Abstract
Computer simulation has been used to identify peptides that mimic the natural target of the SARS-CoV-2 coronavirus spike (S) protein, the angiotensin-converting enzyme type 2 (ACE2) cell receptor. Based on the structure of the complex of the protein S receptor-binding domain (RBD) and ACE2, the design of chimeric molecules consisting of two 22–23-mer peptides linked to each other by disulfide bonds was carried out. The chimeric molecule X1 was a disulfide dimer, in which terminal cysteine residues in the precursor molecules h1 and h2 were connected by the S-S bond. In the chimeric molecule X2, the disulfide bond was located in the middle of each precursor peptide molecule. The precursors h1 and h2 mimic amino acid sequences of α1- and α2-helices of the ACE2 extracellular peptidase domain, respectively, keeping intact most of the amino acid residues involved in the interaction with RBD. The aim of the work was to evaluate the binding efficiency of chimeric molecules and their constituent peptides with RBD (particularly in dependence of the middle and terminal methods of fixing the initial peptides h1 and h2). The proposed polypeptides and chimeric molecules were synthesized by chemical methods, purified to 95–97% purity, and characterized by HPLC and MALDI-TOF mass spectrometry. Binding of these peptides to the SARS-CoV-2 RBD was evaluated by microthermophoresis with recombinant domains corresponding in sequence to the original Chinese (GenBank ID NC_045512.2) and the British (B. 1.1.7, GISAID EPI_ISL_683466) variants. The original RBD of the Chinese variant bound to three synthesized peptides: linear h2 and both chimeric variants. Chimeric peptides were also bound to the RBD of the British variant. The antiviral activity of the proposed peptides was evaluated in Vero cell line.
Keywords: SARS-CoV-2, RBD, ACE2, peptide inhibitor, computer simulation
INTRODUCTION
Over the past 20 years, coronaviruses (SARS-CoV, MERS-CoV and SARS-CoV-2) have caused a number of severe respiratory diseases with alarmingly high mortality rates. The World Health Organization (WHO) has included these infections in the priority list of diseases requiring scientific research. The COVID-19 pandemic stimulated studies on the search for vaccines, the development of diagnostic tools , and specific antivirals. In the context of development of the coronavirus infection a glycoprotein known as the spike protein (S), significantly protruding from the surface of the SARS-CoV-2 virion envelope, attracts much attention [1]. Protein S is a typical type I fusion protein responsible for binding to the cell receptor and mediating the fusion of the viral and cellular membranes. In the virion it exists in the form of a precursor, which is proteolytically cleaved by cellular serine proteases (particularly by furin and TMPRSS2) into S1 and S2 subunits; this cleavage induces conformational changes in the S2 subunit, leading to the exposure of the fusion peptide and attack of the cellular membrane. This triggers the fusion of the viral and cellular membranes. The penetration of the virus into the cell occurs through a mechanism common to many viruses: formation of a coiled-coil complex between two heptad repeats HR1 and HR2 of the S2 subunit.
The main target required for the penetration of SARS-CoV-2 [2] into a human cell is a transmembrane receptor protein known as angiotensin-converting enzyme type 2 (ACE2). The related SARS-CoV-2 coronaviruses SARS-CoV (the causative agent of atypical pneumonia) [3, 4] and HCoV-NL63 (the causative agent of respiratory diseases in children) [5] also use the ACE2 receptor to enter the cell. ACE2 is an enzyme found in cell membranes of many organs of the body: lungs, cardiovascular system, intestine, kidneys, etc. [6]. The ACE2 expression in the cells of the structural elements of the central nervous system determines their susceptibility to the SARS-CoV-2 infection, which leads to anosmia and ageusia during the course of the disease (these symptoms are currently considered as the key symptoms of COVID-19). The binding of the virion to the receptor occurs through the interaction of the protein S RBD with the ACE2 molecule. Inhibition of binding of SARS-CoV-2 RBD (and related viruses) to ACE2 by competing with specifically designed peptide and protein-based drugs is one of the strategies for inhibiting virus entry into human cells. For example, the authors [7] found that a soluble recombinant protein corresponding to the human ACE2 sequence (hrsACE-2) was capable of dose-dependent inhibition of the reproduction of SARS-CoV-2 isolated from the nasopharynx of a patient with confirmed COVID-19.
