Abstract
Background
DNA from many pathogens can be detected in saliva. However, the presence and quantity of Treponema pallidum DNA in patients with syphilis in saliva is unknown.
Methods
234 patients with syphilis with different stages and 30 volunteers were enrolled. Paired saliva and plasma samples were collected from all participants. Consecutive saliva samples from 9 patients were collected every 4 hours following treatment. Treponema pallidum DNA in samples was determined by nested polymerase chain reaction (PCR) and droplet digital PCR targeting polA and Tpp47.
Results
Treponema pallidum DNA detection rates in saliva and plasma were 31.0% (9/29) and 51.7% (15/29) in primary syphilis (P = .11), 87.5% (63/72) and 61.1% (44/72) in secondary syphilis (P < .001), 25.6% (21/82) and 8.5% (7/82) in latent syphilis (P = .004), and 21.6% (11/51) and 5.9% (3/51) in symptomatic neurosyphilis (P = .021), respectively. Median (range) loads of Tpp47 and polA in saliva were 627 (0–101 200) and 726 (0–117 260) copies/mL, respectively, for patients with syphilis. In plasma, however, loads of Tpp47 and polA were low: medians (range) of 0 (0–149.6) and 0 (0–176) copies/mL, respectively. Loads of T. pallidum DNA in saliva during treatment fluctuated downward; the clearance time was positively correlated with the loads of T. pallidum DNA before treatment.
Conclusions
Collection of saliva is noninvasive and convenient. The high loads of T. pallidum DNA in saliva and reduction after treatment indicated that saliva can be not only a diagnostic fluid for syphilis but also an indicator of therapeutic effectiveness.
Keywords: Treponema pallidum DNA, PCR, saliva
There were high loads of Treponema pallidum DNA in saliva in patients with syphilis. In addition, the collection of saliva is noninvasive and convenient. Therefore, saliva may be a new specimen for syphilis diagnosis.
(See the Editorial Commentary by Hook on pages e3259–60.)
Despite extensive strategies for the prevention and control of syphilis, this centuries-old sexually transmitted disease remains a public health concern worldwide. The incidence of syphilis continues to increase globally, including alarming surges in developed countries in recent years [1]. China experienced a surge of syphilis epidemic with the number of diagnosed cases of syphilis increasing by 14–19% annually during 2004–2013 [2], and the number reached 494 867 cases in 2018 [3]. Over the past 2 decades, syphilis has been ranked as 1 of the top 3 infectious diseases in China [3].
The confirmatory diagnosis of infectious diseases mainly relies on direct detection of pathogens from clinical specimens. However, the causative agent of syphilis, Treponema pallidum subspecies pallidum (T. pallidum) is difficult to culture in vitro. Dark-field microscopy is only useful for new, moist primary and secondary lesions, and the sensitivity decreases as the lesions heal or following receipt of antibiotics [4, 5]. A rabbit inoculation test is impractical due to the length of time required [4]. To date, it remains a challenge to detect T. pallidum in patients with syphilis.
Polymerase chain reaction (PCR) methods have been developed for the detection of T. pallidum DNA in recent years [5–9]. Treponema pallidum can disseminate to nearly all tissues and organs, and T. pallidum DNA has been detected by PCR from a variety of biospecimens, such as lesions, blood, cerebrospinal fluid, urine, and semen [5–9]. Recently, several cases provided compelling evidence that mouth-to-mouth transfer of prechewed food or kissing could also result in the transmission of T. pallidum [10–12]. A previous study also showed that there was a high prevalence of T. pallidum DNA in oral swabs from patients with syphilis [13]. Surprisingly, almost 40% of oral swabs from patients without visible oral lesions contained T. pallidum DNA [13]. These studies suggested that saliva could also be a useful biospecimen for the detection of T. pallidum DNA.
To comprehensively investigate whether T. pallidum DNA may be present in saliva in patients with syphilis, we enrolled patients with all stages of syphilis. We used nested PCR (nPCR) and droplet digital PCR (ddPCR) assays to detect and quantify T. pallidum DNA in saliva and plasma from all participants and monitored the dynamic changes in T. pallidum DNA following treatment.
METHODS
Ethics Statement and Subjects
The study was approved by the Ethics Committee of the Shanghai Skin Disease Hospital. Eligible patients who visited Sexually Transmitted Disease (STD) of the Shanghai Skin Disease Hospital during March 2018 and April 2019 were invited to participate in this study. A total of 30 volunteers from the STD Department of the Shanghai Skin Disease Hospital were included in this study. Their serological tests for syphilis were negative and they showed no manifestations of syphilis. Medical and sociodemographic information was collected using a questionnaire, including sexual orientation (Table 1). Patients who received treatment for syphilis prior to sample collection were excluded. Written informed consent was obtained from all participants. The diagnosis and treatment of primary, secondary, and latent syphilis and neurosyphilis were according to the guidelines of the STD Association, China Centers for Disease Control [14].
Table 1.
