Abstract
Despite considerable progress in the development of antiviral drugs, among which anti-immunodeficiency virus (HIV) and anti-hepatitis C virus (HCV) medications can be considered real success stories, many viral infections remain without an effective treatment. This not only applies to infectious outbreaks caused by zoonotic viruses that have recently spilled over into humans such as severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2), but also ancient viral diseases that have been brought under control by vaccination such as variola (smallpox), poliomyelitis, measles, and rabies. A largely unsolved problem are endemic respiratory infections due to influenza, respiratory syncytial virus (RSV), and rhinoviruses, whose associated morbidity will likely worsen with increasing air pollution. Furthermore, climate changes will expose industrialized countries to a dangerous resurgence of viral hemorrhagic fevers, which might also become global infections. Herein, we summarize the recent progress that has been made in the search for new antivirals against these different threats that the world population will need to confront with increasing frequency in the next decade.
Keywords: Severe acute respiratory syndrome coronavirus type 2, Influenza viruses, Picornaviruses, Respiratory syncytial virus, Hemorrhagic fever viruses, Rabies virus, Measles virus, Immunodeficiency virus, Hepatitis C virus, Coronaviruses
1. Introduction
Interest in antiviral research has changed focus within the scientific community over the years. After the pioneering work in the 1960s–1970s on herpesviruses that led to the identification of acyclovir as the gold standard for the treatment of herpes simplex virus (HSV) infections, the field subsequently boomed in the 1980s–1990s after the discovery of the immunodeficiency virus (HIV) as the causative agent of acquired immunodeficiency syndrome (AIDS). In the early 2000s, direct acting antivirals (DAAs) were developed for the treatment of hepatitis C (HCV), following a tremendous medicinal chemistry effort that culminated in the discovery of sofosbuvir in 2007. Over a period of approximately 50 years, the focus has shifted from DNA viruses to retroviruses, and further onto RNA viruses as targets for drug discovery. At this moment, worldwide efforts are ongoing to discover new drugs, including nucleoside analogues, against the newly emerged severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2). An important question to answer is which viruses remain to be treated and against which viral infections drug discovery is focusing in 2021. Due to climate change, frequent international travel, profound demographic shift, increasing air pollution, overpopulation in certain regions, changes in the population age distribution, virus infections are in a continuous flux. Global warming might lead to a continuous re-emergence of hemorrhagic fever viruses in the future, bringing these pathogens closer to the northern countries. Globally prevalent respiratory tract infections, such as influenza, respiratory syncytial virus (RSV) and rhinoviruses, have been declining thanks to current containment measures for SARS-CoV-2; however, increase in air pollution is expected to increase mortality and morbidity due to these viral respiratory infections.
Virus infections of zoonotic origin (e.g., rabies, coronaviruses) will never be eradicated, and new viruses might emerge using wild or domesticated animals as vectors. Other infections that were almost eradicated due to vaccination (e.g., measles, polio) may come back given irregular immunization coverage caused by inequitable global access to vaccines and growing vaccine hesitancy across the world. Bioterrorism could, in theory, also make use of deadly viruses as weapons. For those viruses known to be life-threatening, prophylactic treatment can be considered. Likewise, the population may benefit from the development of antivirals against viruses that may cause cancer or neurological diseases; in this way antivirals may contribute to the general fitness of the society. In this review, we describe the efforts that have been conducted during the last 3 years to discover antivirals against several of these treats. This review highlights some important new discoveries, and it is not meant to give a complete survey of every ongoing effort.
2. Severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2)
The ongoing coronavirus disease 2019 (COVID-19) pandemic caused by the highly transmissible and pathogenic severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can be anticipated to be resolved by vaccination; however, it might be important for future prevention and control to develop a pan-coronavirus vaccine (see also discussion in Section 13), as initiated by VBI vaccines Inc. with a trivalent eVLP Vaccine candidate encoding the antigenic spike proteins of SARS-CoV-2, SARS-CoV, and Middle East respiratory syndrome coronavirus (MERS-CoV) in a single particle.1 This expectation is obviously contingent on a number of factors including equitable access to vaccines, widespread acceptance, high uptake, and sustained coverage specially against rapidly emerging variants. Even in the event of SARS-CoV-2 becoming an endemic pathogen, the future emergence of related epidemic or pandemic outbreaks cannot be ruled out, making critical the availability of effective therapeutic strategies for those who are infected.
The diverse repertoire of structural, non-structural, and accessory proteins encoded by the SARS-CoV-2 RNA genome2 reveals multiple opportunities for antiviral therapeutic targeting, which are all being relentlessly pursued as it can be expected that a cocktail of compounds acting at different steps of the viral life cycle will be needed to efficiently suppress viral replication in vivo. These may include inhibitors of viral entry, polyprotein proteolytic processing, and viral RNA synthesis, which can be additionally complemented by other classes of antivirals interfering with host-mediated regulatory functions. More detailed information about this quickly evolving drug discovery panorama can be retrieved from recent literature reviews.3, 4, 5
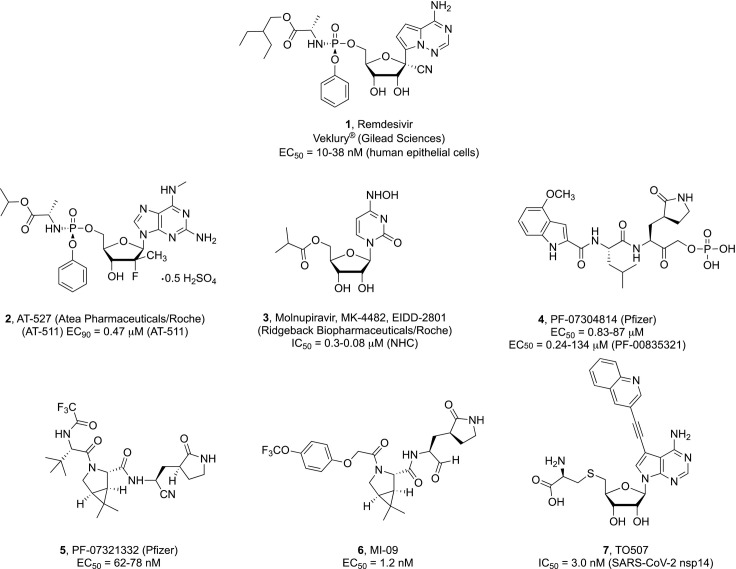
At present, the only investigational drug to have received emergency use authorization for the treatment of adults and children from 12 year old with severe COVID-19 is the pyrrolo[2,1-f][triazin-4-amino]adenine 1’-cyano substituted C-nucleoside ProTide remdesivir 1 (GS-5734, Veklury®, Gilead Sciences, Fig. 1 ),6 an RNA-dependent RNA polymerase (RdRp) inhibitor.7 In a placebo-controlled trial, remdesivir was found to speed the recovery time in hospitalized adult patients with confirmed SARS-CoV-2 lower respiratory tract infection.8 However, the results of a recently published meta-analysis of randomized trials suggested little or no mortality benefit associated with the intravenous administration of remdesivir in COVID-19 patients.9 Being a phosphoramidate prodrug, remdesivir is converted on cell entry to its corresponding modified nucleoside monophosphate by the sequential action of esterases and phosphoramidases; such metabolic step occurs primarily in liver cells where the expression of these hydrolytic enzymes is more pronounced. As this prodrug design was originally not intended for lung delivery, debate is ongoing on whether respiratory viruses such as SARS-CoV-2 that predominantly infect alveolar epithelial cells might be better inhibited by nucleoside prodrugs that could be selectively converted to the active drug in the infected tissue. In a recent study, the nucleoside GS-441524, which turned out to be a major metabolite present in the bloodstream after administration of remdesivir, was demonstrated to significantly reduce viral titers and improve inflammation in the lungs of SARS-CoV-2-infected mice,10 suggesting that a more favorable lung tissue distribution could be achieved with this compound.11 Regardless of this consideration, remdesivir remains an intriguing molecule that may inspire future structural optimization programs for the development of potent RdRp inhibitors against SARS-CoV-2 infection. One possibility could be the synthesis of more lipophilic prodrugs of remdesivir or its analogues for the delivery of 5’-monophosphates in the cells.12
Fig. 1.
Selected nucleoside and protease inhibitors against SARS-CoV-2.
The potential impact of the prodrug moiety on the inhibitory effect of nucleoside analogues towards SARS-CoV-2 replication can be further surmised by the in vitro inactivity of sofosbuvir against this virus in Vero cells versus an EC50 of 6.3 μM displayed by its cyclic phosphate prodrug congener.13 Two moderately active nucleoside lead compounds are 2’-methyl-cytidine and 2’-methyl-7-fluoro-7-deazaadenosine that displayed antiviral activity against SARS-CoV-2 in Vero cells at 9.2 and 7.6 μM, respectively.13
AT-527 (Atea Pharmaceuticals/Roche), compound 2 (Fig. 1), is an oral double prodrug of a 2’-fluoro-2’-methyl guanosine analogue that recently entered clinical trials for the treatment of mild or moderate COVID-19 disease in non-hospitalized patients [NCT04396106 and NCT04709835 (Phase II); NCT04889040 (Phase III)]. Its free base, designated as AT-511, was described to exert sub-micromolar activity (EC90 = 0.47 μM) in human airway epithelial cells (HAE) infected with SARS-CoV-214; however, this activity profile was not supported by an independent study using similar cellular assays,15 leaving the in vivo efficacy of this molecule to be ascertained.
Another broad-spectrum polymerase inhibitor currently in Phase II clinical trials for the treatment of COVID-19 is the 5’-isobutyryl ester prodrug of N 4-hydroxycytidine (NHC) known as molnupiravir (3, EIDD-2801; Ridgeback Biotherapeutic in partnership with Merck), which was recently shown to be well tolerated and effectively reduce nasopharyngeal SARS-CoV-2 infectious virus and viral RNA.16 This orally administered antiviral agent was shown to potently inhibit SARS-CoV-2 replication as well as suppress viral transmission in vivo in different animal models.17, 18 Therapeutic and prophylactic administration of 3 in immunodeficient mice implanted with authentic human lung tissue (LoM) revealed a > 4 log reduction of infectious virus titers and effective prevention of SARS-CoV-2 infection, respectively.19 However, the biochemical mechanism mediating the mutagenic effect of N 4-hydroxycytidine has yet to be elucidated and may be a concern for long term treatment.20
At present, there are 36 studies listed in the ClinicalTrials.gov registry to assess the efficacy and safety of the purine nucleoside prodrug analogue favipiravir (T705) as oral treatment against COVID-19. A meta-analysis of all completed clinical trials till December 2020 revealed a positive clinical improvement in patients receiving favipiravir during seven days after hospitalization compared to control groups, but no significant beneficial effect in reducing mortality for patients with mild to moderate COVID-19.21 Electron cryomicroscopy (cryoEM) studies revealed that favipiravir ribonucleoside triphosphate (favipiravir-RTP) interacts with the catalytic site of SARS-CoV-2 RdRp in a nonproductive binding mode that may account for the inefficient incorporation into a RNA primer strand observed in polymerase activity assays.22
At the moment, it is unclear which specific combination of sugar, nucleobase, and prodrug form would be ideal for enabling potent and tissue-specific antiviral activity against SARS-CoV-2 in vivo.
However, an issue to be faced in developing nucleoside-based inhibitors is associated with an increased risk of toxicity compared to other drug classes, as their antiviral activity is exerted by competing with natural substrates for RdRp-catalyzed incorporation.23 Modifications introduced at the 4’-position of the potent antitumor compound gemcitabine to decrease toxic effects have been attempted without success.24
The virally encoded chymotrypsin-like or main protease (3CLpro or Mpro) is a critical component of the SARS-CoV-2 life cycle. Along with another cysteine protease, the papain-like protease (PLpro), the SARS-CoV-2 Mpro processes the virus p1a/p1ab polyproteins translated from the viral RNA into a series of individual non-structural proteins (nsps), including the Mpro itself, which are essential for viral replication and transcription. Of note, no close human analogues of the coronavirus Mpro are known. Thereby, selective SARS-CoV-2 Mpro inhibition represents an attractive approach to COVID-19 treatment that could avoid unintended drug-target interactions. Furthermore, the antiviral efficacy of small molecule Mpro inhibitors is not expected to be affected by spike protein variants, as the SARS-CoV-2 Mpro and the viral spike protein are distinct protein entities within the viral proteome.
Impressive drug discovery efforts from both academia and industry have provided clear evidence of the ability of various Mpro small molecule inhibitors to efficiently suppress SARS-CoV-2 replication in vitro, as recently reviewed.25 Nonetheless, this research is still at an early discovery stage and only two viable clinical candidate have been identified so far.
The Mpro protease inhibitor PF-07304814 (4) is a phosphate prodrug that was shown to undergo in vivo metabolic conversion to its parent derivative designated as PF-00835321.26 PF-00835231 inhibited recombinant SARS-CoV-2 Mpro activity with nanomolar potency (Ki = 0.271 nM) with excellent off-target selectivity, while exhibiting potent and highly selective antiviral activity in both cellular assays and in vivo murine SARS-CoV-2 model either as a single agent or in combination with remdesivir. A Phase I clinical trial in patients hospitalized with SARS-CoV-2 virus infection to assess the safety, tolerability, and pharmacokinetics of intravenously dosed 4 has been completed and is awaiting publication of results (NCT04535167).
PF-07321332 (compound 5) is the latest orally active inhibitor of SARS-CoV-2 main protease to have reportedly entered phase III clinical trials in 2021 and is currently under investigation for its safety in combination with the antiretroviral protease inhibitor ritonavir (RTV) in nonhospitalized COVID-19 patients with increased risk of developing severe illness (NCT04960202).27 Compound 5 strongly inhibited SARS-CoV-2 Mpro biochemical activity with an inhibitory constant (Ki) value of 3.11 nM, while no inhibitory effects were observed against a range of mammalian proteases. In the SARS-CoV-2 infection Vero E6 cell assay, PF-07321332 displayed potent inhibition of viral replication (EC50 = 74.5 nM), which was further confirmed using physiologically relevant in vitro assays in human adenocarcinoma-derived alveolar basal epithelial cells constitutively expressing ACE2 (A549-ACE2 (EC50 = 77.9 nM, EC90 = 215 nM) and differentiated normal human bronchial epithelial (dNHBE) cells (EC50 = 61.8 nM, EC90 = 181 nM). The in vivo oral antiviral potency, safety, and pharmacokinetics profile of PF-07321332 were demonstrated in a mouse-adapted SARS-CoV-2 model and an ongoing Phase I clinical trial in healthy human participants (NCT04756531).
Mpro inhibitors containing a bicyclic proline similar to 5 were also disclosed by Yang et al. Among a set of 32 compounds, two compounds indicated as MI-09 (compound 6) and MI-30 displayed potent SARS-CoV-2 Mpro inhibitory activity (IC50 = 15.2 and 17.2 nM, respectively) combined with nanomolar antiviral activity in SARS-CoV-2 infected human alveolar epithelial cells (HPAEpiC). These compounds were additionally evaluated in a transgenic mouse model of SARS-CoV-2 infection, where their oral or intraperitoneal administration could significantly reduce lung viral load and ameliorate lung pathological damage.28 These latest discoveries are particularly noteworthy as oral formulations of highly efficacious antiviral agents hold significant potential for more convenient and widespread therapeutic intervention.
More recently, the two methyltransferases (MTases), nsp14 and nsp16, employed by SARS-CoV-2 to protect the 5’-end of its newly synthesized mRNA are also receiving attention as potential therapeutic targets.29, 30 RNA capping is crucial to ensure efficient viral translation and evade clearance by host innate immune responses. The nsp14 MTase catalyzes the transfer of a methyl group from S-adenosyl-l-methionine (SAM) to the 7-position of the terminal guanosine residue. The rational modification of the S-adenosyl-l-homocysteine (SAH), which is formed as main product during this process, led to 7-substituted deaza analogues such as 7 with strong binding affinities and substantial inhibitory activity against this enzyme.30
All discussed results are preliminary and in this quickly evolving scenario it is hard to tell which treatment approach(es) may deliver antiviral drugs with substantial clinical efficacy for the treatment of COVID-19.
3. Influenza viruses
Influenza viruses (IVs, types A, B, C, and D) are commonly circulating respiratory pathogens of the Orthomyxoviridae family, whose genome is organized in a number of (-)-ss RNA segments.31 Clinically, influenza A (IAV) strains are the most significant due to evolutionary advantages conferred by the propensity of their genome to undergo reassortment.32 This may contribute to the emergence of highly virulent pandemic subtypes with potential for cross-species transmission and ability to escape vaccine or drug pressure.33 Early classes of antiviral drugs licensed for flu infections, which interfere with the activity of either the viral M2 ion channel protein (e.g., amantadine and rimantadine) or the surface enzyme neuraminidase (NA) [e.g., oseltamivir (Tamiflu), zanamivir (Relenza), peramivir (Rapivab), laninamivir octanoate (Inavir, approved only in Japan)],34 have been associated with various levels of resistance35, 36; thereby, they can be anticipated to be slowly superseded in the mid term by other small-molecules inhibitors with similar or distinct inhibition mechanisms.
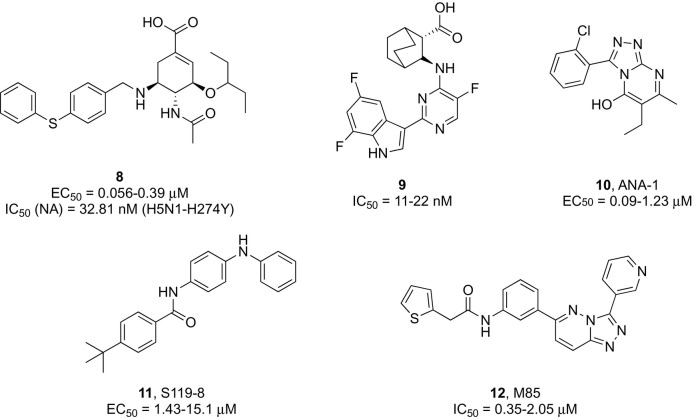
Significant efforts are ongoing to develop second-generation inhibitors targeting highly prevalent M2 proton channel mutants of adamantane-resistant influenza A strains that may raise the genetic barrier to resistance.37, 38 A recently disclosed chemical modification at the 5-position of oseltamivir allowed for an additional interaction within the NA active site that produced new analogues with restored affinity for drug resistant strains.39, 40 Among those, 4-phenylthiobenzyl substituted compound 8 (Fig. 2 ) exhibited potent inhibitory activity against various influenza virus subtypes comparable to oseltamivir in cell based assays, while being 86-fold more active against the predominant H5N1-H274Y mutant strain in a neuraminidase inhibitory assay (IC50 = 32.8 nM).39
Fig. 2.
Examples of mechanistically distinct influenza virus inhibitors.
Alternative approaches for treating the IAV infection sought to abolish the influenza RNA-dependent RNA polymerase (RdRp) activity preventing transcription and replication of the viral genome.41 High-resolution structural analysis revealed this enzyme to be a highly conserved heterotrimeric complex organized in different closely interacting subunits decorated by flexible domains,42 which provided distinctly exploitable targeting sites for antiviral drug discovery. Among selected clinical candidates, only the oral polyheterocyclic ester prodrug baloxavir marboxil (BMX, Xofluza®, Roche and Shionogi) received global approval for the treatment of acute influenza.43 In 2020, this indication was extended to post-exposure prophylaxis for patients 12 years of age or older as the first single-dose preventive treatment of influenza.44 This drug is known to inhibit the cap-dependent endonuclease (CEN) activity of the polymerase acidic (PA) subunit of influenza A and B viruses, thereby blocking initiation of mRNA synthesis.45 However, mutations in the PA subunit associated with BMX resistance were detected during clinical trials,46 prompting further medicinal chemistry efforts towards the exploration of alternative scaffolds.47, 48, 49
Pimodivir (JNJ63623872 or VX787, Janssen Pharmaceutica), an oral azaindole inhibitor of the influenza A polymerase basic 2 (PB2) cap-binding domain,50 was found to significantly decrease viral load in a Phase II study51; however, its development was discontinued after Phase III results in hospitalized patients with influenza A showed no added benefit compared with the standard of care (SOC) (NCT03376321). Notwithstanding, PB2 remains an attractive therapeutic target for its key role in viral transcription, and further lead optimization efforts yielded a 7-fluoroindole bioisostere of pimodivir (compound 9, Fig. 2) with potent anti-IAV activity (EC50 = 11–22 nM, A H1N1 and H3N2 strains), improved metabolic stability, good pharmacokinetics, and in vivo efficacy.52
The purine analogue favipiravir (Avigan®, Toyama Chemical), which blocks influenza virus proliferation either by inducing chain termination during RNA synthesis or through lethal mutagenesis, entered clinical practice in Japan in 2014.53 However, due to teratogenic concerns, approval was granted upon condition of limiting its use against novel and re-emerging influenza virus infections unresponsive to SOC. Results from USA phase III clinical trials in adults with uncomplicated influenza are pending (NCT02026349; NCT02008344).
A parallel approach for inhibiting the RdRP function, which may lead to a next-generation of anti-influenza therapeutics, relies on the disruption of the polymerase assembly process, during which protein-protein interactions (PPIs) occur between the different subunits concurring to the formation of a functional complex.54 From data-driven and biochemical assay-based high throughput screenings of structurally unrelated heterocyclic libraries, numerous hit compounds emerged as potent antiviral agents and strong inhibitors of the viral polymerase activity, which were shown to primarily disrupt the PA-PB1 interaction. Compound 10 (ANA-1, Fig. 2) was effective against multiple IAV subtypes in cell culture, reduced lung virus titers in infected mice, and protected animals from lethal challenge upon intranasal administration.55
The discovery that the small molecule nucleozin can induce loss of viral viability by causing abnormal aggregation of viral nucleoprotein (NP) in the cytoplasm with consequent blockage of its nuclear accumulation56 provided an opportunity for the development of a novel class of antiviral drugs for the treatment of IAV infections.57, 58, 59 Compound S119-8 (11, Fig. 2) was found to exert broad-spectrum micromolar activity against numerous influenza A and B strains and its combination with oseltamivir led to a strong synergism.59
An increasing number of studies have examined direct inhibitors of the viral fusion protein hemagglutinin (HA), a surface glycoprotein that mediates viral attachment and entry, as a mean to complement other classes of anti-influenza agents for achieving pan-subtype efficacy.60, 61, 62 However, doubts remain about the utility of targeting HA for single agent antiviral therapy due to its high susceptibility to mutations.
On the other hand, given the low probability of changes occurring to the host genome during the viral infection cycle, interfering with cellular processes involved in the entry process of influenza is seen as a promising strategy to combat treatment failures due to drug resistance. Two host directed antiviral agents are currently being evaluated in clinical trials. The antiprotozoal agent nitazoxanide (Alinia) is a repurposed thiazolide prodrug of tizoxanide that prevents correct assembly of new viral particles by interfering with cellular factors involved in the maturation of viral hemagglutinin.63 After successfully completing a Phase IIb/III study in adults and adolescents with acute uncomplicated influenza,64 it now moved into Phase 3 (NCT02612922). Fludase (DAS-181, Ansun BioPharma) is a recombinant fusion protein that adheres to the host epithelial cells and removes the sialic acid receptor from the cell-surface preventing viral entry.65 Results from a Phase 2 trial suggested that the drug was well tolerated and significantly reduced viral load and viral shedding.66
Recently, M85 (compound 12, Fig. 2) was reported as an inhibitor of class II PI3K host kinase activity required for IAV internalization into target cells via endocytosis.67 The compound had low cytotoxicity, inhibited viral replication of IAV, IVB, HCV, and human rhinoviruses in various cell types, while exerting a synergistic antiviral action in combination with oseltamivir that protected mice against a lethal challenge of influenza A H1N1 virus.
