Abstract
Trogocytosis is a fast, cell–cell contact-dependent uptake of membrane patches and associated molecules by one cell from another. Here, we report our investigation of trogocytosis of TYRO3, a cell membrane protein, from tumor target cells to natural killer (NK) cells and the associated functional consequences for NK cells. We found that although NK cells did not express endogenous TYRO3 on the cell surface, activated NK cells rapidly acquired TYRO3 from tumor cells via trogocytosis in vitro and in vivo. NK cells that acquired TYRO3, which we termed TYRO3+ NK cells, had significantly enhanced cytotoxicity and IFNγ production as well as higher expression of some activated surface markers compared with TYRO3− NK cells. Furthermore, the activation status of NK cells and TYRO3 expression levels on donor cells, either endogenous or ectopic, positively correlated with trogocytosis levels. When the antigen-presenting cell (APC) K562 leukemia cell line, a feeder cell line to expand NK cells, overexpressed TYRO3, TYRO3 was transferred to NK cells via trogocytosis, which improved NK-cell proliferation ex vivo. This provides a strategy to manufacture NK cells or their engineered counterparts such as chimeric antigen receptor (CAR) NK cells for the treatment of cancer or infectious diseases.
Keywords: TYRO3, trogocytosis, NK cell, tumor cell, proliferation
Introduction
Natural killer (NK) cells belong to the innate immune system and are considered the first line of defense against tumors and viruses (1–3). NK-cell function is regulated by the balance between activating and inhibitory signals. Upon encountering normal cells, NK cells interact with their cognate MHC class I molecules, which provides inhibitory signals to override activating signals (4). In contrast, when NK cells encounter cells lacking self-MHC class I molecules, they exert strong cytotoxic effects via multiple mechanisms, including releasing cytotoxic granules containing perforins and granzymes (5). NK cells can recognize and spontaneously eliminate tumor cells that lack MHC class I molecules, making them an important component of antitumor immunity (5,6).
Trogocytosis is the fast, cell–cell contact-dependent uptake of membrane patches and associated molecules by one cell from another (7). This process is observed for tumor cells (8) as well as several different immune cells, including T cells (9–11), B cells (12), NK cells (13–16), and dendritic cells (17). When an NK cell interacts with a target cell, an immune synapse forms strong enough to allow for the exchange of membrane molecules from one cell to the other. For example, human leukocyte antigen (HLA)-G can be transferred from target cells to NK cells via trogocytosis, and this transfer inhibits the cytolytic function of NK cells (15). The chemokine receptor CCR7 can also be transferred from donor cells onto the surface of NK cells via trogocytosis, enhancing NK-cell homing to lymph nodes (16).
TYRO3 is a receptor tyrosine kinase that belongs to the TAM (TYRO3, AXL, and MERTK) receptor family (18). Many cells of the innate immune system express TAM receptors, including macrophages (19) and dendritic cells (20). Although they are highly expressed on the surface of resting murine NK cells, their surface expression on resting human NK cells is negligible (21,22). TYRO3 also is expressed in different types of cancers and plays important roles in cancer progression. Therefore, the TAM family receptors are considered potential therapeutic targets (23–25).
Here we showed that TYRO3 could be transferred from tumor cells to NK cells with a dependence on cell-cell contact, fast transfer kinetics, and a limited half-life of TYRO3 following its acquisition. We also found NK cells that acquired TYRO3 (TYRO3+) had significantly enhanced cytotoxicity, IFN-γ production, and proliferation in vitro, as well as higher expression of activated surface markers, compared with TYRO3− NK cells. We generated a feeder cell line that expressed TYRO3, thereby allowing human NK cells to acquire TYRO3 via trogocytosis, which improved their proliferation and expansion ex vivo.
Materials and Methods
Isolation and expansion of primary human NK cells
Blood cones were obtained from the City of Hope National Medical Center Blood Bank under institutional review board–approved protocols. All healthy donors provided written informed consent, which followed the ethical guidelines of the Declaration of Helsinki. To isolate NK cells, peripheral blood was mixed and incubated with RosetteSep™ human NK-cell enrichment cocktail (StemCell Technologies, Cat. #15065) for 20 min at room temperature. The blood sample was diluted with DPBS (Gibco, Cat. #14190144) and added into a tube containing Ficoll-Paque (Cytiva, Cat. #17144003). After 18-minute centrifuge without brake, buffer layer was aspirated and treated with red blood cell lysis buffer. The isolated NK cells were directly used for experiments or after being expanded with K562 feeder cells expressing mbIL21 and 4–1BBL (APC K562) with or without co-expressing TYRO3 (APC K562Tyro3) in the presence of IL2 (50 IU/ml, Roche Inc, Cat. #1035–0490). Prior to use for NK expansion, the K562 feeder cells were inactivated by mitomycin (10 ug/ml; Sigma-Aldrich, Cat. #M4287) for 2 hours.
Cell lines
K562 cells were purchase from American Type Culture Collection (ATCC) in 2018. Molm-13, and U937 cells were purchased from Leibniz Institute DSMZ in 2018. Jurkat cells were obtained from Dr. Stephen J. Forman’s lab in 2019. 721.221 cells were a generous gift from Professor J. Miguel Lopez-Botet Arbona at the Universitat Pompeu Fabra, Barcelona. APC K562 cells were obtained from CytoImmune Therapeutics Inc. All above cell lines were cultured in RPMI (Gibco, Cat. #11875119) with 10% heat-inactivated FBS (Gibco, Cat. #16140071). The GP2–293 packaging cell line was purchased from Takara Bio in 2018 and cultured in DMEM supplemented with 1% GlutaMax (Gibco, Cat. #35050061) and 10% FBS. All cells were incubated at 37℃ in a 5% CO2 humidified incubator. No authentication of these cell lines was performed after they were purchased or received. Cell morphology and growth characteristics were monitored during the study and compared with published reports to ensure their authenticity. All cell lines were routinely tested for the absence of mycoplasma using the MycoAlert Mycoplasma Detection Kit from Lonza (Cat. #LT07–318). All cell lines used in experiments were cultured for less than 10 passages.
TYRO3-knockout cell line
TYRO3-knockout K562 cells (K562Tyro3-KO) were generated using CRISPR/Cas9 knockout plasmids (Cat. #sc-401412 and #sc-401412-HDR), UltraCruz transfection reagent (Cat. #sc-395739), and plasmid transfection medium (Cat. #sc-108062), all of which were purchased from Santa Cruz and used according to the manufacturer’s instructions. K562 cells were co-transfected with a homology-directed DNA repair (HDR) plasmid, which incorporates red fluorescent protein (RFP), to select cells containing a successful Cas9-induced site-specific TYRO3 knockout in genomic DNA. The transfected cells were sorted for GFP+RFP+ using FACSAria Fusion (BD Biosciences). Knock-out of TYRO3 clones was confirmed by flow cytometry.
Plasmids generation and retrovirus transduction
The full-length human TYRO3 DNA sequence or its fusion protein with EGFP at the C terminal by a G4S linker (Gly-Gly-Gly-Gly-Ser) were cloned into an RRV plasmid obtained from CytoImmune Therapeutics Inc. to generate the RRV-Tyro3 or the RRV-Tyro3-EGFP plasmids, respectively. The full-length human TYRO3 DNA sequence with K550A mutation was cloned with the Q5 site-directed mutagenesis kit from NEB Inc. (Cat. #E0554) to generate the RRV-Tyro3_K550A plasmid. The sequence with the extracellular domain (ED) of TYRO3 and the PDGFRB transmembrane domain were cloned into the RRV plasmid to generate the RRV-ED-Tyro3 plasmid. The coding sequence of IL15 was cloned to the RRV plasmid to generate the RRV-sIL15 plasmid.
For generating retrovirus to infect cells, GP2–293 cells were cultured until a confluence of 70–80% and then transfected with a retrovirus vector expressing one of the RRV-Tyro3 related plasmids with an envelope plasmid using Lipofectamine 3000 Reagent according to the manufacturer’s instructions (Thermo Fisher Scientific, Cat. #L3000001). The culture supernatant containing the retrovirus was harvested at 48 h post-transfection and filtered. To generate Molm-13Tyro3-OE, JurkatTyro3-OE, APC K562Tyro3, APC K562Tyro3-EGFP, APC K562ED-Tyro3, APC K562Tyro3_K550A, and sIL15 NK cells, retroviral transduction with RetroNectin (Takara Bio, Cat. #T100B) was performed according to a manufacturer protocol.
51Cr-release cytotoxicity assay
51Cr-release cytotoxicity assays were performed as described previously (26). K562 or 721.221 target cells were labeled with 51Cr (Wallac, PerkinElmer, Cat. #NEZ30001MCSBR1) for 1 hour at 37°C. Cells were washed and co-incubated with effector cells (TYRO3− and TYRO3+ NK cells) in triplicates in a 96-well U-bottom plate at multiple effector/target (E/T) ratios for 4 h at 37℃ in 5% CO2. The supernatant was harvested from each well, transferred into a 96-well Luma plate, and analyzed using a Microbeta scintillation counter (Wallac, PerkinElmer).
Reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA was isolated from cells with an RNeasy Mini Kit (Qiagen, Cat. #74034). Complementary DNA (cDNA) was generated from 200 ng RNA using Moloney murine leukemia virus (M-MLV) reverse transcriptase (Invitrogen, Cat. #11756050) and amplified by quantitative (q)RT-PCR with SYBR Green (Thermo Fisher Scientific, Cat. #4472908) or RT-PCR with Q5 PCR Master Mix (NEB, Cat. #M0492) and gene-specific primers. qRT-PCR was performed in triplicates with the reaction protocol of 95°C for 1 min, followed by 40 cycles of 95°C for 10 s, 60°C for 30 s, and 72°C for 30 s, using QuantStudio 12K Flex Real-Time PCR System (Thermo Fisher Scientific). RT-PCR was performed with the reaction protocol of 98°C for 30 s, followed by 25 or 30 cycles of 98°C for 10 s, 60°C for 30 s, and 72°C for 30 s, using C1000 Touch Thermal Cycler (Bio-Rad). Relative amplification values of qRT-PCR were calculated by the 2-ΔΔCt method, normalized to GAPDH or 18S rRNA. Primer sequences are included in Supplemental Table S1.
