ABSTRACT
Plant-pathogenic Ralstonia spp. colonize plant xylem and cause wilt diseases on a broad range of host plants. To identify genes that promote growth of diverse Ralstonia strains in xylem sap from tomato plants, we performed genome-scale genetic screens (random barcoded transposon mutant sequencing screens [RB-TnSeq]) in three strains spanning the genetic, geographical, and physiological range of plant-pathogenic Ralstonia: Ralstonia solanacearum IBSBF1503, Ralstonia pseudosolanacearum GMI1000, and Ralstonia syzygii PSI07. Contrasting mutant fitness phenotypes in culture media versus in xylem sap suggest that Ralstonia strains are adapted to ex vivo xylem sap and that culture media impose foreign selective pressures. Although wild-type Ralstonia grew in sap and in rich medium with similar doubling times and to a similar carrying capacity, more genes were essential for growth in sap than in rich medium. Each strain required many genes associated with envelope remodeling and repair processes for full fitness in xylem sap. These genes were associated with peptidoglycan peptide formation (murI), secretion of periplasmic proteins (tatC), periplasmic protein folding (dsbA), synthesis of osmoregulated periplasmic glucans (mdoGH), and lipopolysaccharide (LPS) biosynthesis. Mutant strains with mutations in four genes had strong, sap-specific fitness defects in all strain backgrounds: murI, thiC, purU, and a lipoprotein (RSc2007). Many amino acid biosynthesis genes were required for fitness in both minimal medium and xylem sap. Multiple mutants with insertions in virulence regulators had gains of fitness in culture media and neutral fitness in sap. Our genome-scale genetic screen identified Ralstonia fitness factors that promote growth in xylem sap, an ecologically relevant condition.
IMPORTANCE Traditional transposon mutagenesis genetic screens pioneered molecular plant pathology and identified core virulence traits like the type III secretion system. TnSeq approaches that leverage next-generation sequencing to rapidly quantify transposon mutant phenotypes are ushering in a new wave of biological discovery. Here, we have adapted a genome-scale approach, random barcoded transposon mutant sequencing (RB-TnSeq), to discover fitness factors that promote growth of three related bacterial strains in a common niche, tomato xylem sap. Fitness of the wild type and mutants show that Ralstonia spp. are adapted to grow well in xylem sap from their natural host plant, tomato. Our screen identified multiple sap-specific fitness factors with roles in maintaining the bacterial envelope. These factors include putative adaptations to resist plant defenses that may include antimicrobial proteins and specialized metabolites that damage bacterial membranes.
KEYWORDS: genetics, plant pathogen, Ralstonia, microbial ecology, xylem
INTRODUCTION
Ralstonia solanacearum, Ralstonia pseudosolanacearum, and Ralstonia syzygii (hereafter, “Ralstonia”) comprise the monophyletic but diverse species complex of plant wilt pathogens (1, 2). Ralstonia strains are adapted to plant hosts belonging to over 50 botanical families and are distributed worldwide in warm tropics and temperate subtropical highlands with year-round precipitation (3–7). Most Ralstonia strains invade plant roots, gain entry to the water-transporting xylem vasculature, and spread systemically, which disrupts xylem function and fatally wilts the host plant (8).
Key molecular pathogenesis, virulence, and fitness traits have been identified by studying model strains like R. pseudosolanacearum GMI1000. However, comparative genomics shows that the Ralstonia species complex is genetically heterogeneous (1, 9–11). Average nucleotide identity (ANI) between strains in the different species ranges between 90.8 and 92.7% (1), which is less than the 95% ANI threshold widely used for species borders. An average Ralstonia genome contains approximately 5,000 genes, but approximately 2,900 core gene families are core to the R. solanacearum species complex (data from Boris Vinatzer and Lowe-Power labs). Accordingly, ecological niches, host ranges, and physiological traits vary widely among Ralstonia strains (1, 12–14). Developing and studying new model strains (15) that capture the phylogenetic diversity of the Ralstonia species complex will provide a more complete view of the biology of these globally important pathogens.
We hypothesized that genes in diverse Ralstonia strains contribute to their growth in xylem sap of tomato plants, a common host. In this study, we used a random barcoded transposon mutant sequencing (RB-TnSeq) mariner transposon library with over 108 possible barcoded Tns (16) to create genome-wide transposon insertion mutant libraries of three tomato-pathogenic Ralstonia strains: R. pseudosolanacearum GMI1000, R. solanacearum IBSBF1503, and R. syzygii PSI07. We mapped the transposon insertion sites and predicted core essential genes. We performed genetic screens to identify fitness factors that influence competitive growth of strains in ex vivo xylem sap from healthy susceptible tomato, as well as minimal and rich culture media. Comparison of genes’ fitness contributions between conditions revealed sap-specific fitness factors and genes that contribute to fitness both in sap and in culture media.
RESULTS
Adapting RB-TnSeq for plant-pathogenic Ralstonia isolates.
To identify genes required for Ralstonia fitness, we constructed barcoded transposon insertion mutant libraries in three Ralstonia species complex backgrounds: R. pseudosolanacearum GMI1000, R. solanacearum IBSBF1503, and R. syzygii PSI07. We selected these isolates because they are all virulent on tomato and because there are high-quality, complete genomes available for each strain. These diverse strains share 76 to 83% of their gene content with average nucleotide identities of 91 to 93% (1), which we hypothesized would allow us to identify strain-specific and conserved fitness factors required by the species complex.
The mutant libraries were created by conjugation with the pKMW3 Escherichia coli donor library (16), which carries a pool of over 108 mariner transposon delivery vectors in which each transposon is marked by a unique 20-bp sequence. These unique sequences function as DNA “barcodes.” After conjugation, we selected and pooled approximately 105 transposon insertion mutants per strain library and mapped the insertion site of each barcoded transposon. Table 1 shows detailed summary statistics for each mutant library. For barcode sequencing (BarSeq) mutant fitness assays, we calculate fitness contributions using transposons that had inserted into the central 80% of the open reading frame (ORF) because insertions at a gene product’s N and C termini are less likely to impair protein function.
TABLE 1.
Properties of barcoded mariner mutant libraries
| Mutant library | Total no. of genes in genome | No. of genes with Fit scores (% total) | Library sizea (no. of mutants) | Median (mean) no. of mutants per gene in fitness calculations | Estimated no. of essential genesb (%) |
|---|---|---|---|---|---|
| GMI1000 | 5,204 | 4,003 (77) | 90,016 | 11 (17.1) | 456–470 (8.7–9.0) |
| PSI07 | 4,968 | 4,057 (82) | 190,155 | 22 (35.7) | 454–487 (9.5–9.8) |
| IBSBF1503 | 4,897 | 3,809 (78) | 104,510 | 12 (20.4) | 454–487 (9.3–10.0) |
Measured by the number of DNA barcodes mapped to a specific insertion site.
Two pipelines (described in Materials and Methods) were used to predict gene essentiality based on the distribution of transposon insertions in open reading frames.
The barcoded mutant libraries cover 79 to 86% of the protein coding genes of each strain. There are several reasons why a given gene may be unrepresented in the mutant libraries. The mariner transposon we used inserts into TA dinucleotide sites. Strains GMI1000, IBSBF1503, and PSI07 have 48, 42, and 101 genes that could not be mutagenized because they lack TA sites. Table S1 in the supplemental material lists genes that lack fitness data and the number of TA sites in the central 80% of each gene. Additionally, it is not possible to uniquely map transposon insertions in repetitive, indistinguishable genomic regions like the 31-kb tandem duplication in the GMI1000 genome. Furthermore, if these genes contribute to fitness in a condition, they would be functionally redundant with their identical paralogs, mitigating any potential impact on fitness. Finally, a subset of the genes without transposon insertions are likely essential or near-essential for growth in rich medium, resulting in their failure to grow during library construction and exclusion from our libraries.
Gene essentiality predictions for strains GMI1000, PSI07, and IBSBF1503. Download Table S1, XLSX file, 0.2 MB (223.4KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Predicting genes essential for Ralstonia growth in rich medium.
We predicted essentiality of genes in each strain (Table S1) using the Bio::TraDIS pipeline (17) and our previously described analysis pipeline (18). Briefly, both methods predict gene essentiality based on genes that lacked transposon insertions in the central 80% of the coding region. After setting cutoffs to exclude short genes (“Above length cutoff” in Table S1), which are more likely to lack transposon insertions, 450 to 500 genes were predicted to be essential per strain by at least one pipeline, while 393 to 396 genes per strain were predicted to be essential by both pipelines (Table S1). To compare results between strains, we identified orthologous genes in strains GMI1000, PSI07, and IBSBF1503 using the Joint Genome Institute (JGI) Integrated Microbial Genomes and Microbiomes (IMG) (19) Genome Gene Best Homologs tool. A core set of 244 genes was predicted to be essential by both pipelines in all three strains (see Fig. S1 in the supplemental material) (20). We inspected the predicted essential gene set for a positive control, the speC ornithine decarboxylase. We previously showed that a GMI1000 speC mutant (RSc2365) is a putrescine auxotroph that cannot grow in rich medium unless it is supplemented with putrescine (8); both pipelines predicted speC was essential in strains GMI1000 and PSI07. Bio::TraDIS, but not our own pipeline, predicted speC to be essential in IBSBF1503.
