Abstract
Activation of Jak tyrosine kinases through hematopoietic cytokine receptors occurs as a consequence of ligand-induced aggregation of receptor-associated Jaks and their subsequent autophosphorylation. Jak kinases consist of a C-terminal tyrosine kinase domain, a pseudokinase domain of unknown function, and Jak homology (JH) domains 3 to 7, implicated in receptor-Jak interaction. We analyzed the functional roles of the different protein domains in activation of Jak2. Deletion analysis of Jak2 showed that the pseudokinase domain but not JH domains 3 to 7 negatively regulated the catalytic activity of Jak2 as well as Jak2-mediated activation of Stat5. Phosphorylation of Stat5 by wild-type Jak2 was dependent on the SH2 domain of Stat5; however, this requirement was lost upon deletion of the pseudokinase domain of Jak2. Investigation of the mechanisms of the pseudokinase domain-mediated inhibition of Jak2 suggested that this regulation did not involve protein tyrosine phosphatases. Instead, analysis of interactions between the tyrosine kinase domain and Jak2 suggested that the pseudokinase domain interacted with the kinase domain. Furthermore, coexpression of the pseudokinase domain inhibited the activity of the single tyrosine kinase domain. Finally, deletion of the pseudokinase domain of Jak2 deregulated signal transduction through the gamma interferon receptor by significantly increasing ligand-independent activation of Stat transcription factors. These results indicate that the pseudokinase domain negatively regulates the activity of Jak2, probably through an interaction with the kinase domain, and this regulation is required to keep Jak2 inactive in the absence of ligand stimulation. Furthermore, the pseudokinase domain may have a role in regulation of Jak2-substrate interactions.
Cytokines regulate the proliferation and differentiation of cells by binding to hematopoietic cytokine receptors and initiating signaling cascades leading to altered gene expression. The cytokine receptors lack intrinsic catalytic activity and rely on receptor-associated cytoplasmic Jak tyrosine kinases for signal transduction (14). Activation of Jak kinases leads to phosphorylation of tyrosine residues in cytokine receptors. This creates docking sites for cytoplasmic signaling molecules, such as the signal transducers and activators of transcription (Stats) that are recruited to the receptor and activated by tyrosine phosphorylation (6). The four different mammalian Jak kinases (Jak1, Jak2, Jak3, and Tyk2) mediate essential and nonredundant functions in cytokine signaling. Mice lacking the Jak2 gene have revealed a critical role for Jak2 in signaling through several receptors, such as the erythropoietin and gamma interferon (IFN-γ) receptors (23, 24).
The Jak kinases are characterized by the presence of seven regions of sequence similarity found among Jak family members and designated Jak homology (JH) domains (41). With the exception of a catalytically active tyrosine kinase domain, JH1, it is not known which of the JH regions form structurally and functionally independent domains. JH2, adjacent to JH1, has sequence similarity to kinase domains but lacks several critical residues, such as the third glycine in the GXGXXG motif in kinase subdomain I and the nearly invariant aspartic acid directly involved in the phosphotransfer reaction in subdomain VI. JH2 is therefore termed the pseudokinase domain and is considered catalytically inactive (33). The N-terminal half of Jaks, encompassing JH domains 3 to 7, has limited sequence similarity to any known protein domains. An SH2-like domain and a 4.1 domain have been predicted around JH domains 3 to 4 and 4 to 7, respectively, but the functionality of these putative domains has not been confirmed (9, 12, 17). The region responsible for association with cytokine receptors has been mapped to JH domains 3 to 7 of Jak kinases, which would be consistent with the role of 4.1 domains in mediating interactions with transmembrane proteins (4, 39).
In most cases Jak kinases are found constitutively associated with cytokine receptors. Activation of Jaks has been considered to result from ligand-induced dimerization or oligomerization of the receptor chains, bringing the receptor-associated Jaks in close proximity and leading to their auto- and transphosphorylation. Apparently Jak kinases do not need other tyrosine kinases for their activation, and induced dimerization of two Jak molecules has been found to be sufficient for activation of Jaks (21). In single-chain receptors, such as the erythropoietin receptor, Jak activation relies on interaction between two similar Jak kinases, and notably, all single-chain receptors utilize Jak2 (34). In receptors composed of multiple chains, initiation of signal transduction is more complex and requires an activation cascade of different Jak kinases (22).
Phosphorylation of Jak kinases is functionally important, since mutation of the activation loop (A-loop) tyrosines in the kinase domain affects activation of Jaks. Although the tyrosine kinase domains of Jaks are highly conserved and all contain the YY motif in the A-loop, the functional role of these tyrosines varies between different Jaks. Phosphorylation of the first tyrosine in the YY motif is critical for activation of Jak1 and Jak2, and it also enhances the activity of Jak3 (8, 19, 40). However, mutation of the second tyrosine in the YY motif greatly increases the kinase activity of Jak3, while similar mutations in Jak1 or Jak2 have no effect (8, 19, 40).
The A-loop tyrosines are potential targets for regulation of Jak activity through dephosphorylation. At least two protein tyrosine phosphatases (PTPases) have been implicated in Jak signaling. SHP-2 has been found to regulate Jak signaling both positively and negatively, whereas SHP-1, which is predominantly expressed in hematopoietic cells, has an important role in downregulation of cytokine receptor signaling (16, 37, 38). The tyrosine-phosphorylated Jaks can also be inhibited by interaction with the SOCS proteins. SOCS-1 and SOCS-3 bind to a phosphorylated tyrosine in the A-loop and to the catalytic center in the kinase domain of Jak2, thus interfering with substrate binding and inhibiting kinase activity (36).
In many cytoplasmic tyrosine kinases, intramolecular interactions form yet another level of regulation of catalytic activity by preventing aberrant autophosphorylation and subsequent activation of the kinases in the absence of a specific activation signal. The Src kinases c-Src and Hck are kept inactive by binding of the Src homology region 2 (SH2) and SH3 domains to target residues in the C-terminal tail and in the linker between the SH2 and kinase domains, respectively (29, 35). In a Tec family tyrosine kinase, Itk, inhibitory interactions are at least partially mediated by the SH3 domain (1). Mutations abrogating the intramolecular regulation may lead to hyperactivation of the kinase, a phenomenon well characterized for v-Src, the oncogenic form of c-Src. Inappropriately regulated forms of Jak kinases have also been identified and shown to have severe biological consequences. A point mutation in the JH2 domain of the Drosophila Jak homolog (Hop) has been found to induce hematopoietic neoplasia in the fly, but the exact mechanism of hyperactivation of the Hop mutant is still unknown (20). In addition, constitutive dimerization of a TEL-Jak2 fusion protein has been found to activate Jak2 and to cause lymphoblastic leukemia (18).
This study was carried out to characterize the mechanisms regulating the activity of the Jak2 tyrosine kinase. Intramolecular interactions play a critical role in the regulation of several cytoplasmic tyrosine kinases, but thus far such intramolecular regulation has not been reported for Jak kinases. We analyzed the roles of the different JH protein domains in activation of Jak2. Our results show that the pseudokinase domain, but not other JH domains, negatively regulated the tyrosine kinase domain of Jak2. Furthermore, deregulation of Jak2 by deletion of JH2 resulted in ligand-independent Stat activation through the IFN-γ receptor. Analysis of the interactions between different Jak2 domains suggested that regulation of Jak2 by the pseudokinase domain is mediated through intramolecular interactions between the tyrosine kinase and pseudokinase domains.
MATERIALS AND METHODS
Reagents, cell culture, and transfections.
