Kumari et al. report two-tiered regulation of mitotic dynein function through the mitosis-specific, cdk1-mediated phosphorylation of its light intermediate chain 1 subunit. Either phosphorylation alone or subsequent phosphorylation-dependent recruitment of the prolyl isomerase Pin1 dictates the assembly of separate dynein-cargo adaptor complexes, which regulate distinct mitotic dynein functions.
Abstract
The dynein motor performs multiple functions in mitosis by engaging with a wide cargo spectrum. One way to regulate dynein’s cargo-binding selectivity is through the C-terminal domain (CTD) of its light intermediate chain 1 subunit (LIC1), which binds directly with cargo adaptors. Here we show that mitotic phosphorylation of LIC1-CTD at its three cdk1 sites is required for proper mitotic progression, for dynein loading onto prometaphase kinetochores, and for spindle assembly checkpoint inactivation in human cells. Mitotic LIC1-CTD phosphorylation also engages the prolyl isomerase Pin1 predominantly to Hook2-dynein-Nde1-Lis1 complexes, but not to dynein-spindly-dynactin complexes. LIC1-CTD dephosphorylation abrogates dynein-Pin1 binding, promotes prophase centrosome–nuclear envelope detachment, and impairs metaphase chromosome congression and mitotic Golgi fragmentation, without affecting interphase membrane transport. Phosphomutation of a conserved LIC1-CTD SP site in zebrafish leads to early developmental defects. Our work reveals that LIC1-CTD phosphorylation differentially regulates distinct mitotic dynein pools and suggests the evolutionary conservation of this phosphoregulation.
Introduction
The microtubule cytoskeleton supports both positive end–directed transport through kinesin motors and negative end–directed transport primarily through the dynein motor (Olenick and Holzbaur, 2019). Vertebrate cells contain mainly one cytoplasmic dynein complex (cytoplasmic dynein 1, henceforth “dynein”) but nearly 50 kinds of plus end–directed kinesins, except for the minus end–directed kinesin 14 family (Kardon and Vale, 2009). Dynein’s remarkable cargo-binding diversity is therefore essential for its multiple interphase and mitotic functions (Reck-Peterson et al., 2018; Vallee et al., 2012). During mitosis, dynein performs several essential functions, including prophase centrosome–nuclear envelope (NE) attachment, NE breakdown (NEB), spindle formation, chromosome congression, spindle orientation, and spindle assembly checkpoint (SAC) inactivation (Bolhy et al., 2011; Goshima et al., 2005; Howell et al., 2001; Mahale et al., 2016a; Mahale et al., 2016b; Raaijmakers et al., 2013; Salina et al., 2002; Varma et al., 2008). Dynein complexes usually require the cofactor dynactin for optimal function (Reck-Peterson et al., 2018; Schroer, 2004); however, some dynein complexes may not (Huang et al., 2012; McKenney et al., 2010; Raaijmakers et al., 2013; Vallee et al., 2012). Several activating adaptor proteins that bridge dynein and dynactin are required for the processivity and cargo-binding of dynein (Lee et al., 2020; Redwine et al., 2017). Of the two copies each of the heavy chains (HCs), intermediate chains (ICs), light intermediate chains (LICs), and multiple light chain subunits present in vertebrate dynein (Pfister et al., 2005; Pfister et al., 2006), the LICs are pivotal for directly engaging adaptors (Celestino et al., 2019; Lee et al., 2020). Among the known dynein adaptors, spindly and Hook2 function exclusively during mitosis (Dwivedi et al., 2019; Griffis et al., 2007).
Dynein’s dramatic interphase-to-mitosis cargo-switching is strongly correlated with cdk1-cyclinB–mediated LIC phosphorylation (Addinall et al., 2001; Dell et al., 2000), but through poorly understood mechanisms. Dynein LIC1 has been studied extensively with respect to the mechanism of cargo adaptor binding (Lee et al., 2020). Human LIC1 (hLIC1) contains four conserved cdk1–cyclin B phosphorylation sites, of which S207 in the N-terminal domain (NTD) remains phosphorylated through interphase and mitosis, while S389, S405, and T408 in the LIC1-CTD are phosphorylated exclusively during mitosis (Daub et al., 2008; Dephoure et al., 2008; Olsen et al., 2010). Importantly, the three clustered LIC1-CTD sites lie upstream of helix-1 (H1, residues 440–455), which binds directly to various cargo-binding adaptor NTDs (Celestino et al., 2019; Lee et al., 2020; Lee et al., 2018). However, the importance of LIC1-CTD phosphorylation in regulating mitotic dynein’s adaptor selectivity and functional repertoire is unknown.
Conformational regulation of phosphorylated proteins can be achieved through the peptidyl prolyl isomerase Pin1 (prolyl isomerase interacting NIMA 1), which binds to phosphorylated Ser/Thr residues in Ser-Pro/Thr-Pro (SP/TP) sites and isomerizes the adjacent proline, thereby regulating a wide variety of cellular functions (Lu et al., 1996; Yaffe et al., 1997). cdk1-phosphorylated SP/TP sites can serve as potential substrates for Pin1 in mitosis (Lu et al., 2002; Shen et al., 1998). The LIC1 cdk1 phosphosites lie in the intrinsically unstructured, adaptor-binding LIC1-CTD (Celestino et al., 2019; Kumari et al., 2021), thus presenting potential targets for Pin1 binding and regulation. However, to our knowledge, no dynein subunit has been reported to be a direct substrate of Pin1.
Here, we report that phosphorylation at the three LIC1-CTD sites regulates mitotic functions in a layered fashion in human cells. LIC1-CTD phosphorylation is required for metaphase-to-anaphase progression by helping dynein-spindly-dynactin complexes localize to mitotic kinetochores and inactivate the SAC. Phosphorylated LIC1-CTD also directly recruits Pin1 preferentially to Hook2-Nde1-Lis1-dynein complexes, but not to dynein-spindly-dynactin complexes. Abrogating LIC1-CTD phosphorylation disengages Pin1 from dynein and causes prophase centrosome-NE detachment as well as chromosome miscongression, suggesting compromised Nde1-Lis1-Hook2-dynein function. Phosphorylation of a corresponding conserved SP site in zebrafish LIC1-CTD is essential for normal embryonic development. Our work reveals discrete mechanisms by which mitotic LIC1 phosphorylation regulates functionally distinct dynein complexes.
Results
Conserved LIC1-CTD phosphorylation is required for normal mitotic progression
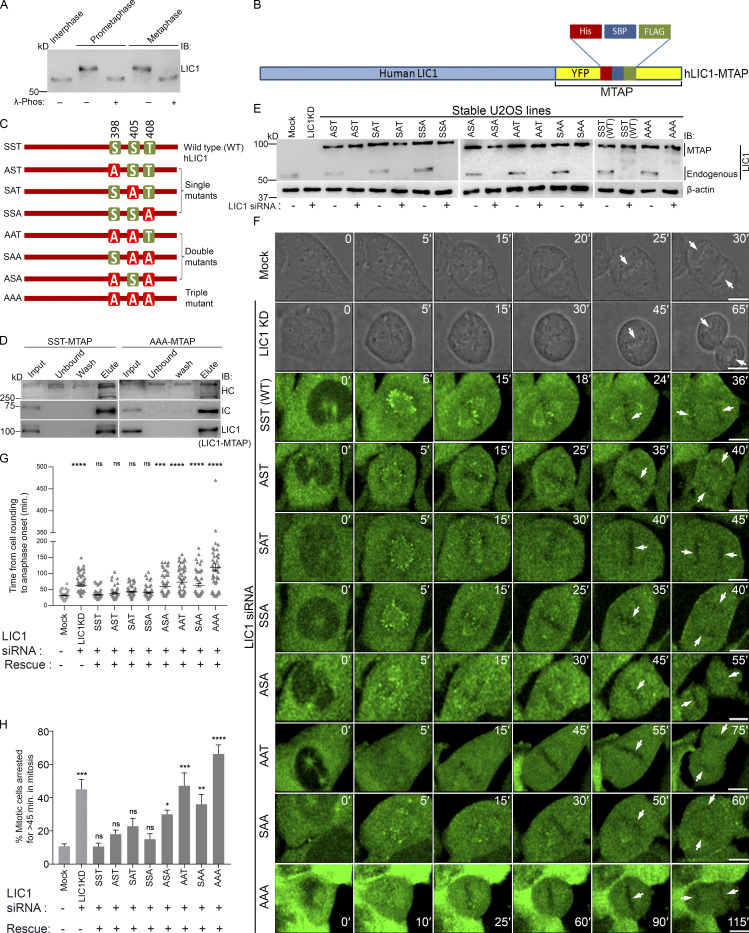
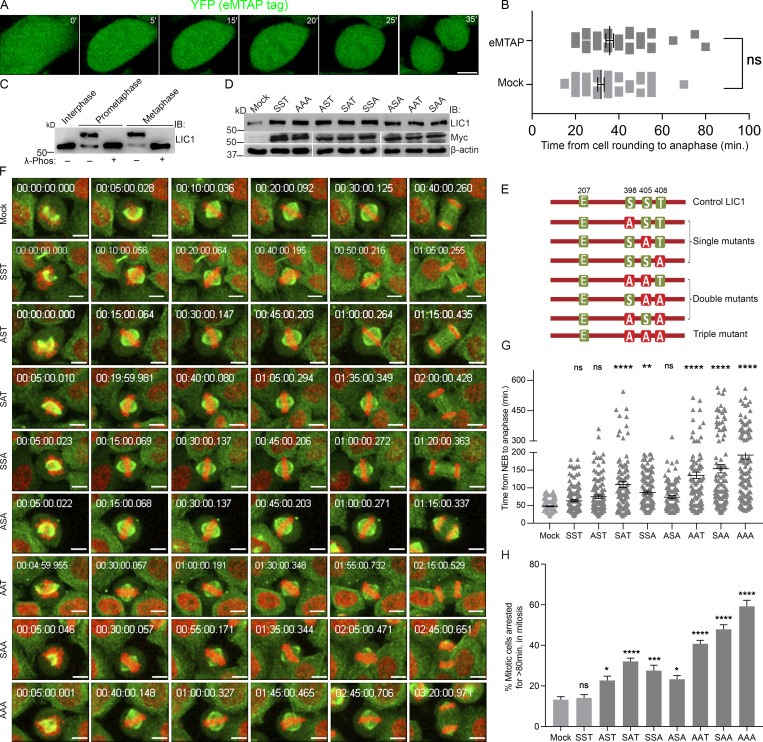
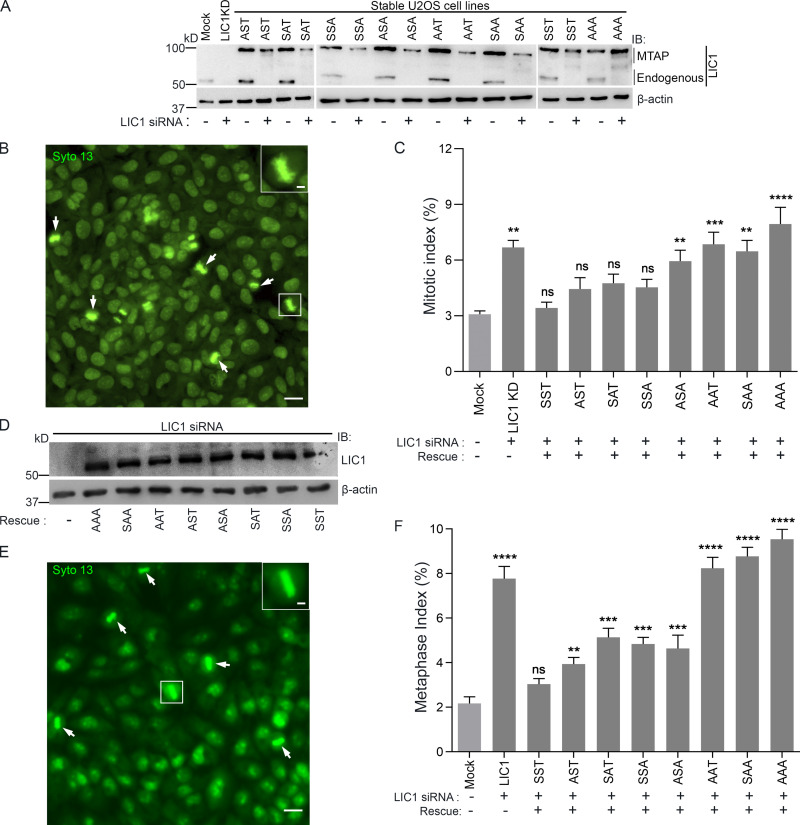
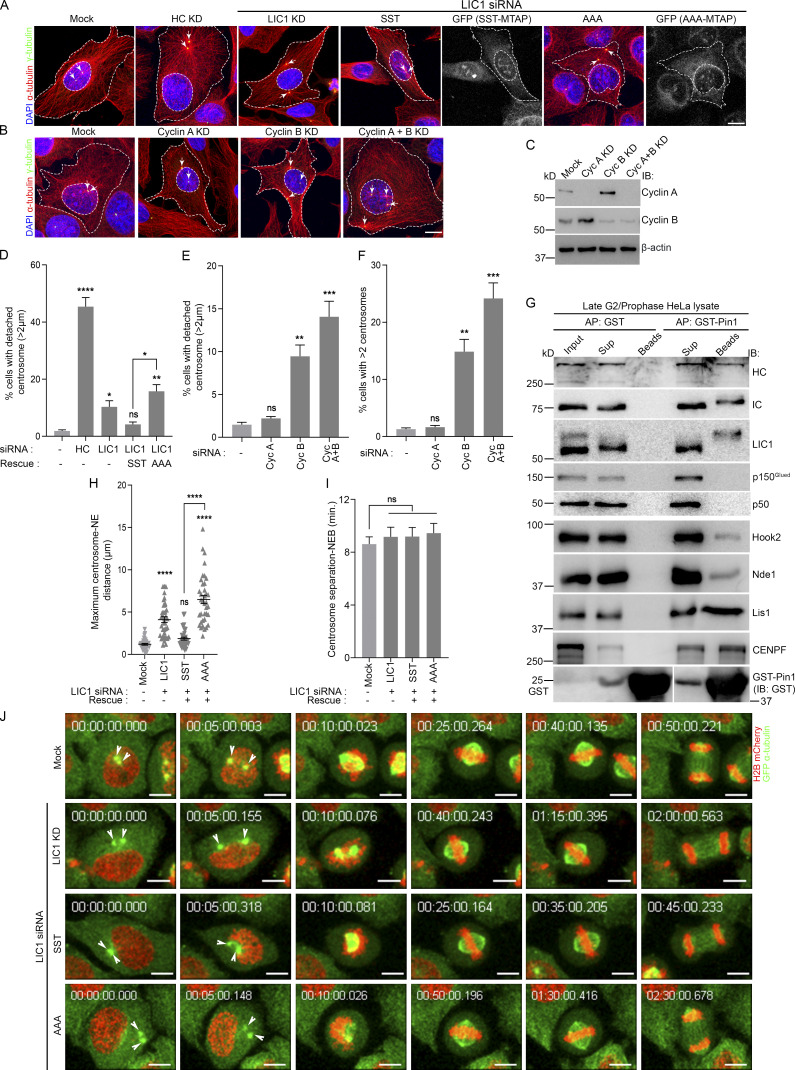
We observed mitotic gel retardation of hLIC1 with respect to interphase in both U2OS and HeLa cells, which was abrogated upon treatment with λ-phosphatase (Figs. 1 A and S1 A), confirming mitotic phosphorylation of LIC1. To assess the role of the three mitosis-specific cdk1 phosphorylation sites in LIC1-CTD, we cloned the single, double, and triple combinatorial hLIC1–multiple tandem affinity purification-tagged (hLIC1-MTAP) phosphodeficient mutants for these sites (S398A, S405A, and T408A) and generated stably expressing U2OS cell lines (Fig. 1, B and C). Both the SST (WT) and AAA (triple mutant) LIC1 proteins could efficiently integrate into the dynein complex (Fig. 1 D). We used suboptimal LIC1 siRNA treatment in high-expression stable lines to simultaneously achieve efficient endogenous hLIC1 depletion and exogenous LIC1-MTAP expression for rescue experiments (Fig. 1, E and F). Time-lapse, live-cell confocal microscopy revealed that SST and the single-site mutants could efficiently rescue hLIC1 depletion–induced delays in anaphase onset; however, the double mutants showed compromised function, and AAA was the least capable (Fig. 1, F–H; and Videos 1, 2, 3, and 4). A stable U2OS cell line expressing just the MTAP tag did not show delays in mitotic progression compared with the mock-transfected control (Fig. S1, A and B). Mitotic index measurements using Syto13-stained cells confirmed the requirement of the LIC1-CTD phosphosites for timely mitotic progression (Fig. S2, A–C). The rat LIC1 orthologue (rLIC1) has been shown to efficiently rescue mitotic phenotypes owing to hLIC1 depletion in HeLa cells (Sivaram et al., 2009). Using time-lapse confocal imaging, we quantified the extent of mitotic delay upon the transient, dominant-negative expression of each of the corresponding rLIC1-CTD mutants (all containing the S207E phosphomimetic mutation) in a GFP-tubulin::histone 2B-mCherry expressing “double-stable” HeLa cell line (Fig. S1, C–H; and Videos 5, 6, and 7). Our observations further supported the requirement of the LIC1-CTD sites for proper mitotic progression, as did metaphase index measurements in hLIC1-depleted HeLa cells rescued with transient rLIC1 mutants expression (Fig. S2, D–F).
