Abstract
The BETA2 (neuroD) gene is expressed in endocrine cells during pancreas development and is essential for proper islet morphogenesis. The objective of this study is to identify potential upstream regulators of the BETA2 gene during pancreas development. We demonstrated that the expression of neurogenin 3 (ngn3), an islet- and neuron-specific basic-helix-loop-helix transcription factor, partially overlaps that of BETA2 during early mouse development. More importantly, overexpression of ngn3 can induce the ectopic expression of BETA2 in Xenopus embryos and stimulate the endogenous RNA of BETA2 in endocrine cell lines. Furthermore, overexpression of ngn3 could cause a dose-dependent activation on the 1.0-kb BETA2 promoter in islet-derived cell lines. Deletion and mutation analyses revealed that two proximal E box sequences, E1 and E3, could bind to ngn3-E47 heterodimer and mediate ngn3 activation. Based on these results, we hypothesize that ngn3 is involved in activating the expression of BETA2 at an early stage of islet cell differentiation through the E boxes in the BETA2 promoter.
The endocrine pancreas, which is organized as the islets of Langerhans, contains at least four distinct types of endocrine cells (α, β, δ, and PP). The differentiation and maturation of islet cells during development is a complex process controlled by a unique network of gene regulation. Recently, it has been demonstrated by gene targeting studies that several tissue-specific transcription factors, such as BETA2 (neuroD) (24, 25), PDX-1 (1, 27), Islet-1 (2), Nkx2.2 (42), PAX-6 (41), and PAX-4 (40), are involved in this process. These factors, alone or in concert, can activate the expression of genes encoding hormones, such as glucagon (9, 44), insulin (9, 25, 28), and somatostatin (3, 31). BETA2 (neuroD), a basic helix-loop-helix (bHLH) transcription factor, was isolated both as a transcriptional activator of the insulin gene (25) and as a differentiation factor of neurogenesis (17). BETA2 is selectively expressed in the developing endocrine pancreas, the small intestine, and the nervous system (17). It has been shown that BETA2 transactivates the insulin (25) and glucagon genes (9) by binding to the E box sequences localized in their promoters. Furthermore, the functional importance of BETA2 to pancreatic islet cell development has been demonstrated by loss-of-function studies (24). BETA2-deficient (BETA2−/−) mice die of severe diabetes caused by a major reduction in the number of β cells and a lack of proper islet formation. These results indicate that BETA2 plays an important role in maintaining the differentiation of endocrine cells and proper islet morphogenesis. Results obtained from BETA2-deficient mice also imply that the upstream factors controlling BETA2 expression are likely to be involved in the early events which determine endocrine cell differentiation. So far, numbers of a novel family of genes, the neurogenin genes (ngn) (19, 39), have been reported to be good candidates for upstream regulators of the BETA2 gene. During neuronal development, neurogenin family members are expressed earlier than BETA2, and they are expressed in either overlapping or adjacent domains (19). When ectopically expressed in Xenopus embryos (19), both mouse neurogenin 1 (ngn1) and Xenopus neurogenin-related-1 are able to induce ectopic expression of Xenopus neuroD, but not vice versa. More importantly, mice lacking ngn1 (18) and ngn2 (12) develop distinct defects in the cranial sensory ganglia and fail to express BETA2. Therefore, it is likely that the BETA2 gene is the direct downstream target of some neurogenin family members in the transcriptional cascade which controls neuronal development. Interestingly, the expression of ngn3 (39) in the developing pancreas also seems to precede and overlap with BETA2 expression, the latter being detected exclusively in the endocrine pancreas (24). Thus, we hypothesize that ngn3 plays a role in activating the transcriptional expression of BETA2 and in determining the cell fate of pancreatic endocrine cells. Very little is known about the cis elements and trans activators regulating the BETA2 promoter. In this study, we describe the cloning of the 5′ genomic sequence of the BETA2 gene and determination of the transcription start site and exon-intron junctions. We found that the 1.7-kb promoter reporter has the highest activity among a series of reporter constructs made from 5′ deletions of the 2.2-kb BETA2 promoter. We also demonstrate that the 2.2-kb promoter is capable of directing proper tissue-specific expression of the lacZ reporter gene in the pancreatic islets and neuronal tissues. In addition, the first 1.0 kb of the BETA2 promoter, which contains nine E box sequences, can confer cell-type-specific activity in vitro. The notion that ngn3 participates in regulating BETA2 gene expression during development is supported by our colocalization studies of ngn3 and BETA2 in the developing pancreas, which indicate that ngn3 expression partially overlaps that of BETA2. More importantly, we also provide evidence that injection of ngn3 mRNA into Xenopus embryos can cause ectopic expression of BETA2, and overexpression of ngn3 can stimulate the endogenous RNA levels of BETA2 in endocrine cells lines. Consistent with these data, transient transfection data indicate that overexpression of ngn3 causes a dose-dependent activation of the BETA2 promoter in islet-derived cell lines. While deletion analysis suggests that the three E boxes (E1, E2, and E3) in the first 419-bp region could be important for ngn3-mediated activation, the mutation analysis and gel shift results clearly indicate that the ngn3-E47 heterodimer transactivates the 419-bp BETA2 promoter by binding to the two E boxes (E1 and E3) in this region specifically. Collectively, our results suggest that ngn3 acts as an upstream regulator of the BETA2 gene and could be involved in the initiation step that switches on the BETA2 gene at early stage of islet cell differentiation.
MATERIALS AND METHODS
Isolation of the 5′ end of the BETA2 cDNA.
A λgt11 cDNA library of β-TC cells (10) was constructed previously (obtained from C. M. M. Stellrecht; M.-J. Tsai, unpublished results). To clone the 5′ end of the BETA2 cDNA from this cDNA library, PCR was performed with an oligonucleotide (B2S [5′-CCTGAGAACTGAGACACT-3′]) that is complementary to the encoding region of the BETA2 gene, combined with either of the two oligonucleotides (λgt5 [5′-CCTGAGAACTGAGACACT-3′] and λgt3 [5′-GACACCAGACCAACTGGTAATG-3′] that are specific for both ends of sequences flanking the EcoRI cloning site. PCRs, using either B2S and λgt5 or B2S and λgt3, were carried out at 95°C for 2 min, followed by 35 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min, followed by 5 min at 72°C. The amplified DNA was subjected to electrophoresis, gel purified, and subcloned into pCR2.1 vector (Invitrogen) using a TA cloning kit (Invitrogen).
RNase protection assay.
To generate the DNA template used in the RNase protection assay, a SacII-HindIII BETA2 genomic fragment, which spans 305 bp of the intron and ∼300 bp upstream of the first exon, was subcloned into pBluescript II KS(+) [pBSII-KS(+); (Stratagene]. The RNA probe was prepared by transcription of SacII-linearized DNA template using T3 polymerase. Followed by electrophoresis and gel purification, ∼105 cpm of probe was hybridized to 5 or 10 μg of total RNA from β-TC cells, yeast, and mouse liver tissues. The hybridization and RNase digestion procedures were performed with an RNase protection assay kit as instructed by the manufacturer (Ambion). Samples without RNase digestion were included as the control. To generate the molecular marker for RNA, 0.5 μg of the Century marker template (Ambion) was in vitro transcribed and labeled with [32P]UTP.
RNA isolation and primer extension.
Total cellular RNA was isolated from cultured β-TC cells and frozen 1-month-old mouse liver using TRIZOL (Gibco BRL) according to the manufacturer's instructions. The yeast RNA was purchased from the manufacturer (Gibco BRL). For the probe used in the assay, a 29-mer oligonucleotide complementary to the sequence located 15 bp upstream of the 3′ end of the first exon of the BETA2 gene was end labeled with T4 polynucleotide kinase. Primer extension was performed by adding 5 × 105 cpm of labeled probe to 20 or 100 μg of total cellular RNA in the hybridization buffer (40 mM PIPES [pH 6.4], 1 mM EDTA, 0.4 M NaCl; 50 μl in volume). Hybridization at 42°C overnight was followed by precipitation with 3 volumes of 1 M ammonium acetate and 4 volumes of isopropanol and a 70% ethanol wash. The precipitated mixture was dissolved in distilled water and subjected to extension using Superscript reverse transcriptase (RT; Gibco BRL). The extension was performed at 42°C for 2.5 h in 50 μl of buffer containing 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 1 mM deoxynucleoside triphosphate, 10 mM dithiothreitol, and 10 U of RNasin (Promega). After isopropanol precipitation, the products were subjected to electrophoresis on a 10% polyacrylamide gel. A dideoxy sequencing reaction was performed with the BETA2 genomic DNA and the same primer, and the reaction was loaded in adjacent lanes to determine the transcription start site.
DNA constructs.
A 14-kb NotI fragment containing the entire BETA2 gene isolated from a 129/Sv mouse genomic library was subcloned into pBSII-KS(+) and mapped as described previously (24). From this construct, a 4.5-kb 5′ flanking sequence-containing BssHII fragment was subcloned into pBSII-KS(+) and named pBSB2 (4.5 kb). The 2.2-kb SacI-EcoRI fragment of pBSB2 (4.5 kb) was then subcloned into a luciferase reporter vector pGL3-basic (Promega). A series of 5′ deletions were made in this plasmid, using the available restriction enzyme XbaI (−1.7 kb), NheI (−1.0 kb), SacII (−0.3 kb), and MluI (−0.1 kb). The resulting promoter reporters are B2 (1.7 kb), B2 (1.0 kb), B2 (0.3 kb), and B2 (0.1 kb). To generate the successive deletions of eight E boxes (E1 to E8) in the 1.0-kb promoter, PCRs were performed with nine lower-strand primers, which correspond the flanking sequences located 3′ from each E box, and an upper-strand primer which spans from −9 to +11. The upper-strand primer sequence is 5′-CCCAAGCTTGAATTCCTCGTGTCCCGGTG-3′. The nine lower-strand primers (and sequences) are E1-8 (−938 to −956), E1-7 (−838 to −858), E1-6 (−820 to −841), E1-5 (−763 to −782), E1-4 (−707 to −726), E1-3 (−400 to −419), E1-2 (−322 to −342), E1 (−251 to −270), and E (−) (−212 to −231). The resulting nine deletion constructs are B2E1-8 (−956 to +11), B2E1-7 (−860 to +11), B2E1-6 (−842 to +11), B2E1-5 (−782 to +11), B2E1-4 (−726 to +11), B2E1-3 (−419 to +11; =B2 (419 bp), B2E1-2 (−341 to +11), B2E1 (−296 to +11), and B2E (−) (−231 to +11). In addition, in the text or figure legends B2 (1.0 kb) is equivalent to B2E1-9 and B2 (231 bp) is equivalent to B2E(−).
