Abstract
Gene expression in mammalian organisms is regulated at multiple levels, including DNA accessibility for transcription factors and chromatin structure. Methylation of CpG dinucleotides is thought to be involved in imprinting and in the pathogenesis of cancer. However, the relevance of methylation for directing tissue-specific gene expression is highly controversial. The cyclin A1 gene is expressed in very few tissues, with high levels restricted to spermatogenesis and leukemic blasts. Here, we show that methylation of the CpG island of the human cyclin A1 promoter was correlated with nonexpression in cell lines, and the methyl-CpG binding protein MeCP2 suppressed transcription from the methylated cyclin A1 promoter. Repression could be relieved by trichostatin A. Silencing of a cyclin A1 promoter-enhanced green fluorescent protein (EGFP) transgene in stable transfected MG63 osteosarcoma cells was also closely associated with de novo promoter methylation. Cyclin A1 could be strongly induced in nonexpressing cell lines by trichostatin A but not by 5-aza-cytidine. The cyclin A1 promoter-EGFP construct directed tissue-specific expression in male germ cells of transgenic mice. Expression in the testes of these mice was independent of promoter methylation, and even strong promoter methylation did not suppress promoter activity. MeCP2 expression was notably absent in EGFP-expressing cells. Transcription from the transgenic cyclin A1 promoter was repressed in most organs outside the testis, even when the promoter was not methylated. These data show the association of methylation with silencing of the cyclin A1 gene in cancer cell lines. However, appropriate tissue-specific repression of the cyclin A1 promoter occurs independently of CpG methylation.
Gene expression in higher organisms is initiated by the interaction of the basal transcriptional machinery with the double-stranded DNA helix of a eukaryotic promoter (for a review, see reference 6). The process of this interaction critically depends on the chromatin structure surrounding the DNA and the abundance and binding ability of the relevant accessory transcription factors (14, 35). Both mechanisms, a nonpermissive chromatin structure as well as either the absence of important transcriptional activators or the presence of repressors, can inhibit gene transcription. A permissive chromatin structure contains acetylated histones that allow unfolding of the nucleosome, making it accessible to transcription factors (48, 56, 59). Conversely, the deacetylation of histones leads to repression of transcriptional activity. A wide variety of transcriptional repressors (e.g., Mad-Max complexes) are known to recruit corepressors, such as mSin3A, N-CoR, and SMRT (2). These corepressors are associated with several other proteins, including histone deacetylases 1 and 2 (HDAC1 and HDAC2), which remove acetyl moieties from specific lysine residues on histones H3 and H4 (50). The positively charged lysine residues interact with the DNA and render it inaccessible to the transcription machinery (57).
Recently, the methylation of CpG dinucleotides has been demonstrated to mediate transcriptional repression by recruiting histone deacetylases (44, 47). HDAC activity is relocated by family members of the methylated CpG binding domain (MBD) proteins (19). Methylation of DNA has long been associated with the regulation of gene expression, and physiological DNA methylation patterns are essential for embryogenesis (36, 67). Abnormalities in CpG methylation are involved in tumorigenesis and senescence (7, 24, 27). The role of methylation in the silencing of genes in cancer cells and especially in cancer cell lines is well documented (1). Methylation of a tumor suppressor gene (e.g., p16ink4A) is closely related to transcriptional repression of its expression, and the inhibition of DNA methylation can lead to reexpression and to altered tumor growth characteristics (3). While studies have implied that methylation is only a secondary event in gene silencing (11), other investigators have suggested that the methylation itself is the primary event in transcriptional repression in cell lines (7).
In contrast to the presumed role of methylation in cell lines, its relevance in directing tissue-specific expression in healthy organisms is highly controversial (53, 60). A correlation between methylation and nonexpression has been documented for several genes (9, 34, 62), but no such correlation exists for other genes (21, 58). A DNA methylation profile for the mouse skeletal α-actin promoter showed that expression of the gene was appropriately repressed in several organs, although the promoter was not methylated (61). On the other hand, consistent with the idea that methylation can suppress transcription, the authors did not find expression in organs with strong CpG methylation of the α-actin promoter. These studies suggest that methylation is not necessary for transcriptional repression in many organs. However, the role of CpG methylation found in organs without expression of the analyzed gene is unclear. Methylation might be critically involved in transcriptional repression, it could act as a secondary layer of repression, or methylation could be an epiphenomenon essentially uninvolved in gene silencing.
We have started to determine the regulation of expression of the cyclin A1 gene, a gene that is physiologically expressed at high levels in pachytene spermatocytes and at much lower levels in hematopoietic progenitor cells (54, 66). Also, very high levels of cyclin A1 have been found in leukemic cells harvested from individuals with acute myeloid leukemia and from myeloid leukemia cell lines (64, 66). In contrast, very low expression at the mRNA level is observed in most nonleukemic, adherent cell lines (64). We have recently cloned the promoter region of cyclin A1 and determined that several GC boxes are essential for promoter activity (42). In addition, the promoter is transactivated by c-myb (43). Surprisingly, in contrast to the highly restricted expression of the cyclin A1 gene in vivo, we found the promoter to be very active in transient transfections in all cell types and cell lines tested so far (42, 43; unpublished data). Since the gene expression pattern is highly restricted in vivo, we concluded that either the isolated promoter fragment (∼1.3 kb) missed an important repressor binding site or mechanisms that alter the chromatin structure are essential for directing tissue-specific expression in vivo.
We therefore carefully analyzed cyclin A1 promoter regulation in cell lines and in mice that were transgenic for a cyclin A1 promoter-enhanced green fluorescent protein (EGFP) construct and obtained the following results. The CpG island of the endogenous cyclin A1 promoter is heavily methylated in adherent cell lines. Methylation of the cyclin A1 promoter in cancer cell lines correlates with transcriptional silencing, and MBD family members are expressed in these cell lines and might be involved in transcriptional repression of the methylated cyclin A1 promoter. Cyclin A1 gene expression could be induced in nonexpressing HeLa cells by inhibitors of deacetylation but not by inhibitors of methylation alone. A fragment containing 1.3 kb upstream of the cyclin A1 transcriptional start site was sufficient to direct tissue-specific expression in the testes of transgenic mice. Interestingly, methylation was not necessary to repress the transgenic promoter in most organs of the transgenic mice. In addition, methylation of the cyclin A1 promoter was not sufficient to inhibit testis-specific gene expression. MeCP2 expression was very low in EGFP-expressing male germ cells. Our study provides evidence that methylation of the cyclin A1 promoter correlates with transcriptional repression in cell lines, but tissue-specific expression of cyclin A1 in vivo is mediated by methylation-independent mechanisms.
MATERIALS AND METHODS
DNA constructs.
The cyclin A1 promoter-EGFP expression plasmid was constructed by inserting a 1,444-bp (−1299 to +145) fragment of the cyclin A1 promoter (42) into the BglII and HindIII sites of the promoterless EGFP-1 plasmid (Clontech). The cyclin A1 promoter-luciferase reporter construct contained a 335-bp fragment of the cyclin A1 promoter (−190 to +145) cloned into the promoter position of PGL3-Basic (Promega). The human MeCP2 expression vector was a kind gift from S. Kudo, Hokkaido Institute, Sapporo, Japan (33).
Generation of transgenic mice.
Transgenic mice were generated by standard techniques (22). The vector sequences were removed from the cyclin A1 promoter-EGFP construct by restriction digestion, and the DNA fragment was purified. Fertilized eggs were harvested from the oviducts of B6D2F1 (C57BL/6J × DBA)F1 females, and the DNA was microinjected into the pronuclei of fertilized eggs. The injected eggs were incubated in KSOM (Specialty Media) at 37°C and 5% CO2 until they were transferred to the oviducts of pseudopregnant females. About 10% of the offspring contained the transgene. The presence of the transgene was confirmed by PCR and Southern blot analysis. The expression of the transgene did not impair either survival or fertility of the mice (data not shown).
Preparation of cells from murine organs for flow cytometric analysis and cell sorting.
The tunica was removed from the testis, and the tubules were teased out and spread in a 5% collagenase solution in HEPES-buffered modified HTF medium (Irvine Scientific) for 15 min at 37°C. The tubules were lifted into 1 ml of enzyme mixture (containing trypsin, collagenase, and DNase)/testis and minced with sterile needles. The minced tubules were incubated at 37°C for 15 min. The undigested tubules were allowed to settle, and the cell suspension was aspirated, washed three times by centrifugation at 400 × g, and resuspended in 6 ml of modified HTF medium. The cells were resuspended in 1 ml of HTF medium and passed through nylon mesh before flow cytometric analysis and cell sorting were performed. The kidney cells were prepared in the same way except that the first collagenase incubation was omitted. Single-cell suspensions from bone marrow and spleen were obtained by pressing the cells through a mesh.
Analysis of CpG methylation.
Bisulfite sequencing was carried out according to the method described by Clark et al. (8) with minor modifications. Ten micrograms of sheared genomic DNA was incubated with the bisulfite-hydroquinone solution for 6 h. A nested PCR was performed (the primers are listed in Table 1), and the final PCR product (356 bp) was gel purified. Also, another set of primers was chosen that exclusively amplified the transgenic cyclin A1 promoter-EGFP construct (Table 1). For analyses of the cell lines, the PCR products were blunt end cloned and 14 to 18 clones were sequenced. Sequences were obtained from two independent batches of bisulfite-treated DNA and at least two independent PCRs. In all other cases, the purified PCR products were directly sequenced using 33P cycle sequencing of nucleotides to obtain an overview of the methylation pattern of the sense strand of the cyclin A1 promoter.
TABLE 1.
