Abstract
Background
Alcohol is an established risk factor for several cancers, but modest alcohol-cancer associations may be missed because of measurement error in self-reported assessments. Biomarkers of habitual alcohol intake may provide novel insight into the relationship between alcohol and cancer risk.
Methods
Untargeted metabolomics was used to identify metabolites correlated with self-reported habitual alcohol intake in a discovery dataset from the European Prospective Investigation into Cancer and Nutrition (EPIC; n = 454). Statistically significant correlations were tested in independent datasets of controls from case-control studies nested within EPIC (n = 280) and the Alpha-Tocopherol, Beta-Carotene Cancer Prevention (ATBC; n = 438) study. Conditional logistic regression was used to estimate odds ratios (ORs) and 95% confidence intervals (CIs) for associations of alcohol-associated metabolites and self-reported alcohol intake with risk of pancreatic cancer, hepatocellular carcinoma (HCC), liver cancer, and liver disease mortality in the contributing studies.
Results
Two metabolites displayed a dose-response association with self-reported alcohol intake: 2-hydroxy-3-methylbutyric acid and an unidentified compound. A 1-SD (log2) increase in levels of 2-hydroxy-3-methylbutyric acid was associated with risk of HCC (OR = 2.54, 95% CI = 1.51 to 4.27) and pancreatic cancer (OR = 1.43, 95% CI = 1.03 to 1.99) in EPIC and liver cancer (OR = 2.00, 95% CI = 1.44 to 2.77) and liver disease mortality (OR = 2.16, 95% CI = 1.63 to 2.86) in ATBC. Conversely, a 1-SD (log2) increase in questionnaire-derived alcohol intake was not associated with HCC or pancreatic cancer in EPIC or liver cancer in ATBC but was associated with liver disease mortality (OR = 2.19, 95% CI = 1.60 to 2.98) in ATBC.
Conclusions
2-hydroxy-3-methylbutyric acid is a candidate biomarker of habitual alcohol intake that may advance the study of alcohol and cancer risk in population-based studies.
In 2016, an estimated 2.8 million deaths, corresponding to 6.8% and 2.2% of age-standardized deaths in men and women, respectively, were attributed to alcohol use worldwide (1). Excessive alcohol consumption is an established risk factor for many acute and chronic health conditions (2), including cancers of the upper aerodigestive tract, female breast, liver, colon, and rectum (3). However, the relationship of alcohol, particularly light-to-moderate alcohol consumption, with other cancer sites remains controversial (4).
Self-reported alcohol intake is, like other dietary factors, prone to underreporting (5). Validation studies have shown larger correlations for alcohol intake measured via dietary questionnaire and 24-hour dietary recall than those many other dietary constituents; however, this information may not reflect the level of accuracy because alcohol is a sensitive exposure, making it susceptible to underreporting across self-reported assessments. Consequently, the extent and distribution of exposure misclassification are unknown (6), and it is likely that observed associations between alcohol use and disease risk in prospective studies are attenuated and that estimates of alcohol-attributable death and disease are underestimated. Biomarkers of liver function and oxidative stress are used to study alcohol-related liver injury and alcoholic liver disease (7,8), but most alcohol consumers, particularly light-to-moderate consumers, will never manifest alcoholic liver disease. There are also biomarkers of recent (eg, ethyl glucuronide) and heavy alcohol use (eg, carbohydrate deficient transferrin and phosphatidylethanol) (9-11). However, biomarkers of habitual alcohol use, including light-to-moderate drinking, are needed to better assess alcohol exposure in epidemiological studies and to improve risk estimates for diseases including cancer where modest associations may exist.
Metabolomics is a powerful tool for discovering dietary biomarkers. When used in an untargeted mode, it can detect a wide range of compounds in biological samples including metabolites formed during digestion, metabolism, and microbial fermentation (12,13), making it well suited for discovering novel biomarkers of exposure or response to habitual alcohol consumption. Herein, we applied a multistage design, using untargeted metabolomics and independent discovery and test datasets, to identify serum metabolites associated with habitual alcohol consumption among free-living individuals with a wide range of intake. We then estimated the associations of these candidate alcohol biomarkers with risk of pancreatic cancer, liver cancers, and liver disease mortality in the European Prospective Investigation into Cancer and Nutrition (EPIC) study and the Alpha-Tocopherol, Beta-Carotene Cancer Prevention Study (ATBC).
Methods
Study Design
EPIC recruitment and study procedures, including dietary assessment methods and blood collection, are described extensively elsewhere (14). Briefly, EPIC is a large cohort study of more than half a million men and women recruited between 1992 and 2000 in 23 European centers. Diet, including average daily alcohol intake, over the 12 months before enrollment was assessed by validated country-specific food frequency questionnaires designed to capture local dietary habits with high compliance. Country-specific self-reported alcohol intake was calculated based on the estimated average glass volume and ethanol content for wine, beer, cider, sweet liquor, distilled spirits, or fortified wines, using information collected in standardized 24-hour dietary recalls from a subset of the cohort (15). The correlation between alcohol intake estimated by food frequency questionnaires and 24-hour dietary recall was 0.79 (16). Blood samples were collected and stored at -196ºC under liquid nitrogen at the International Agency for Research on Cancer (IARC) for all countries except Sweden (-80°C freezers) and Denmark (-150°C, nitrogen vapor).
Our study included a discovery and 2 independent test datasets (see Figure 1). The discovery set (n = 454) was nested in the EPIC cross-sectional study (17,18). The first test set included control subjects from 2 EPIC nested case-control studies of hepatocellular carcinoma (HCC; n = 128) and pancreatic cancer (n = 152) with untargeted metabolomics data (19–21). The second test set included 2 nested case-control studies in the ATBC cohort of male Finnish smokers (22). In ATBC, participants reported on demographics, lifestyle, and medical history via questionnaires and donated a fasting serum sample at baseline, which was stored at -70°C. For this study, we excluded controls (as well as cases) with missing self-reported alcohol intake (n = 72) and those with samples that failed laboratory analysis (n = 18); of the remaining 864 observations, 438 were controls.
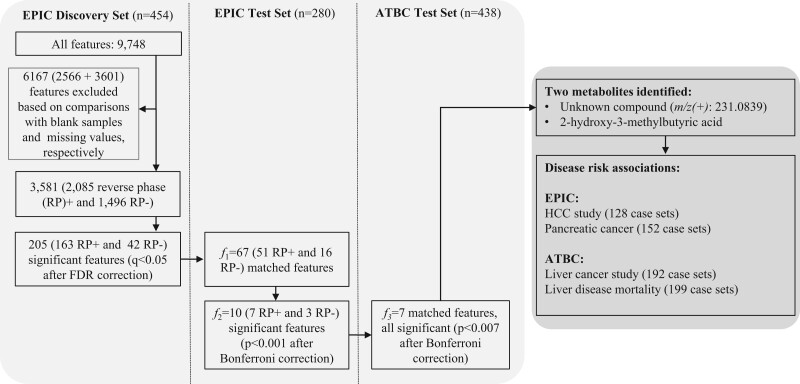
Figure 1.
