Abstract
Major histocompatibility complex class II (MHC-II) molecules occupy a pivotal position in the adaptive immune system, and correct regulation of their expression is therefore of critical importance for the control of the immune response. Several regulatory factors essential for the transcription of MHC-II genes have been identified by elucidation of the molecular defects responsible for MHC-II deficiency, a hereditary immunodeficiency disease characterized by regulatory defects abrogating MHC-II expression. Three of these factors, RFX5, RFXAP, and RFXANK, combine to form the RFX complex, a regulatory protein that binds to the X box DNA sequence present in all MHC-II promoters. In this study we have undertaken a dissection of the structure and function of RFX5, the largest subunit of the RFX complex. The results define two distinct domains serving two different essential functions. A highly conserved N-terminal region of RFX5 is required for its association with RFXANK and RFXAP, for assembly of the RFX complex in vivo and in vitro, and for binding of this complex to its X box target site in the MHC-II promoter. This N-terminal region is, however, not sufficient for activation of MHC-II expression. This requires an additional domain within the C-terminal region of RFX5. This C-terminal domain mediates cooperative binding between the RFX complex and NF-Y, a transcription factor binding to the Y box sequence of MHC-II promoters. This provides direct evidence that RFX5-mediated cooperative binding between RFX and NF-Y plays an essential role in the transcriptional activation of MHC-II genes.
Major histocompatibility complex class II (MHC-II) molecules are heterodimeric (α-chain–β-chain) transmembrane glycoproteins occupying a pivotal position in the adaptive immune system. They play several key roles in the homeostasis of the CD4+-T-cell population. First, MHC-II molecules present antigenic peptides derived from exogenous proteins to the receptors of CD4+ T lymphocytes, thereby leading to T-helper cell activation and to the initiation and propagation of antigen-specific immune responses (14). Second, MHC-II expression in the thymus drives the positive- and negative-selection events that generate and shape the mature CD4+-T-cell repertoire (30). Finally, expression of MHC-II molecules in the periphery affects CD4+-T-cell survival (8, 60, 70). In addition, engagement of MHC-II molecules by the T-cell receptor also participates in the activation of the antigen-presenting cells on which they are expressed (61). Considering these central functions, it is of no surprise that correctly regulated MHC-II expression is of critical importance for the control of the immune response. For instance, the loss of MHC-II expression severely cripples the immune system (20, 34, 54) while inappropriate MHC-II expression is frequently observed in tissues that are attacked during the course of certain CD4+-T-cell-mediated autoimmune diseases (6, 26).
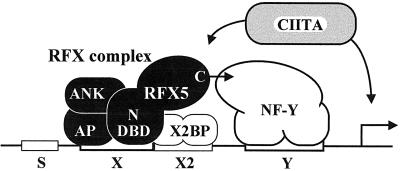
Two modes of MHC-II expression, constitutive and inducible, are generally recognized (5, 25, 39, 71). Constitutive expression is largely restricted to specialized cells of the immune system, including thymic epithelial cells, B cells, macrophages, and dendritic cells. The majority of other cell types lack MHC-II expression but can be induced to express MHC-II by exposure to a variety of agents, of which the most potent and well known is gamma interferon. Both modes of expression are controlled primarily at the level of transcription by a conserved promoter-proximal enhancer consisting of four cis-acting DNA sequences known as the S, X, X2, and Y boxes (Fig. 1) (5, 25, 39, 71). The presence, orientation, and spacing of these regulatory sequences relative to each other are highly conserved in all MHC-II promoters and are critical for activity (5, 25, 39, 71).
FIG. 1.
Schematic representation of a typical MHC-II promoter. The S, X, X2, and Y boxes conserved in all MHC-II promoters and the factors that bind to these sequences are indicated. Elucidation of the molecular defects in MHC-II deficiency complementation groups A, B, C, and D have led, respectively, to the identification of the non-DNA-binding coactivator CIITA and the three subunits (RFXANK, RFX5, and RFXAP) of the RFX complex. The trimeric RFX complex binds cooperatively with the X2 box binding protein X2BP, which has recently been shown to contain CREB, and the Y box binding protein NF-Y. Here we define two distinct functional domains in RFX5. A highly conserved N-terminal region (N) encompassing the DBD is sufficient for assembly and binding of the RFX complex. A less well-conserved C-terminal region (C) mediates cooperative binding with NF-Y.
Identification of the transcription factors that activate MHC-II promoters via the S-X-X2-Y enhancer has been greatly facilitated by the study of cell lines derived from patients suffering from a rare hereditary immunodeficiency disease called MHC-II deficiency (20, 34, 39, 54). This disease is characterized by a complete absence of MHC-II expression, and it results from mutations in transcription factors that are essential for activation of MHC-II promoters. Patients have been classified into four complementation groups (A, B, C, and D) corresponding to four genetic defects (3, 23, 37, 64). The molecular defects in group A patients reside in the gene encoding the class II transactivator (CIITA), a non-DNA-binding coactivator that functions as a molecular switch controlling the cell type specificity and inducibility of MHC-II expression (Fig. 1) (45, 46, 68, 69). In contrast, patients in the remaining three groups are characterized by a deficiency in regulatory factor X (RFX), a heterotrimeric DNA binding complex that binds to the X boxes of all MHC-II promoters (Fig. 1) (19, 29, 42, 56, 67). The molecular defects in groups B, C, and D lie, respectively, in the genes encoding the RFXANK (42, 47), RFX5 (67), and RFXAP (19, 72) subunits of the RFX complex. RFX5 was the fifth member of the RFX family of DNA binding proteins to be identified (21). All members of this family share a characteristic DNA binding domain (DBD) referred to as the RFX motif (21, 22). In contrast to RFX5, RFXANK and RFXAP do not contain the RFX motif and consequently do not formally belong to the RFX family. However, they derive their names from the fact that their association with RFX5 is essential for the binding activity and function of the RFX complex (19, 42).
Although genetic and biochemical studies have demonstrated that all three RFX subunits are essential for the activation of MHC-II promoters and are required for the assembly and binding of the RFX complex (19, 42, 47, 67), the precise roles of the different subunits are not well understood. RFX5 is of special interest because it is the largest subunit (616 amino acids) and contains a DBD that is involved in tethering the RFX complex to the X box of MHC-II promoters (67). However, this DBD covers only 74 amino acids within the N terminus of RFX5, and until now no well-defined function has been mapped to the remainder of the 616-amino-acid protein. Outside of the DBD, RFX5 contains no obvious functional motifs. Besides a proline-rich region reminiscent of certain transcription activation domains, no known protein-protein interaction motifs are evident. Yet RFX5 is known or suspected to interact with several other proteins. These include the other two subunits of the RFX complex (19, 42, 72), CIITA (62), and other transcription factors that are known to bind to MHC-II promoters (Fig. 1). The last include the X2 box binding protein X2BP (28), which was recently shown to contain CREB (43), and the Y box binding protein NF-Y (16, 40, 41, 75, 76). Indeed, in vitro binding studies have previously shown that the RFX complex plays a central role in promoting cooperative binding interactions required for stable occupation of MHC-II promoters by NF-Y and X2BP (18, 38, 44, 55, 57).
To further our understanding of the mode of action of RFX5, we have undertaken a structure-function analysis of the protein. To facilitate this analysis we isolated the mouse RFX5 gene in order to identify conserved regions and optimized a number of genetic and biochemical approaches to dissect the function of the RFX5 protein. Our results allowed us to distinguish between two functionally distinct domains within RFX5. First, there is a highly conserved N-terminal region that encompasses the DBD and is sufficient for assembly of the RFX complex and for its binding to the MHC-II X box target site. This conserved domain is, however, not sufficient to restore expression of the endogenous MHC-II genes in a functional assay relying on the genetic complementation of cells derived from a MHC-II deficiency patient lacking RFX5. To reactivate MHC-II expression in this assay, a second, considerably less strongly conserved region of RFX5 is required. We show that this second poorly conserved domain is essential because it mediates cooperative binding with NF-Y. This finding provides direct evidence that cooperative binding between RFX and NF-Y plays an essential role in the transcriptional activation of MHC-II genes in vivo.
MATERIALS AND METHODS
Isolation of mouse RFX5 cDNA clones.
Full-length mouse RFX5 cDNA clones were isolated from a BALB/c mouse spleen cDNA library (58) by screening with a probe consisting of the DBD of the human RFX5 gene. Several independent cDNA clones were sequenced on both strands.
Construction of RFX5 expression vectors.
A wild-type RFX5 cDNA clone was first subcloned between the ApaI and NotI sites of pBluescript (Stratagene). A sequence encoding a hemagglutinin (HA) tag (MGYPYDVPDYASLGGPHH) was fused to the N terminus of RFX5 via an NdeI site introduced at the ATG initiation codon. N- and C-terminally truncated versions of RFX5 were amplified by PCR from the HA-RFX5 construct by using the following 5′ (N-terminal deletions) and 3′ (C-terminal deletions) primers (coordinates in the nucleotide sequence of the published human RFX5 cDNA [67] are indicated; the N1 and N2 primers contained an NdeI site, while the C1 to C5 primers contained a SalI site [underlined]): N1, 5′-CGAAGAACATATGCCAGGTGGTGCTGAGGCT-3′, nucleotides 215 to 233; N2, 5′-CGAAGGACATATGAAGGCCGTGCAGAACAAAG-3′, nucleotides 279 to 297; C1, 5′-CGTATCTGTCGACTTTGGTATGCTGGGAAC-3′, nucleotides 1819 to 1801; C2, 5′-CCTGAATGTCGACCCCTCCAGCTGAGTTG-3′, nucleotides 1703 to 1688; C3, 5′-CCTCGATGTCGACTAATGCTGTATCCTCTATACT-3′, nucleotides 1541 to 1521; C4, 5′-CAGTAATGTCGACTACCGGGGCTGAGTGAGTCC-3′, nucleotides 1391 to 1372; and C5, 5′-CGTACATGTCGACTACACAGGGCACCTGAAGAAAG-3′, nucleotides 1242 to 1224.