To generate successive mutations in the three most proximal E boxes, a 155-bp promoter fragment which contains the three E boxes (E1 to E3) was amplified by PCR, digested, and subcloned into a pGL3-TATA vector. The primers used were 5′-GGAGTCTCTAACTGGCGA-3′ (lower strand) and 5′-TGCTCCTTCCTCCCCGGCAT-3′ (upper strand). The three E boxes were then mutated using a Quick Change site-directed mutagenesis kit (Stratagene) with primers specific for each E box that change CANNTG to TANNCT. The resulting mE1, mE2, and mE3 constructs were then used as templates to generate m(E1+E2), m(E1+E3), m(E2+E3), and m(E1+E2+E3) by using the same primers and mutagenesis method. The mutated E box oligonucleotide sequences are 5′-CTGGACCGGGAAGACTATACTGCGCATGCCGGGGAG-3′ (mutE1); 5′-AGGCAGGTTACGCTGTTCCCGGCTCTTGGCTGGA-3′ (mutE2); 5′-GTCTCTAACTGGCGATAGACTGGCCACTTTCTTCTG-3′ (mutE3). The mutated nucleotides in the E boxes are underlined. To generate the E47 expression vector, the HindIII-BstEII fragment of pSVE2-5 (14) was subcloned into HindIII-EcoRV sites of pCR3.1 (Invitrogen). To generate the expression plasmid full-length E47 (FL-E47), the full-length cDNA of E47 (37) was amplified using PCR and subcloned into EcoRI and XbaI sites of pCR3.1. The expression plasmid for ngn3 was constructed by subcloning into pCR3.1 the encoding region of ngn3, which was amplified from pSK-ngn3 (39) by PCR using the primers 5′-ACCCAAGCTTGCCACCATGGCGCCTCATCCCTTG-3′ (lower strand) and 5′-CGGGATCCTCACAAGAAGTCTGAG AACAC-3′ (upper strand). To add a FLAG peptide sequence (33) at the C terminus of ngn3, the encoding region of ngn3 was amplified by PCR with the ngn3-specific lower-strand primer mentioned above and an upper-strand primer tagged with the 18 nucleotides encoding the FLAG peptide. The sequence of the upper-strand primer is 5′-CGGGATCC TCAC T TG TCATCG TCATCC T TG TAG TCCAAGAAG TCTGAGAACACCAG-3′. The PCR product was digested with HindIII-BamHI and subcloned into the corresponding sites of pCR3.1. The TATA-luciferase reporters driven by three copies of each E box [(E1)×3-TATA, (E2)×3-TATA, and (E3)×3-TATA] were constructed by inserting three copies of each E box and the surrounding sequences into pGL3-TATA. The E boxes and surrounding sequences used were as follows: E1 (−230 to −247), E2 (−266 to −283), and E3 (−337 to −354). All of the constructs mentioned above were verified by sequencing analysis.
Cell culture and transfections.
The HIT-T15 M2.2.2 (36), β-TC (10), NIH 3T3, and HeLa cell lines were grown in Dulbecco modified Eagle's medium supplemented with 10% fetal bovine serum, 100 U of penicillin/ml, and 100 μg of streptomycin/ml. The HIT and β-TC cells were seeded at 5 × 105 cells per well in Falcon six-well dishes. The HeLa cells were seeded at 105 cells per well. The NIH 3T3 cells were seeded at 2 × 105 cells per well. Transient transfections were performed using FuGENE6 (Boehringer Mannheim) according to the manufacturer's instructions. The amount of each reporter plasmid used in transfection was 300 ng per well. For all other transfections, the amounts of DNA used are noted in the figure legends. Cells were harvested 24 to 36 h after transfections. Cell extracts were assayed for luciferase activity using the Promega luciferase system, and values (relative luciferase units [RLU]) were corrected for protein concentration or Rous sarcoma virus (RSV)-luciferase activity. Data are represented as means of triplicate values obtained from representative experiments. All transfections were repeated at least three times.
BETA2-lacZ transgenic constructs and generation of transgenic mice.
The BETA2 promoter lacZ transgene construct (BPIL-4) was generated by inserting the VspI-XhoI fragment of BP-SBC-1 into the corresponding sites upstream of the lacZ gene in plasmid lacZ-pA-SBC-2. BP-SBC-1 was constructed by inserting a 3.8-kb VspI-EcoRI BETA2 5′ flanking sequence, which includes the first exon and the intron in addition to a 2.2-kb promoter fragment, into the corresponding sites of SBC-1 vector (8). lacZ-pA-SBC-2 contains a 3-kb XbaI-PstI fragment of the lacZ gene (pPD46.21) (24) with a nuclear localization signal (obtained from Eric Olson, University of Texas Southwestern Medical Center, Dallas), fused upstream of a 0.7-kb human growth hormone poly(A) sequence. All the constructs were confirmed by restriction enzyme digestion and partial sequence analysis. The resultant transgene construct was then released from plasmid BPIL-4 by VspI digestion and purified from a low-melting-point gel after electrophoresis. The transgene DNA was microinjected into B6C3FI stud male (Harlan) fertilized one-cell ICR embryos. After microinjection, the fertilized embryos were transferred into pseudo-pregnant ICR recipient mothers (Harlan) to carry the embryos to term. Different batches of DNA were used to generate three lines transgenic mice. Genotypic analysis was performed by Southern analysis of the genomic DNA isolated from mouse tails using a portion of the lacZ gene as a probe.
X-Gal histochemistry and in situ hybridization.
For 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) histochemistry of BETA2-lacZ transgenic mice, the embryonic day 11.5 (E11.5) transgenic embryos or the pancreata from 7-month-old adult transgenic mice were removed, fixed in 2% paraformaldehyde at room temperature for 2 h, and subjected to whole mount X-Gal histochemistry as described elsewhere (6). After X-Gal staining, the tissues were dehydrated, embedded in paraffin, and sectioned at 10 μm. The sections were then counterstained with nuclear fast red and subjected to microscopic analysis. For combined X-Gal histochemistry and in situ hybridization, embryos of different developmental stages were obtained from the wild-type C57BL/6 female mice mated to male BETA2+/−mice which were generated previously (24). The embryos were then fixed in 4% paraformaldehyde at 4°C for 2 h and stained with X-Gal solution at 25°C overnight. The BETA2+/− embryos, which were genotyped by PCR as described previously (24), were then postfixed in 4% paraformaldehyde at 4°C overnight, embedded in paraffin, and sectioned at 6 μm. The sections were then subjected to in situ hybridization with a 35S-labeled antisense RNA probe which is complementary to the 3′ untranslated region (UTR) and the encoding region of ngn3. The procedures for probe preparation and in situ hybridization were described elsewhere (45). After in situ hybridization, the sections were processed for autoradiography using NTB-2 Kodak emulsion and exposed to 4 to 7 days at 4°C. Analyses were carried out using both light- and dark-field optics on a Zeiss microscope.
RT-PCR.
Pancreas total RNA, extracted with TRIZOL (Gibco BRL), was from mouse embryos of different developmental stages (E13.5 to E17.5), postnatal day 2 (P2) mice, and 2-month-old adult mice. First-strand cDNA synthesis was performed with approximately 200 ng of pancreas RNA using the Superscript preamplification system (Gibco BRL). One-tenth of the first-strand cDNA was used for PCR amplification. For BETA2 and the internal control L19 (29), the following parameters were used: 94°C for 1 min, followed by 25 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min. For ngn3, 30 cycles were used with the parameters mentioned above. Following PCR, one-fifth of the reaction products were loaded on a 2% gel and subjected to electrophoresis. The primers used in PCR were 5′-GACAGAGTCTTGATGATCTC-3′ (L19, upper strand), 5′-CTGAAGGTCAAAGGGAATGTG-3′ (L19, lower strand), 5′-AAGCACAGTGGGTTCGTTTC-3′ (BETA2, upper strand), 5′-CATCAATGGCAACTTCTCTTTC-3′ (BETA2, lower strand), 5′-CTTCACAAGAAGTCTGAGAACACCAG-3′ (ngn3, upper strand), and 5′-CTGCGCATAGCGGACCACAGCTTC-3′ (ngn3, lower strand).
Electrophoretic mobility shift assay.
The E1 (−224 to −254), E2 (−260 to −289), and E3 (−331 to −360) double-stranded oligonucleotides were end labeled with [α-32P]dCTP, using the Klenow fragment, to a specific activity of ∼108 cpm/μg. E47 and ngn3 proteins used in gel shift assay were synthesized from plasmids pCR3.1-E47, pCR3.1-ngn3, and pCR3.1-ngn3-FLAG, using the Promega TNT coupled transcription-translation kit according to the manufacturer's instructions. Parallel reactions with [35S]methionine or unlabeled methionine were performed. Labeled proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 10% gel to ensure proper synthesis. Binding reaction mixtures included 10 mM Tris-HCl (pH 7.5), 50 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 5% glycerol, 200 ng of poly(dI-dC), ∼3 × 104 cpm of each probe, and various amounts of in vitro-translated proteins. Protein-DNA complexes were incubated at room temperature for 20 min and then resolved by SDS-PAGE on a 5% polyacrylamide gel. The mutated E box oligonucleotides used for competition studies were 5′-GGACCGGGAAGACTATACTGCGCATGCCGGG-3′ (mutE1, upper strand), 5′-GGACCCGGCATGCGCAGTATAGTCTTCCCGG-3′ (mutE1, lower strand), 5′-GTCTAACTGGCGATAGACTGGCCACTTTCTT-3′ (mutE3, upper strand), and 5′-GGGAAGAAAGTGGCCAGTCTATCGCCAGTTA-3′ (mutE3, lower strand). The mutated nucleotides are underlined. For supershift assay, 1 μl of 1:10-diluted anti-E47 antibody (Santa Cruz Biotechnology) or 1 μl of 1:10 diluted anti-FLAG peptide antibody (Biological Research and Imaging Products) was added to the mixture after the DNA probes were preincubated with proteins for 5 min. The whole mixture was then incubated at room temperature for another 20 min. To obtain cellular extracts from COS-1 cells, COS-1 cells were cotransfected with pCR3.1-E47 and pCR3.1-ngn3-FLAG by the DEAE-dextran suspension method (7), and cellular extracts were collected as described previously. For gel shift assay, 10 μg of cellular extract was used for each reaction. The reaction buffer and conditions used were similar to those described above except that the reaction was carried out at 4°C and 1.2 μg of poly(dI-dC) was added in the reaction buffer.
Xenopus embryo injection and in situ hybridization.
Capped ngn3 RNA was prepared from PCR3.1-ngn3 linearized with AflIII, using a T7 mMessage mMachine kit (Ambion) as recommended by the manufacturer. Xenopus embryos, prepared as described previously (11), were injected with ngnr3 RNA in the animal region of the two prospective left blastomeres of the four-celled embryos. Embryos were cultured in 1× MBS (30) containing 3% Ficoll until 3 to 4 h after fertilization and then transferred to 0.1× MBS containing 3% Ficoll. After overnight incubation, embryos were transferred to 0.1× MBS for subsequent culture. Embryos were collected at approximately stage 26 (26) and fixed for 1 to 2 h in MEMFA (13) prepared using paraformaldehyde and then dehydrated in methanol. In these experiments, green fluorescent protein (GFP) RNA was coinjected as a lineage tracer or injected alone as a control for nonspecific effects due to the microinjection procedure. Before fixation for in situ hybridization, embryos were scored for GFP expression by fluorescence microscopy (data not shown). Embryos not expressing GFP were discarded. In another experiment, embryos were subjected to whole-mount immunostaining using an antibody which could detect GFP expression following in situ hybridization to verify that ectopic BETA2 expression was on the injected side of the embryos (data not shown).
In situ hybridization was performed with a digoxygenin-labeled Xenopus BETA2 probe (see below) as described previously (13) and modified by Turner and Weintraub (43) except that the color reaction substrate solution contained 10-fold less nitroblue tetrazolium, as suggested by Ma et al. (19). To prepare the probe, a fragment of the Xenopus BETA2 cDNA (from nucleotides 501 to 1251, [17]) was isolated by PCR from stage 17 cDNA, using the primers CGGAATTCCAGACCTGGTGTCCTTTGTAC and GCTCTAGAAGTGTCGTATTGGAAGGAGGTG. The resulting 750-bp fragment was digested with EcoRI and XbaI (underlined in the primer sequences) and ligated with similarly digested pBSII KS. Digoxygenin-labeled riboprobe was generated from this plasmid linearized with EcoRI using T7 RNA polymerase (Boehringer Mannheim Biochemicals) as recommended by the manufacturer.
Generation of stably transfected cell line and Northern blotting.
