Abstract
Most COVID-19 convalescents can build effective anti-SARS-CoV-2 humoral immunity, but it remains unclear how long it can maintain and how efficiently it can prevent the reinfection of the emerging SARS-CoV-2 variants. Here, we tested the sera from 248 COVID-19 convalescents around 1 year post-infection in Wuhan, the earliest known epicenter. SARS-CoV-2 immunoglobulin G (IgG) was well maintained in most patients and potently neutralizes the infection of the original strain and the B.1.1.7 variant. However, varying degrees of immune escape was observed on the other tested variants in a patient-specific manner, with individuals showing remarkably broad neutralization potency. The immune escape can be largely attributed to several critical spike mutations. These results suggest that SARS-CoV-2 can elicit long-lasting immunity but this is escaped by the emerging variants.
Key words: SARS-CoV-2, antibody, variant, B.1.617.2, convalescent
Graphical abstract
Public summary
-
•
Sera from a large cohort of COVID-19 convalescents in Wuhan were collected for evaluation of anti-SARS-CoV-2 humoral immunity
-
•
Anti-SARS-CoV-2 IgG was well maintained for 1 year in most convalescents and can potently neutralize the original strain and the B.1.1.7 variant
-
•
Varying degrees of immune escape was observed on the tested variants, especially on B.1.351 and B.1.617.2 variants
-
•
Sera from a few individuals showed remarkably broad neutralization potency against SARS-CoV-2 WT and variants
Introduction
SARS-CoV-2 emerged more than a year ago and rapidly swept across the world, developing into a long-lasting COVID-19 pandemic with devastating impacts.1, 2, 3 The situation has led to perpetual mutation of SARS-CoV-2, with numerous variants emerging around the world. In addition to the initial D614G mutation, the viral spike protein underwent antigenic drift and produced several variants of concern associated with local outbreaks, such as B.1.1.7 (alpha),4 B.1.351 (beta),5 P.1 (gamma, also known as B.1.1.28.1),6 B.1.525 (eta),7 B.1.617.2 (delta),8 and B.1.620, tested in this study.
Most COVID-19 patients can build anti-SARS-CoV-2 humoral immunity against SARS-CoV-2 after recovery.9,10 However, due to the lack of sufficient evidence about how long the protective immunity induced by prior infection or vaccination can maintain, the optimal interval between the vaccinations remains to be determined.11, 12, 13 In addition, increasing evidence showing these variants of concern, especially B.1.617.2 and B.1.351,14, 15, 16, 17, 18, 19, 20 remarkably increased resistance to the neutralizing antibodies, which is largely due to the mutations in the RBD region interacting with human angiotensin-converting enzyme 2 (hACE2).1 It is imperative to figure out the durability and breadth of SARS-CoV-2 neutralizing antibodies elicited by previous infection or vaccination based on the original strain.
Wuhan is the first known epicenter of the SARS-CoV-2 outbreak. Almost all patients in this city were infected by the original strain before the city reopened and the epidemic there had been well controlled before the emergence of variants.21,22 In this study, sera from convalescents infected around 1 year ago in Wuhan were collected to investigate the SARS-CoV-2 antibody durability and the breadth over time. Results from enzyme-linked immunosorbent assay (ELISA) and pseudovirus neutralization assay demonstrated that the SARS-CoV-2 RBD-specific IgG antibodies remain detectable in most patients, which potently inhibits the infection of the SARS-CoV-2 pseudovirus (wild-type [WT]-D614G and B.1.1.7), but to a different extent was compromised in other tested variants (P.1, B.1.620, B.1.525, B.1.617.2, and especially B.1.351) and mutations (K417N, L452R, E484K, and E484Q/L452R). The decline of neutralizing activity was confirmed by authentic viral neutralization assays (WT, B.1.1.7, B.1.617.2, and B.1.351). Of note, we report eight individuals who developed relatively broad immunity against the tested variants, supporting the presence of broadly neutralizing epitopes. Overall, these results provided important implications for the control of the pandemic caused by the constantly emerging SARS-CoV-2 variants.