Figure 1.
Conserved LIC1-CTD phosphorylation is required for mitotic progression. (A) Immunoblots depicting the migration of hLIC1 from U2OS cell lysates at the indicated stages. λ-phos, λ-phosphatase treatment. (B) Schematic of the hLIC1-MTAP construct. (C) Schematic showing various hLIC1-MTAP constructs mutated at the three mitotic cdk1 sites. Green boxes, WT residues; red boxes, phosphodeficient mutation to alanine. (D) Immunoblots of SBP-affinity precipitates from SST-MTAP and AAA-MTAP cell lysates, demonstrating integration into the dynein complex. (E) Immunoblots depicting expression levels of endogenous LIC1 and stably expressing hLIC1-MTAP mutants under the indicated conditions. (F) Stills from representative time-lapse videos of U2OS cells (mock, LIC1 knockdown [KD]), differential interference contrast [DIC]) or the respective U2OS stable cell lines (green, YFP fluorescence from the MTAP tag) as indicated, from cell rounding (mitotic entry) to anaphase onset. Time stamps (min) included in the images. Arrows depict chromosomes. (G and H) Bar graphs quantifying the mitotic timing (G) and fraction of cells showing delayed anaphase onset (H) from the datasets represented in F. n = 3 independent experiments, ≥20 mitotic cells per experiment per construct. IB, immunoblot. Error bars = mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001, ns = not significant vs. mock (G, Kruskal–Wallis test; H, one-way ANOVA). Scale bar = 10 µm.
Figure S1.
LIC1-CTD phosphorylation is required for proper mitotic progression in HeLa cells. (A) Stills from a time-lapse video of a representative U2OS cell line stably expressing the empty MTAP tag (eMTAP, green). Time stamps included in the images. (B) Graph quantifying the average mitotic timing for eMTAP videos. n = 3 experiments, total 60 mitotic cells imaged. (C) Immunoblots (IB) depicting the migration of hLIC1 from HeLa cell lysates at different stages of the cell cycle as indicated. λ-phos, λ-phosphatase treatment to confirm that the gel retardation is due to phosphorylation. (D) Immunoblots depicting the transient expression of the various rLIC1 phosphorylation constructs shown in E, after transfection into a HeLa cell line expressing GFP-α-tubulin::mCherry-histone 2B. (E) Schematic showing various rLIC1 constructs mutated at the three mitotic CTD cdk1 sites in a mammalian expression vector as indicated. All constructs contained the S207E phosphomimetic mutation. Green boxes, WT residues; red boxes, phosphodeficient mutation to alanine. (F) Stills from representative time-lapse videos of HeLa cells stably expressing GFP-tubulin (green) and mcherry-Histone 2B (red) from NEB to anaphase. Time stamps included in the images. (G) Bar graphs representing the average time taken from NEB to anaphase calculated from the videos as described in F. n = 3 experiments, total 150 mitotic cells imaged. (H) Fraction of mitotic cells showing delayed anaphase onset (>80 min after NEB). Scale bar = 10 µm. Error bars = mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001, ns = not significant vs. mock. B, two-tailed Student’s t test; G, Kruskal–Wallis test; H, one-way ANOVA.
Video 1.
Mitotic progression in mock depleted, LIC1-depleted, and eMTAP-expressing stable U2OS cells. Time-lapse confocal videos of U2OS cells after mock transfection (DIC), LIC1 siRNA treatment (DIC), and stable MTAP tag (eMTAP) expression. Green represents YFP fluorescence from the MTAP tag. All videos show the duration from cell rounding to anaphase onset. Time stamps included. Scale bar = 10 µm. Speed is 3 frames per second.
Video 2.
Mitotic progression upon endogenous LIC1 depletion in WT and triple phosphodeficient LIC1 mutant stable U2OS cells. Time-lapse confocal videos of LIC1 siRNA–treated stable U2OS cell lines expressing MTAP-tagged WT LIC1 construct SST and triple site LIC1-CTD mutant AAA. Green represents YFP fluorescence from the MTAP tag. All videos show the duration from cell rounding to anaphase onset. Time stamps included. Scale bar = 10 µm. Speed is 3 frames per second.
Video 3.
Mitotic progression upon endogenous LIC1 depletion in single site phosphodeficient LIC1 mutant stable U2OS cells. Time-lapse confocal videos of LIC1 siRNA treated stable U2OS cell lines expressing MTAP-tagged single site LIC1-CTD mutants AST, SAT and SSA. Green represents YFP fluorescence from the MTAP tag. All videos show the duration from cell rounding to anaphase onset. Time stamps included. Scale bar = 10 µm. Speed is 3 frames per second.
Video 4.
Mitotic progression upon endogenous LIC1 depletion in double site phosphodeficient LIC1 mutant stable U2OS cells. Time-lapse confocal videos of LIC1 siRNA–treated stable U2OS cell lines expressing MTAP-tagged double site LIC1-CTD mutants SAA, ASA, and AAT. Green represents YFP fluorescence from the MTAP tag. All videos show the duration from cell rounding to anaphase onset. Time stamps included. Scale bar = 10 µm. Speed is 3 frames per second.
Figure S2.
LIC1 C-terminal phosphomutants cannot rescue LIC1 depletion–induced mitotic arrest. (A) Immunoblots depicting the expression of endogenous LIC1/stably expressing hLIC1-MTAP constructs under the indicated conditions. (B) Epifluorescence micrograph of U2OS cells stained with the viable DNA dye Syto 13 (green) to visualize mitotic cells (arrows; zoomed inset shows metaphase cell). Scale bar = 50 µm. (C) Mitotic index measurements (fraction of mitotic cells) under the indicated conditions; P values calculated with regard to mock. n = 3, minimum of 500 cells per experiment. (D) Immunoblots depicting the transient expression of the various rLIC1 phosphorylation constructs in HeLa cells. (E) Representative epifluorescence micrograph of HeLa cells stained with the viable DNA dye Syto 13 to visualize metaphase cells. Zoomed inset shows one magnified metaphase cell. Scale bar = 50 µm, inset scale bar = 10 µm. (F) Metaphase index (fraction of metaphase cells) from HeLa cells treated with anti-hLIC1 siRNA and rescued by transient expression of the various rLIC1 constructs as indicated. n = 3, minimum 600 cells per experiment. IB, immunoblot. Error bars = mean ± SEM. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001, ns = not significant vs. mock (one-way ANOVA).
Video 5.
Mitotic progression upon transient expression of SST and AAA rLIC1 proteins in HeLa cells. Confocal fluorescence time-lapse videos of double stable GFP-α-tubulin (green)::histone 2B-mCherry (red) HeLa cells upon mock transfection, and exogenous individual expression of rLIC1(S207E) constructs including SST and triple site rLIC1-CTD mutant AAA. All videos show the duration from NEB to anaphase onset. Time stamps included. Scale bar = 10 µm. Speed is 3 frames per second.
Video 6.
Mitotic progression upon transient expression of single site rLIC1 phosphodeficient proteins in HeLa cells. Confocal fluorescence time-lapse videos of double stable GFP-α-tubulin (green)::histone 2B-mCherry (red) HeLa cells upon exogenous individual expression of rLIC1(S207E) constructs including single-site rLIC1-CTD mutants AST, SAT, and SSA. All videos show the duration from NEB to anaphase onset. Time stamps included. Scale bar = 10 µm. Speed is 3 frames per second.
Video 7.
Mitotic progression upon transient expression of double site rLIC1 phosphodeficient proteins in HeLa cells. Confocal fluorescence time-lapse videos of double stable GFP-α-tubulin (green)::histone 2B-mCherry (red) HeLa cells upon exogenous individual expression of rLIC1(S207E) constructs including double site rLIC1-CTD mutants SAA, ASA, and AAT. All videos show the duration from NEB to anaphase onset. Time stamps included. Scale bar = 10 µm. Speed is 3 frames per second.
LIC1-CTD phosphorylation is required for normal loading of dynein onto unattached prometaphase kinetochores
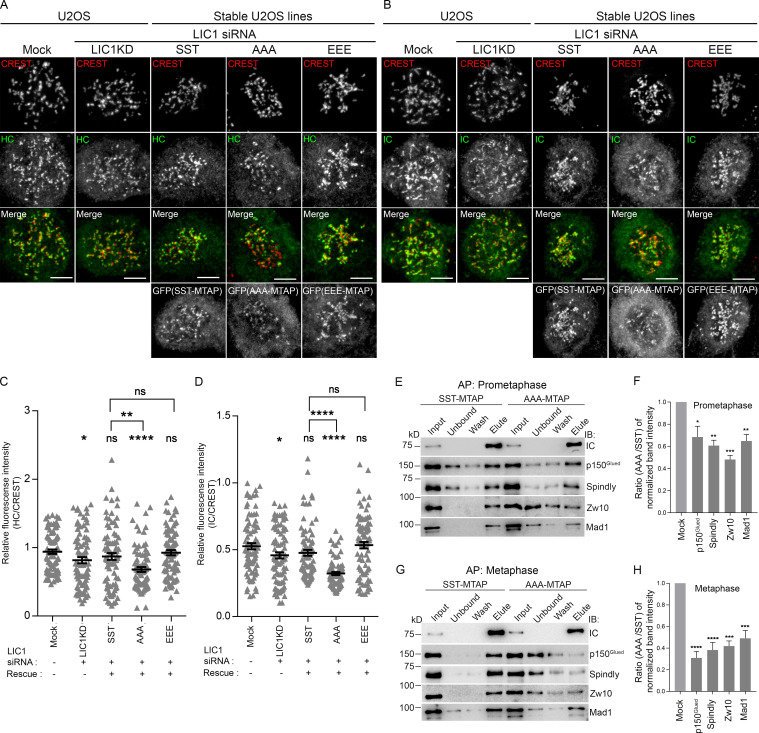
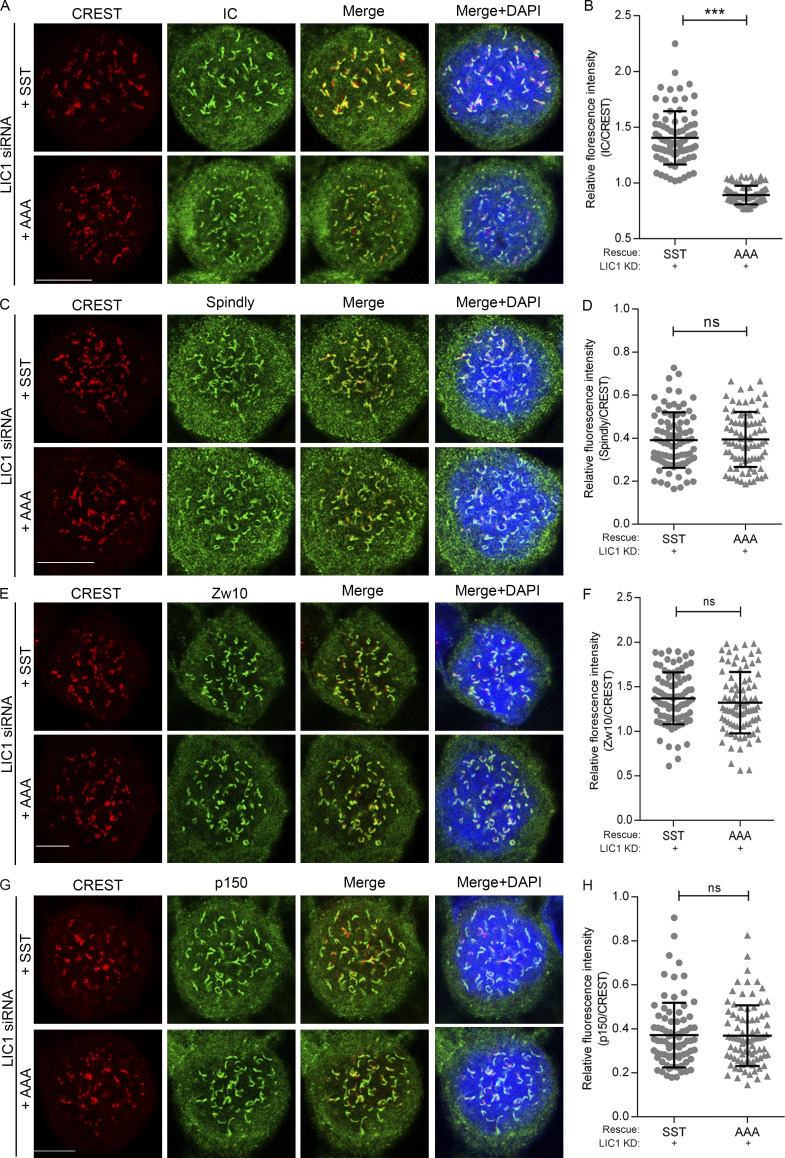
Using quantitative confocal microscopy of the SST, AAA, and phospho-mimetic EEE stable U2OS cell lines following nocodazole treatment, we observed that both HC and IC levels at prometaphase kinetochores were reduced significantly upon rescue of hLIC1 depletion with AAA, while they were restored by rescue with SST or EEE, suggesting suboptimal prometaphase dynein complex recruitment to kinetochores upon LIC1-CTD dephosphorylation (Fig. 2, A–D). We observed similar outcomes for IC loading in hLIC1-depleted HeLa cells rescued with the corresponding rLIC1 mutants (Fig. S3, A and B); however, kinetochore dynein receptors spindly, p150Glued (dynactin subunit), and Zw10 (Gama et al., 2017) remained largely unperturbed (Fig. S3, C–H). Thus, despite proper recruitment of upstream kinetochore receptors, phosphodeficient LIC1-containing dynein failed to optimally load onto mitotic kinetochores.
Figure 2.
LIC1-CTD phosphorylation is required for mitotic kinetochore dynein recruitment and interaction with SAC components. (A and B) Representative confocal micrographs of nocodazole-treated prometaphase cells, immunostained for kinetochores (CREST), HC (A), and IC (B). (C and D) Quantified immunofluorescence intensity of HC (C) and IC (D) normalized to the respective CREST intensity. n = 2 independent experiments, ≥80 prometaphase cells per condition. (E and G) Immunoblots depicting SBP affinity precipitates (APs) obtained from prometaphase (E) and metaphase (G) lysates of SST-MTAP and AAA-MTAP stable cell line lysates, probed (immunoblot) with the indicated antibodies. (F and H) Quantification of AP efficiency from three independent experiments for E and G, respectively. y axes, densitometric band intensity ratios of the elute bands for the indicated proteins (x axes) normalized to the respective IC bands. Error bars, mean ± SEM over three independent AP experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (G, Kruskal–Wallis test; H, one-way ANOVA). Scale bar = 10 µm.
Figure S3.
LIC1-CTD phosphorylation is required for prometaphase kinetochore dynein recruitment in HeLa cells. (A, C, E, and G) Representative confocal immunofluorescence micrographs of prometaphase HeLa cells (1 µM nocodazole treated for 4 h), immunostained to visualize the chromosomes (DAPI, blue), kinetochores (CREST, red), and the indicated kinetochore proteins (green) as follows: IC (A), spindly (C), Zw10 (E), and p150Glued (G). All cells were treated with anti-hLIC1 siRNA and rescued by the transient expression of the indicated rLIC1-CTD constructs. (B, D, F, and H) Scatterplots depicting the quantification of the immunofluorescence signals from A, C, E, and G, respectively, normalized to the respective kinetochore (CREST) intensities, upon rescue of hLIC1 depletion by the indicated rLIC1 constructs (x axis). A minimum of 80 prometaphase cells per condition, over 3 independent experiments, were quantified. Scale bar = 10 µm. Error bars = mean ± SD. ***, P < 0.001 (two-tailed Student’s t test).
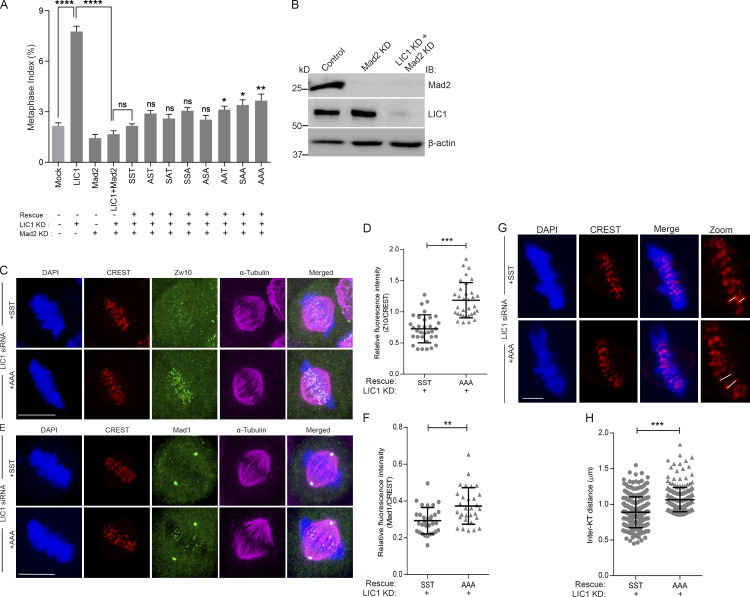
LIC1 is required for the removal of a subset of SAC proteins from mitotic kinetochores to allow anaphase onset (Mahale et al., 2016b; Sivaram et al., 2009). We assessed the ability of the LIC1-CTD mutants to restore normal mitotic progression in the absence of an active SAC in HeLa cells. siRNA-mediated depletion of the SAC effector Mad2, or codepletion of LIC1, did not induce metaphase delay due to an inactive SAC (Fig. S4, A and B). Rescue with the various rLIC1-CTD mutants following Mad2:LIC1 double depletion only mildly increased the metaphase index, suggesting that the SAC is a major target of mitotic LIC1-CTD phosphorylation (Fig. S4 A). We tested whether the AAA-rLIC1, which leads to the strongest metaphase delay, could remove SAC proteins from metaphase kinetochores. Confocal immunofluorescence intensity analysis of congressed metaphase cells (MG132 arrested following nocodazole release) showed that in comparison to rescue with SST, AAA led to significant kinetochore retention of Zw10 and Mad1 (Fig. S4, C–F), despite the cells being in late metaphase (increased interkinetochore distance; Fig. S4, G and H). Thus, mitotic LIC1-CTD phosphorylation is required for efficient dynein-based SAC removal in mitosis. Streptavidin-binding protein (SBP) affinity purification from mitotically enriched lysates of the respective stable cell lines revealed that AAA showed weaker binding with p150Glued, spindly, Zw10, and Mad1 in prometaphase as compared with SST, a difference even more pronounced in metaphase (Fig. 2, E–H). Thus, LIC1-CTD phosphorylation is required for optimal binding of spindly and dynactin and the subsequent removal of SAC components from mitotic kinetochores.
