Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA transmission route was thoroughly investigated in the hospital wastewater, sewage collection network, and wastewater treatment plants. Samples were taken on four occasions from December 2020 to April 2021. The performance of two different wastewater treatment processes of sequencing batch reactor (SBR) and conventional activated sludge (CAS) was studied for virus destruction. For this purpose, liquid phase, solid phase and bioaerosol samples were taken from different units of the investigated wastewater treatment plants (WWTPs). The results revealed that all untreated hospital wastewater samples were positive for SARS-CoV-2 RNA. The virus detection frequency increased when the number of hospitalized cases increased. Detection of viral RNA in the wastewater collection system exhibited higher load of virus in the generated wastewater in areas with poor socioeconomic conditions. Virus detection in the emitted bioaerosols in WWTPs showed that bioaerosols released from CAS with surface aeration contains SARS-CoV-2 RNA posing a potential threat to the working staff of the WWTPs. However, no viral RNA was detected in the bioaerosols of the SBR with diffused aeration system. Investigation of SARS-CoV-2 RNA in WWTPs showed high affinity of the virus to be accumulated in biosolids rather than transporting via liquid phase. Following the fate of virus in sludge revealed that it is completely destructed in anaerobic sludge treatment process. Therefore, based on the results of the present study, it can be concluded that receiving water resources could not be contaminated with virus, if the wastewater treatment processes work properly.
Keywords: Bioaerosol, Biosolids, COVID-19, CAS, RT-qPCR, SBR
Graphical abstract
1. Introduction
The novel Coronavirus Disease 2019 (COVID-19) is a global pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is considered a significant threat to the public health. SARS-CoV-2 has spread worldwide to over 200 countries, and up to now, more than 236 million confirmed cases and 4,831,000 deaths have been reported since the virus outbreak started in Wuhan. In Iran, the total number of cases is 5,674,083 with 122,012 deaths as of October 11, 2021 (WHO, 2021).
Dry cough, loss of taste or smell, tiredness, fever, shortness of breath, diarrhea, muscle pains, sore throat, and conjunctivitis have been reported as the symptoms of this lethal disease (Kitajima et al., 2020; Ahmed et al., 2021; Gonçalves et al., 2021). Although symptoms of SARS-CoV-2 are mainly respiratory diseases, human CoVs are known to cause many gastrointestinal symptoms such as diarrhea and nausea (Kitamura et al., 2021). Previous investigations on SARS-CoV (2002−2003) and MERS-CoV (2012) confirmed the presence of viral RNA in the stool specimens of both symptomatic and asymptomatic cases (Leung et al., 2003; Corman et al., 2016). In the case of SARS-CoV-2, recent studies also confirm the detection of viral RNA in the human stool samples (Parasa et al., 2020; Tang et al., 2020). Furthermore, the detection of viral RNA in the human stool samples with high load (up to 108 genomic copies (GC) per gram of feces) (Kitajima et al., 2020) implies that wastewater can be a pathway to transmit SARS-CoV-2 viral particles into the environment. In addition, respiratory secretions of positive SARS-CoV-2 cases also contain large percentage of infected immune cells that are directly entering into the wastewater collection system. Therefore, probing wastewater for SARS-CoV-2 RNA or wastewater-based epidemiology (WBE) could be a useful tool and early warning system of (re-) emergence and outbreak of SARS-CoV-2 (Gonçalves et al., 2021). In a newly published study, the researchers quantified the SARS-CoV-2 concentration in wastewater to infer viral shedding dynamics, and to find the relationship between SARS-CoV-2 RNA concentration in wastewater and clinically reported positive cases. They found strong correlation between wastewater data and clinically diagnosed new SARS-CoV-2 cases (Wu et al., 2022). In addition, Ai et al. (2021) implemented a quadratic polynomial model to track COVID-19 cases from wastewater surveillance data. The suggested model performed best in predicting COVID-19 cases from wastewater data, which can provide useful information regarding the effectiveness of vaccination in future stage of the pandemic.
To date, several studies worldwide reported the detection of SARS-CoV-2 in hospital wastewater (Achak et al., 2021; Gonçalves et al., 2021), raw and treated municipal wastewater (Sherchan et al., 2020; Hata et al., 2021; Nasseri et al., 2021; Westhaus et al., 2021), wastewater aerosols (Gholipour et al., 2021), and solid fraction of wastewater (D'Aoust et al., 2021; Graham et al., 2021; Kitamura et al., 2021). Survival of coronavirus in wastewater treatment systems depends on some critical factors including the presence of organic matter and oxidizing agents; presence of antagonistic bacteria; pH and temperature values. It has been reported that the presence of the sufficient amount of strong oxidant (e.g., chlorine) improves inactivation of the virus. In contrast, suspended solids and organic matters reduce the virus inactivation efficiency, as they adsorb and protect the virus. The presence of antagonistic bacteria also helps enhance virus inactivation, which is due to the virus removal through enzymatic breakdown and predation (Arslan et al., 2020).
In addition to aforementioned parameters affecting coronaviruses survival/persistence, the types of wastewater treatment processes are also important for virus removal. Therefore, it is of great importance to track the virus fate in various types of biological wastewater treatment processes. Furthermore, WWTP employs various processes that their operation is associated with the production of various waste streams such as biosolids, solid wastes, sand and grits, and bioaerosols, which can also act as a route to transmit SARS-CoV-2 viral particles into the environment. The affinity of SARS-CoV-2 genetic material to be accumulated in biosolids have been proved recently in a study conducted by Balboa et al. (2021). They found primary sludge and mostly sludge thickener as a suitable sampling spot for detecting SARS-CoV-2 genetic materials, since they detected high concentration of accumulated SARS-CoV-2 viral particles in these wastewater fractions. Moreover, SARS-CoV-2 can spread into adjacent environment via generated aerosols from WWTP. Even though SARS-CoV-2 is not an airborne virus, it can attach to particulate matter present in the wastewater aerosols and transport to longer distances. Thereby, this might increase the risk of transmission of viral infections to the plant workers and nearby residents (Kitajima et al., 2020). Although various studies have been conducted worldwide to analyze the presence of SARS-CoV-2 RNA in wastewater, sludge and bioaerosols, no study has been performed on the detection of SARS-CoV-2 RNA in solid wastes and grits separated from the primary units of WWTP. In addition, literature review results revealed that no study was conducted to investigate the effect of wastewater treatment types and aeration types on the spread of SARS-CoV-2 RNA from wastewater.
Accordingly, present study is comprehensively investigating the presence of SARS-CoV-2 RNA in hospital wastewater, sewage collection network and wastewater treatment plant. In fact, this study monitors SARS-CoV-2 from zero-point to the end-point where the virus can enter into the environment (ambient air, soil or receiving water bodies). Moreover, in the present work, we compared the potential of two different biological processes of conventional activated sludge (CAS) and sequencing batch reactor (SBR) processes in case of virus emission in to the environment.
