Abstract
The translocation liposarcoma (TLS) gene is fused to the ETS-related gene (ERG) in human myeloid leukemia, resulting in the generation of a TLS-ERG protein. We demonstrate that both TLS and the TLS-ERG leukemia fusion protein bind to RNA polymerase II through the TLS N-terminal domain, which is retained in the fusion protein; however, TLS recruits members of the serine-arginine (SR) family of splicing factors through its C-terminal domain, whereas the TLS-ERG fusion protein lacks the ability to recruit SR proteins due to replacement of the C-terminal domain by the fusion partner ERG. In transient-transfection assays, the TLS-ERG fusion protein inhibits E1A pre-mRNA splicing mediated by these TLS-associated SR proteins (TASR), and stable expression of the TLS-ERG fusion protein in K562 cells alters the splicing profile of CD44 mRNA. These results suggest that TLS fusion proteins may lead to cellular abnormalities by interfering with the splicing of important cellular regulators.
The TLS (also called FUS) gene was originally identified at the site of the t(12;16) chromosomal translocation in malignant liposarcomas, where it is fused to the CHOP (c/EBP homologous protein) gene (12, 36). In human myeloid leukemias with the t(16;21) chromosomal translocation, the TLS gene is fused to the ERG (ETS-related gene) gene (23). In both instances, the resultant TLS-CHOP and TLS-ERG fusion proteins generated by these translocations retain the N-terminal domain of TLS; however, in both fusion proteins the C-terminal domain of TLS is replaced by the DNA-binding domain from the corresponding transcription factor. The oncogenic potentials of both the TLS-CHOP and the TLS-ERG fusion proteins have been confirmed in transformation assays using mouse cell lines (24, 51) and normal human hematopoietic cells (32), respectively.
The role of TLS in normal cellular function and the mechanisms whereby TLS fusion proteins lead to transformation remain unclear. TLS belongs to a family of closely related proteins, including Ewing's sarcoma protein, EWS (13), and TATA-binding protein-associated factor, TAFII68 (3). Both EWS and TAFII68 interact with components of the RNA polymerase II (Pol II) complex (3, 4, 34), thus implicating TLS in transcriptional activation. The N-terminal domains of the TLS family of proteins are rich in glutamine, serine, and tyrosine, which are amino acid residues commonly found in transcriptional activation domains. Since TLS fusion proteins acquire their C-terminal region from the DNA-binding domains of the corresponding transcription factors, TLS fusion proteins are thought to lead to transformation by transcriptional activation of target genes (33). Moreover, TLS fusion proteins have been shown to transactivate reporter genes in transient-transfection assays (35, 51). However, mutagenesis studies have failed to demonstrate a correlation between the ability to transactivate and the ability to transform (26, 28).
The lack of a correlation between transactivation and transformation with TLS fusion proteins suggested that TLS fusion proteins might transform cells through the disruption of important cellular processes other than transcription. In this regard, there is considerable circumstantial evidence that TLS participates in RNA processing. The C-terminal domain of TLS contains two sequence motifs, ribonucleoprotein consensus sequence (RNP-CS) and arginine-glycine-glycine repeats (RGG), which are signatures of RNA-binding proteins (6). TLS binds to RNA and shuttles between the nucleus and the cytoplasm (52). When overexpressed in erythroid cells, TLS induces the preferential use of the most distal 5′ splice sites during E1A pre-mRNA splicing (19). TLS also associates with splicing factors ribonucleoprotein (RNP) A1 (51) and SF1 (49).
In this study, we demonstrate that TLS and its fusion protein TLS-ERG interact with RNA Pol II through the N-terminal domain of TLS, the domain preserved in the fusion protein. However, wild-type TLS recruits members of the serine-arginine family of splicing factors through the C-terminal domain of TLS, and the TLS-ERG fusion protein lacks this ability due to replacement of its C-terminal domain by the fusion partner. In the E1A pre-mRNA splicing assay, the TLS-ERG fusion protein interferes with E1A pre-mRNA splicing mediated by TLS-associated SR proteins in a dominant-negative manner. These results suggest that TLS protein functions as a docking molecule in the recruitment of SR splicing factors and that the TLS-ERG fusion protein inhibits this function of TLS.
MATERIALS AND METHODS
Two-hybrid screen and cDNA cloning.
A yeast two-hybrid-screen was performed as previously described (47), using the C-terminal domain of TLS as the bait (Fig. 1). To obtain full-length mouse TASR-2 cDNA, the TASR-2 insert identified from the yeast two-hybrid screen was used as a probe in the hybridization screening of a Uni-ZAP phage cDNA library derived from murine EML cells with erythroid-myeloid-lymphoid potentials (42). pBluescript phagemid containing the full-length TASR-2 cDNA was prepared after in vivo excision from the Uni-ZAP XR vector and was used in sequencing reactions with dye terminators (Applied Biosystems). Human TASR-2 cDNA was obtained from K562 leukemia cells by reverse transcription-PCR (RT-PCR) using primers designed on the basis of the mouse TASR-2 sequence.
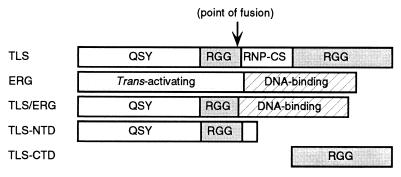
FIG. 1.
Schematic of TLS, TLS-ERG fusion protein, and TLS deletion mutants. Wild-type TLS and ERG proteins are shown with distinct sequence features: QSY, glutamine-, serine-, and tyrosine-rich domain; RGG, regions with multiple Arg-Gly-Gly repeats; RNP-CS, ribonucleoprotein consensus sequence. TLS-NTD indicates the TLS N-terminal domain, and TLS-CTD designates the TLS C-terminal domain (used as a bait in the yeast two-hybrid screen).
In vitro translation.
The TnT coupled reticulocyte lysate system (Promega) was used in the in vitro translation of TASR-2 cDNA. The DNA template was a pBluescript phagemid containing the full-length mouse TASR-2 cDNA along with the empty pBluescript vector as a negative control. The [35S]methionine-labeled protein was prepared as specified by the manufacturer and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 12% polyacrylamide gel.
Plasmid construction.
The cDNAs for TLS, TLS-ERG, and ERG were cloned into the EcoRI-SmaI sites of the pSG5-FL vector for transfection and expression of these proteins with the Flag epitope at the N-terminal end. Plasmid pSG5-FL-TLS-NTD contains a DNA insert corresponding to amino acids 1 to 290 of the TLS sequence, whereas pSG5-FL-TLS-CTD contains a DNA insert corresponding to amino acids 357 to 525 of the TLS sequence. Myc-tagged expression plasmids pCS2-MT-TASR-1 and pCS2-MT-TASR-2 were generated by in-frame cloning of full-length TASR cDNAs into the EcoRI-StuI sites of pCS2-MT vector. For the in vivo splicing assay, TASR cDNAs were inserted into the pMH vector (Boehringer Mannheim) to generate pMH-TASR-1 and pMH-TASR-2 with the influenza virus hemagglutinin (HA) epitope tagged at the C-terminal ends of TASR proteins. Reporter plasmid pCS3-MT-E1A was a kind gift from F. Moreau-Gachelin (19), and reporter plasmid pCS3-MT-E1A-9S was constructed by cloning the cDNA for the 9S E1A splicing isoform into the EcoRI-XbaI sites of pCS3-MT vector.
Immunoprecipitation and Western blotting.