Figure S4.
LIC1-CTD phosphorylation is required for metaphase inactivation of the SAC in HeLa cells. (A) Graph depicting quantification of the metaphase index upon rescue of siRNA-mediated hLIC1 depletion by the various phosphomutant rLIC1 constructs, in the presence (no Mad2 siRNA) and absence (Mad2 siRNA) of a functional SAC. Mock and LIC1 bars are the same data as shown in Fig. S2 C. P values for each of the phosphomutant constructs (sixth bar from the left and beyond) have been calculated with respect to SST. n = 3, ≥500 cells per experiment. (B) Immunoblots (IB) depicting the siRNA-mediated depletion of Mad2 and LIC1 in HeLa cells under the indicated conditions. (C and E) Representative confocal micrographs of congressed metaphase HeLa cells (released from nocodazole into MG132) immunostained to visualize the chromosomes (DAPI, blue), kinetochores (CREST, red), and the indicated SAC proteins, Zw10 or Mad1 (green). All cells were treated with anti-hLIC1 siRNA and rescued by the expression of the indicated rLIC1-CTD constructs. n = 3 independent experiments, ≥30 metaphase cells per condition. Scale bar = 10 µm. (D and F) Scatterplots depicting the quantification of the immunofluorescence signals of SAC proteins Zw10 (D) and Mad1 (F) normalized to the respective kinetochore (CREST) intensities under the indicated conditions. Kinetochore intensities from ≥10 perfectly aligned metaphase cells per experiment over 3 independent experiments were quantified. (G) Representative confocal micrographs of a single z-section of congressed metaphase HeLa cells showing metaphase kinetochores (CREST, red) for calculating interkinetochore distances (white lines) under the indicated conditions. Scale bar = 2 µm. (H) Average interkinetochore distance of cells imaged as in G. n = 3 experiments, a total of 255 kinetochore pairs measured from 10 metaphase cells per experiment for each condition. Error bars = mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ***, P < 0.0001 (two-tailed Student’s t test).
LIC1-CTD phosphorylation recruits the prolyl isomerase Pin1 to the mitotic dynein complex
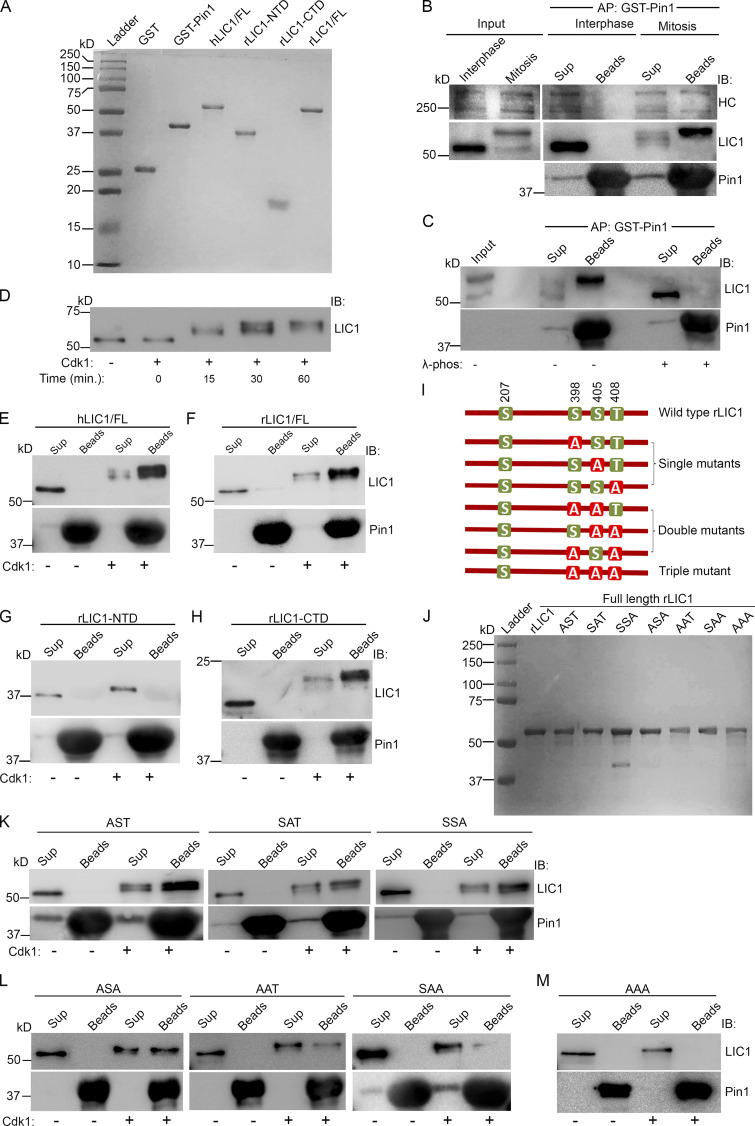
The three LIC1-CTD phosphorylation residues are potential substrates of the phosphorylation-dependent prolyl isomerase Pin1, which regulates multiple mitotically phosphorylated substrates (Cheng and Tse, 2018). Immobilized GST-tagged Pin1 (GST-Pin1) was able to pull down LIC1 and HC from mitotic, but not interphase, lysates (Fig. 3, A and B), suggesting a mitotic dynein–Pin1 interaction. The loss of Pin1-LIC1 binding in λ-phosphatase–treated affinity precipitates confirmed that phosphorylation was imperative for binding (Fig. 3 C). Purified hLIC1 (Fig. 3 A), when phosphorylated in vitro by cdk1-cyclin B, led to saturable gel retardation of hLIC1 (Fig. 3 D). We observed that only phosphorylated hLIC1 (Fig. 3 E) and rLIC1 (Fig. 3 F) could bind directly to GST-Pin1.
Figure 3.
LIC1-CTD phosphorylation is required for Pin1 to interact with mitotic dynein. (A) Coomassie-stained SDS-PAGE (15%) of the indicated recombinantly purified proteins. (B) Immunoblots showing the binding of mitotic dynein, but not interphase dynein, from HeLa cell lysates with purified GST-Pin1. (C) Immunoblots of the affinity precipitates from A after λ-phosphatase (λ-phos) treatment. (D) Anti-LIC1 immunoblot of purified hLIC1 after incubation with purified cdk1-cyclin B for the indicated durations. (E–H) Immunoblots showing the direct binding of purified hLIC1 (E), rLIC1 (F), and rLIC1-CTD (H),but not rLIC1-NTD (G) with purified GST-Pin1 in the presence (+) or absence (–) of cdk1. (I and J) Schematic diagram (I) and Coomassie-stained SDS PAGE (10%) profiles (J) of the indicated purified proteins. (K–M) Immunoblots depicting direct binding assays of the indicated purified rLIC1 proteins with purified GST-Pin1, with (+) and without (–) prior cdk1 treatment. AP, affinity purification; IB, immunoblot, Cdk1, cdk1-cyclin B; beads, affinity precipitate; sup, supernatant.
To identify the domain of LIC1 responsible for Pin1 binding, we purified rLIC1-NTD (containing one cdk-1 site [S207]) and rLIC1-CTD (containing three cdk1 sites [S398, S405, and T408]; Fig. 3 A). In vitro binding assays following cdk1-phosphorylation revealed that only phosphorylated LIC1-CTD, but not phosphorylated LIC1-NTD, could bind to Pin1 (Fig. 3, G and H). To determine the specific cdk1-phospho sites responsible for the Pin1 interaction, we purified all seven possible full-length rLIC1-CTD phosphodeficient combinations (Fig. 3, I and J). Immunoblot analysis showed that all phosphorylated single mutants (AST, SAT, and SSA) and the double mutant ASA were able to bind to Pin1 to various extents (Fig. 3, K and L); AAT and SAA showed reduced binding (Fig. 3 L), and the triple mutant AAA was unable to interact (Fig. 3 M). Thus, no single LIC1-CTD phosphorylation was sufficient for optimal Pin1 binding, and abrogating phosphorylation altogether completely abolished the Pin1 interaction.
Pin1 preferentially binds to the Hook2-Nde1-Lis1-dynein complex but not to the spindly-dynactin-dynein complex
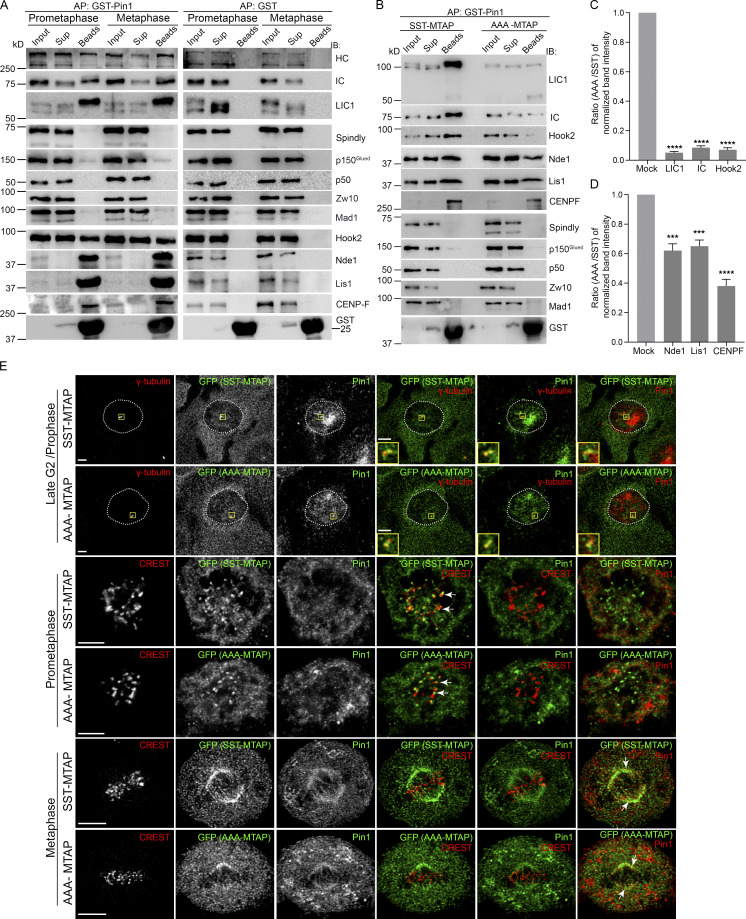
LIC1-CTD is required for engaging directly with activating adaptors of dynein (Lee et al., 2020). We tested whether the kinetochore-localized mitotic adaptor spindly is part of the dynein complex that interacts with Pin1. GST-Pin1 (bait) was able to robustly pull down LIC1 along with IC and HC from both HeLa and U2OS mitotic lysates (Fig. 4, A and B). Surprisingly, spindly, p150Glued, p50, Zw10, and Mad1, all part of the spindly-dynactin-dynein mitotic kinetochore complex, did not coprecipitate (Fig. 4, A and B), suggesting that this spindly-occupied fraction of dynein is unlikely to be regulated by Pin1. Dynein devoid of dynactin can contain the cofactors Nde1 and Lis1 and is known to mediate high load-bearing functions (Huang et al., 2012; McKenney et al., 2010; Raaijmakers et al., 2013; Vallee et al., 2012). Indeed, we found that Nde1 and Lis1 were present in the GST-Pin1 mitotic affinity precipitates, as were centromere protein F (CENPF), a known interactor of Nde1-Lis1-dynein (Bolhy et al., 2011; Vergnolle and Taylor, 2007), and the mitotic dynein adaptor Hook2, which interacts with the late G2/prophase Nde1-Lis1-CENPF-dynein complex (Dwivedi et al., 2019; Fig. 4, A and B). These results suggested that Pin1 binds preferentially to Hook2-bound dynein, but not to spindly-bound dynein, in mitosis. Pin1’s binding to LIC1, IC, and Hook2 was drastically reduced upon making the phosphodeficient AAA mutation (Fig. 4, B–D), suggesting compromised Pin1 binding with the whole dynein complex. Nde1, Lis1, and CENPF also showed reduced Pin1 binding, although significant amounts could still bind (Fig. 4, B–D), perhaps through their phosphorylation-based engagement independently with Pin1. We observed clear localization of both Pin1 and SST/AAA LIC1 at G2/prophase centrosomes and metaphase spindles, but no substantial Pin1 localization at prometaphase kinetochores (Fig. 4 E). Pin1 is known to interact with several mitotic substrates on G2/prophase centrosomes (Lee et al., 2013) and the spindle (St-Denis et al., 2011) apart from dynein, which could explain its localization at these structures independent of LIC1 phosphorylation. Overall, these results revealed the preferential interaction of Pin1 with the Hook2-Nde1-Lis1-CENPF dynein complex, but not with the spindly-dynactin-dynein complex during mitosis, highlighting its ability to differentiate between dynein complexes containing distinct mitotic adaptors.
Figure 4.
Pin1 interacts with the Hook2-Nde1-Lis1 dynein complex, but not with the spindly-dynactin-dynein complex. (A) Immunoblots showing affinity precipitates of GST-Pin1/GST from prometaphase (nocodazole-treated) and metaphase (released into MG132 after nocodazole treatment) HeLa cell lysates, probed for the indicated antigens. (B) Immunoblots showing the affinity precipitates of GST-Pin1 from SST-MTAP and AAA-MTAP prometaphase U2OS stable cell lysates, probed for the indicated antigens. (C and D) Ratios (AAA-MTAP/SST-MTAP) of eluate (beads) fraction band intensities for each indicated antigen (x axes) over three independent affinity purification (AP) experiments. (E) Representative single confocal z-sections of SST- or AAA-MTAP U2OS stable cells showing the localization of the indicated antigens. Boxes are magnified in the insets. The metaphase images are deconvolved to enhance spindle fiber visualization. Arrows indicate colocalization. Scale bar = 10 µm, inset scale bar = 5 µm. IB, immunoblot. Error bars = mean ± SEM. ***, P < 0.001; ****, P < 0.0001 (one-way ANOVA).
LIC1-CTD phosphorylation is required for dynein-mediated prophase centrosome–NE attachment
Hook2-dynein and Nde1-Lis1-CENPF are required to mediate centrosome-NE attachment in prophase (Bolhy et al., 2011; Dwivedi et al., 2019; Raaijmakers et al., 2013). Upon LIC1 siRNA treatment, we observed a small but significant increase in detachment that could be rescued by SST but not by AAA (Fig. 5, A and D), suggesting a requirement of LIC1-CTD phosphorylation. We tested which cdk1 cofactor, cyclin A or B, which are both active at differential levels in late G2/prophase, is required for this function (De Boer et al., 2008; Gavet and Pines, 2010). Upon using cyclin A/B isoform-specific siRNAs (Fig. 5 C), we observed no measurable centrosome-NE detachment upon cyclin A depletion, but significant detachment and centrosome fragmentation upon cyclin B depletion, an effect that was mildly amplified upon codepletion of both cyclins (Fig. 5, B, E, and F), possibly due to the absence of the compensatory action of cyclin A upon cyclin B depletion in early mitosis (Gong and Ferrell, 2010; Hégarat et al., 2020).
Figure 5.
LIC1-CTD phosphorylation is required for prophase centrosome attachment with the NE. (A and B) Representative merged confocal micrographs of G2/prophase synchronized U2OS cells (10 h after double thymidine block and release) showing the relative centrosome (γ-tubulin, arrows) and NE (inner dashed lines) positions upon the indicated siRNA treatments. Outer dashed lines, cell boundaries. Scale bar = 40 µm. (C) Immunoblots showing cyclin A/B depletion upon the respective siRNA treatments. (D–F) Fraction of G2/prophase U2OS cells showing significant centrosome-NE detachment (D and E) and centrosome fragmentation (F) under various conditions. n = 3 independent experiments, ≥150 cells per experiment. (G) Immunoblots showing affinity precipitates of GST-Pin1/GST from late G2/prophase HeLa cell lysate (8 h after double thymidine block and release), probed for the indicated antigens. (H–J) Quantification of the maximal centrosome-NE distance (H) and the timing from inter-centrosome separation to NEB (I) from time lapse, live-cell videos of double-labeled HeLa cells under the indicated treatments (represented in J, sequential still frames with time stamps shown). n = 2 independent experiments, ≥36 mitotic cells per condition. Scale bar = 10 µm. Arrowheads, centrosomes (green); AP, affinity purification; IB, immunoblot. Error bars = mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (one-way ANOVA).
