Abstract
Background
The aim of this study was to quantitatively analysis the diagnostic performance of adenomatous polyposis coli (APC) gene promoter methylation in serum or sputum/bronchoalveolar lavage fluid (BLAF) as a biomarker for lung cancer identification through pooling of open published data.
Methods
The relevant electronic MEDLINE, EMBASE, Ovid, web of science and CNKI databases were systematically searched to identify the studies related to APC gene promoter methylation for lung cancer diagnosis. Data of true positive (tp), false positive (fp), false negative (fn) and true negative (tn) were extracted from the publications included in the study. The pooled diagnostic sensitivity, specificity and area under summary receiver operating characteristic (SROC) curve (AUC‐SROC) of APC gene promoter methylation were calculated. Publication bias was evaluated by Begg's funnel plot and Egger's line regression test.
Results
Fourteen studies associated with APC gene promoter methylation and lung cancer were identified in the databases and finally included in the meta‐analysis. The data was pooled using a random effect model due to significant statistical heterogeneity across the 14 studies (p < 0.05). Using the APC gene promoter methylation as a reference for lung cancer identification, the pooled diagnostic sensitivity and specificity were 0.43 (95% CI: 0.40–0.45), and 0.92 (95% CI: 0.90–0.95), respectively with combined diagnostic positive likelihood ratio (+LR) and negative likelihood ratio (−LR) of 7.15 (95% CI: 3.62–14.12) and 0.63 (95% CI: 0.57–0.71). The pooled diagnostic odds ratio (DOR) and AUC‐SROC of APC gene promoter methylation for lung cancer diagnosis were 9.84 (95% CI: 5.77–16.79) and 0.7, respectively. The Begg's funnel plot and Egger's line regression test both indicated statistical publication bias (t = 5.40, p < 0.05).
Conclusions
APC gene promoter methylation in serum or sputum/BLAF is a potential biomarker for lung cancer diagnosis with high specificity. However, due to its low sensitivity, it may not be suitable for lung cancer screening in the general population.
Keywords: adenomatous polyposis coli, lung cancer, meta‐analysis, methylation, promoter
The summary receiver operating characteristic (SROC) curve of APC gene promoter methylation for lung cancer diagnosis were 0.7 with the diagnostic sensitivity specificity of 0.43 (95% CI: 0.40–0.45) and 0.92 (95% CI: 0.90–0.95) respectively.

INTRODUCTION
Lung carcinoma including non‐small cell lung cancer (NSCLC) and small cell lung cancer (SCLC) is the most common malignant tumor and leading cause of cancer‐related death wordwide. In 2018 it was reported that there was 2 100 000 new lung cancer cases diagnosed and 1800,000 deaths from lung carcinoma globally, accounting for one fifth of all cancer‐related deaths. 1 An epidemiological study indicated that there will be 235 760 lung cancer new cases and 131 880 death of lung cancer in the USA in the year 2021. 2 In China, 787 000 new lung cancer cases and 631 000 death were recorded in 2015. 3 Although lung cancer is clinically the most common carcinoma, the prognosis of this disease is still poor after several decades of surgical progress, radiation and chemotherapy, mainly due to its advanced stage in patients when first diagnosis. Therefore, lung cancer screening and early diagnosis are essential for improving the prognosis of this disease.
Methylation of tumor suppressor genes in promoter regions are common in cancer tissue, which is an important mechanism of tumor suppressor gene inactivation. 4 , 5 Recently, studies have also identified that methylation of tumor suppressor genes of lung cancer patients is also common in body fluid specimens such as serum, sputum or BLAF. 6 , 7 The APC gene encodes a tumor suppressor protein involved in cell migration and adhesion, transcriptional activation, and apoptosis. 8 , 9 Several studies have indicated that the APC gene promoter region is methylated in body fluid specimens such as serum or sputum compared to controls. 10 , 11 However, the diagnostic performance of APC gene promoter methylation as a biomarker for lung cancer identification is unclear. Therefore, we performed this meta‐analysis to further evaluate the feasibility of detection of APC gene promoter methylation in body fluid as a biomarker for lung cancer diagnosis through pooling of open published data.
