Abstract
Src-like adapter protein (Slap) is a recently identified protein that negatively regulates mitogenesis in murine fibroblasts (S. Roche, G. Alonso, A. Kazlausakas, V. M. Dixit, S. A. Courtneidge, and A. Pandey, Curr. Biol. 8:975–978, 1998) and comprises an SH3 and SH2 domain with striking identity to the corresponding Src domains. In light of this, we sought to investigate whether Slap could be an antagonist of all Src functions. Like Src, Slap was found to be myristylated in vivo and largely colocalized with Src when coexpressed in Cos7 cells. Microinjection of a Slap-expressing construct into quiescent NIH 3T3 cells inhibited platelet-derived growth factor (PDGF)-induced DNA synthesis, and the inhibition was rescued by the transcription factor c-Myc but not by c-Jun/c-Fos expression. Fyn (or Src) overexpression overrides the G1/S block induced by both SrcK− and a Slap mutant with a deletion of its C terminus (SlapΔC), but not the block induced by Slap or SlapΔSH3, implying that the C terminus is a noncompetitive inhibitor of Src mitogenic function. Furthermore, a chimeric adapter comprising SrcΔK fused to the Slap C terminus (Src/SlapC) also inhibited Src function during the PDGF response in a noncompetitive manner, as Src coexpression could not rescue PDGF signaling. Slap, however, did not reverse deregulated Src-induced cell transformation, as it was unable to inhibit depolymerization of actin stress fibers while still being able to inhibit SrcY527F-induced DNA synthesis. This was attributed to a distinct Slap SH3 binding specificity, since the chimeric Slap/SrcSH3 molecule, in which the Slap SH3 was replaced by the Src SH3 sequence, substantially restored stress fiber formation. Indeed, three amino acids important for ligand binding in Src SH3 were replaced in the Slap SH3 sequence; Slap SH3 did not bind to the Src SH3 partners p85α, Shc, and Sam68 in vitro, and the chimeric tyrosine kinase Slap/SrcK, composed of SlapΔC fused to the SH2 linker kinase sequence of Src, was not regulated in vivo. Furthermore, the Src SH3 domain is required for signaling during mitogenesis and since Slap/SrcK behaved as a dominant negative in the PDGF mitogenic response when microinjected into quiescent fibroblasts. We conclude that Slap is a negative regulator of Src during mitogenesis involving both the SH2 and the C terminus domains in a noncompetitive manner, but it does not regulate all Src function due to specific SH3 binding substrates.
Growth factors mediate their mitogenic responses via association with cell surface receptors with intrinsic tyrosine kinase activity. Ligand binding induces receptor dimerization and catalytic activation. As a consequence, activated receptors transphosphorylate tyrosine residues within their cytoplasmic domains, creating binding sites for SH2-containing proteins and thereby initiating a signaling cascade (37). The first line of proteins discovered to be involved in this process possessed enzymatic activity: for example, the platelet-derived growth factor beta (PDGFβ) receptor associates with the lipid kinase phosphoinositide 3-kinase, the lipid phosphatase phospholipase Cγ, members of the protein tyrosine kinase family of Src, and the protein phosphatase Shp2, all of which are required for efficient signaling (30, 31, 36). We have been particularly interested in the function of the Src family of tyrosine kinases during growth factor responses. These enzymes comprise eight members in mammals among Src, Fyn, and Yes and are widely expressed. They are associated with the membrane via myristylation at the N terminus and also include an SH3 and SH2 domain, a catalytic sequence, and a short C-terminal sequence involved in enzymatic regulation (26).
Using a microinjection approach, we have previously shown that Src, Fyn, and Yes were required for the mitogenic response in fibroblasts as induced by most growth factors (29, 36). We believe that their function is to activate a signaling cascade that does not involve the small GTP-binding protein Ras but ultimately culminates in expression of the transcription factor c-Myc, which is necessary for DNA synthesis (1). Recently, null mutants for the src, fyn, and yes genes were generated in mice and displayed an embryonic-lethal phenotype, leading the authors to conclude that these kinases serve vital and redundant functions during embryogenesis (14). Using embryo fibroblasts derived from these mice strains that were immortalized with the simian virus 40 large T antigen, the authors demonstrated that Src kinases appeared dispensable for PDGF mitogenic signaling but essential for integrin signaling. While in apparent contradiction with our observations, one could speculate that the simian virus 40 large T somehow overrides the requirement of Src kinases for PDGF signaling, and indeed, recent experiments in our laboratory favor this hypothesis (unpublished observations).
In addition to cytoplasmic enzymes, growth factor receptors also use adapter molecules that are cytoplasmic proteins that have no intrinsic catalytic activity but possess homology domains necessary for protein interaction and signaling events (37). For example, the PDGFβ receptor associates with Grb2, Shc, and Nck (37), which are all necessary for DNA synthesis (31). Early adapters identified were positive regulators of mitogenesis, with some having been transduced by retroviruses and being oncogenic when overexpressed in fibroblasts (37). They are thought to signal by interacting with their effectors and activating them via plasma membrane translocation. The adapter Grb2, for example, is constitutively associated with SOS, the GTP exchange factor for Ras. In quiescent cells, the complex is localized in the cytosol, preventing SOS from activating membrane-bound Ras. However, during growth factor stimulation, the Grb2-SOS complex becomes membrane associated with the activated receptor and enables signaling to proceed (37). More recently, several adapters have been identified that negatively regulate signaling (G. Manes, P. Bello, and S. Roche, submitted for publication). One of the first examples was Cis, an adapter of 27 kDa that contains an SH2 domain with high homology to the transcription factor Stat. When overexpressed, Cis inhibits cell responses induced by various cytokines, probably by preventing Stat transcriptional activity in vivo (41). Other adapters have now been identified, including Cis homologues called SOCS (11), APS (39), Cbl (20), members of the Smad family (10), and FRNK, a dominant negative splice variant of the focal adhesion kinase Fak (24). Most negatively regulate a specific pathway by terminating the signaling cascade. For example, Cis expression is induced following cytokine stimulation, leading to downregulation of Stat activity (40).
We recently identified a new member of this family called Slap (25) that was originally cloned by a yeast two-hybrid genetic screen using the Eck tyrosine kinase receptor cytoplasmic domain as a bait (21). Slap is a 34-kDa protein that contains a short N terminus followed by SH3 and SH2 domains and a unique C terminus of about 100 amino acids. Interestingly, a high homology with the SH2 and SH3 domains of the Src family was observed (about 50% identity), hence the name. It appears to be ubiquitously expressed, as its mRNA was detected in most tissues tested (21). In a previous report, we investigated the function of Slap during mitogenesis and found that it negatively regulates growth factor responses (25). In contrast to other adapters of the subfamily, Slap protein levels increased only moderately during cell cycle progression (unpublished observations), suggesting that it probably does not act by a molecular switching-off mechanism. Rather, a dramatic increase in Slap mRNA level was reported during cell maturation, suggesting a function during the cell differentiation process (19). Slap may therefore define a signaling threshold during mitogenesis, since inhibition of endogenous Slap in fibroblasts led to an increase in cell response induced by growth factors (25). In this report, we investigated the mechanism by which Slap inhibits mitogenesis, and due to its homology with Src, we sought to analyze whether Slap could be a negative regulator of the Src mitogenic function. Using various chimeric Slap and Src constructs and a microinjection approach, we show that Slap is indeed an antagonist of the Src mitogenic pathway, not, however, simply by direct competition involving the SH2 domain, as the unique C-terminal sequence is also required for maximal effect. Our data also suggest that Slap may not regulate all Src functions, as it cannot reverse Src-induced cell transformation due to distinct SH3 binding specificity.
MATERIALS AND METHODS
Plasmid constructs.
