Abstract
We have analyzed the interleukin-4 (IL-4)-triggered mechanisms implicated in cell survival and show here that IL-4 deprivation induces apoptotic cell death but does not modulate Bcl-2 or Bcl-x expression. Since Bcl-x expression is insufficient to ensure cell survival in the absence of IL-4, we speculate that additional molecules replace the antiapoptotic role of Bcl-2 and Bcl-x in an alternative IL-4-triggered pathway. Cell death is associated with Bcl-3 downregulation and Bcl-3 expression blocks IL-4 deprivation-induced apoptosis, suggesting that Bcl-3 acts as a survival factor in the absence of growth factor. To characterize the IL-4-induced regulation of murine Bcl-3 expression, we cloned the promoter of this gene. Sequencing of the promoter showed no TATA box element but did reveal binding sites for AP1, AP1-like, and SP1 transcription factors. Retardation gels showed that IL-4 specifically induces AP1 and AP1-like binding activity and that mutation of these binding sites abolishes the IL-4-induced Bcl-3 promoter activity, suggesting that these transcription factors are important in Bcl-3 promoter transactivation. IL-4 deprivation induces downregulation of Jun expression and upregulation of Fos expression, both of which are proteins involved in the formation of AP1 and AP1-like transcription factors. Overexpression of Jun family proteins transactivates the promoter and restores Bcl-3 expression in the absence of IL-4 stimulation. Taken together, these data describe a new biological role for Bcl-3 and define the regulatory pathway implicated in Bcl-3 expression.
Bcl-3 was originally identified as a putative oncogene and cloned from a chromosomal breakpoint in the t(14;19) translocation, which is found in some cases of chronic B-cell lymphocytic leukemias (31). Bcl-3 is a member of the IκB multigene family, which modulates the activities of NF-κB/Rel transcription factors (2, 3, 41, 43). The Bcl-3 protein contains a proline-rich amino terminus, a series of seven tandem ankyrin repeats, and a proline- and serine-rich carboxyl terminus (22). Bcl-3 can increase transcription from NF-κB responsible promoters, although contradictory data exist concerning this role for Bcl-3 (35). On the other hand, Bcl-3 can dissociate p50-p52 homodimers from DNA (6, 14, 15). The picture is further complicated by the finding that Bcl-3 is phosphorylated and that its phosphorylation status affects its interaction with both p50 and p52 (8, 30).
Physiological functions of Bcl-3 have been revealed by the generation of Bcl-3-deficient mice (13, 38). Bcl-3 is required for T-cell-dependent immunity. Bcl-3-deficient mice are defective in antigen-specific antibody production and germinal-center formation and fail to resist infection (13, 38). Bcl-3 may also contribute to B-cell survival, which may explain its oncogenic potential when expressed at high levels as result of chromosomal translocation.
Bcl-3 overexpression is proposed to contribute to the development of chronic lymphocytic leukemia through dysregulation of genes important in cell proliferation and differentiation (31, 47). Sequence analysis of the human Bcl-3 gene predicted a protein with identity to a number of products of genes involved in cell cycle control and in cell lineage determination. Bcl-3 is the first known oncoprotein containing the SWI6/cdc10 motif, suggesting that this protein may be involved in cellular proliferation. Nonetheless, such motifs have also been observed in membrane-associated proteins that are not necessarily involved in cell cycle (27).
Bcl-3 is detected in different tissues, especially the spleen and other lymphoid organs (30). The regulation of its expression has been inadequately investigated. This gene was shown to be induced by mitogenic stimuli in B and T cells (5, 31). The induction of Bcl-3 by both granulocyte-macrophage colony-stimulating factor (GM-CSF) and erythropoietin (Epo) in proliferating human erythroid precursors involves enhanced expression of Bcl-3 mRNA, as well as an increase in the level of Bcl-3 protein (46). After Epo or GM-CSF stimulation, a gradual translocation of Bcl-3 to the nucleus is observed. In addition, Bcl-3 expression was recently shown to be induced by interleukin-9 (IL-9) (35).
IL-4 is a cytokine produced predominantly by T cells, mast cells, and basophils. It stimulates the proliferation of T and B cells as well as of mast cells and exerts distinctive biologic effects on a variety of cells (32). The biological functions of IL-4 are mediated via its binding to a specific cell surface receptor. This receptor is composed of two chains that are members of the type I cytokine receptor superfamily (10), a ligand-binding chain and the common γ chain, which is shared with the IL-2, IL-7, IL-9, and IL-15 receptors (16, 22–24, 29, 36, 37). IL-4 treatment of cells elicits many distinct biological responses, including an increase in cell proliferation and the transcription of a series of genes (32). Some of these responses are unique to IL-4, whereas others are also elicited by different cytokines. Although the receptors for IL-2 and IL-4 have several features in common, including their use of the γ chain as a receptor component, IL-4 evokes responses that IL-2 does not (9, 18, 26, 34). Many factor-dependent cell lines respond to IL-4 with increased thymidine incorporation into DNA, but only a few lines have been successfully adapted for growth in IL-4 alone. Of these, TS1αβ and LD8 can be propagated indefinitely in IL-4.
We present here the cloning and characterization of the murine Bcl-3 gene promoter. We have delineated a positive regulatory region important for IL-4-inducible promoter activity of the Bcl-3 gene. The AP1 and AP1-like binding sites were critical in Bcl-3 promoter activation, and their mutation abrogates promoter activity. We show that Jun proteins, which are involved in the formation of AP1 and AP1-like transcription factors, play an important role in IL-4-induced Bcl-3 expression. Finally, we demonstrate that Bcl-x expression is not sufficient to ensure cell survival in the absence of IL-4 and that Bcl-3 expression is able to block apoptosis in IL-4-deprived cells. This is the first description of the role of AP1 and AP1-like transcription factors in the control of Bcl-3 promoter activation and protein expression, as well as the antiapoptotic role of Bcl-3 in our cellular model.
MATERIALS AND METHODS
Cells and cultures.
TS1αβ is a murine T-cell line stably transfected with the human IL-2 receptor α and β chains (33). This cell line responds independently to IL-2, IL-4, or IL-9. Cells were cultured in RPMI 1640 (BioWhittaker, Walkersville, Md.) supplemented with 5% heat-inactivated fetal calf serum (Gibco-BRL, Gaithersburg, Md.), 2 mM glutamine, 10 mM HEPES, 0.5 mM arginine, 0.24 mM asparagine, 50 μM 2-mercaptoethanol and 60 U of IL-4 per ml or 5 ng of recombinant IL-2 (rIL-2) per ml.
Lymphokines, antibodies, reagents, plasmids, and probes.
Murine rIL-4 or supernatant of a HeLa subline transfected with pKCRIL-4.neo was used as a source of murine IL-4. Anti-Bcl-3 antibody was from Santa Cruz (Santa Cruz, Calif.) or UBI (Lake Placid, N.Y.). Anti-Jun antibodies were from Oncogene Science (Cambridge, Mass.) or Transduction Laboratories (Lexington, Ky.), and anti-Fos antibody was from Santa Cruz. Bcl-2 and Bcl-x antibodies were from Transduction Laboratories. Anti-histone antibodies were from Chemicon International (Temecula, Calif.). Peroxidase-conjugated goat anti-rabbit or anti-mouse immunoglobulin antibody was from Dako (Glostrup, Denmark). Enhanced chemiluminescence (ECL) and 32P-labeled reagents were from Amersham (Little Chalfont, United Kingdom). NP-40 was from Boehringer, (Mannheim, Germany), DEAE-dextran was from Pharmacia (Uppsala, Sweden), and the Capture-Tec pHook 3 kit was from Invitrogen (San Diego, Calif.). The QuikChange site-directed mutagenesis kit was from Stratagene (La Jolla, Calif.). pAd10SacBII vector was from Genome Systems (St. Louis, Mo.), pGL3 basic vector was from Promega (Palo Alto, Calif.), and the CAPFINDER PCR cDNA synthesis kit was from Clontech (Madison, Wis.). Expression vectors for c-Jun, JunB, and JunD proteins were provided by M. Yaniv, Pasteur Institute, Paris, France.