METHODS
Electronic database searches
MEDLINE, EMBASE, Ovid, web of science and CNKI databases were systematically searched to identify the studies related to APC gene promoter methylation for lung cancer diagnosis. The languages were restricted to English and Chinese. “APC”, “adenomatous polyposis coli”, “non‐small cell lung cancer”, “lung cancer”, “lung neoplasm”, “methylation”, “hypermethylation” were applied for free text word in the process of the publication search. The references of the included studies were also reviewed in order to identify potentially suitable publications.
Inclusion and exclusion criteria
Inclusion and exclusion criteria were restricted to language, study design, patients, methylation detection methods and results. Inclusion criteria: studies published in English or Chinese; study design limited to case–control, cohort or observation study; patients restricted to cases of confirmed diagnosis of lung cancer; methylation detection method was methylation specific PCR (MSP); the results of the originally included studies should provide enough data to calculate the true positive (tp), false positive (fp), false negative (fn) and true negative (tn). Exclusion criteria: case reports, literature review or meta‐analysis were excluded; lung cancer cases included in the original study was not confirmed by pathology or cytology; studies using other methods not MSP for APC gene promoter methylation detection were excluded. Studies which did not provide enough data that could be used for tp, fp, fp and tn calculations were eliminated.
Data extraction
Two reviewers (Fang Liu & He Huang) independently extracted the information and cross checked the data which included first and corresponding authors, time of publication, body fluid specimen type, methylation detection methods, sample size, age of the original cases, histology type, tp, fp, fn and tn of original study.
Statistical analysis
The data was pooled by Meta‐DiSc1.4 (http://www.biomedsearch.com) and STATA12.0 (http://www.stata.com; Stata Corporation) statistical software. Before pooling the diagnostic parameters, the statistical heterogeneity across the 14 original studies was evaluated by chi‐square test and demonstrated by I2. If p < 0.05 (chi‐square test) or I2 > 50%, the data was pooled by random effect model, otherwise by fixed effect model. Publication bias was evaluated using Begg's funnel plot and Egger line regression test. Two‐tailed p < 0.05 was considered statistically significant.
RESULTS
General characteristics
By exclusion of the inappropriate studies, 14 publications were finally included in the meta‐analysis (Figure 1). APC gene promoter methylation was detected by MSP array in all 14 publications. The main features of the 14 original studies are demonstrated in Table 1.
FIGURE 1.
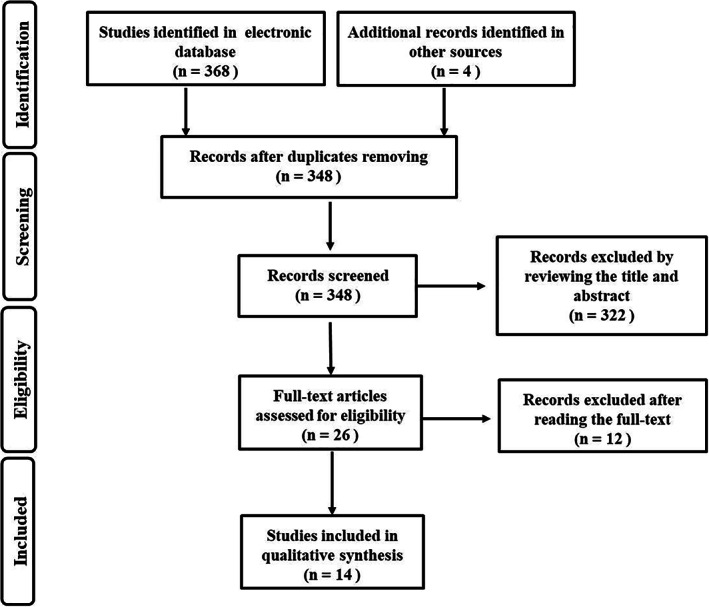
Studies screened and included after searching the relevant databases
TABLE 1.