2. Materials and methods
2.1. Study area
Maragheh is an ancient city and capital of Maragheh County, which is located southwest of the East Azerbaijan province, Iran. Maragheh with a population of more than 185,000 people and an area of 26 Km2 is the second-largest city and the second-most populous city in the province. Maragheh has been situated in longitude 46°9′–46°44′E and latitude 37°1′–37°45′N with a height of 1477 m above sea level.
2.2. Sample collection
Four sampling runs were conducted in four separate days from 28 December 2020 to 13 April 2021. At each sampling run, 12-h composite samples were collected at 2-hourly intervals from the selected points in sterile 1500 mL HDPE bottles, preserved on ice (4 °C) and dispatched to the Maragheh Cellular-Molecular diagnostics laboratory for analysis (Baldovin et al., 2021). The sampling personnel wore standard personal protective equipment during sampling, including face mask, gloves and safety goggles, etc.
2.2.1. Wastewater samples
Municipal wastewater samples were collected from Maragheh wastewater treatment plant (WWTP) located at the study area, as well as from various access points (e.g. manholes) in wastewater collection network. Three points were chosen to take wastewater samples in wastewater collection system in Maragheh city. Sampling points were selected in a way to cover almost the entire wastewater collection system.
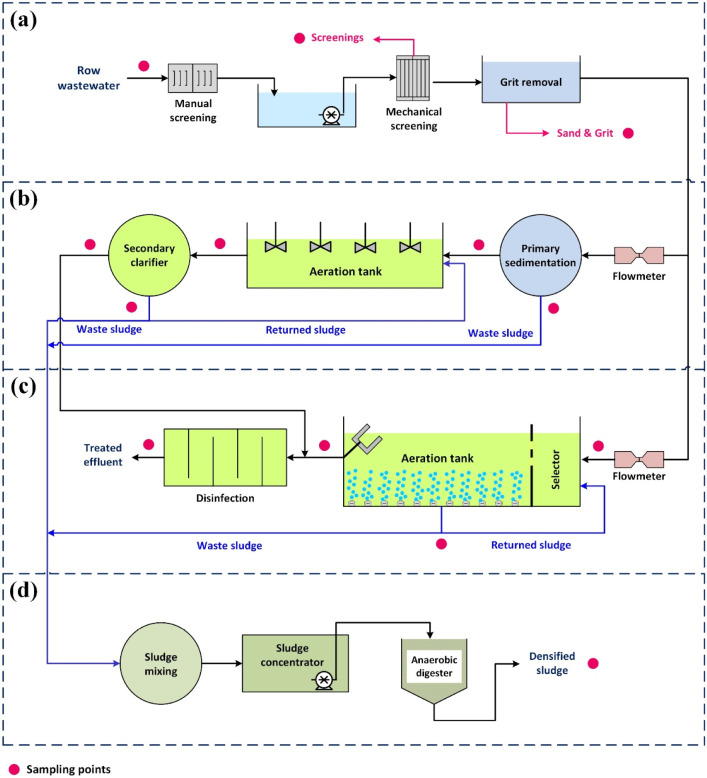
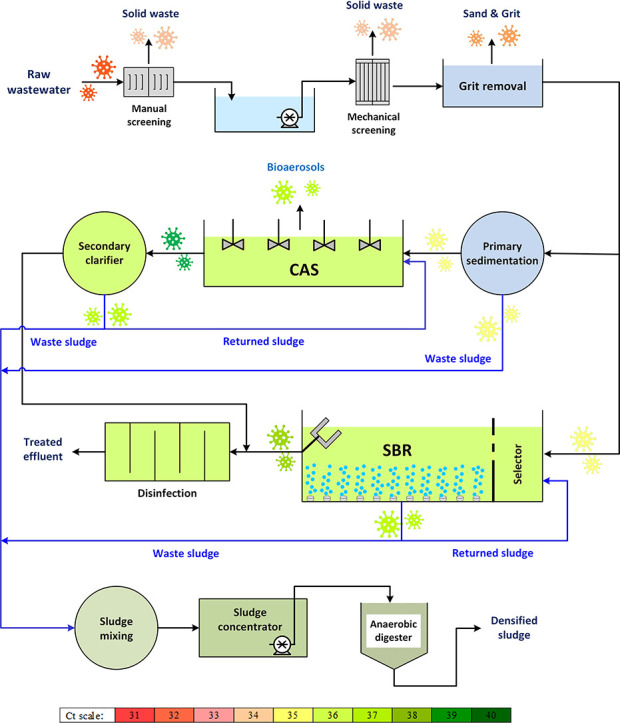
Maragheh municipal WWTP consists of two modules: a conventional activated sludge process and a sequencing batch reactor. These two WWTPs were constructed to treat maximum volumes of 43,000 m3/day of influent wastewater and serve population of 190,000 in total. Fig. 1 illustrates the series of treatment processes implemented in these two WWTPs along with the sampling points.
Fig. 1.
Schematic flow diagram of the studied WWTPs: (a) primary treatment unit, (b) conventional activated sludge process, (c) sequencing batch reactor, and (d) sludge treatment unit.
In this study, untreated hospital wastewater samples were obtained from the main sewer of Amir al-mu'minin Hospital (Maragheh, Iran), which is the main hospital in Maragheh allocated to treat positive COVID-19 patients. As part of the main hospital complex in Maragheh, Amir al-mu'minin Hospital has 5 wards and 236 beds in total, all of which (including 176 beds in the general ward and 32 beds in the intensive care unit) except for the post-angiography ward (20 beds) and the critical care unit (8 beds) are dedicated to positive COVID-19 patients. Totally, during four sampling runs, 44 wastewater samples were collected from Maragheh WWTP, wastewater collection network and hospital wastewater.
2.2.2. Solid waste and grit samples
Solid waste and grits separated from screening and grit removal units were sampled to investigate the presence of SARS-CoV-2 RNA. Time-proportional, 12 h composite samples were taken at 4-hourly intervals. Totally, eight composite samples were collected during four sampling runs. At each sampling day, 2 L of solid waste and grit samples were collected and transported to the laboratory on ice (4 °C).
2.2.3. Sludge samples
Sludge samples were taken from various sections in CAS and SBR processes. In addition, the presence of viral RNA in the anaerobic sludge digester was also monitored. It should be noted that in the studied plant, screening and grit removal sections are the same for SBR and CAS processes. Furthermore, the wasted activated sludge discharged from both SBR and CAS processes follows the same route for digestion.
In each sampling run, 12 h composite sludge samples with 4-h intervals were collected from primary clarifier sludge, secondary clarifier sludge and treated sludge of the CAS process, as well as from settled sludge of the SBR system (Graham et al., 2021). In sum, 16 sludge samples were taken from both CAS and SBR systems. The collected sludge samples were stored on ice and shipped to the Maragheh Cellular-Molecular diagnostics laboratory for analysis. The anaerobic digester was also investigated in case of the virus removal. Samples taken from the anaerobic digester were mixed liquor containing both solid fraction and supernatant.