The STC-1 (34) and β-TC cell lines were grown in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin (Life Sciences-Gibco BRL). Ten-centimeter-diameter dishes of STC-1 and β-TC cells were transfected by the FuGENE6 reagent (Boehringer Mannheim Biochemicals) with either pCR3.1-ngn3 (containing the neomycin resistance gene) or the control pCR3.1 vector without ngn3 insert. The cells were then grown in medium for 2 days. Cells were then split into five 10-cm-diameter plates with medium containing 500 μg of Geneticin (Life Sciences-Gibco BRL) per ml, which was determined earlier to be the effective killing dose for both the STC-1 and β-TC cells. Cells were grown in selection medium for 2 weeks, and colonies were pooled by trypsin-EDTA digestion and expanded. For Northern blotting, total RNA was prepared using RNeasy kit (Qiagen) as recommended by the manufacturer. STC-1 total RNA (10 μg) or β-TC total RNA (5 μg) was denatured, and subjected to electrophoresis in 2.2 M formaldehyde–1% agarose gel in morpholinepropanesulfonic acid (MOPS) buffer at 120 V for 6 h, and transferred to nylon membranes (Hybond-NX; Amersham) overnight. RNA was cross-linked to the membranes by UV irradiation. A 1.1-kb BssHII-SpeI fragment of mouse BETA2 cDNA, a 700-bp HindIII-BamHI fragment of pCR3.1-ngn3 that contained the full-length mouse ngn3, and an RT-PCR-generated clone of mouse L19 mRNA were used as the templates to synthesize [α-32P]dCTP-labeled probes by random priming. Prehybridization and hybridization were carried out in ExpressHyb hybridization solution (Clontech) at 60°C overnight. Unhybridized probes were removed by washing twice in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.5% SDS at 50°C for 20 min and twice in 0.2× SSC–0.1% SDS at 55°C for 15 min. Filters were then processed for autoradiography. Quantitative analysis of the hybridization signals was performed by scanning the phosphor screen (Molecular Dynamics) with a densitometer (Molecular Dynamics).
RESULTS
Analysis of the 5′ flanking sequences of the mouse BETA2 gene.
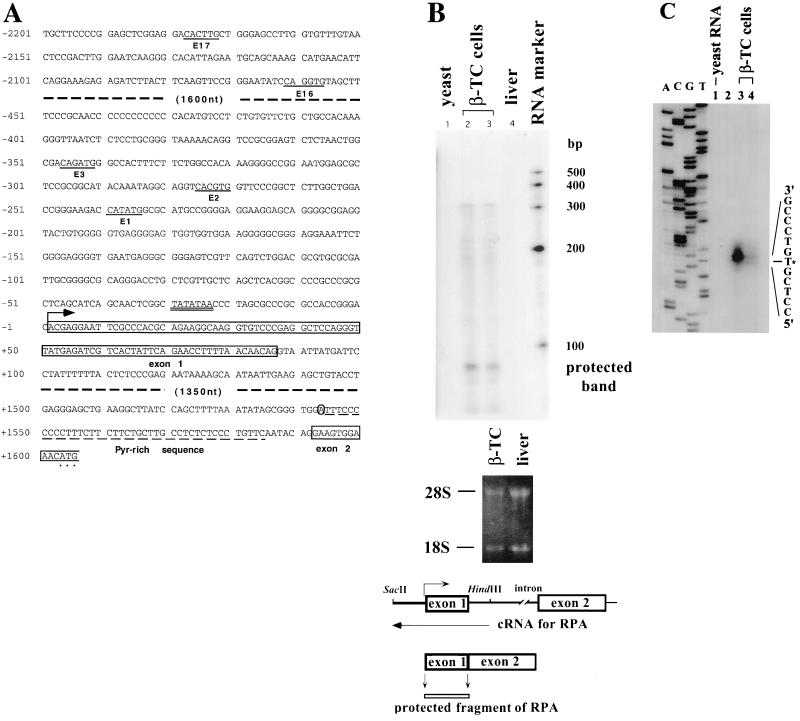
We isolated a 14-kb genomic fragment from a mouse genomic library to characterize the promoter of the mouse BETA2 gene. Preliminary sequence analyses indicated that the fragment contained the BETA2 gene and a ∼3.8-kb region upstream of the first ATG. Recently, two members of BETA2/neuroD family, the mouse NEX-1 (4) and neuroD2 (20) genes, were cloned and analyzed. The results suggest that members of the BETA2/neuroD gene family are composed of two exons; the first exon contains the 5′ UTR, and the second includes the coding region and 3′ UTR. Previously, an incomplete mouse BETA2 cDNA sequence was reported (17), which contains the coding region and a very short 5′ UTR. To identify the full-length cDNA of BETA2, we used PCR to amplify the 5′ UTR of BETA2 from a cDNA library of the β-TC cell line that expresses endogenous mouse BETA2 (25). The longest 5′ UTR sequence that we cloned was 79 bp in size. We then compared the partial 5′ UTR sequence to the known genomic sequence and determined the size of the intron, which is 1.5 kb, and the exon-intron junctions (Fig. 1A). The intron is flanked by consensus sequences for splice donors and acceptors, and the branch point is located 60 nucleotides upstream of the coding sequences (Fig. 1A). An RNase protection assay was performed to determine the exact transcription start site, using a 32P-labeled antisense cRNA probe that spans 305 nucleotides of the intron and 323 nucleotides of the 5′ upstream region (Fig. 1B). A major protected band of approximately 85 bp, which is equivalent to the size of the first exon, was observed when the probe was hybridized to β-TC cell total RNA (Fig. 1B). In contrast, no protected band was observed when the same amount of mouse liver and yeast RNAs was used. In addition to the major protected band, we observed several minor and longer protected bands, indicating the existence of additional minor transcription start sites in this promoter.
FIG. 1.
(A) Partial 5′ flanking nucleotide sequence of the BETA2 gene. The sequence includes the promoter sequence from bp −2201 to −2052, the first exon and surrounding sequences (bp −451 to +150), and both 5′- and 3′-end sequences of the intron. The E box consensus sequences are underlined and numbered (from the transcription start site). The three most proximal E boxes (E1, E2, and E3) and surrounding sequences are included because of their importance in BETA2 promoter activity. The TATA box is double underlined. The first exon sequence, starting from the transcription start site (marked by an arrow) and terminating at the donor site for splicing, is boxed and labeled. The putative branch point, which is an adenine (A), is circled. The pyrimidine-rich region is labeled and underlined by a dash line. The first 14 nucleotides (nt) of the second (encoding) exon are also boxed to indicate the splicing acceptor site. Asterisks mark the first ATG. (B) Determination of transcription start site by RNase protection assay. The upper panel shows the result of RNase protection assay performed with total RNA from β-TC cells (10 μg in lane 2; 5 μg in lane 3), yeast (10 μg, lane 1), and mouse liver (10 μg, lane 4). The middle panel is the RNA electrophoresis gel stained with ethidium bromide. Clear 18S and 28S bands suggest that most RNAs from β-TC cells and liver were not degraded. The lower panel shows a schematic representation of the BETA2 gene and the design of the antisense probe used in the assay. The size of the protected band equals that of the first exon. A major protected band, which is ∼85 bp in size, is observed in the upper panel and labeled. (C) Determination of transcription start site by primer extension assay performed with total cellular RNA from yeast (lane 1, 100 μg) and β-TC cells (lane 3, 100 μg; lane 4, 20 μg). A 29-bp primer complementary to the 3′ end of the first exon was used in the assay. Lane 2 does not contain any reaction. A dideoxy sequencing reaction using the genomic BETA2 DNA and the same primer was performed and loaded in parallel on the left side of the gel for better estimation of the location of the transcription start site. An asterisk marks the mapped transcription start site.
To further confirm the result, we performed a primer extension assay using an oligonucleotide primer selected from the first exon sequence. The result indicated that the putative transcription start site is located at 97 bp upstream of the coding region (Fig. 1C). The RNase protection assay result is consistent with the primer extension result, considering that the second exon contains 11 untranslated nucleotides before the first ATG (Fig. 1A). In addition, there is a typical TATA box located 25 bp upstream of the first exon (Fig. 1A). Therefore, we conclude that the major transcription start site of the BETA2 gene is positioned at 97 bp upstream of the translation start site, and the size of the first exon is 86 bp.
The promoter activity of BETA2 in transient transfection and in transgenic mice.
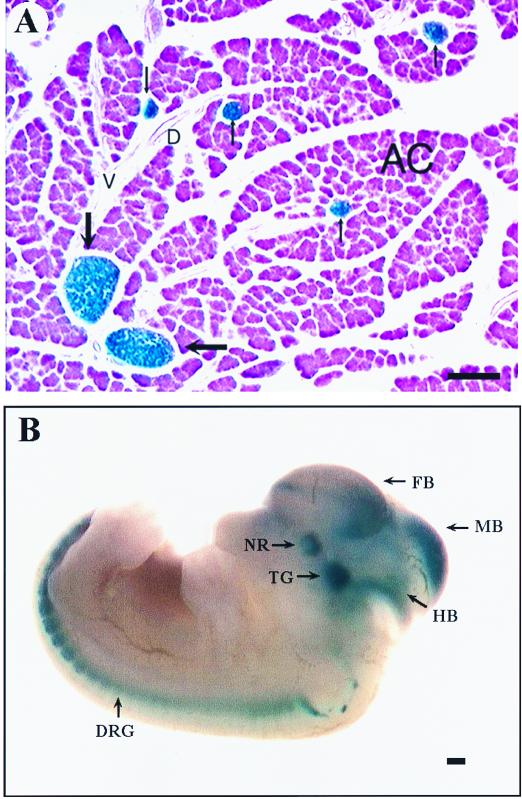
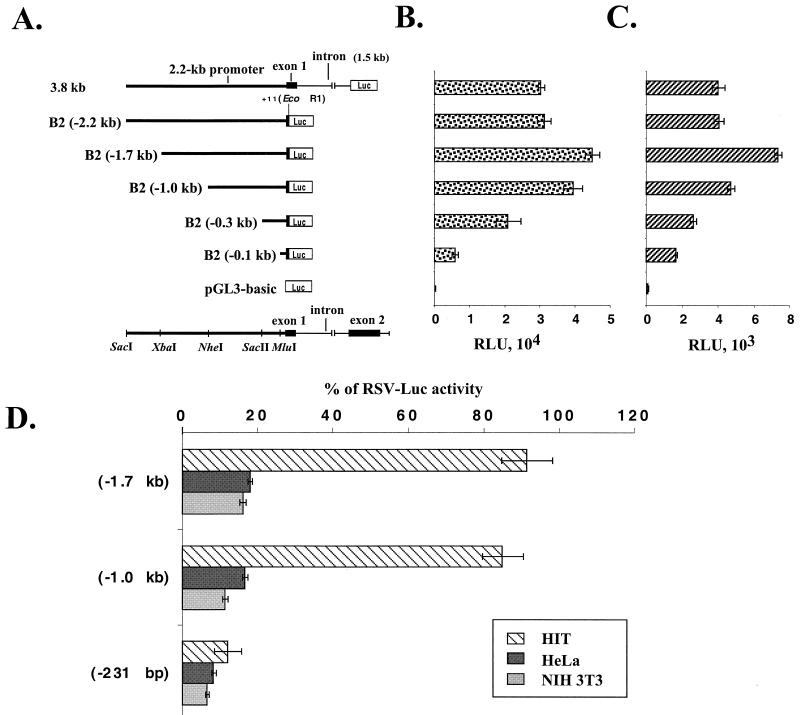
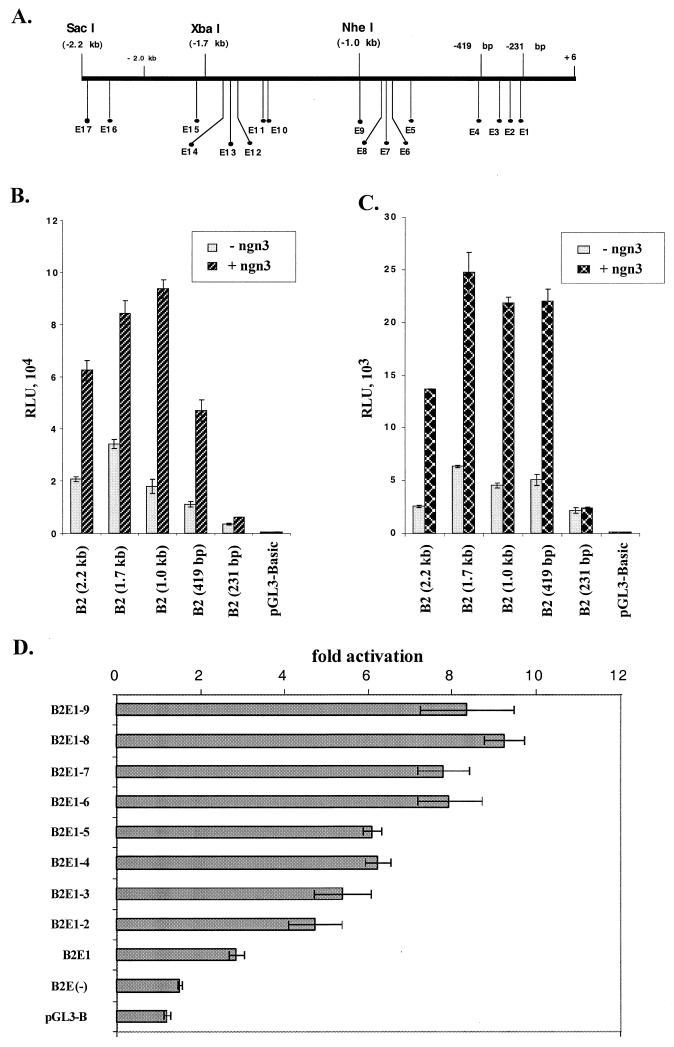
To determine if the 5′ flanking sequence that we isolated is sufficient for islet-specific expression of BETA2, transgenic mice were generated using as a reporter gene lacZ, fused downstream of a 3.8-kb 5′ sequence of the BETA2 gene, including the 2.2-kb BETA2 promoter, the untranslated exon, and the intron. In all three lines of transgenic mice examined by X-Gal histochemistry, the 2.2-kb 5′ flanking region is sufficient to direct appropriate tissue-specific expression of the lacZ gene in the islets of Langerhans of adult mice (Fig. 2A). In addition, strong β-galactosidase (β-Gal) activity can be detected in the adult mouse brain, including the cerebellum, the hippocampus, the pituitary gland, the retina, and the cerebral cortex. Also, transgenic embryos carrying the same transgene were analyzed at E11.5. Expression of LacZ was found only in the pancreas and the nervous system, including the brain, the neural retina, trigeminal ganglia, and dorsal root ganglia (Fig. 2B). Since the expression patterns of the BETA2 promoter-lacZ in embryos and adult mice seem to be indistinguishable from what we observed in BETA2+/− mice and what others have reported, we conclude that the 2.2-kb 5′ flanking sequence contain most if not all cis elements important for tissue-specific expression of BETA2. To identify promoter sequences important for BETA2 transcription, we linked the 2.2-kb BETA2 fragment (−2191 to +11) to a luciferase reporter gene and generated a series of 5′ deletion constructs using available restriction enzyme sites (Fig. 3A). These constructs, together with another 3.8-kb construct that contains additional sequences for the untranslated exon and the intron, were transiently transfected into two cell lines expressing endogenous BETA2. As shown in Fig. 3B and C, the 3.8-kb construct had activity similar to that of the 2.2-kb construct alone in both HIT and β-TC cells. In addition, we consistently observed a slight increase in promoter activity when the fragment between −2.2 and −1.0 kb was deleted (Fig. 3B and C), suggesting the existence of a negative regulatory element(s) in this region. Further deletion from −1.0 to −0.1 kb resulted in a progressive decrease of reporter activities, which indicated that the proximal 1.0-kb region contains cis elements important for the BETA2 transcription (Fig. 3B and C).
