Abstract
Major Histocompatibility Complex Class II (MHCII) is dynamically expressed on intestinal epithelial cells (IECs) throughout the intestine, but its regulation remains poorly understood. We observed that spontaneous upregulation of IEC MHCII in locally-bred Rag1−/− mice correlated with serum Interleukin (IL)-18, was transferrable via cohousing to commercially-bred immunodeficient mice and could be inhibited by both IL-12 and IL-18 blockade. Overproduction of intestinal IL-18 due to an activating Nlrc4 mutation upregulated IEC MHCII via classical inflammasome machinery independently of immunodeficiency or dysbiosis. Immunodeficient dysbiosis increased Il18 transcription, which synergized with NLRC4 inflammasome activity to drive elevations in serum IL-18. This IL-18-MHCII axis was confirmed in several other models of intestinal and systemic inflammation. Elevated IL-18 reliably preceded MHCII upregulation, suggesting an indirect effect on IECs, and mice with IL-18 overproduction showed activation or expansion of type 1 lymphocytes. Interferon Gamma (IFNg) was uniquely able to upregulate IEC MHCII in enteroid cultures and was required for MHCII upregulation in several in vivo systems. Thus, we have linked intestinal dysbiosis, systemic inflammation, and inflammasome activity to IEC MHCII upregulation via an intestinal IL-18-IFNg axis. Understanding this process may be crucial for determining the contribution of IEC MHCII to intestinal homeostasis, host defense, and tolerance.
Keywords: MHCII, Interleukin-18, enterocyte, Interferon-gamma, dysbiosis, inflammasome
Graphical Abstract
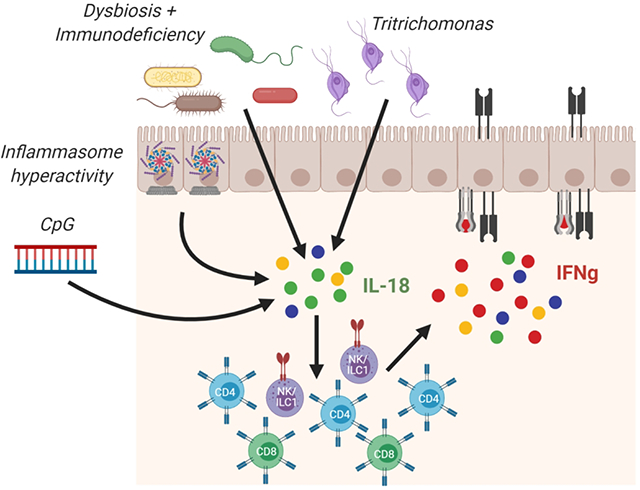
Introduction:
The mammalian intestinal epithelium is a heterogeneous single cell barrier that separates host tissues from a complex microbial ecosystem. It must shepherd vital nutrients systemically from the lumen, exclude commensal microbes, repair itself, and mobilize appropriate immune responses to intestinal pathogens. Among the greatest challenges this barrier faces is negotiating the balance between host defense and immune tolerance. Notably, antigen presentation is critical to both outcomes.
Though the expression of Major Histocompatibility Complex Class II (MHCII) is canonically reserved for “professional” antigen presenting cells, like B- and dendritic cells, Intestinal Epithelial Cells (IECs) are also capable of processing peptides and expressing them on surface MHCII1,2. IEC MHCII appears to be expressed on both luminal and basolateral surfaces, and on a wide variety of IECs from the duodenum through the colon2. Both murine and human studies concur that elevated colonic MHCII expression is observed in gut inflammatory disorders including colitis and Graft versus Host Disease (GvHD), where local production of Interferon Gamma (IFNg) by intestinal T cells appears sufficient to upregulate IEC MHCII1–5.
Despite broad appreciation for the phenomenon, little is known about its regulation or contributions to barrier integrity, host defense, and tolerance. The conventional view of intestinal antigen presentation suggests that dendritic cells (DCs) acquire antigen and migrate to Peyer’s Patches and mesenteric lymph nodes to initiate immune responses to food-, commensal-, and pathogen-derived antigens6,7. Recent studies using conditional deletion of MHCII have largely corroborated this view. For example, intestinal CD11c+ dendritic cells, and not IECs, were found to be required for TH17 responses to Segmented Filamentous Bacteria colonization, as well as bacterial driven-T cell colitis8,9.
However, other studies suggest exceptions to the “professional APC paradigm in the intestine. MHCII expression by Rorc expressing cells was required to prevent development of spontaneous IBD in a microbiota-dependent manner10. The specific role for MHCII expression by IECs remains controversial. Biton et al. showed that IEC MHCII expression was important for epithelial remodeling post infection11. Koyama et al. recently identified that ileal IEC MHCII expression promoted intestinal graft-versus-host responses to cytosolic alloantigens via the microbiota, IL-12, and IFNg1. Consistent with a role for IEC MHCII in gut T-cell activation, mice selectively lacking IEC MHCII developed both more severe infectious (C. rodentium) colitis as well as worsened T-cell transfer or IL-10 receptor blockade colitis (the latter requiring H. hepaticus)4,5.
MHCII expression on traditional APCs can be stimulated by various cytokines and chemokines12. Other than its association with IFNg, little is known about the regulation of IEC MHCII2,12. Specifically, the conditions, chronology, and upstream events leading to IEC MHCII remain largely unknown. Herein, we show that Rag1 deficiency is associated with a transferable dysbiosis that drives MHCII expression on both small and large intestinal epithelial cells. IL-18 and IL-12 are necessary drivers for this IEC MHCII expression. Various genetic models of IL-18 overproduction show that constitutive IL-18 can itself (independently of dysbiosis or immunodeficiency) drive MHCII and supports a role for inflammasome machinery in the regulation of both serum IL-18 and SI IEC MHCII. Specifically, our results support a collaborative model in which microbial induction of Il18 cooperates with inflammasome-mediated proIL-18 maturation to drive changes in serum IL-18, expansion of innate and adaptive type 1 lymphocytes, and IFNg-mediated IEC MHCII. These models, as well as models of systemic inflammation and commensal protist colonization, elucidate a novel mechanism of IEC MHCII regulation via an intestinal IL-18-IL-12/IFNg axis.
Results:
Rag1-deficiency is associated with a microbiota-dependent increase in small and large intestinal epithelial MHCII
Inducible expression of MHCII has been reported on murine small and large intestinal epithelial cells in a variety of circumstances4,5,9. We observed consistently high MHCII expression on Epcam+ colonic and small intestinal (SI) epithelial cells in Rag1−/− mice relative to wild-type (WT) controls (Figure 1a & Supplemental Figure 1). SI IEC MHCII was sometimes observed to be elevated in unstimulated WT mice, suggesting greater sensitivity of SI IECs vs colonic IECs. Treatment of Rag1−/− mice with broad spectrum antibiotics normalized colonic MHCII expression (Figure 1b). Given this microbial association, we compared IEC MHCII in Rag1−/− mice bred in our colony (Pitt) vs mice imported directly from a commercial source (Jackson Laboratory). Imported Rag1−/− mice showed neither colonic nor SI IEC MHCII expression, suggesting a commensal inducer of IEC MHCII present uniquely in our colony (Figure 1c). Though stool from Rag1−/− mice in our colony contained DNA from Segmented Filamentous Bacteria (SFB, a vancomycin-sensitive commensal that can induce IEC MHCII expression), treatment with vancomycin alone did not diminish IEC MHCII (data not shown)13.
Figure 1: Rag1-immunodeficiency is associated with a microbiota-dependent increase in intestinal epithelial cell MHCII.
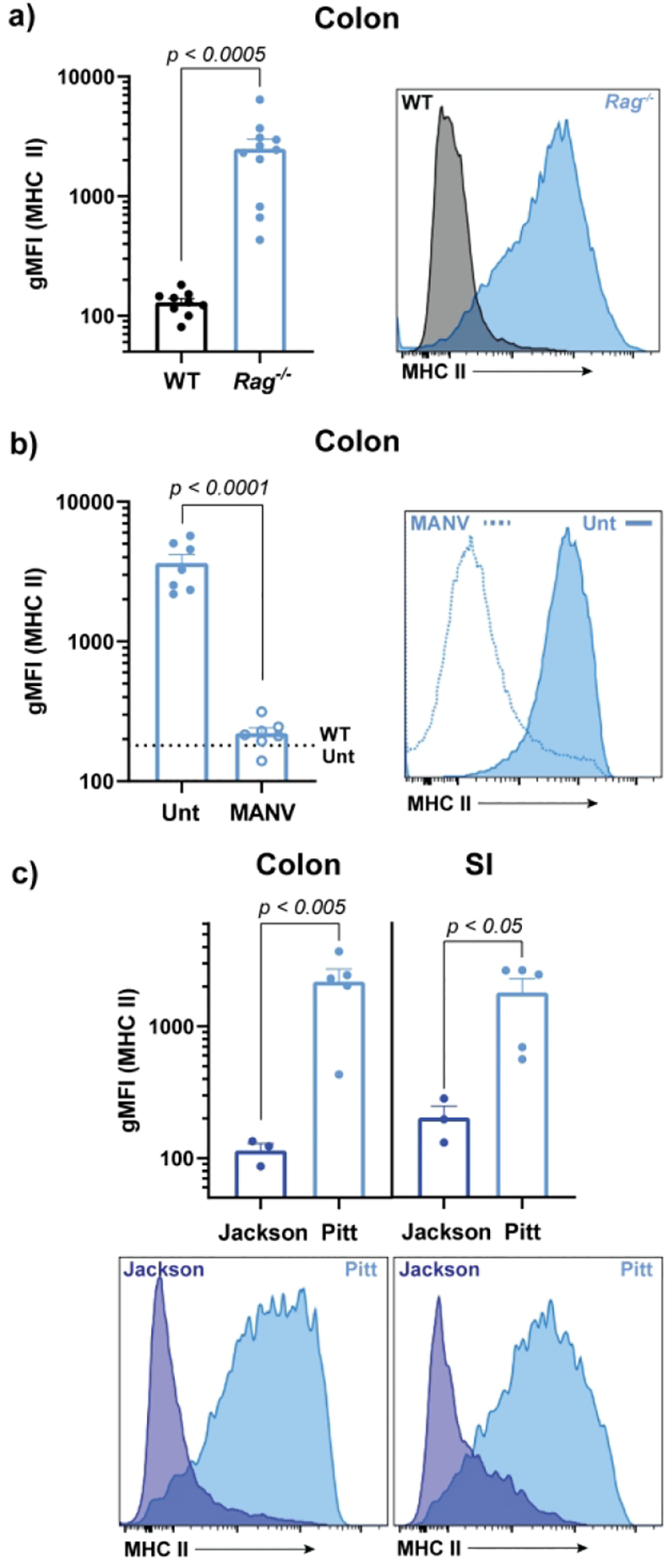
gMFI and representative histograms of colonic and/or SI epithelial cell MHCII in Rag1−/− vs C57Bl/6J mice at baseline (a), following 3-weeks of broad-spectrum antibiotics (b), and in commercially-bred (Jackson) vs locally-bred (Pitt) Rag1−/− mice (c). Combined results of 2-(b) or 3-(a) independent experiments with 3–5 mice per group. Panel c is representative of 3-independent experiments with 2–3 mice per experimental group. Statistical significance is shown on each graph and was determined by unpaired t-tests. Abx=antibiotics, gMFI=geometric mean fluorescent intensity, MANV=antibiotics (metronidazole, ampicillin, neomycin, vancomycin), and Unt=Untreated (no antibiotics)
Elevated IEC MHCII is transferable to immunodeficient, but not immunocompetent hosts
To determine the nature of microbiota-driven IEC MHCII expression in Pitt Rag1−/− mice, we cohoused them for 5 weeks with imported Jackson Rag1−/− mice. Cohousing resulted in a significant increase in IEC MHCII (Figure 2a). To identify unique microbial factors driving changes in MHCII, we performed 16S sequencing on cecal stool. Principal component analysis demonstrated an obvious shift in the Jackson Rag1−/− microbiome towards that of Pitt Rag1−/− cage mates (Figure 2b). Analyzing the specific genera acquired or lost by Rag1−/− mice imported and cohoused (versus imported and housed separately), we found that cohousing was associated with increased abundance of Paraprevotellacea and Helicobacteraceae, and to a lesser extent Deferribacteraceae (Figure 2c). These bacteria are frequently bound by IgA, which is absent in Rag1−/− mice14,15.
