Abstract
In the rapidly growing COVID-19 pandemic, designing of new drugs and evaluating their inhibitory action against main targets of corona virus could be an effective strategy to accelerate the drug discovery process and their efficacy towards corona virus disease. Herein, we design new bis-triazolyl probe for an investigation of inhibitory activity towards COVID-19 main protease by Molecular docking approach. The formulated compound has been thoroughly characterized by elemental analysis, NMR (1H and 13C) and complete structure elucidation was achieved via X-ray crystallography. Docking study reveals that newly synthesized compound confers good inhibitory response to COVID-19 main protease as supported by calculated docking score and binding energy. Strong hydrogen bonding and hydrophobic interactions of the newly synthesized compound with several important amino acids of the main protease also helps to explain the potency of the compound to inhibit the main protease. We hope that the present study would help the researcher in the field of Medicinal chemistry and to develop potential drug against the novel corona virus.
Keywords: COVID-19 main protease, Triazole, Crystallography, Molecular docking
1. Introduction
The ongoing outbreak of novel corona virus (COVID-19) pandemic has claimed more than 2 million human lives worldwide and has posed notable threat to international health and economy [1], [2], [3], but an effective therapeutic intervention has yet to be developed. COVID-19 is caused by new corona virus called severe acute respiratory syndrome corona virus 2 (SARS-CoV-2) and was first reported in December 2019 in Wuhan City, China. The crisis has motivated the researchers from different fields of science towards vaccine finding against this novel disease.
Drug designing in the pharmaceutical industry is largely based on the use of different organic moieties as a constructing unit because it can provide novel libraries of biologically active molecules with improved pharmacokinetic properties [4]. There are many heterocyclic ring structures, which have been designed in such a way that their binding efficiency with the receptors increases after structural modifications [5], [6]. One of the most requisite moieties is triazoles, which have been explored and still its scope is inevitable. These are five-membered heterocyclic frameworks which can be readily obtained in good to excellent yield on the multi-gram scale by click chemistry via reaction of aryl/alkyl halides, alkynes and sodium azide (NaN3) under ambient conditions [7]. These motifs are effective amide surrogates in bioactive molecules because of their strong dipole moments. Triazoles can also be used as a linker and shows bioisosteric effects on peptide linkage, aromatic ring, double bonds and an imidazole ring. Some unique features like hydrogen bond formation, dipole–dipole and pi-stacking interactions of triazole compounds have increased their importance in the field of medicinal chemistry as they bind with the biological target with high affinity due to their improved solubility. It has been an emerging area of interest for many researchers throughout the globe owing to its immense pharmacological scope as it exhibits anti-HIV, anti-malarial, anticancer, antibacterial, antifungal, and anti-TB properties [8], [9], [10], [11], [12], [13], [14]. Some drugs currently in use are based on triazoles especially 1,2,3–triazole moiety such as anti-HIV agent TSAO, Cefatrizine (antibiotic), antibacterial agent Tazobactum, anti-TB agent I-A09 and anti cancer agent CIA [15]. Furthermore, Pantane-3-yl group containing substances play a pivotal role in drug chemistry and also have been reported to display potent binding inhibition activity against a corticotrophin-releasing factor 1 (CRF1) receptor and in vitro antagonistic activity, strong anti-proliferative activity against several tumor cell lines, including multi-drug resistant phenotypes [16].
For many viruses, the protease enzyme plays a critical role in viral protein maturation by cleaning pro-proteins after their translation into the host cell cytosol. As a result, viral proteases are often potential drug targets [17]. The inhibition of viral protease can reduce the assembly of mature viral particles. Molecular docking is one of the best and fast techniques in the scientific community for rational design of drugs. Docking addresses the binding between drugs and proteins via active sites determination. Keeping the above perspectives in mind, herein, we report the synthesis, spectral characterization and crystal structure of new bis-triazolyl probe for an investigation of inhibitory activity towards COVID-19 main protease (Mpro) by Molecular docking approach. The formulated compound has been thoroughly characterized by elemental analysis NMR (1H and 13C) and complete structure elucidation was achieved via X-ray crystallography.
The development of a candidate drug into an approved drug is a long and costly process. Hence, designing a drug molecule on the basis of selected important functional moieties and blind identification of their inhibitory activity against main viral protease with the help of computational methodologies such as molecular docking, virtual screening and binding free energy evaluation serves as a promising alternative for identifying potential drug candidates to combat COVID-19 pandemic.