The pSGT constructs expressing Src, SrcY527F, SrcK−, FynK, FynΔK, and Csk have been described in detail elsewhere (35), as have the pcDNA3 constructs expressing Myc-tagged Shc, Slap, and SlapΔSH3 (25). The plasmids expressing RasN17, c-Myc, c-Fos, and c-Jun have also been described (1). The Slap/SrcK, Slap/SrcSH3, and Src/SlapC chimeras were produced using the four-primer method of Cho and Lemaux (4). Briefly, 34-mer chimeric oligonucleotides were synthesized (Eurogentec) corresponding to 17 bp of each domain and two flanking T7 or SP6 primers. All the PCRs were performed in a 50-μl volume with an MJ Research (Watertown, Mass.) thermocycler. The first PCR comprised the relevant primers (40 pmol of each) and DNA template (1 ng) and was performed in a buffer containing all four deoxynucleoside triphosphates (200 μM each), 1.5 mM MgCl2, and 200 U of Elongase polymerase (Gibco-BRL). A touchdown cycling strategy was used to take into account the differences in melting temperatures. The products of the first PCR were then agarose gel purified (Qiagen) and used for the second-extension PCRs to produce the full-length chimeric sequences. The SlapΔC mutant was also derived by PCR with a customized 3′ antisense primer (Eurogentec), with a 26-bp overlap of the Slap SH2 domain and incorporating both a FLAG epitope and SalI site. These products were then digested with appropriate enzymes, subcloned into pcDNA3.1HYGRO (Invitrogen), and verified by sequencing. The constructs were also translated in a TNT (Promega) reaction or transfected into Cos7 cells to be further characterized by immunoprecipitation and Western blot analyses with appropriate antibodies (see below).
The G-to-A substitution in the myristylation site of Slap was made by using the Transformer site-directed mutagenesis kit (Clontech) according to the manufacturer's instructions and verified by sequencing. The SrcG2A mutant was kindly provided by Giulio Superti-Furga (European Molecular Biology Laboratory) and subcloned into the pcDNA3NEO (Invitrogen) vector.
Cell culture, transient transfection, and microinjection.
NIH 3T3 (clone 7), RS1 (NIH 3T3 stably expressing v-Src), LIA, and Cos7 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing glutamine and antibiotics (penicillin and streptomycin) at 37°C in a humidified 5% CO2 atmosphere. Transient transfection in Cos7 and NIH 3T3 cells was performed with Lipofectamine Plus reagent (Gibco) according to the manufacturer's instructions. Cells were also microinjected as described previously (25). Briefly, cells were seeded onto glass coverslips and grown for 24 h. To render the NIH 3T3 cells quiescent, the medium was replaced with DMEM containing 0.5% serum, and the cells were incubated for at least 30 h. DNA plasmids (0.1 mg/ml) were injected into the nucleus 4 h prior to PDGF (20 ng/ml) or fetal calf serum (10%) stimulation. In the case of SrcY527F plasmid expression, injected cells were incubated for a further 24 h before fixation. DNA synthesis was monitored by adding 0.1 mM bromodeoxyuridine (BrdU) into the medium, and growth factor-stimulated cells were incubated for 18 h and then fixed for immunostaining (see below).
Immunofluorescence.
Coverslips were washed once with phosphate-buffered saline (PBS) and normally fixed for 5 min with ice-cold methanol or with 3% formaldehyde for 10 min in the case of cytoskeletal analysis and ectopic protein localization. In the latter case, cells were permeabilized by a further incubation with PBS containing 0.1% Triton for 1 min. Cells overexpressing Slap-FLAG wild-type and mutant proteins were stained with the monoclonal anti-FLAG antibody (Sigma) (1:30 dilution) for 30 min, followed by fluorescein-conjugated anti-mouse immunoglobulin (Ig) antibody (ICN) incubation. Src wild-type and mutant proteins and the Slap/SrcK chimera-overexpressing cells were stained with affinity-purified anti-cst1 antibody (1:50 dilution) (28), followed by fluorescein-conjugated anti-rabbit Ig antibody (ICN) incubation. Slap/SrcK chimeric kinase- and FynΔK-expressing cells were stained with the 327 monoclonal Src antibody (28) and anti-β-galactosidase antibody (GAL-13; Sigma), respectively, followed by fluorescein-conjugated anti-mouse Ig antibody incubation. To visualize DNA synthesis by BrdU labeling, cells were incubated for 10 min with 1.5 M HCl, washed three times with PBS, and stained with monoclonal anti-BrdU antibody (1:50 dilution) (Pharmingen) followed by rhodamine-conjugated anti-mouse Ig antibody (ICN). Actin was visualized with Texas red-conjugated phalloidin (1:200 dilution) (a generous gift from M. Pucéat, Centre de Recherche de Biochemie Macromoléculaire). All coverslips were finally washed in PBS containing 10 μM Hoechst 33258 (Sigma), rinsed in water, inverted and mounted in Moviol (Hoechst) on glass slides, and viewed with a DMR B microscope (Leica).
The percentage of injected cells that incorporated BrdU for each coverslip was calculated by the following formula: (number of BrdU-positive injected cells/number of injected cells) × 100. Similarly, the percentage of cells with actin stress fibers was determined as follows: (number of stress fiber-positive injected cells/number of injected cells) × 100.
For confocal microscopy, we used a Nikon Optiphot II upright and Bio-Rad 1024 CLSM system with an argon-krypton ion laser (15 mW) with two emission lines at 488 nm for fluorescein isothiocyanate excitation (emission filter, 522/32) and 568 nm for rhodamine (emission filter, 605/32) and 60× (1.4-numerical-aperture or 20× (0.75-numerical-aperture) planopoachromatic objectives (Nikon). Images were collected sequentially to avoid cross-contamination between the fluorochromes. A series of optical sections were collected and projected onto a single image plane using the Laser Sharp 1024 software and processing system. Images were scanned at a 512- by 512-pixel resolution.
Biochemistry.
In vivo labeling assays were performed with lysates from Cos7 cells transfected for 2 days with the plasmids indicated below and then labeled for 24 h with [3H]myristate (Amersham) or 4 h with [35S]methionine (Tran35S-label; ICN) essentially as described before (15).
In vitro binding assays were performed as described previously (25). Glutathione-S-transferase (GST)-SlapΔC, GST-FynSH3, Sf9 insect cell lysates expressing the p85α subunit of phosphoinositide 3-kinase, and Sam68 were all isolated and purified as described before (16, 25, 27, 28). Cell lysates from Cos7 cells transfected for 3 days with a Myc-tagged Shc-encoding construct were used as a source of Shc protein. Bound proteins were detected by Western blotting with anti-p85, (27), anti-Sam68 (16), and 9E10 anti-Myc (Santa Cruz) antibodies. Various Src mutants were detected with EC10 monoclonal antibody (Santa Cruz), which specifically recognizes the chicken Src sequence. Cell lysates were prepared as described previously (27). Briefly, cells were rinsed twice in ice-cold TBS (20 mM Tris [pH 7.5], 150 mM NaCl, 0.1 mM sodium orthovanadate) and then lysed with LB (20 mM Tris [pH 8], 150 mM NaCl, 1% Nonidet P-40, 1% aprotinin, 20 μM leupeptin, 10 mM NaF, 0.1 mM sodium orthovanadate). In vitro kinase assays were performed with acid-denatured enolase (Sigma) as described before (29). In vivo kinase activity was assessed by tyrosine-phosphorylated protein content by Western blotting with the anti-phosphotyrosine 4G10 antibody (UBI) from total cell lysates of Cos7 cells transfected for 3 days with plasmids encoding the indicated kinases (Src, SrcY527F, Slap/SrcK, and Csk). The levels of ectopic protein expressed were assessed by Western blotting of total cell lysates with affinity-purified cst1 (Src, SrcY527F, and Slap/Src) or anti-Csk (Transduction Laboratories) antibodies. Bound antibody was detected with horseradish peroxidase-conjugated protein A (cst1 and αCsk) or horseradish peroxidase-conjugated anti-mouse IgG (4G10) followed by chemiluminescence detection (ECL; Amersham).
RESULTS
Slap is myristylated and colocalizes with Src in vivo.
We first investigated Slap localization within the cell. Cos7 cells were transiently transfected with a Slap construct that was tagged at the C terminus with the FLAG epitope (Fig. 1). As shown in Fig. 2, ectopic Slap was found predominantly in the cytoplasm with some localization at the plasma and perinuclear membranes, a situation reminiscent of that of Src (13). In order to confirm the colocalization with Src, both ectopic proteins were coexpressed in cells and coimmunostaining was performed with specific antibodies. As shown in Fig. 2, Src and Slap localization largely overlaps.