2.2.4. Air samples
In the present study, passive and active methods were used for detecting SARS-CoV-2 RNA in WWTPs aerosols. Sampling points in the studied WWTPs were selected considering the dominant wind direction, which is west-to-east. Active sampling was carried out using impinger containing phosphate buffer solution (Gholipour et al., 2021). Active air samples were collected using a vacuum pump at a flow rate of 24 L/min for 1 h (1440 L). In addition, passive air samples were collected using a petri dishes containing phosphate buffer solution. Sampling was performed in four runs at three points with distances of 1.5, 10, and 50 m from the aeration tanks of the CAS and SBR systems. In sum, a total of 48 air samples were collected and transported to the laboratory in an insulated cooling box.
2.3. Sample processing
Upon arrival to the laboratory, all samples were concentrated prior to the SARS-CoV-2 extraction and detection. Briefly, 200 mL of each wastewater samples was centrifuged at 3500 rpm for 30 min to remove large floating particles. Then, aluminum hydroxide adsorption-precipitation method as described in reference criteria (APHA/AWWA/WEF, 2005) was used to concentrate SARS-CoV-2 in the samples. After concentration, water samples were stored at −80 °C prior to being processed for RNA extraction.
In the case of solid waste and grit samples, considering that no method was found in the literature that illustrates virus recovery from these samples, we used the following method for SARS-CoV-2 RNA recovery from solid waste and grit samples. First, we washed the collected solid waste and grit samples many times with deionized water to transfer viruses from solid phase into the liquid phase. Then, 200 mL of each prepared sample was taken and centrifuged at 3500 rpm for 30 min to eliminate the debris. After that, the rest of concentration process was similar to the water samples.
Sludge samples were concentrated using polyethylene glycol (PEG). The method presented by Balboa et al. (2021) was employed for the separation of SARS-CoV-2 from organic materials present in sludge samples intended for RNA extraction. After which the sludge samples were concentrated and prepared, they were kept at −80 °C for further analysis.
Air samples taken from various points in the studied plants were also concentrated using PEG (Gholipour et al., 2021). In addition in this study, some samples such as air and final effluent samples were concentrated using both PEG and aluminum hydroxide adsorption-precipitation methods in order to enhance virus recovery performance.
2.4. SARS-CoV-2 extraction
SARS-CoV-2 RNA extraction and one-step reverse transcription quantitative polymerase chain reaction (RT-qPCR) was performed at Maragheh Cellular-Molecular diagnostics laboratory, a specialized center dedicated to SARS-CoV-2 diagnosis. 200 μL of each concentrated sample (including wastewater, solid waste, grit, sludge, and air samples) was used for SARS-CoV-2 RNA extraction using the RNJia Virus Kit (ROJETechnologies, Yazd, Iran) (Joukar et al., 2021), according to manufacturer protocol (see the Supplementary Material for details). A negative control was used with SARS-CoV-2 RNA extraction to analyze possible cross-over contamination of samples during viral RNA extraction process. The negative control was prepared using nuclease-free deionized water.
2.5. SARS-CoV-2 detection
The isolated RNA was subjected to RT-qPCR assay for molecular detection of SARS-CoV-2. COVID-19 ONE-STEP RT-PCR kit (Pishtaz Teb Diagnostics, Tehran, Iran) was used for detecting SARS-CoV-2 genes in the samples according to the manufacturer's instructions. The kit's primer and probe mixture is designed based on dual-target gene method targeting two different regions of the SARS-CoV-2 genome, specifically RdRp and N genes. This kit includes a solution containing probe and internal control primer (RNase P) for enhancing the sensitivity of detection and avoiding false negative results. In addition, this kit includes PCR positive and negative (Diethyl pyrocarbonate (DEPC)-treated water) controls. For each PCR run, 10 μL of sample was added to 10 μL of master mix and primer probe mixture. The thermal cycling conditions for the RT-qPCR assay were as follows: reverse transcription at 50 °C for 20 min and cDNA initial denaturation at 95 °C for 3 min, followed by 45 cycles of denaturation at 94 °C for 10 s, and primer annealing and extension reaction at 55 °C for 40 s, and finaly cooling at 25 °C for 10 s.
2.6. Quality control
In this study, for the sake of increasing accuracy and re-checking the samples, we used other SARS-CoV-2 detection kits including Novel Coronavirus (2019-nCoV) Nucleic Acid Diagnostic Kit (Sansure Biotech, China) which targets ORF-1ab and N genes, and Novel Coronavirus 2019-nCoV) Real Time Multiplex RT-PCR Kit (Liferiver Bio-Tech, US) which targets ORF1ab, E, and N genes. Each sample was assayed in triplicate. RT-qPCR results were interpreted as following: the gene target (RdRp, N) with a Ct value lower or equal to 40 was considered positive. When at least two of the three replicates were positive, the corresponding sample was considered positive for SARS-CoV-2. For SARS-CoV-2 quantification, standard curves were constructed using 10-fold dilutions of SARS-CoV-2 positive control of the reference kit according to the manufacturer's instructions. RNA extraction and RTqPCR preparation were carried out in different laboratories in order to avoid cross contamination.
3. Results and discussion
3.1. SARS-CoV-2 detection in hospital wastewater and sewage collection system
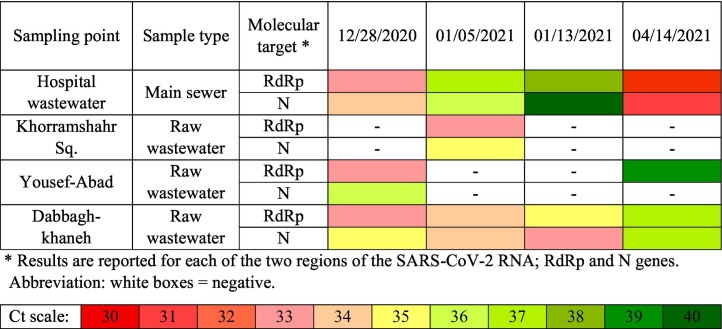
Hospital wastewater is always considered as the source of pathogenic micro-pollutants such as microorganisms, toxic chemicals, antibiotic residual, etc. (Pourakbar et al., 2016; Al Aukidy et al., 2018). However, it is usually discharged into the municipal wastewater collection system without any pre-treatment, which is also true in case of the selected hospital in the present study. In order to identify the fate of released SARS-CoV-2 through wastewater discharge, generated wastewater was monitored by taking samples at four different runs. Table 1 is illustrating the results of this sampling. The molecular detection of SARS-CoV-2 RNA in the collected samples was performed targeting two genes: nucleocapsid (N) gene and RNA-dependent RNA polymerase (RdRp) gene. Although RdRp gene has higher sensitivity than the N gene, N gene is more specific for tracking, screening, and first-line detection of SARS-CoV-2 (Shirato et al., 2020). Furthermore, for additional confirmatory testing, the RdRp gene was used. Based on the results of this study, RT-qPCR amplifications produced almost congruous results between the two assays. However, the N gene, which is specific only for SARS-CoV-2 was used to quantify SARS-CoV-2 RNA in the samples.
Table 1.
Mean amplification cycles of SARS-CoV-2 RNA in hospital wastewater and wastewater collection network in Maragheh city.
