This study describes a unique monomethyl branched-chain fatty acid, C17iso, as the side chain of phospholipids to ensure endoplasmic reticulum integrity for lipid droplet growth in an intact organism, Caenorhabditis elegans.
Abstract
In eukaryote cells, lipid droplets (LDs) are key intracellular organelles that dynamically regulate cellular energy homeostasis. LDs originate from the ER and continuously contact the ER during their growth. How the ER affects LD growth is largely unknown. Here, we show that RNAi knockdown of acs-1, encoding an acyl-CoA synthetase required for the biosynthesis of monomethyl branched-chain fatty acids C15iso and C17iso, remarkably prevented LD growth in Caenorhabditis elegans. Dietary C17iso, or complex lipids with C17iso including phosphatidylcholine, phosphatidylethanolamine, and triacylglycerol, could fully restore the LD growth in the acs-1RNAi worms. Mechanistically, C17iso may incorporate into phospholipids to ensure the membrane integrity of the ER so as to maintain the function of ER-resident enzymes such as SCD/stearoyl-CoA desaturase and DGAT2/diacylglycerol acyltransferase for appropriate lipid synthesis and LD growth. Collectively, our work uncovers a unique fatty acid, C17iso, as the side chain of phospholipids for determining the ER homeostasis for LD growth in an intact organism, C. elegans.
Introduction
Lipid droplets (LDs) are conserved intracellular organelles ubiquitously present in almost all organisms from bacteria to humans (Zhang and Liu, 2017; Murphy, 2012). LDs are composed of a core of neutral lipids, mainly triacylglycerol (TAG) and sterol esters (SE), surrounded by a phospholipid (PL) monolayer containing several hundred proteins. The major function of LDs is to dynamically regulate cellular energy homeostasis (Walther and Farese, 2012; Listenberger et al., 2003). In addition, LDs are involved in the regulation of membrane biosynthesis, lipid-mediated signaling, ER or oxidative stress mitigation, protein maturation, storage and degradation, and so on (Welte and Gould, 2017). Under nutrient-rich conditions, excess energy is transformed mainly into TAG and SE molecules and stored in LDs for later use. However, an aberrant increase of LDs in adipose tissue and nonadipose tissues such as liver, muscle, and heart is highly associated with a variety of metabolic diseases, including obesity, fatty liver, diabetes, and atherosclerosis (Salo et al., 2019; Mori et al., 2001), which are all global public health issues. Thus, understanding LD dynamics provides in-depth insights into the regulation of energy homeostasis and the pathogenesis of metabolic diseases.
It is well known that LDs originate from the ER, where many lipid synthesis enzymes localize and function. The neutral lipids are synthesized and accumulate between the leaflets of the ER membrane, and then they are budded and released toward the cytosol as mature LDs (Walther et al., 2017; Jackson, 2019). Some nascent LDs can grow further by both local lipid synthesis on the LD surface and lipid transfer from the ER (Salo et al., 2016; Wilfling et al., 2013; Jacquier et al., 2011). Among intracellular organelles, the ER has probably the most frequent and prominent contact with LDs (Schuldiner and Bohnert, 2017). Many ER-associated proteins were reported to regulate LD biogenesis, growth, and expansion. For example, fat storage inducing transmembrane (FITM2), long-chain fatty acyl-CoA synthetase (ACSL3), and perilipin protein (PLIN3) have been reported to play important roles in LD lens formation and budding (Choudhary et al., 2016; Kassan et al., 2013; Poppelreuther et al., 2018; Salo and Ikonen, 2019). Seipin is a membrane protein located at LD–ER contact sites and functions to mediate the flow of TAG from the ER to LDs (Salo et al., 2019). The ER-localized ACS-22/FATP1 and LD surface protein DGAT2 (diacylglycerol O-acyltransferase 2) form a conserved protein complex to synthesize TAG for LD expansion (Xu et al., 2012).
In addition, the lipid composition of the ER regulates LD formation and growth. The ER is a major site for PL synthesis, and also accounts for >60% of PL mass in a variety of cell types (Lagace and Ridgway, 2013). The PL content and composition of the ER determine its configuration and functions. It has been shown that PLs affect the ER membrane surface tension to regulate LD formation in vitro (Ben M’barek et al., 2017), in which phosphatidylcholine (PC) specifically promotes the budding of triglyceride LDs, and phosphatidylinositol (PI) or saturated PL promotes the budding of neutral LD formation (Ben M’barek et al., 2017). Defects in PC synthesis, the most abundant lipid of ER membranes, leads to lipid accumulation and LD expansion in Caenorhabditis elegans, mouse liver, and human cells (Jacquemyn et al., 2017; Walker et al., 2011). The lipid saturation profile not only affects LD size and membrane PL composition (Shi et al., 2013), but it is also involved in ER stress in obesity (Fu et al., 2011). Altogether, these reports demonstrate that lipids are crucial for maintaining ER configuration and functions. However, in spite of many ER-associated proteins reported as being involved in LD growth, it is largely unknown how the ER per se affects LD growth. As the membrane is constituted, how certain PLs with specific fatty acids as side chains ensure ER integrity and function for lipid biogenesis or growth is poorly understood.
The genetically tractable organism C. elegans has well-resolved metabolic pathways for lipid biosynthesis and stores neutral lipids in LDs mainly in the intestine and hypodermis (Watts and Ristow, 2017; Zhang et al., 2013). Here, we performed a candidate screen to identify LD growth–associated genes using RNAi. We found that RNAi reduction of acs-1, encoding an acyl-CoA synthetase (ACS), prevented LD growth. ACS-1 functions in the biosynthesis of monomethyl branched-chain fatty acids (mmBCFAs) C15iso and C17iso. Remarkably, C17iso is essential to maintain ER integrity for LD growth.
Results
RNAi screen identified ACS-1 as being required for LD growth
The intracellular organelles LDs dynamically regulate lipid metabolism and energy homeostasis. To identify genes involved in LD growth, we performed a reverse genetic screen by RNAi in the model organism C. elegans. Lipids play a vital role in normal life activities, and dysfunction of lipid metabolism is consequently associated with impaired growth, development, reproduction, etc. Therefore, we collected and integrated 2,343 candidate genes that have been previously reported to display relevant phenotypes in C. elegans through genome-wide RNAi screens by Maeda et al. (2001), Kamath et al. (2003), and Simmer et al. (2003; Fig. 1 A). DHS-3 is a short-chain dehydrogenase/reductase localized on LDs, and the DHS-3::GFP (N2;dhs-3p::dhs-3::gfp) fusion protein is a well-established LD marker in C. elegans (Na et al., 2015; Zhang et al., 2012). Using RNAi knockdown, we initially identified 43 genes displaying significantly altered LD size visualized by both DHS-3::GFP and Nile Red staining of fixed worms (Brooks et al., 2009; Fig. 1 B and Table S3). Interestingly, a majority of genes that caused increased LD size by RNAi knockdown are involved in protein production via ribosome biogenesis, consistent with our previous report (Wu et al., 2018), and aminoacyl-tRNA synthetases (ARS; Webster et al., 2017; Table S3), suggesting that impaired protein production may shift energy to store as fat in LDs.
Figure 1.
RNAi screen identified ACS-1 regulating LD size. (A) Candidate genes (2,343) for RNAi screen from 3 previous reports, in which 259 genes were specifically from Maeda (blue), 273 from Kamath (orange), and 392 from Simmer (green), and 1,419 genes were duplicates from 2 or 3 reports (pink). (B) Schematic workflow of RNAi screen in wild-type N2 and transgenic worms for the regulators of LDs visualized by DHS-3::GFP (N2;dhs-3p::dhs-3::gfp) and Nile Red staining of fixed worms. (C) LDs visualized by DHS-3::GFP, fixed Nile Red staining, and LipidTOX staining. DHS-3::GFP worms were imaged by high-resolution laser confocal microscopy; scale bar represents 2.5 and 0.65 µm (enlarged). For Nile Red– and LipidTOX-stained animals, anterior is left and posterior is right. Scale bar represents 5 and 1.25 µm (enlarged). The pink (DHS-3::GFP-positive LDs) and yellow (Nile Red– and LipidTOX-stained particles) arrows indicate represented LDs. (D and E) The size (mean diameter; D) and distribution (percentage; E) of LDs quantified from five representative Nile Red–stained worms for each worm strain. The data are presented as diameter. (F) The abundance of LDs quantified from Nile Red–stained worms. The data are presented as the number of LDs within a fixed area. n, the number of worms scored for quantification. (G) Percentage of TAG in TL (TAG+ PL) by TLC-GC analysis. Data presented are the mean ± SEM of four biological repeats. Significant difference between the control (EV) and a specific RNAi treatment: ***, P < 0.001.
Among 20 genes that showed reduced LD size by RNAi knockdown, acs-1 and fat-6 were chosen for further analysis, since they are involved in lipid metabolism and displayed more severe reduction of LD size than others on the list (Fig. 1, C–E; and Table S3). fat-6 encodes a stearoyl-CoA desaturase (SCD) that converts saturated fatty acids (SFAs, C16:0 and C18:0) to monounsaturated fatty acids (MUFAs, C16:1(n-7) and C18:1(n-9); Brock et al., 2006, 2007). RNAi knockdown of fat-6 significantly reduced the expression of FAT-6::GFP{lin-15(n765);waEx16[fat-6p::fat-6::gfp;lin-15(+)]}, but inversely activated the expression of both FAT-5::GFP{unc-119(ed3);kunIs161[fat-5p::fat-5::gfp+unc-119(+)]} and FAT-7::GFP{lin-15(n765);waEx15[fat-7p::fat-7::gfp;lin-15(+)]} (Fig. S1, A and B), which could be due to compensation, since FAT-5, -6, and -7 are three SCDs in C. elegans (Brock et al., 2006, 2007). Consistent with our previous report that SCD regulates LD size and PL compositions (Shi et al., 2013), the fat-6RNAi worms showed reduced LD size and abundance and also reduced TAG content, as quantified by thin-layer chromatography (TLC) and gas chromatography (GC; Fig. 1, C–G). Therefore, we used the fat-6RNAi worms as a positive control for the ensuing experiments.
Figure S1.
