Abstract
Variability in SARS-CoV-2 susceptibility and COVID-19 disease severity between individuals is partly due to genetic factors. Here, we identify 4 genomic loci with suggestive associations for SARS-CoV-2 susceptibility and 19 for COVID-19 disease severity. Four of these 23 loci likely have an ethnicity-specific component. Genome-wide association study (GWAS) signals in 11 loci colocalize with expression quantitative trait loci (eQTLs) associated with the expression of 20 genes in 62 tissues/cell types (range: 1:43 tissues/gene), including lung, brain, heart, muscle, and skin as well as the digestive system and immune system. We perform genetic fine mapping to compute 99% credible SNP sets, which identify 10 GWAS loci that have eight or fewer SNPs in the credible set, including three loci with one single likely causal SNP. Our study suggests that the diverse symptoms and disease severity of COVID-19 observed between individuals is associated with variants across the genome, affecting gene expression levels in a wide variety of tissue types.
Keywords: COVID-19, SARS-CoV-2, GWAS, eQTL, colocalization
Graphical abstract
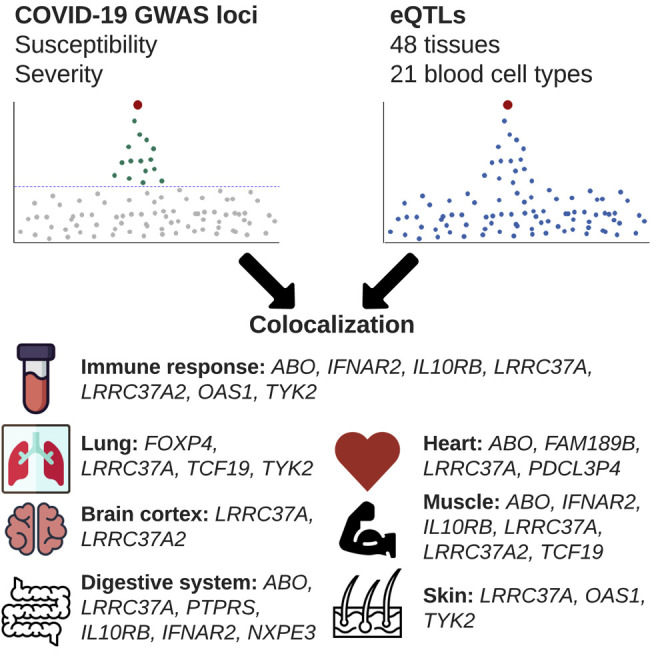
D’Antonio et al. characterize associations between GWAS signals for COVID-19 disease and eQTLs in 69 human tissues to identify causal variants and their underlying molecular mechanisms. They show that diverse symptoms and disease severity of COVID-19 are associated with variants affecting gene expression in a wide variety of tissues.
Introduction
The SARS-CoV-2 virus has infected more than 220 million individuals worldwide between December 2019 and August 2021, and its associated disease (COVID-19) has reportedly caused >4.5 million deaths (Worldometer, 2021). Several risk factors associated with SARS-CoV-2 infection and COVID-19 disease severity have been identified, including older age; sex; ethnicity; blood type; and cardiovascular, respiratory, and kidney diseases (Atkins et al., 2020; Cummings et al., 2020; Guan et al., 2020; Horowitz et al., 2021; Mackey et al., 2021; Richardson et al., 2020; Zhou et al., 2020). Most of these risk factors have a well-known genetic component (Aragam et al., 2018; Kulminski et al., 2018; Sakornsakolpat et al., 2019), suggesting that risk of SARS-CoV-2 infection and/or disease severity may also be influenced by the genetic background of an individual. The COVID-19 Host Genetics Initiative (COVID-19 HGI, https://www.covid19hg.org/) is currently leading a public effort worldwide to analyze COVID-19 information for millions of individuals in conjunction with genotype data in order to identify genetic variants associated with SARS-CoV-2 infection as well as COVID-19 hospitalization and disease severity (COVID-19 Host Genetics Initiative, 2020, 2021). To date, the COVID-19 HGI has conducted meta-analyses on one SARS-CoV-2 susceptibility phenotype (C2, COVID-19 patients versus population) and three COVID-19 disease severity phenotypes (A2, very severe respiratory confirmed COVID-19 versus population; B1, hospitalized versus non-hospitalized COVID-19 patients; and B2, hospitalized versus population) by combining 46 studies worldwide. Of note, B1 and B2 are both COVID-19 hospitalization phenotypes and differ only in their controls. Given that the controls for B1 included only non-hospitalized COVID-19 patients, it has less power to detect associations compared with the A2 and B2 disease severity phenotypes. For each phenotype, four distinct meta-analyses were performed by either including only individuals from European descent (referred to as European-only hereafter) or individuals from multiple ethnicities (referred to as multi-ethnic hereafter) and by either including or excluding individuals from the UK Biobank (UKBB) as controls. Recently published meta-analyses on the A2, B2, and C2 phenotypes by the COVID-19 HGI identified 13 genomic loci that are significantly associated with SARS-CoV-2 infection and/or COVID-19 severity (COVID-19 Host Genetics Initiative, 2021), confirming that this disease has a strong underlying genetic component. While putative target genes for several of the SARS-CoV-2 susceptibility and COVID-19 disease severity genome-wide association study (GWAS) loci have been identified (Pairo-Castineira et al., 2021; Schmiedel et al., 2020; Ellinghaus et al., 2020; Zeberg and Pääbo, 2021; Zhou et al., 2021), a large-scale genetic fine-mapping effort to identify causal regulatory variants, along with the tissue types in which they are functional and their associated genes, has yet to be conducted.
Colocalization is a widely used approach to investigate whether two genetic traits, such as an expression quantitative trait locus (eQTL) for a specific gene located within the same locus as a GWAS signal, share the same causal variants (Giambartolomei et al., 2014; Wallace, 2020). This Bayesian method uses pvalues of the variants tested for two traits to calculate the posterior probabilities (PPs) of five hypotheses at a specific locus: (1) H0, neither trait has a significant association at the tested locus; (2) H1, only the first trait is associated; (3) H2, only the second trait is associated; (4) H3, both traits are associated but the causal variants are different; and (5) H4, both traits are associated and share the same causal variants. Therefore, by conducting colocalization between GWAS and eQTL signals in the same interval, it is possible to detect gene(s) with expression profiles that are associated with SARS-CoV-2 infection or COVID-19 disease severity. Furthermore, this approach can be used to perform genetic fine mapping (Mahajan et al., 2018) of each GWAS locus, which results in the identification of variants that have a high likelihood for causality. In this manner, it is possible to exploit large collections of eQTL analyses, which have assessed the associations between genetic variation and gene expression in many human tissues (Bonder et al., 2021; GTEx Consortium, 2020; DeBoever et al., 2017; Kerimov et al., 2021), to identify causal variants in the COVID-19-associated loci and determine the tissues in which they are functional.
Here, we initially examined the 16 meta-analyses in the COVID-19 HGI dataset and identified 23 loci that we classified as having suggestive associations with either SARS-CoV-2 susceptibility or COVID-19 disease severity. Four of the loci showed heterogeneous GWAS signals consistent with being ethnicity specific. We exploited eQTL data for 48 human tissues in GTEx and for 21 blood-related cell types in the eQTL catalogue (GTEx Consortium, 2020; Kerimov et al., 2021) to investigate the molecular mechanisms underlying the associations between genetic variation and COVID-19 disease status. We performed fine mapping identifying variants associated with the expression of 20 genes in 62 tissues/cell types (range: 1:43 tissues/gene) that strongly colocalize with COVID-19-associated variants in 11 loci. Our study shows that SARS-CoV-2 susceptibility and COVID-19 disease severity GWAS variants are associated with the expression levels of many genes in a wide variety of tissue types.
Results
Genetic architecture of SARS-CoV-2 susceptibility and COVID-19 disease severity
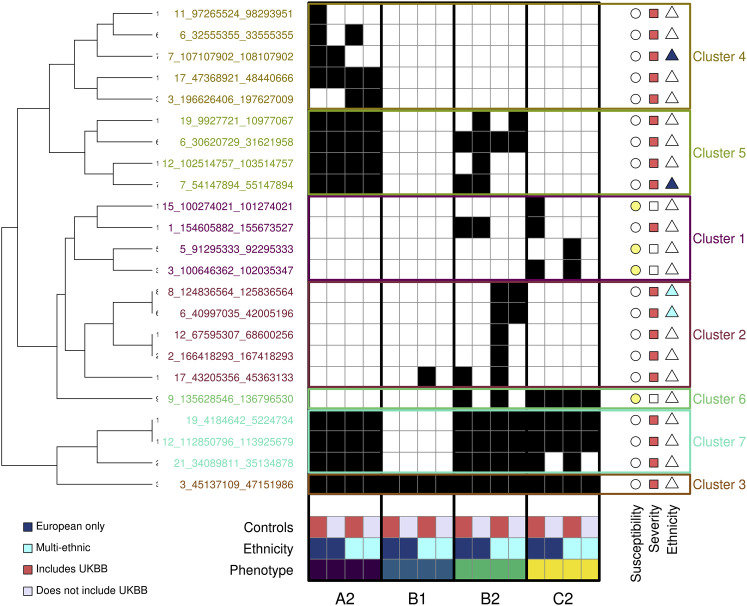
To characterize the similarities and differences of genetic associations between SARS-CoV-2 infection and COVID-19 disease severity phenotypes, we obtained the summary statistics of the 16 HGI COVID-19 meta-analyses from data freeze 5 (COVID-19 Host Genetics Initiative, 2020, 2021). These meta-analyses correspond to four distinct but overlapping phenotypes: C2 with SARS-CoV-2 susceptibility and A2, B1, and B2 with COVID-19 disease severity. For each phenotype, four meta-analyses were conducted, of which two were European-only and two were multi-ethnic (COVID-19 Host Genetics Initiative, 2021). We identified 23 unique loci with suggestive associations (p < 1 × 10−7) in at least one of the 16 meta-analyses (Table S1), of which only one locus on chromosome 3 was associated in all 16 studies (3:45137109-47151986). We assigned three loci as associated with susceptibility because they had p values < 1 × 10−7 only for the C2 phenotype (Figure 1 ). Fourteen loci were assigned as associated with disease severity because they had p values < 1 × 10−7 in one or more of the disease severity phenotypes (A2, B1, or B2) but not the C2 phenotype. Six loci had p values < 1 × 10−7 in both one or more of the disease severity phenotypes (A2, B1, or B2) and the C2 phenotype, of which five were assigned as associated with disease severity because the C2 phenotype had substantially weaker p values and effect sizes (Table S1) and one (9:135628546-136796530) was assigned as associated with susceptibility because it had stronger p values in the C2 meta-analyses (p < 4.9 × 10−16) than in the severity phenotypes (p > 4.4 × 10−9, Table S1). We performed hierarchical clustering using binary association information (p < 1 × 10−7) for each of the 23 loci and identified seven clusters, each of which represents unique information about COVID-19 associations: (1) cluster 1 includes three susceptibility-associated loci significant only for phenotype C2 and one severity-associated locus significant for both B2 and C2 phenotypes, but with substantially weaker p values for the C2 phenotype (Table S1); (2) cluster 2 includes five severity-associated loci significant primarily for phenotype B2; (3) cluster 3 includes only one severity-associated locus (3:45137109-47151986) that was associated with all 16 studies but had substantially weaker p values for the C2 phenotype; (4) cluster 4 includes five severity-associated loci that are significant only for phenotype A2; (5) cluster 5 includes four severity-associated loci significant for both phenotypes A2 and B2; (6) cluster 6 includes one susceptibility-associated locus associated with phenotypes C2 and B2 (9:135628546-136796530) but with weaker p values for the B2 phenotype; and (7) cluster 7 includes three severity-associated loci significant for phenotypes A2, B2, and C2, but with C2 having substantially weaker p values and effect sizes (Table S1). Six severity-associated loci showed heterogeneity in their associations between the European-only and multi-ethnic meta-analyses for one or more phenotypes (Figure 2 ), suggesting the presence of ethnicity-specific signals, but two were barely below the threshold for association in the European-only meta-analyses (p = 2.6 × 10−7 and p = 2.3 × 10−7, respectively) suggesting that they are underpowered and likely not associated with ethnicity. These analyses show that different intervals in the genome are associated with SARS-CoV-2 susceptibility and COVID-19 disease severity, and the genetic associations within four of these intervals are likely ethnicity specific.
Figure 1.
GWAS signals across four COVID-19 phenotypes
Heatmap showing, for each of the 23 COVID-19 loci, the studies with p values < 1 × 10−7 (black squares). The 16 studies are sorted by phenotype (A2, B1, B2, C2), ethnicity (dark blue is for European only, cyan is for multi-ethnic), and type of controls (red indicates studies that included UK Biobank [UKBB] individuals as controls, while gray indicates studies that excluded UKBB individuals). The symbols on the right of each locus indicate whether each locus is associated with susceptibility (yellow circle) or severity (red square) or is ethnicity specific (European only, dark blue triangle; multi-ethnic, cyan triangle). Hierarchical clustering was performed using the hclust and cutree functions in R.
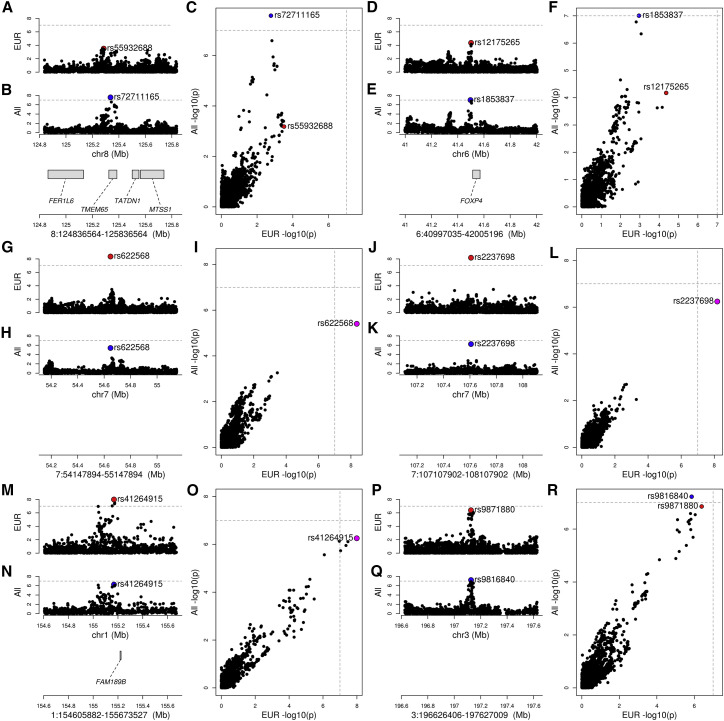
Figure 2.