4. Respiratory syncytial virus (RSV)
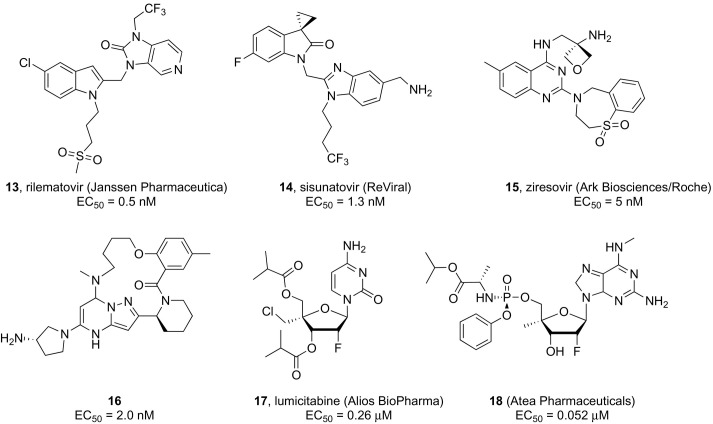
Human respiratory syncytial virus (RSV, major antigenic subtypes A and B) is a Orthopneumovirus of the Pneumoviridae family containing a negative sense, single stranded linear 15 kb RNA genome made of 10 genes encoding 11 proteins.68 RSV is responsible for lower respiratory tract infections that can be life-threatening among patients with impaired immune function like premature infants and the frail elderly.69, 70 Narrow, suboptimal preventive and therapeutic options for RSV infection, confined to monoclonal antibody palivizumab (Synagis) and broad-spectrum nucleoside analogue ribavirin (aerosol formulation),71 along with the potential for nosocomial outbreaks inflict a heavy clinical and economic burden. Significant global efforts are underway by several major pharmaceutical companies to develop RSV fusion (F) protein inhibitors to prevent viral entry into host cells, as recently reviewed in detail elsewhere,72 which seems to suggest that the first marketed drug against RSV infections might be within close reach. Among the latest candidates to advance to late-stage development are 5-chloroindole and spirocyclopropyl azaindole-based analogues rilematovir (13, JNJ-53718678, Janssen Pharmaceutica, Fig. 3 )73 and sisunatovir (14, RV521; ReViral, Fig. 3),74 respectively, which are currently being evaluated for oral administration in hospitalized infants and vulnerable adult populations. Phase II studies are also in progress for ziresovir (15, AK0529; in-licensed by Ark Biosciences from Roche, Fig. 3), which was evolved from a benzoazepine quinoline chemical series.75 On the other hand, despite promising data from preclinical and preliminary clinical studies,76 results from a recently completed Phase II clinical trial examining the efficacy of the pyrazolo[1,5-a]pyrimidines candidate presatovir (GS-5806; Gilead Sciences) revealed a lack of therapeutic benefit in adult hematopoietic cell transplant recipients77, 78 and low barrier to resistance.79 While the clinical utility of RSV fusion inhibitors remains to be fully validated, the identification of new heterocyclic leads binding the viral surface F glycoprotein continues to be of interest to the medicinal chemistry community. A lead optimization strategy based on a machine learning algorithm led to novel compounds integrating the popular benzimidazole scaffold.80 The most active analogue in this series demonstrated subnanomolar in vitro potency comparable to JNJ-53718678 (EC50 = 0.5 vs 0.9 nM) against wild-type RSV with a CC50 > 20 μM; however, it was significantly less potent against the clinically reported viral escape mutant D489E, emphasizing the need for RSV combination therapy rather than monotherapy.
Fig. 3.
Selected RSV fusion and replication inhibitors.
Macrocyclic pyrazolo[1,5-a]pyrimidine derivatives, as exemplified by a 15-membered ring containing compound X, displaying activities in the low nanomolar to subnanomolar range against wild-type RSV A2 were recently disclosed by means of molecular docking studies. Compound 16 combined excellent potency against both wild-type and drug-resistant mutant RSV F proteins owing to productive interactions of the rigid structure, suggesting the potential for developing inhibitors with anti-RSV activities.81
The latest compound to enter the anti-RSV drug discovery pipeline is the post-entry replication inhibitor EDP-938 (Enanta Pharmaceuticals), whose mode of action has been linked to the inhibition of the viral nucleoprotein (N), which serves as a polymerase co-factor in the RSV ribonucleoprotein (RNP) complex responsible for carrying out RNA synthesis. EDP-938 exhibited EC50 values in the 21–64 nM range against a panel of RSV A and B laboratory strains and clinical isolates when using primary human bronchial epithelial cells (HBECs) as in vitro model. Further in vitro studies revealed a high barrier to resistance as well as no cross-resistance between EDP-938 and other classes of RSV inhibitors, but rather a potentially synergistic in vitro antiviral effect. The antiviral efficacy was further confirmed in vivo using an non-human African Green monkey model of RSV infection.82 In current Phase II clinical trials, hospitalized and non-hospitalized infants as well as adult allogeneic hematopoietic cell transplant (HCT) recipients are undergoing treatment with EDP-938.
Several nucleoside polymerase inhibitors have been described with potent in vitro anti-RSV activity72, 83; however, this discovery approach has been delayed by the unavailability of sufficient structural data on the large (L) protein and poor understanding of its interactions (e.g., with the tetrameric phosphoprotein P to form an active RNA polymerase complex) and enzymatic activities.84, 85 The most active nucleoside analogues are prodrugs sharing a 2’-deoxy-2’-fluoro-ribose sugar moiety bearing different 4’-substituents in combination with various nucleobases (e.g., cytosine, amino-pyrrolotriazine, and N 6-methylated diaminopurine). Lumicitabine (17, ALS-8176; Alios BioPharma, Fig. 3), the 3',5'-di-O-isobutyryl prodrug of 2′-fluoro-4′-chloromethyl-cytidine, emerged as a potent (EC50 = 0.26 μM) and selective inhibitor of RSV replication with no concomitant inhibition of human polymerases.86 After achieving clinical proof of concept in human RSV challenge studies, however, the development of this compound was ultimately discontinued due to unclear reasons following the early termination of a study in hospitalized infants (NCT03333317). The potent in vitro anti-RSV activity (EC50 = 0.052 μM) was described for a phosphoramidate prodrug of 2’-deoxy-2’-fluoro-4’-methyl nucleoside analogue bearing a modified purine base (18, Fig. 3) in HAE cultures with no cytotoxicity found in bone marrow stem cell assay.87 Preliminary oral bioavailability data were reported in hamsters along with the demonstration of the conversion to the active triphosphate form in lung tissue. Remdesivir was shown to effectively inhibit RSV A2 replication in vitro with an EC50 of 530 nM in primary human lung cells.88 An in vivo efficacy study was conducted by subjecting RSV A2-infected African green monkeys to intravenous administration of 10 mg/kg delivered once daily, which resulted in a > 2-log10 reduction in the peak lung viral load.
In parallel, non-nucleosides are also being investigated mainly as allosteric inhibitors of the L polymerase along with molecules targeting other viral proteins involved in the virus life cycle such as the attachment G protein, another viral protein involved in the initiation of the RSV infection, as exhaustively discussed by Cockerill et al.72
5. Rhinoviruses
The Picornaviridae is a large and diverse family of (+)-ssRNA viruses whose genome is packed within an icosahedral capsid made of four viral proteins (VP1-4). Notable members include human rhinoviruses (HRVs, type A, B, C),89 which are ubiquitous and persistent pathogens most frequently associated with infections of the upper respiratory tract (i.e., common colds) that can occasionally cause an aggravation of preexisting pulmonary and middle ear inflammatory conditions in vulnerable patient populations.90, 91 Their (+)-ssRNA genome (~ 7.21 kb) is translated into a single polyprotein that is cleaved by virally encoded proteases 2A and 3C to provide 11 structural and non-structural proteins. At present, no antiviral drug has been licensed for therapeutic use in rhinovirus or other enterovirus infections. Numerous active small molecules have been discovered through extensive in vitro screening campaigns that fall mainly into two mechanistic classes, i.e., capsid and 3C protease (3Cpro) inhibitors, with the former being the most advanced avenue for rhinovirus treatment.92, 93 However, lead compounds were either not developed or failed to meet the clinical criteria for their successful use because of their toxicity, poor pharmacology, or rapid resistance development [e.g., pleconaril, pirodavir, vapendavir, and rupintrivir). Moreover, a major challenge will be identifying anti-HRV compounds with a broad activity spectrum due to the circulation of approximately 160 different viral serotypes characterized by high mutation rates.
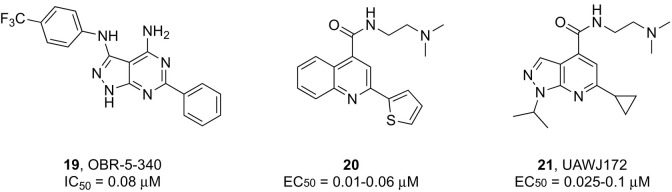
Discovery of novel capsid binders continues through the exploration of structurally diverse heterocyclic scaffolds, some of which showed potent in vitro activities.94, 95 Although their precise uncoating inhibition mechanism has not been fully validated, it is believed that these compounds occupy the hydrophobic pleconaril-binding pocket of VP1, resembling its mode of action. It is of interest to note that pleconaril-resistant viruses feature amino acid substitutions with bulkier residues in this pocket that sterically preclude binding.96 Besides reducing the size of the molecules to fit the narrower pocket of HRV mutants,97, 98 a better strategy to raise the barrier to resistance is identifying inhibitors that stabilize the viral capsid dynamics inducing uncoating by targeting alternative binding sites. Recent cryo-EM studies demonstrated that the pyrazolopyrimidine derivative OB-5-340 (compound 19, Fig. 4 ) binds to a less conserved region of VP1 that only partly overlaps with the attachment site of other capsid inhibitors,99 which could explain its potent inhibitory activity against HRV-B5 (average IC50 = 0.08 μM) and other high-level pleconaril-resistant HRV strains.100
Fig. 4.
Examples of small molecule inhibitors active against HRVs and EV-D68.
The RdRp of rhinoviruses could be a viable target for the development of antiviral agents; however, investigations in this direction have not yet produced any concrete results, except for the report of the inhibitory activity of the nucleoside analogue 7-deaza-2’-methyladenosine (7DMA, MK0608) in HAE cultures infected with several HRV-C strains (EC50 = 1.8–12 nM).101
Despite having distinct cellular tropisms and clinical manifestations, rhinoviruses are structurally and genetically related to other human pathogens of the same genus Enterovirus in the Picornaviridae family. Prior to the advent of genetic sequencing, enterovirus D68 (EV-D68) was misclassified as rhinovirus 87 due to its almost exclusive association with respiratory illnesses, unlike most enteroviruses that infect a variety of cell types.102 EV-D68 is an emerging pathogen that in recent years has been increasingly associated with episodes of neurological disease outbreaks.103
The viral proteins that are being exploited as targets to develop anti-EV-D68 agents are those for which inhibitors were selected in the anti-HRV drug discovery process, since their high degree of sequence conservation among different enteroviruses might allow for broad-spectrum antiviral activity.104 As an example, fluoxetine-modified inhibitors with broad spectrum activity against enteroviruses and rhinoviruses have been reported that target the non-structural 2C protein.105 This protein has been proposed to play multifunctional roles in the viral life cycle of nonpolio enteroviruses, and significant efforts are being made in identifying antivirals against this target.
Structure − activity relationship studies designed to improve the activity of EV-D68 inhibitor dibucaine yielded a series of analogues containing a quinoline scaffold, as represented by compound 20 (Fig. 3), which exhibited EC50 values in the nanomolar range and a high selectivity against different EV-D68 strains.106 Recently, a systematic modification of the pyrazolopyridine scaffold led to several lead compounds including 21 (Fig. 3) endowed with excellent broad-spectrum antiviral potency against several viruses (e.g., EV-D68, EV-A71, and CVB3) coupled and minimal cytotoxicity.107
Apart from targeting specific steps in the viral life cycle via direct-acting antivirals, host factors that are essential for viral replication are equally attractive drug targets for enterovirus inhibition and have been covered in a recent review.93 At the moment, the only small molecule in clinical development (Phase III) for the treatment of common colds caused by enterovirus/rhinovirus infections is the broad-spectrum antiviral nitazoxanide.
6. Ebolavirus (EBOV)
Within the Filoviridae family of negative-sense, single-stranded RNA (ssRNA) viruses, Ebola virus (EBOV) and Marburg virus (MARV) cause clinically similar acute illnesses that can evolve into severe systemic infections culminating in multiorgan failure.108 With reported case-fatality rates as high as 90% (e.g., for the highly pathogenic Zaire EBOV), EBOV stands out as one of the deadliest viruses known to mankind and a potential bioterrorism threat.109 Since 1976, when filoviruses were discovered, more than 25 major Ebola virus disease (EVD) outbreaks have been recorded in several West and Central African countries that claimed thousands of lives. The 2020 regulatory approval of two vaccines [single-dose Ervebo (Merck) and two-component Zabdeno-and-Mvabea (Janssen Pharmaceutica)] validated adaptive immunity as a viable mean to control EBOV transmission,110 however optimal impact on outbreak response remains to be defined mainly due to challenging settings to routine immunization.111
At present, immune therapy is very much on the front line of EVD management following the approval in 2020 of two human monoclonal antibody (mAb) products (i.e., Inmazeb and Ebanga) for intravenous administration112, 113, 114; however, orally small molecule therapeutics are seen as more convenient options in the event of high risk exposure and chronic infections to reach immunologically privileged sites that might harbor the virus. Nonetheless, screening for antifiloviral lead compound identification has been severely hampered by the requirement for biosafety level-4 (BSL-4) containment facilities combined with the limitations associated with surrogate BSL-2 cell-based systems to faithfully model the viral life cycle.115
The 19 kb EBOV genome encodes seven structural proteins and several non-structural proteins, potentially offering multiple druggable targets.115 Nonetheless, the most promising routes towards antiviral treatment have primarily focused either on the inhibition of the viral entry or genome transcription/replication steps, as recently reviewed.114, 115, 116
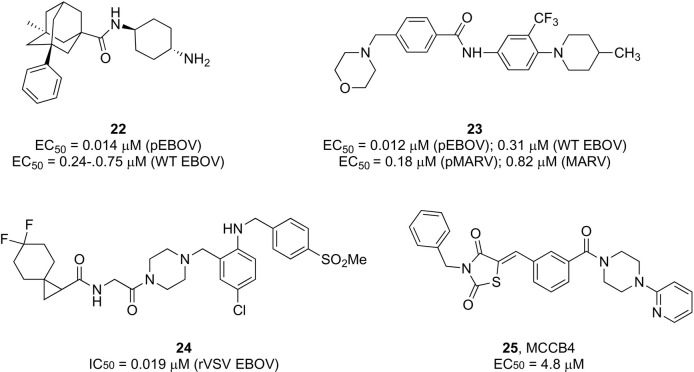
Various small molecules targeting all key aspects of the viral entry process have been actively discovered, however, most have only been studied in preclinical models. Some of the latest examples with the largest potential for further profiling as filovirus entry inhibitors are illustrated in Fig. 5 . Selected adamantane carboxamide derivatives, exemplified by 22, exhibited nanomolar antiviral potency against both BSL2 vesicular stomatitis virus (VSV) pseudotyped EBOV (pEBOV) and wild type EBOV combined with in vitro favorable liver metabolic stability.117 Using various filoviral replication-defective and replicative infectivity assays, Gaisina et al. discovered compounds sharing a common 4-(aminomethyl)benzamide core with broad spectrum activity against both EBOV (Mayinga) and MARV (Angola).118 In addition to low micromolar potency (EC50 < 1 μM), these compounds (exemplified by 23) exhibited favorable pharmacokinetic and pharmacological properties without detected cytotoxicity. Preliminary mechanistic studies suggested that the inhibitory activity of these compounds resulted from the direct binding and inhibition of the EBOV envelope glycoprotein (GP) protein, clearly indicating GP as an attractive target for therapeutic intervention other than the key immunogen for eliciting antibody responses.
Fig. 5.
Selected compounds with antiviral activity against EBOV and MARV.
Besides interfering with the processes controlled by the virus, other viable means of blocking viral entry rely on host-directed inhibitors interfering with cellular regulatory networks. Diamide piperazine derivatives were shown to inhibit EBOV by targeting the endosomal host receptor Niemann-Pick C1 (NPC1) and preventing its subsequent interaction with GP, which is considered an essential step for infection.119 Systematic optimization of the original scaffold facilitated modulation of the inhibitory potency and hydrophobicity of this series of analogues. The most promising antiviral profile against EBOV was observed for compound 24 (EC50 = 0.019 μM in a replication competent VSV vector),120 as supported by in vivo data on oral administration of 24 in combination with ritonavir.
A series of epoxysuccinyl-based derivatives were found to inhibit lysosomal cysteine cathepsins, which are proteases required for cleavage of highly glycosylated sequences from the GP and unmasking of its binding site for further interactions with NPC1.121 The most potent compounds exhibited antiviral activity against EBOV and MARV with EC50 values in the low nanomolar range.
Several nucleoside and nucleotide polymerase inhibitors showed promising antiviral activities in cell-based assays often combined with therapeutic efficacy in animal models; however, none of these agents proved to be clinically useful. Prior to repurposing for SARS-CoV-2, Remdesivir entered clinical trials for EVD treatment following evidence of effective post-exposure protection in EBOV-infected rhesus monkeys.122, 123 However, its experimental use as part of an emergency response to the 2018 EVD outbreak in the Democratic Republic of the Congo (Phase III PALM trial) was associated with inferior survival rates compared to 2 mAb therapies, resulting in treatment discontinuation.124 Favipiravir displayed in vivo efficacy as postexposure prophylaxis (PEP) and early treatment in a therapeutic mouse model; however, survival was reduced at a later stage of the infection.53 The Phase II evaluation of its therapeutic efficacy in patients with Guinea and West African EVD was not conclusive. Another early-stage development candidate is imino-C-nucleoside galidesivir (BXC4430),125, 126 for which a Phase I clinical trial has been recently completed to evaluate its intramuscular administration in healthy subjects (NCT03800173). Based on promising in vitro data,127 the lipid prodrug brincidofovir (CMX001), which was originally developed to treat DNA viral infections and is now approved for treatment of smallpox,128 was authorized for compassionate use during the West African EVD epidemic but the trial was later discontinued due to lack of efficiency in preclinical animal models.129
The potential utility of S-adenosyl-l-homocysteine hydrolase (SAHase) inhibitors to provide protection against a lethal Ebola virus challenge has been demonstrated in vivo for known carbocyclic nucleosides 3-deazaadenosine and 3-deazaneplanocin A.130, 131
MCCB4 (compound 25, Fig. 5) is a non-nucleoside polymerase inhibitor identified by in silico driven design that displayed selective anti-EBOV activity in a minigenome assay. This compound was suggested to interfere with viral RNA synthesis by binding to a hydrophobic pocket on the nucleoprotein (NP), which is an essential component of the ribonucleoprotein complex.132
7. Human immunodeficiency virus (HIV)
In developed countries, access to antiretroviral therapy (ART) has drastically improved the prognosis and life expectancy of human immunodeficiency virus (HIV)-positive individuals. However, HIV transmission remains unabated with an estimated worldwide average of 1.5 million people infected in 2020.133 In addition, an increasing trend in HIV infection rates can be expected as a result of reduced access to HIV testing services and medical care during the COVID-19 pandemic. In the absence of an effective vaccine for HIV, the use of small molecule antiretroviral medications as a prevention strategy represents a potent alternative for preventing transmission among HIV-negative individuals at risk of contracting the infection and a major endeavor in antiviral drug discovery.
In 2012, the HIV medication Truvada® (Gilead Sciences), a fixed dose combination product containing the two nucleoside reverse transcriptase inhibitors (NRTIs) emtricitabine (FTC, 200 mg) and tenofovir disoproxil fumarate (TDF, 300 mg), became the first oral regimen to be approved worldwide for daily use in HIV pre-exposure prophylaxis (PrEP). The safety and efficacy (> 90%) of this HIV prevention regimen has been demonstrated, while resistance was found to be rare (< 0.1%).134 However, one issue is that it is only used use to < 20% of the people in the USA that could benefit from it.134
A second tenofovir-based formulation Descovy® given once-daily (Gilead Sciences, tenofovir alafenamide (TAF)/FTC 25 mg/200 mg) was associated with comparable efficiency in reducing HIV acquisition compared with Truvada® but a higher bone and renal safety in a Phase III study (DISCOVER trial), which led to its FDA approval in 2019 for at-risk uninfected adults and adolescents.135
However, the medication adherence must be high for sustained efficacy, and the potential to induce resistance occurs when PrEP is used during undiagnosed acute HIV infections.136 Obviously, PrEP does not prevent viral infections with strains that are resistant against the selected drugs. Many challenges still remain for prevalence in the African region. The PrEP drug must be cheap, easy to access, easy to administer, allow optimal adherence, while the person should be regularly diagnosed to monitor the effect. The benefits, however, clearly outweigh the disadvantages. A variety of long-acting (LA) antiviral compounds and different extended-release formulations are now under investigation to overcome shortcomings associated with once daily self-administered oral therapy, particularly adherence, non-compliance, and cumulative toxicity.137
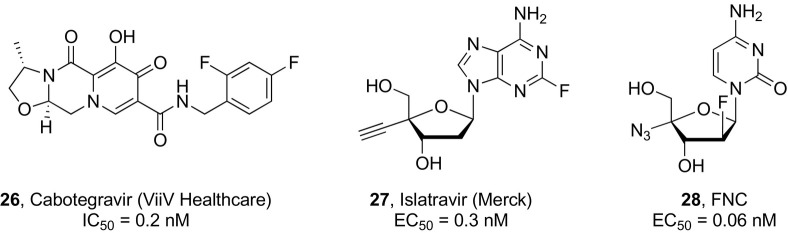
The beginning of 2021 saw the approval of the first long-acting intramuscular injectable anti-HIV treatment for monthly use (Cabenuva®), which was obtained from the coformulation of the integrase inhibitor cabotegravir (200 mg, CAB, ViiV Healthcare, 26, Fig. 6 ) and the non-nucleoside reverse transcriptase inhibitor (NNRTI) rilpivirine (300 mg, RPV, Janssen Pharmaceutica).138 At the same time, a CAB long-acting (CAB-LA) nanosuspension is being clinically evaluated as HIV PrEP monotherapy.139 Two phase III studies (HPTN083 and HPTN084) enrolling different HIV-uninfected risk populations were prematurely unblinded after establishing the superior efficacy of injectable CAB administered every month or every two months versus once-daily oral Truvada.140, 141 However, during the HPTN 083 trial, infections occurred in four cases in spite of preventive CAB-LA exposure but were detected only at a later stage in the collected blood samples most likely due to long-term viral suppression slowing antibody production. This delay in HIV diagnosis led to the emergence of drug resistance, highlighting the need for more sensitive viral load testing.140, 142
Fig. 6.
Long-acting antivirals under investigation for HIV pre-exposure prophylaxis.
Islatavir (4′-ethynyl-2-fluoro-2′-deoxyadenosine, EFdA, MK-8591, Merck, 27, Fig. 6 ) is a highly potent investigational nucleoside analogue that inhibits HIV reverse transcriptase with an unique mode of action by blocking primer translocation without causing direct chain termination. Its active triphosphate metabolite exhibits an exceptionally long intracellular half-life and is responsible for prolonged virological effects, which enables decreased dosing frequencies making this compound suitable for PrEP. Phase III clinical trials are currently undergoing to compare the efficacy of islatravir administered once monthly for lowering the risk of HIV infection to current daily PrEP. Recent clinical studies demonstrated the feasibility of formulating islatavir as an implantable drug for PrEP purposes along with the potential for a once-yearly administration.143
The low oral absorption of the NNRTI dapivirine led to its formulation for topical delivery as an intravaginal silicone ring (IVR), which is currently in the WHO list of prequalified medicines for HIV-1 prevention. In phase II trials, the slow release of dapivirine over one month was shown to reduce the risk of HIV acquisition by 30% compared with placebo, and high acceptance and adherence were observed among the study population.144
Lenacapavir is a capsid inhibitor that appears to disrupt multiple stages of the HIV life cycle. A subcutaneous injectable formulation of this drug candidate displayed a long half-life and is being investigated as a twice-yearly injectable formulation for HIV PrEP.145
An additional agent that could potentially be suitable for PrEP in the future owing to its high potency and slow systemic clearance is 2′-deoxy-2′-β-fluoro-4′-azidocytidine (FNC, 28, Fig. 6),146 which was recently shown to combine the direct inhibition in HIV-1 reverse transcription with an indirect antiviral effect on the host antiviral factor A3G, which may explain its long-lasting action.