293T (American Type Culture Collection) and γ2A (Jak2-deficient fibrosarcoma cell line [32]) cell lines were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Gibco-BRL) and antibiotics. 293T cells were transfected with the calcium-phosphate transfection kit (Gibco) and γ2A cells using the Fugene 6 transfection reagent (Boehringer Mannheim) according to the manufacturer's instructions. Specific cDNAs (0.5 to 5 μg, depending on the experiment) were used to transfect 10-cm dishes of 60% confluent 293T cells, whereas 100 ng of specific cDNA was used for transfection of a six-well plate well of γ2A cells. The amount of each cDNA transfected was adjusted within a single experiment to obtain similar expression levels of the different cDNA constructs. The cells were harvested 72 h after transfection for immunoprecipitation and after 20 h for luciferase assays. Pervanadate was prepared by mixing equal volumes of Na3VO4 (50 mM) and H2O2 (250 mM). The following antibodies were used: antiphosphotyrosine 4G10 (Upstate Biotechnology), antihemagglutinin (anti-HA) 16B12 (Berkeley Antibody Company), anti-Stat5 ST5a-2H2 (Zymed) and polyclonal anti-Jak2, a kind gift from James Ihle (30).
DNA constructs.
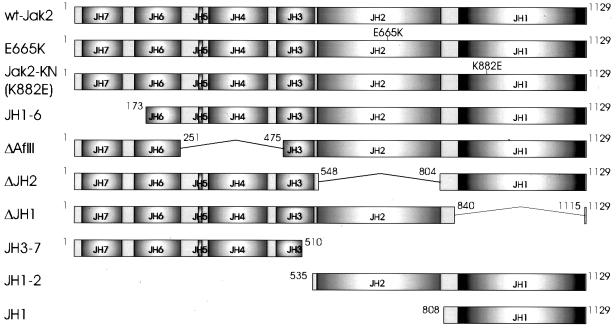
The expression vector for Jak2-HA has been described (26). The amino acids encoded by the Jak2 constructs are shown in Fig. 1. ΔAflII-HA is an internal AflII deletion mutant of Jak2-HA. Other Jak2 constructs were prepared by PCR and cloned into the pCIneo expression vector (Promega). New translational initiation codons were introduced by PCR into Jak2-HA to create JH1-6-HA and JH1-HA. ΔJH2-HA was constructed by replacing a fragment of Jak2-HA with a ligation product of PCR fragments coding for amino acids 259 to 548 and 809 to 1129 of Jak2 with additional SacI sites at the 3′ and 5′ ends, respectively. ΔJH1 is a SanDI-EcoNI deletion mutant of untagged Jak2 cDNA (wt-Jak2) (30) created by filling in with the Klenow fragment. Jak2-KN is identical to wt-Jak2 except for the K882E substitution. All PCR products were confirmed by sequencing (Applied Biosystems). Expression vectors for Stat5A and the R618L mutant of Stat5A were kind gifts from Tim Wood. The luciferase reporter construct containing the Stat1 binding site from the promoter of the IFN regulatory factor 1 gene was a kind gift from Richard Pine (25).
FIG. 1.
Protein products encoded by Jak2 cDNA constructs. Schematic presentation of the proteins encoded by Jak2 deletion constructs. Amino acids encoded by the constructs are indicated; numbers refer to the murine Jak2 sequence. An HA tag, when present, is located in the C terminus and indicated by the suffix HA in the text. The full-length HA-tagged Jak2 is termed Jak2-HA, and the untagged form of Jak2 is termed wt-Jak2.
Immunoblotting and immunoprecipitation.
Immunoblotting and immunoprecipitation in Triton buffer (50 mM Tris [pH 7.5], 10% glycerol, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 50 mM NaF, 1 mM Na3VO4) have been described (26), except that lysates were precleared using normal mouse sera, followed by incubation with protein G-Sepharose. For coimmunoprecipitation, cells were lysed in Brij 58 buffer (10 mM Tris-HCl [pH 7.5], 0.9% Brij 58, 0.1% NP-40, 150 mM NaCl, 50 mM NaF, 1 mM Na3VO4) supplemented with protease inhibitors (phenylmethylsulfonyl fluoride, approtinin, leupeptin, and pepstatin A). Before immunoprecipitation, bovine serum albumin was added to the precleared lysates (final concentration, 1%). Immunoprecipitates were washed once with Brij 58 buffer, twice with Triton buffer, once with high-salt buffer (Brij 58 buffer with 350 mM NaCl), and twice with NP-40 buffer (50 mM Tris-HCl [pH 7.5], 10% glycerol, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 50 mM NaF, 1 mM Na3VO4). Equal amounts of protein from cell lysates were always used for immunoprecipitations and Western blotting of cell lysates. Protein concentrations were determined using the Bio-Rad protein assay system (Bio-Rad Laboratories). Immunodetection was performed with specific primary antibodies, biotinylated anti-mouse immunoglobulin (Ig) or anti-rabbit Ig secondary antibodies (Dako A/S), and streptavidin-biotin-horseradish peroxidase conjugate (Pharmacia-Amersham) followed by enhanced chemiluminescence.
Kinase and luciferase assays.
For the kinase assay, cells were lysed in kinase lysis buffer (10 mM Tris-HCl [pH 7.5], 1% Triton X-100, 20% glycerol, 5 mM EDTA, 50 mM NaCl, 50 mM NaF, 1 mM Na3VO4) supplemented with protease inhibitors. Immunoprecipitates were washed four times with kinase lysis buffer and twice with kinase assay buffer (10 mM HEPES [pH 7.4], 50 mM NaCl, 5 mM MgCl2, 5 mM MnCl2, 50 mM NaF, 0.1 mM Na3VO4). The immunoprecipitates were suspended in kinase assay buffer containing 1 mM dithiothreitol and Stat1 (GPKGTGYIKTELISVS) or Stat5 (AKAADGYVKPQIKQVV) peptides (1 mg/ml), and 10 μCi of [γ-32P]ATP was added. The reaction mixtures were incubated at room temperature for 5 to 10 min, boiled in reducing Laemmli sample buffer, and separated by sodium dodecyl sulfate–20% polyacrylamide gel electrophoresis (SDS–20% PAGE) followed by autoradiography or quantitation of radioactivity with a PhosphorImager (Fuji).
For the luciferase assay, cells were transfected with a Stat-dependent luciferase reporter construct together with a pRLTK control vector constitutively expressing Renilla luciferase (Promega). Cells were lysed in passive lysis buffer (Promega), and the luciferase activity of the lysates was determined using the dual luciferase reporter assay system (Promega) according to the manufacturer's instructions. Stat-dependent luciferase activity was normalized to the activity of the constitutively expressed Renilla luciferase.
Binding of histidine-tagged proteins.
Cells were lysed in kinase lysis buffer without EDTA. After clearing by centrifugation, equal amounts of protein from the lysates were diluted 10-fold with urea binding buffer (8 M urea, 50 mM NaH2PO4, 10 mM Tris [pH 8], 150 mM NaCl) and incubated with Talon metal affinity resin (Clontech) for 30 min at room temperature. The resin was washed four times with urea washing buffer (8 M urea, 50 mM NaH2PO4 [pH 7], 150 mM NaCl) before elution with 100 mM EDTA and boiling in reducing Laemmli sample buffer.
RESULTS
Effect of JH domains on catalytic activity of Jak2.