FIG. 2.
The 2.2-kb promoter sequence of the BETA2 gene can direct tissue-specific expression of LacZ in pancreatic islets and neurons of transgenic mice. (A) In the adult transgenic mouse pancreas, β-Gal activity is restricted to well-formed islets of Langerhans (thick arrows) and in small clusters of islet cells (thin arrows). No β-Gal activity is found in exocrine acini (AC), vessels (V), and ducts (D). (B) Whole-mount X-Gal staining of an E11.5 transgenic embryo carrying the BETA2 promoter–lacZ transgene. High levels of β-Gal activity are seen in the dorsal root ganglia (DRG), trigeminal ganglia (TG), neural retina (NR), forebrain (FB), midbrain (MB), and hindbrain (HB). Scale bars equal 200 μm (A) and 40 μm (B).
FIG. 3.
Activities of BETA2 promoter deletion constructs in HIT and β-TC cells. (A) Schematic diagram of the BETA2 promoter deletion constructs subcloned in pGL3-basic. At the top is the 3.8-kb construct which contains a 2.2-kb promoter (thick line), the first exon (solid box), and the intron (thin line). All other constructs are named according to the location (kilobase) of the 5′ end of each promoter fragment relative to the transcription start site. The restriction enzymes used to make the deletions are indicated on the bottom. (B and C) Comparison of activities of different deletion constructs in HIT (B) and β-TC (C) cells. Luciferase (Luc) activities (expressed as RLU) were corrected by protein concentrations and presented as means of triplicate values (± standard deviation of the mean). All transfection assays were performed in triplicate and repeated at least three times. (D) Both the 1.7- and 1.0-kb BETA2 promoter fragments can confer cell-type-specific activity. Transient transfections were performed with HIT, HeLa, and NIH 3T3 cells. For comparison of luciferase activities in different cell lines, an RSV promoter-driven luciferase construct (RSV-Luc) was transfected to each cell line in parallel wells. The activities of deletion constructs from a representative experiment were presented as percentage of the RSV-Luc activities (±standard deviation of the mean). Transfections were performed in triplicate and repeated three times.
In addition, we tested the cell-type-specific activity of different deletion constructs in both BETA2-expressing and -nonexpressing cell lines. Interestingly, both the 1.7- and 1.0-kb fragments had significantly higher activity in HIT cells than in BETA2-nonexpressing cell lines such as HeLa and NIH 3T3 (Fig. 3D). However, further deletion to −231 seemed to decrease the cell-type-specific activity in HIT cells compared to the activity in HeLa and NIH 3T3 cells (Fig. 3D). This result suggested the existence of a cell-type-specific element(s) between −1.0 kb and −230 bp in the BETA2 promoter.
The spatial expression of ngn3 partially overlaps that of BETA2 during early islet cell differentiation.
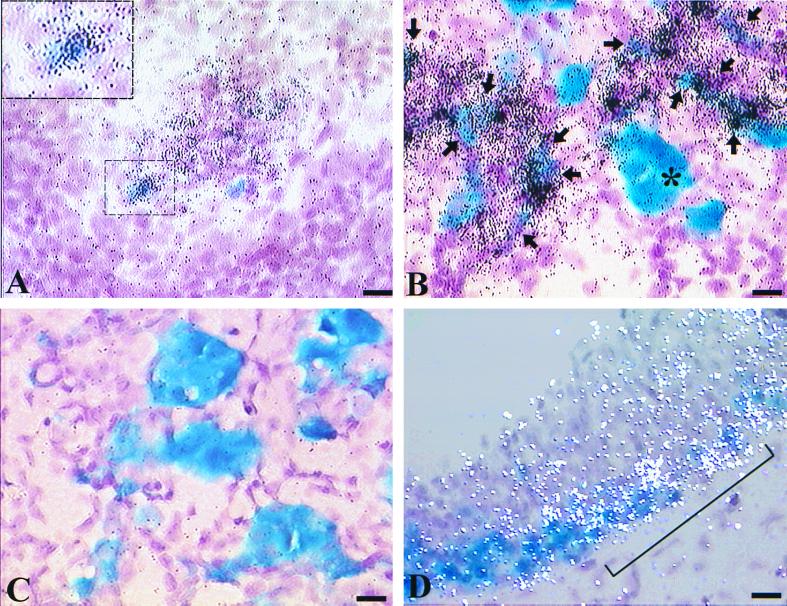
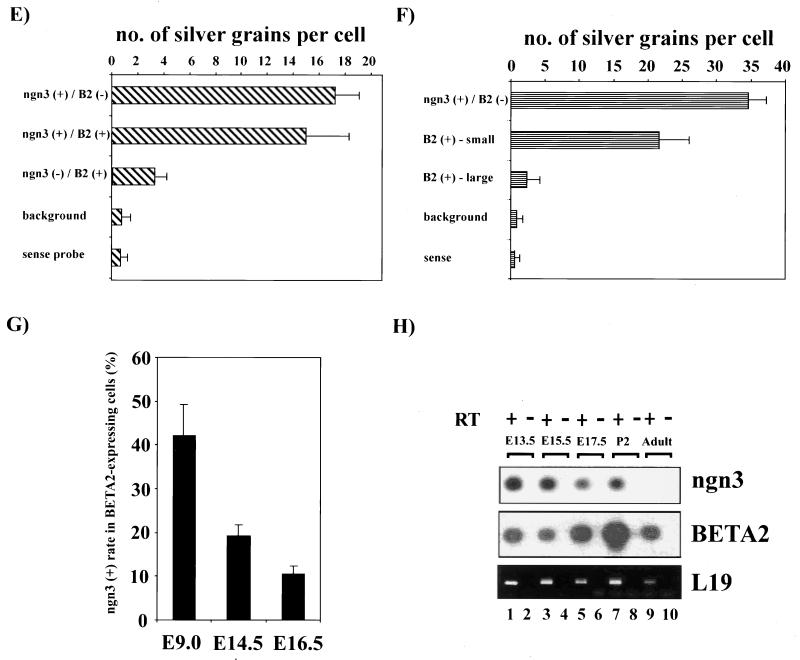
Sommer et al. (39) have demonstrated that ngn3 expression seems to precede that of BETA2 in the mouse endocrine pancreas. In that study, a comparison of in situ hybridization on adjacent sections of the pancreas indicates that ngn3-expressing cells are localized closely to the area where BETA2 is detected. Thus, it is likely that ngn3, like ngn1 and ngn2 in the nervous system (12, 18), is an upstream activator of BETA2 in the developing pancreas. However, the study did not address the question of whether ngn3 and BETA2 are indeed expressed in the same cells during development. To address this question, we performed colocalization studies on the embryonic tissues from the previously generated BETA2+/− mouse (24), in which the lacZ gene replaces one copy of the BETA2 gene, so that the expression of BETA2 could be followed by X-Gal staining. BETA2+/− embryos of different stages were first subjected to standard whole-mount X-Gal staining. The embryonic sections were then hybridized with either the antisense (Fig. 4A, B, and D) or sense control (Fig. 4C) probes for ngn3. As shown in Fig. 4A, at E9.0, when the first pancreatic tissue starts to arise from the foregut, ngn3 mRNA can be easily detected in the foregut region where a small number of BETA2-positive cells (Fig. 4A) can also be seen. Interestingly, at this stage, about 40% of BETA2-positive cells coexpressed the ngn3 transcript (Fig. 4A, E, and G). At later stages such as E14.5 (Fig. 4B and G) and E16.5 (Fig. 4G and data not shown), ngn3 expression still partially colocalized with BETA2. Interestingly, most of those cells coexpressing BETA2 and ngn3 were weakly positive for β-Gal activity and existed either as a single isolated cell or as small clusters (Fig. 4B and F), suggesting that these cells were at the earlier differentiation stage, when the BETA2 gene was just switched on. In contrast, most strongly X-Gal-positive cells were grouped in larger clusters and expressed little ngn3 (Fig. 4B). Moreover, the percentage of BETA2-positive cells which coexpress ngn3 decreased gradually from E9.0 to E16.5 (Fig. 4G), suggesting that as more endocrine cells aggregate and differentiate at later developmental stages, they coexpress less BETA2 and ngn3. Taken together, the observation that ngn3 expression partially overlaps that of BETA2 suggests that ngn3 is a pro-endocrine cell-specific gene and is likely involved in switching on BETA2 expression at the early stage of islet cell differentiation. The expression of ngn3 in the pancreas seems to persist until the newborn stage and disappears in adults, since ngn3 RNA could still be detected using RT-PCR in the pancreas of postnatal day 2 mice but not in the adult mice (Fig. 4H). In addition, we also observed intermediate regions coexpressing ngn3 and BETA2 in the hypothalamus and the spinal cord at E10.5 (Fig. 4D). Again, this suggested that ngn3 could be an upstream trans activator of the BETA2 gene in these regions. Based on these observations, we propose that at the early stage of islet cell differentiation, there exists a transitional cell population that coexpresses BETA2 and ngn3. The function of ngn3 in these cells can be potentially involved in switching on the initial expression of BETA2. Once BETA2 expression is initiated in these cells, BETA2 itself, or other transcription factors, may replace ngn3 and maintain the expression of BETA2.