Venn diagram of putative essential genes in Ralstonia strains GMI1000, IBSBF1503, and PSI07. Orthologous predicted essential genes are represented proportionally in Venn diagram intersections. Genes were included in the Venn diagram only if they were predicted to be essential by both analysis pipelines used in this study (17, 18). Download FIG S1, TIF file, 0.5 MB (559.2KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
To visualize which cellular processes are predicted essential for growth in rich medium, we used KEGG Mapper (21) to query the KEGG (Kyoto Encyclopedia of Genes and Genomes) database with the GMI1000 locus tags of the 244 genes predicted to be essential in all three strains. As expected, many of these genes are involved in central cellular functions, such as DNA replication, transcription, translation, mRNA degradation, and lipid and peptidoglycan biosynthesis. Several genes from central carbon metabolism and respiration are predicted to be essential, including components of the Entner-Doudoroff pathway, the pentose phosphate pathway, the tricarboxylic acid (TCA) cycle, NADH dehydrogenase, and ATP synthase. Many genes involved in cofactor and vitamin biosynthesis (ubiquinone, coenzyme A, thiamine, heme, and folate) are predicted to be essential. Genes that encode biosynthetic steps of several amino acids, including threonine, arginine, lysine, and histidine, also appear to be essential for growth in rich medium.
Growth of Ralstonia species representatives in xylem sap.
To design an RB-TnSeq experiment with a sufficient number of cell doublings to quantify mutant fitness defects, we empirically determined the carrying capacity and doubling rate of Ralstonia in xylem sap. We compared growth of R. pseudosolanacearum GMI1000 in rich medium, minimal medium, or xylem sap harvested from greenhouse-grown Moneymaker tomato (Fig. S2A and S2B). The carrying capacities of xylem sap and rich medium were higher than the carrying capacity of minimal medium: 3.1 × 109 to 3.5 × 109 CFU/ml in sap, 2.4 × 109 to 2.6 × 109 CFU/ml in rich medium, and 2.2 × 108 to 4.6 × 108 CFU/ml in minimal medium. Both xylem sap and rich medium cultures exceeded 1 × 108 CFU within 24 h, while minimal medium cultures had an extended lag time. We were surprised by the high carrying capacity supported by xylem sap based on our previous experience growing diverse Ralstonia in xylem sap from a different susceptible tomato (Bonny Best) grown in environmentally controlled growth chambers (8, 22). To determine whether the growing conditions or plant cultivar influenced the saps’ carrying capacity, we collaborated with researchers at the University of Wisconsin—Madison to repeat this experiment with sap from growth chamber-grown Moneymaker and Bonny Best tomato in all three of our study strains. Each Ralstonia isolate grew to a higher carrying capacity in xylem sap from Moneymaker plants (3.3 × 109 to 6.6 × 109 CFU/ml) than sap from Bonny Best plants (3.6 × 107 to 5.5 × 108 CFU/ml) (Fig. S2C to S2E).
Growth of Ralstonia in rich medium, minimal medium, and xylem sap from multiple susceptible tomato cultivars. (A and B) To determine the carrying capacity of sap and culture media, Ralstonia strain GMI1000 was inoculated into rich CPG medium, quarter strength M63 medium, or xylem sap from Moneymaker tomato at 104, 105, or 106 CFU/ml starting density. (C to E) Wild-type Ralstonia strains GMI1000, PSI07, and IBSBF1503 were grown in sap from Moneymaker and Bonny Best tomato. All strains were inoculated in biological triplicate into three pools of xylem sap incubated at 28°C with shaking. Cell density was measured by dilution plating. Download FIG S2, TIF file, 1.0 MB (1,016.5KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
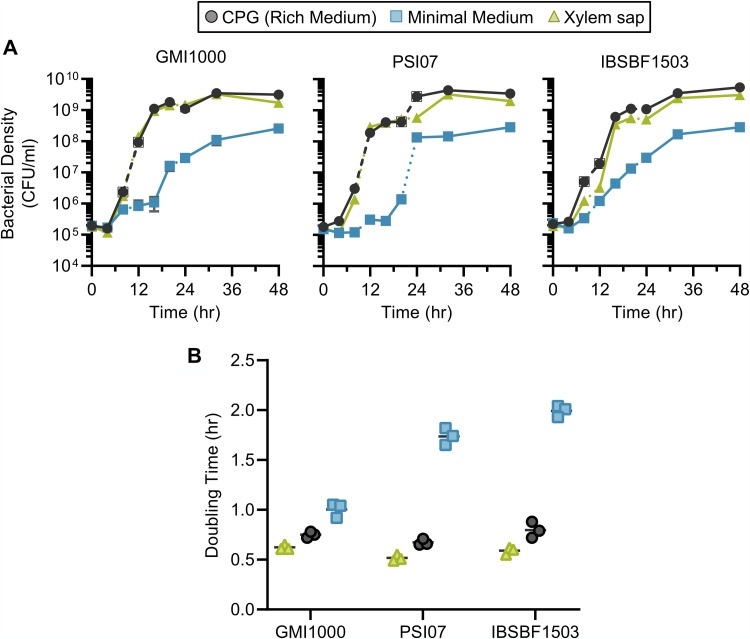
To estimate the doubling rate in xylem sap and rich and minimal culture media, we compared the growth of strains GMI1000, PSI07, and IBSBF1503 with frequent sampling intervals (every 4 h [Fig. 1A]). Strains grew slowest in minimal medium and fastest in xylem sap (Fig. 1B). The maximum doubling rate in xylem sap was 120 to 131% higher than in rich medium (P < 0.005 by analysis of variance [ANOVA] with Tukey’s multiple-comparison test). The higher growth rate and carrying capacity in xylem sap compared to rich medium indicate that Ralstonia strains GMI1000, IBSBF1503, and PSI07 are well adapted to growth in xylem sap from Bonny Best tomato plants.
FIG 1.
Growth of Ralstonia strains in xylem sap is similar to growth in rich medium. Ralstonia strain GMI1000, strain PSI07, or strain IBSBF1503 was grown in rich medium (CPG), quarter-strength M63 minimal medium (MM), or xylem sap from Moneymaker tomato plants. (A) Strains were inoculated at 105 CFU/ml into CPG, MM, or xylem sap. All cultures were set up with three biological replicates (individual overnight cultures and individual batches of xylem sap) and were performed in technical duplicate. Solid lines indicate time points sampled from the same culture; the 12- to 24-h time points were from an independent culture, but 0- to 8-h and 24- to 48-h time points were taken from the same culture. Error bars indicate standard deviations. (B) The minimum doubling time between contiguous 4-h measurements was calculated per biological replicate. The short horizontal lines indicate the means. Per strain, the doubling times under all conditions were significantly different from each other (adjusted P < 0.05 by ANOVA with Tukey’s multiple-comparison test).
RB-TnSeq experiments identify mutants with altered fitness in xylem sap and minimal medium.
We used RB-TnSeq to identify genes that influence fitness in ex vivo xylem sap, minimal medium, and rich medium. Mutant abundance before and after selective competitive growth was measured by BarSeq. In BarSeq, each gene is assigned a fitness score (“Fit” score), which is a weighted average of the log2 ratios of each barcoded mutant’s final abundance divided by its abundance before growth in the selective condition (the “time zero” abundance) (16). In total, our BarSeq assays measured fitness contributions for approximately 4,000 genes per strain. Multiple independent transposon mutants in the same gene are used to calculate each gene’s Fit score (Table 1). We calculated Fit scores and t-like test statistics for each gene under each condition (Table S2). Fitness data are also publicly available on the interactive Fitness Browser (http://fit.genomics.lbl.gov/). Because Fit scores are on a log2 scale, a score of −5 shows in principle that those mutants replicated for five fewer generations than the population. In practice, we found that mutants with the strongest fitness defects had Fit scores that exceeded the number of generations that we measured by dilution plating. For example, GMI1000 murI (RSc1956) had a mean Fit score of −8.6 in xylem sap even though the population grew for only seven generations. However, most Fit scores were in the expected ranges for each TnSeq trial.
Genome-wide Fit scores for strains GMI1000, PSI07, and IBSBF1503. Download Table S2, XLSX file, 3.5 MB (3.5MB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
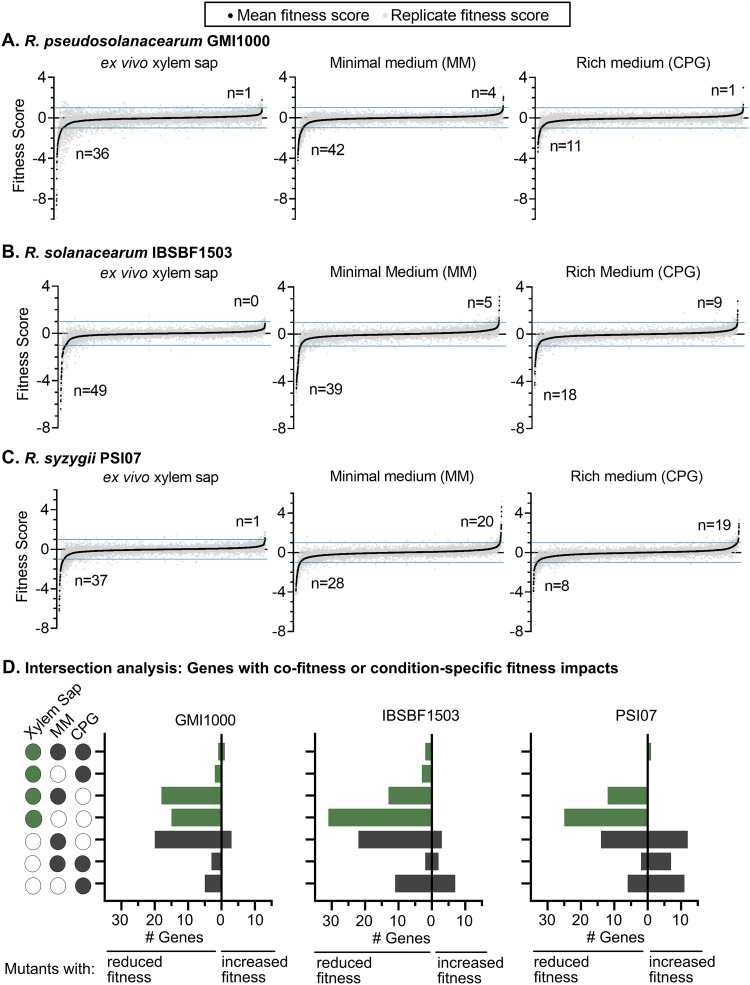
There were trends in the number of genes associated with gain or loss of fitness between growth conditions (Fig. 2A to C). There were more mutants with fitness defects in xylem sap (n = 37 to 55) and minimal medium (n = 28 to 42) than in rich medium (n = 8 to 28) (Fit score of less than −1; t-like statistic less than −2.5 in all trials). Logically, mutants with strong fitness defects in rich medium are excluded from the libraries because they would not have grown on the selection plates. Interestingly, we observed that more mutants had gain-of-fitness phenotypes in minimal (n = 4 to 20) and rich (n = 2 to 19) media than in xylem sap (n = 0 to 1). We next determined which genes influence fitness in a single condition and which genes have intersecting fitness contributions in two or more conditions (Fig. 2D). The intersection analysis reveals that each strain has many genes that contribute to fitness in xylem sap only, in minimal medium only, or in both xylem sap and minimal medium.