General characteristics of included studies
| Author | Time | Case/control | Cancer | Control | TNM (I, II/)III, IV | Specimen | Histology (Ad/Sq/other) | Age (case/control) | Methods |
|---|---|---|---|---|---|---|---|---|---|
| (M+/M‐) | (M+/M‐) | ||||||||
| Ma et al. 12 | 2016 | 254/150 | 65/180 | 4/146 | 84/161 | Serum | 108/100/37 | NA | MSP |
| Xie et al. 13 | 2008 | 58/31 | 11/47 | 0/31 | NA | Serum | NA | NA | MSP |
| Zhu et al. 14 | 2015 | 70/40 | 18/52 | 1/39 | 24/46 | Serum | 39/30/11 | 63.96/59.08 | MSP |
| Kang et al. 15 | 2011 | 47/24 | 14/33 | 0/24 | NA | Sputum | 12/29/6 | 52.9/59.7 | MSP |
| Zhong et al. 16 | 2018 | 50/50 | 19/31 | 0/50 | NA | Sputum | NA | NA | MSP |
| Luo et al. 17 | 2018 | 79/40 | 28/51 | 1/39 | 27/52 | Serum | 40/37/0 | 63.27/57.23 | MSP |
| Ali et al. 10 | 2017 | 160/70 | 84/76 | 10/60 | 0/160 | Serum | 22/48/90 | 57.4/46.6 | MSP |
| Zhang et al. 18 | 2011 | 110/50 | 52/58 | 5/45 | 110/0 | Serum | NA | NA | MSP |
| Pan et al. 19 | 2009 | 40/31 | 19/21 | 0/31 | NA | Serum | 12/9/19 | 53.0/48.0 | MSP |
| Usadel et al. 20 | 2002 | 89/50 | 42/45 | 0/50 | 76/23 | Serum | 35/4717 | NA | MSP |
| Grote et al. 11 | 2004 | 155/67 | 110/45 | 28/39 | NA | BALF | 40/47/68 | 64/65 | MSP |
| Rykova et al. 21 | 2004 | 9/16 | 3/6 | 0/16 | NA | Serum | NA | NA | MSP |
| Liu et al. 22 | 2017 | 120/46 | 38/82 | 5/41 | 53/67 | Serum | 52/36/32 | 56/ | MSP |
| Wang & Song 23 | 2020 | 85/15 | 57/28 | 1/14 | NA | BALF | NA | NA | MSP |
Abbreviations: BALF, bronchoalveolar lavage fluid; NA, not available.
Statistical heterogeneity
Chi‐square test demonstrated significant statistical heterogeneity across the 14 studies in the effect size of sensitivity, specificity, +LR, −LR and DOR (Pall < 0.05). The heterogentity test results indicated that the data should be pooled in a random effect model.
Meta‐analysis
Pooled diagnostic sensitivity and specificity
Using the APC gene promoter methylation as a reference for lung cancer identification, the pooled diagnostic sensitivity and specificity were 0.43 (95% CI: 0.40–0.45) (Figure 2) and 0.92 (95% CI: 0.90–0.95) (Figure 3), respectively by random effect model.
FIGURE 2.
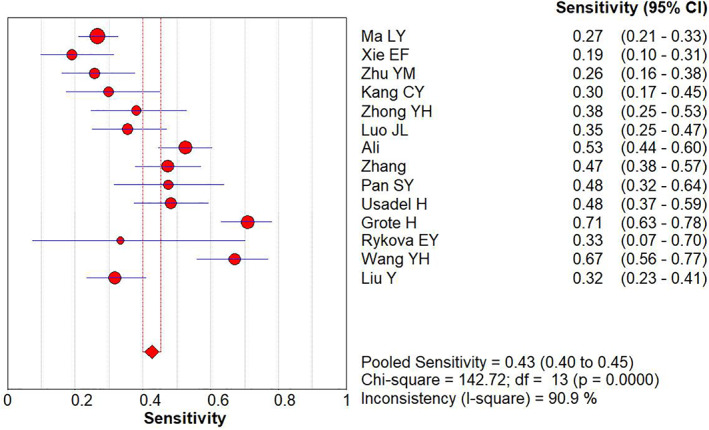
Forest plot of sensitivity for APC gene promoter methylation in the diagnosis of lung cancer
FIGURE 3.
