Abstract
The potato tuber starch trait is changed depending on the composition of amylose and amylopectin. The amount of amylopectin is determined by the activity of the starch branching enzymes SBE1, SBE2, and SBE3 in potato. SBE3, a homolog of rice BEI, is a major gene that is abundant in tubers. In this study, we created mutants of the potato SBE3 gene using CRISPR/Cas9 attached to the translation enhancer dMac3. Potato has a tetraploid genome, and a four-allele mutant of the SBE3 gene is desired. Mutations in the SBE3 gene were found in 89 of 126 transformants of potato plants. Among these mutants, 10 lines contained four mutant SBE3 genes, indicating that 8% efficiency of target mutagenesis was achieved. These mutants grew normally, similar to the wild-type plant, and yielded sufficient amounts of tubers. The potato starch in these tubers was similar to that of the rice BEI mutant. Western blot analysis revealed the defective production of SBE3 in the mutant tubers, suggesting that these transformants were loss-of-function mutants of SBE3.
Keywords: amylopectin, CRISPR/Cas9, potato (Solanum tuberosum L.), starch, starch-branching enzyme (SBE)
Introduction
Storage starch in plants consists of amylose and amylopectin, the composition ratio of which alters starch properties. In many plant species, mutants showing altered amylose contents are known. Granule-bound starch synthase (GBSS) is a major enzyme involved in amylose biosynthesis (Nelson and Rines 1962). Mutants lacking this enzyme produce amylose-free starch. Mutants lacking the GBSS gene in rice show a waxy phenotype (Sano 1984). Similar mutants are also known in other plant species, such as maize (Shure et al. 1983), barley (Rohde et al. 1988), and sorghum (Pedersen et al. 2005).
Amylopectin is synthesized in concert by the activities of soluble starch synthase (SS) and starch branching enzyme (BE) (Guan et al. 1995). Mutants deficient in starch branching enzymes show aberrations in amylopectin. A rice mutant lacking the starch branching enzyme BEIIb, known as ae, shows elongated amylose chains with an increase in amylose content (Nishi et al. 2001; Yano et al. 1985). The mutant lacking BEI showed no significant change in the apparent amylose content, but the structure of amylopectin was altered because of a decrease in long chains and an increase in short chains of amylopectin. In addition, the endosperm starch of this mutant displayed altered properties of gelatinization (Satoh et al. 2003).
Potato, Solanum tuberosum L., is one of the most important crops in the world. There have been many attempts to breed potato mutants showing altered starch traits. However, the traditional breeding of potato is quite difficult because potato has a strong vegetative reproduction phase in addition to its polyploidy. Especially, use of recessive alleles is challenging because of its polyploid nature. To date, some genetically modified potatoes showing desired traits have been established. To obtain amylose-free potato starch, a knockdown transformant was created by introduction of the antisense gene of GBSSI (Visser et al. 1991).
Recently, genome editing technology has been applied to potato. A mutant lacking the production of a glycoalkaloid has been created (Sawai et al. 2014). However, it is difficult to establish a deficient potato mutant because potato has a tetraploid genome. Therefore, to introduce mutations in all four alleles for each gene in the genome, a powerful genome editing tool is required. We found a translational enhancer, dMac3, that strongly elevates the translation efficiency of the downstream ORF (Aoki et al. 2014). We established an improved CRISPR/Cas9 system using dMac3 and applied it to create a potato mutant lacking GBSS activity (Kusano et al. 2018). We confirmed that this mutant shows the trait of low amylose content in tuber starch (Kusano et al. 2018).
Many kinds of starch branching enzymes are found in potato. Among them, SBE1, SBE2, and SBE3 are known as the major enzymes involved in the biosynthesis of amylopectin (Larsson et al. 1996; Van Harsselaar et al. 2017). SBE2 is the counterpart of rice BEIIb and is considered to be involved in short chain amylopectin. A transformant harboring an antisense gene to SBE2 has been produced, and it exhibits an amylose extension trait similar to the rice ae mutant (Jobling et al. 1999). A mutant in which genes encoding SBE1 and SBE2 have been disrupted was reported and resulted in the severe amylose extension phenotype (Tuncel et al. 2019).
Three genes were identified as homologs of the rice BEI gene. Two genes, SBE1.1 and SBE1.2, have been reported as potato homologs due to their homology to Arabidopsis SBE1. Sequences of the potato SBE1 and SBE3 genes also have been published. However, potato SBE1 is not homologous to rice BEI because of low sequence similarity. Potato SBE3 shows high similarity to rice BEI. In addition, SBE1.1 and SBE1.2 were identical to that of potato SBE3 except for their C-terminal regions. Currently, SBE3 is considered the homolog of rice BEI and is a major starch branching enzyme gene that is commonly expressed in developing potato tuber (Van Harsselaar et al. 2017).
In this study, to elucidate the function of potato SBE3, we created a genome-edited potato mutant in which mutations occur in all four alleles of the SBE3 gene and the evaluation of the traits of the obtained mutant.
Materials and methods
Plant material, tissue culture and plant growth conditions
The Solanum tuberosum L. cv. Sayaka was used. Potato plants were cultured on medium containing an MS basal salt mixture (Fuji Film Wako, Tokyo, Japan) (Murashige and Skoog 1962), 3% sucrose, and 0.3% Gelrite (Fuji Film Wako) with the pH adjusted to 6.0. Plants were grown under long-day conditions with 16 h light and 8 h dark at 23°C in a growth chamber. The regeneration of potato plants was performed as described previously (Ohnuma et al. 2020). Regenerated potato plants were cultivated using a growth room. They were grown under 16 h light (200–400 µmol m−2 s−1) and 8 h dark conditions at 23°C in a growth chamber. Tubers were harvested from wild-type and transformed potatoes, which were grown at 22°C under 16 h light and 8 h dark conditions in a growth chamber.