RNAi efficiency and the effect of ACS family on LD size. (A and B) Fluorescence intensity (A) and quantification (B) of FAT-5::GFP{unc-119(ed3);kunIs161[fat-5p::fat-5::gfp,unc-119(+)]}, FAT-6::GFP {lin-15A(n765);waEx16[fat-6p::fat-6::gfp+lin15(+)]}, and FAT-7::GFP{lin-15(n765); waEx15[fat-7p::fat-7::gfp;lin-15(+)]}. Imaged by fluorescent microscopy (Olympus); scale bar represents 100 µm. The data are presented as mean ± SEM of 10 imaged worms. Significant difference between the control (EV) and a specific RNAi treatment: ***, P < 0.001. (C and D) Fluorescence intensity (C) and quantification (D) of ACS-1::GFP {unc-119(ed3);kunEx202[vha-6p::acs-1::gfp+unc-119(+), myo-2p::mcherry]}, and ACS-2::GFP {N2;[acs-2p::acs-2::gfp+rol-6(su1006)]} under acs-1RNAi treatment. The white arrows indicate the transgenic marker of MYO-2::mCherry (myo-2p::mcherry), and the purple triangle indicates ACS-1::GFP. Imaged by fluorescent microscopy (Olympus); scale bar represents 100 µm. The data are presented as mean ± SEM of 10 imaged worms. Significant difference between the control (EV) and a specific RNAi treatment: ***, P < 0.001. (E) Cluster analysis of ACS family in C. elegans and Homo sapiens. (F and H) LDs by Nile Red staining of fixed mutant worms (F) and RNAi-treated worms (H), cultured with E. coli OP50 and HT115, respectively. For all representative animals, posterior is left and anterior is right. Scale bar represents 5 µm. (G and I) The diameter (mean ± SEM) of LDs by quantification of five representative Nile Red–stained worms.
acs-1 encodes an orthologue of human ACSF2 (ACS family member 2) and is required for the synthesis of mmBCFAs (Kniazeva et al., 2008). Disruption affects both lumenal membrane homeostasis (Zhang et al., 2011) and early embryogenesis (Kniazeva et al., 2012). However, its role in regulating LD size has not been reported thus far. The significance and specificity of RNAi knockdown of acs-1 was confirmed by the obviously reduced fluorescent expression of ACS-1::GFP{unc-119(ed3);kunEx202[vha-6p::acs-1::gfp+unc-119(+)]} that is mainly expressed in the intestine, but not for ACS-2::GFP{N2;[acs-2p::acs-2::gfp+rol-6(su1006)]} (Fig. S1, C and D). ACS-1 and ACS-2 showed the closest protein sequence homology (Fig. S1 E). Compared with the control (empty vector [EV]; and similarly to fat-6RNAi but to a greater extent), RNAi depletion of acs-1 resulted in significantly decreased size and abundance of LDs, as indicated by DHS-3::GFP, Nile Red staining, and LipidTOX staining of fixed worms (Fig. 1, C–F), as well as decreased TAG content (Fig. 1 G), suggesting a dramatic reduction of LD growth and lipid storage. Collectively, these results indicate that ACS-1, like FAT-6, regulates LD growth.
The enzyme ACS catalyzes the formation of a thioester bond between a fatty acid and coenzyme A to activate fatty acids for dehydrogenation, elongation, degradation, or incorporation into complex lipids. C. elegans has 21 members of the ACS family in total (Fig. S1 E), some of which have been reported to be involved in lipid metabolism and other biological processes (Watts and Ristow, 2017). To explore whether other ACS members function like ACS-1 and are required for LD growth, we examined the LD size of 16 acs genes with available mutant strains or RNAi clones. However, when compared with the control (either N2 or EV), none of the mutants or acs RNAi other than acs-1 caused an obvious change in LD size and lipid accumulation (Fig. S1, F–I), suggesting that ACS-1 may be the only ACS regulating LD growth.
ACS-1–derived mmBCFA C17iso is necessary for LD growth
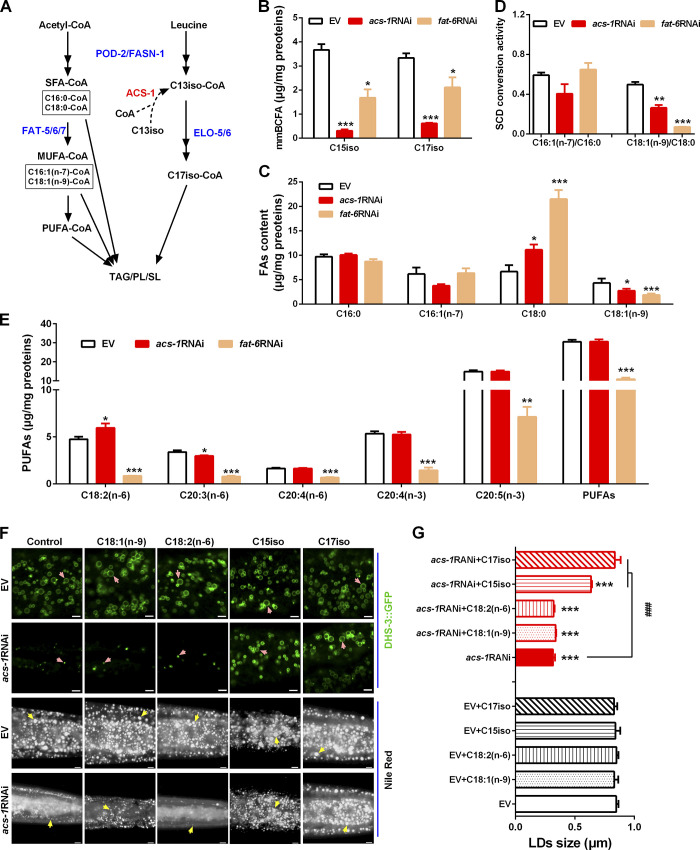
C. elegans contains saturated (SFAs) and unsaturated fatty acids, as well as leucine-derived mmBCFAs, which are major substrates for the synthesis of membrane structural PLs, sphingolipids (SLs), and storage lipid TAG (Fig. 2 A; Watts and Ristow, 2017). ACS-1 has been reported to be mainly involved in mmBCFA biosynthesis (Fig. 2 A; Kniazeva et al., 2008, Zhang et al., 2011, Kniazeva et al., 2012). Consistent with these reports, our analysis of the fatty acid profile by GC further confirmed that the content (micrograms per milligrams of protein) of mmBCFAs C15iso and C17iso were indeed significantly decreased in the acs-1RNAi worms when compared with the control (EV) worms (Fig. 2 B). In addition, RNAi knockdown of acs-1, like fat-6RNAi, displayed a similar shifting profile, but to different degrees, of the SFAs and MUFAs: both had increased content of C18:0 but decreased content of C18:1(n-9) (Fig. 2 C), leading to a decreased ratio of C18:1(n-9) to C18:0, an indicator of SCD conversion activity (Fig. 2 D). As a control, fat-6RNAi greatly reduced the contents of various and total polyunsaturated fatty acids (PUFAs; Fig. 2 E). Although RNAi knockdown of acs-1 did not affect the content of PUFAs, it slightly but significantly increased the content of C18:2(n-6) but decreased the content of C20:3(n-6) (Fig. 2 E). Taken together, these results suggest that ACS-1 may be involved mainly in the synthesis of both C15iso and C17iso mmBCFAs and C18:1(n-9).
Figure 2.
Inactivation of ACS-1 impairs C17iso synthesis and LD growth. (A) Schematic pathways of fatty acid biosynthesis in C. elegans. ACS-1, acyl-CoA synthase; ELO-5/6, elongases; FASN-1, fatty acid synthase; FAT-5/6/7, SCDs; POD-2, acetyl-CoA carboxylase. (B and C) The contents of mmBCFAs C15iso and C17iso (B), SFAs (C16:0 and C18:0; C), and MUFAs (C16:1(n-7) and C18:1(n-9); C), analyzed by GC and further quantified by total proteins. The data are presented as mean ± SEM of three to four biological repeats. (D) SCD conversion activity presented as the ratio of C16:1(n-7)/C16:0 and C18:1(n-9)/C18:0 from C. The data are presented as mean ± SEM of three to four biological repeats. (E) The contents of various and total PUFAs analyzed by GC and further quantified by total proteins. The data are presented as mean ± SEM of three to four biological repeats. (F) LDs visualized by DHS-3::GFP and fixed Nile Red staining with dietary fatty acids. DHS-3::GFP worms were imaged by high-resolution laser confocal microscopy; scale bar represents 2.5 µm; pink arrows indicate represented LDs. For Nile Red–stained animals, anterior is left and posterior is right; scale bar represents 5 µm; yellow arrows indicate represented LDs. (G) The mean diameters of LDs quantified from five representative Nile Red–stained worms. Significant differences between the EV and a specific RNAi treatment: *, P < 0.05; **, P < 0.01; ***, P < 0.001. Significant differences between the acs-1RNAi without and with a specific dietary fatty acid treatment: ###, P < 0.001.
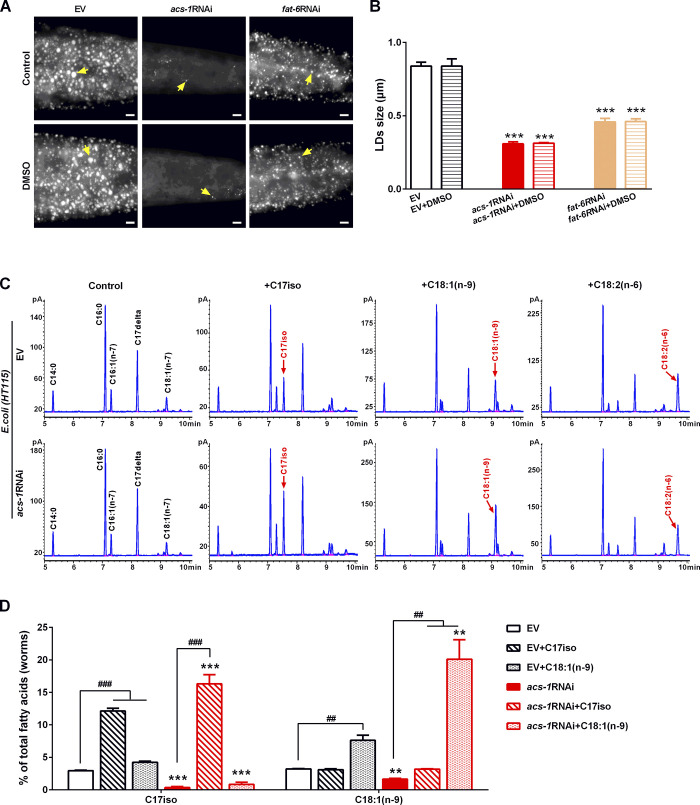
As mentioned above, inactivation of ACS-1 altered the fatty acid profile and reduced LD size, raising the question of whether the reduced LD size was due to the altered fatty acid profile. To examine the role of mmBCFAs or unsaturated fatty acids in LD growth, the acs-1RNAi worms were fed with individual dietary fatty acid C18:1(n-9), C18:2(n-6), C15iso, or C17iso, which were dissolved in DMSO. This treatment had no obvious effect on LD size or lipid storage (Fig. S2, A and B). The GC profiles confirmed that all these dietary fatty acids were successfully taken up by the host Escherichia coli strain HT115 (Fig. S2 C) as well as the C. elegans (Fig. S2 D) containing either the EV or the acs-1RNAi clone. Remarkably, compared with the control, dietary C17iso could fully restore LD size, as indicated by DHS-3::GFP and Nile Red staining of fixed worms in acs-1RNAi (Fig. 2, F and G). In addition, dietary C15iso, like C17iso but to a lesser extent, also partially rescued the LD size of acs-1RNAi worms (Fig. 2, F and G). In contrast, dietary C18:1(n-9) and C18:2(n-6) had no effect at all on the acs-1RNAi worms (Fig. 2, F and G), indicating that ACS-1 may indirectly affect the biosynthesis of C18:1(n-9) and other PUFAs. Taken together, these lines of evidence demonstrate that ACS-1 may specifically function for the biosynthesis of mmBCFA C17iso, which is essential for LD growth.
