Abstract
Background
Entamoeba histolytica infection is a sexually transmitted disease in some developed countries. Asymptomatic infection often occurs and can be a source of transmission; however, limited data are available regarding the pathogenesis of E. histolytica.
Methods
This was a single-center, cross-sectional study. Specimens were prospectively collected from patients with clinically suspected cases. Entamoeba histolytica infection was defined as a case in which the identification of E. histolytica was confirmed by polymerase chain reaction (PCR) of a clinical specimen. Data from asymptomatic cases were compared with those from symptomatic invasive cases.
Results
Sixty-four E. histolytica–infected cases, including 13 asymptomatic cases, were identified during the study period. Microbiological diagnosis was made by endoscopic sampling in 26.6% of these cases (17/64). Endoscopy identified macroscopically visible lesions in all cases; however, the sensitivity of histopathology on biopsy samples was low (45.5%) compared with PCR (94.7%). In asymptomatic cases, infection sites were limited around the proximal colon; moreover, trophozoites were frequently identified at infection sites whereas cystic forms were commonly detected in stools. Gut microbiome analyses showed more uniform composition in asymptomatic cases than in symptomatic invasive cases, which were represented by a relatively high abundance of Ruminococcaceae, Coriobacteriaceae, and Clostridiaceae, and a low abundance of Streptococcaceae.
Conclusions
These results indicate that the encystation and attenuation of E. histolytica are highly affected by the intestinal contents, including the gut microbiome.
Keywords: parasitology, sexually transmitted infections, amebiasis, microbiome
During asymptomatic infection, Entamoeba histolytica caused endoscopically visible ulcers around the cecum. Trophozoites were detected at infection sites, whereas cystic forms were commonly detected in stools, indicating that encystation is induced by gut environmental factors, such as the gut microbiota.
Entamoeba histolytica, the causative agent of invasive amebiasis, is the second most common parasitic cause of mortality worldwide [1]. Over the past 2 decades, it was reported that invasive amebiasis is prevalent not only in developing countries where food and water are frequently contaminated by feces, but also in some developed countries in Asia and Europe [2–5]. In these areas, the pathogen spreads as a sexually transmitted infection (STI), especially among men who have sex with men (MSM) and people living with human immunodeficiency virus (HIV) [2, 5, 6]. Furthermore, recent data indicate that this pathogen is spreading among HIV-uninfected men and women in Japan [7, 8].
The severity of E. histolytica infection varies. Only 10% of individuals who are exposed to the pathogen develop “symptomatic” invasive amebiasis; the majority of cases are asymptomatic or display self-limiting mild diarrhea at an early phase [9]. Therefore, E. histolytica infection is often overlooked in clinical settings. Furthermore, it is known that some infections persist asymptomatically. The half-life of asymptomatic infection is reported to be about 1 year [10], which can result in a transmissible pathogen reservoir among the community, and E. histolytica infection has been unexpectedly diagnosed by endoscopy in developed countries [11–13]. Data from the National Surveillance of Japan indicated an increase in the number of asymptomatic infections from 39 patients in 2010 to 170 patients in 2013 [8]. However, these numbers only accounted for around 10%–20% of all reported cases, indicating that asymptomatic cases of E. histolytica infection are currently underestimated in Japan. For future disease control of E. histolytica infection, it is important to understand the pathogenesis of this microorganism, which involves identifying the determinant factors of disease severity. Previously reported human cohort data indicated that the gut microbiome plays an important role. Cross-sectional studies from India reported that the burden of some bacterial species in the gut microbiota was altered by the presence of E. histolytica [14], and differences in the gut microbiota between asymptomatic colonization and liver abscess during E. histolytica infection have also been reported [15]. The presence of Prevotella copri in the gut flora was shown to be associated with susceptibility to E. histolytica–induced diarrheal disease in 2 geographically distinct areas [16, 17]. It is of interest to better understand the impact of the gut microbiome on the severity of E. histolytica infection.
Herein, we compared the clinical features of asymptomatic E. histolytica infection with those of invasive diseases in our cohort and sought to identify the features of the gut microbiota during asymptomatic infection.
MATERIALS AND METHODS
Study Design and Sampling
This was a single-center, cross-sectional study carried out between 2014 and 2019. Clinical specimens were prospectively collected from patients with suspected E. histolytica infection after obtaining written informed consent. The choice of sampling method was completely dependent on the physicians’ decision; however, samples were examined not only by approved in vitro diagnostic methods in Japan (stool ova and parasite examinations [O&P] and tissue histopathology) but also by other unapproved laboratory diagnostic methods, as detailed below. Stool samples were examined by O&P, which consisted of direct microscopic examination for trophozoites and formalin-ether sedimentation for cyst forms, and polymerase chain reaction (PCR) using E. histolytica–specific primers. For patients undergoing endoscopy, we collected aspirated intestinal fluid samples by washing macroscopically identifiable lesions with 5 mL of saline [18] for O&P and PCR, in addition to tissue biopsy for histopathology with hematoxylin and eosin staining and periodic acid-Schiff staining. Aspirated pus from abscesses was evaluated by O&P and PCR. Residual samples, if any, were immediately frozen at −80°C until further experimental use. This study was approved by the ethics committee of the National Center for Global Health and Medicine (approval number NCGM-G-001566-02 and NCGM-G-003333-00) and was implemented in accordance with the provisions of the Declaration of Helsinki.