2. Experimental section
2.1. Material and methods
9-Bromopentane (Aldrich), 4-bromo-2-hydroxybenzaldehyde (Aldrich), 1,2-dibromoethane (Avra), Sodium azide (Aldrich), Propargyl Bromide (80% in toluene, Aldrich), potassium carbonate (Avra), dibromoethane (Aldrich), NaN3 (Aldrich), bromotris (triphenylphosphine) copper (I) (Sigma Aldrich) were used as received. Solvent were purified and dried before use. 1H and 13C NMR spectra were recorded on a JEOL (AL 400 MHz) spectrometer using CDCl3 as an internal reference and chemical shifts were reported relative to TMS, values are given in Hz. The FT-IR spectra were recorded on a Thermo Scientific NICOLET IS50 spectrophotometer. Melting points were measured in a Mel Temp II device using sealed capillaries and were uncorrected
2.2. X-ray diffraction analysis and data collection
Single crystals of compound 6 suitable for X-ray diffraction were obtained by slow evaporation from a saturated chloroform solution at room temperature. A suitable crystal was carefully selected under polarizing microscope in order to perform its structural analysis by X-ray diffraction. Diffraction data were collected on a Super Nova, Single source at offset/far, HyPix3000 diffractometer. The crystal was kept at 298(2) K during data collection, using Olex2 [18]. The structure was solved with the SHELXT [19] structure solution program using direct methods and refined with the SHELXL [20] refinement package using Least Squares minimization. The drawings were made with Diamond and Mercury 4.0.0. Crystal Data for compound 6 C32H38N6O4 (M = 570.68 g/mol): monoclinic, space group P21/c (no. 14), a = 5.498(3) Å, b = 20.526(4) Å, c = 13.2792(16) Å, β = 93.06(3)°, V = 1496.3(10) Å3, Z = 2, T = 298(2) K, μ(Mo Kα) = 0.085 mm−1, Dcalc = 1.267 g/cm3, 971 reflections measured (6.458° ≤ 2Θ ≤ 46.542°), 741 unique (R int = 0.0466, Rsigma = 0.1137) which were used in all calculations. The final R 1 was 0.0972 (I > 2σ (I) and wR 2 was 0.3035 (all data). The goodness-of-fit was 1.000. CCDC 2094938 contains the supplementary crystallographic data for compound 6. Crystallographic parameters and basic information pertaining to data collection and structure refinement for the compound are summarized in Table 1 .
Table 1.
Crystal data and structure refinement for compound 6.
| Identification code | 6 |
| Empirical formula | C32H38N6O4 |
| Formula weight | 570.68 |
| Temperature/K | 298(2) |
| Crystal system | monoclinic |
| Space group | P21/c |
| a/Å | 5.498(3) |
| b/Å | 20.526(4) |
| c/Å | 13.2792(16) |
| α/° | 90 |
| β/° | 93.06(3) |
| γ/° | 90 |
| Volume/Å[3] | 1496.3(10) |
| Z | 2 |
| ρcalcg/cm[3] | 1.267 |
| μ/mm-1 | 0.085 |
| F(000) | 608.0 |
| Crystal size/mm3 | 0.19 × 0.15 × 0.08 |
| Radiation | Mo Kα (λ = 0.71073) |
| 2Θ range for data collection/° | 6.458 to 46.542 |
| Index ranges | −4 ≤ h ≤ 1, -15 ≤ k ≤ 13, -12 ≤ l ≤ 13 |
| Reflections collected | 971 |
| Independent reflections | 741 [Rint = 0.0466, Rsigma = 0.1137] |
| Data/restraints/parameters | 741/42/192 |
| Goodness-of-fit on F2 | 1.053 |
| Final R indexes [I>=2σ (I)] | R1 = 0.0971, wR2 = 0.2457 |
| Final R indexes [all data] | R1 = 0.1374, wR2 = 0.3035 |
| Largest diff. peak/hole / e Å−3 | 0.25/−0.22 |
2.3. Synthesis
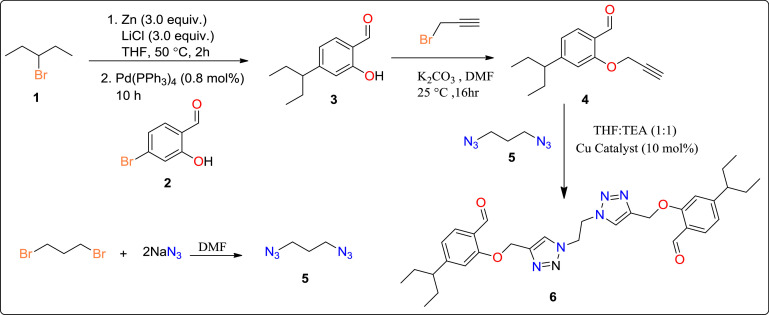
Initially 3-bromopentane (1) was made to react with Zinc and lithium chloride in minimum amount of THF at 50 °C for 2 h followed by addition of 4-bromo-2-hydroxybenzaldehyde (2) and Pd(PPh3)4 and continued to stir for 10 h (scheme 1 ). The reaction was monitored by TLC and on complete consumption of starting materials, the resulting reaction mixture was allowed to cool and filtered to get 2-hydroxy-4-(pentan-3-yl)benzaldehyde (3). Further, compound 3 was dissolved in minimum amount of DMF along with 1.5 equiv of anhydrous K2CO3 and allowed to react with propargyl bromide, which was added slowly and allowed to stir at room temperature for 16 h to get 2-hydroxy-4-(pentan-3-yl)benzaldehyde (4). The progress of the reaction was monitored by TLC. Finally, to get the desired product 6, compound 4 was allowed to react with diazidoethane (5) in presence of copper catalyst (10 mol%) using THF:TEA (1:1) as solvent. The crude product was further purified by column chromatography to get pure compound 6 and thoroughly characterised by NMR and IR. 1,2-diazidoethane (5) was synthesized according to the reported literature by reaction of 1,3-dibromopropane with sodium azide in DMF [21].