FIG 2.
Summary of RB-TnSeq data after growth of mutant libraries in xylem sap, minimal medium (MM), or rich medium (CPG). (A to C) Ranked Fit scores for all measured genes in CPG, MM, and xylem sap for R. pseudosolanacearum GMI1000 (A), R. solanacearum IBSBF1503 (B), and R. syzygii PSI07 (C). The number of genes that influence fitness under each condition is indicated (average Fit score of >1 or less than −1; t-like test statistic > |2.5| all three RB-TnSeq trials). (D) Intersection analysis quantifying number of genes that influence fitness in a single condition or that have cofitness in multiple conditions. Green bars indicate genes that influence fitness in sap, and gray bars indicate genes that influence fitness only in culture media. The intersection visualization is modeled on UpSet Plots (66), an alternative to Venn diagrams.
Ralstonia require multiple envelope-associated genes for fitness in xylem sap.
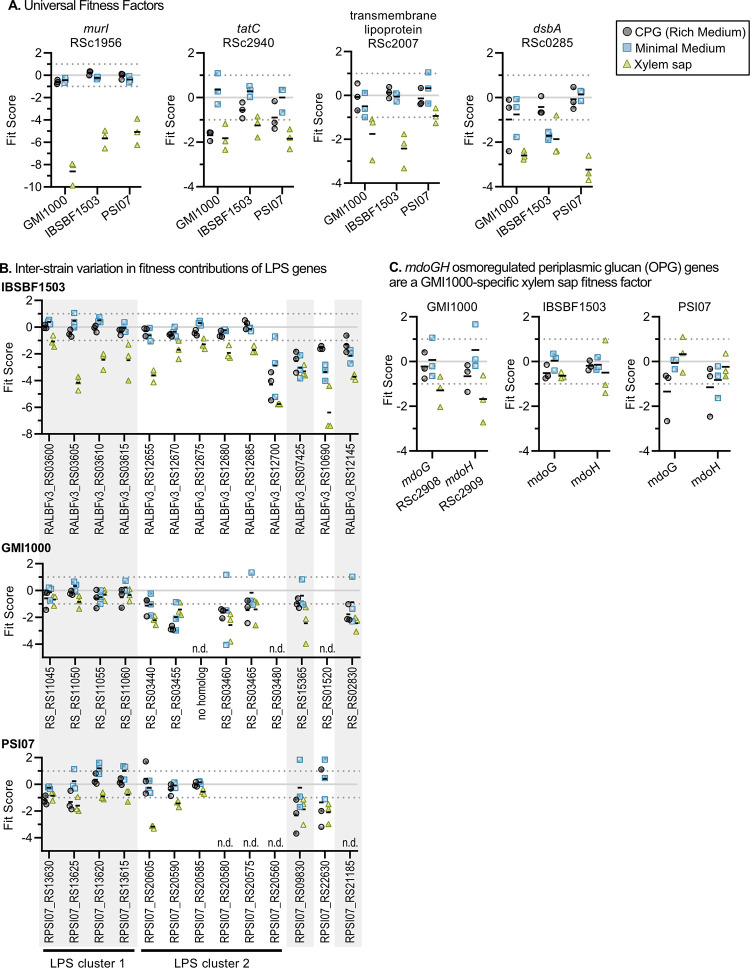
Our intersection analysis identified envelope-associated genes as fitness factors in xylem sap in all three strains, in addition to other strain-specific xylem sap fitness factors (Fig. 3). The strongest xylem sap fitness factor in all strains was the murI gene (RSc1956) with a Fit score of less than −5 in all strains. MurI is a glutamate racemase that converts l-glutamate to d-glutamate, which is incorporated into peptide cross-linkages in peptidoglycan. The murI gene is required for multiple Ralstonia strains to grow in tomato stems (23, 24). For all three study strains, other xylem sap fitness factors included tatC (involved in Tat-dependent protein secretion), dsbA which catalyzes disulfide bonds in periplasmic proteins, and a gene encoding a lipoprotein of unknown function (RSc2940, RSc0285, and RSc2007, respectively). BLAST analysis reveals that this lipoprotein gene is present in the genomes of Ralstonia, Cupriavidus, Paraburkholderia, and Burkholderia, which are all genera in the Burkholderiaceae family.
FIG 3.
Envelope-associated genes contribute to fitness in xylem sap. Fit scores of xylem sap fitness factors in one or more Ralstonia strains are shown. (A to C) Graphs show fitness contributions of four envelope-associated genes that are sap fitness factors in all three strains (A), LPS genes that are xylem sap fitness factors for at least one strain (B), and periplasmic glucan biosynthesis genes that are xylem sap fitness factors for GMI1000 (C). Graphs show Fit scores of n = 3 TnSeq trials, and bars show the means. Full Fit scores and t-like statistics are shown in Table S2 in the supplemental material. n.d., not determined.
IBSBF1503 requires 13 different lipopolysaccharide (LPS) genes for full fitness in xylem sap, and several of these genes are important for fitness in xylem sap in strains PSI07 and GMI1000 as well (Fig. 3B). Though conserved in all three strains, LPS genes colocalized in LPS cluster 1 had stronger fitness phenotypes in strain IBSBF1503 (−4.2 < Fit score < −1.1) than in strains GMI1000 (−0.9 < Fit score < −0.3) and PSI07 (−1.6 < Fit score < −0.8). These results corroborate previous work in Ralstonia strain Pss4 indicating that mutants with structural defects in LPS also have reduced growth in planta (25) while highlighting subtle interstrain differences in the exact orthologs involved.
Other strain-specific envelope proteins identified in our analysis include the finding that strain GMI1000 requires mdoGH (RSc2908 and RSc2909) during growth in xylem sap (Fig. 3C), which putatively encode enzymes that synthesize periplasmic branched oligoglucans. Since periplasmic oligoglucans can act as osmoprotectants to Gram-negative bacteria (26), and mdoGH contributes to the growth of several plant pathogens of the host (27), these enzymes may play a role in protecting GMI1000 from environmental stress posed during growth in xylem sap.
Metabolic functions required for full fitness in xylem sap.
Many of the xylem sap fitness factor genes are associated with bacterial metabolism. Consistent with previous studies that showed Ralstonia scrA sucrose catabolism mutants (RSp1285) have growth defects in tomato stems (28, 29), scrB (RSp1284) sucrose catabolism mutants also had reduced fitness in xylem sap in our TnSeq experiments. The fitness defect of scrB mutants was stronger in the IBSBF1503 background than in the GMI1000 or PSI07 background (Fit scores of −2.0, −1.3, and −0.9, respectively). Overall, our findings validate that sucrose is an important carbon source in xylem sap although Ralston can catabolize other carbon sources present in sap (8, 30, 31).