FIG. 4.
Partial colocalization of ngn3 and BETA2 in the developing pancreas and the brain. (A) Sagittal section through the foregut area of an E9.0 BETA2+/− embryo. BETA2 expression (blue, X-Gal staining) and ngn3 expression (black silver grains, in situ hybridization) are partially colocalized (bracket). The region marked by a bracket is magnified in the inset, showing a cell coexpressing BETA2 and ngn3. (B) Sagittal section of the BETA2+/− pancreas at E14.5. Colocalization of BETA2 (blue) and ngn3 (black grains) is seen in some cells (arrows) that have weaker β-Gal activity and in smaller clusters (fewer than five cells). The larger clusters (more than five) of BETA2-positive cells (∗) are strongly X-Gal positive and express little ngn3. (C) The in situ hybridization of E14.5 pancreas using the sense probe for ngn3. (D) Sagittal section of the hypothalamus region of an E10.5 embryo. The region coexpressing BETA2 (blue) and ngn3 (white silver grains) is shown (bracket). Scale bars equal 10 μm. (E) Distribution of ngn3 signal (average number of silver grains per cell ± standard deviation) in BETA2-positive cells and other cells in the foregut region at E9.0. The number in each column represents the average of 50 cells from four embryos. The ngn3 (+) cell is arbitrarily defined as a cell with more than 10 silver grains. (F) Distribution of ngn3 signal (average number of silver grains per cell ±standard deviation) in BETA2-positive cells and other cells in the pancreas at E14.5. The number of each column represents the average of 200 cells from three embryos. The BETA2-positive [BETA2(+)] cell clusters were classified as small when fewer than five cells were found; otherwise, they were classified as large. (G) The percentage of BETA2-positive cells which express ngn3 decreases in later development. The data (average ± standard deviation) were obtained by counting 50 BETA2-positive cells from four E9.0 embryos and 300 cells from three E14.5 or E16.5 embryos. (H) Expression of ngn3 and BETA2 in the pancreas during development. Total RNA was extracted from the pancreas from different stages of mouse embryos. After DNase I treatment, RT-PCR was performed with approximately same amount of RNA and primer pairs specific to ngn3, BETA2, and the housekeeping gene L19. As a control, the same amount of RNA was subjected to PCR directly without RT treatment. After electrophoresis, the gels (except L19) were transferred to nylon membranes and subjected to Southern blotting using ngn3- or BETA2-specific probes complementary to the central region of the specific fragments amplified.
Induction of ectopic BETA2 expression by ngn3 mRNA in Xenopus embryos.
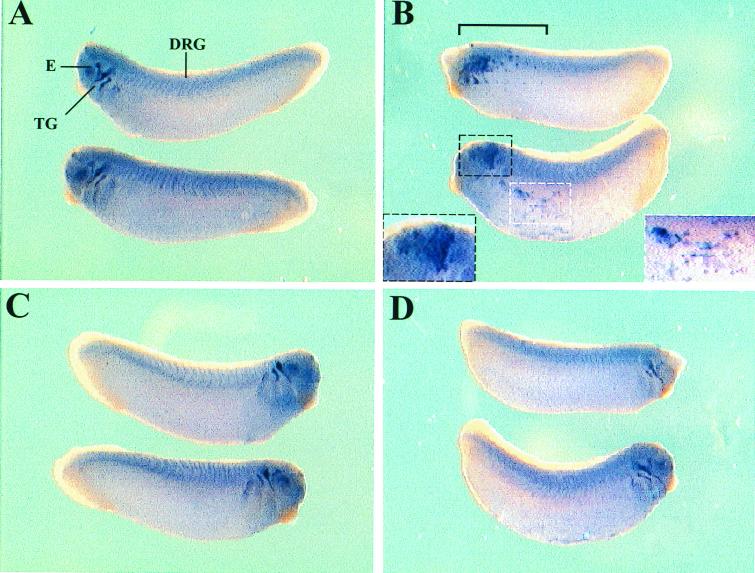
The observation that the ngn3 expression pattern partially overlap that of BETA2 prompted us to clarify whether ectopic ngn3 expression can induce BETA2 expression during embryonic development. Xenopus embryos at the four-cell stage were injected with either ngn3 mRNA or control GFP mRNA and allowed to develop until approximately stage 26. Whole-mount in situ hybridization of BETA2 was performed to examine the expression pattern of BETA2. In addition to GFP-injected embryos (Fig. 5A and C), the uninjected sides of ngn3-injected embryos were included as the control (Fig. 5D). On the ngn3-injected side of embryos, ectopic BETA2 expression was found in the superficial layer of the trunk (Fig. 5B; 68% of embryos examined; 28 embryos from three independent experiments). The ectopic expression of BETA2 was not seen on the uninjected side of the same embryos or in the GFP-injected embryos (0% of embryos examined; 30 embryos from three independent experiments). Moreover, ngn3 injection also caused abnormal expansion of BETA2-expressing area in the head region (Fig. 5B), which obscured the normal expression of BETA2 in the eyes and trigeminal ganglia seen in the uninjected embryos (Fig. 5A, C, and D). These data suggest that ngn3 is able to induce the expression of BETA2 mRNA in vivo.
FIG. 5.
Injection of ngn3 can induce ectopic expression of BETA2 in Xenopus embryos. (A) The injected sides of GFP-injected embryos, which were included as the control. Normal expression of BETA2 is seen in the eyes (E), trigeminal ganglia (TG), and dorsal root ganglia (DRG). (C) Uninjected side of the same GFP-injected embryos. (B) The injected sides of ngn3-injected embryos. In the lower embryo, the two areas that express ectopic BETA2 are marked by black (left) and white (right) dashed lines and are magnified in two insets at the left and right corners, respectively. In the upper embryo, the region expressing an ectopic or abnormal amount of BETA2 is also marked (bracket). In addition, in both embryos the expression of BETA2 in the head region is expanded and obscures the normal expression of BETA2 in the eyes and trigeminal ganglia. (D) The uninjected side of ngn3-injected embryos. Original magnification, ×3.125.
ngn3 up-regulates BETA2 gene transcription in endocrine cells.
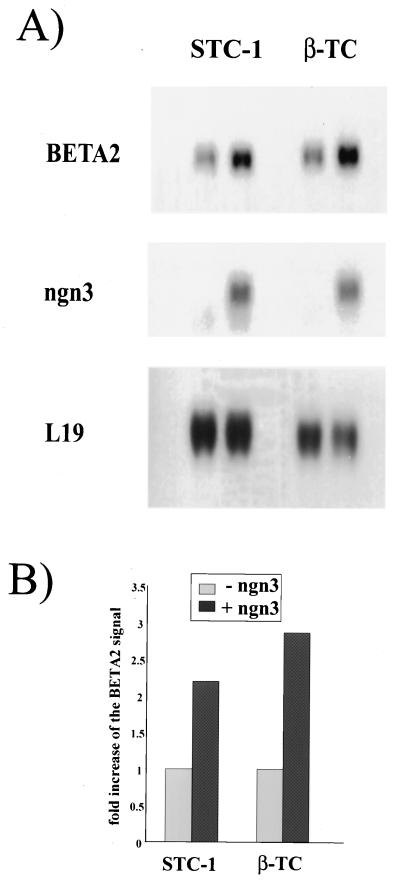
To determine whether overexpression of ngn3 can alter the level of endogenous BETA2 transcript, we established cell lines overexpressing ngn3 by stably transfecting STC-1 (34) and β-TC cells with the expression vector containing ngn3 cDNA (Fig. 6A, lane 2 and 4). As a control, cell lines stably transfected with control expression vector without the ngn3 insert (Fig. 6A, lane 1 and 3) were also established. Northern blotting was performed with total RNA from cells, and the same blots were hybridized with probes for BETA2, ngn3, and L19 (29), which is an RNA control to indicate equal loading in the filters. As shown in Fig. 6A, overexpression of ngn3 in both STC-1 and β-TC cells can lead to a two- to threefold increase (Fig. 6B) of BETA2 RNA signal compared to control cells. Thus, the results suggest that ngn3 up-regulates the level of BETA2 gene expression.
FIG. 6.
Stimulation of BETA2 transcription by overexpression of ngn3 in endocrine cells. (A) Both STC-1 and β-TC cells were stably transfected with either pCR3.1-ngn3 (lanes 2 and 4) or control pCR3.1 empty vector (lanes 1 and 3). After selection with G418, the surviving colonies were pooled and expanded in medium containing G418. Northern blotting was performed with total RNAs from transfected cells. Ten and 5 μg of total RNAs were used for STC-1 and β-TC cells, respectively. The same blots were hybridized with probes for BETA2, ngn3, and control L19 separately. (B) Fold increase of the BETA2 RNA signal in ngn3-overexpressing cells. The levels of BETA2 RNA signals shown in the graph were normalized against those of L19, which was used as a loading control. Quantitative analyses of Northern blot signals were performed by scanning the phosphor screen using a densitometer.
ngn3 up-regulates BETA2 promoter activity in islet-derived and non-islet cell lines.
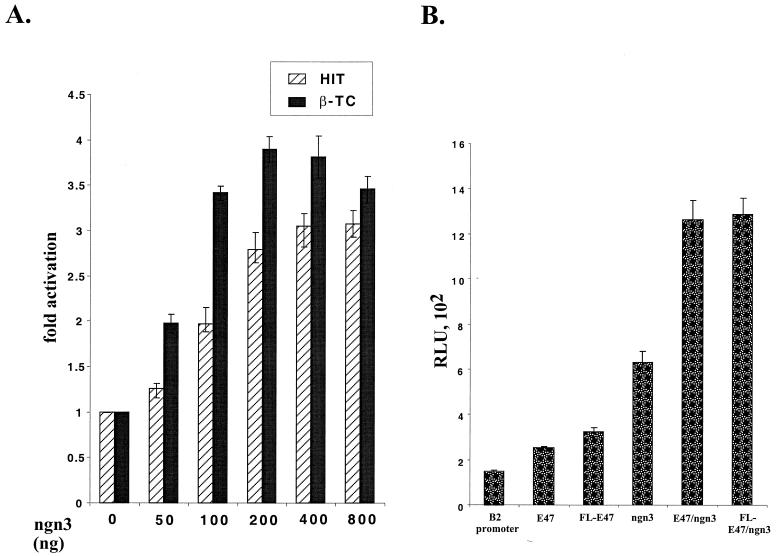
Since overexpression of ngn3 can induce ectopic expression of BETA2 both in Xenopus embryo injection and in endocrine cell lines, we next examined whether the BETA2 promoter contains potential binding sites for bHLH factors. A preliminary analysis of the 2.2-kb 5′ flanking region revealed the existence of 17 putative E boxes, 9 of them located in the proximal 1.0-kb sequence (see Fig. 8A). In addition, a computer-based analysis indicated that two of nine E boxes in the 1-kb promoter have the flanking sequences homologous to the known MyoD binding sites (21). To determine whether ngn3 is capable of stimulating the BETA2 promoter, ngn3 was transiently overexpressed in two BETA2-expressing cell lines, HIT and β-TC, with the BETA2 promoter reporter constructs. As shown in Fig. 7A, in both cell lines ngn3 clearly exerted a dose-dependent activation effect on the 1.0-kb promoter reporter. We also cotransfected these cells with the same amount of both cytomegalovirus (CMV)-ngn3 and CMV-E47; however, CMV-E47 did not further increase the activation ability of ngn3 in these cells (data not shown), possibly because HIT and β-TC cells already had an abundant amount of E47 (23) and other ubiquitous bHLH factors, such as HEB/BETA1 (32, 37, 47). In contrast, in HeLa cells, coexpression of CMV-E47 further increased the ability of ngn3 to activate the BETA2 promoter (Fig. 7B). Since the initial construct of E47 contained a truncated form of E47 that lacked ∼200 bp at the N terminus, we also included a CMV expression vector containing the full-length cDNA for E47 (Fig. 7B, FL-E47). When transfected alone, it had a slightly higher activation effect (2.2-fold versus 1.7-fold) on the BETA2 promoter than E47. However, when cotransfected with CMV-ngn3, both FL-E47 and E47 had similar activation effects on the BETA2 promoter (Fig. 7B).