For expression of Flag- or Myc-tagged proteins, 10 μg of the pSG5-Flag-expression construct and 10 μg of the pCS2-Myc-expression construct were introduced into 3 × 106 COS-7 cells by electroporation. At 48 h after electroporation, the cells were lysed with 0.6 ml of lysis buffer A (10 mM Tris-HCl [pH 7.4], 2.5 mM MgCl2, 100 mM NaCl, 0.5% Triton X-100). Prior to cell lysis, 30 μl of D8 polyclonal rabbit anti-Flag antibody (Santa Cruz Biotechnology), 3 μl of 9E10 monoclonal mouse anti-Myc antibody (Sigma), or 10 μl of 8WG16 monoclonal mouse anti-RNA Pol II antibody (Research Diagnostics, Inc.) was incubated with 30 μl of protein A/G agarose (Santa Cruz Biotechnology) for 50 min at 4°C in 0.3 ml of buffer A, and the antibody-protein A/G-agarose complex was then incubated with 0.2 ml of fresh cell lysate for 20 min at 4°C with gentle rocking. After the samples were washed with RIPA buffer four times, 50 μl of SDS-PAGE sample buffer was added to the agarose beads. The samples were heated at 100°C for 3 min, 20 μl of the sample was separated by SDS-PAGE in a 10% polyacrylamide gel, and the proteins were detected with M2 monoclonal mouse anti-Flag antibody (Sigma) or the 9E10 monoclonal mouse anti-Myc antibody as described in Results. Protein bands were visualized using the enhanced chemiluminescence Western blotting analysis system (Amersham).
E1A pre-mRNA splicing assay.
For in vivo splicing of E1A pre-mRNA, 2 μg of pCS3-MT-E1A and 2 μg of pMH-TASR plus 6 μg of pSG5-Flag construct were mixed with 60 μl of N-[1-(2,3-dioleoxy)propyl]-N,N,N-trimethylammonium (DOTAP; Boeringer Mannheim). The total amount of DNA for each 60-mm dish was kept at 5 μg by the addition of the corresponding empty vector. The DNA-DOTAP mixture was then added to two duplicate dishes with 65% confluent HeLa cells in 4 ml of Dulbecco modified Eagle medium containing 1% fetal bovine serum. After 7 h of incubation with the DNA-DOTAP mixture, the cells were replenished with Dulbecco modified Eagle medium containing 10% fetal bovine serum and further incubated for 40 h. The cells from one dish were then lysed with 0.25 ml of RIPA buffer for Western blot analysis, while the cells from the other dish were used for RNA isolation with an RNeasy column (Qiagen). Total RNA from transfected HeLa cells in each 60-mm dish was eluted with 50 μl of H2O from the RNeasy column.
For RT-PCR amplification of various E1A isoforms, 10 μl of total HeLa RNA was used for overnight hybridization to 10 pmol of T7 primer. After RT with SuperScript H− reverse transcriptase (GIBCO-BRL), the RNA was degraded by RNase H in a 40-μl reaction mixture. A 1.5-μl volume of the reaction mixture was then used as the DNA templates for PCR amplification of various E1A splicing isoforms. The forward primer was 5′GAGCTTGGGCGACCTCA3′ (RR67), and the reverse primer was 5′AATACGACTCACTATAG3′ (T7). The reactions were carried out in 50 μl containing 10 pmol of each primer, 0.2 mM each deoxynucleoside triphosphate, 1.5 mM MgCl2, 1× Taq buffer, and 2.5 U of Platinum Taq polymerase (GIBCO-BRL). PCR was performed by a 2-min incubation at 94°C followed by 35 cycles of 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C, with a final extension for 7 min at 72°C. To eliminate mismatches between different E1A isoforms, 3 μl of 0.1 M EDTA (pH 8.0) was added to each 50 μl of PCR products and the annealing was carried out at 80°C for 10 h. The PCR products were then separated on a 1.5% agarose gel, denatured with NaOH, transferred to a nylon membrane, and detected with a 32P-labeled E1A cDNA probe.
For the RNase protection assay, 10 μl of the total RNA was hybridized to 1.5 × 106 cpm of 32P-labeled RNA probe antisense to the E1A genomic sequence (covering bases 499 to 1316 of the E1A gene). After overnight hybridization, excessive RNA probe was digested with a mixture of RNase A plus RNase T1 supplied with the RNase protection assay system (PharMingen). The protected antisense E1A RNA fragments were isolated as specified by the manufacturer and were separated on a 6% QuickPoint precast denaturing gel (Novex).
CD44 splicing assay.
For the generation of retroviral constructs that express Flag-tagged proteins, Flag-TLS, Flag-TLS-ERG, and Flag-ERG cDNA cassettes were cloned into the HpaI-BamHI sites of retroviral vector LXSN. The retroviral particles were first packaged in PG13 cells and then transduced into K562 cells. After selection with G418 at a concentration of 1 mg/ml of medium, the G418-resistant cells were collected for Western blotting analysis and RNA isolation.
For RT-PCR analysis of CD44 splicing, 10 μg of total RNA from K562 cells was hybridized overnight to 10 pmol of oligo(dT) adapter primer 5′GGCCACGCGTCGACTAGTACTTTTTTTTTTTTTTTTT3′. After RT by SuperScript H− reverse transcriptase, the total RNA was degraded by RNase H in a 40-μl reaction mixture. A 1.5-μl sample of the RT mixture was then used as the DNA templates for PCR amplification of various CD44 splicing isoforms with the Platinum Taq DNA polymerase. To detect subtle changes of CD44 alternative splicing after expression of the Flag-TLS-ERG cassette in K562 cells, two pairs of CD44 primers were used in the PCR. The first primer pair consisted of 5′TCCCAGTATGACACATATTGC3′ (P1) and 5′CAGCTGTCCCTGTTGTCGAA3′ (V6D), and the second primer pair consisted of 5′GTACGTCTTCAAATACCATC3′ (V3U) and 5′CAAGATGATCAGCCATTCTGG3′ (P2). PCR amplification was performed by a 2-min incubation at 94°C followed by 35 cycles of 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C, with a final extension for 7 min at 72°C. To eliminate mismatches between different CD44 splicing isoforms, 3 μl of 0.1 M EDTA (pH 8.0) was added to each 50 μl of PCR products and the annealing was carried out at 80°C for 10 h. The PCR products were then separated on a 1.5% agarose gel, denatured with NaOH, transferred to a nylon membrane, and detected with a 32P-labeled CD44H cDNA probe spanning constitutive exons 3, 4, 5, 16, 17, and 18 (see Fig. 7B).
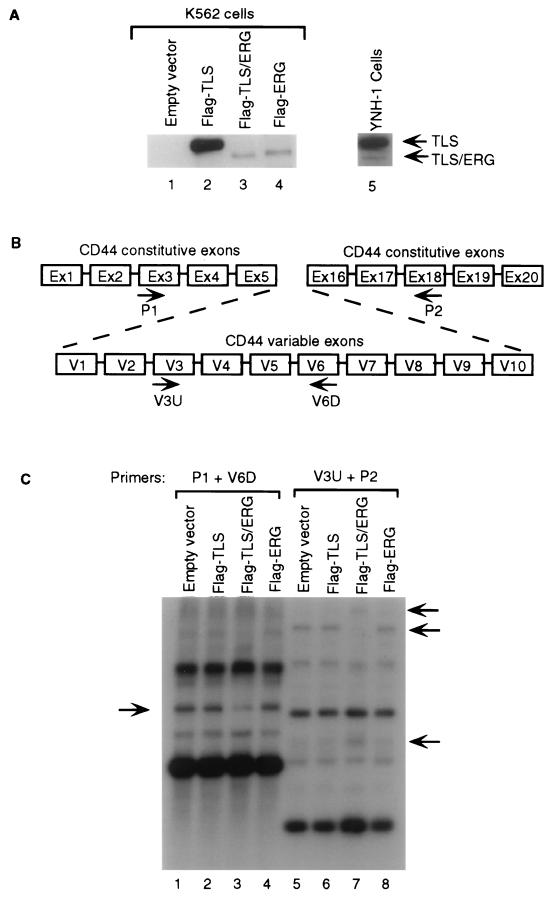
FIG. 7.
Effects of TLS-ERG stable expression on CD44 splicing in K562 cells. (A) Western blot analyses of lysates from K562 cells harboring the empty retroviral vector LXSN (lane 1) or K562 cells harboring the retroviral construct with Flag-TLS (lane 2), Flag-TLS-ERG (lane 3), or Flag-ERG (lane 4) were carried out with a mouse anti-Flag antibody. Lysate from the human YNH-1 myeloid leukemia cells (which express endogenous TLS-ERG fusion protein) was blotted with a rabbit anti-TLS antibody that recognizes the N-terminal region of TLS (lane 5). (B) Genomic structure of the CD44 gene. Regions of constitutive exons and variable exons are indicated. The sizes of the exons are not drawn to scale. Arrows designate primers used for RT-PCR amplification. (C) Southern blot analysis of RT-PCR products for CD44 splicing isoforms. Lanes 1 to 4 represent CD44 RT-PCR products using primer P1 and V6D. Lanes 5 to 8 represent CD44 RT-PCR products using primer 3VU and P2. All RT-PCR products were separated on a 1.5% agarose gel and probed with a 32P-labeled CD44 DNA probe. Arrows indicate positions of CD44 splicing isoforms that are affected by exogenous Flag-TLS-ERG in K562 cells.
