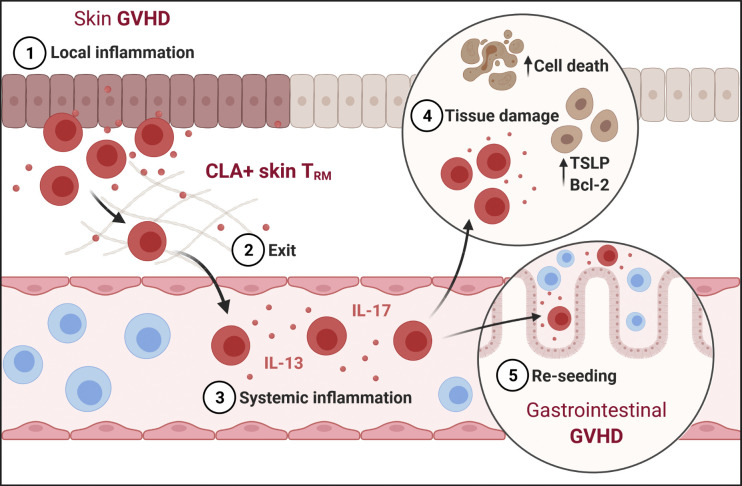
Circulating skin-derived T cells with TRM phenotype (cTRMs) form a distinct population in blood after hematopoietic stem cell transplantation. cTRMs maintain their skin identity, are increased in GVHD, and display a Th2-like cytokine profile contributing to systemic GVHD inflammation.
Abstract
Emigration of tissue-resident memory T cells (TRMs) was recently introduced in mouse models and may drive systemic inflammation. Skin TRMs of patients undergoing allogeneic hematopoietic stem cell transplantation (HSCT) can coexist beside donor T cells, offering a unique human model system to study T cell migration. By genotyping, mathematical modeling, single-cell transcriptomics, and functional analysis of patient blood and skin T cells, we detected a small consistent population of circulating skin-derived T cells with a TRM phenotype (cTRMs) in the blood and unveil their skin origin and striking resemblance to skin TRMs. Blood from patients with active graft-versus-host disease (GVHD) contains elevated numbers of host cTRMs producing pro-inflammatory Th2/Th17 cytokines and mediating keratinocyte damage. Expression of gut-homing receptors and the occurrence of cTRMs in gastrointestinal GVHD lesions emphasize their potential to reseed and propagate inflammation in distant organs. Collectively, we describe a distinct circulating T cell population mirroring skin inflammation, which could serve as a biomarker or therapeutic target in GVHD.
Graphical Abstract
Introduction
Tissue-resident memory T cells (TRMs) play a vital role for the local immune milieu, facilitating immune homeostasis within the tissue and rapid recall responses in recurring infections (Mueller and Mackay, 2016). TRMs are largely independent from the circulatory T cell pool and are identified by the expression of residency markers, including the S1P1-suppressor CD69 and the Integrin αE (CD103; Clark et al., 2006; Kumar et al., 2017; Watanabe et al., 2015).
Originally, TRMs were regarded as a terminally differentiated subset of memory T cells that, once generated in the tissue, were permanently located there, with limited motility and local proliferation (Beura et al., 2018; Park et al., 2018). However, this concept was challenged in murine experimental models that described a retrograde migration of cells with a TRM phenotype to draining lymph nodes, the blood, and secondary tissue sites (Behr et al., 2020; Beura et al., 2018; Fonseca et al., 2020; Klicznik et al., 2019). Klicznik et al. (2019) employed a humanized skin transplant mouse model to demonstrate that human skin–derived CD4+ T cells with a TRM signature, identified by expression of the cutaneous lymphocyte-associated antigen (CLA) and CD103, can down-regulate CD69, exit the skin into the circulation, and reseed a distant skin site. Fonseca et al. (2020) recently revealed an unexpected developmental plasticity of TRMs in a murine transplant model. Upon recall antigen challenge, CD8+ TRMs reentered the circulation and trans-differentiated into central memory–like T cells while maintaining the propensity to home back to their tissue of origin and regain a TRM phenotype.
Despite recent reports of T cells with a skin TRM signature in human lymph and blood (Klicznik et al., 2019), definitive proof of (re)circulating TRMs as a separate entity in the human system remains elusive. As TRMs have been shown to play a pathognomonic role in numerous inflammatory diseases (Chen and Shen, 2020), potential skin exit, trans-differentiation, and entry into distant tissues by disease-driving TRMs may represent a novel systemic factor contributing to these diseases. It is therefore eminent that the presence and role of circulating TRM-like cells warrant further investigation in the human setting.
Recently, we and others demonstrated in patients receiving allogeneic hematopoietic stem cell transplant (HSCT) that host-derived skin TRMs are maintained in the skin for at least 10 yr, whereas the circulating T cell pool is rapidly replaced by the donor T cells (Divito et al., 2020; Strobl et al., 2020b). Importantly, activated host T cells were detected in lesional skin and in the gut of graft-versus-host disease (GVHD) patients, suggesting that they play a role in GVHD and drive an inflammatory host-versus-graft reaction. Based on those findings, the unique clinical setting of allogeneic HSCT provides an ideal model to study (re)entry of human skin TRMs into the circulation.
Results
Circulating CD4+ T cells with a skin-resident phenotype are found in the blood of immune cell–reconstituted patients after HSCT
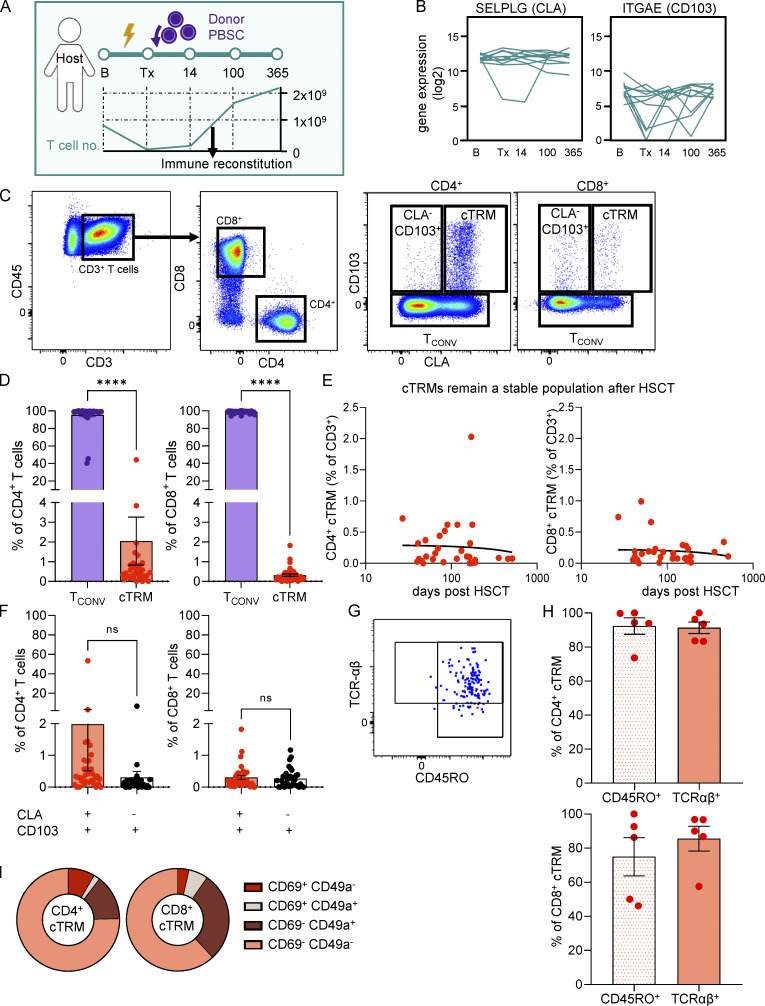
In the course of HSCT, peripheral blood T cells are largely eliminated by conditioning treatments and return to baseline levels weeks to months after transplantation of donor peripheral blood stem cells (PBSCs; Strobl et al., 2020b; Fig. 1 A). To investigate marker genes for circulating T cells with a skin TRM phenotype (cTRMs; Klicznik et al., 2019) in HSCT, we FACS-sorted T cells of 11 patients undergoing HSCT at five time points: B, baseline; Tx, hours before transplantation after completion of conditioning treatment; 14, day 14 after Tx; 100, day 100 after Tx; and 365, 1 yr after Tx; (Fig. S1, Cohort 1). RNA sequencing (RNA-seq) of 100 bulk T cells per time point revealed stable expression of SELPLG, the gene coding for PSLG-1, which can be post-translationally modified to harbor the E-selectin–binding epitope of CLA on skin-homing T cells (Fuhlbrigge et al., 1997) and continuous expression of ITGAE (CD103) at a low level (Fig. 1 B). In contrast, ITGA1 (CD49a), a marker for cytotoxic TRMs (Cheuk et al., 2017), was not consistently detectable by RNA-seq (Fig. S2 A). To evaluate CLA and CD103 on protein level, we checked for surface expression of these cTRM markers in T cells in the blood after immune reconstitution. In accordance with RNA expression, we detected a small population of CLA- and CD103-expressing cells among CD4+ and CD8+ T cells (Fig. 1, C and D; and Fig. S1, Cohort 2). The vast majority of CD4+ T cells were CD103 negative and were termed conventional T cells (TCONV cells), and a minor faction of CD4+ CD103+ T cells were CLA− (Fig. 1, C, D, and F). In contrast, the proportions of CLA+CD103+ and CLA−CD103+ CD8+ T cells were equal, indicating a preferential egress of CD4+ TRMs from the skin in these patients (Fig. 1 F). Skin TRMs commonly express S1PR1-antagonist CD69, which is used as an activation marker of peripheral blood T cells (Cibrián and Sánchez-Madrid, 2017; Mackay et al., 2015). We found only a minor fraction of CD4+ or CD8+ cTRMs expressing CD69 (Fig. 1 I), which is consistent with Klicznik et al. (2019) reporting that CD69 down-regulation is required for skin egress. CD4+ cTRMs were also largely CD49a−, whereas a higher percentage of CD8+ cTRMs expressed CD49a, in line with their cytotoxic function (Cheuk et al., 2017; Fig. 1 I). Pearson correlation analysis found no association between percentage of cTRMs and time after Tx in CD4+ or CD8+ T cells (Fig. 1 E), arguing for a stable subset of cTRMs over time. To confirm that cTRMs represent memory αβT cells, we stained for CD45RO as a memory marker and TCR-αβ (Fig. 1, G and H). The vast majority of cTRMs from both CD4+ and CD8+ T cells were positive for both markers (Fig. 1 H). Since CD4+ cTRMs seem to be enriched in our patients compared with reported values in healthy controls (<0.2% of CD4+ T cells; Klicznik et al., 2019) and compared with CD8+ cTRMs, we focused further experiments on CD4+ cTRMs and refer to them as cTRMs throughout the manuscript.