8. Yellow fever virus (YFV)
Yellow fever virus (YFV) is a vector-borne (+)-ssRNA virus that shares the same genomic structure and geographical distribution as other pathogens of the genus Flavivirus (Flaviviridae family) responsible for causing severe dengue, Zika virus disease, Japanese encephalitis, and West Nile fever.147 Most individuals contracting YFV infection do not experience symptoms, however in symptomatic patients a wide spectrum of clinical manifestations has been documented ranging from a mild acute febrile illness to a severe viral hemorrhagic fever associated with hepatic dysfunction and renal failure, which is fatal in approximately 50% of cases.148
YFV transmission occurs through two main coexisting cycles involving non-human primates (NHPs) or humans and infected mosquito species of the Aedes and Haemagogus genus.149 In the jungle cycle, infected NHPs serve as sylvatic reservoir of YFV, virtually dismissing the possibility of eradicating this virus. Viremic humans who encroached on NHP forest habitats can introduce the virus into densely populated urbanized areas, where transmission can be perpetuated by competent local vector populations (e.g., Aedes aegypti).
The disease is preventable; a single subcutaneous injection of YF 17D live-attenuated vaccine administered from the age of 9 months appears to confer lifelong immunity.150 While vaccination campaigns and vector-control efforts have concurred to reduce the incidence of YFV infections in many high-risk areas, endemic sylvatic circulation continues in tropical and subtropical regions of Africa and South America where immunization coverage is insufficient.151, 152 Recent outbreaks of YFV-related hemorrhagic disease in rural areas of Angola, Democratic Republic of Congo, and Brazil have renewed fears of a resurgence of urban transmission of this old viral threat.153 Although YFV is absent in the Asia-Pacific region, more frequent exchanges between Africa and Asia caused imported yellow fever cases, which are seen as a growing public health concern since suitable ecological and climatic conditions are present in these non-endemic areas that could support local transmission.154 Notably, the increasing geographic distribution of Aedes aegypti due to climate change is expected to exacerbate the risk of global spread and transmission of YFV across previously unaffected countries such as North America, China, and southern Europe by 2050.155 By 2100, even parts of southern Canada and northern Europe may offer suitable ecological niches to sustain its survival.156
In this scenario, antiviral-based treatments are an important complement to the YF vaccine in building a healthcare response to counteract this potential threat, especially for individuals unwilling to be vaccinated or for whom vaccination is contraindicated, such as those experiencing YF vaccine-associated viscerotropic (YEL-AVD) and neutropic (YEL-AND) diseases similar to those caused by the wild-type virus.157
Repurposing compounds previously investigated or marketed for other therapeutic purposes has been largely pursued for yellow fever disease treatment. Through in vitro screening of a small library of nucleoside analogues, 2’-methyl-7-deazaadenosine, 2’-methyl-7-fluoro-7-deazaadenosine, and a ProTide of 2’,2”-dichlorouridine were identified as effective inhibitors of YFV replication with EC50 values in the 0.25–2.1 μM range and no cytotoxicity up to 100 μM.158
Recent in vitro and preclinical studies in a murine model revealed a potential therapeutic use of the HCV approved nucleoside phosphoramidate prodrug sofosbuvir in the treatment of YFV infections. This prodrug has the advantage of being metabolically activated primarily in the liver, which is a major site of YFV infection. Sofosbuvir was shown to bind to a conserved region of the YFV RNA polymerase (NS5) and exert its antiviral activity (EC50 = 4.2–4.8 μM) against both vaccine and wild-type strains of YFV when tested in human hepatoma cell-based assays. In addition, prophylactic therapeutic treatment with sofosbuvir 1 day prior to infection enhanced survival of YFV-infected animals and reduced liver injury.159 The off-label, compassionate use of Sofosbuvir to treat two YFV-infected patients with acute liver failure led to a reduction in viremia and disease improvement.160
Another RdRp nucleoside inhibitor to be tested as potential anti-YFV candidate is the C-nucleoside galidesivir (BCX4430). Despite displaying a moderate in vitro anti-YFV activity (EC90 = 27.6 μM), galidesivir exhibited potent preclinical efficacy in a Syrian golden hamster model of yellow fever infection. Complete survival of infected animals and improved disease parameters were observed upon intraperitoneal treatment with this nucleoside for 4 or 7 days, which also primed an immune response resulting in protection against a secondary YFV challenge. A phase Ib study to determine the safety and antiviral effects of intravenous galidesivir in YF patients was prematurely terminated (NCT03891420).161
The active 5’-triphosphate metabolite of remdesivir, which accumulates intracellularly and serves as substrate for the RdRp of several RNA viruses, was shown to be an efficient in vitro inhibitor of YFV polymerase with an IC50 of 0.26 μM.162 The YFV polymerase emerged as a superior target for remdesivir among other flaviviral polymerases, while being less efficiently inhibited than coronavirus polymerases.
In addition, some broad-spectrum compounds built on different heterocyclic scaffolds have shown inhibition efficacy against YFV in vitro, however their targets remain undefined.163, 164, 165
9. Poliovirus (PV)
At the launch of the Global Polio Eradication Initiative in 1988, paralytic poliomyelitis was an endemic disease in more than 125 countries.166 Today, the global incidence of wild poliovirus infections has plunged by over 99% thanks to effective vaccination programs. However, while wild poliovirus type 2 (WPV2) and type 3 (WPV3) were declared eradicated in 2015 and 2019, respectively, sporadic cases associated with the circulation of wild type poliovirus type 1 are still being recorded in Pakistan and Afghanistan, despite sustained efforts to eliminate this virus.167
Besides potential breaches of poliovirus containment in research laboratories and vaccine manufacturing facilities, a major menace to global eradication is posed by circulating vaccine-derived polioviruses (cVDPVs) evolved from the use of live attenuated oral poliovirus vaccine (OPV).168 These variants are genetically distinct from wild polioviruses but cannot be distinguished in terms of pathogenicity and transmissibility.169 At present, cVDPVs, particularly strains of serotype 2, are responsible for the majority of paralytic polio outbreaks worldwide.170
In addition, individuals with a primary immunodeficiency disorder (PID) exposed to OPV may excrete immunodeficiency-related vaccine-derived polioviruses (iVDPVs) for prolonged periods of time, which are likely to constitute a reservoir of virus after eradication.171 The development of effective antiviral drugs seems to be the best option to halt chronic excretion of iVDPVs and minimize the likelihood of potential polio outbreaks post eradication.172 The viral proteins considered as targets for anti-polio drug development are those exploited for the identification of anti-rhinovirus agents, due to their high degree of conservation among enteroviruses. In the past, many small molecules capable of inhibiting poliovirus replication have been described (e.g., ribavirin, pleconaril, vapendavir, and pirodavir); however, none progressed further than Phase II evaluation.
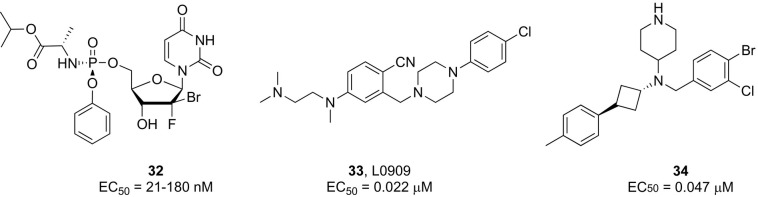
Since a potential poliovirus antiviral drug is considered to have little or no commercial value for its development to be pursued by pharmaceutical companies, a partnership known as the Poliovirus Antiviral Initiative (PAI) was established to advance the discovery of effective poliovirus antiviral drugs. Two antiviral agents with different modes of action, the capsid binder pocapavir (V-073, compound 29, Fig. 8)173, 174 and the peptidomimetic inhibitor of 3C protease V-7404 (compound 30, Fig. 7 ),175 are now under clinical characterization (ViroDefense Inc. & the Bill & Melinda Gates Foundation), for potential use as combination therapy.
Fig. 8.
Structures of anti-HCV lead candidates with different modes of action.
Fig. 7.
Examples of PV small molecule inhibitors.
Recently, some pyrazolopyridine-based compounds were reported to suppress WPV-1 replication in cell culture with EC50 values as low as 5 μM by targeting the viral 2C protein.176 Another report indicated the anticancer nucleoside gemcitabine as an active anti-polio compound in vitro (IC50 = 0.3 μM) able to completely protect HeLa cells against the cytopathic effects of WPV1 at 25 μM.177 Suppression of poliovirus replication was shown to be correlated with the direct inhibition of RNA chain elongation by gemcitabine triphosphate.
Screening of a compound library through a series of in vitro assays against wild-type and vaccine-derived PV strains identified a lead compound (31, Fig. 7) with broad spectrum antiviral activity in the nanomolar range. On the basis of time-of-addition studies, it was hypothesized that 31 could be targeting multiple stages of poliovirus replication; however, other related structural analogues appeared to interfere with the viral entry step.178
10. Hepatitis C virus (HCV)
Hepatis C virus (HCV) is a bloodborne, hepatotropic RNA virus and the only member of the genus Hepacivirus within the Flaviviridae family causing both acute and chronic infections in humans.179 Its genome consists of a 9.6 kb (+)-ss-RNA that is translated into a single large polyprotein precursor. Subsequent polyprotein processing requires two viral proteases (NS2 and NS3/4A), which are autocatalytically excised from the polyprotein, and at least two host cellular peptidases to yield three HCV structural (S) and seven nonstructural (NS) proteins. Several direct-acting antiviral agents (DAAs) have been licensed for clinical use that target three of these NS proteins implicated in different steps of the HCV infectious cycle, i.e., the RdRp (NS5B), NS3/4A protease, and NS5A, a multifunctional RNA-binding protein involved in viral replication, assembly, and regulation of cellular pathways.180 Various fixed-dose combination regimens comprising DAAs with synergistic modes of action have further been approved to offset compensatory evolutionary mechanisms leading to resistance development and viral rebound. In addition, this strategy can reduce treatment cost and duration, improve adherence, and limit dose-limiting toxicity.
The pharmacokinetics and pharmacodynamics of DAA regimens has been recently reviewed.181 Most of them consist of two drugs, either an NS5B and NS5A inhibitor [e.g., sofosbuvir + ledipasvir (Harvoni®), sofosbuvir + velpatasvir (Epclusa®)] or an NS5A and NS3/4A inhibitor [e.g., elbasvir + grazoprevir (Zepatier®), glecaprevir + pibrentasvir (Maviret®, Mavyret®)]. Also a triple combination targeting NS5B, NS5A, and NS3/4A [e.g., Sofosbuvir + Velpatasvir + Voxilaprevir, Vosevi®]182 as well as a multiple regimen (Paritaprevir + Ritonavir + Ombitasvir and Dasabuvir)183 have been approved for the treatment of chronic HCV genotype (GT)1-6 and GT1 infection in adults, respectively. With the availability of these oral combination therapies, the standard of care for chronic HCV infection has dramatically improved. Today, most patients, including those previously refractory to treatment such as individuals with HCV and HIV co-infection, decompensated liver disease, and renal impairment, can be successfully cured, as indicated by sustained virologic response (SVR) rates (defined as aviremia 24 weeks after therapy completion) up to 95%.184 Recently, an 8-week regimen of glecaprevir and pibrentasevir given to untreated patients with chronic HCV genotype 1–6 infection and compensated cirrhosis was shown to afford a virological cure rate as high as 99.7%, irrespective of any pre-treatment patient or viral characteristics.185 While a short treatment of acute HCV infection with sofosbuvir and ribavirin was demonstrated to be suboptimal,186 high efficacy was instead obtained with a 6-week regimen of sofosbuvir and ledipasvir in patients with HCV GT1 monoinfection187 as well as GT1 or 4 HCV infection and HIV-1 coinfection.188 DAA treatment may also lead to partial restoration of HCV-specific immune responses in chronically infected patients, an effect that might be improved with early administration.189 DAA re-treatment of chronic HCV infections has also been shown to be highly effective.190 Re-treatment regimens based on a combination of sofosbuvir, velpatasvir, and voxilaprevir191 or glecaprevir and pibrentasvir192 for DAA-experienced patients have been approved. In addition, the beneficial clinical effects of DAAs has led to a significant reduction in the waitlist for liver transplant due to chronic HCV infection, while increasing the survival of patients with HCV-associated hepatocellular carcinoma.193
Overall, this scenario offers good prospects that the WHO program to eliminate viral hepatitis C as a public health problem can meet its goals by 2030.194 Nonetheless, at present, there are still approximately 58 million people worldwide living with chronic HCV infection and 1.5 million new infections annually,195 reflecting the fact that significant challenges remain to be addressed including accurate diagnosis of HCV infected patients, treatment of patients with decompensated cirrhosis or renal impairment, serious complications due to drug-drug interactions, high cost of treatment for underdeveloped countries, and occurrence of resistance associated variants (RAVs) to DAAs.181 In particular, the development of new curative HCV drugs that may be effective against DAA-resistant mutants and broadly active on multiple HCV genotypes remains an important to optimize current therapeutic regimens and facilitate the global eradication of HCV, especially in the absence of a prophylactic vaccine.
Drug discovery efforts are actively ongoing to discover new pan-genotypic inhibitors of NS5A that are expected to decrease the incidence of treatment failure associated with low barrier to resistance of most marketed NS5A inhibitors for genotype 1 subtype a (GT1a)196, 197, 198 as well as improve the pharmacokinetic profiles of existing drugs by prodrug synthesis to enhance solubility properties and simplify drug dosage formulations (e.g., pibrentasvir).199
The macrocyclic scaffold structure shared by approved HCV NS3/4A protease inhibitors has been modified to modulate both potency and drug-like properties of the inhibitors as well as prevent resistance due to pre-existing resistance-associated substitutions (RASs) and/or baseline polymorphisms among diverse genotypes.200, 201
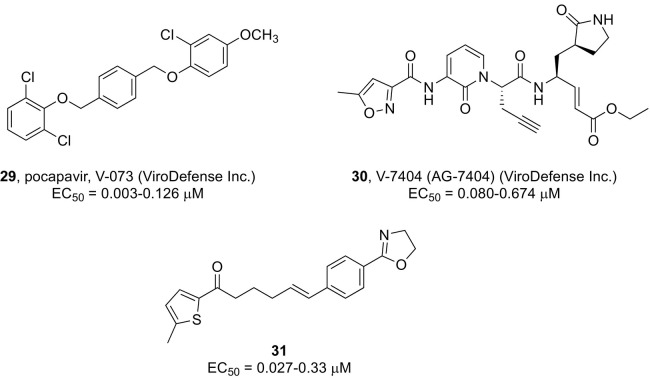
Aiming at identifying alternative uridine-containing sugar-modified NS5B inhibitors, chemical modifications were applied to the sugar moiety of sofosbuvir, such as the introduction of a fluorine atom at the 4′ position of a 2′-methyl-uridine phosphoramidate prodrug (AL-335)202 or the replacement of the 2’-methyl-2’-fluoro moiety with a dihalogenated functional group, such as in compound 32 (Fig. 8 ).203 This Sp phosphoramidate diastereomer of 2′-bromo,2′-fluoro-uridine exhibited potent inhibition of HCV in cell culture (EC50 = 21–180 nM) combined with pan-genotypic anti-HCV replicon activity. Its corresponding 5′-triphosphate was selective for HCV NS5B polymerase of GT1-6 with no concomitant inhibition of human or cellular mitochondrial RNA polymerases, while exhibiting favorable liver accumulation according to a preliminary liver pharmacokinetic study in beagle dogs. Alternatively, guanosine containing nucleoside phosphoramidate prodrug analogues also displayed potent activity against HCV infection and in some cases potential value as new clinical anti-HCV agents. This include a 2’,4’-fluoro substituted guanosine nucleotide analogue (AL-611)204 and AT-527 (compound 2 in Fig. 1), which entered clinical development owing to the attractive broad spectrum coverage of major HCV genotypes.205 Second-generation non-nucleoside inhibitors of NS5B, such as N-benzoxaborole benzofuran analogues, are also undergoing structural optimization programs.206
Recently, there has also been a surge in research efforts to find anti-HCV compounds with different modes of action, i.e., targeting stages of HCV infection either preceding or following viral replication. Although the full understanding of these complex multifactorial processes remains elusive, several related factors and processes have been investigated as potential therapeutic targets. These compounds could have an important synergist effect with DAAs towards HCV elimination or serve as preventive measures against HCV infection and reinfection.
The design of preclinical candidate L0909 (compound 33, Fig. 8), featuring a 2-((4-arylpiperazin-1-yl)methyl)benzonitrile scaffold, was the result of a structure − activity relationship (SAR) study that optimized a modestly potent anti-HCV 2-(arylamino)methylbenzonitrile lead inhibitor by introducing a piperazine moiety. L0909, which displayed nanomolar antiviral activity in a HCVcc system (EC50 = 0.022 μM), appeared to act on the HCV entry stage. A synergistic effect against HCV was observed for L0909 in combination with DAAs together with a high sensitivity to clinical resistant HCV mutants. Further in vivo pharmacological and preclinical results supported the favorable anti-HCV profile of this compound.207
A series of polyheterocyclic derivatives featuring a 4-aminopiperidine scaffold as common structural element have been shown to be involved with the modulation of the HCV assembly process.208 Antiviral synergy was observed for the combination of a starting screening hit with various marketed HCV therapeutics in cell-based assays of HCV, and an advantageous hepatotropic distribution was confirmed by in vivo pharmacokinetic studies. Lead optimization studies defined compound 34 (Fig. 8) as a promising candidate with nanomolar anti-HCV activity for further in vivo and and preclinical efficacy profiling.
In principle, broad spectrum antiviral activity against HCV can only be achieved with compounds acting by multiple modes of action (e.g., ribavirin) or through host-cell directed strategies, such as those targeting the human enzyme dihydroorotate dehydrogenase (DHODH).209
11. Rabies virus (RABV)
Rabies virus (RABV) is the archetypal species of the genus Lyssavirus (Rhabdoviridae family), a group of neurotropic viruses that carry a (-)-ss RNA genome and can infect both humans and animals.210 Rabies disease is the deadliest among all viral infections with a 100% fatality rate after the onset of clinical symptoms.211 Upon entering the peripheral nerves, RABV reaches the central nervous system (CNS) by means of axonal transport, where it preferentially replicates evading the host immune system and causing a severe and incurable neurological disease.212
Effective vaccines are available for pre-exposure immunization as well as human rabies immune globulin (HRIG) for post-exposure prophylaxis (PEP) intervention early after infection, which can successfully prevent the disease from progressing towards symptoms.213, 214 However, the risk of acquiring rabies infections is perpetuated through the low level of routine immunization coverage in high risk countries, manifesting in an estimated 59,000 human deaths claimed by RABV each year worldwide. RABV can be expected to persist globally as a pervasive public health threat because of its zoonotic transmission, which occurs in approximately 99% of cases through the transfer of infectious saliva from bites of infected dogs.
Antiviral molecules that may be able to suppress viral replication in an infected individual during the symptomatic phase of encephalitic rabies will be the next major step in fighting human RABV infections, however direct dosing in the cerebrospinal fluid is still considered an unmet challenge.215 The development of antiviral compounds is also desirable as cost-effective and easily storable alternatives to rabies immunoglobulins currently used for PEP, especially considering the heavy economic burden associated with RABV prophylaxis and the fact that most infections (95%) occur in Africa and Asia where the supply of these biologics is limited.216
The promising in vitro inhibitory activity of the broad-spectrum RNA polymerase inhibitor ribavirin against RABV did not translate into in vivo antiviral efficacy in preclinical animal models and clinical trials in humans.217 Some ribavirin analogues named EICAR and EICNR displayed increased in vitro activity (EC50 = 0.9–3.8 μM) as inhibitors of viral replication and transcription, most likely via IMPDH inhibition, compared to ribavirin (EC50 = 18.6 μM); however, a more systematic evaluation is needed to define their anti-RABV potential.218
Independent studies demonstrated the capability of favipiravir to effectively reduce RABV replication in vitro219, 220; however, little or no effect on survival in a mouse model of RABV infection was observed. Recently, in vivo imaging studies demonstrated that favipiravir treatment 1 h or 2 days after intramuscular inoculation of RABV in mice could suppress viral replication in the periphery, but double or triple doses were needed to suppress replication in the CNS.221
Two adamantane derivatives, 1-adamantyl-(5-bromo-2-methoxybenzyl) amine (ABMA) and its 1-dimethyl analogue DABMA, were described to prevent RABV viral gene expression and genome replication by interfering with different steps of the late endosomal pathway.222 When delivered 2 h pre-infection, these early-stage post-entry inhibitors exhibited in vitro micromolar activities against RABV.
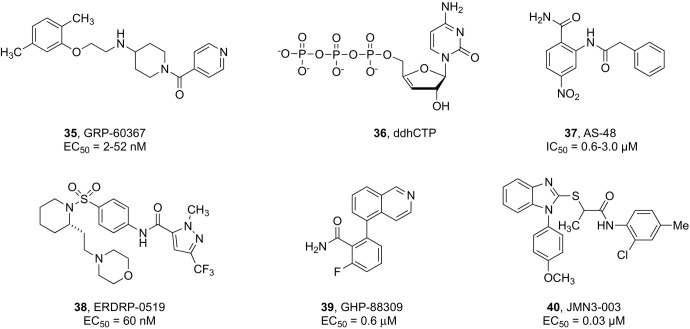
More recently, the implementation of a high-throughput screening (HTS) protocol based on a single-cycle RABV reporter strain allowed to identify a first-in-class, highly potent (EC50 = 2–52 nM) and selective small molecule inhibitor of RABV, designated as GRP-60367 (compound 35, Fig. 9 ). Mechanistic characterization demonstrated that GRP-60367 blocks RABV entry by specifically targeting a previously unknown druggable site on the G protein, representing a suitable lead compound for the design of next-generation antivirals for RABV treatment and prevention.223
Fig. 9.
Small molecules active against RABV and MeV infection.
Notably, a broadly conserved radical S-adenosyl-l-methionine (SAM) enzyme known as viperin appeared to be implicated in the inhibition of the replication of many RNA viruses, including RABV.224 Recently, it was demonstrated that this enzyme synthesizes a broad spectrum antiviral nucleotide, i.e., 3′-deoxy-3′,4′-didehydro-cytidine triphosphate (ddhCTP, 36 in Fig. 9), by catalyzing the dehydration of CTP via a radical mechanism. This study established ddhCTP as a naturally occurring chain terminator encoded by the human genome.225 Additional studies suggested that viperin exerts its antiviral effect by activating a component of the protein ubiquitination machinery with subsequent degradation of cellular and viral proteins essential for viral replication.226
Host-targeted therapies to combat RABV infections have not been very successful to date.216
12. Measles virus (MeV)
Measles virus (MeV) is a (-)-ssRNA virus classified in the genus Morbillivirus within the Paramyxoviridae family that is recognized as the causative agent of one of the most contagious respiratory acute disease affecting humans. Its basic reproduction number (i.e., average cases caused by an infected individual during the infectious period) is estimated to vary between 12 and 18, surpassing by far that of SARS-CoV-2 (i.e., 2.5–3.5).227 A safe, affordable, and effective life-attenuated vaccine is available to prevent outbreaks and ensure immunity against MeV; however, this virus is still prevalent in Third World countries where vaccine access and distribution are inefficient. Recently, developed countries where endemic MeV transmission was considered eliminated experienced a re-emergence of this virus following an alarming decline in immunization coverage due to vaccination hesitancy. In the period between 2017 and 2019, an approximate 130% increase of deaths directly attributed to measles was documented worldwide (207,500 in 2019), mostly among children under 5 years of age.228 The risk of globally recurring measles outbreaks is anticipated to raise due to the COVID-19 pandemic as many countries suspended childhood immunization measles campaigns leaving dangerous immunity gaps.228, 229 Cost-effective and readily distributable small molecule antivirals are urgently needed for outbreak control especially among under-vaccinated populations in vulnerable countries where the clinical need is highest and to aid in the long term measles eradication by synergizing with vaccination to close gaps in herd immunity due to vaccine refusal.