Figure S2.
DMSO effects on LD size and detection of dietary C18:1(n-9), C18:2(n-6), or C17iso in E. coli and C. elegans. (A) LDs by Nile Red staining of fixed worms treated with or without DMSO (1%). Yellow arrows indicate represented LDs. For all representative animals, anterior is left and posterior is right. Scale bar represents 5 µm. (B) The diameter of LDs by quantification of five representative Nile Red–stained worms from A. Data presented are the mean ± SEM. Significant difference between the control (EV) and a specific RNAi treatment: ***, P < 0.001. (C) GC analysis of the presence (peak) of C18:1(n-9), C18:2(n-6), or C17iso, indicated by red arrows, in E. coli HT115 diet. (D) Percentage of C17iso and C18:1(n-9) in total fatty acids in worms analyzed by GC and further quantified by the peak-area normalization method (% of the peak area of a specific fatty acid in the peak areas of total fatty acids). Significant differences between the EV and acs-1RNAi treatment with or without dietary lipid: **, P < 0.01; ***, P < 0.001. Significant differences between a specific dietary fatty acid treatment and not in same worm strain: ##, P < 0.01; ###, P < 0.001.
mmBCFA C17iso ensures the integrity of the ER for LD growth
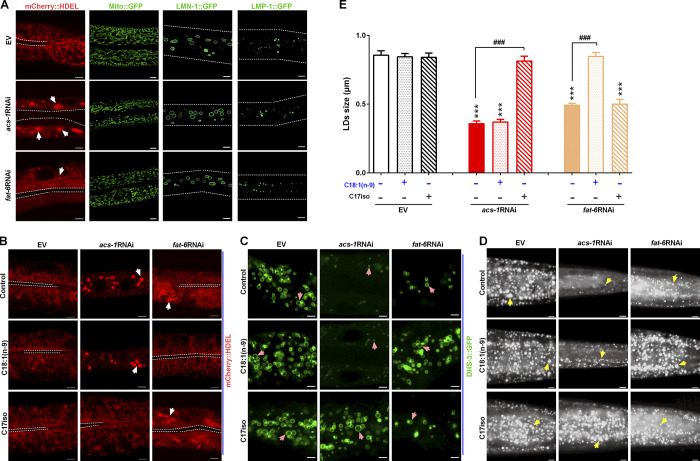
From previous reports, acs-1 is required for maintaining the luminal surface membrane and complex membrane system in C. elegans embryos (Kniazeva et al., 2012; Zhang et al., 2011). mmBCFA C17iso can be used as a substrate for biosynthesis of other complex lipids that constitute the cellular membrane system. LDs originate from the ER and maintain continuous contact with the ER and other intracellular organelles during their growth, raising the question of whether dysfunction of ACS-1 affects the homeostasis of these organelles, consequently preventing LD growth. To examine the morphology of several membrane systems, we treated transgenic strains of SEL(1–79)::mCherry::HDEL (ER-resident protein) {hjSi158[vha-6p::sel-1(1–79)::mcherry::hdel::let-858 3′ utr]}, Mito::GFP (mitochondrial marker) {N2;mito::gfp}, LMP-1::GFP (lysosome-associated membrane protein) {unc-119(ed3);pwIs50[lmp-1p::lmp-1::gfp+unc-119(+)]}, and LMN-1::GFP (nuclear lamina protein) {ccIs4810[(lmn-1p::lmn-1::gfp::lmn-1 3′ utr +dpy-20(+)]} (Table S1) with acs-1RNAi (Fig. 3 A).
Figure 3.
ACS-1 derived C17iso is necessary for maintaining the ER integrity for LD growth. (A) Visualization of the ER morphology by SEL(1–79)::mCherry::HDEL {N2;hjSi158[vha-6p::sel-1(1–79)::mcherry::hdel::let-858 3′ utr]}, imaged by two-photon confocal microscopy, scale bar represents 5 µm. The white dashed lines indicate the lumen of worms, and the white arrows indicate the aggregation of the ER. The morphology of mitochondria, nuclear, and lysosome was visualized by Mito::GFP (N2;mito::gfp), LMN-1::GFP {ccIs4810[lmn-1p::lmn-1::gfp::lmn-1 3′utr+dpy-20(+)]}, and LMP-1::GFP {unc-119(ed3);pwIs50[lmp-1p::lmp-1::gfp+unc-119(+)]}, respectively, imaged by high resolution laser confocal microscopy. Scale bar represents 5 µm for Mito::GFP, and 10 µm both LMN-1::GFP and LMP-1::GFP. The white dotted lines indicate the worm boundaries. (B) Visualization of ER morphology by mCherry::HDEL with dietary C18:1(n-9) or C17iso treatment, imaged by two-photon confocal microscopy, scale bar represents 5 µm. The white dashed lines indicate the lumen of worms, and the white arrows indicate the aggregation of the ER. (C) LDs visualized by DHS-3::GFP with dietary C18:1(n-9) or C17iso treatment, imaged by high resolution laser confocal microscopy, scale bar represents 2.5 µm. Pink arrows indicate represented LDs. (D) LDs by Nile Red staining of fixed worms with dietary C18:1(n-9) or C17iso treatment. Yellow arrows indicate represented LDs. For all representative animals, anterior is left and posterior is right. Scale bar represents 5 µm. (E) The mean diameter of LDs quantified from five representative Nile Red–stained worms for each worm strain. The data are presented as mean ± SEM. Significant differences between the EV and a specific RNAi treatment: ***, P < 0.001. Significant differences between a specific dietary fatty acid treatment and not in same RNAi treatment: ###, P < 0.001.
In cells, the ER is an interconnected network of flattened sacs or tubes encased in membranes. As visualized by SEL(1–79)::mCherry::HDEL, the ER displayed interconnected sheets and tubules in the control (EV) worms. However, the morphology of the ER was severely disrupted with formation of aggregation in the acs-1RNAi worms (Fig. 3, A and B). Similar to the acs-1RNAi worms but to a much lesser extent, the fat-6RNAi worms showed impaired ER integrity as filamentous (Fig. 3, A and B). In addition, the morphology of mitochondria visualized by Mito::GFP displayed somewhat slight division and granularity in both acs-1RNAi and fat-6RNAi worms, while it presented as an interconnected tubule network in the control (EV) worms. In contrast, the morphology of the lysosome and nuclear membrane visualized by LMP-1::GFP and LMN-1::GFP, respectively, did not display an obvious change in the acs-1RNAi worms compared with the control (EV) worms (Fig. 3 A). Thus, these results suggest that inactivation of ACS-1 may more specifically affect the integrity of the ER than other organelles.
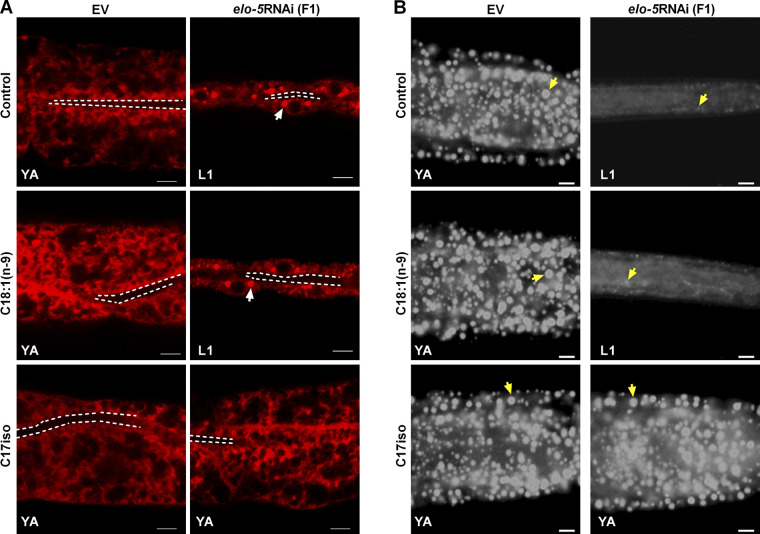
Because the ER plays a critical role in LD biogenesis and growth, and inactivation of ACS-1 severely impairs its integrity, we focused on this organelle. We asked whether the disrupted ER integrity was due to the loss of mmBCFA C17iso or other fatty acids in the acs-1RNAi worms. We found that the abnormal ER integrity was fully rescued by dietary C17iso but not C18:1(n-9) (Fig. 3 B). Simultaneously, dietary C17iso also completely restored the LD size of these worms, as indicated by DHS-3::GFP and Nile red staining of fixation (Fig. 3, C–E). On the other hand, dietary C18:1(n-9) but not C17iso recovered the ER integrity and LD size in fat-6RNAi worms (Fig. 3, B–E). The fatty acid elongase ELO-5 was reported to be required for the synthesis of mmBCFAs C15iso and C17iso in C. elegans (Fig. 2 A), and RNAi of elo-5 resulted in a growth arrest at the first larval stage (L1) in the next (F1) generation (Kniazeva et al., 2004). Consistent with the acs-1RNAi, RNAi knockdown of elo-5 led to worms arrested in L1 (F1 generation) with similarly impaired ER integrity, which was completely rescued to normal by dietary C17iso but not C18:1(n-9) (Fig. S3). Taken together, these lines of evidence demonstrate that mmBCFA C17iso is necessary for maintaining ER integrity for LD growth.
Figure S3.
Dietary supplementation of C18:1(n-9) or C17iso in elo-5RNAi worms. (A) Visualization of ER morphology by SEL(1–79)::mCherry::HDEL with dietary C18:1(n-9) or C17iso treatment, imaged by two-photon confocal microscopy; scale bar represents 5 µm. The white dashed lines indicate the lumen of worms, and the white arrows indicate the aggregation of the ER. (B) LDs by Nile Red staining of fixed worms with dietary C18:1(n-9) or C17iso treatment. Yellow arrows indicate represented LDs. For all representative animals, anterior is left and posterior is right. Scale bar represents 5 µm. F1, F1 generation; YA, young adult stage.