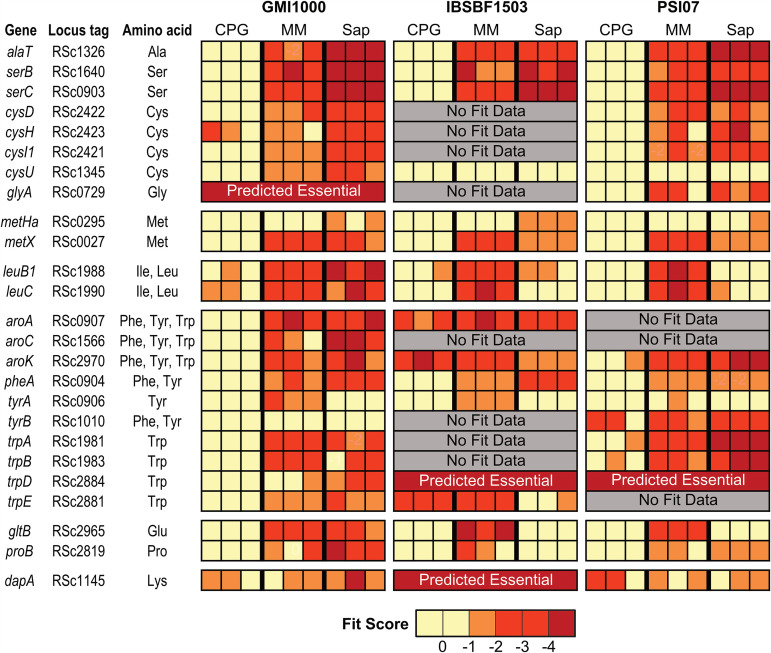
All strains required amino acid biosynthesis genes for fitness in xylem sap and minimal medium (Fig. 4). Mutants lacking Ser, Cys, Met, Phe, and Trp biosynthesis genes had fitness defects in both xylem sap and minimal medium. Although all strains required several Glu, Pro, and branched-chain amino acid (AA) (Leu/Ile) biosynthesis genes for growth in minimal medium, only strain GMI1000 consistently required these genes for full growth in xylem sap. Our TnSeq results are consistent with xylem sap metabolomic studies that indicate that multiple tomato cultivars lack Cys and Trp in sap (8, 31). Although Ser, Met, and Phe are all present in xylem sap, our TnSeq results indicate that Ralstonia requires many de novo amino acid synthesis enzymes for full fitness in xylem sap.
FIG 4.
Amino acid auxotroph mutants have fitness defects in minimal medium (MM) and/or xylem sap. Fit scores of amino acid biosynthesis mutants with reduced fitness in sap, MM, or both. Gene name and locus tag for the GMI1000 gene are displayed. Full Fit scores and t-like test statistics are shown in Table S2.
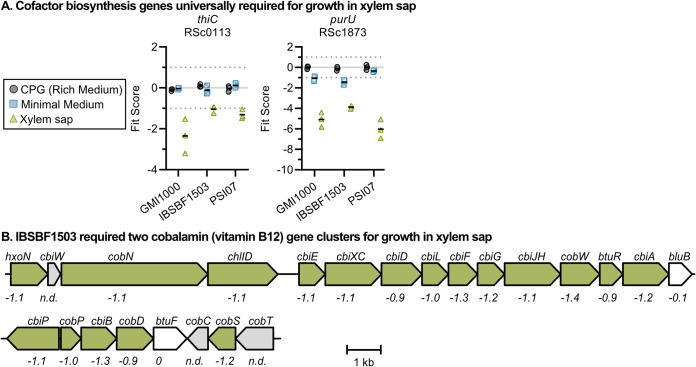
Multiple cofactor biosynthesis mutants had xylem sap fitness defects in one or more strains (Fig. 5). All three strains required the thiamine biosynthesis gene thiC for full growth in xylem sap (Fit score of −1.0 to −2.4). Similarly, all strains strongly required purU (RSc1873) for growth in xylem sap (Fit score of −3.9 and −6.0), with weaker fitness defects in minimal medium (Fit score of −0.3 to −1.4). PurU is a putative formyltetrahydrofolate deformylase that hydrolyzes 10-formyltetrahydrofolate into formate and tetrahydrofolatic acid (THFA). Because THFA is a cofactor for biosynthesis of many amino acids and nucleic acids, it is possible that PurU is necessary for Ralstonia to recycle THFA in order to synthesize amino acids. Interestingly, only one of the three strains required cobalamin biosynthesis to grow in xylem sap. Eighteen IBSBF1503 cobalamin biosynthesis mutants had subtle but consistent fitness defects in xylem sap with Fit scores that ranged from −0.9 to −1.4 (Fig. 5B).
FIG 5.
Mutants lacking cofactor biosynthesis genes have fitness defects in xylem sap. (A) A thiamine and tetrahydrofolic acid gene were both xylem sap fitness factors in all three strains. Graphs show Fit scores of n = 3 TnSeq trials, and bars show the means. (B) Ralstonia IBSBF1503 required cobalamin biosynthesis for growth in xylem sap. The xylem sap fit scores of each gene are written below the genes. “n.d.” indicates fitness was not determined because mutants were not present in the RB-TnSeq library. Full Fit scores and t-like test statistics are shown in Table S2.
Mutants with improved fitness.
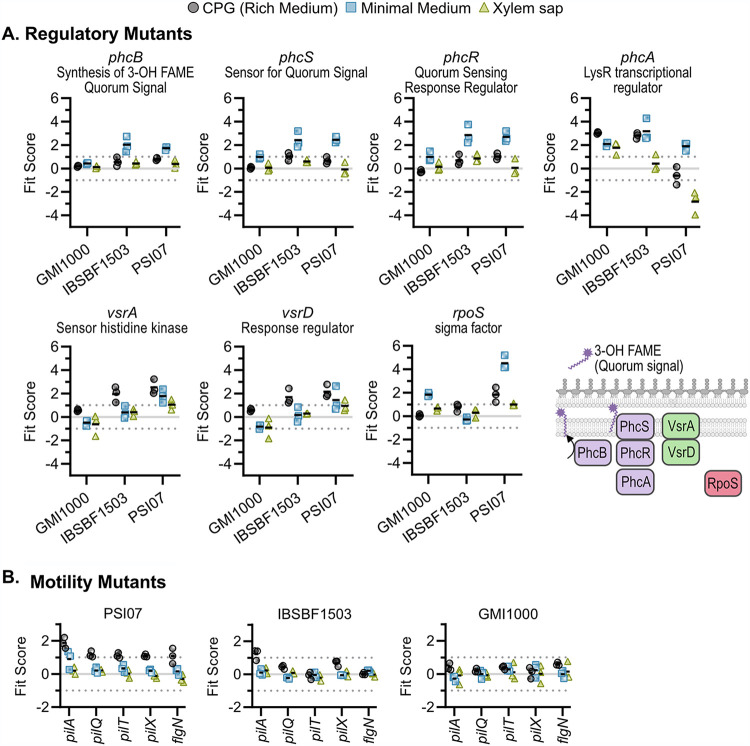
Many mutants with gain-of-fitness phenotypes had transposon insertions in regulatory genes (Fig. 6). Consistent with previous results showing that the PhcA quorum sensing regulator mediates a trade-off between fast growth at low cell density and expression of in planta fitness traits at high cell density (32, 33), GMI1000 and IBSBF1503 phcA mutants had improved growth under all tested conditions (Fig. 6A). Unexpectedly, PSI07 phcA mutants behaved differently. Although PSI07 phcA mutants had increased fitness in minimal medium (Fit score of +1.9), they had strongly reduced growth in xylem sap (Fit score of −2.8). Contrasting phenotypes of phcA mutants in different wild-type backgrounds highlight the genetic and regulatory diversity within plant-pathogenic Ralstonia. At high cell densities, PhcA positively regulates production of an energetically costly extracellular polysaccharide (EPS) among other traits (33, 34). Transposon insertion in other regulatory genes that activate expression of eps and other virulence genes (phcBSR, vsrAD, and rpoS) (35, 36) also increased fitness in one or more strains (Fig. 6A).
FIG 6.
Mutants with gain of fitness in one or more strains and conditions. (A) Regulatory mutants with known defects in extracellular polysaccharide (EPS) production relative to the wild type: phc quorum sensing mutants, vsrAD mutants, and rpoS mutants. (B) Motility mutants lacking type IV pilus genes and the flgN flagellar chaperone genes had increased fitness in rich media in one or more strains. Graphs show Fit scores of n = 3 TnSeq trials, and bars show the means. Full Fit scores and t-like test statistics are shown in Table S2.
Strain PSI07 had the most mutants with gain-of-fitness phenotypes in culture media. Four of these genes were associated with the type IV pilus, and another gene was associated with a flagellum-associated chaperone, FlgN (Fig. 6B). Three IBSBF1503 type IV pilus mutants showed the same trend as PSI07, but with weaker phenotypes. This result suggests that these wild-type strains may express energy-consuming motility systems in liquid rich medium but not in sap or minimal medium. PSI07 had additional regulators that had gain-of-fitness phenotypes in culture media: a MarR-family transcriptional regulator nested in a hydroxycinnamic acid degradation cluster (RSp0224/RPSI07_RS00780) (13), a transcriptional regulator colocalized with an ABC transport system (RSp1579/RPSI07_RS07405) (37), a TetR family transcriptional regulator (RSc3207/RPSI07_RS15470), a GGDEF/EAL domain-encoding gene (RSp1208/RPSI07_RS05560), and the RelA (p)ppGpp synthase (RSc1576/RPSI07_RS15685).
DISCUSSION
Why might envelope genes contribute to Ralstonia fitness in ex vivo xylem sap? Recent studies have shown that clinical antibiotic use can select for pathogens that remodel their envelopes, enabling them to exclude and resist antibiotics (38). Because many envelope-associated genes were xylem sap fitness factors, we hypothesize that xylem sap contains preformed antimicrobial chemicals and proteins. Future studies should test the hypothesis that periplasmic branched oligoglucans, LPS, and other envelope genes protect Ralstonia from in planta stresses like reactive oxygen species and antimicrobial peptides (38).
There are benefits and limitations of predicting gene essentiality using transposon mutagenesis. Here, we identified sets of 454 to 487 essential genes in three Ralstonia strains, of which more than half (n = 244) were conserved in all three strains and predicted essential by both analysis pipelines. Unsurprisingly, many of the essential genes are involved in carbon metabolism, amino acid and cofactor biosynthesis, and central dogma processes. Identifying absolutely essential genes of human pathogens aids in antibacterial drug development research (39), but precision antimicrobial treatment for most plant pathogens is not financially viable and poses environmental concerns. Nonetheless, identifying putative essential genes has fundamental value, particularly to the Ralstonia research community. Creating mutants lacking these genes might be impossible or require specialized selection media as was required for the ΔspeC::Sm mutant that had a putrescine auxotrophy (8). The essentiality of multiple amino acid biosynthesis genes in the peptone- and Casamino Acid-containing CPG rich medium suggests Ralstonia may lack effective transporters for some amino acids.