Primers and probesa
| Transcript or assay | Forward primer | Reverse primer | Probe and/or amplicon size (bp) |
|---|---|---|---|
| Human cyclin A1 | CTC CTG TCT GGT GGG AGG A | CTG ATC CAG AAT AAC ACC TGA | AGA GTG GAG TTG TGC TGG CT, 382 |
| Human β-actin | TAC ATG GCT GGG GTG TTG AA | AAG AGA GGC ATC CTC ACC CT | ATC GAG CAC GGC ATC CTC AC, 218 |
| Bisulfite cyc-A1 1.PCR | GTA GGA GAY GTT AGA GGG GGT TGT TAG | CCC CAA TAA AAA ATC CAA AAT ACA TAA TTA | 772 |
| Bisulfite cyc-A1 2. PCR | GAT TTA ATA GAY GYG GGT GGG TAG TTT AGT | ATC CCA AAT AAC RAA CAA AAT ACT AC | 356 |
| Bisulfite GFP-cycA1 1. PCR | Similar to bisulfite cyclin A1 1.PCR | ATA ATA CAA ATA AAC TTC AAA ATC AAC TTA | 961 |
| Bisulfite GFP-cyc-A1 2. PCR | Similar to bisulfite cyclin A1 1.PCR | CAA CTC CTC RCC CTT ACT CAC CAT AAT AA | 470 |
| Human cyclin A1 (TaqMan) | GGG CTC CCA GAT TTC GTC T | CTG CAG TGC ATT GCT TCA GA | CCA GCA GCA GCC CGT GGA, 58 |
| Human MeCP2 | ACT CCC CAG AAT ACA CCT TGC TT | TGA GGC CCT GGA GGT CCT | TGT TAG GGC TCA GGG AAG AAA AGT CAG AAG A, 112 |
| Human MBD-1 | CCT GGG TGC TGT GAG AAC TGT | TTG AAG GCA ATT CTC TGT GCT C | CAG CTT CTC AGG GGA TGG CAC CCA, 106 |
| Human MBD-2 | AGT GAA ATC AGA CCC ACA ACG AA | CAT CTG ATG CAC TAA GTC CTT GTA GC | AAT GAA CAG CCA CGT CAG CTT TTC TG, 85 |
| Human MBD-3 | GTA TGG CTC AAC ACC ACG CA | AGC ATG TCG GCC ATC AGC | ACG AGG ACA TCA GGA AGC AGG AAG AGC T, 127 |
| Human MBD-4 | TCT AGT GAG CGC CTA GTC CCA G | TTC CAA TTC CAT AGC AAC ATC TTC T | CCG CCG AAT GAC CTC CGC A, 68 |
| Human GAPDH | GAA GGT GAA GGT CGG AGT C | GAA GAT GGT GAT GGG ATT TC | CAA CGT TCC CGT TCT CAG CC, 226 |
| Murine MeCP2 | GAA GTC TGG CCG ATC TGC TG | ATG CAA TCA ATT CTA CTT TAG AGC GAA | 88 |
| Murine MBD-1 | TTG TGG AAT CCA CTT TTC ATG G | CAG CCG ACT CGC TTA AAC ATT | 122 |
| Murine MBD-2 | GCA GAG ACT CCG GAA TGA CC | CTG GTT GCT TGA AAA TTG ATG C | 100 |
| Murine MBD-3 | CGG CCA CAG GGA TGT CTT T | TCA TCA ACA TCT TTC CGG TGC | 130 |
| Murine MBD-4 | AAA TGC CCA GCT GCT CAC A | AGC TTC TTT GTT GTA CTT GCT GGA | 130 |
| Murine MBD-2-testis specific | Identical to murine MBD-2 forward primer | GGA GCA GAG AGG TGC ACA CA | 97 |
| Murine GAPDH | ACT GGC ATG GCC TTC CG | CAG GCG GCA CGT CAG ATC | TTC CTA CCC CCA ATG TGT CCG TCG T, 62 |
All oligonucleotides are shown in 5′-to-3′ direction. Probes for RT-PCR Southern blotting were labeled with digoxigenin. Probes used for the 5′ nuclease (TaqMan) assay were labeled with VIC (both GAPDH probes) or with FAM (all others) at the 5′ end and with the quencher dye TAMRA at the 3′ end. Real-time quantitative RT-PCR for murine MBD proteins was performed with SybrGreen. In these cases, the presence of a single specific band was demonstrated by electrophoresis on a 3% agarose gel (data not shown).
In vitro methylation and luciferase assay.
The cyclin A1 promoter-luciferase reporter construct was in vitro methylated by SssI (New England Biolabs) following the recommendations of the manufacturer (33). S2 Drosophila cells were transfected as described previously (42) using 1 μg of either methylated or mock-methylated luciferase-reporter plasmid, 100 ng of Sp1 expression plasmid, and 1 μg of a cytomegalovirus–β-galactosidase expression plasmid used for standardization purposes. One microgram of either human MeCP2 expression vector or empty vector control was cotransfected. For the experiments using trichostatin A (TSA), the drug was added 24 h after transfection and analyses were performed 24 h later. Luciferase experiments were performed in duplicate and independently repeated three times.
Reverse transcription (RT)-PCR and Southern blotting.
Total RNA was prepared using Trizol (Gibco-Life Technologies). The RNA samples were reverse transcribed using Superscript II (Gibco-Life Technologies) and random hexamers following the recommendations of the manufacturer. Samples without the addition of Superscript II served as controls. The cyclin A1 PCR was performed for 28 cycles, and the β-actin PCR was performed for 20 cycles. The PCR products were run on a 1.5% agarose gel and blotted on a positively charged nylon membrane. After being cross-linked, the blots were hybridized with internal oligonucleotides for cyclin A1 and β-actin. Both were labeled with digoxigenin that was nonradioactively detected using digoxigenin antibodies coupled to alkaline phosphatase (Boehringer Mannheim). A subsequent chemiluminescence reaction (CDP-Star; Tropix, Bedford, Mass.) was visualized on X-ray film.
Quantitative real-time PCR.
The quantitation of mRNA levels for cyclin A1 and the MBD family members was carried out by a real-time fluorescence detection method (13, 17). The cDNA was prepared as described above and amplified by PCR in the ABI Prism 7700 sequence detector (PE Biosystems, Foster City, Calif.). The initial concentrations of template of murine MBD mRNA were determined by real-time analyses of the amount of PCR product present in the samples at each PCR cycle as measured by SybrGreen incorporation. All primer combinations were positioned to span an intron from two different exons. The primer combinations produced a single product of the appropriate length as visualized by electrophoresis in a 3% agarose gel. When genomic DNA was used as a template, no bands were seen after PCR amplification. Cyclin A1, murine and human glyceraldehyde-3-phosphate-dehydrogenase (GAPDH), human MBD proteins 1 to 4, and MeCP2 were detected by the 5′ nuclease assay (38). In brief, oligonucleotide probes annealed to the PCR products during the annealing and extension steps. The probes were labeled at the 5′ end with VIC (GAPDH probes) or with FAM (all others) and at the 3′ end with TAMRA, which served as a quencher (38). The 5′-to-3′ nuclease activity of the Taq polymerase cleaved the probe and released the fluorescent dyes (VIC or FAM) which were detected by the laser detector of the sequence detector. After the detection threshold was reached, the fluorescence signal was proportional to the amount of PCR product generated. The initial template concentration could be calculated from the cycle number when the amount of PCR product passed a threshold set in the exponential phase of the PCR. All probes were positioned across exon-exon junctions. Primer and probe sequences (Table 1) were either supplied by PE Biosystems (human GAPDH) or designed with Primer Express software (PE Biosystems) using published sequences (18, 42). Relative gene expression levels were calculated using standard curves generated by serial dilutions of U937 cDNA (human samples) or 32D cell cDNA (murine samples). The relative amounts of gene expression were calculated by using the expression of GAPDH as an internal standard. At least three independent analyses were performed for each gene, and data are presented as the mean ± standard error (SE). For analyses of mRNA expression levels of murine MBD proteins, testis cells from transgenic mice were sorted by flow cytometry as described above, and RNA was prepared with Trizol reagent (Gibco). The cDNA from the somatic organs was obtained from Clontech.
Exposure of HeLa cells to TSA and 5-aza-C.
HeLa cells were seeded at low density in 100-mm-diameter plates. Twenty-four hours after being seeded, the cells were exposed to 1 μM 5-aza-cytidine (5-aza-C). After 72 h, TSA (final concentration, 1 μM) was added to the appropriate dishes and incubation was continued for another 24 h. The cells were directly lysed in Trizol to extract RNA.
RESULTS
Methylation of the cyclin A1 promoter correlates with nonexpression in somatic cell lines.