Clinical Characteristics of 234 Patients With Syphilis Enrolled in This Study
| Variable | All Cases (n = 234) | Primary Syphilis (n = 29) | Secondary Syphilis (n = 72) | Latent Syphilis (n = 82)a | Symptomatic Neurosyphilis (n = 51)b |
|---|---|---|---|---|---|
| Age, median (IQR), years | 52 (34–61) | 39 (28.5–54) | 35(28–53) | 57 (43–65) | 56 (50–63) |
| Male gender, n (%) | 154 (65.8) | 29 (100) | 38 (52.8) | 44 (53.7) | 43 (84.3) |
| Blood-reactive TPPA, n (%) | 234 (100) | 29 (100) | 72 (100) | 82 (100) | 51 (100) |
| Blood-reactive RPR, n (%) | 232 (99.1) | 27 (93.1) | 72 (100) | 82 (100) | 51 (100) |
| Plasma RPR titer, median (range) | 32 (0–512) | 16 (0–128) | 64 (2–512) | 32 (1–256) | 64 (4–512) |
| HIV infection, n (%) | 19 (8.1) | 1 (3.4) | 18 (25) | 0 (0) | 0 (0) |
| MSM, n (%) | 27 (11.5) | 1 (3.4) | 21 (29.2) | 5 (6.1) | 0 (0) |
| Oral sex, n (%) | 38 (17.1)c | 1 (3.4) | 28 (40.0)d | 8 (10.7)e | 1 (2.1)f |
| Oral lesions, n (%) | 18 (7.7) | 0 (0) | 18 (25)g | 0 (0) | 0 (0) |
Abbreviations: HIV, human immunodeficiency virus; IQR, interquartile range; MSM, men who have sex with men; Oral lesions, mucosal ulcer and/or leukoplakia; RPR, rapid plasma reagin; TPPA, Treponema pallidum particle agglutination.
aIncluding 5 early latent syphilis, 8 late latent syphilis, and 69 unknown duration latent syphilis.
bIncluding 5 meningovascular neurosyphilis, 32 general paresis, 10 ocular neurosyphilis, 1 tabetic neurosyphilis, 1 patient with both tabetic and paretic neurosyphilis, 1 patient with both ocular and paretic neurosyphilis, and 1 patient with both meningovascular and paretic neurosyphilis.
cThe number of all cases was 222 because 12 cases were unknown.
dThe number of all cases was 70 because 2 cases were unknown.
eThe number of all cases was 75 because 7 cases were unknown.
fThe number of all cases was 48 because 3 cases were unknown.
gIncluding 3 patients with oral ulcer, 14 patients with oral leukoplakia, and 1 patient with both oral ulcer and leukoplakia.
Sample Collection
Paired saliva and plasma samples were collected from all participants. Five milliliters of blood with anticoagulant EDTA (ethylenediaminetetraacetic acid) was centrifuged (3000 rpm, 10 minutes) and plasma was collected. Saliva samples (at least 1 mL) were collected in the Omnigene Oral kit (Genoteke, Ottawa, Canada) or 50-mL sterile centrifuge tubes without inducing salivation or using any form of mouthwash. In addition, saliva samples at 4-hour intervals after treatment were collected from 9 patients (1 with secondary syphilis, 1 with secondary syphilis with human immunodeficiency virus [HIV] coinfection, 3 with secondary syphilis with central nervous system [CNS] involvement, 2 patients with relapse of secondary syphilis, 1 with unknown duration of latent syphilis with CNS involvement, and 1 with ocular neurosyphilis). All samples were frozen at −80 °C.
DNA Extraction and Nested PCR Assay
Samples from patients with syphilis and volunteers were extracted under the same experimental condition. One milliliter of plasma and 1 mL of saliva were extracted in a DNA clean environment using the QiaAmp DNA Mini blood kit (Qiagen, Inc, Valencia, CA) according to the manufacturer’s instructions. DNA was dissolved in 100 µL TE buffer. Amplification of polA and Tpp47 was performed by nPCR as previously described [6]. Positive (Nichols strain of T. pallidum) and negative (distilled water) controls were applied for each set of reactions.
Quantity of polA and Tpp47 by Droplet Digital PCR Assay
Droplet digital PCR reaction mixture consisted of 11 μL 2 × ddPCR supermix (Bio-Rad, Pleasanton, CA), 1 μL primers (0.9 pmol/μL)/probe (0.25 pmol/μL) mix, and 10 μl of sample DNA. Twenty microliters of this mix was loaded into a DG8 droplet generator to generate droplets. Droplets were transferred to a PCR plate and amplified under the following settings for polA: 95°C for 5 minutes, 40 cycles at 94°C for 30 seconds, 60°C for 30 seconds, and 72°C for 30 seconds, and followed by 4°C for 5 minutes and 90°C for 5 minutes. The cycling conditions for Tpp47 were as follows: 95°C for 5 minutes, 40 cycles at 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 30 seconds, and followed by 4°C for 5 minutes and 90°C for 5 minutes. After amplification, products were read automatically on a QX200 droplet reader. Data were analyzed with QuantaSoft analysis software, and the final load (copies/mL) of each sample was determined by multiplying the average copy number per microliter of PCR mixture in each well and the sample dilution factor. The sequences of polA and Tpp47 primers were as follows—polA forward primer: 5′-CAGGATCCGGCATATGTCC-3′; and reverse primer: 5′-AAGTGTGAGCGTCTCATCATTCC-3′, probe FAM-CTGTCATGCACCAGCTT CGACGTCTT-MGB; Tpp47 forward primer: 5′-CAACACGGTCCGCTACGACTA-3′, and reverse primer: 5′-TGCCATAACTCGCCATCAGA-3′, probe FAM-CGGTGATGACGCGAGCTACACCA-MGB.