RESULTS
TLS N-terminal domain mediates interactions with RNA Pol II.
The structural features of TLS, TLS-ERG, ERG, TLS-NTD (N-terminal domain), and TLS-CTD (C-terminal domain) are shown in the schematic (Fig. 1). The N-terminal domain of TLS is rich in glutamine, serine, and tyrosine residues and has been suggested to function as a transcriptional activation domain (25, 35, 51). In addition, EWS and TAFII68, two proteins highly homologous to TLS, interact with components of the RNA Pol II complex (3, 4, 34). These observations suggested that TLS might physically interact with RNA Pol II.
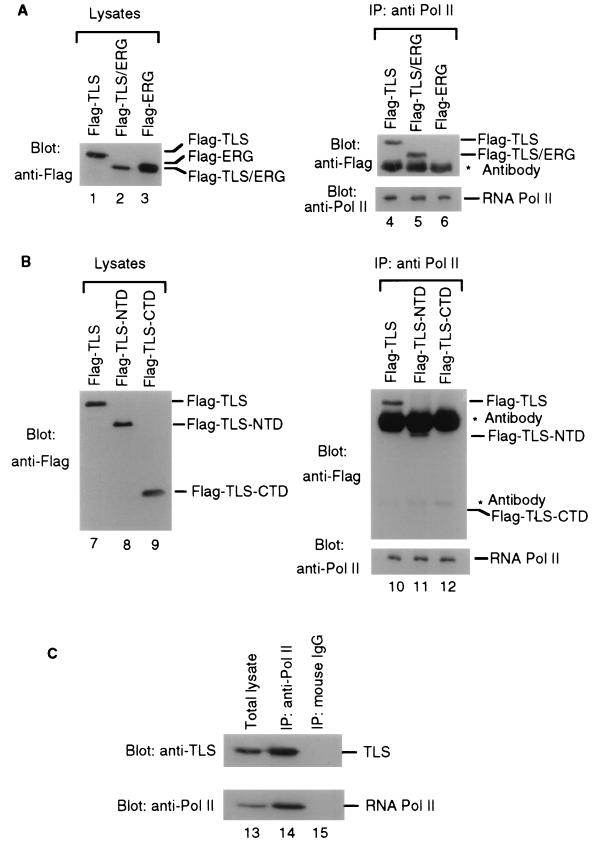
To examine the potential interaction of TLS and RNA Pol II, COS-7 cells were transfected with plasmids expressing Flag-tagged TLS, TLS-ERG, and ERG proteins (Fig. 2A, lanes 1 to 3). Lysates from the transfected cells were then immunoprecipitated with a mouse monoclonal antibody recognizing the C-terminal heptapeptide repeats on the largest subunit of RNA Pol II. Western blotting analysis showed that both Flag-TLS and Flag-TLS-ERG, but not Flag-ERG, were present in the immunocomplexes brought down by the anti-RNA Pol II antibody (lanes 4 to 6). Interestingly, more Flag-TLS-ERG than Flag-TLS was coimmunoprecipitated by the anti-RNA Pol II antibody despite the demonstration that Flag-TLS-ERG was less abundant than Flag-TLS in the total-cell lysates. These results suggest that TLS-ERG leukemia fusion protein may have a higher affinity toward RNA Pol II than does wild-type TLS or that TLS-ERG may localize preferentially to the nucleus.
FIG. 2.
Association of TLS, TLS-ERG, and TLS-NTD with RNA Pol II. (A) COS-7 cells were transfected with Flag-tagged TLS, TLS-ERG, or ERG expression plasmid, and the cell lysates were probed with the M2 anti-Flag antibody to confirm protein expression (lanes 1 to 3). The same lysates were immunoprecipitated with a mouse monoclonal 8WG16 antibody (Research Diagnostics, Inc.) recognizing the C-terminal heptapeptide repeat on the largest subunit of RNA Pol II (lanes 4 to 6), and the immunoprecipitates were blotted with an M2 anti-Flag antibody or a rabbit polyclonal anti-RNA Pol II antibody (Santa Cruz Biotechnology). (B) COS-7 cells were transfected with Flag-tagged TLS deletion constructs, and the lysates were probed with the M2 anti-Flag antibody to confirm protein expression (lanes 7 to 9). The same lysates were immunoprecipitated with the 8WG16 anti-RNA Pol II antibody and blotted with an anti-Flag antibody or an anti-RNA Pol II antibody (lanes 10 to 12). (C) HeLa cell lysate (lane 13) was immunoprecipitated (IP) with the 8WG16 anti-RNA Pol II antibody (lane 14) or a mouse immunoglobulin G (IgG) (lane 15) and then blotted with a rabbit anti-TLS antibody and a rabbit anti-Pol II antibody.
To test our hypothesis that the interaction between TLS and RNA Pol II is mediated through the N-terminal domain of TLS, two additional plasmids expressing the Flag-tagged N-terminal domain of TLS (Flag-TLS-NTD) or the Flag-tagged C-terminal domain of TLS (Flag-TLS-CTD) were constructed (Fig. 1A). After transfection, both Flag-TLS-NTD and Flag-TLS-CTD were expressed in COS-7 cells and migrated faster than Flag-TLS on SDS-PAGE (Fig. 2B, lanes 7 to 9). Along with Flag-TLS, Flag-TLS-NTD was detectable in the anti-RNA Pol II immunocomplex (lanes 10 and 11) whereas Flag-TLS-CTD was absent from the immunocomplex (lane 12). These results suggested that the N-terminal domain of wild-type TLS protein interacts with RNA Pol II and that this ability is retained by the TLS-ERG fusion protein.
To confirm that endogenous TLS associates with endogenous RNA Pol II, we carried out coimmunoprecipitation experiments with the same anti-RNA Pol II antibody using human cell lysates. In these experiments, we used a rabbit anti-TLS antibody (a kind gift from F. Moreau-Gachelin) that recognizes the TLS N-terminal region to detect endogenous TLS. We found that TLS is abundantly expressed in a variety of human cell lines including HeLa (Fig. 2C, lane 13). Endogenous TLS was also present in the anti-Pol II immunocomplex (lane 14) but not in the immunocomplex with the mouse IgG (lane 15), indicating that endogenous TLS associates with RNA Pol II.
TASR-2 is a serine-arginine splicing factor.
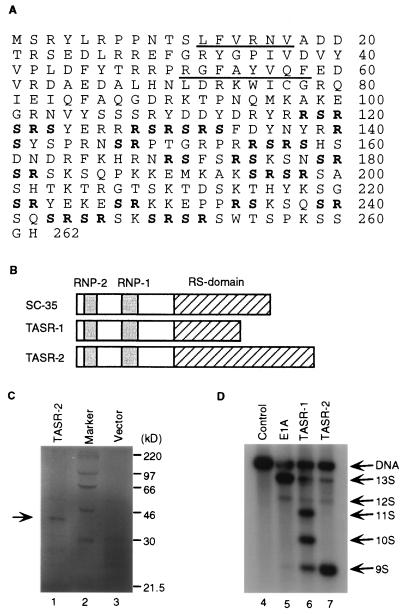
We recently demonstrated through yeast two-hybrid screening that TLS interacts with a well-known SR protein SC35 and a novel SR protein termed TASR-1 (47). To obtain a more complete picture of the proteins interacting with TLS, we carried out a second yeast two-hybrid screen of a mouse hematopoietic cDNA library using the C-terminal domain of TLS as the bait (Fig. 1A). We isolated a cDNA encoding a second novel TLS-associated SR protein termed TASR-2. Mouse TASR-2 cDNA (GenBank accession number AF060490) is 2.7 kb long and encodes a protein of 262 amino acids with a calculated molecular mass of 31 kDa (Fig. 3A). The cDNA encoding human TASR-2 (GenBank accession number AF067730) was isolated from K562 cells by RT-PCR. Mouse and human TASR-2 are identical at the amino acid level, suggesting that TASR-2 is evolutionarily conserved. Similar to SC35 and TASR-1, TASR-2 also has typical RNP2 and RPN1 motifs in its N-terminal region and multiple serine-arginine repeats in its C-terminal region (Fig. 3B) that are characteristic of prototype SR proteins. Even though the calculated molecular mass of TASR-2 is 31 kDa, in vitro-translated TASR-2 protein migrated as a band of 37 kDa (Fig. 3C, lane 1) when analyzed by SDS-PAGE. This discrepancy between the calculated molecular size and that determined by SDS-PAGE is typical of SR proteins such as SC35 and SF2/ASF (16) and results from phosphorylation of the serine residues and perhaps regions of alternating charge.