Results
Durability and neutralizing activity
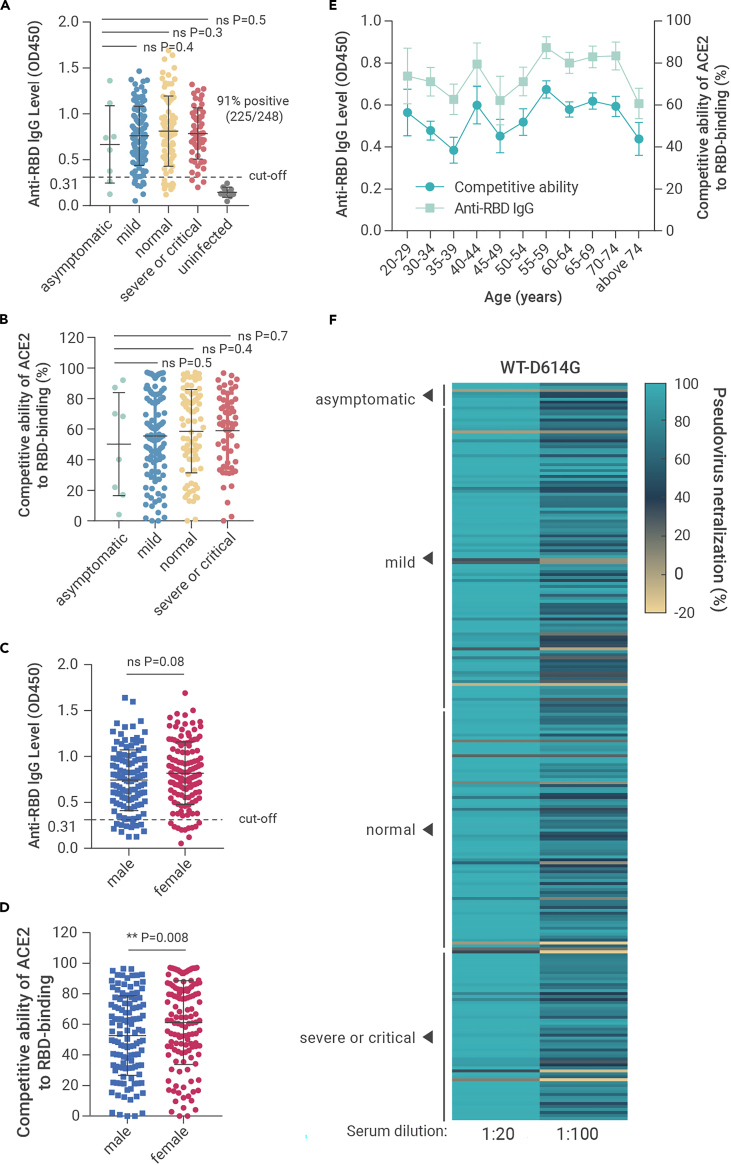
To evaluate the durability and neutralizing activity of SARS-CoV-2 antibodies after infection, we collected sera and clinical records from 248 COVID-19 patients infected around 1 year ago (11–12 months after symptom onset) in Wuhan. None of the convalescents had been administered with SARS-CoV-2 vaccine before sample collection according to the official record. Firstly, a rapid ELISA (colloidal gold) test for SARS-CoV-2-S IgM/IgG antibody was applied. Seventy-nine percent (197/248) of the sera tested positive for anti-SARS-CoV-2 IgG, and 2.0% (5/248) tested positive for anti-SARS-CoV-2 IgM (Table S1). Conventional ELISA assays were then conducted to determine the anti-RBD IgG level and the competitive ability of ACE2 to RBD binding of the sera. As we set OD450 of 0.31 (the mean OD450 value +3 SD of uninfected people's sera) as the cut-off value, 91% (225/248) tested positive for anti-SARS-CoV-2-RBD IgG (Figure 1A). Our results also showed that the competitive ability is highly consistent with the anti-RBD IgG level (Figures 1A–1E, Table S1), indicating that most RBD-targeting antibodies in the patients can interfere with the interaction between ACE2 and RBD. However, the anti-RBD IgG level and the neutralizing activity of the sera showed no statistically significant difference in patients with different severity (Figures 1A and 1B), gender (Figure 1C), and age (Figure 1E) except for the higher sera competitive ability of ACE2 to RBD binding was found in females (Figure 1D).
Figure 1.
The anti-RBD IgG level and neutralizing activity of sera from COVID-19 patients in Wuhan 1 year after recovery
(A–E) Results of ELISA measuring the 248 convalescent COVID-19 patients' sera reactivity to SARS-CoV-2-RBD. The anti-RBD IgG level of the indicated groups (A and C). The competitive ability of ACE2 to RBD binding of the indicated groups (B and D). Comparison of the anti-RBD IgG and the competitive ability of ACE2 to RBD binding in patients with different age (E).
(F) The neutralizing activity (WT-D614G) of the sera from convalescents with different severity was tested with the indicated dilution folds. Mean ± SD are shown in (A–D). Mean ± SEM are shown in (E). Statistical significance was determined using two-tailed Mann-Whitney U tests. ns, no significant difference was found (p > 0.05); ∗∗p < 0.01. The horizontal dotted lines in (A and C) indicate the cut-off value (0.31, the mean OD450 value +3 SD of uninfected people's sera).
The neutralizing activities of the sera were tested with SARS-CoV-2 pseudovirus (WT-D614G) produced based on a replication-deficient VSV pseudotyping system (VSV-dG-Luc) and a BHK-21 cell line stably expressing hACE2 (BHK-21-hACE2). A total of 234 sera (94%) showed more than 50% neutralization when diluted 20-fold (NT50 > 20), and 195 samples (79%) showed 50% neutralization under 100-fold dilution (NT50 > 100). No statistically significant difference was observed between pseudovirus neutralizing activity and disease severity (Figure 1F). These results indicate that most patients could build and maintain an effective humoral immunity against SARS-CoV-2 for at least 1 year.