Statistical Analysis
All statistical analyses were performed using SPSS software, version 19.0 (SPSS, Inc, Chicago, IL). Descriptive statistics were used to calculate the median and interquartile range (IQR) or range. Differences between the groups were analyzed using the nonparametric Mann-Whitney U test. Spearman correlation analysis was performed between the clearance time and the loads of polA and Tpp47 before treatment. The chi-square test and Fisher’s exact test were performed to compare the proportion between groups. Factors with P < .1 were included in the logistical regression. Differences were considered to be statistically significant at a 2-sided P < .05.
RESULTS
Clinical Characteristics of the Enrolled Patients With Syphilis
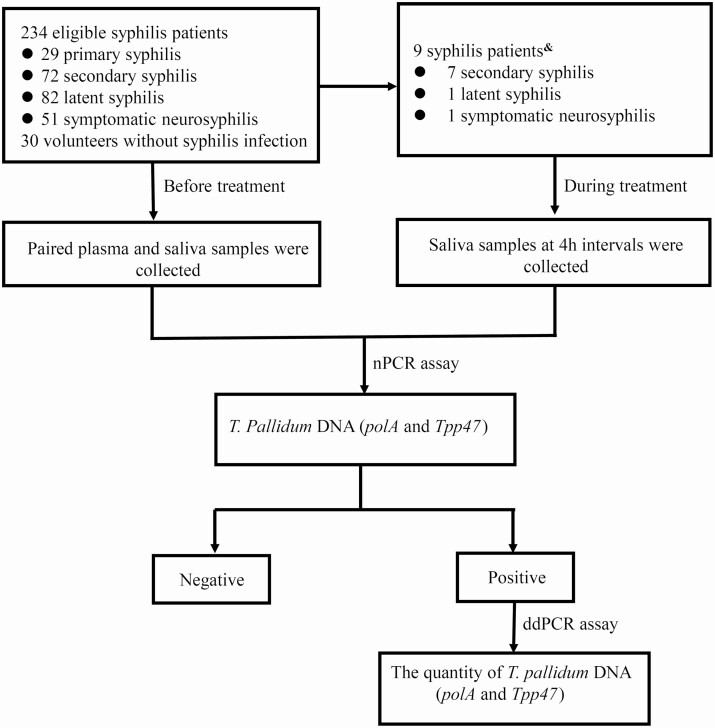
A total of 234 patients were enrolled, including 29 (12.4%) with primary syphilis, 72 (30.8%) with secondary syphilis, 82 (35.0%) with latent syphilis, and 51 (21.8%) with symptomatic neurosyphilis (Figure 1). Among them, there were 14 patients with secondary syphilis and 46 patients with latent syphilis with no neurological/psychiatric symptoms or signs but with cerebrospinal fluid abnormalities. As shown in Table 1, 19 (8.1%) patients were coinfected with HIV, 27 (11.5%) patients were men who have sex with men, and 38 (17.1%) patients had oral sex. All 18 patients who had oral lesions were secondary syphilis (of which 6 reported oral sex).
Figure 1.
Flowchart of participant enrollment and study design. “Negative” indicates that PolA and Tpp47 were negative by nPCR assay; “Positive” indicates that PolA and/or Tpp47 were positive by nPCR assay. &PolA and Tpp47 were positive in saliva from 9 patients. Abbreviations: ddPCR, droplet digital polymerase chain reaction; nPCR, nested polymerase chain reaction.
Treponema pallidum DNA Can Be Detected in Saliva at Any Stage of Syphilis and Saliva Possesses a Higher Detection Rate of T. pallidum DNA Compared With Plasma
To detect the presence of T. pallidum DNA in saliva and plasma, and to compare the detection rate, paired saliva and plasma samples were collected from all participants. Samples were considered positive if nPCR was positive for polA and/or Tpp47. As shown in Table 2, the overall T. pallidum DNA detection rate in saliva was 44.4% (104/234), which was significantly higher than that in plasma (29.5%, 69/234) (P = .001). However, T. pallidum DNA detection rates in saliva (31.0%, 9/29) and plasma (51.7%, 15/29) were not significantly different in primary syphilis (P = .11); however, the T. pallidum DNA detection rates in saliva increased with blood rapid plasma reagin (RPR) titers (P = .013) (Supplementary Table 1). When blood RPR titers ranged from 64 to 128, the detection rate in saliva reached 71.4% (5/7). Treponema pallidum DNA detection rates in saliva were all significantly higher those in plasma among patients with secondary syphilis (87.5% [63/72] vs 61.1% [44/72], P < .001), latent syphilis (25.6% [21/82] vs 8.5% [7/82], P = .004), and symptomatic neurosyphilis (21.6% [11/51] [2 meningovascular, 3 general paralysis, 5 ocular syphilis, 1 tabes dorsalis] vs 5.9% [3/51] P = .021).
Table 2.
Treponema pallidum DNA Detection Rates in Saliva and Plasma From Patients With Different Stages of Syphilis
| Stage | Saliva, % (n/N) | Plasma, % (n/N) | P | ||||
|---|---|---|---|---|---|---|---|
| polA(+) | Tpp47(+) | polA and Tpp47(+) | polA(+) | Tpp47(+) | polA and Tpp47(+) | ||
| Primary syphilis (n = 29) | 20.7 (6/29) | 27.6 (8/29) | 31.0 (9/29) | 37.9 (11/29) | 37.9 (11/29) | 51.7 (15/29) | .11 |
| Secondary syphilis (n = 72) | 87.5 (63/72) | 84.7 (61/72) | 87.5 (63/72) | 52.8 (38/72) | 50 (36/72) | 61.1 (44/72) | <.001 |
| Latent syphilis (n = 82) | 24.4 (20/82) | 18.3 (15/82) | 25.6 (21/82) | 3.7 (3/82) | 7.3 (6/82) | 8.5 (7/82) | .004 |
| Symptomatic neurosyphilis (n = 51) | 17.6 (9/51) | 19.6 (10/51) | 21.6 (11/51) | 5.9 (3/51) | 3.9 (2/51) | 5.9 (3/51) | .021 |
| All cases (n = 234) | 41.9 (98/234) | 40.2 (94/234) | 44.4 (104/234) | 23.5 (55/234) | 23.5 (55/234) | 29.5 (69/234) | .001 |
All of the saliva and plasma samples from 30 uninfected volunteers were negative for both polA and Tpp47. polA and Tpp47(+) indicates positive for polA and/or Tpp47; P values refer to the statistical difference between the percentage of polA and Tpp47 positive in saliva and the percentage of polA and Tpp47 positive in plasma at the same stage.