To analyze the roles of the different protein domains in the regulation of Jak2 activity, we constructed a series of Jak2 expression plasmids (Fig. 1). The JH1 and JH2 domain boundaries were predicted by using the Smart program (28). ΔJH1 lacks the kinase domain, and ΔJH2 lacks the pseudokinase domain. ΔAflII lacks JH domains 4 and 5 and small fragments of domains 3 and 6. JH1-2 contains only the pseudokinase and tyrosine kinase domains, and JH1-6 lacks domain 7 and a short sequence of domain 6. JH1 encodes a single tyrosine kinase domain.
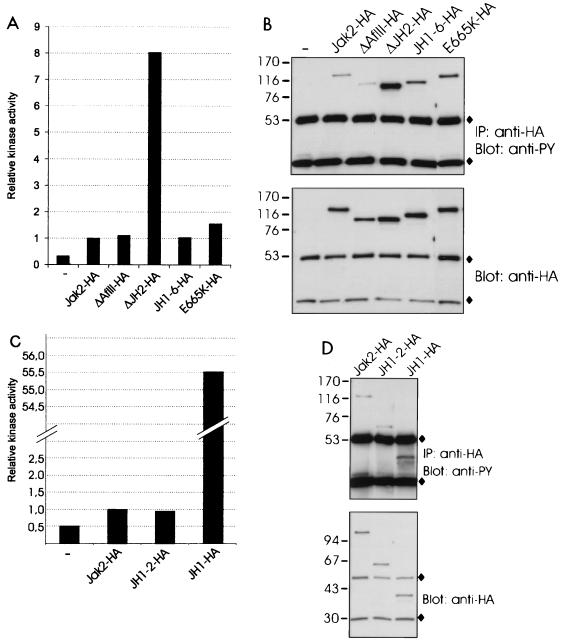
The Jak2-HA expression plasmid and the HA-tagged Jak2 deletion constructs were transiently expressed in mammalian 293T cells, in which expression of Jak2 leads to autoactivation of the kinase. The different Jak2 proteins were immunoprecipitated with anti-HA antibody and subjected to an in vitro kinase assay (Fig. 2A). Aliquots of the same immunoprecipitates were also subjected to antiphosphotyrosine and anti-HA immunoblots (Fig. 2B). As shown in Fig. 2A, deletion of the JH2 domain increased the kinase activity of Jak2 nearly 10-fold, while other deletions had no or only moderate effects. The E665K point mutation in JH2, similar to that found to cause hematopoietic neoplasia in Drosophila (20), increased the kinase activity of Jak2 by 50%. The levels of tyrosine phosphorylation (Fig. 2B) correlated with the kinase activities of the Jak2 proteins, ΔJH2-HA having the highest level of tyrosine phosphorylation. JH1-6-HA showed slightly increased tyrosine phosphorylation without an increase in kinase activity. The anti-HA immunoblot confirmed similar expression of all Jak2 proteins. To directly assess the effect of JH2 on the activity of JH1, we prepared a construct containing only domains JH1 and JH2. The analysis of this construct in an in vitro kinase assay showed that the catalytic activity of the JH1-2-HA protein was equal to that of Jak2-HA (Fig. 2C). When JH2 was deleted from the JH1-2-HA construct, the catalytic activity increased more than 50-fold (JH1-HA). Tyrosine phosphorylation of the expressed proteins (Fig. 2D) correlated with the results from the kinase assays shown in Fig. 2C, although the differences in tyrosine phosphorylation were less profound, suggesting the presence of phosphorylation sites outside JH1. Similar results were obtained when the in vitro kinase assays presented in Fig. 2 were repeated two to four times.
FIG. 2.
Deletion of JH2 increases the catalytic activity of Jak2. (A) 293T cells were transfected with expression plasmids for Jak2-HA, ΔAflII-HA, ΔJH2-HA, JH1-6-HA, and E665K-HA or left untransfected (—). The lysates were immunoprecipitated with anti-HA antibody, and aliquots were subjected to in vitro kinase assays with Stat1-derived peptide and [γ-32P]ATP as substrates. The peptides were separated by SDS–20% PAGE followed by quantification with a PhosphorImager. Relative catalytic activities are shown. Radioactivity incorporated in the Stat1 peptide by the different Jak2 proteins was normalized to the radioactivity incorporated in the Stat1 peptide by Jak2-HA, which was set at 1. (B) Aliquots of the same immunoprecipitates (IP) were analyzed by SDS–4 to 15% PAGE and blotted with antiphosphotyrosine (anti-PY) or anti-HA antibodies. (C) 293T cells were transfected with expression plasmids for Jak2-HA, JH1-2-HA, and JH1-HA or left untransfected. The lysates were immunoprecipitated with anti-HA antibody and analyzed in in vitro kinase assays as in panel A. (D) Aliquots of the same immunoprecipitates were analyzed by SDS–4 to 15% PAGE and blotted with antiphosphotyrosine or anti-HA antibodies. (B and D) ⧫, immunoglobulin chains. The mobilities of the molecular mass markers (in kilodaltons) are shown on the left.
Effect of JH domains on Jak2-induced activation of Stat5.
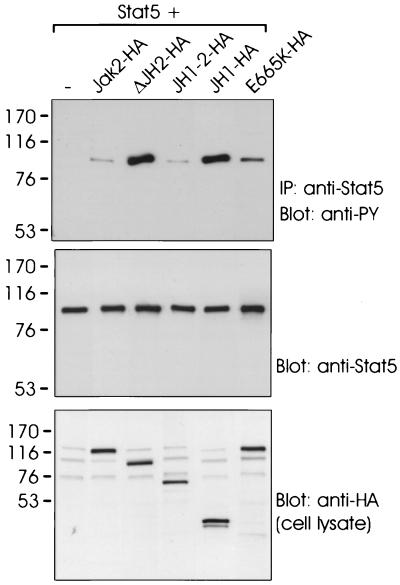
To analyze whether deletion of JH2 also resulted in enhanced Jak2 signaling, we analyzed the ability of the different Jak2 mutants to activate Stat5. For this purpose, Stat5 and the HA-tagged Jak2 plasmids were coexpressed in 293T cells, and Stat5 was immunoprecipitated and subjected to antiphosphotyrosine immunoblotting. JH1-2-HA-induced tyrosine phosphorylation of Stat5 comparable to that with Jak2, while ΔJH2-HA- and JH1-HA-induced phosphorylation of Stat5 was greater than that induced by Jak2-HA (Fig. 3). The E665K mutant of Jak2-HA also phosphorylated Stat5 better than did Jak2-HA, but not as well as ΔJH2-HA or JH1-HA. Similar results were obtained when activation of Stat5-mediated transcription was analyzed in 293T cells coexpressing different Jak2 mutants and the Stat5-dependent luciferase reporter (not shown). Taken together, the results (Fig. 2 and 3) indicate that JH2, but not other JH domains, negatively regulates the activity of Jak2.
FIG. 3.
Deletion of JH2 enhances Jak2-mediated activation of Stat5. 293T cells were cotransfected with expression plasmids for Stat5 alone (—) or Stat5 plus Jak2-HA, ΔJH2-HA, JH1-2-HA, JH1-HA, or E665K-HA. Stat5 was immunoprecipitated with anti-Stat5 antibody, and the immunoprecipitates (IP) were analyzed by SDS–7.5% PAGE followed by antiphosphotyrosine (anti-PY) or anti-Stat5 immunoblotting. Aliquots of cell lysates were analyzed by SDS–4 to 15% PAGE followed by anti-HA immunoblotting (lowest panel). The mobilities of the molecular mass markers (in kilodaltons) are shown on the left.