Case Definitions
In the present study, “E. histolytica infection” was defined as a case in which the identification of E. histolytica was confirmed by PCR in any clinical specimens (stool, aspirated intestinal fluid, and/or aspirated pus). Clinical forms were defined as follows:
• Asymptomatic infection: Abdominal symptoms were not the reason for the hospital visit, and the frequency of defecation and the morphology of stools were the same as usual;
• Liver abscess: Compatible liver lesions identified by computed tomography and/or sonography that responded to nitroimidazole treatment; and
• Colitis: E. histolytica infection that was not categorized as an asymptomatic infection or a liver abscess.
Serum Antibody Testing
Indirect fluorescent antibody assays using a slide precoated with fixed E. histolytica were performed for the detection of anti–E. histolytica antibody in serum [19, 20]. The commercial kit, Amoeba-Spot IF (bioMérieux SA), which was previously (until the end of 2017) approved for the diagnosis of E. histolytica infection in Japan, was carried out in accordance with the instructions enclosed with the kit. Seropositivity was defined as a positive response in a serum sample diluted at 1:100, and the anti–E. histolytica titer was determined by the highest dilution that produced a positive response.
Endoscopic Assessment
The features identified by endoscopy were assessed by lesion site and distinctive macroscopic appearance as follows: aphthae or erosion, slight damage to the mucosa; ulcer, a clear deep mucosal defect; exudate, mucosal epithelial attachment white or yellow in color, accompanied by aphthae, erosion, and an ulcer; bump, edematous swollen mucosa caused by acute inflammation [11].
Polymerase Chain Reaction
Clinical specimens (0.2 g) were subjected to DNA extraction using a QIAamp Fast DNA Stool Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Genomic DNA (5.0 μL) was subjected to PCR using different groups of primers (N-K2 and R-R) in this study, which were used to amplify E. histolytica HM-1:IMSS transfer RNA gene sequences as previously described [21–23].
Gut Microbiome Analysis
DNA extraction and 16S ribosomal RNA (rRNA) sequencing were conducted according to a previously described method [24]. The V3–V4 regions of bacterial 16S rRNA were PCR amplified using the 341f/806r primers and the dual-index method [25–27]. Barcoded amplicons were sequenced using the paired-end, 2 × 284-bp cycle run on the MiSeq system with MiSeq Reagent Kit version 3 (600 Cycle) chemistry. Paired-end sequencing reads were merged using the fastq-join program with default settings [28]. Only reads that had quality value scores of 20 for >99% of the sequences were extracted, and chimeric sequences were removed using usearch 6.1 [29]. Nonchimeric reads were identified using the TechnoSuruga Laboratory, Japan database DB-BA 13.0 [30, 31]. Operational taxonomic units (OTUs) were aligned based on open-reference picking using the USEARCH version 6.1 of QIIME [29, 32]. The OTUs with a 97% similarity level were identified using the Greengenes database version 13.8 [33]. The α- and β-diversity were implemented in the QIIME pipeline [32].
Statistical Methods
The patients’ characteristics and the laboratory results from each diagnostic test for E. histolytica infection were compared between cases of asymptomatic infection and symptomatic invasive infection. Analysis of variance tests were used for comparisons of patients’ demographic data and the sensitivities of diagnostic tests. Statistical significance was defined as a 2-sided P value < .05. All statistical analyses were performed using GraphPad Prism 7.0 software (GraphPad, San Diego, California). The microbiome composition of asymptomatic infections was compared with symptomatic invasive infections by performing permutational multivariate analysis of variance. The α-diversity and β-diversity were assessed based on both unweighted and weighted UniFrac distance metrics [32].
RESULTS
Patients’ Characteristics
During the study period, E. histolytica infection was suspected in 116 patients, for which 125 specimens were evaluated by PCR. Finally, we identified 64 patients with E. histolytica infection. The patients’ characteristics are summarized in Table 1. Forty-five patients (70.3%) were seropositive for at least 1 of the following infections: HIV type 1, hepatitis B virus, hepatitis C virus, or syphilis, indicating that E. histolytica infection is mainly diagnosed as an STI in this study cohort, whereas imported infections from tropical countries were suspected in 20.3% (13/64) of the patients. Based on the clinical forms of E. histolytica infection described in the Methods, 13 (20.3%) and 51 (79.7%) of the E. histolytica–infected patients were categorized as having an asymptomatic infection and symptomatic invasive infection (45 cases of colitis and 6 cases of liver abscess), respectively. These epidemiological data were consistent with the national surveillance data in Japan [8]. Clinical presentations at diagnosis were varied from asymptomatic to unstable vital changes requiring immediate surgical intervention. Although diarrhea was the most common symptom among symptomatic invasive infections, 14 patients presented with acute abdomen. From the laboratory test results, the white blood counts and C-reactive protein levels in patients with symptomatic invasive infection were found to be elevated, whereas these inflammatory markers were rarely elevated in asymptomatic infected individuals. Medical treatment of E. histolytica infection was performed for all patients according to global guidelines [9]. However, surgical intervention (appendectomy, colectomy, and percutaneous drainage) was required in 6 patients. Death from E. histolytica infection was not reported in the study population. Interestingly, serum anti–E. histolytica antibody was detected in 80% of the patients with asymptomatic infection, whereas only 38% and 25% of the patients with colitis and liver abscesses, respectively, showed positive serology, which probably resulted from the fact that seropositivity depends on the time interval from infection to blood testing [9, 34, 35]. Microbiological diagnosis was made by endoscopic sampling in 38.5% and 23.5% of asymptomatic infection and symptomatic invasive infections, respectively. These results indicated that asymptomatic E. histolytica infection is not a rare comorbidity with STIs, but its diagnosis is sometimes difficult in clinical settings in Japan.