We then investigated the T. pallidum DNA in saliva and plasma from the same patient. As shown in Table 3, 22.6% (53/234) of all patients were positive for both saliva and plasma (Saliva+/Plasma+), and 21.8% (51/234) of patients were Saliva+ and negative for plasma (Plasma−); only 6.8% (16/234) of patients were negative for saliva (Saliva−) and Plasma+ (P < .0001). However, there was no significant difference between Saliva+/Plasma− (13.8%, 4/29) and Saliva−/Plasma+ (34.5%,10/29) in primary syphilis (P = .123). The percentage of patients with Saliva+/Plasma− was significantly higher than that with Saliva−/Plasma+ among patients with secondary syphilis, latent syphilis, and symptomatic neurosyphilis (all P < .05). Overall, these results demonstrate a significantly higher detection rate for T. pallidum DNA in saliva than in plasma, except for primary syphilis.
Table 3.
Presence of Treponema pallidum DNA in Saliva and Plasma From the Same Patient
| Variables | All Casesa (N = 234) | Primary Syphilis (n = 29) | Secondary Syphilisa (n = 72) | Latent Syphilisa (n = 82) | Symptomatic Neurosyphilisa (n = 51) |
|---|---|---|---|---|---|
| Saliva+/Plasma+ | 22.6 (53/234) | 17.2 (5/29) | 58.3 (42/72) | 4.9 (4/82) | 3.9 (2/51) |
| Saliva+/Plasma− | 21.8 (51/234) | 13.8 (4/29) | 29.2 (21/72) | 20.7 (17/82) | 17.6 (9/51) |
| Saliva−/Plasma+ | 6.8 (16/234) | 34.5 (10/29) | 2.8 (2/72) | 3.7 (3/82) | 2.0 (1/51) |
| Saliva−/Plasma− | 48.7 (114/234) | 34.5 (10/29) | 9.7 (7/72) | 70.7 (58/82) | 76.5 (39/51) |
Data are presented as % (n/N). Saliva+/Plasma+, patients who were T. pallidum DNA positive in saliva and plasma; Saliva+/Plasma−, patients who were T. pallidum DNA positive in saliva and negative in plasma; Saliva−/Plasma+, patients who were T. pallidum DNA negative in saliva and positive in plasma; Saliva−/Plasma−, patients who were T. pallidum DNA negative in saliva and plasma.
aThere were significant differences between the percentage of Saliva+/Plasma− and the percentage of Saliva−/Plasma+ at the same stage (P < .05).
Treponema pallidum DNA Was Abundant in Saliva Among Patients With Syphilis, and the Loads Were Significantly Higher Than That in Plasma
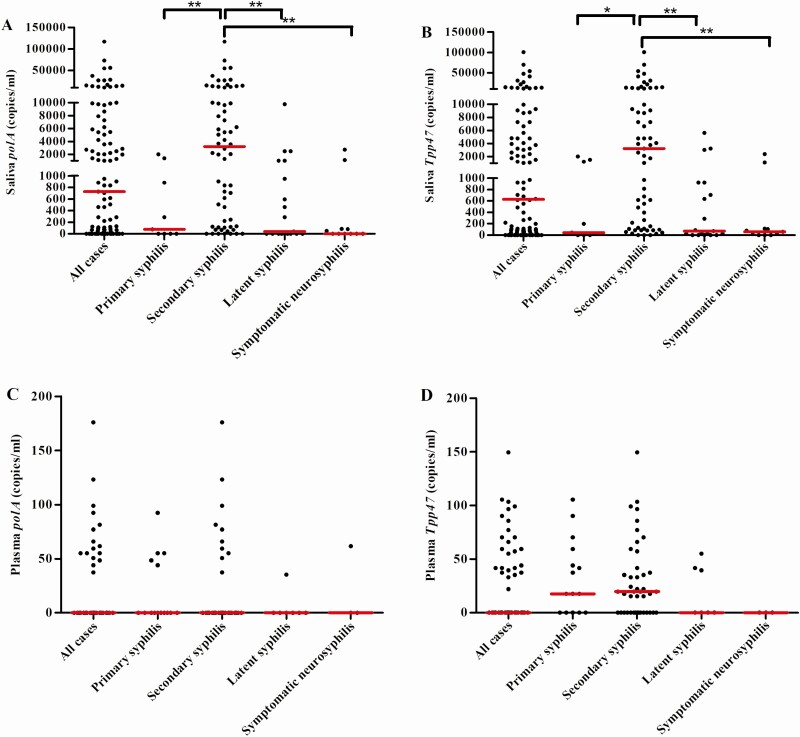
To determine the loads of T. pallidum DNA in saliva and plasma, positive samples were included for ddPCR analysis. As the sensitivity of ddPCR is lower than nPCR, some samples were undetectable by ddPCR and their readings were set as zeros. As shown in Figure 2, the median (range) loads of Tpp47 and polA in saliva were 627 (0–101 200) copies/mL and 726 (0–117 260) copies/mL, respectively. In plasma, however, the loads of Tpp47 and polA were very low, with a median (range) of 0 (0–149.6) copies/mL and 0 (0–176) copies/mL, respectively. The loads of T. pallidum DNA in saliva were significantly higher than that in plasma (both P < .001). The loads of T. pallidum DNA were the highest in saliva from patients with secondary syphilis. As shown in Figure 2A and 2B, the median (range) loads of Tpp47 and polA were 3212 (0–101 200) copies/mL and 3190 (0–117 260) copies/mL in secondary syphilis and were significantly higher than in primary syphilis, latent syphilis, and symptomatic neurosyphilis (all P < .05).