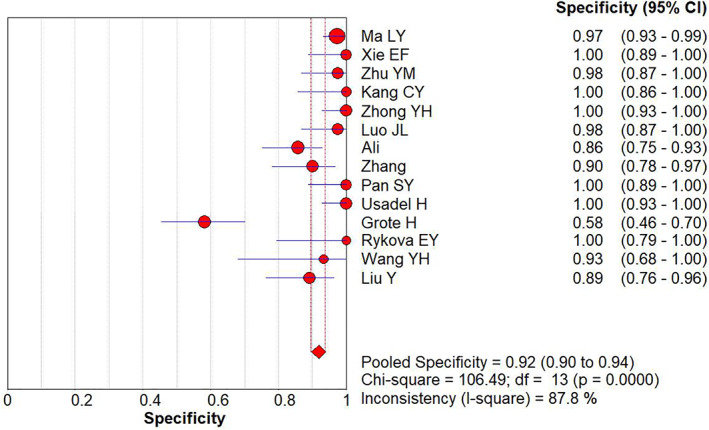
Forest plot of specificity for APC gene promoter methylation in the diagnosis of lung cancer
Pooled diagnostic +LR and –LR
The combined diagnostic +LR and –LR for APC gene promoter methylation as a reference for lung cancer identification were 7.15 (95% CI: 3.62–14.12) (Figure 4) and 0.63 (95% CI: 0.57–0.71) (Figure 5), respectively by random effect model.
FIGURE 4.
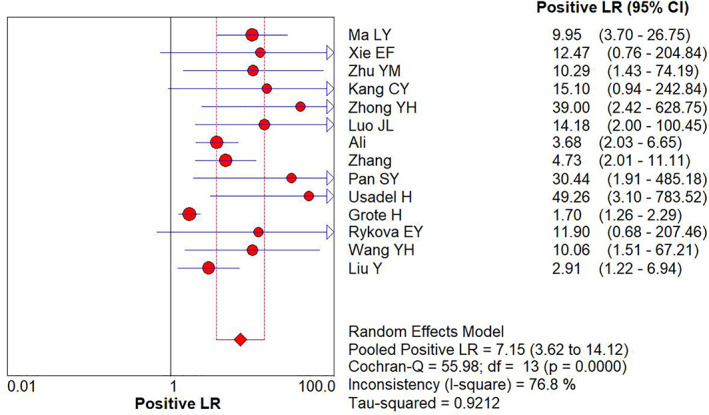
Forest plot of +LR for APC gene promoter methylation in the diagnosis of lung cancer
FIGURE 5.
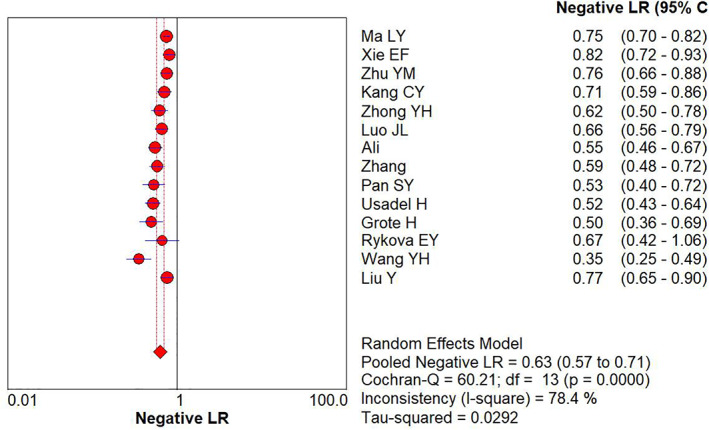
Forest plot of ‐LR for APC gene promoter methylation in the diagnosis of lung cancer
Pooled DOR and SROC
The pooled diagnostic odds ratio (DOR) and summary receiver operating characteristic (SROC) curve of APC gene promoter methylation for lung cancer diagnosis were 9.84 (95% CI: 5.77–16.79) (Figure 6) and 0.7 (Figure 7), respectively.