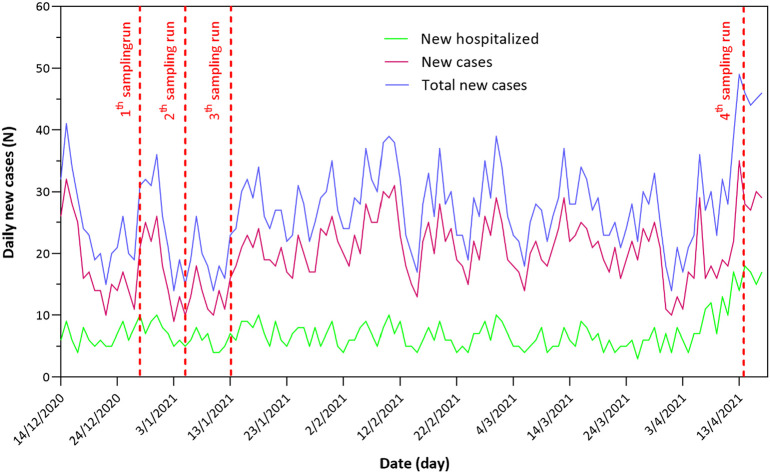
As it shown in Table 1, SARS-CoV-2 RNA has been detected in all hospital wastewater samples. Quantification results revealed that SARS-CoV-2 genome concentration in the hospital wastewater ranged from (0.49–7.3) × 103 gene copies/L. The Ct values also indicate that there was a high load of viral RNA in the hospital wastewater during the fourth run of sampling. This fact could be associated to the number of infected patients in the hospital which was higher than the other investigated runs. To prove this fact, the number of new cases and also new hospitalized cases are illustrated in Fig. 2 . As it is shown in the figure, the lowest new cases of hospitalized patients are in the second run of sampling. However, the forth sampling run was conducted when there were a high number of positive COVID-19 cases in the hospital. Various studies have been conducted to track SARS-CoV-2 RNA in the hospital wastewater. Zhang et al. (2020) investigated the presence of SARS-CoV-2 in medical wastewater treated in a septic tank and high loads of viral RNA have been reported. They have also reported that the recommended disinfection strategy by WHO needs to be re-evaluated to effectively remove virus from wastewater. Gonçalves et al. (2021) also investigated the SARS-CoV-2 presence in hospital wastewater in Slovenia. They detected SARS-CoV-2 RNA in the hospital wastewater one day after hospitalization of the first confirmed positive COVID-19 case. In another study conducted in Japan, 45 raw wastewater samples were taken from the inlet of WWTP. They have reported that 21 out of 45 (46.6%) samples were positive in case of SARS-CoV-2 RNA. It has also been reported that the detection frequency of the positive samples strongly correlates with the number of total confirmed SARS-CoV-2 cases (Hata et al., 2021). Therefore, based on the results of our study and similar reported studies, it can be deduced that wastewater monitoring can be considered as a potential complementary tool for public health monitoring.
Fig. 2.
Daily new positive SARS-CoV-2 cases in the studied area.
As it is shown in the Table 1, SARS-CoV-2 RNA was also detected in wastewater collection system. The samples taken from Dabbagh-khaneh sampling point were tested to be positive in all cases with high level of SARS-CoV-2 RNA concentration ranging between 0.38 × 104–2.8 × 104 gene copies/L. However, only one of the samples taken from Khorramshahr Sq. and Yousef-Abad vicinities was positive for SARS-CoV-2 RNA with genome concentration of 15.4 × 103 and 3.9 × 103 gene copies/L, respectively. This difference could be due to the difference in the socioeconomic features of the investigated households. Dabbagh-khaneh sampling point is collecting wastewater from densely populated area with lower wealth of properties. On the other hand, Khorramshahr Sq. and Yousef-Abad vicinities are among the old areas of the city with small family members. Therefore, it can be deduced that collected wastewater from densely populated areas contain higher loads of virus which is well-confirmed based on our results.
3.2. Fate of SARS-CoV-2 in wastewater treatment plants
Viral RNA of the SARS-CoV-2 has been reported in stool samples of infected patients. In the next chain of the virus transformation into the receiving water bodies, the viral RNA was also detected in the hospital wastewater as well as municipal wastewater collection system. Therefore, wastewater treatment plants and various treatment processes are the last barrier to cease virus entrance into the environment. To better illustrate the fate of SARS-CoV-2 RNA in the WWTP, liquid, solid, and air samples were taken from both CAS and SBR processes.
3.2.1. Virus track in liquid phase of the wastewater treatment plants
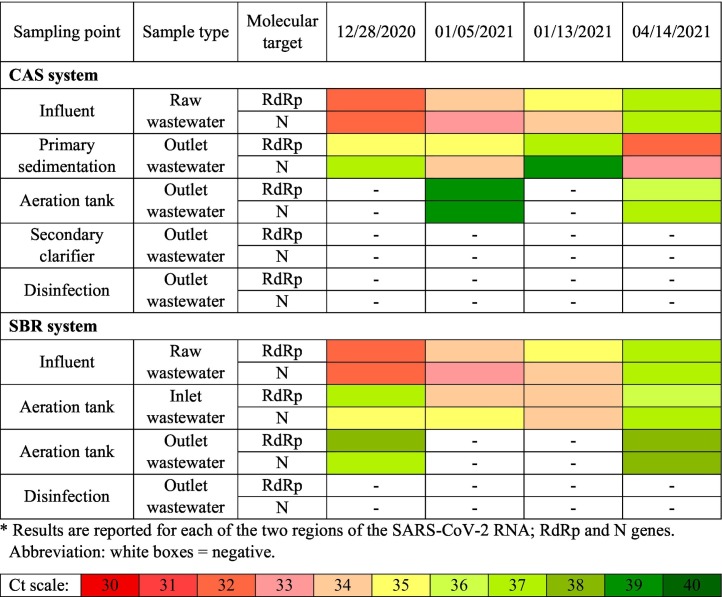
Table 2 is illustrating the mean amplification cycles of SARS-CoV-2 RNA in the liquid phase of the CAS and SBR processes. Both treatment plants receive the same wastewater. As can be seen in Table 2, SARS-CoV-2 RNA was detected in raw wastewater in all sampling runs. Based on calculations, viral RNAs with concentrations in the range of 1.8 × 104–22.4 × 104 gene copies/L were consistently detected in raw wastewater. In a similar study conducted by Kumar et al. (2021a), the average SARS-CoV-2 RNA concentration in raw wastewater samples of the CAS system was calculated about 1.25 × 103 gene copies/L. In CAS system, SARS-CoV-2 RNA was also detected in the effluent of the aeration tank. However, in CAS process which is using surface aeration of the wastewater for biological degradation of the organic contaminants, a reduction in the concentration of SARS-CoV-2 RNA (0.3 × 103–2.1 × 103 gene copies/L) was observed and samples taken in the first and third runs were negative for SARS-CoV-2 RNA. Further investigation of the fate of SARS-CoV-2 RNA in the liquid phase revealed that there were no viral RNAs in the secondary clarifier effluent and also final chlorinated effluent.