In the absence of a specific antiviral agent approved for treating MeV infections, various small molecule inhibitors targeting viral components as well as cellular factors have been evaluated in vitro and in animal models; however, further clinical development has been delayed by the low commercial value of a potential measles drug, ethical challenges associated with the design of clinical trials among a prevailing pediatric patient population, and the limited opportunities for intervention associated with the predominantly acute nature of the disease.230 In rare cases, MeV may invade the central nervous system (CNS) during or after infection leading to severe neurological manifestations that complicate potential antiviral treatments.231
A homology model of the MeV fusion (F) protein trimer guided the rational design of a class of nonpeptidic fusion protein inhibitors featuring an anilide scaffold that appeared to exert a stabilizing effect on the prefusion state of F trimers.232, 233 These compounds, as exemplified by AS-48 (compound 37, Fig. 9), exhibited broad spectrum activity across various MeV strains as well as closely related pathogens of the Morbillivirus genus such as the canine distemper virus (CDV), however their moderate antiviral potency (EC50 values in the low-micromolar range) observed in cellular assays hampered their further evaluation in animal models.
Among nucleoside polymerase inhibitors, remdesivir was evaluated against various paramyxoviruses by employing reporter virus assays and emerged as a potentially viable candidate to treat MeV infections with an EC50 of 0.037 μM.234 The introduction of a fluorine substituent at the 2'-position of 4’-azido-cytidine (R1479) yielded two additional analogues, namely 2’-fluoro and 2’-difluoro-4’-azido-cytidine, which displayed low micromolar activities (EC50 = 0.37 and 0.34 μM, respectively) superior to the parent compound (1.9 μM) when tested in a recombinant measle virus model).235
The investigation of non-nucleoside inhibitors of MeV RdRp has delivered molecules with promising antiviral profiles that could advance the path towards the development of effective measles therapeutics. ERDRP-0519 (compound 38, Fig. 9) is an orally bioavailable pan-morbillivirus inhibitor derived from an optimization process of hits containing the pyrazole carboxamide scaffold that were identified by a cell-based HTS campaign.236 Compound 38 showed potent in vitro inhibitory activity against MeV and CDV strains. Initial animal in vivo characterization focused on testing the efficacy of 38 in a lethal CDV-ferret surrogate model of human MeV. CDV is known to cause lethal measles-like disease in ferrets. This study showed that oral treatment of CDV-infected ferrets 3 days after exposure completely suppressed disease clinical signs. All treated animals survived and developed a robust immune response against the virus providing protection against reinfection when challenged with a later lethal virus dose.237
Recently, the in vivo oral efficacy of this compound was further validated in non-human primates known to develop a human measles-like disease.238 Prophylactic and post-exposure therapeutic treatment of squirrel monkeys (Saimiri sciureus) reduced virus shedding and prevented measles disease. Mechanistically, ERDRP-0519 was demonstrated to target the MeV polymerase and specifically inhibit all phosphodiester bond formation during both the de novo initiation of RNA synthesis and RNA elongation steps.239
A high-throughput screen for inhibitors of human parainfluenza virus type-3 (HPIV3) led to the identification of another broad-spectrum paramyxovirus allosteric inhibitor, i.e., GHP-88309 (compound 39, Fig. 9), which was also active against MeV in well-differentiated human airway organoid cultures.240 This compound was also shown to act as an allosteric inhibitor of viral polymerase activity by interfering with the initiation phase of viral synthesis. In vivo proof-of-concept efficacy was established in a lethal Sendai virus mouse surrogate model of human HPIV3 disease. Oral administration of 39 48 h after infection provided complete protection against infection and led to immunoprotection acquisition against reinfection.
A series of hit compounds emerging from a large scale HTS was shown to constitute a mechanistically novel class of MeV inhibitors able to interfere with host factors required for viral RdRp activity. Among these, JMN3-003 (compound 40, Fig. 9) showed potent and broad activity against MeV as well as a selection of clinically significant members of the para- and orthomyxovirus families. A high barrier against rapid viral escape from inhibition for this class of compounds was demonstrated by in vitro adaptation studies.241 Other efforts to target host factors as a strategy to develop anti-measle compounds include among others repurposing kinase inhibitors.242
13. New coronaviruses (CoVs)
Coronaviruses (CoVs) are diverse (+)-ssRNA viruses classified in the Coronaviridae family and Orthocoronavirinae subfamily, where they are further taxonomically clustered into four genera designated as α, β, γ, and δ.243 Seasonal coronavirus infections are caused in humans by a total of four α-CoVs (HCoV-NL63 and HCoV-229E) and β-CoVs (HCoV-HKU1 and HCoV-OC43), which circulate endemically and are typically responsible for mild to moderate gastrointestinal and upper-tract respiratory illnesses.244 However, the last two decades have witnessed the alarming epidemic and pandemic impact of this class of human pathogens with the emergence of three viral pneumonia outbreaks caused by previously unknown β-CoVs (SARS-CoV in 2003, MERS-CoV in 2012, and SARS-CoV-2 in 2019).245, 246 While new cases of SARS have not been reported since 2004, MERS continues to circulate in the Middle East with the latest confirmed cases and deaths dating between March and July 2021.247 As of the 8 september 2021, the underway SARS-CoV-2 pandemic has affected more than 221 million people and claimed 4.5 million lives.248 The pathogenesis of these infections may vary from mild symptoms to severe respiratory distress and in some cases multiple organ failure.249 Upon binding specific receptors on the surface of respiratory epithelial cells [i.e., angiotensin-converting enzyme 2 (ACE2) for SARS-CoV and SARS-CoV-2, dipeptidyl peptidase 4 (DPP4) for MERS-CoV], these viruses enter the airways and start replicating primarily in the lower respiratory tract where they can trigger an aberrant host immune response.
These three highly pathogenic CoVs are alleged to have originated in bats, which are known to be primary natural hosts for human β-CoVs.250 While adaptation from bats in an intermediary host was shown to have facilitated transfer to humans in the case of SARS-CoV and MERS-CoV (i.e., via infected palm civets and dromedary camels, respectively),246 no clear evidence has been found to support this transmission mode for the new virus.245 Phylogenetic analysis suggested that the SARS-CoV-2 lineage persisted for decades in bats where it evolved as to be capable of direct transmission and replication in human cells.251
Given the zoonotic origin of these viruses as well as their rapid mutation frequency often resulting in variants of higher infectivity, new CoVs related to SARS, MERS, and SARS-CoV-2 may not be excluded in the future and demand urgent action. The development and stockpiling of antiviral therapeutics with broad anti-coronavirus coverage against the currently known pathogenic CoVs must be prioritized for pandemic preparedness and prevention.252
The most logic strategy towards the discovery of drugs that would inhibit a wide range of closely and distantly related viral pathogens is targeting viral proteins that feature the highest degree of sequence conservation among different species and thus may be less likely to mutate. A recent patent literature search indicates that several potential targets can be considered for pan-coronavirus drug design (e.g., RdRp, Mpro, PLpro, RNA helicase, spike glycoprotein).253 A map of conserved regions among the main human coronavirus proteins has been recently reported based on the analysis of a large set of viral genomic sequences.254
The viral main protease Mpro is an ideal target for broad spectrum antiviral inhibition as Mpro structural and functional similarities across different coronaviruses are known. For instance, SARS-CoV and SARS-CoV-2 Mpro sequences share 96% amino acid identity and a 100% sequence homology occur at the level of their substrate binding active site.255
In an initial study, a structure-based approach led to the identification of a series of α-ketoamide-based peptidomimetic Mpro inhibitors with equipotent activity against α-CoVs (HCoV-229E) and β-CoVs (SARS-CoV and MERS-CoV) in cell cultures.256 Scaffold optimization yielded compound 41 (Fig. 10 ) featuring a pyridone ring between the P3 and P2 amide bonds that inhibited SARS-CoV-2 Mpro with an IC50 of 0.67 μM, while displaying IC50 values corresponding to 0.90 and 0.58 μM for inhibiting the SARS-CoV Mpro and the MERS-CoV Mpro, respectively.255 Compound 41 was further tested in a SARS-CoV replicon where it showed the ability to inhibit RNA replication at EC50 = 1.75 μM. In human Calu-3 cells infected with SARS-CoV-2, an EC50 of 4 to 5 μM was observed.
Fig. 10.
Structures of some potential pan-coronavirus antivirals.
Boceprevir and the peptidomimetic GC376 (42, Fig. 10) were also found to possess a broad spectrum multitargeting profile against SARS-CoV-2, SARS-CoV, MERS-CoV, and HCoV.257 Deuterated variants of GC376 were recently reported to have improved potency compared to the parent compound in vitro, while in vivo efficacy was demonstrated by the observation of enhanced survival rates in infected mice following treatment at 24 h postinfection.258
PF-00835231 (the parent of clinical candidate PF-07304814 4, Fig. 1) was originally developed as a SARS-CoVMpro inhibitor in response to the 2002-2003 SARS outbreak.259 Pan-coronavirus antiviral activity was also observed with PF-00835231 against a variety of α-CoVs (NL63-CoV, PEDV, FIPV), β-CoVs (HKU4-CoV, HKU5-CoV, HKU9-CoV, MHV-CoV, OC43-CoV, HKU1- CoV), and γ-CoVs (IBV-CoV) with Ki values ranging from 30 pM to 4 nM in in fluorescence resonance energy transfer (FRET) protease activity assays, suggesting a potential therapeutic use against emerging coronaviruses.26
The orally bioavailable Mpro inhibitor PF-07321332 5 (Fig. 1) currently in clinical trials for COVID-19 could inhibit the proteolytic activity of MPro of various β-CoVs (SARS-CoV, HCoV-HKU1, HCoV-OC43, MERS-CoV) as well as endemic α-CoVs (HCoV-229E, HCoV-NL63) in FRET assays with Ki values ranging from 4.94 to 226 nM.27 The potent pan-human coronavirus antiviral activity of PF07321332 was further validated in cytopathic effect (CPE) cellular assays against SARS-CoV (EC90 = 317 nM), MERS-CoV (EC90 = 351 nM), and HCoV-229E (EC90 = 620 nM).
Since the RdRp (also named nsp 12) and its mode of substrate recognition during viral RNA synthesis are highly conserved among diverse coronaviruses (e.g., RdRp of SARS-CoV-2 shares 99.1% similarity and 96% amino acid identity with that of SARS-CoV),260 a broad antiviral activity spectrum can likewise be expected from nucleoside analogues that are incorporated into the growing RNA chain after intracellular phosphorylation and either induce delayed chain termination or lead to base-pair mismatches resulting in lethal mutation rates, as epitomized by remdesivir and molnupiravir, respectively. Besides SARS-CoV-2, the in vitro antiviral activity spectrum of remdesivir (1, Fig. 1) was revealed to further cover SARS-CoV (EC50 = 0.069 μM) and MERS-CoV (EC50 = 0.025–0.12 μM) as well as endemic HCoV-OC43 and HCoV-229E and porcine CoVs.261 Further in vivo efficacy studies conducted in a rhesus macaque and mouse model of MERS-CoV and SARS-CoV infection, respectively, demonstrated the prophylactic and therapeutic value of this drug.
The parent nucleoside of mutagenic SARS-CoV-2 antiviral molnupiravir 3 (Fig. 1) was shown to be highly active in vitro against SARS-CoV-2, while also conferring a potent inhibition of MERS-CoV (IC50 = 0.024 μM) and SARS-CoV (IC50 = 0.14 μM) in primary human epithelial cell cultures. Interestingly, the antiviral activity of NHC was extended to zoonotic SARS- and MERS-like CoVs (SHC014, HKU3, and HKU5), suggesting that an increase (up to 20%) in the natural amino acid variation of the RdRp did not affect the activity of this nucleoside prodrug.17
AT-511, the free base congener of clinical candidate AT-527 (2, Fig. 1), was also shown to have a broad antiviral activity spectrum among α-CoVs (HCoV-229E) and β-CoVs (HCoV-OC43, SARS-CoV) in different cell lines with EC50 values in the micromolar and sub-micromolar range. Surprisingly, only moderate activity was observed against MERS-CoV (EC50 = 26-27 μM), suggesting that the mode of action of this nucleoside analogue might not be limited to RdRp activity inhibition and subsequent non-obligate chain termination but implicate the additional interaction with another functional domain of the polymerase complex [i.e., nidovirus RdRp-associated nucleotidyltransferase (NiRAN)] crucial for viral replication.14
Further potential drug candidates for the treatment of multiple coronavirus infections could target the viral entry step, and specifically the spike (S) protein-mediated membrane fusion. Among a series of lipopeptide analogues, EK1C4 exhibited the most potent inhibitory activity against membrane fusion and pseudovirus infection of SARS-CoV-2 with IC50 values of 1.3 and 15.8 nM, respectively, while also effectively inhibiting the replication of SARS-CoV, MERS-CoV, and other HCov.262 A protective effect against HCoV-OC43 infection was observed upon intranasal application of EK1C4 in mice before or after viral challenge, suggesting that EK1C4 could be useful for both prevention and treatment of infection by the currently circulating SARS-CoV-2 and other potentially emerging CoVs.
Other broad spectrum antivirals targeting coronaviruses with an unknown mode of actions have been identified by screening programs263 and drug repurposing campaign.264 Targeting host factors such as kinase pathways,265 cysteine protease like cathepsin L,266 or compounds that deplete cellular polyamines by inhibiting their biosynthesis (e.g., fluoromethylornithine) and in this way reduce viral attachment and entry267 maybe also represent cellular approaches towards broad spectrum antivirals.
14. Conclusion
The fight against virus infections is based upon the prophylactic (or preventive) administration of vaccines or therapeutic use of antiviral drugs, although, as testified by PrEP, antivirals (e.g., Truvada® and Descovy®) have also been formally approved for the prevention of HIV infections. Generally, when a virus has economic implications but is mostly harmless, like in the case of rhinovirus infections, a drug that reduces symptoms and infectivity may suffice. By contrast, for viral infections with an enormous impact on social (e.g., HCV-related cirrhosis and hepatocellular carcinoma) or healthcare (e.g., SARS-CoV-2) systems, it is vital to develop drugs that can eradicate the causative pathogen. The development of drugs for antiviral prophylaxis is particularly significant for viral infections leading to severe illnesses and high case fatality rates, and especially those for which the production of a vaccine is problematic. As corroborated in the past by the development of beta-lactam antibiotics, an infection can most effectively be treated using compounds whose mode of action either triggers the formation of covalent bonds or avoids that these bonds are broken. Accordingly, several nucleoside analogues and protease inhibitors have proven their value as antiviral agents so far, but we can envisage more broad-spectrum therapeutic applications for these classes of drugs in the future. It should also be appreciated that heterocyclic-based antiviral drugs may unforeseeably interact with proteins of the host, leading to pharmacological toxicity that can only be discovered during clinical Phase III and IV studies. These compounds need to bind to a viral target protein with high affinity, thus easily leading to the selection of resistant viruses. Targeting host factors remains a challenging approach as the virus may hijack other pathways once the cellular receptors or enzymes that are normally exploited for its in vivo infection, replication, and transmission are blocked.
Overall, the availability of a vaccine for a given viral infection may also dictate the priorities for antiviral drug discovery research. Smallpox (variola virus) was the first virus infection to be eradicated owing to vaccination. Poliovirus maybe the next example, however the final steps towards its global eradication appear to pose a few unique challenges. In the foreseeable future, we may witness the elimination of mumps and measles viruses providing that a continued effort in vaccination coverage is ensured, while rubella virus is virtually eradicated. With the availability of DAAs, HCV can be expected to be eradicated within a few years, while for hepatitis B virus (HBV) both vaccines and antivirals are applicable. The continuously emerging new influenza virus mutants may also be counteracted by vaccines and antivirals, whereas for RSV antiviral drug treatment seems to become increasingly imminent. For several other virus infections, such as rhino or hemorrhagic fever infections, vaccines and antivirals are anxiously awaited. Finally, several successful vaccines have been launched to prevent SARS-CoV-2 infections, and effective antivirals will likely follow.
Depending on their nature, viral infectious diseases may be amenable to either prophylaxis, drug treatment or both. As amply demonstrated in this review, these are important times for the field and many approaches are being pursued in parallel to combat viral infections with a continuing impact on human health. The pace of antiviral drug discovery needs to move well beyond a single target, and it can be anticipated that the next decade will deliver antiviral cocktails against most of these infections.