We observed that IC, HC, phosphorylated LIC1, Hook2, Nde1, Lis1, and CENPF all interacted with GST-Pin1 in late G2/prophase–enriched HeLa cell lysates, while the dynactin subunits p150Glued and p50 did not (Fig. 5 G), suggesting that the interaction of the dynein-Nde1-Lis1-CENPF complex with Pin1 might occur as early as late G2/prophase. We tested whether the AAA mutation, which drastically reduces Pin1-dynein binding (Fig. 4), impacted centrosome-NE attachment. From live-cell videos of hLIC1-depleted GFP-α-tubulin::H2B-mcherry HeLa cells, we observed visible centrosome-NE detachment upon LIC1 depletion or rescue with rLIC1-AAA (Fig. 5, H and J; and Video 8). Dynein also facilitates NEB at late G2/prophase (Salina et al., 2002). However, we observed that, at the temporal resolution of our live imaging experiments (5 min between successive time points), neither LIC1 depletion nor the LIC1-CTD phosphorylation status in the rescue experiments significantly perturbed the timing of NEB onset following intercentrosome separation in late G2/prophase (Fig. 5 I). Overall, these results functionally implicated mitotic LIC1-CTD phosphorylation in maintaining dynein-dependent centrosome-NE attachment in late G2/prophase.
Video 8.
Centrosome–NE detachment dynamics in HeLa cells. Confocal fluorescence time-lapse videos of double stable GFP-α-tubulin (green)::histone 2B-mCherry (red) HeLa cells under the following conditions: mock transfection, hLIC1 depletion, hLIC1 depletion + rLIC1-SST expression, and hLIC1 depletion + rLIC1-AAA expression. All videos show the duration from centrosome separation to anaphase onset. Time stamps included. Scale bars = 10 µm. Speed is 3 frames per second.
LIC1-CTD phosphorylation is required for chromosome congression
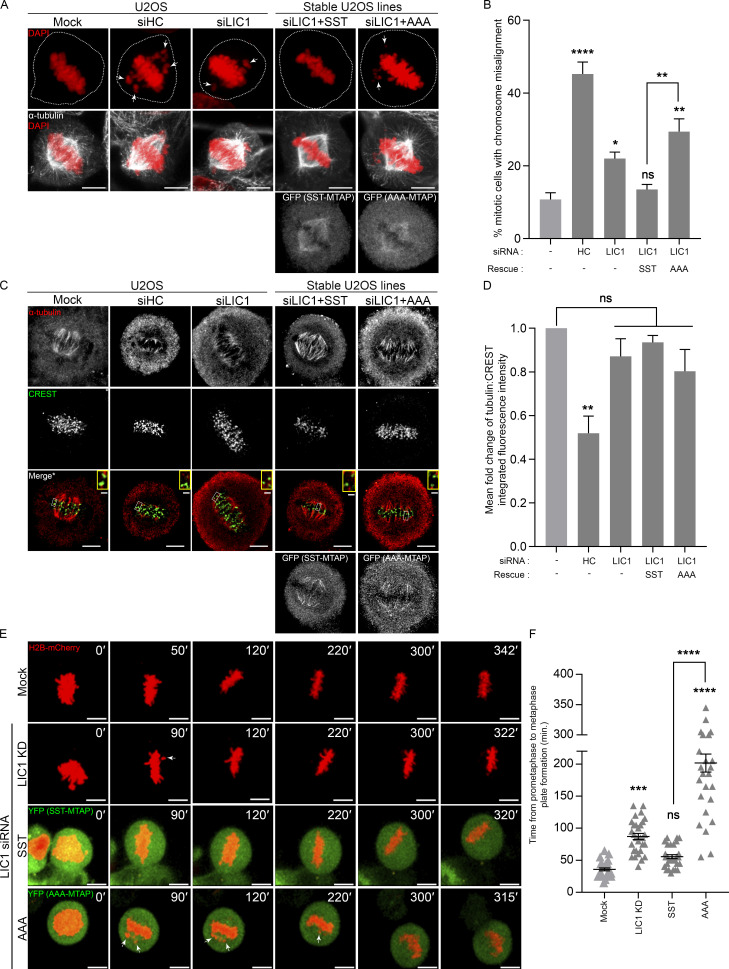
We assessed whether LIC1-CTD mitotic phosphorylation is required for chromosome congression, a known function of dynein, where roles of Hook2, Nde1, and Lis1 have also been reported (Dwivedi et al., 2019; Moon et al., 2014; Vergnolle and Taylor, 2007). LIC1 siRNA treatment led to an increase in the fraction of miscongressed metaphase U2OS cells, which could be rescued by stably expressed SST, but not by AAA (Fig. 6, A and B), implicating mitotic LIC1-CTD phosphorylation in this function. Given the prolonged metaphase arrest observed with AAA (Fig. 1, F–H; and Fig. S1), we measured the stability of kinetochore–microtubule (Kt-MT) attachments by quantifying α-tubulin fluorescence intensity after cold treatment, which positively correlates with the levels of stable attachments (DeLuca et al., 2016). The normalized α-tubulin intensity was significantly reduced upon HC siRNA treatment, but not upon LIC1 siRNA treatment or rescue with SST/AAA, suggesting that LIC1 function or phosphorylation is dispensable (Fig. 6, C and D). Time-lapse imaging of H2B-mCherry–transfected U2OS cells revealed that LIC1 siRNA treatment led to delayed chromosome congression, but most cells eventually congressed and maintained an aligned metaphase plate for prolonged durations (Fig. 6, E and F; and Video 9) when measured up to time points preceding typical cohesion fatigue (Daum et al., 2011; Stevens et al., 2011). Stably expressed SST rescued the congression delay but did not impact subsequent plate integrity, while rescue with stably expressed AAA led to drastic miscongression, while not significantly impacting subsequent plate integrity (Fig. 6, E and F). Overall, these results suggested a role for LIC1-CTD mitotic phosphorylation in initial metaphase chromosome capture and alignment but not in maintaining the stability of Kt-MT attachments.
Figure 6.
LIC1-CTD phosphorylation is required for chromosome congression. (A) Representative confocal micrographs showing metaphase chromosome (mis)alignment in U2OS (mock, HC knockdown) or SST-/AAA-U2OS stable cell lines under the indicated conditions. (B) Fraction of miscongressed metaphase cells. n = 3 experiments, 50 cells per experiment. (C) Representative confocal maximum projections from deconvolved planes showing cold-stable microtubules attached to metaphase kinetochores under the indicated conditions. *, Merge: Single z-plane shown from the stack to clearly visualize kinetochore–microtubule attachment; selected kinetochore pairs magnified in insets. (D) Mean fold-change (vs. mock, first bar) of the microtubule (α-tubulin):kinetochore (CREST) integrated fluorescence intensity from metaphase cells upon cold treatment. n = 3 experiments, 20 cells per experiment. (E) Stills from representative time-lapse videos of mitotically synchronized U2OS cells (mock, LIC1 knockdown [KD]) or SST-/AAA-MTAP (green, YFP) stable U2OS cells expressing H2B-mCherry (red), recording the time taken from prometaphase to metaphase plate formation. Time stamps (min) included in the images. (F) Quantification of the timing from prometaphase to metaphase plate formation. n = 2 experiments, 15 mitotic cells per experiment. Arrows, misaligned chromosomes. Cells were released into MG132 after nocodazole treatment for 2 h (fixed cells), and live video imaging was started immediately after MG132 addition. Scale bar = 10 µm, inset scale bar = 1 µm. Error bars = mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (B and D, one-way ANOVA; F, Kruskal–Wallis test).
Video 9.
Metaphase chromosome alignment and metaphase plate integrity in U2OS cells. Time-lapse confocal videos of U2OS cells transfected with H2B-mCherry (red, chromosomes) upon mock transfection or LIC1 siRNA treatment (top two cells); and LIC1 siRNA-treated stable MTAP-tagged WT LIC1 SST and LIC1-CTD triple mutant AAA U2OS cells (treated with MG132 following nocodazole treatment and release), transfected with H2B-mCherry (red) showing the timing from prometaphase to metaphase plate formation and further metaphase plate integrity. Time stamps included. Scale bars = 10 µm. Speed is 3 frames per second.
LIC1-CTD phosphorylation and Pin1 regulate mitotic Golgi fragmentation but not interphase membrane transport
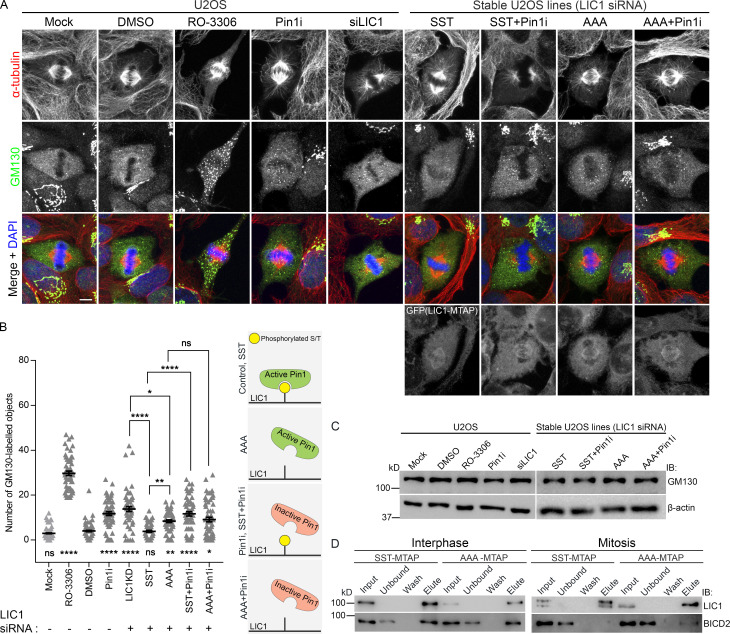
The dramatic fragmentation of interphase Golgi stacks to a “Golgi haze” in metaphase is a characteristic feature of Golgi dynamics during the cell cycle (Misteli and Warren, 1995; Shorter and Warren, 2002). Golgi fragmentation in mitosis requires the dissociation of dynein from Golgi membranes, with cdk1-mediated LIC1 phosphorylation postulated as a potential trigger (Addinall et al., 2001; Yadav and Linstedt, 2011). As expected, treatment with the cdk1 inhibitor RO-3306 (9 µM) dramatically inhibited Golgi fragmentation (Fig. 7, A and B). Upon either LIC1 depletion or rescue with AAA but not with SST, we observed a small but significant persistence of GM130-positive Golgi punctae at metaphase without impacting GM130 levels, indicating incomplete Golgi fragmentation upon preventing mitotic hLIC1-CTD phosphorylation (Fig. 7, A–C). Interestingly, when treated with a potent and specific Pin1 inhibitor BJP-06-005-3 (20 µM; henceforth “Pin1i”; Pinch et al., 2020), both control and SST-rescued cells showed incomplete Golgi fragmentation, while no additional loss of fragmentation was observed in AAA-rescued cells (Fig. 7, A and B). AAA could partially rescue LIC1 depletion-induced Golgi fragmentation as compared with SST (Fig. 7, A and B). Together, these observations suggest that Pin1 activity on the hLIC1-CTD phosphorylation sites likely plays a partial but significant role in enabling complete mitotic Golgi fragmentation, possibly by preventing dynein disengagement from Golgi membranes (Yadav et al., 2012). The interphase dynein adaptor BICD2 is essential for Golgi binding and transport (Hoogenraad et al., 2001; Matanis et al., 2002), but is unlikely to have mitotic functions (Raaijmakers et al., 2013). Using SBP affinity elution, we observed robust binding of both SST and AAA to BICD2 in interphase, as expected (Fig. 7 D). Interestingly, during mitosis, while the phosphorylatable SST expectedly showed complete loss of BICD2 binding, the nonphosphorylatable AAA still retained some BICD2 binding (Fig. 7 D). These results suggested that cdk1-mediated mitotic hLIC1-CTD phosphorylation could regulate dynein adaptor selectivity during the interphase-to-mitosis transition. Correlated with the incomplete Golgi fragmentation observed with AAA but not with SST, these observations suggest the possibility of incomplete dynein-Golgi membrane disengagement upon preventing mitotic LIC1-CTD phosphorylation.
Figure 7.
LIC1-CTD phosphorylation and Pin1 function regulate mitotic Golgi fragmentation. (A) Representative confocal images showing the localization of the Golgi protein GM130 in metaphase (after single thymidine block and release into MG132 for 2 h) under the indicated conditions. (B) Quantification of the number of GM130-positive punctae. n = 3 experiments, 20 cells per experiment. (C) Immunoblots showing the expression of GM130 under the indicated conditions. (D) Immunoblots from SBP-affinity precipitates of SST-MTAP or AAA-MTAP cell lysates at the indicated stages. IB, immunoblot. Scale bar = 10 µm. Error bars = mean ± SEM. *, P < 0.05; **, P < 0.01; ****, P < 0.0001 (Kruskal–Wallis test).
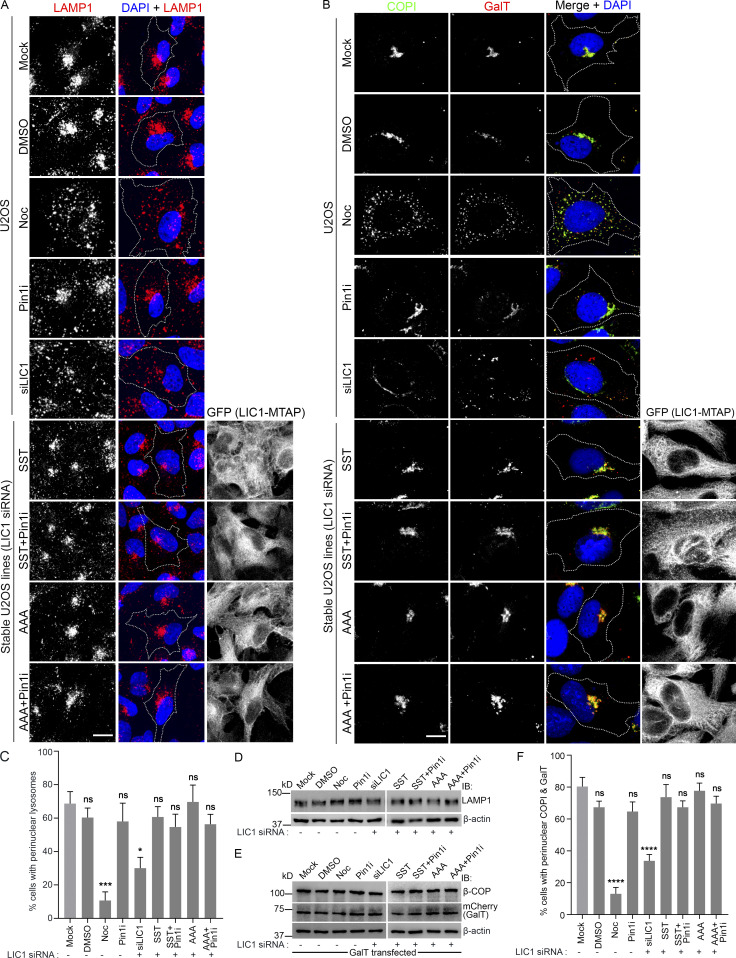
We probed whether LIC1-CTD phosphorylation is required for dynein’s role in interphase membrane transport. LIC1 is required for the perinuclear enrichment of LAMP1-positive lysosomes/late endosomes (Scherer et al., 2014; Tan et al., 2011). SST and AAA were equally competent at rescuing the LIC1 depletion-induced dispersal of perinuclear lysosomes (Fig. 8, A and C). Even treatment with Pin1i did not impede this rescue or perturb LAMP1 levels in either case (Fig. 8, A, C, and D). LIC1 is also known to maintain perinuclear interphase Golgi complex organization and optimal ER-Golgi transport (Palmer et al., 2009). The focused juxtanuclear localization of the Golgi marker COPI (coatomer complex subunit, a marker for Golgi integrity and ER-Golgi transport; Palmer et al., 2009) was disrupted upon LIC1 depletion; however, it was restored by both SST and AAA, even with Pin1i treatment (Fig. 8, B, E, and F). We also observed that the strong perinuclear colocalization of the exogenously expressed Golgi cargo β-1,4-galactosyltransferase I (GalT) with COPI in control cells was drastically disrupted upon LIC1 depletion, but could be rescued by both SST and AAA; Pin1i treatment did not affect this outcome (Fig. 8, B, E, and F). Together, these observations suggest that LIC1-CTD phosphorylation and Pin1 activity are not required for maintaining Golgi integrity or regulating ER-Golgi transport. Overall, these results demonstrate that neither cdk1-mediated LIC1-CTD phosphorylation nor Pin1 activity are involved in dynein-mediated interphase membrane transport, thus confirming their exclusive contributions to mitotic dynein functions.
Figure 8.
LIC1-CTD phosphorylation and Pin1 function are not required for interphase membrane transport. (A and B) Representative confocal images showing the localization of lysosomes (LAMP1, A), Golgi (COPI), and GalT (Golgi cargo) under the indicated conditions. NOC, nocodazole; Pin1i, Pin1 inhibitor. (C) Fraction of cells showing prominent perinuclear lysosomal localization under the indicated conditions. n = 3 experiments, 120 cells per experiment. (D and E) Immunoblots showing the expression of the indicated proteins; GalT is exogenously expressed in E. (F) Fraction of cells showing prominent perinuclear colocalization of COPI and GalT. n = 3 experiments, 75 cells per experiment. IB, immunoblot; dotted lines, cell boundaries. Scale bar = 30 µm. Error bars = mean ± SEM. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001 (one-way ANOVA).