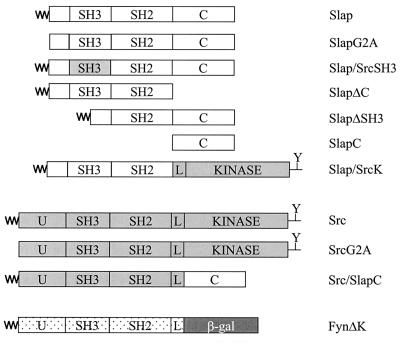
FIG. 1.
Schematic representation of the Slap, Src, and Fyn protein constructs used in this study. The unique (U), Slap C terminus (C), SH3 and SH2 domains, and Src SH2-CD linker region (L) are indicated. The myristyl group is denoted by ∼. β-gal, β-galactosidase; Y, negative regulatory tyrosine (referred to as Y527 in chicken c-Src).
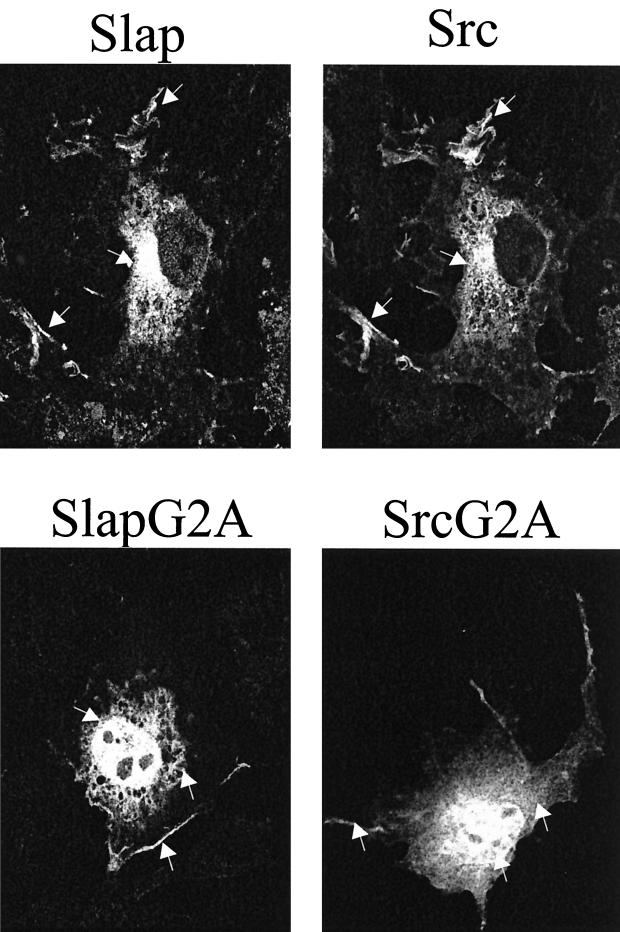
FIG. 2.
Slap colocalizes with Src in Cos cells. Immunofluorescence analyses of Cos7 cells transfected with Src and Slap FLAG wild type (upper panels) and G2A nonmyristylated mutants (lower panels) are shown. The ectopic proteins from the same transfected cell were immunostained with the cst1 (Src) and FLAG (Slap) antibodies and visualized by confocal microscopy as described in Materials and Methods. The arrows indicate specific Src and Slap localizations.
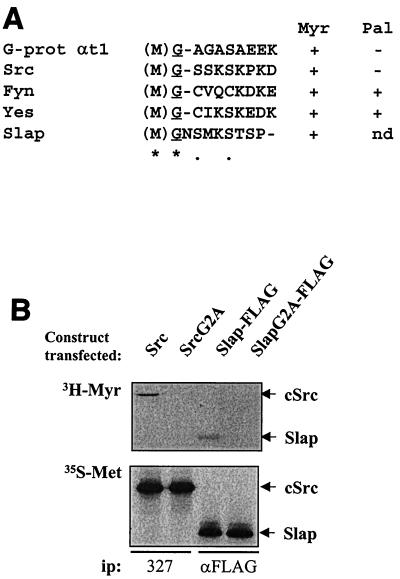
We then asked whether Slap was myristylated in order to explain the colocalization with Src. Supporting this notion, the sequence (M)GNSMKS was observed in the Slap N terminus, which closely resembles the consensus myristylation motif (M)G1–4T/S (Fig. 3A) (17, 23). Cos7 cells transiently transfected with either Src or the Slap-FLAG construct were labeled in vivo with [3H]myristic acid and [35S]methionine, and ectopic proteins were immunoprecipitated with specific antibodies and subsequently analyzed for labeling. As shown in Fig. 3B, both Src and Slap had incorporated myristic acid. However, while equally expressed, no such labeling was detected for either the Src or SlapG2A mutants, which were predicted not to attach lipid (Fig. 3A). The importance of such a posttranslational modification was next assessed. The potential effects on protein localization were first tested and are shown in the lower panels of Fig. 2A. SlapG2A was found in the cytoplasm as expected, but surprisingly, a strong nuclear localization was also seen. Similarly, nonmyristylated Src (SrcG2A) was strongly detected in the nucleus, in the cytosol, and at the plasma membrane (Fig. 2, lower panels), suggesting that some common mechanism(s) exists for Src and Slap cellular localization. Similar results were obtained in murine fibroblasts, indicating that the observed protein distribution was not due to the cell line used (unpublished observation).
FIG. 3.
Slap is myristylated in vivo. (A) Fatty acid recognition motifs for G-protein (G-prot) αt1, the Src family of tyrosine kinases, and Slap using ClustalW alignment. The single-amino-acid code is used, with the initiating methionine in parentheses and the myristylated glycine underlined. Identical residues (∗) and conserved residues and semiconserved substitutions (●) are indicated. The consensus myristylation sequence is (M)GX1–4S/T (17, 23). While not determined (nd), palmitoylation of Slap is unlikely to be due to the absence of cysteine residues in the N terminus. Myr, myristylation; Pal, palmitoylation. (B) Cos7 cells transiently transfected with Src, Slap-FLAG, SrcG2A, and SlapG2A-FLAG mutants as indicated were in vivo labeled with [3H]myristate (Myr) for 24 h or with [35S]methionine (Met) for 3 h. Labeled proteins were then immunoprecipitated (ip) from cell lysates with the indicated antibodies, followed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography.
Like SrcK−, the Slap G1 block is overcome by c-Myc expression.
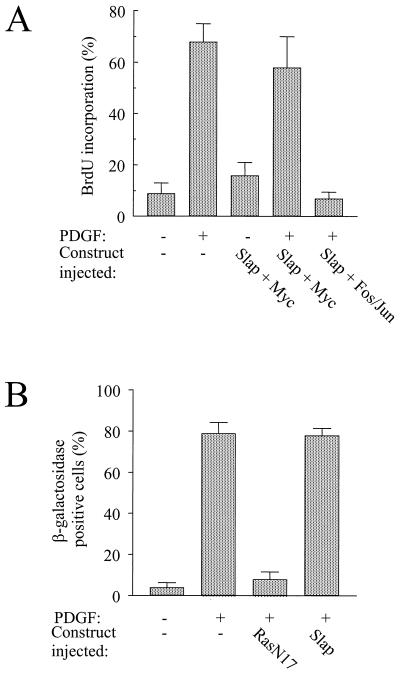
We next investigated which signaling pathway is affected by Slap. c-Myc expression was shown to bypass the G1-to-S block induced by a kinase-inactive form of Src (SrcK−) (1), and therefore we tested whether a similar scenario occurs for Slap. Ectopic proteins were expressed in quiescent cells by microinjection of the corresponding plasmids, and the cells were then stimulated with PDGF for 18 h. DNA synthesis was monitored by adding BrdU to the medium. BrdU is incorporated into the nuclear DNA during the S phase of the cell cycle. Statistical analysis of these experiments is shown in Fig. 4A. Slap strongly inhibited the PDGF response that was overcome by c-Myc coexpression. This was growth factor specific, as c-Myc did not drive cells into S phase in the absence of any extracellular stimulus. Furthermore, this rescue was specific to c-Myc, since coexpression of the transcriptional complex Fos/Jun with Slap was unable to restore PDGF signaling (Fig. 4A). We concluded that Slap inhibits a Src/Myc-dependent pathway and that the absence of rescue observed with Fos/Jun suggests that Slap does not affect the Ras-dependent pathway. In order to confirm this hypothesis further, we investigated the effect of Slap on the expression of the transcription factor c-Fos, an effector of the Ras pathway in early G1 (34). Since c-Fos levels are quite low in cells and the protein is highly labile, we decided to use a fibroblast cell line (LIA) that expresses β-galactosidase under the control of the c-Fos promoter. We have previously used this cell line to show that Src kinases do not affect PDGF-induced c-Fos promoter activation (31). In these cells, Slap overexpression did not affect the c-Fos response, while it was abrogated by dominant negative RasN17 (Fig. 4B).
