Abstract
Imprinted genes are expressed from one allele according to their parent of origin, and many are essential to mammalian embryogenesis. Here we show that the ɛ-sarcoglycan gene (Sgce) and Zac1 (Lot1) are both paternally expressed imprinted genes. They were identified in a subtractive screen for imprinted genes using a cDNA library made from novel parthenogenetic and wild-type fibroblast lines. Sgce is a component of the dystrophin-sarcoglycan complex, Zac1 is a nuclear protein inducing growth arrest and/or apoptosis, and Zac1 is a potential tumor suppressor gene. Sgce and Zac1 are expressed predominantly from their paternal alleles in all adult mouse tissues, except that Zac1 is biallelic in the liver and Sgce is weakly expressed from the maternal allele in the brain. Sgce and Zac1 are broadly expressed in embryos, with Zac1 being highly expressed in the liver primordium, the umbilical region, and the neural tube. Sgce, however, is strongly expressed in the allantoic region on day 9.5 but becomes more widely expressed throughout the embryo by day 11.5. Sgce is located at the proximal end of mouse chromosome 6 and is a candidate gene for embryonic lethality associated with uniparental maternal inheritance of this region. Zac1 maps to the proximal region of chromosome 10, identifying a new imprinted locus in the mouse, homologous with human chromosome 6q24-q25. In humans, unipaternal disomy for this region is associated with fetal growth retardation and transient neonatal diabetes mellitus. In addition, loss of expression of ZAC has been described for a number of breast and ovarian carcinomas, suggesting that ZAC is a potential tumor suppressor gene.
The normal development of the mammalian embryo requires both the maternal and the paternal genomes (4, 27). Mouse embryos in which the entire genome is either of maternal origin (parthenotes) or of paternal origin (androgenotes) usually die at or shortly after implantation. Rarely do these embryos develop to E10, and none have developed to term. The basis for this failure in development is that some genes are imprinted and are expressed from only one parental allele. Thus, loss of the expressed allele can render the embryo null for the gene's function. For some imprinted genes, such as the maternally expressed Igf2/mannose-6-phosphate receptor allele (Igf2r) (3), this results in embryonic lethality, whereas mutation of the paternally inherited allele, which is not expressed, has no effect on viability (23).
In the mouse, most imprinted genes are found in clusters distributed among 10 regions over six chromosomes (C. V. Beechey and B. M. Cattanach, 1998, mouse imprinting data and references [http://www.mgu.har.mrc.ac.uk/imprinting/implink.html]). These regions were first defined by the elegant use of chromosomal translocations to derive embryos uniparental for specific chromosomal regions, with their inheritance having overt phenotypic effects on embryogenesis, postnatal growth, and behavior (6). Some of the imprinted genes responsible for these phenotypes have been identified (3, 8, 15, 24, 47, 48). Though these translocation studies may have been exhaustive in defining regions which when inherited uniparentally result in a severe phenotype, not all imprinted genes in the mouse map to these regions as defined above. Five imprinted genes have been localized outside imprinted loci (Ins1 on chromosome 19 [14], Grf1 on chromosome 9 [32], Peg1 [Mest] on chromosome 6 [24], Nnat on chromosome 2 [46], and Impact on chromosome 18 [16]). Two of these genes, Grf1 and Peg1, when mutated result in the newborns exhibiting subtle postnatal growth and/or behavioral defects (J. M. Itier, G. L. Tremp, J. F. Leonard, M. C. Multon, G. Ret, F. Schweighoffer, B. Tocque, M. T. Bluet-Pajot, V. Cormier, and F. Dautry, Letter, Nature 393:125–126, 1998). These results reveal that imprinted genes are more widely distributed in the mouse genome than previously anticipated and that their mutation can result in subtle phenotypes. However, genes mapping to other imprinted regions, such as the proximal region of chromosome 6 and for the middle and distal regions of chromosome 12, both of which are associated with embryonic lethality, remain to be identified. Consequently, the identification of other imprinted genes in chromosomes 6 and 12 or other unknown imprinted regions is of major importance, as this may provide further insights into the role(s) of imprinting in mammalian development, its contribution to various disease processes in humans, and, ultimately, why and how this form of gene regulation evolved in mammals.
Here we describe a procedure by which androgenetic (AG) and parthenogenetic (PG) mouse embryonic fibroblast (MEF) lines that stably retain the parent-of-origin pattern of imprinted gene expression were established in culture. We used them as a source of mRNA for a suppressive subtractive screen for paternally expressed genes. We identified two novel imprinted genes, one of which, the gene for ɛ-sarcoglycan (Sgce), maps to proximal chromosome 6, while the other, Zac1, maps to chromosome 10 in a region previously not known to be imprinted in the mouse. The importance of Zac1 and Sgce imprinting is discussed in the context of mouse development and human disease. Furthermore, these cell lines will be an important source of material for searching for other imprinted genes and for studying various questions related to imprinting.
MATERIALS AND METHODS
Embryos and the derivation of fibroblasts.
MEFs were derived by explanting and culturing day 13 (d13) (day of plug ≡ d1) embryos after removing the head and internal organs. PG MEFs were generated from 13-day PG↔chimeric embryos made by aggregating PG embryos, constitutively expressing the Neor gene (36), with wild-type (WT) embryos. The WT MEFs from the chimeras were selected against by culturing the primary explants in medium supplemented with G418 for the first three passages, allowing only the PG cells carrying the neomycin gene to survive (37). AG MEFs were generated from chimeras made by injecting AG embryonic stem (ES) cells (25), transfected with the PgkNeo cassette so that they constitutively expressed a Neor gene, into blastocysts (35) with their subsequent isolation by the same procedure used to derive the PG MEFs.
cDNA subtraction and differential screening.