Cross-variant protection
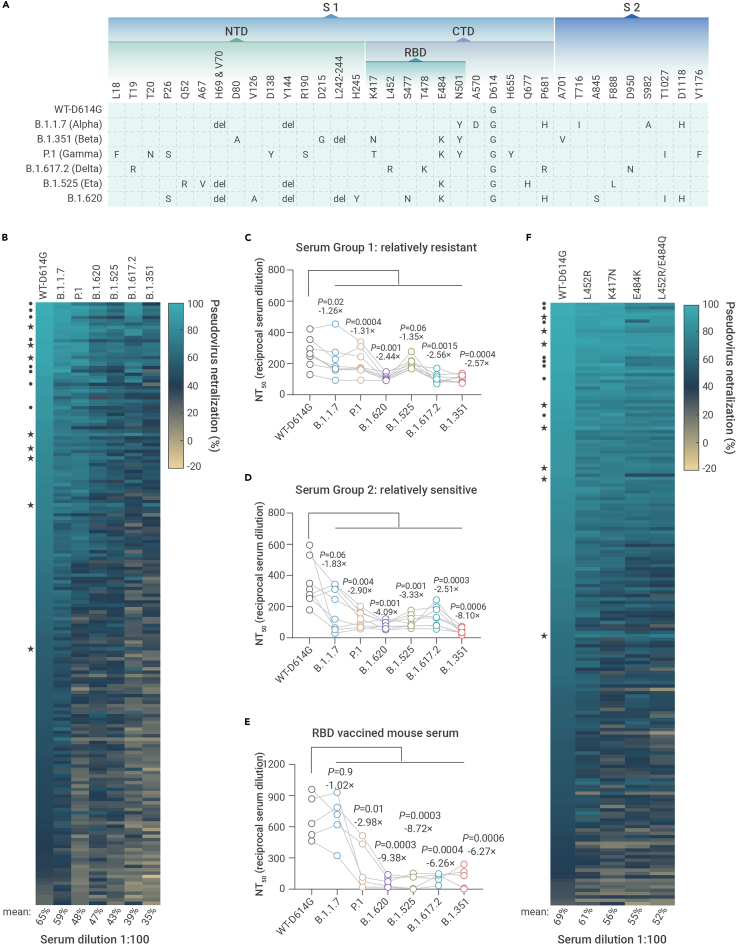
Recently, SARS-CoV-2 variants with multiple mutations in S protein have emerged in the United Kingdom (B.1.1.7), Brazil (P.1), Lithuania (B.1.620), Nigeria (B.1.525), South Africa (B.1.351), the United States (B.1.427/B.1.429),19,23 India (B.1.617.2), etc. We first tested the neutralizing activity of the top 180 potent sera in Figure 1F by the pseudoviruses of four variants, with the WT-D614G as a control (Figure 2A). When tested at 100-fold dilution, the mean neutralization efficiency of the sera against WT-D614G was 65%. However, the efficiency reduced to 59% for B.1.1.7, 48% for P.1, 47% for B.1.620, 43% for B.1.525, 39% for B.1.617.2, and 35% for B.1.351 (Figure 2B). Interestingly, we found that a small fraction of the patients showed cross-variant neutralizing activity. According to the reduction of neutralization to B.1.351, we selected eight representative sera (group 1, relatively resistant) from the 180 patients for further verification by NT50 determination, with another eight sera (group 2, relatively sensitive) for comparison. As expected, the sera in group 1 showed only a slight decrease in NT50 against the tested variants compared with WT-D614G (1.26- to 2.57-fold), while the sera in group 2 showed much higher sensitivity (1.83- to 8.10-fold). The NT50 data further confirmed that B.1.351 is the top immune escape variant among the tested strains, followed by B.1.617.2, B.1.525, B.1.620, P.1, and B.1.1.7, respectively (Figures 2C, 2D, and S1).
Figure 2.
The neutralizing activity of the convalescents' sera to SARS-CoV-2 variants
(A) The amino acid alterations in the S protein of SARS-CoV-2 variants tested in this study.
(B) The neutralizing activity of convalescents' sera at 100-fold dilution against pseudoviruses bearing S proteins from the indicated variants with WT-D614G as a control.
(C–E) Paired analysis of NT50 values of convalescents' sera or RBD-vaccinated mice sera against WT-D614G and variants. Serum group 1, relatively resistant (C). Serum group 2, sensitive (D). RBD-vaccinated mice sera (E). Serum groups 1 and 2 were selected according to their tolerance to B.1.351 pseudovirus. Statistical significance was determined using paired two-tailed t tests. p values and mean fold changes in NT50 values compared with WT-D614G are indicated.
(F) The neutralizing activity of convalescents' sera at 100-fold dilution against pseudoviruses bearing different S with the indicated mutations. (★) The relatively resistant serum sample; (•) the sensitive serum sample.
We further tested whether the immune escape of the tested variants is mainly attributed to the reduced neutralizing activity of SARS-CoV-2 RBD-specific antibodies. Five C57BL/6 mice were immunized three times with WT SARS-CoV-2 RBD human IgG-Fc recombinant protein. Seven weeks later, the mice sera were collected and tested using a pseudovirus neutralization assay. Similar to most of the convalescents' sera, the sera from RBD protein-immunized mice showed reduced neutralization to P.1, B.1.620, B.1.525, B.1.617.2, and B.1.351, but not to B.1.1.7, which is reasonable as B.1.1.7 only has one mutation (N501Y) in the RBD region, a mutation without a prominent immune escape effect15,24,25 (Figure 2E). These results showed that the immune escape ability of the variants can be largely attributed to mutations in the RBD region.
To define the contribution of the critical mutations in the RBD region to immune escape, we produced pseudovirus bearing SARS-CoV-2 S protein (D614G) with K417N, L452R, E484K, and L452R/E484Q mutations, respectively. Notably, L452R and L452R/E484Q are two critical mutations in the B.1.427/B.1.429 (epsilon) and B.1.167.1 (kappa) variants, respectively. We tested the neutralizing activity of the 180 sera by these mutants with WT-D614G as a control. As shown in Figure 2F, when tested at 100-fold dilution, the mean neutralization efficiency of the sera against WT-D614G was 69%. However, the efficiency reduced to 61% for L452R, 55% for K417N, 54% for E484K, and 52% for L452R/E484Q. This result indicates that the RBD mutations are responsible for the reduction of cross-variant neutralization among the variants of concern.