Analysis of RNA expression.
Total RNA was isolated using the Trizol reagent from Gibco-BRL. For Northern blot analysis, RNA samples (15 μg) were electrophoresed in a 1% agarose gel in the presence of formaldehyde and then transferred to a nitrocellulose filter. After hybridization to a 32P-labeled EcoRI DNA fragment containing 1.9 kb of the Bcl-3 gene, the filter was washed and exposed to X-ray film at −70°C with intensifying screens.
Cloning, sequencing, and mutagenesis of the Bcl-3 promoter.
A mouse genomic library constructed in the pAd10SacBII vector was screened by PCR using oligonucleotides specific for the Bcl-3 5′ untranslated region. DNA was extracted from one positive clone and digested with EcoRI, and the resulting fragments were subcloned into pTZ19R. Subclones were screened by colony hybridization using a Bcl-3 probe spanning the first five exons of the Bcl-3 cDNA. Positive clones were isolated and sequenced using plasmid-derived primers. One clone contained a 7-kb fragment showing identity to the 5′ end of the Bcl-3 gene. A 1.5-kb NotI-SstII DNA fragment containing the Bcl-3 promoter and part of exon 1 was subcloned into the pGL3 basic vector at the SmaI site. A shorter DNA fragment containing the promoter was generated by PCR amplification from the latter clone, in which the initiation codon of Bcl-3 gene has been deleted and cloned in the pGL3 vector in front of the reporter luciferase gene. The Bcl-3 promoter was sequenced on both strands with an automatic sequencer (Applied Biosystems, Foster City, Calif.).
Bcl-3 promoter 5′ deletion constructs were generated from pGL3 containing the full-length promoter. The plasmid was linearized, and deletions were generated by NotI, StuI, BamHI, and XcmI digestions, the last two generated by partial digestions. The deleted ends were treated with Klenow fragment and then ligated.
Site-directed mutagenesis was performed using the QuikChange site-directed mutagenesis kit. The primers used for mutation were as follows: AP1 5′ AGATGGCTGAATGATCAAGAAATACTAAAGG 3′ and for AP1-like: 5′ GAGAATCTCAAGGAGCTCGACCCAGACAGAGT 3′. Putative binding sites are underlined, and point mutations are shown in bold type.
Transcription start site mapping.
Transcription start sites were mapped by PCR using the CAPFINDER PCR cDNA synthesis kit (Clontech), in which only cDNA derived from capped mRNA is exponentially amplified by PCR. The oligonucleotides used in defining the start sites were the 5′ PCR oligonucleotide provided and the internal primer from the Bcl-3 gene, 5′ GTGCGGCGAGCTCGGCACG. PCR fragments were electrophoresed in 4% MetaPhor agarose, extracted, purified, and sequenced using the Bcl-3 gene primer.
Transient transfection.
Ts1αβ cells were transiently transfected using the DEAE-dextran method. Cells (10 × 106) in exponential growth were washed with TS buffer (25 mM Tris HCl, 137 mM NaCl, 5 mM KCl, 0.7 mM CaCl2, 0.5 mM MgCl2, 0.6 mM Na2HPO4 [pH 7.4]). A total of 5 μg of plasmid, 750 μl of TS buffer, and 750 μl of freshly prepared DEAE-dextran (1 mg/ml) in TS buffer were mixed successively with the cells and incubated for 20 min at room temperature, after which 13 ml of RPMI 1640–5% fetal calf serum was added. Cells were incubated (1 h at 37°C), centrifuged, and resuspended in 12 ml of RPMI 1640–5% FCS alone or supplemented with 60 U of IL-4 per ml or 5 ng of IL-2 per ml. When Jun or Bcl-3 expression vectors were transfected, cells were cotransfected with the pHook3 plasmid. The pHook3 vector drives the expression of a hapten-specific single-chain antibody (sFv) on the surface of transfected cells. Cells expressing the sFv were isolated from the culture by binding to hapten-coated (pHox) magnetic beads.
Luciferase assay.
After transfection, cells were unstimulated or stimulated with 60 U of IL-4 per ml or 5 ng of IL-2 per ml for different periods (12 to 24 h), washed in cold phosphate-buffered saline, and lysed in Luc buffer (25 mM Tris phosphate [pH 7.8], 8 mM MgCl2, 1 mM dithiothreitol, 1 mM EDTA, 1% Triton X-100, 1% bovine serum albumin, BSA, 15% glycerol) at 4°C. Extracts were diluted in Luc buffer, and the reaction mixture was prepared with 91 μl of 25 mM luciferin, 330 μl of 20 mM ATP, and 4.606 ml of Luc buffer supplemented with 0.5 mM coenzyme A. Luciferase activity in protein extracts was measured using a Berthold LB9501 luminometer.
Western blot analysis.
Nuclear proteins from IL-4-stimulated or -deprived cells were isolated as described below. Alternatively, IL-4-stimulated or -deprived cells were lysed in Laemmli sample buffer and protein extracts were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose, blocked with 5% nonfat milk in Tris-buffered saline (TBS) (20 mM Tris-HCl [pH 7.5], 150 mM NaCl), and incubated with primary antibody in TBS–0.5% nonfat dry milk. Membranes were washed in 0.05% Tween 20 in TBS and incubated with peroxidase-conjugated second antibody. After the washing step, proteins were developed using the ECL system. When stripping was required, the membranes were incubated with 62.5 mM Tris-HCl (pH 6.8)–2% SDS–0.1 M 2-mercaptoethanol for 1 h at 56°C and washed extensively with TBS before being subjected to reblocking and probing.
Nuclear extracts, electrophoretic mobility shift, and supershift assay.
IL-4-stimulated or -deprived cells (3 × 107) were resuspended for 2 min in 1 ml of buffer A (50 mM NaCl, 10 mM HEPES [pH 8], 0.5 mM sucrose, 1 mM EDTA, 0.5 mM spermidine, 0.15 mM spermine, 0.2% Triton X-100). Lysates were centrifuged (4,500 × g for 3 min at 4°C). Nuclei were resuspended in 1 ml of buffer B (50 mM NaCl, 10 mM HEPES [pH 8], 25% glycerol, 0.1 mM EDTA, 0.5 mM spermidine, 0.15 mM spermine) and centrifuged (4,500 × g for 3 min at 4°C). Nuclear proteins were extracted at 4°C for 30 min in 60 μl of buffer C (350 mM NaCl, 10 mM HEPES [pH 8], 25% glycerol, 0.1 mM EDTA, 0.5 mM spermidine, 0.15 mM spermine). Supernatants were cleared by centrifugation and stored at −80°C. The protein concentration was determined using the Bradford method (Bio-Rad). All buffers were supplemented with protease inhibitors. 32P-end-labeled oligonucleotides (0.2 ng) were incubated (at room temperature for 15 min) with 3 μg of nuclear proteins in the presence of 0.25 ng of single-stranded DNA, 60 mM KCl, 0.01% NP-40, 0.1 mg of bovine serum albumin per ml, and 4% Ficoll in a final volume of 10 μl. Protein-DNA complexes were separated from free probe by electrophoresis on 5% polyacrylamide gels (PAGE) in 0.5× TBE (1× TBE is 90 mM Tris, 90 mM boric acid, and 1.5 mM EDTA [pH 8]) at room temperature, and the gels were dried and exposed to X-ray film. A 20-fold molar excess of cold oligonucleotide was used to compete for protein binding to the radiolabeled probe. The following probes were used: SP1 binding site, 5′ GGATTCGATCGGGGCGGGGCGAGC 3′; AP1 binding site, 5′ GGCTTGATGAGTCAGCCG; and AP1-like 5′ GAATCTCAAGGACTCAGACCCAG 3′. For supershift experiments, IL-4-stimulated or -deprived nuclear extracts were preincubated with the corresponding antibody on ice prior to addition of the 32P-labeled probe. The shifted complexes were resolved as above.
Nucleotide sequence accession number.
The Bcl-3 promoter sequence data have been submitted to the EMBL database (accession no. AJ249641).
RESULTS
IL-4 induces Bcl-3 expression in TS1αβ cells.