Plasmid construction
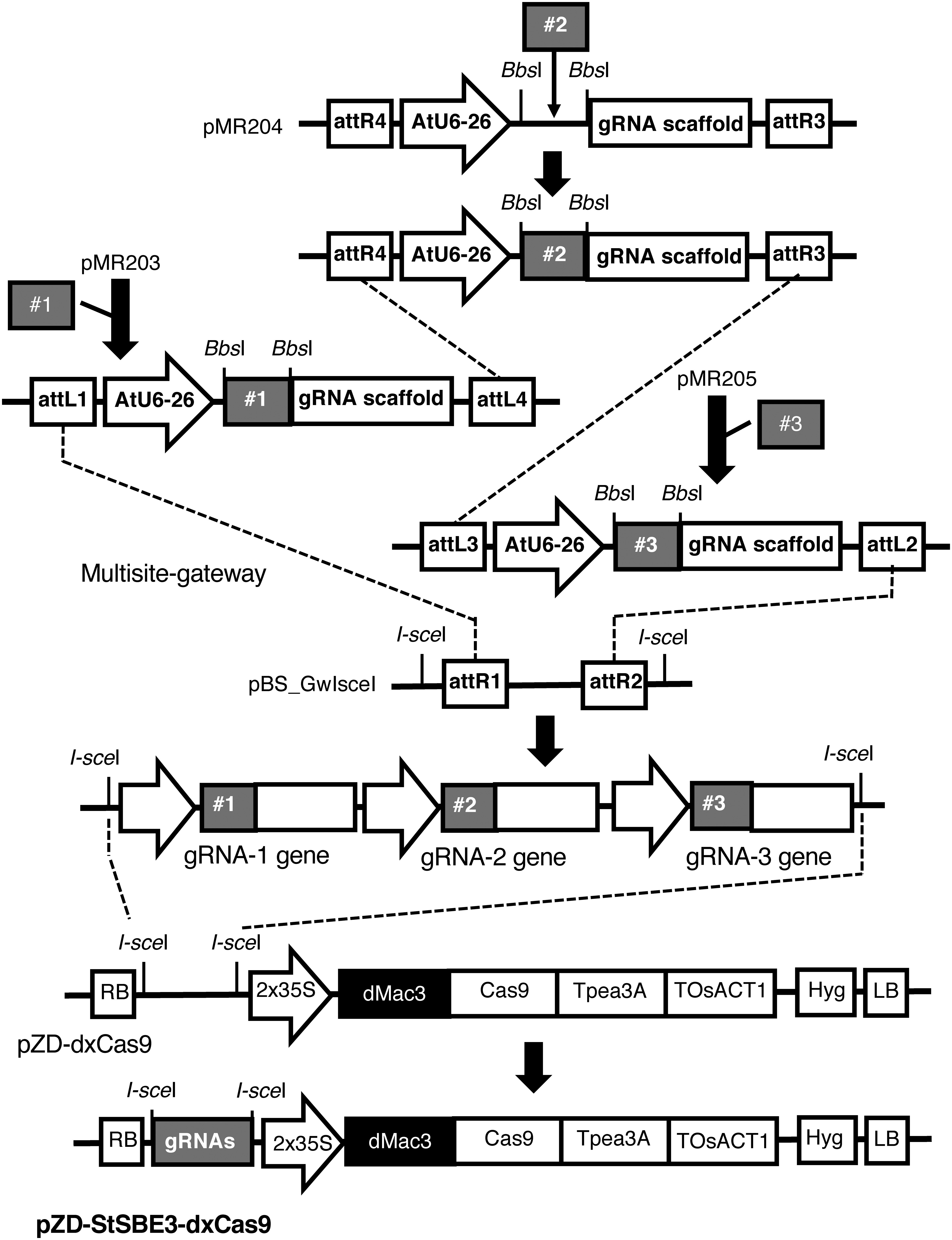
The plasmid for CRISPR/Cas9 was constructed essentially according to Kusano et al. (2018). For construction of the gRNA genes, the DNA fragments corresponding to the target sequences were chemically synthesized. The fragments were inserted into the BbsI sites of the appropriate guide RNA vector, pMR203, pMR204, and pMR205 (kindly provided by M. Endo). The resultant gRNA genes were located between the AtU6-26 promoter and the gRNA scaffold sequence. They were introduced into pBS_GwIsceI using the multisite-Gateway system (Invitrogen, Carlsbad, CA, USA). The pBS_GwIsceI plasmid had the regions for the Gateway attR1 and attR2 sites lying between the two I-SceI sites (Kusano et al. 2018). The resultant plasmid contained the fragments corresponding to the tandem gRNA genes between two I-SceI sites. The gene fragments were digested with I-SceI and then introduced into the I-SceI sites of the pZD-dxCas9 plasmid (Kusano et al. 2018) to generate CRISPR/Cas9 vectors containing three gRNA genes.
Plant transformation
Potato was transformed using the Agrobacterium-mediated procedure using the A. tumefaciens EHA105 strain according to Yamada et al. (2004) and Kusano et al. (2018). Stem internodes were isolated from germ-free potato plants grown in a clean environment for 3 weeks and cut into approximately 1 cm pieces. The pieces were infected with Agrobacterium harboring an appropriate plasmid. Callus induction and plant regeneration were performed on a plate of 3C5ZR medium (Sheerman and Bevan 1988) containing 0.3% Gelrite (Fuji Film Wako) supplemented with 5 mg l−1 hygromycin B (Fuji Film Wako) and cultured for 2 months. Regenerated plants were examined by detection of the transgene by PCR using the Cas9-S (5′-GGC GTA AGA ATA GAA TCT GTT AT-3′) and Cas9-T (5′-GAC AGC GCT ATC AGA TTT CCA A-3′) primers, which amplified part of the Cas9 gene sequence. Genome edited potato plants were cultivated in a green room.
Cleaved amplified polymorphic sequence (CAPS) analysis
Genomic DNA was prepared from potato leaves using the RED Extract N-Amp™ Plant PCR kit (Sigma-Aldrich, St. Louis, USA). The 1.0-kb region in the potato SBE3 gene, in which the sequences of the target sites of the guide RNAs were contained, was PCR-amplified using the StSBE3_CAPS_Fw (5′-CTA TGA GAC GGA TAG TTG AGA ATG TG-3′) and StSBE3_CAPS_Rv (5′-GGC CAG GTG CCA CCT AAA TG-3′) primers that were derived from the regions in introns 1 and 3. The amplified fragment was digested with BamHI. The wild-type gene has a BamHI site in the corresponding region. Therefore, we identified mutant alleles as those where the amplified fragment was no longer digested by BamHI. The number of mutant alleles among the four genes was estimated by the ratio of undigested fragments to digested fragments.
DNA sequence analysis
The 0.5-kb region of the potato SBE3 gene that contained the target sites of the guide RNAs was PCR-amplified using the StSBE3_seq_Fw (5′-TGA TAT AAA CTA ACT GTG GTG CA-3′) and StSBE3_seq_Rv (5′-GGA GTG TCA GTG GCA AAG CAA-3′) primers. Then, the amplified fragment was inserted into the pTA2 vector (TOYOBO, Osaka, Japan). The resultant plasmids were used for nucleotide sequence analysis.