FIG. 3.
Amino acid sequence and characterization of TASR-2. (A) The predicted amino acid sequence of TASR-2 protein is shown. Mouse TASR-2 (GenBank accession number AF060490) and human TASR-2 (GenBank accession number AF067730) are identical at the amino acid level. RNP2 and RNP1 consensus sequences are underlined. Arg-Ser or Ser-Arg dipeptide repeats are shown in bold type. (B) Schematic comparison of SC35, TASR-1, and TASR-2. The RNP consensus sequences are shown in gray boxes. The RS domains are in hatched boxes. (C) In vitro translation reaction products with pBluescript-TASR-2 as the template (lane 1) are separated by SDS-PAGE on a 12% polyacrylamide gel along with protein molecular markers (lane 2) and control reaction products with the pBluescript empty vector (lane 3). The position of the TASR-2 protein is indicated by an arrow. (D) A 2.5-μg sample of pCS3-MT-E1A reporter plasmid was cotransfected with 2.5 μg of pCR3 empty vector (lanes 4 and 5) or with 2.5 μg of pCR3-TASR-1 (lane 6) and 2.5 μg of pCS3-TASR-2 (lane 7) expression plasmid into HeLa cells. RT-PCR was carried out as described in Materials and Methods, and the E1A splicing products were confirmed by hybridization to a 32P-labeled E1A DNA probe. Lane 4 contains control RT-PCR products from a reaction carried out in the absence of reverse transcriptase to show the presence of residual pCS3-MT-E1A template DNA in the total RNA.
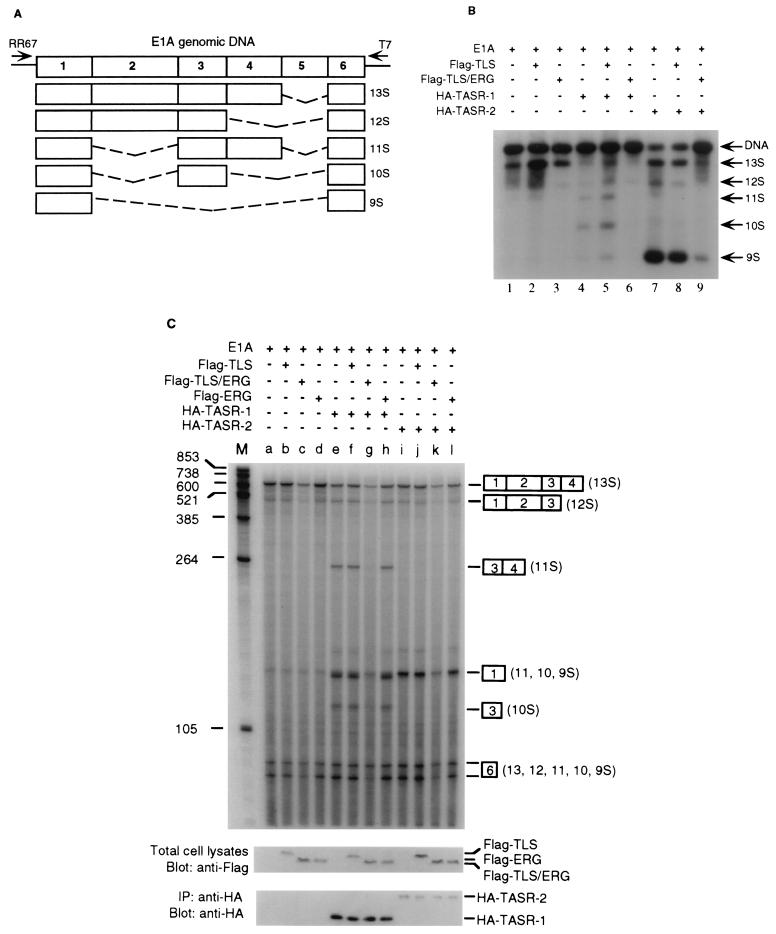
To confirm that TASR-2 functions as a splicing factor, its effect on the alternative splicing of an E1A reporter gene was tested. The alternative splicing of E1A pre-mRNA leads to the generation of five different splicing isoforms designated 13S, 12S, 11S, 10S, and 9S mRNA (see Fig. 5A). For these studies of E1A pre-mRNA splicing, HeLa cells were transfected with the pCS3-MT-E1A reporter containing the E1A genomic sequence, and the alternative splicing of the E1A pre-mRNA was examined by RT-PCR. In HeLa cells the dominant splicing products of E1A pre-mRNA are 13S and 12S isoforms (Fig. 3D, lane 5). TASR-1 promotes multiple splice site selections leading to 11S, 10S, and 9S isoforms (lane 6) whereas coexpression of TASR-2 promotes 5′ splice site selection, leading primarily to the 9S isoform (lane 7). Due to the presence of residual plasmid DNA in the total RNA, the E1A genomic sequence could be amplified by PCR (lane 4).
FIG. 5.
TLS-ERG inhibition of TASR-mediated E1A pre-mRNA splicing. (A) Diagram of individual E1A pre-mRNA splicing isoforms. Numbers indicate individual exons, and dashed lines represent spliced sequences. (B) Alternative splicing of exogenous E1A pre-mRNA was analyzed by RT-PCR using RNA from HeLa cells transfected with the pCS3-MT-E1A reporter construct plus various expression plasmids. DNA combinations for all samples are indicated on the top. (C) In vivo alternative splicing of E1A pre-mRNA in HeLa cells was analyzed by an RNase protection assay. DNA combinations for all samples are indicated at the top, positions of RNA markers are labeled at the left, protected E1A RNA fragments are shown on the right with exons designated by numerals in boxes, and levels of protein expression are shown at the bottom. Since HA-tagged TASR proteins (which promote identical E1A pre-mRNA splicing to the untagged TASR proteins) were not detectable from total-cell lysates, they were concentrated by immunoprecipitation (IP) with a rabbit anti-HA antibody prior to Western blotting.
The TLS C-terminal domain mediates interaction with SR proteins.