Verification by authentic viruses
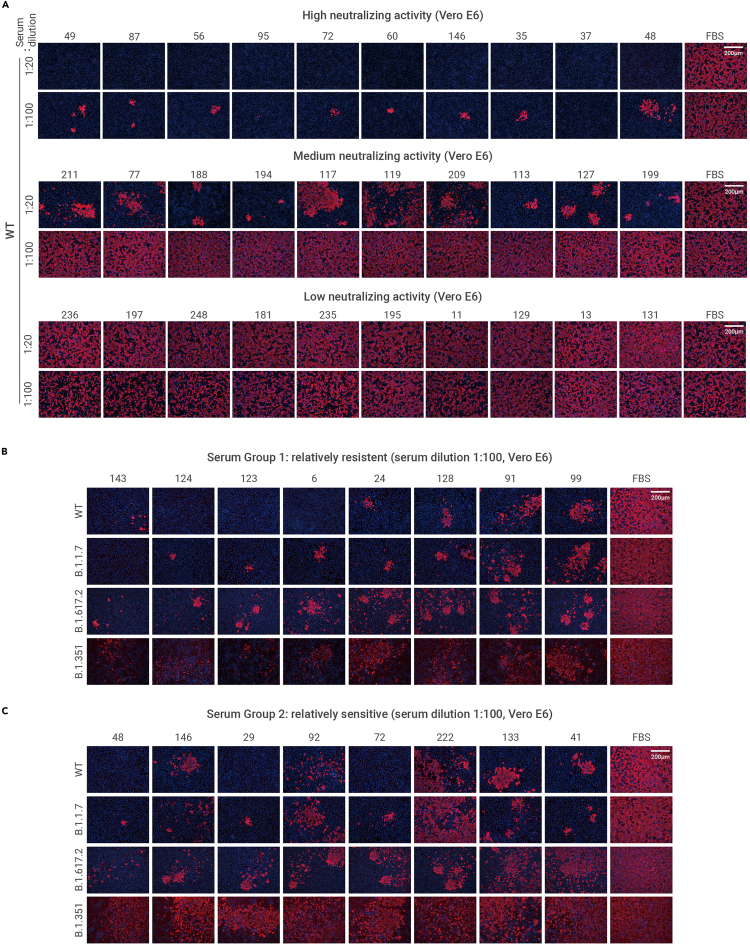
To confirm whether the neutralization results are consistent between the SARS-CoV-2 pseudovirus and the authentic virus, we first conducted the authentic SARS-CoV-2 virus (WT) neutralization assay to verify the 30 representative sera showing high, medium, and low neutralizing activities in Figure 1F, respectively. The SARS-CoV-2 nucleocapsid (N) protein immunofluorescence assay showed consistent results with the pseudovirus neutralization assay (Figure 3A).
Figure 3.
The neutralizing activity of the convalescents' sera to authentic SARS-CoV-2 viruses
(A) The neutralizing activity of 30 representative sera against authentic SARS-CoV-2 WT strain at 20- and 100-fold dilution. The sera were grouped based on their neutralizing activity to WT-D614G pseudovirus.
(B and C) The neutralizing activity of serum group 1 (B) and serum group 2 (C) against the indicated authentic SARS-CoV-2 viruses. SARS-CoV-2 N protein (red) in the infected cells was detected through immunofluorescence assay at 24 h post-infection. The nucleus was stained blue. Scale bar, 200 μm. The serum sample numbers are indicated above the pictures.
We further verified the neutralizing activity of two groups of patients in Figures 2C and 2D (relatively resistant and relatively sensitive) by four authentic SARS-CoV-2 strains, including the WT strain and the B.1.1.7, B.1.617.2, and B.1.351 variant strains. The sera were diluted 100-fold, pre-incubated with these viruses, and then applied to Vero E6 cells for infection neutralization assay. As shown in Figure 3B, the neutralizing activity of the sera against the B.1.1.7 strain is similar to the WT strain but with a slight decrease. As for the B.1.617.2 and B.1.351 strains, the group 1 sera (relatively resistant) efficiently neutralized the infection of both the WT and the B.1.351 strains. In contrast, the group 2 sera (relatively sensitive) showed a remarkable reduction of neutralizing activity against the B.1.617.2 and the B.1.351 strains compared with the WT strain (Figure 3C). Again, these results are also consistent with the results from pseudovirus neutralization assays.
Discussion
Previous studies show the protective immunity against seasonal coronaviruses (HCoV-NL63, HCoV-229E, HCoV-OC43, and HCoV-HKU1) may last only 6–12 months,26 and the titer of SARS-CoV-2 antibody has been reported to decline rapidly in the first several months,11 raising concerns about the durability of SARS-CoV-2 neutralizing antibodies elicited by either previous infection or vaccination. In addition, SARS-CoV-2 variants are constantly emerging during the COVID-19 pandemic, greatly challenging the development and application of effective neutralizing antibodies and vaccines.27,28 It is imperative to investigate the durability and breadth of the anti-SARS-CoV-2 humoral immunity. To this end, we collected convalescent sera in Wuhan in consideration of the several unique advantages over samples collected elsewhere: (1) Wuhan is the earliest reported epicenter of COVID-19 with most patients infected more than 1 year ago; (2) the patients were all infected by the original WT strain without the interference of potential multi-variant infections before sample collection,21 and (3) so far, most administered vaccines globally were designed based on the sequences of the original strain from Wuhan.