Figure 1.
Circulating CD4+ T cells with a skin-resident phenotype are enriched in the blood of immune cell–reconstituted patients after HSCT. (A) Graphical representation of sampling timeline and blood T cell no./liter before initiation of conditioning (B); hours before transplantation after completion of conditioning treatment (Tx); and 14, 100, and 365 d after Tx (14, 100, and 365; n = 11). (B) RNA-based progression scores of SELPLG and ITGAE in FACS-sorted bulk T cells. Data shown as mean expression in 100 T cells per patient per time point (n = 11). (C) Representative gating strategy and sorting gates for cTRMs and TCONV. Cells were pregated as live single leukocytes. (D) Mean percentage (±SEM) of cTRMs and TCONV among CD4+ and CD8+ T cells (n = 36). Statistical analysis was performed with paired Student’s t test (****, P < 0.0001). (E) Linear regression of CD4+ and CD8+ cTRMs (percentage of all T cells) to time after Tx (days; n = 31; CD4+ R2 = 0.0003755, CD8+ R2 = 0.01269). Data plotted as percentage cTRM of total T cells versus days after Tx. One dot indicates one patient. (F) Mean percentage (±SEM) of CLA+ CD103+ and CLA− CD103+ cells among CD4+ and CD8+ T cells (n = 36). Statistical analysis was performed with paired Student’s t test. (G and H) Representative gating plot (G) and mean percentage (±SEM) of CD45RO and TCR-αβ expression of CD4+ and CD8+ cTRMs (n = 5; H). (I) CD69 and CD49a surface molecule expression on cTRMs assessed by flow cytometry of PBMCs. Data shown as mean percentage of CD103+CLA+ T cells positive for the respective markers (n = 21).
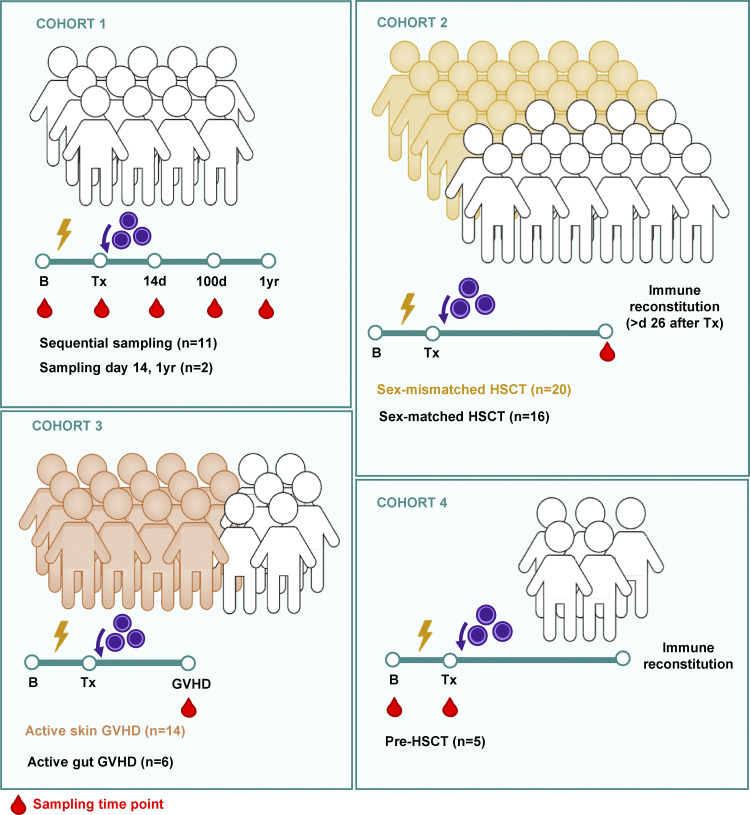
Figure S1.
Graphical representation of patient cohorts .
Figure S2.
RNA expression, T cell subtypes and cross-shared clones. (A) RNA-based progression scores of ITGA1 in FACS-sorted bulk T cells. Data shown as mean expression in 100 T cells per patient per time point (n = 11). (B) Representative sorting strategy of T cell subsets (Tnaive = blue, TCM = green, TEM = orange, TEMRA = red) for XY-FISH (Fig. 2 C), pregated on live single CD3+ cells, and quantification (n = 5). Data shown as percentage ± SEM of live CD3+ T cells. (C) Cross-shared clones of sorted T cells from pre-Tx skin and post-Tx blood at days 14, 100, and 365 after transplantation as identified by Smart-seq2/TRUST pipeline. Data shown as mean percentage ± SEM of all detected TCR clones in each sample (n = 11 patients). SSC, side scatter.
cTRMs are host derived and detectable in skin before transplantation
In solid organ transplantation, resident T cells of gut, kidney, or skin may remain stationary in their respective tissue and are thus cotransplanted with the donor organ into the recipient’s body (Bartolomé-Casado et al., 2019; de Leur et al., 2019; Lian et al., 2014). Inversely, in the unique setting of allogeneic HSCT, TRMs survive conditioning treatment due to altered metabolism and senescent features and coexist with donor T cells, forming a human chimeric host (Divito et al., 2020; Strobl et al., 2020b). To our knowledge, the similarity of human cTRMs to their counterparts in tissues has exclusively been identified by phenotypic features, but proof of the relation between the two compartments in humans is missing. We therefore tracked XX/XY genotypes of T cells after sex-mismatched transplantation to distinguish host and donor cells (Fig. 2 A; and Fig. S1, Cohort 2). As expected after successful HSCT, the overwhelming majority (mean, 88.01%) of TCONV cells were of donor origin (Fig. 2 B). Patients with higher levels of host-derived TCONV (Fig. 2 B, triangle symbols) suffered from skin or mucosal infections at the time of blood sampling (skin infection with HSV or varicella zoster virus and viral sinus infection), providing insights into mobilization mechanisms of TRMs into peripheral blood. Strikingly, analysis of sorted cTRMs revealed that a mean of 50% of cTRMs exhibited a host genotype (Fig. 2 B).
Figure 2.
Host-derived cTRMs share clones detectable in the skin before transplantation. (A) Graphical representation of sample processing after sex-mismatched HSCT. (B and C) Host/donor genotype of TCONV (n = 18) and cTRMs (n = 20; B) and Tnaive, TCM, TEM, and TEMRA (n = 5; C) as determined by XX/XY–FISH. Data shown as mean percentage (±SEM) of detected T cells per sample after sorting of minimum 5,000 cells. One dot represents one patient; statistical analysis was performed with repeated measures one-way ANOVA with multiple comparisons (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). (D) Correlation trend of host genotypes among cTRMs to the time after HSCT in a two-phase exponential decay model (n = 20; decay span 1: ∼46.87 d, decay span 2: 49.10 d, ns). Following initial decay estimated at K1 ∼5.344e−015, half-life of host cTRMs was determined at 39 d (K2 = 0.01776). Data shown as percentage of host cTRMs among all cTRMs and days after Tx. (E) Sorted bulk T cells from 11 patients from pre- and post-transplant time points from skin and blood were RNA sequenced (Smart-seq2) and analyzed for their TCR repertoire by TRUST. Graphical representation of definition of blood and skin TCR clones that are shared between pre- and post-transplant time points and percentage of shared TCR clones of pre-Tx blood versus post-Tx blood (“blood”) and pre-Tx skin versus post-Tx skin (“skin”). (F) Cross-shared clones from pre-Tx skin to post-Tx blood. Graphical representation and percentage of cross-shared clones from all clones (”SxB”) and percentage of cross-shared clones in blood of different individuals at day 100 after transplantation (“neg. ctrl”). Data shown as mean percentage ± SEM of all detected TCR clones in each sample (n = 11). Statistical analysis in E and F was performed with paired (E) and unpaired (F) Student’s t test (*, P < 0.05).
To address the chimerism in other memory populations in the peripheral blood, we sorted T cells according to their naive (Tnaive = CD45RA+ CD62L+), central memory (TCM = CD45RO+ CD62L+), effector memory (TEM = CD45RO+ CD62L−), and terminally differentiated effector memory (TEMRA = CD45RA+ CD62L−) phenotypes (Fig. S2) and analyzed the XX/XY genotype. As expected, the vast majority of Tnaive cells were of donor origin (Fig. 2 C). Importantly, the proportion of host-derived cells among memory T cell subsets (TCM and TEMRA cells) was minor (Fig. 2 C). In contrast to overall cTRMs (Fig. 1 E), host-derived cTRMs showed a trend to slowly decline in two phases, with a faster decay within the first 100 d (Fig. 2 D).
To investigate emergence of TCR clones in contrast to persistence of TRM and cTRM clones, we performed TCR sequencing from matched pre- and post-transplantation skin and blood samples (Fig. 2 E). In both skin and blood, a mean of 1% of clones were shared between pre- and post-transplantation. Interestingly, 0.4% of clones present in blood after transplantation were found in pretransplant skin, thus cross-shared between tissues and time points (Fig. 2 F and Table S4). This fraction of cross-shared host-derived clones insignificantly declined over time (Fig. S2 C).
Figure S3.
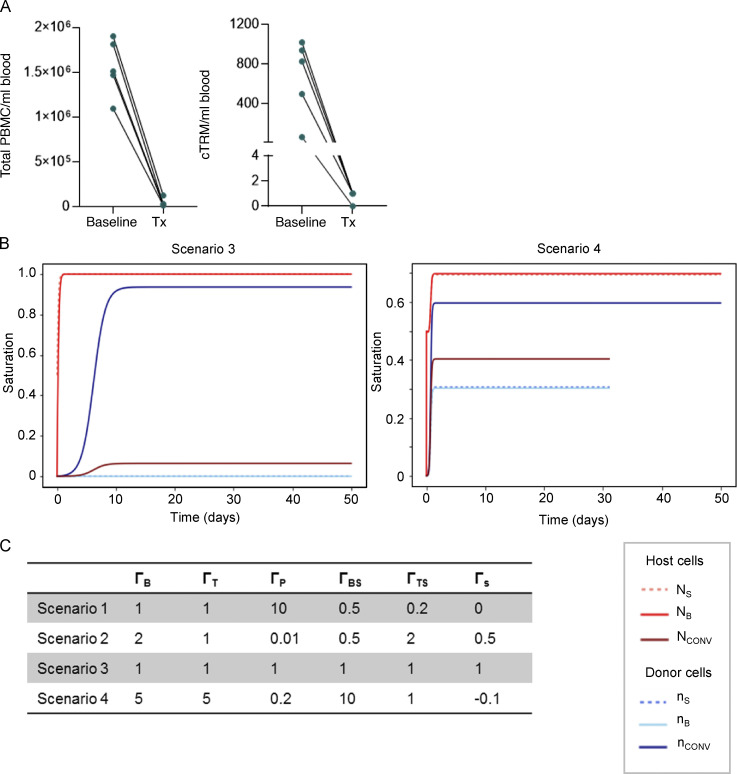
Mathematical model. (A) Quantification of total blood PBMCs (left) and total number of cTRMs (right) per milliliter of blood in patients at B and at Tx (n = 5). (B) Scenarios 3 and 4 using cell proliferation rates shown in Fig. S3 C. (C) Tested proliferation rates for scenarios 1–4.