Recently, Su et al. predicted 464 essential genes in Ralstonia GMI1000 (40) using a TnSeq approach, but our libraries contain mutants with transposon insertions in 126 of these genes. Of these 126 genes, only three genes contributed to fitness more than twofold (Fit score = −1) in any of our experimental conditions, including rich medium. These discrepancies highlight the limitations of using randomly generated mutant libraries to predict gene essentiality. Targeted knockdown experiments using techniques like CRISPR interference (CRISPRi) (41) could test essentiality more robustly. In a subsequent TnSeq experiment using their mutant library in living tomato plant hosts, Su et al. identified 131 genes as being important for in planta fitness (42). Of the 36 genes we identified as important for xylem sap fitness (average | Fit score | > 1, | t | > 2.5 in three trials), Su et al. identified 16 as also being important for in planta fitness. This overlap suggests that Ralstonia experiences some of the same selective pressures in ex vivo xylem sap as it does in living tomato hosts, though additional genes are necessary for full fitness in the more complex in planta environment.
RB-TnSeq reveals interstrain differences in fitness factors. Although model strains such as GMI1000 are well characterized and extensively studied, there are known physiological and genetic differences between strains (1, 28, 43). To more fully capture the diversity of Ralstonia, we constructed barcoded transposon mutant libraries in strains from each of the three plant-pathogenic Ralstonia species: R. solanacearum (GMI1000), R. solanacearum (IBSBF1503), and R. syzygii (PSI07). While RB-TnSeq found specific metabolism- and envelope-related fitness factors in all three strains, the orthologs found to be fitness factors varied from strain to strain. For instance, strain GMI1000 requires mdoGH genes for periplasmic oligoglucan production in xylem sap, while these genes do not produce strong fitness phenotypes in the other two strains. Additionally, several amino acid biosynthesis genes were required by some by not all strains when grown in minimal media or xylem sap, including cysU, leuB1, leuC, gltB, and proB (Fig. 4). These strain-to-strain differences in gene fitness profiles reveal subtle differences in the genetics that underlie the ability of all these strains to robustly grow in tomato xylem sap. Considering our results in ex vivo xylem sap, different genes may be important for virulence of these diverse strains in living hosts as well.
Novel environments provide strong selective pressures for bacterial evolution (44). We suggest that the number of gain-of-fitness mutations in a TnSeq screen may be an indicator of how closely the test conditions match the environment to which the bacterial isolate is adapted. For instance, transposon insertions in over two dozen E. coli genes increased its growth on cheese agar (45), a foreign growth medium. Similarly, serial passage of Ralstonia GMI1000 in Medicago nodules (a foreign environment) selected for more gain-of-fitness mutations than serial passage in tomato stems (46, 47). In our study, few mutations increased fitness in sap, but many mutations increased fitness in artificial culture media, suggesting that the Ralstonia isolates are well adapted to grow in tomato xylem sap. We observed that the model strain Ralstonia GMI1000 had fewer gain-of-function mutations in culture media than the less-studied IBSBF1503 and PSI07 strains, suggesting that GMI1000 may be more domesticated and adapted to culture media since its isolation in 1978. Meta-analyses of TnSeq studies across multiple bacteria in novel and naturalistic conditions can test the hypothesis that gain-of-fitness phenotypes are more common when mutants grow in novel conditions.
Traditional transposon mutagenesis genetic screens pioneered molecular plant pathology and identified core virulence traits like the type III secretion system (48–50). TnSeq approaches leverage next-generation sequencing to rapidly quantify mutant phenotypes. These genome-wide fitness assays are a powerful approach to rapidly investigate basic bacterial biology and identify pathogen, commensal, and mutualist fitness factors. TnSeq studies have identified genes that promote bacterial fitness in their native environments, including plant hosts (51–57) animal hosts (58–60), soft-rind cheese (45), and lake water (61). The RB-TnSeq technique that we use here is a powerful TnSeq methodology that has a low cost and technical barrier per sample, which facilitates profiling mutant fitness across in multiple strain backgrounds across many conditions (16, 62). Future studies will profile fitness of Ralstonia mutants in planta to test whether sap fitness factors also contribute to fitness in living hosts.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
This study uses three Ralstonia strains, one representative per plant-pathogenic species: R. pseudosolanacearum GMI1000 (phylotype I sequevar 18), R. syzygii PSI07 (phylotype IV sequevar 10), and R. solanacearum IBSBF1503 (phylotype IIB sequevar 4). All isolates are pathogenic on tomato (11).
Ralstonia was routinely grown in CPG rich medium (per liter: 1 g Casamino Acids, 10 g Bacto peptone, 5 g glucose, 1 g yeast extract) at 28°C. Agar plates were supplemented with 1% tetrazolium chloride to confirm colony morphology. For minimal medium growth curves and fitness assays, strains were grown in quarter-strength M63 [per liter: 3.4 g KH2PO4, 0.5 g (NH4)2SO4, 10 μl of 1.25 mg/ml FeSO4·7H2O, 51.7 μl of 1 M MgSO4; adjusted to pH 7] with 10 mM glucose.
Construction of barcoded transposon mutant libraries.
Barcoded mariner transposon mutant libraries were created in three wild-type strain backgrounds: GMI1000, PSI07, and IBSBF1503. The barcoded transposons were introduced via conjugation. Recipient strains (Ralstonia isolates) were grown overnight in 5 ml CPG medium at 28°C with shaking at 200 rpm. A 2-ml portion of the overnight culture was subcultured into 25 ml CPG and grown for 2 h at 28°C. The donor E. coli strain (WM3064 background, a synthetic auxotroph requiring 300 μM diaminopimenlate [DAP]) carrying the pKMW3 (Kanr) barcoded mariner transposon vector library (16) was thawed on ice, and 1 ml was inoculated into 20 ml LB with 25 μg/ml kanamycin and 300 μM DAP and grown to mid-log phase at 37°C with shaking. Conjugations were carried out as previously described (63). Donor and recipient strains were centrifuged at 5,000 × g for 10 min and resuspended in CPG plus 300 μM DAP. Recipient strains and the donor strain were adjusted to an optical density at 600 nm (OD600) of 3.0 and 1.0, respectively, and mixed in equal volumes. In total, 1.2 ml of the strain mixture was spotted onto 12 nitrocellulose filters (82-mm discs with 0.45-μm pore size cut into sixths) overlaid on four plates of CPG with 300 μM DAP and without glucose. Mating was allowed to occur for 3 to 4 h at 28°C. Filters were pooled in 20 ml CPG broth and vortexed in a 50-ml tube for 30 s to dislodge bacteria. The suspension was adjusted to 80 ml and evenly spread over 400 CPG plates (200 μl per plate) with 25 μg/ml kanamycin. To calculate transformation efficiencies, a portion of each suspension was dilution plated on CPG with kanamycin (to quantify transformants) and without kanamycin (to quantify total Ralstonia cells). Transformation efficiencies were as follows: 3.9 × 10−5 for GMI1000, 8.6 × 10−3 for PSI07, and 1.5 × 10−4 for IBSBF1503. Selection plates were incubated for 2 days at 28°C. Cells were harvested by scraping and pooling approximately 2 × 106 colonies in CPG broth with kanamycin. Density was adjusted to an OD600 of 0.25 in 200 ml and grown at 28°C with shaking until OD600 reached 1.0. Individual 1-ml aliquots were preserved in 20% (vol/vol) glycerol (final concentration) at −80°C.
Mapping transposon insertion sites.
We used a previously described protocol for mapping the transposon insertion locations and to link these mutants to their unique DNA barcode sequences (16). Full details describing this protocol are available (16). Briefly, genomic DNA was extracted using the Qiagen DNeasy blood and tissue kit, with RNase A treatment, according to the manufacturer’s recommendations. The DNA was sheared to ∼300 bp using a Covaris S220, and the DNA was repaired and A-tailed using the NEBNext DNA Library preparation kit for Illumina (New England Biolabs). After ligating Illumina compatible adapters to the DNA fragments, we PCR amplified the transposon insertion junctions using primers with Illumina P5 and P7 sequences. The final PCR product was purified using AMPure XP beads and assessed for quantity and size using an Agilent Bioanalyzer DNA1000 chip. Samples were sequenced on a HiSeq2500 instrument at the QB3 Berkeley Genomics Center using 2 × 150 reads. TnSeq reads were analyzed with a custom Perl script, MapTnSeq.pl, which assigns each unique barcode sequence to a corresponding location in the genome. Each barcode was mapped to a single insertion site by identifying barcodes that consistently map to a unique location in the genome using DesignRandomPool.pl. All scripts used are available at https://bitbucket.org/berkeleylab/feba/src/master/bin/ (16, 62).
Essential gene calculations.