Some authors [8–10] used molecular dynamics modeling to design potential peptide inhibitors that would restrict the attachment and penetration of the SARS-CoV-2 into the cell. Based on modeling of the crystal structure of the ACE2 and RBD SARS-CoV-2 complex authors of [11] found that the most significant for the entry of SARS-CoV and SARS-CoV-2 coronaviruses into the cell was protein S RBD binding to the peptide sequence of the spike-binding peptide 1 (SBP1, Table 1); they synthesized a 23-membered SBP1 peptide and using bio-layer interferometry showed that SBP1 was tightly bound to RBD with a dissociation constant (Kd) of 47 nM (this value was comparable to that (14.7 nM) for full-length water-soluble ACE2 [12]). In the present work, based on the analysis of the structure of the complexes, an attempt has been undertaken to design peptides that mimic the natural target of the SARS-CoV-2 protein S, the ACE2 cell receptor. The most promising peptides according to the prediction data have been synthesized and the efficiency of their binding to RBD and also their antiviral activity have been investigated in vitro. The newly synthesized SBP1 peptide was used as a control compound.
Table 1.
Characteristics of synthesized peptides
| Code | Sequence | Mol. Mass, g/mol | MALDI-TOF, m/z | Purity HPLC, % |
|---|---|---|---|---|
| SPB1 | IEEQAKTFLDKFNHEAEDLFYQS-NH2 (control) | 2802.01 | 2801.4 | 97.6 |
| h1 | IEEQAKTFLDKFNHEAEDLFYk | 2716.0 | 2715.4 | 97.26 |
| h1-D-Cys | cEEQAKTFLDKFNHEAEDLFYk | 2705.95 | 2705.3 | 96.8 |
| h1-Mpa | IEEQAKTFLDK(Mpa)FNHQAEDLFYk | 2802.4 | 2802.5 | 96.3 |
| h2 | DKWSAFLKEQSTIAQNleYPLQECI | 2712.1 | 2711.4 | 95.9 |
| h2-Cys | DKWSAFLKECSTIAQIYPLQEI-OH | 2584.0 | 2583.1 | 96.4 |
| Х1 |

|
5412.9 | 5413.9 2711.5* 2705.4* | 95.0 |
| X2 |

|
5383.9 | 5382.8 2583.4* 2802.5* | 96.2 |
Modifications are shown in bold; lowercase letters designate D-amino acids; *m/z correspond to the molecular weights of the daughter peptides constituting the disulfide; Mpa—3-mercaptopropionic acid.
