Abstract
The increase of HIV infection in macrophages results in HIV proteins being released, like HIV Tat which impairs the function of monoamine transporters. HIV-infected patients have displayed increased synaptic levels of dopamine (DA) due to reduced binding and function of monoamine transporters such as norepinephrine transporter (NET) and dopamine transporter (DAT). Development of a three-dimensional model of the HIV-1 Tat-human NET (hNET) binding complex would help reveal how HIV-1 Tat causes toxicity in the neuron by affecting DA uptake. Here we use computational techniques such as molecular modeling to study microscopic properties and molecular dynamics of the HIV-1 Tat-hNET binding. These modeling techniques allow us to analyze non-covalent interactions and observe residue-residue contacts to verify a model structure. The modeling results studied here show that HIV-1 Tat-hNET binding is highly dynamic and that HIV-1 Tat preferentially binds to hNET in its outward-open state. In particular, HIV-1 Tat forms hydrogen bond interactions with side chains of hNET residues Y84, K88, and T544. The favorable hydrogen bonding interactions of HIV-1 Tat with the hNET side chain residues Y84 and T544 have been validated by our subsequently performed DA uptake activity assays and site-directed mutagenesis, suggesting that the modeled HIV-1 Tat-hNET binding mode is reasonable. These mechanistic and structural insights gained through homology models discussed in this study are expected to encourage the pursuit of pharmacological and biochemical studies on HIV-1 Tat interacting with hNET mechanisms and detailed structures.
Keywords: Molecular docking, molecular dynamics, protein-protein interaction, norepinephrine transporter, trans-activator of transcription, dopamine
Graphical Abstract
Introduction
Within the Human immunodeficiency virus (HIV) is a gene that encodes the transactivator of transcription (Tat) which is involved in the regulation of proteins that control how HIV infects the cells of its host.1–5 HIV-1 Tat causes viral toxicity in neurons by interacting with monoamine neurotransmitters that are found in the central nervous system (CNS). These transmitters, known as the norepinephrine transporter (NET) and dopamine transporter (DAT), are also targets for widely abused drugs (psychostimulants). More specifically, when HIV-1 Tat binds to these neurotransmitters, it can dysregulate the dopamine (DA) uptake process.6–28 There have been reports showing that HIV viral replication within the CNS leads to the development and progression of HIV-associated neurocognitive disorder (HAND).18 Common symptoms of HAND include difficulty with cognitive functions like learning and making decisions, loss of attention and concentration, deficits in memory and executive function, and tremors or loss of coordination.29,30 Most HIV therapies use highly active antiretroviral therapy (HAART) medications which allow their patients to live longer.31 Majority of HAART, especially protease inhibitors, have difficulty passing the blood-brain barrier (BBB) because of their low permeability, but the virus that infects macrophages does in fact possess the ability to cross the BBB.32 The progression of HIV infection leads to decreased levels of DA and causes selective damage to DA-rich areas of the brain in advanced stages of HIV infection.8,13,14,33
DAT is a monoamine transporter found on the presynaptic axonal membrane and its function is to regulate the neuronal synapse DA concentration. DA is released from the post-synaptic receptors into the synapse, at which point DAT will attempt to transfer DA into the neuron through the pre-synaptic terminal.34–36 HIV-1 Tat has been reported to impact the function of DAT through in vitro studies.37–39 HIV proteins and controlled substances, such as methamphetamine and cocaine, will increase DA levels in a synergistic manner inside of the synapse. DA can bind to the microglia, primarily in the brain, through DA receptors found on the postsynaptic terminal and stimulate HIV replication in HIV-infected cells. Previous work highlighted the severity of HIV infection in DA-rich areas of the brain, dorsal and ventral areas, suggesting a connection between DA levels and neuronal degeneration in HIV cases. These reports included experimental and computational studies, which examined how HIV-1 Tat binds to the human DAT (hDAT) in the extracellular regions.40–43
NET possess the ability to transport DA as well as norepinephrine (NE).44,45 In certain areas of the brain, like the prefrontal cortex (PFC), DA uptake relies on NET for its transport.46 Carboni et al. explains certain situations arise where temporary DAT deletion occurs, DA will be cleared from extracellular space through NET.47 Another important area is the hippocampus, which resembles the PFC where the uptake of DA into the terminal occurs through the more densely expressed NET.46,48–50 Straus et al showed that HIV-1 Tat expression influences DA transmission by decreasing uptake through both NET and DAT in the PFC of HIV-1 Tat transgenic mice.51
This study attempts to provide a computational model describing how HIV-1 Tat interacts with human NET (hNET). It is our hypothesis that HIV-1 Tat binding is similar in NET as DAT. State-of-the-art molecular modeling techniques were used to study the HIV-1 Tat-hNET complex as the RCSB Protein Data Bank (PDB) only hosts crystal structure of the following transporters, Aquifex aeolicus leucine transporter (LeuT)52, Drosophila DAT (dDAT)53, and human serotonin transporter (hSERT).54,55 Currently, the X-ray crystal structure for hNET and hDAT has not been resolved. Previous computational studies have laid the ground work for modelling such structures, this includes work by our group which based hDAT models on the previous LeuT structure using homology modeling techniques.55 The LeuT X-ray crystal structure was used as a template to reasonably model hDAT and hNET structures for their interactions with small-molecule ligands.56–59 The template LeuT shares 20.4% sequence identity and 42% sequence similarity with hDAT. More recently, our group has previously modeled hDAT from dDAT, and offered great insight on conserved residues and inhibitor interactions.42,60 In these studies, hDAT was modeled after dDAT because there was sequence similarity of 59% (identity: 46%). The previously obtained hDAT models allowed the investigation of the interactions between hDAT with dopamine, cocaine, and other inhibitors.61,62
Homology modeling can be quite useful when the model template has a high evolutionary homology sequence and identity percentage when compared to the modeled protein. Recent studies have modeled NET using DAT as a template and showed a high conservation of topological domains. The study shows most of the differences between NET and DAT are found towards the C-terminus.63 The hNET and hDAT sequences are quite similar with sequence similarity of 82% and sequence identity of 71%. The hDAT modelled structures were found to be consistent with known structural insight gained from previously reported wet-experimental hDAT validation. In this present study, the HIV-1 Tat–hNET binding mode has been resolved using protein–protein docking64 and molecular dynamic (MD) simulations.65 The computationally resolved binding mode is supported by subsequently obtained experimental data from DA uptake assays and site-directed. Insights from this study will hopefully establish a basis for further studies on the relationship between HIV-1 infection and DA uptake inhibition as well as HIV-1 Tat molecular mechanism and its effect in neurons.
Results and Discussion
In determining the bound structure of HIV-1 Tat–hNET, we need to investigate and obtain the individual structural details of hNET and HIV-1 Tat. The structure of hNET can be categorized in three different conformational states (including the outward-occluded, outward-open, and inward-open) which are similar to hDAT and built from homology modeled structures discussed in previous reports.42 The modeled structure of hNET is consistent with structural insights discussed in previously reported wet-experimental validations and previous modeling studies of hNET.66,67 The hNET model exhibits very similar features of hDAT which include conserved residues and a binding site for dopamine.