FIG. 8.
The proximal three E boxes are important for ngn3-mediated activation of the BETA2 promoter. (A to C) Effects of E box deletions on ngn3-mediated activation of the BETA2 promoter in HIT and β-TC cells. (A) Schematic diagram of locations of 17 E boxes in the 2.2-kb BETA2 promoter. (B) Deletion analyses in HIT cells cotransfected with 300 ng of deletion constructs and 600 ng of CMV-ngn3. (C) Deletion analyses in β-TC cells cotransfected with 300 ng of deletion constructs and 200 ng of CMV-ngn3. The size of each promoter fragment is given in parentheses. The effects are shown as fold activation of each reporter construct by ngn3 (±standard deviation of the mean). Transfections were performed in triplicate and repeated three times. (D) Effects of a progressive deletion of E boxes ngn3-E47-mediated activation of the 1.0-kb BETA2 promoter. HeLa cells were cotransfected with 300 ng of each deletion construct and an equal amount (600 ng) of the expression vectors for ngn3 and E47. The numbers following B2E represent the E boxes included in each construct. The B2E(−) construct has no E box. The effects are shown as fold activation of each reporter construct by ngn3-E47 (±standard deviation of the mean). Transfections were performed in triplicate and repeated three times.
FIG. 7.
The BETA2 promoter can be activated by ngn3. (A) Dose-dependent activation of the BETA2 promoter by ngn3 in HIT and β-TC cells. In both HIT and β-TC cells, 300 ng of the 1.0-kb BETA2 promoter reporter was cotransfected with increased amounts (50 to 800 ng) of CMV-ngn3 expression vector. Activation effects of different amounts of CMV-ngn3 on the promoter are shown as fold activation (±standard deviation of the mean). The transfections were performed in triplicate and repeated three times. (B) Cotransfection of E47 and ngn3 together had a higher activation effect on the BETA2 promoter in non-islet cells. HeLa cells were cotransfected with 300 ng of the 1.0-kb BETA2 promoter reporter and 600 ng of E47 (or FL-E47) and/or ngn3 expression vectors. The luciferase activities (expressed as RLU) were corrected by protein concentrations and presented as the average of triplicate values (±standard deviation of the mean). The transfection were repeated three times.
Characterization of ngn3 binding sites in the BETA2 promoter.
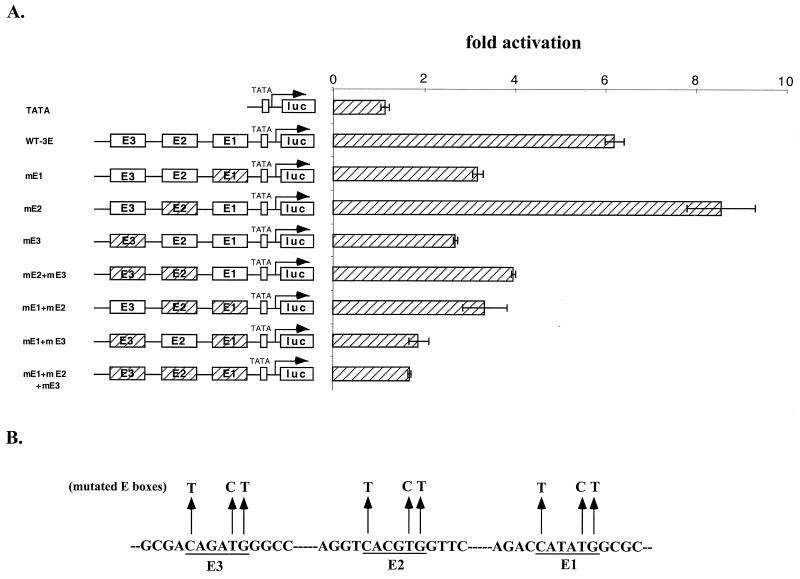
As shown in Fig. 8A, there are 17 E boxes in the 2.2-kb BETA2 promoter. To further define those E boxes important for ngn3-mediated activation of the BETA2 promoter, we assayed the effect of ngn3 on a series of reporter constructs bearing different deletions of the BETA2 promoter. Among the five constructs tested, we consistently found that the 1.0-kb promoter reporter [B2 (1.0 kb)] had the highest level of activation by ngn3 in both HIT and β-TC cells (Fig. 8B and 6C). Moreover, the 419-bp promoter reporter [B2 (419 bp)], which contains the three most proximal E boxes, could still be stimulated by ngn3 to a reasonable level. However, a further deletion of this promoter to eliminate the three proximal E boxes [B2 (231 bp)] resulted in the loss of ngn3-mediated activation (Fig. 8B and 6C). Interestingly, by alignment of the human and mouse BETA2 promoter sequences, we found that only four of the nine E boxes in the 1.0-kb fragment, including the three most proximal ones and another located at bp −754 and −749, are conserved between the human and mouse sequences. To further support the assumption that the proximal three E boxes are important, we generated 10 promoter reporter constructs with stepwise deletions of one E box each time from the 5′ end of the 1.0-kb promoter. Transfection assay of these constructs indicated that, in general, deletion of each E box from the distal end caused either no effect or a slight reduction of ngn3-mediated activation (Fig. 8D). However, deletion of the three proximal E boxes from the 419-bp construct [Fig. 8D, 2E1-3) abolished most of the effect of ngn3 and E47 [Fig. 8D, 2E(−)]. To confirm the importance of the three most proximal E boxes (E1 to E3), we subcloned a 155-bp promoter sequence containing these three E boxes in either a wild-type (CANNTG) or a mutated (TANNCT) form (Fig. 9A and B) into a TATA-luciferase reporter. Wild-type and mutated constructs were cotransfected into HeLa cells with expression vectors for ngn3 and E47. The results indicated that mutation of either E1 or E3 caused an ∼50% decrease of ngn3-mediated fold activation compared to the wild type. However, mutation of E2 did not decrease but rather slightly increased of ngn3-E47-mediated activation. In addition, the reporter constructs bearing mutations of E1 and E3 or all three mutations decreased ngn3-E47-mediated activation to the level close to that of the control vector (TATA). Further, we did not observe any synergistic effect between E1 and E3.
FIG. 9.
E1 and E3 E boxes are important for ngn3-E47-mediated activation of the BETA2 promoter. (A) Mutational analyses of the three proximal E boxes in the BETA2 promoter in the presence of ngn3 and E47. All constructs (except the control) derived from a TATA-luciferase reporter driven by a 155-bp BETA2 promoter fragment containing three E boxes in either a wild type (WT-3E; open box) or mutated (hatched box) form. (B) Illustration of mutated E box sequences, in each of which the sequence CANNTG was replaced by TANNCT. Each construct was cotransfected into HeLa cells with both ngn3 and E47 expression vectors. The activation of each construct by ngn3-E47 is represented as fold activation (±standard deviation of the mean). (C) Synergistic activation of individual E box reporters by ngn3 and E47. Three copies of each E box (E1, E2, and E3) were cloned into pGL3-TATA vector. The resultant reporters (300 ng) were then cotransfected into HeLa cells with ngn3 and/or E47 expression vectors (100 ng). The transfections were repeated three times. Top four rows, E1 results; middle four rows, E3 results; bottom four rows, E2 results.
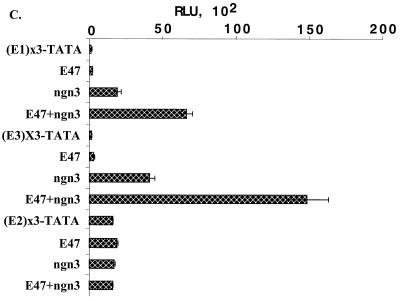
To determine whether an individual E box can, indeed, mediate the activating effect of ngn3-E47, we cloned three copies of each E box into a TATA box-containing luciferase reporter (pGL3-TATA). The reporters and the expression vectors for ngn3 and E47 were then cotransfected into HeLa cells. The activities of all E box reporters (Fig. 9C) were only slightly increased by overexpression of E47 alone. Overexpression of ngn3 alone resulted in significant increases on both the E1 [(E1)×3-TATA]- and E3 [(E3)×3-TATA]-driven reporter activities (Fig. 9C, ngn3 compared to reporters). In addition, coexpression of E47 and ngn3 exerted synergistic activation on both the E1- and E3-driven reporters (Fig. 9C, E47+ngn3 compared to ngn3 and E47 alone). In sharp contrast, the reporter activity of E2 was not affected by either ngn3 or ngn3-E47. Thus, both E1 and E3 E boxes can act as the target of ngn3-E47. In agreement with previous results (Fig. 9A and B), these data suggest that E1 and E3, but not E2, can mediate the activation effect of ngn3-E47.
ngn3-E47 heterodimer can bind to E1 and E3 but not E2 in the BETA2 promoter.
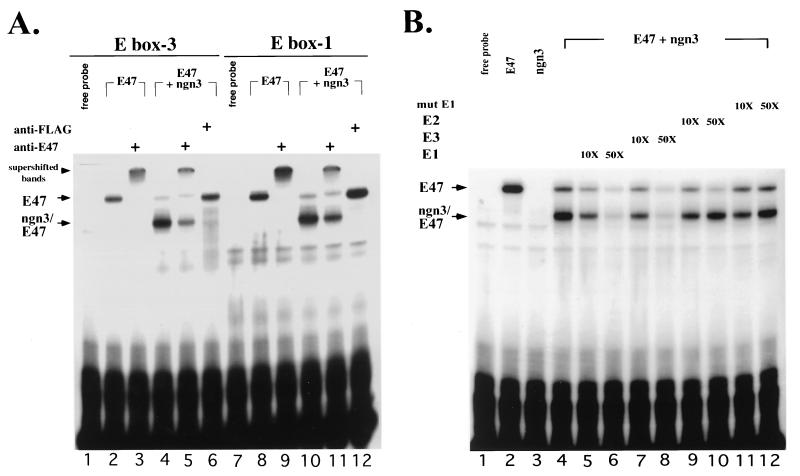
For the next series of experiments, we addressed the question of whether ngn3 can bind specifically to the three proximal E boxes in vitro. The cDNAs of ngn3 and E47 were transcribed and translated in vitro, and the proteins, individually or in combination, were incubated with 32P-labeled E1, E2, and E3 oligonucleotides individually. While incubation of E47 alone with either E1 or E3 gave rise to a single binding complex (Fig. 10A, lanes 2 and 8; Fig. 10B, lane 2), incubation of ngn3 alone generated no visible complex (Fig. 10B, lane 3). Coincubation of ngn3 and E47 with either E1 or E3 generated two complexes: a major, faster-migrating ngn3-E47 heterodimer and a minor, slower-migrating E47 homodimer (Fig. 10). The identities of these complexes were confirmed by supershift assay (Fig. 10A) with anti-E47 antibody and an antibody against a FLAG peptide sequence (33) that was tagged at the C terminus of ngn3. As expected, in the supershift assay using the E1 or E3 oligonucleotide as a probe, anti-E47 antibody could supershift not only the E47 homodimer (Fig. 10A, lanes 3 and 9) but also the major complexes formed by ngn3 and E47 (Fig. 10A, lanes 5 and 11). In contrast, the presence of anti-FLAG antibody seemed to prevent the formation of the major fast-migrating complexes and enhanced the formation of slower-migrating complexes (Fig. 10A, lanes 6 and 12), suggesting that the faster-migrating complexes indeed contain both ngn3 and E47.