Complex lipids containing C17iso determine ER integrity and LD growth
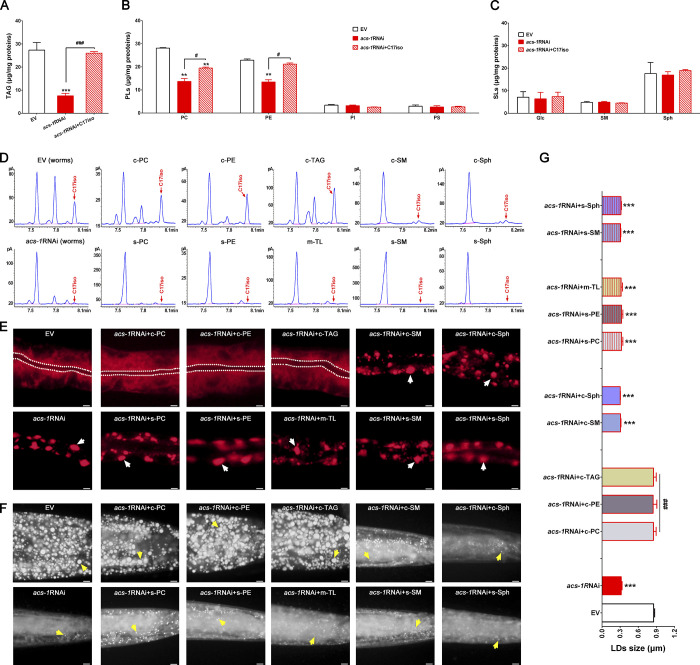
In C. elegans, mmBCFAs can be incorporated into complex lipids including PLs and SLs for membrane scaffolding and TAG for energy storage in LDs (Watts and Ristow, 2017). Therefore, using the same strategy of dietary supplementation, we studied which complex lipid–containing mmBCFAs are required for maintaining ER integrity and LD growth. The lipid profile by TLC-GC analysis revealed that, compared with the control (EV), RNAi knockdown of acs-1 in C. elegans significantly reduced the content (micrograms per milligrams of protein) of TAG, PC, and phosphatidylethanolamine (PE), which could be recovered by dietary C17iso (Fig. 4, A and B). Interestingly, the content of phosphatidylserine (PS) and PI as well as sphingomyelin (SM), sphingosine (Sph), and glucosylceramide (Glc) did not show an obvious change in the control (EV), acs-1RNAi, and acs-1RNAi+C17iso worms (Fig. 4, B and C). C17iso has been reported to be present in total lipids (TL), and in PL PC, PE, PI, and PS to different degrees, with a particularly higher amount in PE (Watts and Ristow, 2017; Kniazeva et al., 2012). Consistently, the fatty acid profiles by GC analysis also revealed a similar phenomenon in the control (EV) worms (Table S4). Moreover, the content of C17iso was significantly reduced in TL, TAG, PC, PE, PI, and PS in the acs-1RNAi worms compared with the control (EV) worms (Table S4). Remarkably, dietary C17iso could largely increase the C17iso content in all these lipids up to two- to threefold in the acs-1RNAi worms over the control (EV) worms (Table S4).
Figure 4.
Complex lipids containing C17iso determine ER integrity and LD growth. (A–C) The contents of TAG, PLs (PC, PE, PI, and PS), and SLs (Glc, SM, and Sph) analyzed by TLC-GC and further quantified by total proteins. The data are presented as mean ± SEM of three to four biological repeats. (D) GC analysis of C17iso in various complex lipids. Red arrows point to the C17iso peaks. (E) Visualization of ER morphology by mCherry::HDEL cultured with different complex lipids and imaged by fluorescent microscopy (Olympus); scale bar represents 5 µm. The white dashed lines indicate the lumen of worms, and the white arrows indicate the aggregation of the ER. (F) LDs by Nile Red staining of fixed worms cultured with different complex lipids. Yellow arrows indicate represented LDs; scale bar represents 5 µm. (G) The mean diameters of LDs quantified from five representative Nile Red–stained worms for each worm strain. For all representative animals, anterior is left and posterior is right. Significant differences between the EV and the acs-1RNAi with or without a specific dietary lipid treatment; **, P < 0.01; ***, P < 0.001. Significant differences between the acs-1RNAi without and with a specific dietary lipid treatment: #, P < 0.05; ###, P < 0.001. c, C. elegans; m, mouse liver; s, sigma.
To further examine whether TAG, PC, and PE were sufficient for ER integrity and LD growth, c-TAG (c meaning derived from C. elegans), c-PC, and c-PE, as well as c-SM and c-Sph, were extracted and separated by TLC from wild-type (N2) worms (Fig. S4, A–C). Each was then individually fed to the acs-1RNAi worms. GC analysis of fatty acid profiles revealed that C17iso was present sparsely in c-SM and c-Sph and generously in c-TAG, c-PC and c-PE (Fig. 4 D), which could be absorbed by the E. coli as diet and then taken by worms (Fig. S4 D). Similar to dietary C17iso alone, supplementation of c-TAG, c-PC, and c-PE, but not c-SM and c-Sph, completely restored ER integrity (Fig. 4 E), as well as LD size and lipid accumulation (Fig. 4, F and G), in the acs-1RNAi worms compared with the control (EV). This can be explained in that, instead of using C16:0 (palmitic acid) as in mammals, C. elegans specifically uses C15iso as the backbone of SLs (Watts and Ristow, 2017), and also that the C. elegans–derived c-SM and c-Sph may not contain enough C17iso for the rescue effect. Intriguingly, dietary supplementation of TL extracted from mouse liver (m-TL), and commercial PC and PE (s-PC and s-PE) obtained from Sigma-Aldrich (Fig. S4 C), which contain C16:0 rather than C17iso as side chains (Fig. 4 D), had no effect at all on the acs-1RNAi worms (Fig. 4, E–G). Altogether, these results indicate that C17iso is probably present as the side chain of complex lipids to maintain ER integrity and LD growth.
Figure S4.
Separation of complex lipids by TLC and detection of C17iso in E. coli HT115 diet and C. elegans. (A–C) Separation of c-SM and c-Glc (A), c-Sph (B), and PL (c-PC, c-PE, c-PS, and c-PI; C) from TL by TLC analysis. (D) GC analysis indicates the presence (peak) of C17iso in E. coli HT115 diet (upper panels) and C. elegans N2 worms (lower panels) under different processing. The red arrows indicated the peak of C17iso. The black arrows indicate the flow direction of solvent. s-PC, s-PE, s-SM, and s-Sph are the standard lipids obtained from Sigma-Aldrich. c-TAG, c-PC, c-PE, c-SM, and c-Sph were isolated and separated from C. elegans wild type (N2).
Deficiency of C17iso impairs ER function in lipid and protein biosynthesis
Among intracellular organelles, the ER acts as a hub of lipid synthesis and is also involved in protein biosynthesis, folding, processing, and so on. Disruption of ER integrity may affect these functions. To get a global view of gene expression, we performed a transcriptome analysis by RNA sequencing of the control (EV) and acs-1RNAi worms. Gene ontology enrichment analysis revealed that RNAi reduction of acs-1, which definitely decreased in expression, led to down-regulation of many genes, particularly those involved in lipid and protein biosynthesis, but also up-regulation of genes involved in the ER unfolded protein response (UPR; Fig. 5, A–C), suggesting that deficiency of ACS-1/C17iso impairs not only ER integrity but also its biological function. Quantitative PCR (QPCR) further confirmed the altered expression of some genes, and more importantly, their expression could be reversed by dietary C17iso (Fig. 5 D), suggesting that C17iso is required for maintaining ER function.
Figure 5.
Deficiency of ACS-1/C17iso impairs ER function for lipid and protein biosynthesis. (A and B) Gene ontology enrichment analysis of down- or up-regulated genes from RNA-sequencing data from the acs-1RNAi worms compared with the control (EV) worms. (C) Transcriptional expression of lipid and protein biosynthesis genes and ER UPR genes using transcriptome profile analysis between the control (EV) worms and the acs-1RNAi worms. (D) Relative mRNA expression of genes in the control (EV) worms and the acs-1RNAi worms with or without C17iso by QPCR analysis. The data are presented as the mean ± SEM of three to four biological repeats. (E and G) Fluorescence intensity of GFP::DGAT-2 {hjSi56[vha-6p::3×flag-tev-gfp::dgat-2::let-858 3′utr]} (E) and ACS-22::GFP {hjSi29[vha-6p::acs-22 cDNA::gfp-tev-3×flag::let-858 3′ utr]} (G) under treatment of dietary C18:1(n-9) or C17iso. Imaged by fluorescent microscopy; scale bar represents 100 µm. (F and H) Quantitation of GFP fluorescence intensity of GFP::DGAT-2 from E and ACS-22::GFP from G. Data presented are the mean ± SEM of 10 representative worms in each treatment. Significant differences between the EV and a specific RNAi treatment with or without dietary lipid: *, P < 0.05; **, P < 0.01; ***, P < 0.001. Significant differences between a specific dietary fatty acid treatment and not in same worm strain: #, P < 0.05; ##, P < 0.01; ###, P < 0.001.
As well, the ER is the organelle most frequently and prominently having contact with LDs (Schuldiner and Bohnert, 2017). The biogenesis, growth, and expansion of LDs are inseparable from the ER, where many lipid synthesis enzymes localize and are involved in these processes. Abnormalities in ER integrity usually affect lipid synthesis and eventually LD growth (Jacquemyn et al., 2017). Thus, we asked whether the impaired ER integrity resulting from ACS-1/C17iso deficiency caused the dysfunction of lipid synthesis enzymes for LD growth. In C. elegans, the FATP1/ACS-22 and DGAT2/diacylglycerol acyltransferase complex acts at the ER–LD interface to facilitate LD expansion/growth (Xu et al., 2012). In light of the evidence that genetic silencing of acs-1 impaired the synthesis of mmBCFA C17iso, which consequently damaged ER integrity and LD growth, we next examined the involvement of ACS-22 and DGAT-2. The RNA sequencing results showed that the transcriptional expression of both acs-22 and dgat-2 were down-regulated in the acs-1RNAi worms compared with the control (EV) worms (Fig. 5 C). Consistently, significantly decreased GFP fluorescent expression of ACS-22::GFP {hjSi29[vha-6p::acs-22 cDNA::gfp-tev-3×flag::let-858 3′ utr]} or GFP::DGAT-2 {hjSi56[vha-6p::3×flag-tev-gfp::dgat-2::let-858 3′ utr]}, was detected in acs-1RNAi or fat-6RNAi worms (Fig. 5, E–H), or both. Similarly, the expression of ACS-22::GFP and GFP::DGAT-2 was fully restored by dietary C17iso in acs-1RNAi worms but not fat-6RNAi worms (Fig. 5, E–H), and likewise by dietary C18:1(n-9) in fat-6RNAi worms but not acs-1RNAi worms (Fig. 5, E–H). Additionally, fat-6RNAi could eliminate the rescue effect of dietary C17iso on the expression of ACS-22::GFP and GFP::DGAT-2 in acs-1RNAi worms (Fig. 5, E–H), suggesting a synergistic role of ACS-1 and SCD in regulating LD growth. Altogether, these lines of evidence consistently reinforce that C17iso, as the product of ACS-1, plays an essential role in maintaining ER integrity for the function of ER-resident lipid synthesis enzymes involved in LD growth.