Scheme 1.
Synthetic route for compound 6.
NMR of compound 6: Yield 96% (dark brown solid), Melting pt. 99–101 °C. Anal. Calcd. For C32H38N6O4: C, 67.11; H, 7.04; N, 14.67; Found: C, 67.11; H, 7.02; N, 14.50. IR (cm−1): 1660 (C O), 1258 (O—CH2), 1560 (C C), 2876, 2925, 2964 (CH). 1H NMR (500 MHz, CDCl3): δ 1.19–1.22 (t, 12 H, J = 7.08), 2.88 (s, 2H), 3.41–3.44 (m, 8H), 4.9 (s, 4H), 5.2 (s, 4 H), 6.22–6.28 (m, 4H), 7.42 (s, 2H), 7.63–7.65 (d, 2H, J = 8.93), 10.01 (s, 2H); 13C NMR (126 MHz, DMSO): δ 190.97, 190.92, 162.84, 160.62, 142.07, 131,65, 129.82, 124.98, 115.51, 114.54, 61.39, 55.74, 49.03, 39.42, 39.25. Experimental characterization of compound 6 is reported in supplementary information.
2.4. Molecular docking protocol
Required PDB file for the crystal structure of COVID-19 main protease (PDB ID: 6LU7) downloaded from Protein Data Bank (https://www.rcsb.org) and the Ligand (compound 6) for screening was synthesized according to scheme 1. The ligand was drawn in the Avogadro molecule editor, and then their stable structure obtained by energy minimization with the MMFF94 Force Field and converted into PDB format with the help of 3D-chemdraw. The 6LU7 protein structure was prepared by removing all water molecules, assignment of Gasteiger partial charges and adding polar hydrogen. Ligand position in Mpro protein was set at grid coordinates x = −18.955, y = −5.188 and z = 8.617 Genetic algorithm (GA) parameters were assigned at 100 GA run and population size of 150. Molecular docking calculations were performed by using Autodock 4.2 with the aid of Auto Dock Tools 1.5.6 [22]. Docked structures and the active sites inter-action were visualized by Discovery Studio Visualizer v20.1.0.19295 [23].
3. Results and discussion
3.1. Synthesis and characterization
A three step synthetic route, shown in scheme 1, was employed to synthesize compound 6. Firstly, 3-bromopentane (1) was made to undergo Negishi coupling with 4-bromo-2-hydroxybenzaldehyde (2) in THF to yield 2-hydroxy-4-(pentan-3-yl)benzaldehyde (3). The obtained compound 3 was then stirred at room temperature with propargyl bromide in DMF using potassium carbonate as base to give 2-hydroxy-4-(pentan-3-yl)benzaldehyde (4). Finally, desired product 6 was synthesized by reaction between compound 4 and 1,2-diazidoethane (5) using copper catalyst in THF: TEA (1:1). The structure and purity of all the compounds were inferred from their infrared and NMR (1H and 13C) spectra. The FT-IR spectra of the newly synthesized compounds were recorded as neat spectra in the range of 4000–400 cm−1. The absorption frequencies recorded were found to be in close agreement with the structure of the synthesized compounds. The typical bands observed for the carbonyl group appeared near 1660 cm−1 and 1258 cm−1 corresponding to OCH2. The absorption band for the aromatic C C vibrations and C—H stretching appears in the region of 1500–1600 cm−1 and for alkyl chain C—H stretching gives a medium absorption band around 2850–2900 cm−1 and aromatic C—H stretching gives absorption band in the region of 3076–3060 cm−1. The 1H NMR spectra of compound 6 have demonstrated the symbolic peaks of aldehydic (HC O) protons at 10.01 ppm. Signals in the region of 6.22–6.28 ppm are assigned for aryl ring protons. Peak at 5.22 ppm are assigned for alkyl protons (-O-CH2-) and peak at 7.42 ppm are observed for triazole ring protons. In 13C NMR spectra, peak corresponds to –C O group at 190.97 ppm, peak at 142.07 ppm and 128.04 ppm to the -C C- of triazole ring and at 55.63 ppm to alkyl protons validate the product formation. Moreover, the aromatic carbon peaks were seen in the region of 162.95 ppm to 115.08 ppm.
