Abstract
RNA viruses cause many routine illnesses, such as the common cold and the flu. Recently, more deadly diseases have emerged from this family of viruses. The hepatitis C virus has had a devastating impact worldwide. Despite the cures developed in the U.S. and Europe, economically disadvantaged countries remain afflicted by HCV infection due to the high cost of these medications. More recently, COVID-19 has swept across the world, killing millions and disrupting economies and lifestyles; the virus responsible for this pandemic is a coronavirus. Our understanding of HCV and SARS CoV-2 replication is still in its infancy. Helicases play a critical role in the replication, transcription and translation of viruses. These key enzymes need extensive study not only as an essential player in the viral lifecycle, but also as targets for antiviral therapeutics. In this review, we highlight the current knowledge for RNA helicases of high importance to human health.
Keywords: SARS CoV-2, Helicase, NSP13, NS3, DEAD-box, DDX3, Hepatitis C, HCV, Inhibitors
1. Introduction
Helicases are molecular motor proteins that manipulate DNA and RNA [1]. They hydrolyze ATP and transduce chemical energy to move along the DNA, separating the double-stranded (ds) DNA, or removing proteins that might block progress of replication, transcription, or repair of DNA [2], [3], [4]. RNA metabolism such as splicing and ribosome assembly requires multiple RNA helicases. One or more helicases are necessary for virtually all aspects of nucleic acid metabolism [5]. Helicases are categorized into superfamilies (SFs) based on sequence, with SF1 and SF2 harboring the largest membership [6]. They are found in most viruses, including those that threaten human health such as Hepatitis C virus and SARS CoV-2. SF1 and SF2 are further categorized as moving along DNA or RNA in a 5′-to-3′ (SF1B) or 3′-to-5′ (SF1A) direction.
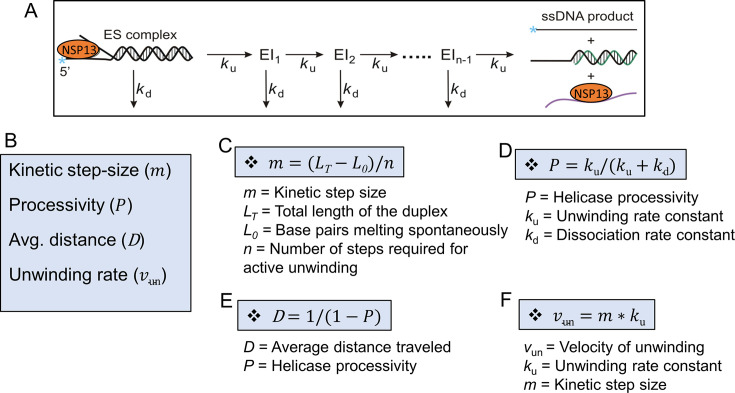
Helicases are characterized by kinetic parameters that define their activity. The number of base pairs (bp) unwound per second is one measure, as is the number of bases traveled per binding event, or processivity. They can unwind DNA at rates of over 1000 bp/s, and some helicases can unwind tens of thousands of bp in a single binding event. The most commonly used helicase assay is performed by creating a partial duplex oligonucleotide (Fig. 1 ). A helicase is incubated with the duplex substrate, and the reaction is started by addition of ATP and Mg+ 2. The enzyme can move along the translocase strand of the substrate, in steps of one or more base pairs. The single-stranded (ss) DNA product can reanneal with the substrate, so a DNA trapping strand is added to the reaction mixture. If a single cycle of activity is preferred, a helicase trap can also be added at the same time as the DNA trap. The helicase trap prevents the enzyme from binding to another substrate molecule. It is possible to determine the kinetic step size by varying the length of the duplex, for example, 16 bp, 20 bp, 24 bp, and 28 bp. Kinetic values that can emerge from these studies are the kinetic step size, the unwinding rate, the dissociation rate, and the processivity. Fig. 1 summarizes the approach developed by the Lohman laboratory [7].
Fig. 1.
Diagram showing the most frequently used assay for characterizing the kinetic parameters that govern helicase activity. (A) The kinetic constants ku and kd are obtained by fitting the data to a model of stepwise unwinding [7], [8]. (B) Kinetic constants that define helicase activity. (C) Kinetic step-size which is determined from a fit to the data. (D) Processivity with brief explanation of its calculation. (E) Distance that helicase travels on DNA or RNA. (F) Velocity of unwinding.
The kinetic step size is one expression of the term step size. Another characteristic is the physical step size, which is the number of base pairs unwound in a simultaneous event. Step size can also refer to the chemical step size, the number of ATPs hydrolyzed per base pair unwound. For some helicases, such as S. cerevisiae Pif1, the different measures of step size resulted in a composite determination of one bp unwound for the kinetic, physical, and chemical step size [8].
Measuring the rate of duplex unwinding as well as the translocation rate on ssDNA or ssRNA can help provide a mechanistic characterization of a helicase. The rate of unwinding has been compared to the rate of translocation on ssDNA as a way to classify helicases as being active or passive. Active helicases are considered to pry apart base pairs actively, whereas passive helicases capture base pairs as they open due to fraying [9]. Croquette proposed using the ratio of the unwinding rate to the translocation rate to discern active from passive helicases. For example, Dda helicase from bacteriophage T4 translocates on ssDNA at a rate of ~ 267 nt/s, and unwinds dsDNA at a rate of 257 bp/s. [10]. The ratio of 0.96 indicates that this is an active helicase in terms of separation of base pairs. However, both an active or passive helicase can be highly processive.
Superfamily 1 and 2 helicases have similar structural features, including two RecA-like domains in which all conserved helicase motifs reside. RecA is an E. coli protein involved in DNA recombination. The domain interacts in a head-to-tail manner to form a filament along DNA that catalyzes strand exchange. In helicases, two RecA domains make up the sites for ATP binding and hydrolysis, which drives the conformational changes that lead to directional translocation on DNA or RNA. Helicase motifs are sequences that are responsible for specific interactions with ATP and RNA. ATP can bind to helicases, but is not rapidly hydrolyzed until RNA also binds. Specific helicase motifs are able to drive conformational changes as a function of specific steps in the ATP hydrolysis mechanism. For example, release of phosphate has been proposed to contribute to the rate-limiting step for unwinding by NS3 helicase [11].
NS3 helicase from the Hepatitis C virus is a SF2A RNA helicase that has been studied extensively due to the large impact of HCV on human health. An estimated 2.4 million people in the United States were living with hepatitis C during 2013–2016 [12]. The function of NS3 helicase activity in the viral life cycle is essential, but its specific role is uncertain. Possible functions include replication of the viral genome, transcription, or packaging. One section of this review highlights NS3 and its possible biological functions and inhibitors.
The SARS CoV-2 (SARS2) helicase, NSP13, is a SF1B helicase, that translocates in the 5′-to-3′ direction. SARS2 is responsible for the COVID-19 pandemic, responsible for over 600,000 deaths in the U.S., and 3.55 million deaths worldwide as of June 2021. The SARS2 helicase is very similar to SARS1 helicase, with only one amino acid change. Therefore, biochemical properties of SARS2 helicase are likely the same as for SARS1 helicase. As with NS3, the specific function for NSP13 is uncertain, but replication, transcription, translation, and/or RNA packaging are possible functions. We review the biochemical characterization, including binding to RNA, ATPase activity, and helicase unwinding activity. A smaller number of inhibitors are reported for NSP13 than for NS3 due to the fact that NS3 has been more heavily studied.
The DEAD-box helicases are a sub-family from SF2. These helicases do not translocate, but undergo conformational changes upon ATP binding and hydrolysis, resulting in local duplex unwinding. DEAD-box helicases such as DDX3X have been shown to interact with viral genomes and/or viral proteins to facilitate viral genome replication. DDX3X helicase is also considered as an anti-cancer target as well as an anti-viral target. Thus, viruses can encode critical helicases for genome replication, or they can hi-jack an endogenous helicase to do so. Here, we provide a review of RNA helicases that are targets to inhibit replication of viruses that are of high importance to human health.
2. Hepatitis C virus NS3 helicase
Hepatitis C virus (HCV) is the major cause of liver-related complications, such as fibrosis, cirrhosis, and hepatocellular carcinoma [13]. According to the 2017 report from the World Health Organization, approximately 400,000 people die each year of liver-related complications from Hepatitis C infection. It is currently estimated that over 71 million people in the world are chronically-infected with HCV [13], and to date, there is no effective vaccine. Current anti-HCV drugs (also known as Direct Acting Antiviral Drugs or DAA), which are a combination of drugs that target viral proteins, are extremely expensive and not accessible to the majority of HCV-infected patients. For instance, in western countries such as Germany and France less than 10% of HCV-infected patients have access to them [13]. As a result, a number of HCV patients are still dependent on interferon-based therapies. In addition to the issue of accessibility and high cost of DAA regimens, drug resistance due to the high mutation rate of the HCV genome can be a significant challenge for DAA based therapies. Moreover, data regarding the efficacy and the safety of DAA regimens across various HCV genotypes and subtypes is limited [14]. Therefore, development of new anti-HCV drugs is needed. Over the last decade, a number of small molecules that target HCV NS3 helicase have emerged as novel potential anti-HCV agents.
The life cycle of HCV is well studied. The entry of the virus to hepatocytes (common HCV targets) is followed by the release of the positive sense, ssRNA genome to the cytoplasm [13]. The HCV RNA genome consists of a single open reading frame (ORF) that is flanked by untranslated regions at the 5′ and 3′ ends [15]. The internal ribosomal entry site (IRES) located within the 5' UTR is important for cap-independent translation [15]. The ORF encodes a single polyprotein which is then cleaved by both host and viral proteases into three structural (core, E1, & E2) and seven non-structural viral proteins (p7, NS2, NS3, NS4A, NS4B, NS5A, & NS5B) [13]. The non-structural proteins NS3, NS4A, NS4B, NS5A, and NS5B are sufficient for successful replication of the virus [16]. Viral replication takes place inside a vesicle derived from the endoplasmic reticulum (ER) double membrane, also known as a membranous web. Inside the ER membranous web, the positive sense-strand is copied to a negative-strand RNA, which is then used as a template to make more of the positive sense-strand RNA [13], [16].
NS3 has a serine protease domain and a helicase domain at the amino and carboxyl termini, respectively. NS4A serves as a cofactor for the protease domain of NS3. NS4A-NS3 is responsible for polyprotein processing. Why the protease and the helicase domains are connected is unknown, though, studies suggest a functional communication between the two domains [16]. Results from experiments conducted in whole animal models or cell-based assays, the helicase activity of NS3 is essential for HCV genome replication; however, it is involved in HCV replication is not well understood [17], [18]. The helicase could (1) unwind the nascent and template strands of the RNA genome during viral replication, (2) unfold RNA secondary structures making the RNA sequences available for replication machineries, or (3) displace proteins that bind to the RNA genome and could potentially block the replication process [13].