Figure 2: Elevated MHCII in Rag1−/− mice is driven by a transferable microbial dysbiosis.
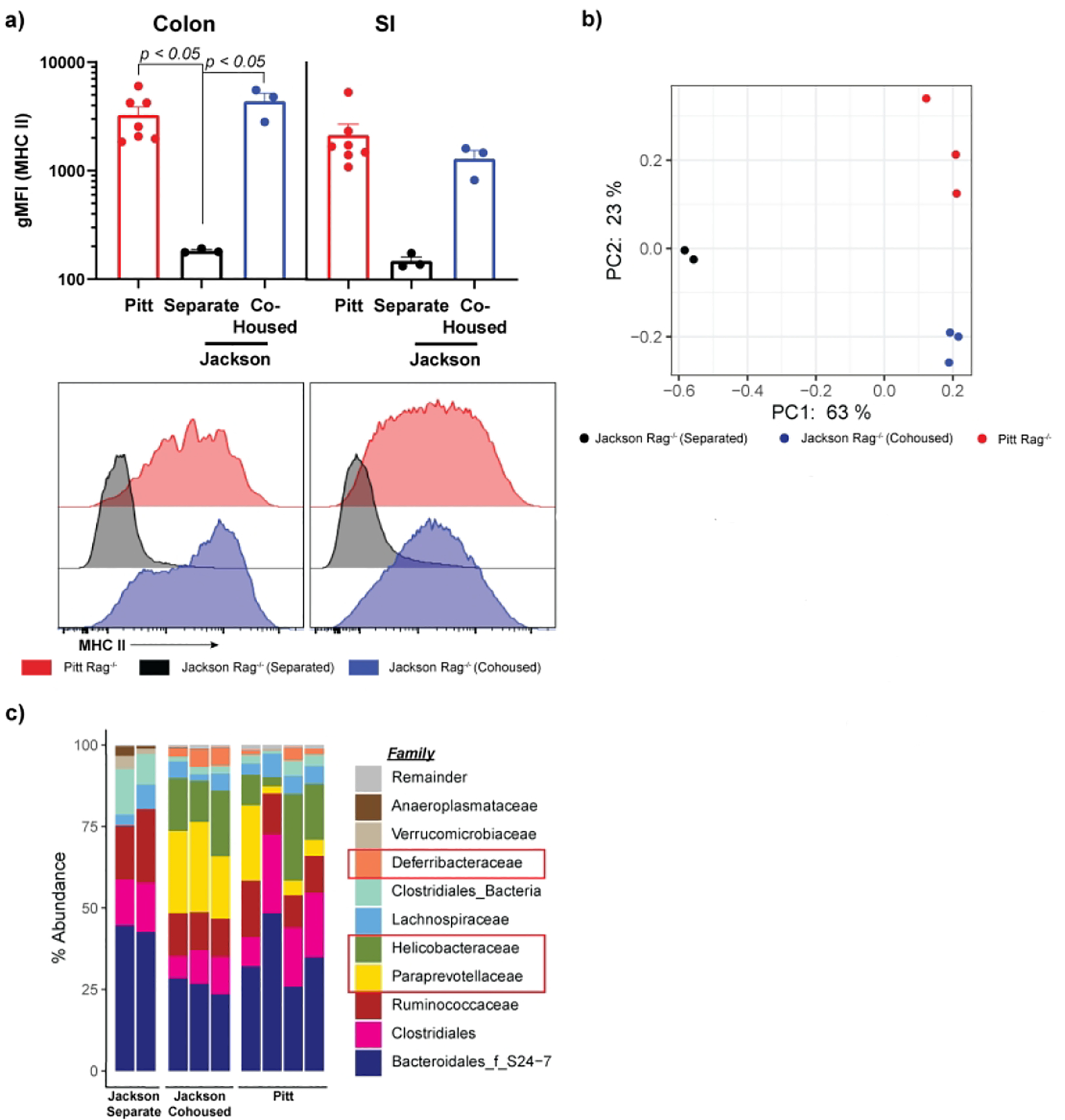
Locally-(Pitt) and commercially-bred (Jackson) Rag1−/− mice were kept isolated or cohoused for 5 weeks. At sacrifice mice were analyzed for epithelial MHCII expression by flow cytometry (a). Each dot represents a single mouse and is representative of two independent experiments with 2–3 mice per condition. Cecal stool was collected and its DNA analyzed by 16S analysis. Jaccard principal coordinate analysis plots of cecal fecal diversity (b) and family diversity (c), for each mouse and is representative of a single experiment. gMFI=geometric mean fluorescent intensity, PC=Principal Coordinate.
To investigate whether the transfer of the MHCII phenotype, and its microbial correlates, were unique to Rag1−/− mice, we housed locally-bred WT mice separately or with Pitt Rag1−/− mice. After five weeks, WT mice had normal colonic MHCII regardless of their housing partners, suggesting induction of IEC MHCII by transferred commensal bacteria occurred only in immunodeficient hosts (Supplemental Figure 2a). This effect was obscured in the SI by spontaneous MHCII upregulation in some WT mice. Though cohoused WT mice mice did not upregulate colonic MHCII, 16s analysis of their cecal contents demonstrated a shift toward the Rag1−/− colonization pattern, with increases in the relative abundance of Helicobacteraceae, Paraprevotellacea and Deferribacteracea upon cohousing (Supplemental Figure 2b and c). Overall, these data suggest that colonization of Rag1-deficient (but not WT) mice with certain commensal bacteria stably upregulates colonic MHCII.
MHCII upregulation correlates with and requires IL-18
We have previously shown a connection between IEC IL-18 production and SI IEC MHCII transcription16. Therefore, we assessed serum IL-18 in Rag1−/− mice and noted elevated serum IL-18 relative to WT controls (Figure 3a). Consistent with our MHCII findings, treatment of Rag1−/− mice with broad spectrum antibiotics returned serum IL-18 to baseline, suggesting that microbial dysbiosis drove both IEC MHCII and elevated IL-18 (Figure 3b). Congruent with our MHCII findings, we also observed that locally-bred Rag1−/− mice had elevated levels of serum IL-18 relative to imported Jackson mice (Figure 3c). Upon cohousing, serum IL-18 in imported Rag1−/−, but not WT mice, rose to that of locally-bred mice, suggesting that IL-18 was induced by a combination of dysbiosis and immunodeficiency and was necessary for MHCII expression (Figure 3d and Supplemental Figure 2d). To confirm a direct effect of IL-18 on MHCII, we treated Rag1−/− mice with an IL-18 receptor blocking antibody and found reduced colonic MHCII expression (Figure 3e). IL-18 canonically acts in synergy with other cytokines, particularly IL-12, to promote type I cytokine production and cytotoxicity17. Consistent with this interaction, a similar downregulation of colonic MHCII was seen following IL12 blockade (Figure 3f). SI MHCII was altered by neither IL-18R nor IL-12 blockade (Supplemental Figure 3a–b). Thus, IL-18 and IL-12 are necessary contributors for microbially-driven colonic IEC MHCII expression in Rag1−/− mice.
Figure 3: Colonic IEC MHCII correlates with serum IL-18 in Rag1−/− mice and can be blunted by IL-18 and IL-12 blockade.