FIG. 4.
Functional interaction between Slap and c-Myc during mitogenesis. (A) c-Myc but not c-Fos/c-Jun overrides the Slap G1 block. NIH 3T3 cells seeded onto coverslips were microinjected with Slap-encoding constructs in the presence or absence of c-Myc or c-Fos/c-Jun, as indicated, and stimulated or not with PDGF. Cells were fixed and processed for immunofluorescence as described in Materials and Methods. The percentage of injected cells that incorporated BrdU for each coverslip was calculated as described in the text. The means and standard deviations of several independent experiments are shown. (B) Slap does not affect PDGF-induced c-Fos promoter activation. Quiescent LIA cells which express β-galactosidase under the control of a c-Fos promoter were microinjected with the constructs encoding Slap or RasN17, as shown, and stimulated or not with PDGF for 90 min. β-Galactosidase activity and ectopic protein expression were assessed as described previously (6). For the microinjected cells, c-Fos expression was calculated as follows: percentage of β-galactosidase-positive cells = [(number of injected cells showing β-galactosidase activity)/(total number of injected cells)] × 100. The means and the standard deviations from three independent experiments are shown.
Src expression does not overcome the Slap G1 block.
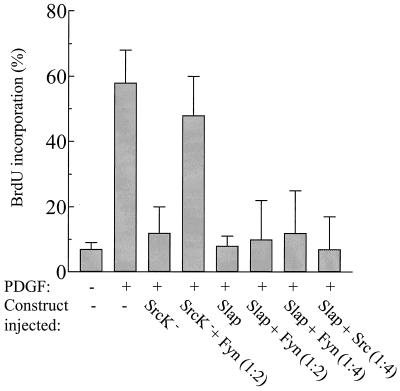
Is Slap a natural antagonist for Src? We first postulated that Slap acts by a competitive mechanism based on the following data: Slap and Src share the same binding site for PDGF receptor association (25), the Slap SH2 was needed for in vivo activity (25), and Slap largely colocalized with Src. Accordingly, Src overexpression should overcome the Slap G1 block. As a positive control, SrcK− inhibited the PDGF mitogenic response, which was then rescued by Fyn expression at a ratio of 2 to 1 (Fig. 5). This confirmed functional redundancy between Src family members, as previously reported (2, 28, 29). However, no such rescue was observed in the case of Slap, even when Fyn was expressed in a fourfold excess. As one can argue that it is due to lower homology between Slap and Fyn at the level of SH2 domains, similar experiments were performed with Src, but again, no rescue was observed (Fig. 5). Therefore, we conclude that Slap does not act solely in a competitive manner towards inhibiting mitogenic Src function.
FIG. 5.
Src kinases do not rescue the Slap G1 block. NIH 3T3 cells seeded onto coverslips were microinjected with Slap-encoding constructs in the presence or absence of various molar amounts of Src or Fyn and stimulated or not with PDGF. Cells were fixed and processed for immunofluorescence as described in Materials and Methods. The percentage of injected cells that incorporated BrdU for each coverslip was scored as described in the legend to Fig. 4A.
The Slap block involves the unique C-terminal sequence.
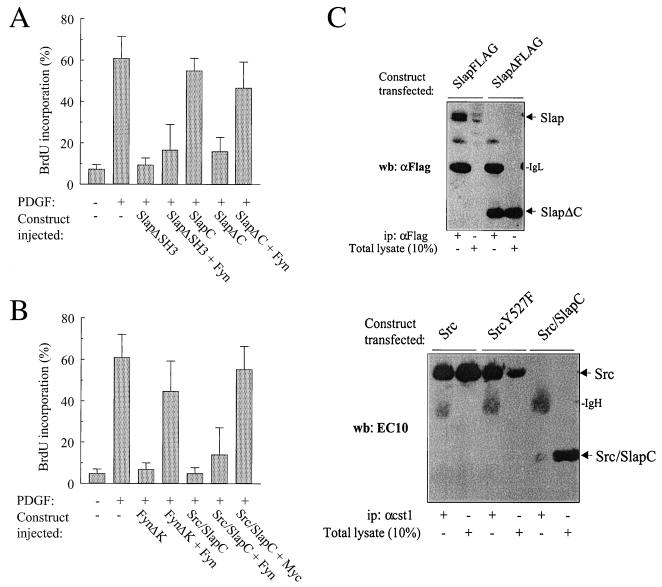
The absence of Src (or Fyn) rescue suggested that a further sequence within Slap is responsible for the noncompetitive inhibitory effect observed. We then turned to the SH3 and the C terminus within this context. As previously reported, SlapΔSH3 was inhibitory when overexpressed in quiescent cells (25). However, the G1 block could not be rescued by Fyn coexpression (Fig. 6A). In contrast, a version of Slap with a C-terminal deletion (SlapΔC) was still inhibitory, but in this case, Fyn coexpression did rescue the PDGF mitogenic response, implying that the Slap C terminus is necessary for Slap function. Since the Fyn rescue could be due to aberrant expression and/or stability of the adapters, the protein levels of Slap versus SlapΔC were analyzed by Western blotting lysates from cells transiently transfected with the corresponding constructs. As shown in Fig. 6C (upper panel), ectopic proteins were equally well immunoprecipitated by the anti-FLAG antibody, but interestingly, a higher level of SlapΔC was observed, suggesting that the SlapΔC protein is more stable and/or more highly expressed than Slap. Importantly, this rules out the absence of Fyn rescue due to a higher level of Slap expression. Since the Slap C terminus was evidently important for Slap function, we also investigated whether this sequence alone was sufficient to induce a G1 block. Microinjection of the Slap C terminus construct, however, had no effect on mitogenesis, suggesting that this unique domain apparently needs to be in the context of the whole Slap molecule to function (Fig. 6A). Pursuing this idea further, we next constructed an adapter comprising the N terminus of Src (U-SH3-SH2-linker) fused to the Slap C terminus (Src/SlapC, Fig. 1), and its effect on the mitogenic response was analyzed. As expected, microinjection of the Src/SlapC chimera inhibited the PDGF mitogenic response; however, Fyn (or Src; data not shown) did not overcome the inhibition (Fig. 6B), thus involving the C terminus sequence in noncompetitive inhibition. Once again, the inability of Src to restore signaling cannot be due to a difference in protein content, since Src was found to be more stably expressed than Src/SlapC when analyzed by Western blotting (Fig. 6C, lower panel). As a positive control for a competitive mechanism, we found that the adaptor FynΔK (U plus SH3 plus SH2) inhibited PDGF-induced signaling but this could be rescued by wild-type Fyn coexpression (Fig. 6B). In order to confirm specificity for the Src pathway, we also analyzed the effect of c-Myc on Src/SlapC inhibition and observed that c-Myc was able to rescue mitogenesis (Fig. 6B). Taken together, these data strongly suggest that Slap acts in a noncompetitive manner that sequentially involves the SH2 domain and the C terminus.
FIG. 6.
The Slap G1 block involves the C terminus. (A) Fyn overcomes the G1 block induced by SlapΔC but not by SlapΔSH3. Quiescent NIH 3T3 cells seeded onto coverslips were microinjected with the SlapΔC or SlapΔSH3 constructs alone or with Fyn and stimulated or not with PDGF as shown. The percentage of injected cells that incorporated BrdU for each coverslip was scored as in the legend to Fig. 4A. (B) Fyn does not override the G1 block induced by the Src/SlapC chimeric adapter. FynΔK or the Src/SlapC adapter constructs were microinjected into seeded quiescent NIH 3T3 cells in the presence or absence of Fyn- or Myc-encoding vectors and stimulated or not with PDGF. Cells were then fixed and processed for immunofluorescence as described in Materials and Methods. The percentage of injected cells that incorporated BrdU for each coverslip was calculated as in the legend to Fig. 4A. (C) Levels of ectopic Slap, SlapΔC, Src, Src527F, and Src/SlapC proteins. Constructs encoding the indicated proteins were transiently transfected into Cos7 cells, and ectopic protein levels were analyzed by Western blotting (wb) of the total cell lysate (10%) and after immunoprecipitation (ip) with the indicated antibodies. The positions of Slap, SlapΔC, Src, Src/SlapC, Ig heavy chain (IgH), and Ig light chain (IgL) are also indicated.