ACS-1–derived C17iso ensures the function of ER-resident enzyme SCD for LD growth
SCD is an ER-resident protein that converts SFAs (C16:0 and C18:0) to MUFAs (16:1(n-7) and C18:1(n-9)). In C. elegans, it is encoded by three genes, fat-5, fat-6, and fat-7 (Paton and Ntambi, 2009, Brock et al., 2007; Fig. 2 A), and it regulates LD size and PL composition (Shi et al., 2013). The RNA sequencing and QPCR results showed that RNAi reduction of acs-1 reduced the expression of fat-6 and fat-7 (Fig. 5, C and D). Meanwhile, as mentioned above, RNAi reduction of acs-1 decreased the content of C18:1(n-9) but increased the content of C18:0 (Fig. 2 C), consequently impairing SCD conversion activity of C18:1(n-9) to C18:0 (Fig. 2 D). These observations raised the question of whether ACS-1/C17iso might also affect SCD function involved in LD growth.
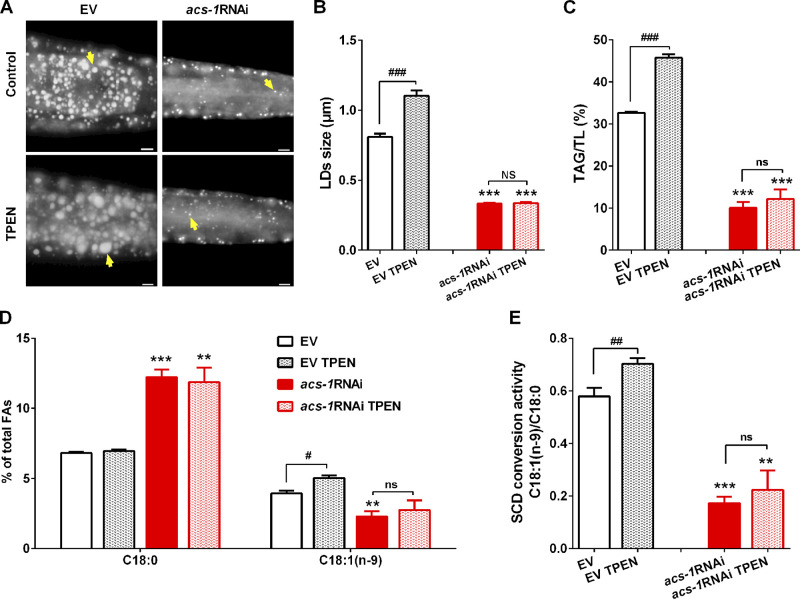
Consistently, the fluorescence intensity of FAT-5::GFP, FAT-6::GFP, and FAT-7::GFP was greatly reduced in acs-1RNAi worms when compared with control (EV) worms (Fig. 6, A and B). Dietary C17iso could fully restore expression levels to normal in acs-1RNAi worms, although it had no obvious effect on control (EV) worms (Fig. 6, A and B). More importantly, dietary C17iso significantly increased the level of C18:1(n-9), but decreased the level of C18:0, thereby up-regulating the SCD conversion activity in acs-1RNAi worms (Fig. 6, C and D). In addition, we previously showed that zinc antagonizes iron to regulate SCD activity (Zhang et al., 2017). Consistently, compared with control (EV) worms, acs-1RNAi worms were completely resistant to zinc deficiency–induced LD expansion, TAG accumulation, altered fatty acids profile, and SCD activity (Fig. S5), suggesting that ACS-1 may act upstream to affect SCD activity. Taken together, these results reinforce that ACS-1–derived C17iso is necessary to maintain SCD expression and activity.
Figure 6.
C17iso ensures the function of ER-resident enzyme SCD for LD growth. (A and B) Fluorescence intensity (A) and quantification (B) of FAT-5::GFP{unc-119(ed3);kunIs161[fat-5p::fat-5::gfp,unc-119(+)]}, FAT-6::GFP {lin-15A(n765);waEx16[fat-6p::fat-6::gfp+lin15(+)]}, and FAT-7::GFP{lin-15(n765); waEx15[fat-7p::fat-7::gfp;lin-15(+)]}. Imaged by fluorescent microscopy (Olympus); scale bar represents 100 µm. (C) Percentage of SFAs C18:0 and MUFAs C18:1(n-9) in total fatty acids, analyzed by GC and further quantified by the peak-area normalization method (% of the peak area of a specific fatty acid in the peak areas of total fatty acids). (D) SCD conversion activity presented as the ratio of C18:1(n-9)/C18:0. (E) Visualization of ER morphology by mCherry::HDEL with the combination of dietary C17iso and C18:1(n-9), imaged by high-resolution laser confocal microscopy; scale bar represents 5 µm. The white dashed lines indicate the lumen of worms, and the white arrows indicate the aggregation of the ER. (F) LDs visualized by DHS-3::GFP and fixed Nile red staining. DHS-3::GFP worms were imaged by high-resolution laser confocal microscopy; scale bar represents 2 µm. For Nile Red–stained animals, anterior is left and posterior is right; scale bar represents 5 µm. The pink (DHS-3::GFP positive LDs) and yellow (Nile Red–stained particles) arrows indicate represented LDs. (G) The mean diameter of LDs quantified from five representative Nile Red–stained worms for each worm strain from F. Data presented are the mean ± SEM. Significant differences between the EV and a specific RNAi treatment with or without dietary lipid: **, P < 0.01; ***, P < 0.001. Significant differences between a specific dietary fatty acid treatment and not in same worm strain: ##, P < 0.01; ###, P < 0.001.
Figure S5.
Inactivation of ACS-1 resistant to TPEN induced SCD activity and LD growth. (A) LDs by Nile Red staining of fixed worms with or without TPEN treatment. Yellow arrows indicate represented LDs. For representative animals, anterior is left and posterior is right. Scale bar represents 5 µm. (B) The mean diameter of LDs by quantification of five representative Nile Red–stained worms for each worm from A. (C) Percentage of TAG in TL (TAG+ PL) by TLC-GC analysis. (D) Percentage of C18:0 and MUFA C18:1(n-9) in total fatty acids analyzed by GC and further quantified by the peak-area normalization method (% of the peak area of a specific fatty acid in the peak areas of total fatty acids). (E) SCD conversion activity presented as the ratio of C18:1(n-9)/C18:0. The data are presented as mean ± SEM of four biological repeats. Significant difference between the EV and acs-1RNAi treatment with or without TPEN: **, P < 0.01; ***, P < 0.001. Significant difference between TPEN treatment and not in same worm strain: #, P < 0.05; ##, P < 0.01; ###, P < 0.001.
Although fat-6RNAi worms also displayed abnormal ER integrity (Fig. 3, A and C), as well as reduced LD size and abundance and lipid accumulation (Fig. 1, C–G; and Fig. 3, D–F) similarly to acs-1RNAi, simultaneous RNAi knockdown of both acs-1 and fat-6 gave rise to effects similar to acs-1RNAi alone in terms of ER integrity and LD size (Fig. 6, E–G), which could be partially recovered by C17iso but not by C18:1(n-9) (Fig. 6, E–G). Meanwhile, dietary C17iso could fully restore LD size and ER integrity in acs-1RNAi worms (Fig. 3), whereas additional fat-6RNAi reduced this effect (Fig. 6, E–G). Furthermore, the combination of dietary C18:1(n-9) with C17iso successfully rescued ER integrity as well as LD size in fat-6;acs-1RNAi double-silenced worms (Fig. 6, E–G). Altogether, these lines of evidence suggest that ACS-1–derived C17iso ensures the function of ER-resident enzyme SCD for LD growth.
ACS-1–derived C17iso may play a central role in LD growth and lipid accumulation
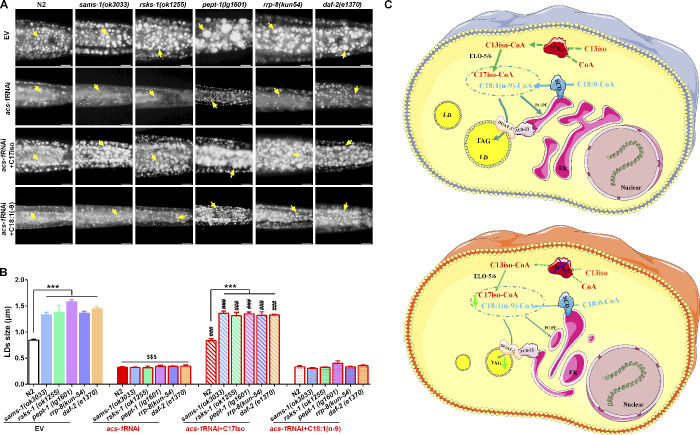
In C. elegans, several signaling pathways play critical roles in regulating LD growth and lipid accumulation. Inactivation of several well-known genes, such as sams-1 (the PC synthesis pathway; Walker et al., 2011), rsks-1 (the mTOR pathway; Shi et al., 2013; Wu et al., 2018), pept-1 (the absorption pathway; Benner et al., 2011; Spanier et al., 2009), rrp-8 (nucleolar stress; Wu et al., 2018), and daf-2 (the insulin-like growth factor pathway; Svensson et al., 2011; Kimura et al., 1997), led to increased LD size and lipid accumulation (Fig. 7 A). Compared with the control (EV), acs-1RNAi completely prevented LD growth and lipid accumulation in all tested gene mutants, which were similar to the acs-1RNAi worms alone (Fig. 7, A and B). Consistently, dietary C17iso was sufficient to fully restore their lipid phenotypes (Fig. 7, A and B), while dietary C18:1(n-9) had no effect. These results further strengthen the dominant role of ACS-1 and its product C17iso in the regulation of LD growth in C. elegans.
Figure 7.