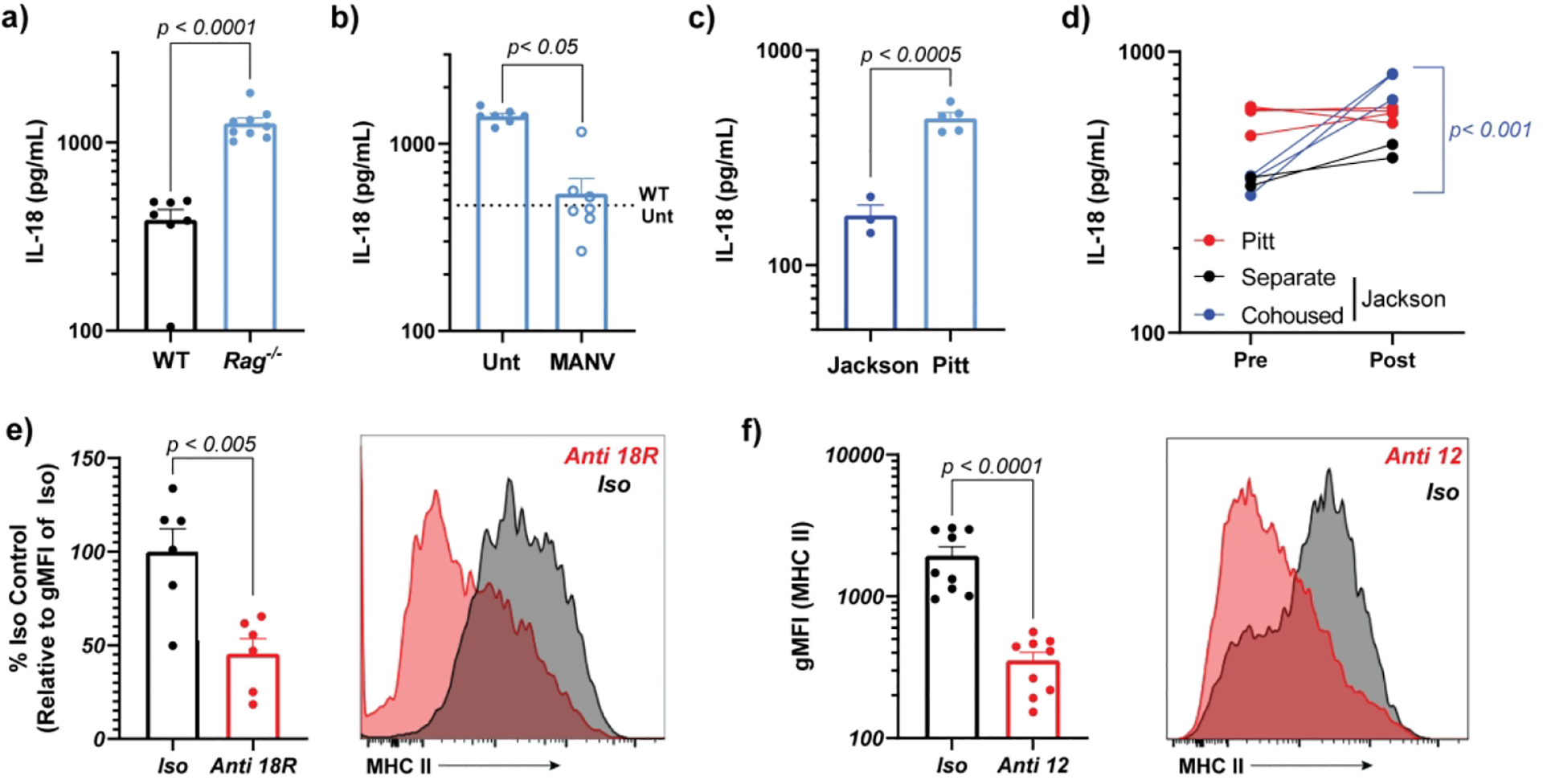
Serum IL-18 in Rag1−/− mice at baseline (a), following antibiotic treatment (b) and in commercially-(Jackson) vs locally-bred (Pitt) mice without (c) or with (d) co-housing (paired analysis). Rag−/− mice were injected every three days with IL-18R (e) or IL-12 (f) blocking antibodies or isotype control and colons analyzed for epithelial MHCII expression. Figures are combined results from 3 of 5 experiments (a), n = 2 experiments (b, e, f) or representative of n=2 experiments (c) with 3–5 mice per condition. To correct for inter-experiment variation in e, gMFI was normalized for each sample to the mean of that experiment’s isotype control group. Statistical significance is shown on each graph and was determined by unpaired t-test (n = 2 groups) or one-way ANOVA (n = 3 groups). Abx=antibiotics, gMFI=geometric mean fluorescent intensity, MANV=antibiotics (Metronidazole, ampicillin, neomycin, vancomycin) and Unt=Untreated (no antibiotics).
Genetic models of IL-18 overproduction cause SI MHCII upregulation independently of the microbiome or immunodeficiency
We have previously shown that mice bearing an activating mutation in the NLRC4 inflammasome (Nlrc4TS/TS) exhibited increased serum IL-18 derived from Intestinal Epithelial Cells (IECs)16. SI IECs from these mice also showed increased proliferation and transcription of genes related to MHCII antigen presentation. We readily detected spontaneous IEC MHCII upregulation on Epcam+ SI IECs (and to a lesser extent colonic IECs) from Nlrc4TS/TS mice (Figure 4a). Contrasting with Rag1−/− mice, Nlrc4TS/TS mice treated with broad spectrum antibiotics (MANV) showed neither diminution of their serum IL-1816 nor a decrease in SI MHCII expression (Figure 4b). As these mice have neither immunodeficiency nor increased IEC Il18 transcription16, this suggests that IEC inflammasome activation, and resultant IL-18 maturation, is sufficient for SI IEC MHCII upregulation.
Figure 4: NLRC4 inflammasome hyperactivity, IL-18bp deficiency, and transgenic IL-18 expression increase SI MHCII expression.
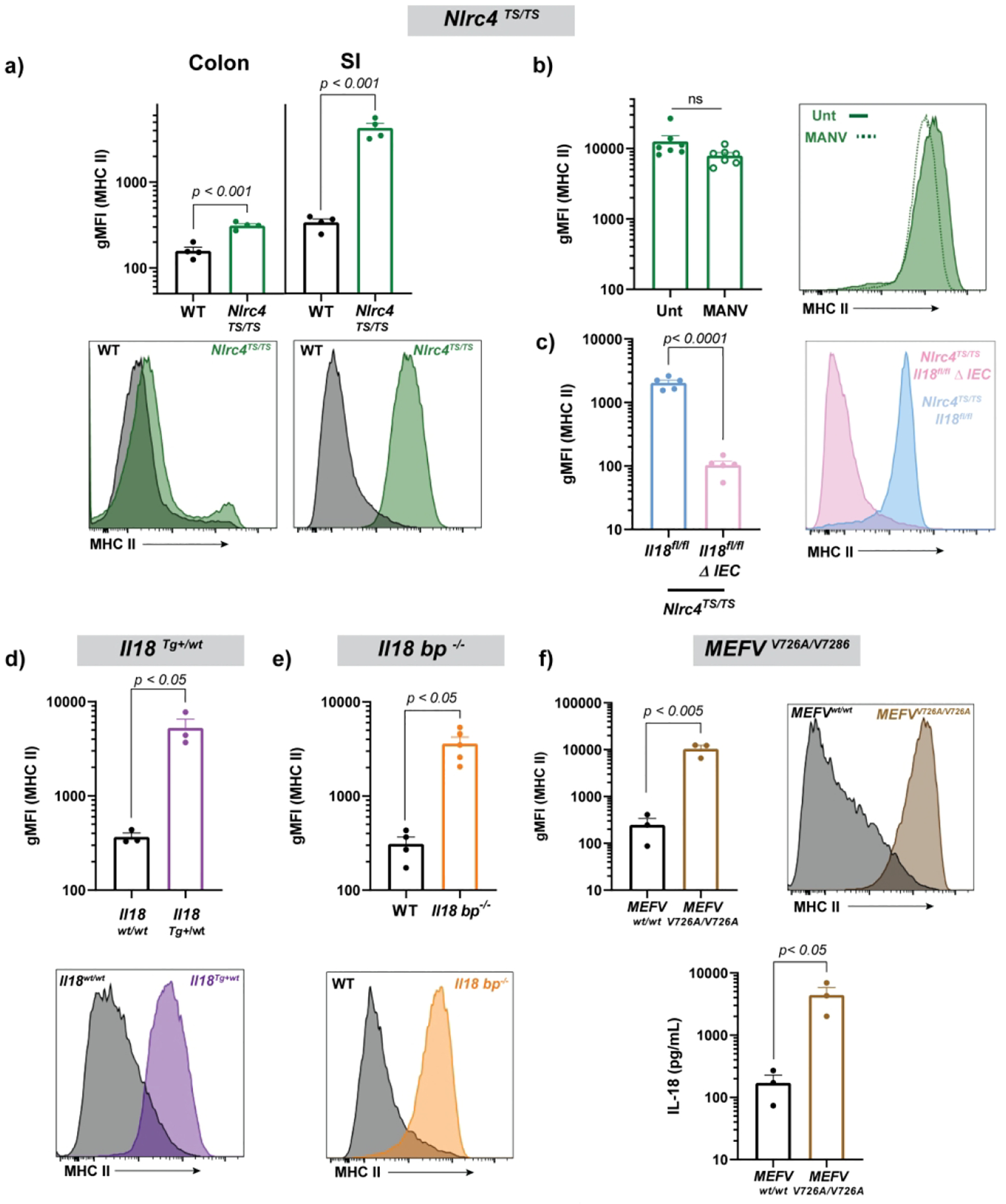
Epithelial MHCII including representative histograms in Nlrc4TS/TS mice at baseline (a), following treatment with broad spectrum antibiotics (b), and with or without deletion of intestinal epithelial Il18 (c). SI MHCII in Il18Tg/wt mice (d), Il18bp−/− mice (e), and MEFV V726A/V726A mice (including serum IL-18) (f). MEFVV726A/V726A mice were assessed at 4 weeks of age due to spontaneous systemic inflammation arising in adulthood. Each symbol represents a single mouse. Representative of n ≥ 3 experiments (a,c), combined results of n = 2 experiments (b, d-f) and representative of a single experiment (h). Statistical significance is shown on each graph and is determined by unpaired t-test (N=2 groups). MANV=antibiotics (Metronidazole, ampicillin, neomycin, vancomycin), gMFI=geometric mean fluorescent intensity, and Unt=Untreated (no antibiotics).
Since IL-18 in Nlrc4TS mice is intestinally-derived16, we directly tested this link by assessing MHCII expression in mice specifically lacking the Il18 gene in Vil1-expressing IECs. Loss of intestinal Il18 normalized both serum IL-1816 and MHCII (Figure 4c). As in Rag1−/− mice, antibody-based blockade of the IL-18R or IL-12 failed to affect SI IEC MHCII expression in Nlrc4TS mice (Supplemental Figure 3c–d).
We hypothesized that other models associated with excess IL-18 would also increase IEC MHCII. We found that mice with transgenic expression of IL-1818, mice lacking IL-18’s endogenous inhibitor IL-18 binding protein (Il18bp−/− mice)19, and mice with genetic hyperactivity of the Pyrin inflammasome (MEFVV726A/V726A)20 all developed elevation of SI MHCII expression, again solidifying the link between IL-18 overproduction and elevated epithelial MHCII (Figure 4d–f).