Ethnicity-associated signals
Four plots are shown for each of the six loci with heterogeneous signals between European-only and multi-ethnic GWAS: (1) GWAS signal in European-only (lead variant highlighted in red); (2) GWAS signal in multi-ethnic (lead variant highlighted in blue); (3) genes with eQTLs (Table S2) overlapping GWAS locus (genes without eQTLs are not shown); and (4) scatterplot showing the -log10 (p value) calculated in the European-only (x axis) and multi-ethnic (y axis) GWAS. If the lead variant is the same between the two studies, it is colored in magenta; otherwise, the lead variant for each study is shown (red, European only; blue, multi-ethnic). For all six loci designated as heterogeneous between the ethnicities, at least two studies on the same ethnicity and phenotype (susceptibility or severity) had p values < 1 × 10−7, and GWAS for the other ethnicity was not significant. For each locus, the following are reported: locus coordinates, phenotype, and the two studies (study names as reported by the COVID-19 HGI).
(A–F) Two cases where only the multi-ethnic studies have p values passing the 1 × 10−7 threshold, suggesting that these two GWAS signals are likely associated with non-European populations. (A–C) 8:124836564-125836564 locus; B2 phenotype (hospitalized COVID-19 patients versus population): COVID19_HGI_B2_ALL_eur_leave_23andme and COVID19_HGI_B2_ALL_leave_23andme; (D–F) 6:40997035-42005196 locus; B2 phenotype (hospitalized COVID-19 patients versus population): COVID19_HGI_B2_ALL_eur_leave_ukbb_23andme and COVID19_HGI_B2_ALL_leave_UKBB_23andme.
(G–L) Three cases where European-only and multi-ethnic studies share the same lead variant, but only the European-only passes the threshold, suggesting that although the multi-ethnic studies include more individuals, they are less powered to detect the COVID-19 association. Therefore, these signals are likely associated with Europeans, rather than other ethnicities. (G–I) 7:54147894-55147894 locus; B2 phenotype (hospitalized COVID-19 patients versus population): COVID19_HGI_B2_ALL_eur_leave_23andme and COVID19_HGI_B2_ALL_leave_UKBB_23andme; (J–L) 7:107107902-108107902 locus; A2 phenotype (very severe respiratory confirmed COVID-19 versus population): COVID19_HGI_A2_ALL_eur_leave_ukbb_23andme and COVID19_HGI_A2_ALL_leave_UKBB_23andme.
(M–R) Two loci (1:154605882-155673527 and 3:196626406-197627009) for which the multi-ethnic study passes the threshold, but the European-only GWAS is barely below the threshold (p = 2.6 × 10−7 and p = 2.3 × 10−7, respectively) suggesting that they are underpowered; therefore, these are likely false negative associations for the European-only, and the locus is not associated with ethnicity. (M–O) 1:154605882-155673527 locus; B2 phenotype (hospitalized COVID-19 patients versus population): COVID19_HGI_B2_ALL_eur_leave_ukbb_23andme and COVID19_HGI_B2_ALL_leave_UKBB_23andme; (P–R) 3:196626406-197627009 locus; A2 phenotype (very severe respiratory confirmed COVID-19 versus population): COVID19_HGI_A2_ALL_eur_leave_23andme and COVID19_HGI_A2_ALL_leave_23andme.
Colocalization with eQTLs and genetic fine mapping of COVID-19 GWAS signals
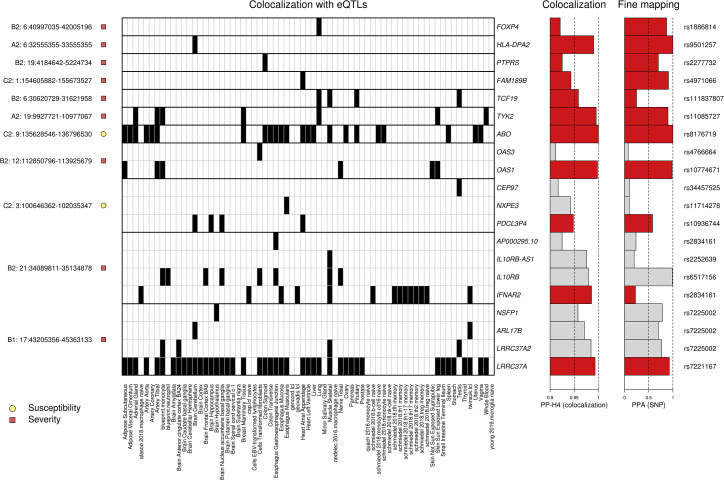
To identify genes whose expression is associated with SARS-CoV2 infection and COVID-19 disease severity, we examined 692 genes that map to the 23 GWAS loci. We obtained eQTL data for the 692 genes in 48 GTEx tissues (GTEx Consortium, 2020) and 21 eQTL datasets from blood-derived cell types (Alasoo et al., 2018; Buil et al., 2015; Chen et al., 2016; Gutierrez-Arcelus et al., 2013; Kerimov et al., 2021; Lappalainen et al., 2013; Nédélec et al., 2016; Quach et al., 2016; Schmiedel et al., 2018; Theusch et al., 2020; Young et al., 2021) and performed a colocalization analysis (Giambartolomei et al., 2014) between each eQTL and GWAS signal. Eleven of the 23 loci had GWAS signals in one or more of the four COVID-19 phenotypes that colocalized (PP-H4 > 0.9) with eQTLs for 20 genes in total (Figure 3 ; Table S2). The GWAS signals in four loci colocalized (PP-H4 > 0.9) with an eQTL signal present in a single tissue for a single gene (FOXP4, HLA-DPA2, PTPRS, FAM189B); three loci colocalized with eQTL signals present in multiple tissues for a single gene (TCF19, TYK2, ABO); and four loci colocalized with eQTL signals for multiple genes (range: 2–4 per locus), of which some eQTLs were present in a single tissue (OAS3, CEP97, NXPE3, AP000295.10, IL10RB-AS1, NSFP1, ARL17B) and others present in multiple tissues (OAS1, PDCL3P4, IL10RB, IFNAR2, ALR17B, LRRC37A2, and LRRC37A) (range: 2–43 per gene). Thus, for 11 of the 20 genes, the colocalized signal was present in a single tissue, while for 9 genes, the colocalized signal was present in multiple tissues. These findings show that causal variants underlying 11 COVID-19 GWAS signals colocalize with genetic variants associated with the expression levels of 20 gene(s), of which some show tissue-specific associations and others show associations across many different tissue types.
Figure 3.
Colocalization of COVID-19 GWAS signals and eQTLs
Heatmap showing tissue eQTLs that colocalized with GWAS signals in 11 loci. Shown are (1) on the left, each locus and its association with susceptibility or severity; (2) in the middle, a heatmap showing the posterior probability of association (PP-H4) between eQTLs in the indicated tissue and the GWAS signal in each locus (black = PP-H4 ≥ 0.9); (3) on the right, barplots indicating the PP-H4 of the indicated eQTL and GWAS signals and of the lead variant tested for each colocalization. “PP-H4” indicates the PP of the GWAS and eQTL signals to have the same underlying causal variant: since all associations shown have PP-H4 > 0.9, the x-axis scale is from 0.9 to 1. PPA (SNP) indicates the PPA for the indicated SNP to be causal for each colocalization. The corresponding RS ID is shown on the right. For each of the four loci with eQTL signals for more than one gene, and only the associations for the gene with the highest PP-H4 (indicated by red bars) were used to fine map the locus. The colocalization data are shown for one phenotype (C2, A1, B1, or B2) per locus and are indicated on the left before the chromosome number and locus coordinates. At one severity-associated locus (1:154605882-155673527), we found that FAM189B colocalized with high PPA (PPA = 0.94) only with the C2 phenotype, but not with any of the severity phenotypes (PPA < 0.67; Table S2).
Of the 48 tissues and 21 blood-related cell types examined for colocalization between eQTLs and COVID-19 GWAS signals, 62 had eQTLs that colocalized with at least one GWAS locus, of which 31 colocalized with two or more (range: 2–7). These include several tissues and cell types that are associated with COVID-19 symptoms, such as lung (associated with cough and shortness of breath: FOXP4, LRRC37A, TCF19, and TYK2), whole blood (immune response: LRRC37A and TYK2), macrophages (immune response: IFNAR2), neutrophils (immune response: IL10RB), B cells (immune response: IFNAR2), lymphoblastoid cell line (LCL; immune response: ARL17B, LRRC37A, and IFNAR2), monocytes (immune response: ABO, IL10RB, LRRC37A, LRRC37A2, OAS1, and TYK2), T cells (immune response: IFNAR2), skeletal muscle (muscle fatigue and body aches: ABO, IFNAR2, IL10RB, IL10RB-AS1, LRRC37A, LRRC37A2, and TCF19), digestive system (nausea and diarrhea: ABO, LRRC37A, PTPRS, AP000295.10, IL10RB, IFNAR2, and NXPE3), brain cortex (loss of smell and taste, cognitive defects: LRRC37A and LRRC37A2; Table S2), heart (myocarditis, cardiomyopathy, and heart failure: ABO, FAM198B, LRRC37A, and PDCL3P4), and skin (skin rashes: LRRC37A, OAS1, and TYK2). Surprisingly, 5 of the 11 (44.4%) GWAS signals did not colocalize with eQTLs in blood or lung, the two tissues that have been described as being the most affected by COVID-19.
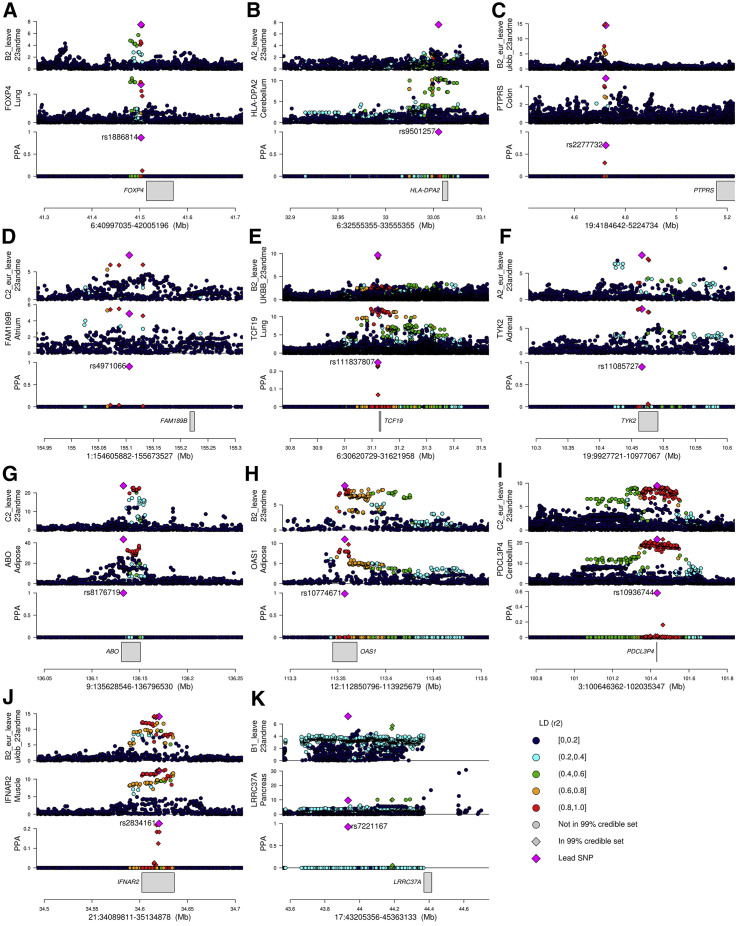
To identify putative causal variants, we performed genetic fine mapping (Mahajan et al., 2018) in each of the 11 loci using the colocalization results between the GWAS signal and the gene with the best colocalizing eQTL signal (i.e., highest PP-H4). We calculated the posterior probability of each SNP tested in both the GWAS and the eQTL analysis to be causal (PP of association [PPA]). We next combined all the SNPs with the highest PPAs to compute 99% credible sets, defined as the SNPs whose sum of PPA is >99% (Figure 4 ; Tables S3 and S4). For 10 GWAS loci, we were able to determine 99% credible sets that included eight or fewer SNPs, including three with a single likely causal SNP. For the remaining GWAS locus, the 99% credible set included 57 SNPs; the most likely causal SNP had PPA = 0.58.
Figure 4.
Colocalizations between COVID-19 GWAS and tissue eQTL signals
For each of the 11 fine-mapped loci, four panels are presented: (1) GWAS p values for indicated COVID-19 phenotype; (2) eQTL p values for indicated gene; (3) posterior probability of colocalization of each variant (coloc PP; calculated using coloc) and color-coded based on linkage disequilibrium in individuals of European descent (r2) calculated using LDlinkR (Myers et al., 2020); and (4) location of gene in locus. The p values of all variants tested in the GWAS and eQTL studies are shown; however, only variants tested in both studies were used for colocalization.
(A) Locus 6:40997035-42005196, phenotype B2 GWAS (B2_ALL_leave_23andme), and FOXP4 eQTL in lung.
(B) Locus 6:32555355-33555355, phenotype A2 GWAS (A2_ALL_leave_23andme), and HLA-DPA2 eQTL in brain cerebellum.
(C) Locus 19:4184642-5224734, phenotype B2 GWAS (B2_ALL_eur_leave_ukbb_23andme), and PTPRS eQTL in colon sigmoid.
(D) Locus 1:154605882-155673527, phenotype C2 GWAS (C2_ALL_eur_leave_23andme), and FAM189B eQTL in heart atrial appendage; although this signal is severity associated, only the C2 phenotype colocalized with an eQTL.
(E) Locus 6:30620729-31621958, phenotype B2 GWAS (B2_ALL_leave_UKBB_23andme), and TCF19 eQTL in lung.
(F) Locus 19:9927721-10977067, phenotype A2 GWAS (A2_ALL_eur_leave_23andme), and TYK2 eQTL in adrenal gland.
(G) Locus 9:135628546-136796530, phenotype C2 GWAS (C2_ALL_leave_23andme), and ABO eQTL in adipose visceral omentum.
(H) Locus 12:112850796-113925679, phenotype B2 GWAS (B2_ALL_leave_23andme), and OAS1 eQTL in adipose subcutaneous.
(I) Locus 3:100646362-102035347, phenotype C2 GWAS (C2_ALL_eur_leave_23andme), and PDCL3P4 eQTL in cerebellum; since the FOXP4 locus was associated with non-European individuals, LD is shown for East Asians.
(J) Locus 21:34089811-35134878, phenotype B2 GWAS (B2_ALL_eur_leave_ukbb_23andme), and IFNAR2 eQTL in muscle skeletal.
(K) Locus 17:43205356-45363133, phenotype B1 GWAS (B1_ALL_leave_23andme), and LRRC37A eQTL in pancreas.
Genetic fine-mapped GWAS signals
Here, we report a description of each of the 11 fine-mapped GWAS signals.