PCR was performed with the Expand high-fidelity PCR system (Boehringer Mannheim). The two N-terminal deletions were amplified by PCR using C5 as the 3′ primer. The resulting PCR products were digested with NdeI (5′ primer) and XhoI (nucleotide 748 in RFX5) and subcloned into the pBluescript HA-RFX5 construct between NdeI and XhoI. The C1 to C5 deletions were amplified by PCR using the M13 reverse primer of pBluescript as the 5′ primer. The PCR products were digested with XhoI (nucleotide 748 in RFX5) and SalI (3′ primer) and subcloned into the pBluescript HA-RFX5 construct between XhoI and SalI. The C6 deletion was generated by deleting the coding region downstream of the XhoI site (nucleotide 748 in RFX5) in pBluescript HA-RFX5. All constructs were verified by sequencing. The wild-type and C-terminally deleted HA-tagged RFX5 constructs were then subcloned either into the episomal EBS expression vector or into a bicistronic lentiviral vector (see Fig. 4). To construct the latter vector, the internal ribosomal entry site (IRES) from the encephalomyocarditis virus (EMCV) (31) was first inserted in front of the mouse CD8 gene from p1704-Lyt2 (mCD8) (generous gift from J.-K. Wang) and the IRES-mCD8 construct was introduced between the BamHI and XhoI sites of pHR′CMV (49, 50). HA-RFX5 constructs were then inserted in front of the IRES-mCD8 with an adapter.
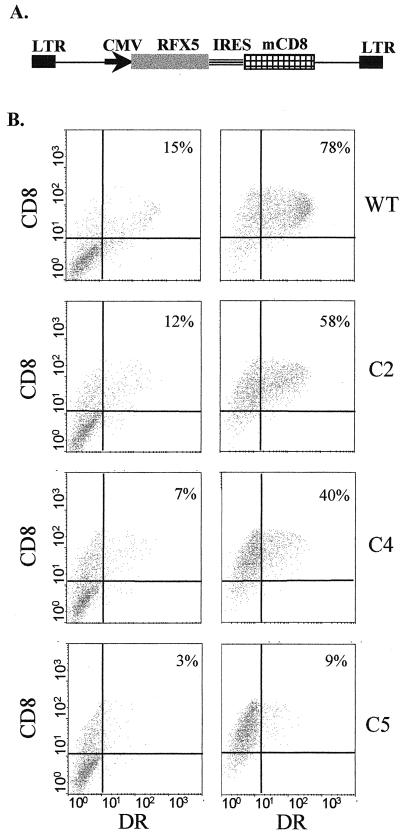
FIG. 4.
Complementation of SJO cells with bicistronic lentiviral constructs. (A) Schematic representation of the vectors used. The vectors contain a cytomegalovirus (CMV) promoter driving an expression cassette encoding wild-type or truncated RFX5 in the first cistron and mCD8 in the second cistron. Translation of mCD8 is under the control of the IRES of EMCV. LTR, long terminal repeat. (B) SJO cells transduced with bicistronic lentiviral vectors encoding wild-type (WT) or truncated (C2, C4, and C5) RFX5 were analyzed by FACS for HLA-DR and mCD8 expression. The analysis was done before (left) and after (right) the transduced CD8-positive cells were sorted out. The percentage of cells that are CD8+ and DR+ is indicated in each case.
Cell culture and complementation assays.
The B-lymphoblastoid cell line Raji and the Epstein-Barr virus-transformed B-cell line SJO derived from an RFX5-deficient patient (10, 11, 67) were grown in RPMI 1640. HeLa cells used for the production of recombinant RFX subunits and 293T cells used for the production of virus were grown in Dulbecco modified Eagle medium. All culture media were supplemented with glutamine, 10% heat-inactivated fetal calf serum, and antibiotics. Cells were grown at 37°C in 5% CO2.
For complementation with EBS-based expression vectors (see Fig. 3), 10 μg of plasmid was transfected by electroporation into SJO cells with a Bio-Rad electropulser using a 300-V and 960-μF pulse. Transfected cells were selected with 50 to 150 μg of hygromycin per ml for 10 to 15 days and were then analyzed by fluorescence-activated cell sorting (FACS) as described previously (68).
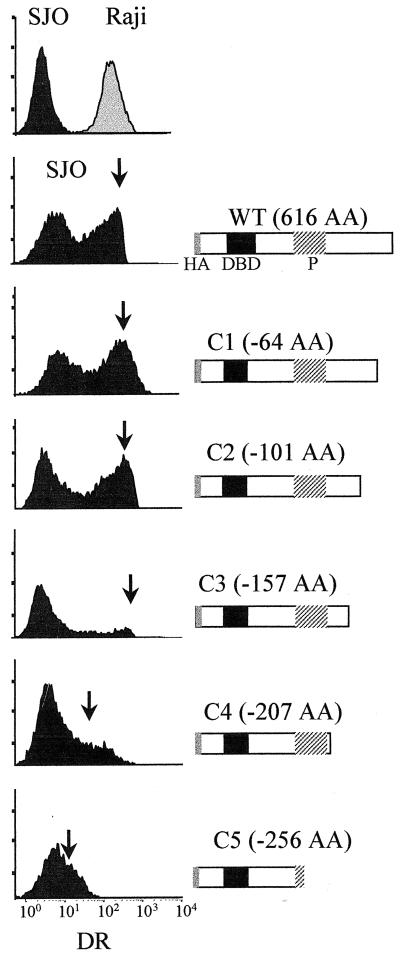
FIG. 3.
Complementation of SJO cells with episomal expression vectors encoding wild-type (WT) and C-terminally truncated (C1 to C5) versions of RFX5. Nontransfected SJO cells (top left, black profile), the MHC-II-positive control cell line Raji (top right, grey profile), and the transfected SJO cells were analyzed for HLA-DR expression by FACS. Transfected cells were selected for prolonged times (up to 20 days) with hygromycin. Arrows indicate the position of complemented MHC-II-positive SJO cells. Schematic maps of the wild-type and truncated versions of RFX5 are indicated at the right. The sizes of the deletions (in amino acids) are indicated. HA, HA tag; P, proline-rich region.
Production of virus from the bicistronic plasmids was done as follows. The packaging plasmid (pCMVΔR8.91) (49, 79) was used to provide all of the viral proteins except for the envelope protein. A third plasmid was used to provide the heterologous envelope G glycoprotein of vesicular stomatitis virus (49, 51). Virus was generated by cotransfection of 293T cells in 10-cm-diameter plates with 5 μg of the pHR′CMV-IRES-mCD8 vectors encoding the wild-type construct or RFX5 deletion constructs, 3 μg of pCMVΔR8.91, and 1 μg of the vesicular stomatitis virus glycoprotein pseudotyped envelope plasmid (48–50). FUGENE 6 (Boehringer Mannheim) was used for transfection. Supernatants were collected 24 and 48 h after transfection, filtered, and concentrated by ultracentrifugation at 20,000 rpm for 90 min in a SW 28 rotor. Virus pellets were then resuspended in a 1/50 volume of RPMI 1640. One hundred thousand SJO cells were infected by incubation with 1 ml of the concentrated supernatants in culture dishes that had been previously coated with recombinant fibronectin fragments (Retronectin; Takara) as described previously (27). One week after infection, transduced cells were washed twice in phosphate-buffered saline and stained with the HLA-DR monoclonal antibody 2.06 (12) and then with fluorescein isothiocyanate-conjugated rabbit anti-mouse immunoglobulin G (Serotec) and with phycoerythrin-conjugated rat anti-mouse-CD8a (Ly-2) monoclonal antibody (Pharmigen). After being washed in phosphate-buffered saline, the cells were analyzed by FACS. For purification of transduced cells, 20 × 106 cells were stained with biotin anti-mouse CD8a (Ly-2) antibodies (Pharmigen) and sorted with streptavidin-coated Dynabeads (Dynal). Sorted cells were reanalyzed by FACS 3 to 5 days later.
Production and analysis of recombinant proteins.
R. Mantovani provided recombinant NF-Y. The three subunits of NF-Y (NF-YA, NF-YB, and NF-YC) were produced in Escherichia coli and purified as described previously (36, 41). Recombinant RFX subunits were produced as follows. HeLa cell monolayers were infected in 5-cm-diameter dishes with 2 to 5 PFU of a vaccinia virus recombinant expressing T7 RNA polymerase per cell as described previously (24). At 1 h postinfection, medium was replaced with a transfection mix consisting of 2 μg of pBluescript plasmids containing the wild type or truncated RFX5, RFXAP, and RFXANK cDNAs cloned downstream of the T7 promoter and 10 μl of transfectase as previously described (15). After 24 h, whole-cell extracts were prepared as described previously (35). Isolated RFX subunits were synthesized separately, and the RFX complex was then reconstituted by mixing the extracts.
Western blotting experiments were used to monitor the synthesis of recombinant RFX proteins. Whole-cell extracts were prepared from 5 × 106 to 10 × 106 cells as described previously (35). Ten to 50 μg of extract was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 7.5% polyacrylamide gels. Proteins were transferred to polyvinylidene difluoride membranes (Immobilon-P; Millipore) using a Trans-Blot semidry transfer cell (Bio-Rad) for 30 min at 15 V. Membranes were incubated with the diluted antibodies indicated below in 1× blocking solution overnight at 4°C and washed in 1× washing buffer (digoxigenin buffer set; Boehringer Mannheim). The monoclonal anti-HA antibody 12CA5 (Babco) was used at a 1/8,000 dilution. A polyclonal rabbit anti-RFX5 antibody (19) was used at a 1/5,000 dilution. A polyclonal rabbit anti-RFXAP antibody (19) was used at a 1/1,000 dilution. A polyclonal rabbit antibody directed against two N-terminal peptides of RFXANK (QTPASELGDPEDPGEEC and CTPEPVNPEPDASVSS) was prepared by Eurogentec and used at a 1/5,000 dilution. Signals were revealed using a peroxidase-labeled anti-rabbit antibody diluted 1/5,000, followed by enhanced chemoluminescence detection (Amersham).
For the immunoprecipitation experiments, the RFX complex was assembled by mixing equal amounts of recombinant RFX5, RFXAP, and RFXANK. The mixture was first preincubated with protein A-Sepharose beads for 30 min and cleared by centrifugation. Supernatants were then immunoprecipitated with an anti-RFXAP or a control antibody (polyclonal VP16 antiserum; Clontech) by classical procedures. The immunoprecipitates were analyzed by Western blotting as described above by using a monoclonal anti-HA antibody (12CA5; Babco) at a 1/800 dilution to detect HA-RFX5 and HA-RFXAP and a monoclonal anti-FLAG antibody (Sigma) at a 1/200 dilution to detect FLAG-RFXANK.
EMSA.