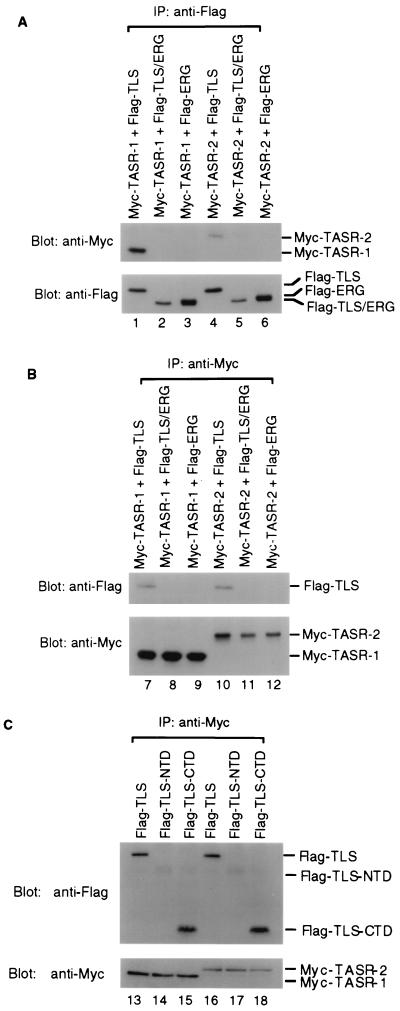
We have previously shown through coimmunoprecipitation that TLS interacts with SC35 and TASR-1 (47). Since the C-terminal domain of TLS is replaced by the DNA-binding domain of ERG in the TLS-ERG leukemia fusion protein, we investigated the interaction between TLS and TLS-ERG and the TASR-1 and TASR-2 proteins. Plasmids expressing Flag-tagged TLS, TLS-ERG, or ERG were cotransfected into COS-7 cells with plasmids expressing Myc-tagged TASR-1 or TASR-2, and lysates from the cotransfected cells were used for immunoprecipitation. In immunoprecipitates brought down using an anti-Flag antibody, Myc-TASR-1 and Myc-TASR-2 were found to associate with the wild-type Flag-TLS (Fig. 4A, lanes 1 and 4) but not with the Flag-TLS-ERG leukemia fusion protein or wild-type Flag-ERG (lanes 2, 3, 5, and 6). In the reciprocal immunoprecipitation, only Flag-TLS was coimmunoprecipitated with Myc-TASR-1 and Myc-TASR-2 using an anti-Myc antibody (Fig. 4B, lanes 7 and 10) whereas Flag-TLS-ERG and Flag-ERG were not immunoprecipitated (lanes 8, 9, 11, and 12). Since Flag-TLS and Flag-TLS-ERG differ only in their C-terminal domains, these results suggested that the TLS C-terminal domain was responsible for interaction with TLS-associated SR proteins. In further experiments, the Flag-tagged TLS C-terminal domain alone was shown to coimmunoprecipitate with both Myc-TASR-1 and Myc-TASR-2 (Fig. 4C, lanes 15 and 18) whereas the Flag-tagged TLS N-terminal domain did not interact with either TASR protein (lanes 14 and 17).
FIG. 4.
Association of TLS-CTD with TASR proteins in COS-7 cells. (A) Plasmids expressing Myc-tagged TASR-1 (lanes 1 to 3) and TASR-2 (lanes 4 to 6) were cotransfected into COS-7 cells with plasmids expressing Flag-tagged TLS, TLS/ERG, or ERG. The cell lysates were immunoprecipitated (IP) with a rabbit polyclonal D8 anti-Flag antibody (Santa Cruz Biotechnology). The immunoprecipitates were blotted with a mouse monoclonal M2 anti-Flag antibody (Sigma) or a mouse monoclonal 9E10 anti-Myc antibody (Sigma). (B) The reciprocal immunoprecipitation described for panel A was carried out with the 9E10 anti-Myc antibody using the same lysates from cells coexpressing Myc-TASR-1 (lanes 7 to 9) or Myc-TASR-2 (lanes 10 to 12), along with Flag-tagged TLS, TLS-ERG, or ERG. (C) Plasmids expressing Myc-tagged TASR-1 (lanes 13 to 15) and TASR-2 (lanes 16 to 19) were cotransfected into COS-7 cells with plasmids expressing Flag-tagged TLS, TLS-NTD, or TLS-CTD. The cell lysates were immunoprecipitated with the 9E10 anti-Myc antibody, and the immunoprecipitates were separated by SDS-PAGE on a 12% polyacrylamide gel and blotted with the anti-Flag antibody.
The TLS-ERG leukemia fusion protein inhibits E1A pre-mRNA splicing.
It is now increasingly recognized that gene transcription by RNA Pol II can be coupled to RNA processing (5, 14, 15, 22, 29, 30, 38, 43, 45, 50). As transcriptional elongation by RNA Pol II is taking place, the nascent pre-mRNA is processed by the splicing machinery associated with RNA Pol II. Because TLS interacts with both RNA Pol II and SR splicing factors and because TLS-ERG lacks the ability to recruit SR proteins due to the replacement of the C-terminal domain by the fusion partner, the fusion protein might cause alterations in RNA splicing mediated by the SR proteins.
The adenovirus E1A gene was used as a reporter gene to analyze splicing, since the alternative splicing of E1A pre-mRNA by SR proteins has been well characterized (7, 44). The five different E1A splicing isoforms designated 13S, 12S, 11S, 10S, and 9S mRNA are illustrated (Fig. 5A). The effect of TLS or the TLS-ERG leukemia fusion protein on TASR-mediated alternative splicing of adenovirus E1A pre-mRNA was tested in HeLa cells and analyzed by RT-PCR. In the absence of exogenous SR protein expression, the major splicing products of E1A pre-mRNA were 13S and 12S isoforms (Fig. 5B, lanes 1 to 3). TASR-1 overexpression resulted in an increase in the 11S, 10S, and 9S isoforms (lane 4), whereas TASR-2 overexpression predominantly increased the 9S isoform (lane 7). Although coexpression of wild-type TLS did not significantly alter TASR-mediated E1A pre-mRNA splicing (lanes 5 and 8), most probably due to the high levels of endogenous TLS, coexpression of TLS-ERG resulted in nearly complete inhibition of E1A pre-mRNA splicing mediated by both TASR proteins (lanes 6 and 9).
To confirm that these results reflected changes in E1A pre-mRNA splicing patterns and were not due to selective amplification by PCR, we used an RNase protection assay to measure the steady-state levels of the various E1A splicing isoforms. After hybridization to a 32P-labeled antisense E1A RNA probe followed by RNase digestion and gel electrophoresis, each E1A splicing product could be identified according to the distinct sizes of the protected RNA fragments. In the absence of exogenous SR protein expression, alternative splicing of E1A pre-mRNA in HeLa cells normally generated 13S, 12S, and 9S isoforms (Fig. 5C, lanes a to d). TASR-1 overexpression resulted in an increase in the levels of 11S, 10S, and 9S isoforms (lane e), whereas TASR-2 overexpression favored 5′ splice site selection, leading primarily to the 9S isoform (lane i). Although coexpression of Flag-TLS and Flag-ERG did not significantly alter TASR-mediated E1A pre-mRNA splicing (lanes f, h, j, and l), coexpression of Flag-TLS-ERG led to a decrease in the levels of 13S and 12S isoforms and an almost complete inhibition of the 11S, 10S, and 9S isoforms mediated by both TASR proteins (lanes g and k). By Western blotting, the levels of Flag-tagged TLS, TLS-ERG, ERG and HA-tagged TASR proteins were shown to remain consistent in all samples, demonstrating that the observed inhibition of E1A splicing was not due to variation in transfection or decrease in TASR expression caused by the TLS-ERG fusion protein. Unspliced E1A pre-mRNA was not detected by RNase protection, consistent with previous reports (7, 44) indicating that unprocessed E1A pre-mRNA is labile under the in vivo experimental conditions. The SR splicing factor SC35 was not assessed in this study, because endogenous SC35 is abundant in HeLa cells and overexpression of SC35 has minimal effect on E1A pre-mRNA splicing (44, 47).
To rule out the possibility that the observed TLS-ERG inhibition of TASR-mediated E1A pre-mRNA splicing was due to repression of transcription from the pCS3-MT-E1A reporter or preferential destabilization of the 11S, 10S, and 9S E1A splicing isoforms, we constructed an additional reporter plasmid, pCS3-MT-E1A-9S, which was generated from the same vector as pCS3-MT-E1A but contains a cDNA insert corresponding to the 9S E1A splicing isoform. When this construct was analyzed under the same conditions, the resultant 9S E1A mRNA levels were not decreased by coexpression of TLS-ERG or altered by TASR-1 or TASR-2 (data not shown). These results indicated that TLS-ERG did not suppress transcription from the pCS3-MT-E1A vector or selectively destabilize the 9S E1A splicing isoform.
TLS deletion mutants are unable to inhibit E1A pre-mRNA splicing.
