Abstract
The second wave of COVID-19 caused by severe acute respiratory syndrome virus (SARS-CoV-2) is rapidly spreading over the world. Mechanisms behind the flee from current antivirals are still unclear due to the continuous occurrence of SARS-CoV-2 genetic variants. Brazil is the world's second-most COVID-19 affected country. In the present study, we identified the genomic and proteomic variants of Brazilian SARS-CoV-2 isolates. We identified 16 different genotypic variants were found among the 27 isolates. The genotypes of three isolates such as Bra/1236/2021 (G15), Bra/MASP2C844R2/2020 (G11), and Bra/RJ-DCVN5/2020 (G9) have a unique mutant in NSP4 (S184N), 2′O-Mutase (R216N), membrane protein (A2V) and Envelope protein (V5A). A mutation in RdRp of SARS-CoV-2, particularly the change of Pro-to Leu-at 323 resulted in the stabilization of the structure in BRA/CD1739-P4/2020. NSP4, NSP5 protein mutants are more virulent in genotype 15 and 16. A fast protein folding rate changes the structural stability and leads to escape for current antivirals. Thus, our findings help researchers to develop the best potent antivirals based on the new mutant of Brazilian isolates.
Keywords: Phylogenomic study, SARS-CoV-2, Genotypic variants, Protein mutations, Brazilian isolates
Abbreviation: hACE2, Human-Angiotensin converting enzyme 2 receptor; SARS-CoV-2, Severe acute respiratory syndrome coronavirus 2; COVID-19, Coronavirus disease-2019; NSP, Non-structural protein; RdRp, RNA-dependent RNA polymerase
1. Introduction
1.1. SARS-CoV-2 genome
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is a single strand positive RNA genome that causes coronavirus disease (COVID-19). World Health Organization (WHO) has declared the COVID-19 pandemic as a global public health emergency (Cucinotta and Vanelli, 2020). On 15th July 2021, WHO reported a total of 72,528 deaths out of 3.7 million new weekly affected cases. Brazil is the second-largest COVID-19 affected place, globally. So far 17, 296, 118 affected cases have been reported in Brazil, of which 4, 54, 710 new cases have been reported in the last seven days as of 6th June 2021. Overall 4, 84, 235 death cases have been reported so far (World Health Organization WHO, 2021), https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---20-july-2021).
Several SARS-CoV-2 variants were found to be associated with increased infectivity rate, pathogenesis and displayed a considerably faster transmission through droplets than the wild-type strain (Prathiviraj et al., 2020; Rajeev et al. 2020; Prathiviraj et al., 2021). A study realized with the primary human airway epithelial (hACE) cell culture reveals that enhanced transmission rate of SARS-CoV-2 is linked with the effective replication of the viral particle in the upper respiratory tract temperature of 33 °C when compared with the lower respiratory tract temperature of 37 °C (V'kovski et al., 2021). RNA-dependent RNA polymerase (RdRp), the enzyme in charge of viral genome replication and transcription (Posthuma et al., 2017) facilitates mutations at various locations in the genome. Errors in the replication and repair systems of the virus lead to the accumulation of additional mutations in the genome (Banoun, 2021; Pachetti et al., 2020). It must be noted that the mutations described at particular locations do not individually provide fitness to the virus, but a combination of favourable mutations at different locations in the genome, resulting in the improved or diminished functionalities have been classified as clades of mutant viruses harbouring certain moderations (Mercatelli and Giorgi, 2020).
It is important to analyze and interpret the changes at both genotypic and phenotypic levels, as these changes help in understanding the structural changes, pathogenesis, replication and transmission of SARS-CoV-2 (Mousavizadeh and Ghasemi, 2021) in a different environmental niche. Genotyping approach employing single nucleotide polymorphism is essential to identify the genetic changes accumulating during the transmission of SARS-CoV-2 and helps to determine the changes in the property of viral transmission and virulence. Thus, helps to track genotypes associated with topography and epidemiology. The identification of genotypes associated with topographical and contagious groups helps in tracking and monitoring the single nucleotide polymorphism (Yin, 2020). Genotypic studies help in identifying the phenotypical changes caused due to the replacements of amino acids (Morais et al., 2020). The protein folding information plays a key role in the therapeutic intervention of many viral diseases (Broglia et al., 2005; Prathiviraj et al., 2021). The folding process of a protein determines the structural stability and drug binding specificity (Prisilla et al., 2016; Prathiviraj et al., 2016) thus it's most important to identify the folding rate of mutants (Murugan et al., 2019; Prathiviraj et al., 2021). Analysis of the genotype and phenotype is essential as it aids in understanding the missense, synonymous, and non-synonymous mutations in the virus. In the present study, we identify the number of genotypic variants and proteomic mutants that have recently evolved in Brazilian SARS-CoV-2 isolates. Thus the identified genotypic variants would be useful for developing a new antivirals depending upon the stability of mutants.
2. Methodology
2.1. Dataset preparation and sequence alignment
The complete genomic/nucleotide sequences of Brazilian SARS-CoV-2 isolates and reference Wuhan-Hu-1 SARS-CoV-2 genome were retrieved from the NCBI Virus data repository (https://www.ncbi.nlm.nih.gov/sars-cov-2/). Initial multiple sequence alignment was performed using the PartTree (Katoh and Toh, 2007) algorithm (systematic of large scale alignment) in the MAFFT online server (Katoh et al., 2019).
2.2. Analysis of evolutionary imprints
The aligned file was further imported in MEGA Version X (Kumar et al., 2018) to perform the molecular phylogenomic analysis using the neighbor-joining algorithm (Saitou and Nei, 1987). Kimura two-parameter (Nishimaki and Sato, 2019) evolutionary substitution model was carried out for estimation of genetic diversity. The phylogenomic tree was computed with 1000 bootstrapping iteration to identify replication among selected isolates. Finally, a huge scale phylogenomic supertree was drawn using the Interactive Tree of Life (iTOL) v4 (Letunic and Bork, 2019). Nodes of each isolate in the cluster are grouped, colored and classified according to its features.
2.3. Analysis of genomic and proteomic mutation
Each open reading frame (ORFs) of nucleotide and protein sequences of retrieved Brazilian isolates were separated, correspond to the genomic and proteomic positions of the reference genome (Wuhan-Hu-1). The alignment of each ORFs was carried out to identify the mutated residues (mismatch region) compared with Wuhan-Hu-1 genome using the BioEdit v7.2 software (Hall, 1999). Each mutation in the genomic bases of corresponding isolates were cross verified with the Virus pathogen database and analysis resource (ViPR) (Pickett et al., 2012).
2.4. Prediction of virulence mechanism and protein folding rate
Virulence mechanism of identified mutation of each ORFs was predicted using the VICMpred (Saha and Raghava, 2006) and VirulentPred server (Garg and Gupta, 2008). Identified prediction score of each mutated residue was compared with the reference proteome (Wuhan-Hu-1), to identify its near-native state. Prediction score was computed based on the support vector machine learning algorithm (Rashid et al., 2007) and the final sub-cellular localization score was determined by its functional motifs. The protein folding rate among all the identified mutants was calculated using the FOLD-RATE server (Gromiha et al., 2006).
3. Results
3.1. Analysis of evolutionary footprint
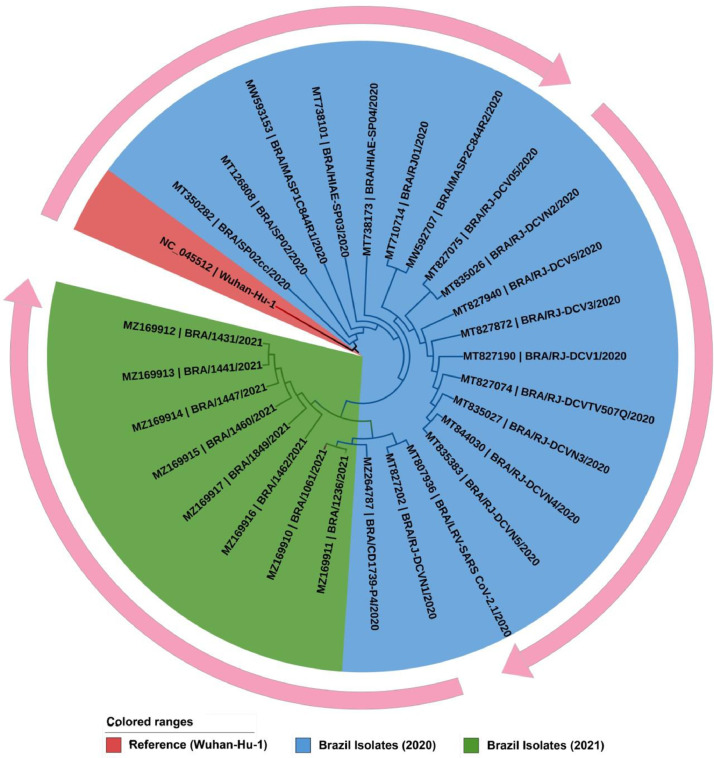
We performed the phylogenomic discrepancy among the 27 full-length genomic sequences of Brazilian SARS-CoV-2 isolates, so far yet deposited in the NCBI-virus repository (Fig 1 ). Out of 27 genomic isolates 19 samples were collected from the year 2020 and 8 from 2021. Genomic features of the selected isolates are represented in Supplementary Table S1. Phylogenetic analysis demonstrated that the tree was separated into two major clades. First clade shows the origin and replication of Brazilian isolate from the reference Wuhan-Hu-1 genome (red color). Cluster in blue indicates Brazilian isolates of 2020, whereas the second clade represents isolates from 2021 (green).