Total RNA from MEFs, PG MEFs, and AG MEFs was isolated using the RNeasy procedure (Qiagen), and poly(A)+ RNA was purified with the poly(A) Track mRNA isolation system (Promega). Suppressive subtractive hybridization was carried out using the PCR-Select cDNA subtraction kit (Clontech) according to the manufacturer's protocol. MEF cDNA was used as the tester and PG MEF cDNA was used as the driver in the forward reaction which was designed to enrich the tester, MEF cDNA, for paternally expressed genes not present in the PG MEF cDNA. The subtraction was also performed in reverse by using PG MEF RNA as tester and MEF RNA as driver, to generate a probe lacking paternally expressed genes. Differential screening was performed by high-throughput cDNA array analysis using the PCR-Select differential screening kit (Clontech). Clones were blotted simultaneously on two membranes, hybridized with the forward probe and the reverse probes, and quantified by phosphorimager analysis.
Reverse transcriptase (RT) PCR.
Total RNA was extracted with the RNeasy columns (Qiagen) and treated with RNase-free DNase I (Promega) to eliminate residual genomic DNA. Amplification consisted of 30 cycles at 94°C for 30 s, 58°C for 30 s, and 72°C for 60 s followed by one cycle at 72°C for 7 min. Primers for Igf2, Igf2r, H19, and Snrpn were as described previously (41). Other primers were as follows: Ndn, 5′ CAGCCGAGGTCCCCGACTGTGAG and 3′ GCAGCCCGAACACTCTGGCGAGG; p57kip2, 5′ CCGCGCAAACGTCTGAGATGAG and 3′ CACCTTGGGACCAGCGTACTCC; Grb10, 5′ CAACGATATTAACTCGTCCGTGG and 3′ CCACTTCTCACATCTGCCACAATG; Peg1 (Mest), 5′ AGCTCAGTGGTAGTGTGCCTGCC and 3′ TCCACGTCAGCCCTGGAGGAGCT; Sgce, 5′ GGGGTGGCAGAGTGCCGCTTCC and 3′ GGCAGCACATGATATAAGCGAG; Zac1, 5′ ATCCTGTTCCTACCTCATATGC and 3′ CTGGATCTGCAACTGAAACTGTGG; Gas2, 5′ CACAGAGAAGCTGTGTTTAGGATGATC and 3′ GATATGTCCTGGGTATACAGTCTGT; Igfbp5, 5′ GCAAGGGCTAAGGAGACACTCCCC and 3′ GGCTAGAGCTGAAAGCAAAAGGGC; and Rpl19, 5′ CTGAAGGTCAAGGGGAATG and 3′ GGACAGAGTTTTGATGATCTC.
Virtual Northern blotting.
For virtual Northern blot analysis, double-stranded cDNA was synthesized by using the SMART cDNA synthesis kit (Clontech). Double-stranded cDNA (0.5 μg) was electrophoresed on a 1.2% agarose gel, transferred to a nylon membrane, and hybridized with the indicated labeled probe.
Whole-mount in situ hybridization.
Dissected embryos were processed for in situ hybridization as described previously (18). Zac1 (1 to 1425) and Sgce (872 to 1422) sense and antisense riboprobes were synthesized from the appropriate mouse cDNA clones.
Chromosomal mapping.
Interspecific backcross progeny were generated by mating (C57BL/6J × Mus spretus)F1 females and C57BL/6J males as described previously (7). A total of 205 N2 mice were used to map the Sgce and Zac1 loci. DNA isolation, restriction enzyme digestion, agarose gel electrophoresis, Southern blot transfer, and hybridization were performed essentially as described previously (19). By use of the Sgce probe, an ∼500-bp fragment of mouse cDNA, fragments of 8.6, 3.4, 2.2, and 1.6 kb were detected in ScaI-digested C57BL/6J DNA and fragments of 8.6, 3.7, 2.8, and 1.6 kb were detected in ScaI-digested M. spretus DNA. The Zac1 probe, an ∼1.2-kb fragment of mouse cDNA, detected an ∼20.0-kb SacI fragment in C57BL/6J DNA and a 14.0-kb SacI fragment in M. spretus DNA. The presence or absence of the M. spretus-specific fragments was monitored in backcross mice. A description of the probes and restriction fragment length polymorphisms (RFLPs) for the loci linked to Sgce including Calcr, Met, and Cpa has been reported previously (17, 44); those linked to Zac1 include Estra, Myb, and Lama2 (20, 29). Recombination distances were calculated using Map Manager, version 2.6.5. Gene order was determined by minimizing the number of recombination events required to explain the allele distribution patterns.
RESULTS
Establishment of uniparental primary MEFs.
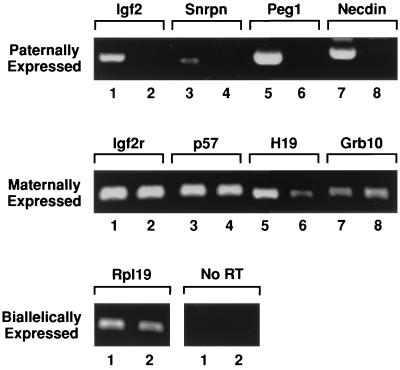
To circumvent the difficulties of producing uniparental embryos in sufficient numbers, AG and PG primary MEFs were established from chimeras made between WT embryos and AG ES cells or PG embryos, respectively (25). The AG ES cells and the PG embryos used to make the chimeras both constitutively expressed the Neor gene and so were resistant to neomycin. AG and PG fibroblasts were isolated from the WT cells by culturing the explanted chimeras in high concentrations of neomycin for the first three passages. To ensure that these lines had stably retained their imprinted status, the expression of eight known imprinted genes, together with biallelically expressed genes, was analyzed by RT-PCR in WT MEFs, isolated from fertilized embryos, and PG MEFs. Paternally expressed genes such as Igf2, Snrpn, Peg1 (Mest) (referred to as Peg1), and Ndn were detected in WT MEFs (Fig. 1, lanes 1, 3, 5, and 7) but not in PG MEFs (lanes 2, 4, 6, and 8). In contrast, maternally expressed genes such as p57kip2, Igf2r, H19, and Grb10 were expressed in both PG MEFs and WT MEFs (Fig. 1, middle panel). The biallelic genes Rpl19 and G3pdh (data not shown) were expressed in both lines at similar levels (Fig. 1, bottom panel). These results clearly demonstrate that PG MEFs retained the appropriate expression pattern of several known imprinted genes. These lines were then used as a source of mRNA for cDNA screening of paternally expressed genes using a suppressive subtractive hybridization procedure.