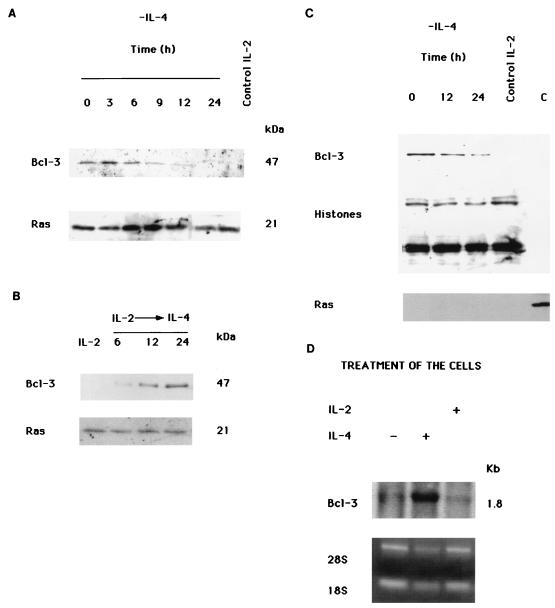
We have described that Bcl-2 is differentially regulated by IL-2 and IL-4 (19) and asked whether other genes might be differentially regulated by IL-2 and IL-4 in TS1αβ cells. Bcl-3 was recently shown to be induced by IL-9 and IL-4 in mouse T helper cells (35). We identified Bcl-3 as a gene expressed in IL-4- but not IL-2-stimulated TS1αβ cells (Fig. 1A). When IL-4-maintained cells were deprived of lymphokine, Bcl-3 expression was downregulated. The amount of Bcl-3 decreased throughout the period of IL-4 deprivation, reaching undetectable levels at 24 h of deprivation. Cells maintained in IL-2 showed no Bcl-3 expression. To determine whether IL-4 modulates Bcl-3 expression, IL-2-maintained cells were switched to IL-4-containing medium and Bcl-3 expression was analyzed by Western blotting. Under these conditions, stimulation with IL-4 induced Bcl-3 expression (Fig. 1B) but IL-2-stimulated cells did not express Bcl-3 (Fig. 1A and B).
FIG. 1.
Regulation of Bcl-3 expression at the mRNA and protein level in TS1αβ cells. (A) TS1αβ cells were IL-4 stimulated (60 U/ml) or lymphokine deprived for the times indicated and then lysed. Protein extracts were separated by SDS-PAGE, transferred to nitrocellulose, and probed with anti-Bcl-3 antibody. Cells maintained in IL-2 (5 ng/ml) were used as a negative control for Bcl-3 expression. Protein bands were detected using the ECL system. The blot was stripped and reprobed with pan-Ras antibody as an internal control of protein loading. Similar results were obtained in two independent experiments. Molecular weights of the corresponding proteins are shown. (B) IL-2-stimulated TS1αβ cells were switched to IL-4 for the times indicated and then lysed. Protein extracts were separated by SDS-PAGE, transferred to nitrocellulose, and probed with anti-Bcl-3 antibody. IL-2-stimulated cells were used as a negative control for Bcl-3 expression. Protein bands were detected using the ECL system. The blot was probed with pan-Ras antibody as an internal control of protein loading. The molecular masses of the corresponding proteins are shown. (C) Nuclear proteins were isolated from IL-4-stimulated (60 U/ml) or -deprived cells and from IL-2-stimulated (5 ng/ml) cells. After quantification, proteins were resolved by SDS-PAGE, transferred to nitrocellulose, and immunoblotted with anti-Bcl-3 antibody. As a nuclear marker of protein loading, the blot was probed with anti-histone H1, H2a, H2b, H3, and H4 antibodies. Protein bands were detected using the ECL system. Similar results were obtained in two independent experiments. As an internal control of protein fractionation, nuclear or cytoplasmic proteins were blotted and probed with pan-Ras antibody (cytoplasmic marker). (D) Total RNA was isolated from 20 × 106 IL-2-stimulated (5 ng/ml), IL-4-stimulated (60 U/ml), and IL-4-deprived cells (24 h). RNA (15 μg) was electrophoresed in 1% agarose with formaldehyde, blotted, and hybridized under high-stringency conditions to a 32P-labeled Bcl-3 probe. The DNA probe was labeled by random priming. Both 28S and 18S are shown to estimate RNA levels. The size of the mRNA for Bcl-3 is indicated. Data are representative of two independent experiments.
Given that Bcl-3 is predominantly a nuclear protein, we analyzed its expression in nuclear extracts under IL-4 stimulation or deprivation conditions (Fig. 1C). High Bcl-3 levels were detected in nuclear extracts of IL-4-stimulated cells by Western blot analysis. The amount of Bcl-3 decreased at 12 h in IL-4-deprived cells, with minimum Bcl-3 levels detected after 24 h of lymphokine deprivation, suggesting an IL-4-induced modulation of nuclear Bcl-3 protein.
Since IL-4 regulates Bcl-3 expression, it was of interest to determine whether IL-4 could modulate Bcl-3 mRNA levels. Total mRNA was isolated from IL-4- or IL-2-stimulated and IL-4-deprived cells, electrophoresed, and hybridized with a Bcl-3 probe. The result shows that the absence of IL-4 downmodulates the Bcl-3 mRNA level (Fig. 1D). In the presence of IL-2, as well as in the absence of IL-4, we were unable to detect mRNA for Bcl-3.
Structure of the Bcl-3 promoter.
To characterize the regulation of the gene encoding Bcl-3, we have isolated, sequenced, and characterized the promoter of this gene. To isolate the Bcl-3 promoter region, we screened by PCR a mouse genomic library using oligonucleotides specific for the 5′ untranslated region. Two clones containing sequences located upstream of the transcribed Bcl-3 gene were isolated. One of these contained a 1.5-kb NotII-SstII fragment of the Bcl-3 promoter, including the initiation codon. A shorter fragment of the promoter without the initiation codon was isolated by PCR amplification. Sequence analysis of this fragment demonstrated that it contained 1,458 bp upstream of the ATG. Analysis of this DNA sequence using the TF sites data bank showed potential binding sites for AP1, AP1-like, and SP1 transcription factors, although no TATA box could be identified (Fig. 2). Transcription start site mapping was done using a PCR base method, in which the cDNA population is enriched in 5′ capped mRNA. DNA amplification of a thymus cDNA using an anchoring primer (5′ PCR, see Materials and Methods) and a Bcl-3 cDNA-specific primer produced three major DNA bands. The predicted transcription start sites (101, 176, and 239 bp upstream of the ATG codon) are shown in Fig. 2.
FIG. 2.
Nucleotide sequence of the Bcl-3 promoter. The nucleotide sequence of the 5′-flanking region of the Bcl-3 gene is shown. The ATG start codon is in bold type. The three transcription start sites are in bold and underlined. The AP1 binding site is in italic and underlined. The SP1 site is double underlined. The AP1-like sites are underlined. The primer used for determination of the transcription start site mapping is boxed. Restriction sites are shown.