Fig. 1.
A circular view of the phylogenomic tree represents the evolutionary transmittance of Brazilian SARS-CoV-2 isolates along with the reference genome (Wuhan-Hu-1). Each color of nodes and clades are represented a corresponding genomic isolated year. The lengths of the branches are proportional to the evolutionary distances. The outer circular arrow indicated the evolutionary flow of Brazilian SARS-CoV-2 isolates.
3.2. Analysis of genotypic and proteomic variants
The selected isolates were aligned and compared with Wuhan-Hu-1 genome to identify the mutations. Each ORFs (including non-structural protein from ORF1ab) is pair-wisely aligned separately based on the genomic organization and its position of a reference genome (Table S2). Interestingly we found 16 genotypic variants among the 27 Brazilian isolates, based upon the mutation analysis.
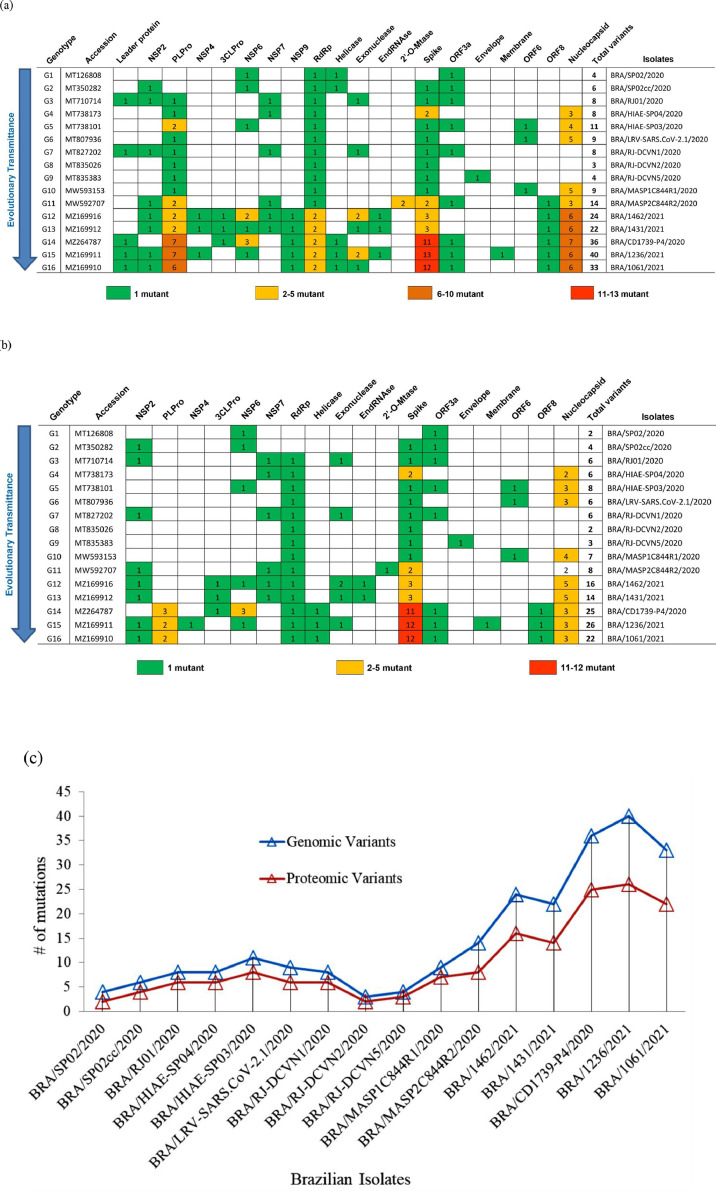
The first wave of SARS-CoV-2 genome of 2020 has 3–14 mutations, whereas the second wave SARS-CoV-2 genome has 15–20 mutants (Fig 2 a; Table S3). 3 to 7 mutants were identified in the Nucleocapsid genes with all genotypes (G1 to G16). Numerous mutations were found in PLPro and spike genes of the isolates BRA/CD1739-P4/2020 (11 base change), Bra/1236/2021 (13 base change), and Bra/1061/2021 (12 base change).
Fig. 2.
The heat map representation indicates the identified mutants in genomic (a) and proteomic (b) regions among the selected 16 genotypic variants from Brazilian SARS-CoV-2 isolates. (c) A graphical representation of mutation rates was predicted from the total hotspot of Brazilian isolates (c). Note: The number inside the cell indicated the total number of mutations.
Comparatively, the identified mutant in the proteomic regions (Fig 2b; Table 1 ) show a huge variation from isolates BRA/CD1739-P4/2020 (11 base changes), BRA/1236/2021, and BRA/1061/2021 (12 base changes) with a higher number of mutations in the spike genes are more virulent. Two to five mutations were identified in the nucleocapsid protein of genotypes G1 to G16. Gene NSP4 (S184N) and membrane protein (A2V) from the isolate BRA/1236/2021 (G15), envelop protein (V5A) from BRA/RJ-DCVN5/2020 (G9), 2′O-mutase (R216N) from BRA/MASP2C844R2/2020 (G11) had the single mutation. No mutations were observed in NSP1, NSP8-NSP10 genes of ORFab. The second wave of Brazilian isolates especially spike protein of the genotypes G14-G16 had four times higher mutation than the first wave (Table 1). Out of the 27 isolates, eight isolates of 2021 second wave had five times higher mutation than the first wave of 2020 isolates (Fig 2C).
Table 1.
Identification of proteomic variants of Brazilian SARS-CoV-2 isolates. The variants were represented among each open reading frame (ORFs).
| Genotype | Accession | Isolates | Genes | Mutation |
|---|---|---|---|---|
| G1 | MT126808 | BRA/SP02/2020 | NSP6 | L37F |
| ORF3a | G251V | |||
| G2 | MT350282 | BRA/SP02cc/2020 | NSP2 | T528I |
| NSP6 | L37F | |||
| Spike | N74K | |||
| ORF3a | G251V | |||
| G3 | MT710714 | BRA/RJ01/2020 | NSP2 | T85I |
| NSP7 | S25L | |||
| RdRp | P323L | |||
| Exonuclease | A320V | |||
| Spike | D614G | |||
| ORF3a | Q57H | |||
| G4 | MT738173 | BRA/HIAE-SP04/2020 | NSP7 | L71F |
| RdRp | P323L | |||
| Spike | D614G, V1176F | |||
| Nucleocapsid | R203K, G204R | |||
| G5 | MT738101 | BRA/HIAE-SP03/2020 | NSP6 | T17I |
| RdRp | P323L | |||
| Spike | D614G | |||
| ORF3a | S171L | |||
| ORF6 | I33T | |||
| Nucleocapsid | R203K, G204R, I292T | |||
| G6 | MT807936 | BRA/LRV-SARS.CoV-2.1/2020 | RdRp | P323L |
| Spike | D614G | |||
| ORF6 | I33T | |||
| Nucleocapsid | R203K, G204R, I292T | |||
| G7 | MT827202 | BRA/RJ-DCVN1/2020 | NSP2 | T85I |
| NSP7 | S25L | |||
| RdRp | P323L | |||
| Exonuclease | A320V | |||
| Spike | D614G | |||
| ORF3a | Q57H | |||
| G8 | MT835026 | BRA/RJ-DCVN2/2020 | RdRp | P323L |
| Spike | D614G | |||
| G9 | MT835383 | BRA/RJ-DCVN5/2020 | RdRp | P323L |
| Spike | D614G | |||
| Envelope | V5A | |||
| G10 | MW593153 | BRA/MASP1C844R1/2020 | RdRp | P323L |
| Spike | D614G | |||
| ORF6 | I33T | |||
| Nucleocapsid | R203K, G204R, I292T, P383L | |||
| G11 | MW592707 | BRA/MASP2C844R2/2020 | NSP2 | V577F |
| NSP7 | L71F | |||
| RdRp | P323L | |||
| Ribose | R216N | |||
| Spike | D614G, V1176F | |||
| Nucleocapsid | R203K, G204R | |||
| G12 | MZ169916 | BRA/1462/2021 | NSP2 | G88E |
| 3CLPro | L205V | |||
| NSP6 | L37F | |||
| NSP7 | L71F | |||
| RdRp | P323L | |||
| Exonuclease | T16A, Y235N | |||
| EndRNAse | K110R | |||
| Spike | E484K, D614G, V1176F | |||
| Nucleocapsid | A119S, S202T, R203K, G204R, M234I | |||
| G13 | MZ169912 | BRA/1431/2021 | NSP2 | G88E |
| 3CLPro | L205V | |||
| NSP7 | L71F | |||
| RdRp | P323L | |||
| Exonuclease | T16A | |||
| EndRNAse | K110R | |||
| Spike | E484K, D614G, V1176F | |||
| Nucleocapsid | A119S, S202T, R203K, G204R, M234I | |||
| G14 | MZ264787 | BRA/CD1739-P4/2020 | PLPro | S375L, K982Q, S1740F |
| 3CLPro | P96H | |||
| NSP6 | S106T, G107S, F108L | |||
| RdRp | P323L | |||
| Helicase | E341D | |||
| Spike | L18F, T20N, P26S, D138Y, R190S, E484K, N501Y, D614G, H655Y, T1027I, V1176F | |||
| ORF3a | S253P | |||
| ORF8 | E92K | |||
| Nucleocapsid | P80R, R203K, G204R | |||
| G15 | MZ169911 | BRA/1236/2021 | NSP2 | S138L |
| PLPro | S375L, K982Q | |||
| NSP4 | S184N | |||
| NSP6 | F108L | |||
| RdRp | P323L | |||
| Helicase | E341D | |||
| Exonuclease | N256S | |||
| Spike | L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I, V1176F | |||
| ORF3a | S253P | |||
| Membrane | A2V | |||
| ORF8 | E92K | |||
| Nucleocapsid | P80R, R203K, G204R | |||
| G16 | MZ169910 | BRA/1061/2021 | NSP2 | K534N |
| PLPro | S375L, K982Q | |||
| RdRp | P323L | |||
| Helicase | E341D | |||
| Spike | L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I, V1176F | |||
| ORF3a | S253P | |||
| ORF8 | E92K | |||
| Nucleocapsid | P80R, R203K, G204R |
3.3. Analysis of virulence mechanism and protein folding state
The virulence mechanism of a protein can be resolute by its amino acid composition and the physicochemical properties of substituted amino acids. Our findings show that most of the proteins are involved in the cellular and metabolic process and none are identified from information molecules. The mutant (P323L) identified in RdRp of BRA/CD1739-P4/2020 (G14) transmits its biological function from cellular process to virulence mechanism (Table 2 ).