References
- 1.VBI Vaccines . 2020. VBI-2901: Pan-Coronavirus Vaccine Candidate Overview, VBI Vaccines; pp. 6–9. [Google Scholar]
- 2.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E.C., Zhang Y.-Z. A New Coronavirus Associated With Human Respiratory Disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Su H., Zhou F., Huang Z., Ma X., Natarajan K., Zhang M., Huang Y., Su H. Molecular Insights into Small-Molecule Drug Discovery for SARS-CoV-2. Angew. Chem. Int. Ed. 2021;60:9789–9802. doi: 10.1002/anie.202008835. [DOI] [PubMed] [Google Scholar]
- 4.Namchuk M.N. Early Returns on Small Molecule Therapeutics for SARS-CoV-2. ACS Infect. Dis. 2021;7:1298–1302. doi: 10.1021/acsinfecdis.0c00874. [DOI] [PubMed] [Google Scholar]
- 5.Shang Z., Chan S.Y., Liu W.J., Li P., Huang W. Recent Insights Into Emerging Coronavirus: SARS-CoV-2. ACS Infect. Dis. 2021;7:1369–1388. doi: 10.1021/acsinfecdis.0c00646. [DOI] [PubMed] [Google Scholar]
- 6.U.S. Food & Drug Administration FDA Approves First Treatment for COVID-19. 2020. https://www.fda.gov/news-events/press-announcements/fda-approves-first-treatment-covid-19
- 7.Kokic G., Hillen H.S., Tegunov D., Dienemann C., Seitz F., Schmitzova J., Farnung L., Siewert A., Höbartner C., Cramer P. Mechanism of SARS-CoV-2 Polymerase Stalling by Remdesivir. Nat. Commun. 2021;12:279. doi: 10.1038/s41467-020-20542-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., Lopez de Castilla D., Finberg R.W., Dierberg K., Tapson V., Hsieh L., Patterson T.F., Paredes R., Sweeney D.A., Short W.R., Touloumi G., Lye D.C., Ohmagari N., Oh M.-D., Ruiz-Palacios G.M., Benfield T., Fätkenheuer G., Kortepeter M.G., Atmar R.L., Creech C.B., Lundgren J., Babiker A.G., Pett S., Neaton J.D., Burgess T.H., Bonnett T., Green M., Makowski M., Osinusi A., Nayak S., Lane H.C. Remdesivir for the Treatment of Covid-19—Final Report. New Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan H., Peto R., Henao-Restrepo A.M., Preziosi M.P., Sathiyamoorthy V., Abdool Karim Q., Alejandria M.M., Hernández García C., Kieny M.P., Malekzadeh R., Murthy S., Reddy K.S., Roses Periago M., Abi Hanna P., Ader F., Al-Bader A.M., Alhasawi A., Allum E., Alotaibi A., Alvarez-Moreno C.A., Appadoo S., Asiri A., Aukrust P., Barratt-Due A., Bellani S., Branca M., Cappel-Porter H.B.C., Cerrato N., Chow T.S., Como N., Eustace J., García P.J., Godbole S., Gotuzzo E., Griskevicius L., Hamra R., Hassan M., Hassany M., Hutton D., Irmansyah I., Jancoriene L., Kirwan J., Kumar S., Lennon P., Lopardo G., Lydon P., Magrini N., Maguire T., Manevska S., Manuel O., McGinty S., Medina M.T., Mesa Rubio M.L., Miranda-Montoya M.C., Nel J., Nunes E.P., Perola M., Portolés A., Rasmin M.R., Raza A., Rees H., Reges P.P.S., Rogers C.A., Salami K., Salvadori M.I., Sinani N., Sterne J.A.C., Stevanovikj M., Tacconelli E., Tikkinen K.A.O., Trelle S., Zaid H., Røttingen J.A., Swaminathan S. Repurposed Antiviral Drugs for Covid-19—Interim WHO Solidarity Trial Results. N. Engl. J. Med. 2021;384:497–511. doi: 10.1056/NEJMoa2023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y., Cao L., Li G., Cong F., Li Y., Sun J., Luo Y., Chen G., Li G., Wang P., Xing F., Ji Y., Zhao J., Zhang Y., Guo D., Zhang X. Remdesivir Metabolite GS-441524 Effectively Inhibits SARS-CoV-2 Infection in Mouse Models. J. Med. Chem. 2021 doi: 10.1021/acs.jmedchem.0c01929. [DOI] [PubMed] [Google Scholar]
- 11.Yan V.C., Muller F.L. Advantages of the Parent Nucleoside GS-441524 Over Remdesivir for Covid-19 Treatment. ACS Med. Chem. Lett. 2020;11:1361–1366. doi: 10.1021/acsmedchemlett.0c00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schooley R.T., Carlin A.F., Beadle J.R., Valiaeva N., Zhang X.-Q., Clark A.E., McMillan R.E., Leibel S.L., McVicar R.N., Xie J., Garretson A.F., Smith V.I., Murphy J., Hostetler K.Y. Rethinking Remdesivir: Synthesis, Antiviral Activity and Pharmacokinetics of Oral Lipid Prodrugs. bioRxiv. 2021 doi: 10.1128/AAC.01155-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zandi K., Amblard F., Musall K., Downs-Bowen J., Kleinbard R., Oo A., Cao D.D., Liang B., Russell O.O., McBrayer T., Bassit L., Kim B., Schinazi R.F. Repurposing Nucleoside Analogs for Human Coronaviruses. Antimicrob. Agents Chemother. 2021;65 doi: 10.1128/AAC.01652-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Good S.S., Westover J., Jung K.H., Zhou X.-J., Moussa A., Colla P.L., Collu G., Canard B., Sommadossi J.-P. AT-527, a Double Prodrug of a Guanosine Nucleotide Analog, is a Potent Inhibitor of SARS-CoV-2 In Vitro and a Promising Oral Antiviral for Treatment of COVID-19. Antimicrob. Agents Chemother. 2021;65:e02420–e02479. doi: 10.1128/AAC.02479-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Do T.N.D., Donckers K., Vangeel L., Chatterjee A.K., Gallay P.A., Bobardt M.D., Bilello J.P., Cihlar T., De Jonghe S., Neyts J., Jochmans D. A Robust SARS-CoV-2 Replication Model in Primary Human Epithelial Cells at the Air Liquid Interface to Assess Antiviral Agents. Antiviral Res. 2021;192 doi: 10.1016/j.antiviral.2021.105122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer W., Eron J.J., Holman W., Cohen M.S., Fang L., Szewczyk L.J., Sheahan T.P., Baric R., Mollan K.R., Wolfe C.R., Duke E.R., Azizad M.M., Borroto-Esoda K., Wohl D.A., Loftis A.J., Alabanza P., Lipansky F., Painter W.P. Molnupiravir, an Oral Antiviral Treatment for COVID-19. medRxiv. 2021 [Google Scholar]
- 17.Sheahan T.P., Sims A.C., Zhou S., Graham R.L., Pruijssers A.J., Agostini M.L., Leist S.R., Schäfer A., Dinnon K.H., Stevens L.J., Chappell J.D., Lu X., Hughes T.M., George A.S., Hill C.S., Montgomery S.A., Brown A.J., Bluemling G.R., Natchus M.G., Saindane M., Kolykhalov A.A., Painter G., Harcourt J., Tamin A., Thornburg N.J., Swanstrom R., Denison M.R., Baric R.S. An orally Bioavailable Broad-Spectrum Antiviral Inhibits SARS-CoV-2 in Human Airway Epithelial Cell Cultures and Multiple Coronaviruses in Mice. Sci. Transl. Med. 2020:12. doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cox R.M., Wolf J.D., Plemper R.K. Therapeutically Administered Ribonucleoside Analogue MK-4482/EIDD-2801 Blocks SARS-CoV-2 Transmission in Ferrets. Nat. Microbiol. 2021;6:11–18. doi: 10.1038/s41564-020-00835-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wahl A., Gralinski L.E., Johnson C.E., Yao W., Kovarova M., Dinnon K.H., Liu H., Madden V.J., Krzystek H.M., De C., White K.K., Gully K., Schäfer A., Zaman T., Leist S.R., Grant P.O., Bluemling G.R., Kolykhalov A.A., Natchus M.G., Askin F.B., Painter G., Browne E.P., Jones C.D., Pickles R.J., Baric R.S., Garcia J.V. SARS-CoV-2 Infection is Effectively Treated and Prevented by EIDD-2801. Nature. 2021;591:451–457. doi: 10.1038/s41586-021-03312-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Menéndez-Arias L. Decoding Molnupiravir-Induced Mutagenesis in SARS-CoV-2. J. Biol. Chem. 2021;297:100867. doi: 10.1016/j.jbc.2021.100867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hassanipour S., Arab-Zozani M., Amani B., Heidarzad F., Fathalipour M., Martinez-de-Hoyo R. The efficacy and Safety of Favipiravir in Treatment of COVID-19: A Systematic Review and Meta-Analysis of Clinical Trials. Sci. Rep. 2021;11:11022. doi: 10.1038/s41598-021-90551-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naydenova K., Muir K.W., Wu L.-F., Zhang Z., Coscia F., Peet M.J., Castro-Hartmann P., Qian P., Sader K., Dent K., Kimanius D., Sutherland J.D., Löwe J., Barford D., Russo C.J. Structure of the SARS-CoV-2 RNA-Dependent RNA Polymerase in the Presence of Favipiravir-RTP. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2021946118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian L., Qiang T., Liang C., Ren X., Jia M., Zhang J., Li J., Wan M., YuWen X., Li H., Cao W., Liu H. RNA-Dependent RNA Polymerase (RdRp) Inhibitors: The Current Landscape and Repurposing for the COVID-19 Pandemic. Eur. J. Med. Chem. 2021;213:113201. doi: 10.1016/j.ejmech.2021.113201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng Z., Groaz E., Snoeck R., De Jonghe S., Herdewijn P., Andrei G. Influence of 4′-Substitution on the Activity of Gemcitabine and Its ProTide Against VZV and SARS-CoV-2. ACS Med. Chem. Lett. 2021;12:88–92. doi: 10.1021/acsmedchemlett.0c00485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang H., Yang J. A Review of the Latest Research on Mpro Targeting SARS-COV Inhibitors. RSC Med. Chem. 2021;12:1026–1036. doi: 10.1039/d1md00066g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boras B., Jones R.M., Anson B.J., Arenson D., Aschenbrenner L., Bakowski M.A., Beutler N., Binder J., Chen E., Eng H., Hammond H., Hammond J., Haupt R.E., Hoffman R., Kadar E.P., Kania R., Kimoto E., Kirkpatrick M.G., Lanyon L., Lendy E.K., Lillis J.R., Logue J., Luthra S.A., Ma C., Mason S.W., McGrath M.E., Noell S., Obach R.S., O’Brien M.N., O’Connor R., Ogilvie K., Owen D., Pettersson M., Reese M.R., Rogers T.F., Rossulek M.I., Sathish J.G., Shirai N., Steppan C., Ticehurst M., Updyke L.W., Weston S., Zhu Y., Wang J., Chatterjee A.K., Mesecar A.D., Frieman M.B., Anderson A.S., Allerton C. Discovery of a Novel Inhibitor of Coronavirus 3CL Protease for the Potential Treatment of COVID-19. bioRxiv. 2021 doi: 10.1038/s41467-021-26239-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Owen D.R., Allerton C.M.N., Anderson A.S., Aschenbrenner L., Avery M., Berritt S., Boras B., Cardin R.D., Carlo A., Coffman K.J., Dantonio A., Di L., Eng H., Ferre R., Gajiwala K.S., Gibson S.A., Greasley S.E., Hurst B.L., Kadar E.P., Kalgutkar A.S., Lee J.C., Lee J., Liu W., Mason S.W., Noell S., Novak J.J., Obach R.S., Ogilvie K., Patel N.C., Pettersson M., Rai D.K., Reese M.R., Sammons M.F., Sathish J.G., Singh R.S.P., Steppan C.M., Stewart A.E., Tuttle J.B., Updyke L., Verhoest P.R., Wei L., Yang Q., Zhu Y. An Oral SARS-CoV-2 Mpro Inhibitor Clinical Candidate for the Treatment of COVID-19. medRxiv. 2021 doi: 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- 28.Qiao J., Li Y.-S., Zeng R., Liu F.-L., Luo R.-H., Huang C., Wang Y.-F., Zhang J., Quan B., Shen C., Mao X., Liu X., Sun W., Yang W., Ni X., Wang K., Xu L., Duan Z.-L., Zou Q.-C., Zhang H.-L., Qu W., Long Y.-H.-P., Li M.-H., Yang R.-C., Liu X., You J., Zhou Y., Yao R., Li W.-P., Liu J.-M., Chen P., Liu Y., Lin G.-F., Yang X., Zou J., Li L., Hu Y., Lu G.-W., Li W.-M., Wei Y.-Q., Zheng Y.-T., Lei J., Yang S. SARS-CoV-2 Mpro Inhibitors With Antiviral Activity in a Transgenic Mouse Model. Science. 2021;371:1374–1378. doi: 10.1126/science.abf1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bobileva O., Bobrovs R., Kanepe I., Patetko L., Kalnins G., Sisovs M., Bula A.L., Grinberga S., Boroduskis M., Ramata-Stunda A., Rostoks N., Jirgensons A., Tars K., Jaudzems K. Potent SARS-CoV-2 mRNA Cap Methyltransferase Inhibitors by Bioisosteric Replacement of Methionine in SAM Cosubstrate. ACS Med. Chem. Lett. 2021;12:1102–1107. doi: 10.1021/acsmedchemlett.1c00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otava T., Sala M., Li F.L., Fanfrlik J., Devkota K., Perveen S., Chau I., Pakarian P., Hobza P., Vedadi M., Boura E., Nencka R. The Structure-Based Design of SARS-CoV-2 nsp14 Methyltransferase Ligands Yields Nanomolar Inhibitors. ACS Infect. Dis. 2021;7:2214–2220. doi: 10.1021/acsinfecdis.1c00131. [DOI] [PubMed] [Google Scholar]
- 31.Krammer F., Smith G.J.D., Fouchier R.A.M., Peiris M., Kedzierska K., Doherty P.C., Palese P., Shaw M.L., Treanor J., Webster R.G., García-Sastre A. Influenza. Nat. Rev. Dis. Primers. 2018;4:3. doi: 10.1038/s41572-018-0002-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim H., Webster R.G., Webby R.J. Influenza Virus: Dealing with a Drifting and Shifting Pathogen. Viral Immunol. 2018;31:174–183. doi: 10.1089/vim.2017.0141. [DOI] [PubMed] [Google Scholar]
- 33.Harrington W.N., Kackos C.M., Webby R.J. The Evolution and Future of Influenza Pandemic Preparedness. Exp. Mol. Med. 2021;53:737–749. doi: 10.1038/s12276-021-00603-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mtambo S.E., Amoako D.G., Somboro A.M., Agoni C., Lawal M.M., Gumede N.S., Khan R.B., Kumalo H.M. Influenza Viruses: Harnessing the Crucial Role of the M2 Ion-Channel and Neuraminidase Toward Inhibitor Design. Molecules. 2021;26:880. doi: 10.3390/molecules26040880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He W., Zhang W., Yan H., Xu H., Xie Y., Wu Q., Wang C., Dong G. Distribution and Evolution of H1N1 Influenza A Viruses With Adamantanes-Resistant Mutations Worldwide From 1918 to 2019. J. Med. Virol. 2021;93:3473–3483. doi: 10.1002/jmv.26670. [DOI] [PubMed] [Google Scholar]
- 36.Bloom J.D., Gong L.I., Baltimore D. Permissive Secondary Mutations Enable the Evolution of Influenza Oseltamivir Resistance. Science. 2010;328:1272–1275. doi: 10.1126/science.1187816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu Y., Hau R.K., Wang Y., Tuohy P., Zhang Y., Xu S., Ma C., Wang J. Structure–Property Relationship Studies of Influenza A Virus AM2-S31N Proton Channel Blockers. ACS Med. Chem. Lett. 2018;9:1111–1116. doi: 10.1021/acsmedchemlett.8b00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y., Hu Y., Xu S., Zhang Y., Musharrafieh R., Hau R.K., Ma C., Wang J. In Vitro Pharmacokinetic Optimizations of AM2-S31N Channel Blockers Led to the Discovery of Slow-Binding Inhibitors With Potent Antiviral Activity Against Drug-Resistant Influenza A Viruses. J. Med. Chem. 2018;61:1074–1085. doi: 10.1021/acs.jmedchem.7b01536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J., Murugan N.A., Tian Y., Bertagnin C., Fang Z., Kang D., Kong X., Jia H., Sun Z., Jia R., Gao P., Poongavanam V., Loregian A., Xu W., Ma X., Ding X., Huang B., Zhan P., Liu X. Structure-Based Optimization of N-Substituted Oseltamivir Derivatives as Potent Anti-Influenza A Virus Agents with Significantly Improved Potency Against Oseltamivir-Resistant N1-H274Y Variant. J. Med. Chem. 2018;61:9976–9999. doi: 10.1021/acs.jmedchem.8b01065. [DOI] [PubMed] [Google Scholar]
- 40.Zhang J., Poongavanam V., Kang D., Bertagnin C., Lu H., Kong X., Ju H., Lu X., Gao P., Tian Y., Jia H., Desta S., Ding X., Sun L., Fang Z., Huang B., Liang X., Jia R., Ma X., Xu W., Murugan N.A., Loregian A., Huang B., Zhan P., Liu X. Optimization of N-Substituted Oseltamivir Derivatives as Potent Inhibitors of Group-1 and -2 Influenza A Neuraminidases, Including a Drug-Resistant Variant. J. Med. Chem. 2018;61:6379–6397. doi: 10.1021/acs.jmedchem.8b00929. [DOI] [PubMed] [Google Scholar]
- 41.Yuan S., Wen L., Zhou J. Inhibitors of Influenza A Virus Polymerase. ACS Infect. Dis. 2018;4:218–223. doi: 10.1021/acsinfecdis.7b00265. [DOI] [PubMed] [Google Scholar]
- 42.te Velthuis A.J.W., Fodor E. Influenza Virus RNA Polymerase: Insights Into the Mechanisms of Viral RNA Synthesis. Nat. Rev. Microbiol. 2016;14:479–493. doi: 10.1038/nrmicro.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hayden F.G., Sugaya N., Hirotsu N., Lee N., de Jong M.D., Hurt A.C., Ishida T., Sekino H., Yamada K., Portsmouth S., Kawaguchi K., Shishido T., Arai M., Tsuchiya K., Uehara T., Watanabe A. Baloxavir Marboxil for Uncomplicated Influenza in Adults and Adolescents. N. Engl. J. Med. 2018;379:913–923. doi: 10.1056/NEJMoa1716197. [DOI] [PubMed] [Google Scholar]
- 44.Ikematsu H., Hayden F.G., Kawaguchi K., Kinoshita M., de Jong M.D., Lee N., Takashima S., Noshi T., Tsuchiya K., Uehara T. Baloxavir Marboxil for Prophylaxis against Influenza in Household Contacts. N. Engl. J. Med. 2020;383:309–320. doi: 10.1056/NEJMoa1915341. [DOI] [PubMed] [Google Scholar]
- 45.Yang T. Baloxavir Marboxil: The First Cap-Dependent Endonuclease Inhibitor for the Treatment of Influenza. Ann. Pharmacother. 2019;53:754–759. doi: 10.1177/1060028019826565. [DOI] [PubMed] [Google Scholar]
- 46.Hashimoto T., Baba K., Inoue K., Okane M., Hata S., Shishido T., Naito A., Wildum S., Omoto S. Comprehensive Assessment of Amino Acid Substitutions in the Trimeric RNA Polymerase Complex of Influenza A Virus Detected in Clinical Trials of Baloxavir Marboxil. Influenza Other Respir. Viruses. 2021;15:389–395. doi: 10.1111/irv.12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ivashchenko A.A., Mitkin O.D., Jones J.C., Nikitin A.V., Koryakova A.G., Ryakhovskiy A., Karapetian R.N., Kravchenko D.V., Aladinskiy V., Leneva I.A., Falynskova I.N., Glubokova E.A., Govorkova E.A., Ivachtchenko A.V. Non-Rigid Diarylmethyl Analogs of Baloxavir as Cap-Dependent Endonuclease Inhibitors of Influenza Viruses. J. Med. Chem. 2020;63:9403–9420. doi: 10.1021/acs.jmedchem.0c00565. [DOI] [PubMed] [Google Scholar]
- 48.Credille C.V., Morrison C.N., Stokes R.W., Dick B.L., Feng Y., Sun J., Chen Y., Cohen S.M. SAR Exploration of Tight-Binding Inhibitors of Influenza Virus PA Endonuclease. J. Med. Chem. 2019;62:9438–9449. doi: 10.1021/acs.jmedchem.9b00747. [DOI] [PubMed] [Google Scholar]
- 49.Miyagawa M., Akiyama T., Taoda Y., Takaya K., Takahashi-Kageyama C., Tomita K., Yasuo K., Hattori K., Shano S., Yoshida R., Shishido T., Yoshinaga T., Sato A., Kawai M. Synthesis and SAR Study of Carbamoyl Pyridone Bicycle Derivatives as Potent Inhibitors of Influenza Cap-dependent Endonuclease. J. Med. Chem. 2019;62:8101–8114. doi: 10.1021/acs.jmedchem.9b00861. [DOI] [PubMed] [Google Scholar]
- 50.Clark M.P., Ledeboer M.W., Davies I., Byrn R.A., Jones S.M., Perola E., Tsai A., Jacobs M., Nti-Addae K., Bandarage U.K., Boyd M.J., Bethiel R.S., Court J.J., Deng H.B., Duffy J.P., Dorsch W.A., Farmer L.J., Gao H., Gu W.X., Jackson K., Jacobs D.H., Kennedy J.M., Ledford B., Liang J.L., Maltais F., Murcko M., Wang T.S., Wannamaker M.W., Bennett H.B., Leeman J.R., McNeil C., Taylor W.P., Memmott C., Jiang M., Rijnbrand R., Bral C., Germann U., Nezami A., Zhang Y.G., Salituro F.G., Bennani Y.L., Charifson P.S. Discovery of a Novel, First-in-Class, Orally Bioavailable Azaindole Inhibitor (VX-787) of Influenza PB2. J. Med. Chem. 2014;57:6668–6678. doi: 10.1021/jm5007275. [DOI] [PubMed] [Google Scholar]
- 51.Finberg R.W., Lanno R., Anderson D., Fleischhackl R., van Duijnhoven W., Kauffman R.S., Kosoglou T., Vingerhoets J., Leopold L. Phase 2b Study of Pimodivir (JNJ-63623872) as Monotherapy or in Combination With Oseltamivir for Treatment of Acute Uncomplicated Seasonal Influenza A: TOPAZ Trial. J. Infect. Dis. 2019;219:1026–1034. doi: 10.1093/infdis/jiy547. [DOI] [PubMed] [Google Scholar]
- 52.McGowan D.C., Balemans W., Embrechts W., Motte M., Keown J.R., Buyck C., Corbera J., Funes M., Moreno L., Cooymans L., Tahri A., Eymard J., Stoops B., Strijbos R., Van den Berg J., Fodor E., Grimes J.M., Koul A., Jonckers T.H.M., Raboisson P., Guillemont J. Design, Synthesis, and Biological Evaluation of Novel Indoles Targeting the Influenza PB2 Cap Binding Region. J. Med. Chem. 2019;62:9680–9690. doi: 10.1021/acs.jmedchem.9b01091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Łagocka R., Dziedziejko V., Kłos P., Pawlik A. Favipiravir in Therapy of Viral Infections. J. Clin. Med. 2021;10 doi: 10.3390/jcm10020273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Massari S., Desantis J., Nizi M.G., Cecchetti V., Tabarrini O. Inhibition of Influenza Virus Polymerase by Interfering With Its Protein–Protein Interactions. ACS Infect. Dis. 2021;7:1332–1350. doi: 10.1021/acsinfecdis.0c00552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yuan S., Chu H., Zhao H., Zhang K., Singh K., Chow B.K.C., Kao R.Y.T., Zhou J., Zheng B.-J. Identification of a Small-Molecule Inhibitor of Influenza Virus via Disrupting the Subunits Interaction of the Viral Polymerase. Antiviral Res. 2016;125:34–42. doi: 10.1016/j.antiviral.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 56.Kao R.Y., Yang D., Lau L.S., Tsui W.H., Hu L., Dai J., Chan M.P., Chan C.M., Wang P., Zheng B.J., Sun J., Huang J.D., Madar J., Chen G., Chen H., Guan Y., Yuen K.Y. Identification of Influenza A Nucleoprotein as an Antiviral Target. Nat. Biotechnol. 2010;28:600–605. doi: 10.1038/nbt.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Woodring J.L., Lu S.-H., Krasnova L., Wang S.-C., Chen J.-B., Chou C.-C., Huang Y.-C., Cheng T.-J.R., Wu Y.-T., Chen Y.-H., Fang J.-M., Tsai M.-D., Wong C.-H. Disrupting the Conserved Salt Bridge in the Trimerization of Influenza A Nucleoprotein. J. Med. Chem. 2020;63:205–215. doi: 10.1021/acs.jmedchem.9b01244. [DOI] [PubMed] [Google Scholar]
- 58.Sethy B., Hsieh C.-F., Lin T.-J., Hu P.-Y., Chen Y.-L., Lin C.-Y., Tseng S.-N., Horng J.-T., Hsieh P.-W. Design, Synthesis, and Biological Evaluation of Itaconic Acid Derivatives as Potential Anti-Influenza Agents. J. Med. Chem. 2019;62:2390–2403. doi: 10.1021/acs.jmedchem.8b01683. [DOI] [PubMed] [Google Scholar]