With the exception of the following modifications, electrophoretic mobility shift assays (EMSA) were performed essentially as described previously (55, 57). EMSA were performed with 8 μg of whole-cell extract per sample. Whole-cell extracts were prepared as described previously (35) from 5 × 106 to 10 × 106 transfected SJO cells or from HeLa cells overexpressing the three RFX subunits. To reconstitute the RFX complex in vitro, HeLa whole-cell extracts containing the isolated RFX subunits were mixed in equal quantities prior to EMSA. To generate the RFX–NF-Y complexes, 1 ng (see Fig. 8D) or 10 ng (see Fig. 8C and 9) of recombinant NF-Y was added to the EMSA reaction mixtures set up with the HeLa cell extracts containing the reconstituted RFX complex. Binding mixtures were preincubated for 30 min prior to the addition of 40,000 cpm of the suitable 32P-labeled oligonucleotide probes and then incubated for a further 30 min to allow binding to proceed to completion. The probes used for binding of RFX (DRA-XX2 probe) or for binding of NF-Y and RFX–NF-Y (DRA-XY probe) and the wild-type and mutated X box competitor oligonucleotides have been described previously (29). For the dissociation rate experiments, the reaction mixtures were supplemented after binding was completed with a 500-fold molar excess of specific unlabeled fragment and then incubated at room temperature for various times prior to gel electrophoresis. For the supershift experiments, 20-μl binding reaction mixtures were set up as usual, reactions were allowed to proceed for 30 min at 0°C, and then the mixtures were supplemented with appropriate dilutions of the anti-HA, anti-RFX5, anti-RFXAP, or anti-RFXANK antibody described above.
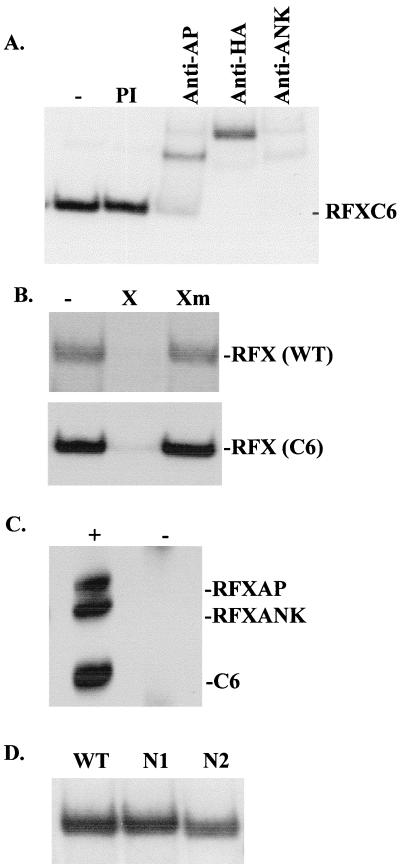
FIG. 8.
Delimitation of the region of RFX5 that is sufficient for assembly and binding of the RFX complex. (A) Recombinant RFX complexes were assembled as described for Fig. 7 with the C6 deletion of RFX5. Binding activity was analyzed by EMSA. The presence of the three subunits was confirmed by the addition of anti-HA (to detect HA-C6), anti-RFXAP, and anti-RFXANK antibodies. Preimmune serum (PI) was used as a negative control. (B) The specificities of wild-type RFX and of the RFX complex assembled with C6 were analyzed by EMSA using competitor oligonucleotides containing a wild-type (X) or mutated (Xm) X box sequence. (C) RFX complexes were assembled in solution from recombinant C6, RFXAP, and RFXANK and were immunoprecipitated with anti-RFXAP (+) or control (−) antibodies. The immunoprecipitates were analyzed by Western blotting for the presence of the three subunits. (D) RFX complexes were assembled with the wild-type, N1, or N2 version of RFX5 and were tested for binding activity by EMSA.
FIG. 9.
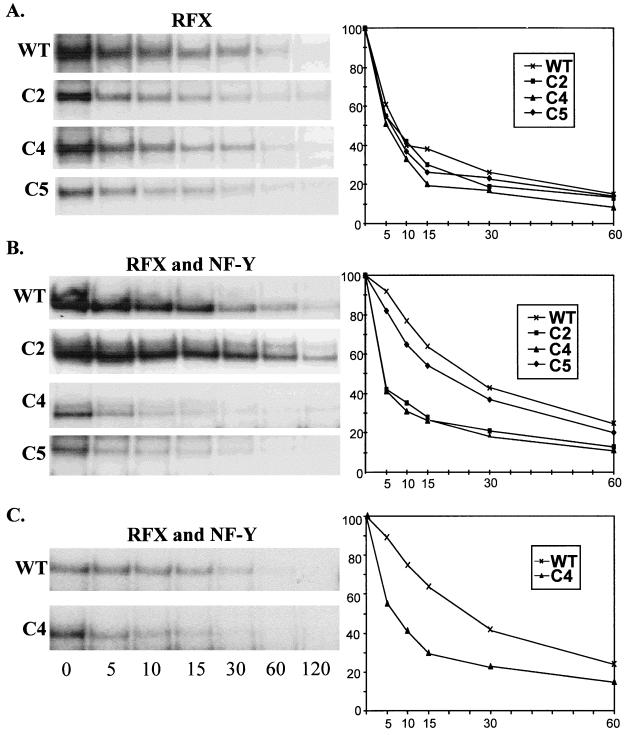
Deletions C4 and C5 eliminate the enhanced stability observed for cooperatively bound RFX and NF-Y. Dissociation rates were determined for protein-DNA complexes containing recombinant RFX complexes produced as described for Fig. 7B (A), recombinant RFX–NF-Y complexes produced as described for Fig. 7C (B), and RFX–NF-Y complexes formed in extracts from SJO cells transduced as described for Fig. 4 (C). As indicated at the left, the complexes analyzed contained the wild-type, C2, C4, or C5 version of RFX5. Reaction mixtures were first incubated to allow binding to proceed to completion and then supplemented with an excess of unlabeled competitor DNA; the reactions continued for 0, 5, 10, 15, 30, 60, and 120 min prior to gel electrophoresis. The gels shown at the left were quantified by phosphorimager analysis. The percentages of the protein-DNA complexes remaining are plotted as a function of time (graphs at right). The amount of complex bound at time zero (addition of unlabeled competitor DNA) was considered 100%.
Nucleotide sequence accession number.
Nucleotide and amino acid sequences of mouse RFX5 have been submitted to GenBank under accession number AF209854.
RESULTS
The C-terminal moiety of RFX5 is poorly conserved yet essential for its function.
The sequence of the mouse RFX5 gene has not yet been reported. We therefore isolated the mouse gene in order to facilitate the identification of conserved regions within RFX5. Full-length RFX5 cDNA clones were isolated from a mouse spleen cDNA library, and several independent clones were sequenced. All exons in the genomic mouse RFX5 gene were also isolated and sequenced. Identity between the human and mouse RFX5 genes was confirmed on the basis of sequence homology (Fig. 2), Southern blotting experiments (data not shown), and the ability of the mouse RFX5 cDNA to complement the genetic defect in RFX5-deficient cells derived from MHC-II deficiency patients (data not shown). The last factor confirmed that the mouse gene could fully substitute for human RFX5 in restoring expression of all the endogenous MHC-II genes (data not shown).
FIG. 2.
Sequence conservation between human and mouse RFX5. (A) Alignment of the human (top) and mouse (bottom) amino acid sequences. Arrowheads indicate positions at which the protein is truncated in patients P1 (and sibling P2), P3, P4, P5, and P6. The DBD is boxed. The proline-rich region is underlined. Arrows indicate endpoints of the N (N1 and N2)- and C (C1 to C5)-terminal deletions. Asterisks indicate amino acid identity. Dashes represent gaps introduced to maximize homology. (B) Schematic maps of the human and mouse RFX5 proteins indicating the same features as those described above. The percentages of amino acid identity in relevant regions are indicated below.
Mouse RFX5 cDNAs contain a 657-amino-acid open reading frame (Fig. 2). Alignment between the human and mouse sequences is quite informative concerning conservation of RFX5 (Fig. 2). The N-terminal third of the protein exhibits very high conservation (>90% amino acid identity), particularly within the region encompassing the DBD (99%), which is to date the only domain in RFX5 of which the function is known. In striking contrast, the C-terminal two-thirds of the protein is considerably less well conserved. This is evident at the level of overall amino acid identity (only 55%). Moreover, a 12-amino-acid deletion and a 53-amino-acid insertion in the mouse gene interrupt the proline-rich region that was previously noted in RFX5 (67). Conserved amino acids are clustered in discreet blocks exhibiting greater (70 to 90%) homology.
Loss-of-function mutations affecting the RFX5 genes in five MHC-II deficiency patients have been characterized (Fig. 2). In all of the patients, the mutations lead to a severe truncation of RFX5 (17, 52, 53, 67, 73). In two patients, P5 and P6, the mutations lead to the synthesis of truncated RFX5 proteins which retain intact the highly conserved N-terminal region but lack the poorly conserved C-terminal moiety (52, 67). Together with the finding that the mouse RFX5 gene can complement the genetic defect in cells from RFX5-deficient patients, the mutations identified in P5 and P6 indicate that the conserved N-terminal region is not sufficient for function. Clearly, although very divergent, the C-terminal region must also contain domains with essential functions.
Functional dissection of the C-terminal moiety of RFX5.
To study the function of the poorly conserved C-terminal region of RFX5, we prepared a series of C-terminal deletions (C1 to C5). The endpoints were chosen such that the deletions would progressively remove blocks of sequence exhibiting greater than average homology between the human and mouse genes (Fig. 2). To study the repercussions of these C-terminal deletions on the function of RFX5, we used a genetic approach relying on the complementation of RFX5-deficient SJO cells. In this system, the ability of the mutated RFX5 proteins to reactivate expression of the endogenous MHC-II genes was evaluated. This complementation approach was chosen for three reasons. First, the readout is the ability to activate expression of the endogenous genes in their native genomic context. In terms of biological relevance, the complementation approach is greatly superior to classical transient-transfection experiments in which the trans-activation of reporter gene constructs is assayed. Second, the effect on all MHC-II genes (including the α- and β-chain genes encoding the HLA-DR, HLA-DQ, and HLA-DP molecules) can be analyzed simultaneously. Finally, activation of MHC-II genes can be scored simply, reliably, and quantitatively by FACS analysis of cell surface MHC-II expression.