Determination of the properties of tuber starch
Starch granules were prepared from potato tubers according to Noda et al. (2004). Sliced sections of potato tuber were dipped into 0.4% potassium iodide-0.12% iodide solution. Photographs were taken after washing with water. The amylose contents in the potato tubers were analyzed according to a previous paper (Noda et al. 2004). The data were statistically analyzed using Dunnett’s multiple comparison test.
Protein extraction, SDS-PAGE and western blot analysis
Sectioned tissues of potato tuber (150 mg) were frozen in liquid nitrogen and ground into a fine powder using an SK mill (Tokken Inc., Kashiwa, Japan). Then, 400 µl of the protein extraction buffer (50 mM Tris-HCl, pH 6.8, 8 M urea, 4% SDS, 20% glycerol, 1 mM DTT, 0.01% bromophenol blue) was added to the powdered tuber. The mixture was well combined and centrifuged at 20,000 g for 15 min at room temperature. The supernatant solution was collected as a crude protein fraction. This fraction (10 µl) was used for 7.5% SDS-polyacrylamide gel electrophoresis and subjected to western blot analysis using antiserum raised against rice BE1, which was prepared from developing rice seeds (Kawasaki et al. 1996). SDS-PAGE and western blot analysis were performed according to Wakasa et al. 2020. For this analysis, a 5,000-fold dilution of the antiserum was used. Protein interaction with the antibody was detected using the anti-rabbit IgG HPR-linked secondary antibody (1 : 10,000 dilution) (GE Healthcare, Chalfont Saint Giles, UK). Signals were detected with Pierce™ ECL Plus western blotting Substrate (Thermo Fishers Scientific, Waltham, MA, USA).
Results
Determination of the nucleotide sequences of the potato SBE3 gene and construction of the CRISPR/Cas9 gene targeted to this gene
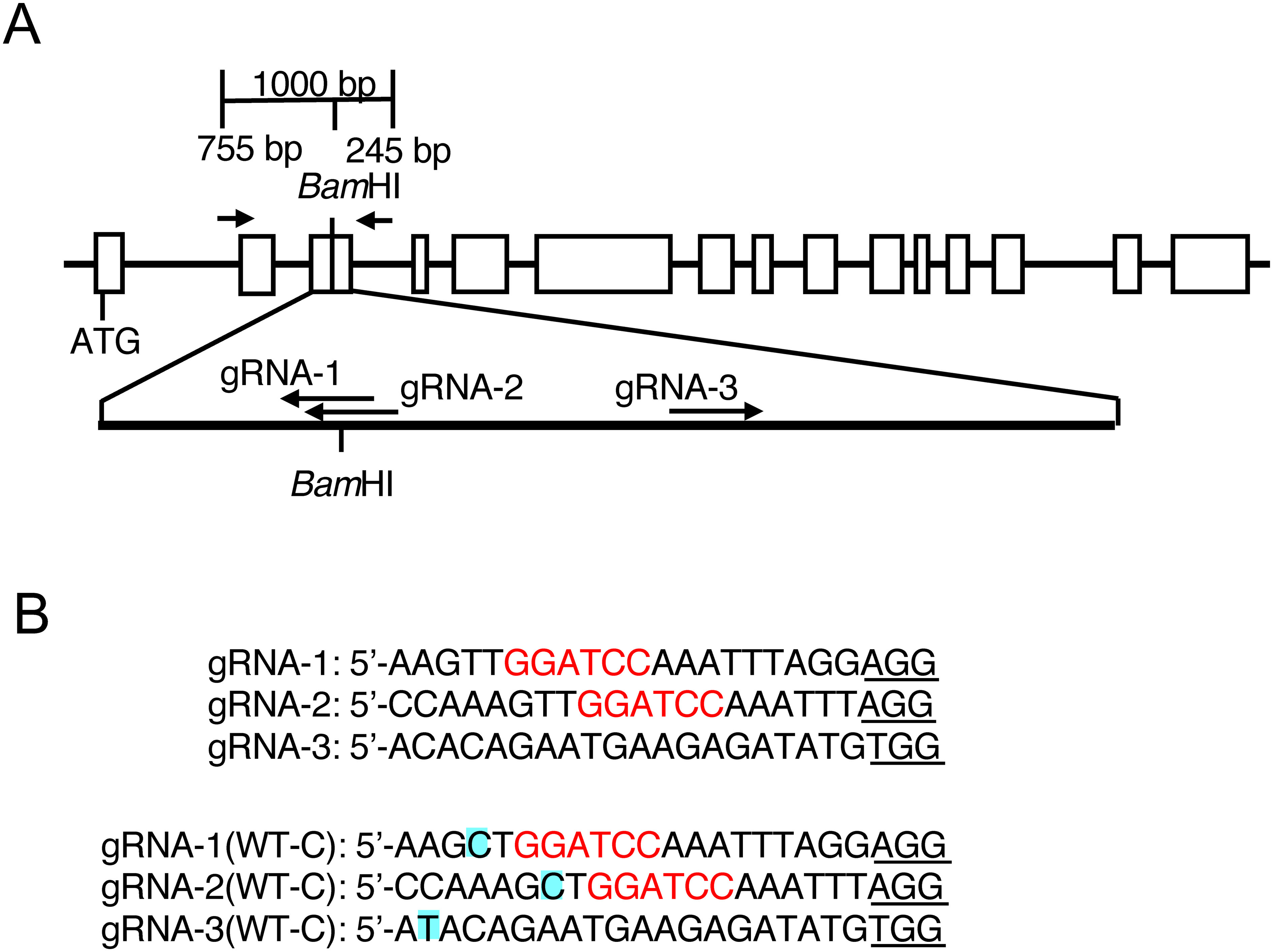
The nucleotide sequence of the potato SBE3 gene of Solanum phureja has been registered in the genome database SpudDB (Acc. no. PGSC0003DMG400009981). This gene consists of 15 exons and is predicted to encode a protein containing the transit peptide for translocation to amyloplasts (Van Harsselaar et al. 2017). The region encoding the mature protein started from the third exon. For target mutagenesis to the SBE3 gene of S. tuberosum, we selected three regions, gRNA-1, gRNA-2 and gRNA-3, corresponding to the sequences within the third exon of the SBE3 gene (Figure 1A). These regions were used for the generation of CRISPR/Cas9 guide RNAs. In the region of the target sequences, there was a BamHI site, which facilitated the detection of targeted mutations by CAPS analysis (Figure 1A).