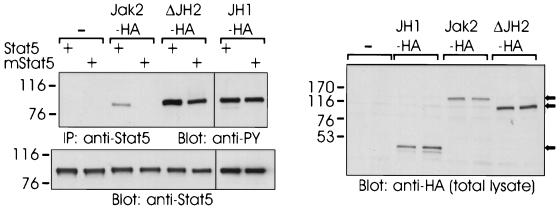
The SH2 domains of Stats have been found to be critical for their activation by Jak kinases (10). To test whether ΔJH2 had the same structural requirement for activation of its substrate proteins, we coexpressed Stat5 and its SH2 mutant carrying an R618L substitution (mStat5) with Jak2-HA, ΔJH2-HA, or JH1-HA. Stat5 was immunoprecipitated from cell lysates and analyzed by antiphosphotyrosine immunoblotting. As shown in Fig. 4, Jak2-HA induced tyrosine phosphorylation of Stat5, but phosphorylation of the mutant Stat5 could not be detected even in longer exposures. In contrast, ΔJH2-HA and JH1-HA readily activated both Stat5 and its SH2 mutant, as detected by induction of tyrosine phosphorylation. The JH1-2-HA construct was unable to induce tyrosine phosphorylation of the mutant Stat5, indicating that deletion of JH domains 3 to 7 of Jak2 did not change the requirement for the SH2 domain for activation of Stat5 (not shown). The high catalytic activity of ΔJH2 might explain phosphorylation of the SH2 mutant Stat5 by ΔJH2. Therefore, we used increasing amounts of JH1-2-HA cDNA in cotransfections with the SH2 mutant of Stat5. However, although JH1-2-HA showed higher tyrosine phosphorylation than ΔJH2-HA, it was not able to phosphorylate the SH2 mutant Stat5 (not shown). Phosphorylation of tyrosine 694 in Stat5 is required for dimerization and activation of Stat5. To test whether ΔJH2 specifically phosphorylated this residue in Stat5, we coexpressed ΔJH2-HA with Y694F-Stat5 in 293T cells. ΔJH2HA, as well as Jak2-HA, was unable to phosphorylate the Y694F mutant of Stat5, indicating that despite its high activity, ΔJH2 retained its specificity by inducing phosphorylation of only the critical tyrosine 694 in Stat5 (not shown). Taken together, these results suggest that the pseudokinase domain affects interactions of Jak2 with target proteins.
FIG. 4.
SH2 domain of Stat5 required for activation by Jak2 but not by ΔJH2 or JH1. 293T cells were transfected with expression plasmids for Stat5 or SH2 mutant of Stat5 (mStat5). In addition, the cells were transfected with expression plasmids for Jak2-HA, ΔJH2-HA, or JH1-HA or left untransfected (—). Stat5 was immunoprecipitated (IP) with anti-Stat5 antibody. The immunoprecipitates were separated by SDS–7.5% PAGE and blotted with antiphosphotyrosine (anti-PY) or anti-Stat5 antibodies. Aliquots of cell lysates were analyzed by SDS–4 to 15% PAGE followed by anti-HA immunoblotting. The mobilities of the molecular mass markers (in kilodaltons) are shown on the left. Arrows on the right indicate the mobilities of the Jak2 proteins.
Effect of JH domains on activity of the Jak2 tyrosine kinase domain.
We next wanted to analyze the mechanism of JH2-mediated inhibition of Jak2. Consistent with previous reports, ΔJH1-HA had no kinase activity when expressed in 293T cells, indicating that JH2 does not possess catalytic activity (7, 8, 33). Therefore, inhibition of Jak2 by JH2 was considered to depend on other than phosphotransfer-dependent mechanisms. JH2 could bind to and recruit a PTPase, which would dephosphorylate and thus inactivate JH1. Alternatively, inhibition by JH2 could be based on a molecular interaction with JH1, keeping JH1 in a conformation that would not allow substrates to bind and/or be phosphorylated. Analogous intramolecular regulation has been found in members of the Src and Tec/Btk tyrosine kinase families (1, 29, 35).
To test the hypothesis that JH2 bound a PTPase required to inhibit Jak2, we expressed different HA-tagged Jak2 constructs in 293T cells. Before lysis, the cells were treated with the cell membrane-permeating PTPase inhibitor pervanadate. The lysates were immunoprecipitated with anti-HA antibody, and aliquots were subjected to antiphosphotyrosine immunoblotting. As shown in Fig. 5, inhibition of PTPases with pervanadate similarly enhanced tyrosine phosphorylation of Jak2-HA, ΔJH2-HA, and JH1-HA. Therefore, we concluded that the increased kinase activity of ΔJH2 was not due to defective dephosphorylation of Jak2.
FIG. 5.
Effect of PTPase inhibitors on tyrosine phosphorylation of Jak2. 293T cells were transfected with expression plasmids for Jak2-HA, JH1-HA, and ΔJH2-HA. The cells were treated for 30 min with the cell membrane-permeating PTPase inhibitor pervanadate (100 μM) (lanes +) or left untreated (lanes −). The Jak2 proteins were immunoprecipitated (IP) with anti-HA antibody, separated by SDS–4 to 15% PAGE, and immunoblotted with antiphosphotyrosine (anti-PY) or anti-HA antibodies. ⧫, Ig chains. The mobilities of the molecular mass markers (in kilodaltons) are shown on the left.
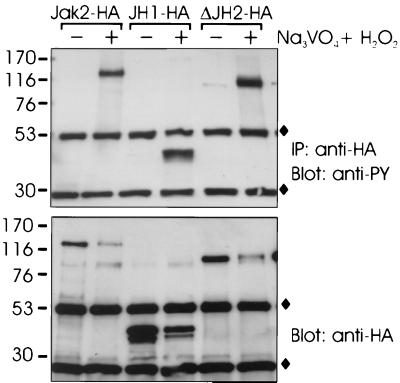
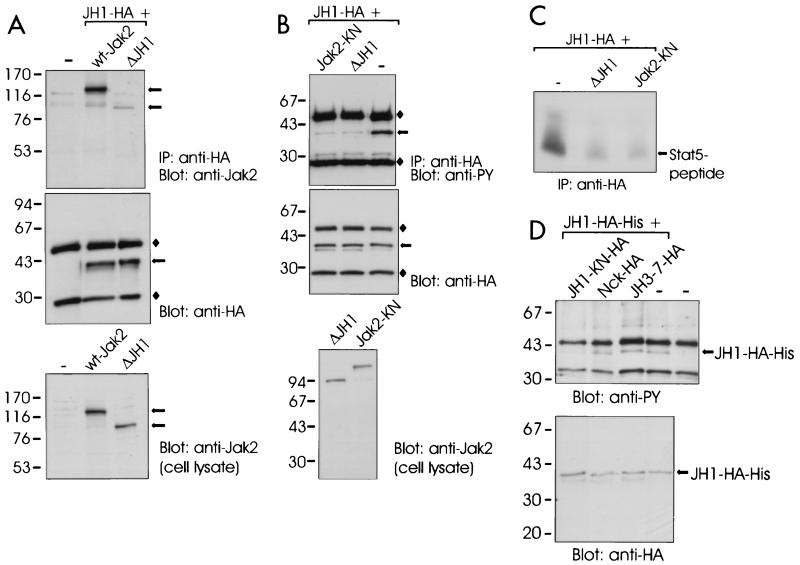
We next tested whether inhibition of Jak2 activity might be caused by an interaction of the JH2 domain with JH1. We created several JH2 constructs with different N and C termini but failed to express JH2 alone in 293T cells at levels comparable to the other Jak2 constructs. Therefore, we first addressed the question of whether JH1-HA was able to interact with Jak2 lacking JH1 (ΔJH1 construct), which would mimic intramolecular interactions of JH1-HA with domains 2 to 7 in Jak2.