HIV-1 Tat subtype B is more prevalent in the North American hemisphere and is composed of 86 amino acids. It contains a +12e net charge with a physiological pH of 7.4. In 2001, experimentalists were successful in using multinuclear NMR spectroscopy to resolve the structure of the HIV-1 Tat protein. From the NMR study, there were 11 conformations presented to the protein databank (RCSB PDB ID: 1JFW)68, the structures are similar with an average root-mean square deviation (RMSD) of 1.3 Å on the backbone heavy atoms.42 HIV-1 Tat is composed of a majority of coil and loop conformations with a few β-turns. Transient folding appears to only occur in two regions composed of residues 38–48 and residues 60–72, while the main functional region is the cysteine-rich region which includes residues 22–37.68
Rigid-body protein–protein docking methods can yield accurate results but has difficulty predicting systems that exhibit conformational flexibility. The complex structures shown in this study were determined from previous works that utilized protein–protein docking, a refined series of energy minimizations, and molecular dynamics (MD) simulations that captured the most favorable HIV-1 Tat–hDAT binding structure. There are three different conformational states of hDAT (outward-occluded, outward-open, and inward-open). Due to significant homology similarity, hNET samples these conformations as well, but the outward-open state is more suited and favored for binding with HIV-1 Tat. The reason HIV-1 Tat prefers binding with hNET in the outward-open structure is due to the favorable binding energy. According to the binding energies calculated by using the Molecular Mechanics/Generalized Born Surface Area (MM/GBSA) approach, the HIV-1 Tat binding with hNET in the outward-open state has the lowest binding energy (which means the highest binding affinity). The binding energies of HIV-1 Tat with hNET in the outward-occluded and inward-open states are much higher (see Supporting Information for the binding structures and their relative binding energies). The most favorable HIV Tat–hNET binding mode (outward-open state) was refined further by performing MD simulations for a total of 2 μs, which included 200 independent MD trajectories each at 10 ns. The computationally modeled HIV-1 Tat-hNET binding interactions in the outward-open state are discussed below in detail.
Key intermolecular interactions in HIV-1 Tat-hNET binding
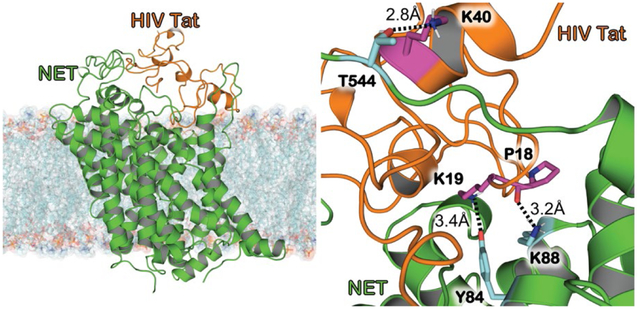
Figure 1A shows HIV-1 Tat binding to hNET in the outward-open confirmational state after energy minimization. The bound complex shows HIV-1 Tat embedded in the extracellular domain of hNET (green) due to electrostatic attraction. HIV-1 Tat is bound above hNET in the vestibule on the extracellular side, forming a contacting interface. This interface between the proteins due to electrostatic complementarity can prevent the uptake of neurotransmitters, because the favorable binding of HIV-1 Tat with hNET in the outward-open state effectively blocks the conformational change required to uptake a neurotransmitter such as DA.
Figure 1.
Modeled structure of HIV-1 Tat bound to hNET in the outward-open state after the molecular docking and energy minimization. (A) HIV-1 Tat–hNET binding structure developed from homology modeling. HIV-1 Tat is (orange) shown here with the cartoon representation, hNET (green) is represented using the cartoon representation. The lipid membrane is shown using semitransparent surface and stick representation colored according to heavy atoms. (B) HIV-1 Tat (orange) and hNET (green) are shown here with the surface representation showing the shape complementarity. (C) Highlighting HIV-1 Tat bound to hNET with critical residues important for maintaining the formed complex. HIV-1 Tat is (orange) shown here with the cartoon representation. Dopamine (red), residues Y84, Y88, and T544 of hNET (purple) are all shown in surface representation. (D) Internuclear distances are shown between the binding interface of HIV-1 Tat–hNET binding structure. HIV-1 Tat (orange) and hNET (green) are represented in the cartoon representation. Residues Y84, Y88, and T544 of hNET (cyan) as well as P18, K19, and K40 of Tat (purple) shown as sticks which are important for maintaining the binding between the two proteins. Intermolecular HBs are indicated as dashed lines, and distances are labeled next to the respected lines.
The most critical interactions include hNET residues Y84 and K88 as well as T544 shown in Figure 1C. There are weak hydrogen-bonding (HB) interactions between the positively charged butylammonium group of HIV-1 Tat K19 side chain and the hydroxyl group of hNET Y84 side chain. Another HB interaction exists between the oxygen atom of HIV-1 Tat P18 backbone and the positively charged butylammonium group of hNET K88 side chain. Lastly, there is a HB interaction between the positively charged butylammonium of HIV-1 Tat K40 side chain and the hydroxyl group of hNET T544 side chain.
We highlight these interactions and show the distances for the modeled structure in Figure 1D. The weak HB interactions between HIV-1 Tat K19 and hNET Y84 exhibit a distance of 3.4 Å between the hydroxyl oxygen of hNET Y84 side chain and the positively charged N atom of HIV-1 Tat K19 side chain, with a distance of 3.2 Å between the positively charged N atom of hNET K88 side chain and the backbone oxygen of HIV-1 Tat P18, and lastly a distance of 2.8 Å between the hydroxyl oxygen of hNET T544 and the positively charged N atom of HIV-1 Tat K40. These HB interactions along with the electrostatic complementarity help to maintain this bound complex.