Potential G-quadruplex forming sequences exist within the genome of various human viruses [19]. G-quadruplex structures are secondary nucleic acid structures that form when a single-stranded guanine-rich DNA or RNA sequence folds into a four-stranded structure through Hoogsteen hydrogen bonding [20]. If they remain folded, G4 structures can block replication, transcription, and translation machineries. As a result, cells need to regulate the folding and unfolding of these stable secondary structures in a controlled manner [21]. By performing a bioinformatics analysis on a number of HCV strains, Wang et al. reported that the positive sense HCV RNA genome contains conserved G4 forming sequences within the gene that encodes the core protein [22]. Jaubert et al. have also shown the presence of conserved G4 forming sequences at the 3′ end of the HCV negative strand RNA [23]. The formation of G4 secondary structures within the HCV genome could inhibit viral replication; as a result, the virus could use its helicase activity to unfold these structures. In our laboratory, we have shown that NS3 (Con 1b strain) unfolds HCV-G4 in vitro, suggesting a novel role of NS3 in viral replication (data not shown; unpublished data). Cell-based assays are needed to define clearly the role of the helicase activity in synthesis of the positive or negative strands of the HCV genome.
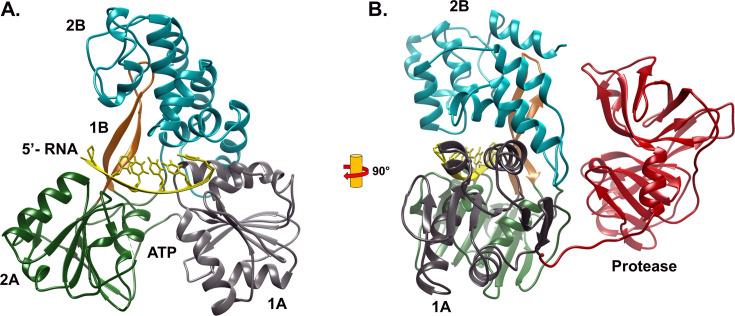
NS3 helicase belongs to the SF2 helicases, specifically within the DExH helicase family. It unwinds RNA or DNA duplex substrates by translocating along the loading strand in the 3′ to 5′ direction, by which it belongs to subfamily A, sub-type α. Therefore, HCV NS3 is classified into the family of SF2Aα [24]. The helicase domain of NS3 has three sub-domains: [17] (Fig. 2 ). Domain 1 and 2 (the motor domains) resemble the RecA-like domain, and are composed of both α helices and β pleated sheets. On the other hand, domain 3 consists of mainly α helices [25]. NS3 helicase binds to ssRNA with a higher affinity than it does to its ssDNA counterpart [26].
Fig. 2.
Structure of the Hepatitis C Virus NS3 helicase. (A) The helicase domains complexed with RNA are highlighted, and the protease domain located behind the structure omitted for clarity. The cleft between the RecA-like domains 1A (gray) and 2B (green) contain the site of ATP binding and hydrolysis. Single-stranded RNA and DNA enter at the 2A domain, bind in a channel along the 1A -2A interface and the 2B domain (cyan) and emerges from the 1A domain to allow for 3′ – > 5′ translocation. The 1B tether region (orange) interacts with the 2B domain to allow for the protease domain to exert conformational control of substrate binding and the helicase activity. (B) Full-length HCV NS3 helicase rotated 90° from (A). The HCV protease (red) located on the N-terminal region of the enzyme is shown.
Despite the fact that the structure of the helicase domain of NS3 has been well studied, the development of helicase-targeting small molecules has been slower in comparison to the extensive work done and success achieved over the years with the molecules that target the protease domain of NS3. To our knowledge, there is no helicase inhibitor available for patient use. The major challenge in developing inhibitors against NS3 helicase is that the helicase domain has structural similarity with cellular motor proteins; as a result, inhibitors that target the HCV helicase could also affect the normal function of cellular proteins, causing toxicity [27]. Most of these compounds exhibit their inhibitory effect by preventing the helicase (1) to bind to DNA or RNA, (2) to unwind dsDNA or dsRNA, or (3) to hydrolyze ATP. In addition, because the helicase domain is connected to the protease domain in NS3, a small molecule that inhibits the activity of the protease domain could also affect the helicase activity.
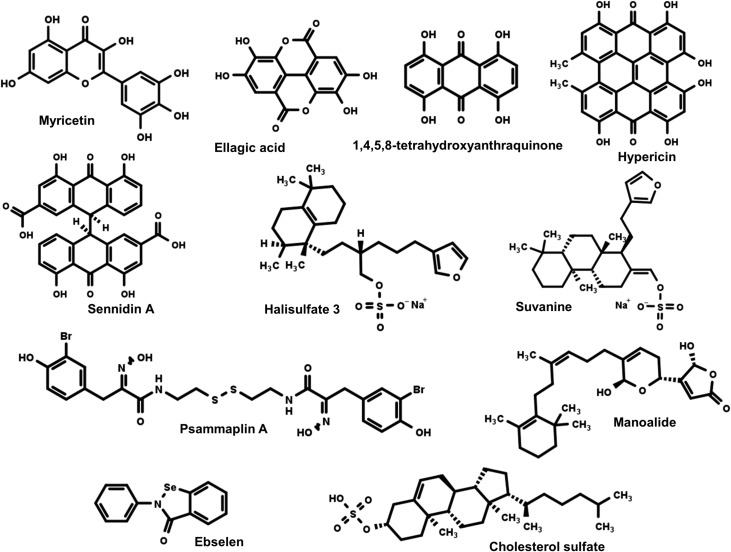
Lim et al. conducted structure-based virtual screening (SBVS) on 13,420 compounds with a benzopyran scaffold to search for benzopyran-based inhibitors that target HCV NS3 from genotype 3. HCV genotype 3 has been associated with high rate of liver-related issues and reduced resistance to direct-acting anti-HCV drugs; as a result, it is difficult to treat patients infected with this type of HCV strain. From the virtual screening method, Lim et al. were able to identify two compounds, ellagic acid and myricetin, as putative protease and helicase dual-target inhibitors of HCV NS3 (Fig. 3 ). Ellagic acid inhibits the unwinding, ATPase, and protease activity of NS3 with IC50 value of 6.58, 68.42, and 40.37 μM, respectively. In addition, ellagic acid reduces viral replication in Huh7.5 cells containing genotype 3a HCV replicon with EC50 and selectivity index (IS) values of 32.37 μM and > 6.18, respectively. Its potency and high selectivity index make it a good candidate for anti-HCV therapeutics. Myricetin inhibits well the dsRNA unwinding activity of NS3; however, it has poor potency against HCV replicon-containing Huh7.5 cells. Future optimization is likely needed to increase its efficacy [18].
Fig. 3.
Chemical structures of the HCV NS3 helicase inhibitors discussed in this review. See the text for discussion of the inhibitors targeted to NS3.
Given the data that shows hydroxyanthraquinone moiety-containing chemicals inhibit NS3 helicase activity, Furuta et al., proposed that the hydroxyanthraquinone moiety alone could also inhibit the helicase activity of NS3 (Fig. 3). To test their hypothesis, they performed fluorescence-based dsRNA helicase unwinding assay in the presence of a series of hydroxyanthraquinone compounds containing varying number of phenolic hydroxyl groups. Of all the hydroxyanthraquinones with varying numbers and positions of phenolic hydroxyl groups, 1,4,5,8-tetrahydroxyanthraquinone inhibited NS3 helicase activity with the lowest IC50 value (6 μM). Moreover, the researchers examined the effect of linking two hydroxyanthraquinone moieties on the helicase activity of NS3. Hypericin and Sennidin A, chemicals containing two hydroxyanthraquinones, exhibited stronger inhibition to the activity of HCV NS3, as evidenced by their low IC50 values, 3 and 0.8 μM, respectively. Interestingly, despite Sennidin A having a low IC50, its ability to suppress the replication of HCV subgenomic replicon in Huh-7 cells was rather small (EC50 > 80 μM). It is possible that the compound has poor permeability across the cell membrane. Future optimization of the drug is likely needed to improve its potency. In contrast, hypericin reduces the replication of the HCV genome with an EC50 value of 3.5 μM. The selectivity index of hypericin is 11.7, which suggests minimal side effects. Hypericin could also inhibit the ATPase and binding activity of NS3. Therefore, its inhibitory effect on the helicase activity could be due to its ability to inhibit binding or/and hydrolysis of ATP [28].
Halisulfate 3 and suvanine inhibit HCV NS3 helicase activity with IC50 values of 4 μM and 3 μM, respectively. In addition, halisulfate 3 and suvanine inhibit the ATPase, RNA binding, and protease activity of HCV NS3. Therefore, it is possible that the inhibitory effect these compounds have on the helicase activity could be associated with their effect on these activities. Interestingly, neither halisulfate 3 nor suvanine have a significant inhibitory effect on NS3 helicase derived from dengue virus, despite its structural similarity to HCV NS3 helicase. Halisulfate 3 and suvanine could be binding to a unique allosteric site on HCV NS3 and inducing conformational changes, which in turn brings about the inhibitory effect on the helicase, protease, ATPase, and the RNA binding activities [29]. Future studies should explore the effect of these compounds on HCV genome-containing cells, to evaluate their ability to suppress viral replication.
Mukherjee et al. performed fluorescence polarization DNA binding assays to screen for compounds that were already known to suppress HCV replication in hepatocytes, with the aim of identifying HCV NS3 helicase inhibitors [25]. Of all the compounds screened, ebselen inhibits the ability of NS3 helicase to bind to DNA with IC50 value of 1.4 μM. Moreover, ebselen prevents NS3 helicase to unwind dsDNA and hydrolyze ATP. However, ebselen is relatively toxic to hepatocytes containing HCV subgenomic replicons [25].
Researchers from Akimitsu's laboratory identified psammaplin A as HCV NS3 helicase inhibitor after screening 54 marine organism extracts using a fluorescence-based, high-throughput screening system to assay dsRNA helicase unwinding. Psammaplin A inhibits HCV NS3 helicase, ATPase, and ssRNA binding activities with IC50 values of 17, 32, and 40 μM, respectively. It did not change the Michaelis constant (Km) for ATP; therefore, it is a non-competitive inhibitor. When cells containing a HCV subgenomic replicon derived from genotype 1b are treated with psammaplin A, viral replication is reduced in a dose-dependent manner (EC50 6.1 μM); however, cytotoxicity was relatively high as indicated by a low selectivity index of 3.7. Therefore, future optimization of psammaplin A could improve potency and efficacy [30].
Cholesterol sulfate (CS) inhibits the helicase activity of NS3 with an IC50 value of 1.7 +/− 0.2 μM. The sulfate group, the sterol backbone, and the alkyl side chain are essential components of CS to maintain high inhibitory potency towards NS3 helicase. It does not inhibit ATP hydrolysis or protease activity. However, it does inhibit NS3 in binding to a ssRNA substrate. Cell- based assays are needed to test its ability to suppress viral replication with minimal cytotoxicity [31].