Caspase-1 and Gasdermin-D are required for basal serum IL-18 and Nlrc4-inflammasome induced elevations of serum IL-18 and IEC MHCII
Elevated serum IL-18 in Nlrc4TS mice is derived from the intestinal epithelium16. To better understand the mechanisms driving epithelial release of IL-18 and subsequent IEC MHCII induction, we evaluated several strains lacking either intestinal Il18 or key components of the inflammasome pathway. Notably, mice with WT Nlrc4 specifically lacking Il18 in intestinal epithelia (Il18fl/fl-ΔIEC) show lower IL-18 levels than Crenegative “WT” controls, suggesting that basal serum IL-18 is intestinally derived (Figure 5a).
Figure 5: Classical inflammasome components are required and synergize with immunodeficiency to modulate Nlrc4-inflammasome induced elevations of serum IL-18 and IEC MHCII.
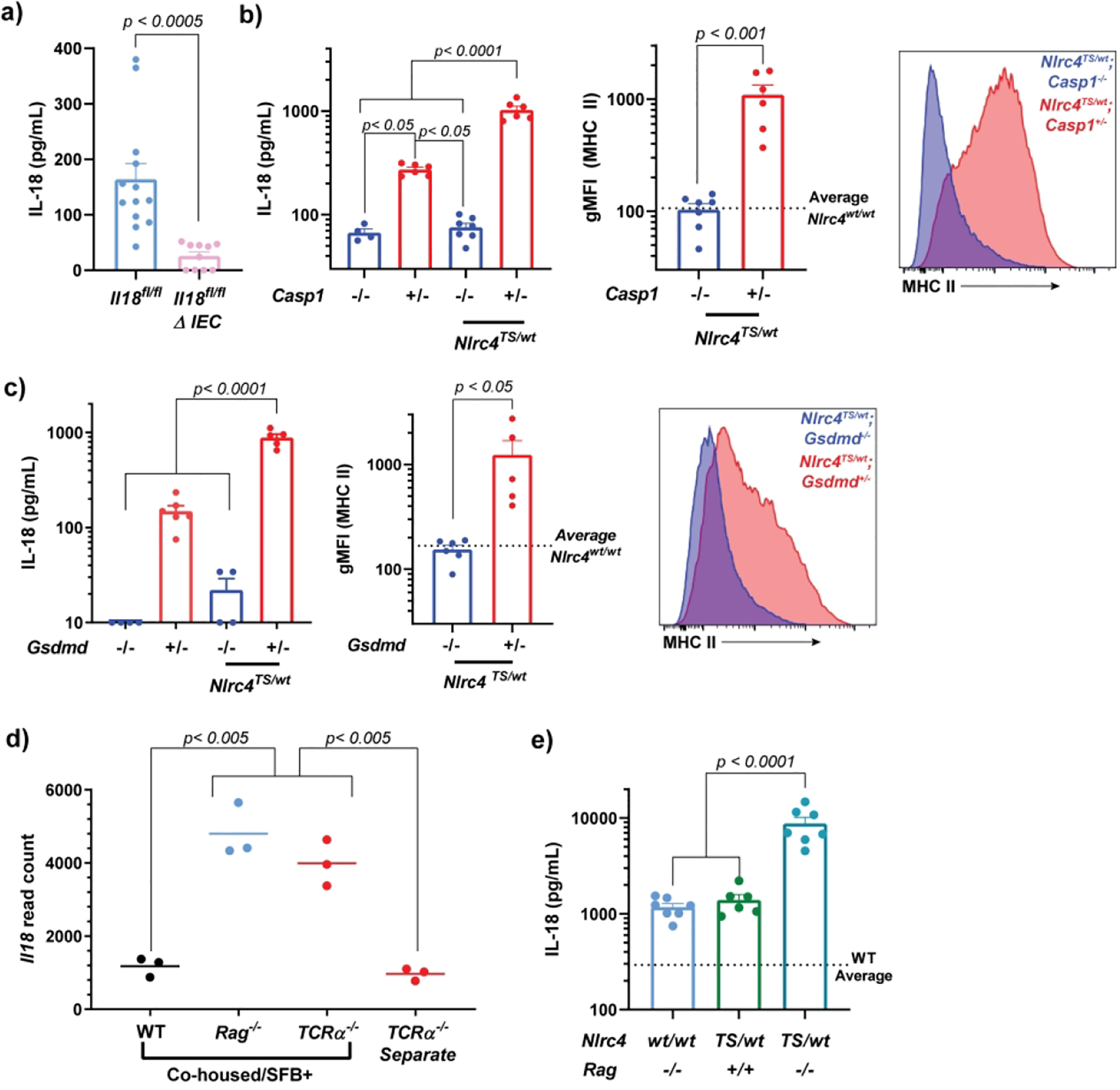
Serum IL-18 and epithelial MHCII including representative histograms in adult mice of the indicated genotypes (a-c). The dotted lines represent the average gMFI of Gasdermin-D or Caspase-1 heterozygous littermate controls. Terminal ileum Il18 read counts in co-housed, Segmented Filamentous Bacteria (SFB)-colonized WT, Rag−/− and TCRa−/− mice compared to separately-housed TCRa−/− mice. Data are derived from Mao et al.24 and accessed from GSE86780 (d). Serum IL-18 in mice bearing combined Nlrc4 hyperactivating and Rag1 mutations (e). Combined results of n=4 (a) or n=2 (e) independent experiments with n > 2 mice per condition. Panels b and c are representative of n=2 independent experiments with n =4–6 mice per genotype. Statistical significance is shown on each graph and is determined by unpaired t-test (N=2 groups) or one-way ANOVA (N=3 groups).gMFI=geometric mean fluorescent intensity, Δ IEC= Villin-Cre+.
Multiple inflammasomes, caspases, and cell death pathways are present in IECs21, and activation of the NLRC4 inflammasome in IECs can trigger Caspase-1 and Caspase-8, both of which can cleave inflammasome substrates22. Serum IL-18 in Casp1−/− mice, even in the presence of the NLRC4TS mutation, was below WT levels, and such mice did not upregulate IEC MHCII (Figure 5b). This suggests there is little/no contribution of Caspase-8 (or any other protease) to either basal or Nlrc4TS-enhnaced IL-18 production or subsequent IEC MHCII induction. Identical results were obtained in mice lacking the pore-forming inflammasome substrate Gasdermin-D23 (Figure 5c). Thus, IEC IL-18 production, basal and NLRC4-driven serum IL-18, and spontaneous IL-18 dependent MHCII upregulation require the classical inflammasome components Caspase-1 and Gasdermin-D.
Rag1-deficiency synergizes with NLRC4 hyperactivity in the elevation of serum IL-18.
Paradoxically, IEC Il18 transcription is slightly decreased in Nlrc4TS mice16. As such, their elevated IL-18 arises due to increased inflammasome-mediated proIL-18 cleavage and export. In locally-bred Rag1−/− mice, we suspected that microbial dysbiosis drove elevated Il18 transcription. Indeed, re-analysis of ileal transcription from Mao et al.24 showed that when SFB-colonized WT, TCRa−/−, and Rag1−/− mice were co-housed, only immunodeficient mice upregulated Il18; separately-housed (SFB-naïve) TCRa−/− mice did not upregulate Il18 (Figure 5d)24. We reasoned that if increased serum IL-18 in Rag1−/− mice is the product of increased Il18 transcription, then combining this mechanism with Nlrc4TS-mediated proIL-18 cleavage should result in (at least) additive IL-18 overproduction. Indeed, Nlrc4TS/WT; Rag1−/− mice had approximately 10-fold higher serum IL-18 than either parental strain, supporting the complementary nature of these mechanisms (Figure 5e).
IL-18 dependent upregulation of IEC MHCII occurs broadly in response to systemic inflammation and Tritrichomonas colonization
We postulated that other stimuli associated with changes in local intestinal and systemic IL-18 would also influence IEC MHCII expression. Repeated systemic stimulation through Toll-Like Receptor (TLR) 9 results in an inflammatory phenotype reminiscent of human macrophage activation syndrome (MAS), which increases serum IL-18 and is dependent on an IL-12-IFNg axis16,25–27. Repeated CpG administration induced high expression of MHCII on colonic and SI IECs (Figure 6a).
Figure 6: Tritrichomonas colonization and systemic TLR9 activation induce IL-18 dependent upregulation of colonic and SI IEC MHCII.