FIG. 10.
ngn3-E47 heterodimer can specifically bind to E1 and E3. (A) The E47 and FLAG-tagged ngn3 proteins were synthesized in vitro in rabbit reticulocyte lysate. Two specific protein-DNA complexes were observed when E47 and/or ngn3 were incubated with E1 or E3 probe: the faster-migrating one (arrow labeled ngn3/E47) and the slower-migrating one (arrow labeled E47). Supershift assay was performed with 1 μl of 1:10-diluted anti-E47 antibody and 1 μl of 1:10 diluted anti-FLAG peptide, respectively. Both antibodies were added after completion of binding reactions. The positions of supershifted complexes (lanes 3, 5, 9, and 11) are indicated (arrowhead). Also notice that the E47 homodimer-DNA complexes were enhanced (lanes 6 and 12) in the presence of anti-FLAG antibody. Nonimmune rabbit serum was not found to have supershift activity and is not shown. (B) ngn3-E47 heterodimer bound specifically to E1. The binding reaction was performed as described for panel A. The homodimer- and heterodimer-DNA complexes are labeled as in panel A. Binding specificity was determined by competition with 10- and 50-fold molar excess of unlabeled double-stranded E1, E2, E3, and mutated E1 (mut E1) oligonucleotides.
To test the specificity of the bindings between E boxes and ngn3-E47 heterodimer, we performed a competition assay using unlabeled E1, E2, E3, mutated E1 (Fig. 10B), and mutated E3 oligonucleotides (data not shown). The results indicated that the complexes formed by ngn3-E47 with either E1 or E3 could be competed by the addition of unlabeled E1 or E3 oligonucleotides in a dose-dependent manner. In contrast, unlabeled E2, mutated E1, or mutated E3 (data not shown) oligonucleotide, in which three nucleotides were changed in the core E box (CANNTG to TANNCT), was unable to compete with the binding of the ngn3-E47 heterodimer to E1 (Fig. 10B) or E3 (data not shown). The inability of E2 to compete for the binding of ngn3-E47 to E1 (Fig. 10B, lanes 9 and 10) or E3 (data not shown) suggested that the ngn3-E47 heterodimer was not able to bind to E2 efficiently. This result may explain why E2 was not important in mediating the ngn3 regulation of the BETA2 promoter as shown in the transfection experiments (Fig. 9). In a similar gel shift assay using the extract from COS-1 cells transfected with expression vectors for E47 and FLAG-tagged ngn3, we found similar bands formed by E47-ngn3 and E boxes, which could be supershifted by anti-FLAG tag antibody (data not shown). Taken together, the data suggest that ngn3-E47 activates the BETA2 promoter by binding specifically to both E1 and E3 but not to E2.
DISCUSSION
Recent studies on cell fate determination during vertebrate neuronal development have led to the identification of several tissue-specific bHLH factors, including members of the neurogenin family (19, 39) and the BETA2 (neuroD) family (17, 24, 25). The early expression of ngns in undifferentiated and dividing neuronal progenitors makes them better candidates than BETA2 (neuroD) members as neuronal determination factors (19). This notion is further supported by the findings that in ngn1- and ngn2-deficient mice (12, 18), the expression of BETA2 is abolished in neuronal cells, possibly because the BETA2 promoter cannot be switched on in the absence of potential transcription activators like ngns. Interestingly, the molecular basis of endocrine pancreas development is in many respects similar to that of neuronal development (38), and both ngn3 and BETA2 are expressed in the developing pancreas and neurons (39). Therefore, by extension, the same cascade relationship may likely hold true for ngn3 and BETA2 in the endocrine pancreas.
In this study, we describe the cloning and characterization of the mouse BETA2 promoter. Our work indicates that the 2.2-kb promoter fragment is sufficient to direct proper expression of the transgene in the mouse pancreas and neuronal tissues. During the preparation of this paper, Xu and Murphy (46) reported the isolation of the mouse BETA2 (neuroD) gene. The transcription start site that we found is 2 bp upstream of the one defined using 5′ RACE PCR alone by Xu and Murphy (46), and the untranslated exon (exon 1) that we identified is 86 bp in length, not 82 bp. In addition to the in vitro transfection assay, we have shown that in transgenic mice, the 3.8-kb BETA2 5′ flanking sequence, which includes the 2.2-kb promoter, the first exon, and the intron, is able to direct proper islet-specific expression of the β-Gal gene. Interestingly, in collaboration with P.-L. Herrera, we obtained preliminary data also indicating that the 2.2-kb promoter with a heterologous intron sequence was able to direct proper islet-specific expression of the reporter gene in the transgenic mice. Therefore, the untranslated exon (exon 1) and the intron do not seem to be required for tissue-specific expression of BETA2 in the islets of Langerhans.
In this paper, we have demonstrated that the BETA2 gene has a tissue- or cell-type-specific promoter. The sequence important for the cell type specificity probably resides between bp −231 and kb −1.0, where nine E boxes exist. It is likely that these E boxes may contribute to the cell-type-specific activity observed. Indeed, for both 1.0-kb and 419-bp reporter constructs, overexpression of ngn3 and E47 in HeLa cells seemed to compensate for the low activity of the BETA2 promoter in these cells (data not shown). However, deletion of the three proximal E boxes (E1 to E3) abolished the activation effect in HeLa cells (data not shown), suggesting that these three E boxes may also contribute to the cell-type-specific activities.
The gel shift analyses demonstrated that ngn3 alone cannot form a visible protein-DNA complex with E boxes (Fig. 10). This is consistent with previous studies (16, 22) showing that tissue-specific or class B bHLH factors cannot form stable homodimers binding to E boxes. However, when heterodimerized with E47, ngn3 could bind to E1 and E3 but not E2. Sequence comparison indicated that the E2 sequence is indeed more homologous to the consensus binding site of Myc (5). Further experiments are under way to determine whether any bHLH-leucine zipper or class C bHLH factor (15) can bind to E2. The interactions between ngn3-E47 and E1 or E3 were significantly weakened when only three nucleotides in the core E boxes were changed. This is in agreement with our mutational analyses showing that the activation effect of ngn3-E47 was decreased when these E boxes were mutated in the same way (Fig. 9). It is also noteworthy that both E1 and E3 contain an E box sequence, CANATG, that is slightly different from the identified BETA2-responsive E box sequence (CANCTG) in the insulin and secretin promoters (23, 25). The significance of this subtle difference is not known. Taken together, these results indicate that the ngn3-E47 heterodimer could discriminate against the slight variances in the E box sequences. Therefore, the interactions that we observed between ngn3-E47 and E1 or E3 are specific and involved in activation of the BETA2 promoter. We should emphasize, however, that other E boxes localized in the distal promoter may be also bound by ngn3-E47 and contribute to maximal activation of the BETA2 promoter. Whether these two E boxes (E1 and E3) that we describe here are essential for BETA2 expression during development requires further experimentation. In addition, it will be interesting to determine whether other transcription factors, such as BETA2 itself, can also bind to these two E boxes and regulate the expression of BETA2.
In the supershift experiments, we do not know exactly whether the anti-FLAG antibody simply prevented the formation of the ngn3-E47 complex on E boxes or formed the supershift complexes migrating at a position indistinguishable from that of the slower-migrating complexes containing E47 homodimer (Fig. 10A). For the former possibility, it is likely that the antibody caused a conformational change of ngn3 and hence interfered with or physically prevented the interaction between ngn3 and E47. Nevertheless, both possibilities suggest that the faster-migrating complex contained both ngn3 and E47.
The role of ngn3 as one of the physiologically relevant upstream regulators of the BETA2 gene is strongly suggested by two biological studies. First, injection of ngn3 mRNA into Xenopus embryos caused the ectopic expression of BETA2 in the trunk of embryos (Fig. 5). Second, overexpression of ngn3 in stably transfected endocrine cell lines up-regulated the endogenous levels of BETA2 RNA (Fig. 6). Furthermore, the observation that the expression domains of ngn3 partially overlaps those of BETA2 in both the developing pancreas (especially at earlier stages) and neuronal tissues (Fig. 4A, B, and D) further supports the conclusion that BETA2 gene expression can be regulated by ngn3. While our results clearly indicate that ngn3 is an upstream regulator of BETA2 gene expression, they do not distinguish whether the effect of ngn3 is a direct one or is mediated by other ngn3 downstream bHLH factors that in turn bind to the E boxes and activate BETA2 transcription. However, the ability of ngn3-E47 heterodimer to directly bind to the BETA2 promoter and transactivate it supports the notion that the activation of BETA2 by ngn3 is a direct effect.
Although we were able to activate BETA2 gene expression in β-TC and STC cells by overexpression of ngn3, we were not able to do so in several other cell lines such as ARIP and Panc1. This result is consistent with data presented in Fig. 5 showing that injection of ngn3 mRNA into Xenopus embryos can ectopically induce BETA2 gene expression in several but not all areas receiving ngn3 mRNA. Thus, ngn3 is not able to stimulate BETA2 gene expression in all cell types. Other factors may also participate in BETA2 gene expression. For example, many factors such as coactivators (i.e., CBP/p300), ngn3 modification (i.e., phosphorylation and dephosphorylation), and other transcription factors may be needed to synergize with ngn3 to induce BETA2 gene expression. If these factors or modification systems are required and not expressed in some cells, we expect that ngn3 will not be able to stimulate BETA2 gene expression in all cells. Indeed, all of these factors have been shown to play a role in modifying the transcriptional activity of other bHLH transcription factors.
Interestingly, at later stages of islet cell differentiation, BETA2 and ngn3 were frequently found to be expressed in separate cell populations, suggesting that ngn3 was no longer required for the expression of BETA2. Therefore, other transcription factors are necessary to maintain the expression of BETA2 in differentiated endocrine cells. Recently, a novel neuron-specific bHLH factor, NeuroM (35), was cloned from the chicken retina. It is possible that the putative intermediate bHLH factors, such as NeuroM or NeuroM-like molecules, play a role in maintaining the expression of BETA2 after the expression of ngn3 is switched off. However, it has not been shown that NeuroM or NeuroM-like molecules are expressed in the developing pancreas or that they can regulate the BETA2 promoter. Alternatively, after being turned on by ngn3 at an early stage, BETA2 may play an autoregulatory role in maintaining its own expression in the developing pancreas. This is more likely, since we have preliminary data indicating that BETA2 could also activate the E1- and E3-driven reporter constructs (data not shown). Intriguingly, the expression of ngn3 (Fig. 4H) could still be detected in the pancreas during later development at E17.5 and P2, suggesting that ngn3-expressing cells may represent a stem cell population in the endocrine pancreas and may be involved in renewal of islet cells.
In conclusion, ngn3 is likely to be one of the transcription factors involved in regulation of the BETA2 promoter during early islet cell differentiation. Future isolation of other transcriptional factors involved in this context and generation of gene-disrupted animals, including ngn3-deficient mice, will help to elucidate the transcriptional cascade of islet-specific transcription factors and its physiological significance during endocrine pancreas development.
ACKNOWLEDGMENTS
We are grateful to David J. Anderson for the gift of plasmid pSK-ngn3. We thank Francesco J. DeMayo's laboratory for microinjections. We are grateful to Roland Stein for providing the plasmid containing full-length E47 cDNA, Francisco J. Naya for providing the BETA2 genomic DNA clone, Christine M. M. Stellrecht for the β-TC cell cDNA library, Fabrice Petit for providing the pGL3-TATA plasmid and valuable help with gel shift assays, and Fred A. Pereira and Cheng Zhou for technical advice on in situ hybridization. We are grateful to Sophia Y. Tsai, Eric Nemoz-Gaillard, and Carlos Pipaon for useful discussions. We thank Debra Bramblett and members in Tsai laboratory for critical reading of the manuscript.
This work was supported by a postdoctoral fellowship (M.L.) and grants (M.-J.T.) from the National Institutes of Health.
ADDENDUM IN PROOF
Following submission of this paper, G. Gradwohl et al. (Proc. Natl. Acad. Sci. USA 97:1607–1611, 2000) reported that expression of BETA2 is lost and all four islet cell types are lacking in ngn3-deficient mice.