Dynamics of HIV-1 Tat-hNET interactions
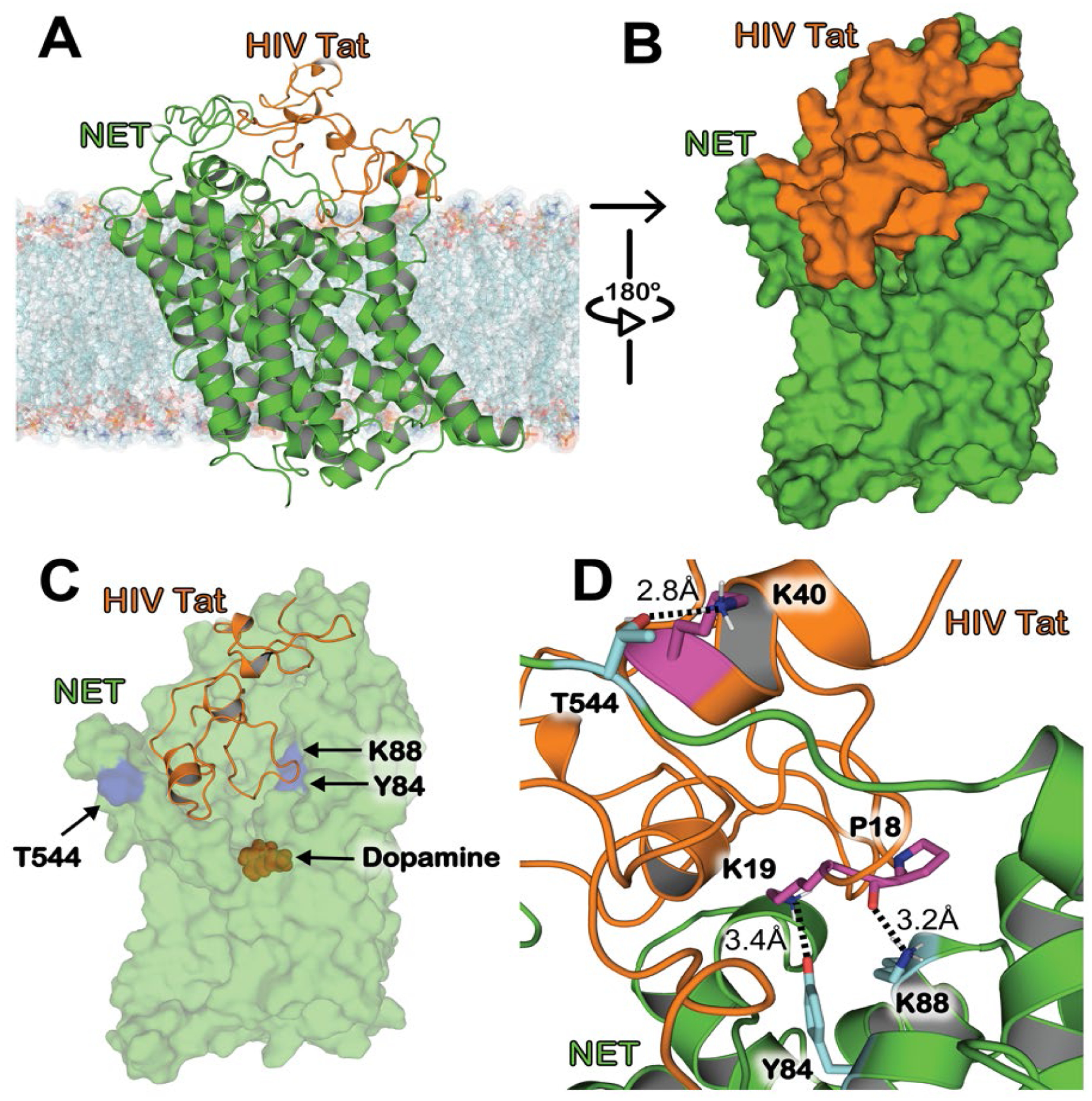
In Figure 2, we show the RMSD for the positions of heavy backbone atoms and distances of the three critical intermolecular interactions of the HIV-1 Tat–hNET binding complex over 200 MD trajectories for a total of 2 μs. In Figure 2A, the RMSD values are shown for 2.0 μs MD trajectories which average 2.99 ± 0.76 Å. This suggests that the HIV-1 Tat-hNET binding structure has a high conformational stability.
Figure 2.
RMSD and critical distances for the HIV-1 Tat–hNET binding structure based on the MD production simulations. (A) RMSD values (black) for the Tat–hNET binding structures backbone heavy atoms (Cɑ, C, O, N) in the 2.0 μs MD trajectories. (B) Critical distance (red) of the interaction between the nitrogen of hNET K88 side chain and the backbone oxygen of HIV-1 Tat P18. (C) Critical distance (blue) of the interaction between the side chain oxygen of hNET Y84 and side-chain nitrogen of HIV-1 Tat K19. (D) Critical distance (green) of the interaction between the oxygen of hNET T544 side chain and the nitrogen of HIV-1 Tat K40 side chain.
Based on the internuclear distances related to the HBs obtained from MD-simulations mentioned above, the intermolecular interactions between HIV-1 Tat and hNET are highly dynamic. For example, the MD-simulated distance between the positively charged nitrogen of hNET K88 side chain and the backbone oxygen of HIV-1 Tat P18 ranged from 2.49 Å to 19.03 Å (see Figure 2B), with an average distance of 4.19 Å (or 4.19 ± 1.90 Å in terms of the standard fluctuation), and 52.13% of the snapshots were shorter than 3.5 Å (the cutoff distance for a hydrogen bond used in this study). In other words, based on the MD simulations, the hydrogen bond between the positively charged nitrogen of hNET K88 side chain and the backbone oxygen of HIV-1 Tat P18 existed in 52.13% of the snapshots (with the distance cutoff of 3.5 Å). The distance shown in Figure 2C represents the HB interaction between the hydroxyl oxygen of hNET Y84 side chain and the positively charged nitrogen of HIV-1 Tat K19 side chain, ranging from 2.57 Å to 18.33 Å, with an average distance of 5.31 ± 1.33 Å, and the hydrogen bond existed in only 8.81% of the snapshots (with the distance cutoff of 3.5 Å). The distance shown in Figure 2D represents the HB interaction between the hydroxyl oxygen of hNET T544 side chain and the positively charged nitrogen of HIV-1 Tat K40 side chain, ranging from 2.55 Å to 19.68 Å, with an average distance of 7.46 ± 2.49 Å, and the hydrogen bond existed in only 5.22% of the snapshots (with the distance cutoff of 3.5 Å). Overall, all the tracked distances depicted in Figure 2 suggest that the HIV-1 Tat-hNET binding mode is dynamically stable. Notably, the occasional long distances were due to the secondary structure characteristics of the HIV-1 Tat protein (which is made up of 86 residues and its secondary structural composition includes a majority of coils and 3 short alpha helices). There was no major structural change during the MD simulation, just the mild stochastic conformational switching of solvent exposed coils.