Table 2.
Mean amplification cycles of SARS-CoV-2 RNA in the liquid phase of Maragheh WWTPs (CAS and SBR systems).
On the other hand, second wastewater treatment plant (one basin cyclic activated process) shows almost the same results. Treated wastewater in the SBR system also showed a reduction in the viral RNA concentration in the liquid phase (1.8 × 103–2.9 × 103 gene copies/L). However, positive samples were detected in the first and forth sampling runs. The final chlorinated effluent samples tested negative for SARS-CoV-2 RNA, proving that the virus was removed via biological and chemical disinfection processes to undetectable level. Sherchan et al. (2020) investigated the presence of SARS-CoV-2 RNA in two WWTPs using CAS process located in Louisiana, USA. They have also reported that the secondary-treated wastewater and the final effluent tests were negative in case of SARS-CoV-2 RNA. In another study in Milano Metropolitan Area, Italy, the presence of SARS-CoV-2 has also been reported in raw wastewater, but the effluent of three investigated WWTP were free of the virus (Rimoldi et al., 2020). In another similar study conducted in United Arab Emirates, SARS-CoV-2 RNA has been detected in 85% of raw wastewater samples taken from different locations across the country. However, none of the 11 WWTPs' liquid effluents was positive during the entire sampling period, indicating the effectiveness of treatment processes for virus removal (Hasan et al., 2021). Considering the results of our study and the similar reported studies, it can be concluded that no virus is released into the environment through the liquid phase of the WWTP effluents. In addition, various investigated biological processes such as CAS and SBR are acting equally in case of virus removal from liquid phase of the WWTP. Therefore, the final effluent is probably safe for discharging into the receiving water bodies or reusing in the plant.
3.2.2. Virus track in solid phase of the wastewater treatment plants
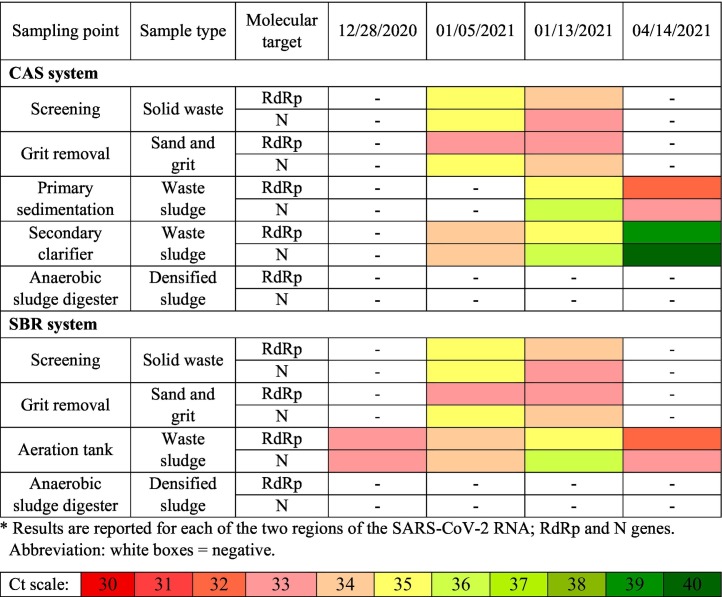
The presence of SARS-CoV-2 RNA in the solid phase of the two investigated WWTPs was also investigated. Table 3 is illustrating the mean amplification cycles of SARS-CoV-2 RNA in the solid phase in both CAS and SBR processes. As it is shown in Table 3, the physically separated solids from screening and grit chamber were positive for SARS-CoV-2 RNA in half of the taken samples. Positive solid waste and grit samples were found at SARS-CoV-2 genome concentration of (1.54–3.9) × 104 and (0.8–2.3) × 104 gene copies/L, respectively. It shows that some portion of the inlet SARS-CoV-2 RNA are removed by attaching to the solids.
Table 3.
Mean amplification cycles of SARS-CoV-2 RNA in the solid phase of Maragheh WWTPs (CAS and SBR systems).
The waste sludge from the primary sedimentation tank is also showing that half of the samples are positive for SARS-CoV-2 RNA (Table 3). Comparing the results of SARS-CoV-2 RNA tracking in solid phase and liquid phase (Table 2, Table 3) shows that only a small portion of the viral RNAs in the influent are removed through unit operation in WWTP. This is due to the presence of SARS-CoV-2 RNA in the aqueous outlet of the primary sedimentation tank.
Tracking SARS-CoV-2 RNA in the aeration tank showed that the liquid outlet of the aeration tanks in both CAS and SBR were not always positive for SARS-CoV-2 RNA (Table 2). However, the separated biological solids at this stage were found positive in all cases for SBR process and three cases for CAS process. Our findings show that SARS-CoV-2 RNA has higher affinity toward the biological solids in activated sludge processes. Therefore, solid lines of the WWTP can be considered as a suitable spot for viral RNA detection.
In addition, the higher concentrations of viral RNAs were observed in the secondary sludge (0.71 × 104–3.1 × 104 gene copies/L) than the primary sludge (0.32 × 104–1.3 × 104 gene copies/L) of the CAS process. This could be due to the higher solids retention time (SRT) in the aerobic section which was about 16–20 days in the investigated process. The higher SRT may lead to the attachment of the viral RNA to the biomass. Furthermore, extracellular polymeric substances (EPS) generated in the biological process are capable of absorption of contaminants in the bioreactor (Lu et al., 2021). Therefore, presence of EPS in the bioreactor could also be considered as another reason for higher virus detection in the sludge. Balboa et al. (2021) also investigated the fate of SARS-CoV-2 RNA in WWTP. They have also reported that most SARS-CoV-2 RNA are retained by the sludge line and they cannot be detected in water effluent of the WWTP. In another study conducted in one of the regions of Japan, the adsorption of SARS-CoV-2 onto the solid fraction of wastewater has also been reported (Kitamura et al., 2021).
Waste activated sludge in the investigated processes goes into the anaerobic sludge digester with SRT value of about 30 days. All the samples taken from the solid phase of the digester were also tested negative in case of SARS-CoV-2 RNA. This finding proves that higher retention times and anaerobic biological processes are able to effectively destruct the viral RNA. This also could be due to the higher temperature of the anaerobic digester (35–40 °C) which can provide a thermal inactivation and removal of the viral inputs. This is also in accordance with the findings of other studies. Balboa et al. (2021) reported that thermal hydrolysis or similar thermal treatment of sludge such as anaerobic digester is capable of destruction of the SARS-CoV-2 RNA which makes the sludge disposal safer. Serra-Compte et al. (2021) also reported that sludge processing units such as sludge thickeners are effective for virus removal, whereas viral RNA was note detected after applying thermal processes for sludge treatment. Kumar et al. (2021b) also reported a considerable reduction in SARS_CoV-2 RNA concentration during anaerobic wastewater treatment process.
Based on the results obtained from tracking SARS-CoV-2 RNA in the liquid and solid phase of the WWTP, it can be concluded that the SARS-CoV-2 RNA transmission into the environment through liquid and solid phase of wastewater could be ceased if the biological processes work properly.