Immunoblotting assay
Cells were harvested and suspended in RIPA lysis buffer (Thermo Fisher Scientific, Cat. # 89900) on ice for 20 min. An equal amount of protein was resolved by a 5–15% Criterion TGX gel (Bio-Rad) and then transferred onto the nitrocellulose (NC) or polyvinylidene difluoride (PVDF) membrane (Thermo Fisher Scientific). The membrane was incubated with a primary antibody at 4℃ overnight and an IRDye secondary antibody (Li-COR Biosciences) for one hour at room temperature. The immunoblots were visualized with Odyssey CLx Imager. Densitometric analysis was performed to quantify the intensity of gel bands with Image J. Primary antibodies used were anti-GZMB (Cell Signaling, Cat. #17215), anti-pSTAT3Ser727 (Cell Signaling, Cat. #9134), anti-pAKTSer473 (Cell Signaling, Cat. #4060), anti-P-p44/42 (Cell Signaling, Cat. #4370), anti-Perforin (R&D systems, Cat. #MAB8011) and anti-Actin (Millipore Sigma, Cat. #MAB1501). Secondary antibodies were IRDye 800CW goat anti-Rabbit IgG (LI-COR Biosciences, Cat. #926–32211) and IRDye 680RD goat anti-Mouse IgG (LI-COR Biosciences, Cat. #926–687070).
Degranulation assays
Primary NK cells were co-cultured with unmodified K562 cells and IL2 (150 IU/ml) for 24 h. Then, NK cells were plated in a 96-well round-bottom plate with anti-human CD107a (BD Biosciences) and 1 mg/ml GolgiPlug (BD Biosciences, Cat. #555029) for a 4-h incubation in RPMI with 10% heat-inactivated FBS at 37℃ in 5% CO2. Then, cells were stained with anti-CD56 and analyzed by flow cytometry. Information about the antibodies used can be found in Supplemental Table S1.
Staining for flow cytometry or sorting
Information on the antibodies used for flow cytometry can be found in Supplemental Table S1. Cells were stained with monoclonal antibodies at room temperature for 20 min and washed with FACS buffer prior to analysis using a Fortessa X 20 flow cytometry (BD Biosciences). For IFNγ intracellular flow cytometric analysis, 1 mg/ml GolgiPlug was added for 4 h before cell harvest. Then, cells were permeabilized and fixed using a Cytofix/Cytoperm Fixation/Permeabilization Solution Kit (BD Biosciences, Cat. #554714). Data were analyzed using Flowjo V10 software (Tree Star, Ashland, OR, USA).
NSG xenograft model
NOD-SCID-IL2Rγ–/– (NSG) mice were purchased from the Jackson Laboratory and housed at the City of Hope Animal Facility. Primary human NK cells were incubated with APC K562 cells or APC K562Tyro3 cells, which were inactivated by mitomycin (10 ug/ml) prior to use, in the presence of IL2 (50 IU/ml) with R10 medium (RPMI + 10% FBS) for 4 days. The NK cells were then transduced with soluble IL15 (sIL15) retrovirus for 48 h, followed by being incubated with inactivated APC K562 cells or APC K562Tyro3 cells in the presence of IL2 (50 IU/ml) for another 7 days prior to being harvested and frozen for mouse injection. For in vivo studies, on day 0, mice were intravenously (i.v.) injected with 1×106 K562-luciferase (K562_Luc) cells (27) and then i.v. treated with 10×106 aforementioned sIL15 NK cells expanded with APC K562 (APC K562_sIL15 NK) or APC K562Tyro3 (APC K562Tyro3_sIL15 NK). On day 1, mice were i.v. treated with a 2nd dose of the NK cells. Bioluminescence imaging was performed on days 9 and 14 by using Lago imaging system (Spectral Instruments Imaging). All animal experiments were approved by the City of Hope Animal Care and Use Committee.
Statistical analysis
Student’s t test and paired t test were used to compare two independent and matched/paired groups, respectively. One-way ANOVA was used to compare three or more independent groups. A linear mixed model or one-way ANOVA model with repeated measures was used to account for the variance-covariance structure due to repeated measures from the same subject. All tests were two-sided and p-values were adjusted for multiple comparisons by Turkey’s or Holm-Sidak’s procedure. A p-value less than 0.05 was defined as statistically significant. GraphPad 9.1.0 was used for the statistical analyses.
Results
TYRO3 expression on primary human NK cells is rapidly induced by TYRO3+ tumor cells
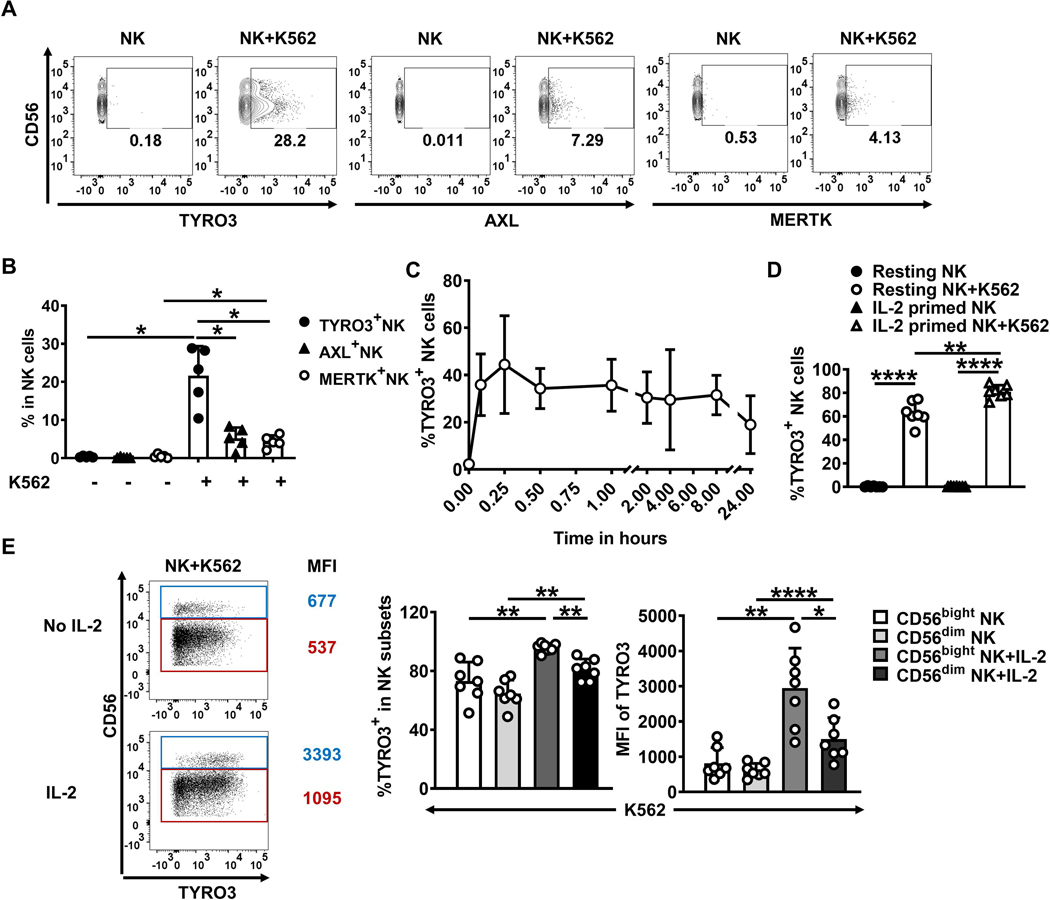
Peeters et al. have reported that T-cell receptor (TCR)-activated primary human CD8+ T cells express Mertk, which subsequently acts as a co-stimulatory receptor on CD8+ T cells (28). To investigate whether activated NK cells express TAM family receptors, we incubated primary human NK cells with different cytokines or with the K562 myeloid leukemia cell line for 24 h. Flow cytometric analysis showed that a substantial proportion of NK cells expressed TYRO3, but very few expressed AXL and MERTK, after co-culture with K562 cells (Fig. 1A and 1B). Resting NK cells did not express any TAM family receptors on surface, which was consistent with our previous study (29), and this did not change when the NK cells were primed with cytokines in the absence of K562 cells (Supplementary Fig. S1A and S1B).
Figure 1. TYRO3 expression on primary human NK cells is rapidly induced by TYRO3+ tumor cells.
(A) TYRO3, AXL, and MERTK expression was determined after primary human NK cells were co-cultured with or without K562 cells at an effector (E)/tumor (T) ratio of 10:1 for 24 h. Representative flow cytometry plots from 4 different donors. Summary data of A are shown in (B). (C) Primary human NK cells were co-cultured with K562 at an E/T ratio of 1:1 for the indicated times, and TYRO3 expression was determined on NK cells by flow cytometry. Data are summarized from 4 different donors. (D)Primary human NK cells were pre-treated without or with IL2 (150 IU/ml) overnight and then co-cultured with K562 cells at an E/T ratio of 1:1, followed by analysis of TYRO3 expression on NK cells by flow cytometry. Data are summarized from 7 different donors. (E) NK cells in (D) were divided into CD56bright and CD56dim NK-cell subsets. Representative flow cytometry plots and summary data of TYRO3 expression in these two subsets from 7 different donors are shown. One-way ANOVA was used for B, D and E. P values were adjusted by the Holm-Sidak’s method. *, P<0.05; **, P<0.01; ****, P<0.0001. Data are presented as mean ± SD.
Next, we measured the expression of TYRO3 at different time points when primary human NK cells were co-cultured with K562 cells. TYRO3 expression was detected on NK cells within 5 minutes of co-culture with K562 cells and reached a plateau at 15 min of co-incubation (Fig. 1C). The rapid detection of TYRO3 on the surface of NK cells suggested that upregulation of TYRO3 may not depend on gene transcription and translation. Although IL2 alone could not induce TYRO3 expression on NK cells (Fig. 1D, and Supplementary Fig. S1B), NK cells that were first primed with IL2 and then co-cultured with K562 cells, expressed more TYRO3 than resting NK cells incubated with K562 cells but without prior IL2 priming (Fig. 1D and Supplementary Fig. S1C). Among the NK cells, IL2-primed CD56bright NK cells expressed more TYRO3 than IL2-primed CD56dim NK cells after incubation with K562 cells for 1 h (Fig. 1E). Collectively, these results indicate that IL2-activated NK cells expressed more TYRO3 than resting NK cells after encountering K562 cells. Moreover, there was no difference in K562-induced TYRO3 expression between enriched NK cells and highly purified (>97%) NK cells (Supplementary Fig. S1D).
We next addressed whether TYRO3 induction on NK cells was related to TRYO3 expression on tumor cells. Flow cytometric analysis showed that K562 and U937 cell lines had high expression levels of TYRO3, whereas Molm-13 and Jurkat cell lines barely expressed TYRO3 (Supplementary Fig. S1E). After NK cells were co-cultured with the different tumor cell lines in the presence of IL-2, NK cells expressed higher levels of TYRO3 after being incubated with K562 or U937 cells than with Molm-13 or Jurkat cells, suggesting that the tumor-cell TYRO3 expression level correlated with the level of TYRO3 induction in NK cells (Supplementary Fig. S1F).