In initial complementation experiments, episomal Epstein-Barr virus-based expression vectors were used. Figure 3 shows the results obtained with the EBS vector (4), in which the strong Srα promoter drives expression. Identical results have also been obtained with the EBO vector (68), in which expression is controlled by the weaker simian virus 40 promoter, indicating that expression levels are not limiting (data not shown). As judged by the mean fluorescence intensity of the complemented cells, the C1, C2, and C3 constructs retained the ability to restore HLA-DR expression to levels that were identical to that observed with wild-type RFX5 (Fig. 3). The last 156 C-terminal amino acids of RFX5 are therefore not essential. In contrast, the C4 and C5 deletions clearly affected the ability to restore HLA-DR expression. Reactivation of HLA-DR expression was strongly reduced for C4 and practically lost for C5 (Fig. 3). The same results were obtained when the deletions were tested for their ability to reactivate expression of HLA-DQ and HLA-DP (data not shown). The proline-rich region (removed in C5) and a 51-amino-acid sequence situated immediately downstream of it (removed in C4) are thus essential for the function of RFX5.
A number of problems were encountered during the course of complementation experiments performed with the episomal expression vectors. First, most cell lines derived from MHC-II deficiency patients, including the SJO cell line used here, grow poorly, exhibit high sensitivity to antibiotics, and are exceedingly difficult to transfect efficiently (67). Second, the episomal RFX5 expression vectors appeared to be unstable because it was difficult to maintain transfected cell populations exhibiting a stable complemented phenotype, even following prolonged hygromycin selection (Fig. 3). Consequently, the fraction of cells that were complemented was variable and rarely greater than 50%, even for the wild-type RFX5 construct. Because of this problem, it was not possible to determine whether differences observed in the fractions of complemented cells (compare results for examples C2 and C3 in Fig. 3) were significant. We therefore developed a second complementation system relying on stable transduction of SJO cells with a bicistronic lentivirus expression vector (49, 50) (Fig. 4A). This system has several advantages. First, the use of a retroviral vector permits stable integration of the expression constructs, thus avoiding the instability observed with the episomal vectors. Second, we designed the vector such that an mCD8 selectable marker cistron was inserted downstream of the RFX5 expression cassette. The IRES of EMCV controls translation of this mCD8 cistron so that cell surface expression of mCD8 represents an excellent internal control for expression of the RFX5 protein encoded by the first cistron. Finally, selection with antibiotics is avoided and purification of the transduced cell populations can be achieved readily by sorting with one round of anti-CD8 antibodies and magnetic beads.
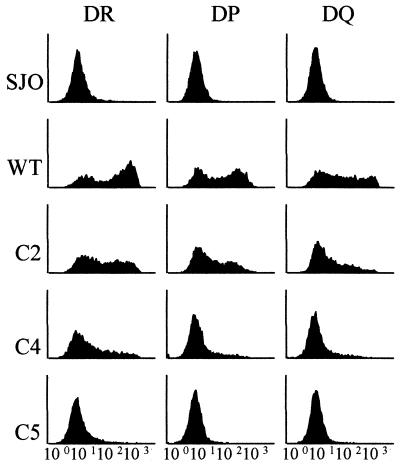
Complementation of SJO cells with the bicistronic lentiviral vectors was very efficient. Sorting of the transduced SJO cells allowed the isolation of populations in which almost all of the cells expressed mCD8 at the cell surface (Fig. 4B). When the construct encoding wild-type RFX5 was used, the majority of the CD8-positive cells also exhibited restored MHC-II expression (Fig. 4B). Moreover, in the complemented cells, expression of all three MHC-II isotypes was restored, although reactivation of HLA-DR appeared to be somewhat more efficient than that of HLA-DQ and HLA-DP (Fig. 5).
FIG. 5.
Complementation of SJO cells with bicistronic lentiviral constructs encoding wild-type (WT) or truncated (C2, C4, and C5) RFX5. Transduced cells were sorted for expression of CD8 and then analyzed by FACS for expression of HLA-DR, HLA-DP, and HLA-DQ. FACS profiles for noncomplemented SJO cells transduced with a negative-control construct are included at the top.
Transduction of SJO cells with bicistronic lentiviral vectors encoding the truncated RFX5 constructs allowed us to confirm and refine the results obtained with the episomal expression vectors (Fig. 4B and 5). Experiments with C3 did not give consistent results, and this deletion was therefore excluded from further analysis. Complementation with C2 was clear, albeit slightly less efficient than with wild-type RFX5. A stronger reduction in complementation efficiency was observed with C4 and C5. This reduction was particularly evident when the mean fluorescence intensity of the transduced cell population was examined. Although less evident, it was also observed at the level of the percentage of cells that scored positive for MHC-II expression. As observed with the episomal vectors, the loss in efficiency was severe for C4 and almost complete for C5. The results obtained were identical irrespective of whether expression of HLA-DR, HLA-DQ, or HLA-DP was examined (Fig. 5).
The RFX5 C-terminal region is dispensable for assembly and binding of the RFX complex.
To confirm that the wild-type and truncated RFX5 proteins were produced in the transduced SJO cells, we performed EMSA with extracts prepared from the sorted CD8-positive cells. These binding experiments revealed not only that the different RFX5 proteins were indeed synthesized but also that they were in each case incorporated into functional RFX complexes that were capable of binding to the X box (Fig. 6A). As expected, the mobility of the RFX-DNA complex progressively increased (wild type < C2 < C4 < C5) as a function of the size of the deletion introduced into RFX5. The RFX-DNA complex observed with cells transduced with wild-type RFX5 comigrated with that formed by the native RFX complex found in normal MHC-II-positive cells (data not shown).
FIG. 6.
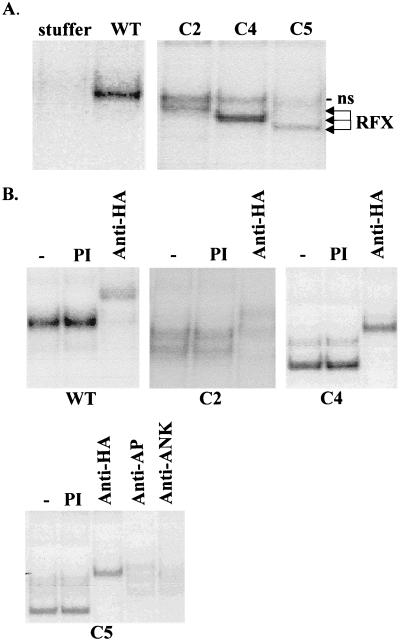
Assembly of RFX complexes exhibiting DNA binding activity in transduced SJO cells. (A) RFX complexes are restored in SJO cells transduced with the bicistronic lentiviral constructs encoding wild-type (WT) and truncated (C2, C4, and C5) versions of RFX5. No RFX complex is detected in SJO cells transduced with a negative-control construct (stuffer). Binding of the RFX complexes was analyzed by EMSA with an oligonucleotide probe containing the X box of the DRA gene. The whole-cell extracts used for the experiments were prepared from the transduced cells after purification of these cells by sorting for CD8 expression. Only the region of the gel containing specific RFX-DNA complexes is shown. All lanes are from the same gel and at the same exposure. A weak nonspecific (ns) complex migrating just below the band due to wild-type RFX is indicated. (B) The mixtures of binding reactions performed as described above were supplemented with preimmune serum (PI), antibodies specific for the HA tag, or antibodies specific for the two other subunits of the RFX complex, RFXANK and RFXAP. The anti-HA antibody supershifts complexes containing the transduced wild-type, C2, C4, and C5 RFX5 proteins. The complex containing C5 is also supershifted by the antibodies directed against RFXAP and RFXANK.
The RFX complexes regenerated in the transduced cells contained all three RFX subunits (Fig. 6B). Since all RFX5 constructs contained an N-terminal HA tag, the presence of the transduced RFX5 proteins could be demonstrated by supershift experiments with an anti-HA antibody. The RFXAP and RFXANK subunits were also shown to be present by means of supershift experiments performed with the appropriate antibodies. Their presence is shown in Fig. 6B for the RFX complex generated with C5 but has also been shown for the complexes containing the less severely deleted C2 and C4 proteins (data not shown). Thus, complete RFX complexes are assembled in the transduced cells by the association of the transduced RFX5 proteins with the endogenous RFXAP and RFXANK subunits.
Although a certain amount of variability was evident in the levels of the RFX complexes that were generated by transduction of SJO cells with the different truncated versions of RFX5 (Fig. 6A), these variations in abundance are unlikely to account for the observed differences in complementation efficiency (Fig. 4 and 5). In the case of C2 it cannot be excluded that the lower abundance of the RFX complex was responsible for the mild reduction in complementation efficiency. However, this possibility cannot be the case for C4 and C5. For example, the wild-type and C4 complexes occurred in equal levels of abundance yet complementation was severely impaired with C4. Similarly, although the C2 and C5 complexes occurred in similar levels of abundance, complementation with C2 was only mildly affected while complementation with C5 was almost completely abolished.
The binding experiments described above imply that the C-terminal region of RFX5 is not required for its association with RFXAP and RFXANK, for assembly of the RFX complex, or for binding of this complex to DNA. The availability of cDNAs encoding all three subunits of RFX allowed us to test this directly with the wild-type and truncated RFX complexes reconstituted in vitro from recombinant proteins. We employed a vaccinia virus-T7 RNA polymerase (Vac-T7) expression system to synthesize recombinant RFX subunits in HeLa cells (see Materials and Methods). The Vac-T7 system was chosen rather than the approach we reported previously when we used in vitro-translated subunits (42). The reason for this was that the recombinant RFX complexes prepared with in vitro-translated subunits do not behave normally in EMSA (42). In contrast, the RFX complex reconstituted from wild-type subunits synthesized with the Vac-T7 system is indistinguishable from the native RFX complex found in extracts from normal cells (see below).