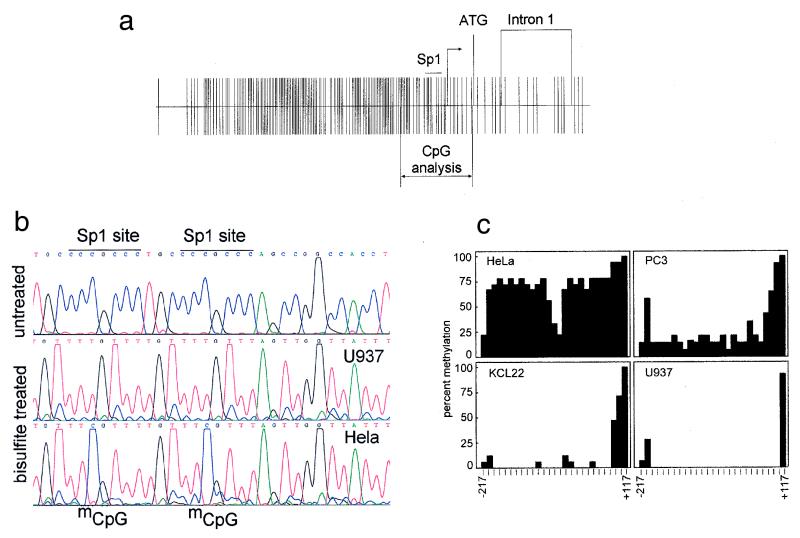
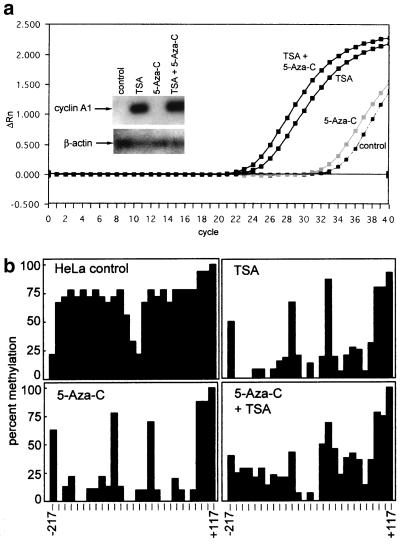
Tissue-specific expression of the cyclin A1 gene is tightly regulated. High expression levels of cyclin A1 in mice and healthy humans are restricted to the testis. To understand the regulation of cyclin A1 expression, we recently cloned and characterized the human cyclin A1 promoter. The 5′ upstream region of the cyclin A1 gene contains a CpG island, and the GC content exceeds 90% in a region 60 bp upstream of the transcriptional start site (Fig. 1a). We analyzed the endogenous human cyclin A1 promoter for CpG methylation at all 27 CpG dinucleotides located between −217 and +117 relative to the transcriptional start site (Fig. 1b and c). Four cell lines with different concentrations of cyclin A1 mRNA were chosen for these analyses. The myeloid leukemia cell lines U937 and KCL22 express levels of cyclin A1 that are detectable by Northern blotting, whereas PC3 and HeLa do not express cyclin A1 (43). Interestingly, we found a close correlation between cyclin A1 nonexpression and a high degree of CpG methylation. No methylation was seen in highly expressing U937 and KCL22 myeloid leukemia cells. Hypomethylation in the leukemic cell lines was clearly restricted to the CpG island, since a CpG dinucleotide at +117, which is outside of the CpG island, was found to be completely methylated in each of the cell lines (Fig. 1c).
FIG. 1.
Methylation of the CpG island in the cyclin A1 promoter correlates with loss of expression in somatic cell lines. (a) Distribution of CpG dinucleotides in the cyclin A1 gene, bp −1299 to +633. The CpG island of the cyclin A1 promoter extends over 1,200 bp and includes four critically important Sp1 binding sites (marked Sp1). The main transcriptional start site and the area analyzed by bisulfite sequencing are also indicated. (b) Bisulfite sequencing was carried out according to the method described by Clark et al. (8) with minor modifications (see Materials and Methods). Shown are the sequencing results for unmodified DNA (U937) and for bisulfite-treated DNA from a cyclin A1-expressing (U937) and a nonexpressing (HeLa) cell line. The methylated CpG dinucleotides (mCpG) are indicated in HeLa cells in the area surrounding the two most proximal Sp1 sites of the cyclin A1 promoter. (c) A total of 27 CpG dinucleotides (14 to 18 independent clones) were analyzed for methylation in each of the four cell lines, which differ in their levels of cyclin A1 expression. The percentage of methylation at each CpG is indicated. The last CpG, at +117, is outside the CpG island and was found to be completely methylated in all of the cell lines. The order of the degree of cyclin A1 expression in these cell lines is U937>KCL22>PC3>HeLa, and cyclin A1 expression in PC3 and HeLa is not detectable by Northern blot analysis (43).
Silencing of a transgenic cyclin A1 promoter is associated with de novo methylation in MG63 osteosarcoma cells.
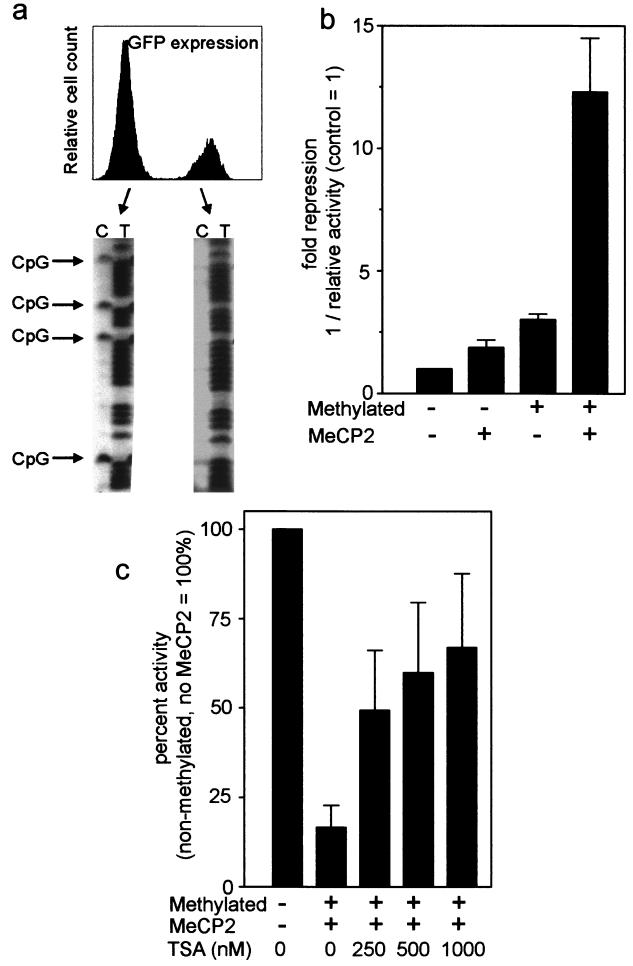
Silencing of transgenes in cell lines involves de novo methylation and histone deacetylation (51). To analyze whether a transgenic cyclin A1 promoter would be silenced in cell lines, we constructed a cyclin A1 promoter-EGFP transgene that was transfected into MG63 osteosarcoma cells. A population of stable transfected cells was established by continuous selection in neomycin-containing medium. These cells were cultured for several months, and the majority of cells lost expression of the transgene over time (Fig. 2a). EGFP-expressing and nonexpressing cells were sorted by flow cytometry, and the DNAs of these cells were subjected to analysis of the methylation status of the transgenic promoter. To avoid amplification and analysis of the endogenous cyclin A1 gene, a different set of primers that included the EGFP gene was used in these experiments (Table 1). Interestingly, no CpG methylation of the transgenic promoter was observed in the EGFP-expressing cells, whereas a high degree of methylation of all analyzed CpG dinucleotides was present in the cell fraction containing the silenced transgene (Fig. 2a). These findings suggest that methylation of the cyclin A1 promoter is associated with gene silencing in cell lines.
FIG. 2.
Methylation is associated with silencing of a cyclin A1 promoter-EGFP transgene in MG63 cells, and MeCP2 represses the methylated cyclin A1 promoter by an HDAC-dependent mechanism. (a) MG63 osteosarcoma cells were stably transfected with a cyclin A1 promoter-EGFP construct. After 2 months of continuous culture, a fraction of neomycin-resistant cells had lost expression of EGFP. Both populations were sorted by flow cytometry and analyzed for CpG methylation of the cyclin A1 promoter transgene. Specific primers were designed for the promoter-EGFP construct to avoid analysis of the endogenous cyclin A1 locus. Strong methylation was observed in nonexpressing cells, whereas negligible or very low levels of methylation were observed in expressing cells. (b) Methylated (+) or mock-methylated (−) reporter constructs were transfected into S2 Drosophila cells along with an Sp1 expression plasmid and either an empty vector (−) or an MeCP2 expression vector (+). Fold repression was calculated as 1/relative activity, with the activity of the control set as 1. The results are shown as the mean + SE of three independent experiments. (c) TSA relieves MeCP2-mediated repression of the methylated cyclin A1. Twenty-four hours after transfection of the methylated construct and the MeCP2 expression vector, TSA was added in different concentrations as indicated. The next day, luciferase activities were analyzed.
MeCP2 can repress transcription from the methylated cyclin A1 promoter in Drosophila cells.
Transcriptional repression of CpG methylation can be mediated by members of the MBD family of proteins (19). The methyl-CpG binding protein 2 (MeCP2) was the first member of this family that was shown to repress transcription by recruiting HDAC activity to methylated promoters (44, 45). For example, the human leukosialin gene is negatively regulated by MeCP2 when the leukosialin promoter is methylated (33). Leukosialin (similar to cyclin A1) is expressed in a tissue-specific manner, and its transcriptional activity depends on the Sp1 transcription factor. S2 Drosophila cells do not express endogenous MeCP2, and transfection of DNA into these cells leads to rapid chromosomal integration. Using these cells, we analyzed whether the methylated cyclin A1 promoter could be suppressed by cotransfected MeCP2 (Fig. 2b). Upon transfection of in vitro-methylated cyclin A1 promoter constructs into Drosophila cells, threefold repression of reporter activity was detected in the absence of MeCP2. When MeCP2 was coexpressed with the methylated cyclin A1 promoter construct, promoter activity was inhibited by 12-fold, indicating that MeCP2 strongly suppressed transcriptional activation of the methylated cyclin A1 promoter (Fig. 2b). The effects of MeCP2 on the methylated cyclin A1 promoter were shown to depend on HDAC activity, since inhibition of histone deacetylase activity restored promoter activity in a dose-dependent manner (Fig. 2c).