MATERIALS AND METHODS
Synthesis of Peptides
Automatic synthesis of linear peptides was carried out on a Tribute-UV synthesizer (Protein Technologies Inc., USA) on a scale of 0.15 mmol in accordance with the program of single coupling of Fmoc-amino acids (Novabiochem, Germany) using a polymer carrier with 2-chlorotrityl chloride anchor group (Iris Biotech, Germany) containing 1.2 eq. Cl/g. The control peptide SPB1 was synthesized on a Rink amide polymer (Novabiochem). To create an amide bond, N,N,N',N'-tetramethyl-O-(benzotriazol-1-yl)uronium hexafluorophosphate (HBTU) was used in the presence of 2 eq. N-methylmorpholine. At the end of the synthesis, the peptides were cleaved from the polymer with simultaneous removal of protecting groups by trifluoroacetic acid (TFA) with scavengers for 1.5 h. The products of the solid-phase synthesis were purified by reverse phase high performance liquid chromatography (HPLC) to 95–97% purity on a Knauer chromatograph (Germany) with a column (250 × 20 mm) Eurospher 100-10 C18 (Knauer). Buffer A (0.1% aqueous TFA) and buffer B—(80% acetonitrile (Carl Roth GmbH, Germany) in buffer A) were used as eluents. Elution was performed from 100% buffer A in a concentration gradient of 0.5%/min buffer B at a flow rate of 10 mL/min, peptides were detected at 220 nm. Analytical HPLC was carried using a Knauer chromatograph and the columns Kromasil 100-5 C18 (4.6 × 250 mm) (AkzoNobel, Sweden), sorbent particle size 5 μm, pore size 100 Å and Vydac 300-5 C18 (Supelco, USA), sorbent particle size 5 μm, pore size 300 Å. The following solvents were used as eluents: buffer A (0.05 M KH2PO4, pH 3.0), buffer B (70% acetonitrile in buffer A). The elution was carried out at a flow rate of 1 mL/min in the concentration gradient of buffer B in buffer A from 20% to 80% in 30 min. Chimeric molecules were obtained by directed disulfide bond formation using temporary 2-pyridine sulfenyl (PyS) [13] protection of cysteine residues involved in the disulfide formation and purified by HPLC on a column Vydac Protein & Peptide 300-5 С18 (10 × 250 mm; Grace/Alltech, USA). The eluents used were: buffer A—0.1% aqueous TFA and buffer B—0% acetonitrile in water. The elution was performed from 100% buffer A in a linear gradient of buffer B concentration by 0.5% per min at a flow rate of 3 mL/min (with detection carried out at 220 nm). The structure of the peptides was confirmed using matrix assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF). Mass spectra were recorded on a Bruker Autoflex speed mass spectrometer (Bruker Daltonics Inc., Germany). The amino acid sequences and characteristics of the peptides are shown in Table 1.
Microthermophoresis
The peptide binding was evaluated by microthermophoresis with recombinant RBD molecules corresponding to sequence of two SARS-CoV-2 variants: the original strain (GenBank ID NC_045512.2, the protein S amino acid residues 331-524) and the British variant B.1.1.7 with the N501Y mutation (GISAID EPI_ISL_683466) (Table 2). We used fluorescently labeled recombinant proteins obtained in the E. coli expression system. Fluorescent labeling of each protein by lysine residue (1 label per protein) and subsequent purification from excess dye was performed by flash chromatography using the NT™ Protein Labeling Kit RED-NHS (Nanotemper, Germany) according to the manufacturer’s protocol. The final protein solution in 10 mM potassium phosphate buffer (pH 7.4) containing 0.05% Tween-20 was titrated with peptide solutions. In this case, the final concentrations of peptides varied in the range from 0 to 175 μM, and the protein concentration was maintained constant (20 nM). Microscale thermophoresis curves were registered by fluorescence label detection at 22°C on a Monolith NT.115 instrument using standard capillaries (NanoTemper). The proportion of the complex was calculated from the change in the normalized fluorescence in the initial part of the thermophoretic curve and then the binding curves were obtained and analyzed using the MO. Affinity Analysis software (Nan-oTemper). The experimental data were approximated by the Hill’s model and the dissociation constants (Kd) were calculated as the effective peptide concentrations corresponding to 50% of the complex fraction.
Table 2.
Binding of the studied peptides with RBD and their antiviral activity in vitro
| Peptide | Binding to RBD (Kd, µM) | Antiviral activity in vitro (EС50, µM) | ||
|---|---|---|---|---|
| RBD (Wuhan) |
RBD (B.1.1.7) |
(1) Preincubation of peptides with the virus |
(2) Preincubation of cells with peptides followed by subsequent viral infection |
|
| SBP1 | >1000 | –* | >250** | >250 |
| h1 | >1000 | – | >250 | >250 |
| h1-Mpa | >1000 | – | >250 | >250 |
| h2 | 40 ± 8 | > 300 | >250 | >250 |
| h2-Cys | >1000 | – | >250 | >250 |
| Chimera X1 | 4.2 ± 0.5 | 0.9 ± 0.2 | >250 | >250 |
| Chimera X2 | 12 ± 5 | 1.8 ± 0.54 | >250 | >250 |
* Not studied; ** inacitve.