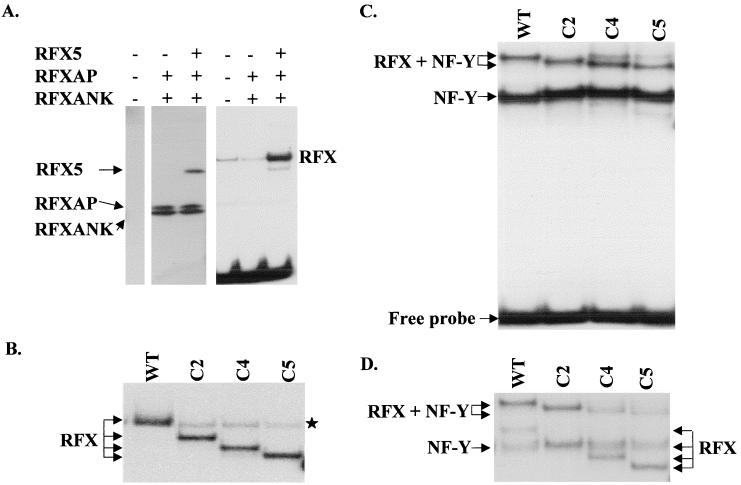
RFXANK, RFXAP, RFX5, C2, C4, and C5 proteins were all synthesized separately using the Vac-T7 system. Western blotting was used to monitor and quantify synthesis of the recombinant proteins (Fig. 7A and data not shown). To reconstitute RFX complexes, extracts containing intact or truncated RFX5 were mixed in equal amounts with extracts containing RFXAP and RFXANK. The binding activities of these in vitro-reconstituted RFX complexes were then studied by EMSA (Fig. 7). In this system, the reconstitution of functional RFX complexes capable of binding required all three subunits (Fig. 7A). The RFX-DNA complex obtained with wild-type RFX5 comigrated with the band formed by the native HeLa cell RFX (Fig. 7A and B). As observed with extracts from complemented SJO cells (Fig. 6), the mobility of the RFX-DNA complex progressively increased (wild type < C2 < C4 < C5) as a function of the size of the deletion introduced into RFX5 (Fig. 7B). The truncated C2, C4, and C5 complexes were generated as efficiently as the wild-type complex (Fig. 7B). Moreover, dissociation rate measurements indicated that the wild-type and truncated complexes had indistinguishable affinities for the X box target site (see Fig. 9A). We conclude that removal of 256 amino acids from the C terminus of RFX5 (deletion C5) has no adverse effect on either the assembly or the binding activity of the RFX complex.
FIG. 7.
Binding of RFX complexes assembled in vitro from recombinant proteins produced in HeLa cells using a Vac-T7 expression system. (A) Extracts from HeLa cells programmed to synthesize the RFX subunits indicated at the top were mixed in equal amounts and analyzed by Western blotting (left) or EMSA (right). Positions of the RFX subunits detected by Western blotting are indicated at the left. The band resulting from binding of the RFX complex is indicated at the right. A weak band resulting from binding of the native RFX complex is visible in the HeLa cell extract (left lane) and the extract lacking RFX5 (middle lane). This native complex comigrates with the band resulting from binding of the RFX complex that is assembled from the three recombinant subunits (right lane). Unbound DNA is visible at the bottom of the gel. (B) EMSA was performed with RFX complexes that were assembled as described above using equal amounts of wild-type (WT), C2, C4, and C5 versions of RFX5. Only the region of the gel containing specific RFX-DNA complexes is shown. Binding of the native RFX complex derived from the HeLa cells is indicated by a star. (C) RFX complexes were assembled as described above and then mixed with an excess (10 ng) of recombinant NF-Y. Binding of the resulting NF-Y and RFX–NF-Y complexes were analyzed by EMSA using an oligonucleotide probe containing both the X box and the Y box. Under these conditions, where NF-Y is in large excess, most RFX complexes are driven into the higher-order protein-DNA complexes containing both RFX and NF-Y. (D) Binding of higher-order RFX–NF-Y complexes was examined as described above, except that a 10-fold lower excess (1 ng) of recombinant NF-Y was added. Under these conditions, formation of the higher-order RFX–NF-Y complex is less efficient for complexes containing C4 and C5.
The conserved N-terminal domain of RFX5 is sufficient for assembly and binding of the RFX complex.
To delimit more precisely the region of RFX5 that is sufficient for assembly and binding of the RFX complex, we prepared additional deletions using the Vac-T7 expression system. A deletion (C6) lacking essentially all of the C-terminal region downstream of the DBD retained its ability to associate with RFXAP and RFXANK and to generate an RFX complex capable of binding specifically to the X box in EMSA. Supershift experiments demonstrated that this complex contains both C6 and the other two RFX subunits (Fig. 8A). Specificity was maintained, because binding of the C6 complex could be eliminated by an X box competitor oligonucleotide but not by an oligonucleotide containing a mutated X sequence (Fig. 8B). Finally, C6, RFXAP, and RFXANK could be efficiently coimmunoprecipitated in the absence of DNA, indicating that the three subunits assemble into a stable complex prior to binding (Fig. 8C).
The analysis of two additional N-terminal deletions (N1 and N2) allowed us to narrow down further the region required for assembly and binding. Both of these deletions could be incorporated efficiently into a functional RFX complex (Fig. 8D). Taken together, these results define a 155-amino-acid segment (amino acids 39 to 194) of RFX5 that is sufficient both for stable assembly of the RFX complex and for its X box-specific binding activity. This region of RFX5 encompasses the DBD and is embedded in the most conserved segment of the protein. In complementation experiments, N1 was fully functional while N2 showed no complementation. However, we cannot exclude the possibility that the lack of complementation observed with N2 was due to a problem with the stability of the protein rather than the loss of a functionally important domain (data not shown).
The C terminus of RFX5 mediates cooperative binding between RFX and NF-Y.
Taken together, the results of the complementation assays and binding experiments indicated that the C-terminal region must mediate a crucial function that is distinct from the association of RFX5 with the other two RFX subunits and from binding of the resulting RFX complex to the MHC-II X box. One potential function that was likely to be affected was the ability of RFX to bind cooperatively with other MHC-II promoter binding factors such as NF-Y. In vitro binding experiments have indicated that cooperative binding between RFX and NF-Y leads to the generation of multiprotein-DNA complexes exhibiting strongly enhanced stability (18, 38, 57). We therefore examined the effect of truncating the C terminus of RFX5 on the stability of the RFX–NF-Y–DNA complex.
RFX–NF-Y–DNA complexes were formed in EMSA by using a probe containing both the X and Y boxes and by mixing in vitro-reconstituted RFX complexes with recombinant E. coli-produced NF-Y (Fig. 7C). When high concentrations of NF-Y were added, most of the RFX was driven into the higher-order complex. At these high NF-Y concentrations, the higher-order RFX–NF-Y–DNA complex could be formed equally well with all of the C-terminal deletions (Fig. 7C). However, at a 10-fold lower excess of NF-Y, the RFX–NF-Y–DNA complexes were found to form less efficiently with the C4 and C5 complexes than with the wild-type and C2 complexes (Fig. 7D). These results suggested that deletions C4 and C5 might affect the stability of the higher-order complex. The relative stabilities of the RFX–NF-Y–DNA complexes formed with the different deletions were therefore compared by examining their dissociation rates (Fig. 9B). In this assay, the C2 deletion had no significant adverse effect. The C4 deletion, on the other hand, clearly led to a major reduction in stability, which is evident from the fact that the 25-min half-life observed for complexes containing wild-type RFX5 was reduced over fivefold, to less than 5 min, for C4. The half-life that was observed for complexes containing C5 was identical to that observed for C4, indicating that the C5 deletion does not lead to a further loss in stability.
The half-life of less than 5 min obtained for RFX–NF-Y–DNA complexes containing C5 and C4 is identical to the half-life observed for RFX bound on its own in the absence of NF-Y (compare Fig. 9A and B). This indicates that the C4 and C5 deletions abolish stabilization of RFX by NF-Y, such that dissociation of the complexes containing these truncated RFX molecules is independent of the adjacently bound NF-Y.
The effect of the C4 deletion on cooperative binding with NF-Y could also be observed by comparing extracts prepared from SJO cells transduced with the wild-type RFX5 and C4 constructs (Fig. 9C). In this case, the multiprotein-DNA complexes were assembled from RFX and NF-Y proteins that were synthesized in vivo at physiological concentrations in the transduced cells. As observed for complexes assembled with recombinant proteins, the C4 deletion abolished the enhanced stability of the RFX–NF-Y–DNA complex (Fig. 8C).
DISCUSSION
Genetic evidence has established that RFX5 is absolutely essential for activation of MHC-II promoters. Mutations of RFX5 that abrogate MHC-II expression have been identified in at least five unrelated MHC-II deficiency patients and in one in vitro-generated mutant cell line (7, 17, 52, 53, 67, 73). RFX5 knockout mice are also characterized by a severe deficiency in MHC-II expression (13). Genetic and biochemical evidence has also demonstrated that RFX5 combines with two other unrelated proteins, RFXAP and RFXANK, to form RFX, a heterotrimeric DNA binding protein that binds to the X box cis-acting element of MHC-II promoters (19, 42, 56). However, until now little has been known about how RFX5 contributes to assembly of the RFX complex and how it mediates activation of MHC-II promoters. We have therefore undertaken a systematic study of the structure and function of RFX5. In this report, we define two domains serving two different essential functions of RFX5. A highly conserved N-terminal region of RFX5 is sufficient for its association with RFXANK and RFXAP, for assembly of the RFX complex in vivo and in vitro, and for binding of this complex to the MHC-II X box (see the model in Fig. 1). However, this N-terminal region is not sufficient for activation of MHC-II expression. This requires an additional, considerably less well-conserved C-terminal region. One of the functions of this C-terminal domain is to promote cooperative binding between the RFX complex and NF-Y, a transcription factor that binds to MHC-II promoters (see the model in Fig. 1).
The conserved N-terminal domain of RFX5 encompasses a 74-amino-acid segment that has previously been identified as the DBD of the protein (39, 67). This DBD is called the RFX motif because it was first identified in other members (RFX1 to RFX4) of the same family of DNA binding proteins (21, 22). That this DBD is implicated in specific binding of RFX5 to the MHC-II X box has been demonstrated previously (67). However, the RFX5 DBD is not by itself sufficient for tethering of the RFX complex to the X box. This insufficiency is demonstrated by the observation that DNA binding activity requires the association of RFX5 with RFXAP and RFXANK (Fig. 7A) (42). The region of RFX5 that is required for assembly of an RFX complex containing all three subunits has been reduced here to a 155-amino-acid segment (amino acids 39 to 194) encompassing the DBD (amino acids 94 to 167) (Fig. 8). The DBD of RFX5 and the regions situated immediately upstream and downstream of it are thus necessary and sufficient for the association with RFXANK and RFXAP, for correct assembly of the RFX complex in solution, and for specific binding to the X box target site. These results are consistent with the finding that amino acid substitutions in a leucine-rich segment situated just upstream of the DBD (amino acids 62 to 68) destroy the function of RFX5 and eliminate the binding activity of the RFX complex (7).