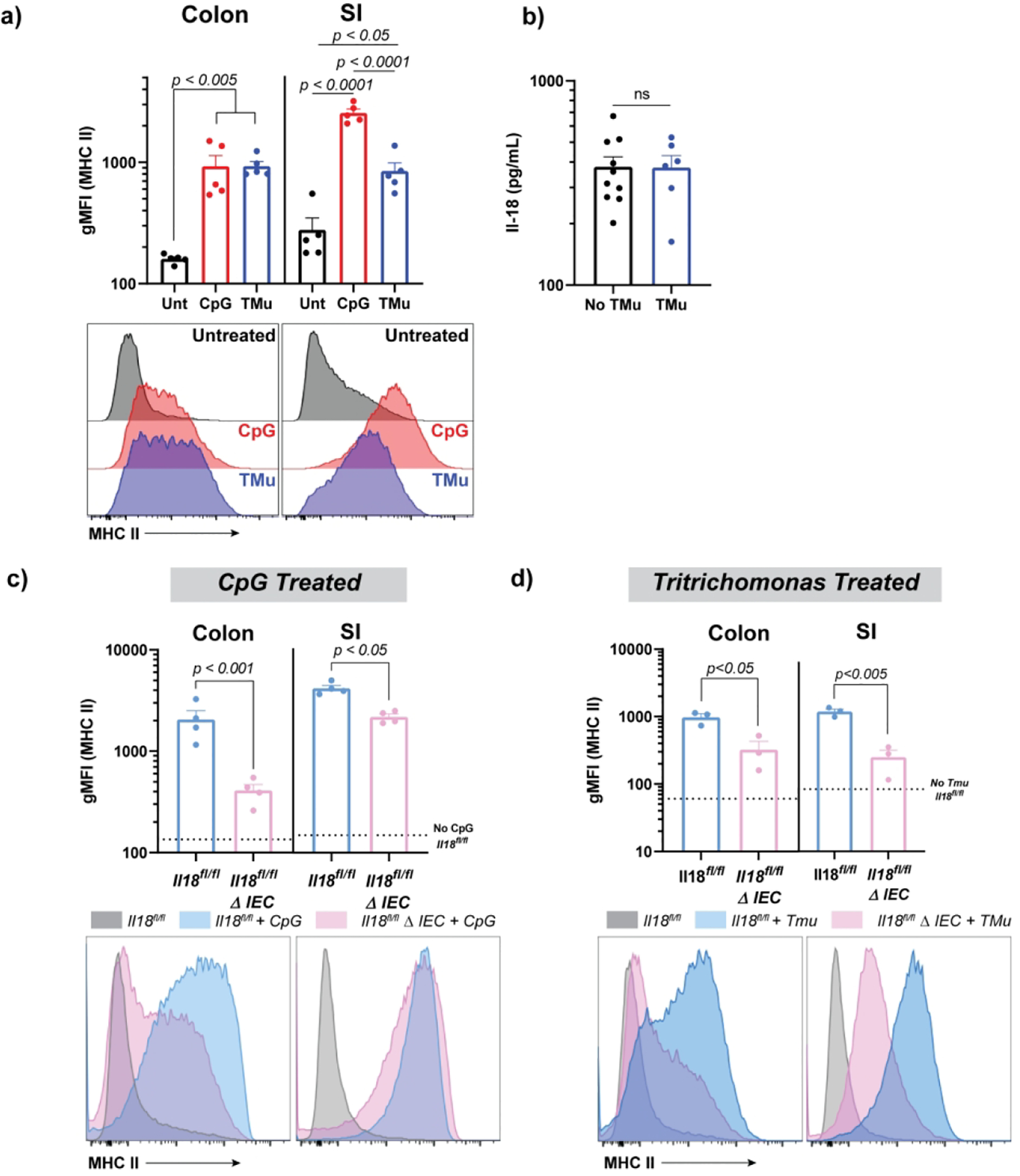
Serum IL-18 and gMFI of epithelial MHCII, including representative histograms in CpG or Tritrichomonas-treated mice (a). WT mice were injected every other day with CpG (ip) or actively gavaged with Tritrichomonas-enriched stool. Two weeks post treatment initiation, mice were sacrificed and analyzed for colonic and small intestinal epithelial MHCII expression by flow cytometry (a) and serum IL-18 by cytometric bead array (b). MHCII expression on small and large intestinal epithelial cells in littermates with or without intestinal epithelial IL-18 treated with two CpG injections (c) or colonized with Tritrichomonas for two weeks (d). Dotted lines represent unstimulated MHCII in Il18fl/fl controls. Each figure is a representative of two independent experiments and each individual mouse is denoted by a datapoint with 3–5 mice per treatment/genotype. Statistical significance is shown on each graph and was determined by unpaired t-test (N = 2 groups) or one-way ANOVA (N=3 groups). TLR=Toll-like receptor, gMFI=geometric mean fluorescent intensity, Unt=untreated, TMu=Tritrichomonas colonized, ΔIEC=Villin-Cre+.
Colonization with the gut-specific commensal protist Tritrichomonas has been associated with intestinal inflammasome activation and lamina propria T-cell IFNg production28,29 as well as Tuft cell activation and induction of type 2 cytokines30. We gavaged WT mice with Tritrichomonas enriched from the cecal contents of a colonized mouse and noted both extensive establishment of Tritrichomonas colonization in recipient animals and robust upregulation of intestinal (particularly colonic) MHCII (Figure 6a). Interestingly, Tritichomonas colonization did not appreciably elevate serum IL-18 (Figure 6b), supporting a remarkably gut-specific immune response. Specific deletion of Il18 in IECs also resulted in decreased IEC MHCII upregulation with either TLR9 stimulation or Tritrichomonas colonization (Figures 6c and d), particularly in the colon.
Elevated IL-18 precedes induction of IEC MHCII
Interestingly, elevated IL-18 occurred around weaning in Rag1−/− mice (~3–4 weeks of age), preceding MHCII induction (6 weeks of age) (Supplemental Figure 4a–b). Despite Nlrc4TS/TS mice elevating serum IL-18 as early as 2-days of age16, they too did not upregulate MHCII until 3–4 weeks of age, suggesting an important role for time/weaning in IL-18-induced MHCII expression (Supplemental Figure 4c). This was largely replicated in ll18bp−/− mice (Supplemental Figure 4d), although they took slightly longer to achieve stable and high MHCII elevation. In addition, 10-day old WT pups treated with CpG showed elevated serum IL-18, but not IEC MHCII (Supplemental Figure 4e).
IFNg is a unique and potent driver of MHCII expression on IECs in vitro
As IECs have little/no expression of the IL-18 or Il-12 receptors (Supplemental Figure 5a) and elevated IL-18 in many of our models precedes the upregulation of MHCII, we hypothesized that IL-18 did not act directly on epithelial cells to upregulate MHCII31. Stimulation of WT small intestinal organoids with high-dose IL-18 (50ng/mL) failed to upregulate MHCII, whereas IFNg stimulation did, even at doses as low as 0.1 ng/mL (Supplemental Figure 5b and data not shown). To determine whether any other stimuli could induce IEC organoid MHCII, we stimulated with various cytokines and pathogen-associated molecules chosen based on their receptor expression in WT IECs16 or their presence within the intestinal microenvironment. Strikingly, only IFNg was able to upregulate MHCII expression on IECs (Supplemental Figure 5b). Similar stimulation of human fetal small intestine organoids showed upregulation of MHCII with IFNg, but not IL-18, LPS, or CpG stimulation (Supplemental Figure 5c) supporting IFNg as a primary driver of MHCII expression in both murine and human IECs.
IEC MHCII expression requires IFNg signaling in vivo
In vivo, T-cell transfer colitis and irradiation1,5 require IFNg to upregulate IEC MHCII. We sought to determine whether IFNg was an absolute requirement for IEC MHCII induction in the variety of IL-18-dependent systems described above. Using Ifng−/− bone marrow chimeras and Ifngr1−/− mice, we found that induction of either SI or colonic IEC MHCII in Rag1−/− (Figure 7a), Nlrc4TS (Figure 7b), TLR9-stimulated (Figure 7c), and Tritrichomonas (Figure 7d) colonized mice required IFNg or the IFNg receptor. Though not comprehensive, these data suggest IFNg is likely to be required for IEC MHCII upregulation regardless of whether the stimulus is acute or chronic, local or systemic.
Figure 7: Diverse in vivo triggers of MHCII expression all require IFNg.
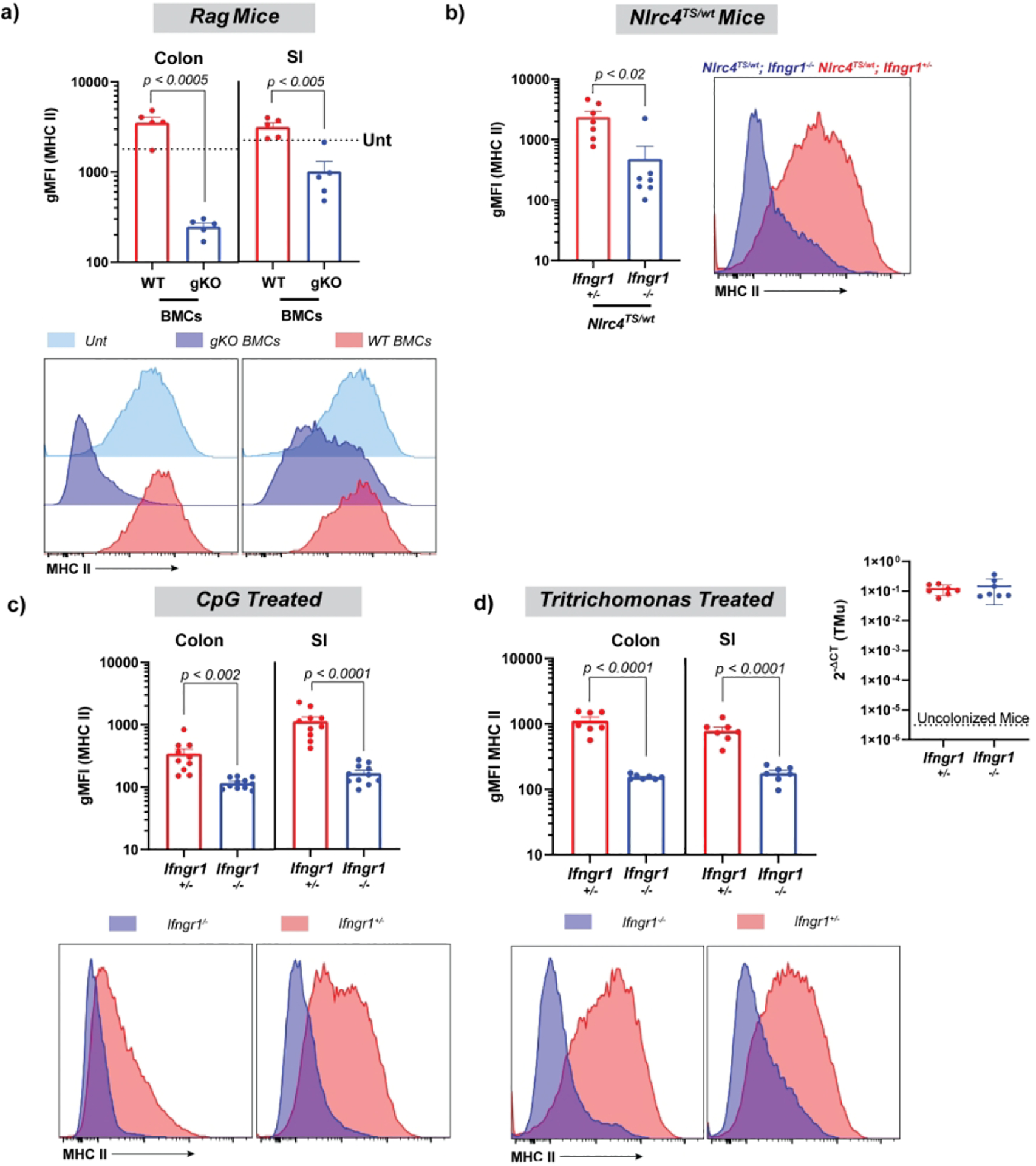
Epithelial cell gMFI and representative histograms in lethally-irradiated Rag1−/− mice reconstituted with either WT or Ifng−/− (gKO) bone marrow (a), NLRC4TS mice with or without Ifngr1 (b), and Ifngr1+/− and Ifngr1−/− littermates treated with CpG (c) or gavaged with Tritrichomonas-enriched stool (d). qPCR showing equal colonization with Tritrichomonas (d, inset). Data shows combined results for two independent experiments, except for b, which shows a representative experiment from n = 2, with 3–5 mice per condition. Statistical significance is shown on each graph and was determined by unpaired t-test. BMCs=bone marrow chimeras, gMFI=geometric mean fluorescent intensity, gKO= Ifng−/−, TMu= Tritrichomonas, Unt=Untreated (no irradiation or reconstitution).