Figure 1. Structure of the potato SBE3 gene and location of the gRNAs used for targeted mutagenesis. (A) Schematic representation of the SBE3 gene. Exons are indicated by boxes. Arrows in the upper panel indicate the position of the PCR primers that were used for amplification of the fragment for the CAPS analysis. The positions of the gRNAs are shown in the lower panel. (B) Nucleotide sequences of the regions corresponding to the gRNAs. The upper panel shows the nucleotide sequences of gRNA-1, gRNA-2, and gRNA-3 used for this experiment. The BamHI site is shown in red letters. The lower panel shows the corresponding nucleotide sequences in WT-C. Polymorphic nucleotides found in the WT-C genome are highlighted. PAM sequences are underlined.
It is considered that the nucleotide sequences of the SBE3 genes are largely different depending on the potato variety. In this study, we used the potato cultivar Sayaka, which is commonly used for genome editing. To determine the nucleotide sequence of the SBE3 gene of Sayaka, we analyzed the sequences around the target sites in the SBE3 gene. The corresponding region was PCR-amplified, and the nucleotide sequences were determined. We found that this region contained a polymorphism in three different nucleotide sequences, named WT-A, WT-B, and WT-C (Supplementary Figure S1). Among the sequences, the nucleotide sequence of WT-A was identical to the registered sequence in the database.
In 18 nucleotide sequences, WT-A, WT-B, and WT-C were repeatedly detected 2, 10, and 6 clones, respectively, suggesting that they may exist in a 1 : 2 : 1 proportion in the four alleles contained in the tetraploid genome of Sayaka. Of the three gRNA target sites, a single nucleotide difference was present in each of the gRNAs in WT-C (Figure 1B), whereas no difference was found in those in WT-B.
We have reported that dMac3 significantly enhanced the translation efficiency of downstream ORFs composed of 161 nt derived from the 5′UTR of OsMac3 mRNA (Aoki et al. 2014). Using this system, we established an improved CRISPR/Cas9 vector system, pZD-dxCas9, which enables effective genome editing of the potato GBSS gene (Kusano et al. 2018). For targeted mutagenesis of the potato SBE3 gene, guide RNAs were chemically synthesized and placed between the AtU6-26 promoter and the chimeric single-guide RNA (gRNA) scaffold (Li et al. 2013; Mali et al. 2013) (Figure 2). The resultant gRNA genes were introduced into the pZD-dxCas9 vector, resulting in the formation of CRISPR/Cas9 vectors containing tandemly located gRNA genes, pZD-StSBE3-dxCas9 (Figure 2).
Figure 2. Schematic representation of the construction of the CRISPR/Cas9 gene for SBE3. Chemically synthesized gRNAs, shown by #1, #2, and #3, were inserted into the vector plasmids pMR203, pMR204, and pMR205, respectively. The resultant gRNA genes were introduced into the intermediate vector pBS_GwIsceI, and they were transferred into the CRISPR/Cas9 vector pZD-dxCas9. AtU6-26: Arabidopsis thaliana U6 promoter, gRNA scaffold: the region for the gRNA scaffold, dMac3: dMac3 enhancer sequence, Cas9: coding region of the Cas9 nuclease, Tpea3A and TOsAct1: terminators of pea 3A and rice actin 1 genes, Hyg: hygromycin resistance gene, att: Gateway recombination sites, and RB and LB: right and left borders.
Creation of a genome edited potato
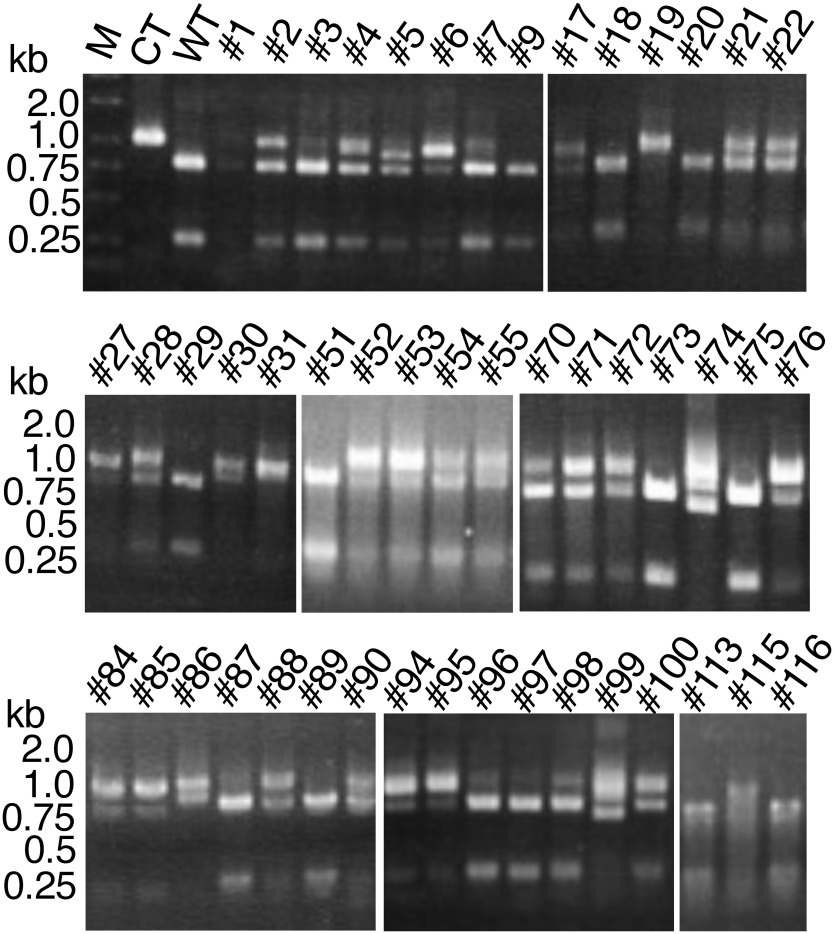
Using pZD-StSBE3-dxCas9, the CRISPR/Cas9 gene was introduced into the stems of potato plants. Among the regenerated potato transformants, mutants of the SBE3 gene were investigated. There is a BamHI site in or near the target sites, and therefore, the occurrence of targeted mutations can be detected by disruption of this BamHI site. The DNA fragment containing the target site was PCR-amplified, and CAPS analysis was carried out by BamHI cleavage.
Potato, S. tuberosum, has a tetraploid genome; thus, genome-editing events may occur on each of the four alleles. Introducing CRISPR/Cas9 obtained 126 potato transformants. Mutant genes were detected by the occurrence of a BamHI-uncleaved PCR-amplified fragment containing the target sites (Figure 3). The frequency of generated mutations was estimated by the ratio of the uncleaved fragments to the amplified fragments. CAPS analysis indicated that 89 (71%) transformants contained mutant genes that exhibited the uncleaved fragment after BamHI digestion (Table 1). Among them, ten transformants (8%) were suggested to be mutants that had mutations in all four alleles of the SBE3 gene (Table 1). This result suggests that site-specific mutation occurred at the target site in the potato cells in planta.