Experimental validation through side-directed mutagenesis
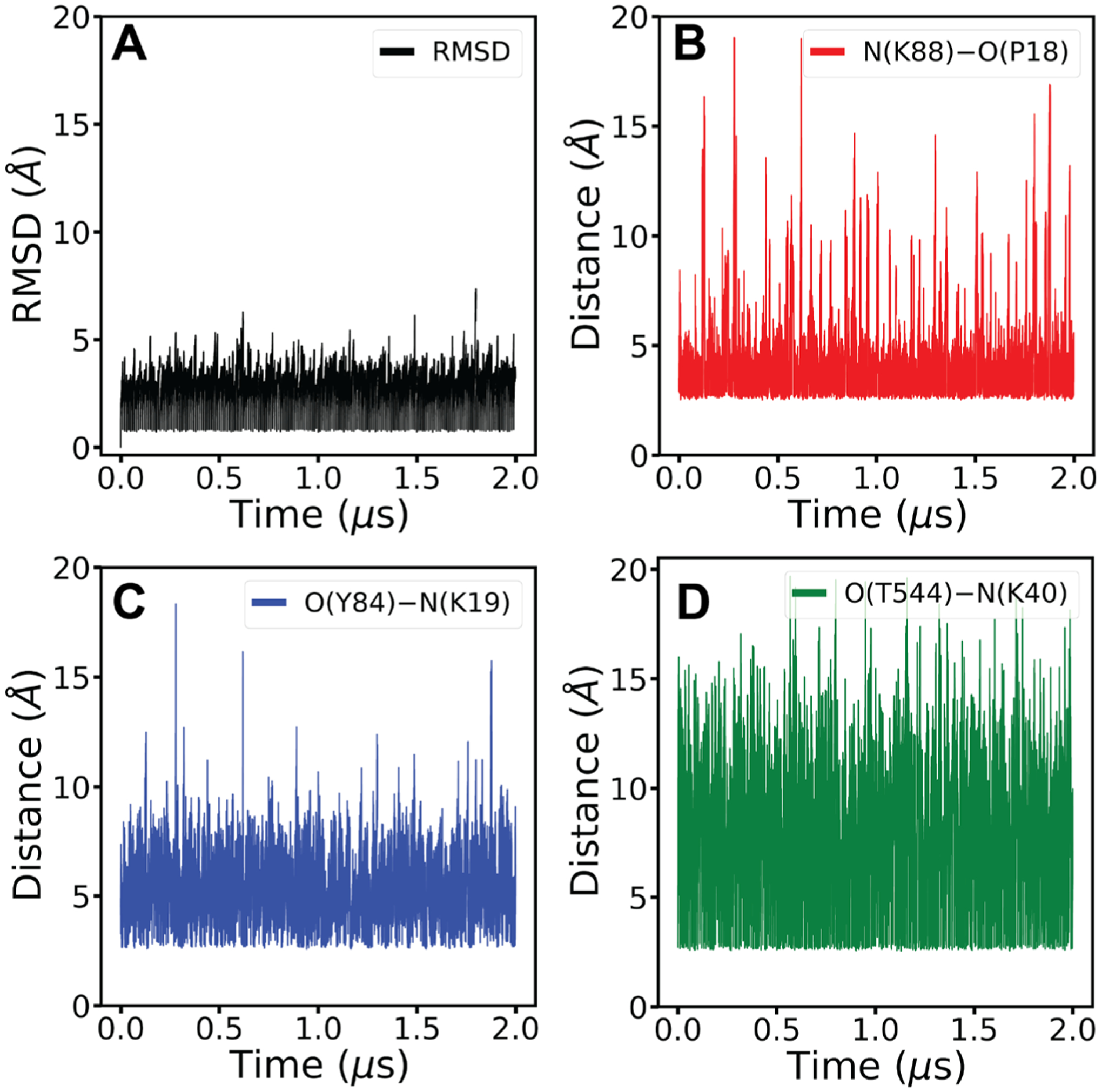
According to the modeled HIV-1 Tat-hNET binding structure in Figure 1, three hNET residues (Y84, K88, and T544) are predicted to have favorable HB interactions with HIV-1 Tat. To validate the predicted HIV-1 Tat-hNET binding mode, three mutants (Y84F, K88M, and T544A) were designed to disrupt the favorable HB interactions of hNET with HIV-1 Tat (as illustrated in Figure 3). These mutants were prepared through site-directed mutagenesis and tested for their DA uptake activity in comparison with wild-type hNET (WT-hNET). It turned out that the Y84F and T544A mutants did have the desirable DA update activity of the WT-hNET. Unfortunately, the K88M mutant was inactive, suggesting that the K88M mutation might have resulted in some major structural change of the protein. So, without detectable DA update activity for the K88M mutant of hNET, we were unable to examine whether HIV-1 Tat can inhibit the DA uptake activity of the K88M mutant. Nevertheless, we were able to test whether HIV-1 Tat can inhibit the DA uptake activity of the Y84F and T544A mutants in comparison with WT-hNET. It turned out that 140 nM recombinant Tat1–86 did not significantly decrease the DA uptake activity of the Y84F and T544A mutants, while decreasing the DA uptake activity of the WT-hNET, suggesting that the favorable HIV-1 Tat-hNET interactions were disrupted by the Y84F and T544A mutations. So, these results of the site-directed mutagenesis support the favorable HB interactions of HIV-1 Tat with the side chains of hNET residues Y84 and T544.
Figure 3.
Critical interactions shown between important residues and mutated residues. Mutants of hNET were modeled from the WT-hNET obtained from the HIV-1 Tat–hNET binding structure followed by energy minimizations. (A) Distances are shown between the side chain oxygen of hNET T544 (cyan) and side chain nitrogen of HIV-1 Tat K40 (purple) as well as the side chain oxygen of hNET Y84 (cyan) and side chain nitrogen of HIV-1 Tat K19 (purple). (B) Y84F mutation shows the disrupted interaction between HIV-1 Tat residue K19 (purple) and hNET residue F84 (cyan) due to a loss of a hydroxyl group from hNET residue Y84. (C) T544A mutation shows the disrupted interaction between HIV-1 Tat residue K40 (purple) and hNET residue A544 (cyan) due to a loss of a hydroxyl group from hNET residue T544.
Conclusion
In this work, we displayed a three-dimensional modeled structure of hNET binding with HIV-1 Tat through the use of protein–protein docking methods and molecular dynamics simulations. The obtained computational data suggests that HIV-1 Tat binds to the outward-open state of hNET, which is similar to the binding of HIV-1 Tat with hDAT. Based on the modeled HIV-1 Tat–hNET binding structure, we have identified hNET residues Y84, K88, and T544 as important residues for hNET interacting with HIV-1 Tat. These interactions were analyzed over 2 μs of MD simulations. The newly obtained HIV-1 Tat–hNET binding mode reveals novel mechanistic insights describing how HIV-1 Tat interacts with hNET. Further, the favorable HB interactions of HIV-1 Tat with the side chains of hNET residues Y84 and T544 have been validated by our subsequently obtained experimental data from the site-directed mutagenesis and DA uptake activity assays, suggesting that the computationally modeled HIV-1 Tat-hNET binding mode is reasonable. In conclusion, this study depicts how molecular dynamics and molecular modeling can be used to study protein-protein interactions and address questions regarding mechanisms and complex structures.
Methods
Molecular Modeling
Although there is a reported PDB structure of dDAT in the RCSB Protein Databank, no x-ray crystal structure of NET has been reported, preventing a direct structural comparison. To understand the impact of the binding between HIV-1 Tat and hNET, we performed a Basic Local Alignment Search Tool (BLAST) search of hNET to identify sequence similarity to hDAT.69,70 Our search determined that, hNET and hDAT have 82% sequence similarity and 71% sequence identity. Mega X was used to generate a fasta file for hNET and hDAT based on the BLAST alignment.71 The Modeller software was then used to model hNET using hDAT as a template.72 After the model was constructed, the Procheck software was used to check the stereochemistry of the model. HIV-1 Tat structure was obtained from a previous study and placed into the extracellular domain with the AmberMD software package.73
Molecular Dynamics
The modeled complex structure of HIV-1 Tat-hNET was imported into the tLeap module of the Amber1865 suite of programs74 to add hydrogens using the ff14SB75,76 modified version of the Cornell et al.77 force field. A series of energy minimization (EM) were conducted on the modeled structure of HIV-1 Tat-hNET complex to address the steric clashes. A second series of energy minimizations were conducted on the entire protein bound structure. The HIV-1 Tat-hNET complex system was energy-minimized with 4,000 steps using the deepest decent EM method and 4,000 steps with the conjugate gradient EM method using the Sander module from the Amber18 package. This series of EM stages were carried out by using a harmonic constraint on the protein atoms and gradually decreasing the force constant from 300, 200, 100, 75, 50, and 25 kcal/mol/Å2. A final energy-minimization stage was carried out without any constraint.
A series of EM stages were carried out prior to the MD production run. A 5-ns MD simulation was used to relax residues involved in steric clashes in vacuum. Then another 5-ns MD simulation was used to relax the binding interface of residues within 5 Å of the bound complex in vacuum. Another 5-ns MD simulation was used to relax the binding interface of residues within 5 Å of the either HIV-1 Tat or hNET in a solvated cubic box. A final 5-ns MD simulation was carried out on the HIV-1 Tat–hNET complex to relax the entire system in a solvated periodic octahedron box of TIP3P78 water model with a box cutoff at 10 Å.