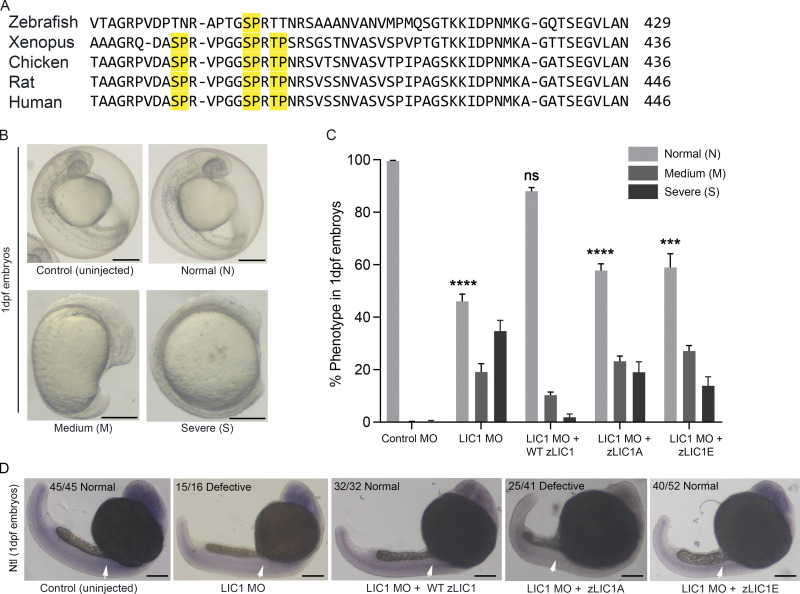
Evolutionarily conserved LIC1-CTD phosphorylation regulates early vertebrate development
Primary sequence alignment shows high conservation of the vertebrate LIC1-CTD cdk1 phosphorylation sites (Fig. 9 A). Morpholino (MO)-based depletion of zebrafish LIC1 (zLIC1) causes mitotic defects in blastomeres, similar to human cells (Mahale et al., 2016a). In comparison to the three LIC1-CTD cdk1 sites found in the more complex vertebrates, only the SP site at S388 (corresponding to hLIC1-S405) is conserved in zebrafish LIC1 (zLIC1; Fig. 9 A). We therefore assessed the ability of WT (S388, zLIC1), phosphodeficient (S388A, zLIC1A), and phosphomimetic (S388E, zLIC1E) mRNA constructs to functionally rescue zLIC1-MO–induced phenotypes in early zebrafish development. About 55% of the zLIC1 MO-treated embryos showed delayed embryonic development of varying severity at 1 day post fertilization (dpf; Fig. 9, B and C; 35%, severe axis elongation and notochord defects [severe, S]; 20%, mild anterior-posterior axis elongation defects [medium, M]). Introduction of WT zLIC1 mRNA led to a significant rescue of the phenotypes; however, zLIC1A or zLIC1E led to only a partial rescue, even though zLIC1E was slightly more competent (Fig. 9 C). These complementation results revealed that locking zLIC1 into either the A or E form, with no scope for phospho-switching, was detrimental for proper early zebrafish development.
Figure 9.
LIC1-CTD phosphomutants are unable to efficiently rescue LIC1 depletion phenotypes in zebrafish embryos. (A) Multiple sequence alignment of representative vertebrate LIC1-CTDs. Yellow, conserved SP/TP cdk1 sites. (B) Bright-field images (5×) of 1-dpf zebrafish embryos. Representative control (uninjected) and zLIC1 MO–injected and/or rescued embryos showing three categories of phenotypes: normal (N), medium (M), or severe (S). (C) Fraction of embryos showing the phenotypes defined in E, upon rescue of zLIC1 depletion with the indicated zLIC1 mRNA constructs. P values calculated with regard to control MO. (D) Representative bright-field images of 1-dpf zebrafish embryos (5×), stained by ISH using a riboprobe against the zebrafish Ntl gene (purple). Numbers indicate the fraction of embryos showing the depicted phenotype. Arrows depict Ntl mRNA expression, which is weakened upon zLIC1 depletion or rescue with zLIC1A. Scale bar = 500 µm. Error bars = mean ± SEM. ***, P < 0.001; ****, P < 0.0001 (one-way ANOVA).
We performed in situ hybridization (ISH) of the No tail (Ntl) gene (TbxTa in zebrafish) in rescued embryos that were visibly normal. Ntl marks the mesoderm, notochord, and tail bud at 1 dpf and is a marker for notochord development (Morley et al., 2009; Schulte-Merker et al., 1994; Zhu et al., 2016). zLIC1 MO treatment resulted in defects of mesodermal origin, as shown by the Ntl ISH (Fig. 9 D). Interestingly, rescue with either zLIC1 or zLIC1E showed a largely normal Ntl expression pattern in the notochord, comparable to control (uninjected) embryos; in contrast, rescue with zLIC1A showed significant reduction in Ntl expression in the notochord and tail bud (Fig. 9 D). Thus, despite showing similar gross morphologies following rescue, zLIC1A was markedly deficient in rescuing Ntl gene expression and notochord development compared with zLIC1E. Overall, these observations emphasized the functional importance of phosphoregulation at this highly conserved zLIC1-CTD residue for early development in the vertebrate zebrafish.
Discussion
Cdk1-mediated mitotic LIC1 phosphorylation has been correlated with dynein’s detachment from interphase membrane cargoes (Addinall et al., 2001), potentially enabling a switch to mitotic cargoes. However, its role in regulating mitotic dynein functions has not been delineated. Our study reveals that LIC1-CTD phosphorylation regulates mitotic dynein function both directly and through selective Pin1 engagement with a subset of dynein complexes. The incomplete mitotic Golgi fragmentation seen with AAA is likely due to a prolonged dynein–Golgi association, a possibility consistent with the sustained binding of the interphase Golgi adaptor BICD2 up to metaphase (Fig. 7). The lack of other significant dynein-dependent interphase membrane trafficking defects (Fig. 8) establishes the exclusively mitotic contributions of LIC1-CTD cdk1 phosphorylation. In addition, this phosphorylation is required for binding to the mitotic kinetochore adaptor spindly (Fig. 2), and thereby for efficient prometaphase dynein loading (Figs. 2 and S3) and subsequent SAC component binding and inactivation (Figs. 2 and S4). The requirement of the solely conserved zLIC1 phosphosite for proper zebrafish embryonic development (Fig. 9) emphasizes the evolutionary importance of LIC1-CTD cdk1 phosphorylation. Importantly, the embryonic developmental defects with either zLICA or zLIC1E, which are constrained in the “off” (possibly interphase) and “on” (possibly mitotic) phosphoisoforms in zebrafish, respectively, suggest the possibility that temporally regulated phosphorylation-dephosphorylation toggling at this site might be required for normal cell cycle progression of blastomeres during embryonic development.
A novel observation from this study is the association of the prolyl isomerase Pin1, a master mitotic regulator, with dynein in mitosis, mediated through the direct Pin1:phospho-LIC1-CTD interaction (Figs. 3 and 4), suggesting interesting possibilities for dynein’s functional modulation. The virtual absence of dynactin, spindly, Mad1, and Zw10 from the mitotic Pin1 affinity precipitates (Fig. 4), while surprising, prompted us to look for the dynactin-free, high load-bearing dynein complexes containing Nde1 and Lis1. Dynactin and Nde1 are known to be mutually exclusive binding cofactors for a common binding site on the IC (McKenney et al., 2011; Nyarko et al., 2012). Indeed, Nde1, Lis1, CENPF, and the mitotic dynein adaptor Hook2 could bind robustly with Pin1, along with dynein subunits (Fig. 4), suggesting that Hook2 can be a part of both dynactin-free and dynactin-containing dynein complexes (Dwivedi et al., 2019). The significant defects in prophase centrosome-NE tethering (Fig. 4) and chromosome congression (Fig. 6) observed with AAA suggested an essential role for LIC1-CTD phosphorylation in these high load-bearing dynein functions (Li et al., 2007; Splinter et al., 2010), wherein Nde1, Lis1, and Hook2, but not dynactin, play important roles (Dwivedi et al., 2019; Raaijmakers et al., 2013). Dynactin is dispensable for chromosome congression and acts only as a recruiter of dynein to the NE for centrosome-NE attachment, with Nde1 proposed as the activator (Raaijmakers et al., 2013). We postulate that CTD-phosphorylated LIC1, with subsequent help from Pin1, could help in the exclusive engagement of Hook2 (but not spindly), and thereby possibly favor the assembly and/or stability of high load-bearing dynein complexes. The prominent Pin1 localization on late G2/prophase centrosomes and the mitotic spindle, but not explicitly at the mitotic kinetochores (Fig. 4), is mirrored by the mutually exclusive localization of the mitotic dynein adaptors, spindly (at kinetochores; Gama et al., 2017; Griffis et al., 2007) and Hook2 (at centrosomes; Dwivedi et al., 2019; Szebenyi et al., 2007). Our results are consistent with a hypothesis in which Pin1 acts on mitotically phosphorylated LIC1 at nonkinetochore dynein pools, possibly on the G2/prophase centrosomes and the mitotic spindle. The interaction of Pin1 with the Hook2-Nde1-Lis1-CENPF dynein complex from prophase through metaphase (Figs. 4 and 5) suggests the possibility of Pin1-mediated regulation of one or more functions of this complex and merits a detailed independent investigation. It is also noteworthy that the mutually exclusive LIC2-dynein complex is independently required for most of dynein’s mitotic functions, often redundantly (Mahale et al., 2016a; Raaijmakers et al., 2013), which could explain the apparently small effects seen upon LIC1 depletion alone. Interestingly, depletion of one LIC subunit has been shown to cause a compensatory increase in levels of the other (our unpublished observations; Raaijmakers et al., 2013; Scherer et al., 2014); rescue of LIC1 depletion with AAA may possibly suppress this feedback loop, leading to a lack of compensatory rescue by LIC2-dynein. In this background, the sequestering of the natural interacting partners by the nonfunctional AAA-LIC1-dynein could result in the dominant-negative effects that further exacerbate the LIC1 depletion phenotypes.
The conservation and functional importance of the single S388 zLIC1-CTD site (analogous to hLIC1 S405; Fig. 9), which has expanded to three sites in the more complex vertebrates, suggests that phosphorylation at a minimum number of the three phosphosites in the 398–408 region could be vital to achieve a minimum threshold of mitotic function. Beyond this threshold, the exact permutation of phosphorylated sites in a given dynein complex could modulate the potency of the downstream cellular response, an idea proposed for clustered phosphosites (Schweiger and Linial, 2010), with the cluster potentially serving as a “cooperative module [that is] resistant to selective pressures” (Yachie et al., 2009).
LIC1-CTD exhibits the architecture of an intrinsically disordered protein region, characterized by helical binding motifs H1 and H2 and flanked by flexible, unstructured linkers L1 and L2 (Kumari et al., 2021). Nuclear magnetic resonance studies have revealed multiple points of contact with adaptors in LIC1-CTD, primarily in H1 (440–455), H2 (493–502), and linker L1 (418–421; Celestino et al., 2019), although H1 is the minimum indispensable binding motif (Lee et al., 2020). Phosphorylation-dependent Pin1-LIC1 binding could help achieve conformational rearrangement of LIC1-CTD through prolyl isomerization, thus resulting in the optimal engagement with specific dynein adaptors (e.g., Hook2), while precluding binding with others (e.g., spindly; Fig. 6), with obvious functional implications. For instance, in spindly (a CC1-box adaptor), the two LIC1 interaction sites are located adjacent on the same coiled coil, whereas in Hook2 (a Hook-domain adaptor), they are positioned farther away from each other (Lee et al., 2018). Remarkably, however, the same helix H1 on LIC1-CTD forms the primary interaction interface with both adaptor NTD clefts (Celestino et al., 2019; Lee et al., 2020; Lee et al., 2018). It is attractive to imagine that the large conformational changes that would be necessary for LIC1-CTD to bind to such conformationally diverse adaptors could be enabled by selective Pin1-mediated isomerization, so as to position H1 in the correct register within divergent adaptor NTD clefts.
During the short span of mitosis, dynein subpopulations could be required to dynamically switch between diverse functions, necessitating the rapid and finely tuned association with a diverse set of adaptors, cofactors, and cargoes, which could be dictated by conformational modulation of the LIC1-CTD. Interestingly, there are also several non-cdk1 mitotic phosphosites in the LIC1-CTD (Dephoure et al., 2008; Olsen et al., 2010), which, in combination with the cdk1 sites, could exponentially increase the potential number of unique phosphorylated permutations to finely regulate the interactions and functions of mitotic dynein (Kumari et al., 2021). Interestingly, the hydrophobic adaptor NTD clefts that interact directly with the hydrophobic H1 of LIC1-CTD are positioned in close proximity to several conserved negatively charged residues (present in all three classes of adaptors; Lee et al., 2020; Schroeder and Vale, 2016) and could therefore be electrostatically repelled due to LIC1-CTD phosphorylation around the points of contact with the adaptor, possibly inhibiting the binding of specific adaptors. For example, such repulsion could be envisioned to help completely dissociate the interphase Golgi adaptor BICD2 upon LIC1-CTD phosphorylation at mitotic entry, but remains partially bound in mitosis upon abrogating LIC1-CTD phosphorylation (Fig. 7 D).
In our immunoblots, we observed a retarded IC band (Fig. 4, A and B; and Fig. 5 G), suggesting the possibility of mitotic IC phosphorylation. Mitotic IC phosphorylation by Plk-1 at T89 is required for Zw10 binding, dynein’s kinetochore recruitment, and chromosome alignment (Bader et al., 2011; Whyte et al., 2008). It is possible that both phospho-LIC1 and phospho-IC could cooperate to enhance dynein binding to RZZ, although this possibility needs to be experimentally tested. S84 phosphorylation of IC strengthens Nde1 binding at the expense of dynactin (Jie et al., 2017), while phosphorylation at its mitotic equivalent T89 reduces dynactin binding but increases Zw10 binding (Bader et al., 2011; Whyte et al., 2008). These correlated observations suggest the possibility of a coordinated regulation of selective dynein complex formation through the phosphorylation of different dynein subunits, perhaps to generate a variety of dynein complexes. This study reveals the dynein motor as a potential new mitotic substrate of Pin1.
Indeed, multiple prolyl isomerases feature in the interphase interactomes of dynein subunits, including IC and LIC (Redwine et al., 2017), raising the possibility of prolyl isomerization-based conformational modulation of dynein as an important means of regulating its interactions and functions. It would be interesting to study whether other mitotically phosphorylated dynein subunits and cofactors, such as IC and Nde1 (Wynne and Vallee, 2018), could also be potential Pin1 substrates, and thus possibly provide considerably higher biochemical diversity to support the vast functional spectrum of mitotic dynein.
Materials and methods
Plasmid constructs and site-directed mutagenesis
The GST-Pin1 plasmid was procured from Addgene (19027). Full-length rat LIC1 cloned in the pCMV 3Tag 3B vector (mammalian expression vector) and in pGEX-6P-1 vector (bacterial expression vector) was used as the template to generate all the rLIC1 phosphomutant constructs using a standard site-directed mutagenesis protocol with the respective primers containing the desired mutations. Similarly, full-length human LIC1 cloned in the MTAP vector (containing a C-terminal triple-TAP tag [His8:SBP:FLAG] embedded in a loop of YFP; Ma et al., 2012) was used as the template to generate the hLIC1 phosphomutant constructs with primers containing the desired mutations. The desired mutations were confirmed by sequencing. The N-terminal (aa 41–386) and C-terminal (aa 389–523) rLIC1 constructs were PCR amplified from a full-length rLIC1 construct in the pGEX-6P-1 vector using their respective primers, followed by cloning in the pGEX-6P-1 vector between the ECoRI and XhoI restriction sites. All the primers used in this study are listed in Table S1. All the plasmids used in this study were transfected using Lipofectamine 2000 (Invitrogen)/X-tremeGENE HP DNA (Roche Applied Science/Sigma-Aldrich) as per manufacturer’s instructions.