MBD gene expression in human cancer cell lines.
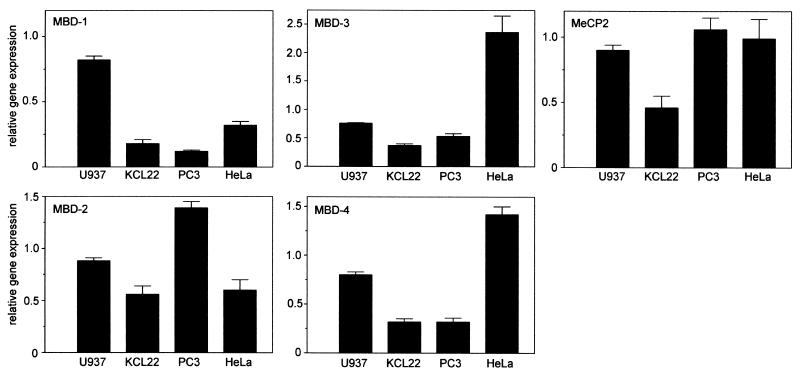
We analyzed whether the known MBD family members were expressed in the human cell lines and would be able to be involved in regulation of the methylated cyclin A1 promoter. Protein expression of MeCP2 in HeLa cells has been reported to be very low (46, 47). We tested protein expression of MeCP2 in HeLa cells using a polyclonal rabbit antibody, and we consistently detected a weak band at 84 kDa (data not shown). Since several other bands were present on the Western blot, we decided to analyze the expression of MeCP2 and MBD-1 to -4 by quantitative real-time PCR (Fig. 3). MeCP2 mRNA was clearly present in all four cell lines, and the differences in the MeCP2 mRNA levels were very small. In addition, we were able to detect expression of MBD-1 to -4 mRNA in the four cell lines. The greatest differences (up to sixfold) in expression were detected for MBD-1 and MBD-3, whereas expression of MBD-2 and MBD-4 was very similar in the different cell lines.
FIG. 3.
mRNA expression levels of MBD protein family members in human cancer cell lines. Expression levels of the indicated MBD family members were determined by quantitative real-time PCR as described in Materials and Methods. The indicated relative gene expression shows expression levels that were standardized using expression of GAPDH as a standard. Expression levels were determined independently at least three times and are shown as the mean + SE.
Inhibitors of histone deacetylation but not inhibitors of methylation alone induce cyclin A1 mRNA in nonexpressing HeLa cells.
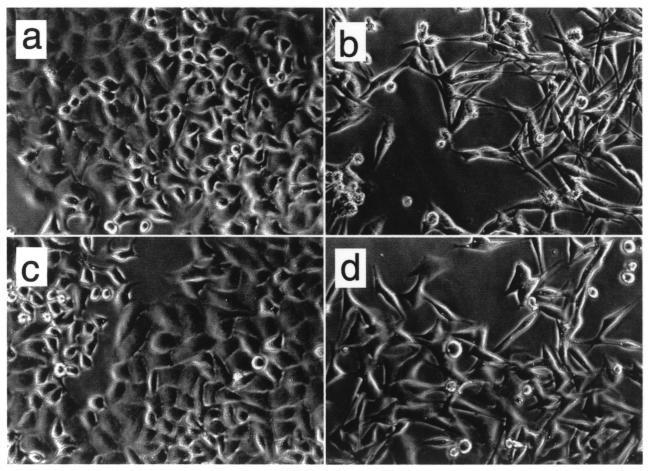
Next, we analyzed whether changes in either the degree of methylation or the acetylation status of histones could induce cyclin A1 expression in nonexpressing cells. HeLa cells were exposed to either the demethylation agent 5-aza-C (1 μM) for 96 h and/or to the inhibitor of histone deacetylases, TSA (1 μM) for 24 h. Whereas 5-aza-C did not have significant effects on the morphology of HeLa cells, TSA led to an obvious change in cell shape, and the cells showed signs of differentiation (Fig. 4). Exposure of HeLa cells to 5-aza-C alone did not lead to significant induction of endogenous cyclin A1 expression (Fig. 5a), although cyclin A1 promoter methylation decreased significantly after HeLa cell exposure to 5-aza-C (Fig. 5b). These results were confirmed using concentrations of 5-aza-C as high as 10 μM (data not shown). Also, 5-aza-deoxy-cytidine, another demethylation reagent, failed to induce cyclin A1 in HeLa cells as well (data not shown). TSA (1 μM) led to a strong induction of endogenous cyclin A1 expression (Fig. 5a). Similar but weaker induction was detected with 250 nM TSA (data not shown). Only slightly stronger induction of cyclin A1 was observed when the cells had been preexposed to 5-aza-C. Interestingly, demethylation also occurred in HeLa cells exposed to either TSA alone or the combination of TSA and 5-aza-C (Fig. 5b). However, demethylation itself was not associated with the increase in cyclin A1 expression, since demethylation by 5-aza-C alone did not lead to induction of cyclin A1 expression.
FIG. 4.
Inhibitors of histone deacetylases alter the phenotypes of HeLa cells. HeLa cells were exposed to 5-aza-C for 96 h and to TSA for 24 h. Exposure of cells to 5-aza-C (c) did not induce morphological changes compared to control cells (a). TSA alone (b) or in combination with 5-aza-C (d) led to cell death and signs of differentiation, such as spindle cell formation.
FIG. 5.
(a) Induction of endogenous cyclin A1 expression by TSA. Induction of cyclin A1 gene expression by 5-aza-C and TSA was analyzed by quantitative real-time PCR as well as by conventional RT-PCR followed by hybridization. The plot for the amplification of cyclin A1 cDNA from HeLa cells that were exposed to the indicated drugs is shown. The x axis shows the cycle number, and the y axis shows the changes in fluorescence compared to baseline values. These samples contained similar amounts of cDNA, as demonstrated by analysis of GAPDH expression (not shown). The inset shows conventional RT-PCR for cyclin A1 (28 cycles) and β-actin (20 cycles). PCR was followed by Southern blotting and nonradioactive detection with an internal oligonucleotide. TSA, but not 5-aza-C, led to a strong induction of cyclin A1 in HeLa cells. (b) 5-aza-C and TSA lead to demethylation of the cyclin A1 promoter in HeLa cells. To analyze the effects of 5-aza-C and TSA on the methylation status of the endogenous cyclin A1 promoter in HeLa cells, genomic DNA was prepared, bisulfite treated, PCR amplified, and sequenced (see Materials and Methods). The percentage of methylation at 27 CpG dinucleotides of the cyclin A1 promoter is indicated. Two independent DNA preparations and PCR amplifications were performed and cloned, and at least 10 independent clones were sequenced.
A 1.3-kb fragment of the cyclin A1 promoter directs tissue-specific expression in testes of transgenic mice.
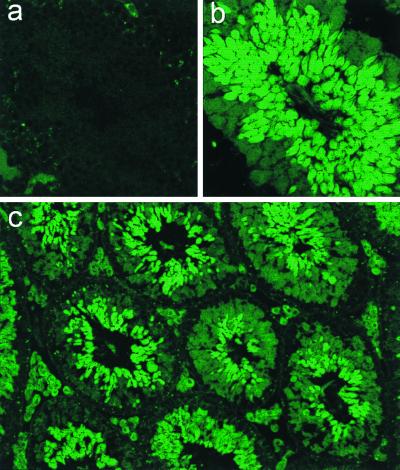
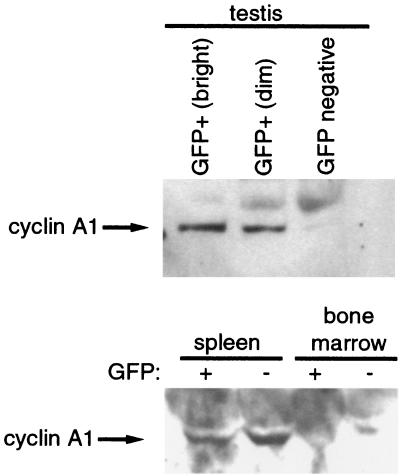
Methylation of the cyclin A1 gene was associated with nonexpression in mammalian cell lines, but these observations did not elucidate whether methylation had a causal role in transcriptional repression and in controlling the tissue specificity of the promoter. This led us to investigate whether the 1.3-kb fragment of the cyclin A1 promoter could direct tissue-specific expression in vivo and whether expression in differentiated tissues in vivo would be influenced by CpG methylation. We established four lines of transgenic mice that carried the cyclin A1 promoter-EGFP reporter construct previously used to generate the stable MG63 cell line. Expression of the transgene was analyzed by fluorescence microscopy and flow cytometry in a wide variety of cell types and tissues. Strong EGFP expression was seen in the testes of all transgenic murine lines. EGFP expression in the testis was confined to male germ cells during spermatogenesis (Fig. 6). Neither Leydig cells nor Sertoli cells expressed significant levels of EGFP. During spermatogenesis, type I spermatogonia at the basal membrane did not express cyclin A1. Intermediate expression was seen in later stages of spermatogonia and in spermatocytes in the early phases of the first meiotic division. A sharp increase in EGFP expression was observed during the first meiotic division, and levels of EGFP subsequently remained very high. No leaky expression was noted in any of the other cell types in the testis, indicating that the 1.3-kb fragment directed very strong and highly specific expression. This pattern of expression in testes was identical in all four transgenic lines. The observed expression pattern matches murine cyclin A1 RNA in situ hybridization results (54). Also, protein expression of the endogenous cyclin A1 gene in the testis was found only in cells expressing EGFP (Fig. 7), verifying the specificity of the cyclin A1 promoter in vivo. Furthermore, the identical transgene expression pattern in testes of different murine lines indicated that the cyclin A1 promoter was able to direct tissue-specific expression in the testis independently of the chromosomal integration site.