We transiently expressed JH1-HA together with wt-Jak2 or ΔJH1 in 293T cells. JH1-HA was immunoprecipitated from cell lysates, and the immunoprecipitates were analyzed by immunoblotting with anti-Jak2 and anti-HA antibodies. As shown in Fig. 6A, both wt-Jak2 and ΔJH1 coimmunoprecipitated with JH1-HA, but the interaction of JH1-HA with ΔJH1 was weaker than with wt-Jak2. The interactions were stable in stringent washing conditions (1% Triton X-100, 1% NP-40, and 350 mM NaCl). As a control, we expressed wt-Jak2 or ΔJH1 without JH1-HA. Anti-HA immunoprecipitates of these lysates did not show precipitation of wt-Jak2 or ΔJH1, indicating that coimmunoprecipitation was specific (not shown). In another experiment in which the amount of coexpressed JH1-HA was increased, coimmunoprecipitation of ΔJH1 with JH1-HA was more evident (not shown). These results indicated that the tyrosine kinase domain could interact with Jak2 in two different ways. First, JH1-HA interacted with another kinase domain, since deletion of the Jak2 kinase domain reduced the ability of the coexpressed JH1-HA to interact with Jak2. This type of interaction is supposed to be important during activation of Jak2. Second, JH1 interacted with JH domains 2 to 7. To further specify the interaction of JH1 with Jak2, we analyzed the association of JH1 with JH1-2 (JH domains 3 to 7 deleted) by coimmunoprecipitation. We found that deletion of domains 3 to 7 did not affect the interaction of JH1 with Jak2, suggesting that JH1 does not interact with domains 3 to 7 in Jak2 (not shown).
FIG. 6.
Effect of JH domains on activity of the Jak2 tyrosine kinase domain. (A) 293T cells were transfected with expression plasmids for JH1-HA and untagged wt-Jak2 or ΔJH1 or left untransfected (—). The cells were lysed in Brij 58 buffer, and the lysates were immunoprecipitated (IP) with anti-HA antibody. The immunoprecipitates were separated by SDS–4 to 15% PAGE and immunoblotted with anti-Jak2 or anti-HA antibodies. Aliquots of the cell lysates were analyzed by SDS–7.5% PAGE followed by anti-Jak2 immunoblotting. (B) 293T cells were transfected with JH1-HA expression plasmid alone (lane —) or together with expression plasmids for untagged ΔJH1 or Jak2-KN. JH1-HA was immunoprecipitated (IP) with anti-HA antibody, analyzed by SDS–4 to 15% PAGE, and blotted with antiphosphotyrosine (anti-PY) or anti-HA antibodies. Aliquots of the cell lysates were analyzed by SDS–4 to 15% PAGE followed by anti-Jak2 immunoblotting. ⧫, Ig chains. (C) Aliquots of immunoprecipitates from panel 6B were subjected to in vitro kinase assays with Stat5-derived peptide and [γ-32P]ATP as substrates. The peptides were separated by SDS–20% PAGE followed by autoradiography. (D) 293T cells were transfected with an expression vector for HA- and histidine-tagged JH1 (JH1-HA-His) alone or together with expression plasmids for JH3-7-HA, JH1-KN-HA (K882E in JH1), or Nck-HA. JH1-HA-His was isolated by metal affinity, analyzed by SDS–4 to 15% PAGE, and blotted with antiphosphotyrosine (anti-PY) or anti-HA antibodies. The mobilities of the molecular mass markers (in kilodaltons) are shown on the left in all panels. The arrows on the right indicate the mobilities of the Jak2 proteins.
We (unpublished results) and others (3, 8) have found that a kinase-inactive form of Jak2 is able to inhibit activation of Jak2 when coexpressed, most probably by competing in dimerization with wild-type Jak2, which is required for trans- or autophosphorylation and subsequent activation. Similarly, we reasoned that if JH1 was inhibited by an interaction with JH2 in Jak2, coexpression of JH2 might result in inhibition of JH1. To analyze whether JH2 could inhibit activation of JH1 in trans, we first coexpressed JH1-HA with an excess of ΔJH1 (domains 2 to 7). A kinase-inactive (K882E) mutant of Jak2 (Jak2-KN) was used as a control. JH1-HA was immunoprecipitated and analyzed by antiphosphotyrosine immunoblotting (Fig. 6B) and in an in vitro kinase assay (Fig. 6C). As shown in Fig. 6B and C, coexpression of either Jak2-KN or ΔJH1 with JH1-HA resulted in reduction of tyrosine phosphorylation and kinase activity of JH1-HA.
To exclude the participation of domains 3 to 7 (included in the ΔJH1 construct) in inhibition of JH1, we coexpressed the JH3-7-HA construct with JH1 tagged with HA and with six histidine residues (JH1-HA-His) (Fig. 6D) and analyzed activation of the latter. As controls, we coexpressed HA-tagged JH1-KN (JH1 carrying the K882E mutation) and Nck, a cytoplasmic adapter protein, with JH1-HA-His. JH1-HA-His was isolated from the cell lysates with metal affinity resin followed by antiphosphotyrosine immunoblotting. As shown in Fig. 6D, tyrosine phosphorylation of JH1-HA-His was reduced by coexpression of JH1-KN-HA but not by coexpression of the JH3-7-HA or Nck-HA proteins. The expression levels of the proteins were controlled using anti-HA immunoblotting of cell lysates (not shown). Since JH1 was inhibited by the simultaneous expression of ΔJH1 including domains 2 to 7 but not by coexpression of domains 3 to 7, we concluded that JH2 was required for inhibition of JH1. Taken together, the results shown in Fig. 6 indicated that the JH2 domain inhibited the activity of the Jak2 tyrosine kinase domain in trans, suggesting that the JH2 domain interacts with the tyrosine kinase domain.
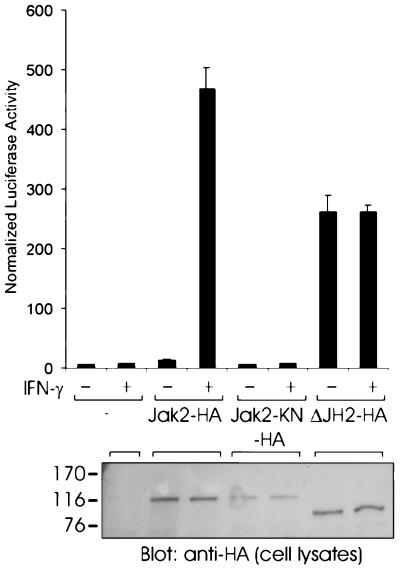
Effect of JH2 on Jak2-mediated signaling through cytokine receptors.
We next examined the functional role of JH2 in cytokine receptor signaling. We set up a cotransfection system in a previously characterized Jak2-deficient cell line, γ2A (32), in which activation of Jak2 was dependent on stimulation with IFN-γ (Fig. 7). The cells were transfected with different HA-tagged Jak2 constructs, pRLTK control vector, and a Stat1-dependent luciferase reporter. Stimulation with IFN-γ had no effect on Stat1 activation, since signaling through the IFN-γ receptor requires the presence of both Jak1 and Jak2 kinases. Expression of Jak2-HA resulted in nearly 500-fold induction of the Stat1 reporter by IFN-γ while having no effect on Stat1 activation in the absence of IFN-γ. In striking contrast, transfection of the ΔJH2-HA expression vector resulted in nearly 300-fold induction of the Stat1 reporter, and this induction was independent of IFN-γ stimulation. The expression levels of the expressed Jak2 proteins were controlled by using anti-HA immunoblotting of cell lysates. These results indicated that JH2 inhibits the basal activity of Jak2, since expression of ΔJH2 resulted in significantly increased basal activity of the Stat1 reporter, which suggests that activation of Jak2 by deletion of JH2 results in constitutive activation of Jak2-dependent cytokine receptor signaling pathways.