C17iso is necessary for LD growth. (A) LDs by Nile Red staining of fixed worms. Yellow arrows indicate represented LDs. For all representative animals, anterior is left and posterior is right. Scale bar represents 10 µm. (B) The mean diameter of LDs quantified from five representative Nile Red–stained worms for each worm strain from A. The data are presented as mean ± SEM. Significant differences between the wild-type N2 and a specific mutant strain under same condition: ***, P < 0.001. Significant differences between the EV and acs-1RNAi treatment in a specific worm strain: $$$, P < 0.001. Significant differences between dietary C17iso treatment and not in a specific worm strain with acs-1RNAi treatment: ###, P < 0.001. (C) A working model of ACS-1–derived C17iso in regulating LD growth. ACS-1–derived mmBCFA C17iso is incorporated into complex PLs for maintaining appropriate ER integrity for the function of ER-resident proteins such as SCD, ACS-22, and DGAT-2 for lipid synthesis and LD growth. Inactivation of ACS-1 impairs C17iso synthesis, which consequently disrupts the ER integrity and thereby leads to dysfunction of SCD, ACS-22, and DGAT-2, eventually preventing lipid synthesis and LD growth.
Discussion
The intracellular organelles LDs not only host the key machinery of lipid manufacturing and storage in their membrane-bound compartment but also play a role in other life processes such as organism growth, development, and reproduction. Through a candidate screen using the model organism C. elegans, we successfully identified 43 genes required for LD growth. Through RNAi silencing and rescue experiments with dietary administration, ACS-1 and its downstream product mmBCFA C17iso were identified as the determining factors controlling ER integrity for LD growth. Inactivation of ACS-1 and another enzyme involved in mmBCFA synthesis, ELO-5, consistently resulted in impaired integrity of the ER and LD growth. Although RNAi knockdown of acs-1 resulted in impaired synthesis of mmBCFA C17iso and C18:1(n-9), only dietary C17iso but not C18:1(n-9) could recover the ER morphology and LD growth, suggesting that ACS-1 may be specifically required for the synthesis of C17iso to maintain ER integrity and LD growth. To the best of our knowledge, this is the first evidence to show that a unique fatty acid, C17iso, distinct from other well-known lipids and proteins, plays an essential role in ER homeostasis and LD growth in an intact organism, C. elegans.
mmBCFAs are widely present in many organisms from bacteria to mammals. The mmBCFAs serve as precursors for the synthesis of complex lipids including SLs, PLs, and TAG (Watts and Ristow, 2017). Specifically, C. elegans uses C15iso, rather than palmitic acid (C16:0) as in mammals, to condense with serine to form a 17-carbon branched-chain sphingoid base for SL synthesis. The importance of the mmBCFAs C15iso and C17iso, and also their derived SLs, manifests in growth and development (Kniazeva et al., 2004; Zhu et al., 2013) and also in the proper polarity of intestinal cells and lumen formation (Zhang et al., 2011). Phospholipids are major membrane components. TLC-GC analysis revealed that the contents of three complex lipids, TAG, PC, and PE, but not others including PI, PS, Glc, SM, and Sph, were significantly reduced in acs-1RNAi worms, and that the effect could be recovered by dietary C17iso incorporated into these lipids. Furthermore, c-TAG, c-PC, and c-PE, but not c-SM and c-Sph or other complex lipids lacking C17iso, had the ability to rescue the impaired ER integrity and LD growth of the acs-1RNAi worms. The dietary lipids c-TAG, c-PC, and c-PE contain C17iso, which can be released by lipases and used by other enzymes yet to be characterized, for further synthesis of PL for membrane structure and TAG for storage in LDs in C. elegans. Thus, this work demonstrates that it is actually the C17iso per se as the side chain of PL that determines ER integrity and LD growth in C. elegans. On the other hand, it may also be possible that C17iso per se or a specific complex lipid containing C17iso functions as a signaling molecule involved in ER integrity and LD growth.
The ER has a number of functions such as lipid biosynthesis, LD biogenesis and growth, and protein biosynthesis, folding, and processing. The transcriptome profile revealed that both lipid and protein biosynthesis pathways were down-regulated, but the ER UPR was up-regulated, which could be reversed by dietary C17iso. Many lipid metabolic enzymes, such as SCD, ACS-22, and DGAT-2, are present in the ER and function in lipid synthesis and LD formation and growth. Deficiency of C17iso by ACS-1 inactivation impairs ER integrity, which consequently leads to the dysfunction of SCD, ACS-22, and DGAT-2. In acs-1RNAi worms, dietary C17iso restored not only ER morphology but also the expression of FAT-5::GFP, FAT-6::GFP, FAT-7::GFP, ACS-22::GFP, and GFP::DGAT-2, in addition to SCD conversion activity. These observations consistently support that C17iso is necessary for ER integrity and functions.
The decreased content of C18:1(n-9) in acs-1RNAi worms may be due to a dysfunction of SCD as the result of impaired ER integrity and function. We previously reported that SCD regulates LD size in C. elegans (Shi et al., 2013). Our current study shows that inactivation of fat-6 also led to impaired ER integrity in addition to reduced LD size, although not to the same extent as acs-1RNAi. Although dietary C17iso can successfully restore the ER morphology and LD growth of acs-1RNAi worms, it fails to do so when FAT-6 is inactivated, suggesting that FAT-6 may act downstream of ACS-1 to regulate LD growth. Consistently, dietary combination of C17iso with C18:1(n-9) greatly recovered the LD size of acs-1;fat-6RNAi double-silenced worms. Therefore, both ACS-1 and SCD play essential roles, not only for maintaining ER integrity and function, but also for determining LD growth.
Recently, it was reported that mmBCFAs are synthesized de novo via mitochondrial branched-chain amino acid (BCAA) catabolism in adipose tissues (Wallace et al., 2018). mmBCFAs levels were significantly decreased in white adipose tissues of obese animals (Wallace et al., 2018) and humans (Su et al., 2015), and also in the plasma of morbidly obese humans (Mika et al., 2016). In contrast, increased levels of BCAAs are linked to obesity and diabetes (Wang et al., 2018). Since mmBCFAs are derived from BCAAs (Wallace et al., 2018), the reduced level of mmBCFAs may be a result of an induced level of BCAAs. As in mammals, the C. elegans mmBCFAs were thought to be derived from the BCAA leucine (Kniazeva et al., 2004), and an inverse correlation between C17iso level in TAG and fat storage level was previously reported in wild-type worms raised on different E. coli diets (Brooks et al., 2009). Additionally, our present work found that inhibition of mmBCFA C17iso biosynthesis prevents LD growth due to impaired ER integrity. Thus, all these works, including our current study, have converged to highlight the crucial role of mmBCFAs in LD growth and obesity.
In summary, we propose a working model as follows: ACS-1–derived mmBCFA C17iso is incorporated into complex PLs to maintain appropriate ER integrity for the function of ER-resident proteins such as SCD, ACS-22, and DGAT-2 in lipid synthesis and LD growth. Inactivation of ACS-1 impairs C17iso synthesis, which consequently disrupts ER integrity and thereby leads to dysfunction of SCD, ACS-22, and DGAT-2, eventually preventing lipid synthesis and LD growth (Fig. 7 C).
Materials and methods
C. elegans strains, RNAi, and culture conditions
C. elegans strains were maintained on nematode growth media plates with E. coli OP50 or HT115 under standard culture conditions, unless otherwise specified. RNAi was performed using the feeding method (Wu et al., 2018), using bacterial strains from the Ahringer C. elegans RNAi library. The wild-type strain was Bristol N2. The organisms and strains used in this study are shown in Table S1.
Construction of transgenic strain and primers
The acs-1::gfp {unc-119(ed3);kunEx202[vha-6p::acs-1::gfp+unc-119(+),myo-2p::mcherry]} transgenic strain was constructed using methods previously described (Frøkjaer-Jensen et al., 2008); the transgenes were generated by microinjection. Briefly, the injection mixture was prepared with 5 ng/ml pCFJ151 inserted with the target DNA fragment and 2.5 ng/ml pCFJ90 (myo-2p::mcherry). DNA mixtures were injected into the gonads of EG4322 (ttTi5605;unc-119(ed3) young adult worms. The primer sequences are listed in Table S2.
Nile Red staining and LipidTOX Red staining of fixed worms and quantification of LD size
Nile Red and LipidTOX staining of fixed worms was performed as previously described (Liang et al., 2010; Brooks et al., 2009). The young adult worms were collected and suspended in 1 ml of water on ice, and then 50 µl of freshly prepared 10% PFA solution was added and mixed. Worms were immediately frozen briefly in liquid nitrogen, subjected to two or three incomplete freeze/thaw cycles, and washed with M9 buffer several times to remove the PFA solution. Nile Red (10 µg/ml) or LipidTOX (1:1,000) was added to the worms and incubated for 30 min at room temperature. Next, the worms were placed onto 2% agarose pads for microscopic observation and photography. Images were obtained with an Olympus BX53 fluorescence microscope (Shi et al., 2013).
For the quantification of LD size, regions of similar location and size were selected for each worm. LD diameter was measured using CellSens Standard software (Olympus). Three or four biological repeats were performed. In each biological repeat, >20 individual worms were selected and observed, and 60–90 worms in total were observed for phenotype determination. Afterward, 5 representative worms were selected for the quantification of LDs in each worm strain or treatment, in which ∼100 LDs were randomly measured from each representative worm. The average diameter of each worm strain or treatment was calculated as mean ± SEM. For the abundance of LDs, a fixed region of similar location and size (60 × 60 µm) was selected for each worm, and the LD number was measured using ImageJ. More than 20 individual worms were scored for each worm strain.
Analysis of fatty acid composition
Approximately 2,000 young adults (without or with 2–3 eggs) were harvested for fatty acid extraction and analysis. To extract fatty acids and form methyl esters, 1 ml of MeOH + 2.5% H2SO4 was added to the harvested worms. They were then heated for 60 min at 70°C. Determination of the fatty acids was done with an Agilent 7890 series gas chromatograph equipped with a 15 m × 0.25 mm × 0.25 µm DB-WAX column (Agilent), with helium as the carrier gas at 1.4 ml/min and a flame ionization detector. Three to four biological replicates were performed for GC analysis.
Analysis of TAG
Young adults were harvested for TL extraction. TAG and PLs were separated by TLC in hexane/ether/acetic acid (80:20:2, vol/vol/vol), and the fatty acids were determined by GC using an Agilent 7890A. Methylated C15:0 was used as a standard for quantitative analysis.
Separation and analysis of SLs and PLs
Standard substance SLs and PLs including PC (Sigma-Aldrich; P3556), PE (Sigma-Aldrich; P7693), SM (Sigma-Aldrich; S0756), Sph (Avanti; 860490P), and TL of mouse liver were dissolved in DMSO and mixed with cultured E. coli bacteria to 1 mM final concentration before spotting plates.