It remains unclear exactly how RFXANK and RFXAP contribute to the binding activity of the RFX complex. Neither of these two subunits contains a recognizable DBD, and only RFX5 has been shown to exhibit site-specific DNA binding activity (67). Yet efficient binding of RFX requires the combination of all three subunits. Several non-mutually exclusive explanations may reconcile these findings. First, it is possible that RFXAP and/or RFXANK does not mediate sequence specificity but that it contributes only by providing nonspecific protein-DNA interactions required for stable binding. This possibility would be consistent with the observation that all three RFX subunits can be cross-linked to DNA in the vicinity of the X box (74). Second, perhaps RFXAP and/or RFXANK mediates dimerization of RFX5. All other known members (RFX1 to RFX4) of the RFX family bind as dimers to palindromic recognition sites conforming to a well-defined consensus sequence (21). Surprisingly, the MHC-II X box target site of RFX5 also conforms to the same palindromic consensus sequence, yet RFX5 lacks the conserved domain that is known to mediate dimerization of the RFX1 to RFX4 proteins. This finding raises the interesting possibility that RFXAP and/or RFXANK is there to replace the missing dimerization domain. Finally, a third possibility is that interactions with RFXAP and/or RFXANK facilitates binding because it induces a conformational change in RFX5. In this respect it may be relevant that RFXANK contains ankyrin repeats (42). In the heterodimeric DNA binding protein GABP, an ankyrin repeat-containing subunit, GABPβ, greatly enhances the affinity of the DNA binding subunit GABPα for its binding site (2). Delimitation of the minimal region of RFX5 required for the assembly and binding of the RFX complex will certainly contribute to the elucidation of the respective roles of the three RFX subunits.
The C-terminal moiety of RFX5, although dispensable for assembly and binding of RFX, is nevertheless essential for activation of MHC-II expression. Removal of a protein segment between amino acids 410 (deletion C4) and 515 (deletion C2) leads to a severe reduction in the function of RFX5 (Fig. 3 to 5). We have shown here that this domain within the C terminus of RFX5 mediates cooperative binding with NF-Y. Elimination of this domain abolishes the stabilization that is observed in vitro when RFX and NF-Y bind together to the same DNA fragment (Fig. 9). The finding that a functionally important domain within RFX5 plays a key role in cooperative binding between RFX and NF-Y provides the first direct evidence that this cooperative binding interaction is critical for activation of MHC-II promoters. Until now, the importance of this cooperative binding in the activity of MHC-II promoters in vivo could only be inferred from two indirect lines of evidence. First, cell lines from MHC-II deficiency patients lacking RFX are characterized by bare MHC-II promoters in which all cis-acting sequences, including the Y box, remain unoccupied (32, 33). Second, disruptions of the Y box in stably transfected MHC-II promoter constructs leads to a reduction in the occupation of the X box (75).
Cooperative binding between RFX and NF-Y suggests the existence of a protein-protein interaction between these two complexes. The C terminus of RFX5 may contribute in a number of ways to this protein-protein interaction. First, it may provide the major contact with NF-Y. Second, it may provide only one of several weak contacts acting together in synergy. Third, it may be required to generate an RFX complex having the correct stoichiometry for the interaction to occur. Finally, it may be required to induce a suitable conformational change in RFXAP and/or RFXANK. In the last two cases, direct contacts with NF-Y may in fact be provided by RFXAP and/or RFXANK rather than by RFX5. To take all of these possibilities into account, we have performed protein-protein interaction studies with intact RFX and NF-Y complexes rather than with isolated subunits. We have used coimmunoprecipitation experiments both with native RFX and NF-Y complexes in crude cell extracts and with recombinant RFX and NF-Y complexes assembled in vitro from Vac-T7- and E. coli-produced proteins. These experiments have not allowed us to detect a direct interaction between RFX and NF-Y, even when high concentrations of recombinant proteins were used to force the interaction and when the immunoprecipitations were performed under very mild conditions (unpublished data).
A number of explanations may account for our inability to detect direct protein-protein interactions between RFX and NF-Y. First of all, these interactions may be too weak in solution to be detected readily in the absence of DNA. In this respect it should be mentioned that a stable interaction between RFX and NF-Y in the absence of DNA would in any case not be expected to occur, because this would defeat the purpose of the synergistic and combinatorial control resulting from cooperative binding. It may also be relevant that the region of RFX5 that is implicated here does not contain any motifs known to mediate high-affinity protein-protein interactions, such as leucine zippers (1), ankyrin repeats (63), and leucine-rich repeats (9). A second possibility is that the protein-protein interaction interfaces need to be unmasked by conformational changes that are induced by binding of RFX and/or NF-Y to their target sites. Finally, it is possible that cooperative binding is mediated by a conformational change of the DNA rather than by direct protein-protein contacts. For instance, binding of RFX may be enhanced by a structural alteration induced in the DNA by NF-Y. In this respect it may be relevant that binding of NF-Y is known to distort DNA (40). If the last explanation is correct, our results suggest that the C-terminal region of RFX5 may render the RFX complex sensitive to the conformation of the DNA. It may do this either by contacting the DNA or by conferring a suitable conformation on the RFX complex.
NF-Y is a ubiquitously expressed transcription factor that has been reported to be involved in the regulation of a wide variety of eukaryotic genes, including MHC-II genes (16, 40, 65). Previous evidence for the role of NF-Y in the expression of MHC-II genes was derived from in vitro binding studies, classical transactivation assays, and in vitro transcription experiments (40, 41, 75, 76). However, direct genetic evidence such as that available for RFX and CIITA was lacking for NF-Y. The data presented here now provide indirect genetic evidence by demonstrating the existence of a physical link between NF-Y and a domain within RFX5, a factor that has been defined genetically to be an essential component of the molecular machinery regulating MHC-II expression. Our findings make a strong argument in favor of a functional role of NF-Y at MHC-II promoters. NF-Y is a heterotrimeric transcription factor consisting of three subunits, NF-YA, NF-YB, and NF-YC. Domains required for DNA binding and transcriptional activation have been mapped within these NF-Y subunits (36, 66, 77). The experiments presented here now also pave the way for the identification of the subunits and regions of NF-Y that mediate cooperative binding with RFX.
It is likely that cooperative binding with NF-Y is not the only function that is mediated by the C terminus of RFX5. This possibility is implicit in the fact that the function of RFX5 is lost progressively rather than suddenly as C-terminal deletions of increasing size are introduced. In particular, a comparison between the effects of deletions C4 and C5 argues for a distinct role of the proline-rich region situated between the endpoints of these two deletions. Thus, C4 eliminates cooperative binding with NF-Y yet it retains residual activity in the functional assays. This residual activity is abolished by C5, although the effect of C5 on cooperative binding with NF-Y is not greater than that observed for C4. One candidate function for the proline-rich region is a protein-protein interaction with the X2 box binding protein X2BP. As described for NF-Y, X2BP binds cooperatively with RFX to MHC-II promoters (18, 38, 44, 55). Until recently, it has been difficult to address this possibility directly because X2BP remained a binding activity detected in nuclear extracts. However, the recent finding that X2BP contains CREB (43) will help to resolve this question. A second important function that may be mediated by the C terminus of RFX5 is recruitment of CIITA. While the function of CIITA as a key transactivator of MHC-II expression is now well established, its mode of action remains obscure. Although CIITA is believed to function as a non-DNA-binding coactivator that is recruited to MHC-II promoters by protein-protein interactions with DNA-bound factors such as RFX (39, 59, 78), it remains to be formally demonstrated that this is indeed the case. One study has reported an interaction between the C-terminal moiety of RFX5 and CIITA, but the region implicated in RFX5 was not precisely defined (62). It will be of key importance to explore further the possibility of an interaction between RFX5 and CIITA, because whether CIITA is actually recruited physically to MHC-II promoters and how this may be achieved remain major unresolved questions. Finally, additional functions of the C-terminal region of RFX5 must also be considered. It may for instance be involved in nuclear import, be the target of modifying activities such as phosphorylation, be implicated in the dimerization of RFX5, or influence the stoichiometry of the assembled RFX complex.
The bicistronic lentiviral vectors we describe here have proved to be efficient for the mapping of functional domains within RFX5. They will certainly also be very useful for similar studies of CIITA and the other two subunits of RFX. In addition, their usefulness extends to a more clinically relevant application, namely, the development of gene therapy for MHC-II deficiency. Gene therapy for MHC-II deficiency is a valid and attractive alternative to bone marrow transplantation, which has a low rate of success with this disease. Lentiviral vectors of the type used here are currently among those that are the best suited for gene therapy. In contrast to other retroviral vectors, such as those based on murine leukemia virus, lentiviral vectors are efficient at transducing quiescent and nondividing cells, including the hematopoietic stem cells that would have to be corrected in MHC-II deficiency (48–50, 79). Vectors such as those described here might be used to deliver RFX5, RFXAP, RFXANK, or CIITA to hematopoietic stem cells derived from MHC-II deficiency patients. The use of bicistronic vectors encoding a selectable marker such as green fluorescent protein in the second cistron would be very useful for enriching the transduced stem cells. Stably corrected stem cells could then be anticipated to reconstitute the entire hematopoietic system with cells capable of reexpressing MHC-II molecules.
ACKNOWLEDGMENTS
We are grateful to D. Trono for providing the plasmids and advice that were needed to set up the lentivirus vector system. We thank M. Zufferey for expert technical assistance. We are indebted to B. Mach, who provided the scientific environment in which this work was first initiated.
This work was supported by the Louis-Jeantet Foundation and by the Swiss National Science Foundation (grants NFP 4037-46197 and 3100-056991.99/1). Marie Peretti was the beneficiary of a studentship stipend from the Yamanouchi Research Institute.