Table 1.
Characteristics of Patients With Entamoeba histolytica Infection
| Characteristic | Asymptomatic Infection (n = 13) |
Symptomatic Invasive Infection (n = 51)a | P Value |
|---|---|---|---|
| Age, y, median Range | 45 (29–68) | 41 (21–67) | .093 |
| Male sex | 12 (92.3) | 45 (88.2) | >.999 |
| MSM | 7 (53.8) | 39 (76.5) | .165 |
| Positive serology of STIs | |||
| HIV (4th generation) | 6 (46.2) | 34 (66.7) | .208 |
| With current OIs | 1 (7.7) | 3 (5.9) | >.999 |
| On ART | 3 (23.1) | 20 (39.2) | .346 |
| Syphilis (TPHA) | 3 (23.1) | 16 (31.4) | .739 |
| Hepatitis B (HBc-Ab) | 5 (38.5) | 22 (43.1) | >.999 |
| Hepatitis C (HCV-Ab) | 2 (15.4) | 2 (3.9) | .181 |
| Travel history to developing countries within 1 y | 4 (30.8) | 9 (17.7) | .439 |
| Clinical symptoms | |||
| Diarrhea | 0 (0.0) | 32 (62.8) | |
| Fever | 0 (0.0) | 20 (39.2) | |
| Abdominal pain | 0 (0.0) | 18 (35.3) | |
| Bloody stool | 0 (0.0) | 15 (29.4) | |
| No complaint | 13 (100) | 0 (0.0) | |
| Laboratory results, median Range | |||
| WBC count, ×103/μL | 5.38 (2.12–8.05) | 7.94 (2.21–30.38) | <.001 |
| CRP, mg/dL | 0.04 (0.01–3.35) | 2.56 (0.02–24.94) | .008 |
| Eosinophils, /μL | 188 (16–718) | 116 (0–1009) | .266 |
| Anti–Entamoeba histolytica Ab | 8/10 (80.0) | 12/33 (36.4) | .028 |
| CD4 count, cells/μLb | 379 (125–507) | 341 (34–1112) | .683 |
| CD4 percentage, %b | 25.3 (9.7–41.9) | 23.7 (3.2–52.1) | .694 |
| HIV RNA, copies/mLb | 8000 (TND–600 000) | 21 (TND–680 000) | .691 |
| Diagnosis method | .265 | ||
| Stool test | 8 (61.5) | 32 (62.7) | |
| Endoscopy | 5 (38.5) | 12 (23.5) | |
| Aspiration/drainage | 0 (0.0) | 7 (13.7) |
Data are presented as no. (%) unless otherwise indicated.
Abbreviations: Ab, antibody; ART, antiretroviral therapy; CRP, C-reactive protein; HBc, hepatitis B core; HCV, hepatitis C virus; HIV, human immunodeficiency virus; MSM, men who have sex with men; OI, opportunistic infection; RNA, ribonucleic acid; STI, sexually transmitted infection; TND, target not detected; TPHA, Treponema pallidum hemagglutination assay; WBC, white blood cell.
aSymptomatic invasive infections included 45 patients with colitis and 6 patients with liver abscess. Acute appendicitis (n = 3), perianal abscess (n = 2), and a fulminant colitis case (n = 1) were included in the colitis group.
bThe peripheral blood tests for CD4+ T cells and HIV viral load were performed only for patients with HIV.