Table 2.
. Comparative analysis of predicted virulence properties of protein mutants identified from Brazilian SARS-CoV-2 isolates. The virulence properties were predicted for both reference and variant proteins. The blue shadow specifies the same functional properties compared with reference ORFs. The orange shadow specifies the functional variants between the reference and mutated proteins.
| Genotype | Isolates | Genes | Mutation | Cellular Process | Metabolism | Virulence factors | |||
|---|---|---|---|---|---|---|---|---|---|
| Ref. | Var. | Ref. | Var. | Ref. | Var. | ||||
| G1 | BRA/SP02/2020 | NSP6 | L37F | 1.988548 | 1.988548 | −0.29145 | −0.29145 | −0.91113 | −0.91113 |
| ORF3a | G251V | 1.59006 | 1.952861 | 0.0081 | 0.104934 | −0.3168 | −0.83548 | ||
| G2 | BRA/SP02cc/2020 | NSP2 | T528I | −2.72165 | −2.72165 | 3.304706 | 3.304706 | −3.39956 | −3.39956 |
| NSP6 | L37F | 1.988548 | 1.988548 | −0.29145 | −0.29145 | −0.91113 | −0.91113 | ||
| Spike | N74K | 1.507647 | 1.492872 | −1.38428 | −1.25244 | −0.67481 | −0.69898 | ||
| ORF3a | G251V | 1.59006 | 1.952861 | 0.0081 | 0.104934 | −0.3168 | −0.83548 | ||
| G3 | BRA/RJ01/2020 | NSP2 | T85I | −2.72165 | −2.72165 | 3.304706 | 3.304706 | −3.39956 | −3.39956 |
| NSP7 | S25L | 1.227022 | 1.227022 | −2.16893 | −2.16893 | 0.960535 | 0.960535 | ||
| RdRp | P323L | −0.54011 | −0.54011 | 0.931417 | 0.931417 | 1.304757 | 1.304757 | ||
| Exonuclease | A320V | 0.205947 | 0.703741 | −1.91813 | −0.93051 | 1.812892 | 0.810148 | ||
| Spike | D614G | 1.507647 | 1.492872 | −1.38428 | −1.25244 | −0.67481 | −0.69898 | ||
| ORF3a | Q57H | 1.59006 | 1.952861 | 0.0081 | 0.104934 | −0.3168 | −0.83548 | ||
| G4 | BRA/HIAE-SP04/2020 | NSP7 | L71F | 1.521723 | 1.521723 | 0.519135 | 0.519135 | −1.40537 | −1.40537 |
| RdRp | P323L | 1.227022 | 1.227022 | −2.16893 | −2.16893 | 0.960535 | 0.960535 | ||
| Spike | D614G, V1176F | 1.507647 | 1.492872 | −1.38428 | −1.25244 | −0.67481 | −0.69898 | ||
| Nucleocapsid | R203K, G204R | 0.623184 | 0.766355 | 0.766728 | 0.788503 | −0.26696 | −0.85619 | ||
| G5 | BRA/HIAE-SP03/2020 | NSP6 | T17I | 1.988548 | 1.988548 | −0.29145 | −0.29145 | −0.91113 | −0.91113 |
| RdRp | P323L | 1.227022 | 1.227022 | −2.16893 | −2.16893 | 0.960535 | 0.960535 | ||
| Spike | D614G | 1.507647 | 1.492872 | −1.38428 | −1.25244 | −0.67481 | −0.69898 | ||
| ORF3a | S171L | 1.59006 | 1.736869 | 0.0081 | 0.319992 | −0.3168 | −0.61861 | ||
| ORF6 | I33T | 1.347941 | 1.492923 | 0.091501 | 0.946338 | −0.55216 | −1.3189 | ||
| Nucleocapsid | R203K, G204R, I292T | 0.623184 | 0.766355 | 0.766728 | 0.788503 | −0.26696 | −0.85619 | ||
| G6 | BRA/LRV-SARS.CoV-2.1/2020 | RdRp | P323L | 1.227022 | 1.227022 | −2.16893 | −2.16893 | 0.960535 | 0.960535 |
| Spike | D614G | 1.507647 | 1.492872 | −1.38428 | −1.25244 | −0.67481 | −0.69898 | ||
| ORF6 | I33T | 1.347941 | 1.492923 | 0.091501 | 0.946338 | −0.55216 | −1.3189 | ||
| Nucleocapsid | R203K, G204R, I292T | 0.623184 | 0.766355 | 0.766728 | 0.788503 | −0.26696 | −0.85619 | ||
| G7 | BRA/RJ-DCVN1/2020 | NSP2 | T85I | −2.72165 | −2.72165 | 3.304706 | 3.304706 | −3.39956 | −3.39956 |
| NSP7 | S25L | 1.521723 | 1.521723 | 0.519135 | 0.519135 | −1.40537 | −1.40537 | ||
| RdRp | P323L | 1.227022 | 1.227022 | −2.16893 | −2.16893 | 0.960535 | 0.960535 | ||
| Exonuclease | A320V | 0.205947 | 0.703741 | −1.91813 | −0.93051 | 1.812892 | 0.810148 | ||
| Spike | D614G | 1.507647 | 1.492872 | −1.38428 | −1.25244 | −0.67481 | −0.69898 | ||
| ORF3a | Q57H | 1.59006 | 1.952861 | 0.0081 | 0.104934 | −0.3168 | −0.83548 | ||
| G8 | BRA/RJ-DCVN2/2020 | RdRp | P323L | 1.227022 | 1.227022 | −2.16893 | −2.16893 | 0.960535 | 0.960535 |
| Spike | D614G | 1.507647 | 1.492872 | −1.38428 | −1.25244 | −0.67481 | −0.69898 | ||
| G9 | BRA/RJ-DCVN5/2020 | RdRp | P323L | 1.227022 | 1.227022 | −2.16893 | −2.16893 | 0.960535 | 0.960535 |
| Spike | D614G | 1.507647 | 1.492872 | −1.38428 | −1.25244 | −0.67481 | −0.69898 | ||
| Envelope | V5A | 0.736976 | 0.727204 | 0.567783 | 1.294167 | −0.08788 | −1.05017 | ||
| G10 | BRA/MASP1C844R1/2020 | RdRp | P323L | 1.227022 | 1.227022 | −2.16893 | −2.16893 | 0.960535 | 0.960535 |
| Spike | D614G | 1.507647 | 1.492872 | −1.38428 | −1.25244 | −0.67481 | −0.69898 | ||
| ORF6 | I33T | 1.347941 | 1.492923 | 0.091501 | 0.946338 | −0.55216 | −1.3189 | ||
| Nucleocapsid | R203K, G204R, I292T, P383L | 0.623184 | 1.516895 | 0.766728 | −0.32718 | −0.26696 | −0.71549 | ||
| G11 | BRA/MASP2C844R2/2020 | NSP2 | V577F | −2.72165 | −2.11747 | 3.304706 | 3.081117 | −3.39956 | −2.99917 |
| NSP7 | L71F | 1.521723 | 1.521723 | 0.519135 | 0.519135 | −1.40537 | −1.40537 | ||
| RdRp | P323L | 1.227022 | 1.227022 | −2.16893 | −2.16893 | 0.960535 | 0.960535 | ||
| Ribose | R216N | 1.464238 | 1.464238 | −0.14461 | −0.14461 | −0.21774 | −0.21774 | ||
| Spike | D614G, V1176F | 1.507647 | 1.492872 | −1.38428 | −1.25244 | −0.67481 | −0.69898 | ||
| Nucleocapsid | R203K, G204R | 0.623184 | 0.766355 | 0.766728 | 0.788503 | −0.26696 | −0.85619 | ||