Cell culture, stable cell line generation, siRNA transfection, and cell synchronization
HeLa cells were procured from the European Collection of Authenticated Cell Cultures (through Sigma-Aldrich), while U2OS cells were kind gift from Prof. Stephen J. Doxsey (University of Massachusetts Medical School, Worcester, MA) and were cultured in DMEM (Invitrogen/Gibco) supplemented with 10% FBS and antibiotic solution of penicillin and streptomycin (HiMedia). Cells were cultured at 37°C with 5% carbon dioxide and 95% humidity. To generate stably expressing cell lines, U2OS cells (human bone osteosarcoma epithelial cell line) were transfected with the respective hLIC1-MTAP construct (SST/AST/SAT/SSA/AAT/SAA/AAT/AAA) cloned into the mammalian expression vector pcDNA4-TO-hygromycin-mVenus-MAP (MTAP), as described earlier (Mahale et al., 2016a). Briefly, this construct was transfected using X-tremeGENE HP DNA, and cells stably expressing the hLIC1-MTAP were selected over 4 wk under antibiotic selection (hygromycin B, 300 µg/ml; TOKU-E) and sorted using flow cytometry based on varying degrees of YFP (a part of the MTAP tag) fluorescence. The stable cell line expressing the YFP fluorescent tag was visualized under a TCS SP8 confocal microscope (Leica), and the level of expression of the transgene was confirmed by immunoblotting.
siRNAs against different human genes were procured from Dharmacon. The siRNA sequences and their respective working concentrations used in the study are as follows: LIC1 (100 nM), 5′-GAAAGUUUGUACAUGAGAA-3′; and Mad2 (100 nM), 5′-GAGUCGGGACCACAGUUU-3′. Both siRNA sequences were previously used and documented (Mahale et al., 2016b; Sivaram et al., 2009). ON-TARGETplus Human DYNC1H1 siRNA- SMARTpool (L-006828-00-0005), ON-TARGETplus human CCNA2 siRNA-SMARTpool (L-003205-00-0005) and ON-TARGETplus human CCNB1 siRNA-SMARTpool (L-003206-00-0005) for dynein heavy chain, cyclin A2, and cyclin B1 siRNAs (Hégarat et al., 2020) were used, respectively. Individual cyclin A2 and cyclin B1 siRNAs (80 nM each) were transfected using Dharmafect1 (Dharmacon) for 72 h. For codepletion experiments, 100 nM each of LIC1 and Mad2 siRNAs were used. For cyclin A2 and cyclin B1 codepletion, 80 nM each of the siRNAs were used for 72 h before immunofluorescence sample preparation. Rescue experiments in cell lines were performed by transfecting the plasmids for 24 h, followed by siRNA transfection for another 48 h. Rescue in the stable U2OS cell lines was performed using 100 nM of LIC1 siRNA for 24 h to be able to achieve endogenous LIC1 siRNA depletion along with expression of MTAP-tagged hLIC1 in the stable lines (SST and various mutants; Fig. 1 E). The mitotic/metaphase index was calculated by counting the number of SYTO 13 (Invitrogen) stained mitotic/metaphase cells as a fraction of total cells.
To prepare mitotic lysates, cells were treated with nocodazole (100 nM HeLa and 300 nM U2OS) for 12–14 h and either harvested immediately (for preparing prometaphase lysates) or released in to nocodazole-free medium containing MG132 (10 µM) for 2 h (for preparing metaphase lysates). For late G2/prophase enrichment, HeLa and U2OS cells were arrested at the G1/S boundary by two consecutive rounds of thymidine treatment and release (treatment with 2.5 mM thymidine for 18 h, followed by 8 h release each time for HeLa cells and 10 h for U2OS cells). After harvesting, cells were flash frozen in liquid nitrogen and physically ground under cryogenic conditions as detailed earlier (Cheeseman and Desai, 2005; Domanski et al., 2012).
Antibodies and chemicals
The following primary antibodies were used: LIC1 (Thermo Fisher Scientific; PA531644), Mad1 (Thermo Fisher Scientific; PA5-28185), Mad2 (Thermo Fisher Scientific; PA5-21594), Zw10 (Abcam; ab21582), dynein IC (IC74) monoclonal antibody (Abcam; ab23905), dynein HC (ProteinTech; 12345-1-AP); Nde1 (36-T Santa Cruz, Sc100328 [NudE1]; and ProteinTech, 10233-1-AP), Lis1 (Abcam, ab68598; and clone LIS1-338, Sigma-Aldrich, L7391), CENP-F (Abcam; ab5), Pin1 (G-8 Santa Cruz, sc-46660; and Pierce, PA1-10013), p150Glued (BD Biosciences; 610474), Hook2 (Abcam; ab154109), spindly (OD 157 and OD 173, kind gifts from Reto Gassman and Arshad Desai), GST (Sigma-Aldrich; G7781), α-tubulin (DM1α, Sigma-Aldrich; T9026), β–actin (Sigma-Aldrich; A3835), anti-GFP (Abcam; ab6556 and ab1218), cyclin A2 (Abcam; ab38), cyclin B1 (Abcam; ab32053), GM130 (Abcam; ab 52649), LAMP1 (Sigma-Aldrich; L1418), γ-tubulin (Abcam; ab11316), mCherry (Abcam; ab167453), β-COP (Abcam; ab2899), BICD2 (Abcam; ab117818), p50 (BD Biosciences; 611002), and CREST (Antibodies Inc.). The following antibodies were procured from Jackson Immunoresearch USA: HRP-conjugated anti-mouse (715-035-150) and anti-rabbit (711-035152) secondary antibodies for immunoblotting and fluorophore-attached anti-mouse and anti-rabbit Alexa Fluor 488–conjugated secondary antibodies (715-545-150 and 711-545-152) and anti-mouse, anti-rabbit Alexa Fluor 594–conjugated secondary antibodies (715-585-150 and 711-585-152), Cy3-conjugated anti-rabbit (111-165-144), biotin-conjugated anti-chicken secondary antibody (103-065-155), and human Cy5 (109-175-008) secondary antibody as well as streptavidin-conjugated Cy3 antibody (016-160-084) for immunofluorescence staining. Thymidine, nocodazole, and the cdk1 inhibitor RO-3306 were purchased from Sigma-Aldrich. The Pin1 inhibitor (BJP-06-005-3) was a kind gift from Dr. Nathanael S. Gray, (Harvard Medical School, Boston, MA; and Stanford University, Palo Alto, CA).
Immunoblotting
2× Laemmli buffer was used to lyse the cells, followed by boiling at 95°C for 10 min. Boiled samples were resolved by SDS-PAGE followed by transfer onto polyvinylidenedifluoride or nitrocellulose membrane (Millipore). After transfer, the membrane was blocked with 5% skim milk followed by overnight incubation in primary antibody at 4°C. Following a 1-h wash after primary antibody incubation, HRP-conjugated secondary antibody was incubated for 1 h at room temperature, and the blot was washed before being developed for chemiluminescence signal using the Luminata Forte reagent (Millipore; WBLUF0500) and captured in the ImageQuant LAS-4000 gel documentation system (GE Healthcare). The dilutions used for the various primary and secondary antibodies were as follows: LIC1, 1:1,000; IC74, 1:1,000; HC, 1:500; Mad1, 1:500; Mad2, 1:250; p150Glued, 1:2,000; p50, 1:500; cyclin A, 1:200; cyclin B, 1:3,000, OD 157, 1:5,000, Zw10, 1:500; Hook2, 1:2,000, NudE1, 1:1,000; Nde1, 1:2,000; Lis1, 1:1,000; GST, 1:1,000; pin1, 1:1,000; β-actin, 1:2,000; myc, 1:2,000; BICD2, 1:2,500; LAMP1, 1:1,000; mCherry, 1:1,000; β-COP, 1:1,000; GM130, 1:1,000; anti-rabbit HRP, 1:10,000; and anti-mouse HRP, 1:10,000. The ImageJ software platform was used to perform all the densitometric analyses of immunoblot band intensities.
Immunofluorescence staining
HeLa and U2OS cells grown on glass coverslips were washed with 1× PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4, pH 7.2) before fixing them in chilled methanol. Fixed coverslips were transferred to a humidified chamber and rehydrated using 1× PBS for 10 min, followed by incubation in blocking solution (1% BSA, 1× PBS, and Triton X-100] for 1 h at room temperature. Primary antibody incubation for 1 h at room temperature was followed by washing and incubation with the appropriate secondary antibodies for 1 h. For lysosomal staining, cells were fixed in 2.5% PFA for 15 min. After washing, the cells were blocked with 1% BSA and 0.1% saponin in 1× PBS for 1 h, followed by the staining steps as described above. For Kt-MT attachment stability assays, immunofluorescence staining was performed as described in DeLuca et al. (2016). Briefly, cells were arrested in prometaphase using 300 nM nocodazole for 14–16 h and then released in MG132-containing medium for 2 h. Cells were washed with 1× PHEM buffer (60 mM Pipes, 25 mM Hepes, 10 mM EGTA, and 4 mM MgSO 4, pH 7.0), put in cold medium (4°C) on an ice tray in a cold room for 10 min, washed with 1× PHEM buffer, and fixed in chilled methanol. Fixed cells were washed three times in 1× PHEM buffer followed by incubation in lysis buffer (0.5% Triton X-100 in 1× PHEM) for 4 min. Cells were blocked for 1 h (blocking solution = 10% BSA in 1× PHEM) followed by addition of the primary antibody mix prepared in 5% BSA solution. The remaining staining steps were the same as described above. For pin1 staining, a chicken Pin1 primary antibody was used, with a biotin-conjugated anti-chicken secondary antibody at 1:500 dilution. After 1 h of incubation followed by washing, a streptavidin-conjugated Cy3 antibody (1:1,000 dilution) was added for 30 min, while the rest of the staining steps were the same as described above. Primary antibody dilutions used for immunofluorescence staining were as follows: GFP, 1:1,000; Mad1, 1:100; Zw10, 1:100; IC74, 1:500; p150Glued, 1:150; OD 173, 1:5,000; α-tubulin, 1:1,000; CREST, 1:50; pin1, 1:500; γ-tubulin, 1:500; DHC, 1:200; LAMP1, 1:1,000; GM130, 1:100; mCherry, 1:500; and β-COP, 1:800. All secondary antibodies were used at a dilution of 1:800. After washing, DAPI was added at 1:10,000 dilution (5 mg/ml stock solution) for 2 min followed by washing with 1× PBS and deionized water (Millipore) and mounted on a frosted glass slide using ProLong Diamond antifade mounting medium (Invitrogen). Coverslips were dried overnight and stored at −20°C until confocal/fluorescence imaging.
Microscopy
Fluorescence imaging and analysis
For live-cell imaging, HeLa cells stably expressing GFP-α-tubulin and histone 2B mCherry (H2B-mCherry; kind gift from Daniel Gerlich) were maintained in DMEM supplemented with puromycin and G418. Cells were grown on glass coverslips and transfected with control and C-terminal phosphomutant LIC1 plasmids. After 36 h of transfection, cells were set up for live-cell imaging using a customized aluminum slide containing 12-mm chambers as described earlier (Mahale et al., 2016a). Sterilized coverslips were used to seal one side of the chamber using VALAP (a 1:1:1 mixture of vaseline, lanolin, and paraffin). Conditioned medium was filled in the well from the opposite side and sealed with a coverslip containing adhered cells using sterile silicone grease. Time-lapse images with z-stacks containing planes 0.5 µm apart were acquired every 5 min for 12 h in conditioned DMEM on a Leica TCS SP8 laser scanning optical confocal microscope using an HCX PL APO CS 40×-1.3 NA/63×-1.4 NA oil-immersion objective fitted in a humidified heating chamber (OkoLab) maintained at 37°C. Similarly, the MTAP-tagged phosphomutant U2OS stable cell lines, maintained in DMEM supplemented with hygromycin B (300 µg/ml), were transfected with LIC1 siRNA. 24 h after transfection, cells were set up for time-lapse live-cell imaging (5- or 3-min [SST] intervals between time points for 12 h) on a Leica TCS SP8 laser scanning optical confocal microscope as described above. Live-cell fluorescence images (stained with SYTO13) were obtained on a Nikon Eclipse TiE epifluorescence microscope using a 20× 0.5 NA Plan Fluor ELWD lens and a DS Qi2 monochrome complementary metal-oxide-semiconductor camera. Fixed-cell immunofluorescence images were acquired on a Leica TCS SP8 laser scanning optical confocal microscope using an HCX PL APO CS 63×-1.4 NA oil-immersion objective and a HyD (hybrid) or PMT detector, with z-stacks containing planes 0.3 µm apart, unless otherwise indicated. All image acquisition settings were kept identical for control and test samples. Leica LASX software was used to control various imaging parameters during image acquisition as well as for postacquisition image analysis. Deconvolved images, where indicated, were acquired using the HyVolution feature of the Leica TCS SP8 confocal microscope. Fluorescence images were analyzed for various parameters on the Imaris software suite (v5.7, Bitplane), Leica offline image analysis software (LASX), and ImageJ software (National Institutes of Health). All representative immunofluorescence images shown are maximum-intensity projections, unless otherwise indicated in the figure legend, and were imported into Adobe Photoshop CS6/Photoshop v22.0 at 600-dpi resolution. Whole-image brightness and contrast were adjusted, where required, for final figure preparation.
Quantification of proteins at kinetochores during prometaphase and metaphase and interkinetochore distance measurement during metaphase
Cells were synchronized using 1 µM nocodazole for 4 h before fixation in chilled methanol and stained for IC (dynein), GFP (hLIC1-MTAP and hLIC1-AAA-MTAP), p150Glued (dynactin), OD 173 (spindly), Zw10, kinetochores (CREST), DNA (DAPI), Zw10, and Mad1 for prometaphase. An additional treatment with 10 µM MG132 for 2 h before fixation was performed for imaging metaphase cells. Images were opened in the Imaris suite for 3D reconstruction from z-stacks, and the integrated fluorescence intensities of various proteins at kinetochores in prometaphase and metaphase cells were quantified. A sphere of diameter 0.45 µm (average kinetochore size) was drawn around each kinetochore to measure the intensity of the respective kinetochore proteins as described earlier, normalized to the intensity of kinetochores (CREST) from the same sphere and plotted as a ratio in scatterplots using the Prism software. Local background was subtracted using features of Imaris software. For SAC protein intensities at metaphase kinetochores, the 20 brightest kinetochores were analyzed as reported earlier (Mahale et al., 2016b). Interkinetochore distance measurement was performed in the Imaris suite (Bitplane) from these images in the “slice” mode, and the distance was calculated between the centers of two sister kinetochore spots using the line tool for ≥10 kinetochore pairs per cell. Measurements were plotted as scatterplots using the Prism software.
Quantification of GM130-positive objects in metaphase
Immunofluorescence images of metaphase cells with 0.3-µm z-sections were analyzed with ImageJ software in the maximum-value projection mode, followed by thresholding using the autothreshold feature. The number of Golgi objects (GM130-positive green fluorescent punctae) per metaphase cell was determined using the “analyze objects” feature after setting a minimum size threshold of 20 pixels. A total of 60 metaphase cells per condition were quantified over three independent experiments.
Quantification of cold-stable tubulin intensity
Spindle intensity measurements after cold-induced depolymerization were performed as described previously (DeLuca et al., 2016), with slight modifications. Briefly, immunofluorescence images of cold-depolymerized metaphase cells with z-sections 0.2 µm apart were analyzed in maximum-value projection mode in ImageJ software. A rectangle was drawn around the spindle including the two spindle poles. The total integrated intensity of α-tubulin and CREST in red and green channels, respectively, along with the area of the rectangle were logged. Background fluorescence in both the channels was calculated by logging the total integrated intensities of an equal area rectangle outside the spindle or kinetochores. Finally, the background-corrected intensity for both α-tubulin and CREST was calculated and plotted as the ratio of the microtubule (α-tubulin)/kinetochore (CREST) integrated fluorescence intensity.
SBP affinity purification
The SBP tag present in the MTAP vector was used for affinity purification using a Streptactin-HP (GE) affinity column. Cellular lysates (“grindates”) of hLIC1-MTAP and hLIC1(AAA)-MTAP mutant cell lines were prepared by cryogenic grinding of harvested mitotic cells/late G2-prophase cells for 40 min followed by resuspension for 20 min in lysis buffer (50 mM Tris, pH 7.5, 125 mM sodium chloride, 1 mM EGTA, 0.2% NP-40, 5% glycerol, protease inhibitors, and phosphatase inhibitors [Roche/Pierce]). The cell grindate was centrifuged for 30 min at 13,000 rpm at 4°C. The supernatant fraction was loaded onto an equilibrated (in lysis buffer) Streptactin-HP affinity column at a flow rate of 0.3 ml/min. The flow-through was collected, and the column was washed using wash buffer (containing 50 mM Tris, pH 7.5, 250 mM sodium chloride, 0.2% NP-40, protease inhibitors, and phosphatase inhibitors). The eluate was collected by passing elution buffer (25 mM Tris, pH 7.5, 125 mM sodium chloride, 2.5 mM desthio-biotin, protease inhibitors, and phosphatase inhibitors) through the column at a flow rate of 0.5 ml/min. The concentrated eluate and other fractions were analyzed by immunoblotting.
Protein purification and pulldown assays
For recombinant protein purification, GST or GST-tagged constructs cloned in pGEX-6P-1 bacterial expression vectors were transformed into Escherichia coli Rosetta cells. Single colonies were inoculated in Luria-Bertani broth containing 100 µg/ml ampicillin at 37°C overnight to prepare the primary culture. Using 1% of the primary inoculum, secondary culture was set up in Luria-Bertani broth, induced with IPTG (0.5 mM) at 18°C overnight, and centrifuged at 5,000 g for 10 min, and the separated pellet fraction was resuspended in ice-cold lysis buffer containing 1× PBS, 400 mM KCl, 5 mM 2 β-mercaptoethanol [β-ME], phenylmethylsulfonyl fluoride (500 µl/50 ml), pepstatin (1 µl/ml), and 5% glycerol. The pellet was properly homogenized before lysis by sonication, followed by centrifugation at 16,000 g for 20 min at 4°C. The supernatant (containing the expressed protein) was loaded onto a preequilibrated GSTPrep FF 16/10 column (GE Healthcare) in the FPLC (AKTA Explorer; GE Healthcare). The column was equilibrated with 10 column volumes of equilibration buffer containing 1× PBS, 400 mM KCl, 5 mM β-ME, phenylmethylsulfonyl fluoride (500 µl/50 ml), and pepstatin (1 µl/ml). After binding, the column was washed with 5 column volumes of wash buffer I (1× PBS, 400 mM KCl, and 5 mM β-ME). 5 column volumes of wash buffer II containing 1× PBS, 400 mM KCl, 5 mM β-ME, 2 mM ATP, and 10 mM MgCl2 were used followed by a repeat of 5 column volume washes with wash buffer I. Bound protein fraction was eluted with five column volumes of elution buffer containing 50 mM Tris (pH 8), 500 mM NaCl, 20 mM reduced glutathione, and 5 mM β-ME. By observing the UV absorption at 280 nm, fractions containing the GST-tagged protein were collected in a prechilled tube. The GST tag was removed from the protein of interest using incubation with PreScission protease (GE Healthcare) and dialysis against a buffer containing 1× PBS, 400 mM KCl, 5 mM β-ME, and 5% glycerol overnight at 4°C, with buffer changes at regular intervals. After overnight dialysis, the protein sample was again loaded on to the equilibrated GSTprep column to remove the cut and unbound GST tag from the protein of interest. The protein of interest (flowthrough) was collected in a prechilled tube and concentrated using Amicon-ultracentrifugal filters (Millipore) with a buffer exchange with 50 mM Tris, 150 mM NaCl, and 5 mM β-ME. The concentrated pool was subjected to size-exclusion chromatography on an equilibrated (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 5 mM β-ME) Superdex 200 16/60 GL column (GE Healthcare) to segregate any aggregated/precipitated pools of the desired protein. The desired fractions were collected and analyzed on SDS-PAGE for purity, pooled, and concentrated. Final protein concentration was estimated using bicinchoninic acid assay kit (Thermo Fisher Scientific; 23227). Finally, 10% glycerol was added to the final concentrated fraction before storage at −80°C after snap freezing in liquid nitrogen as aliquots. GST and GST-Pin1 protein were not subjected to PreScission protease treatment since the GST tag had to be retained on these purified proteins.