Flowchart of the multistage study. The figure shows the features and samples size of the EPIC cross-sectional study that was used as a discovery set (stage 1) and the independent sets of cancer-free controls from EPIC (stage 2) and ATBC (stage 3), as well as of the etiological analyses in nested-case-control studies. ATBC = Alpha-Tocopherol, Beta-Carotene Cancer Prevention; EPIC = European Prospective Investigation into Cancer and Nutrition; FDR = false discovery rate; HCC = hepatocellular carcinoma; m/z = monoisotopic mass divided by the charge state values; RP = reverse phase.
In EPIC, nonmetastatic incident HCC (n = 128) and pancreatic cancer (n = 152) cases were matched 1:1 with cancer-free controls on study center, sex, age at blood collection (± 1 year), date (± 6 months), and time of the day (± 2 h) of blood collection, fasting status, and, for women, exogenous hormone use. Follow-up was based on a combination of methods, including health insurance records, registries, and active follow-up (14). Approval for the EPIC study was obtained from the IARC ethics review board (Lyon, France) and local review bodies of participating institutions. In ATBC, participants were passively followed during the postintervention period via linkage with the Finnish Cancer Registry and death registry. Liver cancer (n = 229) and liver disease mortality (n = 248) cases were individually matched 1:1 with controls, selected by incidence density sampling, on baseline age (± 5 years) and serum draw date (± 30 days) (23). After excluding ATBC cases and controls with missing data, 192 and 199 complete liver cancer and liver disease mortality case-control sets remained. Approval for the ATBC study was obtained from the institutional review boards of the National Cancer Institute (Bethesda, MD) and the National Public Health Institute of Finland. EPIC and ATBC studies were conducted according to the guidelines of the Declaration of Helsinki; all participants provided written informed consent.
Metabolomics Analyses
Sample analysis, data preprocessing, matching of features across datasets, and compound identification are described in detail in the Supplementary Methods (available online). Briefly, all samples were analyzed by the same laboratory at IARC with a UHPLC-QTOF-MS system (1290 Binary Liquid chromatography (LC) system, 6550 quadrupole time-of-flight (QTOF) mass spectrometer; Agilent Technologies, Santa Clara, CA) using reversed phase chromatography and electrospray ionization. Raw data were processed using Agilent MassHunter Qualitative analysis B.06.00, ProFinder B.08.00, and Mass Profiler Professional B.12.1 software with Agilent’s recursive feature finding procedure. The m/z (mass to charge ratio) values of the features of interest were searched against the Human Metabolome Database (HMDB) (24) and METLIN (25). Compound identity was confirmed by comparison of chemical standards and representative samples.
Statistical Analyses
We used an integrated workflow for metabolomics data analysis (26). Features detected in less than 50% of the discovery set samples and background features, (ie, feature intensities present in all blanks with ratio of geometric mean intensities of nonblank: blank samples <5) were excluded. Feature intensities were log2-transformed. Study participants with more than 50% missing features and those identified as outliers by a principal component analysis (PCA)-based approach were excluded (27). Missing values were imputed within each plate by a K-nearest neighbors method, with K = 10 (28). Last, feature intensities measured across plates within any single batch were normalized by applying a random forest-based approach to correct for unwanted variation (29). In the EPIC discovery set and test sets, these steps were applied on feature matrices acquired in positive and negative modes separately. In ATBC, these steps were applied on each batch.
In the discovery and test sets, self-reported alcohol intake (g/day) was adjusted for age, sex, country (in EPIC only), body mass index (kg/m2), smoking status and intensity, and coffee consumption (g/day, log-transformed) via the residual method in linear regression models (30). Coffee drinking and coffee-associated metabolites have been strongly associated with lower risk of liver cancer and liver disease mortality in ATBC (23,31); for consistency, coffee drinking was considered a potential confounder across discovery and test sets. Residuals for feature intensities were also adjusted for well-plate number within the analytical batch, position within the plate (row and column indexes), and the study (EPIC HCC or pancreatic cancer) or batch indicator (ATBC) as random effects. We used the principal component partial-R2 method (32) to quantify the contribution of alcohol and potential confounders to the variability of the 67 feature intensities that were statistically significantly associated with self-reported alcohol intake in the discovery set (33).
We calculated Pearson correlation coefficients using the residuals for self-reported alcohol intake and for feature intensities; correlations with a false discovery rate–corrected P value of less than .05 were considered statistically significant, and each feature in this set (f1) was carried forward for testing in our multistage design. After the discovery stage, f1 residual-adjusted correlation coefficients were computed and corrected by the more conservative Bonferroni method. The correlations between f1 features and self-reported alcohol with a P value of less than .05/f1 were considered statistically significant comprised a second set of features (f2) that were carried forward to the next stage in ATBC. Again, correlations between the residuals of self-reported alcohol intake and of feature intensities were calculated. The linearity of the association between standardized residuals of 2-hydroxy-3-methylbutyric acid and self-reported alcohol intake was evaluated with cubic regression splines with 5 knots (34), by comparing the log-likelihood of models with and without the nonlinear terms to a χ distribution with 2 degrees of freedom.
We estimated odds ratios (OR) and 95% confidence intervals (CI) for candidate features and HCC and pancreatic cancer in EPIC and liver cancer and fatal liver disease in ATBC using conditional logistic regression models. In crude models (conditioned on the matching criteria only), multivariable models, and multivariable models additionally adjusting for self-reported alcohol intake, log2-transformed feature intensities were centered and scaled (ie, mean = 0 [ 1]) to ensure comparability of odds ratio across different endpoints.
All statistical analyses were performed using the Statistical Analysis Software, release 9.4 (SAS Institute Inc, Cary, NC) and R version 3.6.0 (35), and statisical tests were 2-sided.
Results
Population Characteristics
Baseline participant characteristics are presented in Table 1. In the EPIC discovery set, most participants were women (57.5%) and never (52.2%) or former (26.4%) smokers. In the set of EPIC HCC and pancreatic cancer controls, there was a higher percentage of men (52.7%) and a lower percentage of never smokers (46.2%) than in the discovery set. In the set of ATBC liver cancer and liver disease death controls, all participants were Finnish men and current smokers. Median self-reported alcohol intake was 10.0 g/day, 6.6 g/day, and 11.5 g/day in the EPIC discovery, EPIC, and ATBC test sets, respectively.
Table 1.