Firstly, we report that the effective neutralizing antibodies against SARS-CoV-2 could last at least 1 year in most COVID-19 convalescents without vaccination. Our results indicate that most people, including asymptomatic and elderly patients, could build and maintain effective anti-SARS-CoV-2 humoral immunity, with a few exceptions in each group for unknown reasons (Figure 1). We speculate that this phenotype might be attributed to the individual immune differences that deserve further study.
Another particular concern is the breadth of the anti-SARS-CoV-2 humoral immunity. Our study provides additional evidence that most sera collected from convalescents in Wuhan, even for 1 year post-infection, can still efficiently neutralize infection of the B.1.1.7 variant. However, a significant decline of efficacy was observed on P.1, B.1.620, B.1.525, B.1.617.2, and B.1.351 (Figure 2B). B.1.351 displayed the most prominent immune escape ability among these tested variants, consistent with previous reports.15,16,29 Encouragingly, sera from a few individuals showed resistance to all tested variants (Figure 2C), indicating the presence of effective and broadly neutralizing antibodies. This brings hope for the identification of ideal broad neutralization epitopes essential for the development of antibody therapy and next-generation vaccines.
We next sought to characterize the critical factors that contribute to the immune escape. Based on previous studies and our neutralization results from RBD-vaccinated mouse sera, we focused on the four mutation sites in RBD of the tested variants in this study (Figure 2A). N501Y is the only mutation in B.1.1.7 RBD, but this variant did not show remarkable immune escape in both our study (Figure 2E) and previous studies.30,31 Therefore, we tested several mutations in the other three critical sites, including K417N, L452R, E484K, and L452R/E484Q. The neutralization assay demonstrated that L452R/E484Q showed the strongest immune escape, followed by E484K, K417N, and L452R. The mean sera neutralization efficiency (Figures 2B and 2F) demonstrated that the critical RBD mutations play a major role in the immune escape of the variants. Importantly, the cross-variant protective sera from the group 1 convalescents can also tolerate these critical mutations (Figure 2F).
We do acknowledge some limitations of our studies. First, we did not determine the NT50 of all the sera because of the limitation of the sample volume, especially for those with lower neutralizing activity. Secondly, we did not include all the possible mutations that can appear in other emerging variants. Thus, future studies could be done to investigate whether other mutations on spike proteins can result in a leap of the immune escape of other variants.
To our knowledge, this study provides strong evidence that potent anti-SARS-CoV-2 humoral immunity can last for at least 1 year in most convalescents to protect them against the original virus (WT or WT-D614G), which means annual re-administration might be a feasible vaccination strategy to maintain anti-SARS-CoV-2 humoral immunity. However, convalescents and vaccinated people gradually lose protection against the constantly emerging immune escape variants. It remains to be determined whether re-administration of the variant-based or broadly neutralizing epitope-based vaccines can achieve effective cross-variant immunity. Overall, our study suggests that the timely update of vaccines rather than the durability of the SARS-CoV-2 humoral immunity should be more of our concern.
Method
Sample collection
COVID-19 convalescents from ten communities in five different streets (Chezhan Street, Danshuichi Street, Huaqiao Street, Yiyuan Street, and Dazhi Street) in Jiang'an District in Wuhan, whose symptom onset was between January 1, 2020, and March 26, 2020, were randomly recruited to the cohort according to the criteria of the easy sampling principle. Convalescents who had mental disorders, dementia, difficulty in moving freely, or refused to enroll or were not contactable were excluded from the cohort. According to these sampling criteria, all 248 eligible individuals were recruited and their blood samples were collected from 19 January, 2021, to 26 January, 2021, and this collection was carried out through Hubei Provincial Centre For Disease Control and Prevention and Hubei Provincial Academy Of Preventive Medicine (HBCDC) with written consent under appropriate institutional review Boards approval (2021-012-01) and were deidentified. The recovery time, illness severity, gender, and age of donors were recorded (for extended data, see Table S1). Sera of the RBD-hFc vaccinated mice (three immunizations at days 0, 14, and 28, respectively) were collected 7 weeks after the last immunization from the suborbital venous plexus (WP20210007).
ELISA detection of SARS-CoV-2 S protein antibodies in the sera
The rapid IgG/IgM test was applied using the 2019-nCoV IgG/IgM detection kit (colloidal gold-based, Vazyme Biotech), which is based on the reactivity of IgG/IgM against SARS-CoV-2 S protein. The anti-RBD IgG level in sera was tested using the SARS-CoV-2 (2019-nCoV) Spike RBD Antibody Titer Assay Kit (Sino Biological), which is based on the reactivity of IgG against SARS-CoV-2 RBD. The competitive ability of ACE2 to RBD binding of sera was tested through SARS-CoV-2 Neutralizing Antibodies Test Kit (ELISA, Wuxi BioHermes Bio & Medical Technology), which is based on the competition between the neutralizing antibodies in sera and ACE2 to horseradish peroxidase-labeled RBD protein (HRP-RBD). These ELISA detection tests were applied following the manufacturer's instructions.