FIGURE 6.
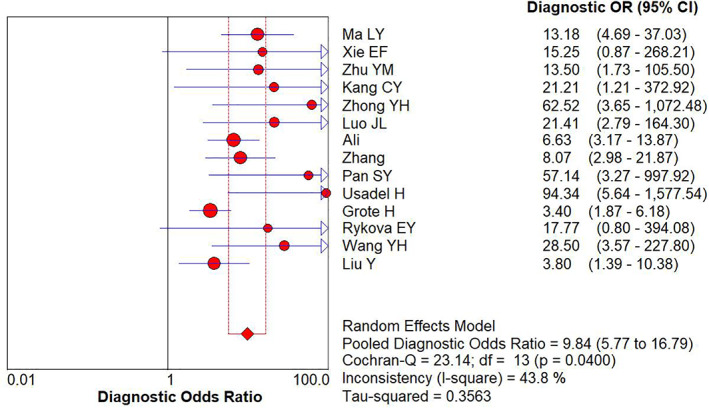
Forest plot of DOR for APC gene promoter methylation in the diagnosis of lung cancer
FIGURE 7.
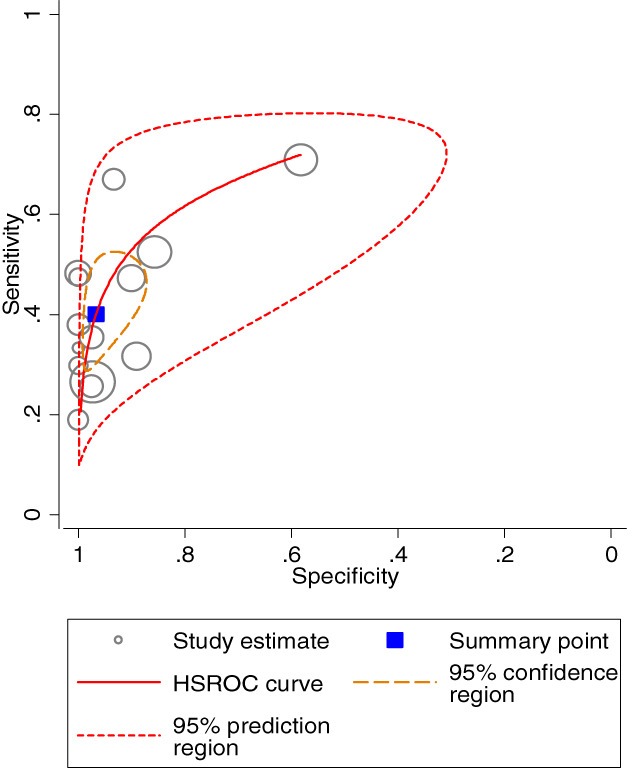
Summary receiver operating characteristic (SROC) cure of APC gene promoter methylation for lung cancer diagnosis
Subgroup analysis
According to specimen type, the diagnostic efficacy was evaluated in the serum or sputum/BALF subgroups. The detailed diagnostic performance is shown in Table 2.
TABLE 2.
Subgroup analysis of APC gene promoter methylation for lung cancer
| Group | SEN | SPE | +LR | –LR | DOR | AUC |
|---|---|---|---|---|---|---|
| Serum | 0.40 (0.37–0.43) | 0.96 (0.93–0.97) | 6.81 (4.22–10.98) | 0.62 (0.54–0.71) | 10.82 (6.91–16.95) | 0.731 |
| BLAF/ Sputum | 0.49 (0.43–0.54) | 0.82 (0.76–0.88) | 3.87 (1.25–12.01) | 0.66 (0.56–0.79) | 5.52 (2.12–14.38) | 0.701 |
Abbreviations: AUC, area under the curve; DOR, diagnostic odds ratio; +LR, positive likelihood ratio; −LR, negative likelihood ratio; SEN, sensitivity; SPE, specificity.