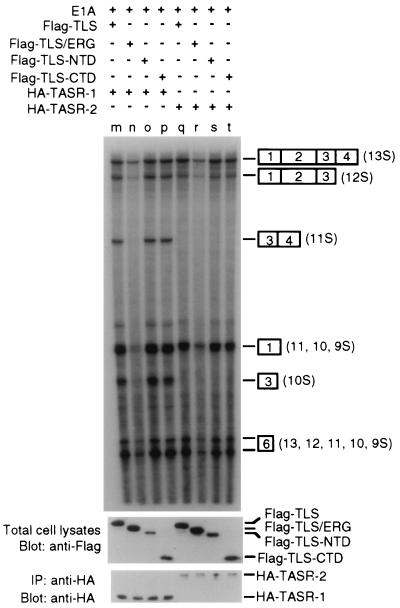
Since the TLS N-terminal domain mediates the interaction with RNA Pol II and the C-terminal domain binds and recruits SR splicing factors, TLS appears to have two functionally distinct domains. To investigate whether the TLS N-terminal or C-terminal domain alone inhibited TASR-mediated E1A pre-mRNA splicing, TLS deletion plasmids and TASR expression plasmids were cotransfected into HeLa cells along with the pCS3-MT-E1A reporter construct and the alternative splicing of E1A pre-mRNA was analyzed by the RNase protection assay. The Flag-TLS-ERG leukemia fusion protein again inhibited TASR-mediated E1A pre-mRNA splicing (Fig. 6, lanes n and r). However, neither the Flag-tagged TLS N-terminal domain nor the C-terminal domain alone affected TASR-mediated E1A pre-mRNA splicing (lanes o, p, s, and t). These results indicated that neither TLS deletion mutant was sufficient to disrupt E1A pre-mRNA splicing mediated by TLS-associated SR proteins and that both the TLS N-terminal domain (RNA Pol II-interaction domain) and the ERG C-terminal domain (DNA-binding domain) were required for the TLS-ERG leukemia fusion protein to inhibit E1A pre-mRNA splicing.
FIG. 6.
Effects of TLS deletion mutants on TASR-mediated E1A pre-mRNA splicing. The TLS deletion mutant containing the N-terminal or C-terminal domain was transfected into HeLa cells, and its effect on TASR-mediated E1A pre-mRNA splicing was examined by an RNase protection assay. Bottom panels represent Western blots of TLS and TLS deletion mutants as well as immunoprecipitation (IP) followed by Western blotting to demonstrate the presence of TASR-1 and TASR-2. Labeling is as in Fig. 5C.
Stable expression of the TLS-ERG fusion protein affects CD44 alternative splicing.
We have shown that coexpression of TLS-ERG inhibits TASR-mediated E1A pre-mRNA splicing in transient-transfection experiments. Since these results were obtained through overexpression of both TLS-ERG and the E1A reporter gene, overexpression of the TLS-ERG fusion protein might displace binding to RNA Pol II by other splicing factors, resulting in a generic decrease in RNA splicing.
To investigate the effect of exogenous TLS-ERG on endogenous RNA splicing when TLS-ERG is expressed at a level comparable to the endogenous protein, we subcloned the Flag-TLS, Flag-TLS-ERG, and Flag-ERG cDNA cassettes into the retroviral vector LXSN. Through retroviral transduction, K562 cells harboring Flag-TLS, TLS-ERG, and ERG retroviral constructs were obtained using G418 selection. From these transduced K562 cells, Flag-tagged proteins were readily detectable by Western blotting with an anti-Flag antibody (Fig. 7A, lanes 1 to 4). The protein level and the relative ratio between the exogenous Flag-TLS and Flag-TLS-ERG in retrovirus-transduced K562 cells were comparable to those between the endogenous TLS and TLS-ERG in YNH-1 cells (compare lanes 2 and 3 to lane 5). YNH-1 is a cell line from a myeloid leukemia patient with the t(16;21) translocation that expresses the TLS-ERG fusion protein (46).
To examine whether stable expression of the exogenous Flag-TLS-ERG fusion protein affects the splicing of an important endogenous RNA, we investigated the alternative splicing profiles of CD44 mRNA in K562 cells harboring different LXSN retroviral constructs. The CD44 gene encodes a cell adhesion molecule consisting of 10 constitutive exons and 10 variable exons (Fig. 7B). Different combinations of the variable exons lead to a variety of CD44 splicing isoforms that differ in their extracellular domains. Abnormal splicing of CD44 mRNA has been found both in solid tumors and in leukemia (10), and recently it has been suggested that stage-specific changes in SR proteins may be linked to changes in CD44 splicing during different stages of mammary cancer (41).
The predominant CD44 mRNA in K562 cells is the CD44H isoform which lacks any of the variable exons (data not shown). To detect subtle changes in variable-exon-containing CD44 splicing isoforms, we used RT-PCR and primers P1 and V6D to amplify CD44 isoforms that contain the variable V6 exon and primers V3U and P2 to amplify CD44 isoforms that contain the variable V3 exon. The amplified CD44 products were then separated by gel electrophoresis and probed with a 32P-labeled CD44 DNA probe. As shown in Fig. 7C, expression of the Flag-TLS or Flag-ERG protein in K562 cells did not alter the profile of CD44 isoforms containing the V6 exon (compare lane 1 with lanes 2 and 4) or the profile of CD44 isoforms containing the V3 exon (compare lane 5 with lanes 6 and 8). However, expression of the Flag-TLS-ERG fusion protein in K562 cells changed the profile of endogenous CD44 isoforms containing the V6 exon (compare lane 1 with lane 3) and the V3 exon (compare lane 5 with lane 7).
DISCUSSION
These studies indicate that TLS interacts with RNA Pol II through the N-terminal domain of TLS and interacts with members of the SR family of splicing factors through the C-terminal domain of TLS. In contrast, the TLS-ERG fusion protein binds RNA Pol II but is unable to bind SR proteins, due to replacement of its C-terminal domain by the fusion partner. The TLS-ERG fusion protein not only lacks the ability to bridge the binding of RNA Pol II to SR proteins but also inhibits E1A pre-mRNA splicing mediated by these SR proteins in transfected HeLa cells. When Flag-TLS-ERG was introduced through retroviral transduction into K562 cells and expressed at a level comparable to the endogenous protein, it changed the splicing profile of the endogenous CD44, a cell adhesion molecule whose abnormal splicing is associated with cellular transformation and tumor metastasis (10).
RNA splicing, a critical step in gene expression, is increasingly recognized as a cotranscriptional event (reviewed in references 31 and 40). Experimental evidence now indicates that transcription, capping, and polyadenylation are also intimately linked by RNA Pol II through its association with RNA-processing factors (9, 21). Among different components of the transcriptional machinery, the C-terminal domain of the largest subunit of RNA Pol II complex is thought to be especially critical for recruitment of RNA-processing factors (29, 30). Even though a novel set of C-terminal domain associated SR-like proteins directly interact via their C-terminal domain-interacting domains with RNA Pol II (11, 48), prototypical SR proteins including SC35 and ASF/SF2 lack such C-terminal domain-interacting domains. At present, the way in which these SR proteins associate with RNA Pol II remains unclear. Recently, a novel transcription coactivator, p52, was found to interact specifically with ASF/SF2 and was suggested to act as an adapter to coordinate transcription and splicing (17). Based on our observation that TLS interacts with both RNA Pol II and SR proteins, we propose that TLS functions as a docking molecule by recruiting SR splicing factors to RNA Pol II, thus coupling gene transcription with RNA splicing.
The TLS gene is ubiquitously expressed, suggesting that TLS may be essential for cell function (1). In myeloid leukemia cells with the t(16;21) translocation and in liposarcoma cells with the t(12;16) translocation, only one TLS allele is interrupted by chromosomal translocation while the remaining TLS allele is intact (12, 36, 46). This indicates that the TLS fusion protein functions in a dominant-negative manner.
The TLS-ERG fusion protein not only inhibits TASR-mediated E1A pre-mRNA splicing, leading to 11S, 10S, and 9S isoforms, but also decreases the expression of the 13S and 12S E1A splicing isoforms. The generation of 13S and 12S isoforms may be mediated by additional splicing factors present in HeLa cells, which in turn implies that the splicing pathway via TLS potentially includes other splicing factors. Based on the observation that expression of the TLS-ERG leukemia fusion protein, despite the presence of endogenous TLS protein, is sufficient to inhibit E1A pre-mRNA splicing in HeLa cells and to alter CD44 splicing in K562 cells, the TLS-ERG fusion protein appears to function in a dominant-negative manner to interfere with RNA splicing. Our finding that neither the N-terminal nor the C-terminal deletion mutant of TLS was sufficient to block TASR-mediated E1A splicing suggests that inclusion of the ERG DNA-binding domain is required for the dominant-negative function of the TLS-ERG leukemia fusion protein, possibly by conferring a higher affinity of the fusion protein for RNA Pol II and/or by localizing the fusion protein to the nucleus.