Clinical Features of Asymptomatic E. histolytica Infection
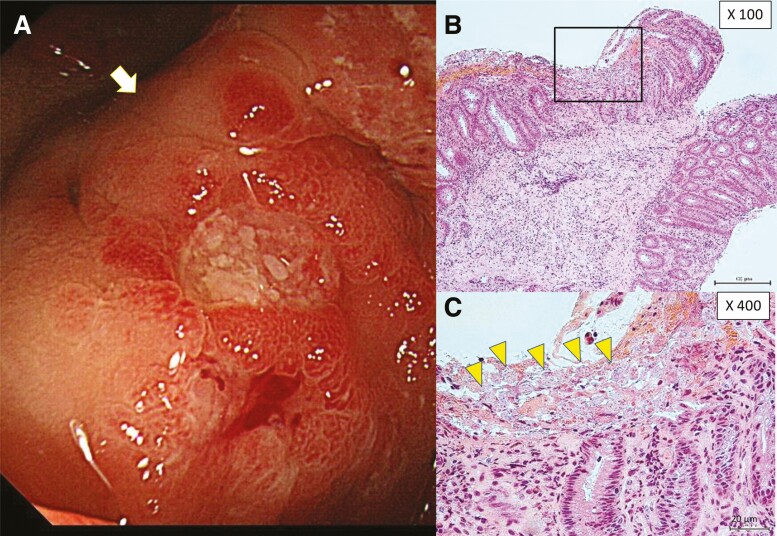
Next, to investigate the clinical features of asymptomatic infection, we summarized the data from 13 patients with asymptomatic E. histolytica infection (Table 2). As per the case definition stated above, abdominal symptoms were not the reason for the hospital visit for any of these asymptomatic patients. The major opportunities to diagnose asymptomatic E. histolytica infection were at cancer screening in 6 patients (fecal occult blood [FOB] positive: 3 patients and complete medical check: 3 patients), and at screening for other STIs in 4 patients (seropositive result for serum anti–E. histolytica antibody). Among 13 patients, stool examination was performed for 9 patients but not for the other 4 patients who were occasionally diagnosed with colonic lesions during endoscopy (2 were FOB positive, 1 underwent a complete medical check, and 1 underwent staging for Kaposi sarcoma). Stool examination identified Entamoeba by O&P in 6 patients (67%), whereas stool PCR identified E. histolytica in 8 patients (89%). Visible colonic ulcers and/or erosions were identified in all 6 patients who underwent endoscopy, even in 1 patient with a negative result for stool PCR (case identification number 11). Interestingly, compared with symptomatic invasive infection, infection sites were limited around the proximal colon, especially in the cecum (Table 3); moreover, trophozoites were frequently identified at infection sites, whereas cystic forms were commonly detected in stools. Unexpectedly, the sensitivity of histopathologic examination of biopsy samples during endoscopy was significantly lower than that of PCR for the same samples (45.5% [10/22] vs 94.7% [18/19], respectively; P < .001), even though all biopsy samples were obtained from the edge of macroscopically visible lesions (Figure 1 and Supplementary Figure 1). This was probably because biopsy samples were small and Entamoeba often resided only on the surface of the mucous layer. Additionally, the antigen detection test (E. HISTOLYTICA II, Techlab, Blacksburg, Virginia) performed on frozen samples (stool and aspirated intestinal fluid samples) had lower sensitivities for pathogen identification compared with PCR.
Table 2.
Summary of the Laboratory Data for 13 Patients With Asymptomatic Intestinal Entamoeba histolytica Infection
| Case ID | Age, y | Sex | Chief Complaint or Reason for Referral | Endoscopy | Serum Anti–Entamoeba histolytica AbTitera | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Stool Examination | Macroscopic Findings | Intestinal Fluid Examination | Histopathology of Biopsied Sample | |||||||
| O&P | PCR | O&P | PCR | |||||||
| 11 | 41 | M | Anti–E. histolytica Ab positive | Negative | Negative | Erosions in the cecum | Trophozoite | Positive | Negative | 400 |
| 104 | 40 | F | FOB test positive | Negative | Positive | Ulcer in the cecumb | NA | NA | Positiveb | NA |
| 71 | 45 | M | Extrapulmonary tuberculosis | Negative | Positive | NA | NA | NA | NA | Negative |
| 77 | 68 | F | Complete medical examination | Cyst | Positive | NA | NA | NA | NA | 400 |
| 92 | 45 | M | Anti–E. histolytica Ab positive | Cyst | Positive | NA | NA | NA | NA | 400 |
| 93 | 42 | M | Anti–E. histolytica Ab positive | Cyst | Positive | NA | NA | NA | NA | 800 |
| 99 | 29 | M | Anti–E. histolytica Ab positive and LFT elevation | Cyst | Positive | NA | NA | NA | NA | 1600 |
| 120 | 45 | M | LFT elevation | Cyst | Positive | NA | NA | NA | NA | NA |
| 124 | 56 | M | Complete medical examination | Cyst | Positive | NA | NA | NA | NA | NA |
| 53 | 38 | M | FOB test positive | NA | NA | Ulcer with exudates in the cecum | Trophozoite | Positive | Positive | 100 |
| 73 | 58 | M | FOB test positive | NA | NA | Multiple erosions with exudates in the cecum | Trophozoite | Positive | Negative | Negative |
| 105 | 52 | M | Complete medical examination | NA | NA | Multiple erosions in the cecum | Trophozoite | Positive | Positive | NA |
| 54 | 49 | M | Kaposi sarcoma | NA | NA | Multiple ulcers and bumpsc with exudates from the cecum to the transverse colon | Negative | Positive | Positive | 100 |
Abbreviations: Ab, antibody; F, female; FOB, fecal occult blood; ID, identification number; LFT, liver function test; M, male; NA, not available; O&P, ova and parasite examination; PCR, polymerase chain reaction.
aAnti–E. histolytica antibody was measured by an indirect immunofluorescence assay. This commercial antibody test was only available until 2017 in Japan, so it could not be used for the samples from 2018 (cases 104–124).
bIn case 104, endoscopy was performed by the previous doctor and only a stool test was performed at our hospital.
cBump was defined as edematous swollen mucosa caused by acute inflammation.
Table 3.