Mathematical modeling confirms concept of cTRMs relating to skin TRMs
The observation that cutaneous host TRMs form a population of cTRMs raises the question of how the cTRM population is maintained after transplantation. Trans-differentiation from host TCONV cells is rather unlikely due to the low numbers of host TCONV in the blood at Tx (Fig. 2 B). Similarly, the novel donor cTRM pool cannot be formed from preexisting donor skin TRMs as donor T cells are only transferred on Tx and need to form a skin TRM population first. This leaves three possible routes of origin for the renewal of the host cTRM population (Fig. 3 D). Either the cTRM pool is maintained from the cutaneous TRM reservoir or it stems from self-renewal of cTRMs or trans-differentiation from TCONV within the peripheral blood.
Figure 3.
Mathematical modeling postulates relation of cTRMs to skin TRMs. (A) TRM number in skin before and after conditioning therapy as determined by immunofluorescence staining. Data shown as mean number (±SEM) of CD103+CD3+ cells per cubic centimeter of skin at B and Tx (n = 11). (B) T cell number in peripheral blood before and after conditioning therapy as determined by flow cytometry. Data shown as mean number (±SEM) of CD3+ lymphocytes per liter of blood at B and Tx, n = 11. (C) Total number of T cells in adult human skin and blood after conditioning before infusion of donor PBSCs. Data shown as mean number and range of cells at Tx calculated for an average human with a body surface area (BSA) of ≥1.63 m2 and a blood volume of 65 ml/kg using the Du Bois formula (BSA = 0.007184 × W0.425 × H0.725). Statistical analysis in B and C was performed with paired Student’s t test; **, P < 0.01; ****, P < 0.0001. (D) Graphical representation of potential routes for maintenance of host/donor cTRMs and equation terms. (E) Modeling of symmetrical differential equation model by applying different cell proliferation and migration rates shown in Fig. S3 C. Scenario 1 (left panel) results in realistic cell saturation values; Scenario 2 (right panel) shows rapid saturation of the cTRM and TRM niches with host-derived cells. Saturation represents the sum of relative host and donor cells (1 = 100%). Time shown in days.
To assess the contribution of skin versus blood T cells to the cTRM pool after transplantation, we measured the number of skin and blood T cells surviving radio-chemotherapy (Fig. 3, A and B; and Fig. S1, Cohort 1). While >1010 TRMs were residing in the skin of a patient at transplantation, <4 × 108 T cells were present in peripheral blood (Fig. 3 C). Consistently, donor-derived peripheral blood mononuclear cells (PBMCs), including cTRMs, were efficiently depleted by conditioning therapy (Fig. S3 A, Cohort 4). We hypothesized that due to the high number of host TRMs in the skin compartment, host cTRMs are maintained by continuous low-level emigration of skin TRMs. To analyze this in detail and determine host and donor T cell cycles needed to maintain the cTRM pool, we created a symmetrical model system based on the measured saturation densities of cTRMs and TCONV cells of host and donor origin (Fig. 3 D). We first established nonlinear differential equations to describe the change in T cell populations. The equations consider exponential growth for small populations and saturation for large populations. The host cTRM cycle comprises the change of the host skin TRM population (NS) and the host cTRM population in blood (NB), as well as the small circulating host TCONV population (Nconv). It depends on (i) the diffusion of skin TRMs to blood cTRMs (and vice versa) with a rate of ΓBS; (ii) the growth of respective skin and blood populations with rates ΓS and ΓB, the self-renewal rate of TCONV (ΓT); and (iii) trans-differentiation from conventional T cells, which acquire a cTRM phenotype (ΓTS). This results in three equation terms:
As opposed to the host cycle, the donor T cell population also grows by differentiation from PBSCs to TCONV with rate ΓP. Therefore, we expect changes in donor T cell populations of transplanted PBSCs (nPBSC), conventional T cells (nconv), skin TRMs (nS), and cTRMs (nB) by differentiation, egress from/to tissue, and proliferation rate.
The two cycles are coupled by requiring that the growth saturates when the total number of TCONV cells, TRMs, and cTRMs equals that of a healthy individual given by NCONVsat, NSsat, and NBsat, respectively. By solving the differential equations for various rates and by analytical approximations, we find that the experimental observation of an equal number of host and donor cTRMs (Fig. 2 C) is met when the diffusion from host skin TRMs to host cTRMs is faster than the proliferation of host cTRMs in blood.
We next tested our equation system by adopting potential cell proliferation and migration rates and plotting them using Mathlib package for Jupyter Notebook. When plotting relative cell proliferation rates based on the proliferation of conventional T cells (ΓT = 1), where ΓB = ΓT, ΓP = 10, ΓBS = 0.5, ΓTS = 0.5, and ΓS = 0, we saw achievement of the experimentally observed relative cell numbers within the first weeks after transplantation (Fig. 3 E, Scenario 1). Other scenarios presuming higher cTRM proliferation rates (ΓB ≥ 2) or rapid adoption of a cTRM phenotype by TCONV (ΓTS ≥ 2) result in unrealistic growth curves (Fig. S3 B, Scenarios 3 and 4). Tested cell cycling rates are shown in Fig. S3 C. Interestingly, due to high skin cell numbers, proliferation of cutaneous TRMs (ΓB > 0) would result in rapid saturation of the cTRM pool with host-derived T cells (Fig. 3 E, Scenario 2), which is not the case according to our genotyping experiments (Fig. 2 B).
Thus, we can conclude that while the donor cTRM pool is formed from PBSCs via TCONV and consequently developed donor TRMs, the host cTRM pool is likely maintained by self-renewal from nonproliferating skin TRMs.
Single-cell RNA-seq unravels gene expression of host- and donor-derived circulating T cells after HSCT
To further characterize circulating host-derived T cells over time, we performed index-sorted single-cell flow cytometry and RNA-seq of T cells isolated from blood after HSCT (Fig. S1, Cohort 1). To increase the likelihood of isolating host-derived T cells, we sequenced peripheral blood T cells of one patient with slow donor T cell recovery and a mixed T cell chimerism at days 14 and 250 after HSCT (Fig. 4, A–D, patient 1; and Table S2) and one patient with normal donor T cell recovery at day 14 only, who had a full donor chimerism of T cells at later time points (Fig. S4, A, patient 2; and Table S3). At both time points, we saw two transcriptionally distinct populations of T cells (Fig. 4, A and C; and Fig. S4 A). Based on unsupervised t-distributed stochastic neighbor embedding (t-SNE) clustering of cells and confirmation by single-nucleotide polymorphisms (SNPs) identified in pretransplant samples, we were able to determine host and donor origin of T cells. SNP calling revealed that host cells were present in all samples and that cells clustered according to host/donor origin (Fig. 4, C and D). Notably, fewer SNPs of host cells were qualified (Fig. 4, C and D) due to low overall numbers of host-derived cells in combination with strict selection criteria (SNPs present in three or more host cells and absent in all donor-derived cells). SNP-based distinction of cells allowed differential gene expression analysis of donor versus host cell transcriptomes, enabling us to investigate differential expression of TRM marker genes from the two patients at day 14 (Fig. 4 E). Interestingly, we found host cells to have a gene expression profile relating to a cTRM phenotype, with a trend of higher expression of cTRM markers ITGAE (CD103), SELPLG (encoding for CLA), LGALS3 (encoding for Galectin-3), and PRDM1, which have been associated with TRMs (de Almeida et al., 2020 Preprint; Mackay et al., 2016; Strobl et al., 2020b). Host cells also displayed significantly higher levels of FABP5, encoding for the fatty acid–binding protein 5 known to be crucial for skin TRM metabolism (Pan et al., 2017). RUNX3, a transcription factor associated with TRM (Milner et al., 2017) and cytotoxic function (Shan et al., 2017), and markers of circulating cells (S1PR1 and CCR7; Mueller and Mackay, 2016) were expressed similarly between host and donor cells.
Figure 4.
Single-cell RNA-seq unravels gene expression of host and donor-derived circulating T cells after HSCT. (A and B) Representative t-SNE plot of single blood T cells of patient 1 at day 14 (A) and day 250 (B) after HSCT as determined by RNA-seq according to the Smart-seq2 protocol. Unsupervised clustering resulted in two distinct clusters. One dot represents one T cell. (C and D) Allele frequency heat map of host- and donor-specific SNPs in cells of clusters 1 and 2 from A and B. (E) Expression of marker genes of interest by donor and host blood T cells at day 14 after HSCT (patients 1 and 2; see Fig. S4 A). Data shown as normalized expression (log2) value per cell. Solid bars indicate the median, and dashed lines show quartiles; one dot represents one cell. Statistical analysis was performed with Mann-Whitney test; P values are indicated in plots. MAF, mean allele frequency.
Figure S4.
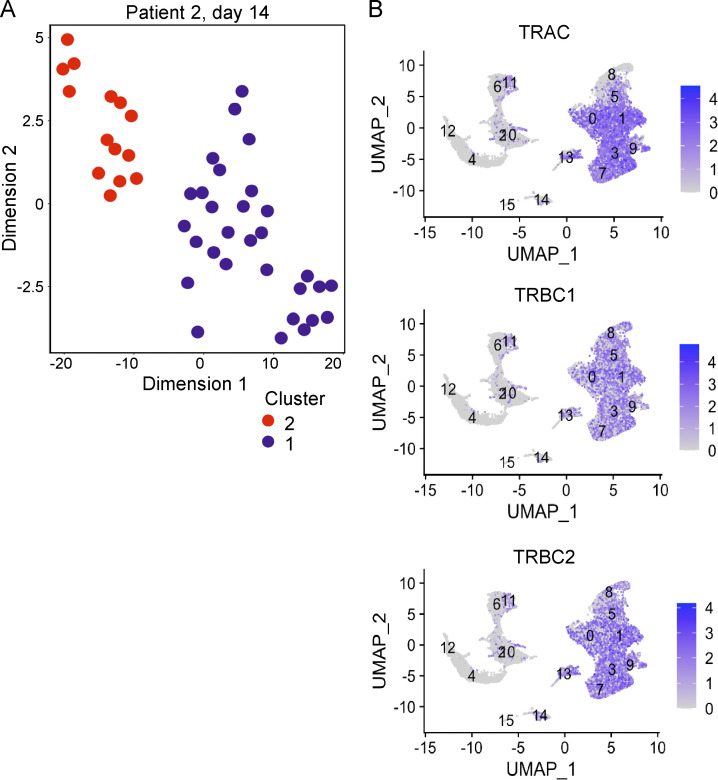
Single-cell RNA expression. (A) Representative t-SNE plot of single blood T cells from patient 2 at day 14 after HSCT as determined by single-cell RNA-seq (Smart-seq2). Unsupervised clustering resulted in two distinct clusters. One dot represents one T cell. (B) UMAP feature plots of log2 gene expression of TCR α and β genes (TRAC, TRBC1, and TRBC2) in blood cells in GVHD determined by single-cell RNA-seq (10X Genomics; n = 2). Cluster 13 represents cells with cTRM phenotype.