The last frame of the solvated HIV-1 Tat–hNET complex was selected to conduct the MD simulation of the system in a bilayer membrane. The bilayer was built using Membrane Builder in the CHARMM-GUI.79,80 The lipid and HIV-1 Tat–hNET complex were built using CHARMM36 and CHARMM36m force field respectfully.81,82 The HIV-1 Tat–hNET complex was solvated using a CHARMM-compatible TIP3P water model.78 The MD simulations of all the solvated HIV-1 Tat–hNET complexes were carried out using the CUDA version of the pmemd module from the Amber18 package74 with a constant pressure (1 bar) and constant temperature (300 K). These MD simulations used the SHAKE algorithm83 to constrain all covalent bonds that involve a hydrogen and the Langevin thermostat to regulate the temperature84 such that it can maintain 300 K with a collision frequency of 1.0 ps–1. During the MD simulations, a 9 Å cutoff was used to calculate the short-range non-bonded interactions, the particle mesh Ewald (PME) method85 was used to evaluate all long-range electrostatic interactions, and a time step of 2 fs was applied to the integration of the Langevin equation of motion. This MD production was simulated in 2000 independent runs each at 10 ns, which contributed to a total of 2 μs.86
Site-directed mutagenesis and DA uptake activity assays
All hNET mutants were prepared with insight gained from the computational model using site-directed mutagenesis starting from the wild-type (WT)-hNET sequence (NCBI, cDNA clone MGC: 190603 IMAGE: 100062757). The combination of hNET mutations on Tyr84, Lys88, and Thr544 (Y84F, tyrosine 84 to phenylalanine; K88M, lysine 88 to methionine; T544A, threonine 544 to alanine) were predicted to eliminate hydrogen bonding interactions of HIV-1 Tat with hNET. GENEWIZ (South Plainfield, NJ) performed site-directed mutagenesis on synthetic cDNA used to encode hNET and subcloned into pcDNA3.1+ as a template. GENEWIZ also performed DNA sequencing conducted on mutant constructs to confirm the hNET sequences. Plasmid DNA was purified and propagated using a Qiagen Hi-speed maxi prep plasmid DNA isolation kit (Qiagen, Valencia, CA, USA). All cell cultures and transfection were conducted as described previously.87,88 PC12 cells were maintained in a 5% CO2 incubator at 37 °C within a Dulbecco’s modified eagle medium (DMEM, Life Technologies, Carlsbad, CA) and supplemented with 2.5% fetal bovine serum, 2 mM glutamine, 15% horse serum, and antibiotics (100 U/ml penicillin and 100 μg/mL streptomycin). Transient transfection was conducted on cells that were seeded into 24-well plates with a density of 1×105 cells/cm2, then allowed to reach 100% confluence, and lastly transfected 24 h later with WT or mutant hNET plasmid DNA using Lipofectamine 2000. These cells were used for transfection after 24 h.
The Michaelis-Menten constant (Km) and maximal velocity (Vmax) of [3H]DA uptake were examined in intact PC12 cells transiently expressing mutant or WT-hNET described previously.89 The cells were washed twice in 1× Krebs-Ringer-HEPES (1× KRH) buffer, then preincubated at room temperature for 10 min in buffer and/or nomifensine (at a final concentration of 10 μM), and then incubated with the addition of unlabeled DA at one of six concentrations (the final DA concentrations: 0.03–5 μM) and a fixed concentration of [3H]DA (500,000 DPM/well, specific activity, 21.2 Ci/mmol) at room temperature for 8 min. Specific uptake was calculated by subtracting nonspecific uptake from total uptake in the presence of 10 μM nomifensine and desipramine. The cells were washed twice with ice-cold 1× KRH buffer, then lysed in 500 μl of 1% SDS for an hour. To determine the effects of HIV-1 Tat inhibition on [3H]DA uptake, cells were harvested then resuspended in culture medium and allowed to incubate for 10 min at room temperature. Dissociated cells were pelleted by centrifugation at 400 × g at 4°C for 5 min, then washed once and resuspended with PBS (phosphate-buffered saline). The dissociated cells underwent an additional centrifugation at 400 × g at 4°C for 5 min, and resuspended in 1X KRH buffer. [3H]DA uptake was determined in cell suspensions prepared from mutant and WT-hDAT in the absence or presence of recombinant Tat1–86 (at a final concentration of 140 nM). Cell suspensions were preincubated with HIV-1 Tat at room temperature for 20 min, and then incubated for 8 min after adding [3H]DA (0.05 μM, final concentration). 10 μM nomifensine and desipramine (in terms of the final concentrations) were used to determine non-specific [3H]DA uptake. The incubation was terminated after immediate filtration through Whatman GF/B glass filters (presoaked with 1× KRH buffer containing 1 mM pyrocatechol for at least 3 h). Glass filters were washed for three times with 3 ml ice-cold KRH buffer containing pyrocatechol using a Brandel cell harvester M-48 (Brandel Inc., Gaithersburg, MD). The radioactivity was measured by using a liquid scintillation counter Tri-Carb 2900TR (PerkinElmer Life and Analytical Sciences, Waltham, MA).
Supplementary Material
Acknowledgments
This work was supported in part by the National Institutes of Health (R01 DA035714, P20 GM130456, and UL1 TR001998) and the National Science Foundation (NSF grant CHE-1111761). The authors also thank the University of Kentucky Computer Center for providing supercomputing time on a Dell Supercomputer Cluster consisting of 388 nodes or 4,816 processors.
Footnotes
Supporting Information
Docked structures of bound complex HIV-1 Tat-hNET shown in three different conformational states (outward-open state, outward-occluded state, and inward-open state).