The separation of SLs was performed as follows: ∼200,000 synchronized young adult N2 worms were harvested for TL extraction. TL (content 150 µg fatty acids) were loaded in TLC silica plates (Merck) and developed to the top of the plate in chloroform/methanol/water/acetic acid (65:25:4:3, vol/vol/vol/vol) for separated Glc and SM, chloroform/methanol/water (65:25:4, vol/vol/vol; Hayashi et al., 2018) for separated Sph, and chloroform:ethanol:water:triethylamine (30:35:7:35, vol/vol/vol/vol; Lee et al., 2008) for separated PC, PE, PI, and PS. Individual SM, Sph, PC, and PE bands were scraped from the TLC plates and dissolved in chloroform/methanol (1:1, vol/vol), and ultrasound was performed for 30 min. The samples were centrifuged, and the liquid was transferred to a new glass tube, immediately blown dry with nitrogen, and redissolved with chloroform. All recycled SLs and PLs were dissolved in 100% DMSO and mixed with 1 ml cultured bacteria before spotting plates.
Dietary supplementation of fatty acids
C18:1(n-9) and C18:2(n-6) (ANPEL), were supplemented into nematode growth media plates at a final concentration 0.2 mM. C15iso and C17iso (Larodan) were dissolved in 100% DMSO and mixed with cultured bacteria to 1 mM working concentration. The mixtures were spotted onto plates for culturing worms. Experimental and control plates for a given assay were prepared with a single source of fatty acids or DMSO-supplemented bacterial food (Kniazeva et al., 2008). To ensure that fatty acids were absorbed by C. elegans from E. coli, the fatty acid compositions of E. coli and worms were analyzed by GC as previously described (Zhang et al., 2016).
mRNA isolation, QPCR, and mRNA sequencing
Synchronized L4 worms were harvested for total RNA extraction as previously described (Wu et al., 2018, Zhang et al., 2017). The cDNA was synthesized using the PrimeScript RT reagent kit (Takara). For QPCR, mRNA levels of each sample were quantified in three biological triplicates on a real-time PCR instrument 7900HT (ABI). The relative expression of mRNA was determined using the ΔΔCt method, and tbb-2 was used as a reference gene. The sequences of all the primers used in this study are listed in Table S2. For mRNA sequencing, total RNA was first isolated, qualified, and quantified, and sequencing libraries were generated and sequenced on an Illumina HiSeq 4000 platform by the Novogene Company.
Visualization of GFP and mCherry
The GFP- or mCherry-positive worms (late L4s and young adults) were washed and plated on agarose gel pad with 10 mM sodium azide for anesthesia and visualized under a high-resolution laser confocal microscope with Airyscan (Zeiss; Axio-Imager_LSM-800), a two-photon confocal microscope (Nikon; A1MP+), and a fluorescent microscope (Olympus; BX53). Images were captured and postprocessed by each device, except for GFP fluorescence intensity, which was quantified by Photoshop CS5.
Data analysis
Data are presented as the mean ± SEM except when specifically indicated. Statistical analysis was performed using unpaired two-tailed t test between two groups or one-way ANOVA with more than two conditions. All figures were made using Prism 6 (GraphPad Software) and Photoshop CS5.
Online supplemental material
Table S1 shows the organisms/strains used in this study. Table S2 shows the primers for construction of transgenic strains and QPCR. Table S3 lists the genes regulating LD size by RNAi screen. Table S4 shows the fatty acid profile in the isolated lipid classes in C. elegans. Fig. S1 shows the RNAi efficiency and the effect of ACS family on LD size. Fig. S2 shows the DMSO effects on LD size and detection of dietary C18:1(n-9), C18:2(n-6), or C17iso in E. coli and C. elegans. Fig. S3 shows the dietary supplementation of C18:1(n-9) or C17iso in elo-5RNAi worms. Fig. S4 shows the separation of complex lipids by TLC and detection of C17iso in E. coli HT115 diet and C. elegans. Fig. S5 shows that acs-1RNAi resists N,N,N′,N′-tetrakis (2-pyridylmethyl) ethyl (TPEN)-induced SCD activity and LD growth.
Supplementary Material
lists the organisms and strains in the experimental models.
lists primers.
lists genes regulating LD size by RNAi screen.
shows profiles of fatty acids in C. elegans isolated lipid classes.
Acknowledgments
We thank Metabo-Profile Biotechnology Co., Ltd. (Shanghai, China) for lipid analysis, Dr. H.H. Zhu (ShanghaiTech University) for generously providing commercial C17iso, and Dr. Junjun Hao and Dr. Leonard Krall for the help with RNA sequence analysis and English copyediting, respectively.
This work was supported by the Ministry of Science and Technology of the People’s Republic of China (2018YFA0800700 and 2018YFA0800703), the National Natural Science Foundation of China (U1702288, 91857113, 31801001, 31671230, 81700520, 31860323, 32071281, 32000818, 32160155, and U1702287), the Yunnan Applied Basic Research Projects (2018FB117, 2019FB046, 2019FB048, and 2019FY003021), and West-light Foundation of the Chinese Academy of Sciences to J.J. Zhang.
The authors declare no competing financial interests.
Author contributions: B. Liang, J. Zhang, and Y. Hu conceived and designed the experiments and wrote the paper. J. Zhang, Y. Hu, and Y. Wang performed most of the experiments and data analysis. L. Zhang and Chengbin Li provided the mouse materials and some technical methods. X. Xu did partial RNAi screening. X. Zou, L. Fu, J. Xu, R. Yang, X. Jiang, Chunxia Li, Y. Wu, and P. Liu provided guidance for some experiments, contributed reagents/materials/analysis tools, and revised the manuscript. B. Liang initiated the project. All authors reviewed the manuscript.
Data availability
The data that support the findings of this study are available from the corresponding authors upon reasonable request.
References
- Ben M’barek, K., Ajjaji D., Chorlay A., Vanni S., Forêt L., and Thiam A.R.. 2017. ER membrane phospholipids and surface tension control cellular lipid droplet formation. Dev. Cell. 41:591–604.e7. 10.1016/j.devcel.2017.05.012 [DOI] [PubMed] [Google Scholar]
- Benner, J., Daniel H., and Spanier B.. 2011. A glutathione peroxidase, intracellular peptidases and the TOR complexes regulate peptide transporter PEPT-1 in C. elegans. PLoS One. 6:e25624. 10.1371/journal.pone.0025624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock, T.J., Browse J., and Watts J.L.. 2006. Genetic regulation of unsaturated fatty acid composition in C. elegans. PLoS Genet. 2:e108. 10.1371/journal.pgen.0020108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock, T.J., Browse J., and Watts J.L.. 2007. Fatty acid desaturation and the regulation of adiposity in Caenorhabditis elegans. Genetics. 176:865–875. 10.1534/genetics.107.071860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks, K.K., Liang B., and Watts J.L.. 2009. The influence of bacterial diet on fat storage in C. elegans. PLoS One. 4:e7545. 10.1371/journal.pone.0007545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary, V., Golden A., and Prinz W.A.. 2016. Keeping FIT, storing fat: Lipid droplet biogenesis. Worm. 5:e1170276. 10.1080/21624054.2016.1170276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjaer-Jensen, C., Davis M.W., Hopkins C.E., Newman B.J., Thummel J.M., Olesen S.P., Grunnet M., and Jorgensen E.M.. 2008. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40:1375–1383. 10.1038/ng.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, S., Yang L., Li P., Hofmann O., Dicker L., Hide W., Lin X., Watkins S.M., Ivanov A.R., and Hotamisligil G.S.. 2011. Aberrant lipid metabolism disrupts calcium homeostasis causing liver endoplasmic reticulum stress in obesity. Nature. 473:528–531. 10.1038/nature09968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, Y., Nemoto-Sasaki Y., Matsumoto N., Hama K., Tanikawa T., Oka S., Saeki T., Kumasaka T., Koizumi T., Arai S., et al. 2018. Complex formation of sphingomyelin synthase 1 with glucosylceramide synthase increases sphingomyelin and decreases glucosylceramide levels. J. Biol. Chem. 293:17505–17522. 10.1074/jbc.RA118.002048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, C.L. 2019. Lipid droplet biogenesis. Curr. Opin. Cell Biol. 59:88–96. 10.1016/j.ceb.2019.03.018 [DOI] [PubMed] [Google Scholar]
- Jacquemyn, J., Cascalho A., and Goodchild R.E.. 2017. The ins and outs of endoplasmic reticulum-controlled lipid biosynthesis. EMBO Rep. 18:1905–1921. 10.15252/embr.201643426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier, N., Choudhary V., Mari M., Toulmay A., Reggiori F., and Schneiter R.. 2011. Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Sci. 124:2424–2437. 10.1242/jcs.076836 [DOI] [PubMed] [Google Scholar]
- Kamath, R.S., Fraser A.G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., et al. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 421:231–237. 10.1038/nature01278 [DOI] [PubMed] [Google Scholar]
- Kassan, A., Herms A., Fernández-Vidal A., Bosch M., Schieber N.L., Reddy B.J.N., Fajardo A., Gelabert-Baldrich M., Tebar F., Enrich C., et al. 2013. Acyl-CoA synthetase 3 promotes lipid droplet biogenesis in ER microdomains. J. Cell Biol. 203:985–1001. 10.1083/jcb.201305142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, K.D., Tissenbaum H.A., Liu Y., and Ruvkun G.. 1997. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 277:942–946. 10.1126/science.277.5328.942 [DOI] [PubMed] [Google Scholar]
- Kniazeva, M., Crawford Q.T., Seiber M., Wang C.Y., and Han M.. 2004. Monomethyl branched-chain fatty acids play an essential role in Caenorhabditis elegans development. PLoS Biol. 2:e257. 10.1371/journal.pbio.0020257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniazeva, M., Euler T., and Han M.. 2008. A branched-chain fatty acid is involved in post-embryonic growth control in parallel to the insulin receptor pathway and its biosynthesis is feedback-regulated in C. elegans. Genes Dev. 22:2102–2110. 10.1101/gad.1692008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniazeva, M., Shen H., Euler T., Wang C., and Han M.. 2012. Regulation of maternal phospholipid composition and IP(3)-dependent embryonic membrane dynamics by a specific fatty acid metabolic event in C. elegans. Genes Dev. 26:554–566. 10.1101/gad.187054.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagace, T.A., and Ridgway N.D.. 2013. The role of phospholipids in the biological activity and structure of the endoplasmic reticulum. Biochim. Biophys. Acta. 1833:2499–2510. 10.1016/j.bbamcr.2013.05.018 [DOI] [PubMed] [Google Scholar]