FIG. 7.
Effect of JH2 on cytokine signal transduction. γ2A cells were transfected with the Stat1-dependent luciferase reporter vector, the pRLTK control vector, and Jak2-HA, Jak2-KN-HA, ΔJH2-HA, or an empty vector (—) as a control. At 5 h after transfection, the cells were removed to serum-free medium and starved for 15 h. The cells were stimulated with IFN-γ (1,000 U/ml) for 5 h or left unstimulated. Luciferase activity was measured as described in Materials and Methods. Shown are the means from three independent experiments and the standard errors of the mean. The mobilities of the molecular mass markers (in kilodaltons) are shown on the left.
DISCUSSION
Tyrosine kinases play a critical role in the regulation of cell proliferation, differentiation, and functional activation. The activity of tyrosine kinases is tightly controlled by autoregulatory mechanisms involving intramolecular interactions or by the action of other regulatory proteins (13). The Jak tyrosine kinases have an important role in the regulation of proliferation and differentiation of hematopoietic cells. The activity of Jak kinases has been found to be under the control of regulatory proteins such as the SOCS proteins and PTPases, while intramolecular regulation of Jaks has not been reported previously. The Jak kinases are characterized by two kinase domains, and thus they differ from most cytoplasmic tyrosine kinases. Only the C-terminal tyrosine kinase domain of Jaks appears to be catalytically active, and therefore the function of the second kinase domain has remained unknown.
We found several lines of evidence indicating an inhibitory role for the JH2 domain in regulation of Jak2 activity. First, deletion of JH2, but not other JH domains, increased the catalytic activity and tyrosine phosphorylation of Jak2. Second, deletion of JH2 enhanced Jak2-mediated activation of Stat5 and resulted in ligand-independent, constitutive activation of Stat proteins in signaling through the IFN-γ receptor. Third, coexpression of the JH2 domain, but not JH domains 3 to 7, inhibited activation of the single tyrosine kinase domain of Jak2 in trans. Studies with deletion mutants of Jak2 have previously suggested that regions outside the kinase domain might contain negative regulatory functions (7, 27). Consistent with our present findings, Stat5 activation in a yeast coexpression system has been found to be enhanced by deletion of JH2 from Jak2, but the mechanism of this enhanced activation was not analyzed (2). However, in the yeast system, the single kinase domain was found to activate Stat5 less efficiently than wt-Jak2 (2), which is in contrast to our results. We found that JH1 activated Stat5 better than wt-Jak2 did and that JH1 possessed even higher catalytic activity than ΔJH2. The JH1 construct used in the yeast system also contained parts of JH2 and JH7, leaving the possibility that the placement of N-terminal domains next to JH1 in the resulting recombinant protein impaired the activity of the kinase domain (2).
Although our results do not indicate any direct regulation of JH1 by JH domains 3 to 7, we cannot totally rule out the possibility that these domains would have a regulatory function in Jak2. Mutations in the N-terminal JH regions have also been found to interfere with regulation of Jak kinases. A point mutation in JH4 of Hop has been reported to activate the kinase, but this residue is not conserved in mammalian Jaks (11). It is noteworthy that mutations in JH7 and JH2 of Jak3 have been reported to cause increased basal tyrosine phosphorylation of Jak3, although at the same time resulting in severe combined immunodeficiency (SCID) and failure to activate Stat5 (4, 5). Exactly how these mutations affect the kinases is currently unknown, but it is plausible that the N-terminal domains 3 to 7 have an important role in activation of Jaks by regulating the association of Jaks with cytokine receptors.
A regulatory role for JH2 in activation of Jak kinases has been suggested by studies in which a mutation in JH2 (E695K in Hop and E665K in Jak2) was found to increase tyrosine phosphorylation of Jak2 and Hop (20). In line with these results, we found that the E665K mutation increased the catalytic activity of Jak2, but deletion of the whole JH2 domain resulted in much higher activation of Jak2. This indicates that the single point mutation in Jak2 did not completely abolish the JH2-mediated regulation of Jak2. The E695K substitution in Hop was also able to deregulate Hop signaling and induce hematopoietic neoplasia in the fly. We also found that deletion of JH2 deregulated cytokine receptor signaling by increasing the basal activity of Jak2 and resulted in constitutive, ligand-independent Stat activation. Therefore, it would be of interest to analyze whether deletion of JH2 in Jak2 has an oncogenic effect in mammalian cells.
Although the E695K mutant of Hop suggested a role for JH2 in negatively regulating the kinase activity, deletion of the JH2 domain in Hop as well as in Tyk2 has been found to cause a loss of enzymatic activity, which is different from our findings with Jak2 (20, 31). This may be due to several reasons, one being that the Jak kinases are differentially regulated and that the role of JH2 varies between different Jaks. In support of this, the role of A-loop tyrosines in regulating the kinase activity appears to be different among the four Jaks. Alternatively, the JH2 deletion in Hop and Tyk2 may have incidentally impaired the activity of the kinase domain. We found that the length of the N-terminal region outside of the JH1 domain boundary critically affected the activity of the kinase domain; e.g., a 40-residue-shorter protein than encoded by our JH1 construct failed to show any catalytic activity (unpublished results).
The SH2 domains of Stats mediate multiple interactions with cytokine receptors, other Stat proteins, and Jak kinases (6). The SH2 domains of Stat1 and Stat2 are required for their activation by Jak1, although the role of the SH2 domain in this interaction is not clearly defined (10). We found that phosphorylation of Stat5 by wt-Jak2 was strictly dependent on the functional SH2 domain of Stat5. Surprisingly, we found that deletion of JH2 allowed Jak2 to phosphorylate Stat5 independently of its SH2 domain, suggesting that JH2 regulates interactions of Jak2 with target proteins.
PTPases play a critical role in regulation of Jak kinases through dephosphorylation, and it has been found that PTPase inhibitors can induce activation of Jak2 in the absence of ligand. We found that in 293T cells, Jak2 as well as ΔJH2 was under the control of protein phosphatases. This suggests that activation of Jak2 by JH2 deletion is not due to defective downregulation of Jak2 through dephosphorylation. However, our results clearly indicate that the PTPase-mediated regulation of Jak2 is very important, since inhibition of PTPase activity resulted in a prominent increase in the tyrosine phosphorylation of Jak2. The regulation by PTPases may be very complex, involving both negative and positive effects on kinase activity. Therefore, we cannot totally rule out the possibility that some PTPases might function through JH2. Thus, it is reasonable to think that PTPases as well as the JH2 domain contribute to inhibition of Jak2 in receptor monomers. Cytokine signaling is also negatively regulated by the SOCS proteins. SOCS-1 and SOCS-3, which have been found to inhibit Jak kinases, appear to bind directly to the tyrosine kinase domain of Jak2 (36). Therefore, the SOCS proteins are not likely to be involved in JH2-mediated regulation of Jak2 in our experimental system.