| G12 | BRA/1462/2021 | NSP2 | G88E | −2.72165 | −2.72165 | 3.304706 | 3.304706 | −3.39956 | −3.39956 |
| 3CLPro | L205V | 0.521603 | 0.521603 | −0.90669 | −0.90669 | 1.521026 | 1.521026 | ||
| NSP6 | L37F | 1.988548 | 1.988548 | −0.29145 | −0.29145 | −0.91113 | −0.91113 | ||
| NSP7 | L71F | 1.521723 | 1.521723 | 0.519135 | 0.519135 | −1.40537 | −1.40537 | ||
| RdRp | P323L | 1.227022 | 1.227022 | −2.16893 | −2.16893 | 0.960535 | 0.960535 | ||
| Exonuclease | T16A, Y235N | 0.205947 | 0.212552 | −1.91813 | −1.27247 | 1.812892 | 1.925776 | ||
| EndRNAse | K110R | 0.886765 | 0.975383 | −0.18029 | 0.012195 | 0.194409 | −0.08423 | ||
| Spike | E484K, D614G, V1176F | 1.507647 | 1.492872 | −1.38428 | −1.25244 | −0.67481 | −0.69898 | ||
| Nucleocapsid | A119S, S202T, R203K, G204R, M234I | 0.623184 | 0.379777 | 0.766728 | 1.258901 | −0.26696 | −0.97281 | ||
| G13 | BRA/1431/2021 | NSP2 | G88E | −2.72165 | −2.72165 | 3.304706 | 3.304706 | −3.39956 | −3.39956 |
| 3CLPro | L205V | 0.521603 | 0.521603 | −0.90669 | −0.90669 | 1.521026 | 1.521026 | ||
| NSP7 | L71F | 1.521723 | 1.521723 | 0.519135 | 0.519135 | −1.40537 | −1.40537 | ||
| RdRp | P323L | 1.227022 | 1.227022 | −2.16893 | −2.16893 | 0.960535 | 0.960535 | ||
| Exonuclease | T16A | 0.205947 | 0.212552 | −1.91813 | −1.27247 | 1.812892 | 1.925776 | ||
| EndRNAse | K110R | 0.886765 | 0.975383 | −0.18029 | 0.012195 | 0.194409 | −0.08423 | ||
| Spike | E484K, D614G, V1176F | 1.507647 | 1.492872 | −1.38428 | −1.25244 | −0.67481 | −0.69898 | ||
| Nucleocapsid | A119S, S202T, R203K, G204R, M234I | 0.623184 | 0.379777 | 0.766728 | 1.258901 | −0.26696 | −0.97281 | ||
| G14 | BRA/CD1739-P4/2020 | PLPro | S375L, K982Q, S1740F | −3.81331 | −3.93465 | −3.09635 | −3.2623 | 0.135194 | 0.258528 |
| 3CLPro | P96H | 0.521603 | 0.521603 | −0.90669 | −0.90669 | 1.521026 | 1.521026 | ||
| NSP6 | S106T, G107S, F108L | 1.988548 | 2.186767 | −0.29145 | −0.68079 | −0.91113 | −1.26714 | ||
| RdRp | P323L | −0.54011 | 1.227022 | 0.931417 | −2.16893 | 1.304757 | 0.960535 | ||
| Helicase | E341D | −0.54011 | −0.54011 | 0.931417 | 0.931417 | 1.304757 | 1.304757 | ||
| Spike | L18F, T20N, P26S, D138Y, R190S, E484K, N501Y, D614G, H655Y, T1027I, V1176F | 1.507647 | 1.75435 | −1.38428 | −1.31278 | −0.67481 | −1.06524 | ||
| ORF3a | S253P | 1.59006 | 2.069778 | 0.0081 | 0.103932 | −0.3168 | −1.04161 | ||
| ORF8 | E92K | 0.70646 | 0.947547 | −0.46479 | 0.551035 | 0.633644 | −0.30211 | ||
| Nucleocapsid | P80R, R203K, G204R | 0.623184 | 0.766355 | 0.766728 | 0.788503 | −0.26696 | −0.85619 | ||
| G15 | BRA/1236/2021 | NSP2 | S138L | −2.72165 | −1.73529 | 3.304706 | 2.882199 | −3.39956 | −2.7792 |
| PLPro | S375L, K982Q | −3.81331 | −3.93465 | −3.09635 | −3.2623 | 0.135194 | 0.258528 | ||
| NSP4 | S184N | −0.47381 | −0.47381 | 2.584585 | 2.584585 | −1.82286 | −1.82286 | ||
| NSP6 | F108L | 1.988548 | 1.969257 | −0.29145 | −0.3559 | −0.91113 | −0.76909 | ||
| RdRp | P323L | 1.227022 | 1.227022 | −2.16893 | −2.16893 | 0.960535 | 0.960535 | ||
| Helicase | E341D | −0.54011 | −0.54011 | 0.931417 | 0.931417 | 1.304757 | 1.304757 | ||
| Exonuclease | N256S | 0.205947 | 0.205947 | −1.91813 | −1.91813 | 1.812892 | 1.812892 | ||
| Spike | L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I, V1176F | 1.507647 | 1.75435 | −1.38428 | −1.31278 | −0.67481 | −1.06524 | ||
| ORF3a | S253P | 1.59006 | 2.069778 | 0.0081 | 0.103932 | −0.3168 | −1.04161 | ||
| Membrane | A2V | 1.772062 | 1.758567 | 0.905378 | 1.061507 | −1.40208 | −2.02498 | ||
| ORF8 | E92K | 0.70646 | 0.947547 | −0.46479 | 0.551035 | 0.633644 | −0.30211 | ||
| Nucleocapsid | P80R, R203K, G204R | 0.623184 | 0.766355 | 0.766728 | 0.788503 | −0.26696 | −0.85619 | ||
| G16 | BRA/1061/2021 | NSP2 | K534N | −2.72165 | −2.72165 | 3.304706 | 3.304706 | −3.39956 | −3.39956 |
| PLPro | S375L, K982Q | −3.81331 | −3.93465 | −3.09635 | −3.2623 | 0.135194 | 0.258528 | ||
| RdRp | P323L | 1.227022 | 1.227022 | −2.16893 | −2.16893 | 0.960535 | 0.960535 | ||
| Helicase | E341D | −0.54011 | −0.54011 | 0.931417 | 0.931417 | 1.304757 | 1.304757 | ||
| Spike | L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I, V1176F | 1.507647 | 1.75435 | −1.38428 | −1.31278 | −0.67481 | −1.06524 | ||
| ORF3a | S253P | 1.59006 | 2.069778 | 0.0081 | 0.103932 | −0.3168 | −1.04161 | ||
| ORF8 | E92K | 0.70646 | 0.947547 | −0.46479 | 0.551035 | 0.633644 | −0.30211 | ||
| Nucleocapsid | P80R, R203K, G204R | 0.623184 | 0.766355 | 0.766728 | 0.788503 | −0.26696 | −0.85619 | ||
The protein folding rate is most important to determine the stability of structure and function. It is most imperative to predict the folding rate of protein to identify the various structural changes in the drug binding region. We also predicted the folding mechanism based on its structural classes (Table 3 ). A fast-fold rate was identified in all mutated proteins exactly in β-classes. And slow fold rate was identified in other classes (All-α, α+β:α/β and unknown).
Table 3.