Endoscopic Findings in Patients With Entamoeba histolytica Infection
| Finding | Asymptomatic Infections (n = 6)a |
Colitis (n = 13)a | P Value |
|---|---|---|---|
| Visible intestinal lesions | 6 (100) | 13 (100) | >.999 |
| Intestinal site of infection | |||
| Proximal sites | 6 (100) | 12 (92.3) | >.999 |
| Cecum | 6 (100) | 12 (92.3) | |
| Ascending | 1 (16.7) | 7 (53.8) | |
| Transverse | 1 (16.7) | 6 (46.2) | |
| Distal sites | 0 (0.0) | 9 (69.2) | .011 |
| Descending | 0 (0.0) | 4 (30.8) | |
| Sigmoid | 0 (0.0) | 4 (30.8) | |
| Rectum | 0 (0.0) | 8 (61.5) | |
| Macroscopic appearanceb | |||
| Erosion | 3 (50.0) | 6 (46.2) | >.999 |
| Ulcer | 3 (50.0) | 7 (53.8) | >.999 |
| Exudate | 3 (50.0) | 12 (92.3) | .071 |
| Bump | 1 (16.7) | 2 (15.4) | >.999 |
| Identification of Entamoeba by histopathology | 4 (66.7) | 6 (46.2) | .629 |
Data are presented as no. (%) unless otherwise indicated.
aEach case in both infection groups was diagnosed with E. histolytica infection by polymerase chain reaction only of the stool sample, because both cases had been evaluated by endoscopy by a previous doctor. A detailed evaluation of the clinical specimens collected by endoscopy was not available, except for the histopathologic findings.
bDefinition of each macroscopic appearance during endoscopy is described in the Methods.
Figure 1.
Endoscopic and histopathologic findings from asymptomatic Entamoeba histolytica infections. A, Aphthae and slightly exudated lesions were observed in the cecum by endoscopy. Biopsy was performed at the edge of the aphthous lesion (white arrow). B and C, Trophozoite forms with phagocytosis of red blood cells [yellow arrowheads (C)] were detected on the membrane surface by hematoxylin and eosin staining. Trophozoites could not be identified in the submucosa.
Gut Microbiome Analyses of Asymptomatic E. histolytica Infection
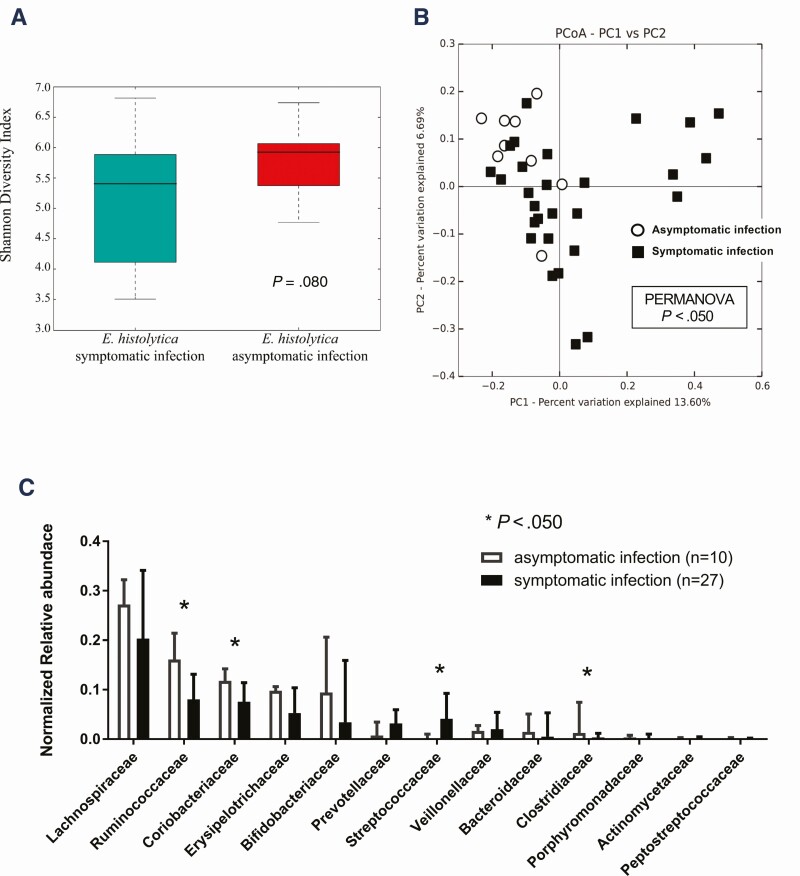
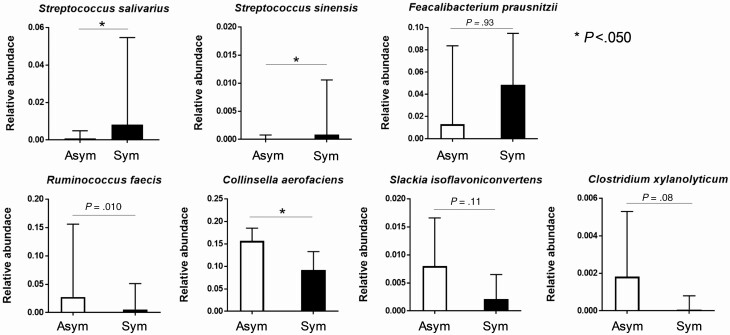
As shown in Table 2, morphological examination by O&P revealed that the cystic form of Entamoeba was frequently observed in stools, whereas the trophozoite form was observed in aspirated intestinal fluid samples of asymptomatically E. histolytica–infected individuals. This result provided a hypothesis that exposure to intestinal contents, such as the gut microbiome, may attenuate the virulence of E. histolytica and induce encystation in asymptomatically infected individuals. We compared the gut microbiotas of asymptomatically infected individuals with those of patients who developed symptomatic invasive infections using 37 stool samples (Supplementary Table 1). First, we assessed the α- and β-diversity in the gut microbiota composition of asymptomatic patients, then compared them with symptomatic patients. Regarding α-diversity, there was no significant difference between the 2 groups (Figure 2A). The β-diversity calculated by principle components analysis revealed that a symptomatic infection (red dots in Figure 2B) provided strong separation along the primary axis of variation of the multidimensional scaling plots, whereas an asymptomatic infection (blue dots in Figure 2B) showed more uniform microbiome composition. These results indicated that the diversity of the gut microbiota was consistent between the 2 groups, but its composition was significantly different, with asymptomatically infected individuals showing specific features of the gut microbiome. Next, to identify the specific bacterial families in prevalence and abundance during asymptomatic infection, we compared the relative abundance of each bacterial family by OTU analysis (Figure 2C). The proportion of Streptococcaceae (potentially exacerbating bacteria) was significantly lower in asymptomatically infected individuals. By contrast, the proportions of Ruminococcaceae, Coriobacteriaceae, and Clostridiaceae (potentially protective bacteria) were significantly higher in asymptomatically infected individuals. We also used Vitcomic2 software (http://vitcomic.org/) to compare the gut microbiome composition between our asymptomatic and symptomatic E. histolytica–infected cases and E. histolytica–uninfected healthy Japanese individuals, using previously published shotgun sequence data for the healthy group [36]; statistical analyses were not applied to these comparisons because of concerns about the inaccuracy resulting from comparing gut microbiome data that were calculated differently (16S rRNA sequencing and shotgun sequencing) (Supplementary Figure 2). These preliminary analyses revealed that some of the symptomatic and uninfected control cases lack 3 “potentially protective” bacterial families (Ruminococcaceae, Coriobacteriaceae, and Clostridiaceae), whereas all of the asymptomatic cases contain these bacterial families (Supplementary Figure 2B). Moreover, 5 symptomatic cases (NA-95, -118, -59, -23, and -66) and 1 healthy control case (DRR042402-DRR042403) showed no (or extremely low) amounts of each of these potentially protective bacterial families in their stool samples (red arrows in Supplementary Figure 2B). In contrast, Streptococcaceae (potentially exacerbating bacteria) was relatively lower in most of the asymptomatic cases, except NA-104, whereas the abundance of Streptococcaceae varied from low to high among healthy control subjects. Next, we investigated prevalence at the genus and species levels for each of the families in asymptomatically infected individuals using Metagenome@KIN software (detailed in the Methods). At the species levels for the Streptococcaceae, the proportions of Streptococcus salivarius and Streptococcus sinensis were significantly lower in asymptomatically infected individuals. At the species level for Ruminococcaceae, Coriobacteriaceae, and Clostridiaceae, only Collinsella aerofaciens was significantly higher in asymptomatically infected individuals (Figure 3). These results suggested that the compositions of the gut microbiome are related to the disease severity of E. histolytica infection, which was represented by low abundance in S. salivarius and S. sinensis and high abundance in C. aerofaciens among asymptomatically infected individuals.
Figure 2.
Composition of the gut microbiota between asymptomatic and symptomatic invasive infections of Entamoeba histolytica. A, Comparison of α-diversity using the Shannon diversity index. B, Principal component analysis (PCoA) of each stool from asymptomatically (white circle) or symptomatically (black square) infected individuals. The first 2 principal components (PC1 and PC2) are shown. The variation (%) explained by each PCoA axis is given. C, Normalized relative abundance of 13 taxa that occurred in at least 0.1% abundance at the family level. The microbiomes (family level) were compared between symptomatic invasive and asymptomatic infections. *P < .05.
Figure 3.
Comparison of the gut microbiotas at the species level. Normalized relative abundance of bacteria that showed significant differences at the family level between asymptomatic (Asym, white bars) and symptomatic invasive infections (Sym, black bars) (see Figure 2C) were compared at the species level. The relative abundance of gene expression levels of bacteria were normalized according to the predicted 16S ribosomal RNA gene abundances (*P < .05, Mann-Whitney U test).