One can envisage at least two potential mechanisms whereby the TLS-ERG fusion protein alters CD44 splicing. In the first scenario, if splicing into a specific CD44 isoform requires docking by TLS, the TLS-ERG fusion protein might block this pathway, leading to degradation of the unfinished CD44 pre-mRNA. In the second scenario, if one CD44 splicing pathway is blocked and alternative splicing pathways exist, the inhibition of the first pathway by the TLS-ERG fusion protein might push the splicing of CD44 pre-mRNA through alternative routes, thus increasing the chance of aberrant splicing.
SR proteins appear to possess the capacity to alter gene expression by influencing RNA splicing. The important regulatory roles of SR proteins in cellular processes have been demonstrated by their functions in Drosophila development and sex determination (2, 20, 37), their involvement in T-cell activation (39), and their close association with cell cycle control (27). Even though transcriptional deregulation by oncogenic fusion proteins has attracted much of the attention, aberrant RNA splicing is also frequently detected in cancer cells and is associated with cellular transformation (8, 18) and tumor metastasis (10). The mechanism underlying aberrant splicing has yet to be elucidated. This study links a specific chromosomal translocation, and the resultant leukemia fusion protein, to disruption of RNA splicing. Although our observation is made from cells expressing exogenous Flag-TLS-ERG fusion protein and further experiments with cancer cells harboring TLS translocations are needed, these results suggest that TLS fusion proteins may lead to abnormal splicing of genes critical for cell growth and differentiation.
Ewing's sarcoma protein EWS and TATA-binding protein-associated factor TAFII68 share sequence homology with TLS, and it is likely that both EWS and TAFII68 also interact with SR splicing factors. The potential disruption of coupling between transcription and RNA splicing by EWS fusion proteins especially deserves further investigation. In addition to fusion with the Fli-1 protein in 90% of cases of Ewing's sarcoma, EWS is involved in chromosomal translocations with a variety of transcription factors including ERG, ETV1, E1A-F, FEV, ATF-1, WT1, and TEC1 (4). In all of these fusions involving EWS, the N-terminal domain of EWS is retained. EWS fusion proteins may thus function in a manner analogous to the TLS fusion proteins by interfering with the recruitment of RNA-processing factors such as the SR proteins.
ACKNOWLEDGMENTS
We thank R. S. Morrison, M. B. Roth, and Y.-C. Yang for helpful discussions; T. R. Bauer, Jr., S. Collins, and B. Kwiatkowski for critical reading of the manuscript; and S. L. Danner and S. M. Stray for DNA sequencing.
REFERENCES
- 1.Aman P, Panagopoulos I, Lassen C, Fioretos T, Mencinger M, Toresson H, Hoglund M, Forster A, Rabbits T H, Ron D, Mandahl N, Mitelman F. Expression patterns of the human sarcoma-associated genes FUS and EWS and genomic structure of FUS. Genomics. 1996;37:1–8. doi: 10.1006/geno.1996.0513. [DOI] [PubMed] [Google Scholar]
- 2.Amrein H, Hedley M L, Maniatis T. The role of specific protein-RNA and protein-protein interactions in positive and negative control of pre-mRNA splicing by Transformer-2. Cell. 1994;76:735–746. doi: 10.1016/0092-8674(94)90512-6. [DOI] [PubMed] [Google Scholar]
- 3.Bertolotti A, Lutz Y, Heard D J, Chambon P, Tora L. hTAFII68, a novel RNA/ss DNA-binding protein with homology to the pro-oncoprotein TLS/FUS and EWS is associated with both TFIID and RNA polymerase II. EMBO J. 1996;15:5022–5031. [PMC free article] [PubMed] [Google Scholar]
- 4.Bertolotti A, Melot T, Acker J, Vigneron M, Delattre O, Tora L. EWS, but not EWS–FLI-1, is associated with both TFIID and RNA polymerase II: interactions between two members of the TET family, EWS and hTAFII68, and subunits of TFIID and RNA polymerase II complexes. Mol Cell Biol. 1998;18:1489–1497. doi: 10.1128/mcb.18.3.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beyer A L, Osheim Y N. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988;2:754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- 6.Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 7.Caceres J F, Stamm S, Helfman D M, Krainer A R. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- 8.Carstens R P, Eaton J V, Krigman H R, Walther P J, Garcia-Blanco M A. Alternative splicing of fibroblast growth factor receptor 2 (FGF-R2) in human prostate cancer. Oncogene. 1997;15:3059–3065. doi: 10.1038/sj.onc.1201498. [DOI] [PubMed] [Google Scholar]
- 9.Cho E J, Takagi T, Moore C R, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper D L, Dougherty G J. To metastasize or not? Selection of CD44 splice sites. Nat Med. 1995;1:635–637. doi: 10.1038/nm0795-635. [DOI] [PubMed] [Google Scholar]
- 11.Corden J L, Patturajan M. A CTD function linking transcription to splicing. Trends Biochem Sci. 1997;22:413–416. doi: 10.1016/s0968-0004(97)01125-0. [DOI] [PubMed] [Google Scholar]
- 12.Crozat A, Aman P, Mandahl N, Ron D. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature. 1993;363:640–644. doi: 10.1038/363640a0. [DOI] [PubMed] [Google Scholar]
- 13.Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, Kovar H, Houbert I, DeJong P, Rouleau G, Aurias A, Thomas G. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumors. Nature. 1992;359:162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 14.Du L, Warren S L. A functional interaction between the carboxy-terminal domain of RNA polymerase II and pre-mRNA splicing. J Cell Biol. 1997;136:5–18. doi: 10.1083/jcb.136.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fakan S, Leser G, Martin T E. Immunoelectron microscope visualization of nuclear ribonucleoprotein antigens within spread transcription complexes. J Cell Biol. 1986;103:1153–1157. doi: 10.1083/jcb.103.4.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu X D, Maniatis T. Isolation of a complementary DNA that encodes the mammalian splicing factor SC35. Science. 1992;256:535–538. doi: 10.1126/science.1373910. [DOI] [PubMed] [Google Scholar]
- 17.Ge H, Si Y, Wolffe A P. A novel transcriptional coactivator, p52, functionally interacts with the essential splicing factor ASF/SF2. Mol Cell. 1998;2:751–759. doi: 10.1016/s1097-2765(00)80290-7. [DOI] [PubMed] [Google Scholar]
- 18.Ge K, DuHadaway J, Du W, Herlyn M, Rodeck U, Prendergast G C. Mechanism for elimination of a tumor suppressor: aberrant splicing of a brain-specific exon causes loss of function of Bin1 in melanoma. Proc Natl Acad Sci USA. 1999;96:9689–9694. doi: 10.1073/pnas.96.17.9689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hallier M, Lerga A, Barnache S, Tavitian A, Moreau-Gachelin F. The transcription factor Spi-1/PU.1 interacts with the potential splicing factor TLS. J Biol Chem. 1998;273:4838–4842. doi: 10.1074/jbc.273.9.4838. [DOI] [PubMed] [Google Scholar]
- 20.Heinrichs V, Baker B S. In vivo analysis of the functional domains of the Drosophila splicing regulator RBP1. Proc Natl Acad Sci USA. 1997;94:115–120. doi: 10.1073/pnas.94.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirose Y, Manley J L. RNA polymerase II is an essential mRNA polyadenylation factor. Nature. 1998;395:93–96. doi: 10.1038/25786. [DOI] [PubMed] [Google Scholar]