Evaluation of the Antiviral Activity
The antiviral activity of peptides was studied in vitro using the SARS-CoV-2 strain PIK35 (GISAID EPI_ISL_428852) [14]. The RBD sequence of the SARS-CoV-2 strain used corresponded to the RBD sequence of the original Wuhan strain, so that the in vitro model was similar to the binding of the studied peptides to the recombinant RBD molecule considered above. The antiviral activity was determined according to a previously published method [15] in two versions: (1) peptide dilutions were mixed with equal volumes of a viral suspension containing 100 TCID50 per well, incubated for 1 h at 37°C, then the virus-peptide mixtures were added to the confluent monolayers of Vero cells (in duplicates); or (2) peptides were added to the confluent monolayers of Vero cells in (duplicates), incubated for 1 h at 37°C, then the cells were infected with a viral suspension containing 100 TCID50 per well. Each experiment contained a positive control—N-hydroxycytidine (NHC) [16]—and virus titration for dose control.
The peptides were dissolved in 10% DMSO (Sigma, Germany) in water to a concentration of 10 mg/mL, and this solution was used in experiments with a maximum concentration in the culture medium of 1.25 mg/mL. (At this concentration we did not observe no precipitation for any of the studied peptides.)
RESULTS AND DISCUSSSION
Design of Peptides
Figure 1 shows a fragment of the spatial structure of the complex formed by the SARS-CoV-2 protein S RBD and the ACE2 cell receptor (Protein Data Bank ID: 7DX9 [17]); it was elucidated by cryoelectron microscopy. Based on this structure, the design of precursor peptides and chimeric molecules was carried out. The aim of the design phase of potential peptide inhibitors was to achieve maximal possible structural similarity between the original (ACE2 receptor) and the model (peptide inhibitor). During peptide structure construction, we have made two assumptions. First, the length of peptide fragments was limited to 22–25 residues; this was due to not only the technical capabilities of peptide synthesis and the practical feasibility of developing prototypes of peptide drugs, but also by the fact that polypeptides of this length could have their own spatial structure. In our case they can simulate the elements of the ACE2 spatial structure. Second, the options for creating intermolecular covalent bonds in chimeric peptides were limited to disulfide bonds. Linear peptides h1 and h2, used to construct chimeric molecules, corresponded to the fragments 21–42 and 64–88 of amino acid sequences of helices α1 and α2 of the ACE2 extracellular peptidase domain, respectively; they maintained unchanged most of the amino acid residues (a.a.) involved in the interaction with RBD. For example, the h1 peptide included most of the amino acids critical for binding to RBD a.a. from helix α1 ACE2 [8]: Q24, T27, D30, K31, H34, E35, E37, D38, Y41.
Fig. 1.
