply as a result of their smaller size and shorter life span, perhaps not allowing enough time for the full spectrum of effects of WS to manifest themselves.
Another potential reason for the discrepancy between the behavior of WRN mutants in mice and humans is that the nature of the WRN target may different. One such target of the WRN protein may be the telomeres. In primary human WS cells, telomeres shorten more rapidly than in wild-type cells, though WS cells ultimately senesce with longer telomeres than do wild-type cells (22). One explanation for the latter observation is that telomeres may be more recombinogenic and unstable in WS cells than in normal cells; hence, there may be more variation in telomere length in WS cells than in wild-type cells. This may occasionally produce a single very short telomere in WS cells which overall retain long telomeres; this could lead to senescence in cells which, for the most part, still possess long telomeres. Data consistent with telomeric instability in WS have been obtained in studies of lymphoblastoid cells (24). Recent studies in our laboratory suggest that introduction of telomerase into primary WS cells can rescue their premature senescence (B. Johnson, personal communication). Mice, unlike humans, express telomerase constitutively in multiple somatic tissues and possess very long telomeres (9, 17); thus, if telomeres are an important target of WRN, many of the effects of WS might not be evident in the mouse. One critical test of this model will be to cross WRN mutant mice with mice lacking the telomerase RNA component to determine whether these double-mutant animals show any synthetic phenotypes. Such experiments are underway.
In summary, we have generated and characterized a murine mutant in the WRN locus. Further studies in both mice and in human cells are necessary to elucidate the role of WRN in normal cellular physiology and its possible role in aging.
ACKNOWLEDGMENTS
We thank all the members of the Guarente and Jaenisch laboratories for helpful discussions. M. Lebel and P. Leder are acknowledged for their generous gift of antiserum directed against the N terminus of the mouse WRN protein.
The Guarente laboratory is supported by grants from the NIH, The Ellison Medical Foundation, The Seaver Institute, and The Linda and Howard Stern Foundation. D.B.L. was supported by an MSTP grant to Harvard Medical School.
REFERENCES
- 1.Blander G, Kipnis J, Leal J F, Yu C E, Schellenberg G D, Oren M. Physical and functional interaction between p53 and the Werner's syndrome protein. J Biol Chem. 1999;274:29463–29469. doi: 10.1074/jbc.274.41.29463. [DOI] [PubMed] [Google Scholar]
- 2.Fukuchi K, Martin G M, Monnat R J., Jr Mutator phenotype of Werner syndrome is characterized by extensive deletions. Proc Natl Acad Sci USA. 1989;86:5893–5897. doi: 10.1073/pnas.86.15.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fukuchi K, Tanaka K, Kumahara Y, Marumo K, Pride M B, Martin G M, Monnat R J., Jr Increased frequency of 6-thioguanine-resistant peripheral blood lymphocytes in Werner syndrome patients. Hum Genet. 1990;84:249–252. doi: 10.1007/BF00200569. [DOI] [PubMed] [Google Scholar]
- 4.Gebhart E, Bauer R, Raub U, Schinzel M, Ruprecht K W, Jonas J B. Spontaneous and induced chromosomal instability in Werner syndrome. Hum Genet. 1988;80:135–139. doi: 10.1007/BF00702855. [DOI] [PubMed] [Google Scholar]
- 5.Hoehn H, Bryant E M, Au K, Norwood T H, Boman H, Martin G M. Variegated translocation mosaicism in human skin fibroblast cultures. Cytogenet Cell Genet. 1975;15:282–298. doi: 10.1159/000130526. [DOI] [PubMed] [Google Scholar]
- 6.Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the mouse embryo: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 7.Imamura O, Ichikawa K, Yamabe Y, Goto M, Sugawara M, Furuichi Y. Cloning of a mouse homologue of the human Werner syndrome gene and assignment to 8A4 by fluorescence in situ hybridization. Genomics. 1997;41:298–300. doi: 10.1006/geno.1997.4661. [DOI] [PubMed] [Google Scholar]
- 8.Jacks T, Remington L, Williams B O, Schmitt E M, Halachmi S, Bronson R T, Weinberg R A. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 9.Kipling D, Cooke H J. Hypervariable ultra-long telomeres in mice. Nature. 1990;347:400–402. doi: 10.1038/347400a0. [DOI] [PubMed] [Google Scholar]