3.2.3. Virus track in the off-gas released from wastewater treatment plants
Wastewaters contain various microorganisms such as viruses, bacteria, fungi, etc., which originate from the household, commercial, and hospital sewage. These microorganisms can easily become airborne bioaerosols during operational processes of the WWTP including aeration and mechanical agitation of wastewater (Sánchez-Monedero et al., 2008; Uhrbrand et al., 2011). Nevertheless, very limited studies have investigated the presence of airborne SARS-CoV-2 RNA in WWTP (Corpuz et al., 2020; Gholipour et al., 2021). Gholipour et al. (2021) have reported the pesence of SARS-CoV-2 RNA in the aerosols released from WWTP, which can pose a potential threat to the plant workers. However, they have not investgated the various types of aeration processes which may affect the presence of viral RNA in the aerosols.
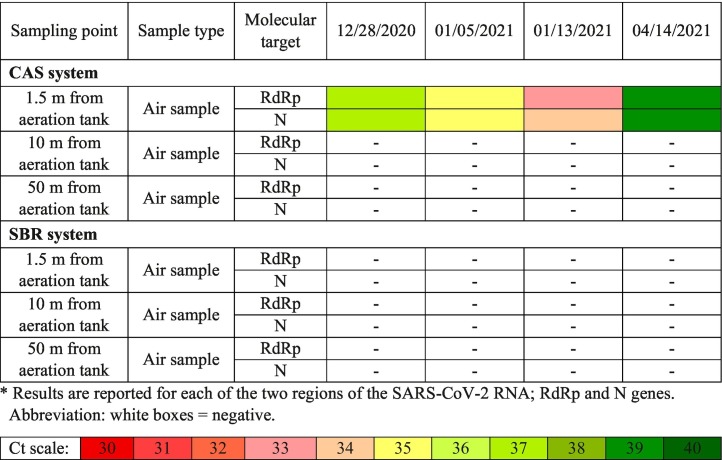
Table 4 gives the mean amplification cycles of SARS-CoV-2 RNA in air samples taken from various units of the studied WWTPs involving CAS (surface aeration) and SBR (diffused aeration) processes. According to Table 4, air samples (active and passive) collected with 1.5 m distance from the aeration tank of CAS system were positive for SARS-CoV-2 RNA. However, other collected samples including samples with 10 and 50 m distances from the aeration tank of CAS system were negative for SARS-CoV-2 RNA.
Table 4.
Mean amplification cycles of SARS-CoV-2 RNA in the air phase of Maragheh WWTPs (CAS and SBR systems).
Gholipour et al. (2021) reported the presence of SARS-CoV-2 RNA in 40% of air samples of WWTPs. Also, Courault et al. (2017) investigated the presence of different viruses in the wastewater reused for irrigation proposes. Their results proved the presence of at least one type of virus in 23% of the collected air samples.
In addition, based on the findings of this study, no SARS-CoV-2 viral RNA was observed in all air samples collected at 1.5, 10, and 50 m distances from the aeration tank of SBR system. The possible reason for detecting SARS-CoV-2 RNA in the air samples of CAS system compared to SBR system can be related to the type of aeration implemented in these two modules. The implemented aeration systems in the studied CAS and SBR processes are surface mechanical aeration and diffused aeration, respectively. The surface mechanical aeration system can create high turbulence that lead to the generation of more bioaerosols containing microorganisms (Brandi et al., 2000; Chen et al., 2021). Han et al. (2020) reported that surface mechanical aeration could cause the transmission of the microorganisms from wastewater into the ambient air. In addition, the results of similar studies (Sánchez-Monedero et al., 2008; Li et al., 2011) showed that surface mechanical aeration such as surface turbines could generate more bioaerosols than diffused aeration system which is in agreement with the results of this study.
The quantification results revealed that the concentration of SARS-CoV-2 RNA in the wasted activated sludge of the CAS system (0.71 × 104–3.1 × 104 gene copies/L) is lower than that of the SBR system (0.75 × 104–4.2 × 104 gene copies/L). This could also be speculated that some portion of the viral RNA in the CAS process (surface aeration) is released into the atmosphere; consequently, leading to lower concentration of SARS-CoV-2 RNA in the wasted activated sludge than SBR process (diffused aeration). However, in general, given the approximately low occurrence of SARS-CoV-2 RNA in the secondary treatment units (e.g. aeration tank), the dispersion potential of SARS-CoV-2 via generated aerosols during aeration is low (0.15 × 102–2.4 × 102 gene copies/L) but still important. Based on our finding, it can be suggested that application of diffused aeration for biological wastewater treatment can be considered as a safe method to cease the transmission of not only SARS-CoV-2 RNA, but also other infectious microorganisms. In addition, it is highly recommended that personal protective equipment and regular disinfection are essential for the epidemic prevention among the WWTP staff.
4. Conclusion
The following conclusions can be pointed out in the present study:
-
•
Wastewater monitoring could be considered as a complicated and fast method for the presence and prevalence of SARS-CoV-2 in the communities.
-
•
SARS-CoV-2 RNA detection in wastewater collection system differs based on the socioeconomic condition of the communities.
-
•
SARS-CoV-2 RNA was detected in all untreated wastewater samples. However, it was not found in the liquid and solid effluent of the WWTPs.
-
•
The large portion of the viral RNA in the WWTPs has higher affinity to biosolids rather than liquid phase.
-
•
SARS-CoV-2 RNA release through gaseous phase as a result of surface aeration has been proved in the present study posing a threat to the WWTP staff.
-
•
Comparing the fate of SARS-CoV-2 RNA in conventional activated sludge (with surface aeration) and cyclic activated sludge (with diffused aeration) show that diffused aeration process is reliable due to the lower gaseous emission of the viral RNAs.
CRediT authorship contribution statement
Mojtaba Pourakbar: Conceptualization, Methodology, Data curation and writing the manuscript. Ali Abdolahnejad: Sampling and writing the manuscript. Saber Raeghi: Conducting PCR analysis. Farhad Ghayourdoust: Sampling and writing manuscript. Roghayeh Yousefi: methodology and conducting experiments. Ali Behnami: Conceptualization, Methodology, Acquisition of the financial support for the project, writing the manuscript.
Declaration of competing interest
The authors declare no conflicts of interest associated with this study.
Acknowledgments
The authors would like to thank the financial support of Maragheh University of Medical Sciences for this research under grant number of A-10-1329-1. The authors also wish to express their sincere appreciation for the help and support provided by Maragheh Water and Wastewater Company for permitting sampling and providing required data.
Editor: Damia Barcelo
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2021.151391.