References
- 1.Carey AN; Sypek EI; Singh HD; Kaufman MJ; McLaughlin JP, Expression of HIV-Tat protein is associated with learning and memory deficits in the mouse. Behav Brain Res 2012, 229 (1), 48–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li W; Li G; Steiner J; Nath A, Role of Tat Protein in HIV Neuropathogenesis. Neurotoxicity Research 2009, 16 (3), 205–220. [DOI] [PubMed] [Google Scholar]
- 3.Ernst T; Yakupov R; Nakama H; Crocket G; Cole M; Watters M; Ricardo-Dukelow ML; Chang L, Declined neural efficiency in cognitively stable human immunodeficiency virus patients. Ann Neurol 2009, 65 (3), 316–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wallace DR; Dodson S; Nath A; Booze RM, Estrogen attenuates gp120- and tat1–72-induced oxidative stress and prevents loss of dopamine transporter function. Synapse 2006, 59 (1), 51–60. [DOI] [PubMed] [Google Scholar]
- 5.Silvers JM; Aksenov MY; Aksenova MV; Beckley J; Olton P; Mactutus CF; Booze RM, Dopaminergic marker proteins in the substantia nigra of human immunodeficiency virus type 1-infected brains. Journal of neurovirology 2006, 12 (2), 140–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silverstein PS; Shah A; Weemhoff J; Kumar S; Singh DP; Kumar A, HIV-1 gp120 and drugs of abuse: interactions in the central nervous system. Curr HIV Res 2012, 10 (5), 369–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scheller C; Arendt G; Nolting T; Antke C; Sopper S; Maschke M; Obermann M; Angerer A; Husstedt IW; Meisner F et al. , Increased dopaminergic neurotransmission in therapy-na < ve asymptomatic HIV patients is not associated with adaptive changes at the dopaminergic synapses. Journal of Neural Transmission 2010, 117 (6), 699–705. [DOI] [PubMed] [Google Scholar]
- 8.Sardar AM; Czudek C; Reynolds GP, Dopamine deficits in the brain: the neurochemical basis of parkinsonian symptoms in AIDS. Neuroreport 1996, 7 (4), 910–912. [DOI] [PubMed] [Google Scholar]
- 9.Purohit V; Rapaka R; Shurtleff D, Drugs of abuse, dopamine, and HIV-associated neurocognitive disorders/HIV-associated dementia. Molecular neurobiology 2011, 44 (1), 102–110. [DOI] [PubMed] [Google Scholar]
- 10.Nath A; Maragos WF; Avison MJ; Schmitt FA; Berger JR, Acceleration of HIV dementia with methamphetamine and cocaine. Journal of NeuroVirology 2001, 7 (1), 66–71. [DOI] [PubMed] [Google Scholar]
- 11.Meade CS; Lowen SB; MacLean RR; Key MD; Lukas SE, fMRI brain activation during a delay discounting task in HIV-positive adults with and without cocaine dependence. Psychiatry Res 2011, 192 (3), 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meade CS; Conn NA; Skalski LM; Safren SA, Neurocognitive impairment and medication adherence in HIV patients with and without cocaine dependence. J Behav Med 2011, 34 (2), 128–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar AM; Ownby RL; Waldrop-Valverde D; Fernandez B; Kumar M, Human immunodeficiency virus infection in the CNS and decreased dopamine availability: Relationship with neuropsychological performance. Journal of NeuroVirology 2011, 17 (1), 26–40. [DOI] [PubMed] [Google Scholar]
- 14.Kumar AM; Fernandez JB; Singer EJ; Commins D; Waldrop-Valverde D; Ownby RL; Kumar M, Human immunodeficiency virus type 1 in the central nervous system leads to decreased dopamine in different regions of postmortem human brains. Journal of Neurovirology 2009, 15 (3), 257–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koutsilieri E; ter Meulen V; Riederer P, Neurotransmission in HIV associated dementia: a short review. J Neural Transm (Vienna) 2001, 108 (6), 767–775. [DOI] [PubMed] [Google Scholar]
- 16.Koutsilieri E; Czub S; Scheller C; Sopper S; Tatschner T; Stahl-Hennig C; ter Meulen V; Riederer P, Brain choline acetyltransferase reduction in SIV infection. An index of early dementia? Neuroreport 2000, 11 (11), 2391–2393. [DOI] [PubMed] [Google Scholar]
- 17.Heaton RK; Clifford DB; Franklin DR Jr.; Woods SP; Ake C; Vaida F; Ellis RJ; Letendre SL; Marcotte TD; Atkinson JH et al. , HIV-associated neurocognitive disorders persist in the era of potent antiretroviral therapy: CHARTER Study. Neurology 2010, 75 (23), 2087–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaskill PJ; Calderon TM; Luers AJ; Eugenin EA; Javitch JA; Berman JW, Human immunodeficiency virus (HIV) infection of human macrophages is increased by dopamine: A bridge between HIV-associated neurologic disorders and drug abuse. American Journal of Pathology 2009, 175 (3), 1148–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferris MJ; Mactutus CF; Booze RM, Neurotoxic profiles of HIV, psychostimulant drugs of abuse, and their concerted effect on the brain: Current status of dopamine system vulnerability in NeuroAIDS. Neuroscience and Biobehavioral Reviews 2008, 32 (5), 883–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferris MJ; Frederick-Duus D; Fadel J; Mactutus CF; Booze RM, Hyperdopaminergic tone in HIV-1 protein treated rats and cocaine sensitization. Journal of neurochemistry 2010, 115 (4), 885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferrarese C; Aliprandi A; Tremolizzo L; Stanzani L; De Micheli A; Dolara A; Frattola L, Increased glutamate in CSF and plasma of patients with HIV dementia. Neurology 2001, 57 (4), 671–675. [DOI] [PubMed] [Google Scholar]
- 22.Ernst T; Jiang CS; Nakama H; Buchthal S; Chang L, Lower brain glutamate is associated with cognitive deficits in HIV patients: a new mechanism for HIV-associated neurocognitive disorder. Journal of magnetic resonance imaging : JMRI 2010, 32 (5), 1045–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang L; Wang GJ; Volkow ND; Ernst T; Telang F; Logan J; Fowler JS, Decreased brain dopamine transporters are related to cognitive deficits in HIV patients with or without cocaine abuse. Neuroimage 2008, 42 (2), 869–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buckner CM; Luers AJ; Calderon TM; Eugenin EA; Berman JW, Neuroimmunity and the blood-brain barrier: molecular regulation of leukocyte transmigration and viral entry into the nervous system with a focus on neuroAIDS. Journal of neuroimmune pharmacology : the official journal of the Society on NeuroImmune Pharmacology 2006, 1 (2), 160–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beuming T; Kniazeff J; Bergmann ML; Shi L; Gracia L; Raniszewska K; Newman AH; Javitch JA; Weinstein H; Gether U et al. , The binding sites for cocaine and dopamine in the dopamine transporter overlap. Nature Neuroscience 2008, 11 (7), 780–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berger JR; Arendt G, HIV dementia: The role of the basal ganglia and dopaminergic systems. Journal of Psychopharmacology 2000, 14 (3), 214–221. [DOI] [PubMed] [Google Scholar]