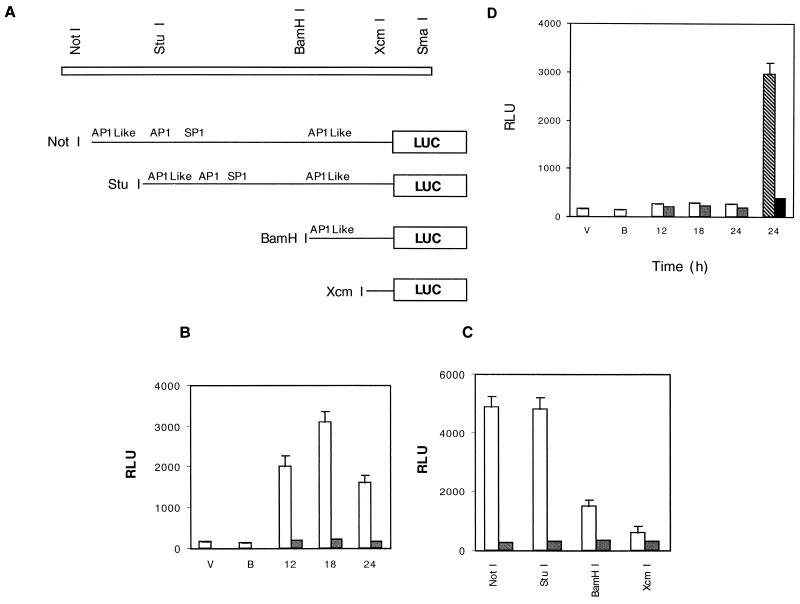
Since the Bcl-3 promoter has no classical TATA box sequence, it was important to perform a functional analysis of the sequence. We cloned the 1.4-kb fragment in front of the luciferase reporter gene into the pGL3 basic vector. Figure 3A shows the restriction map of the full length Bcl-3 promoter and the nested 5′ deletion promoter-luciferase constructs, showing binding sites for some transcription factors such as AP1, AP1-like, and SP1. The full-length Bcl-3 promoter (NotI) showed maximal luciferase activity 18 h after transient transfection of IL-4-stimulated cells, which decreased markedly by 24 h after transfection (Fig. 3B); this period (18 h) was used in further experiments. The fragment in the reverse orientation was unable to drive luciferase reporter gene expression (data not shown). Cells deprived of IL-4 for 12 to 24 h after transfection did not exhibit luciferase activity. The result suggests that the cloned Bcl-3 promoter responds to stimulation by IL-4.
FIG. 3.
Deletion analysis of the Bcl-3 promoter region. (A) Schematic diagram of the 5′ regulatory region showing the restriction map of the Bcl-3 promoter. Deletion mutants with the 5′ endpoints and putative nuclear protein binding sites are indicated. (B) Luciferase assay at different time points after transient transfection of full-length Bcl-3 promoter construct. V, empty vector; B, buffer; open bars, IL-4 stimulation; shaded bars, IL-4 deprivation. Relative light units (RLU) were normalized to β-galactosidase activity. Standard deviation (SD) is shown for n = 3. (C) Summary of the luciferase assay using the 5′ deletion mutants in panel A. After transient transfection, cells were IL-4 stimulated (60 U/ml) (open bars) or deprived (shaded bars) for 18 h, collected, washed, and assayed for luciferase activity. RLU were normalized to β-galactosidase activity. SD is shown for n = 3. (D) Luciferase assay at 24 h or different times after transfection of full-length Bcl-3 promoter. V, empty vector; B, buffer; open bars, IL-2 stimulation; shaded bars, IL-2-deprivation; hatched bar, IL-4 stimulation; solid bar, IL-4 deprivation. RLU were normalized to the β-galactosidase activity. SD is shown for n = 3.
To delineate the cis-acting elements and trans-acting factors that regulate Bcl-3 expression, we examined nested 5′ deletion promoter-luciferase constructs (Fig. 3C) in IL-4-stimulated or -deprived TS1αβ cells by using transient-transfection assays. Control full-length Bcl-3 promoter (NotI-Luc) showed luciferase activity. StuI-Luc deletion does not significantly modify the IL-4-induced luciferase activity, compared to the level observed in control transfected cells. Promoter activity was strongly reduced in the BamHI-Luc deletion and was almost undetectable in the XcmI-Luc deletion. Finally, no luciferase activity was observed with any of the constructs when cells were IL-4 deprived after transfection (Fig. 3C). This result allowed us to delineate the minimum Bcl-3 region with promoter activity, the StuI-Luc construct. IL-2 was not able to transactivate the full-length Bcl-3 promoter at any time after transfection (Fig. 3D). IL-4-stimulated cells after transfection with the full-length Bcl-3 promoter were used as a positive control of Bcl-3 promoter transactivation. This result correlates with the absence of Bcl-3 expression in IL-2-stimulated TS1αβ cells.
Characterization of IL-4-induced proteins binding to the Bcl-3 promoter.
Since the StuI-Luc deletion appears to be the shortest fragment with promoter activity and since additional deletion of this fragment (BamHI-Luc) shows a strong reduction in luciferase activity, we focused our attention on the identification of putative binding sites for transcription factors in the StuI-BamHI promoter fragment. We performed bandshift assays and competition using double-stranded oligonucleotides corresponding to the AP1, AP1-like, and SP1 sites, which were identified in the StuI-BamHI fragment, and nuclear proteins derived from IL-4-stimulated or -deprived TS1αβ cells.
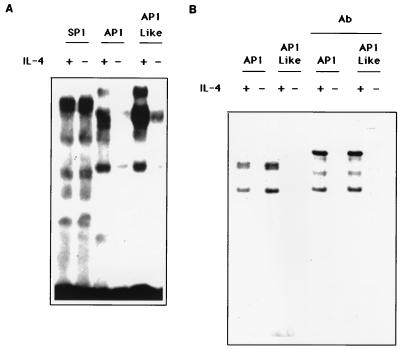
We detected protein binding activity to AP1, AP1-like, and SP1 oligonucleotides when using nuclear extracts from IL-4-stimulated cells (Fig. 4A). When cells were IL-4 deprived for 24 h, protein binding to the AP1 and AP1-like sites was markedly reduced while binding to SP1 site was not affected. Specific DNA–nuclear-protein interaction was confirmed by competition with unlabeled oligonucleotides (data not shown). This suggests that IL-4 specifically induces AP1 and AP1-like activation and that these transcription factors may be responsible for the IL-4 inducibility of the Bcl-3 promoter. Supershift assays were performed to determine whether the nuclear binding activities contained AP1 and AP1-like proteins. Preincubation of the IL-4-stimulated TS1αβ nuclear extracts with c-Jun and JunB antibodies resulted in a supershift of the AP1 and AP1-like complexes (Fig. 4B). No supershift was observed when the antibodies were preincubated with IL-4-deprived nuclear extracts. This result suggests that the complexes contain Jun proteins.
FIG. 4.
Effect of IL-4 on induction of nuclear factor activities. (A) Nuclear proteins from IL-4-stimulated (60 U/ml) or -deprived cells were incubated with the indicated 32P-end-labeled oligonucleotide. For the oligonucleotide sequence, see Materials and Methods. Protein-DNA complexes were separated from free oligonucleotide by PAGE (50% polyacrylamide), dried, and exposed to X-ray film. The specificity for each site was tested using a 20-fold molar excess of specific cold oligonucleotide. Data are representative of three independent experiments. (B) Nuclear proteins from IL-4-stimulated (60 U/ml) or -deprived cells were preincubated with c-Jun and JunB antibodies on ice and then incubated with the indicated 32P-end-labeled oligonucleotide. Protein-DNA complexes were resolved by PAGE (5% polyacrylamide), dried, and exposed to X-ray film. Data are representative of two independent experiments.
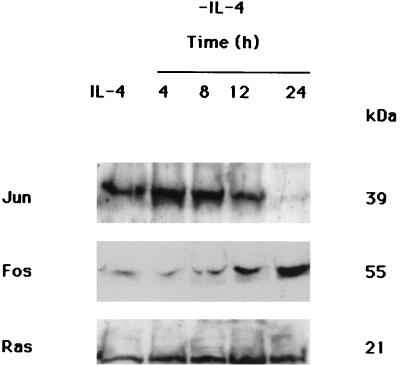
Since AP1 and AP1-like transcription factors are composed of Jun and of Fos protein family members, we asked whether IL-4 deprivation could modulate Jun and Fos expression. TS1αβ cells were stimulated or deprived of IL-4 for different periods and total Jun and Fos protein expression was analyzed by Western blotting. Jun and Fos expression was detected in control IL-4-stimulated cells. Progressive increase of Fos expression was detected throughout the starvation period, reaching maximum at 24 h after IL-4 deprivation (Fig. 5). In contrast, Jun expression decreased following IL-4 deprivation, reaching almost undetectable levels at 24 h after lymphokine deprivation (Fig. 5). This deprivation period corresponds to the period of maximum Fos level detected. The result suggests that absence of AP1 and AP1-like activity in IL-4-deprived cells may be due to the lack of expression of Jun, one of the proteins involved in the formation of AP1 and AP1-like transcription factors.