REFERENCES
- 1.Alber T. Structure of the leucine zipper. Curr Opin Genet Dev. 1992;2:205–210. doi: 10.1016/s0959-437x(05)80275-8. [DOI] [PubMed] [Google Scholar]
- 2.Batchelor A H, Piper D E, de la Brousse F C, McKnight S L, Wolberger C. The structure of GABPalpha/beta: an ETS domain-ankyrin repeat heterodimer bound to DNA. Science. 1998;279:1037–1041. doi: 10.1126/science.279.5353.1037. [DOI] [PubMed] [Google Scholar]
- 3.Benichou B, Strominger J L. Class II-antigen-negative patient and mutant B-cell lines represent at least three, and probably four, distinct genetic defects defined by complementation analysis. Proc Natl Acad Sci USA. 1991;88:4285–4288. doi: 10.1073/pnas.88.10.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bontron S, Ucla C, Mach B, Steimle V. Efficient repression of endogenous major histocompatibility complex class II expression through dominant negative CIITA mutants isolated by a functional selection strategy. Mol Cell Biol. 1997;17:4249–4258. doi: 10.1128/mcb.17.8.4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boss J M. Regulation of transcription of MHC class II genes. Curr Opin Immunol. 1997;9:107–113. doi: 10.1016/s0952-7915(97)80166-5. [DOI] [PubMed] [Google Scholar]
- 6.Bottazzo G F, Todd I, Mirakian R, Belfiore A, Pujol-Borrell R. Organ-specific autoimmunity: a 1986 overview. Immunol Rev. 1986;94:137–169. doi: 10.1111/j.1600-065X.1986.tb01168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brickley W, Wright K, Zhu X, Ting J. Analysis of the defect in INF-gamma induction of MHC class II genes in G1B cells: identification of a novel and functionally critical leucine-rich motif (62-LYLYLQL-68) in the RFX5 transcription factor. J Immunol. 1999;163:6622–6630. [PubMed] [Google Scholar]
- 8.Brocker T. Survival of mature CD4 T lymphocytes is dependent on major histocompatibility complex class II-expressing dendritic cells. J Exp Med. 1997;186:1223–1232. doi: 10.1084/jem.186.8.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buchanan S G, Gay N J. Structural and functional diversity in the leucine-rich repeat family of proteins. Prog Biophys Mol Biol. 1996;65:1–44. doi: 10.1016/s0079-6107(96)00003-x. [DOI] [PubMed] [Google Scholar]
- 10.Bull M, van Hoef A, Gorski J. Transcription analysis of class II human leukocyte antigen genes from normal and immunodeficient B lymphocytes, using polymerase chain reaction. Mol Cell Biol. 1990;10:3792–3796. doi: 10.1128/mcb.10.7.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casper J T, Ash R A, Kirchner P, Hunter J B, Havens P L, Chusid M J. Successful treatment of an HLA-deficient SCID (“bare lymphocyte syndrome”) patient with an unrelated bone marrow transplant. J Pediatr. 1990;116:262–265. doi: 10.1016/s0022-3476(05)82885-9. [DOI] [PubMed] [Google Scholar]
- 12.Charron D J, McDevitt H O. Analysis of HLA-D region-associated molecules with monoclonal antibody. Proc Natl Acad Sci USA. 1979;76:6567–6571. doi: 10.1073/pnas.76.12.6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clausen B, Waldburger J, Schwenk F, Barras E, Mach B, Rajewski K, Förster I, Reith W. Residual MHC class II expression on mature dentritic cells and activated B cells in RFX5-deficient mice. Immunity. 1998;8:143–155. doi: 10.1016/s1074-7613(00)80467-7. [DOI] [PubMed] [Google Scholar]
- 14.Cresswell P. Assembly, transport, and function of MHC class II molecules. Annu Rev Immunol. 1994;12:259–293. doi: 10.1146/annurev.iy.12.040194.001355. [DOI] [PubMed] [Google Scholar]
- 15.Curran J, Boeck R, Lin-Marq N, Lupas A, Kolakofsky D. Paramyxovirus phosphoproteins form homotrimers as determined by an epitope dilution assay, via predicted coiled coils. Virology. 1995;214:139–149. doi: 10.1006/viro.1995.9946. [DOI] [PubMed] [Google Scholar]
- 16.Currie R A. Biochemical characterization of the NF-Y transcription factor complex during B lymphocyte development. J Biol Chem. 1998;273:18220–18229. doi: 10.1074/jbc.273.29.18220. [DOI] [PubMed] [Google Scholar]
- 17.DeSandro A, Nagarajan U M, Boss J M. The bare lymphocyte syndrome: molecular clues to the transcriptional regulation of major histocompatibility complex class II genes. Am J Hum Genet. 1999;65:279–286. doi: 10.1086/302519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durand B, Kobr M, Reith W, Mach B. Functional complementation of MHC class II regulatory mutants by the purified X box binding protein RFX. Mol Cell Biol. 1994;14:6839–6847. doi: 10.1128/mcb.14.10.6839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Durand B, Sperisen P, Emery P, Barras E, Zufferey M, Mach B, Reith W. RFXAP, a novel subunit of the RFX DNA binding complex is mutated in MHC class II deficiency. EMBO J. 1997;16:1045–1055. doi: 10.1093/emboj/16.5.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elhasid R, Etzioni A. Major histocompatibility complex class II deficiency: a clinical review. Blood Rev. 1996;10:242–248. doi: 10.1016/s0268-960x(96)90008-9. [DOI] [PubMed] [Google Scholar]
- 21.Emery P, Durand B, Mach B, Reith W. RFX proteins, a novel family of DNA binding proteins conserved in the eukaryotic kingdom. Nucleic Acids Res. 1996;24:803–807. doi: 10.1093/nar/24.5.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emery P, Strubin M, Hofmann K, Bucher P, Mach B, Reith W. A consensus motif in the RFX DNA binding domain and binding domain mutants with altered specificity. Mol Cell Biol. 1996;16:4486–4494. doi: 10.1128/mcb.16.8.4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fondaneche M C, Villard J, Wiszniewski W, Jouanguy E, Etzioni A, Le Deist F, Peijnenburg A, Casanova J L, Reith W, Mach B, Fischer A, Lisowska-Grospierre B. Genetic and molecular definition of complementation group D in MHC class II deficiency. Hum Mol Genet. 1998;7:879–885. doi: 10.1093/hmg/7.5.879. [DOI] [PubMed] [Google Scholar]
- 24.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glimcher L H, Kara C J. Sequences and factors: a guide to MHC class-II transcription. Annu Rev Immunol. 1992;10:13–49. doi: 10.1146/annurev.iy.10.040192.000305. [DOI] [PubMed] [Google Scholar]
- 26.Guardiola J, Maffei A. Control of MHC class II gene expression in autoimmune, infectious, and neoplastic diseases. Crit Rev Immunol. 1993;13:247–268. [PubMed] [Google Scholar]
- 27.Hanenberg H, Xiao X, Dillo D, Hashino K, Kato I, Williams D. Colocalization of retrovirus and target cells on specific fibronectin fragments increases genetic transduction of mammalian cells. Nat Genet. 1996;2:876–882. doi: 10.1038/nm0896-876. [DOI] [PubMed] [Google Scholar]
- 28.Hasegawa S L, Boss J M. Two B cell factors bind the HLA-DRA X box region and recognize different subsets of HLA class II promoters. Nucleic Acids Res. 1991;19:6269–6276. doi: 10.1093/nar/19.22.6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herrero Sanchez C, Reith W, Silacci P, Mach B. The DNA-binding defect observed in major histocompatibility complex class II regulatory mutants concerns only one member of a family of complexes binding to the X boxes of class II promoters. Mol Cell Biol. 1992;12:4076–4083. doi: 10.1128/mcb.12.9.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janeway C A. Thymic selection: two pathways to life and two to death. Immunity. 1994;1:3–6. doi: 10.1016/1074-7613(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 31.Jang S K, Wimmer E. Cap-independent translation of encephalomyocarditis virus RNA: structural elements of the internal ribosomal entry site and involvement of a cellular 57-kD RNA-binding protein. Genes Dev. 1990;4:1560–1572. doi: 10.1101/gad.4.9.1560. [DOI] [PubMed] [Google Scholar]
- 32.Kara C J, Glimcher L H. In vivo footprinting of MHC class II genes: bare promoters in the bare lymphocyte syndrome. Science. 1991;252:709–712. doi: 10.1126/science.1902592. [DOI] [PubMed] [Google Scholar]
- 33.Kara C J, Glimcher L H. Three in vivo promoter phenotypes in MHC class II deficient combined immunodeficiency. Immunogenetics. 1993;37:227–230. doi: 10.1007/BF00191890. [DOI] [PubMed] [Google Scholar]
- 34.Klein C, Lisowska Grospierre B, LeDeist F, Fischer A, Griscelli C. Major histocompatibility complex class II deficiency: clinical manifestations, immunologic features, and outcome. J Pediatr. 1993;123:921–928. doi: 10.1016/s0022-3476(05)80388-9. [DOI] [PubMed] [Google Scholar]
- 35.Kumar V, Chambon P. The estrogen receptor binds tightly to its responsive element as a ligand-induced homodimer. Cell. 1988;55:145–156. doi: 10.1016/0092-8674(88)90017-7. [DOI] [PubMed] [Google Scholar]
- 36.Liberati C, di Silvio A, Ottolenghi S, Mantovani R. NF-Y binding to twin CCAAT boxes: role of Q-rich domains and histone fold helices. J Mol Biol. 1999;285:1441–1455. doi: 10.1006/jmbi.1998.2384. [DOI] [PubMed] [Google Scholar]
- 37.Lisowska-Grospierre B, Fondaneche M C, Rols M P, Griscelli C, Fischer A. Two complementation groups account for most cases of inherited MHC class II deficiency. Hum Mol Genet. 1994;3:953–958. doi: 10.1093/hmg/3.6.953. [DOI] [PubMed] [Google Scholar]
- 38.Louis-Plence P, Moreno C, Boss J. Formation of a regulatory factor X/X2 box-binding protein/nuclear factor-Y multiprotein complex on the conserved regulatory regions of HLA class II genes. J Immunol. 1997;159:3899–3909. [PubMed] [Google Scholar]
- 39.Mach B, Steimle V, Martinez-Soria E, Reith W. Regulation of MHC class II genes: lessons from a disease. Annu Rev Immunol. 1996;14:301–331. doi: 10.1146/annurev.immunol.14.1.301. [DOI] [PubMed] [Google Scholar]
- 40.Mantovani R. The molecular biology of the CAAT-binding factor NF-Y. Gene. 1999;239:15–27. doi: 10.1016/s0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]
- 41.Mantovani R, Pessara U, Tronche F, Li X Y, Knapp A M, Pasquali J L, Benoist C, Mathis D. Monoclonal antibodies to NF-Y define its function in MHC class II and albumin gene transcription. EMBO J. 1992;11:3315–3322. doi: 10.1002/j.1460-2075.1992.tb05410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Masternak K, Barras E, Zufferey M, Conrad B, Corthals G, Aebersold R, Sanchez J C, Hochstrasser D F, Mach B, Reith W. A gene encoding a novel RFX-associated transactivator is mutated in the majority of MHC class II deficiency patients. Nat Genet. 1998;20:273–277. doi: 10.1038/3081. [DOI] [PubMed] [Google Scholar]