A population of proliferating cTRMs is found in blood of GVHD patients
Increasing evidence points toward a pivotal role of skin TRMs in GVHD pathogenesis (Divito et al., 2020; Strobl et al., 2020b). Studies in mice previously described that activation of skin TRMs during infection leads to skin exit and recirculation (Behr et al., 2020). Based on this, we stratified our patient cohort (Fig. S1, Cohort 2, sex-mismatched HSCT) into patients without GVHD or infection, with active GVHD, and with active viral skin infection at the time of sampling (Fig. 5 A). While percentages of host cTRMs in patients without GVHD development were maintained at ∼25% of total cTRMs, patients with GVHD showed markedly higher percentages of host cTRMs of >60% (Fig. 5 B). Interestingly, patients with viral skin infections such as varicella zoster virus or HSV had equally high percentages of host-derived cTRMs, which may indicate a recall response of long-term skin-resident T cells in the event of antigen encounter.
Figure 5.
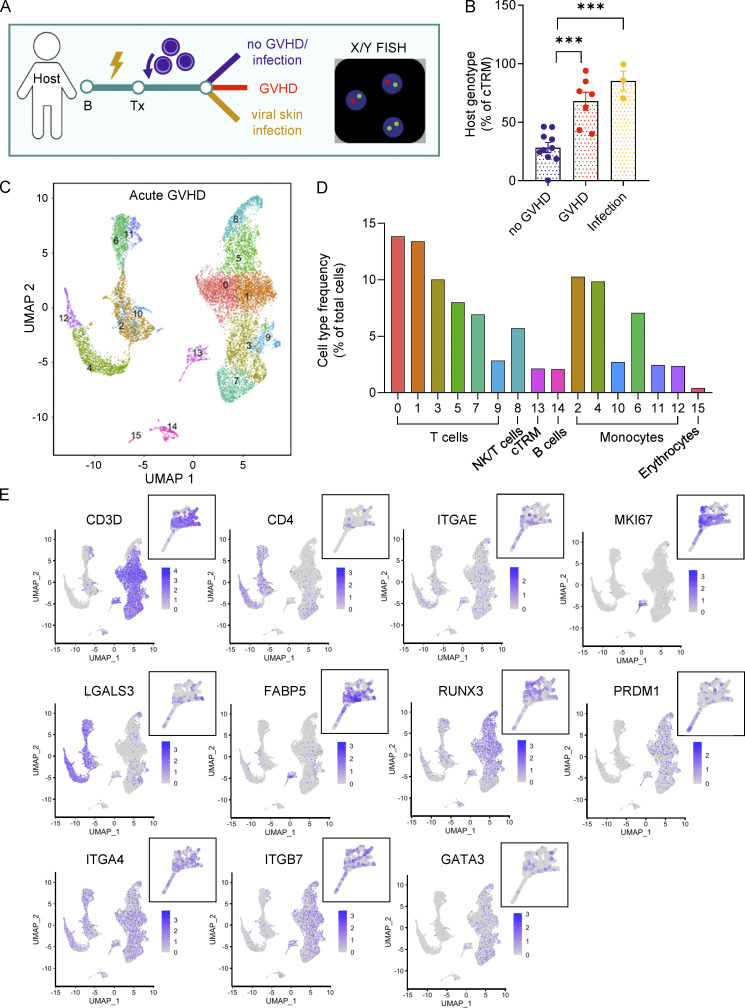
Characterization of cTRMs in GVHD patients. (A) Graphical representation of patient sampling and sample processing for B. (B) cTRM host genotype after HSCT in patients with/without GVHD and viral skin infections. Data shown as mean percentage (±SEM) of host cTRMs of all cTRMs. One dot represents one patient, no GVHD (n = 10), GVHD (n = 7), infection (n = 3). Statistical analysis was performed by one-way ANOVA and Tukey’s multiple comparisons (***, P < 0.001). (C) Combined UMAP plot of lymphoid and nonlymphoid PBMC populations in two patients with acute GVHD as determined by RNA-seq (10X Genomics). (D) Representation of immune cell populations in GVHD. Data shown as percentage of all cells. (E) Feature plots of marker expression of blood cells in GVHD. Data shown as log2 expression for each cell. Zoom-in: cTRM population. NK, natural killer.
To better characterize the cTRM population in the blood, we sampled PBMCs of two patients with active GVHD (Fig. S1, Cohort 3) and performed high-throughput single-cell RNA-seq using the 10x Genomics protocol (Fig. 5 C). The overwhelming majority of cells were T cells, followed by subsets of myeloid cells, non–T lymphocytes (B cells, natural killer cells), and erythrocytes (Fig. 5 D). As cTRMs show a transcriptome distinct from TCONV (Fig. 4) and express CD103 on their surface (Fig. 1 C), we searched for T lymphocytes with high expression of ITGAE (coding for CD103). Indeed, a small population of T cells (Fig. 5 C, Cluster 13) fit this characteristic (Fig. 5 E). Importantly, these cells showed high expression for the proliferation marker MKI67. We also found increased expression of protein-coding genes for fatty acid–binding protein 5 (FABP5) and LGALS3, as well as the transcription factors RUNX3 and PRDM1 (Fig. 5 E), all previously described in TRMs (de Almeida et al., 2020 Preprint; Milner et al., 2017; Pan et al., 2017; Strobl et al., 2020b). The majority of cells in this cluster expressed genes encoding for the αβ-TCR (Fig. S4 B), confirming our previous results (Fig. 1, G and H). The integrins ITGA4 and ITGB7 were both highly expressed in the cTRM cluster (Fig. 5 E). ITGB7 codes for the β7 integrin, which can either form a heterodimer with CD103 to bind E-cadherin or with ITGA4 to form the integrin α4β7, which is important for gut homing. Expression of ITGA4 within the cTRM cluster indicates the potential of the cells to home to other organs than the skin. Moreover, we detected expression of transcription factor GATA3 in cTRMs, which regulates transcription in T helper 2 (Th2) cells. Thus, in GVHD, cTRMs form a proliferative population of Th2-type cells while maintaining a classical TRM transcriptome. These findings suggest that cTRMs are actively proliferating in the blood of patients with GVHD and could contribute to the disease phenotype.
cTRMs are functionally competent in GVHD inflammation
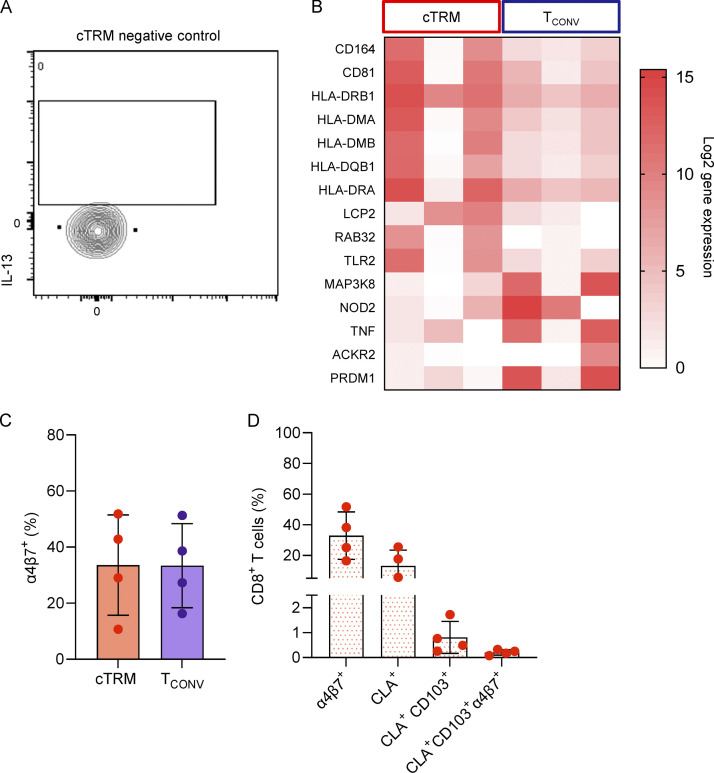
In acute GVHD, skin infiltrate is dominated by Th2 cells, while in both acute and chronic GVHD, tissue damage is described to be largely mediated by Th1 and Th17 donor T cells (Brüggen et al., 2014; Hill and Koyama, 2020). To investigate the contribution of cTRMs to systemic GVHD inflammation, we sampled the blood of patients with active GVHD (Fig. 6 A). In line with our findings on the RNA level (Fig. 5 E), we detected higher percentages of GATA3+ cTRMs compared with TCONV cells (Fig. 6, B and C). The CD4/CD8 ratio was high among cTRMs and <1 in TCONV, indicating a relative increase of CD4+ compared with CD8+ T cells in cTRMs (Fig. 6 D). We next measured cytokine production by cTRMs in patients with active GVHD (Fig. 6, E and F; and Fig. S5 A). Both IL-13– and IL-17–producing T cells were increased among cTRMs, while we observed a trend toward lower IFN-γ production by cTRMs (Fig. 6 F). To confirm the GVHD-promoting phenotype of cTRMs, we sorted cTRMs and TCONV cells from three patients with active acute GVHD (Fig. S1, Cohort 3) and performed bulk RNA-seq. Pathway enrichment analysis of differentially expressed genes (DEGs) of cTRMs versus TCONV revealed that among the top 25 enriched pathways was GVHD itself, as well as several terms relating to GVHD and Th2/Th17-type inflammation, such as allograft rejection, asthma, inflammatory bowel disease, Th1 and Th2 cell differentiation, Th17 cell differentiation, or Staphylococcus aureus infection (Fig. 6 G). Moreover, we found up-regulation of genes linked to T cell activation (HLA Class II genes, LCP2), Th2 cell proliferation (CD81; Maecker, 2003), and memory formation and effector function (TLR2; Komai-Koma et al., 2004; Fig. S5 B). Interestingly, CD164, a marker that is highly expressed on circulating and skin-tropic CD4+ T cells in patients with Sezary syndrome (Benoit et al., 2017), a form of cutaneous T cell lymphoma with a Th2 signature (Saed et al., 1994), was also up-regulated in GVHD cTRMs (Fig. S5 B). Genes previously associated with repression of Th17 responses (NOD2, ACKR2; Hansell et al., 2015; Napier et al., 2020) were down-regulated compared with TCONV (Fig. S5 B). These findings indicate that systemic type 2 and type 17 responses in GVHD are at least partially driven by cTRMs.