The Jak kinases do not contain classical SH2 or SH3 domains, which have been found to mediate intramolecular regulation of, e.g., Src and Tec family tyrosine kinases (1, 29, 35). We analyzed the possibility that JH2-mediated inhibition of Jak2 was based on the inherent interaction of the kinase and pseudokinase domains. We found that the JH1 domain of Jak2 could associate with JH domains 2 to 7 (ΔJH1), although less efficiently than with the full-length Jak2. Association with ΔJH1 also inhibited the activity of JH1, and this inhibition required the presence of JH2. Our results therefore support a model in which the JH2 and JH1 domains interact, resulting in inhibition of the kinase activity of JH1, and that deletion of JH2 relieves this inhibition by abolishing the interaction between JH1 and JH2. The three-dimensional structure of Jak kinases has not been solved but will eventually be important in proving whether Jaks are regulated through JH1-JH2 interaction, as suggested by our results with Jak2, and how such an interaction might result in inhibition of JH1. However, one can envision that an interaction of JH2 with JH1 might induce a conformational change in the JH1 domain, inactivating the kinase activity, or alternatively JH2 might block the access of substrates or ATP by itself interacting with the substrate- or ATP-binding site of JH1. Autoinhibitory domains containing either a phosphorylatable substrate-like sequence or a so-called pseudosubstrate sequence have been identified in many protein kinases, such as protein kinase C (15).
Our results indicated that JH1 is preferentially involved in an interaction with another JH1 domain, supporting the concept that interaction between two kinase domains is important during activation of Jak2. Furthermore, our results suggest that juxtapositioning of Jaks upon receptor dimerization would lead to activation of Jak2, since the “activating” intermolecular JH1-JH1 interaction would be preferred over the weaker “inhibitory” intramolecular JH1-JH2 interaction. The Jak kinases are associated with the cytoplasmic tails of cytokine receptors and are therefore regulated through ligand-induced dimerization of the receptor chains. The results obtained in the IFN-γ-dependent reporter system indicated that activation of ΔJH2 and following downstream signaling was not strictly dependent on cytokine-induced receptor dimerization. This suggests that inhibitory functions possessed by the JH2 domain are important for maintaining Jak2 in a low-activity state in the absence of ligand stimulation.
ACKNOWLEDGMENTS
This study was supported by the Academy of Finland, the Alfred Kordelin Foundation, the Finnish Cancer Organization, the Ida Montin Foundation, the Instrumentarium Science Foundation, the Oskar Öflund Foundation, the Sigrid Juselius Foundation, and the Medical Research Fund of Tampere University Hospital.
We thank Kari Alitalo for comments on the manuscript and Tim Wood, Richard Pine, and James Ihle for kindly providing the reagents specified in Materials and Methods.
REFERENCES
- 1.Andreotti A H, Bunnell S C, Feng S, Berg L J, Schreiber S L. Regulatory intramolecular association in a tyrosine kinase of the Tec family. Nature. 1997;385:93–97. doi: 10.1038/385093a0. [DOI] [PubMed] [Google Scholar]
- 2.Barahmand-Pour F, Meinke A, Groner B, Decker T. Jak2-Stat5 interactions analyzed in yeast. J Biol Chem. 1998;273:12567–12575. doi: 10.1074/jbc.273.20.12567. [DOI] [PubMed] [Google Scholar]
- 3.Briscoe J, Rogers N C, Witthuhn B A, Harpur A G, Wilks A F, Stark G R, Ihle J N, Kerr I M. Kinase-negative mutants of JAK1 can sustain interferon-gamma-inducible gene expression but not an antiviral state. EMBO J. 1996;15:799–809. [PMC free article] [PubMed] [Google Scholar]
- 4.Cacalano N A, Migone T, Bazan F, Hanson E P, Chen M, Candotti F, O'Shea J J, Johnston J A. Autosomal SCID caused by a point mutation in the N-terminus of Jak3: mapping of the Jak3-receptor interaction domain. EMBO J. 1999;18:1549–1558. doi: 10.1093/emboj/18.6.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Candotti F, Oakes S A, Johnston J A, Giliani S, Schumacher R F, Mella P, Fiorini M, Ugazio A G, Badolato R, Notarangelo L D, Bozzi F, Macchi P, Strina D, Vezzoni P, Blaese R M, O'Shea J J, Villa A. Structural and functional basis for JAK3-deficient severe combined immunodeficiency. Blood. 1997;90:3996–4003. [PubMed] [Google Scholar]
- 6.Darnell J E, Jr, Kerr I M, Stark G R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 7.Duhe R J, Farrar W L. Characterization of active and inactive forms of the Jak2 protein-tyrosine kinase produced via the baculovirus expression vector system. J Biol Chem. 1995;270:23084–23089. doi: 10.1074/jbc.270.39.23084. [DOI] [PubMed] [Google Scholar]
- 8.Feng J, Witthuhn B A, Matsuda T, Kohlhuber F, Kerr I M, Ihle J N. Activation of Jak2 catalytic activity requires phosphorylation of Y1007 in the kinase activation loop. Mol Cell Biol. 1997;17:2497–2501. doi: 10.1128/mcb.17.5.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Girault J, Labesse G, Mornon J, Callebaut I. The N-termini of FAK and JAKs contain divergent band 4.1 domains. Trends Biochem Sci. 1999;24:54–57. doi: 10.1016/s0968-0004(98)01331-0. [DOI] [PubMed] [Google Scholar]
- 10.Gupta S, Yan H, Wong L H, Ralph S, Krolewski J, Schindler C. The SH2 domains of Stat1 and Stat2 mediate multiple interactions in the transduction of IFN-α signals. EMBO J. 1996;15:1075–1084. [PMC free article] [PubMed] [Google Scholar]
- 11.Harrison D A, Binari R, Nahreini T, Gilman M, Perrimon N. Activation of Drosophila Janus kinase (JAK) causes hematopoietic neoplasia and developmental defects. EMBO J. 1995;14:2857–2865. doi: 10.1002/j.1460-2075.1995.tb07285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Higgins D G, Thompson J D, Gibson T J. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- 13.Hubbard S R, Mohammadi M, Schlessinger J. Autoregulatory mechanisms in protein-tyrosine kinases. J Biol Chem. 1998;273:11987–11990. doi: 10.1074/jbc.273.20.11987. [DOI] [PubMed] [Google Scholar]
- 14.Ihle J N, Witthuhn B A, Quelle F W, Yamamoto K, Silvennoinen O. Signaling through the hematopoietic cytokine receptors. Annu Rev Immunol. 1995;15:563–593. doi: 10.1146/annurev.iy.13.040195.002101. [DOI] [PubMed] [Google Scholar]
- 15.Kemp B E, Parker M W, Hu S, Tiganis T, House C. Substrate and pseudosubstrate interactions with protein kinases: determinants of specificity. Trends Biochem Sci. 1994;19:440–444. doi: 10.1016/0968-0004(94)90126-0. [DOI] [PubMed] [Google Scholar]
- 16.Klingmuller U, Lorenz U, Cantley L C, Neel B G, Lodish H F. Specific recruitment of SH-PTP1 to the erythropoietin receptor causes inactivation of JAK2 and termination of proliferative signals. Cell. 1995;80:729–738. doi: 10.1016/0092-8674(95)90351-8. [DOI] [PubMed] [Google Scholar]