Descriptive statistics of the EPIC and ATBC samples used to identify and confirm associations of metabolite features with self-reported alcohol intake
| Variable | EPIC discovery, stage 1a | EPIC controls, stage 2b | ATBC controls, stage 3c |
|---|---|---|---|
| Total No. | 454 | 280 | 438 |
| Men, % | 42.5 | 52.7 | 100 |
| BMI, median (10th%-90th%), kg/m2 | 25.8 (20.9-31.6) | 26.6 (20.7-34.1) | 26.2 (22.5-31.3) |
| Age, median (10th%-90th%), y | 55.2 (42.5-63.9) | 59.4 (49.0-68.6) | 56.0 (51.0-63.0) |
| Smoking status, % | |||
| Current | 18.5 | 19.2 | 100 |
| Former | 26.4 | 33.5 | — |
| Never | 52.2 | 46.2 | |
| Unknown | 2.9 | 1.1 | |
| Smoking intensity, median (10th%-90th%), cig/day | 11.5 (2-26) | 15 (4-30) | 20 (10-30) |
| Country, % | |||
| France | 14.5 | 0.4 | — |
| Italy | 34.8 | 18.5 | — |
| Spain | — | 10.0 | — |
| United Kingdom | — | 17.1 | — |
| The Netherlands | — | 10.3 | — |
| Greece | 12.3 | 10.7 | — |
| Germany | 38.3 | 24.9 | — |
| Denmark | — | 8.2 | — |
| Finland | — | — | 100 |
| Alcohol nondrinkers, %d | 8 | 14 | 9 |
| Alcohol intake, median (10th%-90th%), g/day | |||
| Men | 21.4 (1.3-50.4) | 14.9 (1.0-51.7) | 11.5 (0.2-42.1) |
| Women | 5.2 (0.02-24.9) | 2.0 (0.01-23.3) | — |
| Coffee intake, median (10th%-90th%), g/day | 146.3 (21.4-580.2) | 190 (3-857) | 550 (220-1100) |
aEPIC cross-sectional sample. ATBC = Alpha-Tocopherol, Beta-Carotene Cancer Prevention; EPIC = European Prospective Investigation into Cancer and Nutrition.
bControls from both liver and pancreatic cancer EPIC nested case-control studies.
cControls from liver cancer and liver disease mortality ATBC nested case-control studies excluding those with missing data on alcohol intake.
dAlcohol nondrinkers are considered as those with alcohol intake ≤0.1 g/day.
Biomarker Discovery Analysis
After excluding participant samples identified as outliers or as having too many missing values, the final discovery set (stage 1) comprised 451 and 452 study participants in positive and negative ionization mode datasets, respectively. The final EPIC test set (stage 2) comprised 271 and 277 study participants in positive and negative ionization datasets, respectively. Residuals of 205 features in the discovery set were statistically significantly correlated with residuals of self-reported alcohol intake (163 features in positive and 42 features in negative ionization mode; Figure 1), with correlation coefficients ranging from -0.29 to 0.50 in log-log plots (Supplementary Table 1, available online).
Of the 205 features in the discovery set, 51 features in positive and 16 features in negative ionization mode (f1 = 67) matched by mass and retention time with equivalent features in the EPIC test set and principal component partial-R2 analyses showed that self-reported alcohol intake explained more than 7% of variability in the feature intensities (f1 = 67; Figure 2). Residuals of f2 = 10 features were statistically significantly correlated with residuals of self-reported alcohol intake (Table 2). The first 2 features corresponded to a compound that could not be unequivocally identified but had an identical mass, isotope pattern, ion formation (mostly [M+Na]+ and [M+HCOOH-H]-) and retention time to ethyl glucoside (HMDB0029968) (37). However, chromatograms (Supplementary Methods, available online) indicated a lack of specificity, and although fragmentation of the [M+Na]+ ion could not be induced, our results suggest the unknown is a combination of ethyl-α-D-glucoside, ethyl-β-D-glucoside, and an additional structural isomer. The remaining 8 features corresponded to a single compound, which was confirmed by comparison with an authentic standard as 2-hydroxy-3-methylbutyric acid (HMDB0000407). Residuals of all 7 positive ionization mode features selected in the EPIC test set were positively correlated with residuals of self-reported alcohol in the ATBC test set (stage 3; Table 2).
Figure 2.
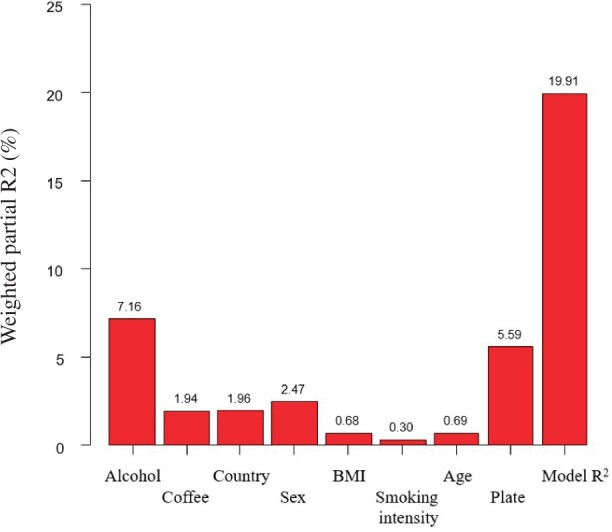
Principal component partial R2 analysis to quantify the contribution of potential confounder variables to the variability of the set of f1 = 67 feature intensities that were statistically significantly associated with alcohol intake in the discovery set. BMI = body mass index.
Table 2.