Figure 3. CAPS analysis of the regenerated potato plants. BamHI-digested PCR-amplified fragments of the representative lines are shown. Numbers with the prefix # indicate the names of transformant lines. M: size marker, CT: PCR-amplified fragment of the region around the target site in the wild-type gene, and WT: PCR-amplified and BamHI-digested WT fragment. Gaps are introduced between the lanes of the gel.
Table 1. Estimated allele numbers of mutations in the SBE3 gene in the transformants.
| Number of mutant alleles estimated | Transformants | |
|---|---|---|
| Number of mutants | Ratio of mutants (%) | |
| 0 | 37 | 29 |
| 1 | 25 | 20 |
| 2 | 34 | 27 |
| 3 | 20 | 16 |
| 4 | 10 | 8 |
| Total | 126 | 100 |
Sequence analysis of the target sites in the mutant alleles of the SBE3 gene
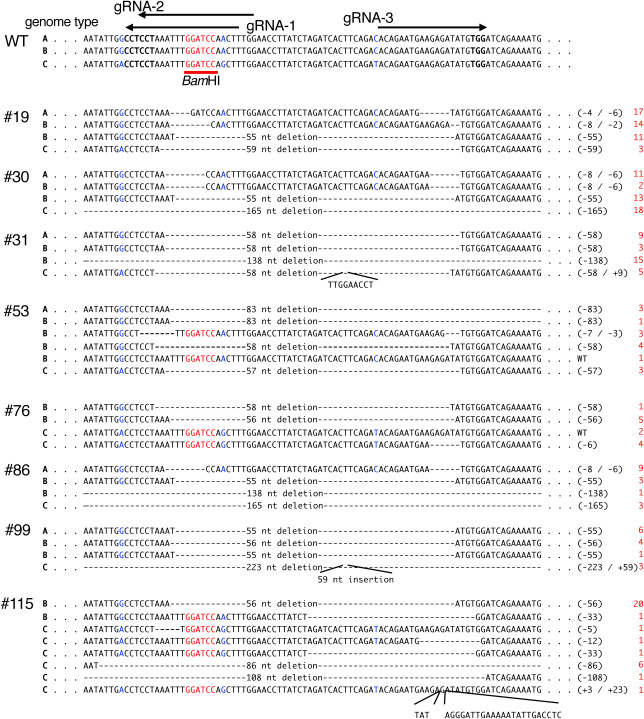
We analyzed the nucleotide sequences of the representative mutants that were considered to contain four or three mutant alleles. We detected four different nucleotide sequences in the transformed lines #19, #30, #31, #86, and #99, whereas no wild-type nucleotide sequence was detected in these lines, suggesting that they were four-allele mutants of SBE3 (Figure 4).
Figure 4. Nucleotide sequences around the target site in the potato SBE3 gene of the representative mutants. The wild-type nucleotide sequence (WT) is shown in the upper panel of the figure. Genome types A, B, and C indicate those from WT-A, WT-B, and WT-C, respectively. Nucleotide sequences containing the outside region are shown in Supplementary Figure S1. The gRNAs (gRNA-1, gRNA-2, and gRNA-3) are indicated above the figure. The region corresponding to the BamHI site is indicated by red letters. Polymorphic nucleotides between three genomes are shown in blue letters. Numbers with the prefix # indicate the individual transformants. Multiple DNA sequences that were detected were aligned. Gaps indicate a nucleotide deletion in the mutants. Nucleotide insertions are indicated in the appended lines. The nucleotide sequences that are indicated by gaps throughout the entire region in #30, #31, #86, #99, and #115 are shown in Supplementary Figure S2. Nucleotide numbers of deletions and insertions are summarized on the right side. Red-colored numbers in the right side showed the frequency of the nucleotide sequences detected.
Line #19 contained four different types of mutant sequences, which were considered to be derived from WT-A, WT-B, WT-B, and WT-C because these mutant sequences contained single nucleotide polymorphisms corresponding to these genomes. One mutant sequence contained the two deletion sites of 4 nt and 6 nt, resulting in a 10-nt deletion in total. The other sequences contained 8-nt and 2-nt (10-nt in total) deletions, a 55-nt deletion, and a 59-nt deletion. These results showed that line #19 contained mutations in four alleles of the SBE3 gene (Figure 4). Among them, two sequences were considered derived from the WT-B sequence, which was predicted to have two copies in the genome (Figure 4). These results suggest that frameshift mutations occurred in these genes (Figure 4).
Similarly, frameshift mutations were found in the SBE3 gene of the mutant lines #30, #31, #86, and #99. These lines also contained four different mutant genes corresponding to each of the three types of SBE3 gene alleles (Figure 4; Supplementary Figure S2). Thus, it is suggested that these lines contained four-allele mutations of the SBE3 gene.
Line #76 contained a wild-type nucleotide sequence along with the three mutant SBE3 sequences, indicating that this transformant was a three-allele mutant (Figure 4). Line #53 contained five mutant sequences in addition to a wild-type sequence (Figure 4). Line #115 showed 8 mutant sequences, but no wild-type sequence was detected (Figure 4; Supplementary Figure S2).