FIG. 6.
Testis-specific expression of the cyclin A1 promoter in transgenic mice. Transgenic mouse lines were established using the cyclin A1 promoter-EGFP reporter plasmid described in the legend to Fig. 2. Frozen sections from testes of adult mice were cut, rinsed in phosphate-buffered saline for 10 min, and analyzed by confocal laser scanning microscopy. (a) No fluorescence was observed in testicular tubuli of control mice. (b and c) In contrast, strong and highly specific expression of EGFP was detected in the testes of the transgenic mice. Maximal EGFP expression was observed during and after the first meiotic division, and a weaker staining was present in spermatogonia. The fluorescence of the interstitial Leydig cells in panel c is unrelated to EGFP expression but depends on the accumulation of lipids. The wavelength of the emitted light is different from that of EGFP. Magnifications are ×380 (a and b) and ×95 (c).
FIG. 7.
EGFP expression in testes but not in somatic cells of transgenic mice correlates with endogenous cyclin A1 expression. Testis, spleen, and bone marrow cells from transgenic animals were sorted by flow cytometry into expressing (+) and nonexpressing (−) populations. Testis cells were further divided into highly (bright) and weakly (dim) expressing populations. A total of 2 × 105 cells of each population were lysed in sodium dodecyl sulfate sample buffer, and samples were run on a 10% Tris-HCl gel. The blots were probed with anti-cyclin A1 rabbit polyclonal antibody as described previously (52). The strong background seen for the spleen and bone marrow cells relates to the much longer exposure necessary to visualize the bands. Results similar to those for the spleen and bone marrow cells were obtained for EGFP-expressing kidney cells.
Methylation does not suppress testis-specific expression of the cyclin A1 promoter.
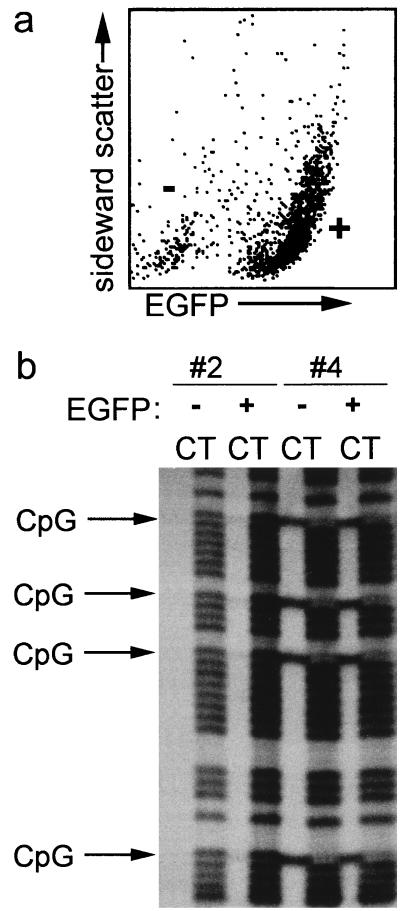
The methylation and activity status of a transgene is thought to be largely determined by either the chromatin structure at the site of integration, the cis-acting sequences in the transgene, and/or the influence of a locus control region (5, 31, 49). Transgene activity has also been reported to be associated with hypomethylation (51). Therefore, we tested whether methylation of the promoter would differ in expressing and nonexpressing cells. Analysis of methylation of the CpG dinucleotides showed that all CpGs were either strongly methylated or nonmethylated in a particular organ. Also, the variation in methylation patterns between different organs was very small (Table 2). In addition, methylation patterns were stable in adult mice from the same line (data not shown). Analysis of the methylation status of the human cyclin A1 promoter in the testes of the transgenic mice showed that the promoter and the transgene were not methylated in the testes of two lines (no. 2 and 3) (Table 2). However, the promoter and the transgene were heavily methylated in the testes of the two other lines (no. 1 and 4). No difference could be found between the EGFP expression patterns in the testes of the murine lines with and without CpG methylation. To confirm that EGFP was highly expressed despite methylation in these male germ cells, testis cells were disaggregated and sorted by flow cytometry (Fig. 8a). Bisulfite sequencing confirmed that methylation of the cyclin A1 promoter in germ cells did not inhibit expression of the transgene in the testis (Fig. 8b). The degree of methylation was similar in EGFP-expressing and non-EGFP-expressing cells of the same transgenic line. Accordingly, nonexpression of EGFP appropriately occurred in the absence of methylation. More surprisingly, strong methylation of all analyzed CpG dinucleotides (total, 27) surrounding the transcriptional start site neither suppressed nor altered transcriptional activity of the transgenic cyclin A1 promoter (Fig. 8b).
TABLE 2.
CpG methylation of the human cyclin A1 promoter and expression of EGFP in transgenic murine linesa
| Line | Testis
|
Kidney
|
Liver
|
Bone marrow
|
Spleen
|
Muscle
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mCpG | EGFP (%) | mCpG | EGFP (%) | mCpG | EGFP (%) | mCpG | EGFP (%) | mCpG | EGFP (%) | mCpG | EGFP (%) | |
| 1 | + | 70 | + | 0 | + | 0 | + | 0 | ND | 0 | ND | 0 |
| 2 | − | 70 | − | 25 | − | 0 | − | 10 | − | 16 | − | 0 |
| 3 | − | 70 | + | 0 | − | 0 | + | 0 | ND | 0 | ND | 0 |
| 4 | + | 70 | + | 0 | + | 0 | + | 0 | + | 0 | + | 0 |
Organs from adult mice of the four transgenic murine lines (no. 1 to 4) were obtained, and the methylation pattern of the transgenic cyclin A1 promoter was determined by direct sequencing of PCR products derived from bisulfite-treated genomic DNA. A total of 23 CpG dinucleotides (−217 to +29) were analyzed. The CpG dinucleotides within one organ from one line were found to be either completely methylated (+) or nonmethylated (−). Methylation patterns were consistent among mice from the same transgenic line. The percentages indicate the fraction of EGFP-expressing cells in the respective organs of the murine lines. Expression outside the testis was seen only when the promoter of the transgene was not methylated. ND, not determined.
FIG. 8.
Expression of EGFP in germ cells of transgenic mice is independent of methylation of the transgenic cyclin A1 promoter. (a) To analyze whether the CpG methylation status differed in EGFP-expressing and non-EGFP-expressing cells in the testis, the testes of two murine lines (no. 2 and 4) were enzymatically disaggregated and sorted by flow cytometry in expressing (+) and nonexpressing (−) cells. Shown is a representative flow cytometry analysis of testicular cells from murine line no. 2. (b) Genomic DNA of the sorted testicular cells was analyzed for CpG methylation. In murine line no. 2, CpG methylation was not seen in EGFP-expressing (+) or non-EGFP-expressing testicular cells (−). In murine line no. 4, EGFP-expressing cells were completely methylated at all CpG dinucleotides. The methylation status did not differ in EGFP-expressing (+) and non-EGFP-expressing (−) testicular cells.
Methylation is not necessary to suppress transcription from the cyclin A1 promoter in nonexpressing cells from most organs.
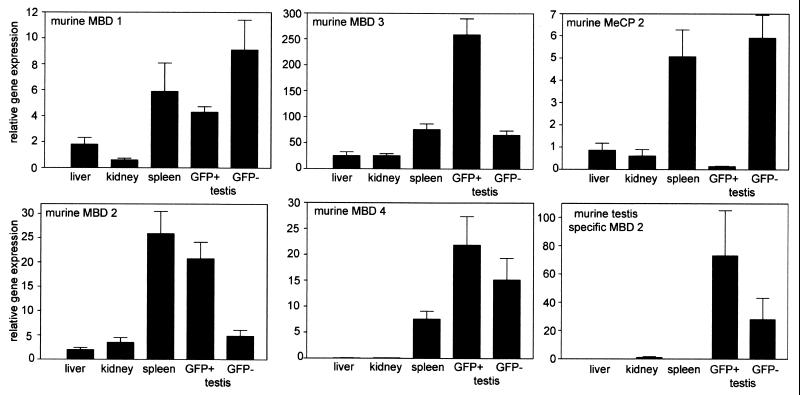
One of the murine transgenic lines without methylation in the testis (no. 3) showed promoter methylation in the kidney and bone marrow but not in the liver. This line showed EGFP expression exclusively in the testis (Table 2). The transgenic murine line no. 2 did not show a significant degree of methylation of the transgenic cyclin A1 promoter anywhere and expressed EGFP in a subset of cells in the kidney (25%), spleen (10%), and bone marrow (16%). Expression of EGFP in these organs was clearly restricted to certain cell types, such as the podocytes and collecting ducts in the kidneys and B-cell subpopulations in the spleen and bone marrow (Fig. 9). A minor percentage of myeloid progenitors expressed EGFP as well. Expression of EGFP in these organs did not reflect expression of the endogenous murine cyclin A1 gene, as evidenced by Western blotting (Fig. 7) and by RT-PCR of sorted cell populations (data not shown). The correct sorting into EGFP-expressing and non-EGFP-expressing cells was confirmed by Western blot analysis with anti-EGFP antibody (not shown). Although a fraction of the cells in the kidneys, spleen, and bone marrow expressed EGFP, the majority of cells did not. In addition, no EGFP expression was found in many other organs (e.g., skin, muscle, lung, and gut), indicating that methylation was not necessary to suppress cyclin A1 promoter activity, at least in most cells. On the other hand, expression outside the testis was restricted to the transgenic mouse line, which did not have methylation of the CpG island of the cyclin A1 promoter. One of the reasons that methylation of the cyclin A1 promoter did not repress promoter activity in the testes could be that relevant MBD family members are not expressed in germ cells. To test this hypothesis, we analyzed expression levels of MeCP2 and MBD family members in EGFP-expressing male germ cells as well as in somatic organs from mice (Fig. 10). While mRNA from MBD-1 to -4 and the testis-specific splice variant of MBD-2 were present in EGFP-expressing testis cells, MeCP2 expression was very low in male germ cells. The testis-specific variant of MBD-2 showed higher expression in EGFP+ testis cells than in EGFP− testis cells.