Slap does not inhibit all Src cellular functions.
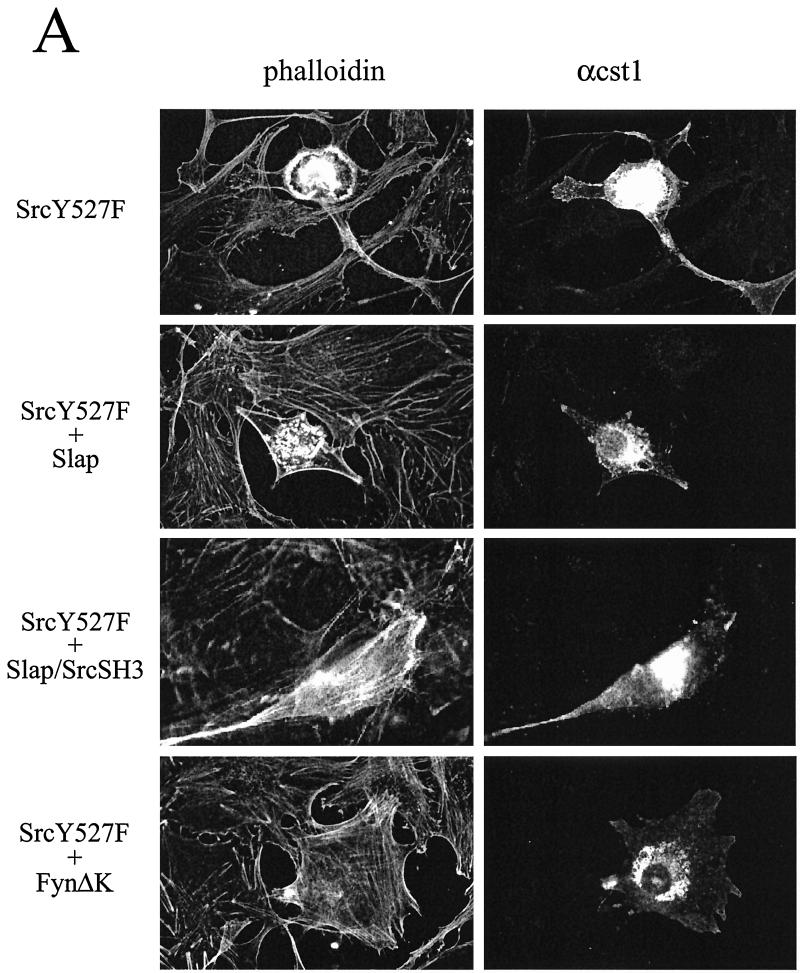
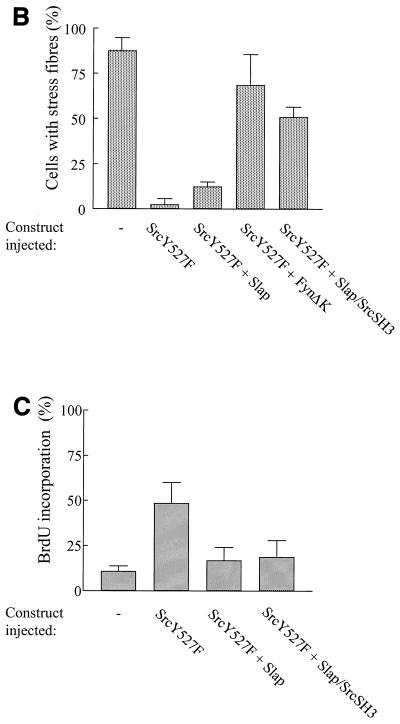
Since Slap inhibits Src signaling during mitogenesis, we sought to investigate whether it could inhibit other Src functions. The effect of Slap was analyzed in v-Src-transformed fibroblasts (RS1 cells). After Slap transfection, however, no reversion of cell morphology was observed (data not shown). For more detailed characterization, a microinjection approach was performed with NIH 3T3 cells. Oncogenic SrcY527F was expressed in quiescent cells, and the subsequent cytoskeletal rearrangements were analyzed. As shown in Fig. 7A, active SrcY527F induced prominent cytoskeletal rearrangements, including the disappearance of stress fibers, aggregation of F-actin into podosome structures, and, in some cases, formation of filopodes and lamelipodes, in agreement with previous observations (26). We next focused our attention on the downregulation of actin polymerization, as it was the most consistent and measurable response observed, and a statistical analysis is shown in Fig. 7B. Approximately 85% of quiescent cells showed actin bundles which were abrogated by SrcY527F expression. Actin polymerization was largely restored by FynΔK expression (Fig. 7A and B), but Slap coexpression poorly reverted the Src-induced cytoskeletal rearrangement while still inhibiting S-phase entry (Fig. 7A and C). Under our experimental conditions, SrcY527F (100 μg/ml) drove 50% of cells into S phase, which was reduced approximately twofold by Slap. Therefore, Slap is a specific regulator of Src during mitogenesis but does not affect Src-induced cell transformation.
FIG. 7.
Slap inhibits DNA synthesis induced by SrcY527F but does not affect Src-induced cytoskeletal rearrangement. (A) Slap does not reverse cytoskeletal rearrangement induced by Src527F. Quiescent NIH 3T3 cells seeded onto coverslips were microinjected with the Src527F construct alone or in the presence of Slap, Slap/SrcSH3, or FynΔK vectors, as shown. Cells were fixed 24 h later and immunostained for Src527F expression with cst1 antibody (right panels). The presence of actin in the cytoskeleton was visualized by phalloidin staining (left panels). The immunofluorescence pattern for a representative cell is shown for each case. (B) Reversion of cell transformation as scored by the presence of actin stress fibers calculated as described in the text. The means and standard deviations of several independent experiments are shown. (C) Slap inhibition of Src527F-induced S-phase entry. Quiescent NIH 3T3 cells seeded onto coverslips were injected with Src527F alone or in the presence of Slap or Slap/SrcSH3 and incubated with BrdU for 20 h. Cells were then fixed and processed for immunofluorescence as described in Materials and Methods. The percentage of injected cells that incorporated BrdU for each coverslip was calculated as described in the legend to Fig. 4A.
Slap and Src SH3 domains have distinct binding specificities.
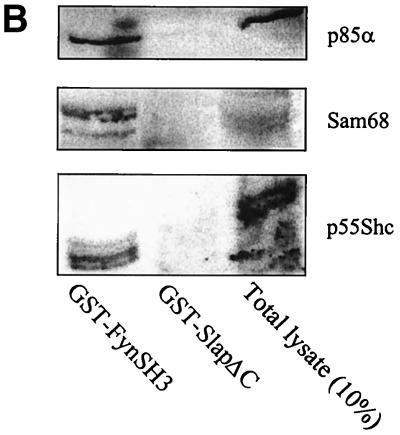
Since Src-induced cytoskeletal rearrangement involves its SH3 domain (12), we sought to determine whether the Src SH3 and Slap SH3 domains were functionally equivalent. A chimeric molecule in which the Slap SH3 was replaced by the Src SH3 sequence (Slap/SrcSH3, Fig. 1) was generated and tested for its ability to revert Src-induced cell transformation. As shown in Fig. 7, this chimeric adapter substantially restored actin polymerization, suggesting that the Slap SH3 is indeed implicated in the inability of Slap to counteract the aberrant cytoskeletal effects induced by deregulated Src expression. This would therefore predict distinct binding specificities between the Src SH3 and Slap SH3 domains. Indeed, despite the strong homology observed in the primary sequences (50% identity), a more careful examination revealed the existence of three nonconservative substitutions in the Slap SH3 sequence (Fig. 8A) that are known to be crucial for intra- and intermolecular interactions for the Src SH3 domain (7). Src residues Tyr-90, Asp-99, and Tyr-136 are replaced by Thr (or Ser in the human Slap sequence), Pro, and Cys, respectively. The ability of Slap SH3 to associate with known Src SH3 ligands was addressed first in an in vitro binding assay (Fig. 8B). While Sam68, Shc, and the p85α regulatory subunit of phosphatidylinositol 3-kinase bound to GST-FynSH3, they did not associate with GST-SlapΔC. This was not due to incorrect protein folding, as the GST-SlapΔC fusion protein has previously been shown to associate with the activated PDGF receptor (25).