Analysis of storage starch in the mutant tubers
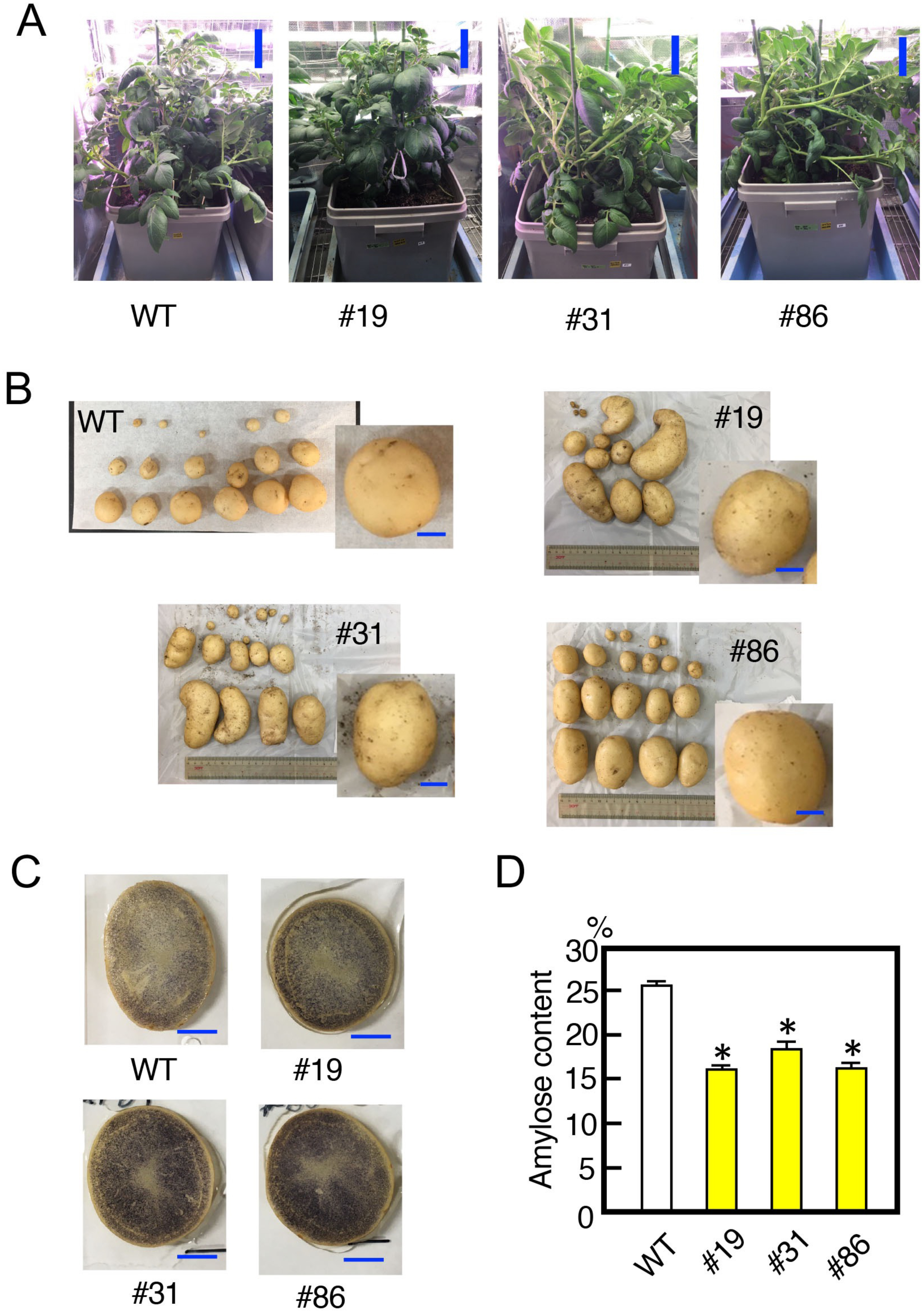
The mutant lines were cultivated. These plants grew normally and showed a morphology similar to that of the wild type during vegetative growth (Figure 5A). They produced normally shaped tubers similar to the wild-type plant (Figure 5B). Iodine staining assays showed that the tubers of these mutants stained strongly, similar to the wild-type plants (Figure 5C). From these mutant tubers starch was prepared. An analysis of the amylose content revealed a significant reduction in the amylose content of the mutants compared with that of wild-type starch (Figure 5D).
Figure 5. Characteristics of potato SBE3 mutants. (A) Shapes of the sbe3 mutants grown in a greenhouse. Plants were cultured for two months. Numbers with the prefix # indicate the individual transformants. WT, wild-type plant. Bars indicate 20 cm. (B) Shapes of the tubers of the mutant lines. Tubers generated from each mutant line are shown. A representative image is highlighted on the right side of each panel. Bars indicate 2 cm. (C) Iodine staining of potato tuber sections. Images were taken at 1.5 h after staining treatment. Bars indicate 2 cm. (D) Amylose content in tuber starch prepared from sbe3 mutants and wild-type plants. Values show the average of triplicate measurements. Error bars indicate the standard errors (n=3). Asterisks indicate significant differences in the values of transformants compared to that of wild type at p<0.05.
Detection of SBE3 in the mutant lines
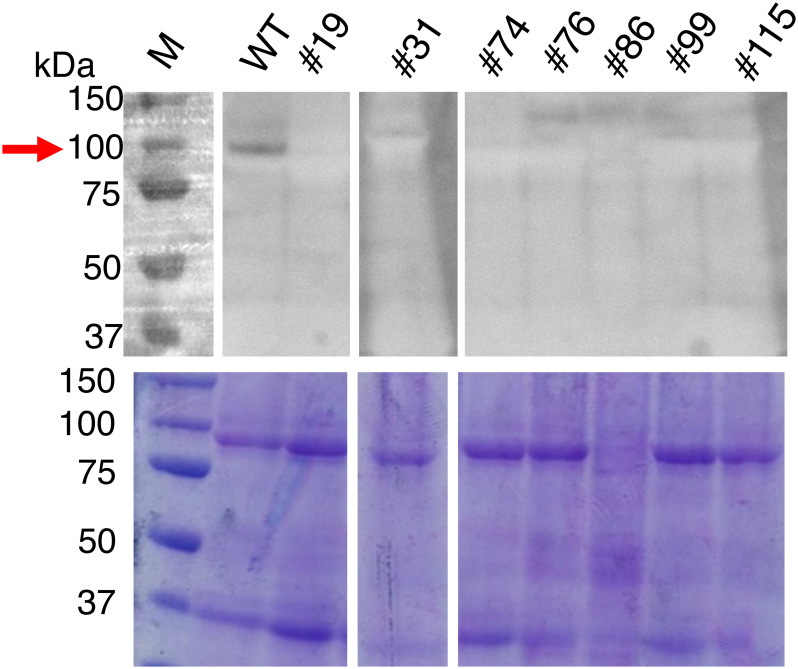
Potato SBE3 is considered the homolog of rice BEI (Supplementary Figure S3) and could be immunologically detected by the antiserum against rice BEI. To detect the SBE3 protein in tubers, we prepared crude protein fractions from the tubers that were the four-allele mutant lines and compared them with those from the wild-type plant. When the rabbit antiserum raised against rice BEI was used for western blot analysis, a protein of approximately 100 kD in molecular size was specifically detected in the protein fraction of the wild-type tuber (Figure 6), suggesting that this protein corresponded to potato SBE3. In the tubers of the four allele mutant lines #19, #31, #99, and #115, no 100-kD protein was detected. This suggests that SBE3 was deficient in these mutants (Figure 6). These results indicated that these mutants lacked the production of SBE3.