- 59.White K.M., Abreu P., Jr., Wang H., De Jesus P.D., Manicassamy B., García-Sastre A., Chanda S.K., DeVita R.J., Shaw M.L. Broad Spectrum Inhibitor of Influenza A and B Viruses Targeting the Viral Nucleoprotein. ACS Infect. Dis. 2018;4:146–157. doi: 10.1021/acsinfecdis.7b00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.White K., Esparza M., Liang J., Bhat P., Naidoo J., McGovern B.L., Williams M.A.P., Alabi B.R., Shay J., Niederstrasser H., Posner B., García-Sastre A., Ready J., Fontoura B.M.A. Aryl Sulfonamide Inhibits Entry and Replication of Diverse Influenza Viruses via the Hemagglutinin Protein. J. Med. Chem. 2021;64:10951–10966. doi: 10.1021/acs.jmedchem.1c00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu G., Yu G., Yu Y., Yang S., Duan Z., Wang W., Liu Y., Yu R., Li J., Zhu T., Gu Q., Li D. Chemoreactive-Inspired Discovery of Influenza A Virus Dual Inhibitor to Block Hemagglutinin-Mediated Adsorption and Membrane Fusion. J. Med. Chem. 2020;63:6924–6940. doi: 10.1021/acs.jmedchem.0c00312. [DOI] [PubMed] [Google Scholar]
- 62.Gaisina I.N., Peet N.P., Cheng H., Li P., Du R., Cui Q., Furlong K., Manicassamy B., Caffrey M., Thatcher G.R.J., Rong L. Optimization of 4-Aminopiperidines as Inhibitors of Influenza A Viral Entry That Are Synergistic With Oseltamivir. J. Med. Chem. 2020;63:3120–3130. doi: 10.1021/acs.jmedchem.9b01900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rossignol J.F., La Frazia S., Chiappa L., Ciucci A., Santoro M.G. Thiazolides, a New Class of Anti-Influenza Molecules Targeting Viral Hemagglutinin at the Post-Translational Level. J. Biol. Chem. 2009;284:29798–29808. doi: 10.1074/jbc.M109.029470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haffizulla J., Hartman A., Hoppers M., Resnick H., Samudrala S., Ginocchio C., Bardin M., Rossignol J.F. Effect of Nitazoxanide in Adults and Adolescents With Acute Uncomplicated Influenza: A Double-Blind, Randomised, Placebo-Controlled, Phase 2b/3 Trial. Lancet Infect. Dis. 2014;14:609–618. doi: 10.1016/S1473-3099(14)70717-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Malakhov M.P., Aschenbrenner L.M., Smee D.F., Wandersee M.K., Sidwell R.W., Gubareva L.V., Mishin V.P., Hayden F.G., Kim D.H., Ing A., Campbell E.R., Yu M., Fang F. Sialidase Fusion Protein as a Novel Broad-Spectrum Inhibitor of Influenza Virus Infection. Antimicrob. Agents Chemother. 2006;50:1470–1479. doi: 10.1128/AAC.50.4.1470-1479.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moss R.B., Hansen C., Sanders R.L., Hawley S., Li T., Steigbigel R.T. A Phase II Study of DAS181, a Novel Host Directed Antiviral for the Treatment of Influenza Infection. J. Infect. Dis. 2012;206:1844–1851. doi: 10.1093/infdis/jis622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.O'Hanlon R., Leyva-Grado V.H., Sourisseau M., Evans M.J., Shaw M.L. An Influenza Virus Entry Inhibitor Targets Class II PI3 Kinase and Synergizes With Oseltamivir. ACS Infect. Dis. 2019;5:1779–1793. doi: 10.1021/acsinfecdis.9b00230. [DOI] [PubMed] [Google Scholar]
- 68.Collins P.L., Fearns R., Graham B.S. Respiratory Syncytial Virus: Virology, Reverse Genetics, and Pathogenesis of Disease. Curr. Top. Microbiol. Immunol. 2013;372:3–38. doi: 10.1007/978-3-642-38919-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Falsey A.R., Hennessey P.A., Formica M.A., Cox C., Walsh E.E. Respiratory Syncytial Virus Infection in Elderly and High-Risk Adults. N. Engl. J. Med. 2005;352:1749–1759. doi: 10.1056/NEJMoa043951. [DOI] [PubMed] [Google Scholar]
- 70.Nair H., Nokes D.J., Gessner B.D., Dherani M., Madhi S.A., Singleton R.J., O'Brien K.L., Roca A., Wright P.F., Bruce N., Chandran A., Theodoratou E., Sutanto A., Sedyaningsih E.R., Ngama M., Munywoki P.K., Kartasasmita C., Simões E.A., Rudan I., Weber M.W., Campbell H. Global Burden of Acute Lower Respiratory Infections Due to Respiratory Syncytial Virus in Young Children: A Systematic Review and Meta-Analysis. Lancet. 2010;375:1545–1555. doi: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Domachowske J.B., Anderson E.J., Goldstein M. The Future of Respiratory Syncytial Virus Disease Prevention and Treatment. Infect. Dis. Ther. 2021;10:47–60. doi: 10.1007/s40121-020-00383-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cockerill G.S., Good J.A.D., Mathews N. State of the Art in Respiratory Syncytial Virus Drug Discovery and Development. J. Med. Chem. 2019;62:3206–3227. doi: 10.1021/acs.jmedchem.8b01361. [DOI] [PubMed] [Google Scholar]
- 73.Vendeville S., Tahri A., Hu L.L., Demin S., Cooymans L., Vos A., Kwanten L., Van den Berg J., Battles M.B., McLellan J.S., Koul A., Raboisson P., Roymans D., Jonckers T.H.M. Discovery of 3-({5-Chloro-1- 3-(methylsulfonyl)propyl -1H-indol-2-yl}methyl)-1-(2,2,2 -trifluoroethyl)-1,3-dihydro-2H-imidazo 4,5-c pyridin-2-one (JNJ-53718678), a Potent and Orally Bioavailable Fusion Inhibitor of Respiratory Syncytial Virus. J. Med. Chem. 2020;63:8046–8058. doi: 10.1021/acs.jmedchem.0c00226. [DOI] [PubMed] [Google Scholar]
- 74.Cockerill G.S., Angell R.M., Bedernjak A., Chuckowree I., Fraser I., Gascon-Simorte J., Gilman M.S.A., Good J.A.D., Harland R., Johnson S.M., Ludes-Meyers J.H., Littler E., Lumley J., Lunn G., Mathews N., McLellan J.S., Paradowski M., Peeples M.E., Scott C., Tait D., Taylor G., Thom M., Thomas E., Barber C.V., Ward S.E., Watterson D., Williams G., Young P., Powell K. Discovery of Sisunatovir (RV521), an Inhibitor of Respiratory Syncytial Virus Fusion. J. Med. Chem. 2021;64:3658–3676. doi: 10.1021/acs.jmedchem.0c01882. [DOI] [PubMed] [Google Scholar]
- 75.Zheng X.F., Gao L., Wang L.S., Liang C.G., Wang B.X., Liu Y.F., Feng S., Zhang B., Zhou M.W., Yu X., Xiang K.L., Chen L., Guo T., Shen H.C., Zou G., Wu J.Z., Yun H.Y. Discovery of Ziresovir as a Potent, Selective, and Orally Bioavailable Respiratory Syncytial Virus Fusion Protein Inhibitor. J. Med. Chem. 2019;62:6003–6014. doi: 10.1021/acs.jmedchem.9b00654. [DOI] [PubMed] [Google Scholar]
- 76.Mackman R.L., Sangi M., Sperandio D., Parrish J.P., Eisenberg E., Perron M., Hui H., Zhang L., Siegel D., Yang H., Saunders O., Boojamra C., Lee G., Samuel D., Babaoglu K., Carey A., Gilbert B.E., Piedra P.A., Strickley R., Iwata Q., Hayes J., Stray K., Kinkade A., Theodore D., Jordan R., Desai M., Cihlar T. Discovery of an Oral Respiratory Syncytial Virus (RSV) Fusion Inhibitor (GS-5806) and Clinical Proof of Concept in a Human RSV Challenge Study. J. Med. Chem. 2015;58:1630–1643. doi: 10.1021/jm5017768. [DOI] [PubMed] [Google Scholar]
- 77.Chemaly R.F., Dadwal S.S., Bergeron A., Ljungman P., Kim Y.J., Cheng G.S., Pipavath S.N., Limaye A.P., Blanchard E., Winston D.J., Stiff P.J., Zuckerman T., Lachance S., Rahav G., Small C.B., Mullane K.M., Patron R.L., Lee D.G., Hirsch H.H., Waghmare A., McKevitt M., Jordan R., Guo Y., German P., Porter D.P., Gossage D.L., Watkins T.R., Marty F.M., Chien J.W., Boeckh M. A Phase 2, Randomized, Double-blind, Placebo-Controlled Trial of Presatovir for the Treatment of Respiratory Syncytial Virus Upper Respiratory Tract Infection in Hematopoietic-Cell Transplant Recipients. Clin. Infect Dis. 2020;71:2777–2786. doi: 10.1093/cid/ciz1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marty F.M., Chemaly R.F., Mullane K.M., Lee D.G., Hirsch H.H., Small C.B., Bergeron A., Shoham S., Ljungman P., Waghmare A., Blanchard E., Kim Y.J., McKevitt M., Porter D.P., Jordan R., Guo Y., German P., Boeckh M., Watkins T.R., Chien J.W., Dadwal S.S. A Phase 2b, Randomized, Double-Blind, Placebo-Controlled Multicenter Study Evaluating Antiviral Effects, Pharmacokinetics, Safety, and Tolerability of Presatovir in Hematopoietic Cell Transplant Recipients With Respiratory Syncytial Virus Infection of the Lower Respiratory Tract. Clin. Infect Dis. 2020;71:2787–2795. doi: 10.1093/cid/ciz1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Porter D.P., Guo Y., Perry J., Gossage D.L., Watkins T.R., Chien J.W., Jordan R. Assessment of Drug Resistance during Phase 2b Clinical Trials of Presatovir in Adults Naturally Infected With Respiratory Syncytial Virus. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.02312-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pribut N., Kaiser T.M., Wilson R.J., Jecs E., Dentmon Z.W., Pelly S.C., Sharma S., Bartsch P.W., Burger P.B., Hwang S.S., Le T., Sourimant J., Yoon J.-J., Plemper R.K., Liotta D.C. Accelerated Discovery of Potent Fusion Inhibitors for Respiratory Syncytial Virus. ACS Infect. Dis. 2020;6:922–929. doi: 10.1021/acsinfecdis.9b00524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yamaguchi-Sasaki T., Tokura S., Ogata Y., Kawaguchi T., Sugaya Y., Takahashi R., Iwakiri K., Abe-Kumasaka T., Yoshida I., Arikawa K., Sugiyama H., Kanuma K. Discovery of a Potent Dual Inhibitor of Wild-Type and Mutant Respiratory Syncytial Virus Fusion Proteins. ACS Med. Chem. Lett. 2020;11:1145–1151. doi: 10.1021/acsmedchemlett.0c00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rhodin M.H.J., McAllister N.V., Castillo J., Noton S.L., Fearns R., Kim I.J., Yu J., Blaisdell T.P., Panarese J., Shook B.C., Or Y.S., Goodwin B., Lin K. EDP-938, a Novel Nucleoprotein Inhibitor of Respiratory Syncytial Virus, Demonstrates Potent Antiviral Activities In Vitro and in a Non-Human Primate Model. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jordan P.C., Stevens S.K., Deval J. Nucleosides for the Treatment of Respiratory RNA Virus Infections. Antivir. Chem. Chemother. 2018;26 doi: 10.1177/2040206618764483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cao D., Gao Y., Roesler C., Rice S., D’Cunha P., Zhuang L., Slack J., Domke M., Antonova A., Romanelli S., Keating S., Forero G., Juneja P., Liang B. Cryo-EM Structure of the Respiratory Syncytial Virus RNA Polymerase. Nat. Commun. 2020;11:368. doi: 10.1038/s41467-019-14246-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gilman M.S.A., Liu C., Fung A., Behera I., Jordan P., Rigaux P., Ysebaert N., Tcherniuk S., Sourimant J., Eléouët J.F., Sutto-Ortiz P., Decroly E., Roymans D., Jin Z., McLellan J.S. Structure of the Respiratory Syncytial Virus Polymerase Complex. Cell. 2019;179:193–204.e114. doi: 10.1016/j.cell.2019.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang G.Y., Deval J., Hong J., Dyatkina N., Prhavc M., Taylor J., Fung A., Jin Z.N., Stevens S.K., Serebryany V., Liu J.W., Zhang Q.L., Tam Y.E., Chanda S.M., Smith D.B., Symons J.A., Blatt L.M., Beigelman L. Discovery of 4'-Chloromethyl-2'-deoxy-3',5'-di-O-isobutyryl-2'-fluorocytidine (ALS-8176), A First-in-Class RSV Polymerase Inhibitor for Treatment of Human Respiratory Syncytial Virus Infection. J. Med. Chem. 2015;58:1862–1878. doi: 10.1021/jm5017279. [DOI] [PubMed] [Google Scholar]
- 87.Sommadossi J.-P., Moussa A. 2018. β-D-2′-Deoxy-2′-Substituted-4′-Substituted-2-Substituted-N6-Substituted-6-Aminopurine Nucleotides for the Treatment of Paramyxovirus and Orthomyxovirus Infections. WO/2018/009623, January 11, 2018. [Google Scholar]
- 88.Mackman R.L., Hui H.C., Perron M., Murakami E., Palmiotti C., Lee G., Stray K., Zhang L., Goyal B., Chun K., Byun D., Siegel D., Simonovich S., Du Pont V., Pitts J., Babusis D., Vijjapurapu A., Lu X., Kim C., Zhao X., Chan J., Ma B., Lye D., Vandersteen A., Wortman S., Barrett K.T., Toteva M., Jordan R., Subramanian R., Bilello J.P., Cihlar T. Prodrugs of a 1'-CN-4-Aza-7,9-dideazaadenosine C-Nucleoside Leading to the Discovery of Remdesivir (GS-5734) as a Potent Inhibitor of Respiratory Syncytial Virus With Efficacy in the African Green Monkey Model of RSV. J. Med. Chem. 2021;64:5001–5017. doi: 10.1021/acs.jmedchem.1c00071. [DOI] [PubMed] [Google Scholar]
- 89.Jacobs S.E., Lamson D.M., St George K., Walsh T.J. Human Rhinoviruses. Clin. Microbiol. Rev. 2013;26:135–162. doi: 10.1128/CMR.00077-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Heikkinen T., Chonmaitree T. Importance of Respiratory Viruses in Acute Otitis Media. Clin. Microbiol. Rev. 2003;16:230–241. doi: 10.1128/CMR.16.2.230-241.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gern J.E. How rhinovirus Infections Cause Exacerbations of Asthma. Clin. Exp. Allergy. 2015;45:32–42. doi: 10.1111/cea.12428. [DOI] [PubMed] [Google Scholar]
- 92.Coultas J.A., Cafferkey J., Mallia P., Johnston S.L. Experimental Antiviral Therapeutic Studies for Human Rhinovirus Infections. J. Exp. Pharmacol. 2021;13:645–659. doi: 10.2147/JEP.S255211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bauer L., Lyoo H., van der Schaar H.M., Strating J.R.P.M., van Kuppeveld F.J.M. Direct-Acting Antivirals and Host-Targeting Strategies to Combat Enterovirus Infections. Curr. Opin. Virol. 2017;24:1–8. doi: 10.1016/j.coviro.2017.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kim J., Shin J.S., Ahn S., Han S.B., Jung Y.-S. 3-Aryl-1,2,4-Oxadiazole Derivatives Active Against Human Rhinovirus. ACS Med. Chem. Lett. 2018;9:667–672. doi: 10.1021/acsmedchemlett.8b00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Da Costa L., Scheers E., Coluccia A., Casulli A., Roche M., Di Giorgio C., Neyts J., Terme T., Cirilli R., La Regina G., Silvestri R., Mirabelli C., Vanelle P. Structure-Based Drug Design of Potent Pyrazole Derivatives Against Rhinovirus Replication. J. Med. Chem. 2018;61:8402–8416. doi: 10.1021/acs.jmedchem.8b00931. [DOI] [PubMed] [Google Scholar]
- 96.Ledford R.M., Patel N.R., Demenczuk T.M., Watanyar A., Herbertz T., Collett M.S., Pevear D.C. VP1 Sequencing of All Human Rhinovirus Serotypes: Insights Into Genus Phylogeny and Susceptibility to Antiviral Capsid-Binding Compounds. J. Virol. 2004;78:3663–3674. doi: 10.1128/JVI.78.7.3663-3674.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lacroix C., Laconi S., Angius F., Coluccia A., Silvestri R., Pompei R., Neyts J., Leyssen P. In Vitro Characterisation of a Pleconaril/Pirodavir-Like Compound With Potent Activity Against Rhinoviruses. Virol. J. 2015;12:106. doi: 10.1186/s12985-015-0330-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Egorova A., Kazakova E., Jahn B., Ekins S., Makarov V., Schmidtke M. Novel pleconaril Derivatives: Influence of Substituents in the Isoxazole and Phenyl Rings on the Antiviral Activity Against Enteroviruses. Eur. J. Med. Chem. 2020;188:112007. doi: 10.1016/j.ejmech.2019.112007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wald J., Pasin M., Richter M., Walther C., Mathai N., Kirchmair J., Makarov V.A., Goessweiner-Mohr N., Marlovits T.C., Zanella I., Real-Hohn A., Verdaguer N., Blaas D., Schmidtke M. Cryo-EM Structure of Pleconaril-Resistant Rhinovirus-B5 Complexed to the Antiviral OBR-5-340 Reveals Unexpected Binding Site. Proc. Natl. Acad. Sci. U.S.A. 2019;116:19109–19115. doi: 10.1073/pnas.1904732116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Makarov V.A., Braun H., Richter M., Riabova O.B., Kirchmair J., Kazakova E.S., Seidel N., Wutzler P., Schmidtke M. Pyrazolopyrimidines: Potent Inhibitors Targeting the Capsid of Rhino- and Enteroviruses. ChemMedChem. 2015;10:1629–1634. doi: 10.1002/cmdc.201500304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mello C., Aguayo E., Rodriguez M., Lee G., Jordan R., Cihlar T., Birkus G. Multiple Classes of Antiviral Agents Exhibit In Vitro Activity Against Human Rhinovirus Type C. Antimicrob. Agents Chemother. 2014;58:1546–1555. doi: 10.1128/AAC.01746-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Blomqvist S., Savolainen C., Råman L., Roivainen M., Hovi T. Human Rhinovirus 87 and Enterovirus 68 Represent a Unique Serotype With Rhinovirus and Enterovirus Features. J. Clin. Microbiol. 2002;40:4218–4223. doi: 10.1128/JCM.40.11.4218-4223.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Holm-Hansen C.C., Midgley S.E., Fischer T.K. Global Emergence of Enterovirus D68: A Systematic Review. Lancet Infect. Dis. 2016;16:E64–E75. doi: 10.1016/S1473-3099(15)00543-5. [DOI] [PubMed] [Google Scholar]
- 104.Hu Y., Musharrafieh R., Zheng M., Wang J. Enterovirus D68 Antivirals: Past, Present, and Future. ACS Infect. Dis. 2020;6:1572–1586. doi: 10.1021/acsinfecdis.0c00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bauer L., Manganaro R., Zonsics B., Hurdiss D.L., Zwaagstra M., Donselaar T., Welter N.G.E., van Kleef R., Lopez M.L., Bevilacqua F., Raman T., Ferla S., Bassetto M., Neyts J., Strating J., Westerink R.H.S., Brancale A., van Kuppeveld F.J.M. Rational Design of Highly Potent Broad-Spectrum Enterovirus Inhibitors Targeting the Nonstructural Protein 2C. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Musharrafieh R., Zhang J., Tuohy P., Kitamura N., Bellampalli S.S., Hu Y., Khanna R., Wang J. Discovery of Quinoline Analogues as Potent Antivirals Against Enterovirus D68 (EV-D68) J. Med. Chem. 2019;62:4074–4090. doi: 10.1021/acs.jmedchem.9b00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hu Y., Kitamura N., Musharrafieh R., Wang J. Discovery of Potent and Broad-Spectrum Pyrazolopyridine-Containing Antivirals Against Enteroviruses D68, A71, and Coxsackievirus B3 by Targeting the Viral 2C Protein. J. Med. Chem. 2021;64:8755–8774. doi: 10.1021/acs.jmedchem.1c00758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jacob S.T., Crozier I., Fischer W.A., Hewlett A., Kraft C.S., Vega M.-A.D.L., Soka M.J., Wahl V., Griffiths A., Bollinger L., Kuhn J.H. Ebola Virus Disease. Nat. Rev. Dis. Primers. 2020;6:13. doi: 10.1038/s41572-020-0147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bray M. Defense Against Filoviruses Used as Biological Weapons. Antiviral Res. 2003;57:53–60. doi: 10.1016/s0166-3542(02)00200-0. [DOI] [PubMed] [Google Scholar]
- 110.Satyanarayana M. Vaccines Could Make Big Ebola Outbreaks a Thing of the Past. C&EN Global Enterprise. 2021;99:16–19. [Google Scholar]
- 111.Bausch D.G. The Need for a New Strategy for Ebola Vaccination. Nat. Med. 2021;27:580–581. doi: 10.1038/s41591-021-01313-w. [DOI] [PubMed] [Google Scholar]
- 112.Satyanarayana M. First Ebola Treatment Approved by FDA. C&EN Global Enterprise. 2020;98:13. [Google Scholar]
- 113.https://www.fda.gov/drugs/drug-safety-and-availability/fda-approves-treatment-ebola-virus
- 114.Hansen F., Feldmann H., Jarvis M.A. Targeting Ebola Virus Replication Through Pharmaceutical Intervention. Expert Opin. Investig. Drugs. 2021;30:201–226. doi: 10.1080/13543784.2021.1881061. [DOI] [PubMed] [Google Scholar]
- 115.Hoenen T., Groseth A., Feldmann H. Therapeutic Strategies to Target the Ebola Virus Life Cycle. Nat. Rev. Microbiol. 2019;17:593–606. doi: 10.1038/s41579-019-0233-2. [DOI] [PubMed] [Google Scholar]
- 116.Edwards M.R., Basler C.F. Current Status of Small Molecule Drug Development for Ebola Virus and Other Filoviruses. Curr. Opin. Virol. 2019;35:42–56. doi: 10.1016/j.coviro.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Plewe M.B., Sokolova N.V., Gantla V.R., Brown E.R., Naik S., Fetsko A., Lorimer D.D., Dranow D.M., Smutney H., Bullen J., Sidhu R., Master A., Wang J.R., Kallel E.A., Zhang L.H., Kalveram B., Freiberg A.N., Henkel G., McCormack K. Discovery of Adamantane Carboxamides as Ebola Virus Cell Entry and Glycoprotein Inhibitors. ACS Med. Chem. Lett. 2020;11:1160–1167. doi: 10.1021/acsmedchemlett.0c00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gaisina I.N., Peet N.P., Wong L., Schafer A.M., Cheng H., Anantpadma M., Davey R.A., Thatcher G.R.J., Rong L. Discovery and Structural Optimization of 4-(Aminomethyl)benzamides as Potent Entry Inhibitors of Ebola and Marburg Virus Infections. J. Med. Chem. 2020;63:7211–7225. doi: 10.1021/acs.jmedchem.0c00463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cote M., Misasi J., Ren T., Bruchez A., Lee K., Filone C.M., Hensley L., Li Q., Ory D., Chandran K., Cunningham J. Small Molecule Inhibitors Reveal Niemann-Pick C1 is Essential for Ebola Virus Infection. Nature. 2011;477:344–U122. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Liu H., Tian Y., Lee K., Krishnan P., Wang M.K., Whelan S., Mevers E., Soloveva V., Dedic B., Liu X., Cunningham J.M. Identification of Potent Ebola Virus Entry Inhibitors With Suitable Properties for in Vivo Studies. J. Med. Chem. 2018;61:6293–6307. doi: 10.1021/acs.jmedchem.8b00704. [DOI] [PubMed] [Google Scholar]
- 121.van der Linden W.A., Schulze C.J., Herbert A.S., Krause T.B., Wirchnianski A.A., Dye J.M., Chandran K., Bogyo M. Cysteine Cathepsin Inhibitors as Anti-Ebola Agents. ACS Infect. Dis. 2016;2:173–179. doi: 10.1021/acsinfecdis.5b00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Warren T.K., Jordan R., Lo M.K., Ray A.S., Mackman R.L., Soloveva V., Siegel D., Perron M., Bannister R., Hui H.C., Larson N., Strickley R., Wells J., Stuthman K.S., Van Tongeren S.A., Garza N.L., Donnelly G., Shurtleff A.C., Retterer C.J., Gharaibeh D., Zamani R., Kenny T., Eaton B.P., Grimes E., Welch L.S., Gomba L., Wilhelmsen C.L., Nichols D.K., Nuss J.E., Nagle E.R., Kugelman J.R., Palacios G., Doerffler E., Neville S., Carra E., Clarke M.O., Zhang L., Lew W., Ross B., Wang Q., Chun K., Wolfe L., Babusis D., Park Y., Stray K.M., Trancheva I., Feng J.Y., Barauskas O., Xu Y., Wong P., Braun M.R., Flint M., McMullan L.K., Chen S.-S., Fearns R., Swaminathan S., Mayers D.L., Spiropoulou C.F., Lee W.A., Nichol S.T., Cihlar T., Bavari S. Therapeutic Efficacy of the Small Molecule GS-5734 Against Ebola Virus in Rhesus Monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Siegel D., Hui H.C., Doerffler E., Clarke M.O., Chun K., Zhang L., Neville S., Carra E., Lew W., Ross B., Wang Q., Wolfe L., Jordan R., Soloveva V., Knox J., Perry J., Perron M., Stray K.M., Barauskas O., Feng J.Y., Xu Y., Lee G., Rheingold A.L., Ray A.S., Bannister R., Strickley R., Swaminathan S., Lee W.A., Bavari S., Cihlar T., Lo M.K., Warren T.K., Mackman R.L. Discovery and Synthesis of a Phosphoramidate Prodrug of a Pyrrolo[2,1-f][triazin-4-amino] Adenine C-Nucleoside (GS-5734) for the Treatment of Ebola and Emerging Viruses. J. Med. Chem. 2017;60:1648–1661. doi: 10.1021/acs.jmedchem.6b01594. [DOI] [PubMed] [Google Scholar]