Feature-specific intensity and reproducibility (coefficient of variation [CV]) in quality control (QC) samples and adjusted Pearson correlation coefficients (r) with alcohol intake in the discovery and independent test sets
| m/z a | Retention time, minb |
Method | Associated metabolite | QC samplesc (n = 38) |
EPIC discovery (stage 1; n = 454)d |
EPIC controls (stage 2; n = 280)e |
ATBC controls (stage 3; n = 438) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean intensity | CV, % | r | P f | Q g | r | P h | r | P i | ||||
| 231.0839j | 0.89 | RP+ | Unknown | 58 378 | 18.5 | 0.41 | 1.2 x 10 -19 | 4.4 x 10-16 | 0.38 | 7.0 x 10-11 | 0.40 | 6.3 x 10-18 |
| 253.0925 | 0.93 | RP- | Unknown | 11 140 | 13.2 | 0.39 | 2.6 x 10-18 | 4.6 x 10-15 | 0.32 | 3.2 x 10-8 | —k | — |
| 203.0227j | 2.78 | RP+ | 2-hydroxy-3-methylbutyric acid | 204 079 | 14.8 | 0.26 | 1.9 x 10-8 | 2.0 x 10-6 | 0.24 | 5.3 x 10-5 | 0.40 | 1.1 x 10-18 |
| 217.9895 | 2.78 | RP+ | 2-hydroxy-3-methylbutyric acid | 36 539 | 11.7 | 0.30 | 9.0 x 10-11 | 2.1 x 10-8 | 0.25 | 2.3 x 10-5 | 0.38 | 2.4 x 10-16 |
| 250.0134 | 2.78 | RP+ | 2-hydroxy-3-methylbutyric acid | 122 838 | 12.5 | 0.28 | 9.0 x 10-10 | 1.6 x 10-7 | 0.27 | 8.2 x 10-6 | 0.40 | 3.5 x 10-18 |
| 221.0605 | 2.78 | RP+ | 2-hydroxy-3-methylbutyric acid | 56 192 | 11.2 | 0.28 | 2.6 x 10-9 | 3.2 x 10-7 | 0.25 | 2.1 x 10-5 | 0.39 | 1.9 x 10-17 |
| 218.9958 | 2.78 | RP+ | 2-hydroxy-3-methylbutyric acid | 115 590 | 11.7 | 0.28 | 1.3 x 10-9 | 2.1 x 10-7 | 0.26 | 1.8 x 10-5 | 0.40 | 1.7 x 10-18 |
| 235.0479 | 2.78 | RP+ | 2-hydroxy-3-methylbutyric acid | 34 447 | 15.5 | 0.20 | 2.3 x 10-5 | 1.0 x 10-3 | 0.26 | 2.1 x 10-5 | 0.38 | 4.2 x 10-16 |
| 117.0559 | 2.78 | RP- | 2-hydroxy-3-methylbutyric acid | 211 842 | 12.1 | 0.28 | 1.3 x 10-9 | 2.2 x 10-7 | 0.28 | 2.0 x 10-6 | —k | — |
| 261.9788 | 2.78 | RP- | 2-hydroxy-3-methylbutyric acid | 15 985 | 11.9 | 0.27 | 7.2 x 10-9 | 8.3 x 10-7 | 0.28 | 2.7 x 10-6 | —k | — |
m/z = monoisotopic mass divided by the charge state values, as observed in the discovery set.
Retention time.
Quality control samples within the discovery set. ATBC = Alpha-Tocopherol, Beta-Carotene Cancer Prevention; EPIC = European Prospective Investigation into Cancer and Nutrition; m/z = monoisotopic mass divided by the charge state values; RP = reverse phase; QC = Quality control.
The analyses of features acquired in positive and negative modes used data from 451 and 452 participants, respectively, after the exclusion of outliers and samples with too many missing values.
The analyses of features acquired in positive and negative modes used data from 271 and 277 participants, respectively, after the exclusion of outliers and samples with too many missing values.
P values for correlations computed in 2-sided tests.
Q values associated to false discovery rate (FDR) procedure to correct for multiple testing (36), alpha = 0.05.
Threshold for statistical significance corrected in 2-sided tests with Bonferroni method for multiple testing, equal to 0.0007463 (0.05/f1, with f1 = 67).
Threshold for statistical significance corrected in 2-sided tests with Bonferroni method for multiple testing, equal to 0.007 (0.05/f3, with f3 = 7).
Feature chosen for analysis of disease, see Table 3.
Feature not available in ATBC.
For subsequent analyses, the feature with the greatest chromatographic intensity (ie, main feature) for each metabolite was used (Table 2). In each of the 3 datasets, the residuals of the main features for the 2 candidate metabolites were statistically significantly correlated, with correlation coefficients ranging from 0.23 in the EPIC discovery set to 0.54 in the ATBC test set. The test for nonlinearity with cubic regression splines using restricted regression spline was marginally statistically significant for residuals of 2-hydroxy-3-methylbutyric acid and self-reported alcohol intake (P = .06; Supplementary Figure 1, available online).
Disease Risk Associations
In multivariable models (Table 3), 2-hydroxy-3-methylbutyric acid was associated with increased odds of HCC (OR1-SD = 2.54, 95% CI = 1.51 to 4.27) and pancreatic cancer (OR1-SD = 1.43, 95% CI = 1.03 to 1.99) in EPIC, as well as liver cancer (OR1-SD = 2.00, 95% CI = 1.44 to 2.77) and fatal liver disease (OR1-SD = 2.16, 95% CI = 1.63 to 2.86) in ATBC; associations remained following adjustment for self-reported alcohol intake. The unknown candidate biomarker was associated with increased odds of liver cancer (OR1-SD = 1.70, 95% CI = 1.29 to 2.25) and liver disease mortality (OR = 1.98, 95% CI = 1.51 to 2.60) in ATBC, and these associations were also independent of self-reported alcohol intake. However, the unknown was not associated with HCC or pancreatic cancer in EPIC. Self-reported alcohol intake was not associated with HCC (OR1-SD = 0.78, 95% CI = 0.56 to 1.09) or pancreatic cancer risk (OR1-SD = 1.03, 95% CI = 0.77 to 1.39) in EPIC but was strongly associated with liver disease mortality (OR1-SD = 2.19, 95% CI = 1.60 to 2.98) in ATBC. The alcohol findings are in line with previously published EPIC and ATBC analyses (37–39).
Table 3.