Figure 6. Detection of potato SBE3 in the tubers. Total protein was prepared from the tubers of the mutant lines and WT. Proteins were separated by SDS-PAGE (lower panel), and SBE3 was detected by western blot analysis using a rabbit antiserum raised against rice BE1 (upper panel). Gaps are introduced between the lanes of the gel. An arrow indicates the band corresponding to SBE3. Size markers are shown to the left. WT: wild-type, Numbers with the prefix #: mutant lines.
Discussion
In this study, we created a potato mutant in which the SBE3 gene was disrupted using a modified CRISPR/Cas9 system containing three gRNAs corresponding to the region in the third exon of the SBE3 gene (Figure 1). Genome editing is a newly developed technology using a sequence-specific artificial nuclease to enable modification of a specific gene (Osakabe and Osakabe 2015). Whereas the conventional breeding method takes a long time to obtain a variety possessing a desired trait, genome editing technology may facilitate rapid establishment of an appropriate mutant with a desired trait (Xiong et al. 2015). CRISPR/Cas9 is a powerful tool to induce a mutation at the target site with a high editing efficiency (Ménoret et al. 2013; Remy et al. 2014).
The transformation of potato has been established using a cultivar, Sayaka, that is commonly used for the creation of genetically modified plants as well as genome-edited plants (Kusano et al. 2016, 2018). Because potato has a tetraploid genome, it is necessary to modify all four alleles of the target gene. Previous studies have reported that efficient genome editing is achieved by a modified CRISPR/Cas9 system assembled with the translational enhancer dMac3 along with three guide RNAs derived from a region close to or inside the target gene (Kusano et al. 2018). We used gRNAs that were derived from the region encoding the mature protein downstream of the transition signal region and expected the loss of function of the SBE3 gene in the mutants.
The nucleotide sequences of the genes of S. tuberosum cv. Sayaka have not been registered in the database. The nucleotide sequences of the guide RNAs were taken based on the registered nucleotide sequences of another Solanum species, S. phureja. The structures of the genes in potato cultivars are known to be largely divergent. To identify these gRNA sequences to be included in the SBE3 gene in the Sayaka genome, we analyzed the nucleotide sequences of the region containing the third exon. We revealed that there were three different nucleotide sequences, WT-A, WT-B, and WT-C (Figure 4; Supplementary Figure S1). Between these genome sequences, WT-C contained many single nucleotide polymorphisms. The nucleotide sequences of the guide RNAs had some sequence differences (Figure 1). Although gRNA-1 and gRNA-3 contained unmatched nucleotides in the 5′-region to the WT-C sequence, other regions were complementary to them. gRNA-2 seemed to be inadequately because it contained unmatched nucleotide in the central region, but the other gRNAs are considered to have functioned to introduce a mutation even in the WT-C genome. We presume that they functioned as target-specific artificial nucleases in this case because mutant genes were generated on the WT-C genome.
We created a CRISPR/Cas9 construct containing these gRNAs and obtained many transformants showing multiple mutations in the SBE3 gene (Figure 3). Ten transformants exhibited mutations in all four alleles. The percent of four allele mutants was 8% of all transformants (Table 1). This result revealed that our CRISPR/dMac3-Cas9 system worked efficiently in the induction of targeted mutagenesis of the SBE3 gene.
The nucleotide sequences of the SBE3 gene revealed multiple mutations. These mutants contained indel mutations in the SBE3 gene. Among them, mutant lines #19, #30, #31, #86, and #99 contained four mutant and no wild-type sequences. These results confirm that they were the four-allele mutants. Because frameshift mutations occurred in these mutant SBE3 genes, these mutant genes were considered to have defective SBE3 activity (Figure 4). In lines #53 and #115, more than four mutant gene sequences were detected (Figure 4), suggesting chimeric mutant lines.
Potato SBE3 is a homolog of rice BEI. The antiserum raised against rice BEI also detected potato SBE3 (Figure 6). Western blot analysis showed defective production of SBE3 in the tubers of four-allele mutants, suggesting that they were mutants with a loss of function of SBE3 (Figure 6).
The rice mutant lacking BEI (sbe1 mutant) shows an aberrant storage starch module, in which the structure of amylopectin is quite different from that of the wild-type strain, as evidenced by an increased degree of iodine staining and alteration of the chain length distribution but no significant difference in the amylose content (Satoh et al. 2003). There could be similarities between the starch trait caused by the potato sbe3 mutant to that caused by the rice sbe1 mutant. The potato sbe3 mutant showed strong iodine staining of the tuber starch along with a significantly reduced amylose content (Figure 5). We have reported that the rice flo2 mutant, which markedly decreased the BE1 activity, showed a decrease in amylose content (Kawasaki et al. 1996). This suggests that properties of starch are influenced by many factors. These results suggest that the potato sbe3 mutant showed an altered storage starch trait as in the rice sbe1 mutant.
To date, there has been no sbe3 mutant potato. The four-allele mutants of the SBE3 gene (sbe3 mutant) that we created grew normally and produced sufficient amounts of tubers with a morphology similar to that of the wild type (Figure 5). Such a mutant potato is expected to contribute determining the functions of the genes involved in starch biosynthesis. It is suggested that the sbe3 mutant may be applied in agriculture as a potato cultivar.
Acknowledgments
We thank Takahiro Asahi, Yukino Okubo, Yuna Akatsu, Karin Hamada, and Ryo Morozumi for technical assistance, and Toshiya Muranaka, Shuhei Yasumoto, Naoyuki Umemoto, and Masaharu Mizutani for valuable discussions. This work was supported in part by the administration of individual commissioned project study (Development of new varieties and breeding materials in crops by genome editing, the Ministry of Agriculture, Forestry and Fisheries, Japan (H.S. and K.A.).