Rapid induction of TYRO3 expression on NK cells by TYRO3+ tumor cells requires cell-cell contact
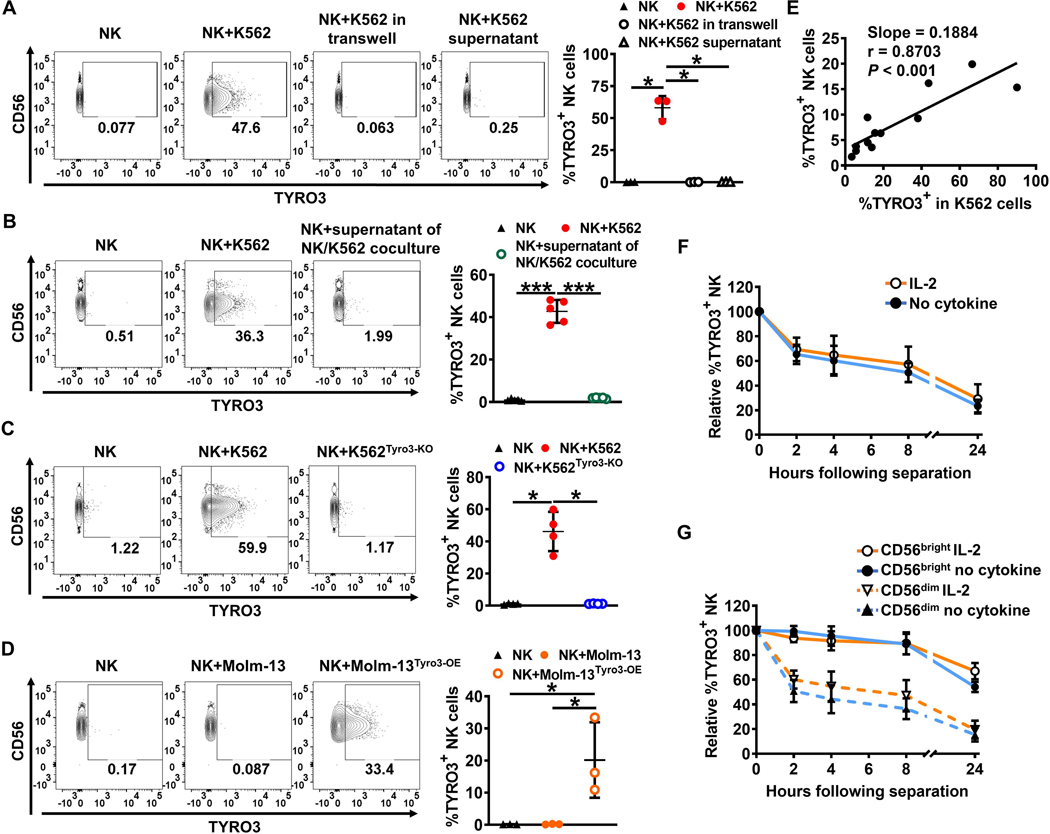
To investigate the mechanism of TYRO3 upregulation on NK cells we tested whether direct cell contact was required for TYRO3 induction. We found K562 cells incubated in transwells did not induce TYRO3 expression on NK cells (Fig. 2A). In addition, supernatant from K562 cells cultured alone or supernatant from K562 cells co-cultured with NK cells was unable to induce TYRO3 expression on NK cells (Fig. 2A and 2B). These data show that direct interaction between NK cells and K562 cells is necessary to induce rapid TYRO3 expression on NK cells.
Figure 2. Rapid induction of TYRO3 expression on NK cells by TYRO3+ tumor cells requires cell–cell contact.
(A) Representative flow cytometry plots and summary data (n = 3) showing TYRO3 expression on NK cells from IL2-stimulated NK cells co-cultured with or without K562 cells in trans-well plates, co-cultured directly with or without K562, or co-cultured with or without supernatant from K562 culture for 1 h at an E/T ratio of 1:1. (B) Representative flow cytometry plots and summary data (n = 5) showing TYRO3 expression on NK cells from IL2-stimulated NK cells co-cultured directly with or without K562, or with or without supernatant from NK cells and K562 co-cultures for 1 h at an E/T ratio of 1:1. (C) Representative flow cytometry plots and summary data (n = 4) showing TYRO3 expression on NK cells from IL2-stimulated NK cells co-cultured with K562 or K562Tyro3-KO for 1 h at an E/T ratio of 1:1. (D) Representative flow cytometry plots and summary data (n = 3) showing TYRO3 expression on NK cells from IL2-stimulated NK cells co-cultured with Molm-13 or Molm-13Tyro3-OE for 1 h at an E/T ratio of 1:1. (E) Nonlinear regression analysis of the correlation between the percentages of acquired TYRO3 on NK cells and their corresponding TYRO3% on K562 cells. The correlation as calculated by Pearson test was statistically significant. Data are summarized from 3 different donors. (F and G) NK cells pre-treated with or without IL2 (150 IU/ml) were co-cultured with K562 for 1 h at an E/T ratio of 1:1. Summarized data show kinetics of persistence of acquired TYRO3 on NK cells (after sorting separation) from 4 independent experiments. One-way ANOVA was used for A–D and Pearson correlation coefficient for E. P values were adjusted by the Holm-Sidak method. *, P<0.05; **, P<0.01; ***, P<0.001. Data are presented as mean ± SD.
Since TYRO3 levels on the surface of tumor cells correlated with the levels of TYRO3 induction on NK cells (Supplementary Fig. S1E and S1F), we generated TYRO3-deficient K562 cells (K562Tyro3-KO) by CRISPR/Cas9-mediated genome editing (Supplementary Fig. S2A). The K562Tyro3-KO cells failed to induce TYRO3 expression on NK cells (Fig. 2C). In contrast, TYRO3 expression on NK cells co-cultured with Molm-13 cells overexpressing TYRO3 (Molm-13Tyro3-OE) was considerably upregulated as compared to NK cells co-cultured with parental Molm-13 cells (Fig. 2D). However, NK cells could not acquire TYRO3 from Jurkat cells overexpressing TYRO3 (JurkatTyro3-OE) (Supplementary Fig. S2B). Since the level of protein expression in the donor cells appeared to affect the amount of transferred protein, different K562 cell lines with different TYRO3 knockout efficiencies were generated using a CRISPR/Cas9-mediated genome editing procedure. There was a positive correlation between the frequency of TYRO3 expressing K562 cells and the frequency of TYRO3 expressing NK cells (r = 0.8703, p < 0.001) (Fig. 2E). Furthermore, approximately half of the NK cells that had acquired TYRO3 from K562 cells no longer displayed it after 8 h when they had been purified and separated from K562 cells (Fig. 2F, and Supplementary Fig. S2C), regardless of IL-2 priming, suggesting that a continuous presence of target cells is required to maintain TYRO3 expression on NK cells. TYRO3 acquired by CD56bright NK cells showed a better persistence than TYRO3 acquired by CD56dim NK cells with or without IL-2 priming (Fig. 2G, and Supplementary Fig. S2C), consistent with the observation that IL-2-primed CD56bright NK cells acquired more TYRO3 than CD56dim cells (Fig. 1E). Collectively, these results demonstrate that induction of TYRO3 expression on NK cells by TYRO3+ tumor cells happens rapidly and requires cell–cell contact.
Tyro3-EGFP can be transferred from tumor cells to human NK cells via trogocytosis
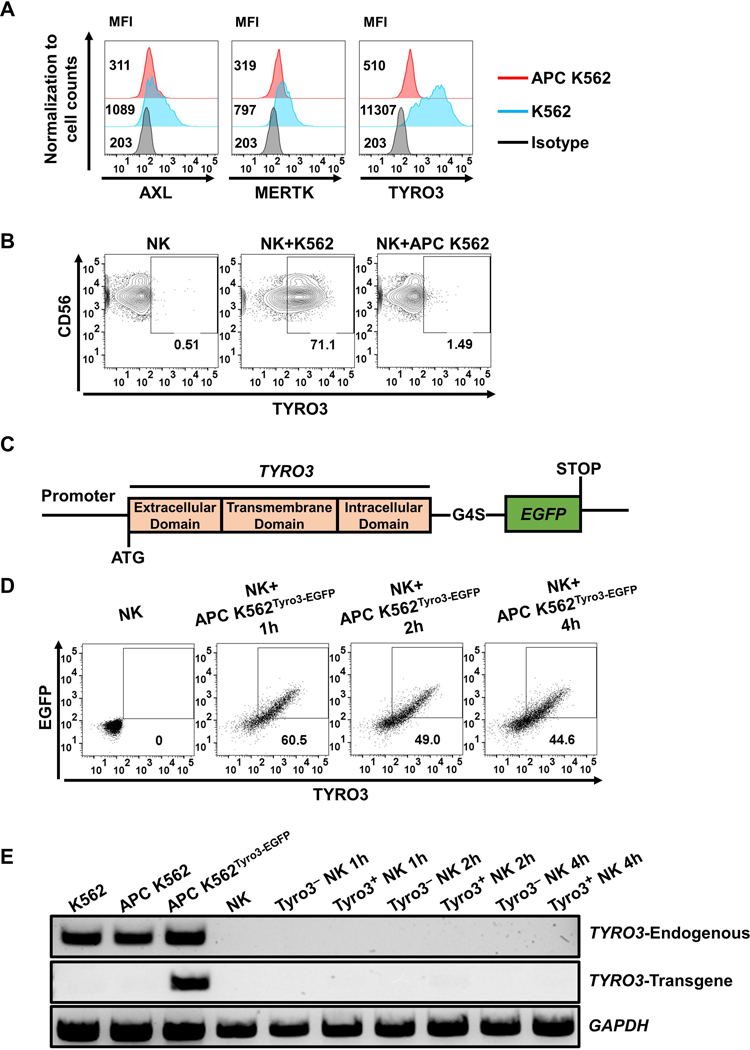
K562 cells expressing mbIL-21 and 4–1BBL (APC K562) are usually used to expand NK cells in vitro (30). We established clinical-grade master and working cell banks of APC K562 cells for NK-cell expansion (Supplementary Table S2). Flow cytometric analysis showed APC K562 cells lacked or had low levels of expression of TYRO3, AXL and MERTK, whereas parental K562 cells expressed all three of these proteins (Fig. 3A). Consistently, co-incubation of primary human NK cells with APC K562 cells did not induce TYRO3 expression on NK cells (Fig. 3B). To further assess whether ectopically expressed TYRO3 can be transferred from APC K562 cells to NK cells, APC K562 cells were transduced with a Tyro3-EGFP fusion protein to generate APC K562Tyro3-EGFP cells (Fig. 3C). EGFP fluorescence and TYRO3 extracellular domains were transferred from APC K562Tyro3-EGFP cells to primary human NK cells during co-culture, indicating that the entire protein and not a part of the protein, such as a shed extracellular domain, was transferred from the tumor cells to the NK cells (Fig. 3D).