FIG. 8.
SlapSH3 and SrcSH3 show distinct binding specificities. (A) ClustalW alignment of the Src and human (h) and mouse (m) Slap SH3 primary sequences. Important residues for the ligand binding surface that are conserved (bold) or replaced (#) in the Slap sequence are highlighted. In the consensus line, identical or conserved residues (∗), conserved substitutions (:), and semiconserved substitutions (.) are indicated. (B) Slap SH3 does not interact with various Fyn SH3 partners. GST-FynSH3 or GST-SlapΔC fusion proteins were bound to glutathione-Sepharose beads and incubated with Sf9 cell lysates expressing p85α or Sam68 and with Cos7 cell lysate transiently transfected with an Shc-encoding construct. After GST preclearing and extensive washing, Western blotting with specific antibodies as described in Materials and Methods revealed the presence of bound proteins. Total cell lysate (10%) was used as a positive control.
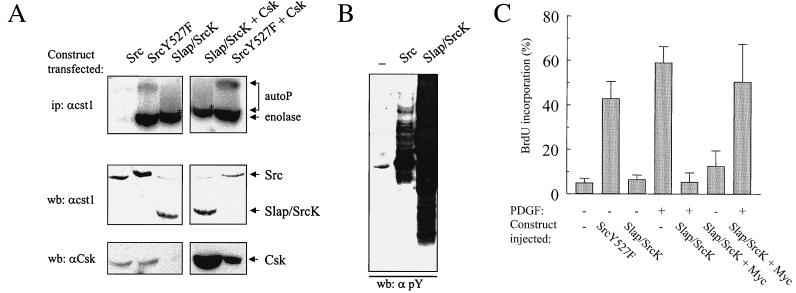
In addition to intermolecular interactions, Src SH3 also associates with the peptide sequence lying between the SH2 and the catalytic domain (SH2-CD linker). Such an interaction, together with the SH2-pY527 association, induces a closed conformation of the protein and repressed kinase activity (22). Whether intramolecular interactions could occur within the Slap SH3 was next investigated. A chimeric Slap-Src tyrosine kinase (Slap/SrcK) was generated that consists of the Slap N-SH3-SH2 sequence fused to the SH2-CD linker, kinase domain, and the regulatory, phosphotyrosine-containing tail of the Src sequence (Fig. 1). To address whether this chimera could be regulated, we performed the following experiment. The Src, SrcY527F, and Slap/SrcK kinases were transiently expressed in Cos7 cells, and kinase activity was assessed in vitro with denatured enolase as an exogenous substrate. Proteins were then immunoprecipitated with the cst1 antibody, which recognizes all the kinases tested, in order to avoid discrepancies due to aberrant antibody effects on kinase activity. Strong kinase activity was detected for SrcY527F and Slap/SrcK, whereas wild-type Src displayed approximately 10-fold less activity (Fig. 9A). Similar data were also obtained when coexpressing Csk, suggesting that the absence of Slap/SrcK regulation is not due to inefficient phosphorylation of the regulatory Y527 residue. In vivo activity was also assessed by total lysate phosphotyrosine content from the same lysates and is shown in Fig. 9B. Slap/SrcK expression induced strong overall tyrosine phosphorylation of numerous endogenous proteins, in agreement with deregulated activity, while Src showed reduced tyrosine phosphorylation, with a major phosphorylated p60 protein that may correspond to the kinase itself. Taken together, these data show that Slap/SrcK is not regulated and that the Slap SH3 is not equivalent to the Src SH3 with respect to intramolecular interactions.
FIG. 9.
Slap/SrcK chimeric tyrosine kinase is not regulated and behaves as a dominant negative during mitogenesis. (A) Slap/SrcK is not regulated. Ectopic tyrosine kinase was immunoprecipitated (ip) with the cst1 antibody from extracts of Cos7 cells transiently transfected with Src, Src527F, and Slap/SrcK alone or in the presence of a Csk-expressing vector, as shown. In vitro kinase assays were then performed with denatured enolase as the substrate. Autoradiography of labeled proteins separated by SDS-PAGE is shown. The positions of labeled enolase and autophosphorylated tyrosine kinases (autoP) are indicated. ∗, autophosphorylated Slap/SrcK which migrates closely with the enolase. The levels of expressed Src, Slap/SrcK, and Csk are also shown and were assessed by Western blotting (wb) of total cell lysates with the indicated antibodies. (B) In vivo activity of Src and Slap/SrcK. The kinase activities of Src and Slap/SrcK were assessed by an antiphosphotyrosine (αpY) Western blot (wb) of cell lysates prepared from Cos7 cells transiently transfected with the appropriate constructs. (C) Slap/SrcK is inhibitory to the PDGF mitogenic response. NIH 3T3 cells seeded onto coverslips were microinjected with Src527F- or Slap/SrcK-encoding constructs alone or in the presence of c-Myc and stimulated or not with PDGF. Cells were fixed and processed for immunofluorescence as described in Materials and Methods. The percentage of injected cells that incorporated BrdU for each coverslip was calculated as described in the legend to Fig. 4A.
Finally, the effect of Slap/SrcK was investigated during cell mitogenesis. We and others have previously shown that Src SH3 was needed for efficient mitogenic signaling, including oncogenic Src-induced cell transformation (7) and PDGF-induced DNA synthesis (3, 6). In contrast to SrcY527F, Slap/SrcK did not drive NIH 3T3 cells into S phase when expressed in quiescent cells, suggesting that it is not oncogenic. In addition, despite being kinase active, Slap/SrcK acted in a dominant negative fashion during PDGF signaling, as previously observed with chimeric Src molecules in which either the Lck SH3 or Spectrin SH3 (6) replaced Src SH3. Moreover, c-Myc was able to overcome the Slap/SrcK inhibitory effect on DNA synthesis, in agreement with a specific inhibition of the Src pathway.
DISCUSSION
Slap as a negative regulator of Src function during mitogenesis.
In this report, we have investigated whether Slap is a negative in vivo regulator of Src function. The mechanism by which Slap inhibits growth factor receptor signaling was first considered, and indeed, our data strongly suggest that Slap is a negative regulator of the Src pathway: (i) Slap is myristylated in vivo and largely colocalizes with Src, (ii) Slap and Src share the same binding site for the activated PDGF receptor (Y579), (iii) the Slap SH2 and Src SH2 domains show similar binding specificities and are both required for their respective function (25, 36), (iv) the G1 block induced by SrcK− and Slap is rescued by constitutive c-Myc expression, and (v) Src (and Fyn) can overcome the G1 block induced by SlapΔC. Evidence of Src effector molecule association with Slap would further support our conclusion. However, the absence of known Src substrates that ensure growth factor signaling precludes such an analysis, and so we cannot exclude the possibility that the adaptor acts farther downstream. However, the fact that Src rescued the SlapΔC block, together with the observed colocalization, would rather suggest that Slap acts directly at the Src level.
The mechanism by which Slap inhibits Src function was further investigated by the use of mutagenesis and chimeric mutant approaches. Our data demonstrated that Slap does not act by direct competition alone, suggesting that it is not just a natural antagonist for Src, as first thought. In addition to the SH2 domain, full Slap inhibitory function requires the C terminus. Interestingly, this domain does not show any homology with any sequences in the current databases but is thought to mediate protein-protein interactions; indeed, we have identified several Slap C terminus interactors with a yeast two-hybrid genetic screen (unpublished data). In light of all this, we propose a scenario in which Slap uses its SH2 domain as a prerequisite for proper localization (by binding to the activated PDGF receptor, for example), enabling the C-terminal sequence to interact with a Src effector(s) important for mitogenic signaling. In agreement with this model, SlapΔC inhibited mitogenesis by competition with Src, probably for receptor association, whereas the whole Slap molecule would in addition titrate Src effectors with the C terminus, thus preventing signaling despite Src overexpression. Involvement of the C terminus in this process is further supported by the similar noncompetitive inhibitory mechanism observed with the chimeric adapter Src/SlapC.