. A comparison of the folding rate and mechanism of protein mutants isolated from Brazilian SARS-CoV-2 isolates. The folding rate was predicted for both reference and variant proteins. The positive value represents fast-folding rate whereas negative indicates slow-folding rate.
| Genotype | Isolates | Genes | Mutation | All-α | All-β | α+β:α/β | Unknown | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ref.. | Var. | Ref.. | Var. | Ref.. | Var. | Ref.. | Var. | ||||
| G1 | BRA/SP02/2020 | NSP6 | L37F | 6.6 | 7.5 | 63.8 | 66.9 | −17.9 | −3.54 | 0.996 | −7.75 |
| ORF3a | G251V | −6.16 | −5.23 | 39.4 | 38.9 | 0.335 | −2.42 | −4.11 | 2.74 | ||
| G2 | BRA/SP02cc/2020 | NSP2 | T528I | 12.3 | 12.5 | 10.5 | 16.7 | −12.7 | −23.7 | 4.41 | −2.22 |
| NSP6 | L37F | 6.6 | 7.5 | 63.8 | 66.9 | −17.9 | −3.54 | 0.996 | −7.75 | ||
| Spike | N74K | 3.4 | 4.1 | 21.5 | 17.7 | −11 | −9.97 | 6.12 | 2.25 | ||
| ORF3a | G251V | −6.16 | −5.23 | 39.4 | 38.9 | 0.335 | −2.42 | −4.11 | 2.74 | ||
| G3 | BRA/RJ01/2020 | NSP2 | T85I | 12.3 | 12.5 | 10.5 | 16.7 | −12.7 | −23.7 | 4.41 | −2.22 |
| NSP7 | S25L | 21.2 | 22.5 | 37.2 | 34.2 | 22.3 | −0.383 | 13.1 | 7.57 | ||
| RdRp | P323L | 13.9 | 13.8 | 21.7 | 21.5 | −13.3 | −0.237 | 4.04 | 4.96 | ||
| Exonuclease | A320V | 9.05 | 9.47 | 16.9 | 22.5 | 4.29 | 2.22 | 0.83 | 2.97 | ||
| Spike | D614G | 3.4 | 4.65 | 21.5 | 24.9 | −11 | −0.137 | 6.12 | 5.89 | ||
| ORF3a | Q57H | −6.16 | −5.22 | 39.4 | 39.2 | 0.335 | −23.9 | −4.11 | 4.27 | ||
| G4 | BRA/HIAE-SP04/2020 | NSP7 | L71F | 21.2 | 20.5 | 37.2 | 33.7 | 22.3 | −13.3 | 13.1 | 14.3 |
| RdRp | P323L | 13.9 | 13.8 | 21.7 | 21.5 | −13.3 | −0.237 | 4.04 | 4.96 | ||
| Spike | D614G, V1176F | 3.4 | 3.49 | 21.5 | 21.4 | −11 | 9.7 | 6.12 | 11.7 | ||
| Nucleocapsid | R203K, G204R | −9.64 | −10.2 | −7.18 | −6.32 | 28.6 | 17.2 | 5.53 | 6.23 | ||
| G5 | BRA/HIAE-SP03/2020 | NSP6 | T17I | 6.6 | 6.89 | 63.8 | 66.6 | −17.9 | −26 | 0.996 | −0.894 |
| RdRp | P323L | 13.9 | 13.8 | 21.7 | 21.5 | −13.3 | −0.237 | 4.04 | 4.96 | ||
| Spike | D614G | 3.4 | 4.65 | 21.5 | 24.9 | −11 | −0.137 | 6.12 | 5.89 | ||
| ORF3a | S171L | −6.16 | −6.21 | 39.4 | 38.7 | 0.335 | 4.01 | −4.11 | −2.7 | ||
| ORF6 | I33T | 8.4 | 8.65 | 53.1 | 51 | −32.1 | −12.5 | 4.37 | 6.66 | ||
| Nucleocapsid | R203K, G204R, I292T | −9.64 | −9.41 | −7.18 | −6.69 | 28.6 | 29.5 | 5.53 | −1.67 | ||
| G6 | BRA/LRV-SARS.CoV-2.1/2020 | RdRp | P323L | 13.9 | 13.8 | 21.7 | 21.5 | −13.3 | −0.237 | 4.04 | 4.96 |
| Spike | D614G | 3.4 | 4.65 | 21.5 | 24.9 | −11 | −0.137 | 6.12 | 5.89 | ||
| ORF6 | I33T | 8.4 | 8.65 | 53.1 | 51 | −32.1 | −12.5 | 4.37 | 6.66 | ||
| Nucleocapsid | R203K, G204R, I292T | −9.64 | −9.41 | −7.18 | −6.69 | 28.6 | 29.5 | 5.53 | −1.67 | ||
| G7 | BRA/RJ-DCVN1/2020 | NSP2 | T85I | 12.3 | 12.5 | 10.5 | 16.7 | −12.7 | −23.7 | 4.41 | −2.22 |
| NSP7 | S25L | 21.2 | 22.5 | 37.2 | 34.2 | 22.3 | −0.383 | 13.1 | 7.57 | ||
| RdRp | P323L | 13.9 | 13.8 | 21.7 | 21.5 | −13.3 | −0.237 | 4.04 | 4.96 | ||
| Exonuclease | A320V | 9.05 | 9.47 | 16.9 | 22.5 | 4.29 | 2.22 | 0.83 | 2.97 | ||
| Spike | D614G | 3.4 | 4.65 | 21.5 | 24.9 | −11 | −0.137 | 6.12 | 5.89 | ||
| ORF3a | Q57H | −6.16 | −5.22 | 39.4 | 39.2 | 0.335 | −23.9 | −4.11 | 4.27 | ||
| G8 | BRA/RJ-DCVN2/2020 | RdRp | P323L | 13.9 | 13.8 | 21.7 | 21.5 | −13.3 | −0.237 | 4.04 | 4.96 |
| Spike | D614G | 3.4 | 4.65 | 21.5 | 24.9 | −11 | −0.137 | 6.12 | 5.89 | ||
| G9 | BRA/RJ-DCVN5/2020 | RdRp | P323L | 13.9 | 13.8 | 21.7 | 21.5 | −13.3 | −0.237 | 4.04 | 4.96 |
| Spike | D614G | 3.4 | 4.65 | 21.5 | 24.9 | −11 | −0.137 | 6.12 | 5.89 | ||
| Envelope | V5A | 7.51 | 7.54 | 55 | 53.9 | 3.46 | −13.5 | 7.44 | 7.83 | ||
| G10 | BRA/MASP1C844R1/2020 | RdRp | P323L | 13.9 | 13.8 | 21.7 | 21.5 | −13.3 | −0.237 | 4.04 | 4.96 |
| Spike | D614G | 3.4 | 4.65 | 21.5 | 24.9 | −11 | −0.137 | 6.12 | 5.89 | ||
| ORF6 | I33T | 8.4 | 8.65 | 53.1 | 51 | −32.1 | −12.5 | 4.37 | 6.66 | ||
| Nucleocapsid | R203K, G204R, I292T, P383L | −9.64 | −9.3 | −7.18 | −3.3 | 28.6 | 27.1 | 5.53 | 7.81 | ||
| G11 | BRA/MASP2C844R2/2020 | NSP2 | V577F | 12.3 | 12.8 | 10.5 | 14.2 | −12.7 | −16 | 4.41 | 6.91 |
| NSP7 | L71F | 21.2 | 20.5 | 37.2 | 33.7 | 22.3 | −13.3 | 13.1 | 14.3 | ||
| RdRp | P323L | 13.9 | 13.8 | 21.7 | 21.5 | −13.3 | −0.237 | 4.04 | 4.96 | ||
| Ribose | R216N | 6.18 | 6.39 | 25.6 | 22.9 | 10.9 | −0.719 | 0.743 | 5.63 | ||
| Spike | D614G, V1176F | 3.4 | 3.49 | 21.5 | 21.4 | −11 | 9.7 | 6.12 | 11.7 | ||
| Nucleocapsid | R203K, G204R | −9.64 | −10.2 | −7.18 | −6.32 | 28.6 | 17.2 | 5.53 | 6.23 | ||
| G12 | BRA/1462/2021 | NSP2 | G88E | 12.3 | 13.7 | 10.5 | 16.6 | −12.7 | −8.3 | 4.41 | 10.9 |
| 3CLPro | L205V | 15.5 | 15.8 | 19.2 | 22.3 | −2.24 | −17.1 | 5.22 | 2.44 | ||
| NSP6 | L37F | 6.6 | 7.5 | 63.8 | 66.9 | −17.9 | −3.54 | 0.996 | −7.75 | ||
| NSP7 | L71F | 21.2 | 20.5 | 37.2 | 33.7 | 22.3 | −13.3 | 13.1 | 14.3 | ||
| RdRp | P323L | 13.9 | 13.8 | 21.7 | 21.5 | −13.3 | −0.237 | 4.04 | 4.96 | ||
| Exonuclease | T16A, Y235N | 9.05 | 8.82 | 16.9 | 17.1 | 4.29 | −8.21 | 0.83 | −0.196 | ||
| EndRNAse | K110R | 6.52 | 7.37 | 18.4 | 22.7 | 27.4 | 2.66 | 5.26 | 5.64 | ||
| Spike | E484K, D614G, V1176F | 3.4 | 4.02 | 21.5 | 22 | −11 | −8.88 | 6.12 | 10.8 | ||
| Nucleocapsid | A119S, S202T, R203K, G204R, M234I | −9.64 | −10 | −7.18 | −3.17 | 28.6 | 22.4 | 5.53 | 9.46 | ||
| G13 | BRA/1431/2021 | NSP2 | G88E | 12.3 | 13.7 | 10.5 | 16.6 | −12.7 | −8.3 | 4.41 | 10.9 |
| 3CLPro | L205V | 15.5 | 15.8 | 19.2 | 22.3 | −2.24 | −17.1 | 5.22 | 2.44 | ||
| NSP7 | L71F | 21.2 | 20.5 | 37.2 | 33.7 | 22.3 | −13.3 | 13.1 | 14.3 | ||
| RdRp | P323L | 13.9 | 13.8 | 21.7 | 21.5 | −13.3 | −0.237 | 4.04 | 4.96 | ||
| Exonuclease | T16A | 9.05 | 8.22 | 16.9 | 21.5 | 4.29 | −3.75 | 0.83 | 0.684 | ||
| EndRNAse | K110R | 6.52 | 7.37 | 18.4 | 22.7 | 27.4 | 2.66 | 5.26 | 5.64 | ||
| Spike | E484K, D614G, V1176F | 3.4 | 4.02 | 21.5 | 22 | −11 | −8.88 | 6.12 | 10.8 | ||
| Nucleocapsid | A119S, S202T, R203K, G204R, M234I | −9.64 | −10 | −7.18 | −3.17 | 28.6 | 22.4 | 5.53 | 9.46 | ||
| G14 | BRA/CD1739-P4/2020 | PLPro | S375L, K982Q, S1740F | 5.63 | 6.62 | 16.6 | 13.1 | −3.42 | −5 | 0.533 | 1.81 |