6:40997035-42005196 and lung-specific eQTL for FOXP4
The 6:40997035-42005196 locus was associated with COVID-19 disease severity in multi-ethnic meta-analyses, but not with European-only meta-analyses (Figures 1 and 2D–2F). Interestingly, the lead GWAS variant in this locus (rs1886814) is less common in Europeans (gnomAD allele frequency = 2.4%) than in East Asians (37.3%), Admixed Americans (18.9%), or South Asians (11.8%) (Karczewski et al., 2020) and was found to be associated with COVID-19 in East Asians by the COVID-19 HGI (COVID-19 Host Genetics Initiative, 2020, 2021). We found that this GWAS signal colocalized with a lung-specific eQTL for FOXP4, a transcription factor associated with neurodevelopmental disorders and congenital abnormalities with lung cancer (Snijders Blok et al., 2021; Yang et al., 2015). Fine mapping with this lung-specific eQTL signal for FOXP4 identified rs1886814 as the variant with the highest likelihood of causality (PPA = 0.870, Figure 4A), which is located ∼10 kb upstream of FOXP4 and is likely a regulatory variant whose alternative allele is associated with increased FOXP4 expression (effect size β = 0.46; Table S3). rs1886814 has previously been described as the lead variant in this locus for COVID-19 hospitalization (COVID-19 Host Genetics Initiative, 2021) and is in linkage disequilibrium (LD) with an emphysema-associated variant (rs2894439, r2 = 0.7) (Manichaikul et al., 2017; Yang et al., 2015). These data suggest increased expression of FOXP4 in lung tissue is causally associated with COVID-19 disease severity.
6:32555355-33555355 and cerebellum-specific eQTL for HLA-DPA2
The 6:32555355-33555355 locus overlapped the major histocompatibility complex (MHC). The GWAS signal in this locus was associated with COVID-19 disease severity and colocalized with a cerebellum-specific eQTL for HLA-DPA2 (Figure 4B), a member of the MHC class II with unknown function (Jones et al., 2006; Roche and Furuta, 2015). Fine mapping identified a single variant (rs9501257, PPA > 0.99) as likely causal for both genetic signals, associated with increased HLA-DPA2 expression (effect size β = 1.08; Table S3) and located in the 3′ UTR of the neighboring HLA-DPB1 gene. This variant is in moderate LD with a hepatitis B infection GWAS SNP (rs3128923, R2 = 0.47 in Africans) and is in high Dʹ (Dʹ = 1 in multiple populations) with variants associated with liver cancer following hepatitis B infection (rs2295119) and autoimmune diseases (rs2295119 and rs2281388: immunoglobulin A glomerulonephritis and Graves’ disease) (Chu et al., 2011; Li et al., 2020; Sawai et al., 2018; Zeng et al., 2021; Zhao et al., 2019). These data suggest increased expression of HLA-DPA2 in the cerebellum is causally associated with COVID-19 disease severity.
19:4184642-5224734 and sigmoid-colon-specific eQTL for PTPRS
The 19:4184642-5224734 locus was associated with COVID-19 disease severity and colocalized with a sigmoid colon-specific eQTL (rs2277732) for PTPRS (effect size β = 0.29; Table S3; Figure 4C), which encodes a phosphatase with three immunoglobulin-like domains whose knockdown results in ulcerative colitis in mice (Muise et al., 2007). The eQTL is 481 kb downstream of PTPRS (which is transcribed on the reverse strand) and, hence is likely in a distal regulatory element. Interestingly, the protein product of PTPRS was shown to interact with the membrane (M) protein of SARS-CoV-2 (Laurent et al., 2020; Samavarchi-Tehrani et al., 2020), which likely glycosylates the spike protein (Thomas, 2020). These data suggest that increased expression of PTPRS in the sigmoid colon is causally associated with COVID-19 disease severity.
1:154605882-155673527 and atrial-appendage-specific eQTL for FAM189B
In the 1:154605882-155673527 locus, the GWAS signal colocalized with an eQTL for FAM189B in cardiac tissues (atrial appendage, effect size β = −0.23; Figures 3 and 4D; Table S3). Although this gene’s function has not been well characterized, it has been shown that rare coding variants in its sequence are associated with increased triglyceride levels in the blood (Gilly et al., 2018). Of note, high triglyceride levels prior to infection serve as a prognostic marker for severe COVID-19 outcomes (Fan et al., 2020; Hu et al., 2020; Wang et al., 2020; Wei et al., 2020). Recent evidence showed that the FAM189B protein product can bind to the SARS-CoV-2 envelope (E) protein (Laurent et al., 2020; Samavarchi-Tehrani et al., 2020), which modulates maturation and retention of the spike protein and thereby contributes to virus assembly and pathogenicity (Mandala et al., 2020). These observations suggest that decreased FAM189B expression in cardiac tissues contributes to COVID-19 disease severity potentially through altered FAM189B binding to the SARS-CoV-2 envelope (E) protein.
6:30620729-31621958 and TCF19
The 6:30620729-31621958 locus disease-severity-associated GWAS signal colocalized with TCF19 eQTLs in lung, skeletal muscle, pituitary gland, and testis (effect size β = 0.34; Figures 3 and 4E; Table S3). This transcription factor is ubiquitously expressed, regulates inflammation and DNA damage response (Yang et al., 2021), and is involved in lung cancer (Zhou et al., 2019). A previous study identified COVID-19 GWAS variants located in the TCF19 promoter and associated with altered expression in immune cells (Schmiedel et al., 2020). Our fine mapping identified four variants with PPAs between 0.22 and 0.25, all in high LD (R2 = 1, cumulative PPA = 0.93) located ∼6 kb upstream of TCF19 in an intron of the neighboring CCHCR1 gene. Given the presence of multiple variants in high LD, we were not able to identify one single likely causal variant, but fine mapping was able to reduce the number of potentially causal variants to four. Our results suggest that increased expression of TCF19 in multiple tissues may be associated with COVID-19 disease severity, potentially due to alterations in the immune response regulating inflammation.
19:9927721-10977067 and TYK2
The 19:9927721-10977067 locus disease-severity-associated GWAS signal colocalized with eQTLs for TYK2 in seven GTEx tissues, including adrenal gland, tibial artery, breast, lung, monocytes, skin, and whole blood (effect size β = 0.45; Figures 3 and 4F; Table S3). TYK2 encodes for a tyrosine kinase involved in the immune response, whose inhibition has been described as a potential treatment for multiple autoimmune diseases (Ghoreschi et al., 2021). The variant with the highest likelihood of causality (rs11085727, PPA = 0.89) was also the lead variant in GWAS signals for neutrophil percentage, systemic lupus erythematosus, and thyroid medication (Astle et al., 2016; Bentham et al., 2015; Wu et al., 2019). These findings suggest that increased expression of TYK2 in multiple tissues may be causally associated with inflammation and the cytokine storm observed in severe COVID-19 cases (Huang et al., 2020; Pedersen and Ho, 2020) and further suggest TYK2 inhibitors could have therapeutic potential for treating COVID-19 symptoms.
9:135628546-136796530 and ABO
The ABO gene is expressed in many tissues (Larson et al., 2016; Oriol et al., 1992). Two coding variants in the ABO gene (rs8176746 and rs8176719) confer the O, A, B, and AB blood types (Melzer et al., 2008). Although multiple studies previously described this locus as associated with COVID-19 and blood type as a risk factor (COVID-19 Host Genetics Initiative, 2021; Pairo-Castineira et al., 2021; Ellinghaus et al., 2020), there is still controversy about the causal variant. We observed that eQTLs for the ABO gene in 21 GTEx tissues and two blood-related cell types colocalized with COVID-19 susceptibility (Figures 3 and 4G; Table S3). The variant with the highest likelihood of causality (PPA ≈1) was rs8176719, which distinguishes the O blood type (reference allele) from the A and B blood types (C insertion). This variant is in high LD with rs912805253 (R2 = 0.732), a distal variant previously proposed to be causal at this locus (COVID-19 Host Genetics Initiative, 2021). Our results suggest that the coding variant rs8176719 is likely causal for the association between the 9:135628546-136796530 locus and SARS-CoV-2 susceptibility, which is consistent with previous studies showing that individuals with blood type O have a reduced risk for SARS-CoV-2 infection (Barnkob et al., 2020).
12:112850796-113925679 and OAS1
The 12:112850796-113925679 locus encodes a cluster of three 2′–5′-oligoadenylate synthetases (OAS1, OAS2, and OAS3) and harbors eQTLs for OAS1 in six tissues including adipose, tibial artery, tibial nerve, skin, and monocytes (effect size β = −0.25 in adipose tissue; Figures 3 and 4H; Table S3) and for OAS3 in transformed fibroblasts. All three OAS genes play a critical role in cellular innate antiviral response (Birdwell et al., 2016), and OAS1 is known to inhibit viral infection by activating RNase L, which promotes viral RNA degradation (Banerjee et al., 2019). While fine mapping with OAS1 results in a single likely causal variant (rs10774671) with PPA = 0.997, fine mapping with OAS3 is unresolved because we found 59 variants in the 99% credible set, and the variant with the highest likelihood of causality (rs6489879) had a PPA = 0.051. Our data suggest that decreased expression levels of OAS1 are likely causally implicated in COVID-19 disease severity and are in agreement with previous studies showing that high plasma OAS1 levels are associated with reduced susceptibility and COVID-19 severity (Zhou et al., 2021).
3:100646362-102035347 and PDCL3P4, CEP97 and NXPE3
The GWAS signal for SARS-CoV-2 susceptibility in locus 3:100646362-102035347 colocalized with an eQTL signal of three genes in six distinct tissues (PDCL3P4: cerebellum, hippocampus, nucleus accumbens basal ganglia, and atrial appendage; CEP97: testis; and NXPE3: esophagus muscularis [effect size β = 0.74 for PDCL3P4 in cerebellum]; Figures 3 and 4I; Table S3). Although we observed large 99% credible sets for each of these three genes (57, 73, and 28, respectively), the lead variant for PDCL3P4 (rs10936744) had a moderately high PPA (PPA = 0.579). The lead variants for the other two genes were in high LD with rs10936744 (rs34457525 for CEP97 and rs11714278 for NXPE3, R2 > 0.97), suggesting that a single eQTL signal likely affects the expression of all three genes. Although we were able to determine associations between the GWAS and eQTL signals in the 3:100646362-102035347 locus, further experiments will be needed to understand how increased expression of one or more of the genes in this interval is implicated in SARS-CoV-2 susceptibility.
21:34089811-35134878 and IFNAR2, IL10RB, IL10RB-AS1, and AP000295.10
The 21:34089811-35134878 locus was associated with COVID-19 disease severity and colocalized with eQTLs for four genes (IFNAR2, IL10RB, IL10RB-AS1, and AP000295.10) in seven GTEx tissues (skeletal muscle, esophagus mucosa, gastresophageal junction, frontal cortex, nucleus accumbens basal ganglia, tibial nerve, transformed fibroblasts) and 14 blood-related cell types (Figures 3 and 4J; Table S2). IFNAR2, IL10RB, and IL10RB-AS1 are associated with immune response (Duncan et al., 2015; Shouval et al., 2014), while the function of AP000295.10 is unknown. While IFNAR2 had a slightly higher colocalization PPA than IL10RB (Figure 3), its fine mapping resulted in five variants with posterior probability of being causal between 0.1 and 0.3, whereas IL10RB resulted in one single variant as likely causal (rs6517156, PPA = 0.99). The lead eQTL variants for all four genes are in high LD (R2 > 0.75), and their effect sizes were negative, suggesting that one single eQTL signal likely reduces the expression of all four genes. Both IFNAR2 and IL10RB have been previously described as associated with COVID-19 (Ahn and Prince, 2020; Meyts and Casanova, 2021). While our genetics analysis determined that decreased expression of at least one of the four genes in this locus is likely causally associated with COVID-19 disease severity, we are unable to pinpoint the exact gene(s).
17:43205356-45363133 and LRRC37A
The 17:43205356-45363133 locus showed colocalization between eQTLs in multiple tissues and COVID-19 disease severity (Figures 3 and 4K). This GWAS signal colocalized with LRRC37A eQTLs in 41 GTEx tissues and two blood-derived cell types; with LRRC37A2 in anterior cingulate cortex, monocytes, skeletal muscle, and testis; with ARL17B in cerebellum and LCL; and with NSFP1 in hypothalamus (Table S2). All eQTLs had positive effect sizes (Table S3), indicating that they were associated with increased expression. Although the credible sets for each of the four genes included three or four variants, the lead variant for LRRC37A (rs7221167) had the highest PPA (PPA = 0.93) and was in moderate LD (R2 = 0.49) with the lead variant for the other three genes (rs7225002) only in Europeans. These findings suggest the presence of a complex relationship among the genotypes of multiple variants, the expression of four genes, and their effects on COVID-19 disease severity. Understanding the associations between this locus and COVID-19 is further complicated by the fact that multiple structural polymorphisms (deletions and inversions) are common, difficult to assay, and have been proposed to potentially affect both gene expression and predisposition to other diseases including Koolen-De Vries syndrome, mental challenge, progressive supranuclear palsy, Parkinson’s disease, and Alzheimer’s disease (Baker et al., 1999; Koolen et al., 2006; Myers et al., 2005; Skipper et al., 2004; Webb et al., 2008). Therefore, although our genetic analyses suggest that increased expression of one of the four genes in this locus is associated with COVID-19 disease severity, the interval is genetically complex, and the association could be due to molecular mechanisms that we did not assay.
Discussion
The COVID-19 pandemic has united the focus of the broad biomedical scientific community to improve our understanding of the mechanisms underlying disease susceptibility and severity. The COVID-19 HGI has collected genetic and COVID-19 status information from ∼2 million individuals across 46 studies (COVID-19 Host Genetics Initiative, 2020, 2021), making COVID-19 one of the best-powered GWAS studies ever conducted. Standard approaches to characterize GWAS summary statistics focus on obtaining all of the genomic loci where variants have p value < 5 × 10−8 and identifying the most likely causal gene by proximity to the lead variant. Here, we compared 16 meta-analyses for four COVID-19 disease phenotypes to investigate the similarities and differences in their genetic signals. We identified four genomic loci with suggestive associations for SARS-CoV-2 susceptibility and 19 for COVID-19 disease severity. Loci where the European-only or the multi-ethnic meta-analyses, but not both, had p values < 1 × 10−7 were labeled as “ethnicity specific.” Using these annotations, we were able to confirm the findings by the COVID-19 HGI at multiple loci and discover novel associations at previously uncharacterized loci. Overall, our analyses showed that different intervals in the genome are associated with SARS-CoV-2 susceptibility and COVID-19 disease severity, and the genetic associations within four of these intervals are likely ethnicity specific. We next applied a colocalization method (Giambartolomei et al., 2014) to compare the COVID-19 GWAS and eQTLs signals in the same intervals, based on the assumption that the molecular underpinning of genetic variants associated with disease could be that they alter the expression of specific genes. GTEx and the eQTL catalogue (GTEx Consortium, 2020; Kerimov et al., 2021) provided us with the unique opportunity to investigate the associations among genotype, gene expression, and COVID-19 in 48 tissues and 21 blood-related cell types. While the association between COVID-19 and altered gene expression in blood or lung may seem obvious, this disease affects many other tissues and organs, suggesting that associations between genetic variation and gene expression in a wide variety of tissues may result in differential disease susceptibility and severity. Our colocalization analysis identified 11 GWAS loci whose causal variants overlapped regulatory and coding variants associated with the expression of 20 genes, of which some showed tissue-specific associations and others showed associations across many different tissue types. By exploiting the colocalization between GWAS and eQTL signals, we were able to perform genetic fine mapping (Mahajan et al., 2018) at all 11 COVID-19 loci and prioritize variants based on their likelihood for being causal.