Figure 2.
The quantity of Treponema pallidum DNA in saliva and plasma from patients with different stages of syphilis. The loads of polA (A) and Tpp47 (B) in saliva from all cases (n = 101), primary syphilis (n = 9), secondary syphilis (n = 62), latent syphilis (n = 19), and symptomatic neurosyphilis (n = 11). The loads of polA (C) and Tpp47 (D) in plasma from all cases (n = 68), primary syphilis (n = 15), secondary syphilis (n = 43), latent syphilis (n = 7), and symptomatic neurosyphilis (n = 3). The red horizontal lines show the medians.*P < .05; **P < .001. Three saliva-positive samples and 1 plasma-positive sample were not further quantified because there was not enough DNA template.
Secondary Syphilis, Blood Rapid Plasma Reagin Titer, and Positive Plasma Treponema pallidum DNA Were Associated With Positive Saliva T. pallidum DNA
We further analyzed the factors that were associated with positive saliva T. pallidum DNA by logistic regression. As shown in Table 4, blood RPR titer equal to or higher than 1:32 was significantly associated with positive detection of saliva T. pallidum DNA, with an adjusted odds ratio (OR) of 11.21 (95% confidence interval [CI], 3.78–33.25) (P < .001). Compared with patients with symptomatic neurosyphilis, secondary syphilis was significantly associated with positive detection of saliva T. pallidum DNA, with an adjusted OR of 13.2 (95% CI, 3.88–44.84) (P < .001). Positive plasma T. pallidum DNA was also associated with positive detection of saliva T. pallidum DNA, with an adjusted OR of 4.30 (95% CI, 1.59–11.59) (P = .004).
Table 4.
Factors Associated With Saliva Treponema pallidum DNA Positive Findingsin Patients With Syphilis
| Variables | Saliva T. pallidum DNA Positive (n = 104) | Saliva T. pallidum DNA Negative (n = 130) | Adjusted OR | 95% CI | P |
|---|---|---|---|---|---|
| Age, median (range), years | 39 (17–75) | 56 (20–84) | .96 | .94–1 | .007 |
| MSM, % (n/N) | 21.4 (22/104) | 3.8 (5/130) | 3.62 | .34–39.09 | .289 |
| Oral sex, % (n/N) | 29.0 (29/100)a | 7.4 (9/122)b | .50 | .09–2.82 | .433 |
| Plasma RPR titer ≥1:32, % (n/N) | 91.3 (95/104) | 53.1 (69/130) | 11.21 | 3.78–33.25 | <.001 |
| Plasma RPR titer, median (range) | 64 (2–512) | 32 (0–512) | 1.00 | 1.0–1.01 | .694 |
| Plasma DNA(+), % (n/N) | 51.0 (53/104) | 12.3 (16/130) | 4.30 | 1.59–11.59 | .004 |
| Stages, % (n/N) | |||||
| Symptomatic neurosyphilis | 10.7 (11/104) | 30.5 (40/130) | … | … | … |
| Primary | 8.7 (9/104) | 15.3 (20/130) | 1.40 | .35–5.67 | .636 |
| Secondary | 60.6 (63/104) | 6.9 (9/130) | 13.20 | 3.88–44.84 | <.001 |
| Latent | 20.4 (21/104) | 46.6 (61/130) | 1.38 | .54–3.52 | .505 |
| HIV infection, % (n/N) | 15.5 (16/104) | 2.3 (3/130) | .15 | .01–1.54 | .109 |
P values refer to the statistical difference of the association between the presence of T. pallidum DNA in saliva and the variables using the logistic regression. Abbreviations: CI, confidence interval; MSM, men who have sex with men; OR, odds ratio; RPR, rapid plasma reagin.
aThe number of all cases was 100 because 4 cases were unknown.
bThe number of all cases was 122 because 8 cases were unknown.