Abbreviations
- PCR
polymerase chain reaction
- gRNA
guide RNA
Supplementary Data
References
- Aoki H, Teramura H, Schepetilnikov M, Ryabova LA, Kusano H, Shimada H (2014) Enhanced translation of the downstream ORF attributed to a long 5′ untranslated region in the OsMac1 gene family members, OsMac2 and OsMac3. Plant Biotechnol 31: 221–228 [Google Scholar]
- Guan H, Kuriki T, Sivak M, Preiss J (1995) Maize branching enzyme catalyzes synthesis of glycogen-like polysaccharide in glgB-deficient Escherichia coli. Proc Natl Acad Sci USA 92: 964–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobling SA, Schwall GP, Westcott RJ, Sidebottom CM, Debet M, Gidley MJ, Jeffcoat R, Safford R (1999) A minor form of starch branching enzyme in potato (Solanum tuberosum L.) tubers has a major effect on starch structure: Cloning and characterization of multiple forms of SBE A. Plant J 18: 163–171 [DOI] [PubMed] [Google Scholar]
- Kawasaki T, Mizuno K, Shimada H, Satoh H, Kishimoto N, Okumura S, Ichikawa N, Baba T (1996) Coordinated regulation of the genes participating in starch biosynthesis by the rice Floury-2 locus. Plant Physiol 110: 89–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano H, Onodera H, Kihira M, Aoki H, Matsuzaki H, Shimada H (2016) A simple Gateway-assisted construction system of TALEN genes for plant genome editing. Sci Rep 6: 30234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano H, Ohnuma M, Mutsuro-Aoki H, Asahi T, Ichinosawa D, Onodera H, Asano K, Noda T, Horie T, Fukumoto K, et al. (2018) Establishment of a modified CRISPR/Cas9 system with increased mutagenesis frequency using the translational enhancer dMac3 and multiple guide RNAs in potato. Sci Rep 8: 13753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson CT, Hofvander P, Khoshnoodi J, Ek B, Rask L, Larsson H (1996) Three isoforms of starch synthase and two isoforms of branching enzyme are present in potato tuber starch. Plant Sci 117: 9–16 [Google Scholar]
- Li JF, Norville JE, Aach J, McCormack M, Zhang D, Bush J, Church GM, Sheen J (2013) Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol 31: 688–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM (2013) RNA-guided human genome engineering via Cas9. Science 339: 823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménoret S, Fontanière S, Jantz D, Tesson L, Thinard R, Rémy S, Usual C, Ouisse LH, Fraichard A, Anegon I (2013) Generation of Rag1-knockout immunodeficient rats and mice using engineered meganucleases. FASEB J 27: 703–711 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog FA (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15: 473–497 [Google Scholar]
- Nelson OE, Rines HW (1962) The enzymatic deficiency in the waxy mutant of maize. Biochem Biophys Res Commun 9: 297–300 [DOI] [PubMed] [Google Scholar]
- Nishi A, Nakamura Y, Tanaka N, Satoh H (2001) Biochemical and genetic analysis of the effects of Amylose-Extender mutation in rice endosperm. Plant Physiol 127: 459–472 [PMC free article] [PubMed] [Google Scholar]
- Noda T, Tsuda S, Mori M, Takigawa S, Matsuura-Endo C, Saito K, Mangalika WHA, Hanaoka A, Suzuki Y, Yamauchi H (2004) The effect of harvest date on starch properties in various potato cultivars. Food Chem 86: 119–125 [Google Scholar]
- Ohnuma M, Teramura H, Shimada H (2020) A simple method to establish an efficient medium suitable for potato regeneration. Plant Biotechnol 37: 25–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe Y, Osakabe K (2015) Genome editing with engineered nucleases in plants. Plant Cell Physiol 56: 389–400 [DOI] [PubMed] [Google Scholar]
- Pedersen JF, Bean SR, Graybosch RA, Park SH, Tilley M (2005) Characterization of waxy grain sorghum lines in relation to granule-bound starch synthase. Euphytica 144: 151–156 [Google Scholar]
- Remy S, Tesson L, Menoret S, Usal C, Cian AD, Thepenier V, Thinard R, Baron D, Charpentier M, Renaud JB, et al. (2014) Efficient gene targeting by homology-directed repair in rat zygotes using TALE nucleases. Genome Res 24: 1371–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde W, Becker D, Salamini F (1988) Structural analysis of the waxy locus from Hordeum vulgare. Nucleic Acids Res 16(14B): 7185–7186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano Y (1984) Differential regulation of waxy gene expression in rice endosperm. Theor Appl Genet 68: 467–473 [DOI] [PubMed] [Google Scholar]
- Satoh H, Nishi A, Yamashita K, Takemoto Y, Tanaka Y, Hosaka Y, Sakurai A, Fujita N, Nakamura Y (2003) Starch-branching enzyme I-deficient mutation specifically affects the structure and properties of starch in rice endosperm. Plant Physiol 133: 1111–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawai S, Ohyama K, Yasumoto S, Seki H, Sakuma T, Yamamoto T, Takebayashi Y, Kojima M, Sakakibara H, Aoki T, et al. (2014) Sterol side chain reductase 2 is key enzyme in the biosynthesis of cholesterol, the common precursor of toxic steroidal glycoalkaloids in potato. Plant Cell 26: 3763–3774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheerman S, Bevan MW (1988) A rapid transformation method for Solanum tuberosum using binary Agrobacterium tumefaciens vectors. Plant Cell Rep 7: 13–16 [DOI] [PubMed] [Google Scholar]
- Shure M, Wessler S, Fedoroff N (1983) Molecular identification and isolation of the waxy locus in maize. Cell 35: 225–233 [DOI] [PubMed] [Google Scholar]
- Tuncel A, Corbin KR, Ahn-Jarvis J, Harris S, Hawkins E, Smedley MA, Harwood W, Warren FJ, Patron NJ, Smith AM (2019) Cas9-mediated mutagenesis of potato starch-branching enzymes generates a range of tuber starch phenotypes. Plant Biotechnol J 17: 2259–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Harsselaar JK, Lorenz J, Senning M, Sonnewald U, Sonnewald S (2017) Genome-wide analysis of starch metabolism genes in potato. BMC 18: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser RGF, Somhorst I, Kuipers GJ, Ruys NJ, Feenstra WJ, Jacobsen E (1991) Inhibition of the expression of the gene for granule-bound starch synthase in potato by antisense constructs. Mol Gen Genet 225: 289–296 [DOI] [PubMed] [Google Scholar]
- Wakasa Y, Kasai A, Yamazaki M, Tabei Y, Tsuyama M, Igarashi T, Okazaki T, Yamamoto K, Fujihara H, Kanno A, et al. (2020) Rapid analysis of GBSS1 and Vinv genes expressed in potato tubers using microtubers produced in liquid culture medium. Plant Cell Rep 39: 1415–1424 [DOI] [PubMed] [Google Scholar]
- Xiong JS, Ding J, Li Y (2015) Genome-editing technologies and their potential application in horticultural crop breeding. Hortic Res 2: 15019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Tozawa Y, Hasegawa H, Terakawa T, Ohkawa Y, Wakasa K (2004) Use of a feedback-insensitive α subunit of anthranilate synthase as a selectable marker for transformation of rice and potato. Mol Breed 14: 363–373 [Google Scholar]
- Yano M, Okuno K, Kawakami J, Satoh H, Omura T (1985) High amylose mutants of rice, Oryza sativa L. Theor Appl Genet 69: 253–257 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.