Cell culture
HEK293T (ATCC, CRL-1168), BHK-21(ATCC, CCL-10), and Vero E6 cells (ATCC, CRL-1586) were cultured in Dulbecco's modified Eagle’s medium (DMEM) (Gibco) supplemented with 10% fetal bovine serum (FBS), 2.0 mM L-glutamine, 110 mg/L sodium pyruvate, and 4.5 g/L D-glucose. BHK-21-ACE2 (clone 7), which stably expresses human ACE2 was generated based on BHK-21 through lentiviral transduction and maintained with puromycin at 1 μg/mL. I1-Hybridoma (CRL-2700) secreting a monoclonal antibody targeting VSV glycoprotein was cultured in minimum essential medium with Earle's salts and 2.0 mM L-glutamine (MEM; Gibco) supplemented with 10% FBS. All cells were cultured at 37°C in 5% CO2 with regular passage every 2–3 days.
Plasmids and protein
The DNA sequences of human codon-optimized S proteins from SARS-COV-2 variants (B.1.1.7, GISAID: EPI-ISL-601443; B.1.351, GISAID: EPI_ISL_678597; P.1, GISAID: EPI_ISL_906075; B.1.617.2, GISAID: EPI_ISL_2378732; B.1.525, GISAID: EPI_ISL_1093472; B.1.620, GISAID: EPI_ISL_1661662) and S protein mutations were commercially synthesized or generated by overlapping PCR-based mutagenesis using pCAGGS-SARS-CoV-2-S-C9 (a gift from Dr. Wenhui Li, National Institute of Biological Science, Beijing, China) as the template and cloned into pCAGGS vector with C-terminal 18 amino acid (aa) truncation to improve VSV pseudotyping efficiency.32 The plasmid pCAGGS-SARS-CoV-2-RBD-hFc was constructed by inserting the RBD sequence (aa: 331–525) into the pCAGGS vector for the expression of RBD-hFc recombinant protein in HEK293T cells. The proteins were purified by protein A resin (GenScript) following the manufacturer's instructions.
SARS-CoV-2 pseudovirus production and titration
The pseudovirus packaged with spike proteins from the WT SARS-CoV-2 and the SARS-CoV-2 variants were produced according to a published protocol with minor modifications.33 In brief, Vero-E6 cells were transfected with plasmids expressing different S proteins through lipofectamine 2000 (Biosharp, China). After 24 h, the transfected cells were inoculated with VSV-dG-fLuc (1 × 106 50% tissue culture infectious dose [TCID50]/mL) diluted in DMEM for 5 h at 37°C and then replenished with growth medium (DMEM with 10% FBS) containing anti-VSV-G monoclonal antibody (I1-hybridoma, cultured supernatant, 1:20). Twenty-four hours later, the SARS-CoV-2 pseudovirus-containing supernatant was harvested and clarified at 3,000 rpm for 10 min, aliquoted, and frozen at −80°C for storage. The TCID50 of the pseudovirus was determined using a serial dilution-based infection assay on BHK-21-hACE2 cells and calculated according to the Reed-Muench method.33,34
SARS-CoV-2 pseudovirus neutralization assay
The SARS-CoV-2 pseudoviruses (3 × 105 TCID50/well) were incubated with serial-diluted sera at room temperature for 30 min in 96-well white flat-bottom culture plates and then mixed with trypsinized BHK-21-hACE2 cells at a density of 2 × 104/well. After 16 h, the medium of the infected cells was removed, and the cells were lysed with 1× Bright-Glo Luciferase Assay reagent (Promega) for chemiluminescence detection using a SpectraMax iD3 multi-well luminometer (Molecular devices). The 50% neutralization dilution titer (NT50) was calculated by GraphPad Prism 7 software with nonlinear regression curve fitting (normalized response, variable slope).
Authentic SARS-CoV-2 neutralization assay
The SARS-CoV-2 WT strain (IVCAS 6.7512) and B.1.351 strain (NPRC 2.062100001)35 were provided by the National Virus Resource, Wuhan Institute of Virology, Chinese Academy of Sciences, the SARS-CoV-2 B.1.1.7 strain (240108) and B.1.617.2 strain (YJ20210707-01) was provided by Hubei Provincial Centre for Disease Control and Prevention. All SARS-CoV-2 authentic virus-related experiments (S01321010A) were approved by the Biosafety Committee Level 3 (ABSL-3) of Wuhan University, Wuhan Institute of Virology and Hubei Provincial Centre for Disease Control and Prevention. In brief, sera were serially diluted in culture medium and mixed with 200 TCID50 SARS-CoV-2 for 30 min at room temperature. The mixture was then added to Vero E6 cells in 96-well plates and incubated for 24 h, and the cells were fixed with 4% paraformaldehyde in PBS at room temperature for 1 h, permeabilized with 0.2% Triton X-100 for 10 min, and then blocked with 1% BSA/PBS at 37°C for 1 h. Cells were subsequently incubated with a mouse monoclonal antibody targeting SARS-CoV/SARS-CoV-2 nucleocapsid (40143-MM05, Sino Biological) at 1:500 dilution and 37°C for 1 h, and then incubated with 2 μg/mL of Alexa Fluor 594-conjugated goat anti-mouse IgG antibody (A-11032, Thermo Fisher Scientific) at 37°C for 1 h. The nucleus was stained with Hoechst 33342. Images were acquired with an inverted fluorescence microscope (DMi8, Leica).