3.2. Crystal structure
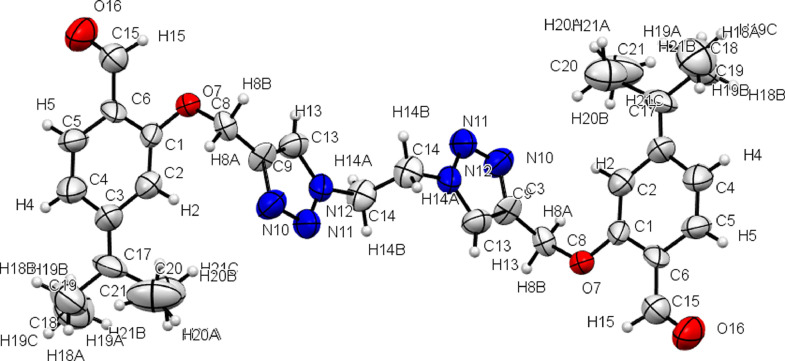
Single crystal X-ray diffraction studies disclose that the compound 6 crystallized in monoclinic crystal system containing eight molecules per unit cell with P21/c space group and has comparable structure, as is evident from the bond angles and bond parameters (Table S2). Crystallographic data and structure determination parameters are summarized in Table 1. The ORTEP diagram and atomic labeling for empirical formula C32H38N6O4 (6) is shown in Fig. 1 (Thermal ellipsoid have been drawn at 40% probability level). In the molecules both the triazoles ring are in the parallel plane and Pent-3-yl substituted aromatic ring assume to lie above and below (anti-position) the plane at an angle of 112.57(13)˚ with an extended conformation which can be seen from the C1-O7-C8-C9 torsion angles of 75.4(17)˚. The pent-3-yl group in crystal structure lies in twisted conformation and the whole structure looks like a dumbbell (Fig. 2 ).
Fig. 1.
ORTEP diagram of compound 6 with thermal ellipsoids at 40% probability, selected bond length (Å) and angle (˚) O7-C1 = 1.347(13), O7-C8 = 1.43(2), N11-N12 = 1.34(3), N12-C13 = 1.354(18), C15-C6 =1.45(3), N12-C14 = 1.460(15), N10-C9 = 1.36(2), C1-O7-C8 = 118.7(13), N10-N11-N12 = 107.1(14), C13-N12-C14 =127.4(17), O7-C1-C6 = 115.3(16), O7-C8-C9 = 112.7(13), C19-C18-C17 = 113.7(2).
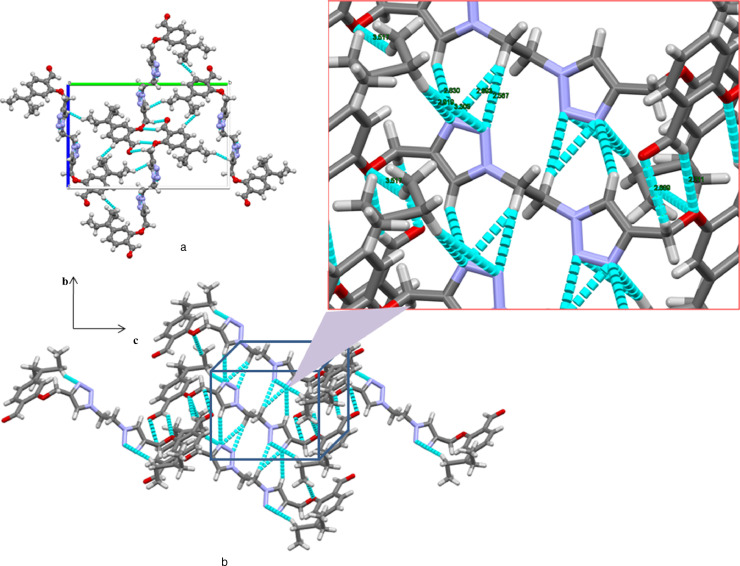
Fig. 2.
(a) Packing arrangements of compound 6 in the unit cell along a-axis (b) Non-classical interactions and hydrogen bonding interaction with zoomed portion.