- Lee, H.C., Inoue T., Imae R., Kono N., Shirae S., Matsuda S., Gengyo-Ando K., Mitani S., and Arai H.. 2008. Caenorhabditis elegans mboa-7, a member of the MBOAT family, is required for selective incorporation of polyunsaturated fatty acids into phosphatidylinositol. Mol. Biol. Cell. 19:1174–1184. 10.1091/mbc.e07-09-0893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, B., Ferguson K., Kadyk L., and Watts J.L.. 2010. The role of nuclear receptor NHR-64 in fat storage regulation in Caenorhabditis elegans. PLoS One. 5:e9869. 10.1371/journal.pone.0009869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listenberger, L.L., Han X., Lewis S.E., Cases S., Farese R.V. Jr., Ory D.S., and Schaffer J.E.. 2003. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc. Natl. Acad. Sci. USA. 100:3077–3082. 10.1073/pnas.0630588100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda, I., Kohara Y., Yamamoto M., and Sugimoto A.. 2001. Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr. Biol. 11:171–176. 10.1016/S0960-9822(01)00052-5 [DOI] [PubMed] [Google Scholar]
- Mika, A., Stepnowski P., Kaska L., Proczko M., Wisniewski P., Sledzinski M., and Sledzinski T.. 2016. A comprehensive study of serum odd- and branched-chain fatty acids in patients with excess weight. Obesity (Silver Spring). 24:1669–1676. 10.1002/oby.21560 [DOI] [PubMed] [Google Scholar]
- Mori, M., Itabe H., Higashi Y., Fujimoto Y., Shiomi M., Yoshizumi M., Ouchi Y., and Takano T.. 2001. Foam cell formation containing lipid droplets enriched with free cholesterol by hyperlipidemic serum. J. Lipid Res. 42:1771–1781. 10.1016/S0022-2275(20)31503-0 [DOI] [PubMed] [Google Scholar]
- Murphy, D.J. 2012. The dynamic roles of intracellular lipid droplets: from archaea to mammals. Protoplasma. 249:541–585. 10.1007/s00709-011-0329-7 [DOI] [PubMed] [Google Scholar]
- Na, H., Zhang P., Chen Y., Zhu X., Liu Y., Liu Y., Xie K., Xu N., Yang F., Yu Y., et al. 2015. Identification of lipid droplet structure-like/resident proteins in Caenorhabditis elegans. Biochim. Biophys. Acta. 1853(10, 10 Pt A):2481–2491. 10.1016/j.bbamcr.2015.05.020 [DOI] [PubMed] [Google Scholar]
- Paton, C.M., and Ntambi J.M.. 2009. Biochemical and physiological function of stearoyl-CoA desaturase. Am. J. Physiol. Endocrinol. Metab. 297:E28–E37. 10.1152/ajpendo.90897.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppelreuther, M., Sander S., Minden F., Dietz M.S., Exner T., Du C., Zhang I., Ehehalt F., Knüppel L., Domschke S., et al. 2018. The metabolic capacity of lipid droplet localized acyl-CoA synthetase 3 is not sufficient to support local triglyceride synthesis independent of the endoplasmic reticulum in A431 cells. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 1863:614–624. 10.1016/j.bbalip.2018.03.003 [DOI] [PubMed] [Google Scholar]
- Salo, V.T., and Ikonen E.. 2019. Moving out but keeping in touch: contacts between endoplasmic reticulum and lipid droplets. Curr. Opin. Cell Biol. 57:64–70. 10.1016/j.ceb.2018.11.002 [DOI] [PubMed] [Google Scholar]
- Salo, V.T., Belevich I., Li S., Karhinen L., Vihinen H., Vigouroux C., Magré J., Thiele C., Hölttä-Vuori M., Jokitalo E., and Ikonen E.. 2016. Seipin regulates ER-lipid droplet contacts and cargo delivery. EMBO J. 35:2699–2716. 10.15252/embj.201695170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salo, V.T., Li S., Vihinen H., Hölttä-Vuori M., Szkalisity A., Horvath P., Belevich I., Peränen J., Thiele C., Somerharju P., et al. 2019. Seipin facilitates triglyceride flow to lipid droplet and counteracts droplet ripening via endoplasmic reticulum contact. Dev. Cell. 50:478–493.e9. 10.1016/j.devcel.2019.05.016 [DOI] [PubMed] [Google Scholar]
- Schuldiner, M., and Bohnert M.. 2017. A different kind of love - lipid droplet contact sites. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 1862(10, 10 Pt B):1188–1196. 10.1016/j.bbalip.2017.06.005 [DOI] [PubMed] [Google Scholar]
- Shi, X., Li J., Zou X., Greggain J., Rødkær S.V., Færgeman N.J., Liang B., and Watts J.L.. 2013. Regulation of lipid droplet size and phospholipid composition by stearoyl-CoA desaturase. J. Lipid Res. 54:2504–2514. 10.1194/jlr.M039669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer, F., Moorman C., van der Linden A.M., Kuijk E., van den Berghe P.V., Kamath R.S., Fraser A.G., Ahringer J., and Plasterk R.H.. 2003. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 1:e12. 10.1371/journal.pbio.0000012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanier, B., Lasch K., Marsch S., Benner J., Liao W., Hu H., Kienberger H., Eisenreich W., and Daniel H.. 2009. How the intestinal peptide transporter PEPT-1 contributes to an obesity phenotype in Caenorhabditits elegans. PLoS One. 4:e6279. 10.1371/journal.pone.0006279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, X., Magkos F., Zhou D., Eagon J.C., Fabbrini E., Okunade A.L., and Klein S.. 2015. Adipose tissue monomethyl branched-chain fatty acids and insulin sensitivity: Effects of obesity and weight loss. Obesity (Silver Spring). 23:329–334. 10.1002/oby.20923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson, E., Olsen L., Mörck C., Brackmann C., Enejder A., Faergeman N.J., and Pilon M.. 2011. The adiponectin receptor homologs in C. elegans promote energy utilization and homeostasis. PLoS One. 6:e21343. 10.1371/journal.pone.0021343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, A.K., Jacobs R.L., Watts J.L., Rottiers V., Jiang K., Finnegan D.M., Shioda T., Hansen M., Yang F., Niebergall L.J., et al. 2011. A conserved SREBP-1/phosphatidylcholine feedback circuit regulates lipogenesis in metazoans. Cell. 147:840–852. 10.1016/j.cell.2011.09.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace, M., Green C.R., Roberts L.S., Lee Y.M., McCarville J.L., Sanchez-Gurmaches J., Meurs N., Gengatharan J.M., Hover J.D., Phillips S.A., et al. 2018. Enzyme promiscuity drives branched-chain fatty acid synthesis in adipose tissues. Nat. Chem. Biol. 14:1021–1031. 10.1038/s41589-018-0132-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther, T.C., and Farese R.V. Jr. 2012. Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 81:687–714. 10.1146/annurev-biochem-061009-102430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther, T.C., Chung J., and Farese R.V. Jr. 2017. Lipid droplet biogenesis. Annu. Rev. Cell Dev. Biol. 33:491–510. 10.1146/annurev-cellbio-100616-060608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Xu S., Gao J., Zhang L., Zhang Z., Yang W., Li Y., Liao S., Zhou H., Liu P., and Liang B.. 2018. SILAC-based quantitative proteomic analysis of the livers of spontaneous obese and diabetic rhesus monkeys. Am. J. Physiol. Endocrinol. Metab. 315:E294–E306. 10.1152/ajpendo.00016.2018 [DOI] [PubMed] [Google Scholar]
- Watts, J.L., and Ristow M.. 2017. Lipid and carbohydrate metabolism in Caenorhabditis elegans. Genetics. 207:413–446. 10.1534/genetics.117.300106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster, C.M., Pino E.C., Carr C.E., Wu L., Zhou B., Cedillo L., Kacergis M.C., Curran S.P., and Soukas A.A.. 2017. Genome-wide RNAi screen for fat regulatory genes in C. elegans identifies a proteostasis-AMPK axis critical for starvation survival. Cell Rep. 20:627–640. 10.1016/j.celrep.2017.06.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte, M.A., and Gould A.P.. 2017. Lipid droplet functions beyond energy storage. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 1862(10, 10 Pt B):1260–1272. 10.1016/j.bbalip.2017.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfling, F., Wang H., Haas J.T., Krahmer N., Gould T.J., Uchida A., Cheng J.X., Graham M., Christiano R., Fröhlich F., et al. 2013. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev. Cell. 24:384–399. 10.1016/j.devcel.2013.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J., Jiang X., Li Y., Zhu T., Zhang J., Zhang Z., Zhang L., Zhang Y., Wang Y., Zou X., and Liang B.. 2018. PHA-4/FoxA senses nucleolar stress to regulate lipid accumulation in Caenorhabditis elegans. Nat. Commun. 9:1195. 10.1038/s41467-018-03531-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, N., Zhang S.O., Cole R.A., McKinney S.A., Guo F., Haas J.T., Bobba S., Farese R.V. Jr., and Mak H.Y.. 2012. The FATP1-DGAT2 complex facilitates lipid droplet expansion at the ER-lipid droplet interface. J. Cell Biol. 198:895–911. 10.1083/jcb.201201139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C., and Liu P.. 2017. The lipid droplet: A conserved cellular organelle. Protein Cell. 8:796–800. 10.1007/s13238-017-0467-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Abraham N., Khan L.A., Hall D.H., Fleming J.T., and Göbel V.. 2011. Apicobasal domain identities of expanding tubular membranes depend on glycosphingolipid biosynthesis. Nat. Cell Biol. 13:1189–1201. 10.1038/ncb2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, P., Na H., Liu Z., Zhang S., Xue P., Chen Y., Pu J., Peng G., Huang X., Yang F., et al. 2012. Proteomic study and marker protein identification of Caenorhabditis elegans lipid droplets. Mol. Cell. Proteomics. 11:317–328. 10.1074/mcp.M111.016345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Zou X., Ding Y., Wang H., Wu X., and Liang B.. 2013. Comparative genomics and functional study of lipid metabolic genes in Caenorhabditis elegans. BMC Genomics. 14:164. 10.1186/1471-2164-14-164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Wang H., Zhang J., Hu Y., Zhang L., Wu X., Su X., Li T., Zou X., and Liang B.. 2016. The cytochrome b5 reductase HPO-19 is required for biosynthesis of polyunsaturated fatty acids in Caenorhabditis elegans. Biochim. Biophys. Acta. 1861:310–319. 10.1016/j.bbalip.2016.01.009 [DOI] [PubMed] [Google Scholar]
- Zhang, J.J., Hao J.J., Zhang Y.R., Wang Y.L., Li M.Y., Miao H.L., Zou X.J., and Liang B.. 2017. Zinc mediates the SREBP-SCD axis to regulate lipid metabolism in Caenorhabditis elegans. J. Lipid Res. 58:1845–1854. 10.1194/jlr.M077198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, H., Shen H., Sewell A.K., Kniazeva M., and Han M.. 2013. A novel sphingolipid-TORC1 pathway critically promotes postembryonic development in Caenorhabditis elegans. eLife. 2:e00429. 10.7554/eLife.00429 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists the organisms and strains in the experimental models.
lists primers.
lists genes regulating LD size by RNAi screen.
shows profiles of fatty acids in C. elegans isolated lipid classes.
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon reasonable request.