Discussion
The disease severity associated with E. histolytica infection varies from asymptomatic chronic infection to life-threatening fulminant diseases; however, the pathogenesis of amebiasis remains unclear. Interestingly, in asymptomatic E. histolytica–infected cases, visible ulcer lesions and/or erosions were limited to around the cecum, whereas conversely, in symptomatic invasive patients, lesions were identified in various sites throughout the colon by endoscopy. Moreover, similar to previous studies [10, 37], most of the asymptomatically E. histolytica–infected patients in this study evacuated cyst forms of E. histolytica in their stools. Surprisingly, aspirated intestinal fluid obtained during endoscopy from macroscopic membranous lesions in asymptomatically infected individuals frequently contains the trophozoite form of E. histolytica. Furthermore, we examined the gut microbiomes of the study participants. According to the family level analysis, the composition of the microbiome represented by β-diversity analysis in asymptomatic patients showed a relatively uniform microbial community compared with symptomatic invasive patients, with a significant difference being evident between the 2 groups (Supplementary Figure 3). Further assessment of the gut microbiome at the species level revealed that the gene expression levels of 2 bacteria (S. salivarius and S. sinensis) were significantly lower in patients with asymptomatic infection, and the gene expression levels of 1 bacterium (C. aerofaciens) was significantly higher in patients with asymptomatic infection (Figure 3). It was previously reported that S. salivarius has a protective effect on colitis in an animal model due to the inhibitory effect of NF-κB activation [38, 39]. It was also shown that host NF-κB levels were suppressed by E. histolytica in an in vitro infection model [40]. However, no previous reports assessed the direct interaction between these microbiota and E. histolytica during its infection. Taken together, these findings suggest that the gut microbiota might play an important role in determining the disease pathogenesis of E. histolytica infection; however, further studies are needed to elucidate the pathogenesis of asymptomatic/symptomatic E. histolytica infection.
Some limitations need to be considered for the present study. First, this study was performed in a single institute with only a limited number of patients with confirmed E. histolytica infection. Second, initial recruitment and sampling methods were all dependent on clinical judgments in the present observational study design. The frequency of E. histolytica infection, especially asymptomatic cases, may have been underestimated, although the ratio of disease forms among the study population was consistent with that of the Japanese national surveillance data. Third, the gut microbiome can be influenced by numerous host conditions, such as food consumption and comorbidities. Gut microbiome comparisons using samples from a single timepoint (eg, upon diagnosis) cannot be conclusively correlated with disease pathogenesis. Finally, no experimental evaluation was performed for elucidating the mechanism that explains the effect of the gut microbiota on the clinical outcome of E. histolytica infection. Validation using an experimental model of E. histolytica infection should be performed in a future study.
In conclusion, we revealed clinicopathological features of asymptomatic E. histolytica infection. Also, our data support the previous reports that the gut microbiome may play an important role. Further investigations will be needed for elucidating its molecular mechanisms.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors thank Genki Ozawa of TechnoSuruga Laboratory Co, Ltd, Japan, for the gut microbiome analysis; and Analisa Avila, ELS, and Katie Oakley, PhD, of Edanz Group (www.edanzediting.com/ac) for editing drafts of this manuscript.
Financial support. This work was supported by the Emerging/Re-emerging Infectious Diseases Project of Japan from the Japan Agency for Medical Research and Development (grant number JP20fk0108138s0301); and the National Center for Global Health and Medicine of Japan (grant number 19A2016); A research grant from MSD K.K. (Tokyo, Japan).
Potential conflicts of interest. The authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. Lozano R, Naghavi M, Foreman K, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012; 380:2095–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nagata N, Shimbo T, Akiyama J, et al. Risk factors for intestinal invasive amebiasis in Japan, 2003–2009. Emerg Infect Dis 2012; 18:717–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hung CC, Chang SY, Ji DD. Entamoeba histolytica infection in men who have sex with men. Lancet Infect Dis 2012; 12:729–36. [DOI] [PubMed] [Google Scholar]
- 4. James R, Barratt J, Marriott D, Harkness J, Stark D. Seroprevalence of Entamoeba histolytica infection among men who have sex with men in Sydney, Australia. Am J Trop Med Hyg 2010; 83:914–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Roure S, Valerio L, Soldevila L, et al. Approach to amoebic colitis: epidemiological, clinical and diagnostic considerations in a non-endemic context (Barcelona, 2007–2017). PLoS One 2019; 14:e0212791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Watanabe K, Gatanaga H, Escueta-de Cadiz A, Tanuma J, Nozaki T, Oka S. Amebiasis in HIV-1-infected Japanese men: clinical features and response to therapy. PLoS Negl Trop Dis 2011; 5:e1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yanagawa Y, Nagata N, Watanabe K, et al. Increases in Entamoeba histolytica antibody-positive rates in human immunodeficiency virus-infected and noninfected patients in Japan: a 10-year hospital-based study of 3,514 patients. Am J Trop Med Hyg 2016; 95:604–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ishikane M, Arima Y, Kanayama A, et al. Epidemiology of domestically acquired amebiasis in Japan, 2000–2013. Am J Trop Med Hyg 2016; 94:1008–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Petri WA Jr., Haque R. Entamoeba species, including amebiasis. In: Bennett JE, Dolin R, Blaser MJ, et al. , eds. Mandell, Douglas, and Bennett’s principles and practice of infectious diseases. 9th ed. Vol. 2. Philadelphia: Elsevier/Saunders, 2019:3422–5. [Google Scholar]