Crude and adjusted odds ratios (OR, 95% confidence interval [CI]) of self-reported alcohol intake (12 g/day) and the main features of the unknown compound and 2-hydroxy-3-methylbutyric acid (per 1-SD) with hepatocellular carcinoma (HCC; 129 case-control sets) and pancreatic cancer (152 case-control sets) in EPIC, and with liver cancer (194 case-control sets) and liver disease mortality (201 case-control sets) in ATBC
| Exposure | Crude models |
Adjusted modelsa |
Alcohol-adjusted modelsb |
||||||
|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | ||||
| HCC, EPIC (128 case-control sets) | |||||||||
| Alcohol intake, 12 g/day | 1.13 (1.00 to 1.27) | .05 | 1.04 (0.89 to 1.20) | .65 | — | — | |||
| Alcohol intake, 1-SD (log2) | 0.93 (0.73 to 1.20) | .59 | 0.78 (0.56 to 1.09) | .14 | — | — | |||
| Unknown compound, 1-SD (log2)c | 1.27 (0.92 to 1.76) | .15 | 1.01 (0.66 to 1.52) | .98 | 1.23 (0.75 to 2.01) | .40 | |||
| 2-hydroxy-3-methylbutyric acid, 1-SD (log2)d | 2.28 (1.52 to 3.43) | 7.0 x 10-5 | 2.54 (1.51 to 4.27) | 4.2 x 10-4 | 3.12 (1.74 to 5.56) | 4.2 x 10-4 | |||
| Pancreatic cancer, EPIC (152 case-control sets) | |||||||||
| Alcohol intake, 12 g/day | 1.07 (0.92 to 1.25) | .36 | 1.04 (0.88 to 1.24) | .65 | — | — | |||
| Alcohol intake, 1-SD (log2) | 1.08 (0.83 to 1.40) | .58 | 1.03 (0.77 to 1.39) | .83 | — | — | |||
| Unknown compound, 1-SD (log2)c | 1.15 (0.92 to 1.46) | .22 | 1.10 (0.91 to 1.41) | .48 | 1.10 (0.83 to 1.46) | .50 | |||
| 2-hydroxy-3-methylbutyric acid, 1-SD (log2)d | 1.43 (1.07 to 1.92) | .02 | 1.43 (1.03 to 1.99) | .03 | 1.46 (1.03 to 2.06) | .03 | |||
| Liver cancer, ATBC (192 case-control sets) | |||||||||
| Alcohol intake, 12 g/day | 1.25 (1.09 to 1.43) | .001 | 1.17 (1.01 to 1.36) | .03 | — | — | |||
| Alcohol intake, 1-SD (log2) | 1.33 (1.05 to 1.67) | .02 | 1.23 (0.94 to 1.60) | .13 | — | — | |||
| Unknown compound, 1-SD (log2)c | 1.34 (1.07 to 1.68) | .01 | 1.70 (1.29 to 2.25) | 2.0 x 10-4 | 1.76 (1.28 to 2.41) | 5.0 x 10-4 | |||
| 2-hydroxy-3-methylbutyric acid, 1-SD (log2)d | 2.08 (1.53 to 2.82) | 2.7 x 10-6 | 2.00 (1.44 to 2.77) | 3.4 x 10-5 | 2.07 (1.43 to 2.98) | .01 | |||
| Liver disease mortality, ATBC (199 case-control sets) | |||||||||
| Alcohol intake, 12 g/day | 1.38 (1.22 to 1.55) | 1.1 x 10-7 | 1.32 (1.16 to 1.50) | 1.6 x 10-5 | — | — | |||
| Alcohol intake, 1-SD (log2) | 2.37 (1.78 to 3.14) | 2.8 x 10-8 | 2.19 (1.60 to 2.98) | 8.4 x 10-7 | — | — | |||
| Unknown compound, 1-SD (log2)c | 2.11 (1.63 to 2.72) | 1.0 x 10-8 | 1.98 (1.51 to 2.60) | 8.6 x 10-7 | 1.65 (1.24 to 2.20) | 7.0 x 10-4 | |||
| 2-hydroxy-3-methylbutyric acid, 1-SD (log2)d | 2.26 (1.73 to 2.95) | 2.1 x 10-9 | 2.16 (1.63 to 2.86) | 9.6 x 10-8 | 1.85 (1.38 to 2.48) | 3.9 x 10-5 | |||
Models for hepatocellular carcinoma (HCC) were adjusted for body mass index (BMI; kg/m2), waist circumference (cm), recreational and household physical activity (Met-hours/week), a composite variable for smoking status and intensity (never, current: 1-15 cig/day; current: 16-25 cig/day; current: ≥26 cig/day; former: quit ≤10 years; former: quit 11-20 years; former: quit ≥20 years; current: occasional pipe/cigar/use; current/former: missing, unknown), level of educational attainment, and coffee intake [(log2)grams/day]; models for pancreatic cancer were adjusted for BMI (kg/m2), sex-specific physical activity categories, and the composite variable for smoking status and intensity; ATBC liver cancer and fatal liver disease models were adjusted for age (years), BMI (kg/m2), leisure time physical activity, smoking intensity (cigarettes/day), level of educational attainment, and coffee intake [(log2)grams/day]. ATBC = Alpha-Tocopherol, Beta-Carotene Cancer Prevention; EPIC = European Prospective Investigation into Cancer and Nutrition; m/z = monoisotopic mass divided by the charge state values.
Models were further adjusted for self-reported alcohol intake (log2)grams/day.
Unknown compound (m/z = 231.0839).
2-hydroxy-3-methylbutyric acid (m/z = 203.0227).
Discussion
Using untargeted metabolomics data from a discovery and 2 independent sets of cancer-free controls to validate correlations between candidate metabolite feature and self-reported alcohol, we found 2 serum metabolites that were highly correlated with self-reported habitual alcohol intake. One compound was identified as 2-hydroxy-3-methylbutyric acid; the other remains unknown but is likely a combination of isomers of ethyl glucoside. Of note, ethyl-α-D-glucoside is a known constituent of some alcoholic beverages (40). Notably, 2-hydroxy-3-methylbutyric acid was strongly associated with HCC and pancreatic cancer risks in EPIC and with liver cancer and fatal liver disease in ATBC, and these associations remained after adjustment for self-reported alcohol intake. This suggests that 2-hydroxy-3-methylbutyric acid, which is not a constituent or a by-product of alcohol intake, may reflect a relevant biological response to alcohol intake that potentially plays a role in the etiology of multiple chronic diseases. In contrast, self-reported alcohol intake was only consistently associated with liver disease mortality risk in ATBC. Further research is needed to elucidate the potential metabolic cascade from alcohol drinking to 2-hydroxy-3-methylbutyric acid to disease and to replicate and extend the observed associations. Additionally, targeted metabolomics panels that can simultaneously measure multiple alcohol-related metabolites using authentic standards, including 2-hydroxy-3-methylbutyric acid and related compounds, should be developed to measure absolute concentrations, which will enable comparisons and pooling of data across studies, supporting replication and improving risk estimation; this is especially important for diseases such as pancreatic cancer, for which the literature is suggestive (41) yet inconsistent (42).
Prior population-based studies have used a targeted or semitargeted metabolomics approach to identify alcohol-specific metabolomic profiles of self-reported alcohol intake. Three studies, including 1 in EPIC, used targeted metabolomics, measuring 123 to 163 metabolites, to gain insight into metabolic pathways linking alcohol drinking to human health (43–45); 10 alcohol-metabolite associations were common to all 3 studies and included phosphatidylcholines (PCs), LysoPCs, acylcarnitines, and sphingomyelins. Of note, PCs contribute to the formation of phosphatidylethanol in human tissues (46), which is a known biomarker of recent and heavy alcohol consumption used to diagnose alcohol abuse (47,48). A fourth targeted study used nuclear magnetic resonance to evaluate cross-sectional associations of 76 lipids, fatty acids, amino acids, ketone bodies, and gluconeogenesis-related metabolites with alcohol consumption (49). The endogenous metabolites identified by these targeted platforms did not overlap with the compounds most highly correlated with self-reported alcohol intake in our untargeted study, underscoring the breadth of the metabolome and discovery potential of untargeted metabolomics methods.