FIG. 5.
Effect of IL-4 deprivation on Jun and Fos expression. Control IL-4-stimulated (60 U/ml) or -deprived cells were harvested at different times (from 4 to 24 h). Total-protein extracts were separated by SDS-PAGE, transferred to nitrocellulose, and probed sequentially with anti-Jun and anti-Fos antibodies. The blot was stripped and probed with pan-Ras antibody as an internal control of protein loading. The protein bands were detected using the ECL system. Similar results were obtained in two independent experiments. The molecular masses of the corresponding proteins are shown.
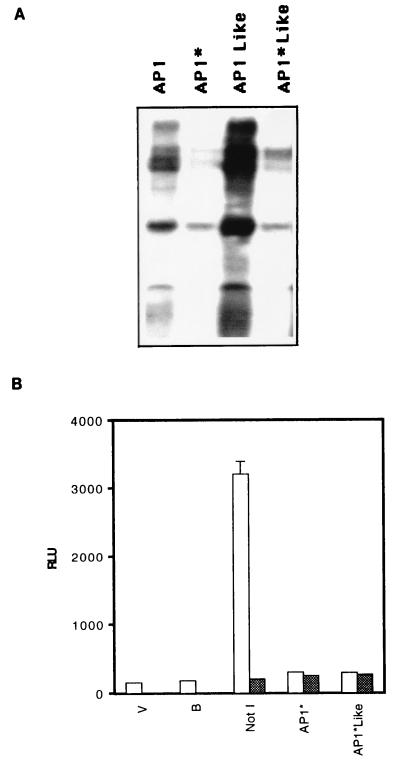
To evaluate the functional role of AP1 and AP1-like transcription factors in the control of Bcl-3 promoter activity, we mutated both binding sites in Bcl-3 promoter so that it could not bind to nuclear proteins. Binding activity for AP1 and AP1-like factors was detected in nuclear extracts of IL-4-stimulated cells. This binding activity was undetectable using oligonucleotides containing mutated AP1* and AP1*-like binding sites (Fig. 6A). The specificity of the DNA-protein interaction was confirmed by competition with unlabeled oligonucleotides (data not shown). Cells transfected with mutated AP1* or AP1*-like full-length Bcl-3 promoter constructs in the presence of IL-4 showed no transactivation of the luciferase reporter gene compared with cells transfected with the wild-type full-length Bcl-3 promoter construct (Fig. 6B). Similarly, no luciferase activity was detected when cells transfected with wild-type or mutated AP1* or AP1*-like constructs were maintained in the absence of IL-4. The results suggest that AP1 and AP1-like factors play a direct, important role in Bcl-3 promoter transactivation.
FIG. 6.
AP1 and AP1-like mutation abolishes nuclear protein binding and promoter activity. (A) Nuclear proteins from IL-4-stimulated (60 U/ml) cells were incubated with 32P-end-labeled oligonucleotide containing the wild-type or mutated AP1* and AP1*-like binding sites. For the oligonucleotide sequence, see Materials and Methods. Protein-DNA complexes were separated from free oligonucleotide by PAGE (5% polyacrylamide), dried, and exposed to X-ray film. Data are representative of two independent experiments. (B) TS1αβ cells were transiently transfected with full-length wild type Bcl-3 promoter (NotI-Luc) or the full-length Bcl-3 promoter containing mutated AP1* and AP1*-like binding sites. After transfection, the cells were IL-4 stimulated or deprived for 18 h, collected, washed, and analyzed for luciferase activity. B, buffer; V, empty vector; AP1* and AP1*-like, full-length Bcl-3 promoter with mutated AP1 and AP1-like binding sites. Relative light units (RLU) were normalized to β-galactosidase activity. SD is shown for n = 3.
Jun proteins induce Bcl-3 expression in the absence of IL-4.
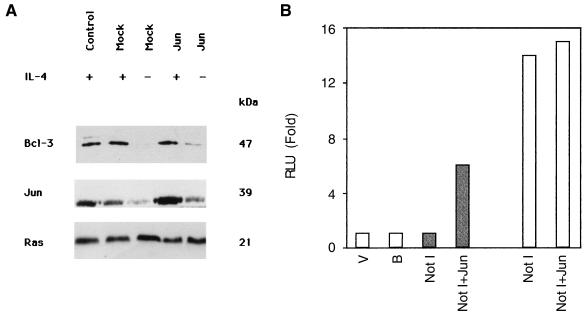
Since mutation of AP1* and AP1*-like binding sites in the Bcl-3 promoter abolishes DNA binding and luciferase activity, and since both Jun and Bcl-3 expression are downregulated in the absence of IL-4, we asked whether Jun proteins are involved in the control of Bcl-3 expression. TS1αβ cells were transiently transfected with a mixture of c-Jun, JunB, and JunD expression vectors and analyzed for Bcl-3 expression (Fig. 7A). After IL-4 stimulation, mock transfectants or cells transfected with Jun proteins showed Bcl-3 expression levels comparable to those of control IL-4-stimulated cells. Mock transfectants maintained in the absence of IL-4 showed no Bcl-3 expression. Interestingly, in cells transfected with the Jun protein mixture, Bcl-3 expression was induced without IL-4 addition (Fig. 7A) whereas independent transfection of c-Jun, JunB, or JunD did not restore Bcl-3 expression in the absence of IL-4 (data not shown). Expression of transiently transfected Jun was confirmed by direct comparison of Jun protein levels in transfected cells and mock controls. Unaltered Ras expression was demonstrated under all transfection conditions as an internal protein loading control. Jun proteins thus appear to induce Bcl-3 expression in the absence of IL-4. We asked whether expression of Jun proteins affects Bcl-3 promoter transactivation. Cotransfection of Bcl-3 promoter and Jun proteins (c-Jun, JunB, and JunD) induced luciferase activity in the absence of IL-4-stimulation (Fig. 7B). Following IL-4-stimulation, cells transfected with the Bcl-3 promoter alone or in combination with the Jun proteins show comparable levels of luciferase activity (Fig. 7B), suggesting that expression of Jun proteins transactivates the Bcl-3 promoter in IL-4-deprived cells.
FIG. 7.
Transfection of Jun proteins induces Bcl-3 expression in IL-4-deprived cells. (A) Cells were transiently cotransfected with pHook3 and a mixture of c-Jun, JunB, and JunD expression vectors using the DEAE-dextran method and then stimulated with or deprived of IL-4 for 24 h. After transfection, the cells were washed, separated from untransfected cells as described in Materials and Methods, and lysed. Protein extracts were separated by SDS-PAGE, transferred to nitrocellulose, and probed with anti-Bcl-3 antibody. IL-4-maintained cells were used as a positive Bcl-3 expression control. Protein bands were developed using ECL. Transfected Jun protein expression was confirmed by comparing Jun protein levels in transfected and mock-transfected controls. Hybridization with pan-Ras is shown as the internal protein loading control. The molecular masses of the corresponding proteins are shown. Similar results were obtained in two independent experiments. (B) Luciferase assay after transient transfection of the full-length Bcl-3 promoter alone or cotransfected with c-Jun, JunB, and JunD. V, empty vector; B, buffer; open bars, IL-4 stimulation; shaded bars, IL-4 deprivation. RLU were normalized to β-galactosidase activity.
Bcl-3 expression prevents apoptosis of IL-4-deprived TS1αβ cells.