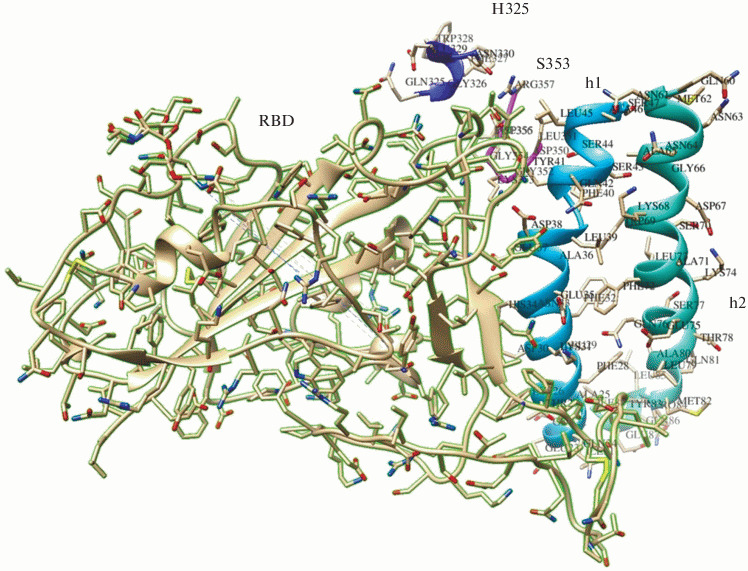
A fragment of the spatial structure (contact zone) of the SARS-CoV-2 receptor-binding domain (RBD) complex with the ACE2 cellular receptor [17] in a ribbon presentation. RBD—beige with a green border. Fragments of ACE2: helix α1 (h1) is shown in blue, helix α2 (h2)—in turquoise, beta hairpin centered on residue 353—in pink, a loop near residue 325—in blue.
Modifications of the precursor peptides included residues that were not involved in the interaction with RBD and were aimed at providing the possibility of synthesizing chimeric molecules on their basis, in which these peptides could be linked by covalent disulfide bonds.
For construction of the X1 chimera, we introduced the substitutions Ile21D-Cys (h1 peptide) and Glu87Cys (h2 peptide) and for construction of the X2 chimera, the Lys31 residue (h1 peptide) was modified with 3-mercaptopropionic acid (Mpa), and Gln76 was replaced with Cys76. The need of Lys31 modification was due to the fact that the length of the linker bond between the h1 and h2 peptides in the case of Cys31-Cys76 was insufficient in the X2 chimera. The distance between the peptides was increased by Lys31 modification. In addition, in the h2 peptide, the Met82 residue, which was not involved in contact with RBD, was replaced with isoleucine or norleucine to avoid oxidation problems. At this stage of the study, we also tried to preserve the stacking motif of aromatic amino acid residues Phe32-Phe72, Phe40-Trp69, although these regions were not involved in the interaction with RBD. In addition, minimal structural modification of the regions directly interacting with RBD was aimed at the increase in binding efficiency mainly due to dimers formation, rather than by other reasons. The chimeric molecules X1 and X2 differed from each other in the localization of disulfide bridges. The chimeric X1 molecule was a disulfide dimer in which the terminal cysteine residues in the h1 and h2 precursor molecules were connected by an S-S bond. In the case of modeling the chimeric X1 molecule, the optimal orientations of the side chains of the N-terminal cysteine residue and the C-terminal lysine residue in the h1 peptide, as well as the configuration of the disulfide bond, were obtained using the Cys21 and Lys42 D-isomers. In addition, the presence of D‑amino acids on the flanks of the h1 peptide should increase its resistance to the action of amino and carboxypeptidases. In the chimeric X2 molecule, the disulfide bond was in the middle of the molecule of each of the precursor peptides. Table 1 shows the structural formulas and characteristics of precursor peptides and chimeric molecules obtained on their basis.