Figure 3. Tyro3-EGFP can be transferred from tumor cells to human NK cells via trogocytosis.
(A) Representative flow cytometry shows TYRO3, AXL and MERTK expression on parental K562 and APC K562 cells, confirming that the APC K562 cells barely express TYRO3. (B) NK cells did not express TYRO3 after co-cultured with APC K562 cells at an E/T ratio of 1:1 for 1 h. Each experiment was performed with 4 different donors. (C) The construct design of the Tyro3-EGFP fusion protein. G4S linker, Gly-Gly-Gly-Gly-Ser. (D) Representative flow cytometry plots showing co-culture of NK cells with APC K562 cells overexpressing TYRO3 with C-terminal fusion EGFP (APC K562Tyro3-EGFP). Each experiment was performed with NK cells from 3 different donors. (E) PCR analysis of TYRO3 expression in NK cells after co-cultured with APC K562Tyro3-EGFP cells. K562 cells were used as positive control for endogenous TYRO3 transcript and served as negative control for transgenic TYRO3 transcript. TYRO3− and TYRO3+ NK cells both lacked the expression of endogenous and transgenic TYRO3 transcript over the entire 4 h period of co-culture. GAPDH was used as control for quality of cDNA synthesis. Each experiment was performed with 3 different donors.
To show that the TYRO3 displayed on NK cells after co-incubation with APC K562Tyro3-EGFP feeder cells was not endogenously produced, we sorted TYRO3+ NK cells and TYRO3− NK cells from co-culture with APC K562Tyro3-EGFP cells at different time points and measured transcription of TYRO3. One pair of primers that we used was specific for native TYRO3 mRNA and lack the ability to amplify TYRO3 mRNA derived from transfected APC K562Tyro3-EGFP cells, whereas the other pair of primers was specific for transfected APC K562Tyro3-EGFP cells and unable to amplify native TYRO3 mRNA. After co-incubation with APC K562Tyro3-EGFP cells, NK cells displayed cell-surface Tyro3 (Fig. 3D), but not TYRO3 mRNA regardless of whether the primers detected endogenous or transduced TYRO3 (Fig. 3E). Collectively, these data show that NK cells acquire TYRO3 from K562 cells via a process of trogocytosis.
NK cells acquiring TYRO3 via trogocytosis possess improved effector functions
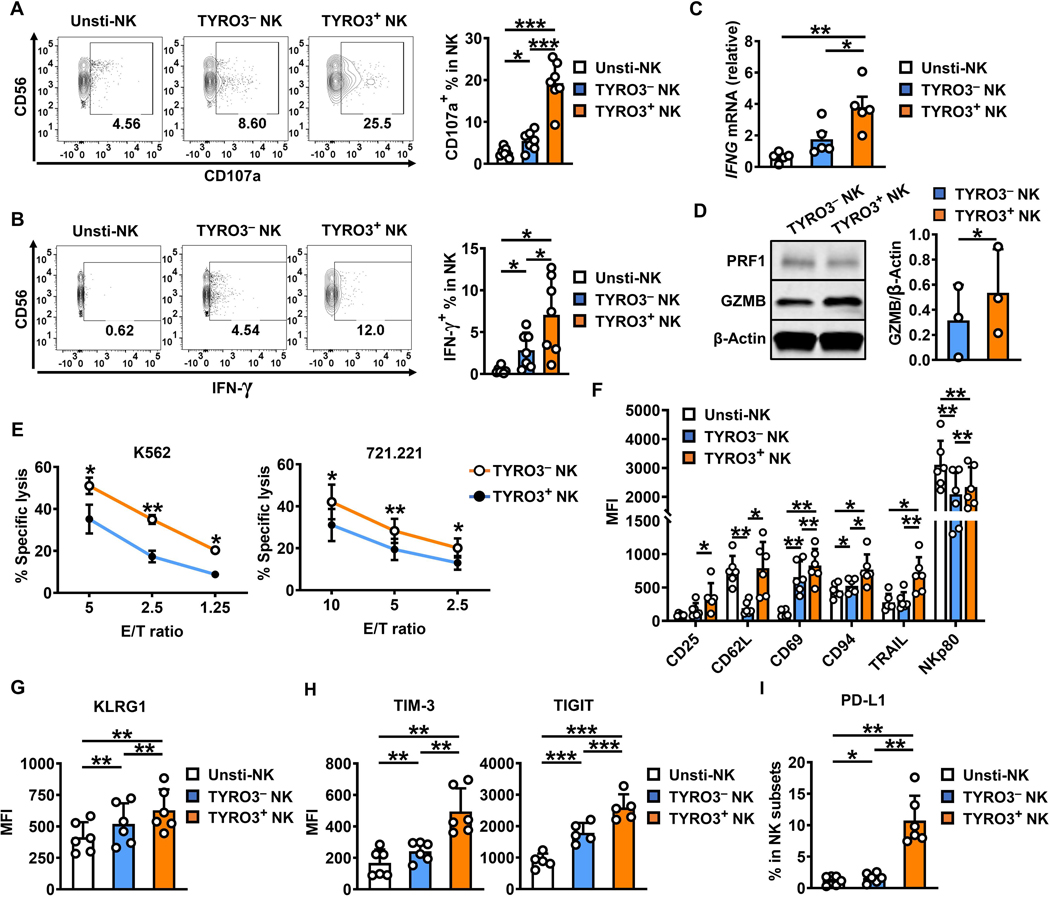
We next investigated the effect of TYRO3 acquisition by NK cells. NK-cell expression of CD107a and IFN-γ are commonly used as functional markers for NK-cell degranulation and cytokine production, respectively, following NK-cell activation. After co-culture of NK cells with K562 cells for 24 h, we found that expression of CD107a and IFN-γ was significantly increased in TYRO3+ NK cells compared with TYRO3− NK cells (Fig. 4A and 4B). IFNG mRNA expression levels also were significantly increased in TYRO3+ NK cells compared with TYRO3− NK cells (Fig. 4C), whereas there were no significant differences in GZMB and PRF1 mRNA expression levels between the TYRO3+ and TYRO3− NK cells (Supplementary Fig. S3A). Moreover, even after co-culture of IL2-primed NK cells with K562 cells for 4 h, we found that expression of CD107a and IFN-γ significantly increased in TYRO3+ NK cells compared with TYRO3− NK cells (Supplementary Fig. S3B and S3C). There was also an increase in protein levels of GZMB but not PRF1 (Fig. 4D). A 51Cr release assay confirmed that the cytotoxicity level of TYRO3+ NK cells was significantly increased compared with TYRO3− NK cells (Fig. 4E). These results indicate that TYRO3+ NK cells are highly activated immune effector cells. Collectively, our data demonstrate that NK cells can acquire TYRO3 upon encountering TYRO3+ tumor cells, and that compared with TYRO3− NK cells, TYRO3+ NK cells possess more robust effector functions.
Figure 4. NK-cells acquiring TYRO3 via trogocytosis possess improved effector functions.
(A and B) Representative flow cytometry plots and summary data from 7 different donors showing the expression of CD107a and IFN-γ in TYRO3− and TYRO3+ NK cells after NK cells were co-cultured with K562 cells for 24 h at an E/T ratio of 10:1. (C) Expression of IFNG was assessed by qRT-PCR in FACS-sorted unstimulated, TYRO3− and TYRO3+ NK cells after NK cells were co-cultured with K562 cells for 24 h at an E/T ratio of 10:1. Summary data are representative from 5 different donors. (D) The protein levels of GZMB and PRF1 were assessed by immunoblotting in FACS-sorted TYRO3− and TYRO3+ NK cells after IL2-stimulated NK cells were co-cultured with K562 cells for 4 h at an E/T ratio of 1:1 with GlogiPlug. Summary data are for 3 different donors. (E) TYRO3− and TYRO3+ NK cells were FACS-sorted after NK cells were co-cultured with K562 cells for 24 h at an E/T ratio of 10:1, then co-cultured with 51Cr labeled-K562 or −721.221 cells for 4 h, followed by quantification of specific K562 or 721.221 cell lysis. Data are summarized from 4 different donors. (F-I) Primary human NK cells were incubated with or without K562 cells for 24 h at an E/T ratio of 10:1. (F) Summary data of activating receptors (CD25, CD62L, CD69, CD94, TRAIL and NKp80) on NK cells from 6 different donors. (G) Summary data of KLRG1 on NK cells from 6 different donors. (H) Summary data of exhaustion markers (TIM-3 and TIGIT) on NK cells from at least 5 different donors. (I) Summary data of PD-L1 on NK cells from 6 different donors. One-way ANOVA was used for A–C and G–I, paired t test for D, multiple t test for E, and two-way ANOVA for F. P values were adjusted by the Holm-Sidak’s method. *, P<0.05; **, P<0.01; ***, P<0.001. Data are presented as mean ± SD for A, B, D and F–I and as mean ± SEM for C and E.
Since changes of NK-cell surface receptor expression are commonly reported in cancers (31–33) and our data show that the functional capacities of TYRO3+ and TYRO3− NK cells are different, we next measured the expression of surface markers on these two distinct NK-cell subsets using flow cytometry. Co-incubation of NK cells with K562 cells led to increased expression of several activation markers CD25, CD69, and TRAIL on TYRO3+ NK cells compared with TYRO3− NK cells and unstimulated NK cells (Fig. 4F), whereas other activation markers such as NKG2D and NKp30 showed no significant difference in expression between TYRO3+ and TYRO3− NK cells (Supplementary Fig. S3D). Moreover, CD62L expression was maintained on TYRO3+ NK cells but decreased on TYRO3− NK cells compared with unstimulated NK cells (Fig. 4F). Cell maturation markers CD94, NKp80, and KLRG1 showed an increase in expression on the TYRO3+ NK-cell subset compared with the TYRO3− NK-cell subset and unstimulated NK cells, whereas the inhibitory receptor NKG2A showed no difference in expression between the TYRO3+ and TYRO3− NK-cell subsets (Fig. 4F-G, and Supplementary Fig. S3E). Furthermore, co-incubation of NK cells with K562 cells induced the expression of two exhaustion-related markers, TIM-3 and TIGIT, with expression levels significantly higher on TYRO3+ NK cells than TYRO3− NK cells (Fig. 4H). Our previous research has revealed that PD-L1 is induced on NK cells after encountering K562 cells (34). Interestingly, PD-L1 expression was significantly upregulated on TYRO3+ NK cells compared with TYRO3− NK cells after NK cell co-culture with K562 cells for 24 h (Fig. 4I).