Quantity of Treponema pallidum DNA in Saliva Declined After Treatment
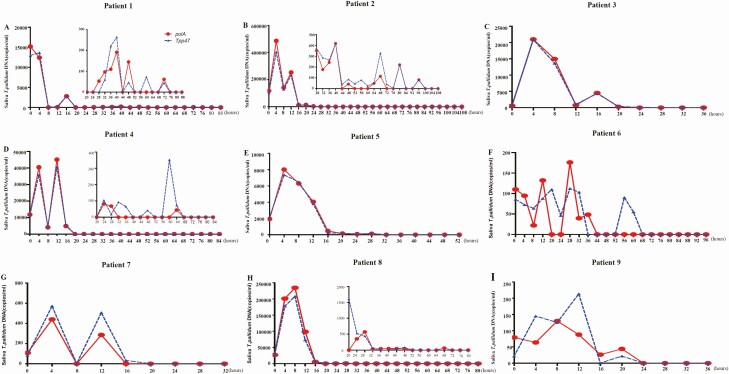
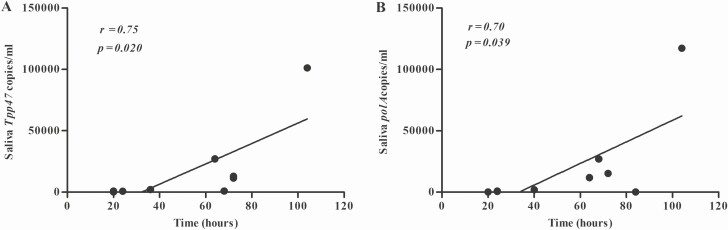
We consecutively collected saliva samples from 9 patients at 4-hour intervals following treatment to investigate the dynamic characteristics of T. pallidum DNA clearance in saliva after treatment initiation. Clearance of T. pallidum DNA was defined as at least 3 consecutive negative detections by nPCR (Supplementary Table 2). The median clearance time for T. pallidum DNA in saliva was 64 hours, for polA was 64 hours (IQR, 24–78 hours), and for Tpp47 was 64 hours (IQR, 22–72 hours). As shown in Figure 3, the loads of T. pallidum DNA fluctuated downward after initiation of treatment. The loads of Tpp47 and polA reached a peak at 4– 8 hours after treatment. The time-to-clearance was correlated with the loads of T. pallidum DNA before treatment (Figure 4), with correlation coefficients of 0.75 and 0.70 for Tpp47 and polA, respectively (both P < .05).
Figure 3.
The quantity of polA and Tpp47 in saliva from 9 patients with syphilis after treatment by ddPCR assay. Patient 2, patient 5, and patient 7 had secondary syphilis with CNS involvement and were treated with aqueous crystalline penicillin G, 4MU intravenously every 4 hours for 14 days (B, E, G). Patient 4 and patient 8 had secondary syphilis relapse and were treated with aqueous crystalline penicillin G, 4MU intravenously every 4 hours for 14 days (D, H). Patient 9 had an ocular neurosyphilis and was treated with aqueous crystalline penicillin G, 4MU intravenously every 4 hours for 14 days (I). Patient 6 had secondary syphilis and was treated with benzathine penicillin 2.4MU/qw intramuscularly for 2 weeks (F). Patient 3 had latent syphilis with CNS involvement and was treated with aqueous crystalline penicillin G, 4MU intravenously every 4 hours for 14 days (C). Patient 1 had secondary syphilis coinfected with HIV and was treated with aqueous crystalline penicillin G, 4MU intravenously every 4 hours for 14 days (A). The loads of polA and Tpp47 were significantly decreased compared with before treatment in patients 1, 2, 4, and 8; insert graphs are shown for these patients to better show the dynamic change in follow-up. Abbreviation: CNS, central nervous system; ddPCR, droplet digital polymerase chain reaction; HIV, human immunodeficiency virus.
Figure 4.
Correlation between the clearance time of Tpp47 (A) and polA (B) in saliva and the loads of Treponema pallidum DNA before treatment.
We continued to collect saliva samples from patients 5, 7, 8, and 9 after 3 consecutive negative detections to rule out the possibility of a late recrudescence. As shown in Supplementary Table 2, for patient 5, saliva polA was negative at 40 hours after treatment, but it turned positive at 56 hours. Similarly, saliva T. pallidum DNA turned positive at 40 hours after treatment for patient 7, at 88 hours for patient 8, and at 68 hours for patient 9.
DISCUSSION
Syphilis remains a severe public health concern globally. Developing a convenient and noninvasive method to collect samples on a large scale is of great importance for syphilis prevention. Saliva samples undoubtedly provide such convenience. Our data revealed that T. pallidum DNA could be detected in saliva at all stages of syphilis, and the detection rates were much higher in saliva than in plasma, except for primary syphilis. These findings were consistent with previous studies, which also showed that an extensive array of pathogens’ nucleic acid was detectable in saliva [15–20]. Saliva testing is believed to be superior in sensitivity than serum testing for human herpesvirus, cytomegalovirus, and varicella-zoster virus [19, 20]. Here, the T. pallidum DNA detection rate was 87.5% in secondary syphilis, which is higher than that in a previous study, which detected T. pallidum DNA in 64.5% of oral swab samples from patients with secondary syphilis [13]. The possibility that saliva was a more sensitive diagnostic fluid is reasonable, because saliva is a reservoir from systemic sources [21].
Pathogens enter saliva mainly through the following ways: (1) direct transfer via saliva or pathogens from other infected individuals, (2) a blood-borne infection via the salivary glands, (3) infection through the oral mucosa, and (4) serum exudates [22]. However, Yang et al [13] showed that, out of 267 participants, 238 had oral sex and 22 cases had oral ulcers, yet 7 patients without reported oral sex also had T. pallidum DNA detected in oral swabs. Our study indicated that T. pallidum in the oral cavity might not only associate with oral sex or oral lesions but also associate with infection route, such as blood-borne infection via the salivary glands, or serum exudates. Of interest, most patients in our study did not report oral sex, and there was prevalent T. pallidum DNA detected in saliva. The findings that secondary syphilis, a blood RPR titer of 1:32 or greater, and positive plasma T. pallidum DNA were significantly associated with the detection rate of T. pallidum DNA in saliva indicated that T. pallidum DNA could be easily detected in the oral cavity in spirochaetaemia. In our study, 18 out of 72 patients with secondary syphilis had oral lesions, and all saliva samples were T. pallidum DNA positive. However, in those who had oral lesions, only 33.33% (6/18) of the patients reported oral sex. Therefore, in addition to the direct transmission of T. pallidum by oral sex, systemic T. pallidum infection may also play a crucial role in oral cavity infection, as in other infectious diseases (eg, protozoan flagellate Leishmania has proven that systemic infections could also affect the oral cavity) [23].