In addition to its role as an anti-inflammatory compound, manoalide could potentially have anti-viral activity. It inhibits the dsRNA unwinding activity of HCV NS3 with an IC50 value of 15 μM, and its ability to hydrolyze ATP in a non-competitive manner (IC50 value of 70 μM). Data from an isothermal denaturation assay indicates that manoalide binds directly to HCV NS3 and induces conformational changes. Therefore, it is possible that its inhibitory effect on HCV NS3 activities stems from its ability to induce structural changes [32].
The anti-HCV drugs available in the market are expensive, and are unavailable for the majority of HCV-infected patients. In addition, there is insufficient data regarding the potency of these drugs against various strains of the virus. Because direct acting anti-HCV drugs target mutation-prone viral proteins, it is possible that drug resistance could still be a challenge for DAA based therapy. Therefore, it is important that the development of novel anti-HCV drugs continue. A number of small molecules that target the helicase activity of NS3 have been proposed as good candidates for anti-HCV infection therapy. Even though none of the HCV NS3 helicase inhibitors studied thus far are being used in the clinic, efforts have been made to examine the helicase domain of NS3 as a potential target for anti-HCV therapeutics. Future optimization of NS3 helicase inhibitors could improve their efficacy and safety for clinical use.
3. Coronaviruses, SARS CoV-1 and SARS CoV-2 NSP13 helicases
The rapid spread of SARS CoV-2 in 2019 to a world-wide pandemic generated intense focus on coronavirus virology and propagation. Previous outbreaks and containment of SARS and the Middle Eastern Respiratory Syndrome (MERS) viruses in 2002 and 2012, respectively, sparked the identification and initial characterization of the non-structural protein 13 (NSP13) helicases, though the functional purpose for the helicase activity during the replication has still not been elucidated (see Lehmann et al. [33] for an extensive review). Nevertheless, the NSP13 sequence conservation and structural homologies within the MERS, SARS CoV-1 and SARS CoV-2 family of viruses suggest that much of what we have come to understand will inform future studies with the NSP13 of SARS CoV-2, as well as newly-emergent coronaviruses.
The Coronaviridae family of viruses have large, positive-stranded RNA genomes. The genomes of the SARS viruses (hereafter referred to as SARS1 and SARS2) are ~ 30 kB, and contain multiple hairpins and regions of secondary structure needed for efficient packaging [34]. During viral replication and the transcription of the polyprotein ORF, stretches of dsRNA must be unwound, while protective RNA-coating proteins must be displaced to prevent pausing or disassociation of the replication transcription complex (RTC). Virally-encoded translocating helicases are particularly well suited to interact both with the viral genome and the RTC, to efficiently remove any impediments and drive the multiplication and propagation of RNA viruses.
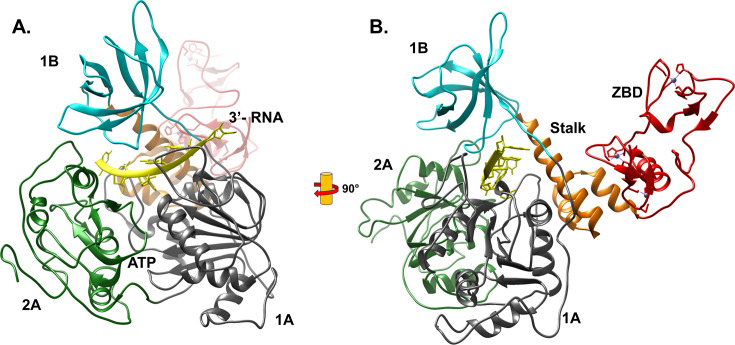
The SARS1 and SARS2 helicases are encoded by the NSP13 gene and are nearly identical, differing by a single isoleucine to valine mutation at position 572 [35]. The five domain structures of NSP13 (Fig. 4 ) are the unique cysteine-rich, zinc-binding domain (ZBD), an α-helical stalk domain that connects the ZBD with the accessory domain (2B), and the RecA-like 1A and 2A domains harboring the ATPase and nucleic acid binding sites. The similarity of NSP13 to human RNA helicase Upf1, a critical component of the non-sense mediated mRNA decay pathways, places the SARS helicases in the super family 1 group. Like Upf1, the SARS NSP13 helicase binds single-stranded nucleic acids and translocates in the 5′ to 3′ direction, further classifying it in the SF1B subgroup [36], [37]. In solution, NSP13 appears to bind to ssDNA and ssRNA as a monomer.
Fig. 4.
Structure of the SARS CoV-2 NSP13 helicase. (A) The helicase domains complexed with RNA are highlighted, and the zinc binding domain (ZBD) lightened for clarity. The cleft between the RecA-like domains 1A (gray) and 2A (green) contains the site of ATP binding and hydrolysis. Single-stranded RNA and DNA enter at the 1A domain, bind in the channel along the 1A-2A interface and the 2B domain (cyan) and emerges from the 2A domain for 5′ - > 3′ translocation. (B) SARS CoV-2 NSP13 helicase rotated 90° from (A). The N-terminal zinc binding domain (red) binds 3 zinc atoms and is essential for helicase activity. The ZBD is connected to the helicase domains through the α-helical-rich Stalk domain (orange).
The N-terminal portion of the protein contains a well-conserved, cysteine-rich domain capable of coordinating three zinc atoms in highly-structured zinc fingers. While the SARS1 NSP13 ZBD has not been well studied, a mutational analysis of the ZBD of the related human coronavirus HCoV-229E demonstrated that substitutions to the zinc-binding histidine and cysteine residues significantly decreased ATPase and helicase activities [38]. In fact, complete removal of the ZBD from the helicase core led to an inactive enzyme, and attempts to reconstitute the holo enzyme using separately-expressed domains also failed to restore activity. Clearly the intrinsic mechanism for the helicase activity is intimately connected with the zinc domain, and may provide a key regulatory mechanism to control ATPase and RNA affinity. The CH domain of Upf1, which is homologous to the ZBD, has a relatively flexible hinge, leading to significantly greater spatial flexibility for the interaction of the regulatory protein Upf2 modulating the helicase activity [39]. In the SARS helicases, the ZBD is connected to the helicase by a series of rigid α-helices making up the stalk domain. This interaction allows the face of the ZBD to pack tightly against the helicase core, while the zinc fingers interact with the accessory protein NSP8 in the large replication complex [40], [41], [42]. Evidence points to the ZBD modulating the movement of the helicase domains to alter its activity; however, little else is known about how it participates with viral or host protein partners to influence viral replication.
Until recently, the limited number of structural studies of coronavirus helicases have hampered our understanding of the coordination of the various domain structures during nucleic acid binding and the mechanisms of translocation. The structure of the helicase from the equine arteritis virus (EAV), a member of the nidovirus family, was solved, providing the initial clues about the structural arrangement of a viral helicase from a positive-stranded RNA virus [43]. While Nsp10 of EAV lacks the stalk domain connecting the ZBD and the 2A domain [43], the crystal structure provided an important framework for the study of helicase from other nidoviruses. By 2017, Hao et al. [44] had reported the crystal structure of a human coronavirus helicase (NSP13) from MERS. They were the first group to crystalize a human coronavirus helicase and utilized the sequence and structural similarities to the yeast Upf1 [44] to establish a model for coronavirus helicase domains. Jia et al. [39] reported the first crystal structure of the SARS CoV-1 NSP13 helicase (PDB:6JYT), though they did not co-crystalize the protein with either an ATP analog or an RNA or DNA substrate. Combined with hydrogen-deuterium exchange and ensemble activity measurements, they were able to establish that the β-sheets of the 1A domain directly participated in the unwinding and translocation activities. This was the most extensive structure-based model for the coordination of the NTP-binding pocket with the nucleic acid binding channel.
Within a short time of the identification and sequencing of SARS CoV-2, two groups provided cryo-EM structures of an assembled replication transcription complex (RTC) including NSP12 (RdRP), NSP13 (helicase), NSP7 and NSP8 (accessory proteins) [40], [41]. Structures from both groups were in good agreement, and provided key insights into the viral protein arrangements in the RTC. Both groups suggested that two helicase proteins bind to the complex, yet not as a dimer. Chen et al. [40] were also able to produce partial EM assignments for NSP12 bound to a template RNA and one of the NSP13 units, though due to the heterogeneity of structures with RNA, the full engagement of a contiguous piece of RNA was not resolved. Interestingly, one of the helicase subunits is situated adjacent to the active center of the RdRP, poised to unwind duplex template strand RNA. However, with the directionality of NSP13 translocation being 5′-to-3′, binding to the template strand would place the helicase in direct opposition to the polymerase during negative strand synthesis. However, the authors hypothesized that during the production of the positive-strand genome, the helicase and the polymerase could coordinate by binding to the separate strands, thus allowing the helicase to separate the duplex RNA ahead of the polymerase. Yan et al. [42] extended the structural information for the complex to include the RNA binding protein Nsp9, to determine how the essential 5′-GpppA cap structure is synthesized on the SARS2 mRNA. They speculate that NSP13 participates in the removal of the 5′-γ-phosphate, an activity previously reported for the SARS1 NSP13 [45], though it is unclear if the special positioning of either NSP13 in the complex would allow this to occur. In addition, the cryo-EM structures point to an interaction of the ZBD with duplex RNA, something not seen in any of the previous cryo-EM or crystallographic structures. As the capacity to visualize the RTC at the molecular level rapidly improves, the structural coordination of the helicase proteins during viral genome transcription and replication will aid with biochemical determinations of the role of the helicase in SARS2.
Much of what we know about coronavirus helicase mechanisms has come from work with helicases from SARS1 and MERS. The single amino acid substitution in the SARS2 NSP13 occurs along the surface of the RecA-like 2A domain, and does not appear to participate directly with the ATPase or unwinding activities. Thus, much of the biochemical and kinetic data for NSP13 from SARS2 governing its activity may be inferred from SARS1, with some caveats and caution. Comparisons of reported measurements for the nucleic acid-dependent NTPase activity showed significantly differing values for ATP turnover. Tanner et al. [46] observed a kcat of 19.1 s− 1 using a polyuridine RNA substrate, while Ivanov et al. [45] reported a 10-fold lower value of 2.3 s− 1. Clues to the mechanisms responsible for differences came from Adedeji et al. who reported two rate constants for independently-purified forms of NSP13. A 6X-histidine-tagged NSP13 hydrolyzed ATP at a rate of 0.22 s− 1, whereas a much more rapid rate of 104.1 s− 1 was measured for the GST-tagged protein [47]. The ATPase rate constant for the native protein lacking any affinity or protein tag was not reported. Notably, Tanner utilized a 6X-histidine-tagged NSP13, while Ivanov expressed a maltose-binding protein (MBP)-fusion. The Sarafianosis group also reported difficulty with the protease-mediated cleavage of the affinity tag in their construct, though others did not seem to experience similar issues. Using uncleaved proteins, a preliminary kinetic comparison was made of protein-tagged NSP13 ATPase rates, which lead to the conclusion that the differences in the catalytic efficiencies resulted from interference in ATP binding [48]. This explanation, however, does not consider the significant role of the N-terminal ZBD on the ATPase activity. Though the rigid stalk domain holds the ZBD close to the helicase subunits, the rotation and mobility of the ZBD during the NTP hydrolysis cycle and nucleic acid translocation may be hampered by the presence of an N-terminal extension, depending on the design of amino acid linkers between the N-terminus of the enzyme and the tag. The initial X-ray crystal structure of SARS1 NSP13 used the 6X-histidine-tagged construct, though the structural data from subsequent cryo-EM studies that utilized tagged proteins that were subsequently fully cleaved and purified as the native protein were in good agreement [40], [41]. Clearly the small 6X-histidine tag has a negative effect on the ATPase and unwinding activities, though not on substrate binding. The much larger MDP and GST tags also seem to disrupt the intrinsic helicase activity, though possibly allow a freer coordinated movement during substrate turnover. A thorough kinetic analysis (see Fig. 1) of the native protein has not been reported, though it would shed considerable light on the intrinsic activity of the enzyme.