Figure 6.
GATA3+ cTRMs are functionally competent in systemic GVHD inflammation. (A) Graphical representation of GVHD patient sampling. (B and C) Representative gating plot (B) and mean percentage (±SEM; C) of intracellular GATA3 expression of cTRMs versus TCONV in GVHD patients (n = 6). (D) Mean (±SEM) CD4/CD8 T cell ratio in cTRMs versus TCONV in GVHD in blood samples determined by flow cytometry. (E and F) Representative gating plots (E) and mean expression (±SEM) of IL-13, IL-17A, and IFN-γ cytokine expression in cTRMs versus TCONV (F) in GVHD. Controls represent isotype or fluorescence-minus-one controls. Statistical analyses in C and F were performed by paired Student’s t test (*, P < 0.05; **, P < 0.01; n = 6). (G) Top 25 enriched KEGG pathways in up-regulated genes (P < 0.05) in bulk RNA-seq of sorted cTRMs versus TCONV. Bars indicate the combined enrichment score per pathway (n = 3).
Figure S5.
Additional flow cytometry data and differential gene expression. (A) Matched negative control of IL-13 expression from cTRMs to plots in Fig. 6 D. (B) Heat map showing log2 gene expression of selected DEGs (adjusted P value <0.05) comparing bulk transcriptomes of sorted cTRMs and TCONV cells (n = 3). (C and D) Mean percentage (±SEM) of α4β7-integrin–expressing CD8+ cTRMs and TCONV (C) and of CD8+ T cells positive for the indicated markers in the blood of GVHD patients (n = 4; D) determined by flow cytometry.
cTRMs can exert damage and home to distant tissue sites during GVHD
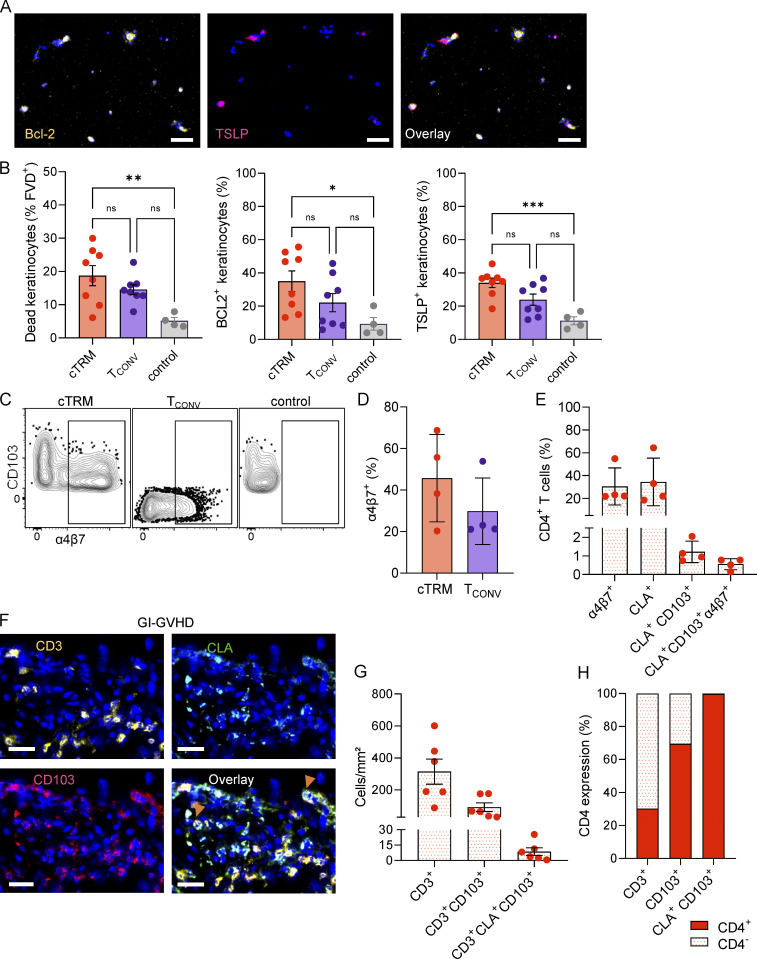
To investigate if cTRMs are a byproduct of increased skin inflammation or can actively mediate damage in tissue GVHD, we sorted cTRMs and TCONV cells from the blood of patients with GVHD (Fig. S1, Cohort 3), activated them with PMA/Ionomycin, and co-cultured them alone or in combination with keratinocytes isolated from healthy donor skin. We then checked for cell viability of keratinocytes, as well as expression of the pro-inflammatory cytokine TSLP and the anti-apoptotic molecule Bcl-2, since these markers were previously described to be elevated in keratinocytes in GVHD skin (Brüggen et al., 2014; Strobl et al., 2020a; Fig. 7 A). We observed that the presence of cTRMs leads to significantly more keratinocyte cell death compared with control, which was not the case for TCONV cells (Fig. 7 B). Moreover, TSLP, as well as Bcl-2, was significantly up-regulated after co-cultures with cTRMs but not with TCONV (Fig. 7 B). These data indicate that cTRMs can elicit tissue damage in the context of cutaneous GVHD.
Figure 7.
cTRMs can promote tissue damage and home to distant organ sites. (A) Representative immunofluorescence images of keratinocytes after co-culture with cTRM, stained with DAPI (blue), Bcl-2–APC (left), TSLP-PE (middle), and overlay (right); scale bar = 100 µM. (B) Mean percentage (±SEM) of dead keratinocytes (determined by LIVE/DEAD staining), Bcl-2, and TSLP expression of keratinocytes cultured alone (control) with cTRMs or with TCONV at a ratio of 20:1, determined by immunofluorescence staining. Statistical analysis was performed by ordinary one-way ANOVA with multiple comparisons (*, P < 0.05; **, P < 0.01; ***, P > 0.001; n = 4). (C and D) Representative gating plots (C) and mean percentage (±SEM) of α4β7-integrin–expressing cTRMs and TCONV from GVHD patients (n = 4; D). Control represents α4β7 fluorescence-minus-one staining. (E) Mean percentage (±SEM) of indicated markers among total peripheral blood-derived CD4+ T cells of GVHD patients (n = 4) determined by flow cytometry. (F) Representative immunofluorescence images of a GI-GVHD tissue sample stained with DAPI (blue), CD3-APC, CLA-FITC, and CD103-PE-Dazzle and overlay. Arrowheads indicate cTRMs; scale bar = 100 µM. (G) Total number (±SEM) of cells with labeled phenotype detected per square millimeter in gut biopsies of patients with acute GI-GVHD (n = 6). (H) Mean percentage of CD4+ and CD4− cells in the indicated parent population in patients with acute GI-GVHD (n = 6), determined by immunofluorescence staining.
Meanwhile, skin GVHD often precedes the involvement of other organs, including the gastrointestinal (GI) tract. Given that we found increased RNA expression of ITGA4 and ITGB7 on cTRMs in GVHD, we assessed in the blood of patients with active GVHD the presence of cTRMs expressing the gut-homing molecule α4β7 integrin (Fig. S1, Cohort 3). We found that a substantial portion (20–70%) of cTRMs coexpress α4β7 (Fig. 7, C and D). Among total CD4+ T cells, CLA+CD103+ α4β7+ cTRMs comprised <1% (Fig. 7 E). CD8+ T cells displayed a similar pattern of α4β7 expression (Fig. S5, C and D).
Based on this, we examined gut biopsies of patients with active GI GVHD (GI-GVHD) for the presence of skin marker molecule-expressing TRMs (Fig. S1, Cohort 3). Indeed, we found that in all samples investigated a fraction of GI mucosa T cells expressed CLA in addition to CD103 (Fig. 7, F and G). Among the T cells coexpressing CLA and CD103, >99% were CD4+ (Fig. 7 H). The presence of T cells with a phenotype consistent with skin-derived cTRMs in GI-GVHD emphasizes the role of cTRMs in systemic GVHD and their involvement in tissue inflammation of different organs.
Collectively, we show that host-derived cTRMs can be tracked in the blood of patients after HSCT and during GVHD, display an activated Th2/Th17-like phenotype, and can elicit keratinocyte damage and home to distant organ sites, thereby contributing to systemic inflammation and propagating disease.
Discussion
Since their discovery, TRMs have been defined as a rather homogeneous and differentiated T cell subset terminally located in the tissue. Recent findings from mouse models and humans have challenged this concept and demonstrated a previously unknown plasticity of TRMs, including the potential to reenter the circulation and reseed distant organ sites (Behr et al., 2020; Fonseca et al., 2020; Milner et al., 2020). This has been proposed to be a mechanism for spreading tissue-specific immunity from the primary site of antigen encounter over large areas and organs. However, it also raises the possibility that, in the case of detrimental effects mediated by TRMs, pathology can be distributed to distant tissue sites. Here, we show for the first time that skin-derived cTRMs can be tracked as a discrete population in human blood over prolonged periods of time. We previously reported that host skin TRMs survive conditioning during HSCT, as opposed to circulating T cells. In the present study, we provide evidence that surviving host TRMs exit the skin after transplantation and give rise to a stable pool of cTRMs, which (i) retains in part a skin-resident transcriptional signature and functional profile, (ii) elicits a GVHD-like phenotype on healthy keratinocytes upon activation, and (iii) homes to other GVHD target organs, highlighting their potential in systemic immune reactions. While cTRMs were present among CD4+ and CD8+ T cells, CD4+ cTRMs were enriched compared with CD8+ cTRMs in our patient cohorts. This is in line with the reports from healthy human individuals (Klicznik et al., 2019) and might represent one important difference with mouse models, where CD8+ TRMs are the dominant recirculating TRM type (Behr et al., 2020; Fonseca et al., 2020).