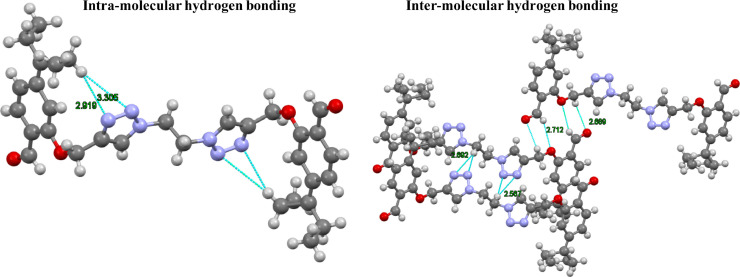
In molecular lattice arrangement of compound 6 along a-axis, molecules are packed via strong intra-molecular hydrogen-bonding interactions (N•••H—C) 2.919 Å and 3.305 Å involving N10 and N11 of triazole ring with H20B of the pent-3-yl chain in the molecule (Fig. 3 , left). Furthermore, N10 of triazole ring of one molecule linked with H13 of the adjacent triazole ring of another molecule and O7 of one molecule linked with the H21 of the adjacent pent-3-yl chain of the another molecule via strong intermolecular hydrogen bonding interaction (N•••H—C) type 2.830 Å and (O•••H—C) 3.517 Å. Similarly O16 of the aldehyde group linked with the H18A and H20A of pent-3-yl chain of another molecule via intermolecular hydrogen bong type (O•••H—C) 2.741 and 2.839 Å. Fig. 3 (right) and overall packing looks like plane sheet (Fig. 4 ).
Fig. 3.
Left image shows intra-molecular hydrogen bonding (N•••H—C); Right image shows inter-molecular hydrogen bonding between (N•••H—C) and (O•••H—C).
Fig. 4.
Molecular lattice arrangements for compound 6 along a-axis shows molecules are joined by non-classical interactions and strong molecular hydrogen bonding.
Moreover, the pair of molecules in unit cell are joined by non-classical interactions of (C—H•••O) type between H15 of one moiety and O7 of another moiety with bond length of 2.711 Å and O16 of the carbonyl group with H8B of another molecules leading to a closed packing structure.
3.3. Molecular docking study
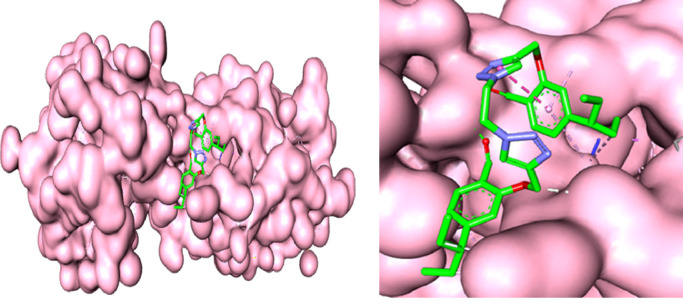
Mpro also termed 3CL protease which is a potential drug target for corona virus infections due to its essential role in processing the poly-proteins that are translated from the viral RNA and mediates the maturation of functional polypeptides involved in the assembly of replication-transcription machinery [24]. Mpro digests the poly-protein at no. less than 11 conserved sites, starting with the autolytic cleavage of this enzyme itself from pp1a and pp1ab. In addition, Mpro has no human homolog and is highly conserved among all corona-virus25. These above features make it an attractive drug target against corona viruses. Therefore, in order to find out the binding conformation of synthesized compound 6 within the active sites of main protease of COVID-19 and to obtain additional validations for experimental results, molecular docking was performed. All single bonds were considered as rotatable and fitness function was given as the score and all the calculations were made by using standard default settings. After Docking several pose was saved for compound 6 out of them best are shown in Fig. 5 .
Fig. 5.
Surface diagram of the main protease along with docked conformations for compound 6 (left); zoomed picture of compound interactions with cavity of protein surface (right).
The compound 6 was found to be most promising molecule and neatly fits in the active site of protein making various hydrogen bonding interactions through lone pair of hetero-atoms and pi-bonds present in compound with the protein active site amino acid residues including Sys156, Tyr154, Ile152, Phe8, Phe294, Asp295 and Arg298 shown in Fig. 6 a and 6b with 3D and 2D diagrams respectively.
Fig. 6.
(a) 3D Interactions of compound 6 docked in active site of protease of COVID-19; (b) 2D diagram showing hydrogen bonding (green), pi-donor hydrogen bond (light green), pi-Alkyl interactions (light pink).
Visual inspection of the highly suitable pose of compound 6 revealed that the methyl part of the pent-3-yl and pi-bond in benzene ring shows strong interaction with oxygen of carboxyl group on the side chain of Ile152, Pro9, Tyr 154 and Phe294 respectively. Moreover, nitrogen atom of triazole ring of the compound 6 is interacting with Phe8 by strong hydrogen bonding and pi-anion interactions between side chain carboxyl oxygen of Phe8. Moreover, carboxyl oxygen of compound 6 is interacting with Arg298 by strong hydrogen bonding interactions (O•••H—N) shown in Fig. 6. Directed bonds between protein and ligand are drawn as black dashed lines and the interacting protein residues and the ligand are visualized as structure diagrams. Hydrophobic contacts are represented more indirectly by means of spline sections highlighting the hydrophobic parts of the ligand and the label of the contacting amino acid. Fig. 7 b On the contrary, all interaction between compound 8 and the protein residues summarized in Table 2 a and 2b.
Fig. 7.