FIG. 1.
Appropriate expression of known imprinted genes in PG MEFs. Total RNA isolated from PG and WT MEFs was analyzed by RT-PCR (lanes 1, 3, 5, and 7 are WT MEFs, and lanes 2, 4, 6, and 8 are PG MEFs).
Identification of novel imprinted genes.
To identify genes exclusively expressed from their paternal allele, cDNA libraries made from WT and PG MEF mRNAs were used in a suppressive subtractive hybridization (9) as tester and driver, respectively. Following subtractive hybridization, a cDNA library was prepared and screened using probes enriched in paternally expressed genes (forward subtracted) versus those lacking paternally expressed genes (reverse subtracted).
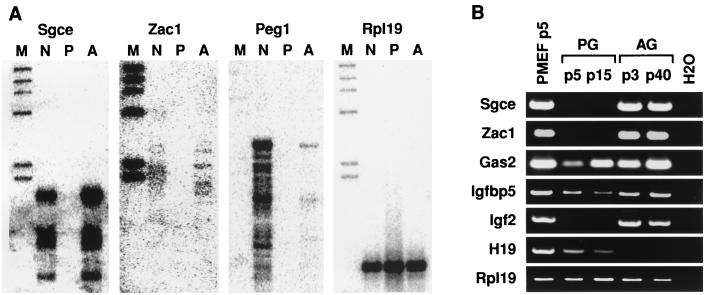
A total of 1,200 clones were screened for differential hybridization with the forward and reverse probes. Approximately 10% of these clones showed strong hybridization with the forward but not the reverse probe and were sequenced. Among these differentially expressed clones, one was identified as Igf2 and six were Peg1 (Mest), these both being known as maternally imprinted genes, and as well there were two previously unidentified candidate imprinted genes, that for ɛ-sarcoglycan (Sgce) and Zac1. The presence of Igf2 and Peg1 (Mest) in the isolated clones provided a clear indication that the screened cDNA library was enriched in paternally expressed genes. The expression of candidate imprinted genes was further analyzed by virtual Northern blotting in which mRNA from the WT, AG, and PG MEFs was reverse transcribed into cDNA and probed with the candidate imprinted genes. AG MEF cDNA was used as a positive control to confirm the paternal expression of candidate genes which should be absent in PG MEFs but present in WT MEFs. As shown in Fig. 2A, Sgce (isolated in four independent clones) and Zac1 (one clone) are expressed in AG and WT MEFs but not in PG MEFs. Peg1, a paternally expressed gene, and Rpl19, a biallelic gene, are shown as controls. The potential imprinted status of Sgce and Zac1 was further analyzed by RT-PCR of the AG, PG, and WT cDNAs. We found high levels of expression of both Sgce and Zac1 in AG MEFs and WT MEFs but no expression in PG MEFs (Fig. 2B). Other genes such as Gas2, Igfbp5, Sarp, and Cnp6, as well as two expressed sequence tags, were differentially expressed but not imprinted as shown by RT-PCR (Fig. 2B and data not shown). These results indicate that about 10% of the clones that were found to be differentially expressed in the enriched cDNA library were imprinted genes. The other nonimprinted genes showing differential expression were probably detected due to differences in the growth rates between our AG and PG fibroblast lines (unpublished observations).
FIG. 2.
Differential expression of clones isolated from the screen by virtual Northern blotting (A) and RT-PCR (B). Rpl19 probe was used as a control for both the virtual Northern blotting and RT-PCR. Both Sgce and Zac1 are expressed only in the AG and WT MEFs by both virtual Northern blotting and RT-PCR. Other genes such as Gas and Igfbp5 are biallelically expressed. The maternally expressed H19 was analyzed by RT-PCR as a control for AG MEFs not expressing maternally expressed imprinted genes (abbreviations: M, markers; N, WT MEFs; P, PG MEFs; A, AG MEF cDNA).
Imprinting of Sgce and Zac1 in vivo.
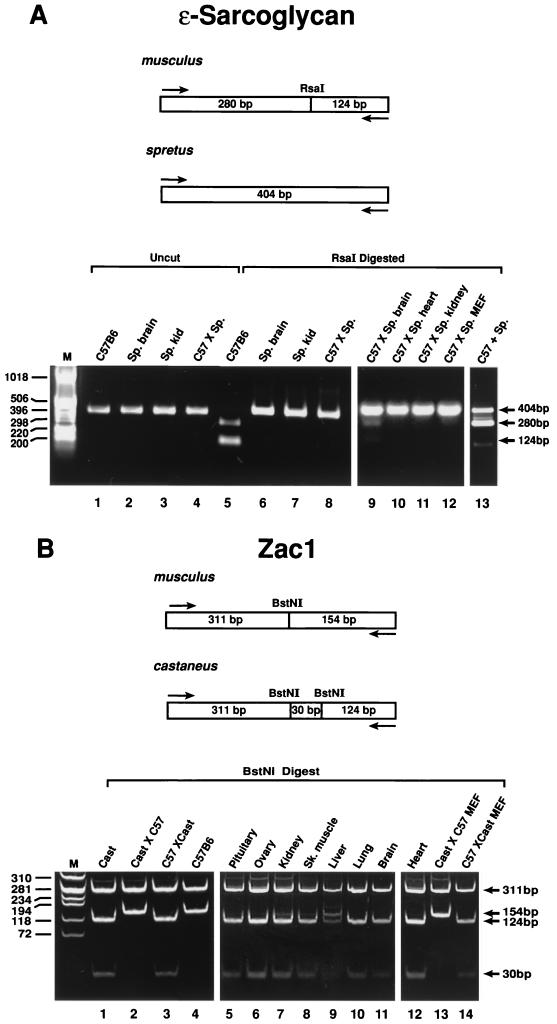
To confirm that Sgce and Zac1 were imprinted, allele-specific expression of the genes was analyzed in interspecific hybrid embryos and adult tissues using restriction enzyme polymorphisms in their cDNA sequences. A polymorphic change from GTAC to ATAC between Mus musculus and M. spretus deletes an RsaI site in the cDNA of Sgce from M. spretus (Fig. 3A). After PCR, a 404-bp fragment was isolated and digested with RsaI. cDNA from M. musculus, following RsaI digestion, yielded two fragments of 280 and 124 bp (Fig. 3A, lane 5). In contrast, only the undigested fragment of 404 bp was found in M. spretus cDNA (Fig. 3A, lanes 6 and 7). When the cDNA from C57BL/6J × M. spretus interspecific hybrid embryos and a variety of adult tissues was analyzed, only the undigested M. spretus paternal 404-bp fragment (Fig. 3A, lane 8) was detected in the majority of these samples. The one exception was the adult brain, where, in addition to the predominant 404-bp paternal band, a signal from the 280- and 124-bp bands indicative of the maternal allele is weakly expressed in this tissue (Fig. 3A, lane 9). M. musculus males mated with M. spretus females do not breed successfully, and reciprocal crosses could not be made. As a control, to ensure that both alleles could be amplified in the same reaction, total RNA from both M. musculus and M. spretus was mixed and subjected to RT-PCR, and equal amplification of both alleles is shown in Fig. 3A, lane 13. This demonstrated that Sgce was transcribed from the paternal allele in the embryo and the majority of adult tissues and that therefore Sgce is maternally imprinted in vivo.