In murine models, cell origin can easily be tracked by congenic strains for generating host/donor chimera. In our system, we use a similar approach for tracking host and donor cells on a genetic level. By creating a mathematical model of T cell cycles and testing cell proliferation scenarios with multiple variables, we demonstrate that a blood origin of cTRMs after HSCT is highly unlikely, as exceedingly high proliferation rates of remaining host cTRMs would be required to repopulate the cTRM reservoir, which is virtually absent at the time of transplantation. We furthermore detected that the equal distribution of host- and donor-derived cTRMs is maintained only by relatively high proliferation rates of donor PBSCs and TCONV cells and low or absent proliferation of host skin TRMs. This concurs with previous studies showing host-derived skin TRMs after HSCT to express markers for cellular senescence (Strobl et al., 2020b).
Naturally, our findings raise the question of how the emigration of TRMs from skin is orchestrated. According to the concept of recently described “outside-in” recall responses (Fonseca et al., 2020), in a HSCT scenario, TRMs may be activated during the tissue-damaging conditioning regimen and subsequently undergo retrograde migration to the circulation. In line with this hypothesis, we found an increased percentage of cTRMs in the blood of patients with GVHD and cutaneous infection. Previous studies proposed outside-in recall responses as a mechanism to promote host-protective immunity, but our results suggest that this mechanism could also lead to a systemic contribution to inflammatory diseases originally manifesting in tissues. The clinical relevance of circulating host-derived T cells in the pathogenesis of GVHD is supported by the association of mixed T cell chimerism with GVHD development (Mousavi et al., 2017). We found that cTRMs display a Th2/Th17 signature with increased expression of GATA3 and IL-13, as well as higher levels of IL-17 compared with TCONV after stimulation. We previously showed that Th2 cytokines are up-regulated in the skin during acute GVHD (Brüggen et al., 2014), and IL-17 has been reported to exacerbate cutaneous GVHD in mouse models (Mousavi et al., 2017). Of note, host-derived skin TRMs in GVHD lesions produce IFN-γ and IL-17 in situ, thereby promoting host-versus-graft immune responses (Divito et al., 2020). Interestingly, we did not observe increased IFN-γ production from cTRMs. In line with our findings, Divito et al. (2020) showed in a humanized mouse model of GVHD that IFN-γ was the only T cell–derived cytokine significantly decreased in the absence of donor T cells, suggesting that IFN-γ–mediated effects in GVHD can be largely attributed to donor T cells.
Keratinocyte damage mediated by cytokine-producing T cells in the skin is a hallmark of GVHD, and keratinocytes in acute GVHD display elevated levels of TSLP, a cytokine that promotes Th2 responses and can in turn be up-regulated by Th2 cytokines (Brüggen et al., 2014). Our finding of increased TSLP levels and enhanced cell death in healthy keratinocytes upon co-culture with activated cTRMs suggests that cTRMs can exacerbate tissue damage in GVHD and promote Th2 skewing in secondary tissue sites via TSLP signaling.
Alloreactive T cells expressing the gut-homing marker α4β7 integrin are known to promote GI-GVHD (Petrovic et al., 2004), and several studies indicate that treatment with the humanized α4β7 antagonist vedolizumab can ameliorate GI-GVHD (Coltoff et al., 2018). Here, we show that cTRMs in the blood of patients with GVHD express high levels of α4β7 and that cells with a cTRM phenotype can be detected in GI-GVHD. This is in line with murine CD8+ TRMs emigrating to the circulation and reseeding different tissues such as the intestine, salivary gland, or female reproductive tract (Fonseca et al., 2020). Similar to this study, where ex-TRMs acquired a central memory phenotype before reexpressing TRM markers in the tissue, our data suggest that cTRMs partially change their TRM profile during recirculation but maintain crucial marker genes like FABP5. It remains to be determined where and by which signals cTRMs acquire gut-homing properties and how long a single cTRM remains in the circulation before tissue reentry.
In contrast to a CD8+ TRM recall-induced tissue exit, it was postulated that human CD4+ CLA+ CD103+ T cells can exit the skin in steady state (Klicznik et al., 2019). Although cTRM numbers were increased during inflammation, we observed that the population in the blood remained stable over several years even in the absence of infection or cutaneous inflammation, indicating that human skin TRMs can undergo both proposed exit routes of recall-induced retrograde migration and steady-state emigration from skin. It will be important to determine in future studies what differences exist between homeostatic and activation-induced exit of cTRMs. Of note, it is likely that plasticity within the skin TRM population is not uniform, but that specific subsets are better equipped to exit the tissue.
Collectively, our study identifies human skin–derived CD4+ cTRMs as a stably maintained low-frequency T cell population in human blood with the potential to contribute to systemic inflammation and tissue damage at secondary target sites in the setting of GVHD. The present work highlights the power of single-cell technologies in addressing these questions, since we could detect rare CD4+ blood cTRMs as a distinct population by their skin TRM-like transcriptome and surface molecules. Combining single-cell approaches for transcriptome, epigenome, and TCR sequencing will be a useful tool for elucidating the functional repertoire of cTRMs in the future.
Materials and methods
Patient sampling
For this study, we included patients presenting with myelodysplastic syndrome or acute or chronic leukemia receiving allogeneic peripheral blood HSCT. All patients participated in the study voluntarily and with fully informed written consent. The research is in line with national law and has been approved by the ethics committee of the Medical University of Vienna (ECS 1087/2016). Peripheral blood was sampled from four patient cohorts according to time to transplantation. For Cohort 1, patients were sampled at hospital admission before start of conditioning (B, n = 11), the day of HSCT (Tx, n = 11), at day 14 (n = 14), and after immune reconstitution between day 100 and day 365 after stem cell transfer (n = 14), and healthy-appearing skin (6-mm punch biopsy) was sampled at B (n = 11) and Tx (n = 11). For flow cytometric analysis and genotyping of cTRMs, patients were sampled after HSCT at the time of immune reconstitution (Cohort 2). From patients presenting with active skin GVHD, blood was sampled at disease onset before start of treatment with corticosteroids (Cohort 3). Additionally, intestinal biopsies of six patients with active gut GVHD were sampled at disease onset (Cohort 3). For cohort 4, we sampled five patients at B and the day of HSCT (Tx). GVHD was diagnosed according to the National Institutes of Health consensus criteria (Filipovich et al., 2005). Patient details are found in Table S5.
Flow cytometry and cell sorting
Single-cell suspensions of blood and skin were stained for surface or intracellular markers as previously described (Strobl et al., 2020a). For intracellular cytokine staining, PBMCs were stimulated for 4 h with 1X Cell Activation Cocktail containing PMA/Ionomycin and Brefeldin A (Biolegend; 423303) in RPMI 1640 (Gibco; 52400025) according to the manufacturers’ protocol. Antibodies used are indicated in Table S1. Stained cells were acquired on a BD Biosciences FACS Aria III using FACS Diva software or a CYTEK Aurora using SpectroFlo software. Acquired flow data were analyzed with FlowJo software v.10.6 or v.10.7 (Tree Star).
T cell–keratinocyte co-culture experiments
T cells were sorted according to the cTRM (CD4+/CD8+ and CLA+CD103+) or TCONV (CD4+/CD8+ and CLA−CD103−) phenotype and rested overnight in RPMI 1640 (Gibco; 52400025) with 10% FBS (Gibco; 10500064) and 1% penicillin-streptomycin (Gibco; 15140122). Keratinocytes isolated from healthy donor skin by explant cultures were seeded in 8-well chamber slides (Ibidi; 80897) at 20,000 cells per well. T cells were stimulated for 2 h with phorbol 12-myristate 13-acetate (Sigma-Aldrich; P1581-1MG) at 10 ng/ml and 1 µg/ml Ionomycin (Sigma-Aldrich; I0634-1MG), and then 1,000 cTRMs or TCONV cells were added per well in X-VIVO 15 Medium (Lonza; BE02-060F). Cells were co-cultured for 16 h before T cells were removed by washing. Keratinocytes were stained with GFP LIVE/DEAD Fixable Green Dead Cell Stain Kit (Invitrogen; L34969) according to the manufacturer’s instructions and subsequently fixed with 4% formaldehyde. Afterward, CD3-AF750, Bcl-2-PE, and purified TSLP with a second-step AF647 were stained as previously described (see Table S1 for antibody details; Brüggen et al., 2014). Briefly, cells were permeabilized with 2% BSA + 0.1% Triton-X-100 and subsequently stained with unlabeled, second-step, and labeled antibodies for 1 h each at 37°C and 5 min with DAPI at room temperature. Mounted slides were imaged on a Z1 Axio Observer microscope equipped with an LD Plan-Neofluar 20×/0.4 objective (Zeiss) and TissueFAXS imaging system and analyzed using TissueQuest software (TissueGnostics GmbH). Data from sorted CD4+ and CD8+ T cells were combined for analysis.
cTRM staining of GI-GVHD biopsies
Cryosection of optimal cutting temperature media–embedded gut biopsies from patients with acute GI-GVHD were acetone-fixed and stained for cTRM markers CD3, CD4, CLA, and CD103 as previously described (Brüggen et al., 2014; antibodies listed in Table S1). Briefly, after blocking with rat and mouse serum (+ 2% BSA), the tissue was stained with fluorochrome-conjugated antibodies for 2 h, Alexa Fluor 488–labeled anti-FITC for 30 min, and DAPI nuclear marker for 5 min. All steps were performed at room temperature. Slides were imaged as described above.
Cytospins and X/Y fluorescence in situ hybridization of T cell subsets
T cells were sorted in Dulbecco’s PBS with 10% FBS. Cytospins of >1,000 cells were harvested onto microscope slides using a cytofunnel system (Shandon) and processed using FAST FISH Prenatal X, Y and 18 Enumeration Probe Kit (Cytocell; LPF 002) as described previously (Strobl et al., 2020b). In brief, the protocol included a digestion step with pepsin/HCl, fixation with formaldehyde, denaturation with formamide, hybridization at 81°C, incubation with the florescence in situ hybridization (FISH) probe overnight at 37°C, and DAPI staining before mounting. Slides were imaged as described above.
Smart-seq2 bulk and single-cell RNA-seq
For Smart-seq2 (Picelli et al., 2014), bulk (Fig. 1, A and B; Fig. 2, E–H; and Fig. 6 G) and single-cell (Fig. 4) RNA-seq libraries were prepared and sequenced at the Biomedical Sequencing Facility of the Center for Molecular Medicine of the Austrian Academy of Sciences as previously described (Strobl et al., 2020b). Bioinformatical analysis, including gene expression analysis, SNP calling, and T cell clonality analysis, was performed as previously described (Strobl et al., 2020b). Determination of single-cell T cell clones from Smart-seq2 sequencing was identified using the TRUST (TCR repertoire utilities for solid tissue) tool version 4.0 (Li et al., 2016). For pathway enrichment (Fig. 6 G), DEGs in cTRMs with adjusted P value <0.05 were loaded in Enrichr (Xie et al., 2021), and the top 25 KEGG 2021 human pathways with adjusted P value <0.01 were plotted.