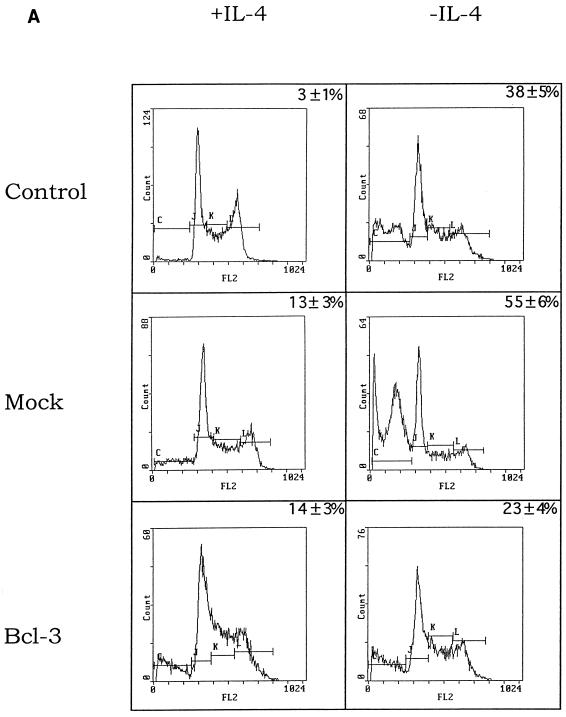
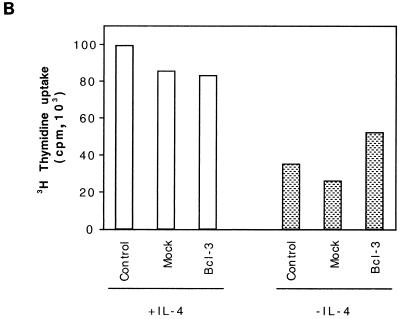
IL-4-stimulated cells do not express Bcl-2, while IL-4-stimulated or -deprived cells express Bcl-x (data not shown). Since IL-4 deprivation in TS1αβ cells correlates with downregulation of Bcl-3 expression and apoptosis, without modification of Bcl-x expression, we hypothesized that Bcl-3 may prevent apoptosis. Mock transfectants or cells transfected with Bcl-3 expression vector were selected from a mixed population of transfected and nontransfected cells. Cells transfected with Bcl-3 and deprived of IL-4 for 24 h showed a strong reduction in the fraction of apoptotic cells compared to IL-4-deprived mock-transfected cells (Fig. 8A). The frequency of apoptotic cells remained similar in all transfected cells in the presence of IL-4. Similar results were obtained for Jun protein expression (data not shown). Taken together, these results suggest that Bcl-3 may act as a survival factor in TS1αβ cells. To analyze the ability of IL-4-deprived Bcl-3-transfected cells to inhibit apoptosis, we performed a proliferation assay (Fig. 8B). Control, mock-transfected, or Bcl-3-transfected cells maintained in the presence of IL-4 after transfection showed thymidine incorporation. Control or mock-transfected IL-4-deprived transfected cells showed strong resuction of thymidine uptake. Interestingly, Bcl-3-transfected cells maintained in the absence of IL-4 showed higher thymidine incorporation than did transfected cells maintained in the absence of IL-4, although they did not reach the level of proliferation detected in IL-4-stimulated cells. This result suggests that IL-4 supplies an additional intracellular signal that complements the Bcl-3 survival signal, allowing cell proliferation.
FIG. 8.
Cell cycle and proliferation analysis of TS1αβ cells transfected with Bcl-3. (A) Cells were transfected with or without Bcl-3 and pHook3, using the DEAE-dextran method, and maintained for 24 h after transfection alone or with IL-4. The cells were washed, selected, permeabilized, stained with propidium iodide, and analyzed by flow cytometry. The most proximal region of the fluorescence scale represents the sub-G1 region. The percentages of apoptotic cells in each sample are superimposed. Nontransfected cells maintained alone or in the presence of IL-4 for 24 h were also used as controls. Similar results were obtained in three independent experiments. SD is shown for n = 3. (B) TS1αβ cells were transfected as for panel A and maintained for 24 h after transfection with [3H]thymidine in the presence or absence of IL-4. The cells were washed and selected, and the proliferation response was analyzed. Nontransfected cells maintained in the presence or the absence of IL-4 for 24 h were used as a control of [3H]thymidine incorporation.
DISCUSSION
For a more complete understanding of Bcl-3 expression regulation by IL-4, we have cloned and characterized the murine Bcl-3 gene promoter region. We have delineated the 5′ regulatory region, which is essential for IL-4-induced promoter activity, and have investigated the role of AP1 and AP1-like nuclear proteins in the control of Bcl-3 expression. Stimulation of TS1αβ cells by IL-4, but not IL-2, induces Bcl-3 expression at the RNA and protein levels. The differential control of gene expression by IL-2 and IL-4 has been previously found in TS1αβ cells; IL-2 induces Bcl-2 and NFAT expression, whereas IL-4 does not (18, 19). IL-9, GM-CSF, and Epo also induce Bcl-3 expression in T cells and erythroid cell precursors, as well as stimulating proliferation (35, 46). It has also been found that Bcl-3 is related to genes implicated in cell lineage determination and cell cycle control (31). Another function described for Bcl-3 is the activation of retinoblastoma expression through interaction with E4TF1 (40). Bcl-3 is located preferentially in the cell nucleus (6, 44, 47), although other reports describe its location in both nuclear and cytoplasmic compartments (46), suggesting that nuclear expression may be regulated under physiological conditions. The cytoplasmic retention may be due to physical association with other proteins or to posttranslational modifications. Other members of the NF-κB inhibitor family have been also observed in the nucleus (4, 11, 28, 45).
A remarkable feature of the 1.3-kb promoter is the absence of a TATA box element. The lack of this motif has also been observed in a number of genes whose products have housekeeping functions (17, 39, 42). Analysis of the Bcl-3 promoter revealed the presence of three transcription start sites; the initiation of gene transcription at multiple sites is consistent with the lack of a canonical TATA box in the promoter. In addition to Bcl-3, the presence of several transcription start sites has been described for other genes (12, 25). The luciferase activity observed after IL-4 stimulation is consistent with the increased level of Bcl-3 expression. In constructs with endpoints at XcmI and BamHI sites, respectively, no activity or nearly undetectable promoter activity was observed. It is interesting that the BamHI deletion retains only one of the AP1-like binding sites and that the XcmI deletion has no binding sites for these transcription factors.
Protein binding to AP1 and AP1-like binding sites was induced by IL-4 stimulation. Antibodies against Jun proteins can supershift both AP1 and AP1-like complexes, suggesting that the DNA-protein complexes observed in gel retardation contain Jun proteins. IL-4 deprivation induces downregulation of Jun expression, suggesting that Jun proteins may be the limiting factor in the formation of AP1 and AP1-like transcription factors. IL-4-deprived cells that receive an additional dose of Jun proteins are able to synthesize significant quantities of Bcl-3, suggesting that the presence of AP1 and AP1-like transcription factors may be essential for IL-4-dependent promoter activity and Bcl-3 expression. We do not exclude the possibility that the AP1 and AP1-like factors interact with other proteins to control Bcl-3 expression or, alternatively, that these factors may cooperate in the induction of Bcl-3 expression. In studies on Bcl-3 expression control by IL-9, the effect of IL-9 is controlled by STAT proteins, suggesting differences between IL-4 and IL-9 signaling or, alternatively, synergy between STAT and Jun proteins (35).
Cell death by IL-2 deprivation has been correlated with a decrease in the level of Bcl-x (7), but in our experimental system, Bcl-x protein levels were constant after IL-4 deprivation (data not shown). Although Bcl-x is expressed after IL-4-stimulation, it appears to be insufficient to promote cell survival, since Bcl-x is also expressed in IL-4-deprived cells. An alternative pathway different of Bcl-2 and Bcl-x may be triggered by IL-4 to prevent cell death and to induce proliferation. IL-4 deprivation induces inhibition of Bcl-3 expression, resulting in apoptotic cell death, which is blocked by Bcl-3 expression, suggesting that Bcl-3 can replace the antiapoptotic role of Bcl-2 and Bcl-x and act as survival factor in IL-deprived TS1αβ cells.
A correlation has been demonstrated between Bcl-3 expression and proliferation of B lymphocytes (5). Similarly, activation of retinoblastoma expression by Bcl-3 protects cells from apoptosis, which may contribute to leukemogenesis following the model suggested for Bcl-2 (20, 40). Bcl-3 downregulation presumably results in a change in the regulation of a gene or genes important in some aspect of cell proliferation, differentiation, or survival. The downregulation of Bcl-3 during IL-4 deprivation-triggered apoptosis, together with the ability of Bcl-3 to act as a cell lineage-specific gene, allowed us to conclude that Bcl-3 may act as a survival gene for Th2 cell differentiation.