Since IEC MHCII induction requires IFNg but is not observed prior to weaning, we first confirmed that neonatal IECs express the IFNg receptor (Supplemental Figure 4f). As murine intestines are not fully populated with lymphocytes until after weaning24,32, we hypothesized that IL-18 acted on IFNg-producing intestinal lymphocytes to increase IEC MHCII. Neither T- nor B-cells are absolutely required for IEC MHCII upregulation, given the robust responses we observed in Rag1−/− mice. Investigation of colonic lamina propria innate lymphoid cells in Rag1−/− mice did not show an increase in the proportion of colonic NK or Innate Lymphoid Cell (ILC)1 cells, but rather a shift in their expression of KLRG1 consistent with an activated phenotype (Figure 8a–c). Ex vivo stimulation with PMA/Ionomycin resulted in dramatic loss of ILC/NK cells, preventing direct confirmation of their IFNg production. By contrast, examination of SI lamina propria of Nlrc4TS mice showed no change in NK/ILC1 populations, but a dramatic increase in both CD4 and CD8 T-cells (Figures 8d–g). These cells showed a non-significant trend towards more type I differentiation/activation. These data suggest that chronic IL-18 is associated with activation and/or expansion of diverse, and likely redundant, IFNg-competent intestinal lymphocyte populations.
Figure 8: Elevated IL-18 alters lamina propria immune cell composition in Rag1−/− and NLRC4TS/TS mice.
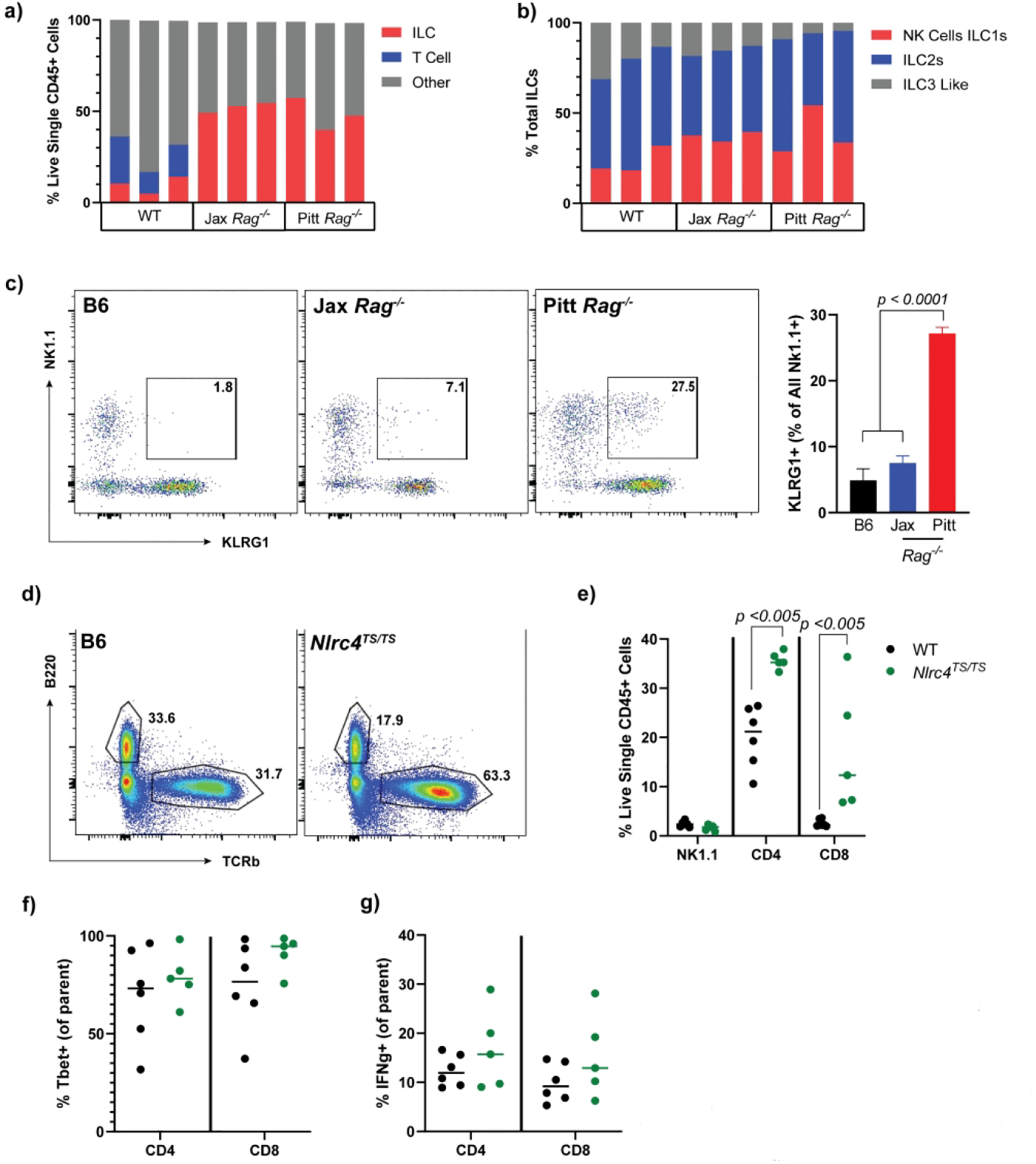
Quantification of lamina propria lymphocytes by flow cytometry. Total live single CD45+ (a) and ILC (b) cell populations in Jackson WT vs Jackson and Pitt Rag1−/− mice. Representative flow plots and quantification showing KLRG1 NK1.1 double positive cells (c). T cell and NK cell composition (d and e) including representative flow plots and Tbet+ cells (f) in in house WT vs NLRC4TS/TS. Isolated ILCs were stimulated with Brefeldin, PMA and ionomycin for 2.5 hours and IFNg expression quantified (g). Panels a-c are representative on n=2 experiments with 3 mice per experimental group. Panels d-g show combined results from n=3 experiments with n=1–2 mice per group. Statistical significance is shown on each graph and was determined by unpaired t-test. Jax= Jackson Rag1−/− mice.
Discussion:
Intestinal epithelial expression of MHCII was described nearly 50 years ago, and has since then largely existed as a phenomenon associated with specific microbial or inflammatory triggers, or its analysis restricted to specific portions of the intestine33. More recent selective deletion and single-cell technologies have ushered a resurgence of interest in the phenomenon. Though many studies implicate dendritic cells as the main sources of productive intestinal antigen presentation8,9,34, others nevertheless highlight a role for IEC MHCII in epithelial remodeling, GvHD, and infectious colitis1,4,5,11. As our appreciation of the functions of IEC MHCII expands, understanding the factors regulating its expression become increasingly important.
We found that Rag1−/− mice in our colony had elevated serum IL-18 and colonic and SI MHCII expression. IL-18 and IEC MHCII were antibiotic-sensitive in Rag1−/− mice, suggesting the activity of specific commensal mediators. Co-housing commercially sourced Rag1−/− mice (low for IL-18 and MHCII) with those locally-bred resulted in a pronounced fecal microbial shift characterized by the increased abundance of Helicobacter, Prevotella, and Mucispirillum (Deferribacteracea) genera. Similar cohousing of WT mice did not induce serum IL-18 or IEC MHCII despite a similar microbial shift, suggesting a role for immunodeficiency in microbial-driven MHCII induction. Though the resolution of this analysis did not permit the identification of specific bacterial species, Helicobacter, Prevotella, and Mucispirillum are all organisms bound strongly by IgA14,15. The interaction of these bacteria and host immunodeficiency may have enabled them to interact more directly with IECs and driven Il18 transcription. A similar induction of Il18 was observed in dysbiotic TCRa−/− mice (Figure 5d), suggesting T-cell functions such as production of barrier-maintaining cytokines, T-cell dependent antibody responses, or limiting the outgrowth of innate lymphoid cells could prevent the dysbiosis and IEC MHCII induction observed in Rag1−/− mice.
Strengthening the link between elevated IL-18 and IEC MHCII, we found that NLRC4 inflammasome hyperactivity drove IEC-dependent IL-18 overproduction and upregulated MHCII and related genes16. We also observed robust IEC MHCII expression on SI, but not colonic cells in other known IL-18 overproducing mouse strains (Il18Tg+/wt, Il18bp−/− and MEFVV726A/V726A), suggesting differential regulation of SI and colon IECs, and confirming a strong link between IL-18 and IEC MHCII. IEC MHCII was IL-18 dependent, as blocking IL-18 signaling or deleting Il18 from IECs significantly decreased MHCII expression in these models.
In NLRC4 hyperactive mice, both elevated serum IL-18 and IEC MHCII depended on IEC production of IL-18, but not microbial factors, suggesting that IL-18 connects intestinal inflammatory stimulation to IEC MHCII16. As in myeloid cells, this intestinal IL-18 was cleaved and released via a classical inflammasome pathway that required Caspase-1 and Gasdermin-D. While other inflammasome components, particularly Caspase-8, have been identified as downstream targets of NLRC421,22, our results leave little room for a contribution by other proteases. Even WT mice depended on Caspase-1, Gasdermin-D, and IEC IL-18 for maintaining basal serum IL-18, demonstrating a requirement for homeostatic intestinal inflammasome activity. Other inflammasome-nucleating proteins present in IECs, like NLRP6 or AIM2, may also be important contributors to basal IL-1835. We also identified an impressive synergy between factors driving IEC Il18 transcription (e.g. dysbiosis and immunodeficiency) and factors promoting IEC inflammasome activation.