Appendix A. Supplementary data
Supplementary material
References
- Achak M., Alaoui Bakri S., Chhiti Y., M'Hamdi Alaoui F.E., Barka N., Boumya W. SARS-CoV-2 in hospital wastewater during outbreak of COVID-19: a review on detection, survival and disinfection technologies. Sci. Total Environ. 2021;761 doi: 10.1016/j.scitotenv.2020.143192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Tscharke B., Bertsch P.M., Bibby K., Bivins A., Choi P., Clarke L., Dwyer J., Edson J., Nguyen T.M.H., O'Brien J.W., Simpson S.L., Sherman P., Thomas K.V., Verhagen R., Zaugg J., Mueller J.F. SARS-CoV-2 RNA monitoring in wastewater as a potential early warning system for COVID-19 transmission in the community: a temporal case study. Sci. Total Environ. 2021;761 doi: 10.1016/j.scitotenv.2020.144216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai Y., Davis A., Jones D., Lemeshow S., Tu H., He F., Ru P., Pan X., Bohrerova Z., Lee J. Wastewater SARS-CoV-2 monitoring as a community-level COVID-19 trend tracker and variants in Ohio,United States. 2021;801 doi: 10.1016/j.scitotenv.2021.149757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Aukidy M., Al Chalabi S., Verlicchi P. Hospital Wastewaters: Characteristics, Management, Treatment and Environmental Risks. Springer International Publishing; 2018. pp. 171–188. [Google Scholar]
- APHA/AWWA/WEF . American Public Health Association; 2005. Standard Methods for the Examination of Water and Wastewater. [Google Scholar]
- Arslan M., Xu B.Gamal, El-Din M. Transmission of SARS-CoV-2 via fecal-oral and aerosols–borne routes: environmental dynamics and implications for wastewater management in underprivileged societies. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balboa S., Mauricio-Iglesias M., Rodriguez S., Martínez-Lamas L., Vasallo F.J., Regueiro B., Lema J.M. The fate of SARS-COV-2 in WWTPS points out the sludge line as a suitable spot for detection of COVID-19. Sci. Total Environ. 2021;772 doi: 10.1016/j.scitotenv.2021.145268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldovin T., Amoruso I., Fonzo M., Buja A., Baldo V., Cocchio S., Bertoncello C. SARS-CoV-2 RNA detection and persistence in wastewater samples: an experimental network for COVID-19 environmental surveillance in Padua, Veneto region (NE Italy) Sci. Total Environ. 2021;760 doi: 10.1016/j.scitotenv.2020.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandi G., Sisti M., Amagliani G. Evaluation of the environmental impact of microbial aerosols generated by wastewater treatment plants utilizing different aeration systems. J. Appl. Microbiol. 2000;88:845–852. doi: 10.1046/j.1365-2672.2000.01024.x. [DOI] [PubMed] [Google Scholar]
- Chen Y.-H., Yan C., Yang Y.-F., Ma J.-X. Quantitative microbial risk assessment and sensitivity analysis for workers exposed to pathogenic bacterial bioaerosols under various aeration modes in two wastewater treatment plants. Sci. Total Environ. 2021;755 doi: 10.1016/j.scitotenv.2020.142615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Albarrak A.M., Omrani A.S., Albarrak M.M., Farah M.E., Almasri M., Muth D., Sieberg A., Meyer B., Assiri A.M., Binger T., Steinhagen K., Lattwein E., Al-Tawfiq J., Müller M.A., Drosten C., Memish Z.A. Viral shedding and antibody response in 37 patients with Middle East respiratory syndrome coronavirus infection. Clin. Infect. Dis. 2016;62:477–483. doi: 10.1093/cid/civ951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpuz M.V.A., Buonerba A., Vigliotta G., Zarra T., Ballesteros F., Campiglia P., Belgiorno V., Korshin G., Naddeo V. Viruses in wastewater: occurrence, abundance and detection methods. Sci. Total Environ. 2020;745 doi: 10.1016/j.scitotenv.2020.140910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courault D., Albert I., Perelle S., Fraisse A., Renault P., Salemkour A., Amato P. Assessment and risk modeling of airborne enteric viruses emitted from wastewater reused for irrigation. Sci. Total Environ. 2017;592:512–526. doi: 10.1016/j.scitotenv.2017.03.105. [DOI] [PubMed] [Google Scholar]
- D'Aoust P.M., Mercier E., Montpetit D., Jia J.-J., Alexandrov I., Neault N., Baig A.T., Mayne J., Zhang X., Alain T., Langlois M.-A., Servos M.R., MacKenzie M., Figeys D., MacKenzie A.E., Graber T.E., Delatolla R. Quantitative analysis of SARS-CoV-2 RNA from wastewater solids in communities with low COVID-19 incidence and prevalence. Water Res. 2021;188 doi: 10.1016/j.watres.2020.116560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gholipour S., Mohammadi F., Nikaeen M., Shamsizadeh Z., Khazeni A., Sahbaei Z., Mousavi S.M., Ghobadian M., Mirhendi H. COVID-19 infection risk from exposure to aerosols of wastewater treatment plants. Chemosphere. 2021;273 doi: 10.1016/j.chemosphere.2021.129701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves J., Koritnik T., Mioč V., Trkov M., Bolješič M., Berginc N., Prosenc K., Kotar T., Paragi M. Detection of SARS-CoV-2 RNA in hospital wastewater from a low COVID-19 disease prevalence area. Sci. Total Environ. 2021;755 doi: 10.1016/j.scitotenv.2020.143226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham K.E., Loeb S.K., Wolfe M.K., Catoe D., Sinnott-Armstrong N., Kim S., Yamahara K.M., Sassoubre L.M., Mendoza Grijalva L.M., Roldan-Hernandez L., Langenfeld K., Wigginton K.R., Boehm A.B. SARS-CoV-2 RNA in wastewater settled solids is associated with COVID-19 cases in a large urban sewershed. Environ. Sci. Technol. 2021;55:488–498. doi: 10.1021/acs.est.0c06191. [DOI] [PubMed] [Google Scholar]
- Han Y., Li L., Wang Y., Ma J., Li P., Han C., Liu J. Composition, dispersion, and health risks of bioaerosols in wastewater treatment plants: a review. Front. Env. Sci. Eng. 2020;15:38. [Google Scholar]