Metabolomics analyses that limit biomarker discovery to previously annotated compounds have also identified several alcohol-related biomarkers. For example, using prediagnostic serum samples from a nested breast cancer case-control study within a US cohort, self-reported alcohol intake was associated with 16 of the 617 annotated metabolites, including 2-hydroxy-3-methylbutyric acid, 2,3-dihydroxyisovaleric acid (ie, 2,3-hydroxy-3-methylbutyric acid), ethyl glucuronide, and several endogenous metabolites related to androgen metabolism (50). Other cross-sectional analyses, measuring hundreds of metabolites, also found associations of 2-hydroxy-3-methylbutyric acid, 2,3-dihydroxyisovaleric acid (ie, 2,3-hydroxy-2-methylbutyric acid), and ethyl glucuronide with self-reported alcohol intake using prediagnostic serum (51,52). However, these studies did not test associations in multiple, independent datasets and estimated correlations in cases and controls combined. One study, which reported using discovery and replication sets, evaluated associations between self-reported alcohol intake and 356 known metabolites among 1500 African Americans and carried statistically significant metabolites forward for testing in a smaller set of 477 African Americans (53). This study found that alcohol was associated with five 2-hydroxybutyrate-related metabolites including 2-hydroxy-3-methylbutyric acid (53). Also using a multistage design, a Japanese study of 107 metabolites identified positive associations between 2-hydroxybutyric acid and self-reported alcohol intake in a discovery set and independent test set (54).
The production of 2-hydroxy-3-methylbutyric acid and other hydroxybutyric acid–related metabolites is linked to the rate of hepatic glutathione synthesis, which can increase considerably in response to oxidative stress or detoxification of xenobiotics in the liver (55). A targeted metabolomics investigation in EPIC found evidence suggesting that glutathione metabolism is involved in the development of HCC (20). Additionally, 2-hydroxy-3-methylbutyric acid is a product of branched-chain amino acid metabolism, which has been linked to alcohol drinking (54,56). Finally, prior research on metabolite variability reported 1-year intraclass correlation coefficients for 2-hydroxy-3-methylbutyric acid (ie, alpha-hydroxyisovalerate) ranging from 0.76 to 0.49 in independent samples of 60 Chinese women and 30 US men and women, respectively (57), suggesting low to moderate within-subject variability (ie, good to moderate reliability) over 1 year.
To our knowledge, this study is unique in its untargeted metabolomics approach without preselected metabolites and its use of a multistage design to test the associations of thousands of metabolite features with self-reported alcohol intake in a large discovery dataset and then to retest candidate metabolite features in 2 independent sets of cancer-free controls. By considering nearly 7000 features, many of which are correlated, we greatly increased the number of potential candidates, but we also incurred stronger penalization for multiple testing. Consequently, our approach may have missed features that did not meet stringent statistical significance thresholds. A strength of our approach was the use of 3 large, independent datasets although matching features across sets may have resulted in the loss of relevant information. Other potential limitations relate to generalizability, measurement error, and changes in alcohol use over time. Circulating metabolite levels reflect environmental exposures as well as host and microbial metabolism (58–60), and identification of candidate biomarkers that are sufficiently specific to ethanol and generalizable to diverse populations is challenging. Measurement error, both systematic and random, is inherent to self-reported assessments (61–63) and likely biases association estimates in etiological studies as well as biomarker discovery studies. Additionally, self-reported alcohol intake and blood measures were assessed in each study at baseline only; therefore, we are unable to account for changes in alcohol intake or metabolites over time. Despite our use of cutting-edge untargeted metabolomics methods, a robust study design, and an etiological component to evaluate the associations of our candidate biomarkers with disease outcomes, we cannot dismiss the possibility that our findings were impacted by measurement error in self-reported alcohol intake.
In summary, we observed robust correlations between self-reported habitual alcohol intake and 2-hydroxy-3-methylbutyric acid and an unidentified compound in a discovery set and 2 independent test sets of cancer-free participants. Associations for 2-hydroxy-3-methylbutyric acid with risk of HCC and pancreatic cancer in the EPIC study and with liver cancer in ATBC were stronger than those for either self-reported alcohol intake or the unidentified compound. Both candidate biomarkers were associated with liver endpoints independent of self-reported alcohol intake, indicating value beyond being correlates of intake. In conclusion, 2-hydroxy-3-methylbutyric acid is a promising candidate biomarker for studying the relationship between habitual alcohol intake and health (50–53), but further research, preferably in the context of a randomized controlled trial, is needed to better characterize the relationship between 2-hydroxy-3-methylbutyric acid and alcohol at varying levels of intake.
Funding
The coordination of EPIC is financially supported by International Agency for Research on Cancer (IARC) and also by the Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, which has additional infrastructure support provided by the NIHR Imperial Biomedical Research Centre (BRC). This work was supported by the Intramural Research Program of the National Cancer Institute at the National Institutes of Health. For EPIC-Oxford, it is Cancer Research UK C8221/A29017 and C8221/A19170, and Medical Research Council MR/M012190/1. RZ-R was supported by the “Miguel Servet” program (CP15/00100) from the Institute of Health Carlos III (Co-funded by the European Social Fund (ESF) - ESF investing in your future). EPIC-Spain received support from the Regional Governments of Andalucía, Asturias, Basque Country, Murcia and Navarra, and the Catalan Institute of Oncology (Spain). This work was supported in part by the French National Cancer Institute (L’Institut National du Cancer; INCA; grant numbers 2009-139 and 2014-1-RT-02-CIRC-1; PI: M. Jenab). For pancreatic cancer in EPIC, the work was supported by internal IARC funds.
Notes
Role of the funders: The funders of this study had no role in the collection, analysis, and interpretation of the data, in the writing of the manuscript, and in the decision to submit the manuscript for publication.
Disclosures: The authors have no potential conflicts of interest to disclose.
Author contributions: MBS, EW, TMS, VK, RK, AT, MJS, ATr, RT, MDC, GM, EA, RV, PB, AS, MJG, MJ, PKR, and PF collected, acquired, and administered study participants’ information on lifestyle exposure and metabolomics within the EPIC study. EL, DA, SJW, NDF, and RS collected, acquired, and administered study participants’ information on lifestyle exposure and metabolomics within the ATBC study. EL, PF, LT, VV, PK, MJG, and RS designed the study. EL, MS, VV, LT, JR, CB, PKR, and PF performed the statistical analyses. EL, MS, VV, IAB, SB, MB, JAS, RZR, THN, ES, BO, FR, CCD, DA, SJW, AS, NDF, MJG, MJ, PKR, and PF interpreted the results and prepared the first versions of the manuscript. All authors actively contributed to the final manuscript.
Disclaimer: Where authors are identified as personnel of the International Agency for Research on Cancer/World Health Organization, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy, or views of the International Agency for Research on Cancer/World Health Organization.
Acknowledgements: EPIC Umeå investigators thank the Västerbotten Intervention Programme and the County Council of Västerbotten for providing data and samples and acknowledge the contribution from Biobank Sweden, supported by the Swedish Research Council (VR 2017-00650). We thank the National Institute for Public Health and the Environment (RIVM), Bilthoven, the Netherlands, for their contribution and ongoing support to the EPIC Study.
Data Availability
For information on how to submit an application for gaining access to EPIC data and/or biospecimens, please follow the instructions at http://epic.iarc.fr/access/index.php.