An alternative explanation would be that the Slap SH2 domain and C terminus act independently. However, this mechanism has been ruled out, since overexpression of the C terminus alone does not affect mitogenesis, indicating that it needs to be in the context of the whole molecule for function. A similar case has been proposed for the Src kinases; Fyn SH2 (FynΔSH3ΔK) but not Fyn SH3 (FynΔSH2ΔK) was found to inhibit the PDGF response, implying that the SH2 domain is required for signaling (36). However, deletion of an SH3 domain alone (SrcΔSH3) also inhibited mitogenic signaling (6), and therefore the Src SH2 domain appears to be important for proper cell localization, subsequently enabling the SH3 domain to associate with Src (or Fyn) effectors. Finally, other functions for the C terminus can be proposed, including protein localization, but this again has been excluded, since this sequence alone showed strong cytosolic localization and its deletion did not affect Slap intracellular localization (unpublished observations). Identification of Slap effectors that associate with the C terminus will be very informative in this regard. With regard to the C terminus affecting protein stability, the results are only preliminary and no firm conclusions can be drawn.
Slap does not regulate all Src functions due to distinct SH3 binding specificity.
In contrast to what was originally thought, Slap does not regulate all Src functions, as was observed for Src-induced cell morphological change and actin depolymerization. The absence of a Slap effect may be attributed to specific Src sequences important for signaling, including the N terminus and/or the SH3 domain. Despite largely overlapping expression patterns, Src displayed some unique localization, signifying specific function. For example, SrcY527F is known to be targeted to focal adhesions (12), whereas Slap was not detected at these sites. In addition to myristylation, specific localization of membrane-bound Src could involve stabilization by three Arg residues located in the unique domain which are not present in most other members of the Src family (23). Rather, the related kinases use a palmitic lipid group on Cys-3 or Cys-5 to further stabilize their membrane attachment. In this context, Slap possesses neither the stretch of Arg residues (only one Arg is present) nor Cys in its N terminus. In addition, our mutagenesis study identified a function for myristylation in cytoplasmic protein retention, preventing nuclear entry or aberrant protein localization. Indeed, both the Src and SlapG2A mutants were expressed predominantly in the nucleus, and interestingly, a Src allele has been reported with an altered N-terminal sequence that displays partial nuclear localization (5). However, we cannot exclude another mechanism for Slap membrane targeting, since substantial amounts of nonmyristylated Src and Slap were evident at the plasma membrane.
Src-induced cell transformation also involves its SH3 domain (33), and the inability of Slap to revert cell transformation is largely due to a distinct SH3 binding specificity, as demonstrated by the SH3 swapping experiment. The SH3 domain is required for mouse fibroblast transformation by SrcY527F (7) and is also involved in SrcY527F cellular distribution and association with a number of substrates, including the cytoskeleton-associated proteins AFAP-110 (9), paxillin (38), p130Cas (18), and Fak (32). While not fully understood, SrcY527F-induced actin depolymerization involves the inhibition of Rho activity, probably via phosphorylation and inactivation of p190RhoGap (8), a process that may not be affected by Slap. The distinct function for SH3 domains was confirmed at the molecular level, as a comparison of Src SH3 and Slap SH3 sequences revealed nonconservative amino acid substitutions for crucial residues in the ligand binding pocket. All of these residues have been implicated in intramolecular interactions that regulate kinase activity (7) and for binding Src to signaling effectors required for mitogenesis (6). Our in vitro and in vivo data demonstrate a distinct specificity for ligand interactions: the chimeric Slap/SrcK could not induce either cell cycle progression or cell transformation, although constitutively active. In fact, it acted as a dominant negative during mitogenesis, a situation reminiscent of SrcΔSH3 or a chimeric Src bearing the Lck SH3 domain (6). From these observations, we postulate that Slap may also have specific functions unrelated to Src for signaling involving specific SH3 ligands. Determination of the Slap SH3 structure as well as specific interactors will be important in this regard. Nevertheless, we cannot exclude the existence of a common subset of partners for the Slap and Src SH3 domains.
In conclusion, our results strongly suggest that Slap is a negative regulator for some but not all Src biological functions. Slap mitogenic inhibition may involve association with Src interactors via its SH2 and unique C-terminal domains through a noncompetitive mechanism. The distinct SH3 binding specificity also confirmed that Slap is not just a natural antagonist of Src, and we propose that Slap may also have independent cellular functions involving specific Slap SH3 ligands.
ACKNOWLEDGMENTS
We thank N. Lautredou for help with confocal microscopy, A. Padilla and C. Dumas for discussion on Slap SH3 structure, G. Superti-Furga for the SrcG2A construct, and our colleagues for discussion and critical reading of the manuscript.
G.M. was a supported by the Fondation pour la Recherche Médicale, P.B. by La Ligue Contre le Cancer, and S.R. by the Institut National de la Santé et de la Recherche Médicale. This work was supported by l'Association pour la Recherche contre le Cancer and the Centre National de la Recherche Scientifique.
REFERENCES
- 1.Barone M V, Courtneidge S A. Myc but not Fos rescue of PDGF signalling G1 block caused by kinase-inactive Src. Nature. 1995;378:509–512. doi: 10.1038/378509a0. [DOI] [PubMed] [Google Scholar]
- 2.Boyer B, Roche S, Denoyelle M, Thiery J P. Src and Ras are involved in separate pathways in epithelial cell scattering. EMBO J. 1997;16:5904–5913. doi: 10.1093/emboj/16.19.5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broome M A, Hunter T. Requirement for c-Src catalytic activity and the SH3 domain in platelet-derived growth factor BB and epidermal growth factor mitogenic signaling. J Biol Chem. 1996;271:16798–16806. doi: 10.1074/jbc.271.28.16798. [DOI] [PubMed] [Google Scholar]
- 4.Cho M J, Lemaux P G. Rapid PCR amplification of chimeric products and its direct application to in vivo testing of recombinant DNA construction strategies. Mol Biotechnol. 1997;8:13–16. doi: 10.1007/BF02762336. . (Erratum, 8:197.) [DOI] [PubMed] [Google Scholar]
- 5.David-Pfeuty T, Bagrodia S, Shalloway D. Differential localization patterns of myristylated and nonmyristylated c-Src proteins in interphase and mitotic c-Src overexpresser cells. J Cell Sci. 1993;105:613–628. doi: 10.1242/jcs.105.3.613. [DOI] [PubMed] [Google Scholar]
- 6.Erpel T, Alonso G, Roche S, Courtneidge S A. The Src SH3 domain is required for DNA synthesis induced by platelet-derived growth factor and epidermal growth factor. J Biol Chem. 1996;271:16807–16812. doi: 10.1074/jbc.271.28.16807. [DOI] [PubMed] [Google Scholar]
- 7.Erpel T, Superti-Furga G, Courtneidge S A. Mutational analysis of the Src SH3 domain: the same residues of the ligand binding surface are important for intra- and intermolecular interactions. EMBO J. 1995;14:963–975. doi: 10.1002/j.1460-2075.1995.tb07077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fincham V J, Chudleigh A, Frame M C. Regulation of p190 Rho-GAP by v-Src is linked to cytoskeletal disruption during transformation. J Cell Sci. 1999;112:947–956. doi: 10.1242/jcs.112.6.947. [DOI] [PubMed] [Google Scholar]