In summary, our study examines the associations among COVID-19 disease phenotypes, genetic variation, and gene expression in 48 human tissues and 21 blood-derived cell types and provides novel insights into the molecular mechanisms underlying inherited predispositions to SARS-CoV-2 infection and COVID-19 disease severity. While previous studies have mostly focused on associations between COVID-19 and lung or immune cell types, here, we emphasize that associations between genetic variation and gene expression in diverse tissue and cell types contribute to the observed differences in susceptibility and COVID-19 disease severity between individuals and indicate that genetic variation likely plays an important role in determining the systemic effects of COVID-19 disease.
Limitations of the study
Although colocalization is a powerful tool to characterize the associations between GWAS and eQTL signals and to fine map, it is limited by the fact that it assumes one single causal variant underlying both genetic signals per region. In regions with multiple independent signals, this approach likely results in an underestimation of the colocalization between GWAS and eQTLs. Therefore, our results do not exclude that multiple GWAS signals with independent molecular underpinnings may be present in some of the 11 fine-mapped loci.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| COVID-19 GWAS summary statistics | (COVID-19 Host Genetics Initiative, 2021) | https://www.covid19hg.org/ |
| GTEx V.7 eQTL summary statistics | (GTEx Consortium, 2020) | https://www.gtexportal.org/home/datasets |
| eQTL Catalogue summary statistics | (Kerimov et al., 2021) | https://www.ebi.ac.uk/eqtl/ |
| Software and algorithms | ||
| liftOver | UCSC Genome Browser | http://hgdownload.soe.ucsc.edu/downloads.html |
| Colocalization | (Giambartolomei et al., 2014) | https://cran.r-project.org/web/packages/coloc/ |
| LDlinkR | (Myers et al., 2020) | https://cran.r-project.org/web/packages/LDlinkR/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Matteo D’Antonio (madantonio@health.ucsd.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
This is a computational study that only used publicly available data. Therefore it does not include any experimental models.
Method details
GWAS summary statistics processing
We downloaded the summary statistics on GRCh37 coordinates of 16 meta-analyses from the COVID-19 HGI data freeze 5 (January 18, 2021, https://www.covid19hg.org/). These studies include four phenotypes: 1) A2 = Very severe respiratory confirmed COVID-19 versus population; 2) B1 = Hospitalized versus non-hospitalized COVID-19 patients; 3) B2 = Hospitalized COVID-19 patients versus population; and 4) C2 = COVID-19 patients versus population. For each phenotype, four meta-analyses were performed, including two on individuals from European descent and two on multiple ethnicities. All the studies from three phenotypes (A2, B2 and C2) include more than 1 million controls, whereas one (B1), which tested hospitalized and non-hospitalized patients, included < 6,000 cases and < 16,000 controls.
For each study, we identified all variants with p value < 10−7 (considered to have “suggestive associations with COVID-19”) from the filtered meta-analysis summary statistics (p < 1 × 10−5) provided by the COVID-19 HGI (https://www.covid19hg.org/results/r5/) and expanded each variant’s position by 500 kb upstream and downstream. We used bedtools merge (Quinlan, 2014) to identify 23 unique loci.
To annotate a locus as associated with ethnicity, we required at least two studies on the same ethnicity to have p value < 1 × 10−7.
eQTL data
We downloaded all SNP-gene association tests (including non-significant test) in all GTEx V.7 tissues and gene-level information from the GTEx Portal (https://www.gtexportal.org/home/datasets). We also obtained eQTL data from 21 blood-related tissues from the eQTL Catalogue (https://www.ebi.ac.uk/eqtl/). The genomic coordinates from the eQTL Catalogue were converted from hg38 to hg19 using liftOver.
In each tissue, we extracted the eQTL information for 692 genes that overlapped the 23 COVID-19 genome-wide significant loci.
Colocalization
For each gene in the 23 COVID-19 loci, we performed colocalization between its eQTL signal in each of 48 GTEx tissues and 21 blood-related tissue and each of the 16 COVID-19 meta-analyses. We used the coloc.abf function from the coloc package in R (Giambartolomei et al., 2014) to compare the p values between each pair of SNPs genotyped in both datasets. To determine the total number of individuals genotyped in each meta-analysis, we used the “all_meta_sample_N” column in each summary statistics file. We used default priors: p1 (prior probability that a SNP is associated with the eQTL) = 1 × 10−4; p2 (prior probability that a SNP is associated with the GWAS trait) = 1 × 10−4; and p12 (prior probability that a SNP is associated with both GWAS trait and eQTL) = 1 × 10−5. We considered as each pair of signals “colocalized” if their PP-H4 was greater than 0.9. At the 11 loci that colocalized with eQTLs, we selected the colocalization with highest PPA and used it for fine mapping. Genetic fine mapping was performed using the coloc.abf function and extracting the PPA of each SNP tested in both COVID-19 and eQTL. We defined 99% credible sets by selecting the variants that contribute to 99% of the cumulative PPA.
LD calculation
To obtain LD information for each of the most likely causal variants in the colocalization between GWAS and eQTLs (Figure 4), we used LDlinkR (Myers et al., 2020). We obtained a token from https://ldlink.nci.nih.gov/?tab=apiaccess and interrogated the 1000 Genomes LD structure using the LDmatrix and LDproxy functions.
All LD information provided in the text and figures was generated using LDlinkR.
Quantification and statistical analysis
As described above, we identified all variants with p value < 10−7 (considered to have “suggestive associations with COVID-19”) from the meta-analysis summary statistics provided by the COVID-19 HGI (https://www.covid19hg.org/results/r5/). To conduct colocalization we expanded each variant’s position by 500 kb upstream and downstream.
We considered an eQTL as colocalizing with a GWAS signal if their colocalization PPA was greater than 0.9.
Consortia
The members of The COVID-19 Host Genetics Initiative are Benjamin M. Neale, Mark Daly, Andrea Ganna, Christine Stevens, Gita A. Pathak, Shea J. Andrews, Masahiro Kanai, Mattia Cordioli, Andrea Ganna, Juha Karjalainen, Gita A. Pathak, Renato Polimanti, Shea J. Andrews, Mattia Cordioli, Matti Pirinen, Masahiro Kanai, Nadia Harerimana, Kumar Veerapen, Brooke Wolford, Huy Nguyen, Matthew Solomonson, Christine Stevens, Rachel G. Liao, Karolina Chwialkowska, Amy Trankiem, Mary K. Balaconis, Caroline Hayward, Anne Richmond, Archie Campbell, Marcela Morris, Chloe Fawns-Ritchie, Joseph T. Glessner, Douglas M. Shaw, Xiao Chang, Hannah Polikowski, Petty E. Lauren, Hung-Hsin Chen, Zhu Wanying, Hakon Hakonarson, David J. Porteous, Jennifer Below, Kari North, Joseph B. McCormick, Paul R.H.J. Timmers, James F. Wilson, Albert Tenesa, Kenton D’Mellow, Shona M. Kerr, Mari E.K. Niemi, Mattia Cordioli, Lindokuhle Nkambul, Kathrin Aprile von Hohenstaufen, Ali Sobh, Madonna M. Eltoukhy, Amr M. Yassen, Mohamed A.F. Hegazy, Kamal Okasha, Mohammed A. Eid, Hanteera S. Moahmed, Doaa Shahin, Yasser M. El-Sherbiny, Tamer A. Elhadidy, Mohamed S. Abd Elghafar, Jehan J. El-Jawhari, Attia A.S. Mohamed, Marwa H. Elnagdy, Amr Samir, Mahmoud Abdel-Aziz, Walid T. Khafaga, Walaa M. El-Lawaty, Mohamed S. Torky, Mohamed R. El-shanshory, Chiara Batini, Paul H. Lee, Nick Shrine, Alexander T. Williams, Martin D. Tobin, Anna L. Guyatt, Catherine John, Richard J. Packer, Altaf Ali, Robert C. Free, Xueyang Wang, Louise V. Wain, Edward J. Hollox, Laura D. Venn, Catherine E. Bee, Emma L. Adams, Mari E.K. Niemi, Ahmadreza Niavarani, Mattia Cordioli, Lindokuhle Nkambul, Bahareh Sharififard, Rasoul Aliannejad, Ali Amirsavadkouhi, Zeinab Naderpour, Hengameh Ansari Tadi, Afshar Etemadi Aleagha, Saeideh Ahmadi, Seyed Behrooz Mohseni Moghaddam, Alireza Adamsara, Morteza Saeedi, Hamed Abdollahi, Abdolmajid Hosseini, Pajaree Chariyavilaskul, Monpat Chamnanphon, Thitima B. Suttichet, Vorasuk Shotelersuk, Monnat Pongpanich, Chureerat Phokaew, Wanna Chetruengchai, Watsamon Jantarabenjakul, Opass Putchareon, Pattama Torvorapanit, Thanyawee Puthanakit, Pintip Suchartlikitwong, Nattiya Hirankarn, Voraphoj Nilaratanakul, Pimpayao Sodsai, Ben M. Brumpton, Kristian Hveem, Cristen Willer, Brooke Wolford, Wei Zhou, Tormod Rogne, Erik Solligard, Bjørn Olav Åsvold, Malak Abedalthagafi, Manal Alaamery, Saleh Alqahtani, Dona Baraka, Fawz Al Harthi, Ebtehal Alsolm, Leen Abu Safieh, Albandary M. Alowayn, Fatimah Alqubaishi, Amal Al Mutairi, Serghei Mangul, Abdulraheem Alshareef, Mona Sawaji, Mansour Almutairi, Nora Aljawini, Nour Albesher, Yaseen M. Arabi, Ebrahim S. Mahmoud, Amin K. Khattab, Roaa T. Halawani, Ziab Z. Alahmadey, Jehad K. Albakri, Walaa A. Felemban, Bandar A. Suliman, Rana Hasanato, Laila Al-Awdah, Jahad Alghamdi, Deema AlZahrani, Sameera AlJohani, Hani Al-Afghani, May Alrashed, Nouf AlDhawi, Hadeel AlBardis, Sarah Alkwai, Moneera Alswailm, Faisal Almalki, Maha Albeladi, Iman Almohammed, Eman Barhoush, Anoud Albader, Salam Massadeh, Abdulaziz AlMalik, Sara Alotaibi, Bader Alghamdi, Junghyun Jung, Mohammad S. Fawzy, Yunsung Lee, Per Magnus, Lill-Iren S. Trogstad, Øyvind Helgeland, Jennifer R. Harris, Massimo Mangino, Tim D. Spector, Duncan Emma, Sandra P. Smieszek, Bartlomiej P. Przychodzen, Christos Polymeropoulos, Vasilios Polymeropoulos, Mihael H. Polymeropoulos, Israel Fernandez-Cadenas, Jordi Perez-Tur, Laia Llucià-Carol, Natalia Cullell, Elena Muiño, Jara Cárcel-Márquez, Marta L. DeDiego, Lara Lloret Iglesias, Anna M. Planas, Alex Soriano, Veronica Rico, Daiana Agüero, Josep L. Bedini, Francisco Lozano, Carlos Domingo, Veronica Robles, Francisca Ruiz-Jaén, Leonardo Márquez, Juan Gomez, Eliecer Coto, Guillermo M. Albaiceta, Marta García-Clemente, David Dalmau, Maria J. Arranz, Beatriz Dietl, Alex Serra-Llovich, Pere Soler, Roger Colobrán, Andrea Martín-Nalda, Alba Parra Martínez, David Bernardo, Silvia Rojo, Aida Fiz-López, Elisa Arribas, Paloma de la Cal-Sabater, Tomás Segura, Esther González-Villa, Gemma