Evaluation of Peptide Binding to SARS-CoV-2 RBD and their Antiviral Activity
The primary screening for binding to the Wuhan RBD revealed 3 active peptides: h2, and also chimeras X1 and X2. Therefore, the binding to variant B.1.1.7 RBD was evaluated only for these peptides. The active peptides demonstrated dose-dependent binding. Chimeric peptides X1 and X2 bound to RBD with the highest affinity (dissociation constants of 1–10 μM). At the same time, the X1 chimera with the terminal localization of the disulfide bond (D-Cys21-Cys87) between the h1 and h2 peptides had a higher (approximately 2 times) affinity for both RBD variants than the X2 chimera with the central localization of the disulfide bond [Lys31 (Mpa)—Cys76] between peptides h1 and h2. The data are presented in Table 2 and Fig. 2. The data obtained suggest that chimeric peptides may have antiviral activity by blocking interaction with the cellular ACE2 receptor. It should be noted that in the studied concentration range the control peptide SBP1 showed very weak binding to the recombinant RBD of the Wuhan virus. Similar results were obtained by the authors of [11]; they used RBDs obtained in various commercially available systems. Despite the micromolar dissociation constants of the SBP1 complex with RBD obtained by Sino Biological (China) in insect cells, this peptide did not bind to other RBD preparations obtained both in insects and in HEK cells. In our experiments, none of the synthesized peptides, including the control peptide SBP1 [11], exhibited any antiviral activity in the Vero cells (Table 2), regardless of the experimental protocol.
Fig. 2.
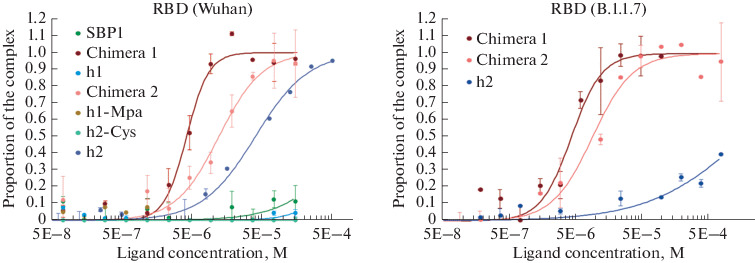
Binding of the studied peptides with the recombinant RBD molecules corresponding in sequence to the original Wuhan strain (Wuhan, GenBank ID NC_045512.2) and the British variant with the N501Y mutation (B.1.1.7, GISAID EPI_ISL_683466) as determined by microthermophoresis.
Similar data were obtained by the authors of the study [18]; they developed a series of stapled peptidomimetics based on the α1 ACE2 helix, in which an intramolecular hydrocarbon bridge was created to fix the α-helical structure. However, none of these 15 peptidomimetics, including SBP1 used by the authors as a control, showed any antiviral activity in vitro [18].
The putative mechanism of action of the proposed peptides is similar to the action of neutralizing antibodies and must be realized before the virion penetration into the cell by means of blocking the RBD binding to the receptor. Thus, the ability of peptides to penetrate the cell should not have a significant impact, at least in cell culture studies. It can be assumed that the lack of in vitro activity of peptides is associated with the RBD conformation in the virion, which does not allow peptides to bind tightly to the protein.
CONCLUSIONS
The proposed chimeric peptides showed binding to RBDs of both the original Wuhan variant and the British variant B.1.1.7 of the SARS-CoV-2 with dissociation constant values ranged from 1 to 10 μM. The obtained experimental results demonstrated the essential role of the localization of covalent crosslinks between monomers for the binding of peptides to SARS-CoV-2 RBD.
ACKNOWLEDGMENTS
The authors are grateful to D. Osolodkin, PhD, for his valuable suggestions on data presentation.
FUNDING
This work was supported by the Russian Foundation for Basic Research (project no. 20-04-60110).