NSP13 binds to ssDNA and RNA with similar nanomolar affinities. Upf1 and other members of the SF1 and SF2 families of helicase also have this characteristic, though the underlying mechanisms are still unknown. In addition, the nucleic acid-stimulated ATPase activity of NSP13 for saturating amounts of DNA is greater than that for RNA [46], suggesting that different structural features of the nucleic acid channel are involved in DNA and RNA binding. The minimal functional length of ssDNA or RNA needed for NSP13 binding is approximately ten nucleotides [47], [49], though the cryo-EM structure only resolved six bases within the channel [41]. Though the SARS genome is RNA, NSP13 does not appear to serve any alternate functions in the nucleus of host cells, or function with host RNA or DNA; this has been a pressing unanswered question in viral helicase biology.
Multiple groups have reported kinetic analyses for uncovering the mechanisms of the unwinding activities of MERS and SARS1 helicases, and by extension due to it high homology, SARS2 NSP13. Due to the dual DNA/RNA binding and unwinding activity, determinations of substrate preferences, processivity, chemical and kinetic stepping and cooperativity have been examined using more stable and less expensive DNA substrates, and minimally confirmed with RNA substrates. Due to the different binding of RNA and DNA, a thorough survey of the viral helicase with RNA is warranted. Nevertheless, Lee et al. [49] synthesized multiple DNA substrates with differing duplex lengths, ssDNA overhangs and gap sizes to measure processivity, cooperativity and binding site size, respectively. They reported that the distance that NSP13 can unwind DNA prior to dissociation from the nucleic acid is significantly lower than that seen with other helicases such as HCV NS3 and the NPH-II RNA helicase. When using DNA substrate with longer ssDNA tails, they also observed an increasing amount of unwound product, suggesting that multiple helicase subunits can bind and work in concert to unwind longer duplexes. When the study was repeated with RNA substrates containing the same duplex and overhang lengths, Jang et al. from the same laboratory observed even lower processivity and significantly less cooperativity, possibly due to differences in affinity and binding to the RNA versus DNA [50]. Because the ATPase activity differs between RNA and DNA and by inference its mode of binding, it is not surprising then that translocation and duplex displacement is also different. Perhaps the presence of a much larger replication complex consisting of the RNA polymerase and accessory proteins NSP7 and NSP8 help tether NSP13 to the vicinity of the RNA genome.
The mechanisms of NSP13 helicase activity were further advanced by the Sarafianos laboratory with the measurement of the kinetic step size of the enzyme, or the apparent number of nucleotides traversed with a single catalytic cycle. Interestingly, they also demonstrated significant differences in the unwinding efficiency of different tagged enzymes. They clearly demonstrate that the GST-tagged-NSP13 unwound a 30 base pair duplex substrate up to 100- fold faster than the purified 6X-histidine-tagged (used by the Kim laboratory) and MBP-tagged helicase [47]. They also observed little difference between DNA and RNA unwinding rates for the same-sized substrate. These data demonstrate that N-terminal modifications and affinity tags that aid in the purification of NSP13 need to be removed prior to in vitro kinetic experiments. Using rapid quench-flow assays to measure the pre-steady state phase of duplex unwinding in a single turnover reaction, unwinding occurred at 30 steps per second, with a step size being approximately 9.3 base pairs. The net unwinding rate was calculated to be ~ 280 base pairs per second, and they demonstrated a much more robust processivity with DNA. In addition, they observed that the interaction with Nsp12, which was subsequently confirmed by SPR [39], enhanced the unwinding rate by nearly 2-fold. These observations agree with the RTC complex structures and provide biochemical confirmation of the cooperative interaction between the polymerase and helicase activity. While the rationale for using the DNA substrates seem sound, kinetic characterization of NSP13 will require the use of native protein, RNA substrates and rapid kinetic experiments to fully characterize the helicase unwinding mechanisms.
Little is known regarding the involvement of the DEAD-box helicases during the progression of the SARS viral life cycle. Based on findings from other RNA viruses such as the hepatitis C virus (HCV) [51], [52], West Nile Virus (WNV) [53], [54], several host-encoded DEAD-box helicases likely play a role in the genome processing, maintenance and replication. The only reported interaction of a DEAD-box helicase with NSP13 is from Chen et al. [55] using a two-hybrid screen in yeast. The interaction was confirmed by co-immunoprecipitation of DDX5 with exogenously expressed SARS1 NSP13 in A549 cells. Further, the siRNA knockdown of DDX5 in a model of SARS1 infection in fetal rhesus kidney cells reduced active production of virus suggesting a critical role for DDX5 during SARS1 infection. However, these experiments did not directly address question regarding the stage of viral infection that may require the interaction of NSP13 with DDX5. It is tempting to consider DDX5 as a potential host factor aiding in the resolution of duplex RNA structures in the large coronavirus genome. Interestingly, Schmidt et al. reported the pull-down of the RNA-binding DEAD-box helicase DDX3X bound to the SARS2 genome [56], though pharmacological knockdown of the DDX3X activity led to a modest reduction in viral replication in Calu3 cells. A large-scale survey of tagged SARS2 proteins using affinity-purification mass spectrometry identified multiple potential host-encoded binding partners for NSP13, though the interaction with either DDX3X or DDX5 was not found [57]. In this study, NSP13 seemed to interact with multiple proteins involved in the Golgi organization, PKA signaling as well as the centrosome, and may have been a result of zinc finger motifs in the ZBD interacting with human proteins. It is unclear whether NSP13 resides solely within the peri-nuclear double membrane replication vesicles associated with the endoplasmic reticulum [58], or can interact with host proteins in the cytoplasm and nucleus.
The critical role in viral multiplication for the helicase activity suggests that NSP13 is a prime target for small-molecule compounds to effectively control viral infection. Viral helicase targets contain a common helicase motif for nucleotide binding; thus, developing SARS-specific small molecule inhibitors without reducing or eliminating endogenous helicase NTPase activity is challenging. Due to the successes with other viral infections such as HIV-1 and HCV, the protease and the core polymerase has been the favored targets for small molecule viral inhibitors. In silico studies utilizing the crystal structures of the SARS1 NSP13 helicase quickly appeared in the literature following the recognition of the remarkable conserved homology of the SARS1 and SARS2 proteins and SARS CoV-2 as a world-wide threat [59], [60], [61], [62], [63], [64], [65]. Importantly, these studies made it clear that NSP13 contains multiple druggable structures and pockets predicted to disrupt the helicase activity outside of the canonical NTP binding site. Ideally, effective antiviral compounds would bind an allosteric site, allow the enzyme to form the RTC and disrupt the duplex displacement activity. Presumably, the stalled RTC complex would dissociate, or be unable to participate in RdRP-mediated genome replication. The speed of discovery is hampered by the necessity for medium to high throughput screens to eliminate compounds that affect ATPase rates as well as nucleic acid-mediated helicase activity [66]. Nevertheless, the rational design and screening of viral helicase-specific small molecule inhibitors is possible.
Molecular dynamic surveys of the crystal structure for SARS1 NSP13 utilized large libraries of FDA-approved compounds [59], [62] as well as natural compounds [67] to screen for potential antiviral drugs. The in vitro and in vivo follow-up studies of potential inhibitors from these studies are more difficult and time consuming, and thus have been slower to appear in the literature. Other investigators have taken the approach of using fragment-based molecular modeling to elucidate binding pockets throughout the structure and build optimized compounds that fully engage the structure while disrupting activity [64]. Increasingly, pockets in and around the intersection of the ZBD with the RecA1 and RecA2 domains adjacent to the stalk domain appear to have the potential to interfere with the substrate binding or the transfer of energy from ATP hydrolysis through domain movements to produce efficient translocation and duplex unwinding. Adedeji et al. reported the screening and helicase activity inhibition of SSYA10–001 (Fig. 5 ), a 1,2,4-triazole compound for SARS1 NSP13 [68], as well as MERS and the mouse hepatitis virus (MHV) [68]. This compound and derivatives, however, suffer from relatively high potency requirements with the EC50 in the micromolar range. Early binding calculations suggested an interaction with amino acids on the surface of the 1B domain, but crystal structure data brought that into question. Thus, the binding site for a better-characterized NSP13 inhibitors, SSYA10–001 still not has been identified.
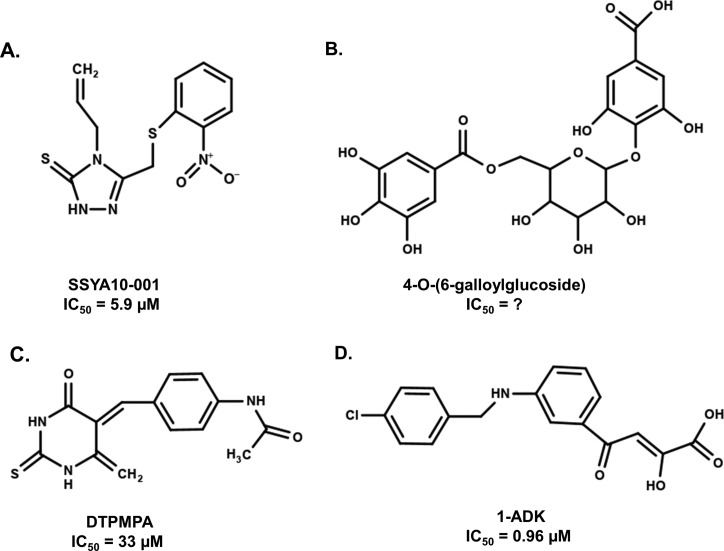
Fig. 5.
Structures of SARS1/2 NSP13 helicase inhibitors. (A) The patented compound SSYA10–001 inhibits the DNA unwinding activity of NSP13 in a FRET-based in vitro assay with an IC50 of 5.9 μM [102]. (B) Docking studies identified 4-O-(6-galloylglucoside) as a potential inhibitor for SARS1/2 NSP13, though no in vitro studies have evaluated its ability to inhibit helicase activity [61]. (C) DTPMPA (N-(4-((4,6-dioxo-2-thioxotetrahydropyrimidin-5(2H)-ylidene)methyl)phenyl)acetamide) was found to be a dual inhibitor of the ATPase activity with an IC50 of 1.2 μM, and the helicase activity with an IC50 of 33 μM [70]. (D) The aryl diketoacid compound ADK-1 inhibits NSP13 in vitro with an IC50 of 0.96 μM [63].