10X Genomics single-cell sequencing
For 10X Genomics single-cell sequencing, CD45+ live cells were sorted into PBS/10% FCS for 10X library preparation with the Single-Cell 3′ Library & Gel Bead Kit v2 (10X Genomics) according to the manufacturer’s instructions. Briefly, sorted cells were partitioned into gel bead-in-emulsions for cDNA synthesis and subsequently amplified and prepared for sequencing. The samples were sequenced at the Biomedical Sequencing Facility on the Illumina HiSeq 3000/4000 platform in the 75-bp paired-end configuration. Raw sequencing data were processed and analyzed using the 10X Genomics Cell Ranger platform and 10X Genomics Loupe Browser.
Mathematical modeling of cTRM origin
Whole T cell numbers and TRM numbers were determined per cubic centimeter of skin as described by Clark et al. (2006) and per milliliter of blood at B and Tx and extrapolated to whole-body surface and whole blood volume for an average human adult (weight: ≥66.1 kg) using the Du Bois formula (BSA = 0.007184 × W0.425 × H0.725; Fig. 3, A–C). T cell cycles (Fig. 3 D) were modeled using a system of ordinary differential equations based on previously published models of T cell dynamics in HIV infection (Reeves et al., 2017). The equation system was modeled in Jupyter Notebook software (Python). Plots were created using Matplotlib visualization tool (Hunter, 2007).
Statistical analysis
Statistical analyses on flow cytometry and imaging data were performed using GraphPad Prism 8.0. To determine statistically significant differences between two groups, we used paired or unpaired, two-tailed Student’s t test. To compare multiple groups, we used parametric one-way ANOVA followed by Tukey’s pairwise comparisons and two-tailed Student’s t test.
Online supplemental material
Fig. S1 shows graphical representation of the patient cohorts. Fig. S2 shows the RNA-based progression score of ITGA1 expression in patient T cells, gating/sorting strategy and FACS-based quantification of Tnaive, TCM, TEM, and TEMRA cells, and quantification cross-shared TCR clones in pre-Tx skin versus post-Tx blood. Fig. S3 shows quantification of total PBMC and cTRM numbers in blood of patients at B and Tx and modeled growth curves for scenarios 3 and 4, as well as tested proliferation rates for scenarios 1–3. Fig. S4 shows a t-SNE plot of Smart-seq2 single T cells of patient 2 at d 14 and uniform manifold approximation and projections (UMAPs) depicting expression of TCR genes in GVHD 10X Genomics single-cell data. Fig. S5 shows negative control for IL-13 staining on cTRMs, heat map of selected DEGs in bulk sequenced cTRMs versus TCONV cells of GVHD patients, flow cytometry quantification of α4β7 expression on cTRMs and TCONV cells, and selected markers on CD8+ T cells in GVHD blood. Table S1 lists antibodies used in the study. Table S2 and Table S3 show host chimerism of blood T cells in patients A and B, corresponding to Fig. 4. Table S4 lists cross-shared TCR clone sequences of pre-Tx skin and post-Tx blood per patient. Table S5 shows patient details, including clinical information.
Supplementary Material
lists antibodies and dyes used for flow cytometry, FACS sorting, and immunofluorescence.
presents host chimerism in peripheral blood CD3+ cells of patient A.
presents host chimerism in peripheral blood CD3+ cells of patient B.
lists shared TCR clone sequences in post-Tx blood and pre-Tx skin for every patient.
presents patient details.
Acknowledgments
We thank the Biomedical Sequencing Facility at Center for Molecular Medicine of the Austrian Academy of Sciences for assistance with next-generation sequencing. The authors acknowledge the Core Facility Flow Cytometry of the Medical University of Vienna, a member of Vienna Life-Science Instruments. The graphical abstract was created with BioRender.com.
This work was supported by funds of the Oesterreichische Nationalbank (Austrian Central Bank, Anniversary Fund, project number: 17872), by the Austrian Science Fund (P31494), and by the Innovation Fund of the Austrian Academy of Sciences (project number: IF_2017_29). J. Strobl was supported by a DOCmed fellowship of the Austrian Academy of Sciences. This work was further supported through a DOC fellowship received by J. Huber from the Austrian Academy of Sciences. V. Smejkal acknowledges financial support by the doctoral college program “TU-D Unravelling advanced 2D materials” funded by Technische Universität Wien. This research was funded in whole or in part by the Austrian Science Fund. For the purpose of Open Access, the author has applied a CC-BY public copyright license to any Author Accepted Manuscript (AAM) version arising from this submission.
Author contributions: J. Strobl and G. Stary designed the study. J. Strobl, L.M. Gail, and L. Kleissl planned and carried out experiments and acquired data. R. Dingelmaier-Hovorka, D. Atzmüller, and T. Krausgruber performed experiments and acquired data. J. Strobl, L.M. Gail, R.V. Pinay, and V. Puxkandl analyzed, interpreted, and visualized the data. V. Smejkal, J. Huber, and J. Strobl designed, analyzed, and visualized the mathematical model. L. Unterluggauer, P. Wohlfarth, and W. Rabitsch provided patient material and performed disease scoring. J. Strobl and L.M. Gail compiled the figures. J. Strobl, L.M. Gail, and G. Stary drafted the manuscript. C. Bock provided resources and critically revised the manuscript. G. Stary provided funding and resources and supervised the study. All authors reviewed the final version of the manuscript.
Data availability
Sequencing data presented in Fig. 1 B and Fig. S2 A are accessible in Gene Expression Omnibus under accession no. GSE146495. All remaining sequencing data are available via Gene Expression Omnibus database accession no. GSE183212.
References
- Bartolomé-Casado, R., Landsverk O.J.B., Chauhan S.K., Richter L., Phung D., Greiff V., Risnes L.F., Yao Y., Neumann R.S., Yaqub S., et al. 2019. Resident memory CD8 T cells persist for years in human small intestine. J. Exp. Med. 216:2412–2426. 10.1084/jem.20190414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behr, F.M., Parga-Vidal L., Kragten N.A.M., van Dam T.J.P., Wesselink T.H., Sheridan B.S., Arens R., van Lier R.A.W., Stark R., and van Gisbergen K.P.J.M.. 2020. Tissue-resident memory CD8+ T cells shape local and systemic secondary T cell responses. Nat. Immunol. 21:1070–1081. 10.1038/s41590-020-0723-4 [DOI] [PubMed] [Google Scholar]
- Benoit, B.M., Jariwala N., O’Connor G., Oetjen L.K., Whelan T.M., Werth A., Troxel A.B., Sicard H., Zhu L., Miller C., et al. 2017. CD164 identifies CD4+ T cells highly expressing genes associated with malignancy in Sézary syndrome: the Sézary signature genes, FCRL3, Tox, and miR-214. Arch. Dermatol. Res. 309:11–19. 10.1007/s00403-016-1698-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beura, L.K., Mitchell J.S., Thompson E.A., Schenkel J.M., Mohammed J., Wijeyesinghe S., Fonseca R., Burbach B.J., Hickman H.D., Vezys V., et al. 2018. Intravital mucosal imaging of CD8+ resident memory T cells shows tissue-autonomous recall responses that amplify secondary memory. Nat. Immunol. 19:173–182. 10.1038/s41590-017-0029-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brüggen, M.C., Klein I., Greinix H., Bauer W., Kuzmina Z., Rabitsch W., Kalhs P., Petzelbauer P., Knobler R., Stingl G., and Stary G.. 2014. Diverse T-cell responses characterize the different manifestations of cutaneous graft-versus-host disease. Blood. 123:290–299. 10.1182/blood-2013-07-514372 [DOI] [PubMed] [Google Scholar]
- Chen, L., and Shen Z.. 2020. Tissue-resident memory T cells and their biological characteristics in the recurrence of inflammatory skin disorders. Cell. Mol. Immunol. 17:64–75. 10.1038/s41423-019-0291-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheuk, S., Schlums H., Gallais Sérézal I., Martini E., Chiang S.C., Marquardt N., Gibbs A., Detlofsson E., Introini A., Forkel M., et al. 2017. CD49a Expression Defines Tissue-Resident CD8+ T Cells Poised for Cytotoxic Function in Human Skin. Immunity. 46:287–300. 10.1016/j.immuni.2017.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibrián, D., and Sánchez-Madrid F.. 2017. CD69: from activation marker to metabolic gatekeeper. Eur. J. Immunol. 47:946–953. 10.1002/eji.201646837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, R.A., Chong B., Mirchandani N., Brinster N.K., Yamanaka K., Dowgiert R.K., and Kupper T.S.. 2006. The vast majority of CLA+ T cells are resident in normal skin. J. Immunol. 176:4431–4439. 10.4049/jimmunol.176.7.4431 [DOI] [PubMed] [Google Scholar]
- Coltoff, A., Lancman G., Kim S., and Steinberg A.. 2018. Vedolizumab for treatment of steroid-refractory lower gastrointestinal acute graft-versus-host disease. Bone Marrow Transplant. 53:900–904. 10.1038/s41409-018-0094-8 [DOI] [PubMed] [Google Scholar]
- de Almeida, G.P., Lichtner P., Mädler S., Chu C.F., and Zielinski C.E.. 2020. Transcriptional profiling and single-cell chimerism analysis identifies human tissue resident T cells in the human skin after allogeneic stem cell transplantation. bioRxiv. (Preprint posted April 13, 2020) 10.1101/2020.04.11.037101 [DOI]
- de Leur, K., Dieterich M., Hesselink D.A., Corneth O.B.J., Dor F.J.M.F., de Graav G.N., Peeters A.M.A., Mulder A., Kimenai H.J.A.N., Claas F.H.J., et al. 2019. Characterization of donor and recipient CD8+ tissue-resident memory T cells in transplant nephrectomies. Sci. Rep. 9:5984. 10.1038/s41598-019-42401-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divito, S.J., Aasebø A.T., Matos T.R., Hsieh P.C., Collin M., Elco C.P., O’Malley J.T., Bækkevold E.S., Reims H., Gedde-Dahl T., et al. 2020. Peripheral host T cells survive hematopoietic stem cell transplantation and promote graft-versus-host disease. J. Clin. Invest. 130:4624–4636. 10.1172/JCI129965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipovich, A.H., Weisdorf D., Pavletic S., Socie G., Wingard J.R., Lee S.J., Martin P., Chien J., Przepiorka D., Couriel D., et al. 2005. National Institutes of Health consensus development project on criteria for clinical trials in chronic graft-versus-host disease: I. Diagnosis and staging working group report. Biol. Blood Marrow Transplant. 11:945–956. 10.1016/j.bbmt.2005.09.004 [DOI] [PubMed] [Google Scholar]