Serrano-Heras, Joan Martí-Fàbregas, Elena Jiménez-Xarrié, Alicia de Felipe Mimbrera, Jaime Masjuan, Sebastian García-Madrona, Anna Domínguez-Mayoral, Joan Montaner Villalonga, Paloma Menéndez-Valladares, Daniel I. Chasman, Julie E. Buring, Paul M. Ridker, Giulianini Franco, Howard D. Sesso, JoAnn E. Manson, Xiao Chang, Joseph R. Glessner, Hakon Hakonarson, Caroline Hayward, Anne Richmond, David J. Porteous, Archie Campbell, Chloe Fawns-Ritchie, Carolina Medina-Gomez, Andre G. Uitterlinden, M. Arfan Ikram, Kati Kristiansson, Sami Koskelainen, Markus Perola, Kati Donner, Katja Kivinen, Aarno Palotie, Samuli Ripatti, Sanni Ruotsalainen, Mari Kaunisto, FinnGen, Tomoko Nakanishi, Guillaume Butler-Laporte, Vincenzo Forgetta, David R. Morrison, Biswarup Ghosh, Laetitia Laurent, Alexandre Belisle, Danielle Henry, Tala Abdullah, Olumide Adeleye, Noor Mamlouk, Nofar Kimchi, Zaman Afrasiabi, Nardin Rezk Branka Vulesevic, Meriem Bouab, Charlotte Guzman, Louis Petitjean, Chris Tselios, Xiaoqing Xue, Erwin Schurr, Jonathan Afilalo, Marc Afilalo, Maureen Oliveira, Bluma Brenner, Pierre Lepage, Jiannis Ragoussis, Daniel Auld, Nathalie Brassard, Madeleine Durand, Michaël Chassé, Daniel E. Kaufmann, G. Mark Lathrop, Vincent Mooser, J. Brent Richards, Rui Li, Darin Adra, Souad Rahmouni, Michel Georges, Michel Moutschen, Benoit Misset, Gilles Darcis, Julien Guiot, Julien Guntz, Samira Azarzar, Stéphanie Gofflot, Yves Beguin, Sabine Claassen, Olivier Malaise, Pascale Huynen, Christelle Meuris, Marie Thys, Jessica Jacques, Philippe Léonard, Frederic Frippiat, Jean-Baptiste Giot, Anne-Sophie Sauvage, Christian Von Frenckell, Yasmine Belhaj, Bernard Lambermont, Mari E.K. Niemi, Mattia Cordioli, Sara Pigazzini, Lindokuhle Nkambule, Michelle Daya, Jonathan Shortt, Nicholas Rafaels, Stephen J. Wicks, Kristy Crooks, Kathleen C. Barnes, Christopher R. Gignoux, Sameer Chavan, Triin Laisk, Kristi Läll, Maarja Lepamets, Reedik Mägi, Tõnu Esko, Ene Reimann, Lili Milani, Helene Alavere, Kristjan Metsalu, Mairo Puusepp, Andres Metspalu, Paul Naaber, Edward Laane, Jaana Pesukova, Pärt Peterson, Kai Kisand, Jekaterina Tabri, Raili Allos, Kati Hensen, Joel Starkopf, Inge Ringmets, Anu Tamm, Anne Kallaste, Pierre-Yves Bochud, Carlo Rivolta, Stéphanie Bibert, Mathieu Quinodoz, Dhryata Kamdar, Noémie Boillat, Semira Gonseth Nussle, Werner Albrich, Noémie Suh, Dionysios Neofytos, Véronique Erard, Cathy Voide, FHoGID, RegCOVID, P-PredictUs, SeroCOVID, CRiPSI, Rafael de Cid, Iván Galván-Femenía, Natalia Blay, Anna Carreras, Beatriz Cortés, Xavier Farré, Lauro Sumoy, Victor Moreno, Josep Maria Mercader, Marta Guindo-Martinez, David Torrents, Manolis Kogevinas, Judith Garcia-Aymerich, Gemma Castaño-Vinyals, Carlota Dobaño, Alessandra Renieri, Francesca Mari, Chiara Fallerini, Sergio Daga, Elisa Benetti, Margherita Baldassarri, Francesca Fava, Elisa Frullanti, Floriana Valentino, Gabriella Doddato, Annarita Giliberti, Rossella Tita, Sara Amitrano, Mirella Bruttini, Susanna Croci, Ilaria Meloni, Maria Antonietta Mencarelli, Caterina Lo Rizzo, Anna Maria Pinto, Giada Beligni, Andrea Tommasi, Laura Di Sarno, Maria Palmieri, Miriam Lucia Carriero, Diana Alaverdian, Stefano Busani, Raffaele Bruno, Marco Vecchia, Mary Ann Belli, Nicola Picchiotti, Maurizio Sanarico, Marco Gori, Simone Furini, Stefania Mantovani, Serena Ludovisi, Mario Umberto Mondelli, Francesco Castelli, Eugenia Quiros-Roldan, Melania Degli Antoni, Isabella Zanella, Massimo Vaghi, Stefano Rusconi, Matteo Siano, Francesca Montagnani, Arianna Emiliozzi, Massimiliano Fabbiani, Barbara Rossetti, Elena Bargagli, Laura Bergantini, Miriana D’Alessandro, Paolo Cameli, David Bennett, Federico Anedda, Simona Marcantonio, Sabino Scolletta, Federico Franchi, Maria Antonietta Mazzei, Susanna Guerrini, Edoardo Conticini, Luca Cantarini, Bruno Frediani, Danilo Tacconi, Chiara Spertilli, Marco Feri, Alice Donati, Raffaele Scala, Luca Guidelli, Genni Spargi, Marta Corridi, Cesira Nencioni, Leonardo Croci, Maria Bandini, Gian Piero Caldarelli, Paolo Piacentini, Elena Desanctis, Silvia Cappelli, Anna Canaccini, Agnese Verzuri, Valentina Anemoli, Agostino Ognibene, Alessandro Pancrazzi, Maria Lorubbio, Antonella D’Arminio Monforte, Federica Gaia Miraglia, Massimo Girardis, Sophie Venturelli, Andrea Cossarizza, Andrea Antinori, Alessandra Vergori, Arianna Gabrieli, Agostino Riva, Daniela Francisci, Elisabetta Schiaroli, Francesco Paciosi, Pier Giorgio Scotton, Francesca Andretta, Sandro Panese, Renzo Scaggiante, Francesca Gatti, Saverio Giuseppe Parisi, Stefano Baratti, Matteo Della Monica, Carmelo Piscopo, Mario Capasso, Roberta Russo, Immacolata Andolfo, Achille Iolascon, Giuseppe Fiorentino, Massimo Carella, Marco Castori, Giuseppe Merla, Gabriella Maria Squeo, Filippo Aucella, Pamela Raggi, Carmen Marciano, Rita Perna, Matteo Bassetti, Antonio Di Biagio, Maurizio Sanguinetti, Luca Masucci, Serafina Valente, Marco Mandalà, Alessia Giorli, Lorenzo Salerni, Patrizia Zucchi, Pierpaolo Parravicini, Elisabetta Menatti, Tullio Trotta, Ferdinando Giannattasio, Gabriella Coiro, Fabio Lena, Domenico A. Coviello, Cristina Mussini, Enrico Martinelli, Sandro Mancarella, Luisa Tavecchia, Lia Crotti, Chiara Gabbi, Marco Rizzi, Franco Maggiolo, Diego Ripamonti, Tiziana Bachetti, Maria Teresa La Rovere, Simona Sarzi-Braga, Maurizio Bussotti, Stefano Ceri, Pietro Pinoli, Francesco Raimondi, Filippo Biscarini, Alessandra Stella, Kristina Zguro, Katia Capitani, Claudia Suardi, Mari E.K. Niemi, Mattia Cordioli, Sara Pigazzini, Simona Dei, Gianfranco Parati, Sabrina Ravaglia, Rosangela Artuso, Mattia Cordioli, Sara Pigazzini, Lindokuhle Nkambule, Giordano Bottà, Paolo Di Domenico, Ilaria Rancan, Antonio Perrella Francesco Bianchi, Davide Romani, Paola Bergomi, Emanuele Catena, Riccardo Colombo, Marco Tanfoni, Antonella Vincenti, Claudio Ferri, Davide Grassi, Gloria Pessina, Mario Tumbarello, Massimo Di Pietro, Ravaglia Sabrina, Sauro Luchi, Chiara Barbieri, Donatella Acquilini, Elena Andreucci, Francesco Paciosi, Francesco Vladimiro Segala, Giusy Tiseo, Marco Falcone, Mirjam Lista, Monica Poscente, Oreste De Vivo, Paola Petrocelli, Alessandra Guarnaccia, Silvia Baroni, Albert V. Smith, Andrew P. Boughton, Kevin W. Li, Jonathon LeFaive, Aubrey Annis, Anne E. Justice, Tooraj Mirshahi, Geetha Chittoor, Navya Shilpa Josyula, Jack A. Kosmicki, Manuel A.R. Ferreira, Joseph B. Leader, Dave J. Carey, Matthew C. Gass, Julie E. Horowitz, Michael N. Cantor, Ashish Yadav, Aris Baras, Goncalo R. Abecasis, David A. van Heel, Karen A. Hunt, Dan Mason, Qin Qin Huang, Sarah Finer, Genes & Health Research Team, Bhavi Trivedi, Christopher J. Griffiths, Hilary C. Martin, John Wright, Richard C. Trembath, Nicole Soranzo, Jing Hua Zhao, Adam S. Butterworth, John Danesh, Emanuele Di Angelantonio, Lude Franke Marike Boezen, Patrick Deelen, Annique Claringbould, Esteban Lopera, Robert Warmerdam, Judith. M. Vonk, Irene van Blokland, Pauline Lanting, Anil P.S. Ori, Brooke Wolford Sebastian Zöllner, Jiongming Wang, Andrew Beck, Gina Peloso, Yuk-Lam Ho, Yan V. Sun, Jennifer E. Huffman, Christopher J. O’Donnell, Kelly Cho, Phil Tsao, J. Michael Gaziano, Michel (M.G.) Nivard, Eco (E.J.C.) de geus, Meike Bartels, Jouke Jan Hottenga, Scott T. Weiss, Elizabeth W. Karlson, Jordan W. Smoller, Robert C. Green, Yen-Chen Anne Feng, Josep Mercader, Shawn N. Murphy, James B. Meigs, Ann E. Woolley, Emma F. Perez, Daniel Rader, Anurag Verma, Marylyn D. Ritchie, Binglan Li, Shefali S. Verma, Anastasia Lucas, Yuki Bradford, Hugo Zeberg, Robert Frithiof, Michael Hultström, Mari E.K. Niemi, Mattia Cordioli, Sara Pigazzini, Miklos Lipcsey, Lindo Nkambul, Nicolas Tardif, Olav Rooyackers, Jonathan Grip, Tomislav Maricic, Tomoko Nakanishi, Guillaume Butler-Laporte, Vincenzo Forgetta, J. Brent Richards, Konrad J. Karczewski, Elizabeth G. Atkinson, Masahiro Kanai, Kristin Tsuo, Nikolas Baya, Patrick Turley, Rahul Gupta, Shawneequa Callier, Raymond K. Walters, Duncan S. Palmer, Gopal Sarma, Matthew Solomonson, Nathan Cheng, Wenhan Lu, Sam Bryant, Claire Churchhouse, Caroline Cusick, Jacqueline I. Goldstein, Daniel King, Wei Zhou, Cotton Seed, Hilary Finucane, Alicia R. Martin, Sam Bryant, F. Kyle Satterstrom, Daniel J. Wilson, Jacob Armstrong, Justine K. Rudkin, Gavin Band, Sarah G. Earle, Shang-Kuan Lin, Nicolas Arning, Derrick W. Crook, David H. Wyllie, Anne Marie O’Connell, Chris C.A. Spencer, Nils Koelling, Mark J. Caulfield, Richard H. Scott, Tom Fowler, Loukas Moutsianas, Athanasios Kousathanas, Dorota Pasko, Susan Walker, Augusto Rendon, Alex Stuckey, Christopher A. Odhams, Daniel Rhodes, Georgia Chan, Prabhu Arumugam, Catherine A. Ball, Eurie L. Hong, Kristin Rand, Ahna Girshick, Harendra Guturu, Asher Haug Baltzell, Genevieve Roberts, Danny Park, Marie Coignet, Shannon McCurdy, Spencer Knight, Raghavendran Partha, Brooke Rhead, Miao Zhang, Nathan Berkowitz, Michael Gaddis, Keith Noto, Luong Ruiz, Milos Pavlovic, Laura G. Sloofman, Shea J. Andrews, Alexander W. Charney, Noam D. Beckmann, Eric E. Schadt, Daniel M. Jordan, Ryan C. Thompson, Kyle Gettler, Noura S. Abul-Husn, Steven Ascolillo, Joseph D. Buxbaum, Kumardeep Chaudhary, Judy H. Cho, Yuval Itan, Eimear E. Kenny, Gillian M. Belbin, Stuart C. Sealfon, Robert P. Sebra, Irene Salib, Brett L. Collins, Tess Levy, Bari Britvan, Katherine Keller, Lara Tang, Michael Peruggia, Liam L. Hiester, Kristi Niblo, Alexandra Aksentijevich, Alexander Labkowsky, Avromie Karp, Menachem Zlatopolsky, Michael Preuss, Ruth J.F. Loos, Girish N. Nadkarni, Ron Do, Clive Hoggart, Sam Choi, Slayton J. Underwood, Paul O’Reilly, Laura M. Huckins, Marissa Zyndorf. A complete list of the members of the Consortium and their affiliations is provided at https://docs.google.com/spreadsheets/d/1cp9pFeFUxXz5WMjRFv4X-AM1Hlc0iXYJa1rorSSj2Dc.
Acknowledgments
This work was supported by Emergency COVID-19 Seed Award from the Tobacco-Related Disease Research Program of the University of California, grant R00RG2716, and by the NHGRI CEGS grant RM1HG011558. J.P.N. and T.D.A. were supported by an NIH training grant (T15LM011271).
Author contributions
K.A.F. and M.D. conceived the study. M.D., A.D.-C., T.D.A., J.P.N., and H.M. conducted analyses. M.D. and K.A.F. prepared the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: November 16, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.110020.