Additional compounds have been suggested to inhibit coronavirus helicase activity including gallic acid-derivatives [61], bismuth compounds [69], pyrimidine derivatives [70] and RNA aptamers [71] (Fig. 5). However, these compounds affect the ATPase activity with low pharmacological potencies as well as little if any pharmacokinetic data for efficacy or safety. The availability of NSP13 cryo-EM structures complexed with NSP12, NSP7 and NSP8 provides new, though more complex opportunities for the development of SARS2 antiviral compounds. The consideration of the dynamics between the helicase, its activity, and the interplay with other components of viral genome replication will need to be included when targeting NSP13 for novel and effective therapeutics.
4. DEAD-Box RNA helicases in viral multiplication not encoded in the viral genome
DEAD-box RNA helicases, characterized by a highly conserved “Asp-Glu-Ala-Asp” (DEAD) motif, are found in all eukaryotes, as well as in many bacteria and archaea [72]. DEAD-box RNA helicases form the largest family of superfamily 2 (SF2) helicases, with 37 members in humans [72]. This family of helicases plays important roles in RNA metabolism regulating gene transcription and expression, intron splicing, mRNA processing and transport, ribosome assembly and formation, mRNA translation, mRNA decay, ribonucleoprotein (RNP) complex remodeling, and stress granule and P-body formation [72], [73]. In humans, DEAD-box RNA helicases can act as cytosolic DNA sensors [74]. They are involved in cellular stress responses, disease (such as cancer) development and progression, viral infection and replication, as well as host innate immune responses. Additionally, the DEAD-box RNA helicases can serve as links among the cellular energy state, gene expression, and metabolism, through their ability to bind ATP and unwind RNA substrates [75], [76].
DEAD-box RNA helicases have a highly conserved SF2 helicase core composed of two RecA-like domains, often accompanied by N- or C-terminal extensions that are not conserved (Fig. 6 ) (comprehensive review in [72]). Inside the helicase core, at least 12 characteristic sequence motifs were distributed into two RecA-like domains. Motifs Q, I, Ia, Ib, Ic, II, and III are located in the N-terminal domain (Domain 1), and motifs IV, IVa, V, Va, VI are located in the C-terminal domain (Domain 2). These two domains form a cleft that harbors the ATP-binding site, which is seated between the two domains. RNA binds perpendicular to and at the base of the ATP-binding site and the interaction spans across both domains. The conserved motifs participate in ATP binding and hydrolysis (motifs Q, I, II, and VI), and RNA binding (motifs Ia, Ib, Ic, IV, IVa, and V). The communication linkage (motifs III, and Va) is located between the ATP-binding site and the RNA binding site.
Fig. 6.
Structure of the human DEAD-box helicase DDX3X. (A) Overall structure of the active helicase D1D2:dsRNA:D1D2 core from DDX3X. The RecA-like D1 domain (gray in core 1 and orange in core 2) binds ATP and the energy from hydrolysis induces a conformational change in the RecA-like D2 domain (green in core 1 and cyan in core 2) leading to RNA strand separation. (B) DDX3X core helicase rotated 90° from (A). The two helicases act on separate strands of the dsRNA leading to complete strand separation.
In the absence of ATP and RNA, these two RecA-like domains of the helicase core move freely and are flexible with respect to each other (open conformation). The collaborative binding of ATP and RNA, stimulates these two domains to form a closed position (closed conformation), to induce ATP hydrolysis [77]. Interestingly, for many DEAD-box RNA helicases, only ATP binding is required for RNA strand separation but not ATP hydrolysis. DEAD-box RNA helicases have extremely tight RNA binding affinity in the presence of ATP ground and transition state analogues. ADP greatly reduces the affinity of DEAD-box proteins for RNA. Thus, the ATP hydrolysis is necessary for the release of DEAD-box helicase from the RNA substrate, and promotes substrate turnover [78]. In contrast with other processive helicases (for example, Pif1) which translocate along DNA or RNA substrate during ATP hydrolysis cycle, DEAD-box RNA helicases are typically non-processive helicases. Thus, their RNA duplex unwinding is more restricted to local strand separation. Many members of this family of helicases can only unwind duplexes with less than 10 to 12 base pairs [73].
Flanking the helicase core, the N-terminal and C-terminal extensions are the auxiliary domains. The auxiliary domains in DEAD-box RNA helicases are not conserved, ranging from a few to several hundred amino acids in length. These auxiliary domains are responsible for interacting with other proteins or with RNA targets, to achieve various functions.
Most viruses do not encode all proteins required for their propagation in their genome. The interactions between virus and host cells are important for viral replication. On one side, viruses need to recruit host proteins and utilize the host cell machinery to help their replication. On the other side, the anti-viral innate immunity by host cells suppresses viral replication. Although DEAD-box RNA helicases are encoded only by host cells, they play essential roles as the driving force across both viral replication and antiviral innate immunity (comprehensive review in [79]).
In the past two decades, there are two strategies primarily used for identifying host factors/proteins involved in viral propagation: (1) using the small-interfering RNA (siRNA) knock-down strategy to screen host genes that affect viral propagation, (2) proteomic screens by using viral proteins (or viral nucleic acid sequences) as bait. In many cases, DEAD-box helicases have been identified as essentially required for several steps in a virus life cycle. Currently, there are five human DEAD-box helicases (DDX1, DDX3X, DDX5, DDX6, and DDX17) that have been found to have important pro-viral (positive) or antiviral (negative) roles in viral propagations [79].
The mechanisms of DEAD-box helicases in viral propagation are newly emerging research topics in the past two decades but have not been fully explored. In general, DEAD-box helicases can participate in viral propagation by several ways [79]. RNA unwinding or unfolding by the helicase and RNA chaperon activities, to reform RNA structures for viral genome replication, gene transcription and translation. For example, DDX5 can interact with RNA-dependent RNA polymerase of HCV (NS5B), and is required for the transcription of negative-strand from positive-strand HCV RNA [80]. DDX1 binds to the 3′(+) untranslated region (3'-UTR) of HCV RNA, and plays important role in the initiation of HCV RNA replication [81]. DEAD-box helicases can interact with virus elements and proteins, to help viral RNA splicing and subcellular exporting. For example, DDX1 binds to HIV Rev-response element (RRE) to form RNA complexes that facilitate the export of RRE-containing HIV RNAs to the cytoplasm from the nucleus via the chromosomal maintenance 1 (CRM1) pathway. RRE is required for maintaining HIV RNA splicing [82], [83]. In HIV-1 infected cells, DDX3X localizes to nuclear pores, interacts with CRM1 to form the CRM1/DDX3X complex at the cytoplasmic side of the nuclear envelope, and exports the RRE-containing HIV-1 RNAs into cytoplasm from the nucleus [84], [85]. DDX19 interacts with Influenza A virus polymerase and promote the nuclear export of viral mRNAs [86].
DEAD-box helicases interact with virus elements and host (or viral) proteins, to mediate translational initiation of viral RNAs. For example, DDX3X binds to the 7-methylguanosine 5′-triphosphate (m7GTP) cap of the HIV-1 RNA, interacts with eukaryotic translation initiation factor 4G (eIF4G) and PolyA-binding protein cytoplasmic 1 (PABPC1), to form a core DDX3X/PABPC1/eIF4G trimeric complex [87], [88]. DDX3X is recruited to the Tat-recognition region of the 5'-UTR of HIV mRNAs by the viral trans-activating regulatory protein Tat, to promote the Tat-dependent translation of HIV-1 mRNAs [89], [90]. DEAD-box helicases can undergo liquid–liquid phase separation (LLPS) to provide cells with spatial and temporal control of various RNA-processing steps involved in viral propagation. Several DEAD-box RNA helicases have the ability to interact with nucleic acids to undergo LLPS as global regulators of phase-separated organelles [91]. For example, in HCV infected cells, DDX3X is involved in P-body formation, stress granules (SGs), and is important for HCV viral propagation [52], [92]. The detailed functions of LLPS in viral propagation remain unknown.
DEAD-box helicases can promote viral particle assembly and incorporation into viral particles. DDX3X is found in several virion particles, such as Hepatitis B virus [93], and human cytomegalovirus (HCMV) [94]. The DEAD-box helicase might bind to viral genome RNA (or DNA) to maintain certain secondary structures to protect the viral genome, or interact with other proteins to maintain the stability of viral particles. DEAD-box helicases are essential sensors, regulators and effectors for the host innate immunity antiviral defense [74].
Although some DEAD-box helicases can be found inside viral particles, none are encoded in viral genomes. One possible explanation is that the DEAD-box helicases have so many important functions, it is evolutionarily a selective advantage for keeping DEAD-box genes inside the host genome, while allowing the viral genome to be as minimized as possible. Rather than encoding their own, viruses hijack host DEAD-box helicases to aid in transcription, translation, and viral packaging.
Based on the possible role of DDX3X and DDX5 in cancer, high throughput screening has been applied to search for inhibitors. It is early for these possible inhibitors to be counter-screened for anti-viral activity. The biological outcomes for DEAD-box helicases, especially DDX3X, makes this class of helicases a logical anti-viral for HCV and SARS2. Added to the mix, DDX3X can also act as an oncogene in the development of different types of cancers, suggesting that small molecule inhibitors developed as antivirals may also act as anticancer drugs [95], [96]. Importantly, DDX3X-specific compounds have the potential to inhibit a key host helicase involved with the replication of multiple co-infecting viruses, such as HIV-1 with HCV with a single, targeted therapeutic. Thus, the development of DEAD-box helicase inhibitors, and their mechanism of action are increasingly recognized as a prime target in viral and cancer treatments.
As with viral-specific helicases such as NS3 and NSP13, compounds that are specific for the DDX3X helicase activity are crucial for reducing cellular toxicities due to off-target effects. Screening of multiple libraries of commercially available compounds against the dsRNA DDX3X helicase activity yielded [97], [98], 3-(5-butyl-1,2-dihydrotriazol-3-yl)-N-(2-hydroxyphenyl)benzenesulfonamide [54] (Fig. 7 ). This compound competitively binds DDX3X along the RNA channel of the helicase [54]. Surprisingly, this compound and derivatives in the series had low toxicities in cell culture. Additionally, a favorable IC50 of 0.3 μM for the DDX3X helicase resulted in a promising replication inhibitor of HIV, HCV, Dengue virus and West Nile virus. The aqueous solubility of the compound is low hampering use in animal studies. Nevertheless, the compounds have shown great promise as a DDX3X-specific compound with pan-antiviral properties.