Interactions established after docking of compound 6 with 6lu7 Mpro protease; (a) Three dimensional representation of intermolecular interaction in complex (compound 6/6lu7) Mpro; (b) 2D plot showing both hydrogen and hydrophobic interactions.
Table 2.
(a) Hydrophobic Interactions (b) Hydrogen Bonds of complex (compound 6/6lu7).
|
2a. Hydrophobic Interactions |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Index | Residue | AA | Distance | Ligand Atom | Protein Atom | ||||
| 1 | 8A | PHE | 3.65 | 2903 | 80 | ||||
| 2 | 9A | PRO | 3.94 | 2900 | 90 | ||||
| 3 | 154A | TYR | 3.20 | 2925 | 1460 | ||||
| 4 | 294A | PHE | 3.64 | 2922 | 2771 | ||||
| 5 | 294A | PHE | 3.25 | 2923 | 2773 | ||||
|
2b. Hydrogen Bonds | |||||||||
| Index | Residue | AA | Distance H-A | Distance D-A | Donor Angle | Protein donor? | Sidechain | Donor Atom | Acceptor Atom |
| 1 | 298A | ARG | 1.96 | 2.87 | 147.23 |  |
 |
2813 [Ng+] | 2911 [O2] |
| 2 | 298A | ARG | 2.07 | 2.96 | 144.67 |  |
 |
2807 [Ng+] | 2911 [O2] |
The binding energy (EB) of compound 6 was −6.43 kcal/mol with rmsd value, 0.00. The more negative binding energy (EB) and smaller value of inhibition constant (Ki) implies best docking score. Hydrogen bonds are primary contributing factor in supporting the binding affinity of drugs with the receptor. Strong hydrogen bonding interaction represents a high binding capability between ligand and protein which points toward the strong inhibitory activity of synthesized compound 6 against the main protease of corona virus (Table 3 ).
Table 3.
The binding affinity and docked structures of the synthesized compound 6 with main protease (6LU7) of COVID-19.
| Comp. | Protein | Compound Docked structure | EB (Kcal/mol) | Ki (uM) | RMSDb | Hydrogen bond (˚A) |
|---|---|---|---|---|---|---|
| 6 | 6LU7 |  |
−6.43 | −51.19 | 0.00 | ARG298A 1.96 |
| ARG298A 2.07 | ||||||
4. Conclusion
In summary, we have successfully synthesized a new bis-triazolyl probe via click chemistry. The synthesis is very simple, high yielding and employs commercially available starting materials. The molecular structure of the compound 6 is authenticated by single crystal X-ray diffraction analysis showing intra and intermolecular hydrogen bonding which has great importance in many biological properties of the compound in medicinal chemistry. Docking analysis showed interactions of synthesized compound with Mpro of COVID-19 with binding affinity of −6.43 kcal/mol and concludes that tested compound can serve as a potential Mpro inhibitor. In brief, the present work opens a promising approach to design new drug and will probably be useful in molecular modeling, drug designing and wider scope in medicinal chemistry.
Declaration of Competing Interest
There is no conflict of interest.
Acknowledgment
The authors are thankful to CSIR (01(2950)/18/EMR-11, New Delhi, DST-PURSE, DST-SERB (SB/FT/CS-132/2014) for providing financial support and University Grant Commission (UGC), New Delhi, for junior research fellowship.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.molstruc.2021.131858.
Appendix. Supplementary materials
Supporting information contains 1H NMR,13C NMR and single X-Ray structure data, bond parameters and CCDC number (2094938) of compound 6 crystal.