We consistently observed differences in the regulation of colonic vs SI IEC MHCII such that genetically-mediated excess of IL-18 or NLRC4 inflammasome activation predominantly affected SI IECs, whereas Tritrichomonas colonization, TLR9 stimulation, and Rag1-deficiency/dysbiosis altered both colonic and SI MHCII. We also observed occasional spontaneous increases in WT SI, but not colonic, IEC MHCII expression. SI MHCII may be more sensitive to transient stimuli (e.g. SFB colonization8,13,36) and also more durable, as blockade of IL-18 or IL-12 via systemic antibody injection was able to alter colonic but not SI IEC expression (Figure 3 and Supplemental Figure 3). Likewise, genetic ablation of IEC Il18 had a more profound effect on colonic vs SI IEC MHCII expression (particularly in the TLR9 and Tritrichomonas systems). These regional differences in IEC MHCII regulation could be for several reasons: a) greater feedback inhibition by microbial products in the colon leading to less sensitivity/durability of MHCII expression, b) a greater abundance of IL-18 responsive/IFNg producing lymphocytes in the SI, or c) other mechanisms36,37.
In various systems, we found that IEC MHCII induction did not occur until about the time of weaning, typically days to weeks after elevation of systemic IL-18. Weaning is a dynamic time in the murine intestine, witnessing an increase in exposure to food antigens, a window where waning luminal IgA from ingested breast milk is not yet replaced by endogenous IgA, a temporary flash of intestinal STAT3 signals, and an influx of intestinal lymphocytes24,32. Food and microbial signals may not be necessary for IEC upregulation of MHCII, as lymphocytes enter the human intestine in utero and human IECs can upregulate MHCII around 18 weeks gestation2,38,39. We speculate that the influx and effector differentiation of lymphocytes competent to produce IFNg may be crucial for driving IEC MHCII in response to local stimuli, particularly IL-18. Obviously, IFNg production by T-cells is not absolutely required given robust IEC MHCII upregulation in dysbiotic Rag1−/− mice. Indeed, we observed activation of colonic NK1.1-expressing ILCs in locally-bred, but not Jackson, Rag-deficient mice. Our analysis was not able to distinguish NK from ILC1 cells, but both express the IL-18 receptor and are competent to produce IFNg. In NLRC4TS mice, the most likely producers of IFNg appeared to be the dramatically-expanded population of (largely) Tbet+ CD4 and CD8 T-cells in the lamina propria. Thus, both T-cells and ILCs may be sufficient IFNg producers for induction of IEC MHCII.
Our mouse and human enteroid studies not only supported an indirect role for IL-18 in driving IEC MHCII, they expanded upon previous findings1,2,5,40 to suggest that IFNg was uniquely capable of inducing IEC MHCII. Why IFNg would drive IEC MHCII even under homeostatic conditions, and why upregulation of IEC MHCII is the unique domain of IFNg, remain mysterious. The latter may relate to a greater diversity of cytokine- and pattern recognition-receptors on classical APCs versus IECs, or to the differential usage of promoters driving the MHC Class II Transactivator (CIITA)5,41. All CIITA promoters are IFNg-responsive, but promoter IV is more commonly used in non-hematopoietic cells and specifically regulates IFNg-inducible CIITA expression42.
Overall, these data link the diverse mechanisms contributing to intestinal IL-18 to local production of IFNg and geographically-diverse upregulation of IEC MHCII. They provide a uniquely broad picture of the regulation of this energetically costly process, but questions pertaining to the significance of IEC MHCII in barrier integrity, host defense, and tolerance induction remain open. As these functions gain greater clarity, understanding IEC MHCII regulation will be critical for identifying and correcting pathology related to its dysregulation and exploiting this process to treat intestinal disease.
Materials and Methods:
Mice:
All mice were bred and manipulated under specific pathogen free conditions in 12-hour light-dark cycles with food and water available ad libitum. All procedures were approved by the University of Pittsburgh animal care and use committee. All mice are on a C57Bl/6 background and originated from Jackson Laboratories, except the Il-18Tg+/wt (Dr. Tomoaki Hoshino, Kurume University), MEFVV726A/V726A mice (Drs. Daniel Kastner and Jae Jin Chae, NHGRI), Il18bp−/− (Knockout Mouse Project), Il18fl/fl (Dr. Richard Flavell, Yale University) and the NLRC4TS/TS16,18–20,43. IFNgKO bone marrow was a generous gift from Drs. Dario Vignali and Warren Shlomchik (University of Pittsburgh). All treatments were conducted on 6 week or older mice, except where noted.
Murine Manipulation Studies:
Epithelial and Lamina Propria Isolations and Flow Cytometry:
To obtain intestinal scrapes, mice were sacrificed and their colon and ~10 cm of small intestine (~5 cm distal to the stomach) were removed, colons were flushed with PBS, and organs flayed open. Sections were rinsed with PBS and the epithelial surfaces scraped with a sterile slide into cold DMEM with 2% FCS and 2mM EDTA. Supplemental Figure 6 shows the flow gating strategy for the colonic epithelium. Lamina propria cells were isolated according to described protocols44 and stimulated for 2.5 hours with PMA/ionomycin and Brefeldin A to study cytokine secretion. Filtered single cells suspensions were stained with anti-mouse antibodies against CD3(OTK3), CD4(RM4–5), CD8b(YTS 156.7.7), CD45.2(104), CD90(53–2.1), B220(RA3–6B2), Epcam(G8.8), Fixable Live Dead Stain (Thermofisher, Waltham, MA), IFNg(XMG1.2), KLRG1(2F1), NK1.1 (pk136), MHCII(M5/114.152), RORgT(2B2), Tbet(4B10), TCRb(H57–597), TCRgd(eBioGL3), and/or unlabeled CD16/32(93) for 30–60 minutes at 4°C. Cells were fixed prior flow and/or the addition of intracellular antibodies. Samples were run on an LSR Fortessa (BD Biosciences, San Jose, CA) and data analyzed using FlowJo v10.6 (Treestar, Ashland, OR) software. All antibodies were purchased from eBiosciences (San Diego, CA) or Biolegend (San Diego, CA).
CpG:
CpG1826 (IDT Technologies) was injected at a dose of 2.5 mg/kg intraperitoneally (ip) as done previously26. A total of 2–5 CpG injections, spaced two days apart, were given with takedown 24 hours after the last injection. For the pup experiments, 5-day old mice were injected daily with 0.125 mg CpG for 4 days and sacrificed on Day 5.
Antibiotic Treatments:
Adult mice were transferred to fresh cage bases and given drinking water supplemented with metronidazole (1 g L−1), ampicillin (1 g L−1), neomycin (1 g L−1) and vancomycin (0.5 g L−1) for three weeks. Control and antibiotic water were sweetened with saccharine 6g/500 mL.
IL-12/IL-18 Blockade:
Adult Rag1−/− and Nlrc4TS mice were injected ip with 0.25–0.5 mg/mouse Anti-IL-18R Clone 9E6 (IL-1F4(mu):10235, Genentech, San Francisco, CA) or 0.2–0.4 mg/mouse (Anti-Mouse IL-12p40 Clone 17.8, Bio X Cell, Lebanon, NH) every 3 days for two weeks. To account for variability in MHCII fluorescence between the IL18-R block experiments, MHCII gMFI was calculated as a percentage of the average of the isotype treated controls in each experiment.
Tritrichomonas Treatment, Isolation, and qPCR:
Tritrichomonas was identified during routine import testing at the University of Pittsburgh and confirmed to be T. muris or musculis based on qPCR amplification45. Cecal stool from colonized mice was isolated and Tritrichomonads were enriched by centrifugation as done previoulsy29 prior to gavage. Mice were sacrificed and analyzed for IEC MHCII and serum IL-18 two weeks post-colonization.
BMCs:
Rag1−/− mice were lethally irradiated with 1100 cGy (split dose, 2 hours apart) in an XRAD320 (Precision Xray Inc) irradiator. The following day mice were reconstituted with 4–6 million WT or IFNg−/− bone marrow cells. Intestinal scrapes were collected 4–6 weeks post treatment initiation.
Serum IL-18 Measurements:
Serum was collected from terminal bleeds and analyzed using a BD Cytometric Bead Array according to the manufacturer’s instructions, run on the BD LSR II, and analyzed using FCAP Array Software (BD Biosciences, v. 3.0, San Jose, CA).
Organoid Cultures:
Murine:
WT organoid cultures were generated from small intestinal samples according to the manufacturer’s protocol (Stem Cell Technologies, Vancouver, Canada) in IntestiCult™ Organoid Growth Medium. Organoids were routinely passaged and maintained in Matrigel domes every 7–10 days. For TLR and cytokine stimulation, mature organoids were seeded at a density of ~300 organoids/well in a 12 well dish. Following organoid budding (3–5 days post passage), the media was removed and replaced with TLR/cytokine supplemented media and the samples incubated at 37°C for 3.5 days. Organoids were removed, stained, and analyzed as described above for the IEC scrapes. All murine cytokines were purchased from Peprotech (Rockyhill, NJ) and used at a concentration of 10 ng/mL, apart from IL-18 (MBL International, 50ng/mL) and IL-2 (Miltenyi, Germany). See Supplemental Table 1 for concentration and source of the TLR ligands analyzed.
Human:
Small intestinal enteroids were isolated from surgically resected fetal (< 24 weeks) small intestinal tissue according to described protocols46,47. Samples were procured from the Health Science Tissue Bank of UPMC Magee-Womens Hospital in accordance with the guidelines set forth by the University of Pittsburgh Ethics Committee. Briefly, 100 crypts were plated in Matrigel in 48 well plates containing a 50:50 vol:vol ratio of human intestinal stem cells media (plus 50 mM EGF)47 and WRN-conditioned media48. Three days post plating the media was removed and replaced with TLR/Cytokines supplemented media (same concentrations as the murine organoids). Organoids were harvested 3.5 days post stimulation according to the methods described in40 and stained and analyzed for IEC MHCII surface expression.