On the basis of the TnSeq data, we used standard computational methods (62) to predict which genes are likely essential for growth in CPG medium. Briefly, this analysis predicts gene essentiality based on genes that lacked transposon insertions in the central 80% of the coding region. We excluded two categories of genes from the essentiality calculation. Genes sharing high nucleotide identity with other genes in the genome (measured via BLAT) (64) were excluded because we could not map transposon insertions to highly similar genes. Second, we excluded genes below a minimum length because they are less likely to be disrupted. Briefly, this cutoff is determined using our previously described analysis pipeline (18) using saturation of mariner transposon insertions to calculate the length at which genes have a 1% chance of being missed due to random chance. Due to differences in insertion mutant coverage depth between the transposon libraries, the minimum gene length considered was 450 bp for GMI1000, 325 bp for PSI07, and 400 bp for IBSBF1503. For comparison, we also used the Bio-Tradis pipeline (github.com/sanger-pathogens/Bio-Tradis) (17) to predict essential genes in these strains, using the same minimum gene length cutoffs and excluding insertions in the first and last 10% of the coding region from analysis.
Orthologous gene predictions.
Orthologous genes were predicted using the “Genome Gene Best Homologs” tool from the JGI IMG database (19). We searched for homologs in strains PSI07 and IBSBF1503 against strain GMI1000 as the reference genome using the JGI IMG default identity threshold of 60%. The results, originally reported with IMG locus tags, were matched to the corresponding NCBI locus tags by gene sequence.
Tomato xylem sap harvesting.
Xylem sap was harvested from susceptible tomato plants (cv. Moneymaker and Bonny Best). Most experiments were performed with sap from Moneymaker tomato plants grown in the Oxford Tract Greenhouse in Berkeley, CA. Bacterial growth curves were independently replicated with sap from tomato plants grown in a growth chamber (28°C with a 12 h day/night cycle) at University of Wisconsin (UW)-Madison.
Xylem sap was harvested by detopping each plant at the cotyledon juncture with a razor blade and allowing sap to pool on the stump (65). To reduce cytoplasmic contamination, the sap that accumulated in the first few minutes was discarded; the stump was washed with distilled water (diH2O) and gently blotted dry. Sap was then pipetted and pooled into a 50-ml cone over a collection period of 3 h. Pooled sap was centrifuged at 5,000 × g and room temperature (RT) for 10 min, and the supernatant was filter sterilized using filters with 0.22-μm pores (catalog no. 725-2520; Thermo Scientific). Sap was aliquoted and stored at −20°C until use. Each batched pool of xylem sap was collected from approximately 70 plants.
R. solanacearum growth in tomato xylem sap.
Colonies of each strain were inoculated into 6 ml of CPG medium and grown overnight at 28°C with shaking at 200 rpm for a total of three biological replicates. Cells from stationary-phase cultures were pelleted at 13,000 × g and washed twice in 1 ml of double-distilled water (ddH2O). The washed pellet was resuspended in ddH2O and adjusted to a final OD600 of 0.02. Each growth condition was inoculated with 7 μl of each culture at a starting cell density of ∼105 CFU/ml in 48-well plates (catalog no. 353078; Corning) for a total of two technical replicates per biological replicate. Growth was measured by dilution plating at 0-, 4-, 8-, 24-, 32-, and 48-h time points. In parallel, measurements for 12-, 16-, and 20-h time points were taken from independent cultures that were started 12 h after the first, using the same cultures, growth media, and technique described above.
Fitness experiments with transposon libraries.
For genome-wide fitness experiments, we adapted established methods (16). Per condition (minimal medium, rich medium, or Moneymaker tomato xylem sap), three 1-ml aliquots of each transposon library (strains GMI1000, PSI07, and IBSBF1503) were thawed on ice and revived in separate 100-ml flasks of CPG medium with 12.5 mg/ml kanamycin with shaking incubation at 28°C for 16 to 20 h. Once cultures reached an OD600 of 0.2 to 0.5, cells were pelleted by centrifuging for 10 min at 5,000 × g and RT and resuspended in 2 ml of rich or minimal medium or sterile H2O (xylem sap experiments). Cells were washed three times in media or xylem sap. An aliquot of the washed cells (>109 cells) was retained (pelleted and frozen) for a time zero control. For rich and minimal medium experiments, cells were seeded into 5 ml at an OD600 of 0.02 (∼2 × 107 to 5 × 107 total cells). Rich medium cultures were grown to saturation (20 to 24 h; 5 or 6 cell doublings) and minimal medium cultures were grown for over 48 h (5 or 6 cell doublings for strains PSI07 and GMI1000 and 4 doublings for strain IBSBF1503). For xylem sap experiments, cells were resuspended in 20 ml of xylem sap at a starting cell density of 106 cells/ml (2 × 107 cells total) and grown for 25 h (7 or 8 doublings for GMI1000 and 8 or 9 doublings for PSI07). Genomic DNA was extracted (Qiagen DNeasy blood and tissue kit) from the time zero control, and cells were harvested after growing in differential media.
BarSeq and fitness calculations.
Prior to PCR, the quality of total genomic DNA isolated from fitness experiments was assessed on an agarose gel, while DNA concentration was determined by NanoDrop. Barcodes were PCR amplified from each sample using the previously described reaction protocol (16). For most experiments, we used indexed P2 oligonucleotides and a mix of nonindexed P1 oligonucleotides of variable length (two to five bases) to stagger the reads. For some experiments, we used indexed versions of both the P1 and P2 oligonucleotides to minimize incorrectly assigned indexes in Illumina HiSeq4000 runs. Following PCR, amplicon mixtures from each sample were pooled in equal volumes, purified over a Zymo DNA Clean and Concentrator column, and eluted with water. Prior to sequencing, the quality of cleaned and concentrated amplicon pools was assessed using a Bioanalyzer. Quantitative PCR (qPCR) was used to determine how much of each sample to load for sequencing. Barcode amplicons were sequenced on a HiSeq4000 at QB3 Berkeley Genomics Center with 96 samples multiplexed (50-bp reads, single end).
Sequencing data were analyzed using the BarSeq pipeline, available at https://bitbucket.org/berkeleylab/feba/src/master/bin/. For each competitive fitness assay, fitness score (“Fit scores”) for each gene were calculated as the log2 ratio between barcode abundance after outgrowth in a condition versus its abundance in the time zero sample. Each Fit score is the weighted average of fitness values for all mutant strains with transposon insertions in a given gene. These Fit scores are normalized across the genome such that a gene with neither a fitness cost nor benefit has a value of 0. Significance was determined based on an absolute t-like test statistic with a threshold of | t | > 2.5 in all three RB-TnSeq trials. The t-like test statistic considers the consistency of the fitness scores for all barcoded mutants for each gene in the experiment as previously described in detail (16).
Data availability.
The raw reads used for TnSeq mapping is available in the NCBI SRA under accession number PRJNA629015. The Fitness Browser (http://fit.genomics.lbl.gov) offers a graphical user interface for exploring the fitness data from these experiments, orthology of the genes between these strains and other Gram-negative bacteria, and cross-references to KEGG, Paperblast, and NCBI databases.
Supplementary Material
ACKNOWLEDGMENTS
We thank Steve Lindow, Caitilyn Allen, and Jeff Flynn for useful discussions. We also thank Boris Vinatzer and Parul Sharma for sharing unpublished data on the size of the Ralstonia core genome. This work used the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by NIH S10 OD018174 Instrumentation Grant. Oxford Tract Greenhouse Staff and the QB3 Berkeley Genomics Center provided technical assistance.
This work was supported by USDA NIFA 2018-67012-31497 awarded to T.M.L.-P.; partial support was provided by a UC Berkeley SURF Rose Hills Independent Fellowship awarded to K.E.S. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Contributor Information
Tiffany M. Lowe-Power, Email: tlowepower@ucdavis.edu.