FIG. 3.
(A) Monoallelic expression of Sgce in interspecific embryos by RFLP analysis. A PCR fragment was amplified from both C57BL/6J and M. spretus and sequenced. In lanes 1 to 4, the 404-bp fragment is amplified in both species. In lanes 5 to 7, only the C57BL/6J product is cut with RsaI. In embryonic cDNAs from the interspecific cross, the 404-bp fragment is undigested by RsaI, showing exclusive paternal (M. spretus) expression (lane 8), as seen also for adult tissues (lanes 10 to 12). Only in the adult interspecific brain is weak expression of the maternal allele detected (lane 9). Lane 13 is a control showing coamplification of M. musculus and M. spretus alleles after they are mixed in the same reaction. (B) Paternal expression of Zac1 in interspecific embryos by RFLP analysis. A 465-bp PCR fragment was amplified from both C57BL/6J and M. m. castaneus and digested with BstNI. M. m. castaneus produces three fragments, of 311, 124, and 30 bp (lane 1), whereas the C57BL/6 allele produces two fragments, of 311 and 154 bp (lane 4). Embryo and MEF interspecific cDNA from reciprocal crosses revealed that only the paternal allele is expressed (lanes 2, 4, 13, and 14). In adult tissues, biparental expression is most apparent in the liver (lane 9) and to a lesser extent in kidney and skeletal muscle (lanes 7 and 8).
A similar analysis was performed to assess the imprinted status of Zac1. A polymorphism was found between M. musculus and Mus musculus castaneus. In both, the primers amplified a 465-bp fragment after PCR. A change in the sequence from TCTGG to CCTGG created an additional BstNI site in M. m. castaneus cDNA. Restriction digestion with BstNI resulted in three fragments (311, 124, and 30 bp) in M. m. castaneus (Fig. 3B, lane 1) versus two fragments (311 and 154 bp) in M. musculus cDNA (Fig. 3B, lane 4). Analysis of cDNA from E13 embryos of both reciprocal crosses (C57BL/6J × M. m. castaneus and M. m. castaneus × C57BL/6J) revealed that the restriction pattern always corresponded to that of the father, demonstrating that Zac1 was expressed from the paternal allele and therefore is maternally imprinted (Fig. 3B, lanes 2 and 3). Subsequent analysis of cDNA from adult C57BL/6J × M. m. castaneus tissues revealed that paternal expression was retained in the pituitary, ovary, lung, brain, and heart tissue (lanes 5, 6, and 10 to 12) but that equal expression of the maternal and paternal alleles occurred in the liver (lane 9).
Expression of Sgce and Zac1 in embryos and adult tissues.
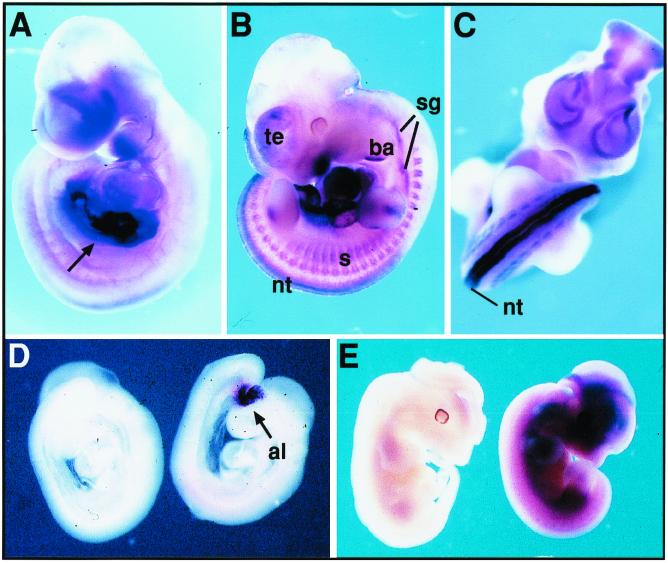
Using RT-PCR (Fig. 3) and Northern blotting (data not shown), we observed that Sgce and Zac1 are expressed in all adult tissues analyzed, including skeletal muscle, kidney, liver, lung, brain, and heart. During embryogenesis, in midgestation (d9.5) embryos, Sgce and Zac1 are both detected by in situ hybridization. Zac1 is strongly expressed in the liver primordium as well as the umbilical region (Fig. 4A). Subsequently, in d11 to d12 embryos, Zac1 showed high levels of localization to the neural tube, with weaker expression in the somites, sympathetic ganglia, distal second brachial arch, and telencephalic vesicles (Fig. 4B and C). Sgce is first detected in the allantoic region (Fig. 4D), and in later stages (d11 to d12), its expression becomes more widespread and diffuse among many tissues (Fig. 4E).
FIG. 4.