Extensive analysis of the response to infections indicated that Bcl-3−/− mice failed to establish a proper antigen-specific Th1 response. In addition, production of IL-12 and gamma interferon, two cytokines necessary for generation of a normal Th1 response, were impaired. The antibody response was also affected in Bcl-3−/− mice. This defect correlates with impaired formation of germinal centers. Such centers are the primary anatomical sites where antigen-specific B cells undergo rapid expansion and finally differentiate into plasma cells or memory cells. These data correlate with our results showing that overexpression of Bcl-3 in T cells, in the absence of complementary signals, allow the survival but not the proliferation of T cells.
Bcl-3 expression is probably controlled by IL-4-regulated transcription factors through binding-site recognition in the promoter region of Bcl-3. Our data demonstrate the significant role of AP1 and AP1-like factors in Bcl-3 promoter transactivation, and their absence provides an explanation for the disruption of Bcl-3 expression in IL-4-deprived cells. Our results also suggest that Bcl-3 can replace the antiapoptotic role of Bcl-2 and Bcl-x in TS1αβ cells. We have established the basis of specific molecular Bcl-3 functions and its integration into regulatory pathways. Further studies are needed to determine whether the findings presented here are applicable to other growth factor signaling systems.
ACKNOWLEDGMENTS
We thank J. L. Barbero for helping with Bcl-3 promoter sequencing; M. Yaniv for the c-Jun, JunB, and JunD expression vectors; and C. Mark for editorial assistance.
V.A. is the recipient of a Pharmacia & Upjohn fellowship. The Department of Immunology and Oncology was founded and is supported by the Spanish Research Council (CSIC) and Pharmacia & Upjohn.
REFERENCES
- 1.Adachi M, Cossman J, Longo D, Croce C M, Tsujimoto Y. Variant translocation of the Bcl-2 gene to immunoglobulin lambda light chain gene in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 1989;86:2771–2774. doi: 10.1073/pnas.86.8.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baeuerle P A, Baltimore D. NF-κB: ten years after. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 3.Baldwin A S., Jr The NF-κB and Iκ-B proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 4.Beg A A, Ruben S M, Scheinman R I, Haskill S, Rosen C A, Baldwin A S. Iκ-B interacts with the nuclear localization sequences of the subunits of NF-κB: a mechanism of cytoplasmic retention. Genes Dev. 1992;6:1899–1913. doi: 10.1101/gad.6.10.1899. [DOI] [PubMed] [Google Scholar]
- 5.Bhatia K, Huppi K, McKeithan T, Siwarski D, Mushinski J F, Magrath I. Mouse Bcl-3: cDNA structure, mapping and stage-dependent expression in B lymphocytes. Oncogene. 1991;6:1569. [PubMed] [Google Scholar]
- 6.Bours V, Franzoso G, Azarenko V, Park S, Kanno T, Brown K, Siebenlist U. The oncoprotein Bcl-3 directly transactivates through κB motifs via association with DNA binding p50 homodimers. Cell. 1993;72:729–739. doi: 10.1016/0092-8674(93)90401-b. [DOI] [PubMed] [Google Scholar]
- 7.Broome H E, Dargan C M, Krajewski S, Reed J C. Expression of Bcl-2, Bcl-x and Bax after T cell activation and IL-2 withdrawal. J Immunol. 1995;155:2311. [PubMed] [Google Scholar]
- 8.Caamaño J H, Perez P, Lira S A, Bravo R. Constitutive expression of Bcl-3 in thymocytes increases the DNA binding of NF-κB homodimers in vivo. Mol Cell Biol. 1996;16:1342–1348. doi: 10.1128/mcb.16.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cerezo A, Martínez-A C, González A, Gómez J, Rebollo A. L-2 deprivation triggers apoptosis which is mediated by c-Jun N-terminal kinase 1 activation and prevented by Bcl-2. Cell Death Differ. 1999;6:87–94. doi: 10.1038/sj.cdd.4400458. [DOI] [PubMed] [Google Scholar]
- 10.Cosman D. The hematopoietin receptor superfamily. Cytokine. 1993;5:95–106. doi: 10.1016/1043-4666(93)90047-9. [DOI] [PubMed] [Google Scholar]
- 11.Davis N, Bargmann W, Lim M Y, Bose H J. Avian reticuloendotheliosis virus-transformed lymphoid cells contain multiple p59 V-rel complexes. J Virol. 1990;64:584–591. doi: 10.1128/jvi.64.2.584-591.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Martin R, Vanhoven B, Chen Q, Hofer E, Csizmadia V, Wincler H, Bach F H. Cytokine-inducible expression in endothelial cells of an Iκ-Bα like gene is regulated by NF-κB. EMBO J. 1993;12:2773–2779. doi: 10.1002/j.1460-2075.1993.tb05938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franzoso G, Carlson L, Scharton T, Kersten T, Shores E W, Epstein S, Grinberg A, Tran T, Shacter E, Leonardi A, Anver M, Love P, Sher A, Siebenlist U. Critical roles for the Bcl-3 oncoprotein in T cell mediated immunity, splenic microarchitecture and germinal centers reactions. Immunity. 1997;6:479–490. doi: 10.1016/s1074-7613(00)80291-5. [DOI] [PubMed] [Google Scholar]
- 14.Franzoso G, Bours V, Park S, Tomita T, Yamaguchi M, Kelly K, Siebenlist U. The candidate oncoprotein Bcl-3 is an antagonist of p50/NF-κB-mediated inhibition. Nature. 1992;359:339–342. doi: 10.1038/359339a0. [DOI] [PubMed] [Google Scholar]
- 15.Fujita T, Nolan G P, Liou H C, Scott M L, Baltimore D. The candidate proto-oncogene Bcl-3 encodes a transcription coactivator that activates through NF-κB p50 homodimers. Genes Dev. 1993;7:1354–1366. doi: 10.1101/gad.7.7b.1354. [DOI] [PubMed] [Google Scholar]
- 16.Giri J, Ahdieh M, Eisenman J, Shanebeck K, Grabstein K, Kumaki S, Namen A, Park L, Cosman D, Anderson D. Utilization of the β and γ chains of the IL-2R by the novel cytokine IL-15. EMBO J. 1994;13:2822–2830. doi: 10.1002/j.1460-2075.1994.tb06576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gnarra J R, Otani H, Wang M, McBride O, Sharon M, Warren J L. Human IL-2Rβ chain gene: chromosomal localization and identification of 5′ regulatory sequences. Proc Natl Acad Sci USA. 1990;87:3440–3444. doi: 10.1073/pnas.87.9.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gómez J, Martinez-A C, Gonzalez A, Garcia A, Rebollo A. The Bcl-2 gene is differentially regulated by IL-2 and IL-4: role of the transcription factor NFAT. Oncogene. 1998;17:1235–1243. doi: 10.1038/sj.onc.1202049. [DOI] [PubMed] [Google Scholar]
- 19.Gómez J, Martínez-A C, Fernandez B, García A, Rebollo A. Ras activation leads to cell proliferation or apoptotic cell death upon IL-2 stimulation or lymphokine deprivation, respectively. Eur J Immunol. 1997;27:1610–1618. doi: 10.1002/eji.1830270704. [DOI] [PubMed] [Google Scholar]
- 20.Gurfinkel N, Unger T, Givol D, Mushinski J F. Expression of the Bcl-2 gene in mouse B lymphocytic cell lines is differentiation stage specific. Eur J Immunol. 1987;17:567–570. doi: 10.1002/eji.1830170421. [DOI] [PubMed] [Google Scholar]