Publication bias
The Begg's funnel plot was obviously left–right asymmetric especially at the bottom which indicates a significant publication bias (Figure 8). The Egger's line regression test also indicated statistical publication bias (t = 5.40, p < 0.05).
FIGURE 8.
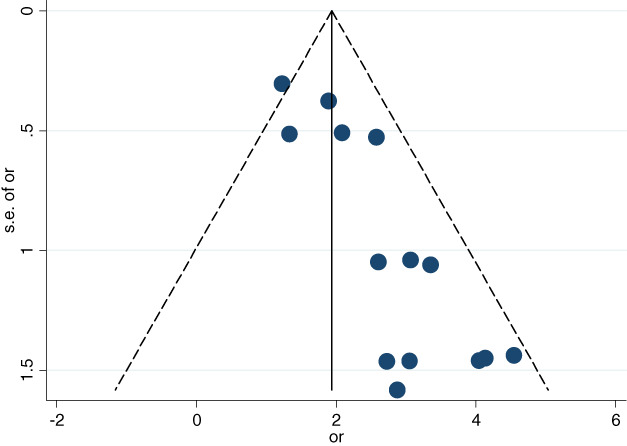
Begg's funnel plot was used for the evaluation of publication bias
DISCUSSION
A total of 14 studies relevant to APC gene promoter methylation and lung cancer were included in this meta‐analysis. The pooled results showed diagnostic sensitivity and specificity were 0.43 (95% CI: 0.40–0.45) and 0.92 (95% CI: 0.90–0.95), respectively. The pooled DOR and SROC curves of APC gene promoter methylation for lung cancer diagnosis were 9.84 (95% CI: 5.77–16.79) and 0.7, respectively. The pooled results indicate that APC gene promoter methylation in serum or sputum/BLAF is a potential biomarker for lung cancer diagnosis with high specificity.
APC gene promoter methylation as a potential biomarker for cancer diagnosis by meta‐analysis has previously been evaluated by studies on breast, colorectal, bladder, prostate and lung cancers. 24 , 25 , 26 , 27 Qian et al.27 performed a meta‐analysis relevant to APC gene promoter aberrant methylation in serum as a biomarker for breast cancer diagnosis. The authors found that APC gene promoter methylation in serum was not suitable for breast cancer screening due to low diagnostic sensitivity, but could be used as potential serological marker for breast cancer confirmation. The low diagnostic sensitivity and high specificity was also identified in our present meta‐analysis for APC methylation in serum or sputum/BLAF body fluid specimen. The low sensitivity limits its clinical value in lung cancer screening. Zhang et al.28 performed a meta‐analysis to evaluate the association between APC promoter methylation and lung cancer. A total of 12 case–control studies from 10 publications were included, which involved 1190 cases and 606 controls. A random effect model was used to merge the data, and the odds ratio (OR) was 9.84 (95% CI: 5.03–19. 27, p < 0.05), which indicates that the risk of lung cancer in a population with APC promoter methylation was 9.84 times higher than that in the general population. The authors indicated that APC promoter methylation might be a potential biomarker for the diagnosis of lung cancer. This conclusion was in accordance with our findings. In our meta‐analysis, we included 14 publications and calculated the exact diagnostic parameter which clearly demonstrates the diagnostic performance of APC promoter methylation detected in serum or sputum/BLAF. Zhang et al. only discussed the methylation frequency in the body fluid of lung cancer cases and controls but didn't provide the diagnostic performance. Compared to the study by Zhang et al., our meta‐analysis provides more information on APC promoter methylation as biomarkers in lung cancer detection and has a more practical clinical application.