Detection of Zac1 and Sgce transcripts in mouse embryos by in situ hybridization. (A) Zac1 is highly expressed in the liver primordium and body wall of the umbilical region (arrow) of a d9.5 embryo. (B) At d11, Zac1 expression is observed in neural tube (nt), somites (s), sympathetic ganglia (sg), distal second branchial arch (ba), and telencephalic vesicles (te). (C) Zac1 showed a strong expression in the neural tube (nt) at d10.5. (D and E) Sgce expression is restricted to the allantoic region (al) at d9.0 (D, right), whereas at d12 (E, right) this transcript is widely distributed (embryos hybridized with a sense probe are shown on the left of each panel).
Mapping of Sgce and Zac1.
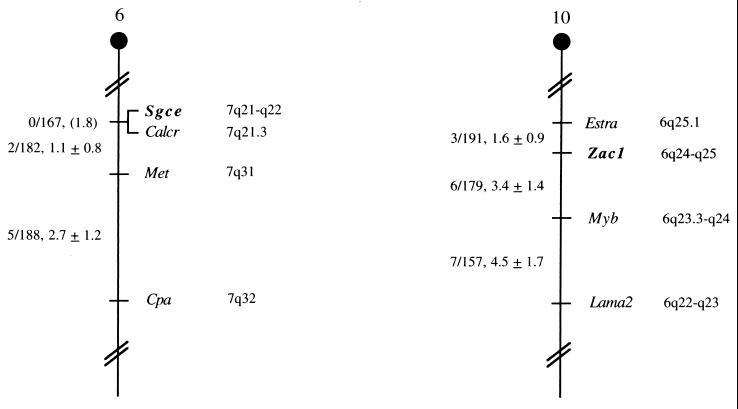
The mouse chromosomal locations of Sgce and Zac1 were determined by interspecific backcross analysis using progeny derived from matings of (C57BL/6J × M. spretus)F1 × C57BL/6J mice (7). Sgce is located in the proximal region of mouse chromosome 6 linked to Calcr, Met, and Cpa. Zac1 mapped to the proximal region of mouse chromosome 10 linked to Estra, Myb, and Lama2 (Fig. 5). The proximal region of mouse chromosome 6 shares a region of homology with human chromosome 7q. Our placement of Scge in this interval is consistent with the assignment of SGCE to 7q21-q22 (28). The proximal region of mouse chromosome 10 shares a region of homology with human chromosome 6q, and ZAC has been mapped to human chromosome 6q24-q25 (43).
FIG. 5.
Murine chromosomal location of Sgce and Zac1. Partial chromosome 6 and 10 linkage maps showing the location of Sgce and Zac1 in relation to linked genes are shown. The number of recombinant N2 animals over the total of N2 animals typed together with the recombination frequency (genetic distance in centimorgans ± standard error) is shown for each pair of loci on the left of the map. Where no recombination was detected between loci, the upper 95% confidence limit of the recombination distance is shown in parentheses. The positions of homologous loci on human chromosomes are shown to the right.
DISCUSSION
Here we report the identification of two novel paternally expressed imprinted genes, Sgce and Zac1, by the subtractive hybridization screen of a cDNA library derived from the WT and PG MEF mRNAs. Previous attempts at deriving ES lines from AG and PG embryos to analyze imprinted gene expression in vitro were only partially successful since the cell lines showed extensive leakiness in retaining the appropriate expression of imprinted genes following their differentiation (2, 40). By making chimeras and then selecting for AG and PG fibroblasts from explanted midgestation embryos, we established lines that are uniform in their differentiation and stable in the expression of known imprinted genes. These lines provide distinct advantages in searching for novel imprinted genes in that they allow workers to avoid having to repeatedly produce AG and PG embryos, they can be expanded in vitro so that sufficient quantities of mRNA can be isolated, and they provide a quick and efficient means to screen for candidate imprinted genes.
We screened 1,200 clones, compared to some 50,000 analyzed in a previous report (21). Our screen resulted in the isolation of two known imprinted genes, Peg1 and Igf2, and two genes that were not known to be imprinted, Sgce and Zac1. Using polymorphisms in the Sgce and Zac1 cDNAs, we showed that both of these genes were imprinted in vivo, in embryonic fibroblasts and the majority of adult tissues.
Sgce is one of five members in the sarcoglycan family, transmembrane proteins which are components of the dystrophin-sarcoglycan complex (10, 28). This complex forms a structural link between the extracellular matrix and cytoskeleton predominantly in the various types of muscle. The α-, β-, γ-, δ-, and ɛ-sarcoglycans are all found in skeletal and cardiac muscle (38). Unlike the other members, Sgce is more widely expressed among adult and embryonic tissues, as shown by our in situ analysis (10), which is consistent with the recent demonstration that it is predominantly localized to smooth muscle, particularly that of the blood vessels (39). Embryonic expression of the other sarcoglycans is first detected during later stages of myogenesis with expression being largely restricted to the various musculatures throughout adulthood. Sgce may therefore have a more widespread role in maintaining cell adhesion and tissue integrity than those of the other sarcoglycans. In the mouse, Sgce maps to the very proximal region of chromosome 6 close to the centromere. This is within the region where, according to the more recently refined imprinting map, maternal uniparental inheritance is associated with embryonic lethality, excluding Peg1 (C. V. Beechey and B. M. Cattanach, 1998, mouse imprinting data and references [http://www.mgu.har.mrc.ak.uk/imprinting/implink.html]). Since Sgce is widely expressed in the embryo, it is a candidate gene for embryonic lethality associated with maternal uniparental isodisomy in this region of the chromosome. The proximal region of mouse chromosome 6 shares homology with human chromosome 7q21-q22 (Fig. 5), and maternal uniparental disomy for chromosome 7 has been sporadically associated with Silver-Russell syndrome (SRS) (45), a condition characterized by pre- and postnatal growth retardation. In mice, mutation of the paternal allele of Peg1 (Mest) resulted in fetal growth retardation and was considered a candidate for SRS (24). However, a recent analysis has suggested that PEG1 has no role in SRS (33). Therefore, Sgce is another candidate for this condition.