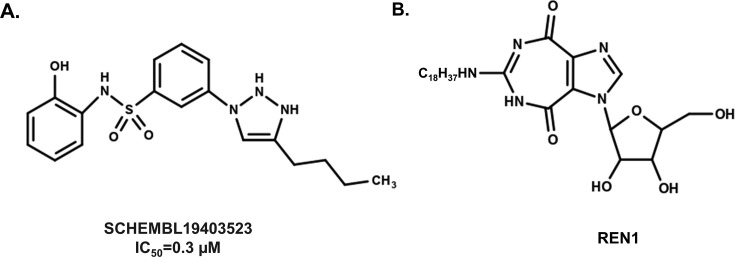
Fig. 7.
Structures of inhibitors of the DEAD-box helicase DDX3X. (A) Structure of the DDX3X inhibitor 3-(5-butyl-1,2-dihydrotriazol-3-yl)-N-(2-hydroxyphenyl)benzenesulfonamide (PubChem Name: SCHEMBL19403523). The IC50 for the RNA-dependent helicase activity for DDX3X is 0.3 μM [54]. (B) Structure of the ring-expanded nucleoside (REN1) ATPase inhibitor for DDX3 and the NS3 helicases from multiple viruses [99], [100], [101].
A separate class of DEAD-box helicase inhibitors is based on a ring-expanded nucleoside (REN) structure that mimics the adenosine nucleoside (Fig. 7) [99], [100], [101]. These compounds inhibit the activity of DDX3X, though an IC50 for DDX3X ATPase activity has not been reported. RENs also bind and inhibit the NS3 helicases from HCV, West Nile and Dengue viruses with estimated IC50 values in the low micromolar range [101]. The mechanism is thought to involve competitive binding at the ATPase site suggesting a pan-helicase activity that spans both a DEAD-box helicase as well as viral-encoded proteins. Simultaneous inhibition of viral helicases as well as DDX3X provides an attractive therapeutic agent for patients with co-infections of multiple viruses.
RNA viruses will continue to cause major health issues for the foreseeable future. The rapid development of vaccines for SARS2 is highly effective and could provide control at least in the short term. The ability of the virus to mutate rapidly indicates that more than one immunogenic target is needed to reduce the risk of additional infection. A major outcome of the fight against RNA virus’ is the extensive knowledge that is accumulating based on increased research funding. We now know much about the mutation frequency due to the low fidelity RNA polymerases in these viruses. However, an example of what we do not know is how SARS2 uses its proofreading capacity to enhance overall fidelity during viral replication. Indeed, we need to understand the enzyme mechanisms of the entire replication and transcription cycle. The role of the helicase needs to be determined to aid in the development of highly effective anti-virals.
References
- 1.Brosh R.M., Matson S.W. History of DNA helicases. Genes (Basel) 2020;11 doi: 10.3390/genes11030255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pyle A.M. RNA helicases and remodeling proteins. Curr. Opin. Chem. Biol. 2011;15:636–642. doi: 10.1016/j.cbpa.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lohman T.M., Tomko E.J., Wu C.G. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat. Rev. Mol. Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- 4.Singleton M.D.D.W.M.R. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 5.Bourgeois C.F., Mortreux F., Auboeuf D. The multiple functions of RNA helicases as drivers and regulators of gene expression. Nat. Rev. Mol. Cell Biol. 2016;17:426–438. doi: 10.1038/nrm.2016.50. [DOI] [PubMed] [Google Scholar]
- 6.Gorbalenya A.E., Koonin E.V. Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 1993;3:419–429. doi: 10.1016/S0959-440X(05)80116-2. [DOI] [Google Scholar]
- 7.Lucius A.L., Maluf N.K., Fischer C.J., Lohman T.M. General methods for analysis of sequential “n-step” kinetic mechanisms: application to single turnover kinetics of helicase-catalyzed DNA unwinding. Biophys. J. 2003;85:2224–2239. doi: 10.1016/S0006-3495(03)74648-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramanagoudr-Bhojappa R., Chib S., Byrd A.K., Aarattuthodiyil S., Pandey M., Patel S.S., Raney K.D. Yeast Pif1 helicase exhibits a one-base-pair stepping mechanism for unwinding duplex DNA. J. Biol. Chem. 2013;288:16185–16195. doi: 10.1074/jbc.M113.470013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manosas M., Xi X.G., Bensimon D., Croquette V. Active and passive mechanisms of helicases. Nucleic Acids Res. 2010;38:5518–5526. doi: 10.1093/nar/gkq273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Byrd A.K., Matlock D.L., Bagchi D., Aarattuthodiyil S., Harrison D., Croquette V., Raney K.D. Dda helicase tightly couples translocation on single-stranded DNA to unwinding of duplex DNA: Dda is an optimally active helicase. J. Mol. Biol. 2012;420:141–154. doi: 10.1016/j.jmb.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Q., Arnold J.J., Uchida A., Raney K.D., Cameron C.E. Phosphate release contributes to the rate-limiting step for unwinding by an RNA helicase. Nucleic Acids Res. 2009;38:1312–1324. doi: 10.1093/nar/gkp1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenberg E.S., Rosenthal E.M., Hall E.W., Barker L., Hofmeister M.G., Sullivan P.S., Dietz P., Mermin J., Ryerson A.B. Prevalence of hepatitis C virus infection in US states and the district of Columbia, 2013 to 2016. JAMA Netw. Open. 2018;1 doi: 10.1001/jamanetworkopen.2018.6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manns M.P., Buti M., Gane E., Pawlotsky J.M., Razavi H., Terrault N., Younossi Z. Hepatitis C virus infection. Nat. Rev. Dis. Prim. 2017;3 doi: 10.1038/nrdp.2017.6. [DOI] [PubMed] [Google Scholar]
- 14.Mei Y.-Y., Chen Y.-M., Wu Y.-K., Zhang X.-H., Xu W.-X. Efficacy and safety of sofosbuvir-based direct-acting antiviral agents treatment for patients with genotype 3/6 Hepatitis C virus infection. Can J Gastroenterol Hepatol. 2020 doi: 10.1155/2020/8872120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Penin F., Dubuisson J., Rey F.A., Moradpour D., Pawlotsky J.-M. Structural biology of hepatitis C virus. Hepatology. 2004;39:5–19. doi: 10.1002/hep.20032. [DOI] [PubMed] [Google Scholar]
- 16.Paul D., Madan V., Bartenschlager R. Hepatitis C virus RNA replication and assembly: living on the fat of the land. Cell Host Microbe. 2014;16:569–579. doi: 10.1016/j.chom.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frick D.N. HCV helicase: structure, function, and inhibition. Hepat. C Viruses Genomes Mol. Biol. 2006:207–244. [PubMed] [Google Scholar]
- 18.Lim S.K., Othman R., Yusof R., Heh C.H. Rational drug discovery: Ellagic acid as a potent dual-target inhibitor against hepatitis C virus genotype 3 (HCV G3) NS3 enzymes. Chem. Biol. Drug Des. 2021;97:28–40. doi: 10.1111/cbdd.13756. [DOI] [PubMed] [Google Scholar]
- 19.Ruggiero E., Richter S.N. Annu. Rep. Med. Chem. Academic Press Inc.; 2020. Viral G-quadruplexes: New frontiers in virus pathogenesis and antiviral therapy; pp. 101–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rhodes D., Lipps H.J. Survey and summary G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015;43:8627–8637. doi: 10.1093/nar/gkv862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sauer M., Paeschke K. G-quadruplex unwinding helicases and their function in vivo. Biochem. Soc. Trans. 2017;45:1173–1182. doi: 10.1042/BST20170097. [DOI] [PubMed] [Google Scholar]
- 22.Wang S.R., Min Y.Q., Wang J.Q., Liu C.X., Fu B.S., Wu F., Wu L.Y., Qiao Z.X., Song Y.Y., Xu G.H., Wu Z.G., Huang G., Peng N.F., Huang R., Mao W.X., Peng S., Chen Y.Q., Zhu Y., Tian T., Zhang X.L., Zhou X. A highly conserved G-rich consensus sequence in hepatitis C virus core gene represents a new anti–hepatitis C target. Sci. Adv. 2016;2 doi: 10.1126/sciadv.1501535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaubert C., Bedrat A., Bartolucci L., Di Primo C., Ventura M., Mergny J.-L., Amrane S., Andreola M.-L. RNA synthesis is modulated by G-quadruplex formation in Hepatitis C virus negative RNA strand OPEN. Sci. Rep. 2018;8:8120. doi: 10.1038/s41598-018-26582-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Byrd A.K., Raney K.D. Superfamily 2 helicases. Front. Biosci. 2012;17:2070–2088. doi: 10.2741/4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mukherjee S., Weiner W.S., Schroeder C.E., Simpson D.S., Hanson A.M., Sweeney N.L., Marvin R.K., Ndjomou J., Kolli R., Isailovic D., Schoenen F.J., Frick D.N. Ebselen inhibits hepatitis C virus NS3 helicase binding to nucleic acid and prevents viral replication. ACS Chem. Biol. 2014;9:2393–2403. doi: 10.1021/cb500512z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rajagopal V., Patel S.S. In: Viral Genome Replication. Raney K., Gotte M., Cameron C., editors. Springer; Boston, MA: 2009. Viral Helicases. [DOI] [Google Scholar]
- 27.Salam K.A., Akimitsu N. Hepatitis C virus NS3 inhibitors: current and future perspectives. Biomed. Res. Int. 2013;2013 doi: 10.1155/2013/467869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Furuta A., Tsubuki M., Endoh M., Miyamoto T., Tanaka J., Salam K.A., Akimitsu N., Tani H., Yamashita A., Moriishi K., Nakakoshi M., Sekiguchi Y., Tsuneda S., Noda N. Identification of Hydroxyanthraquinones as novel inhibitors of hepatitis C virus NS3 helicase. Int. J. Mol. Sci. 2015;16:18440. doi: 10.3390/ijms160818439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Furuta A., Salam K.A., Hermawan I., Akimitsu N., Tanaka J., Tani H., Yamashita A., Moriishi K., Nakakoshi M., Tsubuki M., Peng P.W., Suzuki Y., Yamamoto N., Sekiguchi Y., Tsuneda S., Noda N. Identification and biochemical Charachterization of Halisulfate 3 and Suvanine as novel inhibitors of hepatitis C virus NS3 helicase from a marine sponge. Mar. Drugs. 2014;12:462–476. doi: 10.3390/md12010462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salam K.A., Furuta A., Noda N., Tsuneda S., Sekiguchi Y., Yamashita A., Moriishi K., Nakakoshi M., Tsubuki M., Tani H., Tanaka J., Akimitsu N. Psammaplin a Inhibits Hepatitis C Virus NS3 Helicase. J. Nat. Med. 2013;67:765–772. doi: 10.1007/s11418-013-0742-7. [DOI] [PubMed] [Google Scholar]
- 31.Furuta A., Salam K.A., Akimitsu N., Tanaka J., Tani H., Yamashita A., Moriishi K., Nakakoshi M., Tsubuki M., Sekiguchi Y., Tsuneda S., Noda N. Cholesterol sulfate as a potential inhibitor of hepatitis C virus NS3 helicase. J. Enzyme Inhib. Med. Chem. 2014;29:223–229. doi: 10.3109/14756366.2013.766607. [DOI] [PubMed] [Google Scholar]
- 32.Salam K.A., Furuta A., Noda N., Tsuneda S., Sekiguchi Y., Yamashita A., Moriishi K., Nakakoshi M., Tsubuki M., Tani H., Tanaka J., Akimitsu N. Inhibition of Hepatitis C Virus NS3 helicase by manoalide. J Nat Prod. 2012;75:650–654. doi: 10.1021/np200883s. [DOI] [PubMed] [Google Scholar]