Jack A. Gilbert, University of California San Diego
REFERENCES
- 1.Prior P, Ailloud F, Dalsing BL, Remenant B, Sanchez B, Allen C. 2016. Genomic and proteomic evidence supporting the division of the plant pathogen Ralstonia solanacearum into three species. BMC Genomics 17:90. doi: 10.1186/s12864-016-2413-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Safni I, Cleenwerck I, De Vos P, Fegan M, Sly L, Kappler U. 2014. Polyphasic taxonomic revision of the Ralstonia solanacearum species complex. Int J Syst Evol Microbiol 64:3087–3103. doi: 10.1099/ijs.0.066712-0. [DOI] [PubMed] [Google Scholar]
- 3.Hayward AC. 1991. Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu Rev Phytopathol 29:65–87. doi: 10.1146/annurev.py.29.090191.000433. [DOI] [PubMed] [Google Scholar]
- 4.Safni I, Subandiyah S, Fegan M. 2018. Ecology, epidemiology and disease management of Ralstonia syzygii in Indonesia. Front Microbiol 9:419. doi: 10.3389/fmicb.2018.00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lopes CA, Rossato M. 2018. History and status of selected hosts of the Ralstonia solanacearum species complex causing bacterial wilt in Brazil. Front Microbiol 9:1228. doi: 10.3389/fmicb.2018.01228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang G, Wei Z, Xu J, Chen H, Zhang Y, She X, Macho AP, Ding W, Liao B. 2017. Bacterial wilt in China: history, current status, and future perspectives. Front Plant Sci 8:1549. doi: 10.3389/fpls.2017.01549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lowe-Power T, Chipman K. 2020. A meta-analysis of the known global distribution and host range of the Ralstonia species complex. bioRxiv 10.1101/2020.07.13.189936. [DOI]
- 8.Lowe-Power TM, Hendrich CG, von Roepenack-Lahaye E, Li B, Wu D, Mitra R, Dalsing BL, Ricca P, Naidoo J, Cook D, Jancewicz A, Masson P, Thomma B, Lahaye T, Michael AJ, Allen C. 2018. Metabolomics of tomato xylem sap during bacterial wilt reveals Ralstonia solanacearum produces abundant putrescine, a metabolite that accelerates wilt disease. Environ Microbiol 20:1330–1349. doi: 10.1111/1462-2920.14020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Remenant B, Coupat-Goutaland B, Guidot A, Cellier G, Wicker E, Allen C, Fegan M, Pruvost O, Elbaz M, Calteau A, Salvignol G, Mornico D, Mangenot S, Barbe V, Médigue C, Prior P. 2010. Genomes of three tomato pathogens within the Ralstonia solanacearum species complex reveal significant evolutionary divergence. BMC Genomics 11:379. doi: 10.1186/1471-2164-11-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Remenant B, de Cambiaire JC, Cellier G, Jacobs JM, Mangenot S, Barbe V, Lajus A, Vallenet D, Medigue C, Fegan M, Allen C, Prior P. 2011. Ralstonia syzygii, the blood disease bacterium and some Asian R. solanacearum strains form a single genomic species despite divergent lifestyles. PLoS One 6:e24356. doi: 10.1371/journal.pone.0024356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ailloud F, Lowe T, Cellier G, Roche D, Allen C, Prior P. 2015. Comparative genomic analysis of Ralstonia solanacearum reveals candidate genes for host specificity. BMC Genomics 16:270. doi: 10.1186/s12864-015-1474-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayward AC. 1964. Characteristics of Pseudomonas solanacearum. J Appl Bacteriol 27:265–277. doi: 10.1111/j.1365-2672.1964.tb04912.x. [DOI] [Google Scholar]
- 13.Lowe TM, Ailloud F, Allen C. 2015. Hydroxycinnamic acid degradation, a broadly conserved trait, protects Ralstonia solanacearum from chemical plant defenses and contributes to root colonization and virulence. Mol Plant Microbe Interact 28:286–297. doi: 10.1094/MPMI-09-14-0292-FI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowe-Power TM, Jacobs JM, Ailloud F, Fochs B, Prior P, Allen C. 2016. Degradation of the plant defense signal salicylic acid protects Ralstonia solanacearum from toxicity and enhances virulence on tobacco. mBio 7:e00656-16. doi: 10.1128/mBio.00656-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu H, Deutschbauer AM. 2018. Rapidly moving new bacteria to model-organism status. Curr Opin Biotechnol 51:116–122. doi: 10.1016/j.copbio.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 16.Wetmore KM, Price MN, Waters RJ, Lamson JS, He J, Hoover CA, Blow MJ, Bristow J, Butland G, Arkin AP, Deutschbauer A. 2015. Rapid quantification of mutant fitness in diverse bacteria by sequencing randomly bar-coded transposons. mBio 6:e00306-15. doi: 10.1128/mBio.00306-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barquist L, Mayho M, Cummins C, Cain AK, Boinett CJ, Page AJ, Langridge GC, Quail MA, Keane JA, Parkhill J. 2016. The TraDIS toolkit: sequencing and analysis for dense transposon mutant libraries. Bioinformatics 32:1109–1111. doi: 10.1093/bioinformatics/btw022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rubin BE, Wetmore KM, Price MN, Diamond S, Shultzaberger RK, Lowe LC, Curtin G, Arkin AP, Deutschbauer A, Golden SS. 2015. The essential gene set of a photosynthetic organism. Proc Natl Acad Sci USA 112:e6634–e6643. doi: 10.1073/pnas.1519220112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen I-MA, Chu K, Palaniappan K, Pillay M, Ratner A, Huang J, Huntemann M, Varghese N, White JR, Seshadri R, Smirnova T, Kirton E, Jungbluth SP, Woyke T, Eloe-Fadrosh EA, Ivanova NN, Kyrpides NC. 2019. IMG/M v.5.0: an integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res 47:D666–D677. doi: 10.1093/nar/gky901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hulsen T, De Vlieg J, Alkema W. 2008. BioVenn – a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dalsing BL, Allen C. 2014. Nitrate assimilation contributes to Ralstonia solanacearum root attachment, stem colonization, and virulence. J Bacteriol 196:949–960. doi: 10.1128/JB.01378-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi K, Son GJ, Ahmad S, Lee SY, Lee HJ, Lee S-W. 2020. Contribution of the murI gene encoding glutamate racemase in the motility and virulence of Ralstonia solanacearum. Plant Pathol J 36:355–363. doi: 10.5423/PPJ.OA.03.2020.0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin YM, Chou IC, Wang JF, Ho FI, Chu YJ, Huang PC, Lu DK, Shen HL, Elbaz M, Huang SM, Cheng CP. 2008. Transposon mutagenesis reveals differential pathogenesis of Ralstonia solanacearum on tomato and Arabidopsis. Mol Plant Microbe Interact 21:1261–1270. doi: 10.1094/MPMI-21-9-1261. [DOI] [PubMed] [Google Scholar]
- 25.Li CH, Wang KC, Hong YH, Chu TH, Chu YJ, Chou IC, Lu DK, Chen CY, Yang WC, Lin YM, Cheng CP. 2014. Roles of different forms of lipopolysaccharides in Ralstonia solanacearum pathogenesis. Mol Plant Microbe Interact 27:471–478. doi: 10.1094/MPMI-08-13-0248-R. [DOI] [PubMed] [Google Scholar]
- 26.Bohin JP. 2000. Osmoregulated periplasmic glucans in Proteobacteria. FEMS Microbiol Lett 186:11–19. doi: 10.1111/j.1574-6968.2000.tb09075.x. [DOI] [PubMed] [Google Scholar]
- 27.Bontemps-Gallo S, Bohin J-P, Lacroix J-M. 6 June 2017, posting date. Osmoregulated periplasmic glucans. EcoSal Plus. doi: 10.1128/ecosalplus.ESP-0001-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacobs JM, Babujee L, Meng F, Milling A, Allen C. 2012. The in planta transcriptome of Ralstonia solanacearum: conserved physiological and virulence strategies during bacterial wilt of tomato. mBio 3:e00114-12. doi: 10.1128/mBio.00114-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamilton CD, Steidl O, MacIntyre AM, Allen C. 2021. Ralstonia solanacearum depends on catabolism of myo-inositol, sucrose, and trehalose for virulence in an infection stage-dependent manner. Mol Plant Microbe Interact 34:669−679. doi: 10.1094/MPMI-10-20-0298-R. [DOI] [PubMed] [Google Scholar]
- 30.Xian L, Yu G, Wei Y, Li Y, Zhang H, Xue H, Morcillo RJL, Macho AP. 2020. A bacterial effector protein hijacks plant metabolism to support bacterial nutrition. Cell Host Microbe 28:548−557.e7. doi: 10.1016/j.chom.2020.07.003. [DOI] [PubMed] [Google Scholar]
- 31.Gerlin L, Escourrou A, Cassan C, Maviane Macia F, Peeters N, Genin S, Baroukh C. 2021. Unravelling physiological signatures of tomato bacterial wilt and xylem metabolites exploited by Ralstonia solanacearum. Environ Microbiol doi: 10.1111/1462-2920.15535. [DOI] [PubMed] [Google Scholar]
- 32.Khokhani D, Lowe-Power TM, Tran TM, Allen C. 2017. A single regulator mediates strategic switching between attachment/spread and growth/virulence in the plant pathogen Ralstonia solanacearum. mBio 8:e00895-17. doi: 10.1128/mBio.00895-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peyraud R, Cottret L, Marmiesse L, Gouzy J, Genin S. 2016. A resource allocation trade-off between virulence and proliferation drives metabolic versatility in the plant pathogen Ralstonia solanacearum. PLoS Pathog 12:e1005939. doi: 10.1371/journal.ppat.1005939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brumbley SM, Denny TP. 1990. Cloning of wild-type Pseudomonas solanacearum phcA, a gene that when mutated alters expression of multiple traits that contribute to virulence. J Bacteriol 172:5677–5685. doi: 10.1128/jb.172.10.5677-5685.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang J, Carney BF, Denny TP, Weissinger AK, Schell MA. 1995. A complex network regulates expression of eps and other virulence genes of Pseudomonas solanacearum. J Bacteriol 177:1259–1267. doi: 10.1128/jb.177.5.1259-1267.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flavier AB, Schell MA, Denny TP. 1998. An RpoS (σS) homologue regulates acylhomoserine lactone-dependent autoinduction in Ralstonia solanacearum. Mol Microbiol 28:475–486. doi: 10.1046/j.1365-2958.1998.00804.x. [DOI] [PubMed] [Google Scholar]