Zac1 has been independently identified in two previous screens, the first for genes regulated by neuropeptides (34) and the second for genes whose expression was lost on transformation in a rat ovarian carcinoma model (1). Zac1 is a zinc finger DNA binding protein of the C2H2 family. Its biological functions remain unclear, although in mice (Fig. 4 and data not shown) and humans it is most strongly expressed in the pituitary gland and to a lesser extent in other tissues (43). Zac1 maps to the proximal region of mouse chromosome 10, at a region homologous to 6q24-q25 in the human. Parent-of-origin defects have been reported for chromosome 6 with paternal duplication-isodisomy being associated with fetal growth retardation (often severe) and transient neonatal diabetes mellitus (12, 13, 42). However, maternal duplication or a deletion encompassing this region does not impair growth (22, 31). These observations suggest that Zac1 (ZAC) or some closely linked gene(s) is a candidate gene which, when paternally duplicated, may be responsible for fetal growth retardation and transient neonatal diabetes mellitus.
The distal region of chromosome 6 also shows a high incidence of loss of heterozygosity in the development of a variety of tumors, particularly those of the breast, ovary, and cervix (11, 26, 30). Transfection of mouse and human cells with Zac1 (ZAC) induces proliferative arrest at G1 and apoptosis, suggesting that Zac1 (ZAC) could function as a tumor suppressor gene (34, 43; our unpublished observations). Whether imprinting of ZAC, as a potential tumor suppressor, contributes to tumor formation is unclear. However, in an analysis of 42 primary breast carcinomas, none showed any detectable mutation in the coding sequence of ZAC but 8 failed to express ZAC or expressed it at very low levels (5). Expression could, however, be induced by treatment of the carcinomas with the demethylating reagent 5-azacytidine, suggesting that expression may be epigenetically regulated. It would be of interest to determine whether in these eight lines the paternal allele had been lost, leaving the intact but transcriptionally repressed maternal allele. Furthermore, Zac1 can be induced in our PG fibroblasts following treatment with 5-azacytidine (unpublished observations). Gene targeting and transgenic approaches to manipulating the expression of Zac1 and Sgce should aid in determining the role(s) of these imprinted genes in the development and regulation of both embryonic and cellular growth.
ACKNOWLEDGMENTS
We thank Ruth Wolf, Terry Sulivan, Teresa Shatzer, Lori Sewell, Andreé Reuss, and Deborah B. Householder for excellent technical assistance; Anne Wang, Mark Lewandowski, and John Hagan for critical reading of the manuscript; and Rachel Wevrick for fruitful discussions and advice.
This research was supported, in part, by the National Cancer Institute, DHHS, under contract with ABL.
ADDENDUM IN PROOF
Kamiya et al. (Hum. Mol. Genet. 9:453–460, 2000) reported that ZAC is imprinted in humans.
REFERENCES
- 1.Abdollahi A, Roberts D, Godwin A K, Schultz D C, Sonoda G, Testa J R, Hamilton T C. Identification of a zinc-finger gene at 6q25: a chromosomal region implicated in development of many solid tumors. Oncogene. 1997;14:1973–1979. doi: 10.1038/sj.onc.1201034. [DOI] [PubMed] [Google Scholar]
- 2.Allen N D, Barton S C, Hilton K, Norris M L, Surani M A. A functional analysis of imprinting in parthenogenetic embryonic stem cells. Development. 1994;120:1473–1482. doi: 10.1242/dev.120.6.1473. [DOI] [PubMed] [Google Scholar]
- 3.Barlow D P, Stoger R, Herrmann B G, Saito K, Schweifer N. The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature. 1991;349:84–87. doi: 10.1038/349084a0. [DOI] [PubMed] [Google Scholar]
- 4.Barton S C, Surani M A, Norris M L. Role of paternal and maternal genomes in mouse development. Nature. 1984;311:374–376. doi: 10.1038/311374a0. [DOI] [PubMed] [Google Scholar]
- 5.Bilanges B, Varrault A, Basyuk E, Rodriguez C, Mazumdar A, Pantaloni C, Bockaert J, Theillet C, Spengler D, Journot L. Loss of expression of the candidate tumor suppressor gene ZAC in breast cancer cell lines and primary tumors. Oncogene. 1999;18:3979–3988. doi: 10.1038/sj.onc.1202933. [DOI] [PubMed] [Google Scholar]
- 6.Cattanach B M, Kirk M. Differential activity of maternally and paternally derived chromosome regions in mice. Nature. 1985;315:496–498. doi: 10.1038/315496a0. [DOI] [PubMed] [Google Scholar]
- 7.Copeland N G, Jenkins N A. Development and applications of a molecular genetic linkage map of the mouse genome. Trends Genet. 1991;7:113–118. doi: 10.1016/0168-9525(91)90455-y. [DOI] [PubMed] [Google Scholar]
- 8.DeChiara T M, Robertson E J, Efstratiadis A. Parental imprinting of the mouse insulin-like growth factor II gene. Cell. 1991;64:849–859. doi: 10.1016/0092-8674(91)90513-x. [DOI] [PubMed] [Google Scholar]
- 9.Diatchenko L, Lau Y F, Campbell A P, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov E D, Siebert P D. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci USA. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ettinger A J, Feng G, Sanes J R. Epsilon-sarcoglycan, a broadly expressed homologue of the gene mutated in limb-girdle muscular dystrophy 2D. J Biol Chem. 1997;272:32534–32538. doi: 10.1074/jbc.272.51.32534. . (Erratum, 273:19922, 1998.) [DOI] [PubMed] [Google Scholar]