FIG. 9.
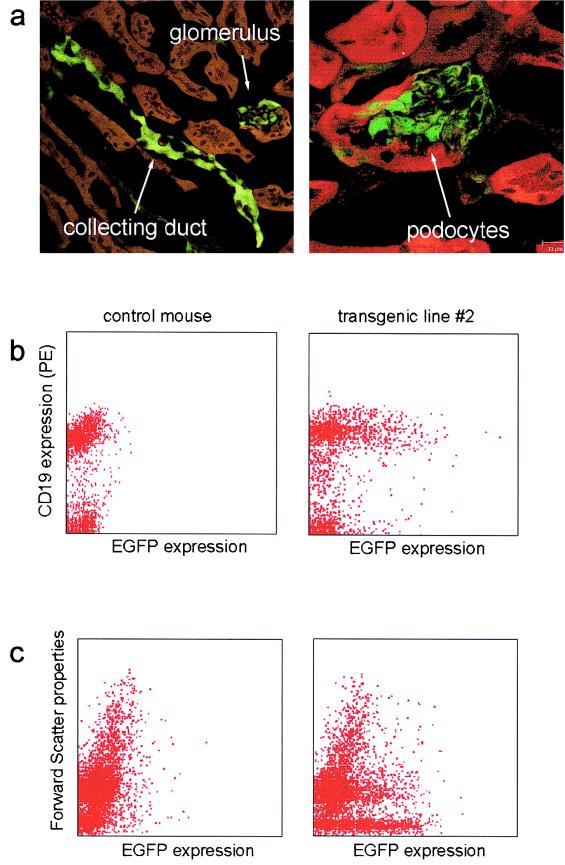
Aberrant expression of EGFP was found in kidneys and B lymphocytes from transgenic murine line no. 2. (a) Expression of EGFP in kidneys of murine line no. 2 is tightly restricted to podocytes in glomeruli and to collecting ducts. Nonfluorescent structures detected by phase-contrast microscopy were colored red to allow a better visualization of the overall structure of the kidney. (b) B lymphocytes from the spleen of a transgenic mouse (line no. 2) and from a negative-control mouse were stained using anti-CD19 monoclonal antibody labeled with phycoerythrin (PE). About 40% of the B cells of murine line no. 2 expressed EGFP. Note that the population that does not express CD19 also does not express EGFP. (c) Bone marrow from a control mouse and a mouse of line no. 2 were analyzed by flow cytometry without antibody staining. The forward scatter properties distinguish cell populations according to their sizes. The very small percentage of hematopoietic stem cells (<1%) precludes us from determining whether EGFP expression in these cells reflects the endogenous cyclin A1.
FIG. 10.
Expression levels of MBD family members in murine tissues. The mRNA levels of murine MBD proteins were determined by quantitative real-time RT-PCR as described in Material and Methods. EGFP-expressing (GFP+) and non-EGFP-expressing (GFP−) testicular cells from transgenic mice were sorted by flow cytometry before the preparation of RNA. MeCP2 expression was very low in EGFP-expressing male germ cells. On the other hand, MeCP2 was robustly expressed in EGFP-negative testis cells from the same mice. Expression levels of MBD-1 to MBD-4 and the testis-specific variant of MBD-2 showed only minor differences in expression between the two testicular cell populations. The error bars represent the SE.
DISCUSSION
Cyclin A1 is essential for spermatogenesis and is expressed at very high levels in pachytene spermatocytes (37). We have previously shown that human cyclin A1 protein directly interacts with E2F-1 and the Rb family of proteins, indicating its potential role in the cell cycle of somatic cells (65). In contrast to other cyclins that regulate the cell cycle during S and G2/M phases, cyclin A1 displays a highly restricted pattern of expression (54, 64). To analyze the mechanisms that determine cyclin A1 expression, we have recently cloned the promoter region of the human cyclin A1 gene (42). Upon transient transfection, the cyclin A1 promoter was active in somatic-cell lines from various tissues, even in those that do not normally express cyclin A1 (including HeLa and PC3 cells) (43).
Cyclin A1 promoter activity critically depends on four Sp1 binding sites that are located between 60 and 120 bp upstream of the main transcriptional start site (42). Transcriptional activity is enhanced by c-myb, which is expressed in hematopoietic progenitors and myeloid leukemia cells (43). The Sp1 transcription factor that is essential for cyclin A1 promoter activity is ubiquitously expressed, and the cyclin A1 promoter does not show tissue specificity in transient transfections. These findings hinted at a role of epigenetic regulation in the tissue-specific expression of cyclin A1.
Methylation of CpG dinucleotides has been shown to lead to transcriptional repression by several mechanisms. Binding of transcription factors can be inhibited by CpG methylation; for example, c-myb did not bind to its consensus binding site when it was methylated (32). On the other hand, Sp1 binding is not inhibited by CpG methylation (16, 23). Members of the MBD family of proteins bind to methylated CpGs in genomic DNA and confer the biological effects of methylation (19, 39). MeCP2 as well as MBD-2, which is a member of the MeCP1 complex, can repress transcription by recruiting corepressors and histone deacetylase activity (28, 45, 47). The MeCP1 complex, identified several years ago, mediates repression of heavily methylated DNA (4, 40). MeCP2 is essential for embryonic development and can bind to single methylated CpG dinucleotides (44, 55). Among the other members of the MBD family of proteins, MBD-4 is involved in DNA repair mechanisms, whereas the functions of the other members are currently unknown (20).
To test the hypothesis that MBD family proteins are involved in transcriptional silencing of the methylated cyclin A1 promoter in tumor cell lines, we examined the effects of MeCP2 on the transcriptional activity of a methylated cyclin A1 promoter construct. Our data demonstrate that the methylated cyclin A1 gene can be repressed by a MeCP2-mediated mechanism. However, MeCP2 has been reported to be expressed at very low levels in HeLa cells that do not express cyclin A1 (47). Our own Western blotting experiments confirmed the low levels of MeCP2 protein expression in HeLa cells (data not shown). Using quantitative real-time PCR with the 5′ nuclease assay, we demonstrated that MeCP2, as well as the other MBD family members, was expressed at the mRNA level in different human cell lines. No significant differences in MBD mRNA expression levels were found between adherent (HeLa and PC3) and leukemic (U937 and KCL22) cell lines. These findings indicate that in the absence of MeCP2, the other MBD family members, especially MBD-2, might be involved in transcriptional repression of genes with methylated promoters, including cyclin A1.
Inhibition of histone deacetylase activity restored cyclin A1 promoter activity in the insect cell transfection assay and induced cyclin A1 gene expression from the endogenous promoter in otherwise nonexpressing HeLa cells. Demethylation of the promoter in HeLa cells had only minor effects on cyclin A1 expression. These findings indicate that stable repression of the cyclin A1 promoter is mediated by histone deacetylation-dependent mechanisms. These data differ from those obtained for several other methylated promoters where demethylation of promoter regions by 5-aza-C was the dominant mechanism that led to expression of silenced genes (7). MBD family members that recruit HDAC activity to methylated promoters are likely to play a role in transcriptional repression of the cyclin A1 gene in solid-tumor cell lines.
The close correlation between methylation and silencing of the cyclin A1 promoter suggests that hypomethylation of the promoter is important for transcriptional activity of the cyclin A1 promoter in the leukemia cell lines. Despite the presence of aberrant methyltransferase activity (25, 30), tumor cells show hypomethylation of the promoter regions of genes that promote growth and inhibit apoptosis (12, 15). Transcriptional activity of a promoter can preserve its hypomethylated status through binding sites for specific transcription factors, such as Sp1 (5). In addition, cyclin A1 induction by TSA in HeLa cells was correlated with a loss of cyclin A1 promoter methylation.
To analyze whether methylation plays a role in directing tissue-specific cyclin A1 expression in vivo, we generated mice that were transgenic for the cyclin A1 promoter driving an EGFP reporter gene. We chose to analyze the association between the status of the transgene methylation and the expression of the reporter gene for several reasons. Since microinjection of foreign DNA leads to random integration, the methylation status of the transgene varies in different transgenic lines. This provided the opportunity to study the effects of methylation versus nonmethylation of the cyclin A1 promoter on gene expression rather than the pure correlation between expression and methylation of the gene. Correlative data between expression and methylation of endogenous genes rarely provide hard evidence either in favor of or against a role for methylation in directing tissue-specific expression (61). A pure correlation is also unable to prove whether methylation is a primary or a secondary event in silencing the expression of a gene.
Using transgenic mice as a model system, we demonstrated that transcriptional repression of cyclin A1 promoter activity could be established in almost all organs without CpG methylation of the transgenic promoter. In addition, the presumed suppressive effect of CpG methylation is specific to the cell type, since the correct patterns of expression in the testis were detected in two murine lines despite high levels of CpG methylation of the transgenic cyclin A1 promoter. Expression patterns in male germ cells did not differ between transgenic lines with and without CpG methylation of the transgenic promoter. To our knowledge, this is the first time that a transgenic promoter in a whole organism has been shown to be active despite strong methylation.