| 3CLPro | P96H | 15.5 | 16.6 | 19.2 | 18.8 | −2.24 | −8.53 | 5.22 | 2.78 | ||
| NSP6 | S106T, G107S, F108L | 6.6 | 6.84 | 63.8 | 61.7 | −17.9 | −22 | 0.996 | 1.26 | ||
| RdRp | P323L | 13.9 | 13.8 | 21.7 | 21.5 | −13.3 | −0.237 | 4.04 | 4.96 | ||
| Helicase | E341D | 3.56 | 3.24 | 17.8 | 16.9 | −3.33 | −19.7 | 11.2 | 7.38 | ||
| Spike | L18F, T20N, P26S, D138Y, R190S, E484K, N501Y, D614G, H655Y, T1027I, V1176F | 3.4 | 4.05 | 21.5 | 24.6 | −11 | −1.44 | 6.12 | 13.3 | ||
| ORF3a | S253P | −6.16 | −6.18 | 39.4 | 37 | 0.335 | −18 | −4.11 | 0.908 | ||
| ORF8 | E92K | 0.827 | 0.363 | 42.2 | 45.3 | −44.6 | −25.5 | 5.47 | 1.2 | ||
| Nucleocapsid | P80R, R203K, G204R | −9.64 | −8.92 | −7.18 | −5.31 | 28.6 | 27.1 | 5.53 | 7.52 | ||
| G15 | BRA/1236/2021 | NSP2 | S138L | 12.3 | 13 | 10.5 | 15.6 | −12.7 | −20.4 | 4.41 | 0.365 |
| PLPro | S375L, K982Q | 5.63 | 5.51 | 16.6 | 10.5 | −3.42 | −17.3 | 0.533 | 3.93 | ||
| NSP4 | S184N | −0.339 | 0.632 | 41.8 | 38.5 | −10.2 | −28.3 | 4.57 | −2.79 | ||
| NSP6 | F108L | 6.6 | 6.66 | 63.8 | 64.3 | −17.9 | −26.2 | 0.996 | 3.07 | ||
| RdRp | P323L | 13.9 | 13.8 | 21.7 | 21.5 | −13.3 | −0.237 | 4.04 | 4.96 | ||
| Helicase | E341D | 3.56 | 3.24 | 17.8 | 16.9 | −3.33 | −19.7 | 11.2 | 7.38 | ||
| Exonuclease | N256S | 9.05 | 7.59 | 16.9 | 22.4 | 4.29 | −0.968 | 0.83 | −2.08 | ||
| Spike | L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I, V1176F | 3.4 | 4.28 | 21.5 | 25.6 | −11 | −3.9 | 6.12 | 8.9 | ||
| ORF3a | S253P | −6.16 | −6.18 | 39.4 | 37 | 0.335 | −18 | −4.11 | 0.908 | ||
| Membrane | A2V | 0.355 | 0.722 | 43.8 | 45.6 | −19.3 | −14.7 | −0.2 | −3.23 | ||
| ORF8 | E92K | 0.827 | 0.363 | 42.2 | 45.3 | −44.6 | −25.5 | 5.47 | 1.2 | ||
| Nucleocapsid | P80R, R203K, G204R | −9.64 | −8.92 | −7.18 | −5.31 | 28.6 | 27.1 | 5.53 | 7.52 | ||
| G16 | BRA/1061/2021 | NSP2 | K534N | 12.3 | 11.7 | 10.5 | 15.3 | −12.7 | −8.76 | 4.41 | 2.65 |
| PLPro | S375L, K982Q | 5.63 | 5.51 | 16.6 | 10.5 | −3.42 | −17.3 | 0.533 | 3.93 | ||
| RdRp | P323L | 13.9 | 13.8 | 21.7 | 21.5 | −13.3 | −0.237 | 4.04 | 4.96 | ||
| Helicase | E341D | 3.56 | 3.24 | 17.8 | 16.9 | −3.33 | −19.7 | 11.2 | 7.38 | ||
| Spike | L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I, V1176F | 3.4 | 4.28 | 21.5 | 25.6 | −11 | −3.9 | 6.12 | 8.9 | ||
| ORF3a | S253P | −6.16 | −6.18 | 39.4 | 37 | 0.335 | −18 | −4.11 | 0.908 | ||
| ORF8 | E92K | 0.827 | 0.363 | 42.2 | 45.3 | −44.6 | −25.5 | 5.47 | 1.2 | ||
| Nucleocapsid | P80R, R203K, G204R | −9.64 | −8.92 | −7.18 | −5.31 | 28.6 | 27.1 | 5.53 | 7.52 | ||
4. Discussion
Our consequence from evolutionary footprints determines that the mean G + C content was found to be low (< 38%) while comparing to the A + T disparity index. It determines that some divergence may occur during the evolutionary transmittance of this genome. Compared with previous research (Prathiviraj et al., 2020), our findings indicate that some genomic variations may occur at the sequence level, and leads to the origination of a new clade within the isolates in a short time (short-term evolution). Host RNA editing systems, APOBEC and ADAR, responsible for C˃U (accompanied by U˃C mutations by the virus’ defense system) and A˃G mutations respectively, account for around 65% of the mutations recorded (Banoun, 2021; Klimczak et al., 2020; Wang et al., 2020). Deletion of one or a few nucleotides has been reported chiefly in the genes encoding proteins involved in host interaction and the spike protein (Banoun, 2021). Lin et al. (2021) designated the ∼ 500–532 coding region of NSP as a deletion hotspot, and the mutants were found to be linked with reduced virulence (mild disease, low IFN-β in serum) and lower viral load (p14). More deletion variants, primarily of structural protein origin are under scrutiny (Banoun, 2021). The transition, C>U in the genome has altered the codons to encode amino acids of more hydrophobic nature, thereby producing intense effects on the properties of the protein. This deamination of cytosine-mediated conversion of C to U has helped the SARS-CoV-2 to prompt evolution and adaptation in the host (Matyášek and Kovaří, 2020).
By analyzing the genomic and proteomic variants of selected isolates we identified both synonymous and non-synonymous substitution mutation evenly occur in all isolates, which leads to drastic changes that may occur in their protein sequences (Chu and Wei, 2019). Several reports have been previously published, that an increased level of mutation in surface glycoprotein may escort a significant role in viral contamination and disease transmission (Yan et al., 2020; Prathiviraj et al., 2021). As same as the previous report, consequently, our study also correlates the mutational rate is moderately increased in 2020 (Zhao et al., 2004; van Dorp et al., 2020) isolate and drastically increased in 2021 isolates (Pachetti et al., 2020; Callaway, 2020; Vilar and Isom, 2021) for relapsing the antivirals against COVID-19.
The spike protein of SARS-CoV-2 is more prone to multiple point mutations (Guruprasad, 2021; Ogawa et al., 2020; Plante et al., 2021), most notable being D614G (Asp614-to-Gly) variant (globally dominant viral form) with increment in transmissibility, infectivity and neutralization susceptibility (Korber et al., 2020; Koyama et al., 2020; Zhang et al., 2020; Weissman et al., 2021). Several studies have implied that the D614G mutation in the spike glycoprotein of SARS-CoV-2, being closely located in the receptor-binding domain. It has an open conformation by changing hydrogen-bond interactions in the spike trimer, consequentially, its binding ability to the hACE2 receptor is enhanced and increased infectivity (Mansbach et al., 2021; Omotuyi et al., 2020; Yurkovetskiy et al., 2020). Certain studies reported the increment in viral load in the upper respiratory tract (URT) of patients infected with the D614G mutant type virus (Korber et al., 2020). Further experimental studies correlated this spike protein mutation to enhanced viral replication in vitro (primary human URT cell lines, animal models), consistent with the URT viral load increment (; Zhou et al., 2021).