The present study also had obvious limitations. (i) Diagnostic performance was pooled by a random effect model due to significant statistical heterogeneity. (ii) A significant publication bias was also identified. (iii) Due to low diagnostic sensitivity, APC promoter methylation may not be suitable for lung cancer screening in the general population. (iv) Clinical heterogeneity such as mixed clinical stages included in the original study may affect the results and decrease the power of the conclusions. (v) For the BALF subgroup, only two studies were included and the results of this subgroup were unstable. In conclusion, APC gene promoter methylation is common in body fluid of serum and sputum/BALF and may be a potential biomarker for lung cancer diagnosis, but may not suitable for lung cancer screening in the general population. Therefore, more relevant studies which meet the requirements should be included to further evaluate the value of APC gene methylation in the diagnosis of lung cancer, so as to provide more sufficient and powerful evidence‐based medical results. Comprehensive diagnosis should be based on a combination of multiple diagnostic approaches such as imaging and cytology to improve the diagnostic accuracy.
CONFLICT OF INTEREST
The authors confirm that there is no conflict of interest.
ACKNOWLEDGEMENTS
This work was supported by Medical and health science of Zhejiang Province (2021KY437).
Liu F, Lu X, Zhou X, Huang H. APC gene promoter methylation as a potential biomarker for lung cancer diagnosis: A meta‐analysis. Thorac Cancer. 2021;12:2907–2913. 10.1111/1759-7714.14151
REFERENCES
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- 2. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33. [DOI] [PubMed] [Google Scholar]
- 3. Zheng RS, Sun KX, Zhang SW, Zeng HM, Zou XN, Chen R, et al. [Report of cancer epidemiology in China, 2015]. Zhonghua Zhong Liu Za Zhi. 2019;41:19–28. [DOI] [PubMed] [Google Scholar]
- 4. Hua F, Fang N, Li X, Zhu S, Zhang W, Gu J. A meta‐analysis of the relationship between RARβ gene promoter methylation and non‐small cell lung cancer. PLoS One. 2014;9:e96163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gu J, Wen Y, Zhu S, Hua F, Zhao H, Xu H, et al. Association between P(16INK4a) promoter methylation and non‐small cell lung cancer: a meta‐analysis. PLoS One. 2013;8:e60107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lissa D, Robles AI. Sputum‐based DNA methylation biomarkers to guide lung cancer screening decisions. J Thorac Dis. 2017;9:4308–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Leng S, Wu G, Klinge DM, Thomas CL, Casas E, Picchi MA, et al. Gene methylation biomarkers in sputum as a classifier for lung cancer risk. Oncotarget. 2017;8:63978–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Benchabane H, Ahmed Y. The adenomatous polyposis coli tumor suppressor and Wnt signaling in the regulation of apoptosis. Adv Exp Med Biol. 2009;656:75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huang X, Guo B. Adenomatous polyposis coli determines sensitivity to histone deacetylase inhibitor‐induced apoptosis in colon cancer cells. Cancer Res. 2006;66:9245–51. [DOI] [PubMed] [Google Scholar]
- 10. Ali A, Kumar S, Kakaria VK, Mohan A, Luthra K, Upadhyay AD, et al. Detection of promoter DNA methylation of APC, DAPK, and GSTP1 genes in tissue biopsy and matched serum of advanced‐stage lung cancer patients. Cancer Invest. 2017;35:423–30. [DOI] [PubMed] [Google Scholar]
- 11. Grote HJ, Schmiemann V, Kiel S, Böcking A, Kappes R, Gabbert HE, et al. Aberrant methylation of the adenomatous polyposis coli promoter 1A in bronchial aspirates from patients with suspected lung cancer. Int J Cancer. 2004;110:751–5. [DOI] [PubMed] [Google Scholar]