Supplementary Material
References
- 1.Global Burden of Disease Collaborators. Alcohol use and burden for 195 countries and territories, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2018;392(10152):1015–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. Alcohol fact sheet. http://www.who.int/mediacentre/factsheets/fs349/en/.
- 3.IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Personal habits and indoor combustions. Volume 100 E. A review of human carcinogens. IARC Monogr Eval Carcinog Risks Hum. 2012;100(pt E):1–538. [PMC free article] [PubMed] [Google Scholar]
- 4.World Cancer Research Fund. Continuous Update Project Expert Report 2018. In Alcoholic Drinks and the Risk of Cancer.
- 5. Klatsky AL, Udaltsova N, Li Y, et al. Moderate alcohol intake and cancer: the role of underreporting. Cancer Causes Control. 2014;25(6):693–699. [DOI] [PubMed] [Google Scholar]
- 6. Kroke A, Klipstein-Grobusch K, Hoffmann K, et al. Comparison of self-reported alcohol intake with the urinary excretion of 5-hydroxytryptophol:5-hydroxyindole-3-acetic acid, a biomarker of recent alcohol intake. Br J Nutr. 2001;85(5):621–627. [DOI] [PubMed] [Google Scholar]
- 7. Das SK, Nayak P, Vasudevan DM.. Biochemical markers for alcohol consumption. Indian J Clin Biochem. 2003;18(2):111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Das SK, Vasudevan DM.. Biochemical diagnosis of alcoholism. Indian J Clin Biochem. 2005;20(1):35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Peterson K. Biomarkers for alcohol use and abuse–a summary. Alcohol Research & Health. 2004;28(1):30–37. [PMC free article] [PubMed] [Google Scholar]
- 10. Torrente MP, Freeman WM, Vrana KE.. Protein biomarkers of alcohol abuse. Expert Rev Proteomics. 2012;9(4):425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Helander A, Bottcher M, Dahmen N, et al. Elimination characteristics of the alcohol biomarker phosphatidylethanol (PEth) in blood during alcohol detoxification. Alcohol Alcohol. 2019;54(3):251–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Scalbert A, Brennan L, Manach C, et al. The food metabolome: a window over dietary exposure. Am J Clin Nutr. 2014;99(6):1286–1308. [DOI] [PubMed] [Google Scholar]
- 13. Edmands WMB, Ferrari P, Rothwell JA, et al. Polyphenol metabolome in human urine and its association with intake of polyphenol-rich foods across European countries. Am J Clin Nutr. 2015;102(4):905–913. [DOI] [PubMed] [Google Scholar]
- 14. Riboli E, Hunt KJ, Slimani N, et al. European prospective investigation into cancer and nutrition (EPIC): Study populations and data collection. Public Health Nutr. 2002;5(6B):1113–1124. [DOI] [PubMed] [Google Scholar]
- 15. Slimani N, Ferrari P, Ocke M, et al. Standardization of the 24-hour diet recall calibration method used in the European Prospective Investigation into Cancer and Nutrition (EPIC): general concepts and preliminary results. Eur J Clin Nutr. 2000;54(12):900–917. [DOI] [PubMed] [Google Scholar]
- 16. Kaaks R, Slimani N, Riboli E.. Pilot phase studies on the accuracy of dietary intake measurements in the EPIC project: overall evaluation of results. European Prospective Investigation into Cancer and Nutrition. Int J Epidemiol. 1997;26(suppl 1):S26–36. [DOI] [PubMed] [Google Scholar]
- 17. Slimani N, Bingham S, Runswick S, et al. Group level validation of protein intakes estimated by 24-hour diet recall and dietary questionnaires against 24-hour urinary nitrogen in the European Prospective Investigation into Cancer and Nutrition (EPIC) calibration study. Cancer Epidemiol Biomarkers Prev. 2003;12(8):784–795. [PubMed] [Google Scholar]
- 18. Rothwell JA, Keski-Rahkonen P, Robinot N, et al. A metabolomic study of biomarkers of habitual coffee intake in four European countries. Mol Nutr Food Res. 2019;63(22):e1900659. [DOI] [PubMed] [Google Scholar]
- 19. Stepien M, Keski-Rahkonen P, Kiss A, et al. Metabolic perturbations prior to hepatocellular carcinoma diagnosis–findings from a prospective observational cohort study. Int J Cancer. 2021;148(3):609–625. [DOI] [PubMed] [Google Scholar]
- 20. Stepien M, Duarte-Salles T, Fedirko V, et al. Alteration of amino acid and biogenic amine metabolism in hepatobiliary cancers: findings from a prospective cohort study. Int J Cancer. 2016;138(2):348–360. [DOI] [PubMed] [Google Scholar]
- 21. Gasull M, Pumarega J, Kiviranta H, et al. Methodological issues in a prospective study on plasma concentrations of persistent organic pollutants and pancreatic cancer risk within the EPIC cohort. Environ Res. 2019;169:417–433. [DOI] [PubMed] [Google Scholar]
- 22. ATBC Cancer Prevention Study Group. The alpha-tocopherol, beta-carotene lung cancer prevention study: Design, methods, participant characteristics, and compliance. Ann Epidemiol. 1994;4(1):1–10. [DOI] [PubMed] [Google Scholar]
- 23. Loftfield E, Rothwell JA, Sinha R, et al. Prospective investigation of serum metabolites, coffee drinking, liver cancer incidence, and liver disease mortality. J Natl Cancer Inst. 2020;112(3):286–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wishart DS, Feunang YD, Marcu A, et al. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018;46(D1):D608–D617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Smith CA, O’Maille G, Want EJ, et al. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;27(6):747–751. [DOI] [PubMed] [Google Scholar]
- 26. Kirpich AS, Ibarra M, Moskalenko O, et al. SECIMTools: a suite of metabolomics data analysis tools. BMC Bioinform. 2018;19(1):151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Edmands WM, Barupal DK, Scalbert A.. MetMSLine: an automated and fully integrated pipeline for rapid processing of high-resolution LC-MS metabolomic datasets. Bioinformatics. 2015;31(5):788–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Do KT, Wahl S, Raffler J, et al. Characterization of missing values in untargeted MS-based metabolomics data and evaluation of missing data handling strategies. Metabolomics. 2018;14(10):128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fan S, Kind T, Cajka T, et al. Systematic error removal using random forest for normalizing large-scale untargeted lipidomics data. Anal Chem. 2019;91(5):3590–3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kleinbaum DG, Kupper LK, Muller KE, Applied Regression Analysis and Other Multivariable Methods. Bellmont, CA: Duxbury Press; 1987. [Google Scholar]
- 31. Lai GY, Weinstein SJ, Albanes D, et al. The association of coffee intake with liver cancer incidence and chronic liver disease mortality in male smokers. Br J Cancer. 2013;109(5):1344–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fages A, Ferrari P, Monni S, et al. Investigating sources of variability in metabolomic data in the EPIC study: the principal component partial R-square (PC-PR2) method. Metabolomics. 2014;10(6):1074–1083. [Google Scholar]
- 33. Perrier F, Novoloaca A, Ambatipudi S, et al. Identifying and correcting epigenetics measurements for systematic sources of variation. Clin Epigenet. 2018;10(1):38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chambers J, Hastie T, Pregibon D, Statistical Models in S: Chapter 7. Generalized additive models. Heidelberg, 1990, p. 317–321. Physica-Verlag HD. [Google Scholar]
- 35.R Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing; 2013.