Acquisition of TYRO3 by NK cells enhances NK-cell ex vivo expansion
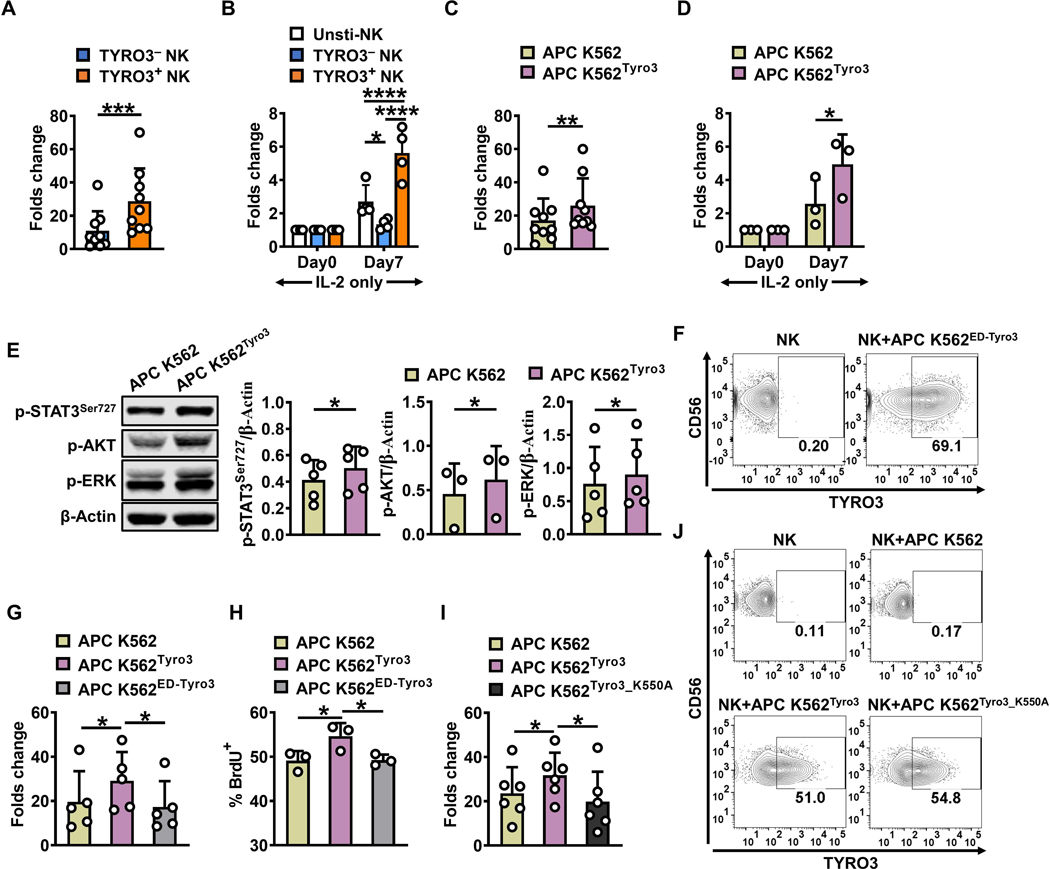
After incubation of NK cells with parental K562 cells for 24 h, we sorted TYRO3+ and TYRO3− NK cells and then expanded the two NK-cell subsets further with the APC K562 cell line, which expresses 4–1BBL and mbIL-21, in the presence of IL2 (50 IU/ml) for 7 days. On day 7, expansion of TYRO3+ NK cells was significantly greater than the expansion of TYRO3− NK cells (Fig. 5A). In addition, we cultured the sorted TYRO3+ and TYRO3− NK cells in the presence of IL-2 (150 IU/ml) alone for 7 days and found that TYRO3+ NK cells expanded significantly more than TYRO3− NK cells (Fig. 5B), suggesting that TYRO3+ NK cells are more responsive to IL2 than TYRO3− NK cells.
Figure 5. Acquisition of TYRO3 by NK cells enhances NK-cell ex vivo expansion.
(A)TYRO3− and TYRO3+ NK cells were FACS-sorted and incubated with inactivated APC K562 cells in the presence of IL2 (50 IU/ml) for 7 days, followed by enumerating live NK cells on day 7 (n = 9). (B) TYRO3− and TYRO3+ NK cells were FACS-sorted and cultured in the presence of IL2 (150 IU/ml) for 7 days, followed by enumerating live NK cells on day 7 (n = 4). (C)Primary human NK cells were incubated with inactivated APC K562 cells or APC K562Tyro3 in the presence of IL2 (50 IU/ml) for 7 days, followed by enumerating live NK cells on day 7 (n = 9). (D) Primary human NK cells were incubated with APC K562 cells or APC K562Tyro3 for 1 h. Then, NK cells were separated from tumor cells, followed by co-culture with IL2 (50 IU/ml) for 7 days, followed by enumerating live NK cells on day 7 (n = 3). (E)Primary human NK cells were incubated with APC K562 cells or APC K562Tyro3 for 1 h. Then NK cells were separated from tumor cells, followed by immunoblotting assay. Summary data of p-STAT3Ser727, p-AKT or p-ERK expression are presented after being normalized to β-Actin expression. Each experiment was repeated with NK cells from at least 3 different donors. (F) TYRO3 expression was determined after IL2-stimulated primary human NK cells co-cultured with APC K562ED-Tyro3 cells for 1 h at an E/T ratio of 1:1 (n = 3). (G) Primary human NK cells were incubated with APC K562 cells, APC K562Tyro3 or APC K562ED-Tyro3 in the presence of IL2 (50 IU/ml) for 7 days, followed by enumerating live NK cells on day 7 (n = 5). (H) NK cells in (G) were followed by BrdU assay with 3 different donors. (I) Primary human NK cells were incubated with APC K562 cells, APC K562Tyro3 or APC K562Tyro3_K550A in the presence of IL2 (50 IU/ml) for 7 days, followed by enumerating live NK cells on day 7 (n = 6). (J) TYRO3 expression was determined after IL2-stimulated primary human NK cells were co-cultured with APC K562Tyro3_K550A cells at an E/T ratio of 1:1 (n = 3). Paired t test was used for A, C and E, two-way ANOVA for B and D, one-way ANOVA for G–I. P values were adjusted by the Turkey’s or Holm-Sidak’s method. *, P<0.05; **, P<0.01; ***, P<0.001; ****, P<0.0001. Data are presented as mean ± SD.
As noted earlier, flow cytometric analysis showed that the APC K562 cell line has very modest expression of TYRO3 compared with the parental K562 cell line (Fig. 3A). We next generated APC K562 cells that stably overexpressed full-length TYRO3 (APC K562Tyro3) or the extracellular domain of TYRO3 with the PDGFRB transmembrane domain (APC K562ED-Tyro3). Since inactivated APC K562 cells are used to expand unmodified primary human NK cells or engineered NK cells, such as chimeric antigen receptor (CAR) NK cells, for clinical use, we tested whether primary human NK cells can acquire TYRO3 from APC K562 cells inactivated by mitomycin. We observed that primary human NK cells could capture TYRO3 from both inactivated APC K562Tyro3 cells and non-inactivated APC K562Tyro3 cells (Supplementary Fig. S4A). The expansion of NK cells co-incubated with inactivated APC K562Tyro3 cells and IL-2 for 7 days was significantly higher than the expansion of NK cells co-incubated with APC K562 cells and IL2 for 7 days (Fig. 5C). With IL2 treatment, NK cells incubated with APC K562Tyro3 cells proliferated significantly more than those incubated with APC K562 cells (Fig. 5D). Furthermore, when NK cells were separated from tumor cells after co-incubation with either APC K562 or APC K562Tyro3 cells for 1 h, we found that levels of p-STAT3, p-AKT and p-ERK in NK cells increased more with APC K562Tyro3 cell stimulation compared with APC K562 cell stimulation (Fig. 5E).
When NK cells were co-cultured with APC K562ED-Tyro3 cells for 1 h, expression of TYRO3 was detected on the surface of NK cells (Fig. 5F). However, APC K562ED-Tyro3 cells did not confer the same benefits on NK-cell expansion as observed with APC K562Tyro3 cells (Fig. 5G). In NK cells expanded with inactivated APC K562Tyro3 cells, the percentage of BrdU incorporated into DNA (Fig. 5H) and the proliferation rate (Supplementary Fig. S4B) was significantly higher than in NK cells expanded with APC K562 or APC K562ED-Tyro3 cells. In contrast, there was no difference in cell apoptosis or cell survival among the three different groups of expanded NK cells (Supplementary Fig. S4C and S4D). We next generated APC K562 cells that stably overexpressed kinase dead full-length TYRO3 (APC K562Tyro3_K550A) with a K550A mutation (35). Similar to APC K562ED-Tyro3 cells, APC K562Tyro3_K550A cells did not confer the same benefits on NK-cell expansion observed with APC K562Tyro3 cells (Fig. 5I), even though expression of TYRO3 was detected on the surface of NK cells when NK cells were co-cultured with the APC K562Tyro3_K550A cells (Fig. 5J).
Taken together, these results suggest that TYRO3 transferred to NK cells via trogocytosis may initiate signaling to control NK-cell activation. Consistent with a previous report (21), overexpression of TYRO3 in NK cells by transduction rather than trogocytosis did not enhance NK-cell expansion regardless of the presence or absence of IL-2 (Supplementary Fig. S4E).