For GST pull-down assays, whole-cell lysates of interphase or mitotic HeLa cells were prepared by thawing the cryogenic grindate and adding the chilled lysis buffer (50 mM Tris, 150 mM NaCl, 0.2% NP-40, 1 mM EDTA, 5% glycerol, Halt protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific), and 5 mM β-ME). After 30-min incubation with shaking, the lysate was subjected to centrifugation at 13,000 g for 20 min, and the supernatant was collected. Meanwhile, glutathione Sepharose beads (GE Healthcare; GE17-0756-01) were washed and incubated with purified GST or GST-Pin1 in binding buffer containing 1× PBS, 400 mM KCl, and 5 mM β-ME for 3 h at 4°C. After incubation, the mixture was centrifuged at 1,000 g for 2 min, and the beads were washed with binding buffer three times to remove any unbound fraction. The GST or GST-Pin1 protein bound to glutathione beads was incubated with the interphase or mitotic cell lysates overnight. After centrifugation at 1,000 g for 2 min, samples were washed three times with lysis buffer and boiled at 95°C for 10 min in 2× Laemmli buffer. The samples were run on SDS-PAGE, transferred to a polyvinylidenedifluoride membrane, and analyzed by immunoblotting.
In vitro cdk1-cyclinB kinase reaction
1 µg bacterially purified LIC1 proteins were incubated with 50 ng of recombinant Cdk1/cyclin B (NEB, P6020S; or Thermo Fisher Scientific, PV3292) with 100 µM ATP (NEB; P0756) and 1× protein kinase buffer (NEB; B6022) in a total volume of 30 µl. The control mixture had every component except the cdk1-cyclin B. The mixture was incubated in a 30°C water bath for 1 h. Phosphorylation was confirmed by an upshift of the phosphorylated protein band, as well as a downshift upon λ-phosphatase (NEB P0753L) treatment, as assessed by immunoblotting, Phosphorylated LIC1 proteins were used in the GST-Pin1 pull-down experiments.
Zebrafish lines, MO injection, and characterization of phenotypes
Tuebingen strain (TU-AB) zebrafish were raised according to standard protocols as described earlier (Westerfield, 2000). All experiments were performed according to protocols approved by the Institutional Animal Ethics Committee of the Council of Scientific and Industrial Research, Centre for Cellular and Molecular Biology, India. Embryos were obtained from natural spawning of adult fish, kept at 28.5°C, and staged according to hours after fertilization (Kimmel et al., 1995). The endogenous zLIC1 levels were depleted by using MO (zLIC1 translation blocker, 5′-GTGTATTTCTGCCCGTCGTCGCCAT-3′, Gene Tools; 10 ng per embryo). p53 MO and standard control MO, 5′-CCTCTTACCTCAGTTACAATTTATA-3′ (both from Gene Tools), were used as negative controls. For expression of the zLIC1 phosphomutant constructs, WT zLIC1 was cloned into pGEM-T easy vector system (Promega; A1360). The phosphomutants zLIC1A and zLIC1E were generated using site-directed mutagenesis. LIC1-A and LIC1-E mRNA were synthesized using the mMESSAGE mMACHINE kit (Ambion; AM1348). For rescue experiments, 50 pg of mRNA was coinjected along with LIC1 MO per embryo at the one-cell stage. The embryos were then analyzed for gross morphological defects at later stages of development (1 dpf).
ISH and gene expression analysis in zebrafish embryos
The Ntl gene (TbxTa) was cloned using 1-dpf embryos of the TU-AB zebrafish strain. For cloning, total RNA was isolated from 100 embryos using an RNA isolation kit (MN; 740955.50), and cDNA was prepared using the iSCRIPT kit (Bio-Rad; 1708891). Sequence-specific primers were used to amplify the 268-bp fragment of the Ntl coding sequence from the cDNA using the following PCR program: 95°C for 30 s; 30 cycles of 95°C for 10 s, 62°C for 30 s, and 72°C for 30 s; and 72°C for 10 min as the final extension. The Ntl fragment was then cloned into the pGEMT easy vector, and the sequence was verified. Antisense digoxigenin (DIG)-labeled riboprobes were synthesized against the Ntl fragment using the Roche DIG RNA labeling kit (11175025910).
For determining Ntl expression, the following ISH protocol was used. Day 1: The embryos were manually dechorionated and fixed in 4% formaldehyde/PBS overnight at 4°C, washed with 1× PBT (1× PBS and 0.1% Tween 20), 50% methanol/1× PBT, and finally 100% methanol and stored at −20°C overnight. Day 2: The embryos were rehydrated to 1× PBT followed by 1:1 1× PBT:prewarmed hybridization wash (50% formamide, 1.3× SSC, 5 mM EDTA, and 0.2% Tween 20 with no heparin and no tRNA) at 65°C. The embryos were then washed with prewarmed hybridization wash for 2 h at 65°C, following by the addition of Ntl riboprobe mixed with hybridization mix (hybridization wash with 100 µg/ml heparin and 50 µg/ml tRNA) at 65°C overnight for effective hybridization. Day 3: The embryos were washed with prewarmed hybridization wash solution twice for 30 min each at 65°C, then with 1× TBST (5 M NaCl, 1 M KCl, 1 M Tris, pH 7.5, and 10% Tween 20) at room temperature and incubated with 10% heat-inactivated sheep serum (Gibco; 16210-064) for blocking at room temperature for 2 h. The embryos were then incubated with the anti-DIG–labeled alkaline phosphatase antibody (Roche; 11093274910) overnight at 4°C. Day 4: The embryos were transferred to 12-well plates, washed twice with 1× NTMT (0.1 M NaCl, 0.1 M Tris, pH 9.5, 0.05 M MgCl2, and 1% Tween 20) for 10 min each, and incubated with NBT/BCIP solution (Roche; 1-383-213 and 1-383-221, respectively). The embryos were monitored during this color reaction and stopped with 1× PBT as a stop solution at the same time to ascertain differences in the levels of gene expression under each condition.
Statistical analysis
The total number of cells counted for each experiment for statistical analysis is mentioned in the respective figure legends. Error bars represent SD or SEM from at least three independent experiments, unless otherwise indicated. Student’s t test, one-way ANOVA, or Kruskal–Wallis test was used to statistically analyze the data and calculate statistical significance through Prism software, as indicated in the figure legends. Data distribution was tested for normality using the D’Agostino–Pearson normality test in the GraphPad Prism software. Parametric tests were used for data that compared the means of multiple independent experiments. Post hoc analysis using Dunn’s/Tukey’s multiple comparison test, as well as generation of graphs. was done using the GraphPad Prism software.
Online supplemental material
Fig. S1 shows that U2OS cells stably expressing the MTAP tag do not delay in mitosis. It also shows that in HeLa cells, LIC1-CTD phosphorylation is required for proper mitotic progression. Fig. S2 shows that in U2OS and HeLa cells, LIC1-CTD phosphomutants cannot efficiently rescue the mitotic arrest caused by LIC1 depletion. Fig. S3 documents the levels of loading of the dynein IC subunit and the kinetochore components Zw10, spindly, and p150Glued (dynactin) at prometaphase kinetochores upon rescuing hLIC1 depletion with the different rLIC1-CTD phosphomutants in HeLa cells, measured through quantitative immunofluorescence imaging. Fig. S4 shows that rLIC1-CTD phosphorylation acts at least in part through an active SAC to ensure timely mitotic progression in HeLa cells. It also shows accumulation (due to a failure of proper removal by dynein) of the SAC proteins Mad1 and Zw10 at congressed metaphase kinetochores upon rescuing hLIC1 depletion with AAA-rLIC1 but not with SST-rLIC1. Table S1 lists all primers used in this study. Video 1 shows representative videos depicting the timing of mitotic progression of live U2OS cells with and without LIC1 siRNA treatment (both by differential interference contrast [DIC]) and stably expressing the MTAP tag (fluorescent). Videos 2, 3, and 4 show the mitotic timing from representative hLIC1 siRNA–treated U2OS cells stably expressing MTAP-tagged SST or the triple mutant AAA (Video 2), the three single mutants (AST, SAT, and SSA; Video 3), and the three double mutants (SAA, ASA, and AAT; Video 4). Videos 5, 6, and 7 show the mitotic timing from representative double stable HeLa cells (stably expressing GFP-α-tubulin and H2B-mCherry) transiently expressing the following myc-tagged rLIC1 constructs (all containing the S207E phospho-mimicking mutation): mock (no transfection), SST (WT) or the triple mutant AAA (Video 5), the three single mutants (AST, SAT, and SSA; Video 6), and the three double mutants (SAA, ASA, and AAT; Video 7). Video 8 shows representative double stable HeLa cells depicting the centrosome-NE detachment phenotypes observed with mock or LIC1 siRNA treatment, as well as rescue with rLIC1-SST or AAA. Video 9 shows representative cells depicting the timing from prometaphase to metaphase plate formation and beyond in U2OS cells with and without LIC1 siRNA treatment or in stable U2OS cell lines expressing SST or AAA after release of nocodazole-arrested prometaphase cells into MG132-containing medium to measure effects on the stability of Kt-MT attachment.
Supplementary Material
lists the primer sequences used.
Acknowledgments
We thank Dr. Sharmishtha Samantaray for help with cloning. We thank Dr. Mahak Sharma (Indian Institute of Science Education and Research, Mohali, Punjab, India) and Dr. Nagaraj Balasubramanian (Indian Institute of Science Education and Research, Pune, Maharashtra, India) for advice regarding specific experiments. We thank the following for providing reagents as indicated: Prof. Stephen J. Doxsey (U2OS cells), Prof. Dannel McCollum (University of Massachusetts Medical School, Worcester, MA; pMTAP-mVenus expression vector), Prof. Daniel W. Gerlich (Institute of Molecular Biotechnology, Vienna, Austria; GFP-tubulin::mCherry-H2B stable HeLa cell line), Prof. Roop Mallik (Indian Institute of Technology Bombay, Mumbai, Maharashtra, India; NudE1, Lis1 and p50 antibodies), Drs. Reto Gassman (Instituto de Biologia Molecular e Celular [IBMC], Universidade do Porto, Porto, Portugal and Instituto de Investigação e Inovação em Saúde [i3S], Universidade do Porto, Porto, Portugal) and Arshad Desai (Department of Cellular and Molecular Medicine and Ludwig Institute for Cancer Research, San Diego, CA; spindly antibodies), Dr. Mahak Sharma (Hook2 antibody and H2B-mCherry plasmid), Dr. Nagaraj Balasubramanian (GalT-RFP plasmid), Drs. Prasenjit Guchhait and Chittur V. Srikanth (Regional Centre for Biotechnology, Faridabad, Haryana, India; LAMP1 antibody), and Dr. Nathanael S Gray (Pin1 inhibitor BJP-06-005-3). We thank Suraj Tewari for assistance with confocal microscopy and the Regional Centre for Biotechnology (RCB) for providing the requisite infrastructure. We are grateful to members of the Laboratory of Cellular Dynamics, Regional Centre for Biotechnology, for critical comments and suggestions during the study.
A. Kumari received fellowship support from the Council of Scientific and Industrial Research, India and the Indian Council of Medical Research, India; R. Pergu and C. Kumar from the Department of Biotechnology, Regional Centre for Biotechnology; R. Pergu and N. Wasnik from the Indian Council of Medical Research and S. Mahale from the Regional Centre for Biotechnology. M. Kumar was supported by a Young Investigator Award from the Regional Centre for Biotechnology, a DST-INSPIRE faculty award from the Department of Science and Technology, Government of India (award number DST/INSPIRE/04/2016/001436) and the Council of Scientific and Industrial Research, Centre for Cellular and Molecular Biology. This work was supported by funding to S.V.S. Mylavarapu from Department of Biotechnology, Regional Centre for Biotechnology (grant number BT/PR6420/GBD/27/435/2012) and the Science and Engineering Research Board (grant number EMR/2016/007842).
The authors declare no competing financial interests.
Author contributions: A. Kumari, C. Kumar, R. Pergu, M. Kumar, S.P. Mahale, and S.V.S. Mylavarapu designed experiments. A. Kumari, C. Kumar, R. Pergu, M. Kumar, and S.P. Mahale performed experiments. A. Kumari, C. Kumar, R. Pergu, M. Kumar, S.P. Mahale, N. Wasnik, and S.V.S. Mylavarapu analyzed data. A. Kumari, C. Kumar, R. Pergu, M. Kumar, N. Wasnik, and S.V.S. Mylavarapu wrote/prepared the manuscript; A. Kumari and S.V.S. Mylavarapu also edited the manuscript. S.V.S. Mylavarapu conceptualized and supervised the entire study and obtained funding.