Contributor Information
The COVID-19 Host Genetics Initiative:
Benjamin M. Neale, Mark Daly, Andrea Ganna, Christine Stevens, Gita A. Pathak, Shea J. Andrews, Masahiro Kanai, Mattia Cordioli, Andrea Ganna, Juha Karjalainen, Gita A. Pathak, Renato Polimanti, Shea J. Andrews, Mattia Cordioli, Matti Pirinen, Masahiro Kanai, Nadia Harerimana, Kumar Veerapen, Brooke Wolford, Huy Nguyen, Matthew Solomonson, Christine Stevens, Rachel G. Liao, Karolina Chwialkowska, Amy Trankiem, Mary K. Balaconis, Caroline Hayward, Anne Richmond, Archie Campbell, Marcela Morris, Chloe Fawns-Ritchie, Joseph T. Glessner, Douglas M. Shaw, Xiao Chang, Hannah Polikowski, Petty E. Lauren, Hung-Hsin Chen, Zhu Wanying, Hakon Hakonarson, David J. Porteous, Jennifer Below, Kari North, Joseph B. McCormick, Paul R.H.J. Timmers, James F. Wilson, Albert Tenesa, Kenton D’Mellow, Shona M. Kerr, Mari E.K. Niemi, Mattia Cordioli, Lindokuhle Nkambul, Kathrin Aprile von Hohenstaufen, Ali Sobh, Madonna M. Eltoukhy, Amr M. Yassen, Mohamed A.F. Hegazy, Kamal Okasha, Mohammed A. Eid, Hanteera S. Moahmed, Doaa Shahin, Yasser M. El-Sherbiny, Tamer A. Elhadidy, Mohamed S. Abd Elghafar, Jehan J. El-Jawhari, Attia A.S. Mohamed, Marwa H. Elnagdy, Amr Samir, Mahmoud Abdel-Aziz, Walid T. Khafaga, Walaa M. El-Lawaty, Mohamed S. Torky, Mohamed R. El-shanshory, Chiara Batini, Paul H. Lee, Nick Shrine, Alexander T. Williams, Martin D. Tobin, Anna L. Guyatt, Catherine John, Richard J. Packer, Altaf Ali, Robert C. Free, Xueyang Wang, Louise V. Wain, Edward J. Hollox, Laura D. Venn, Catherine E. Bee, Emma L. Adams, Mari E.K. Niemi, Ahmadreza Niavarani, Mattia Cordioli, Lindokuhle Nkambul, Bahareh Sharififard, Rasoul Aliannejad, Ali Amirsavadkouhi, Zeinab Naderpour, Hengameh Ansari Tadi, Afshar Etemadi Aleagha, Saeideh Ahmadi, Seyed Behrooz Mohseni Moghaddam, Alireza Adamsara, Morteza Saeedi, Hamed Abdollahi, Abdolmajid Hosseini, Pajaree Chariyavilaskul, Monpat Chamnanphon, Thitima B. Suttichet, Vorasuk Shotelersuk, Monnat Pongpanich, Chureerat Phokaew, Wanna Chetruengchai, Watsamon Jantarabenjakul, Opass Putchareon, Pattama Torvorapanit, Thanyawee Puthanakit, Pintip Suchartlikitwong, Nattiya Hirankarn, Voraphoj Nilaratanakul, Pimpayao Sodsai, Ben M. Brumpton, Kristian Hveem, Cristen Willer, Brooke Wolford, Wei Zhou, Tormod Rogne, Erik Solligard, Bjørn Olav Åsvold, Malak Abedalthagafi, Manal Alaamery, Saleh Alqahtani, Dona Baraka, Fawz Al Harthi, Ebtehal Alsolm, Leen Abu Safieh, Albandary M. Alowayn, Fatimah Alqubaishi, Amal Al Mutairi, Serghei Mangul, Abdulraheem Alshareef, Mona Sawaji, Mansour Almutairi, Nora Aljawini, Nour Albesher, Yaseen M. Arabi, Ebrahim S. Mahmoud, Amin K. Khattab, Roaa T. Halawani, Ziab Z. Alahmadey, Jehad K. Albakri, Walaa A. Felemban, Bandar A. Suliman, Rana Hasanato, Laila Al-Awdah, Jahad Alghamdi, Deema AlZahrani, Sameera AlJohani, Hani Al-Afghani, May Alrashed, Nouf AlDhawi, Hadeel AlBardis, Sarah Alkwai, Moneera Alswailm, Faisal Almalki, Maha Albeladi, Iman Almohammed, Eman Barhoush, Anoud Albader, Salam Massadeh, Abdulaziz AlMalik, Sara Alotaibi, Bader Alghamdi, Junghyun Jung, Mohammad S. Fawzy, Yunsung Lee, Per Magnus, Lill-Iren S. Trogstad, Øyvind Helgeland, Jennifer R. Harris, Massimo Mangino, Tim D. Spector, Duncan Emma, Sandra P. Smieszek, Bartlomiej P. Przychodzen, Christos Polymeropoulos, Vasilios Polymeropoulos, Mihael H. Polymeropoulos, Israel Fernandez-Cadenas, Jordi Perez-Tur, Laia Llucià-Carol, Natalia Cullell, Elena Muiño, Jara Cárcel-Márquez, Marta L. DeDiego, Lara Lloret Iglesias, Anna M. Planas, Alex Soriano, Veronica Rico, Daiana Agüero, Josep L. Bedini, Francisco Lozano, Carlos Domingo, Veronica Robles, Francisca Ruiz-Jaén, Leonardo Márquez, Juan Gomez, Eliecer Coto, Guillermo M. Albaiceta, Marta García-Clemente, David Dalmau, Maria J. Arranz, Beatriz Dietl, Alex Serra-Llovich, Pere Soler, Roger Colobrán, Andrea Martín-Nalda, Alba Parra Martínez, David Bernardo, Silvia Rojo, Aida Fiz-López, Elisa Arribas, Paloma de la Cal-Sabater, Tomás Segura, Esther González-Villa, Gemma Serrano-Heras, Joan Martí-Fàbregas, Elena Jiménez-Xarrié, Alicia de Felipe Mimbrera, Jaime Masjuan, Sebastian García-Madrona, Anna Domínguez-Mayoral, Joan Montaner Villalonga, Paloma Menéndez-Valladares, Daniel I. Chasman, Julie E. Buring, Paul M. Ridker, Giulianini Franco, Howard D. Sesso, JoAnn E. Manson, Xiao Chang, Joseph R. Glessner, Hakon Hakonarson, Caroline Hayward, Anne Richmond, David J. Porteous, Archie Campbell, Chloe Fawns-Ritchie, Carolina Medina-Gomez, Andre G. Uitterlinden, M. Arfan Ikram, Kati Kristiansson, Sami Koskelainen, Markus Perola, Kati Donner, Katja Kivinen, Aarno Palotie, Samuli Ripatti, Sanni Ruotsalainen, Mari Kaunisto, FinnGen, Tomoko Nakanishi, Guillaume Butler-Laporte, Vincenzo Forgetta, David R. Morrison, Biswarup Ghosh, Laetitia Laurent, Alexandre Belisle, Danielle Henry, Tala Abdullah, Olumide Adeleye, Noor Mamlouk, Nofar Kimchi, Zaman Afrasiabi, Nardin Rezk Branka Vulesevic, Meriem Bouab, Charlotte Guzman, Louis Petitjean, Chris Tselios, Xiaoqing Xue, Erwin Schurr, Jonathan Afilalo, Marc Afilalo, Maureen Oliveira, Bluma Brenner, Pierre Lepage, Jiannis Ragoussis, Daniel Auld, Nathalie Brassard, Madeleine Durand, Michaël Chassé, Daniel E. Kaufmann, G. Mark Lathrop, Vincent Mooser, J. Brent Richards, Rui Li, Darin Adra, Souad Rahmouni, Michel Georges, Michel Moutschen, Benoit Misset, Gilles Darcis, Julien Guiot, Julien Guntz, Samira Azarzar, Stéphanie Gofflot, Yves Beguin, Sabine Claassen, Olivier Malaise, Pascale Huynen, Christelle Meuris, Marie Thys, Jessica Jacques, Philippe Léonard, Frederic Frippiat, Jean-Baptiste Giot, Anne-Sophie Sauvage, Christian Von Frenckell, Yasmine Belhaj, Bernard Lambermont, Mari E.K. Niemi, Mattia Cordioli, Sara Pigazzini, Lindokuhle Nkambule, Michelle Daya, Jonathan Shortt, Nicholas Rafaels, Stephen J. Wicks, Kristy Crooks, Kathleen C. Barnes, Christopher R. Gignoux, Sameer Chavan, Triin Laisk, Kristi Läll, Maarja Lepamets, Reedik Mägi, Tõnu Esko, Ene Reimann, Lili Milani, Helene Alavere, Kristjan Metsalu, Mairo Puusepp, Andres Metspalu, Paul Naaber, Edward Laane, Jaana Pesukova, Pärt Peterson, Kai Kisand, Jekaterina Tabri, Raili Allos, Kati Hensen, Joel Starkopf, Inge Ringmets, Anu Tamm, Anne Kallaste, Pierre-Yves Bochud, Carlo Rivolta, Stéphanie Bibert, Mathieu Quinodoz, Dhryata Kamdar, Noémie Boillat, Semira Gonseth Nussle, Werner Albrich, Noémie Suh, Dionysios Neofytos, Véronique Erard, Cathy Voide, FHoGID, RegCOVID, P-PredictUs, SeroCOVID, CRiPSI, Rafael de Cid, Iván Galván-Femenía, Natalia Blay, Anna Carreras, Beatriz Cortés, Xavier Farré, Lauro Sumoy, Victor Moreno, Josep Maria Mercader, Marta Guindo-Martinez, David Torrents, Manolis Kogevinas, Judith Garcia-Aymerich, Gemma Castaño-Vinyals, Carlota Dobaño, Alessandra Renieri, Francesca Mari, Chiara Fallerini, Sergio Daga, Elisa Benetti, Margherita Baldassarri, Francesca Fava, Elisa Frullanti, Floriana Valentino, Gabriella Doddato, Annarita Giliberti, Rossella Tita, Sara Amitrano, Mirella Bruttini, Susanna Croci, Ilaria Meloni, Maria Antonietta Mencarelli, Caterina Lo Rizzo, Anna Maria Pinto, Giada Beligni, Andrea Tommasi, Laura Di Sarno, Maria Palmieri, Miriam Lucia Carriero, Diana Alaverdian, Stefano Busani, Raffaele Bruno, Marco Vecchia, Mary Ann Belli, Nicola Picchiotti, Maurizio Sanarico, Marco Gori, Simone Furini, Stefania Mantovani, Serena Ludovisi, Mario Umberto Mondelli, Francesco Castelli, Eugenia Quiros-Roldan, Melania Degli Antoni, Isabella Zanella, Massimo Vaghi, Stefano Rusconi, Matteo Siano, Francesca Montagnani, Arianna Emiliozzi, Massimiliano Fabbiani, Barbara Rossetti, Elena Bargagli, Laura Bergantini, Miriana D’Alessandro, Paolo Cameli, David Bennett, Federico Anedda, Simona Marcantonio, Sabino Scolletta, Federico Franchi, Maria Antonietta Mazzei, Susanna Guerrini, Edoardo Conticini, Luca Cantarini, Bruno Frediani, Danilo Tacconi, Chiara Spertilli, Marco Feri, Alice Donati, Raffaele Scala, Luca Guidelli, Genni Spargi, Marta Corridi, Cesira Nencioni, Leonardo Croci, Maria Bandini, Gian Piero Caldarelli, Paolo Piacentini, Elena Desanctis, Silvia Cappelli, Anna Canaccini, Agnese Verzuri, Valentina Anemoli, Agostino Ognibene, Alessandro Pancrazzi, Maria Lorubbio, Antonella D’Arminio Monforte, Federica Gaia Miraglia, Massimo Girardis, Sophie Venturelli, Andrea Cossarizza, Andrea Antinori, Alessandra Vergori, Arianna Gabrieli, Agostino Riva, Daniela Francisci, Elisabetta Schiaroli, Francesco Paciosi, Pier Giorgio Scotton, Francesca Andretta, Sandro Panese, Renzo Scaggiante, Francesca Gatti, Saverio Giuseppe Parisi, Stefano Baratti, Matteo Della Monica, Carmelo Piscopo, Mario Capasso, Roberta Russo, Immacolata Andolfo, Achille Iolascon, Giuseppe Fiorentino, Massimo Carella, Marco Castori, Giuseppe Merla, Gabriella Maria Squeo, Filippo Aucella, Pamela Raggi, Carmen Marciano, Rita Perna, Matteo Bassetti, Antonio Di Biagio, Maurizio Sanguinetti, Luca Masucci, Serafina Valente, Marco Mandalà, Alessia Giorli, Lorenzo Salerni, Patrizia Zucchi, Pierpaolo Parravicini, Elisabetta Menatti, Tullio Trotta, Ferdinando Giannattasio, Gabriella Coiro, Fabio Lena, Domenico A. Coviello, Cristina Mussini, Enrico Martinelli, Sandro Mancarella, Luisa Tavecchia, Lia Crotti, Chiara Gabbi, Marco Rizzi, Franco Maggiolo, Diego Ripamonti, Tiziana Bachetti, Maria Teresa La Rovere, Simona Sarzi-Braga, Maurizio Bussotti, Stefano Ceri, Pietro Pinoli, Francesco Raimondi, Filippo Biscarini, Alessandra Stella, Kristina Zguro, Katia Capitani, Claudia Suardi, Mari E.K. Niemi, Mattia Cordioli, Sara Pigazzini, Simona Dei, Gianfranco Parati, Sabrina Ravaglia, Rosangela Artuso, Mattia Cordioli, Sara Pigazzini, Lindokuhle Nkambule, Giordano Bottà, Paolo Di Domenico, Ilaria Rancan, Antonio Perrella Francesco Bianchi, Davide Romani, Paola Bergomi, Emanuele Catena, Riccardo Colombo, Marco Tanfoni, Antonella Vincenti, Claudio Ferri, Davide Grassi, Gloria Pessina, Mario Tumbarello, Massimo Di Pietro, Ravaglia Sabrina, Sauro Luchi, Chiara Barbieri, Donatella Acquilini, Elena Andreucci, Francesco Paciosi, Francesco Vladimiro Segala, Giusy Tiseo, Marco Falcone, Mirjam Lista, Monica Poscente, Oreste De Vivo, Paola Petrocelli, Alessandra Guarnaccia, Silvia Baroni, Albert V. Smith, Andrew P. Boughton, Kevin W. Li, Jonathon LeFaive, Aubrey Annis, Anne E. Justice, Tooraj Mirshahi, Geetha Chittoor, Navya Shilpa Josyula, Jack A. Kosmicki, Manuel A.R. Ferreira, Joseph B. Leader, Dave J. Carey, Matthew C. Gass, Julie E. Horowitz, Michael N. Cantor, Ashish Yadav, Aris Baras, Goncalo R. Abecasis, David A. van Heel, Karen A. Hunt, Dan Mason, Qin Qin Huang, Sarah Finer, Genes & Health Research Team, Bhavi Trivedi, Christopher J. Griffiths, Hilary C. Martin, John Wright, Richard C. Trembath, Nicole Soranzo, Jing Hua Zhao, Adam S. Butterworth, John Danesh, Emanuele Di Angelantonio, Lude Franke Marike Boezen, Patrick Deelen, Annique Claringbould, Esteban Lopera, Robert Warmerdam, Judith.M. Vonk, Irene van Blokland, Pauline Lanting, Anil P.S. Ori, Brooke Wolford Sebastian Zöllner, Jiongming Wang, Andrew Beck, Gina Peloso, Yuk-Lam Ho, Yan V. Sun, Jennifer E. Huffman, Christopher J. O’Donnell, Kelly Cho, Phil Tsao, J. Michael Gaziano, Michel (M.G.) Nivard, Eco (E.J.C.) de geus, Meike Bartels, Jouke Jan Hottenga, Scott T. Weiss, Elizabeth W. Karlson, Jordan W. Smoller, Robert C. Green, Yen-Chen Anne Feng, Josep Mercader, Shawn N. Murphy, James B. Meigs, Ann E. Woolley, Emma F. Perez, Daniel Rader, Anurag Verma, Marylyn D. Ritchie, Binglan Li, Shefali S. Verma, Anastasia Lucas, Yuki Bradford, Hugo Zeberg, Robert Frithiof, Michael Hultström, Mari E.K. Niemi, Mattia Cordioli, Sara Pigazzini, Miklos Lipcsey, Lindo Nkambul, Nicolas Tardif, Olav Rooyackers, Jonathan Grip, Tomislav Maricic, Tomoko Nakanishi, Guillaume Butler-Laporte, Vincenzo Forgetta, J. Brent Richards, Konrad J. Karczewski, Elizabeth G. Atkinson, Masahiro Kanai, Kristin Tsuo, Nikolas Baya, Patrick Turley, Rahul Gupta, Shawneequa Callier, Raymond K. Walters, Duncan S. Palmer, Gopal Sarma, Matthew Solomonson, Nathan Cheng, Wenhan Lu, Sam Bryant, Claire Churchhouse, Caroline Cusick, Jacqueline I. Goldstein, Daniel King, Wei Zhou, Cotton Seed, Hilary Finucane, Alicia R. Martin, Sam Bryant, F. Kyle Satterstrom, Daniel J. Wilson, Jacob Armstrong, Justine K. Rudkin, Gavin Band, Sarah G. Earle, Shang-Kuan Lin, Nicolas Arning, Derrick W. Crook, David H. Wyllie, Anne Marie O’Connell, Chris C.A. Spencer, Nils Koelling, Mark J. Caulfield, Richard H. Scott, Tom Fowler, Loukas Moutsianas, Athanasios Kousathanas, Dorota Pasko, Susan Walker, Augusto Rendon, Alex Stuckey, Christopher A. Odhams, Daniel Rhodes, Georgia Chan, Prabhu Arumugam, Catherine A. Ball, Eurie L. Hong, Kristin Rand, Ahna Girshick, Harendra Guturu, Asher Haug Baltzell, Genevieve Roberts, Danny Park, Marie Coignet, Shannon McCurdy, Spencer Knight, Raghavendran Partha, Brooke Rhead, Miao Zhang, Nathan Berkowitz, Michael Gaddis, Keith Noto, Luong Ruiz, Milos Pavlovic, Laura G. Sloofman, Shea J. Andrews, Alexander W. Charney, Noam D. Beckmann, Eric E. Schadt, Daniel M. Jordan, Ryan C. Thompson, Kyle Gettler, Noura S. Abul-Husn, Steven Ascolillo, Joseph D. Buxbaum, Kumardeep Chaudhary, Judy H. Cho, Yuval Itan, Eimear E. Kenny, Gillian M. Belbin, Stuart C. Sealfon, Robert P. Sebra, Irene Salib, Brett L. Collins, Tess Levy, Bari Britvan, Katherine Keller, Lara Tang, Michael Peruggia, Liam L. Hiester, Kristi Niblo, Alexandra Aksentijevich, Alexander Labkowsky, Avromie Karp, Menachem Zlatopolsky, Michael Preuss, Ruth J.F. Loos, Girish N. Nadkarni, Ron Do, Clive Hoggart, Sam Choi, Slayton J. Underwood, Paul O’Reilly, Laura M. Huckins, and Marissa Zyndorf
Supplemental information
The table shows the coordinates of each of the 23 loci with a minimum p value < 10−7 in at least one of the 16 meta-analyses obtained from the COVID-19 HGI data freeze 5. For each locus and each study, we report the p value and effect size of the variant with the most significant p value.