TYRO3-expressing K562 feeder cells expand NK cells with similar in vivo antitumor activity
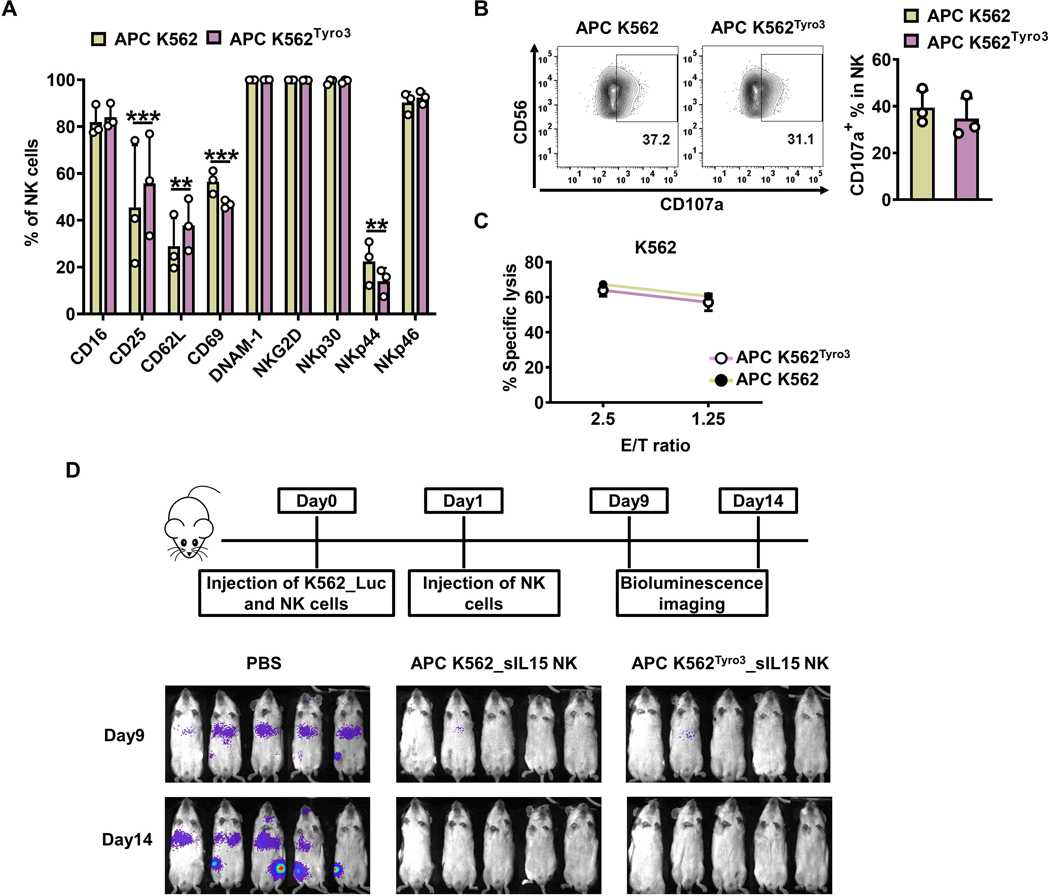
Since acquisition of TYRO3 by NK cells from APC K562 cells expressing TYRO3 enhanced NK-cell ex vivo expansion, we assessed the function of these NK cells both in vivo and in vitro. Primary human NK cells were incubated with inactivated APC K562 or APC K562Tyro3 cells in the presence of IL-2 (50 IU/ml) for 7 days and assessed for expression of surface markers. Expression of the activating surface markers CD16, DNAM-1, NKG2D, NKp30, and NKp46 showed no differences between NK cells expanded in the presence of APC K562Tyro3 or those expanded in the presence of APC K562 cells. NK cells expanded by APC K562Tyro3 cells expressed higher levels of CD25 and CD62L but lower levels of CD69 and NKp44 than NK cells expanded by APC K562 cells (Fig. 6A). We also assessed the function of the expanded NK cells via a CD107a degradation assay and 51Cr cytotoxicity assay, and found that NK cells expanded with APC K562Tyro3 cells and APC K562 cells showed similar levels of CD107a degranulation (Fig. 6B) and cytotoxicity (Fig. 6C).
Figure 6. TYRO3-expressing K562 feeder cells expand NK cells with similar in vivo antitumor activity.
(A-C) Primary human NK cells were incubated with inactivated APC K562 cells or APC K562Tyro3 cells in the presence of IL2 (50 IU/ml) for 7 days. Summary data from 3 different donors showing the expression of surface markers on NK cells expanded with APC K562 cells or APC K562Tyro3 cells (A). Representative flow cytometry plots and summary data from 3 different donors showing the expression of CD107a on NK cells expanded with APC K562 cells or APC K562Tyro3 cells for 4 h at an E/T ratio of 4:1 (B). NK cells expanded with APC K562 cells or APC K562Tyro3 cells were co-cultured with 51Cr-labeled K562 cell line for 4 h, followed by quantification of specific K562 cell lysis. Data are summarized from 3 different donors (C). (D) Primary human NK cells were incubated with APC K562Tyro3 or APC K562 cells in the presence of IL2 (50 IU/ml) for 4 days. Then cells were transduced with a retrovirus vector expressing soluble IL15 (sIL15) for 48 h. On day 6, transduced NK cells were stimulated with APC K562Tyro3 cells or APC K562 cells in the presence of IL2 (50 IU/ml) for another 7 days prior to harvest and storage for following usage. On day 0, mice were injected with 1×106 K562_Luc cells and then treated with 10×106 sIL15 NK cells expanded with APC K562 (APC K562_sIL15 NK) or APC K562Tyro3 (APC K562Tyro3_sIL15 NK). On day 1, mice were treated with a 2nd dose of these NK cells. Bioluminescence imaging was performed on days 9 and 14. Five mice in each group. Two-way ANOVA was used for A. **, P<0.01; ***, P<0.001. Data are presented as mean ± SD.
Next, the ability of NK cells expanded by APC K562Tyro3 cells to control tumors in vivo was investigated by engrafting NSG mice with luciferase-expressing K562 cells (K562_Luc), followed by treating the mice with or without APC K562_sIL15 NK cells or APC K562Tyro3_sIL15 NK cells. APC K562Tyro3_sIL15 NK cells significantly suppressed tumor burden at a level similar to that of APC K562_sIL15 NK cells, which was assessed by whole-body bioluminescence imaging (Fig. 6D). Thus, APC K562Tyro3_sIL15 NK cells display similar functions as APC K562_sIL15 NK cells in vitro and in vivo.
We next tested whether TYRO3 trogocytosis from tumor cells to NK cells occurred in vivo. For this purpose, NSG mice were engrafted with K562_Luc cells for 14 days, followed by treatment with sIL15 NK cells, and TYRO3 expression in human NK cells was assessed in different organs or tissues of mice 2 days post-treatment. We found that TYRO3 was highly expressed in human NK cells from the bone marrow and liver, moderately in the spleen, but barely in the peripheral blood of mice (Supplementary Fig. S5A). To assess if TYRO3 in these human NK cells was due to endogenous expression of the TYRO3 gene, we sorted human NK cells from different organs or tissues of mice, followed by detection of TYRO3 at the mRNA level by RT-PCR. Our data showed that the endogenous expression of TYRO3 in these human NK cells was undetectable (Supplementary Fig. S5B), indicating that TYRO3 on those NK cells are acquired from K562_Luc cells in vivo via trogocytosis.
Discussion
Trogocytosis, which is a rapid process that occurs within minutes, consists of the active transfer of membrane fragments from one donor cell to a recipient cell in a strictly physical cell–cell contact-dependent manner (7). In our current study, we showed that the expression of TYRO3 in NK cells increased via trogocytosis from tumor cells to NK cells. We also found that there were no differences in TYRO3 trogocytosis from tumor cells to CD56bright and CD56dim NK cells in the absence of IL-2; however, when activated by IL-2, CD56bright NK cells captured more TYRO3 from donor cells than CD56dim NK cells. This may be explained by the fact that CD56bright NK cells express high-affinity heterodimeric IL2R, whereas CD56dim NK cells do not (36,37). We also characterized the functional and phenotypical differences between the subsets with and without trogocytosis of TYRO3. Of significance, we generated a novel feeder cell line that enhanced primary human NK-cell expansion, which could be used for future NK cell-based therapy.
NK cells activated by IL-2 acquired more TYRO3 from K562 and U937 cells compared with resting NK cells. Consistent with our results, previous studies have shown that trogocytosis may occur during cell–cell interactions between target cells and NK cells stimulated with IL2 alone (15,38); IL2 and PHA (16); IL2, PHA and irradiated peripheral blood lymphocytes as feeders (39); IL2 with K562-based feeder cells (14,40–42). The functional activity of NK cells is regulated by various inhibitory and activating receptors expressed on NK cells, some of which interact with HLA molecules (43). Target cells expressing MHC class I molecules are protected from NK-cell lysis by interactions between these molecules and inhibitory NK-cell receptors (44–46). In contrast, in the absence of these interactions, target cells become sensitive to NK-cell lysis through triggering of NK cell–activating receptors. Compared with the U937 cell line, the K562 cell line is known as an NK cell–sensitive target due to lack of membrane MHC class I expression (47). Our data showed that more TYRO3 was acquired by NK cells from K562 cells than from U937 cells. Therefore, it seems trogocytosis is a universal process between NK cells and target cells, especially activated NK cells, and the amount of trogocytosis is correlated to the activation level of the NK cells as well the expression level of transferred molecules on target cells.
Phenotypic and functional consequences of trogocytosis in recipient cells can be different (48). The IL2-activated NK cell line NKL becomes suppressive after acquiring HLA-G1 from an HLA-G1–transfected melanoma cell line (15). A reduction in NK-cell cytotoxicity is observed after the intracellular transfer of NKG2D from NK cells to target cells (40). Our results showed that upon encounter and activation by the NK cell–susceptible K562 myeloid leukemia cell line, the acquisition of TYRO3 by NK cells significantly enhanced their cytotoxicity, IFN-γ production and expression density of activated surface proteins compared with TYRO3− NK cells. Expression of PD-L1, which is upregulated on NK cells after encounter with K562 cells (34), was significantly increased in TYRO3+ NK cells compared with TYRO3− NK cells. Our previous study also revealed that PD-L1+ NK cells are more activated than PD-L1− NK cells (33), which is consistent with TYRO3+ NK cells being more activated than TYRO3− NK cells, as observed in our current study.
Trogocytosis is not a transfer of individual molecules or a part of an individual protein, rather it is the transfer of entire membrane patches that contain several proteins or an individual protein as a whole (10,49). Using C-terminal GFP-tagged H2-Dd, Andersson et al. showed that MHC molecules transferred from donor cells to NK cells are whole and include the intact intracellular part (50). Similarly, when we tagged TYRO3 with C-terminal fusion EGFP protein, the EGFP signaling was detected with TYRO3 together after trogocytosis, indicating that the TYRO3 molecules transferred from the tumor cells to the NK cells were likely full-length proteins. Not only can TYRO3 with the extracellular domain be transferred from target cells to NK cells, but also TYRO3 with a K550A mutation that yields a kinase dead TYRO3 protein (35), and the transferred TYRO3 with a kinase-dead domain lacked the functionality of transferred wild-type TYRO3. These data suggest that trogocytosis of TYRO3 from tumor cells to primary human NK cells is independent of TYRO3 kinase function, but the function of TYRO3 acquired by NK cells from tumor cells via trogocytosis is TYRO3-kinase dependent.
GAS6 and PROS1 are ligands of the TAM receptor family (51–53). K562 cells express high levels of GAS6 protein, whereas Molm-13, Jurkat, and U937 cells do not express GAS6 protein (54). Our data showed that IL2-stimulated NK cells could acquire TYRO3 from K562 cells and U937 cells (at a low level) but not from TYRO3-overexpressing Jurkat cells, suggesting that trogocytosis of TYRO3 in NK cells can occur independent of GAS6. According to the ProteinAtlas database, PROS1 is expressed on K562, Jurkat, and U937 cells, whereas our data showed that only K562 cells and U937 cells (at a low level) but not Jurkat overexpressing TYRO3 could perform trogocytosis, further suggesting that trogocytosis is ligand independent.