- 33.Lehmann K.C., Snijder E.J., Posthuma C.C., Gorbalenya A.E. What we know but do not understand about nidovirus helicases. Virus Res. 2015 doi: 10.1016/j.virusres.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huston N.C., Wan H., Strine M.S., de Cesaris Araujo Tavares R., Wilen C.B., Pyle A.M. Comprehensive in vivo secondary structure of the SARS-CoV-2 genome reveals novel regulatory motifs and mechanisms. Mol. Cell. 2021;81:584–598.e5. doi: 10.1016/j.molcel.2020.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshimoto F.K. The proteins of severe acute respiratory syndrome Coronavirus-2 (SARS CoV-2 or n-COV19), the cause of COVID-19. Protein J. 2020;39:198–216. doi: 10.1007/s10930-020-09901-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanaan J., Raj S., Decourty L., Saveanu C., Croquette V., Le Hir H. UPF1-like helicase grip on nucleic acids dictates processivity. Nat. Commun. 2018;9 doi: 10.1038/s41467-018-06313-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fiorini F., Bagchi D., Le Hir H., Croquette V. Human Upf1 is a highly processive RNA helicase and translocase with RNP remodelling activities. Nat. Commun. 2015;6 doi: 10.1038/ncomms8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seybert A., Posthuma C.C., Van Dinten L.C., Snijder E.J., Gorbalenya A.E., Ziebuhr J. A complex zinc finger controls the enzymatic activities of Nidovirus helicases. J. Virol. 2005;79:696–704. doi: 10.1128/JVI.79.2.696-704.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jia Z., Yan L., Ren Z., Wu L., Wang J., Guo J., Zheng L., Ming Z., Zhang L., Lou Z., Rao Z. Delicate structural coordination of the severe acute respiratory syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res. 2019;47:6538–6550. doi: 10.1093/nar/gkz409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen J., Malone B., Llewellyn E., Grasso M., Shelton P.M.M., Olinares P.D.B., Maruthi K., Eng E.T., Vatandaslar H., Chait B.T., Kapoor T.M., Darst S.A., Campbell E.A. Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Cell. 2020;182:1560–1573.e13. doi: 10.1016/j.cell.2020.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan L., Zhang Y., Ge J., Zheng L., Gao Y., Wang T., Jia Z., Wang H., Huang Y., Li M., Wang Q., Rao Z., Lou Z. Architecture of a SARS-CoV-2 mini replication and transcription complex. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-19770-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan L., Ge J., Zheng L., Zhang Y., Gao Y., Wang T., Huang Y., Yang Y., Gao S., Li M., Liu Z., Wang H., Li Y., Chen Y., Guddat L.W., Wang Q., Rao Z., Lou Z. Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis. Cell. 2021;184:184–193.e10. doi: 10.1016/j.cell.2020.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deng Z., Lehmann K.C., Li X., Feng C., Wang G., Zhang Q., Qi X., Yu L., Zhang X., Feng W., Wu W., Gong P., Tao Y., Posthuma C.C., Snijder E.J., Gorbalenya A.E., Chen Z. Structural basis for the regulatory function of a complex zinc-binding domain in a replicative arterivirus helicase resembling a nonsense-mediated mRNA decay helicase. Nucleic Acids Res. 2014;42:3464–3477. doi: 10.1093/nar/gkt1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hao W., Wojdyla J.A., Zhao R., Han R., Das R., Zlatev I., Manoharan M., Wang M., Cui S. Crystal structure of Middle East respiratory syndrome coronavirus helicase. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ivanov K.A., Thiel V., Dobbe J.C., van der Meer Y., Snijder E.J., Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 2004;78:5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanner J.A., Watt R.M., Chai Y.B., Lu L.Y., Lin M.C., Peiris J.S.M., Poon L.L.M., Kung H.F., Huang J.D. The severe acute respiratory syndrome (SARS) coronavirus NTPase helicase belongs to a distinct class of 5′ to 3′ viral helicases. J. Biol. Chem. 2003;278:39578–39582. doi: 10.1074/jbc.C300328200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adedeji A.O., Marchand B., Te Velthuis A.J.W., Snijder E.J., Weiss S., Eoff R.L., Singh K., Sarafianos S.G. Mechanism of nucleic acid unwinding by SARS-CoV helicase. PLoS One. 2012;7 doi: 10.1371/journal.pone.0036521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adedeji A.O., Singh K., Sarafianos S.G. Structural and biochemical basis for the difference in the helicase activity of two different constructs of SARS-CoV helicase. Cell. Mol. Biol. (Noisy-le-Grand) 2012;58:114–121. [PMC free article] [PubMed] [Google Scholar]
- 49.Lee N.R., Kwon H.M., Park K., Oh S., Jeong Y.J., Kim D.E. Cooperative translocation enhances the unwinding of duplex DNA by SARS coronavirus helicase nsP13. Nucleic Acids Res. 2010;38:7626–7636. doi: 10.1093/nar/gkq647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jang K.J., Jeong S., Kang D.Y., Sp N., Yang Y.M., Kim D.E. A high ATP concentration enhances the cooperative translocation of the SARS coronavirus helicase nsP13 in the unwinding of duplex RNA. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-61432-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ariumi Y. Multiple functions of DDX3 RNA helicase in gene regulation, tumorigenesis, and viral infection. Front. Genet. 2014;5 doi: 10.3389/fgene.2014.00423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pène V., Li Q., Sodroski C., Hsu C.-S., Liang T.J. Dynamic interaction of stress granules, DDX3X, and IKK-α mediates multiple functions in hepatitis C virus infection. J. Virol. 2015;89:5462–5477. doi: 10.1128/jvi.03197-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reid C.R., Hobman T.C. The nucleolar helicase DDX56 redistributes to West Nile virus assembly sites. Virology. 2017;500:169–177. doi: 10.1016/j.virol.2016.10.025. [DOI] [PubMed] [Google Scholar]
- 54.Brai A., Martelli F., Riva V., Garbelli A., Fazi R., Zamperini C., Pollutri A., Falsitta L., Ronzini S., Maccari L., Maga G., Giannecchini S., Botta M. DDX3X helicase inhibitors as a new strategy to fight the West Nile virus infection. J. Med. Chem. 2019;62:2333–2347. doi: 10.1021/acs.jmedchem.8b01403. [DOI] [PubMed] [Google Scholar]
- 55.Chen J.Y., Chen W.N., Poon K.M.V., Zheng B.J., Lin X., Wang Y.X., Wen Y.M. Interaction between SARS-CoV helicase and a multifunctional cellular protein (Ddx5) revealed by yeast and mammalian cell two-hybrid systems. Arch. Virol. 2009;154:507–512. doi: 10.1007/s00705-009-0323-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmidt N., Lareau C.A., Keshishian H., Ganskih S., Schneider C., Hennig T., Melanson R., Werner S., Wei Y., Zimmer M., Ade J., Kirschner L., Zielinski S., Dölken L., Lander E.S., Caliskan N., Fischer U., Vogel J., Carr S.A., Bodem J., Munschauer M. The SARS-CoV-2 RNA–protein interactome in infected human cells. Nat. Microbiol. 2021;6:339–353. doi: 10.1038/s41564-020-00846-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Hüttenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Kain A.M., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.P., Liu Y.F., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O’Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu-Ozturk D., Wang H.Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d’Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., García-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.V’kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gorgulla C., Das K.M.P., Leigh K.E., Cespugli M., Fischer P.D., Wang Z.-F., Tesseyre G., Pandita S., Shnapir A., Calderaio A., Gechev M., Rose A., Lewis N., Hutcheson C., Yaffe E., Luxenburg R., Herce H.D., Durmaz V., Halazonetis T.D., Fackeldey K., Patten J.J., Chuprina A., Dziuba I., Plekhova A., Moroz Y., Radchenko D., Tarkhanova O., Yavnyuk I., Gruber C., Yust R., Payne D., Näär A.M., Namchuk M.N., Davey R.A., Wagner G., Kinney J., Arthanari H. A multi-pronged approach targeting SARS-CoV-2 proteins using ultra-large virtual screening. ChemRxiv Prepr. Serv. Chem. 2020;24:102021. doi: 10.26434/chemrxiv.12682316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ahmad S., Waheed Y., Ismail S., Bhatti S., Abbasi S.W., Muhammad K. Structure-based virtual screening identifies multiple stable binding sites at the RecA domains of SARS-CoV-2 helicase enzyme. Molecules. 2021;26 doi: 10.3390/molecules26051446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Umar H.I., Siraj B., Ajayi A., Jimoh T.O., Chukwuemeka P.O. Molecular docking studies of some selected gallic acid derivatives against five non-structural proteins of novel coronavirus. J. Genet. Eng. Biotechnol. 2021;19:16. doi: 10.1186/s43141-021-00120-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gurung A.B. In silico structure modelling of SARS-CoV-2 Nsp13 helicase and Nsp14 and repurposing of FDA approved antiviral drugs as dual inhibitors. Gene Rep. 2020;21 doi: 10.1016/j.genrep.2020.100860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spratt A.N., Gallazzi F., Quinn T.P., Lorson C.L., Sönnerborg A., Singh K. Coronavirus helicases: attractive and unique targets of antiviral drug-development and therapeutic patents. Expert Opin. Ther. Pat. 2021 doi: 10.1080/13543776.2021.1884224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Freidel M.R., Armen R.S. Mapping major SARS-CoV-2 drug targets and assessment of druggability using computational fragment screening: Identification of an allosteric small-molecule binding site on the Nsp13 helicase. PLoS One. 2021;16 doi: 10.1371/journal.pone.0246181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zia M., Muhammad S., Shafiq-urRehman S.B., Abbasi S.W., Al-Sehemi A.G., Chaudhary A.R., Bai F.Q. Exploring the potential of novel phenolic compounds as potential therapeutic candidates against SARS-CoV-2, using quantum chemistry, molecular docking and dynamic studies. Bioorganic Med. Chem. Lett. 2021;43 doi: 10.1016/j.bmcl.2021.128079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marecki J.C., Byrd A.K., Raney K.D. Methods Mol. Biol. Humana Press Inc.; 2021. Identifying RNA helicase inhibitors using duplex unwinding assays; pp. 53–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kousar K., Majeed A., Yasmin F., Hussain W., Rasool N. Phytochemicals from selective plants have promising potential against SARS-CoV-2: investigation and corroboration through molecular docking, MD simulations, and quantum computations. Biomed. Res. Int. 2020;2020 doi: 10.1155/2020/6237160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Adedeji A.O., Singh K., Calcaterra N.E., DeDiego M.L., Enjuanes L., Weiss S., Sarafianos S.G. Severe acute respiratory syndrome coronavirus replication inhibitor that interferes with the nucleic acid unwinding of the viral helicase. Antimicrob. Agents Chemother. 