Statistical analysis
Data are presented as mean values with SEM or SD. All statistical analyses were done using GraphPad Prism 7. Differences between two independent samples were evaluated by two-tailed Mann-Whitney U tests, and p < 0.05 was considered statistically significant. Differences between two related samples were evaluated by paired two-tailed t tests.
Acknowledgments
This study was supported by grants from the National Science and Technology Major Project (2018YFA0900801), China NSFC projects (32041007, 32070160, and 82041004), Fundamental Research Funds for the Central Universities (2042021kf0220 and 2042020kf0024), the Advanced Customer Cultivation Project of Wuhan National Biosafety Laboratory (2021ACCP-MS10) and Special Fund for COVID-19 Research of Wuhan University. We are grateful to Beijing Taikang Yicai Foundation for their great support to this work.
Author contributions
K.L., Y.C., H.Y., and K.C. conceptualized the study design. F.H.M., F.X.Z., X.Y.C., B.H., J.Q.X., Y.Z.J., S.H.Z., and L.T. collected the samples. Q.Y.L., Q.X., F.H.M., C.B.M., Z.Z., X.Y.C., M.G., X.W., Y.H.F., S.S., and C.W.K. did the laboratory tests. Q.Y.L., Q.X., F.H.M., Y.L.L., F.L., L.Z., K.X., C.W.K., F.D., H.Y., and Y.C. analyzed the data. Q.Y.L., Q.X., F.H.M., H.Y., Y.C., and K.L. interpreted the results. Q.Y.L., Q.X., F.H.M., H.Y., and Y.C. wrote the initial drafts of the manuscript. Q.Y.L., K.C., H.Y., Y.C., and K.L. revised the manuscript. Y.L.L., F.L., L.Z., K.X., C.W.K., J.Q.X., Y.Z.J., and F.D. commented on the manuscript. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Published Online: November 3, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xinn.2021.100181.
Contributor Information
Kun Cai, Email: ckreal@163.com.
Huan Yan, Email: huanyan@whu.edu.cn.
Yu Chen, Email: chenyu@whu.edu.cn.
Ke Lan, Email: klan@whu.edu.cn.
Lead contact website
http://www.bio.whu.edu.cn/info/1191/4268.htm.
Supplemental information
Convalescents' sera were serially diluted and then were inoculated with 3 × 105 TCID50 pseudoviruses as indicated for 30 min at room temperature. BHK-21-hACE2 cells were infected by this mixture, followed by a chemiluminescence detection after 16 h. Mean ± SD (n = 2) are shown. The NT50 was calculated by GraphPad Prism 7 software with nonlinear regression curve fitting (normalized response, variable slope). The horizontal dotted lines on each graph indicate 50% and 0% neutralization
Document S2. Article plus supplemental information
References
- 1.Zhou P., Yang X.L., Wang X.G., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grubaugh N.D., Hodcroft E.B., Fauver J.R., et al. Public health actions to control new SARS-CoV-2 variants. Cell. 2021;184:1127–1132. doi: 10.1016/j.cell.2021.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen L., Liu W., Zhang Q., et al. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerging Microbes Infections. 2020;9:313–319. doi: 10.1080/22221751.2020.1725399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Public Health England . 2020. Investigation of Novel SARS-CoV-2 Variant: Variant of Concern 202012/01. Technical Briefing 5. [Google Scholar]
- 5.Tegally H., Wilkinson E., Giovanetti M., et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv. 2020 doi: 10.1101/2020.12.21.20248640. [DOI] [Google Scholar]
- 6.Faria N.R., Claro I.M., Candido D., et al. Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings. Virological. 2021;372:815–821. [Google Scholar]
- 7.Ozer E.A., Simons L.M., Adewumi O.M., et al. High prevalence of SARS-CoV-2 B.1.1.7 (UK variant) and the novel B.1.5.2.5 lineage in Oyo State, Nigeria. medRxiv. 2021 doi: 10.1101/2021.04.09.21255206. [DOI] [Google Scholar]
- 8.Yadav P.D., Nyayanit D.A., Sahay R.R., et al. Isolation and characterization of the new SARS-CoV-2 variant in travellers from the United Kingdom to India: VUI-202012/01 of the B.1.1.7 lineage. J. Trav. Med. 2021;28:taab009. doi: 10.1093/jtm/taab009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ni L., Ye F., Cheng M.L., et al. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity. 2020;52:971–977.e3. doi: 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Juno J.A., Tan H.X., Lee W.S., et al. Humoral and circulating follicular helper T cell responses in recovered patients with COVID-19. Nat. Med. 2020;26:1428–1434. doi: 10.1038/s41591-020-0995-0. [DOI] [PubMed] [Google Scholar]
- 11.Turner J.S., Kim W., Kalaidina E., et al. SARS-CoV-2 infection induces long-lived bone marrow plasma cells in humans. Nature. 2021;595:421–425. doi: 10.1038/s41586-021-03647-4. [DOI] [PubMed] [Google Scholar]