- 11.Fujii H, Zhou W, Gabrielson E. Detection of frequent allelic loss of 6q23-q25.2 in microdissected human breast cancer tissues. Genes Chromosomes Cancer. 1996;16:35–39. doi: 10.1002/(SICI)1098-2264(199605)16:1<35::AID-GCC5>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 12.Gardner R J, Mungall A J, Dunham I, Barber J C, Shield J P, Temple I K, Robinson D O. Localisation of a gene for transient neonatal diabetes mellitus to an 18.72 cR3000 (approximately 5.4 Mb) interval on chromosome 6q. J Med Genet. 1999;36:192–196. [PMC free article] [PubMed] [Google Scholar]
- 13.Gardner R J, Robinson D O, Lamont L, Shield J P, Temple I K. Paternal uniparental disomy of chromosome 6 and transient neonatal diabetes mellitus. Clin Genet. 1998;54:522–525. doi: 10.1111/j.1399-0004.1998.tb03774.x. [DOI] [PubMed] [Google Scholar]
- 14.Giddings S J, King C D, Harman K W, Flood J F, Carnaghi L R. Allele specific inactivation of insulin 1 and 2, in the mouse yolk sac, indicates imprinting. Nat Genet. 1994;6:310–313. doi: 10.1038/ng0394-310. [DOI] [PubMed] [Google Scholar]
- 15.Guillemot F, Caspary T, Tilghman S M, Copeland N G, Gilbert D J, Jenkins N A, Anderson D J, Joyner A L, Rossant J, Nagy A. Genomic imprinting of Mash2, a mouse gene required for trophoblast development. Nat Genet. 1995;9:235–242. doi: 10.1038/ng0395-235. [DOI] [PubMed] [Google Scholar]
- 16.Hagiwara Y, Hirai M, Nishiyama K, Kanazawa I, Ueda T, Sakaki Y, Ito T. Screening for imprinted genes by allelic message display: identification of a paternally expressed gene impact on mouse chromosome 18. Proc Natl Acad Sci USA. 1997;94:9249–9254. doi: 10.1073/pnas.94.17.9249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho H T, Jenkins N A, Copeland N G, Gilbert D J, Winkles J A, Louie H W, Lee F K, Chung S S, Chung S K. Comparisons of genomic structures and chromosomal locations of the mouse aldose reductase and aldose reductase-like genes. Eur J Biochem. 1999;259:726–730. doi: 10.1046/j.1432-1327.1999.00110.x. [DOI] [PubMed] [Google Scholar]
- 18.Hogan B, Beddington R, Costantini F, Lacy L. Manipulating the mouse embryo. 2nd ed. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 19.Jenkins N A, Copeland N G, Taylor B A, Lee B K. Organization, distribution, and stability of endogenous ecotropic murine leukemia virus DNA sequences in chromosomes of Mus musculus. J Virol. 1982;43:26–36. doi: 10.1128/jvi.43.1.26-36.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Justice M J, Siracusa L D, Gilbert D J, Heisterkamp N, Groffen J, Chada K, Silan C M, Copeland N G, Jenkins N A. A genetic linkage map of mouse chromosome 10: localization of eighteen molecular markers using a single interspecific backcross. Genetics. 1990;125:855–866. doi: 10.1093/genetics/125.4.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaneko-Ishino T, Kuroiwa Y, Miyoshi N, Kohda T, Suzuki R, Yokoyama M, Viville S, Barton S C, Ishino F, Surani M A. Peg1/Mest imprinted gene on chromosome 6 identified by cDNA subtraction hybridization. Nat Genet. 1995;11:52–59. doi: 10.1038/ng0995-52. [DOI] [PubMed] [Google Scholar]
- 22.Kumar A, Cassidy S B, Romero L, Schwartz S. Molecular cytogenetics of a de novo interstitial deletion of chromosome arm 6q in a developmentally normal girl. Am J Med Genet. 1999;86:227–231. doi: 10.1002/(sici)1096-8628(19990917)86:3<227::aid-ajmg6>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 23.Lau M M, Stewart C E, Liu Z, Bhatt H, Rotwein P, Stewart C L. Loss of the imprinted IGF2/cation-independent mannose 6-phosphate receptor results in fetal overgrowth and perinatal lethality. Genes Dev. 1994;8:2953–2963. doi: 10.1101/gad.8.24.2953. [DOI] [PubMed] [Google Scholar]
- 24.Lefebvre L, Viville S, Barton S C, Ishino F, Keverne E B, Surani M A. Abnormal maternal behaviour and growth retardation associated with loss of the imprinted gene Mest. Nat Genet. 1998;20:163–169. doi: 10.1038/2464. [DOI] [PubMed] [Google Scholar]
- 25.Mann J R, Gadi I, Harbison M L, Abbondanzo S J, Stewart C L. Androgenetic mouse embryonic stem cells are pluripotent and cause skeletal defects in chimeras: implications for genetic imprinting. Cell. 1990;62:251–260. doi: 10.1016/0092-8674(90)90363-j. [DOI] [PubMed] [Google Scholar]
- 26.Mazurenko N, Attaleb M, Gritsko T, Semjonova L, Pavlova L, Sakharova O, Kisseljov F. High resolution mapping of chromosome 6 deletions in cervical cancer. Oncol Rep. 1999;6:859–863. doi: 10.3892/or.6.4.859. [DOI] [PubMed] [Google Scholar]
- 27.McGrath J, Solter D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell. 1984;37:179–183. doi: 10.1016/0092-8674(84)90313-1. [DOI] [PubMed] [Google Scholar]
- 28.McNally E M, Ly C T, Kunkel L M. Human epsilon-sarcoglycan is highly related to alpha-sarcoglycan (adhalin), the limb girdle muscular dystrophy 2D gene. FEBS Lett. 1998;422:27–32. doi: 10.1016/s0014-5793(97)01593-7. [DOI] [PubMed] [Google Scholar]