2012;56:4718–4728. doi: 10.1128/AAC.00957-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shu T., Huang M., Wu D., Ren Y., Zhang X., Han Y., Mu J., Wang R., Qiu Y., Zhang D.Y., Zhou X. SARS-coronavirus-2 Nsp13 possesses NTPase and RNA helicase activities that can be inhibited by bismuth salts. Virol. Sin. 2020;35:321–329. doi: 10.1007/s12250-020-00242-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee J.M., Cho J.B., Ahn H.C., Jeong Y.J. Selective inhibition of enzymatic activities of severe acute respiratory syndrome coronavirus helicase with a Thioxopyrimidine derivative. Bull. Korean Chem. Soc. 2016;37:2066–2068. doi: 10.1002/bkcs.11002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jang K.J., Lee N.R., Yeo W.S., Jeong Y.J., Kim D.E. Isolation of inhibitory RNA aptamers against severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase. Biochem. Biophys. Res. Commun. 2008;366:738–744. doi: 10.1016/j.bbrc.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Linder P., Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 2011;12:505–516. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- 73.Jarmoskaite I., Russell R. DEAD-box proteins as RNA helicases and chaperones. Wiley Interdiscip. Rev. RNA. 2011;2:135–152. doi: 10.1002/wrna.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Taschuk F., Cherry S. DEAD-box helicases: sensors, regulators, and effectors for antiviral defense. Viruses. 2020;12:181. doi: 10.3390/v12020181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang S., Xing Z., Pascuzzi P.E., Tran E.J. Metabolic adaptation to nutrients involves coregulation of gene expression by the RNA helicase Dbp2 and the Cyc8 corepressor in saccharomyces cerevisiae, G3 genes, genomes. Genet. 2017;7:2235–2247. doi: 10.1534/g3.117.041814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Putnam A.A., Jankowsky E. AMP sensing by DEAD-box RNA helicases. J. Mol. Biol. 2013;425:3839–3845. doi: 10.1016/j.jmb.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hilbert M., Karow A.R., Klostermeier D. The mechanism of ATP-dependent RNA unwinding by DEAD box proteins. Biol. Chem. 2009;390:1237–1250. doi: 10.1515/BC.2009.135. [DOI] [PubMed] [Google Scholar]
- 78.Liu F., Putnam A., Jankowsky E. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc. Natl. Acad. Sci. U. S. A. 2008;105:20209–20214. doi: 10.1073/pnas.0811115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ali M.A.M. DEAD-box RNA helicases: the driving forces behind RNA metabolism at the crossroad of viral replication and antiviral innate immunity. Virus Res. 2021;296:198352. doi: 10.1016/j.virusres.2021.198352. [DOI] [PubMed] [Google Scholar]
- 80.Goh P.-Y., Tan Y.-J., Lim S.P., Tan Y.H., Lim S.G., Fuller-Pace F., Hong W. Cellular RNA helicase p68 Relocalization and interaction with the hepatitis C virus (HCV) NS5B protein and the potential role of p68 in HCV RNA replication. J. Virol. 2004;78:5288–5298. doi: 10.1128/jvi.78.10.5288-5298.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tingting P., Caiyun F., Zhigang Y., Pengyuan Y., Zhenghong Y. Subproteomic analysis of the cellular proteins associated with the 3′ untranslated region of the hepatitis C virus genome in human liver cells. Biochem. Biophys. Res. Commun. 2006;347:683–691. doi: 10.1016/j.bbrc.2006.06.144. [DOI] [PubMed] [Google Scholar]
- 82.Fang J., Kubota S., Yang B., Zhou N., Zhang H., Godbout R., Pomerantz R.J. A DEAD box protein facilitates HIV-1 replication as a cellular co-factor of rev. Virology. 2004;330:471–480. doi: 10.1016/j.virol.2004.09.039. [DOI] [PubMed] [Google Scholar]
- 83.Edgcomb S.P., Carmel A.B., Naji S., Ambrus-Aikelin G., Reyes J.R., Saphire A.C.S., Gerace L., Williamson J.R. DDX1 is an RNA-dependent ATPase involved in HIV-1 rev function and virus replication. J. Mol. Biol. 2012;415:61–74. doi: 10.1016/j.jmb.2011.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yedavalli V.S.R.K., Neuveut C., Chi Y.H., Kleiman L., Jeang K.T. Requirement of DDX3 DEAD box RNA helicase for HIV-1 rev-RRE export function. Cell. 2004;119:381–392. doi: 10.1016/j.cell.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 85.Lorgeoux R.P., Guo F., Liang C. From promoting to inhibiting: diverse roles of helicases in HIV-1 Replication. Retrovirology. 2012;9 doi: 10.1186/1742-4690-9-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Diot C., Fournier G., Dos Santos M., Magnus J., Komarova A., Van Der Werf S., Munier S., Naffakh N. Influenza A virus polymerase recruits the RNA Helicase DDX19 to promote the nuclear export of viral mRNAs. Sci. Rep. 2016;6 doi: 10.1038/srep33763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Soto-Rifo R., Rubilar P.S., Limousin T., De Breyne S., Décimo D., Ohlmann T. DEAD-box protein DDX3 associates with eIF4F to promote translation of selected mRNAs. EMBO J. 2012;31:3745–3756. doi: 10.1038/emboj.2012.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Soto-Rifo R., Rubilar P.S., Ohlmann T. The DEAD-box helicase DDX3 substitutes for the cap-binding protein eIF4E to promote compartmentalized translation initiation of the HIV-1 genomic RNA. Nucleic Acids Res. 2013;41:6286–6299. doi: 10.1093/nar/gkt306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lai M.C., Wang S.W., Cheng L., Tarn W.Y., Tsai S.J., Sun H.S. Human DDX3 Interacts with the HIV-1 Tat Protein to Facilitate Viral mRNA Translation. PLoS One. 2013;8 doi: 10.1371/journal.pone.0068665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yasuda-Inoue M., Kuroki M., Ariumi Y. DDX3 RNA helicase is required for HIV-1 tat function. Biochem. Biophys. Res. Commun. 2013;441:607–611. doi: 10.1016/j.bbrc.2013.10.107. [DOI] [PubMed] [Google Scholar]
- 91.Hondele M., Sachdev R., Heinrich S., Wang J., Vallotton P., Fontoura B.M.A., Weis K. DEAD-box ATPases are global regulators of phase-separated organelles. Nature. 2019;573:144–148. doi: 10.1038/s41586-019-1502-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ariumi Y., Kuroki M., Kushima Y., Osugi K., Hijikata M., Maki M., Ikeda M., Kato N. Hepatitis C virus hijacks P-body and stress granule components around lipid droplets. J. Virol. 2011;85:6882–6892. doi: 10.1128/jvi.02418-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang H., Kim S., Ryu W.-S. DDX3 DEAD-box RNA helicase inhibits hepatitis B virus reverse transcription by incorporation into Nucleocapsids. J. Virol. 2009;83:5815–5824. doi: 10.1128/jvi.00011-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Varnum S.M., Streblow D.N., Monroe M.E., Smith P., Auberry K.J., Paša-Tolić L., Wang D., Camp D.G., Rodland K., Wiley S., Britt W., Shenk T., Smith R.D., Nelson J.A. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 2004;78:10960–10966. doi: 10.1128/jvi.78.20.10960-10966.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kukhanova M.K., Karpenko I.L., Ivanov A.V. DEAD-box RNA helicase DDX3: Functional properties and development of DDX3 inhibitors as antiviral and anticancer drugs. Molecules. 2020;25 doi: 10.3390/molecules25041015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Heerma van Voss M.R., van Diest P.J., Raman V. Targeting RNA helicases in cancer: the translation trap. Biochim. Biophys. Acta - Rev. Cancer. 2017;1868:510–520. doi: 10.1016/j.bbcan.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Brai A., Fazi R., Tintori C., Zamperini C., Bugli F., Sanguinetti M., Stigliano E., Esté J., Badia R., Franco S., Martinez M.A., Martinez J.P., Meyerhans A., Saladini F., Zazzi M., Garbelli A., Maga G., Botta M. Human DDX3 protein is a valuable target to develop broad spectrum antiviral agents. Proc. Natl. Acad. Sci. U. S. A. 2016;113:5388–5393. doi: 10.1073/pnas.1522987113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fazi R., Tintori C., Brai A., Botta L., Selvaraj M., Garbelli A., Maga G., Botta M. Homology model-based virtual screening for the identification of human helicase DDX3 inhibitors. J. Chem. Inf. Model. 2015;55:2443–2454. doi: 10.1021/acs.jcim.5b00419. [DOI] [PubMed] [Google Scholar]
- 99.Yedavalli V.S.R.K., Zhang N., Cai H., Zhang P., Starost M.F., Hosmane R.S., Jeang K.T. Ring expanded nucleoside analogues inhibit RNA helicase and intracellular human immunodeficiency virus type 1 replication. J. Med. Chem. 2008;51:5043–5051. doi: 10.1021/jm800332m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang N., Chen H.M., Koch V., Schmitz H., Minczuk M., Stepien P., Fattom A.I., Naso R.B., Kalicharran K., Borowski P., Hosmane R.S. Potent inhibition of NTPase/helicase of the West Nile virus by ring-expanded (“fat”) nucleoside analogues. J. Med. Chem. 2003;46:4776–4789. doi: 10.1021/jm030277k. [DOI] [PubMed] [Google Scholar]
- 101.Zhang N., Chen H.M., Koch V., Schmitz H., Liao C.L., Bretner M., Bhadti V.S., Fattom A.I., Naso R.B., Hosmane R.S., Borowski P. Ring-expanded (“fat”) nucleoside and nucleotide analogues exhibit potent in vitro activity against Flaviviridae NTPases/helicases, including those of the West Nile virus, hepatitis C virus, and Japanese encephalitis virus. J. Med. Chem. 2003;46:4149–4164. doi: 10.1021/jm030842j. [DOI] [PubMed] [Google Scholar]
- 102.Adedeji A.O., Singh K., Kassim A., Coleman C.M., Elliott R., Weiss S.R., Frieman M.B., Sarafianos S.G. Evaluation of SSYA10-001 as a replication inhibitor of severe acute respiratory syndrome, mouse hepatitis, and Middle East respiratory syndrome coronaviruses. Antimicrob. Agents Chemother. 2014;58:4894–4898. doi: 10.1128/AAC.02994-14. [DOI] [PMC free article] [PubMed] [Google Scholar]