The detection rate of T. pallidum DNA in saliva was not significantly different compared with plasma in primary syphilis. However, an interesting finding is that the detection rates of T. pallidum DNA in saliva were elevated with blood RPR titer in primary syphilis. When blood RPR titers were at a low level (range, 0 to 8), 53.8% of the patients had detectable T. pallidum DNA in plasma, while it was almost undetectable in saliva. When blood RPR titers were at a high level (range, 64 to 128), the detection rate of T. pallidum DNA in saliva was 71.4%. This finding indicates that T. pallidum may enter the bloodstream first and then transmit to the oral cavity after infection. Saliva may concentrate and store T. pallidum like a reservoir.
Currently, the reduction in VDRL or RPR titer 3 to 6 months after treatment is used as an index of therapeutic response. However, long-term follow-up can result in loss to follow-up. The clearance of pathogens’ nucleic acid is a good indicator of treatment response or clinical outcome [24, 25]. We monitored the clearance of T. pallidum DNA from saliva after treatment. The median time of T. pallidum DNA becoming undetectable in saliva was 64 hours after treatment in 9 patients, which indicated that T. pallidum DNA could be quickly cleared. A previous study reported that the mean time of T. pallidum DNA being cleared from blood was 32 hours [26]. However, notably, T. pallidum DNA in saliva turned positive in follow-up among some patients even if it had been negative at multiple successive times. Whether there is a possibility of a late recrudescence is unclear in this study. Relapse of syphilis after treatment is challenging using the current follow-up and treatment strategies [27]. Thus, more patients receiving different therapies, and especially those with treatment failure, should be enrolled, and the time of follow-up should be longer in further studies.
Syphilis could be transmitted through direct lesion contact via broken skin or mucosa during sexual activity. However, the high loads of T. pallidum DNA detected in saliva in patients with syphilis in this study suggested that saliva could be an alternative transmission pathway of syphilis. This is not difficult to understand for secondary syphilis, as previous studies have reported that clinical cases of mouth-to-mouth transfer of prechewed food or kissing with patients with secondary syphilis have resulted in infection with T. pallidum [11, 12]. Surprisingly, the high loads of T. pallidum DNA in saliva were detected in latent syphilis and symptomatic neurosyphilis (the highest values were 3212 copies/mL and 2728 copies/mL, respectively and much higher than those in the blood). If the saliva of a patient in latent or late stages of syphilis could be contagious, then the strategies for syphilis prevention should be substantially reconsidered. At present, only primary and secondary syphilis are considered as “infectious” and targeted in public health control. Although a reported clinical case of an infant who was infected by mouth-to-mouth transfer of prechewed food from a patient with early latent syphilis [10] may support this idea, the existing technology is insufficient to demonstrate active T. pallidum in saliva in this study. Further investigation is required.
In conclusion, our study demonstrates a significantly higher detection rate of T. pallidum DNA in saliva than in plasma. Together with the advantages of convenience and noninvasiveness, saliva samples may not only be a sensitive diagnostic fluid for syphilis but also a promising method for monitoring T. pallidum DNA clearance as an indicator of therapeutic effectiveness.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank Professor Sheila A. Lukehart for her kind help in the review and revision of this manuscript and Professor Lei Zhang for the language improvements.
Financial support. This work was supported by the National Natural Science Foundation of China (grant number 81572039), the Basic Research Project of Shanghai Science and Technology Commission (grant number 15JC1403000), the Shanghai Natural Science Foundation (grant numbers 15ZR1437000, 16411961300, 17DZ2293300), Clinical Research Plan of SHDC (grant number 16CR1029B), the National Megaproject on Key Infectious Diseases (grant number 2017ZX10202102-001–007) and the Science and Technology Commission of Shanghai Municipality (grant number YDZX20193100002868).
Potential conflicts of interest. The authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Rowley J, Toskin I, Ndowa F. World Health Organization: global incidence and prevalence of selected curable sexually transmitted infections—2008. Available at: http://apps.who.int/iris/bitstream/10665/75181/1/9789241503839_eng.pdf.
- 2. Yang S, Wu J, Ding C, et al. Epidemiological features of and changes in incidence of infectious diseases in China in the first decade after the SARS outbreak: an observational trend study. Lancet Infect Dis 2017; 17:716–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. National Health and Family Planning Commission of the People’s Republic of China. General situation of infectious diseases in China, 2018. Available at: http://www.nhc.gov.cn/jkj/s3578/201904/050427ff32704a5db64f4ae1f6d57c6c.shtml.