- 29.Miner J H, Patton B L, Lentz S I, Gilbert D J, Snider W D, Jenkins N A, Copeland N G, Sanes J R. The laminin alpha chains: expression, developmental transitions, and chromosomal locations of alpha1-5, identification of heterotrimeric laminins 8-11, and cloning of a novel alpha3 isoform. J Cell Biol. 1997;137:685–701. doi: 10.1083/jcb.137.3.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noviello C, Courjal F, Theillet C. Loss of heterozygosity on the long arm of chromosome 6 in breast cancer: possibly four regions of deletion. Clin Cancer Res. 1996;2:1601–1606. [PubMed] [Google Scholar]
- 31.Pivnick E K, Qumsiyeh M B, Tharapel A T, Summitt J B, Wilroy R S. Partial duplication of the long arm of chromosome 6: a clinically recognisable syndrome. J Med Genet. 1990;27:523–526. doi: 10.1136/jmg.27.8.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plass C, Shibata H, Kalcheva I, Mullins L, Kotelevtseva N, Mullins J, Kato R, Sasaki H, Hirotsune S, Okazaki Y, Held W A, Hayashizaki Y, Chapman V M. Identification of Grf1 on mouse chromosome 9 as an imprinted gene by RLGS-M. Nat Genet. 1996;14:106–109. doi: 10.1038/ng0996-106. [DOI] [PubMed] [Google Scholar]
- 33.Riesewijk A M, Blagitko N, Schinzel A A, Hu L, Schulz U, Hamel B C, Ropers H H, Kalscheuer V M. Evidence against a major role of PEG1/MEST in Silver-Russell syndrome. Eur J Hum Genet. 1998;6:114–120. doi: 10.1038/sj.ejhg.5200164. [DOI] [PubMed] [Google Scholar]
- 34.Spengler D, Villalba M, Hoffmann A, Pantaloni C, Houssami S, Bockaert J, Journot L. Regulation of apoptosis and cell cycle arrest by Zac1, a novel zinc finger protein expressed in the pituitary gland and the brain. EMBO J. 1997;16:2814–2825. doi: 10.1093/emboj/16.10.2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stewart C L. Production of chimeras between embryonic stem cells and embryos. Methods Enzymol. 1993;225:823–855. doi: 10.1016/0076-6879(93)25053-5. [DOI] [PubMed] [Google Scholar]
- 36.Stewart C L, Schuetze S, Vanek M, Wagner E F. Expression of retroviral vectors in transgenic mice obtained by embryo infection. EMBO J. 1987;6:383–388. doi: 10.1002/j.1460-2075.1987.tb04766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stewart C L, Vanek M, Wagner E F. Expression of foreign genes from retroviral vectors in mouse teratocarcinoma chimaeras. EMBO J. 1985;4:3701–3709. doi: 10.1002/j.1460-2075.1985.tb04138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Straub V, Campbell K P. Muscular dystrophies and the dystrophin-glycoprotein complex. Curr Opin Neurol. 1997;10:168–175. doi: 10.1097/00019052-199704000-00016. [DOI] [PubMed] [Google Scholar]
- 39.Straub V, Ettinger A J, Durbeej M, Venzke D P, Cutshall S, Sanes J R, Campbell K P. Epsilon-sarcoglycan replaces alpha-sarcoglycan in smooth muscle to form a unique dystrophin-glycoprotein complex. J Biol Chem. 1999;274:27989–27996. doi: 10.1074/jbc.274.39.27989. [DOI] [PubMed] [Google Scholar]
- 40.Szabo P, Mann J R. Expression and methylation of imprinted genes during in vitro differentiation of mouse parthenogenetic and androgenetic embryonic stem cell lines. Development. 1994;120:1651–1660. doi: 10.1242/dev.120.6.1651. [DOI] [PubMed] [Google Scholar]
- 41.Szabo P E, Mann J R. Biallelic expression of imprinted genes in the mouse germ line: implications for erasure, establishment, and mechanisms of genomic imprinting. Genes Dev. 1995;9:1857–1868. doi: 10.1101/gad.9.15.1857. [DOI] [PubMed] [Google Scholar]
- 42.Temple I K, Eccles D M, Winter R M, Baraitser M, Carr S B, Shortland D, Jones M C, Curry C. Craniofacial abnormalities, agenesis of the corpus callosum, polysyndactyly and abnormal skin and gut development—the Curry Jones syndrome. Clin Dysmorphol. 1995;4:116–129. [PubMed] [Google Scholar]
- 43.Varrault A, Ciani E, Apiou F, Bilanges B, Hoffmann A, Pantaloni C, Bockaert J, Spengler D, Journot L. hZAC encodes a zinc finger protein with antiproliferative properties and maps to a chromosomal region frequently lost in cancer. Proc Natl Acad Sci USA. 1998;95:8835–8840. doi: 10.1073/pnas.95.15.8835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vaughan K T, Mikami A, Paschal B M, Holzbaur E L F, Hughes S M, Echeverri C J, Moore K J, Gilbert D J, Copeland N G, Jenkins N A, Vallee R B. Multiple mouse chromosomal loci for dynein-based motility. Genomics. 1996;36:29–38. doi: 10.1006/geno.1996.0422. [DOI] [PubMed] [Google Scholar]
- 45.Wakeling E L, Abu-Amero S, Price S M, Stanier P, Trembath R C, Moore G E, Preece M A. Genetics of Silver-Russell syndrome. Horm Res. 1998;49:32–36. [PubMed] [Google Scholar]
- 46.Williamson C M, Beechey C V, Ball S T, Dutton E R, Cattanach B M, Tease C, Ishino F, Peters J. Localisation of the imprinted gene neuronatin, Nnat, confirms and refines the location of a second imprinting region on mouse chromosome 2. Cytogenet Cell Genet. 1998;81:73–78. doi: 10.1159/000014992. [DOI] [PubMed] [Google Scholar]
- 47.Yang T, Adamson T E, Resnick J L, Leff S, Wevrick R, Francke U, Jenkins N A, Copeland N G, Brannan C I. A mouse model for Prader-Willi syndrome imprinting-centre mutations. Nat Genet. 1998;19:25–31. doi: 10.1038/ng0598-25. [DOI] [PubMed] [Google Scholar]
- 48.Yu S, Yu D, Lee E, Eckhaus M, Lee R, Corria Z, Accili D, Westphal H, Weinstein L S. Variable and tissue-specific hormone resistance in heterotrimeric Gs protein alpha-subunit (Gsalpha) knockout mice is due to tissue-specific imprinting of the gsalpha gene. Proc Natl Acad Sci USA. 1998;95:8715–8720. doi: 10.1073/pnas.95.15.8715. [DOI] [PMC free article] [PubMed] [Google Scholar]