- 12.He Z., Ren L., Yang J., et al. Seroprevalence and humoral immune durability of anti-SARS-CoV-2 antibodies in Wuhan, China: a longitudinal, population-level, cross-sectional study. Lancet. 2021;397:1075–1084. doi: 10.1016/S0140-6736(21)00238-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han J., Zhang N., Chen D., et al. Distinct durability of IgM/IgG antibody responses in COVID-19 patients with differing severity. Sci. China Life Sci. 2021:1–4. doi: 10.1007/s11427-020-1947-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Planas D., Veyer D., Baidaliuk A., et al. Reduced sensitivity of SARS-CoV-2 variant delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 15.Wang P., Nair M.S., Liu L., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 16.Hoffmann M., Arora P., Gross R., et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184:2384–2393.e12. doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang R., Zhang Q., Ge J., et al. Analysis of SARS-CoV-2 variant mutations reveals neutralization escape mechanisms and the ability to use ACE2 receptors from additional species. Immunity. 2021 doi: 10.1016/j.immuni.2021.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Planas D., Bruel T., Grzelak L., et al. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat. Med. 2021;27:917–924. doi: 10.1038/s41591-021-01318-5. [DOI] [PubMed] [Google Scholar]
- 19.Shen X., Tang H., Pajon R., et al. Neutralization of SARS-CoV-2 variants B.1.429 and B.1.351. New Engl. J. Med. 2021;384:2352–2354. doi: 10.1056/NEJMc2103740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang L., Tian D., Han J.B., et al. A recombinant receptor-binding domain in trimeric form generates protective immunity against SARS-CoV-2 infection in nonhuman primates. Innovation. 2021;2:100140. doi: 10.1016/j.xinn.2021.100140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao S., Gan Y., Wang C., et al. Post-lockdown SARS-CoV-2 nucleic acid screening in nearly ten million residents of Wuhan, China. Nat. Commun. 2020;11:5917. doi: 10.1038/s41467-020-19802-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen M., Peng Z., Xiao Y., Zhang L. Modeling the epidemic trend of the 2019 novel coronavirus outbreak in China. Innovation. 2020;1:100048. doi: 10.1016/j.xinn.2020.100048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tchesnokova V., Kulakesara H., Larson L., et al. Acquisition of the L452R mutation in the ACE2-binding interface of spike protein triggers recent massive expansion of SARS-Cov-2 variants. bioRxiv. 2021 doi: 10.1101/2021.02.22.432189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou D., Dejnirattisai W., Supasa P., et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell. 2021;184:2348–2361.e6. doi: 10.1016/j.cell.2021.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu C., Ginn H.M., Dejnirattisai W., et al. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell. 2021;184:4220–4236.e13. doi: 10.1016/j.cell.2021.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edridge A.W.D., Kaczorowska J., Hoste A.C.R., et al. Seasonal coronavirus protective immunity is short-lasting. Nat. Med. 2020;26:1691–1693. doi: 10.1038/s41591-020-1083-1. [DOI] [PubMed] [Google Scholar]
- 27.Liu J., Liu Y., Xia H., et al. BNT162b2-elicited neutralization of B.1.617 and other SARS-CoV-2 variants. Nature. 2021;596:273–275. doi: 10.1038/s41586-021-03693-y. [DOI] [PubMed] [Google Scholar]
- 28.Edara V.V., Norwood C., Floyd K., et al. Infection- and vaccine-induced antibody binding and neutralization of the B.1.351 SARS-CoV-2 variant. Cell Host & Microbe. 2021;29:516–521.e13. doi: 10.1016/j.chom.2021.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang G.L., Wang Z.Y., Duan L.J., et al. Susceptibility of circulating SARS-CoV-2 variants to neutralization. New Engl. J. Med. 2021;384:2354–2356. doi: 10.1056/NEJMc2103022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi P.Y., Xie X., Zou J., et al. Neutralization of N501Y mutant SARS-CoV-2 by BNT162b2 vaccine-elicited sera. bioRxiv. 2021 doi: 10.1101/2021.01.07.425740. [DOI] [Google Scholar]
- 31.Xie X., Liu Y., Liu J., et al. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat. Med. 2021;27:620–621. doi: 10.1038/s41591-021-01270-4. [DOI] [PubMed] [Google Scholar]
- 32.Schwegmann-Wessels C., Glende J., Ren X., et al. Comparison of vesicular stomatitis virus pseudotyped with the S proteins from a porcine and a human coronavirus. J. Gen. Virol. 2009;90:1724–1729. doi: 10.1099/vir.0.009704-0. [DOI] [PubMed] [Google Scholar]
- 33.Nie J., Li Q., Wu J., et al. Quantification of SARS-CoV-2 neutralizing antibody by a pseudotyped virus-based assay. Nat. Protoc. 2020;15:3699–3715. doi: 10.1038/s41596-020-0394-5. [DOI] [PubMed] [Google Scholar]
- 34.Matumoto M. A note on some points of calculation method of LD50 by Reed and Muench. Jpn. J. Exp. Med. 1949;20:175–179. [PubMed] [Google Scholar]
- 35.Fengjuan C., Bosheng L., Peter H., et al. A case of new variant COVID-19 first emerging in South Africa detected in airplane pilot—Guangdong province, China, January 6, 2021. China CDC Weekly. 2021;3:28–29. doi: 10.46234/ccdcw2021.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Convalescents' sera were serially diluted and then were inoculated with 3 × 105 TCID50 pseudoviruses as indicated for 30 min at room temperature. BHK-21-hACE2 cells were infected by this mixture, followed by a chemiluminescence detection after 16 h. Mean ± SD (n = 2) are shown. The NT50 was calculated by GraphPad Prism 7 software with nonlinear regression curve fitting (normalized response, variable slope). The horizontal dotted lines on each graph indicate 50% and 0% neutralization
Document S2. Article plus supplemental information