- 27.Bagashev A; Sawaya BE, Roles and functions of HIV-1 Tat protein in the CNS: an overview. Virol J 2013, 10, 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aksenov MY; Aksenova MV; Mactutus CF; Booze RM, D1/NMDA receptors and concurrent methamphetamine+ HIV-1 Tat neurotoxicity. Journal of neuroimmune pharmacology : the official journal of the Society on NeuroImmune Pharmacology 2012, 7 (3), 599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saylor D; Dickens AM; Sacktor N; Haughey N; Slusher B; Pletnikov M; Mankowski JL; Brown A; Volsky DJ; McArthur JC, HIV-associated neurocognitive disorder — pathogenesis and prospects for treatment. Nature Reviews Neurology 2016, 12, 234–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heaton RK; Franklin DR; Ellis RJ; McCutchan JA; Letendre SL; Leblanc S; Corkran SH; Duarte NA; Clifford DB; Woods SP et al. , HIV-associated neurocognitive disorders before and during the era of combination antiretroviral therapy: differences in rates, nature, and predictors. J Neurovirol 2011, 17 (1), 3–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anthony IC; Bell JE, The Neuropathology of HIV/AIDS. Int Rev Psychiatry 2008, 20 (1), 15–24. [DOI] [PubMed] [Google Scholar]
- 32.Rao KS; Ghorpade A; Labhasetwar V, Targeting anti-HIV drugs to the CNS. Expert Opinion on Drug Delivery 2009, 6 (8), 771–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aylward EH; Henderer JD; McArthur JC; Brettschneider PD; Harris GJ; Barta PE; Pearlson GD, Reduced basal ganglia volume in HIV-1-associated dementia: results from quantitative neuroimaging. Neurology 1993, 43 (10), 2099–2104. [DOI] [PubMed] [Google Scholar]
- 34.Seger D, Cocaine, metamfetamine, and MDMA abuse: The role and clinical importance of neuroadaptation. Clinical Toxicology 2010, 48 (7), 695–707. [DOI] [PubMed] [Google Scholar]
- 35.Volz TJ; Hanson GR; Fleckenstein AE, The role of the plasmalemmal dopamine and vesicular monoamine transporters in methamphetamine-induced dopaminergic deficits. Journal of Neurochemistry 2007, 101 (4), 883–888. [DOI] [PubMed] [Google Scholar]
- 36.Camí J; Farré M, Mechanisms of disease: Drug addiction. New England Journal of Medicine 2003, 349 (10), 975–986. [DOI] [PubMed] [Google Scholar]
- 37.Zhu J; Mactutus CF; Wallace DR; Booze RM, HIV-1 Tat Protein-Induced Rapid and Reversible Decrease in [3H]Dopamine Uptake: Dissociation of [3H]Dopamine Uptake and [3H]2 -Carbomethoxy-3- -(4-fluorophenyl)tropane (WIN 35,428) Binding in Rat Striatal Synaptosomes. Journal of Pharmacology and Experimental Therapeutics 2009, 329 (3), 1071–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferris MJ; Frederick-Duus D; Fadel J; Mactutus CF; Booze RM, The human immunodeficiency virus-1-associated protein, Tat1–86, impairs dopamine transporters and interacts with cocaine to reduce nerve terminal function: A no-net-flux microdialysis study. Neuroscience 2009, 159 (4), 1292–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferris MJ; Frederick-Duus D; Fadel J; Mactutus CF; Booze RM, In vivo microdialysis in awake, freely moving rats demonstrates HIV-1 tat-induced alterations in dopamine transmission. Synapse 2009, 63 (3), 181–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu J; Yuan Y; Midde NM; Gomez AM; Sun WL; Quizon PM; Zhan CG, HIV-1 transgenic rats display an increase in [3H]dopamine uptake in the prefrontal cortex and striatum. Journal of NeuroVirology 2016, 22 (3), 282–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan Y; Quizon PM; Sun W-L; Yao J; Zhu J; Zhan C-G, Role of Histidine 547 of Human Dopamine Transporter in Molecular Interaction with HIV-1 Tat and Dopamine Uptake. Scientific Reports 2016, 6, 27314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuan Y; Huang X; Midde NM; Quizon PM; Sun W-L; Zhu J; Zhan C-G, Molecular Mechanism of HIV-1 Tat Interacting with Human Dopamine Transporter. Acs Chemical Neuroscience 2015, 6 (4), 658–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Midde NM; Yuan Y; Quizon PM; Sun W-L; Huang X; Zhan C-G; Zhu J, Mutations at Tyrosine 88, Lysine 92 and Tyrosine 470 of Human Dopamine Transporter Result in an Attenuation of HIV-1 Tat-Induced Inhibition of Dopamine Transport. Journal of Neuroimmune Pharmacology 2015, 10 (1), 122–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horn AS, Structure‐activity relations for the inhibition of catecholamine uptake into synaptosomes from noradrenaline and dopaminergic neurones in rat brain homogenates. British Journal of Pharmacology 1973, 47 (2), 332–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raiteri M; Del Carmine R; Bertollini A; Levi G, Effect of sympathomimetic amines on the synaptosomal transport of noradrenaline, dopamine and 5-hydroxytryptamine. European Journal of Pharmacology 1977, 41 (2), 133–143. [DOI] [PubMed] [Google Scholar]
- 46.Morón JA; Brockington A; Wise RA; Rocha BA; Hope BT, Dopamine uptake through the norepinephrine transporter in brain regions with low levels of the dopamine transporter: Evidence from knock-out mouse lines. Journal of Neuroscience 2002, 22 (2), 389–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carboni E; Silvagni A; Vacca C; Di Chiara G, Cumulative effect of norepinephrine and dopamine carrier blockade on extracellular dopamine increase in the nucleus accumbens shell, bed nucleus of stria terminalis and prefrontal cortex. Journal of Neurochemistry 2006, 96 (2), 473–481. [DOI] [PubMed] [Google Scholar]
- 48.Carboni E; Tanda GL; Frau R; Chiara GD, Blockade of the Noradrenaline Carrier Increases Extracellular Dopamine Concentrations in the Prefrontal Cortex: Evidence that Dopamine Is Taken up In Vivo by Noradrenergic Terminals. Journal of Neurochemistry 1990, 55 (3), 1067–1070. [DOI] [PubMed] [Google Scholar]
- 49.Pozzi L; Invernizzi R; Cervo L; Vallebuona F; Samanin R, Evidence that Extracellular Concentrations of Dopamine Are Regulated by Noradrenergic Neurons in the Frontal Cortex of Rats. Journal of Neurochemistry 1994, 63 (1), 195–200. [DOI] [PubMed] [Google Scholar]
- 50.Yamamoto BK; Novotney S, Regulation of extracellular dopamine by the norepinephrine transporter. Journal of Neurochemistry 1998, 71 (1), 274–280. [DOI] [PubMed] [Google Scholar]
- 51.Strauss M; O’Donovan B; Ma Y; Xiao Z; Lin S; Bardo MT; Ortinski PI; McLaughlin JP; Zhu J, 3H]Dopamine Uptake through the Dopamine and Norepinephrine Transporters is Decreased in the Prefrontal Cortex of Transgenic Mice Expressing HIV-1 Transactivator of Transcription Protein. The Journal of pharmacology and experimental therapeutics 2020, 374 (2), 241–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamashita A; Singh SK; Kawate T; Jin Y; Gouaux E, Crystal structure of a bacterial homologue of Na+/Cl --dependent neurotransmitter transporters. Nature 2005, 437 (7056), 215–223. [DOI] [PubMed] [Google Scholar]
- 53.Penmatsa A; Wang KH; Gouaux E, X-ray structure of dopamine transporter elucidates antidepressant mechanism. Nature 2013, 503 (7474), 85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Coleman JA; Green EM; Gouaux E, X-ray structures and mechanism of the human serotonin transporter. Nature 2016, 532 (7599), 334–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang KH; Penmatsa A; Gouaux E, Neurotransmitter and psychostimulant recognition by the dopamine transporter. Nature 2015, 521 (7552), 322–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang X; Zhan C-G, How dopamine transporter interacts with dopamine: insights from molecular modeling and simulation. Biophysical journal 2007, 93 (10), 3627–3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang X; Gu HH; Zhan C-G, Mechanism for cocaine blocking the transport of dopamine: insights from molecular modeling and dynamics simulations. The Journal of Physical Chemistry B 2009, 113 (45), 15057–15066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hill ER; Huang X; Zhan C-G; Ivy Carroll F; Gu HH, Interaction of tyrosine 151 in