The nonsuppressive effect of methylation in male germ cells might be explained by the presence of specific activating factors that could override the suppressive activity of methylation. Alternatively, corepressors that are necessary for methylation-mediated repression could be either inactive or missing in male germ cells. The overall degree of DNA methylation increases during spermatogenesis and is very high in mature sperm compared to other cells (29, 41). The absence of repressing factors could be associated with the generally less tightly regulated repression of gene expression that occurs during meiosis (10, 26). In addition, MeCP2 activity is barely detectable in adult testes (40). We used quantitative real-time PCR to demonstrate the absence of MeCP2 mRNA expression in EGFP-expressing male germ cells. Interestingly, MeCP2 mRNA was highly expressed in murine testis cells without detectable cyclin A1 promoter activity. This close correlation suggests that the absence of MeCP2 expression is a possible explanation for the nonsuppressive effect of methylation on cyclin A1 promoter activity in male germ cells. Expression of all other MBD family members, including the testis-specific variant of MBD-2, could be demonstrated in EGFP-positive and -negative cells. Taken together, our finding that methylation did not suppress cyclin A1 promoter activity in male germ cells suggests that the suppressive role of methylation for some genes might be cell type specific.
In somatic cells, transcriptional repression of the transgenic promoter appropriately occurred irrespective of the methylation pattern in most of the tissues, showing that methylation is not necessary for transcriptional repression in most differentiated tissues. Since the cyclin A1 promoter was active in transient transfections in each of the cell types examined, including primary cells, the repression of the transgenic cyclin A1 promoter is most likely based on a repressor mechanism that alters the chromatin structure. A specific repressor protein might bind to an unidentified binding site and recruit the relevant corepressors and histone deacetylases. The nature of this presumed repressor is unknown. One repressor protein, c-mos, has been described that directs tissue-specific repression outside of the testis by strong suppression of germ cell-specific promoters in somatic cells (63). However, the c-mos repressor is highly repressive even in transient transfections; thus, this mechanism is unlikely to be relevant for the cyclin A1 promoter.
Aberrant expression of the transgene was seen in kidney podocytes and collecting ducts and B lymphocytes of murine line no. 2, the only transgenic line without CpG methylation in any of the analyzed organs. In contrast to expression in the testis, EGFP expression in these cells did not match expression of the endogenous murine cyclin A1 gene (Fig. 7). The very low levels of expression of cyclin A1 in B cells were confirmed by RT-PCR analysis of several B-lymphocyte subpopulations (data not shown). The expression pattern of the murine line no. 2 might be related to the absence of CpG methylation of the transgenic promoter and/or to the influence of a locus control region that is located close to the chromosomal integration site of the transgene (31, 49). Methylation might have a role in suppression of cyclin A1 promoter activity in these cell populations. On the other hand, methylation is neither necessary nor sufficient to direct tissue-specific gene expression in all other organs that we analyzed (Table 3).
TABLE 3.
Cyclin A1 promoter in relation to chromosomal integration and methylation of the CpG island in somatic cells and in male germ cellsa
| Parameter | Somatic cell lines
|
Murine lines 1, 3, and 4 (methylated)
|
Murine line 2 (nonmethylated)
|
|||||
|---|---|---|---|---|---|---|---|---|
| Myeloid leukemia | Adherent carcinoma | Somatic organs | Male germ cells | Somatic organs | Male germ cells | |||
| Promoter analyzed | Endogenous | Endogenous | Transient transfection | Transgene (stable transfection) | Transgene | Transgene | Transgene | Transgene |
| CpG island | Nonmethylated | Methylated | Nonmethylated | Methylated | Methylated | Methylated | Nonmethylated | Nonmethylated |
| Chromosomal integration | Yes | Yes | No | Yes | Yes | Yes | Yes | Yes |
| Promoter status | Active | Repressed | Active | Repressed | Repressed | Active | Repressed in most organs | Active |
Shown is a summary of the relationship between chromosomal integration, methylation status of the CpG island, and cyclin A1 promoter activity in the different systems analyzed. Chromosomal integration is necessary for cyclin A1 promoter silencing in somatic cells. Silencing can occur independently of CpG methylation. CpG methylation is associated with silencing in somatic cell lines but does not inhibit cyclin A1 promoter activity in male germ cells.
Taken together, these data show that methylation of the cyclin A1 promoter correlates with nonexpression of the endogenous cyclin A1 gene in somatic cell lines. Members of the MBD family of proteins, such as MeCP2, might be involved in transcriptional repression of the methylated cyclin A1 promoter. Tissue-specific repression of the cyclin A1 promoter in differentiated organs can occur independently of CpG methylation. Finally, even strong methylation does not necessarily lead to transcriptional repression, since cyclin A1 promoter activity in male germ cells cannot be repressed by a high degree of methylation.
ACKNOWLEDGMENTS
We are grateful to S. Kudo, Hokkaido Institute, Sapporo, Japan, for the MeCP2 expression vector and to A. Bird, University of Edinburgh, Edinburgh, United Kingdom, for antibodies against MeCP2. We thank Ian Williamson, Danlin Chen, Stephan Rust, Jochen Kremerskothen, Omeni Osian, Xin Min Yan, and Christina Bornemann for technical assistance and Kamlesh Asotra, Scott Fraser, and Mary Dickinson for their assistance with confocal laser scanning microscopy.
This work is supported by grants from NIH and by U.S. Defense Grants as well as grants from the Parker Hughes and C. and H. Koeffler Funds and the Gladys Lichtenstein Trust. C. Müller's work is supported by grants from the Deutsche Forschungsgemeinschaft (Mu 1328/2-1) and the Deutsche Krebshilfe (10-1539-Mü1). G. Idos was supported by the Howard Hughes Institute Undergraduate Program. H. P. Koeffler holds the Mark Goodson Chair in Oncology Research and is a member of the Jonsson Cancer Center. C. Readhead's work is supported by NIH grant 1RO1RR12406.
REFERENCES
- 1.Antequera F, Boyes J, Bird A. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell. 1990;62:503–514. doi: 10.1016/0092-8674(90)90015-7. [DOI] [PubMed] [Google Scholar]
- 2.Ayer D E, Laherty C D, Lawrence Q A, Armstrong A P, Eisenman R N. Mad proteins contain a dominant transcription repression domain. Mol Cell Biol. 1996;16:5772–5781. doi: 10.1128/mcb.16.10.5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bender C M, Pao M M, Jones P A. Inhibition of DNA methylation by 5-aza-2′-deoxycytidine suppresses the growth of human tumor cell lines. Cancer Res. 1998;58:95–101. [PubMed] [Google Scholar]
- 4.Boyes J, Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991;64:1123–1134. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- 5.Brandels M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H. Sp1 elements protect a CpG island from de novo methylation. Nature. 1994;371:435–438. doi: 10.1038/371435a0. [DOI] [PubMed] [Google Scholar]
- 6.Burley S K, Roeder R G. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 7.Cameron E E, Bachman K E, Myohanen S, Herman J G, Baylin S B. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 8.Clark S J, Harrison J, Paul C L, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Smet C, Lurquin C, Lethe B, Martelange V, Boon T. DNA methylation is the primary silencing mechanism for a set of germ line- and tumor-specific genes with a CpG-rich promoter. Mol Cell Biol. 1999;19:7327–7335. doi: 10.1128/mcb.19.11.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eddy E M, O'Brien D A. Gene expression in mammalian meiosis. Curr Top Dev Biol. 1998;37:141–200. [PubMed] [Google Scholar]
- 11.Enver T, Zhang J W, Papayannopoulou T, Stamatoyannopoulos G. DNA methylation: a secondary event in globin gene switching? Genes Dev. 1988;2:698–706. doi: 10.1101/gad.2.6.698. [DOI] [PubMed] [Google Scholar]
- 12.Feinberg A P, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 13.Gibson U E, Heid C A, Williams P M. A novel method for real time quantitative RT-PCR. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- 14.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 15.Hanada M, Delia D, Aiello A, Stadtmauer E, Reed J C. bcl-2 gene hypomethylation and high-level expression in B-cell chronic lymphocytic leukemia. Blood. 1993;82:1820–1828. [PubMed] [Google Scholar]
- 16.Harrington M A, Jones P A, Imagawa M, Karin M. Cytosine methylation does not affect binding of transcription factor Sp1. Proc Natl Acad Sci USA. 1988;85:2066–2070. doi: 10.1073/pnas.85.7.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heid C A, Stevens J, Livak K J, Williams P M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 18.Hendrich B, Abbott C, McQueen H, Chambers D, Cross S, Bird A. Genomic structure and chromosomal mapping of the murine and human Mbd1, Mbd2, Mbd3, and Mbd4 genes. Mamm Genome. 1999;10:906–912. doi: 10.1007/s003359901112. [DOI] [PubMed] [Google Scholar]