The table shows the pairwise colocalization results between eQTLs and COVID-19 GWAS signals for each gene mapping into the 23 COVID-19 genome-wide significant loci. Shown are each locus coordinates (chromosome, start and end positions), gene ID, gene name, tissue tested for eQTLs, GWAS study, the SNP ID of the variant with the highest posterior probability of being causal, the p values of this variant in both studies, the number of SNPs tested for colocalization, the posterior probabilities of the five colocalization hypotheses, and posterior probability of the lead SNP. The colocalization hypotheses are (1) H0, neither study has a significant association at the tested locus; (2) H1, only the first study is associated; (3) H2, only the second study is associated; (4) H3, both studies are associated but the underlying variants are different; and (5) H4, both studies are associated and share the same underlying variants.
We found eQTL signals for 20 genes that colocalized with the COVID-19 GWAS signals at 11 loci (PP-H4 > 0.9). For each of these signals, we performed fine mapping using the coloc.abf function in the R package coloc. This table reports the information about the variant with the highest PPA at each of these signals. Specifically, we show locus, COVID-19 study, gene ID, gene name, eQTL tissue, PPA of each of the five colocalization hypotheses, the variant with highest PPA (including position, reference allele, alternative allele, and RS ID), its p value and effect size in the GWAS and eQTL analyses, its PPA, and the size of the 99% credible set.
For each of the 20 genes whose eQTL signals colocalize with GWAS signals with PPA > 0.9, we selected the phenotype (C2, A1, B1, or B2) that had the eQTL signal with the highest PPA (hypothesis 4) and report fine-mapping results obtained using the coloc.abf function in the R package coloc. The table shows the phenotype with the highest PPA and information about each tested variant: locus, phenotype, gene ID, gene name, variant information (including RS ID), PPA, cumulative PPA, and whether it is included in the 99% credible set. For each colocalization, variants are sorted by their PPA. There are two exceptions to reporting the phenotype with the highest PPA. For the severity-associated 12:112850796-113925679 locus, we report the fine mapping between its eQTL signal in adipose tissue and the B2 phenotype, but PPA is higher with the susceptibility phenotype (C2), although both colocalizations share the same putative causal variant (rs10774671) and their PPA is > 0.99. Additionally, the 1:154605882-155673527 locus is severity-associated, however, the FAM189B eQTL in Heart Atrial Appendage colocalized only with phenotype C2. Therefore, we report the fine mapping between the FAM189B eQTL and the C2 phenotype.
Data and code availability
This paper does not have original code to report.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Ahn D., Prince A. Participation of the IL-10RB Related Cytokines, IL-22 and IFN-λ in Defense of the Airway Mucosal Barrier. Front. Cell. Infect. Microbiol. 2020;10:300. doi: 10.3389/fcimb.2020.00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alasoo K., Rodrigues J., Mukhopadhyay S., Knights A.J., Mann A.L., Kundu K., Hale C., Dougan G., Gaffney D.J., Gaffney D.J., HIPSCI Consortium Shared genetic effects on chromatin and gene expression indicate a role for enhancer priming in immune response. Nat. Genet. 2018;50:424–431. doi: 10.1038/s41588-018-0046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragam K.G., Chaffin M., Levinson R.T., McDermott G., Choi S.H., Shoemaker M.B., Haas M.E., Weng L.C., Lindsay M.E., Smith J.G., et al. GRADE Investigators Phenotypic Refinement of Heart Failure in a National Biobank Facilitates Genetic Discovery. Circulation. 2018 doi: 10.1161/CIRCULATIONAHA.118.035774. Published online November 11, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astle W.J., Elding H., Jiang T., Allen D., Ruklisa D., Mann A.L., Mead D., Bouman H., Riveros-Mckay F., Kostadima M.A., et al. The Allelic Landscape of Human Blood Cell Trait Variation and Links to Common Complex Disease. Cell. 2016;167:1415–1429.e19. doi: 10.1016/j.cell.2016.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins J.L., Masoli J.A.H., Delgado J., Pilling L.C., Kuo C.L., Kuchel G.A., Melzer D. Preexisting Comorbidities Predicting COVID-19 and Mortality in the UK Biobank Community Cohort. J. Gerontol. A Biol. Sci. Med. Sci. 2020;75:2224–2230. doi: 10.1093/gerona/glaa183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M., Litvan I., Houlden H., Adamson J., Dickson D., Perez-Tur J., Hardy J., Lynch T., Bigio E., Hutton M. Association of an extended haplotype in the tau gene with progressive supranuclear palsy. Hum. Mol. Genet. 1999;8:711–715. doi: 10.1093/hmg/8.4.711. [DOI] [PubMed] [Google Scholar]
- Banerjee S., Gusho E., Gaughan C., Dong B., Gu X., Holvey-Bates E., Talukdar M., Li Y., Weiss S.R., Sicheri F., et al. OAS-RNase L innate immune pathway mediates the cytotoxicity of a DNA-demethylating drug. Proc. Natl. Acad. Sci. USA. 2019;116:5071–5076. doi: 10.1073/pnas.1815071116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnkob M.B., Pottegård A., Støvring H., Haunstrup T.M., Homburg K., Larsen R., Hansen M.B., Titlestad K., Aagaard B., Møller B.K., Barington T. Reduced prevalence of SARS-CoV-2 infection in ABO blood group O. Blood Adv. 2020;4:4990–4993. doi: 10.1182/bloodadvances.2020002657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentham J., Morris D.L., Graham D.S.C., Pinder C.L., Tombleson P., Behrens T.W., Martín J., Fairfax B.P., Knight J.C., Chen L., et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat. Genet. 2015;47:1457–1464. doi: 10.1038/ng.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birdwell L.D., Zalinger Z.B., Li Y., Wright P.W., Elliott R., Rose K.M., Silverman R.H., Weiss S.R. Activation of RNase L by Murine Coronavirus in Myeloid Cells Is Dependent on Basal Oas Gene Expression and Independent of Virus-Induced Interferon. J. Virol. 2016;90:3160–3172. doi: 10.1128/JVI.03036-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonder M.J., Smail C., Gloudemans M.J., Frésard L., Jakubosky D., D’Antonio M., Li X., Ferraro N.M., Carcamo-Orive I., Mirauta B., et al. HipSci Consortium. iPSCORE consortium. Undiagnosed Diseases Network. PhLiPS consortium Identification of rare and common regulatory variants in pluripotent cells using population-scale transcriptomics. Nat. Genet. 2021;53:313–321. doi: 10.1038/s41588-021-00800-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buil A., Brown A.A., Lappalainen T., Viñuela A., Davies M.N., Zheng H.F., Richards J.B., Glass D., Small K.S., Durbin R., et al. Gene-gene and gene-environment interactions detected by transcriptome sequence analysis in twins. Nat. Genet. 2015;47:88–91. doi: 10.1038/ng.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Ge B., Casale F.P., Vasquez L., Kwan T., Garrido-Martín D., Watt S., Yan Y., Kundu K., Ecker S., et al. Genetic Drivers of Epigenetic and Transcriptional Variation in Human Immune Cells. Cell. 2016;167:1398–1414.e24. doi: 10.1016/j.cell.2016.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu X., Pan C.M., Zhao S.X., Liang J., Gao G.Q., Zhang X.M., Yuan G.Y., Li C.G., Xue L.Q., Shen M., et al. China Consortium for Genetics of Autoimmune Thyroid Disease A genome-wide association study identifies two new risk loci for Graves’ disease. Nat. Genet. 2011;43:897–901. doi: 10.1038/ng.898. [DOI] [PubMed] [Google Scholar]
- COVID-19 Host Genetics Initiative The COVID-19 Host Genetics Initiative, a global initiative to elucidate the role of host genetic factors in susceptibility and severity of the SARS-CoV-2 virus pandemic. Eur. J. Hum. Genet. 2020;28:715–718. doi: 10.1038/s41431-020-0636-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COVID-19 Host Genetics Initiative Mapping the human genetic architecture of COVID-19. Nature. 2021 doi: 10.1038/s41586-021-03767-x. Published online July 8, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings M.J., Baldwin M.R., Abrams D., Jacobson S.D., Meyer B.J., Balough E.M., Aaron J.G., Claassen J., Rabbani L.E., Hastie J., et al. Epidemiology, clinical course, and outcomes of critically ill adults with COVID-19 in New York City: a prospective cohort study. Lancet. 2020;395:1763–1770. doi: 10.1016/S0140-6736(20)31189-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBoever C., Li H., Jakubosky D., Benaglio P., Reyna J., Olson K.M., Huang H., Biggs W., Sandoval E., D’Antonio M., et al. Large-Scale Profiling Reveals the Influence of Genetic Variation on Gene Expression in Human Induced Pluripotent Stem Cells. Cell Stem Cell. 2017;20:533–546.e7. doi: 10.1016/j.stem.2017.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan C.J., Mohamad S.M., Young D.F., Skelton A.J., Leahy T.R., Munday D.C., Butler K.M., Morfopoulou S., Brown J.R., Hubank M., et al. Human IFNAR2 deficiency: Lessons for antiviral immunity. Sci. Transl. Med. 2015;7:307ra154. doi: 10.1126/scitranslmed.aac4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghaus D., Degenhardt F., Bujanda L., Buti M., Albillos A., Invernizzi P., Fernández J., Prati D., Baselli G., Asselta R., et al. Severe Covid-19 GWAS Group Genomewide Association Study of Severe Covid-19 with Respiratory Failure. N. Engl. J. Med. 2020;383:1522–1534. doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J., Wang H., Ye G., Cao X., Xu X., Tan W., Zhang Y. Letter to the Editor: Low-density lipoprotein is a potential predictor of poor prognosis in patients with coronavirus disease 2019. Metabolism. 2020;107:154243. doi: 10.1016/j.metabol.2020.154243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoreschi K., Augustin M., Baraliakos X., Krönke G., Schneider M., Schreiber S., Schulze-Koops H., Zeißig S., Thaçi D. TYK2 inhibition and its potential in the treatment of chronic inflammatory immune diseases. J. Dtsch. Dermatol. Ges. 2021;19:1409–1420. doi: 10.1111/ddg.14585. [DOI] [PubMed] [Google Scholar]
- Giambartolomei C., Vukcevic D., Schadt E.E., Franke L., Hingorani A.D., Wallace C., Plagnol V. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilly A., Suveges D., Kuchenbaecker K., Pollard M., Southam L., Hatzikotoulas K., Farmaki A.E., Bjornland T., Waples R., Appel E.V.R., et al. Cohort-wide deep whole genome sequencing and the allelic architecture of complex traits. Nat. Commun. 2018;9:4674. doi: 10.1038/s41467-018-07070-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., et al. China Medical Treatment Expert Group for Covid-19 Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Arcelus M., Lappalainen T., Montgomery S.B., Buil A., Ongen H., Yurovsky A., Bryois J., Giger T., Romano L., Planchon A., et al. Passive and active DNA methylation and the interplay with genetic variation in gene regulation. eLife. 2013;2:e00523. doi: 10.7554/eLife.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz J.E., Kosmicki J.A., Damask A., Sharma D., Roberts G.H.L., Justice A.E., Banerjee N., Coignet M.V., Yadav A., Leader J.B., et al. Common genetic variants identify targets for COVID-19 and individuals at high risk of severe disease. medRxiv. 2021 [Google Scholar]
- Hu X., Chen D., Wu L., He G., Ye W. Declined serum high density lipoprotein cholesterol is associated with the severity of COVID-19 infection. Clin. Chim. Acta. 2020;510:105–110. doi: 10.1016/j.cca.2020.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones E.Y., Fugger L., Strominger J.L., Siebold C. MHC class II proteins and disease: a structural perspective. Nat. Rev. Immunol. 2006;6:271–282. doi: 10.1038/nri1805. [DOI] [PubMed] [Google Scholar]
- Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. Genome Aggregation Database Consortium The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerimov N., Hayhurst J.D., Peikova K., Manning J.R., Walter P., Kolberg L., Samoviča M., Sakthivel M.P., Kuzmin I., Trevanion S.J., et al. eQTL Catalogue: a compendium of uniformly processed human gene expression and splicing QTLs. Nat. Genet. 2021;53:1290–1299. doi: 10.1038/s41588-021-00924-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koolen D.A., Vissers L.E., Pfundt R., de Leeuw N., Knight S.J., Regan R., Kooy R.F., Reyniers E., Romano C., Fichera M., et al. A new chromosome 17q21.31 microdeletion syndrome associated with a common inversion polymorphism. Nat. Genet. 2006;38:999–1001. doi: 10.1038/ng1853. [DOI] [PubMed] [Google Scholar]
- Kulminski A.M., Huang J., Loika Y., Arbeev K.G., Bagley O., Yashkin A., Duan M., Culminskaya I. Strong impact of natural-selection-free heterogeneity in genetics of age-related phenotypes. Aging (Albany NY) 2018;10:492–514. doi: 10.18632/aging.101407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen T., Sammeth M., Friedländer M.R., ’t Hoen P.A., Monlong J., Rivas M.A., Gonzàlez-Porta M., Kurbatova N., Griebel T., Ferreira P.G., et al. Geuvadis Consortium Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson N.B., Bell E.J., Decker P.A., Pike M., Wassel C.L., Tsai M.Y., Pankow J.S., Tang W., Hanson N.Q., Alexander K., et al. ABO blood group associations with markers of endothelial dysfunction in the Multi-Ethnic Study of Atherosclerosis. Atherosclerosis. 2016;251:422–429. doi: 10.1016/j.atherosclerosis.2016.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent E.M.N., Sofianatos Y., Komarova A., Gimeno J.-P., Samavarchi Tehrani P., Kim D.-K., Abdouni H., Duhamel M., Cassonnet P., Knapp J.J., et al. Global BioID-based SARS-CoV-2 proteins proximal interactome unveils novel ties between viral polypeptides and host factors involved in multiple COVID19-associated mechanisms. bioRxiv. 2020 doi: 10.1101/2020.08.28.272955. [DOI] [Google Scholar]