- 12. Ma LY, Liang J, Zhou JL. Evaluation of the combination of APC and DCC gene methylation in the early diagnosis of lung cancer. J Mod Lab Med. 2016;1:17–20. [Google Scholar]
- 13. Xie EF, Pan SY, Gao L, Yu TF, Chen D, Zhang LX, et al. Detection of APC gene promoter methylation in plasma of early lung cancer. Chinese J Clin Lab Sci. 2008;26:121–2. [Google Scholar]
- 14. Zhu YM, Li J, Yu LC, Zhu LH, Chen P. Clinical utility of plasma APC and DCC genes promoter hypermethylation in the early diagnosis of lung cancer. J Jiangsu Univ. 2015;25:62–7. [Google Scholar]
- 15. Kang CY, Wang DD, Sha SP, Xiao H. Effect of promoter hypermethylation of FHIT,P16,MGMT,RASSF1A and APC genes in sputum specimens on diagnosis of lung cancer. Chinese J Clin Exp Pathol. 2011;27:869–73. [Google Scholar]
- 16. Zhong YH, Li Y, Lu BT. Correlation between methylation status of APC, p16,CDH13 and RASSF1A and lung cancer. Label Immuno Clin Med. 2018;25:1214–7. +1222. [Google Scholar]
- 17. Luo JL, Li K, Feng X. Value of p16, DAPK and APC gene methylation in early diagnosis of lung cancer. J Minim Invas Med. 2018;13:12–6. [Google Scholar]
- 18. Zhang Y, Wang R, Song H, Huang G, Yi J, Zheng Y, et al. Methylation of multiple genes as a candidate biomarker in non‐small cell lung cancer. Cancer Lett. 2011;303:21–8. [DOI] [PubMed] [Google Scholar]
- 19. Pan SY, Xie EF, Shu YQ, Gao L, Zhang LX, Chen D, et al. [Methylation quantification of adenomatous polyposis coli (APC) gene promoter in plasma of lung cancer patients]. Ai Zheng. 2009;28:384–9. [PubMed] [Google Scholar]
- 20. Usadel H, Brabender J, Danenberg KD, Jerónimo C, Harden S, Engles J, et al. Quantitative adenomatous polyposis coli promoter methylation analysis in tumor tissue, serum, and plasma DNA of patients with lung cancer. Cancer Res. 2002;62:371–5. [PubMed] [Google Scholar]
- 21. Rykova EY, Skvortsova TE, Laktionov PP, Tamkovich SN, Bryzgunova OE, Starikov AV, et al. Investigation of tumor‐derived extracellular DNA in blood of cancer patients by methylation‐specific PCR. Nucleosides Nucleotides Nucleic Acids. 2004;23:855–9. [DOI] [PubMed] [Google Scholar]
- 22. Liu Y, Wang YY, Jia WQ, Ni R, Zhang H, Wang J, et al. Determination of p16, FHIT and APC gene methylation in serum from patients with lung cancer. J Zhengzhou Univ. 2017;52:51–4. [Google Scholar]
- 23. Wang YH, Song YC. The value of combined detection p16,MGMT, FHIT and APC gene methylation in alveolar lavage fluid of patients with pulmonary nodules for lung carcinoma early screening. Zhejiang Med Educ. 2020;19:57–60. [Google Scholar]
- 24. Wang Y, Fan C, Yu J, Wang X. APC methylation predicts biochemical recurrence of patients with prostate cancer: a meta‐analysis. Int J Clin Exp Med. 2015;8:15575–80. [PMC free article] [PubMed] [Google Scholar]
- 25. Bai ZJ, Liu Q, Wang XS, Liu WY. APC promoter methylation is correlated with development and progression of bladder cancer, but not linked to overall survival: a meta‐analysis. Neoplasma. 2019;66:470–80. [DOI] [PubMed] [Google Scholar]
- 26. Zhou X, Jiao D, Dou M, Zhang W, Hua H, Chen J, et al. Association of APC gene promoter methylation and the risk of gastric cancer: a meta‐analysis and bioinformatics study. Med (Baltimore). 2020;99:e19828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Qian X, Ruan L. APC gene promoter aberrant methylation in serum as a biomarker for breast cancer diagnosis: a meta‐analysis. Thorac Cancer. 2018;9:284–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang YQ, Xu N, Zheng P, Wang Q. Association between APC promoter methylation and lung cancer: a meta‐analysis. J Environ Hyg. 2020;10:470–8. [Google Scholar]