- 9.Flynn D C, Leu T H, Reynolds A B, Parsons J T. Identification and sequence analysis of cDNAs encoding a 110-kilodalton actin filament-associated pp60src substrate. Mol Cell Biol. 1993;13:7892–7900. doi: 10.1128/mcb.13.12.7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heldin C H, Miyazono K, ten Dijke P. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 11.Hilton D J, Richardson R T, Alexander W S, Viney E M, Willson T A, Sprigg N S, Starr R, Nicholson S E, Metcalf D, Nicola N A. Twenty proteins containing a C-terminal SOCS box form five structural classes. Proc Natl Acad Sci USA. 1998;95:114–119. doi: 10.1073/pnas.95.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaplan K B, Bibbins K B, Swedlow J R, Arnaud M, Morgan D O, Varmus H E. Association of the amino-terminal half of c-Src with focal adhesions alters their properties and is regulated by phosphorylation of tyrosine 527. EMBO J. 1994;13:4745–4756. doi: 10.1002/j.1460-2075.1994.tb06800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaplan K B, Swedlow J R, Varmus H E, Morgan D O. Association of p60c-src with endosomal membranes in mammalian fibroblasts. J Cell Biol. 1992;118:321–333. doi: 10.1083/jcb.118.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klinghoffer R A, Sachsenmaier C, Cooper J A, Soriano P. Src family kinases are required for integrin but not PDGFR signal transduction. EMBO J. 1999;18:2459–2471. doi: 10.1093/emboj/18.9.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koegl M, Zlatkine P, Ley S C, Courtneidge S A, Magee A I. Palmitoylation of multiple Src-family kinases at a homologous N-terminal motif. Biochem J. 1994;303:749–753. doi: 10.1042/bj3030749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lock P, Fumagalli S, Polakis P, McCormick F, Courtneidge S A. The human p62 cDNA encodes Sam68 and not the RasGAP-associated p62 protein. Cell. 1996;84:23–24. doi: 10.1016/s0092-8674(00)80989-7. [DOI] [PubMed] [Google Scholar]
- 17.Milligan G, Parenti M, Magee A I. The dynamic role of palmitoylation in signal transduction. Trends Biochem Sci. 1995;20:181–187. doi: 10.1016/s0968-0004(00)89004-0. [DOI] [PubMed] [Google Scholar]
- 18.Nakamoto T, Sakai R, Ozawa K, Yazaki Y, Hirai H. Direct binding of C-terminal region of p130Cas to SH2 and SH3 domains of Src kinase. J Biol Chem. 1996;271:8959–8965. doi: 10.1074/jbc.271.15.8959. [DOI] [PubMed] [Google Scholar]
- 19.Ohtsuki T, Hatake K, Ikeda M, Tomizuka H, Terui Y, Uwai M, Miura Y. Expression of Src-like adapter protein mRNA is induced by all-trans retinoic acid. Biochem Biophys Res Commun. 1997;230:81–84. doi: 10.1006/bbrc.1996.5887. [DOI] [PubMed] [Google Scholar]
- 20.Ota Y, Samelson L E. The product of the proto-oncogene c-Cbl: a negative regulator of the Syk tyrosine kinase. Science. 1997;276:418–420. doi: 10.1126/science.276.5311.418. [DOI] [PubMed] [Google Scholar]
- 21.Pandey A, Duan H, Dixit V M. Characterization of a novel Src-like adapter protein that associates with the Eck receptor tyrosine kinase. J Biol Chem. 1995;270:19201–19204. doi: 10.1074/jbc.270.33.19201. [DOI] [PubMed] [Google Scholar]
- 22.Pawson T. New impressions of Src and Hck. Nature. 1997;385:582–583. doi: 10.1038/385582b0. , 585. [DOI] [PubMed] [Google Scholar]
- 23.Resh M D. Myristylation and palmitylation of Src family members: the fats of the matter. Cell. 1994;76:411–413. doi: 10.1016/0092-8674(94)90104-x. [DOI] [PubMed] [Google Scholar]
- 24.Richardson A, Parsons T. A mechanism for regulation of the adhesion-associated protein tyrosine kinase pp125FAK. Nature. 1996;380:538–540. doi: 10.1038/380538a0. . (Erratum, 381:810.) [DOI] [PubMed] [Google Scholar]
- 25.Roche S, Alonso G, Kazlauskas A, Dixit V M, Courtneidge S A, Pandey A. Src-like adaptor protein (Slap) is a negative regulator of mitogenesis. Curr Biol. 1998;8:975–978. doi: 10.1016/s0960-9822(98)70400-2. [DOI] [PubMed] [Google Scholar]
- 26.Roche S, Courtneidge S A. Oncogenic cytoplasmic tyrosine kinases. Vol. 19. Oxford, United Kingdom: Oxford University Press; 1997. [Google Scholar]
- 27.Roche S, Dhand R, Waterfield M D, Courtneidge S A. The catalytic subunit of phosphatidylinositol 3-kinase is a substrate for the activated platelet-derived growth factor receptor, but not for middle-T antigen-pp60c-src complexes. Biochem J. 1994;301:703–711. doi: 10.1042/bj3010703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roche S, Fumagalli S, Courtneidge S A. Requirement for Src family protein tyrosine kinases in G2 for fibroblast cell division. Science. 1995;269:1567–1569. doi: 10.1126/science.7545311. [DOI] [PubMed] [Google Scholar]
- 29.Roche S, Koegl M, Barone M V, Roussel M F, Courtneidge S A. DNA synthesis induced by some but not all growth factors requires Src family protein tyrosine kinases. Mol Cell Biol. 1995;15:1102–1109. doi: 10.1128/mcb.15.2.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roche S, Koegl M, Courtneidge S A. The phosphatidylinositol 3-kinase alpha is required for DNA synthesis induced by some, but not all, growth factors. Proc Natl Acad Sci USA. 1994;91:9185–9189. doi: 10.1073/pnas.91.19.9185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roche S, McGlade J, Jones M, Gish G D, Pawson T, Courtneidge S A. Requirement of phospholipase C gamma, the tyrosine phosphatase Syp and the adaptor proteins Shc and Nck for PDGF-induced DNA synthesis: evidence for the existence of Ras-dependent and Ras-independent pathways. EMBO J. 1996;15:4940–4948. [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas J W, Ellis B, Boerner R J, Knight W B, White G C, 2nd, Schaller M D. SH2- and SH3-mediated interactions between focal adhesion kinase and Src. J Biol Chem. 1998;273:577–583. doi: 10.1074/jbc.273.1.577. [DOI] [PubMed] [Google Scholar]
- 33.Tian M, Martin G S. The role of the Src homology domains in morphological transformation by v-src. Mol Biol Cell. 1997;8:1183–1193. doi: 10.1091/mbc.8.7.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Treisman R. Ternary complex factors: growth factor regulated transcriptional activators. Curr Opin Genet Dev. 1994;4:96–101. doi: 10.1016/0959-437x(94)90097-3. [DOI] [PubMed] [Google Scholar]
- 35.Twamley G M, Kypta R M, Hall B, Courtneidge S A. Association of Fyn with the activated platelet-derived growth factor receptor: requirements for binding and phosphorylation. Oncogene. 1992;7:1893–1901. [PubMed] [Google Scholar]
- 36.Twamley-Stein G M, Pepperkok R, Ansorge W, Courtneidge S A. The Src family tyrosine kinases are required for platelet-derived growth factor-mediated signal transduction in NIH 3T3 cells. Proc Natl Acad Sci USA. 1993;90:7696–7700. doi: 10.1073/pnas.90.16.7696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Geer P, Hunter T, Lindberg R A. Receptor protein-tyrosine kinases and their signal transduction pathways. Annu Rev Cell Biol. 1994;10:251–337. doi: 10.1146/annurev.cb.10.110194.001343. [DOI] [PubMed] [Google Scholar]
- 38.Weng Z, Taylor J A, Turner C E, Brugge J S, Seidel-Dugan C. Detection of Src homology 3-binding proteins, including paxillin, in normal and v-Src-transformed Balb/c 3T3 cells. J Biol Chem. 1993;268:14956–14963. [PubMed] [Google Scholar]
- 39.Yokouchi M, Wakioka T, Sakamoto H, Yasukawa H, Ohtsuka S, Sasaki A, Ohtsubo M, Valius M, Inoue A, Komiya S, Yoshimura A. APS, an adaptor protein containing PH and SH2 domains, is associated with the PDGF receptor and c-Cbl and inhibits PDGF-induced mitogenesis. Oncogene. 1999;18:759–767. doi: 10.1038/sj.onc.1202326. [DOI] [PubMed] [Google Scholar]
- 40.Yoshimura A. The CIS family: negative regulators of JAK-STAT signaling. Cytokine Growth Factor Rev. 1998;9:197–204. doi: 10.1016/s1359-6101(98)00019-7. [DOI] [PubMed] [Google Scholar]
- 41.Yoshimura A, Ohkubo T, Kiguchi T, Jenkins N A, Gilbert D J, Copeland N G, Hara T, Miyajima A. A novel cytokine-inducible gene CIS encodes an SH2-containing protein that binds to tyrosine-phosphorylated interleukin-3 and erythropoietin receptors. EMBO J. 1995;14:2816–2826. doi: 10.1002/j.1460-2075.1995.tb07281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]