- 10.Lebel M, Leder P. A deletion within the murine Werner syndrome helicase induces sensitivity to inhibitors of topoisomerase and loss of cellular proliferative capacity. Proc Natl Acad Sci USA. 1998;95:13097–13102. doi: 10.1073/pnas.95.22.13097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li E, Bestor T H, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 12.Marciniak R A, Lombard D B, Johnson F B, Guarente L. Nucleolar localization of the Werner syndrome protein in human cells. Proc Natl Acad Sci USA. 1998;95:6887–6892. doi: 10.1073/pnas.95.12.6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin G M, Sprague C A, Epstein C J. Replicative life-span of cultivated human cells. Effects of donor's age, tissue, and genotype. Lab Investig. 1970;23:86–92. [PubMed] [Google Scholar]
- 14.Monnat R J, Jr, Hackmann A F, Chiaverotti T A. Nucleotide sequence analysis of human hypoxanthine phosphoribosyltransferase (HPRT) gene deletions. Genomics. 1992;13:777–787. doi: 10.1016/0888-7543(92)90153-j. [DOI] [PubMed] [Google Scholar]
- 15.Moser M J, Oshima J, Monnat R J., Jr WRN mutations in Werner syndrome. Hum Mutat. 1999;13:271–279. doi: 10.1002/(SICI)1098-1004(1999)13:4<271::AID-HUMU2>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 16.Ogburn C E, Oshima J, Poot M, Chen R, Hunt K E, Gollahon K A, Rabinovitch P S, Martin G M. An apoptosis-inducing genotoxin differentiates heterozygotic carriers for Werner helicase mutations from wild-type and homozygous mutants. Hum Genet. 1997;101:121–125. doi: 10.1007/s004390050599. [DOI] [PubMed] [Google Scholar]
- 17.Prowse K R, Greider C W. Developmental and tissue-specific regulation of mouse telomerase and telomere length. Proc Natl Acad Sci USA. 1995;92:4818–4822. doi: 10.1073/pnas.92.11.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salk D. Werner's syndrome: a review of recent research with an analysis of connective tissue metabolism, growth control of cultured cells, and chromosomal aberrations. Hum Genet. 1982;62:1–5. doi: 10.1007/BF00295598. [DOI] [PubMed] [Google Scholar]
- 19.Salk D, Au K, Hoehn H, Stenchever M R, Martin G M. Evidence of clonal attenuation, clonal succession, and clonal expansion in mass cultures of aging Werner's syndrome skin fibroblasts. Cytogenet Cell Genet. 1981;30:108–117. doi: 10.1159/000131597. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 21.Scappaticci S, Cerimele D, Fraccaro M. Clonal structural chromosomal rearrangements in primary fibroblast cultures and in lymphocytes of patients with Werner's Syndrome. Hum Genet. 1982;62:16–24. doi: 10.1007/BF00295599. [DOI] [PubMed] [Google Scholar]
- 22.Schulz V P, Zakian V A, Ogburn C E, McKay J, Jarzebowicz A A, Edland S D, Martin G M. Accelerated loss of telomeric repeats may not explain accelerated replicative decline of Werner syndrome cells. Hum Genet. 1996;97:750–754. doi: 10.1007/BF02346184. [DOI] [PubMed] [Google Scholar]
- 23.Spillare E A, Robles A I, Wang X W, Shen J C, Yu C E, Schellenberg G D, Harris C C. p53-mediated apoptosis is attenuated in Werner syndrome cells. Genes Dev. 1999;13:1355–1360. doi: 10.1101/gad.13.11.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tahara H, Tokutake Y, Maeda S, Kataoka H, Watanabe T, Satoh M, Matsumoto T, Sugawara M, Ide T, Goto M, Furuichi Y, Sugimoto M. Abnormal telomere dynamics of B-lymphoblastoid cell strains from Werner's syndrome patients transformed by Epstein-Barr virus. Oncogene. 1997;15:1911–1920. doi: 10.1038/sj.onc.1201377. [DOI] [PubMed] [Google Scholar]
- 25.Weeda G, Donker I, de Wit J, Morreau H, Janssens R, Vissers C J, Nigg A, van Steeg H, Bootsma D, Hoeijmakers J H J. Disruption of mouse ERCC1 results in a novel repair syndrome with growth failure, nuclear abnormalities and senescence. Curr Biol. 1997;7:427–439. doi: 10.1016/s0960-9822(06)00190-4. [DOI] [PubMed] [Google Scholar]
- 26.Yu C E, Oshima J, Fu Y H, Wijsman E M, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, Martin G M, Mulligan J, Schellenberg G D. Positional cloning of the Werner's syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]