- Hasan S.W., Ibrahim Y., Daou M., Kannout H., Jan N., Lopes A., Alsafar H., Yousef A.F. Detection and quantification of SARS-CoV-2 RNA in wastewater and treated effluents: surveillance of COVID-19 epidemic in the United Arab Emirates. Sci. Total Environ. 2021;764 doi: 10.1016/j.scitotenv.2020.142929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata A., Hara-Yamamura H., Meuchi Y., Imai S., Honda R. Detection of SARS-CoV-2 in wastewater in Japan during a COVID-19 outbreak. Sci. Total Environ. 2021;758 doi: 10.1016/j.scitotenv.2020.143578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joukar F., Yaghubi Kalurazi T., Khoshsorour M., Taramian S., Mahfoozi L., Balou H.A., Jafarinezhad A., Pourkazemi A., Hesni E., Asgharnezhad M., Shenagari M., Jahanzad I., Naghipour M., Maroufizadeh S., Mansour-Ghanaei F. Persistence of SARS-CoV-2 RNA in the nasopharyngeal, blood, urine, and stool samples of patients with COVID-19: a hospital-based longitudinal study. Virol. J. 2021;18:134. doi: 10.1186/s12985-021-01599-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A., Haramoto E., Rose J.B. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura K., Sadamasu K., Muramatsu M., Yoshida H. Efficient detection of SARS-CoV-2 RNA in the solid fraction of wastewater. Sci. Total Environ. 2021;763 doi: 10.1016/j.scitotenv.2020.144587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Kuroda K., Joshi M., Bhattacharya P., Barcelo D. First comparison of conventional activated sludge versus root-zone treatment for SARS-CoV-2 RNA removal from wastewaters: statistical and temporal significance. Chem. Eng. J. 2021;425 doi: 10.1016/j.cej.2021.130635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Kuroda K., Patel A.K., Patel N., Bhattacharya P., Joshi M., Joshi C.G. Decay of SARS-CoV-2 RNA along the wastewater treatment outfitted with upflow anaerobic sludge blanket (UASB) system evaluated through two sample concentration techniques. Sci. Total Environ. 2021;754:142329. doi: 10.1016/j.scitotenv.2020.142329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung W.K., To K.-F., Chan P.K.S., Chan H.L.Y., Wu A.K.L., Lee N., Yuen K.Y., Sung J.J.Y. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology. 2003;125:1011–1017. doi: 10.1016/j.gastro.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Gao M., Liu J. Distribution characterization of microbial aerosols emitted from a wastewater treatment plant using the orbal oxidation ditch process. Process Biochem. 2011;46:910–915. [Google Scholar]
- Lu X., Xu W., Liu C., Zhao Q., Ye Z. Insight into the role of extracellular polymeric substances in denitrifying biofilms under nitrobenzene exposure. Ecotoxicol. Environ. Saf. 2021;222 doi: 10.1016/j.ecoenv.2021.112539. [DOI] [PubMed] [Google Scholar]
- Nasseri S., Yavarian J., Baghani A.N., Azad T.M., Nejati A., Nabizadeh R., Hadi M., Jandaghi N.Z.S., Vakili B., Vaghefi S.K.A., Baghban M., Yousefi S., Nazmara S., Alimohammadi M. The presence of SARS-CoV-2 in raw and treated wastewater in 3 cities of Iran: Tehran, Qom and Anzali during coronavirus disease 2019 (COVID-19) outbreak. J. Environ. Health Sci. Eng. 2021;19:1–12. doi: 10.1007/s40201-021-00629-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parasa S., Desai M., Thoguluva Chandrasekar V., Patel H.K., Kennedy K.F., Roesch T., Spadaccini M., Colombo M., Gabbiadini R., Artifon E.L.A., Repici A., Sharma P. Prevalence of gastrointestinal symptoms and fecal viral shedding in patients with coronavirus disease 2019: a systematic review and meta-analysis. JAMA Netw. Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourakbar M., Moussavi G., Shekoohiyan S. Homogenous VUV advanced oxidation process for enhanced degradation and mineralization of antibiotics in contaminated water. Ecotox. Environ. Saf. 2016;125:72–77. doi: 10.1016/j.ecoenv.2015.11.040. [DOI] [PubMed] [Google Scholar]
- Rimoldi S.G., Stefani F., Gigantiello A., Polesello S., Comandatore F., Mileto D., Maresca M., Longobardi C., Mancon A., Romeri F., Pagani C., Cappelli F., Roscioli C., Moja L., Gismondo M.R., Salerno F. Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Sci. Total Environ. 2020;744 doi: 10.1016/j.scitotenv.2020.140911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Monedero M.A., Aguilar M.I., Fenoll R., Roig A. Effect of the aeration system on the levels of airborne microorganisms generated at wastewater treatment plants. Water Res. 2008;42:3739–3744. doi: 10.1016/j.watres.2008.06.028. [DOI] [PubMed] [Google Scholar]
- Serra-Compte A., González S., Arnaldos M., Berlendis S., Courtois S., Loret J.F., Schlosser O., Yáñez A.M., Soria-Soria E., Fittipaldi M., Saucedo G., Pinar-Méndez A., Paraira M., Galofré B., Lema J.M., Balboa S., Mauricio-Iglesias M., Bosch A., Pintó R.M., Bertrand I., Gantzer C., Montero C., Litrico X. Elimination of SARS-CoV-2 along wastewater and sludge treatment processes. Water Res. 2021;202 doi: 10.1016/j.watres.2021.117435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherchan S.P., Shahin S., Ward L.M., Tandukar S., Aw T.G., Schmitz B., Ahmed W., Kitajima M. First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana, USA. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Nao N., Katano H., Takayama I., Saito S., Kato F., Katoh H., Sakata M., Nakatsu Y., Mori Y., Kageyama T., Matsuyama S., Takeda M. Development of genetic diagnostic methods for detection for novel coronavirus 2019(nCoV-2019) in Japan. Jpn. J. Infect. Dis. 2020;73:304–307. doi: 10.7883/yoken.JJID.2020.061. [DOI] [PubMed] [Google Scholar]
- Tang A., Tong Z.-D., Wang H.-L., Dai Y.-X., Li K.-F., Liu J.-N., Wu W.-J., Yuan C., Yu M.-L., Li P., Yan J.-B. Detection of novel coronavirus by RT-PCR in stool specimen from asymptomatic child,China. 2020;26:1337–1339. doi: 10.3201/eid2606.200301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrbrand K., Schultz A.C., Madsen A.M. Exposure to airborne noroviruses and other bioaerosol components at a wastewater treatment plant in Denmark. Food Environ. Virol. 2011;3:130–137. [Google Scholar]
- Westhaus S., Weber F.-A., Schiwy S., Linnemann V., Brinkmann M., Widera M., Greve C., Janke A., Hollert H., Wintgens T., Ciesek S. Detection of SARS-CoV-2 in raw and treated wastewater in Germany – suitability for COVID-19 surveillance and potential transmission risks. Sci. Total Environ. 2021;751 doi: 10.1016/j.scitotenv.2020.141750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2021. WHO Coronavirus (COVID-19) Dashboard.https://covid19.who.int/ [Google Scholar]
- Wu F., Xiao A., Zhang J., Moniz K., Endo N., Armas F., Bonneau R., Brown M.A., Bushman M., Chai P.R., Duvallet C., Erickson T.B., Foppe K., Ghaeli N., Gu X., Hanage W.P., Huang K.H., Lee W.L., Matus M., McElroy K.A., Nagler J., Rhode S.F., Santillana M., Tucker J.A., Wuertz S., Zhao S., Thompson J., Alm E.J. SARS-CoV-2 RNA concentrations in wastewater foreshadow dynamics and clinical presentation of new COVID-19 cases. Sci. Total Environ. 2022;805 doi: 10.1016/j.scitotenv.2021.150121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Ling H., Huang X., Li J., Li W., Yi C., Zhang T., Jiang Y., He Y., Deng S., Zhang X., Wang X., Liu Y., Li G., Qu J. Potential spreading risks and disinfection challenges of medical wastewater by the presence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) viral RNA in septic tanks of Fangcang Hospital. Sci. Total Environ. 2020;741 doi: 10.1016/j.scitotenv.2020.140445. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material