Cohousing and 16S Sequence Analysis:
Three to five-week old Rag1−/− mice bred locally or purchased commercially (Jackson labs) and/or WT mice were cohoused for 5 weeks prior to sacrifice. Cecal stool samples were collected at time of death and frozen until processing. Fecal DNA was extracted using the QiaAMP Stool DNA Mini Kit (Qiagen, Germantown, MD) according to the manufacturer’s instructions and sequencing performed by Microbiome Insights (University of British Columbia, Canada). Microbiome informatics were performed by AHPB using QIIME2 2020.249. Raw sequences were quality-filtered and denoised with DADA250. Amplicon sequence variants (ASVs) were aligned with mafft and used to construct a phylogeny with fasttree251,52. Alpha diversity metrics (observed OTUs), beta diversity metrics (Jaccard Similarity) and Principal Coordinate Analysis (PCoA) were estimated after samples were rarefied to 63,000 (subsampled without replacement) sequences per sample. Taxonomy was assigned to ASVs using naive Bayes taxonomy classifier against the Greengenes 18_8 99% OTUs reference sequences53. All plots were made with publicly available R packages. Full sequence data is available upon request.
Analysis of Publicly Available Data Sets
Publicly available data sets were obtained from the Gene Expression Omnibus. Briefly, terminal ileum Il18 read counts in co-housed, Segmented Filamentous Bacteria (SFB)-colonized WT, Rag−/− and TCRa−/− mice were obtained from Mao et al. and accessed from GSE8678024. Normalized read counts from bulk RNA-seq of FACS-sorted IECs from neo-natal intestine derived from mice four hours after subcutaneous injection of saline54. t-SNE plot (perplexity=25) colored by inferred cell type or gene expression of select murine cytokine receptors from Haber et al31 visualized in Single Cell Expression Atlas.
Statistics:
Unpaired Student T tests or One-Way ANOVAs with Tukey post correction were performed in GraphPad Prism (Version 8.3.1) as identified in the figure legends. Results were considered significant if p < 0.05.
Supplementary Material
Acknowledgements:
SC, CS, and VD were supported by NICHD R01HD098428. LVDK and AHPB were supported by Autoimmunity and Immunopathology T32 (5T32AI089443-11). TH was supported by the Kenneth Rainin Foundation and R01DK120697. We also thanks Drs. Daniel Kastner and Jae Jin Chae (NHGRI); Dario Vignali, Warren Shlomchik, and Sarah Gaffen (University of Pittsburgh); Tomoaki Hoshino (Kurume University); and Richard Flavell (Yale University) for sharing key reagents.
Disclosures:
SC has received research support for a clinical trial from AB2Bio, Ltd.
Footnotes
No other authors report a COI
References:
- 1.Koyama M et al. MHC Class II Antigen Presentation by the Intestinal Epithelium Initiates Graft-versus-Host Disease and Is Influenced by the Microbiota. Immunity 51, 885–898 e887 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wosen JE, Mukhopadhyay D, Macaubas C & Mellins ED Epithelial MHC Class II Expression and Its Role in Antigen Presentation in the Gastrointestinal and Respiratory Tracts. Front Immunol 9, 2144 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hershberg RM et al. Intestinal epithelial cells use two distinct pathways for HLA class II antigen processing. J Clin Invest 100, 204–215 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jamwal DR et al. Intestinal Epithelial Expression of MHCII Determines Severity of Chemical, T Cell-Induced, and Infectious Colitis in Mice. Gastroenterology 159, 1342–1356 e1346 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thelemann C et al. Interferon-gamma induces expression of MHC class II on intestinal epithelial cells and protects mice from colitis. PLoS One 9, e86844 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bekiaris V, Persson EK & Agace WW Intestinal dendritic cells in the regulation of mucosal immunity. Immunol Rev 260, 86–101 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Grainger JR, Askenase MH, Guimont-Desrochers F, da Fonseca DM & Belkaid Y Contextual functions of antigen-presenting cells in the gastrointestinal tract. Immunol Rev 259, 75–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goto Y et al. Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity 40, 594–607 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maggio-Price L et al. Lineage targeted MHC-II transgenic mice demonstrate the role of dendritic cells in bacterial-driven colitis. Inflamm Bowel Dis 19, 174–184 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hepworth MR et al. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature 498, 113–117 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biton M et al. T Helper Cell Cytokines Modulate Intestinal Stem Cell Renewal and Differentiation. Cell 175, 1307–1320 e1322 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ting JP & Trowsdale J Genetic control of MHC class II expression. Cell 109 Suppl, S21–33 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Umesaki Y, Okada Y, Matsumoto S, Imaoka A & Setoyama H Segmented filamentous bacteria are indigenous intestinal bacteria that activate intraepithelial lymphocytes and induce MHC class II molecules and fucosyl asialo GM1 glycolipids on the small intestinal epithelial cells in the ex-germ-free mouse. Microbiol Immunol 39, 555–562 (1995). [DOI] [PubMed] [Google Scholar]
- 14.Bunker JJ & Bendelac A IgA Responses to Microbiota. Immunity 49, 211–224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palm NW et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158, 1000–1010 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weiss ES et al. Interleukin-18 diagnostically distinguishes and pathogenically promotes human and murine macrophage activation syndrome. Blood 131, 1442–1455 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poznanski SM et al. Combined Stimulation with Interleukin-18 and Interleukin-12 Potently Induces Interleukin-8 Production by Natural Killer Cells. J Innate Immun 9, 511–525 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoshino T et al. Cutting edge: IL-18-transgenic mice: in vivo evidence of a broad role for IL-18 in modulating immune function. J Immunol 166, 7014–7018 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Girard-Guyonvarc’h C et al. Unopposed IL-18 signaling leads to severe TLR9-induced macrophage activation syndrome in mice. Blood 131, 1430–1441 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Chae JJ et al. Gain-of-function Pyrin mutations induce NLRP3 protein-independent interleukin-1beta activation and severe autoinflammation in mice. Immunity 34, 755–768 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vance RE The NAIP/NLRC4 inflammasomes. Curr Opin Immunol 32, 84–89 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rauch I et al. NAIP-NLRC4 Inflammasomes Coordinate Intestinal Epithelial Cell Expulsion with Eicosanoid and IL-18 Release via Activation of Caspase-1 and −8. Immunity 46, 649–659 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Opdenbosch N & Lamkanfi M Caspases in Cell Death, Inflammation, and Disease. Immunity 50, 1352–1364 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mao K et al. Innate and adaptive lymphocytes sequentially shape the gut microbiota and lipid metabolism. Nature 554, 255–259 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Canna SW et al. Interferon-gamma mediates anemia but is dispensable for fulminant toll-like receptor 9-induced macrophage activation syndrome and hemophagocytosis in mice. Arthritis Rheum 65, 1764–1775 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Behrens EM et al. Repeated TLR9 stimulation results in macrophage activation syndrome-like disease in mice. J Clin Invest 121, 2264–2277 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsoukas P et al. Interleukin-18 and cytotoxic impairment are independent and synergistic causes of murine virus-induced hyperinflammation. Blood (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chudnovskiy A et al. Host-Protozoan Interactions Protect from Mucosal Infections through Activation of the Inflammasome. Cell 167, 444–456 e414 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Escalante NK et al. The common mouse protozoa Tritrichomonas muris alters mucosal T cell homeostasis and colitis susceptibility. J Exp Med 213, 2841–2850 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schneider C et al. A Metabolite-Triggered Tuft Cell-ILC2 Circuit Drives Small Intestinal Remodeling. Cell 174, 271–284 e214 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haber AL et al. A single-cell survey of the small intestinal epithelium. Nature 551, 333–339 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Al Nabhani Z et al. A Weaning Reaction to Microbiota Is Required for Resistance to Immunopathologies in the Adult. Immunity 50, 1276–1288 e1275 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Skoskiewicz MJ, Colvin RB, Schneeberger EE & Russell PS Widespread and selective induction of major histocompatibility complex-determined antigens in vivo by gamma interferon. J Exp Med 162, 1645–1664 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stagg AJ Intestinal Dendritic Cells in Health and Gut Inflammation. Front Immunol 9, 2883 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ratsimandresy RA, Indramohan M, Dorfleutner A & Stehlik C The AIM2 inflammasome is a central regulator of intestinal homeostasis through the IL-18/IL-22/STAT3 pathway. Cell Mol Immunol 14, 127–142 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bowcutt R et al. Heterogeneity across the murine small and large intestine. World J Gastroenterol 20, 15216–15232 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olivares-Villagomez D & Van Kaer L Intestinal Intraepithelial Lymphocytes: Sentinels of the Mucosal Barrier. Trends Immunol 39, 264–275 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stras SF et al. Maturation of the Human Intestinal Immune System Occurs Early in Fetal Development. Dev Cell 51, 357–373 e355 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Torow N, Marsland BJ, Hornef MW & Gollwitzer ES Neonatal mucosal immunology. Mucosal Immunol 10, 5–17 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Wosen JE et al. Human Intestinal Enteroids Model MHC-II in the Gut Epithelium. Front Immunol 10, 1970 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pai RK, Askew D, Boom WH & Harding CV Regulation of class II MHC expression in APCs: roles of types I, III, and IV class II transactivator. J Immunol 169, 1326–1333 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Morris AC, Beresford GW, Mooney MR & Boss JM Kinetics of a gamma interferon response: expression and assembly of CIITA promoter IV and inhibition by methylation. Mol Cell Biol 22, 4781–4791 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nowarski R et al. Epithelial IL-18 Equilibrium Controls Barrier Function in Colitis. Cell 163, 1444–1456 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hall JA et al. Essential role for retinoic acid in the promotion of CD4(+) T cell effector responses via retinoic acid receptor alpha. Immunity 34, 435–447 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Howitt MR et al. Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science 351, 1329–1333 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Drummond CG et al. Enteroviruses infect human enteroids and induce antiviral signaling in a cell lineage-specific manner. Proc Natl Acad Sci U S A 114, 1672–1677 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sato T et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 141, 1762–1772 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Miyoshi H & Stappenbeck TS In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat Protoc 8, 2471–2482 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bolyen E et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37, 852–857 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Callahan BJ et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13, 581–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katoh K, Misawa K, Kuma K & Miyata T MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30, 3059–3066 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Price MN, Dehal PS & Arkin AP FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One 5, e9490 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McDonald D et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6, 610–618 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Winkle JA, Constant DA, Li L & Nice TJ Selective Interferon Responses of Intestinal Epithelial Cells Minimize Tumor Necrosis Factor Alpha Cytotoxicity. J Virol 94 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