The function of TYRO3 in humans is still not clear. According to another study, TAM family receptors attenuate NK-cell function and inhibition of TAM-receptor signaling prevents metastasis in a mouse tumor model in an NK cell-dependent manner (21). Mature primary human NK cells show little, if any, TAM receptor expression (22). TYRO3 overexpression in NK cells via viral transduction inhibited NK-cell proliferation under IL2-stimulation, which contrasts with our observation that NK cells acquiring TYRO3 via trogocytosis show enhanced proliferation. These data suggest there are critical differences between engineering NK cells with viral transduction and trogocytosis. This difference can be explained by two possible reasons. First, trogocytosis is not a transfer of individual molecules, but an entire membrane patch that may contain several proteins (10,49). When TYRO3 is transferred from tumor cells to NK cells, one or more related molecules may be transferred in a parallel. It is possible that unidentified molecules on the membrane patch co-transferred with TYRO3 may function as positive regulators. Second, TYRO3 acquired by trogocytosis and genetically expressed TYRO3 may have different post-translational modifications. This is important because the molecular weights of TAM family members vary in different tissues and cells due to posttranslational modifications including glycosylation, phosphorylation, and ubiquitination (55), and these modifications may affect TAM-receptor function.
Because of safety concerns regarding the use of traditional viral transduction methods, several studies recently investigated engineering NK cells via trogocytosis in vitro. Srinivas et al. used trogocytosis as a tool to transiently express CCR7 on expanded human NK cells to enhance their homing to lymph nodes on adoptive transfer (41). Cho et al. generated expanded NK cells that had received an anti-CD19 CAR from a K562 cell line via trogocytosis, and used them to treat B-cell tumors (14). NK cells are complimentary to T cells as a promising tool for use in adoptive immunotherapy against different types of cancer due to their innate ability to recognize and lyse tumor cells through the rapid activation of a series of NK cell–activating receptors without the need for prior sensitization, along with their off-the-shelf potential (3). Thus, optimal expansion of NK cells ex vivo is important for NK-cell adoptive immunotherapy. Currently, APC K562-based feeder cells expressing chimeras of mbIL-21 have been developed into a cell line with full compendial testing for human clinical use to expand NK cells for clinical trials (56–59). Since NK cells that acquired TYRO3 via trogocytosis showed better proliferation, we engineered the APC K562 cell line expressing high levels of TYRO3 to be used as a tool for NK-cell expansion ex vivo. Our results show that the newly generated APC K562 feeder cell line stably expressing TYRO3 is superior to the APC K562 cells line with low to absent TYRO3 for in vitro NK-cell expansion. Since TYRO3 acquired by trogocytosis cannot last very long, at the end of the cell manufacturing, NK cells do not express TYRO3, which is likely because almost all inactivated feeder cells die or are killed by NK cells. Consistent with this, NK cells expanded by the feeder cells expressing TYRO3 have in vitro and in vivo effector functions comparable to an equal number of NK cells expanded by feeder cells without TYRO3 overexpression.
To our knowledge, this is the first report that describes enhanced functions and enhanced expansion of NK cells following the acquisition of TYRO3 from a tumor feeder cell line via trogocytosis. We have developed a new APC K562 cell line as a tool to significantly enhance NK-cell expansion ex vivo. Our data suggest that trogocytosis could be a useful tool to modify human NK cells without genomic integration for adoptive immunotherapy for the treatment of cancer and infectious diseases.
Supplementary Material
Synopsis:
The authors show TYRO3 is transferred to natural killer (NK) cells via trogocytosis from feeder cells, improving NK-cell proliferation ex vivo. This provides a potential novel strategy to manufacture NK cells for adoptive-cell transfer cancer immunotherapy.
Acknowledgments
The 721.221 cells were a generous gift to Dr. Caligiuri from Professor J. Miguel Lopez-Botet Arbona at the Universitat Pompeu Fabra, Barcelona. This work was supported by grants from the NIH (CA210087, CA068458, CA265095, and CA163205 to M.A. Caligiuri; NS106170, AI129582, CA247550, and CA223400 to J. Yu), the Leukemia and Lymphoma Society (6503-17 and 1364-19 to J. Yu), and The California Institute for Regenerative Medicine (DISC2COVID19-11947 to J. Yu).
Financial support: This work was supported by grants from the NIH (CA210087, CA068458, CA265095, and CA163205 to M.A. Caligiuri; NS106170, AI129582, CA247550, and CA223400 to J. Yu), the Leukemia and Lymphoma Society (6503-17 and 1364-19 to J. Yu), and The California Institute for Regenerative Medicine (DISC2COVID19-11947 to J. Yu).
Footnotes
Conflict of Interest Disclosure: M.A. Caligiuri and J. Yu are co-founders of CytoImmune Therapeutics Inc. Other authors declare no direct conflicts of interest.
References
- 1.Sun JC, Lanier LL. NK cell development, homeostasis and function: parallels with CD8(+) T cells. Nat Rev Immunol 2011;11(10):645–57 doi 10.1038/nri3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol 2008;9(5):503–10 doi 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- 3.Spits H, Lanier LL, Phillips JH. Development of human T and natural killer cells. Blood 1995;85(10):2654–70. [PubMed] [Google Scholar]
- 4.Gasser S, Raulet DH. Activation and self-tolerance of natural killer cells. Immunol Rev 2006;214:130–42 doi 10.1111/j.1600-065X.2006.00460.x. [DOI] [PubMed] [Google Scholar]
- 5.Liao NS, Bix M, Zijlstra M, Jaenisch R, Raulet D. MHC class I deficiency: susceptibility to natural killer (NK) cells and impaired NK activity. Science 1991;253(5016):199–202 doi 10.1126/science.1853205. [DOI] [PubMed] [Google Scholar]
- 6.Karre K, Ljunggren HG, Piontek G, Kiessling R. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature 1986;319(6055):675–8 doi 10.1038/319675a0. [DOI] [PubMed] [Google Scholar]
- 7.Ahmed KA, Munegowda MA, Xie Y, Xiang J. Intercellular trogocytosis plays an important role in modulation of immune responses. Cell Mol Immunol 2008;5(4):261–9 doi 10.1038/cmi.2008.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vanherberghen B, Andersson K, Carlin LM, Nolte-’t Hoen EN, Williams GS, Hoglund P, et al. Human and murine inhibitory natural killer cell receptors transfer from natural killer cells to target cells. Proc Natl Acad Sci U S A 2004;101(48):16873–8 doi 10.1073/pnas.0406240101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arnold PY, Davidian DK, Mannie MD. Antigen presentation by T cells: T cell receptor ligation promotes antigen acquisition from professional antigen-presenting cells. Eur J Immunol 1997;27(12):3198–205 doi 10.1002/eji.1830271217. [DOI] [PubMed] [Google Scholar]
- 10.Hudrisier D, Riond J, Mazarguil H, Gairin JE, Joly E. Cutting edge: CTLs rapidly capture membrane fragments from target cells in a TCR signaling-dependent manner. J Immunol 2001;166(6):3645–9 doi 10.4049/jimmunol.166.6.3645. [DOI] [PubMed] [Google Scholar]
- 11.Espinosa E, Tabiasco J, Hudrisier D, Fournie JJ. Synaptic transfer by human gamma delta T cells stimulated with soluble or cellular antigens. J Immunol 2002;168(12):6336–43 doi 10.4049/jimmunol.168.12.6336. [DOI] [PubMed] [Google Scholar]
- 12.Batista FD, Iber D, Neuberger MS. B cells acquire antigen from target cells after synapse formation. Nature 2001;411(6836):489–94 doi 10.1038/35078099. [DOI] [PubMed] [Google Scholar]
- 13.Carlin LM, Eleme K, McCann FE, Davis DM. Intercellular transfer and supramolecular organization of human leukocyte antigen C at inhibitory natural killer cell immune synapses. J Exp Med 2001;194(10):1507–17 doi 10.1084/jem.194.10.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho FN, Chang TH, Shu CW, Ko MC, Liao SK, Wu KH, et al. Enhanced cytotoxicity of natural killer cells following the acquisition of chimeric antigen receptors through trogocytosis. PLoS One 2014;9(10):e109352 doi 10.1371/journal.pone.0109352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caumartin J, Favier B, Daouya M, Guillard C, Moreau P, Carosella ED, et al. Trogocytosis-based generation of suppressive NK cells. EMBO J 2007;26(5):1423–33 doi 10.1038/sj.emboj.7601570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marcenaro E, Cantoni C, Pesce S, Prato C, Pende D, Agaugue S, et al. Uptake of CCR7 and acquisition of migratory properties by human KIR+ NK cells interacting with monocyte-derived DC or EBV cell lines: regulation by KIR/HLA-class I interaction. Blood 2009;114(19):4108–16 doi 10.1182/blood-2009-05-222265. [DOI] [PubMed] [Google Scholar]
- 17.Herrera OB, Golshayan D, Tibbott R, Salcido Ochoa F, James MJ, Marelli-Berg FM, et al. A novel pathway of alloantigen presentation by dendritic cells. J Immunol 2004;173(8):4828–37 doi 10.4049/jimmunol.173.8.4828. [DOI] [PubMed] [Google Scholar]
- 18.Rothlin CV, Carrera-Silva EA, Bosurgi L, Ghosh S. TAM receptor signaling in immune homeostasis. Annu Rev Immunol 2015;33:355–91 doi 10.1146/annurev-immunol-032414-112103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zagorska A, Traves PG, Lew ED, Dransfield I, Lemke G. Diversification of TAM receptor tyrosine kinase function. Nat Immunol 2014;15(10):920–8 doi 10.1038/ni.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carrera Silva EA, Chan PY, Joannas L, Errasti AE, Gagliani N, Bosurgi L, et al. T cell-derived protein S engages TAM receptor signaling in dendritic cells to control the magnitude of the immune response. Immunity 2013;39(1):160–70 doi 10.1016/j.immuni.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paolino M, Choidas A, Wallner S, Pranjic B, Uribesalgo I, Loeser S, et al. The E3 ligase Cbl-b and TAM receptors regulate cancer metastasis via natural killer cells. Nature 2014;507(7493):508–12 doi 10.1038/nature12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park IK, Giovenzana C, Hughes TL, Yu J, Trotta R, Caligiuri MA. The Axl/Gas6 pathway is required for optimal cytokine signaling during human natural killer cell development. Blood 2009;113(11):2470–7 doi 10.1182/blood-2008-05-157073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen D, Liu Q, Cao G, Zhang W. TYRO3 facilitates cell growth and metastasis via activation of the Wnt/beta-catenin signaling pathway in human gastric cancer cells. Aging (Albany NY) 2020;12(3):2261–74 doi 10.18632/aging.102744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsai CL, Chang JS, Yu MC, Lee CH, Chen TC, Chuang WY, et al. Functional Genomics Identifies Hepatitis-Induced STAT3-TYRO3-STAT3 Signaling as a Potential Therapeutic Target of Hepatoma. Clin Cancer Res 2020;26(5):1185–97 doi 10.1158/1078-0432.CCR-18-3531. [DOI] [PubMed] [Google Scholar]