- 19.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hendrich B, Hardeland U, Ng H H, Jiricny J, Bird A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999;40:301–304. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- 21.Hjelle B L, Phillips III J A, Seeburg P H. Relative levels of methylation in human growth hormone and chorionic somatomammatropin genes in expressing and non-expressing tissues. Nucleic Acids Res. 1982;10:3459–3474. doi: 10.1093/nar/10.11.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hogan B, Beddington R, Constantini F, Lacy E. Manipulating the mouse embryo. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 23.Holler M, Westin G, Jiricny J, Schaffner W. Sp1 transcription factor binds DNA and activates transcription even when the binding site is CpG methylated. Genes Dev. 1988;2:1127–1135. doi: 10.1101/gad.2.9.1127. [DOI] [PubMed] [Google Scholar]
- 24.Issa J P, Ottaviano Y L, Celano P, Hamilton S R, Davidson N E, Baylin S B. Methylation of the oestrogen receptor CpG island links aging and neoplasia in human colon. Nat Genet. 1994;7:536–540. doi: 10.1038/ng0894-536. [DOI] [PubMed] [Google Scholar]
- 25.Issa J P, Vertino P M, Wu J, Sazawal S, Celano P, Nelkin B D, Hamilton S R, Baylin S B. Increased cytosine DNA-methyltransferase activity during colon cancer progression. J Natl Cancer Inst. 1993;85:1235–1240. doi: 10.1093/jnci/85.15.1235. [DOI] [PubMed] [Google Scholar]
- 26.Ivell R. ‘All that glisters is not gold’—common testis gene transcripts are not always what they seem. Int J Androl. 1992;15:85–92. doi: 10.1111/j.1365-2605.1992.tb01117.x. [DOI] [PubMed] [Google Scholar]
- 27.Jones P A. DNA methylation errors and cancer. Cancer Res. 1996;56:2463–2467. [PubMed] [Google Scholar]
- 28.Jones P L, Veenstra G J C, Wade P A, Vermaak D, Kass S U, Landsberger N, Strouboulis J, Wolffe A P. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 29.Kafri T, Ariel M, Brandeis M, Shemer R, Urven L, McCarrey J, Cedar H, Razin A. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes Dev. 1992;6:705–714. doi: 10.1101/gad.6.5.705. [DOI] [PubMed] [Google Scholar]
- 30.Kautiainen T L, Jones P A. DNA methyltransferase levels in tumorigenic and nontumorigenic cells in culture. J Biol Chem. 1986;261:1594–1598. [PubMed] [Google Scholar]
- 31.Kioussis D, Festenstein R. Locus control regions: overcoming heterochromatin-induced gene inactivation in mammals. Curr Opin Genet Dev. 1997;7:614–619. doi: 10.1016/s0959-437x(97)80008-1. [DOI] [PubMed] [Google Scholar]
- 32.Klempnauer K H. Methylation-sensitive DNA binding by v-myb and c-myb proteins. Oncogene. 1993;8:111–115. [PubMed] [Google Scholar]
- 33.Kudo S. Methyl-CpG binding protein MeCP2 represses Sp1-activated transcription of the human leukosialin gene when the promoter is methylated. Mol Cell Biol. 1998;18:5492–5499. doi: 10.1128/mcb.18.9.5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kudo S, Fukusato T. Tissue-specific transcriptional regulation of human leukosialin (CD43) gene is achieved by DNA methylation. J Biol Chem. 1995;270:13298–13302. doi: 10.1074/jbc.270.22.13298. [DOI] [PubMed] [Google Scholar]
- 35.Lee D Y, Hayes J J, Pruss D, Wolffe A P. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72:73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- 36.Li E, Beetor T H, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 37.Liu D, Matzuk M M, Sung W K, Guo Q, Wang P, Wolgemuth D J. Cyclin A1 is required for meiosis in the male mouse. Nat Genet. 1998;20:377–380. doi: 10.1038/3855. [DOI] [PubMed] [Google Scholar]
- 38.Livak K J, Flood S J, Marmaro J, Giusti W, Deetz K. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 1995;4:357–362. doi: 10.1101/gr.4.6.357. [DOI] [PubMed] [Google Scholar]
- 39.Meehan R R, Lewis J D, Bird A P. Characterization of MeCP2, a vertebrate DNA binding protein with affinity for methylated DNA. Nucleic Acids Res. 1992;20:5085–5092. doi: 10.1093/nar/20.19.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meehan R R, Lewis J D, McKay S, Kleiner E L, Bird A P. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989;58:499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- 41.Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987;99:371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- 42.Müller C, Yang R, Beck-von-Peccoz L, Idos G, Verbeek W, Koeffler H P. GC boxes are required for cell cycle regulated activation of the cyclin A1 promoter. Cloning of the gene and characterization of the promoter region. J Biol Chem. 1999;274:11220–11228. doi: 10.1074/jbc.274.16.11220. [DOI] [PubMed] [Google Scholar]
- 43.Müller C, Yang R, Idos G, Diederichs S, Verbeek W, Bender T, Koeffler H P. c-myb transactivates the human cyclin A1 promoter and induces cyclin A1 gene expression. Blood. 1999;94:4255–4262. [PubMed] [Google Scholar]
- 44.Nan X, Campoy F J, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 45.Nan X, Ng H H, Johnson C A, Laherty C D, Turner B M, Eisenman R N, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 46.Ng H H, Bird A. DNA methylation and chromatin modification. Curr Opin Genet Dev. 1999;9:158–163. doi: 10.1016/s0959-437x(99)80024-0. [DOI] [PubMed] [Google Scholar]
- 47.Ng H H, Zhang Y, Hendrich B, Johnson A A, Turner B M, Erdjument-Bromage H, Tempst P, Reinberg D, Bird A. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- 48.Nightingale K P, Wellinger R E, Sogo J M, Becker P B. Histone acetylation facilitates RNA polymerase II transcription of the Drosophila hsp26 gene in chromatin. EMBO J. 1998;17:2865–2876. doi: 10.1093/emboj/17.10.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ortiz B D, Cado D, Chen V, Diaz P W, Winoto A. Adjacent DNA elements dominantly restrict the ubiquitous activity of a novel chromatin-opening region to specific tissues. EMBO J. 1997;16:5037–5045. doi: 10.1093/emboj/16.16.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pazin M J, Kadonaga J T. What's up and down with histone deacetylation and transcription? Cell. 1997;89:315–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 51.Pikkaart M J, Recillas-Targa F, Felsenfeld G. Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev. 1998;12:2852–2862. doi: 10.1101/gad.12.18.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Razin A. CpG methylation, chromatin structure and gene silencing—a three way connection. EMBO J. 1998;17:4905–4908. doi: 10.1093/emboj/17.17.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siegfried Z, Eden S, Mendelsohn M, Feng X, Tsuberi B Z, Cedar H. DNA methylation represses transcription in vivo. Nat Genet. 1999;22:203–206. doi: 10.1038/9727. [DOI] [PubMed] [Google Scholar]
- 54.Sweeney C, Murphy M, Kubeika M, Ravnik S E, Hawkins C F, Wolgemuth D J, Carrington M. A distinct cyclin A is expressed in germ cells in the mouse. Development. 1996;122:53–64. doi: 10.1242/dev.122.1.53. [DOI] [PubMed] [Google Scholar]
- 55.Tate P, Skarnes W, Bird A. The methyl-CpG binding protein MeCP2 is essential for embryonic development in the mouse. Nat Genet. 1996;12:205–208. doi: 10.1038/ng0296-205. [DOI] [PubMed] [Google Scholar]
- 56.Tse C, Sera T, Wolffe A P, Hansen J C. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol Cell Biol. 1998;18:4629–4638. doi: 10.1128/mcb.18.8.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ura K, Kurumizaka H, Dimitrov S, Almouzni G, Wolffe A P. Histone acetylation: influence on transcription, nucleosome mobility and positioning and linker histone-dependent transcriptional repression. EMBO J. 1997;16:2096–2107. doi: 10.1093/emboj/16.8.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van der Ploeg L H T, Flavell R A. DNA methylation in the human γδβ-globin locus in erythroid and nonerythroid tissues. Cell. 1980;19:947–958. doi: 10.1016/0092-8674(80)90086-0. [DOI] [PubMed] [Google Scholar]
- 59.Vettese-Dadey M, Grant P A, Hebbes T R, Crane-Robinson C, Allis C D, Workman J L. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J. 1996;15:2508–2528. [PMC free article] [PubMed] [Google Scholar]
- 60.Walsh C P, Bestor T H. Cytosine methylation and mammalian development. Genes Dev. 1999;13:26–34. doi: 10.1101/gad.13.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wamecke P M, Clark S J. DNA methylation profile of the mouse skeletal alpha-actin promoter during development and differentiation. Mol Cell Biol. 1999;19:164–172. doi: 10.1128/mcb.19.1.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Watts G S, Pleper R O, Costello J F, Peng Y M, Dalton W S, Futscher B W. Methylation of discrete regions of the O6-methylguanine DNA methyltransferase (MGMT) CpG island is associated with heterochromatinization of the MGMT transcription start site and silencing of the gene. Mol Cell Biol. 1997;17:5612–5619. doi: 10.1128/mcb.17.9.5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu W, Cooper G M. Identification of a candidate c-mos repressor that restricts transcription of germ cell-specific genes. Mol Cell Biol. 1995;15:5369–5375. doi: 10.1128/mcb.15.10.5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang R, Morosetti R, Koeffler H P. Characterization of a second human cyclin A that is highly expressed in testis and in several leukemia cell lines. Cancer Res. 1997;57:913–920. [PubMed] [Google Scholar]
- 65.Yang R, Müller C, Huynh V, Fung Y K, Yee A S, Koeffler H P. Functions of cyclin A1 in the cell cycle and its interactions with transcription factor E2F-1 and the Rb family of proteins. Mol Cell Biol. 1999;19:2400–2407. doi: 10.1128/mcb.19.3.2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang R, Nakamaki T, Lubbert M, Said J, Sakashita A, Freyaldenhoven B S, Spira S, Huynh V, Müller C, Koeffler H P. Cyclin A1 expression in leukemia and normal hematopoietic cells. Blood. 1999;93:2067–2074. [PubMed] [Google Scholar]
- 67.Yelvin A, Razin A. Pages 523–568. In: Jost J P, Saluz H P, editors. DNA methylation: molecular biology and biological significance. Basel, Switzerland: Birkhauser; 1993. [Google Scholar]