N501Y mutation in the spike protein of B.1.1.7 mutant viruses was experimentally showed to exert enhanced replication rates in cell lines and human respiratory cells (ex-vivo studies), which might occur through the same mechanism (Liu et al., 2021). It enhanced the rate of transmission and virulence probability due to its increased interaction with the human ACE2 receptor (Zhao et al., 2021). Accessory protein ORF3a was found to be four distinct types in India, namely, Type 1–4 and it changed the amino sequences in each type. Type 2 of ORF3a was raised from Type 1 by the change of amino acid mutation at the 171st position. Similarly, the change of amino acids from Asp-to Tyr-and Ser-to Leu-by mutation at 463rd position and 512th position led to the generation of Type 3 and Type 4, respectively from Type 2. These mutations possibly will affect the pathogenesis of the virus by activating the inflammasomes (Hassan et al., 2020a). Type 2 mutation (S171L) of ORF3a was found in BRA/HIAE-SP03/2020, none other type doesn't exist in Brazilian isolates. Non-synonymous mutations are reported in the envelope protein (E) of the novel coronavirus. The SARS-CoV-2 in comparison with SARS-CoV has the E protein with one deletion at 70th position and three-point mutations with Thr-to Ser, Val-to Phe, and Glu-to Arg-at 55th, 56th and 69th positions at its C terminal. These mutations might have effects on the propagation and multiplication of the virus (Hassan et al., 2020b).
BRA/CD1739-P4/2020 (G14) isolates can transmit their cellular and biological process into virulence mechanism by changing its amino acid from Pro-to Leu-at 323rd position in RdRp gene and resulted in the destabilization of its structure (Begum et al., 2020). Similarly, the change of Pro-to Leu-in helicase protein resulted in the enhanced flexibility of the secondary structure. Both of these mutations in SARS-CoV2 helped in the replication of the virus (Begum et al., 2020). A few mutants in NSP4, NSP5 are becoming more virulent in Genotype 15 and 16. All other mutants are remaining the same sub-cellular localization. The mutation is responsible for emerging low-frequency viral variants which are drug-resistant, resulting in therapy failure (Begum et al., 2020). Some mutational events may occur in nucleocapsid-associated proteins with a mid and severe clinical outcome (Nagy et al., 2021). A protein's biophysical properties, such as folding, stability, and functions, are drastically altered by amino acid substitution (Gromiha et al., 2009; Prathiviraj et al., 2016). Hence, the fluctuation in protein folding leads to replication and multiplication of SARS-CoV-2 in the new environmental niche (Prisilla et al., 2016; Prathiviraj et al., 2016, 2021).
It must be noted that the mutations described at particular locations do not individually provide fitness to the virus, but a combination of favourable mutations at different locations in the genome, resulting in the improved or diminished functionalities have been classified as a cluster of mutant viruses harbouring certain moderations (Mercatelli and Giorgi, 2020). For relapse contemporary antiviral treatments, it is crucial because of randomized genomic variants in SARS-CoV-2 isolates at a global level (Prathiviraj et al., 2021). Consequently, the predicted mutations in our study help researchers to determine new antivirulants based on their genomic variants. Thus, computational strategies to assess the side effects of therapeutic drugs are propitious to improve efficiency and accelerate the development of drug discovery (Wu et al., 2020; Murugesan et al., 2021). Recent technological and computational advances in genomics, systems biology and drug repurposing have offered possibilities for antiviral detection and identifying side effects or therapeutic agents by integrating disease proteins and drug-target interactions for the treatment of COVID-19 (Hashimoto 2021; Yousefi et al., 2021).
5. Conclusion
We performed a complete genomic and proteomic investigation of the Brazilian SARS-CoV-2 isolate to identify its genotypic variants. From our study, we identified 16 genotypic variants among 27 isolates. The genotype G14-G16 has a high mutation rate in spike protein. The mutations in NSP4 and NSP5 in genotype (G15 and G16) confer lower stability of the protein structures. Our study revealed the mutational effect on the folding rate of assembly and maturation proteins. The slow and fast folding rate of SARS-CoV-2 protein was observed concerning specific mutations. As a result, the current study will help researchers understand the mutation rate among Brazilian isolates and will aid in the discovery of new drugs.
Funding
Authors acknowledge DST-SERB, New Delhi, India under the NPDF scheme (Sanction No. PDF/2019/002,762/Dated: 23/12/2019), India.
CRediT authorship contribution statement
Ragothaman Prathiviraj: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft. Paulchamy Chellapandi: Conceptualization, Validation, Supervision, Writing – review & editing. Ajima Begum: Visualization, Writing – review & editing. George Seghal Kiran: Visualization, Writing – review & editing. Joseph Selvin: Conceptualization, Validation, Supervision, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no conflict of interest.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.virusres.2021.198618.
Appendix. Supplementary materials
References
- Banoun H. Evolution of SARS-CoV-2: review of mutations, role of the host immune system. Nephron. 2021:1–12. doi: 10.1159/000515417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begum F., Mukherjee D., Thagriki D., Das S., Tripathi P.P., Banerjee A.K., Ray U. Analyses of spike protein from first deposited sequences of SARS-CoV2 from West Bengal, India. F1000Res. 2020;9:371. doi: 10.12688/f1000research.23805.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broglia R.A., Tiana G., Sutto L., Provasi D., Simona F. Design of HIV-1-PR inhibitors that do not create resistance: blocking the folding of single monomers. Protein Sci. 2005;14:2668–2681. doi: 10.1110/ps.051670905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E. The coronavirus is mutating - does it matter? Nature. 2020;585:174–177. doi: 10.1038/d41586-020-02544-6. [DOI] [PubMed] [Google Scholar]
- Chu D., Wei L. Nonsynonymous, synonymous and nonsense mutations in human cancer-related genes undergo stronger purifying selections than expectation. BMC Cancer. 2019;19:359. doi: 10.1186/s12885-019-5572-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucinotta D., Vanelli M. WHO declares COVID-19 a pandemic. Acta Biomed. 2020;91:157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A., Gupta D. VirulentPred: a SVM based prediction method for virulent proteins in bacterial pathogens. BMC Bioinform. 2008;9:62. doi: 10.1186/1471-2105-9-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromiha M.M., Thangakani A.M., Selvaraj S. FOLD-RATE: prediction of protein folding rates from amino acid sequence. Nucl. Acids Res. 2006;34:W70–W74. doi: 10.1093/nar/gkl043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromiha M.M., Yabuki Y., Suresh M.X., Thangakani A.M., Suwa M., Fukui K. TMFunction: database for functional residues in membrane proteins. Nucl. Acids Res. 2009;37:D201–D204. doi: 10.1093/nar/gkn672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guruprasad L. Human SARS CoV-2 spike protein mutations. Proteins. 2021;89:569–576. doi: 10.1002/prot.26042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucl. Acids Symp. Ser. 1999;41(1999):95–98. [Google Scholar]
- Hashimoto K. Repurposing of CNS drugs to treat COVID-19 infection: targeting the sigma-1 receptor. Eur. Arch. Psychiatry Clin. Neurosci. 2021;271:249–258. doi: 10.1007/s00406-020-01231-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan S.S., Choudhury P.P., Basu P., Jana S.S. Molecular conservation and differential mutation on ORF3a gene in Indian SARS-CoV2 genomes. Genomics. 2020;112:3226–3237. doi: 10.1016/j.ygeno.2020.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan S.S., Choudhury P.P., Roy B. SARS-CoV2 envelope protein: non-synonymous mutations and its consequences. Genomics. 2020;112:3890–3892. doi: 10.1016/j.ygeno.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Toh H. PartTree: an algorithm to build an approximate tree from a large number of unaligned sequences. Bioinformatics. 2007;23:372–384. doi: 10.1093/bioinformatics/btl592. [DOI] [PubMed] [Google Scholar]
- Klimczak L.J., Randall T.A., Saini N., Li J.L., Gordenin D.A. Similarity between mutation spectra in hypermutated genomes of rubella virus and in SARS-CoV-2 genomes accumulated during the COVID-19 pandemic. PLoS One. 2020;15 doi: 10.1371/journal.pone.0237689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., Platt D., Parida L. Variant analysis of SARS-CoV-2 genomes. Bull. World Health Organ. 2020;98:495–504. doi: 10.2471/BLT.20.253591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucl. Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.W., Tang C., Wei H.C., et al. Genomic monitoring of SARS-CoV-2 uncovers an Nsp1 deletion variant that modulates type I interferon response. Cell Host Microbe. 2021;29:489–502. doi: 10.1016/j.chom.2021.01.015. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Liu J., Plante K.S., Plante, J.A., Xie, X., Zhang, X., Ku, Z., An, Z., Scharton, D., Schindewolf, C., Menachery, V.D., Shi, P.Y., Weaver, S.C., 2021. The N501Y spike substitution enhances SARS-CoV-2 transmission. bioRxiv [Preprint]. 2021.03.08.434499. doi: 10.1101/2021.03.08.434499. [DOI] [PMC free article] [PubMed]
- Mansbach R.A., Chakraborty S., Nguyen K., Montefiori D.C., Korber B., Gnanakaran S. The SARS-CoV-2 Spike variant D614G favors an open conformational state. Sci Adv. 2021;7:eabf3671. doi: 10.1126/sciadv.abf3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyášek R., Kovařík A. Mutation patterns of Human SARS-CoV-2 and Bat RaTG13 coronavirus genomes are strongly biased towards C>U transitions, indicating rapid evolution in their hosts. Genes. 2020;11:761. doi: 10.3390/genes11070761. (Basel) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercatelli D., Giorgi F.M. Geographic and genomic distribution of SARS-CoV-2 mutations. Front. Microbiol. 2020;11:1800. doi: 10.3389/fmicb.2020.01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais I.J., Polveiro R.C., Souza G.M., Bortolin D.I., Sassaki F.T., Lima A.T.M. The global population of SARS-CoV-2 is composed of six major subtypes. Sci. Rep. 2020;10:18289. doi: 10.1038/s41598-020-74050-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavizadeh L., Ghasemi S. Genotype and phenotype of COVID-19: their roles in pathogenesis. J. Microbiol. Immunol. Infect. 2021;54:159–163. doi: 10.1016/j.jmii.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugan A., Prathiviraj R., Mothay D., Chellapandi P. Substrate-imprinted docking of Agrobacterium tumefaciens uronate dehydrogenase for increased substrate selectivity. Int. J. Biol. Macromol. 2019;140:1214–1225. doi: 10.1016/j.ijbiomac.2019.08.194. [DOI] [PubMed] [Google Scholar]
- Murugesan S., Kottekad S., Crasta I., Sreevathsan S., Usharani D., Perumal M.K., Mudliar S.N. Targeting COVID-19 (SARS-CoV-2) main protease through active phytocompounds of ayurvedic medicinal plants - Emblica officinalis (Amla), phyllanthus niruri Linn. (Bhumi Amla) and tinospora cordifolia (Giloy) - a molecular docking and simulation study. Comput. Biol. Med. 2021;136 doi: 10.1016/j.compbiomed.2021.104683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy Á., Pongor S., Győrffy B. Different mutations in SARS-CoV-2 associate with severe and mild outcome. Int. J. Antimicrob. Agents. 2021;57 doi: 10.1016/j.ijantimicag.2020.106272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimaki T., Sato K. An extension of the Kimura two-parameter model to the natural evolutionary process. J. Mol. Evol. 2019;87:60–67. doi: 10.1007/s00239-018-9885-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa, J., Zhu, W., Tonnu, N., Singer, O., Hunter, T., Ryan, A.L., Pao, G.M., 2020. The D614G mutation in the SARS-CoV2 Spike protein increases infectivity in an ACE2 receptor dependent manner. bioRxiv [Preprint]. 2020.07.21.214932. doi: 10.1101/2020.07.21.214932.