References
- Addinall, S.G., Mayr P.S., Doyle S., Sheehan J.K., Woodman P.G., and Allan V.J.. 2001. Phosphorylation by cdc2-CyclinB1 kinase releases cytoplasmic dynein from membranes. J. Biol. Chem. 276:15939–15944. 10.1074/jbc.M011628200 [DOI] [PubMed] [Google Scholar]
- Bader, J.R., Kasuboski J.M., Winding M., Vaughan P.S., Hinchcliffe E.H., and Vaughan K.T.. 2011. Polo-like kinase1 is required for recruitment of dynein to kinetochores during mitosis. J. Biol. Chem. 286:20769–20777. 10.1074/jbc.M111.226605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolhy, S., Bouhlel I., Dultz E., Nayak T., Zuccolo M., Gatti X., Vallee R., Ellenberg J., and Doye V.. 2011. A Nup133-dependent NPC-anchored network tethers centrosomes to the nuclear envelope in prophase. J. Cell Biol. 192:855–871. 10.1083/jcb.201007118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celestino, R., Henen M.A., Gama J.B., Carvalho C., McCabe M., Barbosa D.J., Born A., Nichols P.J., Carvalho A.X., Gassmann R., and Vögeli B.. 2019. A transient helix in the disordered region of dynein light intermediate chain links the motor to structurally diverse adaptors for cargo transport. PLoS Biol. 17:e3000100. 10.1371/journal.pbio.3000100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman, I.M., and Desai A.. 2005. A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci. STKE. 2005:pl1. 10.1126/stke.2662005pl1 [DOI] [PubMed] [Google Scholar]
- Cheng, C.W., and Tse E.. 2018. PIN1 in Cell Cycle Control and Cancer. Front. Pharmacol. 9:1367. 10.3389/fphar.2018.01367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daub, H., Olsen J.V., Bairlein M., Gnad F., Oppermann F.S., Körner R., Greff Z., Kéri G., Stemmann O., and Mann M.. 2008. Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol. Cell. 31:438–448. 10.1016/j.molcel.2008.07.007 [DOI] [PubMed] [Google Scholar]
- Daum, J.R., Potapova T.A., Sivakumar S., Daniel J.J., Flynn J.N., Rankin S., and Gorbsky G.J.. 2011. Cohesion fatigue induces chromatid separation in cells delayed at metaphase. Curr. Biol. 21:1018–1024. 10.1016/j.cub.2011.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boer, L., Oakes V., Beamish H., Giles N., Stevens F., Somodevilla-Torres M., Desouza C., and Gabrielli B.. 2008. Cyclin A/cdk2 coordinates centrosomal and nuclear mitotic events. Oncogene. 27:4261–4268. 10.1038/onc.2008.74 [DOI] [PubMed] [Google Scholar]
- Dell, K.R., Turck C.W., and Vale R.D.. 2000. Mitotic phosphorylation of the dynein light intermediate chain is mediated by cdc2 kinase. Traffic. 1:38–44. 10.1034/j.1600-0854.2000.010107.x [DOI] [PubMed] [Google Scholar]
- DeLuca, K.F., Herman J.A., and DeLuca J.G.. 2016. Measuring Kinetochore-Microtubule Attachment Stability in Cultured Cells. Methods Mol. Biol. 1413:147–168. 10.1007/978-1-4939-3542-0_10 [DOI] [PubMed] [Google Scholar]
- Dephoure, N., Zhou C., Villén J., Beausoleil S.A., Bakalarski C.E., Elledge S.J., and Gygi S.P.. 2008. A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. USA. 105:10762–10767. 10.1073/pnas.0805139105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domanski, M., Molloy K., Jiang H., Chait B.T., Rout M.P., Jensen T.H., and LaCava J.. 2012. Improved methodology for the affinity isolation of human protein complexes expressed at near endogenous levels. Biotechniques. 0:1–6. 10.2144/000113864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi, D., Kumari A., Rathi S., Mylavarapu S.V.S., and Sharma M.. 2019. The dynein adaptor Hook2 plays essential roles in mitotic progression and cytokinesis. J. Cell Biol. 218:871–894. 10.1083/jcb.201804183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama, J.B., Pereira C., Simões P.A., Celestino R., Reis R.M., Barbosa D.J., Pires H.R., Carvalho C., Amorim J., Carvalho A.X., et al. 2017. Molecular mechanism of dynein recruitment to kinetochores by the Rod-Zw10-Zwilch complex and Spindly. J. Cell Biol. 216:943–960. 10.1083/jcb.201610108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavet, O., and Pines J.. 2010. Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Dev. Cell. 18:533–543. 10.1016/j.devcel.2010.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, D., and Ferrell J.E. Jr. 2010. The roles of cyclin A2, B1, and B2 in early and late mitotic events. Mol. Biol. Cell. 21:3149–3161. 10.1091/mbc.e10-05-0393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, G., Nédélec F., and Vale R.D.. 2005. Mechanisms for focusing mitotic spindle poles by minus end-directed motor proteins. J. Cell Biol. 171:229–240. 10.1083/jcb.200505107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis, E.R., Stuurman N., and Vale R.D.. 2007. Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore. J. Cell Biol. 177:1005–1015. 10.1083/jcb.200702062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hégarat, N., Crncec A., Suarez Peredo Rodriguez M.F., Echegaray Iturra F., Gu Y., Busby O., Lang P.F., Barr A.R., Bakal C., Kanemaki M.T., et al. 2020. Cyclin A triggers Mitosis either via the Greatwall kinase pathway or Cyclin B. EMBO J. 39:e104419. 10.15252/embj.2020104419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogenraad, C.C., Akhmanova A., Howell S.A., Dortland B.R., De Zeeuw C.I., Willemsen R., Visser P., Grosveld F., and Galjart N.. 2001. Mammalian Golgi-associated Bicaudal-D2 functions in the dynein-dynactin pathway by interacting with these complexes. EMBO J. 20:4041–4054. 10.1093/emboj/20.15.4041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell, B.J., McEwen B.F., Canman J.C., Hoffman D.B., Farrar E.M., Rieder C.L., and Salmon E.D.. 2001. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol. 155:1159–1172. 10.1083/jcb.200105093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J., Roberts A.J., Leschziner A.E., and Reck-Peterson S.L.. 2012. Lis1 acts as a “clutch” between the ATPase and microtubule-binding domains of the dynein motor. Cell. 150:975–986. 10.1016/j.cell.2012.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jie, J., Löhr F., and Barbar E.. 2017. Dynein Binding of Competitive Regulators Dynactin and NudE Involves Novel Interplay between Phosphorylation Site and Disordered Spliced Linkers. Structure. 25:421–433. 10.1016/j.str.2017.01.003 [DOI] [PubMed] [Google Scholar]
- Kardon, J.R., and Vale R.D.. 2009. Regulators of the cytoplasmic dynein motor. Nat. Rev. Mol. Cell Biol. 10:854–865. 10.1038/nrm2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel, C.B., Ballard W.W., Kimmel S.R., Ullmann B., and Schilling T.F.. 1995. Stages of embryonic development of the zebrafish. Dev. Dyn. 203:253–310. 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- Kumari, A., Kumar C., Wasnik N., and Mylavarapu S.V.S.. 2021. Dynein light intermediate chains as pivotal determinants of dynein multifunctionality. J. Cell Sci. 134:jcs254870. 10.1242/jcs.254870 [DOI] [PubMed] [Google Scholar]
- Lee, Y.C., Que J., Chen Y.C., Lin J.T., Liou Y.C., Liao P.C., Liu Y.P., Lee K.H., Lin L.C., Hsiao M., et al. 2013. Pin1 acts as a negative regulator of the G2/M transition by interacting with the Aurora-A-Bora complex. J. Cell Sci. 126:4862–4872. 10.1242/jcs.121368 [DOI] [PubMed] [Google Scholar]
- Lee, I.G., Olenick M.A., Boczkowska M., Franzini-Armstrong C., Holzbaur E.L.F., and Dominguez R.. 2018. A conserved interaction of the dynein light intermediate chain with dynein-dynactin effectors necessary for processivity. Nat. Commun. 9:986. 10.1038/s41467-018-03412-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, I.G., Cason S.E., Alqassim S.S., Holzbaur E.L.F., and Dominguez R.. 2020. A tunable LIC1-adaptor interaction modulates dynein activity in a cargo-specific manner. Nat. Commun. 11:5695. 10.1038/s41467-020-19538-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Yu W., Liang Y., and Zhu X.. 2007. Kinetochore dynein generates a poleward pulling force to facilitate congression and full chromosome alignment. Cell Res. 17:701–712. 10.1038/cr.2007.65 [DOI] [PubMed] [Google Scholar]
- Lu, K.P., Hanes S.D., and Hunter T.. 1996. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature. 380:544–547. 10.1038/380544a0 [DOI] [PubMed] [Google Scholar]
- Lu, K.P., Liou Y.C., and Zhou X.Z.. 2002. Pinning down proline-directed phosphorylation signaling. Trends Cell Biol. 12:164–172. 10.1016/S0962-8924(02)02253-5 [DOI] [PubMed] [Google Scholar]
- Ma, H., McLean J.R., Chao L.F., Mana-Capelli S., Paramasivam M., Hagstrom K.A., Gould K.L., and McCollum D.. 2012. A highly efficient multifunctional tandem affinity purification approach applicable to diverse organisms. Mol. Cell. Proteomics. 11:501–511. 10.1074/mcp.O111.016246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahale, S., Kumar M., Sharma A., Babu A., Ranjan S., Sachidanandan C., and Mylavarapu S.V.S.. 2016a. The Light Intermediate Chain 2 Subpopulation of Dynein Regulates Mitotic Spindle Orientation. Sci. Rep. 6:22. 10.1038/s41598-016-0030-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahale, S.P., Sharma A., and Mylavarapu S.V.. 2016b. Dynein Light Intermediate Chain 2 Facilitates the Metaphase to Anaphase Transition by Inactivating the Spindle Assembly Checkpoint. PLoS One. 11:e0159646. 10.1371/journal.pone.0159646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matanis, T., Akhmanova A., Wulf P., Del Nery E., Weide T., Stepanova T., Galjart N., Grosveld F., Goud B., De Zeeuw C.I., et al. 2002. Bicaudal-D regulates COPI-independent Golgi-ER transport by recruiting the dynein-dynactin motor complex. Nat. Cell Biol. 4:986–992. 10.1038/ncb891 [DOI] [PubMed] [Google Scholar]
- McKenney, R.J., Vershinin M., Kunwar A., Vallee R.B., and Gross S.P.. 2010. LIS1 and NudE induce a persistent dynein force-producing state. Cell. 141:304–314. 10.1016/j.cell.2010.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney, R.J., Weil S.J., Scherer J., and Vallee R.B.. 2011. Mutually exclusive cytoplasmic dynein regulation by NudE-Lis1 and dynactin. J. Biol. Chem. 286:39615–39622. 10.1074/jbc.M111.289017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli, T., and Warren G.. 1995. Mitotic disassembly of the Golgi apparatus in vivo. J. Cell Sci. 108:2715–2727. 10.1242/jcs.108.7.2715 [DOI] [PubMed] [Google Scholar]
- Moon, H.M., Youn Y.H., Pemble H., Yingling J., Wittmann T., and Wynshaw-Boris A.. 2014. LIS1 controls mitosis and mitotic spindle organization via the LIS1-NDEL1-dynein complex. Hum. Mol. Genet. 23:449–466. 10.1093/hmg/ddt436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley, R.H., Lachani K., Keefe D., Gilchrist M.J., Flicek P., Smith J.C., and Wardle F.C.. 2009. A gene regulatory network directed by zebrafish No tail accounts for its roles in mesoderm formation. Proc. Natl. Acad. Sci. USA. 106:3829–3834. 10.1073/pnas.0808382106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyarko, A., Song Y., and Barbar E.. 2012. Intrinsic disorder in dynein intermediate chain modulates its interactions with NudE and dynactin. J. Biol. Chem. 287:24884–24893. 10.1074/jbc.M112.376038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olenick, M.A., and Holzbaur E.L.F.. 2019. Dynein activators and adaptors at a glance. J. Cell Sci. 132:jcs227132. 10.1242/jcs.227132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, J.V., Vermeulen M., Santamaria A., Kumar C., Miller M.L., Jensen L.J., Gnad F., Cox J., Jensen T.S., Nigg E.A., et al. 2010. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3:ra3. 10.1126/scisignal.2000475 [DOI] [PubMed] [Google Scholar]
- Palmer, K.J., Hughes H., and Stephens D.J.. 2009. Specificity of cytoplasmic dynein subunits in discrete membrane-trafficking steps. Mol. Biol. Cell. 20:2885–2899. 10.1091/mbc.e08-12-1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister, K.K., Fisher E.M., Gibbons I.R., Hays T.S., Holzbaur E.L., McIntosh J.R., Porter M.E., Schroer T.A., Vaughan K.T., Witman G.B., et al. 2005. Cytoplasmic dynein nomenclature. J. Cell Biol. 171:411–413. 10.1083/jcb.200508078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister, K.K., Shah P.R., Hummerich H., Russ A., Cotton J., Annuar A.A., King S.M., and Fisher E.M.. 2006. Genetic analysis of the cytoplasmic dynein subunit families. PLoS Genet. 2:e1. 10.1371/journal.pgen.0020001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinch, B.J., Doctor Z.M., Nabet B., Browne C.M., Seo H.S., Mohardt M.L., Kozono S., Lian X., Manz T.D., Chun Y., et al. 2020. Identification of a potent and selective covalent Pin1 inhibitor. Nat. Chem. Biol. 16:979–987. 10.1038/s41589-020-0550-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers, J.A., Tanenbaum M.E., and Medema R.H.. 2013. Systematic dissection of dynein regulators in mitosis. J. Cell Biol. 201:201–215. 10.1083/jcb.201208098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reck-Peterson, S.L., Redwine W.B., Vale R.D., and Carter A.P.. 2018. The cytoplasmic dynein transport machinery and its many cargoes. Nat. Rev. Mol. Cell Biol. 19:382–398. 10.1038/s41580-018-0004-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redwine, W.B., DeSantis M.E., Hollyer I., Htet Z.M., Tran P.T., Swanson S.K., Florens L., Washburn M.P., and Reck-Peterson S.L.. 2017. The human cytoplasmic dynein interactome reveals novel activators of motility. eLife. 6:e28257. 10.7554/eLife.28257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salina, D., Bodoor K., Eckley D.M., Schroer T.A., Rattner J.B., and Burke B.. 2002. Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell. 108:97–107. 10.1016/S0092-8674(01)00628-6 [DOI] [PubMed] [Google Scholar]
- Scherer, J., Yi J., and Vallee R.B.. 2014. PKA-dependent dynein switching from lysosomes to adenovirus: a novel form of host-virus competition. J. Cell Biol. 205:163–177. 10.1083/jcb.201307116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder, C.M., and Vale R.D.. 2016. Assembly and activation of dynein-dynactin by the cargo adaptor protein Hook3. J. Cell Biol. 214:309–318. 10.1083/jcb.201604002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroer, T.A. 2004. Dynactin. Annu. Rev. Cell Dev. Biol. 20:759–779. 10.1146/annurev.cellbio.20.012103.094623 [DOI] [PubMed] [Google Scholar]
- Schulte-Merker, S., van Eeden F.J., Halpern M.E., Kimmel C.B., and Nüsslein-Volhard C.. 1994. no tail (ntl) is the zebrafish homologue of the mouse T (Brachyury) gene. Development. 120:1009–1015. 10.1242/dev.120.4.1009 [DOI] [PubMed] [Google Scholar]
- Schweiger, R., and Linial M.. 2010. Cooperativity within proximal phosphorylation sites is revealed from large-scale proteomics data. Biol. Direct. 5:6. 10.1186/1745-6150-5-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, M., Stukenberg P.T., Kirschner M.W., and Lu K.P.. 1998. The essential mitotic peptidyl-prolyl isomerase Pin1 binds and regulates mitosis-specific phosphoproteins. Genes Dev. 12:706–720. 10.1101/gad.12.5.706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter, J., and Warren G.. 2002. Golgi architecture and inheritance. Annu. Rev. Cell Dev. Biol. 18:379–420. 10.1146/annurev.cellbio.18.030602.133733 [DOI] [PubMed] [Google Scholar]
- Sivaram, M.V., Wadzinski T.L., Redick S.D., Manna T., and Doxsey S.J.. 2009. Dynein light intermediate chain 1 is required for progress through the spindle assembly checkpoint. EMBO J. 28:902–914. 10.1038/emboj.2009.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter, D., Tanenbaum M.E., Lindqvist A., Jaarsma D., Flotho A., Yu K.L., Grigoriev I., Engelsma D., Haasdijk E.D., Keijzer N., et al. 2010. Bicaudal D2, dynein, and kinesin-1 associate with nuclear pore complexes and regulate centrosome and nuclear positioning during mitotic entry. PLoS Biol. 8:e1000350. 10.1371/journal.pbio.1000350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Denis, N.A., Bailey M.L., Parker E.L., Vilk G., and Litchfield D.W.. 2011. Localization of phosphorylated CK2alpha to the mitotic spindle requires the peptidyl-prolyl isomerase Pin1. J. Cell Sci. 124:2341–2348. 10.1242/jcs.077446 [DOI] [PubMed] [Google Scholar]
- Stevens, D., Gassmann R., Oegema K., and Desai A.. 2011. Uncoordinated loss of chromatid cohesion is a common outcome of extended metaphase arrest. PLoS One. 6:e22969. 10.1371/journal.pone.0022969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szebenyi, G., Hall B., Yu R., Hashim A.I., and Krämer H.. 2007. Hook2 localizes to the centrosome, binds directly to centriolin/CEP110 and contributes to centrosomal function. Traffic. 8:32–46. 10.1111/j.1600-0854.2006.00511.x [DOI] [PubMed] [Google Scholar]
- Tan, S.C., Scherer J., and Vallee R.B.. 2011. Recruitment of dynein to late endosomes and lysosomes through light intermediate chains. Mol. Biol. Cell. 22:467–477. 10.1091/mbc.e10-02-0129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee, R.B., McKenney R.J., and Ori-McKenney K.M.. 2012. Multiple modes of cytoplasmic dynein regulation. Nat. Cell Biol. 14:224–230. 10.1038/ncb2420 [DOI] [PubMed] [Google Scholar]
- Varma, D., Monzo P., Stehman S.A., and Vallee R.B.. 2008. Direct role of dynein motor in stable kinetochore-microtubule attachment, orientation, and alignment. J. Cell Biol. 182:1045–1054. 10.1083/jcb.200710106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergnolle, M.A., and Taylor S.S.. 2007. Cenp-F links kinetochores to Ndel1/Nde1/Lis1/dynein microtubule motor complexes. Curr. Biol. 17:1173–1179. 10.1016/j.cub.2007.05.077 [DOI] [PubMed] [Google Scholar]
- Westerfield, M. 2000. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio Rerio). 4th Edition. University of Oregon Press, Eugene, OR. [Google Scholar]
- Whyte, J., Bader J.R., Tauhata S.B., Raycroft M., Hornick J., Pfister K.K., Lane W.S., Chan G.K., Hinchcliffe E.H., Vaughan P.S., and Vaughan K.T.. 2008. Phosphorylation regulates targeting of cytoplasmic dynein to kinetochores during mitosis. J. Cell Biol. 183:819–834. 10.1083/jcb.200804114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynne, C.L., and Vallee R.B.. 2018. Cdk1 phosphorylation of the dynein adapter Nde1 controls cargo binding from G2 to anaphase. J. Cell Biol. 217:3019–3029. 10.1083/jcb.201707081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachie, N., Saito R., Sugahara J., Tomita M., and Ishihama Y.. 2009. In silico analysis of phosphoproteome data suggests a rich-get-richer process of phosphosite accumulation over evolution. Mol. Cell. Proteomics. 8:1061–1071. 10.1074/mcp.M800466-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav, S., and Linstedt A.D.. 2011. Golgi positioning. Cold Spring Harb. Perspect. Biol. 3:a005322. 10.1101/cshperspect.a005322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav, S., Puthenveedu M.A., and Linstedt A.D.. 2012. Golgin160 recruits the dynein motor to position the Golgi apparatus. Dev. Cell. 23:153–165. 10.1016/j.devcel.2012.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe, M.B., Schutkowski M., Shen M., Zhou X.Z., Stukenberg P.T., Rahfeld J.U., Xu J., Kuang J., Kirschner M.W., Fischer G., et al. 1997. Sequence-specific and phosphorylation-dependent proline isomerization: a potential mitotic regulatory mechanism. Science. 278:1957–1960. 10.1126/science.278.5345.1957 [DOI] [PubMed] [Google Scholar]
- Zhu, J., Kwan K.M., and Mackem S.. 2016. Putative oncogene Brachyury (T) is essential to specify cell fate but dispensable for notochord progenitor proliferation and EMT. Proc. Natl. Acad. Sci. USA. 113:3820–3825. 10.1073/pnas.1601252113 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists the primer sequences used.