- 124.Mulangu S., Dodd L.E., Davey R.T., Jr., Tshiani Mbaya O., Proschan M., Mukadi D., Lusakibanza Manzo M., Nzolo D., Tshomba Oloma A., Ibanda A., Ali R., Coulibaly S., Levine A.C., Grais R., Diaz J., Lane H.C., Muyembe-Tamfum J.J., Sivahera B., Camara M., Kojan R., Walker R., Dighero-Kemp B., Cao H., Mukumbayi P., Mbala-Kingebeni P., Ahuka S., Albert S., Bonnett T., Crozier I., Duvenhage M., Proffitt C., Teitelbaum M., Moench T., Aboulhab J., Barrett K., Cahill K., Cone K., Eckes R., Hensley L., Herpin B., Higgs E., Ledgerwood J., Pierson J., Smolskis M., Sow Y., Tierney J., Sivapalasingam S., Holman W., Gettinger N., Vallée D., Nordwall J. A Randomized, Controlled Trial of Ebola Virus Disease Therapeutics. N. Engl. J. Med. 2019;381:2293–2303. doi: 10.1056/NEJMoa1910993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Warren T.K., Wells J., Panchal R.G., Stuthman K.S., Garza N.L., Van Tongeren S.A., Dong L., Retterer C.J., Eaton B.P., Pegoraro G., Honnold S., Bantia S., Kotian P., Chen X., Taubenheim B.R., Welch L.S., Minning D.M., Babu Y.S., Sheridan W.P., Bavari S. Protection Against Filovirus Diseases by a Novel Broad-Spectrum Nucleoside Analogue BCX4430. Nature. 2014;508:402–405. doi: 10.1038/nature13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Taylor R., Kotian P., Warren T., Panchal R., Bavari S., Julander J., Dobo S., Rose A., El-Kattan Y., Taubenheim B., Babu Y., Sheridan W.P. BCX4430—A Broad-Spectrum Antiviral Adenosine Nucleoside Analog Under Development for the Treatment of Ebola Virus Disease. J. Infect. Public Health. 2016;9:220–226. doi: 10.1016/j.jiph.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.McMullan L.K., Flint M., Dyall J., Albariño C., Olinger G.G., Foster S., Sethna P., Hensley L.E., Nichol S.T., Lanier E.R., Spiropoulou C.F. The Lipid Moiety of Brincidofovir is Required for In Vitro Antiviral Activity Against Ebola Virus. Antiviral Res. 2016;125:71–78. doi: 10.1016/j.antiviral.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 128.https://www.fda.gov/drugs/drug-safety-and-availability/fda-approves-drug-treat-smallpox
- 129.Dunning J., Kennedy S.B., Antierens A., Whitehead J., Ciglenecki I., Carson G., Kanapathipillai R., Castle L., Howell-Jones R., Pardinaz-Solis R., Grove J., Scott J., Lang T., Olliaro P., Horby P.W., Team, R.-B. T Experimental Treatment of Ebola Virus Disease With Brincidofovir. PLoS One. 2016;11:e0162199. doi: 10.1371/journal.pone.0162199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bray M., Driscoll J., Huggins J.W. Treatment of lethal Ebola Virus Infection in Mice with a Single Dose of an S-Adenosyl-L-Homocysteine Hydrolase Inhibitor. Antiviral Res. 2000;45:135–147. doi: 10.1016/s0166-3542(00)00066-8. [DOI] [PubMed] [Google Scholar]
- 131.Huggins J., Zhang Z.-X., Bray M. Antiviral Drug Therapy of Filovirus Infections: S-Adenosylhomocysteine Hydrolase Inhibitors Inhibit Ebola Virus In Vitro and in a Lethal Mouse Model. J. Infect. Dis. 1999;179:S240–S247. doi: 10.1086/514316. [DOI] [PubMed] [Google Scholar]
- 132.Easton V., McPhillie M., Garcia-Dorival I., Barr J.N., Edwards T.A., Foster R., Fishwick C., Harris M. Identification of a Small Molecule Inhibitor of Ebola Virus Genome Replication and Transcription Using In Silico Screening. Antiviral Res. 2018;156:46–54. doi: 10.1016/j.antiviral.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.https://www.who.int/news-room/fact-sheets/detail/hiv-aids
- 134.Riddell J.t., Amico K.R., Mayer K.H. HIV Preexposure Prophylaxis: A Review. JAMA. 2018;319:1261–1268. doi: 10.1001/jama.2018.1917. [DOI] [PubMed] [Google Scholar]
- 135.Mayer K.H., Molina J.M., Thompson M.A., Anderson P.L., Mounzer K.C., De Wet J.J., DeJesus E., Jessen H., Grant R.M., Ruane P.J., Wong P., Ebrahimi R., Zhong L., Mathias A., Callebaut C., Collins S.E., Das M., McCallister S., Brainard D.M., Brinson C., Clarke A., Coll P., Post F.A., Hare C.B. Emtricitabine and Tenofovir Alafenamide vs Emtricitabine and Tenofovir Disoproxil Fumarate for HIV pre-exposure Prophylaxis (DISCOVER): Primary Results From a Randomised, Double-blind, Multicentre, Active-controlled, Phase 3, Non-Inferiority Trial. Lancet. 2020;396:239–254. doi: 10.1016/S0140-6736(20)31065-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gibas K.M., van den Berg P., Powell V.E., Krakower D.S. Drug Resistance During HIV Pre-Exposure Prophylaxis. Drugs. 2019;79:609–619. doi: 10.1007/s40265-019-01108-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Straubinger T., Kay K., Bies R. Modeling HIV Pre-Exposure Prophylaxis. Front. Pharmacol. 2020;10 doi: 10.3389/fphar.2019.01514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.https://www.fda.gov/news-events/press-announcements/fda-approves-first-extended-release-injectable-drug-regimen-adults-living-hiv
- 139.Engelman K.D., Engelman A.N. Long-Acting Cabotegravir for HIV/AIDS Prophylaxis. Biochemistry. 2021;60:1731–1740. doi: 10.1021/acs.biochem.1c00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Marzinke M.A., Grinsztejn B., Fogel J.M., Piwowar-Manning E., Li M., Weng L., McCauley M., Cummings V., Ahmed S., Haines C.D., Bushman L.R., Petropoulos C., Persaud D., Adeyeye A., Kofron R., Rinehart A., St Clair M., Rooney J.F., Pryluka D., Coelho L., Gaur A., Middelkoop K., Phanuphak N., Cohen M.S., Hendrix C.W., Anderson P., Hanscom B., Donnell D., Landovitz R.J., Eshleman S.H. Characterization of HIV Infection in Cisgender Men and Transgender Women Who Have Sex With Men Receiving Injectable Cabotegravir for HIV Prevention: HPTN 083. J. Infect. Dis. 2021 doi: 10.1093/infdis/jiab152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.https://www.who.int/news/item/09-11-2020-trial-results-reveal-that-long-acting-injectable-cabotegravir-as-prep-is-highly-effective-in-preventing-hiv-acquisition-in-women
- 142.Maxmen A. Achilles Heel Spotted for Promising HIV-Prevention Drug. Nature. 2021;591:357–358. doi: 10.1038/d41586-021-00618-7. [DOI] [PubMed] [Google Scholar]
- 143.Markowitz M., Grobler J.A. Islatravir for the Treatment and Prevention of Infection With the Human Immunodeficiency Virus Type 1. Curr. Opin. HIV AIDS. 2020;15:27–32. doi: 10.1097/COH.0000000000000599. [DOI] [PubMed] [Google Scholar]
- 144.Baeten J.M., Palanee-Phillips T., Mgodi N.M., Mayo A.J., Szydlo D.W., Ramjee G., Gati Mirembe B., Mhlanga F., Hunidzarira P., Mansoor L.E., Siva S., Govender V., Makanani B., Naidoo L., Singh N., Nair G., Chinula L., Parikh U.M., Mellors J.W., Balán I.C., Ngure K., van der Straten A., Scheckter R., Garcia M., Peda M., Patterson K., Livant E., Bunge K., Singh D., Jacobson C., Jiao Y., Hendrix C.W., Chirenje Z.M., Nakabiito C., Taha T.E., Jones J., Torjesen K., Nel A., Rosenberg Z., Soto-Torres L.E., Hillier S.L., Brown E.R. Safety, Uptake, and Use of a Dapivirine Vaginal Ring for HIV-1 Prevention in African Women (HOPE): An Open-Label, Extension Study. Lancet HIV. 2021;8:e87–e95. doi: 10.1016/S2352-3018(20)30304-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Begley R., Lutz J., Rhee M., et al. GS-6207 Sustained Delivery Formulation Supports 6-Month Dosing Interval; Presented at: 23rd International AIDS Conference; July 6–10, 2020; Virtual; 2020. [Google Scholar]
- 146.Sun L., Peng Y., Yu W., Zhang Y., Liang L., Song C., Hou J., Qiao Y., Wang Q., Chen J., Wu M., Zhang D., Li E., Han Z., Zhao Q., Jin X., Zhang B., Huang Z., Chai J., Wang J.-H., Chang J. Mechanistic Insight into Antiretroviral Potency of 2′-Deoxy-2′-β-fluoro-4′-azidocytidine (FNC) With a Long-Lasting Effect on HIV-1 Prevention. J. Med. Chem. 2020;63:8554–8566. doi: 10.1021/acs.jmedchem.0c00940. [DOI] [PubMed] [Google Scholar]
- 147.Pierson T.C., Diamond M.S. The Continued Threat of Emerging Flaviviruses. Nat. Microbiol. 2020;5:796–812. doi: 10.1038/s41564-020-0714-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.https://www.who.int/news-room/fact-sheets/detail/yellow-fever
- 149.Monath T.P., Vasconcelos P.F.C. Yellow fever. J. Clin. Virol. 2015;64:160–173. doi: 10.1016/j.jcv.2014.08.030. [DOI] [PubMed] [Google Scholar]
- 150.Chen L.H., Wilson M.E. Yellow Fever Control: Current Epidemiology and Vaccination Strategies. Trop. Dis. Travel Med. Vac. 2020;6:1. doi: 10.1186/s40794-020-0101-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Shearer F.M., Longbottom J., Browne A.J., Pigott D.M., Brady O.J., Kraemer M.U.G., Marinho F., Yactayo S., de Araújo V.E.M., da Nóbrega A.A., Fullman N., Ray S.E., Mosser J.F., Stanaway J.D., Lim S.S., Reiner R.C., Jr., Moyes C.L., Hay S.I., Golding N. Existing and Potential Infection Risk Zones of Yellow Fever Worldwide: A Modelling Analysis. Lancet Glob. Health. 2018;6:e270–e278. doi: 10.1016/S2214-109X(18)30024-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gaythorpe K.A., Hamlet A., Jean K., Garkauskas Ramos D., Cibrelus L., Garske T., Ferguson N. The Global Burden of Yellow Fever. Elife. 2021;10 doi: 10.7554/eLife.64670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Paules C.I., Fauci A.S. Yellow Fever—Once Again on the Radar Screen in the Americas. N. Engl. J. Med. 2017;376:1397–1399. doi: 10.1056/NEJMp1702172. [DOI] [PubMed] [Google Scholar]
- 154.Lataillade L.D.G.D., Vazeille M., Obadia T., Madec Y., Mousson L., Kamgang B., Chen C.-H., Failloux A.-B., Yen P.-S. Risk of Yellow Fever Virus Transmission in the Asia-Pacific Region. Nat. Commun. 2020;11:5801. doi: 10.1038/s41467-020-19625-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Iwamura T., Guzman-Holst A., Murray K.A. Accelerating Invasion Potential of Disease Vector Aedes aegypti Under Climate Change. Nat. Commun. 2020;11:2130. doi: 10.1038/s41467-020-16010-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Khan S.U., Ogden N.H., Fazil A.A., Gachon P.H., Dueymes G.U., Greer A.L., Ng V. Current and Projected Distributions of Aedes aegypti and Ae. albopictus in Canada and the U.S. Environ. Health Perspect. 2020;128 doi: 10.1289/EHP5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lindsey N.P., Schroeder B.A., Miller E.R., Braun M.M., Hinckley A.F., Marano N., Slade B.A., Barnett E.D., Brunette G.W., Horan K., Staples J.E., Kozarsky P.E., Hayes E.B. Adverse Event Reports Following Yellow Fever Vaccination. Vaccine. 2008;26:6077–6082. doi: 10.1016/j.vaccine.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 158.Zandi K., Amblard F., Amichai S., Bassit L., Tao S.J., Jiang Y., Zhou L.H., Russell O.O., Mengshetti S., Schinazi R.F. Nucleoside Analogs With Antiviral Activity Against Yellow Fever Virus. Antimicrob. Agents Chemother. 2019;63 doi: 10.1128/AAC.00889-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.de Freitas C.S., Higa L.M., Sacramento C.Q., Ferreira A.C., Reis P.A., Delvecchio R., Monteiro F.L., Barbosa-Lima G., James Westgarth H., Vieira Y.R., Mattos M., Rocha N., Hoelz L.V.B., Leme R.P.P., Bastos M.M., Rodrigues G.O.L., Lopes C.E.M., Queiroz-Junior C.M., Lima C.X., Costa V.V., Teixeira M.M., Bozza F.A., Bozza P.T., Boechat N., Tanuri A., Souza T.M.L. Yellow Fever Virus is Susceptible to Sofosbuvir Both In Vitro and In Vivo. PLoS Negl. Trop. Dis. 2019;(13) doi: 10.1371/journal.pntd.0007072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Mendes É.A., Pilger D.R.B.D., Santos Nastri A.C.D.S., Malta F.D.M., Pascoalino B.D.S., Carneiro D’Albuquerque L.A., Balan A., Freitas L.H.G.D., Durigon E.L., Carrilho F.J., Rebello Pinho J.R. Sofosbuvir Inhibits Yellow Fever Virus In Vitro and in Patients With Acute Liver Failure. Ann. Hepatol. 2019;18:816–824. doi: 10.1016/j.aohep.2019.09.001. [DOI] [PubMed] [Google Scholar]
- 161.Julander J.G., Bantia S., Taubenheim B.R., Minning D.M., Kotian P., Morrey J.D., Smee D.F., Sheridan W.P., Babu Y.S. BCX4430, a Novel Nucleoside Analog, Effectively Treats Yellow Fever in a Hamster Model. Antimicrob. Agents Chemother. 2014;58:6607–6614. doi: 10.1128/AAC.03368-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Konkolova E., Dejmek M., Hřebabecký H., Šála M., Böserle J., Nencka R., Boura E. Remdesivir Triphosphate can Efficiently Inhibit the RNA-Dependent RNA Polymerase From Various Flaviviruses. Antiviral Res. 2020;182:104899. doi: 10.1016/j.antiviral.2020.104899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Fioravanti R., Desideri N., Carta A., Atzori E.M., Delogu I., Collu G., Loddo R. Inhibitors of Yellow Fever Virus Replication Based on 1,3,5-Triphenyl-4,5-Dihydropyrazole Scaffold: Design, Synthesis and Antiviral Evaluation. Eur. J. Med. Chem. 2017;141:15–25. doi: 10.1016/j.ejmech.2017.09.060. [DOI] [PubMed] [Google Scholar]
- 164.Cannalire R., Tarantino D., Piorkowski G., Carletti T., Massari S., Felicetti T., Barreca M.L., Sabatini S., Tabarrini O., Marcello A., Milani M., Cecchetti V., Mastrangelo E., Manfroni G., Querat G. Broad Spectrum Anti-flavivirus Pyridobenzothiazolones Leading to Less Infective Virions. Antiviral Res. 2019;167:6–12. doi: 10.1016/j.antiviral.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 165.Gwon Y.-D., Strand M., Lindqvist R., Nilsson E., Saleeb M., Elofsson M., Överby A.K., Evander M. Antiviral Activity of Benzavir-2 Against Emerging Flaviviruses. Viruses. 2020;12:351. doi: 10.3390/v12030351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.https://www.who.int/news-room/fact-sheets/detail/poliomyelitis
- 167.https://www.who.int/news-room/feature-stories/detail/two-out-of-three-wild-poliovirus-strains-eradicated
- 168.Chumakov K., Ehrenfeld E., Agol V.I., Wimmer E. Polio Eradication at the Crossroads. Lancet Glob. Health. 2021;9:e1172–e1175. doi: 10.1016/S2214-109X(21)00205-9. [DOI] [PubMed] [Google Scholar]
- 169.Kew O., Morris-Glasgow V., Landaverde M., Burns C., Shaw J., Garib Z.a., André J., Blackman E., Freeman C.J., Jorba J., Sutter R., Tambini G., Venczel L., Pedreira C., Laender F., Shimizu H., Yoneyama T., Miyamura T., van der Avoort H., Oberste M.S., Kilpatrick D., Cochi S., Pallansch M., de Quadros C. Outbreak of Poliomyelitis in Hispaniola Associated with Circulating Type 1 Vaccine-Derived Poliovirus. Science. 2002;296:356–359. doi: 10.1126/science.1068284. [DOI] [PubMed] [Google Scholar]
- 170.https://www.who.int/emergencies/disease-outbreak-news/item/circulating-vaccine-derived-poliovirus-type-2-global-update
- 171.Macklin G., Diop O.M., Humayun A., Shahmahmoodi S., El-Sayed Z.A., Triki H., Rey G., Avagyan T., Grabovac V., Jorba J., Farag N., Mach O. Update on Immunodeficiency-Associated Vaccine-Derived Polioviruses—Worldwide, July 2018-December 2019. MMWR Morb. Mortal. Wkly Rep. 2020;69:913–917. doi: 10.15585/mmwr.mm6928a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sutter R.W., Modlin J.F., Zaffran M. Completing Polio Eradication: The Case for Antiviral Drugs. J. Infect. Dis. 2016;215:333–334. doi: 10.1093/infdis/jiw547. [DOI] [PubMed] [Google Scholar]
- 173.Oberste M.S., Moore D., Anderson B., Pallansch M.A., Pevear D.C., Collett M.S. In Vitro Antiviral Activity of V-073 Against Polioviruses. Antimicrob. Agents Chemother. 2009;53:4501–4503. doi: 10.1128/AAC.00671-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Collett M.S., Hincks J.R., Benschop K., Duizer E., van der Avoort H., Rhoden E., Liu H., Oberste M.S., McKinlay M.A., Hartford M. Antiviral Activity of Pocapavir in a Randomized, Blinded, Placebo-Controlled Human Oral Poliovirus Vaccine Challenge Model. J. Infect. Dis. 2016;215:335–343. doi: 10.1093/infdis/jiw542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Rhoden E., Liu H.M., Wang-Chern S.W., Oberste M.S. Anti-Poliovirus Activity of Protease Inhibitor AG-7404, and Assessment of In Vitro Activity in Combination With Antiviral Capsid Inhibitor Compounds. Antiviral Res. 2013;98:186–191. doi: 10.1016/j.antiviral.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 176.Xing Y., Zuo J., Krogstad P., Jung M.E. Synthesis and Structure-Activity Relationship (SAR) Studies of Novel Pyrazolopyridine Derivatives as Inhibitors of Enterovirus Replication. J. Med. Chem. 2018;61:1688–1703. doi: 10.1021/acs.jmedchem.7b01863. [DOI] [PubMed] [Google Scholar]
- 177.Zhang Z., Yang E., Hu C., Cheng H., Chen C.Y., Huang D., Wang R., Zhao Y., Rong L., Vignuzzi M., Shen H., Shen L., Chen Z.W. Cell-Based High-Throughput Screening Assay Identifies 2′,2′-Difluoro-2′-deoxycytidine Gemcitabine as a Potential Antipoliovirus Agent. ACS Infect. Dis. 2017;3:45–53. doi: 10.1021/acsinfecdis.6b00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Madia V.N., Messore A., Pescatori L., Saccoliti F., Tudino V., De Leo A., Scipione L., Fiore L., Rhoden E., Manetti F., Oberste M.S., Di Santo R., Costi R. In Vitro Antiviral Activity of New Oxazoline Derivatives as Potent Poliovirus Inhibitors. J. Med. Chem. 2019;62:798–810. doi: 10.1021/acs.jmedchem.8b01482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Manns M.P., Buti M., Gane E., Pawlotsky J.-M., Razavi H., Terrault N., Younossi Z. Hepatitis C Virus Infection. Nat. Rev. Dis. Primers. 2017;3:17006. doi: 10.1038/nrdp.2017.6. [DOI] [PubMed] [Google Scholar]
- 180.Stanciu C., Muzica C.M., Girleanu I., Cojocariu C., Sfarti C., Singeap A.M., Huiban L., Chiriac S., Cuciureanu T., Trifan A. An Update on Direct Antiviral Agents for the Treatment of Hepatitis C. Expert Opin. Pharmacother. 2021:1–13. doi: 10.1080/14656566.2021.1921737. [DOI] [PubMed] [Google Scholar]
- 181.Smolders E.J., Jansen A.M.E., Ter Horst P.G.J., Rockstroh J., Back D.J., Burger D.M. Viral Hepatitis C Therapy: Pharmacokinetic and Pharmacodynamic Considerations: A 2019 Update. Clin. Pharmacokinet. 2019;58:1237–1263. doi: 10.1007/s40262-019-00774-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Heo Y.A., Deeks E.D. Sofosbuvir/Velpatasvir/Voxilaprevir: A Review in Chronic Hepatitis C. Drugs. 2018;78:577–587. doi: 10.1007/s40265-018-0895-5. [DOI] [PubMed] [Google Scholar]
- 183.Martinello M., Bhagani S., Gane E., Orkin C., Cooke G., Dore G.J., Petoumenos K., Applegate T.L., Tu E., Marks P., Pagani N., Grebely J., Nelson M., Matthews G.V. Shortened Therapy of Eight Weeks With Paritaprevir/Ritonavir/Ombitasvir and Dasabuvir is Highly Effective in People With Recent HCV Genotype 1 Infection. J. Viral Hepat. 2018;25:1180–1188. doi: 10.1111/jvh.12917. [DOI] [PubMed] [Google Scholar]
- 184.Do A., Reau N.S. Chronic Viral Hepatitis: Current Management and Future Directions. Hepatol. Commun. 2020;4:329–341. doi: 10.1002/hep4.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Brown R.S., Jr., Buti M., Rodrigues L., Chulanov V., Chuang W.L., Aguilar H., Horváth G., Zuckerman E., Carrion B.R., Rodriguez-Perez F., Urbánek P., Abergel A., Cohen E., Lovell S.S., Schnell G., Lin C.W., Zha J., Wang S., Trinh R., Mensa F.J., Burroughs M., Felizarta F. Glecaprevir/Pibrentasvir for 8 Weeks in Treatment-Naïve Patients With Chronic HCV Genotypes 1-6 and Compensated Cirrhosis: The Expedition-8 Trial. J. Hepatol. 2020;72:441–449. doi: 10.1016/j.jhep.2019.10.020. [DOI] [PubMed] [Google Scholar]
- 186.Martinello M., Gane E., Hellard M., Sasadeusz J., Shaw D., Petoumenos K., Applegate T., Grebely J., Maire L., Marks P., Dore G.J., Matthews G.V. Sofosbuvir and Ribavirin for 6 Weeks is Not Effective Among People With Recent hepatitis C Virus Infection: The DARE-C II Study. Hepatology. 2016;64:1911–1921. doi: 10.1002/hep.28844. [DOI] [PubMed] [Google Scholar]
- 187.Deterding K., Spinner C.D., Schott E., Welzel T.M., Gerken G., Klinker H., Spengler U., Wiegand J., Schulze Zur Wiesch J., Pathil A., Cornberg M., Umgelter A., Zöllner C., Zeuzem S., Papkalla A., Weber K., Hardtke S., von der Leyen H., Koch A., von Witzendorff D., Manns M.P., Wedemeyer H. Ledipasvir Plus Sofosbuvir Fixed-Dose Combination for 6 Weeks in Patients With Acute Hepatitis C Virus Genotype 1 Monoinfection (HepNet Acute HCV IV): An Open-Label, Single-Arm, Phase 2 Study. Lancet Infect. Dis. 2017;17:215–222. doi: 10.1016/S1473-3099(16)30408-X. [DOI] [PubMed] [Google Scholar]
- 188.Rockstroh J.K., Bhagani S., Hyland R.H., Yun C., Dvory-Sobol H., Zheng W., Brainard D.M., Ingiliz P., Lutz T., Boesecke C., Nelson M. Ledipasvir-Sofosbuvir for 6 Weeks to Treat Acute Hepatitis C Virus Genotype 1 or 4 Infection in Patients With HIV Coinfection: An Open-Label, Single-Arm Trial. Lancet Gastroenterol. Hepatol. 2017;2:347–353. doi: 10.1016/S2468-1253(17)30003-1. [DOI] [PubMed] [Google Scholar]