- Li M., Wang L., Shi D.C., Foo J.N., Zhong Z., Khor C.C., Lanzani C., Citterio L., Salvi E., Yin P.R., et al. Genome-Wide Meta-Analysis Identifies Three Novel Susceptibility Loci and Reveals Ethnic Heterogeneity of Genetic Susceptibility for IgA Nephropathy. J. Am. Soc. Nephrol. 2020;31:2949–2963. doi: 10.1681/ASN.2019080799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey K., Ayers C.K., Kondo K.K., Saha S., Advani S.M., Young S., Spencer H., Rusek M., Anderson J., Veazie S., et al. Racial and Ethnic Disparities in COVID-19-Related Infections, Hospitalizations, and Deaths : A Systematic Review. Ann. Intern. Med. 2021;174:362–373. doi: 10.7326/M20-6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan A., Taliun D., Thurner M., Robertson N.R., Torres J.M., Rayner N.W., Payne A.J., Steinthorsdottir V., Scott R.A., Grarup N., et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat. Genet. 2018;50:1505–1513. doi: 10.1038/s41588-018-0241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandala V.S., McKay M.J., Shcherbakov A.A., Dregni A.J., Kolocouris A., Hong M. Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers. Nat. Struct. Mol. Biol. 2020;27:1202–1208. doi: 10.1038/s41594-020-00536-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichaikul A., Wang X.Q., Sun L., Dupuis J., Borczuk A.C., Nguyen J.N., Raghu G., Hoffman E.A., Onengut-Gumuscu S., Farber E.A., et al. Genome-wide association study of subclinical interstitial lung disease in MESA. Respir. Res. 2017;18:97. doi: 10.1186/s12931-017-0581-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzer D., Perry J.R., Hernandez D., Corsi A.M., Stevens K., Rafferty I., Lauretani F., Murray A., Gibbs J.R., Paolisso G., et al. A genome-wide association study identifies protein quantitative trait loci (pQTLs) PLoS Genet. 2008;4:e1000072. doi: 10.1371/journal.pgen.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyts I., Casanova J.L. Viral infections in humans and mice with genetic deficiencies of the type I IFN response pathway. Eur. J. Immunol. 2021;51:1039–1061. doi: 10.1002/eji.202048793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muise A.M., Walters T., Wine E., Griffiths A.M., Turner D., Duerr R.H., Regueiro M.D., Ngan B.Y., Xu W., Sherman P.M., et al. Protein-tyrosine phosphatase sigma is associated with ulcerative colitis. Curr. Biol. 2007;17:1212–1218. doi: 10.1016/j.cub.2007.06.013. [DOI] [PubMed] [Google Scholar]
- Myers A.J., Kaleem M., Marlowe L., Pittman A.M., Lees A.J., Fung H.C., Duckworth J., Leung D., Gibson A., Morris C.M., et al. The H1c haplotype at the MAPT locus is associated with Alzheimer’s disease. Hum. Mol. Genet. 2005;14:2399–2404. doi: 10.1093/hmg/ddi241. [DOI] [PubMed] [Google Scholar]
- Myers T.A., Chanock S.J., Machiela M.J. LDlinkR: An R Package for Rapidly Calculating Linkage Disequilibrium Statistics in Diverse Populations. Front. Genet. 2020;11:157. doi: 10.3389/fgene.2020.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nédélec Y., Sanz J., Baharian G., Szpiech Z.A., Pacis A., Dumaine A., Grenier J.C., Freiman A., Sams A.J., Hebert S., et al. Genetic Ancestry and Natural Selection Drive Population Differences in Immune Responses to Pathogens. Cell. 2016;167:657–669.e21. doi: 10.1016/j.cell.2016.09.025. [DOI] [PubMed] [Google Scholar]
- Oriol R., Mollicone R., Coullin P., Dalix A.M., Candelier J.J. Genetic regulation of the expression of ABH and Lewis antigens in tissues. APMIS. 1992;Suppl 27:28–38. [PubMed] [Google Scholar]
- Pairo-Castineira E., Clohisey S., Klaric L., Bretherick A.D., Rawlik K., Pasko D., Walker S., Parkinson N., Fourman M.H., Russell C.D., et al. GenOMICC Investigators. ISARIC4C Investigators. COVID-19 Human Genetics Initiative. 23andMe Investigators. BRACOVID Investigators. Gen-COVID Investigators Genetic mechanisms of critical illness in COVID-19. Nature. 2021;591:92–98. doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- Pedersen S.F., Ho Y.C. SARS-CoV-2: a storm is raging. J. Clin. Invest. 2020;130:2202–2205. doi: 10.1172/JCI137647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quach H., Rotival M., Pothlichet J., Loh Y.E., Dannemann M., Zidane N., Laval G., Patin E., Harmant C., Lopez M., et al. Genetic Adaptation and Neandertal Admixture Shaped the Immune System of Human Populations. Cell. 2016;167:643–656.e17. doi: 10.1016/j.cell.2016.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A.R. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr. Protoc. Bioinformatics. 2014;47:11.12–11.34. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson S., Hirsch J.S., Narasimhan M., Crawford J.M., McGinn T., Davidson K.W., Barnaby D.P., Becker L.B., Chelico J.D., Cohen S.L., et al. the Northwell COVID-19 Research Consortium Presenting Characteristics, Comorbidities, and Outcomes Among 5700 Patients Hospitalized With COVID-19 in the New York City Area. JAMA. 2020;323:2052–2059. doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche P.A., Furuta K. The ins and outs of MHC class II-mediated antigen processing and presentation. Nat. Rev. Immunol. 2015;15:203–216. doi: 10.1038/nri3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakornsakolpat P., Prokopenko D., Lamontagne M., Reeve N.F., Guyatt A.L., Jackson V.E., Shrine N., Qiao D., Bartz T.M., Kim D.K., et al. SpiroMeta Consortium. International COPD Genetics Consortium Genetic landscape of chronic obstructive pulmonary disease identifies heterogeneous cell-type and phenotype associations. Nat. Genet. 2019;51:494–505. doi: 10.1038/s41588-018-0342-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P., Abdouni H., Knight J.D.R., Astori A., Samson R., Lin Z.-Y., Kim D.-K., Knapp J.J., St-Germain J., Go C.D., et al. A SARS-CoV-2 – host proximity interactome. bioRxiv. 2020 doi: 10.1101/2020.09.03.282103. [DOI] [Google Scholar]
- Sawai H., Nishida N., Khor S.S., Honda M., Sugiyama M., Baba N., Yamada K., Sawada N., Tsugane S., Koike K., et al. Genome-wide association study identified new susceptible genetic variants in HLA class I region for hepatitis B virus-related hepatocellular carcinoma. Sci. Rep. 2018;8:7958. doi: 10.1038/s41598-018-26217-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiedel B.J., Singh D., Madrigal A., Valdovino-Gonzalez A.G., White B.M., Zapardiel-Gonzalo J., Ha B., Altay G., Greenbaum J.A., McVicker G., et al. Impact of Genetic Polymorphisms on Human Immune Cell Gene Expression. Cell. 2018;175:1701–1715.e1716. doi: 10.1016/j.cell.2018.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiedel B.J., Chandra V., Rocha J., Gonzalez-Colin C., Bhattacharyya S., Madrigal A., Ottensmeier C.H., Ay F., Vijayanand P. COVID-19 genetic risk variants are associated with expression of multiple genes in diverse immune cell types. bioRxiv. 2020 doi: 10.1101/2020.12.01.407429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shouval D.S., Biswas A., Goettel J.A., McCann K., Conaway E., Redhu N.S., Mascanfroni I.D., Al Adham Z., Lavoie S., Ibourk M., et al. Interleukin-10 receptor signaling in innate immune cells regulates mucosal immune tolerance and anti-inflammatory macrophage function. Immunity. 2014;40:706–719. doi: 10.1016/j.immuni.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skipper L., Wilkes K., Toft M., Baker M., Lincoln S., Hulihan M., Ross O.A., Hutton M., Aasly J., Farrer M. Linkage disequilibrium and association of MAPT H1 in Parkinson disease. Am. J. Hum. Genet. 2004;75:669–677. doi: 10.1086/424492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijders Blok L., Vino A., den Hoed J., Underhill H.R., Monteil D., Li H., Reynoso Santos F.J., Chung W.K., Amaral M.D., Schnur R.E., et al. Heterozygous variants that disturb the transcriptional repressor activity of FOXP4 cause a developmental disorder with speech/language delays and multiple congenital abnormalities. Genet. Med. 2021;23:534–542. doi: 10.1038/s41436-020-01016-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theusch E., Chen Y.I., Rotter J.I., Krauss R.M., Medina M.W. Genetic variants modulate gene expression statin response in human lymphoblastoid cell lines. BMC Genomics. 2020;21:555. doi: 10.1186/s12864-020-06966-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S. The Structure of the Membrane Protein of SARS-CoV-2 Resembles the Sugar Transporter SemiSWEET. Pathog. Immun. 2020;5:342–363. doi: 10.20411/pai.v5i1.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace C. Eliciting priors and relaxing the single causal variant assumption in colocalisation analyses. PLoS Genet. 2020;16:e1008720. doi: 10.1371/journal.pgen.1008720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Zhang Q., Zhao X., Dong H., Wu C., Wu F., Yu B., Lv J., Zhang S., Wu G., et al. Low high-density lipoprotein level is correlated with the severity of COVID-19 patients: an observational study. Lipids Health Dis. 2020;19:204. doi: 10.1186/s12944-020-01382-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb A., Miller B., Bonasera S., Boxer A., Karydas A., Wilhelmsen K.C. Role of the tau gene region chromosome inversion in progressive supranuclear palsy, corticobasal degeneration, and related disorders. Arch. Neurol. 2008;65:1473–1478. doi: 10.1001/archneur.65.11.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X., Zeng W., Su J., Wan H., Yu X., Cao X., Tan W., Wang H. Hypolipidemia is associated with the severity of COVID-19. J. Clin. Lipidol. 2020;14:297–304. doi: 10.1016/j.jacl.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worldometer . 2021. COVID-19 CORONAVIRUS PANDEMIC.https://www.worldometers.info/coronavirus/ [Google Scholar]
- Wu Y., Byrne E.M., Zheng Z., Kemper K.E., Yengo L., Mallett A.J., Yang J., Visscher P.M., Wray N.R. Genome-wide association study of medication-use and associated disease in the UK Biobank. Nat. Commun. 2019;10:1891. doi: 10.1038/s41467-019-09572-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Li H., Thakur A., Chen T., Xue J., Li D., Chen M. FOXP4 modulates tumor growth and independently associates with miR-138 in non-small cell lung cancer cells. Tumour Biol. 2015;36:8185–8191. doi: 10.1007/s13277-015-3498-8. [DOI] [PubMed] [Google Scholar]
- Yang G.H., Fontaine D.A., Lodh S., Blumer J.T., Roopra A., Davis D.B. TCF19 Impacts a Network of Inflammatory and DNA Damage Response Genes in the Pancreatic β-Cell. Metabolites. 2021;11:513. doi: 10.3390/metabo11080513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A.M.H., Kumasaka N., Calvert F., Hammond T.R., Knights A., Panousis N., Park J.S., Schwartzentruber J., Liu J., Kundu K., et al. A map of transcriptional heterogeneity and regulatory variation in human microglia. Nat. Genet. 2021;53:861–868. doi: 10.1038/s41588-021-00875-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeberg H., Pääbo S. A genomic region associated with protection against severe COVID-19 is inherited from Neandertals. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2026309118. e2026309118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z., Liu H., Xu H., Lu H., Yu Y., Xu X., Yu M., Zhang T., Tian X., Xi H., et al. HBVstudy consortium Genome-wide association study identifies new loci associated with risk of HBV infection and disease progression. BMC Med. Genomics. 2021;14:84. doi: 10.1186/s12920-021-00907-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S.X., Liu W., Liang J., Gao G.Q., Zhang X.M., Yao Y., Wang H.N., Yuan F.F., Xue L.Q., Ma Y.R., et al. China Consortium for the Genetics of Autoimmune Thyroid Disease Assessment of Molecular Subtypes in Thyrotoxic Periodic Paralysis and Graves Disease Among Chinese Han Adults: A Population-Based Genome-Wide Association Study. JAMA Netw. Open. 2019;2:e193348. doi: 10.1001/jamanetworkopen.2019.3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z.H., Chen G., Deng C., Tang J.M., Xie L., Zhou H.Y., Ye X., Zhang D.K., Shi R.Q., Tian D., et al. TCF19 contributes to cell proliferation of non-small cell lung cancer by inhibiting FOXO1. Cell Biol. Int. 2019 doi: 10.1002/cbin.11189. Published online May 29, 2019. [DOI] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Butler-Laporte G., Nakanishi T., Morrison D.R., Afilalo J., Afilalo M., Laurent L., Pietzner M., Kerrison N., Zhao K., et al. A Neanderthal OAS1 isoform protects individuals of European ancestry against COVID-19 susceptibility and severity. Nat. Med. 2021;27:659–667. doi: 10.1038/s41591-021-01281-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The table shows the coordinates of each of the 23 loci with a minimum p value < 10−7 in at least one of the 16 meta-analyses obtained from the COVID-19 HGI data freeze 5. For each locus and each study, we report the p value and effect size of the variant with the most significant p value.
The table shows the pairwise colocalization results between eQTLs and COVID-19 GWAS signals for each gene mapping into the 23 COVID-19 genome-wide significant loci. Shown are each locus coordinates (chromosome, start and end positions), gene ID, gene name, tissue tested for eQTLs, GWAS study, the SNP ID of the variant with the highest posterior probability of being causal, the p values of this variant in both studies, the number of SNPs tested for colocalization, the posterior probabilities of the five colocalization hypotheses, and posterior probability of the lead SNP. The colocalization hypotheses are (1) H0, neither study has a significant association at the tested locus; (2) H1, only the first study is associated; (3) H2, only the second study is associated; (4) H3, both studies are associated but the underlying variants are different; and (5) H4, both studies are associated and share the same underlying variants.
We found eQTL signals for 20 genes that colocalized with the COVID-19 GWAS signals at 11 loci (PP-H4 > 0.9). For each of these signals, we performed fine mapping using the coloc.abf function in the R package coloc. This table reports the information about the variant with the highest PPA at each of these signals. Specifically, we show locus, COVID-19 study, gene ID, gene name, eQTL tissue, PPA of each of the five colocalization hypotheses, the variant with highest PPA (including position, reference allele, alternative allele, and RS ID), its p value and effect size in the GWAS and eQTL analyses, its PPA, and the size of the 99% credible set.
For each of the 20 genes whose eQTL signals colocalize with GWAS signals with PPA > 0.9, we selected the phenotype (C2, A1, B1, or B2) that had the eQTL signal with the highest PPA (hypothesis 4) and report fine-mapping results obtained using the coloc.abf function in the R package coloc. The table shows the phenotype with the highest PPA and information about each tested variant: locus, phenotype, gene ID, gene name, variant information (including RS ID), PPA, cumulative PPA, and whether it is included in the 99% credible set. For each colocalization, variants are sorted by their PPA. There are two exceptions to reporting the phenotype with the highest PPA. For the severity-associated 12:112850796-113925679 locus, we report the fine mapping between its eQTL signal in adipose tissue and the B2 phenotype, but PPA is higher with the susceptibility phenotype (C2), although both colocalizations share the same putative causal variant (rs10774671) and their PPA is > 0.99. Additionally, the 1:154605882-155673527 locus is severity-associated, however, the FAM189B eQTL in Heart Atrial Appendage colocalized only with phenotype C2. Therefore, we report the fine mapping between the FAM189B eQTL and the C2 phenotype.
Data Availability Statement
This paper does not have original code to report.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.