References
- 1.Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of corona virus disease (COVID-19) outbreak. J. Autoimmun. 2020;109 doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sohrabi C., Alsafi Z., O ’Neill N., et al. World Health Organization declares global emergency: a review of the 2019 Novel Coronavirus (COVID-19) Int. J. Surg. 2020;76 doi: 10.1016/j.ijsu.2020.02.034. 71-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shanmugaraj B., Malla A., Phoolcharoen W. Emergence of novel coronavirus 2019 nCoV: need for rapid vaccine and biologics development. Pathogens. 2020;9:148–158. doi: 10.3390/pathogens9020148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El-Dakdouki M.H., Erhardt P.W. Pure Appl. Chem. 2012;84:1479–1542. [Google Scholar]
- 5.a) Baumann M., Baxendale I.R. An overview of the synthetic routes to the best selling drugs containing 6-membered heterocycles. Beilstein J. Org. Chem. 2013;9:2265–2319. doi: 10.3762/bjoc.9.265. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Hopkinson M.N., Richter C., Schedler M., Glorius F. An overview of N-heterocyclic carbenes. Nature. 2014;510:485–496. doi: 10.1038/nature13384. [DOI] [PubMed] [Google Scholar]; c) Selvam T.P., James C.R., Dniandev P.V., Valzita S.K. A mini review of pyrimidine and fuse pyrimidine marketed drugs. Res. Pharm. 2012;2:1–9. [Google Scholar]; d) Shankar R., Chakravarti B., Singh U.S., Ansari M.I., Deshpande S., Dwivedi S.K.D., Bid H.K., Konwar R., Kharkwal G., Chandra V., Dwivedi A., Hajela K. Synthesis and biological evaluation of 3,4,6-triaryl-2-pyranones as a potential new class of anti-breast cancer agents. Bioorg. Med. Chem. 2009;17:3847–3856. doi: 10.1016/j.bmc.2009.04.032. [DOI] [PubMed] [Google Scholar]; e) Shankar R., Jha A.K., Singh U.S., Hajela K. An efficient and improved synthesis of 1, 5-diketones: versatile conjugate addition of nucleophiles to a,b-unsaturated enones and alkynones. Tetrahedron Lett. 2006;47:3077–3079. [Google Scholar]
- 6.a) Welsch M.E., Snyder S.A., Stockwell B.R. Privileged scaffolds for library design and drug discovery. Curr. Opin. Chem. Biol. 2010;14:1–15. doi: 10.1016/j.cbpa.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Dua R., Shrivastava S., Sonwane S.K., Srivastava S.K. Pharmacological significance of synthetic heterocycles scaffold: a review. Adv. Biol. Res. 2011;5:120–144. [Google Scholar]; c) Zhang H., Yang Y., Steinbrecher U.P. Structural requirements for the binding of modified proteins to the scavenger receptor of macrophage. J. Biol. Chem. 1993;268:5535–5542. [PubMed] [Google Scholar]; d) Kumar B., Singh V., Shankar R., Kumar K., Rawal R.K. Synthetic and medicinal prospective of structurally modified curcumins. Curr. Top. Med. Chem. 2016;16:1–14. doi: 10.2174/1568026616666160605050052. [DOI] [PubMed] [Google Scholar]; e) Singh U.S., Shankar R., Kumar A., Trivedi R., Chattopadhyay N., Shakya N., Palne S., Gupta S., Hajela K. Synthesis and biological evaluation of indolyl bisphosphonates as anti-bone resorptive and anti-leishmanial agents. Bioorg. Med. Chem. 2008;16:8482–8491. doi: 10.1016/j.bmc.2008.08.024. [DOI] [PubMed] [Google Scholar]; f) Singh U.S., Shankar R., Yadav G.P., Kharkwal G., Dwivedi A., Keshri G., Singh M.M., Moulik P.R., Hajela K. Synthesis and structure guided evaluation of estrogen agonist and antagonist activities of some new tetrazolyl indole derivatives. Eur. J. Med. Chem. 2008;43:2149–2158. doi: 10.1016/j.ejmech.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 7.a) Da Silva F.D.C., De Souza M.C.B.V., Frugulhetti I.I.P., Castro H.C., Souza S.L.D.O., De Souza T.M.L., Rodrigues D.Q., Souza A.M.T., Abreu P.A., Passamani F., Rodrigues C.R., Ferreira V.F. Synthesis, HIV-RT inhibitory activity and SAR of 1-benzyl-1H-1,2,3-triazole derivatives of carbohydrates. Eur. J. Med. Chem. 2009;44:373–383. doi: 10.1016/j.ejmech.2008.02.047. [DOI] [PubMed] [Google Scholar]; b) Giffin M.J., Heaslet H., Brik A., Lin Y.C., Cauvi G., Wong C.-H., McRee D.E., Elder J.H., Stout C.D., Torbett B.E. A copper(I)-catalyzed 1,2,3-triazole azide- alkyne click compound is a potent inhibitor of a multidrug-resistant HIV-1 protease variant. J. Med. Chem. 2008;51:6263–6270. doi: 10.1021/jm800149m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Cocklin S., Gopi H., Querido B., Nimmagadda M., Kuriakose S., Cicala C., Ajith S., Baxter S., Arthos J., Martin-Garcia J., Chaiken I.M. Broad-spectrum antihuman immunodeficiency virus (HIV) potential of a peptide HIV type 1 entry inhibitor. J. Virol. 2007;81:3645–3648. doi: 10.1128/JVI.01778-06. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Brik A., Alexandratos J., Lin Y.C., Elder J.H., Olson A.J., Wlodawer A., Goodsell D.S., Wong C.-H. 1,2,3-Triazole as a peptide surrogate in the rapid synthesis of HIV-1 protease inhibitors. Chem. Bio. Chem. 2005;6:1167–1169. doi: 10.1002/cbic.200500101. [DOI] [PubMed] [Google Scholar]