- Omotuyi I.O., Nash O., Ajiboye O.B., Iwegbulam C.G., Oyinloye E.B., Oyedeji O.A., Kashim Z.A., Okaiyeto K. Atomistic simulation reveals structural mechanisms underlying D614G spike glycoprotein-enhanced fitness in SARS-COV-2. J. Comput. Chem. 2020;41:2158–2161. doi: 10.1002/jcc.26383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachetti M., Marini B., Benedetti F., Giudici F., Mauro E., Storici P., Masciovecchio C., Angeletti S., Ciccozzi M., Gallo R.C., Zella D., Ippodrino R. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 2020;18:179. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett B.E., Sadat E.L., Zhang Y., Noronha J.M., Squires R.B., Hunt V., Liu M., Kumar S., Zaremba S., Gu Z., Zhou L., Larson C.N., Dietrich J., Klem E.B., Scheuermann R.H. ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res. 2012;40:D593–D598. doi: 10.1093/nar/gkr859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R., Mirchandani D., Scharton D., Bilello J.P., Ku Z., An Z., Kalveram B., Freiberg A.N., Menachery V.D., Xie X., Plante K.S., Weaver S.C., Shi P.Y. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592:116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posthuma C.C., Te Velthuis A.J.W., Snijder E.J. Nidovirus RNA polymerases: complex enzymes handling exceptional RNA genomes. Virus Res. 2017;234:58–73. doi: 10.1016/j.virusres.2017.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prathiviraj R., Kiran G.S., Selvin J. Phylogenomic proximity and comparative proteomic analysis of SARS-CoV-2. Gene Rep. 2020;20 doi: 10.1016/j.genrep.2020.100777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prathiviraj R., Prisilla A., Chellapandi P. Structure-function discrepancy in clostridium botulinum C3 toxin for its rational prioritization as a subunit vaccine. J. Biomol. Struct. Dyn. 2016;34:1317–1329. doi: 10.1080/07391102.2015.1078745. [DOI] [PubMed] [Google Scholar]
- Prathiviraj R., Saranya S., Bharathi M., Chellapandi P. A hijack mechanism of Indian SARS-CoV-2 isolates for relapsing contemporary antiviral therapeutics. Comput Biol Med. 2021;132 doi: 10.1016/j.compbiomed.2021.104315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prisilla A., Prathiviraj R., Sasikala R., Chellapandi P. Structural constraints-based evaluation of immunogenic avirulent toxins from clostridium botulinum C2 and C3 toxins as subunit vaccines. Infect. Genet. Evol. 2016;44:17–27. doi: 10.1016/j.meegid.2016.06.029. [DOI] [PubMed] [Google Scholar]
- Rajeev R., Prathiviraj R., Kiran G.S., Selvin J. Zoonotic evolution and implications of microbiome in viral transmission and infection. Virus Res. 2020;290 doi: 10.1016/j.virusres.2020.198175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid M., Saha S., Raghava G.P. Support Vector Machine-based method for predicting subcellular localization of mycobacterial proteins using evolutionary information and motifs. BMC Bioinform. 2007;8:337. doi: 10.1186/1471-2105-8-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S., Raghava G.P. VICMpred: an SVM-based method for the prediction of functional proteins of gram-negative bacteria using amino acid patterns and composition. Genom. Proteom. Bioinform. 2006;4:42–47. doi: 10.1016/S1672-0229(06)60015-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- van Dorp L., Richard D., Tan C.C.S., Shaw L.P., Acman M., Balloux F. No evidence for increased transmissibility from recurrent mutations in SARS-CoV-2. Nat. Commun. 2020;11:5986. doi: 10.1038/s41467-020-19818-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilar S., Isom D.G. One year of SARS-CoV-2: how much has the virus changed? Biology. 2021;10:91. doi: 10.3390/biology10020091. (Basel) [DOI] [PMC free article] [PubMed] [Google Scholar]
- V'kovski P., Gultom M., Kelly J.N., Steiner S., Russeil J., Mangeat B., Cora E., Pezoldt J., Holwerda M., Kratzel A., Laloli L., Wider M., Portmann J., Tran T., Ebert N., Stalder H., Hartmann R., Gardeux V., Alpern D., Deplancke B., Thiel V., Dijkman R. Disparate temperature-dependent virus-host dynamics for SARS-CoV-2 and SARS-CoV in the human respiratory epithelium. PLoS Biol. 2021;19 doi: 10.1371/journal.pbio.3001158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Hozumi Y., Zheng Y.H., Yin C., Wei G.W. Host immune response driving SARS-CoV-2 evolution. Viruses. 2020;12:1095. doi: 10.3390/v12101095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman D., Alameh M.G., de Silva T., Collini P., Hornsby H., Brown R., LaBranche C.C., Edwards R.J., Sutherland L., Santra S., Mansouri K., Gobeil S., McDanal C., Pardi N., Hengartner N., Lin P.J.C., Tam Y., Shaw P.A., Lewis M.G., Boesler C., Sahin U., Acharya P., Haynes B.F., Korber B., Montefiori D.C. D614G Spike Mutation Increases SARS CoV-2 Susceptibility to Neutralization. Cell Host Microbe. 2021;29:23–31. doi: 10.1016/j.chom.2020.11.012. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (WHO), (2021) Weekly epidemiological update on COVID-19, 15th June 2021. https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---20-july-2021.
- Wu C., Chen X., Cai Y., Xia J., Zhou X., Xu S., Huang H., Zhang L., Zhou X., Du C., Zhang Y., Song J., Wang S., Chao Y., Yang Z., Xu J., Zhou X., Chen D., Xiong W., Xu L., Zhou F., Jiang J., Bai C., Zheng J., Song Y. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern. Med. 2020;180:934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin C. Genotyping coronavirus SARS-CoV-2: methods and implications. Genomics. 2020;112:3588–3596. doi: 10.1016/j.ygeno.2020.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefi H., Mashouri L., Okpechi S.C., Alahari N., Alahari S.K. Repurposing existing drugs for the treatment of COVID-19/SARS-CoV-2 infection: a review describing drug mechanisms of action. Biochem. Pharmacol. 2021;183 doi: 10.1016/j.bcp.2020.114296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L., Wang X., Pascal K.E., et al. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell. 2020;183:739–751. doi: 10.1016/j.cell.2020.09.032. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Jackson C.B., Mou H., et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat. Commun. 2020;11:6013. doi: 10.1038/s41467-020-19808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Lee A., Composto K., et al. A novel diagnostic test to screen SARS-CoV-2 variants containing E484K and N501Y mutations. EmergyMicrobes Infect. 2021;10:994–997. doi: 10.1080/22221751.2021.1929504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Li H., Wu X., Zhong Y., Zhang K., Zhang Y.P., Boerwinkle E., Fu Y.X. Moderate mutation rate in the SARS coronavirus genome and its implications. BMC Evol. Biol. 2004;4:21. doi: 10.1186/1471-2148-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Thao T.T.N., Hoffmann D., et al. SARS-CoV-2 spike D614G change enhances replication and transmission. Nature. 2021;592:122–127. doi: 10.1038/s41586-021-03361-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.