- 21.Hatada E N, Nieters N, Wulczyn F G, Maumann M, Meyer R, Nucifora G, McKeithan T W, Schedereit C. The ankyrin repeat domains of the NF-κB precursor p105 and the protooncogene Bcl-3 act as specific inhibitors of NF-κB DNA binding. Proc Natl Acad Sci USA. 1992;89:2489–2493. doi: 10.1073/pnas.89.6.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimura Y, Takeshita T, Kondo M, Ishii N, Nakamura M, Van Snick J, Sugamura K. Sharing of the IL-2Rγ chain with the functional IL-9 receptor complex. Int Immunol. 1995;7:115–120. doi: 10.1093/intimm/7.1.115. [DOI] [PubMed] [Google Scholar]
- 23.Kondo M, Takeshita T, Higuchi M, Nakamura M, Sudo T, Nishikawa S, Sugamura K. Functional participation of the IL-2Rγ chain in IL-7R complexes. Science. 1994;263:1453–1454. doi: 10.1126/science.8128231. [DOI] [PubMed] [Google Scholar]
- 24.Kondo M, Takeshita T, Ishii N, Nakamura M, Watanabe S, Arai K, Sugamura K. Sharing of the IL-2Rγ chain between receptors for IL-2 and IL-4. Science. 1993;262:1874–1877. doi: 10.1126/science.8266076. [DOI] [PubMed] [Google Scholar]
- 25.Le Bail O, Schmidt-Ulrich R, Israel A. Promoter analysis of the gene encoding the MAD3 inhibitor of NF-κB: positive regulation by members of the rel/NF-κB family. EMBO J. 1993;12:5043–5049. doi: 10.1002/j.1460-2075.1993.tb06197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J X, Migone T S, Tsang M, Friedman M, Watherbee J A, Zhou L, Yamauchi A, Bloom E T, Mietz J, John S, Leonard W J. The role of shared receptor motifs and common Stat proteins in the generation of cytokine pleiotropy and redundancy by IL-2, IL-4, IL-7, IL-13 and IL-15. Immunity. 1995;2:331–339. doi: 10.1016/1074-7613(95)90141-8. [DOI] [PubMed] [Google Scholar]
- 27.Lux S E, John K M, Bennett V. Analysis of cDNA for human erythrocyte ankyrin indicates a repeated structure with homology to tissue-differentiation and cell cycle control proteins. Nature. 1990;344:36–42. doi: 10.1038/344036a0. [DOI] [PubMed] [Google Scholar]
- 28.Morin P J, Gilmore T D. The C-terminus of the NF-κB p50 precursor and a Iκ-B isoform contain transcription activation domains. Nucleic Acid Res. 1992;20:2453–2458. doi: 10.1093/nar/20.10.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noguchi M, Nakamura Y, Russell S M, Ziegler S F, Tsang M, Cao X, Leonard W J. IL-2Rγ chain: a functional component of the IL-4R. Science. 1993;262:1877–1880. doi: 10.1126/science.8266077. [DOI] [PubMed] [Google Scholar]
- 30.Nolan G P, Fujita T, Bhatia K, Huppi C, Liou H C, Scott M L, Baltimore D. The Bcl-3 protooncogene encodes a nuclear IκB like molecule that preferentially interacts with NF-κB p50 and p52 in a phosphorylation dependent manner. Mol Cell Biol. 1993;13:3557–3566. doi: 10.1128/mcb.13.6.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohno H, Takimoto G, McKeithan T W. The candidate proto-oncogene Bcl-3 is related to genes implicated in cell lineage determination and cell cycle control. Cell. 1990;60:991–999. doi: 10.1016/0092-8674(90)90347-h. [DOI] [PubMed] [Google Scholar]
- 32.Paul W. Interleukin 4: a prototypic immunoregulatory lymphokine. Blood. 1991;77:1859–1870. [PubMed] [Google Scholar]
- 33.Pitton C, Rebollo A, Van Snick J, Theze J, Garcia A. High affinity and intermediate affinity forms of the human interleukin 2 receptor expressed in an IL-9-dependent murine T cell line, deliver proliferative signals via differences in their transduction pathways. Cytokine. 1993;5:362–371. doi: 10.1016/1043-4666(93)90069-h. [DOI] [PubMed] [Google Scholar]
- 34.Quelle F W, Shimoda K, Thierfelder W, Fischer C, Kim A, Reuben S M, Cleveland J L, Pierce J H, Keegan A D, Nelms K, Paul W E, Ihle J N. Cloning of murine Stat 6, proteins that are tyrosine phosphorylated in response to IL-4 and IL-13 but are not required for mitogenesis. Mol Cell Biol. 1995;15:3336–3343. doi: 10.1128/mcb.15.6.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richard M, Louahed J, Demoulin J P, Renauld J C. Interleukin 9 regulates NF-κB activity through Bcl-3 gene induction. Blood. 1999;93:4318–4327. [PubMed] [Google Scholar]
- 36.Russell S, Johnston J, Noguchi M, Kawamura M, Bacon C, Friedman M, Berg M, McVicar D, Witthuhn B, Silvenhoinen O, Goldman A, Schmalstieg F, Ihle J, O'Shea J, Leonard W. Interaction of IL-2R β and γ chin with Jak1 and Jak3: implications for XSCID and XCIC. Science. 1994;266:1042–1045. doi: 10.1126/science.7973658. [DOI] [PubMed] [Google Scholar]
- 37.Russell S M, Keegan A D, Harada N, Nakamura Y, Noguchi M, Leland P, Friedmann M C, Miyajima A, Puri R, Paul W E, Leonard W J. IL-2Rγ chain: a functional component of the IL-4R. Science. 1993;262:1880–1883. doi: 10.1126/science.8266078. [DOI] [PubMed] [Google Scholar]
- 38.Schwarz E M, Krimpenfort P, Berns A, Verma I M. Immunological defects in mice with a targeted disruption in Bcl-3. Genes Dev. 1997;11:187–197. doi: 10.1101/gad.11.2.187. [DOI] [PubMed] [Google Scholar]
- 39.Sharon M, Siegel J P, Tosato G, Yodoi J, Gerrard T L, Leonard W. The human IL-2Rβ chain: direct identification, partial purification and patterns of expression on peripheral blood mononuclear cells. J Exp Med. 1988;167:1265–1270. doi: 10.1084/jem.167.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shiio Y, Sawada J, Handa H, Yamamoto T, Inoue J. Activation of the Rb gene expression by Bcl-3: implication for muscle cell differentiation. Oncogene. 1996;12:1837–1845. [PubMed] [Google Scholar]
- 41.Siebenlist U. NF-κB/IκB proteins: their role in the cell growth, differentiation and development. Biochim Biophys Acta. 1997;1332:7–13. doi: 10.1016/s0304-419x(96)00038-8. [DOI] [PubMed] [Google Scholar]
- 42.Siegel J P, Sharon M, Smith P L, Leonard W J. The IL-2Rβ chain: role in mediating signaling for LAK, NK and proliferative activities. Science. 1987;238:75–78. doi: 10.1126/science.3116668. [DOI] [PubMed] [Google Scholar]
- 43.Wulczyn F G, Krappmann D, Scheidereit C. Candidate proto-oncogene Bcl-3 encodes a subunit-specific inhibitor of transcription factor NF-κB. J Mol Med. 1992;74:749–769. doi: 10.1038/358597a0. [DOI] [PubMed] [Google Scholar]
- 44.Wulczyn F G, Naumann M, Scheidereit C. Candidate protooncogene Bcl-3 encodes a subunit specific inhibitor of transcription factor NF-κB. Nature. 1992;358:597–599. doi: 10.1038/358597a0. [DOI] [PubMed] [Google Scholar]
- 45.Zabel U, Henkel T, Silva M D, Baeuerle P A. Nuclear uptake control of NF-κB by MAD-3, an Iκ-B protein present in the nucleus. EMBO J. 1993;12:201–211. doi: 10.1002/j.1460-2075.1993.tb05646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang M, Harhaj E, Bell L, Sun S, Miller B A. Bcl-3 expression and nuclear translocation are induced by granulocyte-macrophage colony-stimulating factor and erythropoietin in proliferating human erythroid precursors. Blood. 1998;92:1225–1234. [PubMed] [Google Scholar]
- 47.Zhang Q, Didonato J A, Karin M, McKeithan T W. Bcl-3 encodes a nuclear protein which can alter the subcellular location of NF-κB proteins. Mol Cell Biol. 1994;14:3915–3926. doi: 10.1128/mcb.14.6.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]