- 37.Brown DG, Allen C. 2004. Ralstonia solanacearum genes induced during growth in tomato: an inside view of bacterial wilt. Mol Microbiol 53:1641–1660. doi: 10.1111/j.1365-2958.2004.04237.x. [DOI] [PubMed] [Google Scholar]
- 38.Powers MJ, Trent MS. 2018. Phospholipid retention in the absence of asymmetry strengthens the outer membrane permeability barrier to last-resort antibiotics. Proc Natl Acad Sci USA 115:E8518–E8527. doi: 10.1073/pnas.1806714115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poulsen BE, Yang R, Clatworthy AE, White T, Osmulski SJ, Li L, Penaranda C, Lander ES, Shoresh N, Hung DT. 2019. Defining the core essential genome of Pseudomonas aeruginosa. Proc Natl Acad Sci USA 116:10072–10080. doi: 10.1073/pnas.1900570116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Su Y, Xu Y, Li Q, Yuan G, Zheng D. 2020. The essential genome of Ralstonia solanacearum. Microbiol Res 238:126500. doi: 10.1016/j.micres.2020.126500. [DOI] [PubMed] [Google Scholar]
- 41.Larson MH, Gilbert LA, Wang X, Lim WA, Weissman JS, Qi LS. 2013. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc 8:2180–2196. doi: 10.1038/nprot.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su Y, Xu Y, Liang H, Yuan G, Wu X, Zheng D. 2021. Genome-wide identification of Ralstonia solanacearum genes required for survival in tomato plants. mSystems 6:e00838-21. doi: 10.1128/mSystems.00838-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kai K, Ohnishi H, Shimatani M, Ishikawa S, Mori Y, Kiba A, Ohnishi K, Tabuchi M, Hikichi Y. 2015. Methyl 3-hydroxymyristate, a diffusible signal mediating phc quorum sensing in Ralstonia solanacearum. Chembiochem 16:2309–2318. doi: 10.1002/cbic.201500456. [DOI] [PubMed] [Google Scholar]
- 44.Lenski RE, Rose MR, Simpson SC, Tadler SC. 1991. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am Nat 138:1315–1341. doi: 10.1086/285289. [DOI] [Google Scholar]
- 45.Morin M, Pierce EC, Dutton RJ. 2018. Changes in the genetic requirements for microbial interactions with increasing community complexity. Elife 7:e37072. doi: 10.7554/eLife.37072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Capela D, Marchetti M, Clérissi C, Perrier A, Guetta D, Gris C, Valls M, Jauneau A, Cruveiller S, Rocha EPC, Masson-Boivin C. 2017. Recruitment of a lineage-specific virulence regulatory pathway promotes intracellular infection by a plant pathogen experimentally evolved into a legume symbiont. Mol Biol Evol 34:2503–2521. doi: 10.1093/molbev/msx165. [DOI] [PubMed] [Google Scholar]
- 47.Guidot A, Jiang W, Ferdy J, Thébaud C, Barberis P, Gouzy J, Genin S. 2014. Multihost experimental evolution of the pathogen Ralstonia solanacearum unveils genes involved in adaptation to plants. Mol Biol Evol 31:2913–2928. doi: 10.1093/molbev/msu229. [DOI] [PubMed] [Google Scholar]
- 48.Arlat M, Gough CL, Zischek C, Barberis PA, Trigalet A, Boucher CA. 1992. Transcriptional organization and expression of the large hrp gene cluster of Pseudomonas solanacearum. Mol Plant Microbe Interact 5:187–193. doi: 10.1094/mpmi-5-187. [DOI] [PubMed] [Google Scholar]
- 49.Boucher CA, Barberis PA, Trigalet APH, Demery DA. 1985. Transposon mutagenesis of Pseudomonas solanacearum: isolation of Tn5-induced avirulent mutants. J Gen Microbiol 131:2449–2457. doi: 10.1099/00221287-131-9-2449. [DOI] [Google Scholar]
- 50.Staskawicz BJ, Dahlbeck D, Keen NT. 1984. Cloned avirulence gene of Pseudomonas syringae pv. glycinea determines race-specific incompatibility on Glycine max (L.) Merr. Proc Natl Acad Sci USA 81:6024–6028. doi: 10.1073/pnas.81.19.6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cole BJ, Feltcher ME, Waters RJ, Wetmore KM, Mucyn TS, Ryan EM, Wang G, Ul-Hasan S, McDonald M, Yoshikuni Y, Malmstrom RR, Deutschbauer AM, Dangl JL, Visel A. 2017. Genome-wide identification of bacterial plant colonization genes. PLoS Biol 15:e2002860. doi: 10.1371/journal.pbio.2002860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Duong DA, Jensen RV, Stevens AM. 2018. Discovery of Pantoea stewartii ssp. stewartii genes important for survival in corn xylem through a Tn-Seq analysis. Mol Plant Pathol 19:1929–1941. doi: 10.1111/mpp.12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gonzalez-Mula A, Lachat J, Mathias L, Naquin D, Lamouche F, Mergaert P, Faure D. 2019. The biotroph Agrobacterium tumefaciens thrives in tumors by exploiting a wide spectrum of plant host metabolites. New Phytol 222:455–467. doi: 10.1111/nph.15598. [DOI] [PubMed] [Google Scholar]
- 54.Helmann TC, Deutschbauer AM, Lindow SE. 2019. Genome-wide identification of Pseudomonas syringae genes required for fitness during colonization of the leaf surface and apoplast. Proc Natl Acad Sci USA 116:18900–18910. doi: 10.1073/pnas.1908858116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Helmann TC, Ongsarte CL, Lam J, Deutschbauer AM, Lindow SE. 2019. Genome-wide transposon screen of a Pseudomonas syringae mexB mutant reveals the substrates of efflux transporters. mBio 10:e02614-19. doi: 10.1128/mBio.02614-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu Z, Beskrovnaya P, Melnyk RA, Hossain SS, Khorasani S, O’Sullivan LR, Wiesmann CL, Bush J, Richard JD, Haney CH. 2018. A genome-wide screen identifies genes in rhizosphere-associated Pseudomonas required to evade plant defenses. mBio 9:e00433-18. doi: 10.1128/mBio.00433-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Royet K, Parisot N, Rodrigue A, Gueguen E, Condemine G. 2019. Identification by Tn-seq of Dickeya dadantii genes required for survival in chicory plants. Mol Plant Pathol 20:287–306. doi: 10.1111/mpp.12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brooks JFI, Gyllborg MC, Cronin DC, Quillin SJ, Mallama CA, Foxall R, Whistler C, Goodman AL, Mandel MJ. 2014. Global discovery of colonization determinants in the squid symbiont Vibrio fischeri. Proc Natl Acad Sci USA 111:17284–17289. doi: 10.1073/pnas.1415957111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hava DL, Camilli A. 2002. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol Microbiol 45:1389–1406. doi: 10.1046/j.1365-2958.2002.t01-1-03106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lewin GR, Stacy A, Michie KL, Lamont RJ, Whiteley M. 2019. Large-scale identification of pathogen essential genes during coinfection with sympatric and allopatric microbes. Proc Natl Acad Sci USA 116:19685–19694. doi: 10.1073/pnas.1907619116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hentchel KL, Ruiz LMR, Curtis PD, Fiebig A, Coleman ML, Crosson S. 2019. Genome-scale fitness profile of Caulobacter crescentus grown in natural freshwater. ISME J 13:523–536. doi: 10.1038/s41396-018-0295-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Price MN, Wetmore KM, Waters RJ, Callaghan M, Ray J, Liu H, Kuehl JV, Melnyk RA, Lamson JS, Suh Y, Carlson HK, Esquivel Z, Sadeeshkumar H, Chakraborty R, Zane GM, Rubin BE, Wall JD, Visel A, Bristow J, Blow MJ, Arkin AP, Deutschbauer AM. 2018. Mutant phenotypes for thousands of bacterial genes of unknown function. Nature 557:503–509. doi: 10.1038/s41586-018-0124-0. [DOI] [PubMed] [Google Scholar]
- 63.Perrier A, Barberis P, Genin S. 2018. Introduction of genetic material in Ralstonia solanacearum through natural transformation and conjugation. Methods Mol Biol 1734:201–207. doi: 10.1007/978-1-4939-7604-1_16. [DOI] [PubMed] [Google Scholar]
- 64.Kent WJ. 2002. BLAT—the BLAST-like alignment tool. Genome Res 12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Khokhani D, Tran TM, Lowe-Power TM, Allen C. 2018. Plant assays for quantifying Ralstonia solanacearum virulence. Bio Protoc 8:e3028. doi: 10.21769/BioProtoc.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lex A, Gehlenborg N, Strobelt H, Vuillemot R, Pfister H. 2014. UpSet: visualization of intersecting sets. IEEE Trans Vis Comput Graph 20:1983–1992. doi: 10.1109/TVCG.2014.2346248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene essentiality predictions for strains GMI1000, PSI07, and IBSBF1503. Download Table S1, XLSX file, 0.2 MB (223.4KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Venn diagram of putative essential genes in Ralstonia strains GMI1000, IBSBF1503, and PSI07. Orthologous predicted essential genes are represented proportionally in Venn diagram intersections. Genes were included in the Venn diagram only if they were predicted to be essential by both analysis pipelines used in this study (17, 18). Download FIG S1, TIF file, 0.5 MB (559.2KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Growth of Ralstonia in rich medium, minimal medium, and xylem sap from multiple susceptible tomato cultivars. (A and B) To determine the carrying capacity of sap and culture media, Ralstonia strain GMI1000 was inoculated into rich CPG medium, quarter strength M63 medium, or xylem sap from Moneymaker tomato at 104, 105, or 106 CFU/ml starting density. (C to E) Wild-type Ralstonia strains GMI1000, PSI07, and IBSBF1503 were grown in sap from Moneymaker and Bonny Best tomato. All strains were inoculated in biological triplicate into three pools of xylem sap incubated at 28°C with shaking. Cell density was measured by dilution plating. Download FIG S2, TIF file, 1.0 MB (1,016.5KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Genome-wide Fit scores for strains GMI1000, PSI07, and IBSBF1503. Download Table S2, XLSX file, 3.5 MB (3.5MB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
The raw reads used for TnSeq mapping is available in the NCBI SRA under accession number PRJNA629015. The Fitness Browser (http://fit.genomics.lbl.gov) offers a graphical user interface for exploring the fitness data from these experiments, orthology of the genes between these strains and other Gram-negative bacteria, and cross-references to KEGG, Paperblast, and NCBI databases.