- 36. Benjamini Y, Hochberg Y.. Controlling the false discovery rate–a practical and powerful approach to multiple testing. J Roy Statl Soc Ser B-Stat Methodol. 1995;57(1):289–300. [Google Scholar]
- 37. Rohrmann S, Linseisen J, Vrieling A, et al. Ethanol intake and the risk of pancreatic cancer in the European Prospective Investigation into Cancer and Nutrition (EPIC). Cancer Causes Control. 2009;20(5):785–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Trichopoulos D, Bamia C, Lagiou P, et al. Hepatocellular carcinoma risk factors and disease burden in a European cohort: a nested case-control study. J Natl Cancer Inst. 2011;103(22):1686–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schwartz LM, Persson EC, Weinstein SJ, et al. Alcohol consumption, one-carbon metabolites, liver cancer and liver disease mortality. PLoS One. 2013;8(10):e78156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mishima T, Harino S, Sugita J, et al. Plasma kinetics and urine profile of ethyl glucosides after oral administration in the rat. Biosci Biotechnol Biochem. 2008;72(2):393–397. [DOI] [PubMed] [Google Scholar]
- 41. Naudin S, Li K, Jaouen T, et al. Lifetime and baseline alcohol intakes and risk of pancreatic cancer in the European Prospective Investigation into Cancer and Nutrition study. Int J Cancer. 2018;143(4):801–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.World Cancer Research Fund/American Institute for Cancer Research. Continuous Update Project Expert Report 2018. Diet, nutrition, physical activity and pancreatic cancer; 2018.
- 43. Jaremek M, Yu Z, Mangino M, et al. Alcohol-induced metabolomic differences in humans. Transl Psychiatry. 2013;3(7):e276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. van Roekel EH, Trijsburg L, Assi N, et al. Circulating metabolites associated with alcohol intake in the European Prospective Investigation into Cancer and Nutrition cohort. Nutrients. 2018;10(5):654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lacruz ME, Kluttig A, Tiller D, et al. Cardiovascular risk factors associated with blood metabolite concentrations and their alterations during a 4-year period in a population-based cohort. Circ Cardiovasc Genet. 2016;9(6):487–494. [DOI] [PubMed] [Google Scholar]
- 46. Brühl A, Faldum A, Löffelholz K.. Degradation of phosphatidylethanol counteracts the apparent phospholipase D-mediated formation in heart and other organs. Biochim Biophys Acta (BBA) - Mol Cell Biol Lipids. 2003;1633(2):84–89. [DOI] [PubMed] [Google Scholar]
- 47. Walther L, de Bejczy A, Lof E, et al. Phosphatidylethanol is superior to carbohydrate-deficient transferrin and gamma-glutamyltransferase as an alcohol marker and is a reliable estimate of alcohol consumption level. Alcohol Clin Exp Res. 2015;39(11):2200–2208. [DOI] [PubMed] [Google Scholar]
- 48. Zheng Y, Beck O, Helander A.. Method development for routine liquid chromatography-mass spectrometry measurement of the alcohol biomarker phosphatidylethanol (PEth) in blood. Clin Chim Acta. 2011;412(15-16):1428–1435. [DOI] [PubMed] [Google Scholar]
- 49. Wurtz P, Cook S, Wang Q, et al. Metabolic profiling of alcohol consumption in 9778 young adults. Int J Epidemiol. 2016;45(5):1493–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Playdon MC, Ziegler RG, Sampson JN, et al. Nutritional metabolomics and breast cancer risk in a prospective study. Am J Clin Nutr. 2017;106(2):637–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Guertin KA, Moore SC, Sampson JN, et al. Metabolomics in nutritional epidemiology: identifying metabolites associated with diet and quantifying their potential to uncover diet-disease relations in populations. Am J Clin Nutr. 2014;100(1):208–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Playdon MC, Sampson JN, Cross AJ, et al. Comparing metabolite profiles of habitual diet in serum and urine. Am J Clin Nutr. 2016;104(3):776–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zheng Y, Yu B, Alexander D, et al. Metabolomic patterns and alcohol consumption in African Americans in the Atherosclerosis Risk in Communities study. Am J Clin Nutr. 2014;99(6):1470–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Harada S, Takebayashi T, Kurihara A, et al. Metabolomic profiling reveals novel biomarkers of alcohol intake and alcohol-induced liver injury in community-dwelling men. Environ Health Prev Med. 2016;21(1):18–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lord RS, Bralley JA.. Clinical applications of urinary organic acids. Part I: detoxification markers. Altern Med Rev. 2008;13(3):205–215. [PubMed] [Google Scholar]
- 56. Pallister T, Jennings A, Mohney RP, et al. Characterizing Blood metabolomics profiles associated with Self-reported food intakes in female twins. PLoS One. 2016;11(6):e0158568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sampson JN, Boca SM, Shu XO, et al. Metabolomics in epidemiology: sources of variability in metabolite measurements and implications. Cancer Epidemiol Biomarkers Prev. 2013;22(4):631–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vipperla K, O’Keefe SJ.. Intestinal microbes, diet, and colorectal cancer. Curr Colorectal Cancer Rep. 2013;9(1):95–105. [Google Scholar]
- 59. Putignani L, Dallapiccola B.. Foodomics as part of the host-microbiota-exposome interplay. J Proteom. 2016;147:3–20. [DOI] [PubMed] [Google Scholar]
- 60. Shin S-Y, Fauman EB, Petersen A-K, et al. ; the Multiple Tissue Human Expression Resource (MuTHER) Consortium. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46(6):543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kipnis V, Subar AF, Midthune D, et al. Structure of dietary measurement error: results of the OPEN biomarker study. Am J Epidemiol. 2003;158(1):14–21. [DOI] [PubMed] [Google Scholar]
- 62. Prentice RL, Mossavar-Rahmani Y, Huang Y, et al. Evaluation and comparison of food records, recalls, and frequencies for energy and protein assessment by using recovery biomarkers. Am J Epidemiol. 2011;174(5):591–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Willett W. Nutritional Epidemiology. Oxford: Oxford University Press; 2013. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
For information on how to submit an application for gaining access to EPIC data and/or biospecimens, please follow the instructions at http://epic.iarc.fr/access/index.php.