- 9.a) Yu W.J., Rao Q., Wang M., Tian Z., Lin D., Liu X.R., Wang J.X. The Hsp90 inhibitor 17-allylamide-17- demethoxygeldanamycin induces apoptosis and differentiation of Kasumi-1 harboring the Asn822Lys KIT mutation and down-regulates KIT protein level. Leukemia Res. 2006;30:575–582. doi: 10.1016/j.leukres.2005.08.028. [DOI] [PubMed] [Google Scholar]; b) Peterson L.B., Brian Blagg S.J. Click chemistry to probe Hsp90: synthesis and evaluation of a series of triazole-containing novobiocin analogs. Bioorg. Med. Chem. Lett. 2010;20:3957–3960. doi: 10.1016/j.bmcl.2010.04.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) Nahrwold M., Bogner T., Eissler S., Verma S., Sewald N. ‘Clicktophycin- 52”: a bioactive cryptophycin-52 triazole analogue. Org. Lett. 2010;12:1064–1067. doi: 10.1021/ol1000473. [DOI] [PubMed] [Google Scholar]; b) Doiron J., Soultan A.H., Richard R., Toure M.M., Picot N., Richard R., Cuperlovic-Culf M., Robichaud G.A., Touaibia M. Synthesis and structure activity relationship of 1- and 2-substituted-1,2,3-triazole letrozole-based analogues as aromatase inhibitors. Eur. J. Med. Chem. 2011;46:4010–4024. doi: 10.1016/j.ejmech.2011.05.074. [DOI] [PubMed] [Google Scholar]
- 11.Fichtali I., Chraibi M., Aroussi F.E., Ben-Tama A., Hadrami E.M.E., Benbrahim K.F., Stiriba S.E. Synthesis of some 1,2,3-triazoles derivatives and evaluation of their antimicrobial activity. Der. Pharma. Chem. 2016;8:236–242. [Google Scholar]
- 12.Abdel-Wahab B.F., Mohamed H.A., Awad G.E.A. Synthesis and biological activity of some new 1,2,3-triazole hydrazone derivatives. Eur. Chem. Bull. 2015;4:106–109. [Google Scholar]
- 13.Raja R., Singh P., Singh P., Gut J., Rosenthal P.J., Kumar V. Azide-alkyne cycloaddition en route to 1H-1,2,3-triazole-tethered 7-chloroquinoline-isatin chimeras: Synthesis and antimalarial evaluation. Eur. J. Med. Chem. 2013;62:590–596. doi: 10.1016/j.ejmech.2013.01.032. [DOI] [PubMed] [Google Scholar]
- 14.Nagesh H.N., Naidu K.M., Rao D.H., Sridevi J.P., Sriram D., Yogeeswari P., Gowri K.V., Sekhar C. Design, synthesis and evaluation of 6-(4-((substituted-1H-1,2,3-triazol-4-yl)methyl)piperazin-1-yl)phenanthridine analogues as antimycobacterial agents. Bioorg. Med. Chem. Lett. 2013;23:6805–6810. doi: 10.1016/j.bmcl.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 15.Mochizuki M., Kojima T., Kobayashi K., Kotani E., Ishichi Y., Kanzaki N., Nakagawa H., Okuda T., Kosugi Y., Yano T., Sako Y., Tanaka M., Aso K. Discovery of 4-chloro-2-(2,4-dichloro-6-methylphenoxy)-1-methyl-7-(pentan-3-yl)-1H-benzimidazole, a novel CRF1 receptor antagonist. Bioorg. Med. Chem. 2017;25:1556–1570. doi: 10.1016/j.bmc.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 16.Cui W., Yang K., Yang H. Front. Mol. Biosci. 2020 doi: 10.3389/fmolb.2020.616341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dolomanov O.V., Bourhis L.J., Gildea R.J., Howard J.A.K., Puschmann H. OLEX2: a complete structure solution, refinement and analysis program. J. Appl. Cryst. 2009;42:339–341. [Google Scholar]
- 18.Sheldrick G.M. Acta Cryst. A. 2015;71:3–8. doi: 10.1107/S2053273314026370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheldrick G.M. Crystal structure refinement with SHELXL. Acta Cryst. C. 2015;71:3–8. doi: 10.1107/S2053229614024218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Betzler F.M., Klapötke T.M., Sproll S.M. Eur. J. Org. Chem. 2013:509–514. [Google Scholar]
- 21.Morris G.M., et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dassault Systèmes; San Diego, CA, USA: 2017. Dassault Systèmes BIOVIA Discovery Studio Modelling Environment, Release 2017. [Google Scholar]
- 23.H. M. Mengist, T. Dilnessa, T Jin, Front. Chem. 9:622898. [DOI] [PMC free article] [PubMed]
- 24.Xue X., Yu H., Yang H., Xue F., Wu Z., Shen W., Li J., Zhou Z., Ding Y., Zhao Q., Zhang X.C., Liao M., Bartlam M., Rao Z. Structures of Two Coronavirus Main Proteases: Implications for Substrate Binding and Antiviral Drug Design. J. Virol. 2008;82:2515–2527. doi: 10.1128/JVI.02114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information contains 1H NMR,13C NMR and single X-Ray structure data, bond parameters and CCDC number (2094938) of compound 6 crystal.