- 17.Kohlhuber F, Rogers N C, Watling D, Feng J, Guschin D, Briscoe J, Witthuhn B A, Kotenko S V, Pestka S, Stark G R, Ihle J N, Kerr I M. A JAK1/JAK2 chimera can sustain alpha and gamma interferon responses. Mol Cell Biol. 1997;17:695–706. doi: 10.1128/mcb.17.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lacronique V, Boureux A, Valle V D, Poirel H, Quang C T, Mauchauffe M, Berthou C, Lessard M, Berger R, Ghysdael J, Bernard O A. A TEL-JAK2 fusion protein with constitutive kinase activity in human leukemia. Science. 1997;278:1309–1312. doi: 10.1126/science.278.5341.1309. [DOI] [PubMed] [Google Scholar]
- 19.Liu K D, Gaffen S L, Goldsmith M A, Greene W C. Janus kinases in interleukin-2-mediated signaling: JAK1 and JAK3 are differentially regulated by tyrosine phosphorylation. Curr Biol. 1997;7:817–826. doi: 10.1016/s0960-9822(06)00369-1. [DOI] [PubMed] [Google Scholar]
- 20.Luo H, Rose P, Barber D, Hanratty W P, Lee S, Roberts T M, D'Andrea A D, Dearolf C R. Mutation in the Jak kinase JH2 domain hyperactivates Drosophila and mammalian Jak-Stat pathways. Mol Cell Biol. 1997;17:1562–1571. doi: 10.1128/mcb.17.3.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mizuguchi R, Hatakeyama M. Conditional activation of Janus kinase (JAK) confers factor independence upon interleukin-3-dependent cells. J Biol Chem. 1998;273:32297–32303. doi: 10.1074/jbc.273.48.32297. [DOI] [PubMed] [Google Scholar]
- 22.Muller M, Briscoe J, Laxton C, Guschin D, Ziemiecki A, Silvennoinen O, Harpur A G, Barbieri G, Witthuhn B A, Schindler C, et al. The protein tyrosine Jak1 complements defects in interferon-alpha/beta and -gamma signal transduction. Nature. 1993;366:129–135. doi: 10.1038/366129a0. [DOI] [PubMed] [Google Scholar]
- 23.Neubauer H, Cumano A, Muller M, Wu H, Huffstadt U, Pfeffer K. Jak2 deficiency defines an essential developmental checkpoint in definitive hematopoiesis. Cell. 1998;93:397–409. doi: 10.1016/s0092-8674(00)81168-x. [DOI] [PubMed] [Google Scholar]
- 24.Parganas E, Wang D, Stravopodis D, Topham D J, Marine J C, Teglund S, Vanin E F, Bodner S, Colamonici O R, van Deursen J M, Grosveld G, Ihle J N. Jak2 is essential for signaling through a variety of cytokine receptors. Cell. 1998;93:385–395. doi: 10.1016/s0092-8674(00)81167-8. [DOI] [PubMed] [Google Scholar]
- 25.Pine R, Canova A, Schindler C. Tyrosine phosphorylated p91 binds to a single element in the ISGF2/IRF-1 promoter to mediate induction by IFN alpha and IFN gamma, and is likely to autoregulate the p91 gene. EMBO J. 1994;13:158–167. doi: 10.1002/j.1460-2075.1994.tb06245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saharinen P, Ekman N, Sarvas K, Parker P, Alitalo K, Silvennoinen O. The Bmx tyrosine kinase induces activation of the Stat signaling pathway, which is specifically inhibited by protein kinase Cδ. Blood. 1997;90:4341–4353. [PubMed] [Google Scholar]
- 27.Sakai I, Kraft A S. The kinase domain of Jak2 mediates induction of Bcl-2 and delays cell death in hematopoietic cells. J Biol Chem. 1997;272:12350–12358. doi: 10.1074/jbc.272.19.12350. [DOI] [PubMed] [Google Scholar]
- 28.Schultz J, Milpetz F, Bork P, Ponting C P. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sicheri F, Moarefi I, Kuriyan J. Crystal structure of the Src family tyrosine kinase Hck. Nature. 1997;385:602–609. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- 30.Silvennoinen O, Witthuhn B A, Quelle F W, Cleveland J L, Yi T, Ihle J N. Structure of the murine Jak2 protein-tyrosine kinase and its role in interleukin 3 signal transduction. Proc Natl Acad Sci USA. 1993;90:8429–8435. doi: 10.1073/pnas.90.18.8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Velazquez L, Mogensen K E, Barbieri G, Fellous M, Uze G, Pellegrini S. Distinct domains of the protein tyrosine kinase Tyk2 required for binding of interferon-α/β and for signal transduction. J Biol Chem. 1995;270:3327–3334. doi: 10.1074/jbc.270.7.3327. [DOI] [PubMed] [Google Scholar]
- 32.Watling D, Guschin D, Muller M, Silvennoinen O, Witthuhn B A, Quelle F W, Rogers N C, Schindler C, Stark G R, Ihle J N, et al. Complementation by the protein tyrosine kinase JAK2 of a mutant cell line defective in the interferon-gamma signal transduction pathway. Nature. 1993;366:166–170. doi: 10.1038/366166a0. [DOI] [PubMed] [Google Scholar]
- 33.Wilks A F, Harpur A G, Kurban R R, Ralph S J, Zurcher G, Ziemiecki A. Two novel protein-tyrosine kinases, each with a second phosphotransferase-related catalytic domain, define a new class of protein kinase. Mol Cell Biol. 1991;11:2057–2065. doi: 10.1128/mcb.11.4.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Witthuhn B A, Quelle F W, Silvennoinen O, Yi T, Tang B, Miura O, Ihle J N. Jak2 associates with the erythropoietin receptor and is tyrosine phosphorylated and activated following stimulation with erythropoietin. Cell. 1993;74:227–236. doi: 10.1016/0092-8674(93)90414-l. [DOI] [PubMed] [Google Scholar]
- 35.Xu W, Harrison S C, Eck M J. Three-dimensional structure of the tyrosine kinase c-Src. Nature. 1997;385:595–602. doi: 10.1038/385595a0. [DOI] [PubMed] [Google Scholar]
- 36.Yasukawa H, Misawa H, Sakamoto H, Masuhara H, Sasaki A, Wakioka T, Ohtsuka S, Imaizumi T, Matsuda T, Ihle J N, Yoshimura A. The JAK-binding protein JAB inhibits Janus tyrosine kinase activity through binding in the activation loop. EMBO J. 1999;18:1309–1320. doi: 10.1093/emboj/18.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yi T, Zhang J, Miura O, Ihle J N. Hematopoietic cell phosphatase associates with erythropoietin (Epo) receptor after Epo-induced receptor tyrosine phosphorylation: identification of potential binding sites. Blood. 1995;85:87–95. [PubMed] [Google Scholar]
- 38.You M, Yu D-H, Feng G-S. Shp-2 tyrosine phosphatase functions as a negative regulator of the interferon-stimulated Jak/STAT pathway. Mol Cell Biol. 1999;19:2416–2424. doi: 10.1128/mcb.19.3.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao Y, Wagner F, Frank S J, Kraft A S. The amino-terminal portion of the Jak2 protein kinase is necessary for binding and phosphorylation of the granulocyte-macrophage colony-stimulating factor receptor β chain. J Biol Chem. 1995;270:13814–13818. doi: 10.1074/jbc.270.23.13814. [DOI] [PubMed] [Google Scholar]
- 40.Zhou Y J, Hanson E P, Chen Y Q, Magnuson K, Chen M, Swann P G, Wange R L, Changelian P S, O'Shea J J. Distinct tyrosine phosphorylation sites in JAK3 kinase domain positively and negatively regulate its enzymatic activity. Proc Natl Acad Sci USA. 1997;94:13850–13855. doi: 10.1073/pnas.94.25.13850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ziemiecki A, Harpur A G, Wilks A F. JAK protein tyrosine kinases: their role in cytokine signaling. Trends Cell Biol. 1994;4:207–212. doi: 10.1016/0962-8924(94)90143-0. [DOI] [PubMed] [Google Scholar]