- 25.Morimoto M, Horikoshi Y, Nakaso K, Kurashiki T, Kitagawa Y, Hanaki T, et al. Oncogenic role of TYRO3 receptor tyrosine kinase in the progression of pancreatic cancer. Cancer Lett 2020;470:149–60 doi 10.1016/j.canlet.2019.11.028. [DOI] [PubMed] [Google Scholar]
- 26.Yu J, Wei M, Becknell B, Trotta R, Liu S, Boyd Z, et al. Pro- and antiinflammatory cytokine signaling: reciprocal antagonism regulates interferon-gamma production by human natural killer cells. Immunity 2006;24(5):575–90 doi 10.1016/j.immuni.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, Zhang Y, Yi P, Dong W, Nalin AP, Zhang J, et al. The IL-15-AKT-XBP1s signaling pathway contributes to effector functions and survival in human NK cells. Nat Immunol 2019;20(1):10–7 doi 10.1038/s41590-018-0265-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peeters MJW, Dulkeviciute D, Draghi A, Ritter C, Rahbech A, Skadborg SK, et al. MERTK Acts as a Costimulatory Receptor on Human CD8(+) T Cells. Cancer Immunol Res 2019;7(9):1472–84 doi 10.1158/2326-6066.CIR-18-0841. [DOI] [PubMed] [Google Scholar]
- 29.Lu T, Chen L, Mansour AG, Yu MJ, Brooks N, Teng KY, et al. Cbl-b Is Upregulated and Plays a Negative Role in Activated Human NK Cells. J Immunol 2021;206(4):677–85 doi 10.4049/jimmunol.2000177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Denman CJ, Senyukov VV, Somanchi SS, Phatarpekar PV, Kopp LM, Johnson JL, et al. Membrane-bound IL-21 promotes sustained ex vivo proliferation of human natural killer cells. PLoS One 2012;7(1):e30264 doi 10.1371/journal.pone.0030264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nieto-Velazquez NG, Torres-Ramos YD, Munoz-Sanchez JL, Espinosa-Godoy L, Gomez-Cortes S, Moreno J, et al. Altered Expression of Natural Cytotoxicity Receptors and NKG2D on Peripheral Blood NK Cell Subsets in Breast Cancer Patients. Transl Oncol 2016;9(5):384–91 doi 10.1016/j.tranon.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kono K, Ressing ME, Brandt RM, Melief CJ, Potkul RK, Andersson B, et al. Decreased expression of signal-transducing zeta chain in peripheral T cells and natural killer cells in patients with cervical cancer. Clin Cancer Res 1996;2(11):1825–8. [PubMed] [Google Scholar]
- 33.Fauriat C, Mallet F, Olive D, Costello RT. Impaired activating receptor expression pattern in natural killer cells from patients with multiple myeloma. Leukemia 2006;20(4):732–3 doi 10.1038/sj.leu.2404096. [DOI] [PubMed] [Google Scholar]
- 34.Dong W, Wu X, Ma S, Wang Y, Nalin AP, Zhu Z, et al. The Mechanism of Anti-PD-L1 Antibody Efficacy against PD-L1-Negative Tumors Identifies NK Cells Expressing PD-L1 as a Cytolytic Effector. Cancer Discov 2019;9(10):1422–37 doi 10.1158/2159-8290.CD-18-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu S, Wurdak H, Wang Y, Galkin A, Tao H, Li J, et al. A genomic screen identifies TYRO3 as a MITF regulator in melanoma. Proc Natl Acad Sci U S A 2009;106(40):17025–30 doi 10.1073/pnas.0909292106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caligiuri MA, Zmuidzinas A, Manley TJ, Levine H, Smith KA, Ritz J. Functional consequences of interleukin 2 receptor expression on resting human lymphocytes. Identification of a novel natural killer cell subset with high affinity receptors. J Exp Med 1990;171(5):1509–26 doi 10.1084/jem.171.5.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagler A, Lanier LL, Phillips JH. Constitutive expression of high affinity interleukin 2 receptors on human CD16-natural killer cells in vivo. J Exp Med 1990;171(5):1527–33 doi 10.1084/jem.171.5.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poupot M, Fournie JJ, Poupot R. Trogocytosis and killing of IL-4-polarized monocytes by autologous NK cells. J Leukoc Biol 2008;84(5):1298–305 doi 10.1189/jlb.0508278. [DOI] [PubMed] [Google Scholar]
- 39.Tabiasco J, Espinosa E, Hudrisier D, Joly E, Fournie JJ, Vercellone A. Active trans-synaptic capture of membrane fragments by natural killer cells. Eur J Immunol 2002;32(5):1502–8 doi . [DOI] [PubMed] [Google Scholar]
- 40.Roda-Navarro P, Vales-Gomez M, Chisholm SE, Reyburn HT. Transfer of NKG2D and MICB at the cytotoxic NK cell immune synapse correlates with a reduction in NK cell cytotoxic function. Proc Natl Acad Sci U S A 2006;103(30):11258–63 doi 10.1073/pnas.0600721103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Somanchi SS, Somanchi A, Cooper LJ, Lee DA. Engineering lymph node homing of ex vivo-expanded human natural killer cells via trogocytosis of the chemokine receptor CCR7. Blood 2012;119(22):5164–72 doi 10.1182/blood-2011-11-389924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robertson MJ, Manley TJ, Donahue C, Levine H, Ritz J. Costimulatory signals are required for optimal proliferation of human natural killer cells. J Immunol 1993;150(5):1705–14. [PubMed] [Google Scholar]
- 43.Bakker AB, Wu J, Phillips JH, Lanier LL. NK cell activation: distinct stimulatory pathways counterbalancing inhibitory signals. Hum Immunol 2000;61(1):18–27 doi 10.1016/s0198-8859(99)00160-3. [DOI] [PubMed] [Google Scholar]
- 44.Moretta A, Bottino C, Vitale M, Pende D, Biassoni R, Mingari MC, et al. Receptors for HLA class-I molecules in human natural killer cells. Annu Rev Immunol 1996;14:619–48 doi 10.1146/annurev.immunol.14.1.619. [DOI] [PubMed] [Google Scholar]
- 45.Long EO. Regulation of immune responses through inhibitory receptors. Annu Rev Immunol 1999;17:875–904 doi 10.1146/annurev.immunol.17.1.875. [DOI] [PubMed] [Google Scholar]
- 46.Lopez-Botet M, Bellon T. Natural killer cell activation and inhibition by receptors for MHC class I. Curr Opin Immunol 1999;11(3):301–7 doi 10.1016/s0952-7915(99)80048-x. [DOI] [PubMed] [Google Scholar]
- 47.Nishimura M, Mitsunaga S, Akaza T, Mitomi Y, Tadokoro K, Juji T. Protection against natural killer cells by interferon-gamma treatment of K562 cells cannot be explained by augmented major histocompatibility complex class I expression. Immunology 1994;83(1):75–80. [PMC free article] [PubMed] [Google Scholar]
- 48.Campana S, De Pasquale C, Carrega P, Ferlazzo G, Bonaccorsi I. Cross-dressing: an alternative mechanism for antigen presentation. Immunol Lett 2015;168(2):349–54 doi 10.1016/j.imlet.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 49.Hwang I, Shen X, Sprent J. Direct stimulation of naive T cells by membrane vesicles from antigen-presenting cells: distinct roles for CD54 and B7 molecules. Proc Natl Acad Sci U S A 2003;100(11):6670–5 doi 10.1073/pnas.1131852100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andersson KE, Williams GS, Davis DM, Hoglund P. Quantifying the reduction in accessibility of the inhibitory NK cell receptor Ly49A caused by binding MHC class I proteins in cis. Eur J Immunol 2007;37(2):516–27 doi 10.1002/eji.200636693. [DOI] [PubMed] [Google Scholar]
- 51.Stitt TN, Conn G, Gore M, Lai C, Bruno J, Radziejewski C, et al. The anticoagulation factor protein S and its relative, Gas6, are ligands for the Tyro 3/Axl family of receptor tyrosine kinases. Cell 1995;80(4):661–70 doi 10.1016/0092-8674(95)90520-0. [DOI] [PubMed] [Google Scholar]
- 52.Varnum BC, Young C, Elliott G, Garcia A, Bartley TD, Fridell YW, et al. Axl receptor tyrosine kinase stimulated by the vitamin K-dependent protein encoded by growth-arrest-specific gene 6. Nature 1995;373(6515):623–6 doi 10.1038/373623a0. [DOI] [PubMed] [Google Scholar]
- 53.Nagata K, Ohashi K, Nakano T, Arita H, Zong C, Hanafusa H, et al. Identification of the product of growth arrest-specific gene 6 as a common ligand for Axl, Sky, and Mer receptor tyrosine kinases. J Biol Chem 1996;271(47):30022–7 doi 10.1074/jbc.271.47.30022. [DOI] [PubMed] [Google Scholar]
- 54.Dirks W, Rome D, Ringel F, Jager K, MacLeod RA, Drexler HG. Expression of the growth arrest-specific gene 6 (GAS6) in leukemia and lymphoma cell lines. Leuk Res 1999;23(7):643–51 doi 10.1016/s0145-2126(99)00075-2. [DOI] [PubMed] [Google Scholar]
- 55.Linger RM, Keating AK, Earp HS, Graham DK. TAM receptor tyrosine kinases: biologic functions, signaling, and potential therapeutic targeting in human cancer. Adv Cancer Res 2008;100:35–83 doi 10.1016/S0065-230X(08)00002-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ciurea SO, Schafer JR, Bassett R, Denman CJ, Cao K, Willis D, et al. Phase 1 clinical trial using mbIL21 ex vivo-expanded donor-derived NK cells after haploidentical transplantation. Blood 2017;130(16):1857–68 doi 10.1182/blood-2017-05-785659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shah N, Martin-Antonio B, Yang H, Ku S, Lee DA, Cooper LJ, et al. Antigen presenting cell-mediated expansion of human umbilical cord blood yields log-scale expansion of natural killer cells with anti-myeloma activity. PLoS One 2013;8(10):e76781 doi 10.1371/journal.pone.0076781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shah N, Li L, McCarty J, Kaur I, Yvon E, Shaim H, et al. Phase I study of cord blood-derived natural killer cells combined with autologous stem cell transplantation in multiple myeloma. Br J Haematol 2017;177(3):457–66 doi 10.1111/bjh.14570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shenouda MM, Gillgrass A, Nham T, Hogg R, Lee AJ, Chew MV, et al. Ex vivo expanded natural killer cells from breast cancer patients and healthy donors are highly cytotoxic against breast cancer cell lines and patient-derived tumours. Breast Cancer Res 2017;19(1):76 doi 10.1186/s13058-017-0867-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.