norepinephrine transporter with the 2β group of cocaine analog RTI-113. Neuropharmacology 2011, 61, 112–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yuan Y; Huang X; Zhu J; Zhan C-G, Computational modeling of human dopamine transporter structures, mechanism and its interaction with HIV-1 transactivator of transcription. Future Med. Chem 2016, 8, 2077–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yuan Y; Zhu J; Zhan C-G, Flipped phenyl ring orientations of dopamine binding with human and drosophila dopamine transporters: Remarkable role of three nonconserved residues. ACS Chem. Neurosci 2018, 9, 1426–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rothman RB; Ananthan S; Partilla JS; Saini SK; Moukha-Chafiq O; Pathak V; Baumann MH, Studies of the Biogenic Amine Transporters 15. Identification of Novel Allosteric Dopamine Transporter Ligands with Nanomolar Potency. Journal of Pharmacology and Experimental Therapeutics 2015, 353 (3), 529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun WL; Quizon PM; Yuan Y; Zhang W; Ananthan S; Zhan CG; Zhu J, Allosteric modulatory effects of SRI-20041 and SRI-30827 on cocaine and HIV-1 Tat protein binding to human dopamine transporter. Scientific Reports 2017, 7 (1), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Góral I; Łątka K; Bajda M, Structure modeling of the norepinephrine transporter. Biomolecules 2020, 10 (1), 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen R; Li L; Weng Z, ZDOCK: An initial-stage protein-docking algorithm. Proteins: Structure, Function, and Bioinformatics 2003, 52 (1), 80–87. [DOI] [PubMed] [Google Scholar]
- 65.Case DA; Ben-Shalom IY; Brozell SR; Cerutti DS; Cheatham TE III; Cruzeiro VWD; Darden TA; Duke RE; Ghoreishi D; Gilson MK et al. , Amber 18. University of California, San Francisco, 2018. [Google Scholar]
- 66.Aggarwal S; Mortensen OV, Overview of Monoamine Transporters. Current Protocols in Pharmacology 2017, 79 (1), 12.16.1–12.16.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Torres GE; Gainetdinov RR; Caron MG, Plasma membrane monoamine transporters: structure, regulation and function. Nature Reviews Neuroscience 2003, 4 (1), 13–25. [DOI] [PubMed] [Google Scholar]
- 68.Péloponèse JM; Grégoire C; Opi S; Esquieu D; Sturgis J; Lebrun É; Meurs É; Collette Y; Olive D; Aubertin AM et al. , 1H-13C nuclear magnetic resonance assignment and structural characterization of HIV-1 Tat protein. Comptes Rendus de l’Academie des Sciences - Serie III 2000, 323 (10), 883–894. [DOI] [PubMed] [Google Scholar]
- 69.Gish W; States DJ, Identification of protein coding regions by database similarity search. Nature Genetics 1993, 3 (3), 266–272. [DOI] [PubMed] [Google Scholar]
- 70.Altschul SF; Gish W; Miller W; Myers EW; Lipman DJ, Basic local alignment search tool. Journal of Molecular Biology 1990, 215 (3), 403–410. [DOI] [PubMed] [Google Scholar]
- 71.Kumar S; Stecher G; Li M; Knyaz C; Tamura K, MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Molecular Biology and Evolution 2018, 35 (6), 1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Webb B; Sali A, Comparative Protein Structure Modeling Using MODELLER. Curr Protoc Bioinformatics 2016, 54, 5.6.1–5.6.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Laskowski RA; MacArthur MW; Moss DS; Thornton JM, PROCHECK: a program to check the stereochemical quality of protein structures. Journal of Applied Crystallography 1993, 26 (2), 283–291. [Google Scholar]
- 74.Case DA; Cheatham TE; Darden T; Gohlke H; Luo R; Merz KM; Onufriev A; Simmerling C; Wang B; Woods RJ, The Amber biomolecular simulation programs. Journal of Computational Chemistry 2005, 26 (16), 1668–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hornak V; Abel R; Okur A; Strockbine B; Roitberg A; Simmerling C, Comparison of multiple amber force fields and development of improved protein backbone parameters. Proteins-Structure Function and Bioinformatics 2006, 65 (3), 712–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Maier JA; Martinez C; Kasavajhala K; Wickstrom L; Hauser KE; Simmerling C, ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J Chem Theory Comput 2015, 11 (8), 3696–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cornell WD; Cieplak P; Bayly CI; Gould IR; Merz KM; Ferguson DM; Spellmeyer DC; Fox T; Caldwell JW; Kollman PA, A 2nd Generation Force-Field for the Simulation of Proteins, Nucleic-Acids, and Organic-Molecules. Journal of the American Chemical Society 1995, 117 (19), 5179–5197. [Google Scholar]
- 78.Jorgensen WL, Revised TIPS for simulations of liquid water and aqueous solutions. Journal of Chemical Physics 1982, 77 (8), 4156–4163. [Google Scholar]
- 79.Wu EL; Cheng X; Jo S; Rui H; Song KC; Dávila-Contreras EM; Qi Y; Lee J; Monje-Galvan V; Venable RM et al. , CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. Journal of Computational Chemistry 2014, 35 (27), 1997–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jo S; Kim T; Iyer VG; Im W, CHARMM-GUI: A web-based graphical user interface for CHARMM. Journal of Computational Chemistry 2008, 29 (11), 1859–1865. [DOI] [PubMed] [Google Scholar]
- 81.Huang J; Rauscher S; Nawrocki G; Ran T; Feig M; de Groot BL; Grubmüller H; MacKerell AD, CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nature Methods 2017, 14 (1), 71–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Klauda JB; Venable RM; Freites JA; O’Connor JW; Tobias DJ; Mondragon-Ramirez C; Vorobyov I; MacKerell AD; Pastor RW, Update of the CHARMM All-Atom Additive Force Field for Lipids: Validation on Six Lipid Types. The Journal of Physical Chemistry B 2010, 114 (23), 7830–7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ryckaert JP; Ciccotti G; Berendsen HJC, Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys 1977, 23 (3), 327–341. [Google Scholar]
- 84.Izaguirre JA; Catarello DP; Wozniak JM; Skeel RD, Langevin stabilization of molecular dynamics. Journal of Chemical Physics 2001, 114 (5), 2090–2098. [Google Scholar]
- 85.Essmann U; Perera L; Berkowitz ML; Darden T; Lee H; Pedersen LG, A smooth particle mesh ewald method. Journal of Chemical Physics 1995, 103 (19), 8577–8593. [Google Scholar]
- 86.Kollman PA; Massova I; Reyes C; Kuhn B; Huo S; Chong L; Lee M; Lee T; Duan Y; Wang W et al. , Calculating Structures and Free Energies of Complex Molecules: Combining Molecular Mechanics and Continuum Models. Accounts of Chemical Research 2000, 33 (12), 889–897. [DOI] [PubMed] [Google Scholar]
- 87.Sun WL; Quizon PM; Yuan Y; Strauss MJ; McCain RJ; Zhan C-G; Zhu J, Mutational effects of human dopamine transporter at tyrosine88, lysine92, and histidine547 on basal and HIV-1 Tat-inhibited dopamine transport. Sci. Rep 2019, 9, 3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Quizon PM; Sun WL; Yuan Y; Midde NM; Zhan C-G; Zhu J, Molecular mechanism: the human dopamine transporter histidine 547 regulates basal and HIV-1 Tat protein-inhibited dopamine transport. Sci. Rep 2016, 6, 39048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Midde NM; Huang X; Gomez AM; Booze RM; Zhan C-G; Zhu J, Mutation of tyrosine 470 of human dopamine transporter is critical for HIV-1 Tat-induced inhibition of dopamine transport and transporter conformational transitions. J. Neuroimmune Pharmacol 2013, 8 (4), 975–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