- 10. Blessmann J, Ali IK, Nu PA, et al. Longitudinal study of intestinal Entamoeba histolytica infections in asymptomatic adult carriers. J Clin Microbiol 2003; 41:4745–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nagata N, Shimbo T, Akiyama J, et al. Predictive value of endoscopic findings in the diagnosis of active intestinal amebiasis. Endoscopy 2012; 44:425–8. [DOI] [PubMed] [Google Scholar]
- 12. Okamoto M, Kawabe T, Ohata K, et al. Amebic colitis in asymptomatic subjects with positive fecal occult blood test results: clinical features different from symptomatic cases. Am J Trop Med Hyg 2005; 73:934–5. [PubMed] [Google Scholar]
- 13. Spinzi G, Pugliese D, Filippi E. An unexpected cause of chronic diarrhea. Gastroenterology 2016; 150:e5–6. [DOI] [PubMed] [Google Scholar]
- 14. Verma AK, Verma R, Ahuja V, Paul J. Real-time analysis of gut flora in Entamoeba histolytica infected patients of northern India. BMC Microbiol 2012; 12:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rani R, Murthy RS, Bhattacharya S, Ahuja V, Rizvi MA, Paul J. Changes in bacterial profile during amebiasis: demonstration of anaerobic bacteria in ALA pus samples. Am J Trop Med Hyg 2006; 75:880–5. [PubMed] [Google Scholar]
- 16. Gilchrist CA, Petri SE, Schneider BN, et al. Role of the gut microbiota of children in diarrhea due to the protozoan parasite Entamoeba histolytica. J Infect Dis 2016; 213:1579–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ngobeni R, Abhyankar MM, Jiang NM, et al. Entamoeba histolytica-encoded homolog of macrophage migration inhibitory factor contributes to mucosal inflammation during amebic colitis. J Infect Dis 2017; 215:1294–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nagata N, Shimbo T, Sekine K, et al. Combined endoscopy, aspiration, and biopsy analysis for identifying infectious colitis in patients with ileocecal ulcers. Clin Gastroenterol Hepatol 2013; 11:673–80.e2. [DOI] [PubMed] [Google Scholar]
- 19. Fotedar R, Stark D, Beebe N, Marriott D, Ellis J, Harkness J. Laboratory diagnostic techniques for Entamoeba species. Clin Microbiol Rev 2007; 20:511–32, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Garcia LS, Bruckner DA, Brewer TC, Shimizu RY. Comparison of indirect fluorescent-antibody amoebic serology with counterimmunoelectrophoresis and indirect hemagglutination amoebic serologies. J Clin Microbiol 1982; 15: 603–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ali IK, Zaki M, Clark CG. Use of PCR amplification of tRNA gene-linked short tandem repeats for genotyping Entamoeba histolytica. J Clin Microbiol 2005; 43:5842–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ali IK, Mondal U, Roy S, Haque R, Petri WA Jr, Clark CG. Evidence for a link between parasite genotype and outcome of infection with Entamoeba histolytica. J Clin Microbiol 2007; 45:285–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zaki M, Meelu P, Sun W, Clark CG. Simultaneous differentiation and typing of Entamoeba histolytica and Entamoeba dispar. J Clin Microbiol 2002; 40:1271–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Takahashi S, Tomita J, Nishioka K, Hisada T, Nishijima M. Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS One 2014; 9:e105592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 1993; 59:695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Caporaso JG, Lauber CL, Walters WA, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 2011; 108(Suppl 1):4516–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hisada T, Endoh K, Kuriki K. Inter- and intra-individual variations in seasonal and daily stabilities of the human gut microbiota in Japanese. Arch Microbiol 2015; 197:919–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. GitHub Pages. Command-line tools for processing biological sequencing data. Available at: https://expressionanalysis.github.io/ea-utils/. Accessed 20 January 2020.
- 29. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011; 27:2194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 2007; 73:5261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kasai C, Sugimoto K, Moritani I, et al. Comparison of the gut microbiota composition between obese and non-obese individuals in a Japanese population, as analyzed by terminal restriction fragment length polymorphism and next-generation sequencing. BMC Gastroenterol 2015; 15:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010; 7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006; 72:5069–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Katzenstein D, Rickerson V, Braude A. New concepts of amebic liver abscess derived from hepatic imaging, serodiagnosis, and hepatic enzymes in 67 consecutive cases in San Diego. Medicine (Baltimore) 1982; 61:237–46. [DOI] [PubMed] [Google Scholar]
- 35. Haque R, Mollah NU, Ali IK, et al. Diagnosis of amebic liver abscess and intestinal infection with the TechLab Entamoeba histolytica II antigen detection and antibody tests. J Clin Microbiol 2000; 38:3235–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nishijima S, Suda W, Oshima K, et al. The gut microbiome of healthy Japanese and its microbial and functional uniqueness. DNA Res 2016; 23:125–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Blessmann J, Tannich E. Treatment of asymptomatic intestinal Entamoeba histolytica infection. N Engl J Med 2002; 347:1384. [DOI] [PubMed] [Google Scholar]
- 38. Kaci G, Goudercourt D, Dennin V, et al. Anti-inflammatory properties of Streptococcus salivarius, a commensal bacterium of the oral cavity and digestive tract. Appl Environ Microbiol 2014; 80:928–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Frick JS, Fink K, Kahl F, et al. Identification of commensal bacterial strains that modulate Yersinia enterocolitica and dextran sodium sulfate–induced inflammatory responses: implications for the development of probiotics. Infect Immun 2007; 75:3490–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kammanadiminti SJ, Chadee K. Suppression of NF-kappaB activation by Entamoeba histolytica in intestinal epithelial cells is mediated by heat shock protein 27. J Biol Chem 2006; 281:26112–20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.