REFERENCES
- 1.Ahlgren U, Jonsson J, Edlund H. The morphogenesis of the pancreatic mesenchyme is uncoupled from that of the pancreatic epithelium in IPF1/PDX1-deficient mice. Development. 1996;122:1409–1416. doi: 10.1242/dev.122.5.1409. [DOI] [PubMed] [Google Scholar]
- 2.Ahlgren U, Pfaff S L, Jessell T M, Edlund T, Edlund H. Independent requirement for ISL1 in formation of pancreatic mesenchyme and islet cells. Nature. 1997;385:257–260. doi: 10.1038/385257a0. [DOI] [PubMed] [Google Scholar]
- 3.Andersen F G, Jensen J, Heller R S, Petersen H V, Larsson L I, Madsen O D, Serup P. Pax6 and Pdx1 form a functional complex on the rat somatostatin gene upstream enhancer. FEBS Lett. 1999;445:315–320. doi: 10.1016/s0014-5793(99)00144-1. [DOI] [PubMed] [Google Scholar]
- 4.Bartholoma A, Nave K A. NEX-1: a novel brain-specific helix-loop-helix protein with autoregulation and sustained expression in mature cortical neurons. Mech Dev. 1994;48:217–228. doi: 10.1016/0925-4773(94)90061-2. [DOI] [PubMed] [Google Scholar]
- 5.Blackwell T K, Kretzner L, Blackwood E M, Eisenman R N, Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990;250:1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- 6.Bonnerot C, Nicolas J F. Application of LacZ gene fusions to postimplantation development. Methods Enzymol. 1993;225:451–469. doi: 10.1016/0076-6879(93)25031-v. [DOI] [PubMed] [Google Scholar]
- 7.Busch K, Martin B, Baniahmad A, Renkawitz R, Muller M. At least three subdomains of v-erbA are involved in its silencing function. Mol Endocrinol. 1997;11:379–389. doi: 10.1210/mend.11.3.9903. [DOI] [PubMed] [Google Scholar]
- 8.Dirks W, Wirth M, Hauser H. Dicistronic transcription units for gene expression in mammalian cells. Gene. 1993;128:247–249. doi: 10.1016/0378-1119(93)90569-o. [DOI] [PubMed] [Google Scholar]
- 9.Dumonteil E, Laser B, Constant I, Philippe J. Differential regulation of the glucagon and insulin I gene promoters by the basic helix-loop-helix transcription factors E47 and BETA2. J Biol Chem. 1998;273:19945–19954. doi: 10.1074/jbc.273.32.19945. [DOI] [PubMed] [Google Scholar]
- 10.Efrat S, Linde S, Kofod H, Spector D, Delannoy M, Grant S, Hanahan D, Baekkeskov S. Beta-cell lines derived from transgenic mice expressing a hybrid insulin gene-oncogene. Proc Natl Acad Sci USA. 1988;85:9037–9041. doi: 10.1073/pnas.85.23.9037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El-Hodiri H M, Perry M. Interaction of the CCAAT displacement protein with shared regulatory elements required for transcription of paired histone genes. Mol Cell Biol. 1995;15:3587–3596. doi: 10.1128/mcb.15.7.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fode C, Gradwohl G, Morin X, Dierich A, LeMeur M, Goridis C, Guillemot F. The bHLH protein NEUROGENIN 2 is a determination factor for epibranchial placode-derived sensory neurons. Neuron. 1998;20:483–494. doi: 10.1016/s0896-6273(00)80989-7. [DOI] [PubMed] [Google Scholar]
- 13.Harland R M. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- 14.Henthorn P, Kiledjian M, Kadesch T. Two distinct transcription factors that bind the immunoglobulin enhancer microE5/kappa 2 motif. Science. 1990;247:467–470. doi: 10.1126/science.2105528. [DOI] [PubMed] [Google Scholar]
- 15.Landschulz W H, Johnson P F, McKnight S L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- 16.Lassar A B, Davis R L, Wright W E, Kadesch T, Murre C, Voronova A, Baltimore D, Weintraub H. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell. 1991;66:305–315. doi: 10.1016/0092-8674(91)90620-e. [DOI] [PubMed] [Google Scholar]
- 17.Lee J E, Hollenberg S M, Snider L, Turner D L, Lipnick N, Weintraub H. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- 18.Ma Q, Chen Z, del Barco Barrantes I, de la Pompa J L, Anderson D J. Neurogenin1 is essential for the determination of neuronal precursors for proximal cranial sensory ganglia. Neuron. 1998;20:469–482. doi: 10.1016/s0896-6273(00)80988-5. [DOI] [PubMed] [Google Scholar]
- 19.Ma Q, Kintner C, Anderson D J. Identification of neurogenin, a vertebrate neuronal determination gene. Cell. 1996;87:43–52. doi: 10.1016/s0092-8674(00)81321-5. [DOI] [PubMed] [Google Scholar]
- 20.McCormick M B, Tamimi R M, Snider L, Asakura A, Bergstrom D, Tapscott S J. NeuroD2 and neuroD3: distinct expression patterns and transcriptional activation potentials within the neuroD gene family. Mol Cell Biol. 1996;16:5792–5800. doi: 10.1128/mcb.16.10.5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murre C, McCaw P S, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 22.Murre C, Voronova A, Baltimore D. B-cell- and myocyte-specific E2-box-binding factors contain E12/E47-like subunits. Mol Cell Biol. 1991;11:1156–1160. doi: 10.1128/mcb.11.2.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mutoh H, Fung B P, Naya F J, Tsai M J, Nishitani J, Leiter A B. The basic helix-loop-helix transcription factor BETA2/NeuroD is expressed in mammalian enteroendocrine cells and activates secretin gene expression. Proc Natl Acad Sci USA. 1997;94:3560–3564. doi: 10.1073/pnas.94.8.3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naya F J, Huang H P, Qiu Y, Mutoh H, DeMayo F J, Leiter A B, Tsai M J. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/neuroD-deficient mice. Genes Dev. 1997;11:2323–2334. doi: 10.1101/gad.11.18.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naya F J, Stellrecht C M, Tsai M J. Tissue-specific regulation of the insulin gene by a novel basic helix-loop-helix transcription factor. Genes Dev. 1995;9:1009–1019. doi: 10.1101/gad.9.8.1009. [DOI] [PubMed] [Google Scholar]
- 26.Nieuwkoop P D, Faber J. Normal table of Xenopus laevis (Daudin) 2nd ed. Amsterdam, The Netherlands: Elsevier/North Holland Publishing Co.; 1967. [Google Scholar]
- 27.Offield M F, Jetton T L, Labosky P A, Ray M, Stein R W, Magnuson M A, Hogan B L, Wright C V. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- 28.Ohlsson H, Karlsson K, Edlund T. IPF1, a homeodomain-containing transactivator of the insulin gene. EMBO J. 1993;12:4251–4259. doi: 10.1002/j.1460-2075.1993.tb06109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orly J, Rei Z, Greenberg N M, Richards J S. Tyrosine kinase inhibitor AG18 arrests follicle-stimulating hormone-induced granulosa cell differentiation: use of reverse transcriptase-polymerase chain reaction assay for multiple messenger ribonucleic acids. Endocrinology. 1994;134:2336–2346. doi: 10.1210/endo.134.6.7514996. [DOI] [PubMed] [Google Scholar]
- 30.Peng H B. Xenopus laevis: practical uses in cell and molecular biology. Solutions and protocols. Methods Cell Biol. 1991;36:657–662. [PubMed] [Google Scholar]
- 31.Peshavaria M, Henderson E, Sharma A, Wright C V, Stein R. Functional characterization of the transactivation properties of the PDX-1 homeodomain protein. Mol Cell Biol. 1997;17:3987–3996. doi: 10.1128/mcb.17.7.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peyton M, Moss L G, Tsai M J. Two distinct class A helix-loop-helix transcription factors, E2A and BETA1, form separate DNA binding complexes on the insulin gene E box. J Biol Chem. 1994;269:25936–25941. [PubMed] [Google Scholar]
- 33.Prickett K S, Amberg D C, Hopp T P. A calcium-dependent antibody for identification and purification of recombinant proteins. BioTechniques. 1989;7:580–589. [PubMed] [Google Scholar]
- 34.Rindi G, Grant S G, Yiangou Y, Ghatei M A, Bloom S R, Bautch V L, Solcia E, Polak J M. Development of neuroendocrine tumors in the gastrointestinal tract of transgenic mice. Heterogeneity of hormone expression. Am J Pathol. 1990;136:1349–1363. [PMC free article] [PubMed] [Google Scholar]
- 35.Roztocil T, Matter-Sadzinski L, Alliod C, Ballivet M, Matter J M. NeuroM, a neural helix-loop-helix transcription factor, defines a new transition stage in neurogenesis. Development. 1997;124:3263–3272. doi: 10.1242/dev.124.17.3263. [DOI] [PubMed] [Google Scholar]
- 36.Santerre R F, Cook R A, Crisel R M, Sharp J D, Schmidt R J, Williams D C, Wilson C P. Insulin synthesis in a clonal cell line of simian virus 40-transformed hamster pancreatic beta cells. Proc Natl Acad Sci USA. 1981;78:4339–4343. doi: 10.1073/pnas.78.7.4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma A, Henderson E, Gamer L, Zhuang Y, Stein R. Analysis of the role of E2A-encoded proteins in insulin gene transcription. Mol Endocrinol. 1997;11:1608–1617. doi: 10.1210/mend.11.11.0004. [DOI] [PubMed] [Google Scholar]
- 38.Slack J M. Developmental biology of the pancreas. Development. 1995;121:1569–1580. doi: 10.1242/dev.121.6.1569. [DOI] [PubMed] [Google Scholar]
- 39.Sommer L, Ma Q, Anderson D J. Neurogenins, a novel family of atonal-related bHLH transcription factors, are putative mammalian neuronal determination genes that reveal progenitor cell heterogeneity in the developing CNS and PNS. Mol Cell Neurosci. 1996;8:221–241. doi: 10.1006/mcne.1996.0060. [DOI] [PubMed] [Google Scholar]
- 40.Sosa-Pineda B, Chowdhury K, Torres M, Oliver G, Gruss P. The Pax4 gene is essential for differentiation of insulin-producing beta cells in the mammalian pancreas. Nature. 1997;386:399–402. doi: 10.1038/386399a0. [DOI] [PubMed] [Google Scholar]
- 41.St-Onge L, Sosa-Pineda B, Chowdhury K, Mansouri A, Gruss P. Pax6 is required for differentiation of glucagon-producing alpha-cells in mouse pancreas. Nature. 1997;387:406–409. doi: 10.1038/387406a0. [DOI] [PubMed] [Google Scholar]
- 42.Sussel L, Kalamaras J, Hartigan-O'Connor D J, Meneses J J, Pedersen R A, Rubenstein J L, German M S. Mice lacking the homeodomain transcription factor Nkx2.2 have diabetes due to arrested differentiation of pancreatic beta cells. Development. 1998;125:2213–2221. doi: 10.1242/dev.125.12.2213. [DOI] [PubMed] [Google Scholar]
- 43.Turner D L, Weintraub H. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- 44.Wang M, Drucker D J. The LIM domain homeobox gene isl-1 is a positive regulator of islet cell-specific proglucagon gene transcription. J Biol Chem. 1995;270:12646–12652. doi: 10.1074/jbc.270.21.12646. [DOI] [PubMed] [Google Scholar]
- 45.Wilkinson D G, Nieto M A. Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods Enzymol. 1993;225:361–373. doi: 10.1016/0076-6879(93)25025-w. [DOI] [PubMed] [Google Scholar]
- 46.Xu W, Murphy L J. Isolation and characterization of the mouse beta 2/neuroD gene promoter. Biochem Biophys Res Commun. 1998;247:814–818. doi: 10.1006/bbrc.1998.8897. [DOI] [PubMed] [Google Scholar]
- 47.Zhuang Y, Cheng P, Weintraub H. B-lymphocyte development is regulated by the combined dosage of three basic helix-loop-helix genes, E2A, E2-2, and HEB. Mol Cell Biol. 1996;16:2898–2905. doi: 10.1128/mcb.16.6.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]