COMPLIANCE WITH ETHICAL STANDARDS
This article does not contain any research involving humans or using animals as experimental objects.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
Footnotes
Translated by A. Medvedev
REFERENCES
- 1.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Cell. 2020;180:281–292. doi: 10.1016/j.cell.2020.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang Ch.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zhen X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Imai Y., Kuba K., Rao S., Huan Y., Guo F., Guan B., Yang P., Sarao R., Wada T., Leong-Poi H., Crackower M.A., Fukamizu A., Hui C.-Ch., Hein L., Uhlig S., Slutsky A.S., Jiang C., Penninger J.M. Nature. 2005;436:112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.NCBI. ACE2 angiotensin I converting enzyme 2 [Homo sapiens (human)] Available from: https://www.ncbi.nlm.nih.gov/gene/59272, 2020
- 6.Hamming I., Timens W., Bulthuis M.L.C., Lely A.T., Navis G.J., van Goor H. J. Pathology. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monteil V., Kwon H., Prado P., Hagelkrüys A., Wimmer R.A., Stahl M., Leopoldi A., Garreta E., Hurtado del Pozo C., Prosper F., Romero J.P., Wirnsberger G., Zhang H., Slutsky A.S., Conder R., Montserrat N., Mirazimi A., Penninger J.M. Cell. 2020;181:905–913. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han Y., Krґal P. ACS Nano. 2020;14:5143–5147. doi: 10.1021/acsnano. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barh, D., Tiwari, S., Silva Andrade, B., Giovanetti, M., Kumavath, R., Ghosh, P., Góes-Neto, A., Carlos Junior Alcantara, L., and Azevedo, V., Preprints, 2020, 2020040347. 10.20944/preprints202004.0347.v1
- 10.Huang X., Pearce R., Zhang Y. Aging. 2020;12:11263–11276. doi: 10.18632/aging.103416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang, G., Pomplun, S., Loftis, A.R., Tan, X., Loas, A., and Pentelute, B.L., bioRxiv, 2020, p. 2020.2003.2019.999318. 10.1101/2020.03.19.999318
- 12.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sidorova M.V., Arefieva T.I., Palkeeva M.E., Molokoedov A.S., Az’muko A.A., Ruleva N.Y., Pylaeva E., Krasnikova T., Bespalova Zh.D. Russ. J. Bioorg. Chem. 2015;41:10–18. doi: 10.1134/S1068162015010124. [DOI] [PubMed] [Google Scholar]
- 14.Kozlovskaya L., Piniaeva A., Ignatyev G., Seliva-nov A., Shishova A., Kovpak A., Gordeychuk I., Ivin Y., Berestovskay A., Prokhortchouk E., Protsenko D., Rychev M., Ishmukhametov A. Int. J. Infect. Dis. 2020;99:40–46. doi: 10.1016/j.ijid.2020.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kozlovskaya L.I., Volok V.P., Shtro A.A., Nikolaeva Y.V., Chistov A.A., Matyugina E.S., Belyaev E.S., Jegorov A.V., Snoeck R., Korshun V.A., Andrei G., Osolodkin D.I., Ishmukhametov A.A., Aralov A.V. Eur. J. Med. Chem. 2021;220:113467. doi: 10.1016/j.ejmech.2021.113467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sheahan, T.P., Sims, A.C., Zhou, S.T., Graham, R.L., Pruijssers, A.J., Agostini, M.L., Leist, S.R., Schafer, A., Dinnon, K.H., Stevens, L.J., Chappell, J.D., Lu, X.T., Hughes, T.M., George, A.S., Hill, S.A., Montgomery, A.J., Brown, G.R., Bluemling, M.G., Natchus, M., Saindane, A.A., Koly-khalov, C.S., Painter, G., Harcourt, J., Tamin, A., Thornburg, N.J., Swanstrom, R., Denison, M.R., and Baric, R.S., Sci. Transl. Med., 2020, vol. 12, no. 541, eabb5883. 10.1126/scitranslmed.abb5883 [DOI] [PMC free article] [PubMed]
- 17.Yan R., Zhang Y., Li Y., Ye F., Guo Y., Xia L., Zhong X., Chi X., Zhou Q. Cell Res. 2021;31:717–719. doi: 10.1038/s41422-021-00490-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morgan D.C., Morris C., Mahindra A., Blair C.M., Tejeda G., Herbert I., Turnbull M.L., Lieber G., Willett B.J., Logan N., Smith B., Tobin A.B., Bhella D., Baillie G., Jamieson A.G. Peptide Science. 2021;2021:e24217. doi: 10.1002/pep2.24217. [DOI] [PMC free article] [PubMed] [Google Scholar]