- 22.Huang S, Spector D L. Intron-dependent recruitment of pre-mRNA splicing factors to sites of transcription. J Cell Biol. 1996;133:719–732. doi: 10.1083/jcb.133.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ichikawa H, Shimizu K, Katsu R, Ohki M. An RNA-binding protein gene, TLS/FUS, is fused to ERG in human myeloid leukemia with t(16;21) chromosomal translocation. Cancer Res. 1994;54:2865–2868. [PubMed] [Google Scholar]
- 24.Ichikawa H, Shimizu K, Katsu R, Ohki M. Dual transforming activities of the FUS (TLS)-ERG leukemia fusion protein conferred by two N-terminal domains of FUS (TLS) Mol Cell Biol. 1999;19:7639–7650. doi: 10.1128/mcb.19.11.7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Immanuel D, Zinszner H, Ron D. Association of SARFH (sarcoma-associated RNA-binding fly homolog) with regions of chromatin transcribed by RNA polymerase II. Mol Cell Biol. 1995;15:4562–4571. doi: 10.1128/mcb.15.8.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaishankar S, Zhang J, Roussel M F, Baker S J. Transforming activity of EWS/Fli is not strictly dependent upon DNA-binding activity. Oncogene. 1999;18:5592–5597. doi: 10.1038/sj.onc.1202940. [DOI] [PubMed] [Google Scholar]
- 27.Jumaa H, Guenet J L, Nielsen P J. Regulated expression and RNA processing of transcripts from the Srp20 splicing factor gene during the cell cycle. Mol Cell Biol. 1997;17:3116–3124. doi: 10.1128/mcb.17.6.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lessnick S L, Braun B S, Denny C T, May W A. Multiple domains mediate transformation by the Ewing's sarcoma EWS/FLI-1 fusion gene. Oncogene. 1995;10:423–431. [PubMed] [Google Scholar]
- 29.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson S D, Wickens M, Bentley D. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 30.Mortillaro M J, Blencowe B J, Wei X, Nakayasu H, Du L, Warren S L, Sharp P A, Berezney R. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc Natl Acad Sci USA. 1996;93:8253–8257. doi: 10.1073/pnas.93.16.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neugebauer K M, Roth M B. Transcription units as RNA processing units. Genes Dev. 1997;11:3279–3285. doi: 10.1101/gad.11.24.3279. [DOI] [PubMed] [Google Scholar]
- 32.Pereira D S, Dorrell C, Ito C Y, Gan O I, Murdoch B, Rao V N, Zou J P, Reddy E S, Dick J E. Retroviral transduction of TLS-ERG initiates a leukemogenic program in normal human hematopoietic cells. Proc Natl Acad Sci USA. 1998;95:8239–8244. doi: 10.1073/pnas.95.14.8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perrotti D, Bonatti S, Trotta R, Martinez R, Skorski T, Salomoni P, Grassilli E, Lozzo R V, Cooper D R, Calabretta B. TLS/FUS, a pro-oncogene involved in multiple chromosomal translocations, is a novel regulator of BCR/ABL-mediated leukemogenesis. EMBO J. 1998;17:4442–4455. doi: 10.1093/emboj/17.15.4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petermann R, Mossier B M, Aryee D NT, Khazak V, Golemis E A, Kovar H. Oncogenic EWS-Fli1 interacts with hsRPB7, a subunit of human RNA polymerase II. Oncogene. 1998;17:603–610. doi: 10.1038/sj.onc.1201964. [DOI] [PubMed] [Google Scholar]
- 35.Prasad D D, Ouchida M, Lee L, Rao V N, Reddy E S. TLS/FUS fusion domain of TLS/FUS-erg chimeric protein resulting from the t(16;21) chromosomal translocation in human myeloid leukemia functions as a transcriptional activation domain. Oncogene. 1994;9:3717–3729. [PubMed] [Google Scholar]
- 36.Rabbitts T H, Forster A, Larson R, Nathan P. Fusion of the dominant negative transcription regulator CHOP with a novel gene FUS by translocation t(12;16) in malignant liposarcoma. Nat Genet. 1993;4:175–180. doi: 10.1038/ng0693-175. [DOI] [PubMed] [Google Scholar]
- 37.Ring H Z, Lis J T. The SR protein B52/SRp55 is essential for Drosophila development. Mol Cell Biol. 1994;14:7499–7506. doi: 10.1128/mcb.14.11.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sass H, Pederson T. Transcription-dependent localization of U1 and U2 small nuclear ribonucleoproteins at major sites of gene activity in polytene chromosomes. J Mol Biol. 1984;180:911–926. doi: 10.1016/0022-2836(84)90263-8. [DOI] [PubMed] [Google Scholar]
- 39.Screaton G R, Caceres J F, Mayeda A, Bell M V, Plebanski M, Jackson D G, Bell J I, Krainer A R. Identification and characterization of three members of the human SR family of pre-mRNA splicing factors. EMBO J. 1995;14:4336–4349. doi: 10.1002/j.1460-2075.1995.tb00108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steinmetz E J. Pre-mRNA processing and the CTD of RNA polymerase II: the tail that wags the dog? Cell. 1997;89:491–494. doi: 10.1016/s0092-8674(00)80230-5. [DOI] [PubMed] [Google Scholar]
- 41.Stickeler E, Kittrell F, Medina D, Berget S M. Stage-specific changes in SR splicing factors and alternative splicing in mammary tumorigenesis. Oncogene. 1999;18:3574–3582. doi: 10.1038/sj.onc.1202671. [DOI] [PubMed] [Google Scholar]
- 42.Tsai S, Bartelmez S, Sitnicka E, Collins S. Lymphohematopoietic progenitors immortalized by a retroviral vector harboring a dominant-negative retinoic acid receptor can recapitulate lymphoid, myeloid, and erythroid development. Genes Dev. 1994;8:2831–2841. doi: 10.1101/gad.8.23.2831. [DOI] [PubMed] [Google Scholar]
- 43.Vincent M, Lauriault P, Dubois M-F, Lavoie S, Bensaude O, Chabot B. The nuclear matrix protein p255 is a highly phosphorylated form of RNA polymerase II largest subunit which associates with spliceosomes. Nucleic Acids Res. 1996;24:4649–4652. doi: 10.1093/nar/24.23.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang J, Manley J L. Overexpression of the SR proteins ASF/SF2 and SC35 influences alternative splicing in vivo in diverse ways. RNA. 1995;1:335–346. [PMC free article] [PubMed] [Google Scholar]
- 45.Xing Y, Johnson C V, Dobner P R, Lawrence J B. Higher level organization of individual gene transcription and RNA splicing. Science. 1993;259:1326–1330. doi: 10.1126/science.8446901. [DOI] [PubMed] [Google Scholar]
- 46.Yamamoto K, Hamaguchi H, Nagata K, Kobayashi M, Tanimoto F, Taniwaki M. Establishment of a novel human acute myeloblastic leukemia cell line (YNH-1) with t(16;21), t(1;16) and 12q13 translocations. Leukemia. 1997;11:599–608. doi: 10.1038/sj.leu.2400594. [DOI] [PubMed] [Google Scholar]
- 47.Yang L, Embree L J, Tsai S, Hickstein D D. Oncoprotein TLS interacts with serine-arginine proteins involved in RNA splicing. J Biol Chem. 1998;273:27761–27764. doi: 10.1074/jbc.273.43.27761. [DOI] [PubMed] [Google Scholar]
- 48.Yuryev A, Patturajan M, Litingtung Y, Joshi R V, Gentile C, Gebara M, Corden J L. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc Natl Acad Sci USA. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang D, Paley A J, Childs G. The transcriptional repressor ZFM1 interacts with and modulates the ability of EWS to activate transcription. J Biol Chem. 1998;273:18086–18091. doi: 10.1074/jbc.273.29.18086. [DOI] [PubMed] [Google Scholar]
- 50.Zhang G, Taneja K L, Singer R H, Green M R. Localization of pre-mRNA splicing in mammalian nuclei. Nature. 1994;372:809–812. doi: 10.1038/372809a0. [DOI] [PubMed] [Google Scholar]
- 51.Zinszner H, Albalat R, Ron D. A novel effector domain from the RNA-binding protein TLS or EWS is required for oncogenic transformation by CHOP. Genes Dev. 1994;8:2513–2526. doi: 10.1101/gad.8.21.2513. [DOI] [PubMed] [Google Scholar]
- 52.Zinszner H, Sok J, Immanuel D, Yin Y, Ron D. TLS (FUS) binds RNA in vivo and engages in nucleo-cytoplasmic shuttling. J Cell Sci. 1997;110:1741–1750. doi: 10.1242/jcs.110.15.1741. [DOI] [PubMed] [Google Scholar]