- Fonseca, R., Beura L.K., Quarnstrom C.F., Ghoneim H.E., Fan Y., Zebley C.C., Scott M.C., Fares-Frederickson N.J., Wijeyesinghe S., Thompson E.A., et al. 2020. Developmental plasticity allows outside-in immune responses by resident memory T cells. Nat. Immunol. 21:412–421. 10.1038/s41590-020-0607-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhlbrigge, R.C., Kieffer J.D., Armerding D., and Kupper T.S.. 1997. Cutaneous lymphocyte antigen is a specialized form of PSGL-1 expressed on skin-homing T cells. Nature. 389:978–981. 10.1038/40166 [DOI] [PubMed] [Google Scholar]
- Hansell, C.A., MacLellan L.M., Oldham R.S., Doonan J., Chapple K.J., Anderson E.J., Linington C., McInnes I.B., Nibbs R.J., and Goodyear C.S.. 2015. The atypical chemokine receptor ACKR2 suppresses Th17 responses to protein autoantigens. Immunol. Cell Biol. 93:167–176. 10.1038/icb.2014.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, G.R., and Koyama M.. 2020. Cytokines and costimulation in acute graft-versus-host disease. Blood. 136:418–428. 10.1182/blood.2019000952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, J.D. 2007. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 9:90–95. 10.1109/MCSE.2007.55 [DOI] [Google Scholar]
- Klicznik, M.M., Morawski P.A., Höllbacher B., Varkhande S.R., Motley S.J., Kuri-Cervantes L., Goodwin E., Rosenblum M.D., Long S.A., Brachtl G., et al. 2019. Human CD4+CD103+ cutaneous resident memory T cells are found in the circulation of healthy individuals. Sci. Immunol. 4:eaav8995. 10.1126/sciimmunol.aav8995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komai-Koma, M., Jones L., Ogg G.S., Xu D., and Liew F.Y.. 2004. TLR2 is expressed on activated T cells as a costimulatory receptor. Proc. Natl. Acad. Sci. USA. 101:3029–3034. 10.1073/pnas.0400171101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, B.V., Ma W., Miron M., Granot T., Guyer R.S., Carpenter D.J., Senda T., Sun X., Ho S.H., Lerner H., et al. 2017. Human Tissue-Resident Memory T Cells Are Defined by Core Transcriptional and Functional Signatures in Lymphoid and Mucosal Sites. Cell Rep. 20:2921–2934. 10.1016/j.celrep.2017.08.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B., Li T., Pignon J.C., Wang B., Wang J., Shukla S.A., Dou R., Chen Q., Hodi F.S., Choueiri T.K., et al. 2016. Landscape of tumor-infiltrating T cell repertoire of human cancers. Nat. Genet. 48:725–732. 10.1038/ng.3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian, C.G., Bueno E.M., Granter S.R., Laga A.C., Saavedra A.P., Lin W.M., Susa J.S., Zhan Q., Chandraker A.K., Tullius S.G., et al. 2014. Biomarker evaluation of face transplant rejection: association of donor T cells with target cell injury. Mod. Pathol. 27:788–799. 10.1038/modpathol.2013.249 [DOI] [PubMed] [Google Scholar]
- Mackay, L.K., Braun A., Macleod B.L., Collins N., Tebartz C., Bedoui S., Carbone F.R., and Gebhardt T.. 2015. Cutting edge: CD69 interference with sphingosine-1-phosphate receptor function regulates peripheral T cell retention. J. Immunol. 194:2059–2063. 10.4049/jimmunol.1402256 [DOI] [PubMed] [Google Scholar]
- Mackay, L.K., Minnich M., Kragten N.A., Liao Y., Nota B., Seillet C., Zaid A., Man K., Preston S., Freestone D., et al. 2016. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science. 352:459–463. 10.1126/science.aad2035 [DOI] [PubMed] [Google Scholar]
- Maecker, H.T. 2003. Human CD81 directly enhances Th1 and Th2 cell activation, but preferentially induces proliferation of Th2 cells upon long-term stimulation. BMC Immunol. 4:1. 10.1186/1471-2172-4-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner, J.J., Toma C., Yu B., Zhang K., Omilusik K., Phan A.T., Wang D., Getzler A.J., Nguyen T., Crotty S., et al. 2017. Runx3 programs CD8+ T cell residency in non-lymphoid tissues and tumours. Nature. 552:253–257. 10.1038/nature24993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner, J.J., Toma C., He Z., Kurd N.S., Nguyen Q.P., McDonald B., Quezada L., Widjaja C.E., Witherden D.A., Crowl J.T., et al. 2020. Heterogenous Populations of Tissue-Resident CD8+ T Cells Are Generated in Response to Infection and Malignancy. Immunity. 52:808–824.e7. 10.1016/j.immuni.2020.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi, S.A., Javadimoghadam M., Ghavamzadeh A., Alimoghaddam K., Sayarifard A., Ghaffari S.H., Chahardouli B., and Basi A.. 2017. The Relationship between STR-PCR Chimerism Analysis and Chronic GvHD Following Hematopoietic Stem Cell Transplantation. Int. J. Hematol. Oncol. Stem Cell Res. 11:24–29. [PMC free article] [PubMed] [Google Scholar]
- Mueller, S.N., and Mackay L.K.. 2016. Tissue-resident memory T cells: local specialists in immune defence. Nat. Rev. Immunol. 16:79–89. 10.1038/nri.2015.3 [DOI] [PubMed] [Google Scholar]
- Napier, R.J., Lee E.J., Davey M.P., Vance E.E., Furtado J.M., Snow P.E., Samson K.A., Lashley S.J., Brown B.R., Horai R., et al. 2020. T cell-intrinsic role for Nod2 in protection against Th17-mediated uveitis. Nat. Commun. 11:5406. 10.1038/s41467-020-18961-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, Y., Tian T., Park C.O., Lofftus S.Y., Mei S., Liu X., Luo C., O’Malley J.T., Gehad A., Teague J.E., et al. 2017. Survival of tissue-resident memory T cells requires exogenous lipid uptake and metabolism. Nature. 543:252–256. 10.1038/nature21379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S.L., Zaid A., Hor J.L., Christo S.N., Prier J.E., Davies B., Alexandre Y.O., Gregory J.L., Russell T.A., Gebhardt T., et al. 2018. Local proliferation maintains a stable pool of tissue-resident memory T cells after antiviral recall responses. Nat. Immunol. 19:183–191. 10.1038/s41590-017-0027-5 [DOI] [PubMed] [Google Scholar]
- Petrovic, A., Alpdogan O., Willis L.M., Eng J.M., Greenberg A.S., Kappel B.J., Liu C., Murphy G.J., Heller G., and van den Brink M.R.. 2004. LPAM (alpha 4 beta 7 integrin) is an important homing integrin on alloreactive T cells in the development of intestinal graft-versus-host disease. Blood. 103:1542–1547. 10.1182/blood-2003-03-0957 [DOI] [PubMed] [Google Scholar]
- Picelli, S., Faridani O.R., Björklund A.K., Winberg G., Sagasser S., and Sandberg R.. 2014. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9:171–181. 10.1038/nprot.2014.006 [DOI] [PubMed] [Google Scholar]
- Reeves, D.B., Peterson C.W., Kiem H.P., and Schiffer J.T.. 2017. Autologous Stem Cell Transplantation Disrupts Adaptive Immune Responses during Rebound Simian/Human Immunodeficiency Virus Viremia. J. Virol. 91:e0095–e17. 10.1128/JVI.00095-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saed, G., Fivenson D.P., Naidu Y., and Nickoloff B.J.. 1994. Mycosis fungoides exhibits a Th1-type cell-mediated cytokine profile whereas Sezary syndrome expresses a Th2-type profile. J. Invest. Dermatol. 103:29–33. 10.1111/1523-1747.ep12388985 [DOI] [PubMed] [Google Scholar]
- Shan, Q., Zeng Z., Xing S., Li F., Hartwig S.M., Gullicksrud J.A., Kurup S.P., Van Braeckel-Budimir N., Su Y., Martin M.D., et al. 2017. The transcription factor Runx3 guards cytotoxic CD8+ effector T cells against deviation towards follicular helper T cell lineage. Nat. Immunol. 18:931–939. 10.1038/ni.3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobl, J., Pandey R.V., Krausgruber T., Kleissl L., Reininger B., Herac M., Bayer N., Krall C., Wohlfarth P., Mitterbauer M., et al. 2020a. Anti-Apoptotic Molecule BCL2 Is a Therapeutic Target in Steroid-Refractory Graft-Versus-Host Disease. J. Invest. Dermatol. 140:2188–2198. 10.1016/j.jid.2020.02.029 [DOI] [PubMed] [Google Scholar]
- Strobl, J., Pandey R.V., Krausgruber T., Bayer N., Kleissl L., Reininger B., Vieyra-Garcia P., Wolf P., Jentus M.M., Mitterbauer M., et al. 2020b. Long-term skin-resident memory T cells proliferate in situ and are involved in human graft-versus-host disease. Sci. Transl. Med. 12:eabb7028. 10.1126/scitranslmed.abb7028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, R., Gehad A., Yang C., Scott L.L., Teague J.E., Schlapbach C., Elco C.P., Huang V., Matos T.R., Kupper T.S., and Clark R.A.. 2015. Human skin is protected by four functionally and phenotypically discrete populations of resident and recirculating memory T cells. Sci. Transl. Med. 7:279ra39. 10.1126/scitranslmed.3010302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z., Bailey A., Kuleshov M.V., Clarke D.J.B., Evangelista J.E., Jenkins S.L., Lachmann A., Wojciechowicz M.L., Kropiwnicki E., Jagodnik K.M., et al. 2021. Gene Set Knowledge Discovery with Enrichr. Curr Protoc. 1:e90. 10.1002/cpz1.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists antibodies and dyes used for flow cytometry, FACS sorting, and immunofluorescence.
presents host chimerism in peripheral blood CD3+ cells of patient A.
presents host chimerism in peripheral blood CD3+ cells of patient B.
lists shared TCR clone sequences in post-Tx blood and pre-Tx skin for every patient.
presents patient details.
Data Availability Statement
Sequencing data presented in Fig. 1 B and Fig. S2 A are accessible in Gene Expression Omnibus under accession no. GSE146495. All remaining sequencing data are available via Gene Expression Omnibus database accession no. GSE183212.