- 43.Moreno C S, Beresford G W, Louis-Plence P, Morris A C, Boss J M. CREB regulates MHC class II expression in a CIITA-dependent manner. Immunity. 1999;10:143–151. doi: 10.1016/s1074-7613(00)80015-1. [DOI] [PubMed] [Google Scholar]
- 44.Moreno C S, Emery P, West J E, Durand B, Reith W, Mach B, Boss J M. Purified X2 binding protein (X2BP) cooperatively binds the class II MHC X box region in the presence of purified RFX, the X box factor deficient in the bare lymphocyte syndrome. J Immunol. 1995;155:4313–4321. [PubMed] [Google Scholar]
- 45.Muhlethaler-Mottet A, Di Berardino W, Otten L A, Mach B. Activation of the MHC class II transactivator CIITA by interferon-gamma requires cooperative interaction between Stat1 and USF-1. Immunity. 1998;8:157–166. doi: 10.1016/s1074-7613(00)80468-9. [DOI] [PubMed] [Google Scholar]
- 46.Muhlethaler-Mottet A, Otten L A, Steimle V, Mach B. Expression of MHC class II molecules in different cellular and functional compartments is controlled by differential usage of multiple promoters of the transactivator CIITA. EMBO J. 1997;16:2851–2860. doi: 10.1093/emboj/16.10.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nagarajan U M, Louis-Plence P, DeSandro A, Nilsen R, Bushey A, Boss J M. RFX-B is the gene responsible for the most common cause of the bare lymphocyte syndrome, an MHC class II immunodeficiency. Immunity. 1999;10:153–162. doi: 10.1016/s1074-7613(00)80016-3. [DOI] [PubMed] [Google Scholar]
- 48.Naldini L. Lentiviruses as gene transfer agents for delivery to non-dividing cells. Curr Opin Biotechnol. 1998;9:457–463. doi: 10.1016/s0958-1669(98)80029-3. [DOI] [PubMed] [Google Scholar]
- 49.Naldini L, Blömer U, Gallay P, Ory D, Mulligan R, Gage F H, Verma I, Trono D. In vivo gene delivery and stable transduction of non dividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 50.Naldini L, Blomer U, Gage F H, Trono D, Verma I M. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc Natl Acad Sci USA. 1996;93:11382–11388. doi: 10.1073/pnas.93.21.11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Page K A, Landau N R, Littman D R. Construction and use of a human immunodeficiency virus vector for analysis of virus infectivity. J Virol. 1990;64:5270–5276. doi: 10.1128/jvi.64.11.5270-5276.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peijnenburg A, Van Eggermond M C, Gobin S J, van den Berg R, Godthelp B C, Vossen J M, van den Elsen P J. Discoordinate expression of invariant chain and MHC class II genes in class II transactivator-transfected fibroblasts defective for RFX5. J Immunol. 1999;163:794–801. [PubMed] [Google Scholar]
- 53.Peijnenburg A, van Eggermond M C, van den Berg R, Sanal O, Vossen J M, van den Elsen P J. Molecular analysis of an MHC class II deficiency patient reveals a novel mutation in the RFX5 gene. Immunogenetics. 1999;49:338–345. doi: 10.1007/s002510050501. [DOI] [PubMed] [Google Scholar]
- 54.Reith W, Steimle V, Lisowska-Grospierre B, Fischer A, Mach B. Molecular basis of major histocompatibility complex class II deficiency. In: Ochs H, Smith C, Puck J, editors. Primary immunodeficiency diseases. New York, N.Y: Oxford University Press; 1999. pp. 167–180. [Google Scholar]
- 55.Reith W, Kobr M, Emery P, Durand B, Siegrist C A, Mach B. Cooperative binding between factors RFX and X2bp to the X and X2 boxes of MHC class II promoters. J Biol Chem. 1994;269:20020–20025. [PubMed] [Google Scholar]
- 56.Reith W, Satola S, Herrero Sanchez C, Amaldi I, Lisowska-Grospierre B, Griscelli C, Hadam M R, Mach B. Congenital immunodeficiency with a regulatory defect in MHC class II gene expression lacks a specific HLA-DR promoter binding protein, RF-X. Cell. 1988;53:897–906. doi: 10.1016/s0092-8674(88)90389-3. [DOI] [PubMed] [Google Scholar]
- 57.Reith W, Siegrist C A, Durand B, Barras E, Mach B. Function of major histocompatibility complex class II promoters requires cooperative binding between factors RFX and NF-Y. Proc Natl Acad Sci USA. 1994;91:554–558. doi: 10.1073/pnas.91.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reith W, Ucla C, Barras E, Gaud A, Durand B, Herrero Sanchez C, Kobr M, Mach B. RFX1, a transactivator of hepatitis B virus enhancer I, belongs to a novel family of homodimeric and heterodimeric DNA-binding proteins. Mol Cell Biol. 1994;14:1230–1244. doi: 10.1128/mcb.14.2.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Riley J L, Westerheide S D, Price J A, Brown J A, Boss J M. Activation of class II MHC genes requires both the X box and the class II transactivator (CIITA) Immunity. 1995;2:533–543. doi: 10.1016/1074-7613(95)90033-0. [DOI] [PubMed] [Google Scholar]
- 60.Rooke R, Waltzinger C, Benoist C, Mathis D. Targeted complementation of MHC class II deficiency by intrathymic delivery of recombinant adenoviruses. Immunity. 1997;7:123–134. doi: 10.1016/s1074-7613(00)80515-4. [DOI] [PubMed] [Google Scholar]
- 61.Scholl P R, Geha R S. MHC class II signaling in B-cell activation. Immunol Today. 1994;15:418–422. doi: 10.1016/0167-5699(94)90271-2. [DOI] [PubMed] [Google Scholar]
- 62.Scholl T, Mahanta S K, Strominger J L. Specific complex formation between the type II bare lymphocyte syndrome-associated transactivators CIITA and RFX5. Proc Natl Acad Sci USA. 1997;94:6330–6334. doi: 10.1073/pnas.94.12.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sedgwick S G, Smerdon S J. The ankyrin repeat: a diversity of interactions on a common structural framework. Trends Biochem Sci. 1999;24:311–316. doi: 10.1016/s0968-0004(99)01426-7. [DOI] [PubMed] [Google Scholar]
- 64.Seidl C, Saraiya C, Osterweil Z, Fu Y P, Lee J S. Genetic complexity of regulatory mutants defective for HLA class-II gene-expression. J Immunol. 1992;148:1576–1584. [PubMed] [Google Scholar]
- 65.Serra E, Zemzoumi K, di Silvio A, Mantovani R, Lardans V, Dissous C. Conservation and divergence of NF-Y transcriptional activation function. Nucleic Acids Res. 1998;26:3800–3805. doi: 10.1093/nar/26.16.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Silvio A, Imbriano C, Mantovani R. Dissection of the NF-Y transcriptional activation potential. Nucleic Acids Res. 1999;27:2578–2584. doi: 10.1093/nar/27.13.2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Steimle V, Durand B, Barras E, Zufferey M, Hadam M R, Mach B, Reith W. A novel DNA binding regulatory factor is mutated in primary MHC class II deficiency (bare lymphocyte syndrome) Genes Dev. 1995;9:1021–1032. doi: 10.1101/gad.9.9.1021. [DOI] [PubMed] [Google Scholar]
- 68.Steimle V, Otten L A, Zufferey M, Mach B. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome) Cell. 1993;75:135–146. [PubMed] [Google Scholar]
- 69.Steimle V, Siegrist C, Mottet A, Lisowska-Grospierre B, Mach B. Regulation of MHC class II expression by interferon-gamma mediated by the transactivator gene CIITA. Science. 1994;265:106–109. doi: 10.1126/science.8016643. [DOI] [PubMed] [Google Scholar]
- 70.Takeda S, Rodewald H R, Arakawa H, Bluethmann H, Shimizu T. MHC class II molecules are not required for survival of newly generated CD4+ T cells, but affect their long-term life span. Immunity. 1996;5:217–228. doi: 10.1016/s1074-7613(00)80317-9. [DOI] [PubMed] [Google Scholar]
- 71.Ting J P Y, Baldwin A S. Regulation of MHC gene expression. Curr Opin Immunol. 1993;5:8–16. doi: 10.1016/0952-7915(93)90074-3. [DOI] [PubMed] [Google Scholar]
- 72.Villard J, Lisowska-Grospierre B, van den Elsen P, Fischer A, Reith A, Mach B. Primary MHC class II deficiency: genetic and molecular definition of a novel complementation group. N Engl J Med. 1997;337:748–753. doi: 10.1056/NEJM199709113371104. [DOI] [PubMed] [Google Scholar]
- 73.Villard J, Reith W, Barras E, Gos A, Morris M A, Antonarakis S E, van den Elsen P J, Mach B. Analysis of mutations and chromosomal localisation of the gene encoding RFX5, a novel transcription factor affected in major histocompatibility complex class II deficiency. Hum Mutat. 1997;10:430–435. doi: 10.1002/(SICI)1098-1004(1997)10:6<430::AID-HUMU3>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 74.Westerheide S D, Boss J M. Orientation and positional mapping of the subunits of the multicomponent transcription factors RFX and X2BP to the major histocompatibility complex class II transcriptional enhancer. Nucleic Acids Res. 1999;27:1635–1641. doi: 10.1093/nar/27.7.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wright K L, Vilen B J, Itoh Lindstrom Y, Moore T L, Li G, Criscitiello M, Cogswell P, Clarke J B, Ting J P. CCAAT box binding protein NF-Y facilitates in vivo recruitment of upstream DNA binding transcription factors. EMBO J. 1994;13:4042–4053. doi: 10.1002/j.1460-2075.1994.tb06721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zeleznik Le N J, Azizkhan J C, Ting J P. Affinity-purified CCAAT-box-binding protein (YEBP) functionally regulates expression of a human class II major histocompatibility complex gene and the herpes simplex virus thymidine kinase gene. Proc Natl Acad Sci USA. 1991;88:1873–1877. doi: 10.1073/pnas.88.5.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zemzoumi K, Frontini M, Bellorini M, Mantovani R. NF-Y histone fold alpha1 helices help impart CCAAT specificity. J Mol Biol. 1999;286:327–337. doi: 10.1006/jmbi.1998.2496. [DOI] [PubMed] [Google Scholar]
- 78.Zhou H, Glimcher L H. Human MHC class II gene transcription directed by the carboxyl terminus of CIITA, one of the defective genes in type II MHC combined immune deficiency. Immunity. 1995;2:545–553. doi: 10.1016/1074-7613(95)90034-9. [DOI] [PubMed] [Google Scholar]
- 79.Zufferey R, Nagy D, Mandel R J, Naldini L, Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]