- 4. Larsen SA, Steiner BM, Rudolph AH. Laboratory diagnosis and interpretation of tests for syphilis. Clin Microbiol Rev 1995; 8:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Grange PA, Gressier L, Dion PL, et al. Evaluation of a PCR test for detection of treponema pallidum in swabs and blood. J Clin Microbiol 2012; 50:546–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang C, Cheng Y, Liu B, et al. Sensitive detection of Treponema pallidum DNA from the whole blood of patients with syphilis by the nested PCR assay. Emerg Microbes Infect 2018; 7:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hagihara M, Yamagishi Y, Kato H, et al. Frequency of Treponema pallidum invasion into cerebrospinal fluid in primary or secondary early-stage syphilis. J Infect Chemother 2018; 24:404–6. [DOI] [PubMed] [Google Scholar]
- 8. Godornes C, Ciccarese G, Drago F, Giacani L. Treponema pallidum subsp. pallidum DNA and RNA in semen of a syphilis patient without genital or anal lesions. Sex Transm Dis 2019; 46:e62–4. [DOI] [PubMed] [Google Scholar]
- 9. Dubourg G, Edouard S, Prudent E, Fournier PE, Raoult D. Incidental syphilis diagnosed by real-time PCR screening of urine samples. J Clin Microbiol 2015; 53:3707–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang RZ, Jin HL. Syphilis in an infant acquired by mouth-to-mouth transfer of prechewed food. Pediatr Dermatol 2016; 33:e344–5. [DOI] [PubMed] [Google Scholar]
- 11. Zhou P, Qian Y, Lu H, Guan Z. Nonvenereal transmission of syphilis in infancy by mouth-to-mouth transfer of prechewed food. Sex Transm Dis 2009; 36:216–7. [DOI] [PubMed] [Google Scholar]
- 12. Yu X, Zheng H. Syphilitic chancre of the lips transmitted by kissing: a case report and review of the literature. Medicine (Baltimore) 2016; 95:e3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang CJ, Chang SY, Wu BR, et al. Unexpectedly high prevalence of Treponema pallidum infection in the oral cavity of human immunodeficiency virus-infected patients with early syphilis who had engaged in unprotected sex practices. Clin Microbiol Infect 2015; 21: 787.e1–7. [DOI] [PubMed] [Google Scholar]
- 14. Centers for Disease Control by the Chinese Center for STD Control. The diagnosis and treatment guidelines of syphilis, gonorrhea, genital herpes and chlamydial trachomatis infection (2014). Chin J Dermatol 2014; 47:365–72. [Google Scholar]
- 15. Pow EH, Law MY, Tsang PC, Perera RA, Kwong DL. Salivary Epstein-Barr virus DNA level in patients with nasopharyngeal carcinoma following radiotherapy. Oral Oncol 2011; 47:879–82. [DOI] [PubMed] [Google Scholar]
- 16. Eventov-Friedman S, Manor H, Bar-Oz B, et al. Saliva real-time polymerase chain reaction for targeted screening of congenital cytomegalovirus infection. J Infect Dis 2019; 220:1790–6. [DOI] [PubMed] [Google Scholar]
- 17. Ikeno R, Yamada E, Yamazaki S, et al. Factors contributing to salivary human immunodeficiency virus type-1 levels measured by a Poisson distribution-based PCR method. J Int Med Res 2018; 46:996–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hooshmand B, Alavian SM, Kouhestani F, Firouzmandi M, Motamedian SR. Detection of hepatitis C virus RNA in blood and saliva of transfusion-dependent thalassemia patients diagnosed with hepatitis C. Contemp Clin Dent 2018; 9:5–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Park SY, Kim JY, Kim JA, et al. Diagnostic usefulness of varicella-zoster virus real-time polymerase chain reaction analysis of DNA in saliva and plasma specimens from patients with herpes zoster. J Infect Dis 2017; 217:51–7. [DOI] [PubMed] [Google Scholar]
- 20. Nefzi F, Ben Salem NA, Khelif A, Feki S, Aouni M, Gautheret-Dejean A. Quantitative analysis of human herpesvirus-6 and human cytomegalovirus in blood and saliva from patients with acute leukemia. J Med Virol 2015; 87:451–60. [DOI] [PubMed] [Google Scholar]
- 21. Pesce MA, Spitalnik SL. Saliva and the clinical pathology laborator. In: Malamud D, Niedbala RS, eds. Oral-based diagnostics. The New; York Academy of Sciences, 2007; 1098:2192–9. [DOI] [PubMed] [Google Scholar]
- 22. Slots J, Slots H. Bacterial and viral pathogens in saliva: disease relationship and infectious risk. Periodontol 2000 2011; 55:48–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bergquist R. Parasitic infections affecting the oral cavity. Periodontol 2000 2009; 49: 96–105. [DOI] [PubMed] [Google Scholar]
- 24. Liu Y, Cathcart AL, Delaney WE 4th, Kitrinos KM. Development of a digital droplet PCR assay to measure HBV DNA in patients receiving long-term TDF treatment. J Virol Methods 2017; 249:189–93. [DOI] [PubMed] [Google Scholar]
- 25. Awungafac G, Amin ET, Fualefac A, et al. Viral load testing and the use of test results for clinical decision making for HIV treatment in Cameroon: an insight into the clinic-laboratory interface. PLoS One 2018; 13:e0198686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tipple C, Jones R, McClure M, Taylor G. Rapid Treponema pallidum clearance from blood and ulcer samples following single dose benzathine penicillin treatment of early syphilis. PLoS Negl Trop Dis 2015; 9:e0003492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhou P, Gu X, Lu H, Guan Z, Qian Y. Re-evaluation of serological criteria for early syphilis treatment efficacy: progression to neurosyphilis despite therapy. Sex Transm Infect 2012; 88:342–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.