- 189.Casey J.L., Feld J.J., MacParland S.A. Restoration of HCV-Specific Immune Responses With Antiviral Therapy: A Case for DAA Treatment in Acute HCV Infection. Cell. 2019;8 doi: 10.3390/cells8040317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Piecha F., Gänßler J.M., Ozga A.K., Wehmeyer M.H., Dietz J., Kluwe J., Laschtowitz A., von Felden J., Sterneck M., Jordan S., Pischke S., Lohse A.W., Schulze Zur Wiesch J. Treatment and Re-treatment Results of HCV Patients in the DAA Era. PLoS One. 2020;15 doi: 10.1371/journal.pone.0232773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Bourlière M., Gordon S.C., Flamm S.L., Cooper C.L., Ramji A., Tong M., Ravendhran N., Vierling J.M., Tran T.T., Pianko S., Bansal M.B., de Lédinghen V., Hyland R.H., Stamm L.M., Dvory-Sobol H., Svarovskaia E., Zhang J., Huang K.C., Subramanian G.M., Brainard D.M., McHutchison J.G., Verna E.C., Buggisch P., Landis C.S., Younes Z.H., Curry M.P., Strasser S.I., Schiff E.R., Reddy K.R., Manns M.P., Kowdley K.V., Zeuzem S. Sofosbuvir, Velpatasvir, and Voxilaprevir for Previously Treated HCV Infection. New Engl. J. Med. 2017;376:2134–2146. doi: 10.1056/NEJMoa1613512. [DOI] [PubMed] [Google Scholar]
- 192.Sezaki H., Suzuki F., Hosaka T., Fujiyama S., Kawamura Y., Akuta N., Kobayashi M., Suzuki Y., Saitoh S., Arase Y., Ikeda K., Kobayashi M., Kumada H. Initial- and Re-treatment Effectiveness of Glecaprevir and Pibrentasvir for Japanese Patients With Chronic Hepatitis C Virus-Genotype 1/2/3 Infections. J. Gastroenterol. 2019;54:916–927. doi: 10.1007/s00535-019-01575-9. [DOI] [PubMed] [Google Scholar]
- 193.Meanwell N.A., Georg G.I., Wang S.T. Nobel Prize in Physiology or Medicine. J. Med. Chem. 2020;2020(63):13197–13204. doi: 10.1021/acs.jmedchem.0c01877. [DOI] [PubMed] [Google Scholar]
- 194.Dore G.J., Bajis S. Hepatitis C Virus Elimination: Laying the Foundation for Achieving 2030 Targets. Nat. Rev. Gastroenterol. Hepatol. 2021;18:91–92. doi: 10.1038/s41575-020-00392-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.https://www.who.int/news-room/fact-sheets/detail/hepatitis-c
- 196.Ramdas V., Talwar R., Banerjee M., Joshi A.A., Das A.K., Walke D.S., Borhade P., Dhayagude U., Loriya R., Gote G., Bommakanti A., Sivaram A., Agarwal G., Goswami A., Nigade P., Mehta M., Patil V., Modi D., Kumar H., Mallurwar S., Dash A., Modi F., Kuldharan S., Srivastava P., Singh M., Narasimham L., Gundu J., Sharma S., Kamboj R.K., Palle V.P. Discovery and Characterization of Potent Pan-Genotypic HCV NS5A Inhibitors Containing Novel Tricyclic Central Core Leading to Clinical Candidate. Journal of Medicinal Chemistry. 2019;62:10563–10582. doi: 10.1021/acs.jmedchem.9b01562. [DOI] [PubMed] [Google Scholar]
- 197.Liu B., Gai K., Qin H., Wang J., Liu X., Cao Y., Lu Q., Lu D., Chen D., Shen H., Song W., Mei J., Wang X., Xu H., Zhang Y. Discovery of a Silicon-Containing Pan-Genotype Hepatitis C Virus NS5A Inhibitor. J. Med. Chem. 2020;63:5312–5323. doi: 10.1021/acs.jmedchem.0c00082. [DOI] [PubMed] [Google Scholar]
- 198.Kazmierski W.M., Baskaran S., Walker J.T., Miriyala N., Meesala R., Beesu M., Adjabeng G., Grimes R.M., Hamatake R., Leivers M.R., Crosby R., Xia B., Remlinger K. GSK2818713, a Novel Biphenylene Scaffold-Based Hepatitis C NS5A Replication Complex Inhibitor with Broad Genotype Coverage. J. Med. Chem. 2020;63:4155–4170. doi: 10.1021/acs.jmedchem.9b02176. [DOI] [PubMed] [Google Scholar]
- 199.Randolph J.T., Voight E.A., Greszler S.N., Uno B.E., Newton J.N., Gleason K.M., Stolarik D., Van Handel C., Bow D.A.J., DeGoey D.A. Prodrug Strategies to Improve the Solubility of the HCV NS5A Inhibitor Pibrentasvir (ABT-530) J. Med. Chem. 2020;63:11034–11044. doi: 10.1021/acs.jmedchem.0c00956. [DOI] [PubMed] [Google Scholar]
- 200.Sun L.Q., Mull E., D'Andrea S., Zheng B., Hiebert S., Gillis E., Bowsher M., Kandhasamy S., Baratam V.R., Puttaswamy S., Pulicharla N., Vishwakrishnan S., Reddy S., Trivedi R., Sinha S., Sivaprasad S., Rao A., Desai S., Ghosh K., Anumula R., Kumar A., Rajamani R., Wang Y.K., Fang H., Mathur A., Rampulla R., Zvyaga T.A., Mosure K., Jenkins S., Falk P., Tagore D.M., Chen C., Rendunchintala K., Loy J., Meanwell N.A., McPhee F., Scola P.M. Discovery of BMS-986144, a Third-Generation, Pan-Genotype NS3/4A Protease Inhibitor for the Treatment of Hepatitis C Virus Infection. J. Med. Chem. 2020;63:14740–14760. doi: 10.1021/acs.jmedchem.0c01296. [DOI] [PubMed] [Google Scholar]
- 201.Nageswara Rao D., Zephyr J., Henes M., Chan E.T., Matthew A.N., Hedger A.K., Conway H.L., Saeed M., Newton A., Petropoulos C.J., Huang W., Kurt Yilmaz N., Schiffer C.A., Ali A. Discovery of Quinoxaline-Based P1–P3 Macrocyclic NS3/4A Protease Inhibitors With Potent Activity Against Drug-Resistant Hepatitis C Virus Variants. J. Med. Chem. 2021;64:11972–11989. doi: 10.1021/acs.jmedchem.1c00554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Wang G., Dyatkina N., Prhavc M., Williams C., Serebryany V., Hu Y., Huang Y., Wan J., Wu X., Deval J., Fung A., Jin Z., Tan H., Shaw K., Kang H., Zhang Q., Tam Y., Stoycheva A., Jekle A., Smith D.B., Beigelman L. Synthesis and Anti-HCV Activities of 4′-Fluoro-2′-Substituted Uridine Triphosphates and Nucleotide Prodrugs: Discovery of 4′-Fluoro-2′-C-methyluridine 5′-Phosphoramidate Prodrug (AL-335) for the Treatment of Hepatitis C Infection. J. Med. Chem. 2019;62:4555–4570. doi: 10.1021/acs.jmedchem.9b00143. [DOI] [PubMed] [Google Scholar]
- 203.Mengshetti S., Zhou L.H., Sari O., De Schutter C., Zhang H.W., Cho J.H., Tao S.J., Bassit L.C., Verma K., Domaoal R.A., Ehteshami M., Jiang Y., Ovadia R., Kasthuri M., Russell O.I., McBrayer T., Whitaker T., Pattassery J., Pascual M.L., Uher L., Lin B.Y., Lee S., Amblard F., Coats S.J., Schinazi R.F. Discovery of a Series of 2'-Alpha-Fluoro,2'-Beta-bromo-ribonucleosides and Their Phosphoramidate Prodrugs as Potent Pan-Genotypic Inhibitors of Hepatitis C Virus. J. Med. Chem. 2019;62:1859–1874. doi: 10.1021/acs.jmedchem.8b01300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Wang G., Dyatkina N., Prhavc M., Williams C., Serebryany V., Hu Y., Huang Y., Wu X., Chen T., Huang W., Rajwanshi V.K., Deval J., Fung A., Jin Z., Stoycheva A., Shaw K., Gupta K., Tam Y., Jekle A., Smith D.B., Beigelman L. Synthesis and Anti-HCV Activity of Sugar-Modified Guanosine Analogues: Discovery of AL-611 as an HCV NS5B Polymerase Inhibitor for the Treatment of Chronic Hepatitis C. J. Med. Chem. 2020;63:10380–10395. doi: 10.1021/acs.jmedchem.0c00935. [DOI] [PubMed] [Google Scholar]
- 205.Good S.S., Moussa A., Zhou X.J., Pietropaolo K., Sommadossi J.P. Preclinical Evaluation of AT-527, a Novel Guanosine Nucleotide Prodrug With Potent, Pan-Genotypic Activity Against Hepatitis C Virus. PLoS One. 2020;15 doi: 10.1371/journal.pone.0227104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Chong P.Y., Shotwell J.B., Miller J., Price D.J., Maynard A., Voitenleitner C., Mathis A., Williams S., Pouliot J.J., Creech K., Wang F., Fang J., Zhang H., Tai V.W.F., Turner E., Kahler K.M., Crosby R., Peat A.J. Design of N-Benzoxaborole Benzofuran GSK8175—Optimization of Human Pharmacokinetics Inspired by Metabolites of a Failed Clinical HCV Inhibitor. J. Med. Chem. 2019;62:3254–3267. doi: 10.1021/acs.jmedchem.8b01719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Jiang X., Tan J., Wang Y., Chen J., Li J., Jiang Z., Quan Y., Jin J., Li Y., Cen S., Li Y., Peng Z., Li Z. 2-((4-Arylpiperazin-1-yl)methyl)benzonitrile Derivatives as Orally Available Inhibitors of Hepatitis C Virus with a Novel Mechanism of Action. J. Med. Chem. 2020;63:5972–5989. doi: 10.1021/acs.jmedchem.0c00232. [DOI] [PubMed] [Google Scholar]
- 208.Rolt A., Talley D.C., Park S.B., Hu Z., Dulcey A., Ma C., Irvin P., Leek M., Wang A.Q., Stachulski A.V., Xu X., Southall N., Ferrer M., Liang T.J., Marugan J.J. Discovery and Optimization of a 4-Aminopiperidine Scaffold for Inhibition of Hepatitis C Virus Assembly. J. Med. Chem. 2021;64:9431–9443. doi: 10.1021/acs.jmedchem.1c00696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Yang Y., Cao L., Gao H., Wu Y., Wang Y., Fang F., Lan T., Lou Z., Rao Y. Discovery, Optimization, and Target Identification of Novel Potent Broad-Spectrum Antiviral Inhibitors. J. Med. Chem. 2019;62:4056–4073. doi: 10.1021/acs.jmedchem.9b00091. [DOI] [PubMed] [Google Scholar]
- 210.Singh R., Singh K.P., Cherian S., Saminathan M., Kapoor S., Manjunatha Reddy G.B., Panda S., Dhama K. Rabies—Epidemiology, Pathogenesis, Public Health Concerns and Advances in Diagnosis and Control: A Comprehensive Review. Vet. Q. 2017;37:212–251. doi: 10.1080/01652176.2017.1343516. [DOI] [PubMed] [Google Scholar]
- 211.https://www.who.int/news-room/fact-sheets/detail/rabies
- 212.Jackson A.C. Diabolical Effects of Rabies Encephalitis. J. Neurovirol. 2016;22:8–13. doi: 10.1007/s13365-015-0351-1. [DOI] [PubMed] [Google Scholar]
- 213.Kessels J.A., Recuenco S., Navarro-Vela A.M., Deray R., Vigilato M., Ertl H., Durrheim D., Rees H., Nel L.H., Abela-Ridder B., Briggs D. Pre-Exposure Rabies Prophylaxis: A Systematic Review. Bull. World Health Organ. 2017;95:210–219c. doi: 10.2471/BLT.16.173039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Kessels J., Tarantola A., Salahuddin N., Blumberg L., Knopf L. Rabies Post-Exposure Prophylaxis: A Systematic Review on Abridged Vaccination Schedules and the Effect of Changing Administration Routes During a Single Course. Vaccine. 2019;37:A107–A117. doi: 10.1016/j.vaccine.2019.01.041. [DOI] [PubMed] [Google Scholar]
- 215.Smith S.P., Wu G., Fooks A.R., Ma J., Banyard A.C. Trying to Treat the Untreatable: Experimental Approaches to Clear Rabies Virus Infection From the CNS. J. Gen. Virol. 2019;100:1171–1186. doi: 10.1099/jgv.0.001269. [DOI] [PubMed] [Google Scholar]
- 216.Du Pont V., Plemper R.K., Schnell M.J. Status of Antiviral Therapeutics Against Rabies Virus and Related Emerging Lyssaviruses. Curr. Opin. Virol. 2019;35:1–13. doi: 10.1016/j.coviro.2018.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Appolinario C.M., Jackson A.C. Antiviral Therapy for Human Rabies. Antivir. Ther. 2015;20:1–10. doi: 10.3851/IMP2851. [DOI] [PubMed] [Google Scholar]
- 218.Anindita P.D., Sasaki M., Okada K., Ito N., Sugiyama M., Saito-Tarashima N., Minakawa N., Shuto S., Otsuguro S., Ichikawa S., Matsuda A., Maenaka K., Orba Y., Sawa H. Ribavirin-Related Compounds Exert In Vitro Inhibitory Effects Toward Rabies Virus. Antiviral Res. 2018;154:1–9. doi: 10.1016/j.antiviral.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 219.Yamada K., Noguchi K., Komeno T., Furuta Y., Nishizono A. Efficacy of Favipiravir (T-705) in Rabies Postexposure Prophylaxis. J. Infect. Dis. 2016;213:1253–1261. doi: 10.1093/infdis/jiv586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Banyard A.C., Mansfield K.L., Wu G., Selden D., Thorne L., Birch C., Koraka P., Osterhaus A., Fooks A.R. Re-Evaluating the Effect of Favipiravir Treatment on Rabies Virus Infection. Vaccine. 2019;37:4686–4693. doi: 10.1016/j.vaccine.2017.10.109. [DOI] [PubMed] [Google Scholar]
- 221.Yamada K., Noguchi K., Kimitsuki K., Kaimori R., Saito N., Komeno T., Nakajima N., Furuta Y., Nishizono A. Reevaluation of the Efficacy of Favipiravir Against Rabies Virus Using In Vivo Imaging Analysis. Antiviral Res. 2019;172:104641. doi: 10.1016/j.antiviral.2019.104641. [DOI] [PubMed] [Google Scholar]
- 222.Kali S., Jallet C., Azebi S., Cokelaer T., Da Fonseca J.P., Wu Y., Barbier J., Cintrat J.C., Gillet D., Tordo N. Broad Spectrum Compounds Targeting Early Stages of Rabies Virus (RABV) Infection. Antiviral Res. 2021;188:105016. doi: 10.1016/j.antiviral.2021.105016. [DOI] [PubMed] [Google Scholar]
- 223.Pont V.D., Wirblich C., Yoon J.-J., Cox R.M., Schnell M.J., Plemper R.K., Dutch R.E. Identification and Characterization of a Small-Molecule Rabies Virus Entry Inhibitor. J. Virol. 2020;94:e00320–e00321. doi: 10.1128/JVI.00321-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Tang H.-B., Lu Z.-L., Wei X.-K., Zhong T.-Z., Zhong Y.-Z., Ouyang L.-X., Luo Y., Xing X.-W., Liao F., Peng K.-K., Deng C.-Q., Minamoto N., Luo T.R. Viperin Inhibits Rabies Virus Replication Via Reduced Cholesterol and Sphingomyelin and is Regulated Upstream by TLR4. Sci. Rep. 2016;6:30529. doi: 10.1038/srep30529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Gizzi A.S., Grove T.L., Arnold J.J., Jose J., Jangra R.K., Garforth S.J., Du Q., Cahill S.M., Dulyaninova N.G., Love J.D., Chandran K., Bresnick A.R., Cameron C.E., Almo S.C. A Naturally Occurring Antiviral Ribonucleotide Encoded by the Human Genome. Nature. 2018;558:610–614. doi: 10.1038/s41586-018-0238-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Patel A.M., Marsh E.N.G. The Antiviral Enzyme, Viperin, Activates Protein Ubiquitination by the E3 Ubiquitin Ligase, TRAF6. J. Am. Chem. Soc. 2021;143:4910–4914. doi: 10.1021/jacs.1c01045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Durrheim D.N. Measles Eradication-Retreating is Not an Option. Lancet Infect. Dis. 2020;20:e138–e141. doi: 10.1016/S1473-3099(20)30052-9. [DOI] [PubMed] [Google Scholar]
- 228.Durrheim D.N., Andrus J.K., Tabassum S., Bashour H., Githanga D., Pfaff G. A Dangerous Measles Future Looms Beyond the COVID-19 Pandemic. Nat. Med. 2021;27:360–361. doi: 10.1038/s41591-021-01237-5. [DOI] [PubMed] [Google Scholar]
- 229.Rana M.S., Alam M.M., Ikram A., Salman M., Mere M.O., Usman M., Umair M., Zaidi S.S.Z., Arshad Y. Emergence of Measles During the COVID-19 Pandemic Threatens Pakistan's Children and the Wider Region. Nat. Med. 2021;27:1127–1128. doi: 10.1038/s41591-021-01430-6. [DOI] [PubMed] [Google Scholar]
- 230.Laventhal N., Tarini B.A., Lantos J. Ethical Issues in Neonatal and Pediatric Clinical Trials. Pediatr. Clin. North Am. 2012;59:1205–1220. doi: 10.1016/j.pcl.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Ferren M., Horvat B., Mathieu C. Measles Encephalitis: Towards New Therapeutics. Viruses. 2019;11:1017. doi: 10.3390/v11111017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Plemper R.K., Doyle J., Sun A., Prussia A., Cheng L.-T., Rota P.A., Liotta D.C., Snyder J.P., Compans R.W. Design of a Small-Molecule Entry Inhibitor With Activity Against Primary Measles Virus Strains. Antimicrob. Agents Chemother. 2005;49:3755–3761. doi: 10.1128/AAC.49.9.3755-3761.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Sun A., Prussia A., Zhan W., Murray E.E., Doyle J., Cheng L.-T., Yoon J.-J., Radchenko E.V., Palyulin V.A., Compans R.W., Liotta D.C., Plemper R.K., Snyder J.P. Nonpeptide Inhibitors of Measles Virus Entry. J. Med. Chem. 2006;49:5080–5092. doi: 10.1021/jm0602559. [DOI] [PubMed] [Google Scholar]
- 234.Lo M.K., Jordan R., Arvey A., Sudhamsu J., Shrivastava-Ranjan P., Hotard A.L., Flint M., McMullan L.K., Siegel D., Clarke M.O., Mackman R.L., Hui H.C., Perron M., Ray A.S., Cihlar T., Nichol S.T., Spiropoulou C.F. GS-5734 and Its Parent Nucleoside Analog Inhibit Filo-, Pneumo-, and Paramyxoviruses. Sci. Rep. 2017;7:43395. doi: 10.1038/srep43395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Lo M.K., Jordan P.C., Stevens S., Tam Y., Deval J., Nichol S.T., Spiropoulou C.F. Susceptibility of Paramyxoviruses and Filoviruses to Inhibition by 2'-Monofluoro- and 2'-Difluoro-4'-Azidocytidine Analogs. Antiviral Res. 2018;153:101–113. doi: 10.1016/j.antiviral.2018.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Ndungu J.M., Krumm S.A., Yan D., Arrendale R.F., Reddy G.P., Evers T., Howard R., Natchus M.G., Saindane M.T., Liotta D.C., Plemper R.K., Snyder J.P., Sun A. Non-nucleoside Inhibitors of the Measles Virus RNA-Dependent RNA Polymerase: Synthesis, Structure–Activity Relationships, and Pharmacokinetics. J. Med. Chem. 2012;55:4220–4230. doi: 10.1021/jm201699w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Krumm S.A., Yan D., Hovingh E.S., Evers T.J., Enkirch T., Reddy G.P., Sun A., Saindane M.T., Arrendale R.F., Painter G., Liotta D.C., Natchus M.G., von Messling V., Plemper R.K. An Orally Available, Small-Molecule Polymerase inhibitor Shows Efficacy Against a Lethal Morbillivirus Infection in a Large Animal Model. Sci. Transl. Med. 2014;6:232ra252. doi: 10.1126/scitranslmed.3008517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Wittwer K., Anderson D.E., Pfeffermann K., Cox R.M., Wolf J.D., Santibanez S., Mankertz A., Plesker R., Sticher Z.M., Kolkykhalov A.A., Natchus M.G., Pfaller C.K., Plemper R.K., von Messling V. Small-Molecule Polymerase Inhibitor Protects Non-Human Primates From Measles and Reduces Shedding. Nat. Commun. 2021;12:5233. doi: 10.1038/s41467-021-25497-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Cox R.M., Sourimant J., Govindarajan M., Natchus M.G., Plemper R.K. Therapeutic Targeting of Measles Virus Polymerase With ERDRP-0519 Suppresses All RNA Synthesis Activity. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Cox R.M., Sourimant J., Toots M., Yoon J.-J., Ikegame S., Govindarajan M., Watkinson R.E., Thibault P., Makhsous N., Lin M.J., Marengo J.R., Sticher Z., Kolykhalov A.A., Natchus M.G., Greninger A.L., Lee B., Plemper R.K. Orally Efficacious Broad-Spectrum Allosteric Inhibitor of Paramyxovirus Polymerase. Nat. Microbiol. 2020;5:1232–1246. doi: 10.1038/s41564-020-0752-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Krumm S.A., Ndungu J.M., Yoon J.J., Dochow M., Sun A., Natchus M., Snyder J.P., Plemper R.K. Potent Host-Directed Small-Molecule Inhibitors of Myxovirus RNA-Dependent RNA-Polymerases. PLoS One. 2011;6 doi: 10.1371/journal.pone.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Grafen A., Schumacher F., Chithelen J., Kleuser B., Beyersdorf N., Schneider-Schaulies J. Use of Acid Ceramidase and Sphingosine Kinase Inhibitors as Antiviral Compounds Against Measles Virus Infection of Lymphocytes In Vitro. Front. Cell Dev. Biol. 2019;7 doi: 10.3389/fcell.2019.00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Perlman S., Poon L.L.M., Samborskiy D.V., Sidorov I.A., Sola I., Ziebuhr J., Coronaviridae Study Group of the International Committee on Taxonomy of V. The Species Severe Acute Respiratory Syndrome-Related Coronavirus: Classifying 2019-nCoV and Naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Hu B., Guo H., Zhou P., Shi Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021;19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Cui J., Li F., Shi Z.L. Origin and Evolution of Pathogenic Coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.https://www.who.int/emergencies/disease-outbreak-news/item/2021-DON333
- 248.https://covid19.who.int/
- 249.Gerges Harb J., Noureldine H.A., Chedid G., Eldine M.N., Abdallah D.A., Chedid N.F., Nour-Eldine W. SARS, MERS and COVID-19: Clinical Manifestations and Organ-System Complications: A Mini Review. Pathog. Dis. 2020;78 doi: 10.1093/femspd/ftaa033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Discovery of Seven Novel Mammalian and Avian Coronaviruses in the Genus Deltacoronavirus Supports Bat Coronaviruses as the Gene Source of Alphacoronavirus and Betacoronavirus and Avian Coronaviruses as the Gene Source of Gammacoronavirus and Deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Boni M.F., Lemey P., Jiang X., Lam T.T.-Y., Perry B.W., Castoe T.A., Rambaut A., Robertson D.L. Evolutionary Origins of the SARS-CoV-2 Sarbecovirus Lineage Responsible for the COVID-19 Pandemic. Nat. Microbiol. 2020;5:1408–1417. doi: 10.1038/s41564-020-0771-4. [DOI] [PubMed] [Google Scholar]
- 252.Dolgin E. The Race for Antiviral Drugs to Beat COVID—and the Next Pandemic. Nature. 2021;592:340–343. doi: 10.1038/d41586-021-00958-4. [DOI] [PubMed] [Google Scholar]
- 253.Nascimento Junior J.A.C., Santos A.M., Quintans-Júnior L.J., Walker C.I.B., Borges L.P., Serafini M.R. SARS, MERS and SARS-CoV-2 (COVID-19) Treatment: A Patent Review. Expert Opin. Ther. Pat. 2020;30:567–579. doi: 10.1080/13543776.2020.1772231. [DOI] [PubMed] [Google Scholar]
- 254.Artese A., Svicher V., Costa G., Salpini R., Di Maio V.C., Alkhatib M., Ambrosio F.A., Santoro M.M., Assaraf Y.G., Alcaro S., Ceccherini-Silberstein F. Current Status of Antivirals and Druggable Targets of SARS CoV-2 and Other Human Pathogenic Coronaviruses. Drug Resist. Updat. 2020;53:100721. doi: 10.1016/j.drup.2020.100721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal Structure of SARS-CoV-2 Main Protease Provides a Basis for Design of Improved α-Ketoamide Inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Zhang L., Lin D., Kusov Y., Nian Y., Ma Q., Wang J., von Brunn A., Leyssen P., Lanko K., Neyts J., de Wilde A., Snijder E.J., Liu H., Hilgenfeld R. α-Ketoamides as Broad-Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, Synthesis, and Activity Assessment. J. Med. Chem. 2020;63:4562–4578. doi: 10.1021/acs.jmedchem.9b01828. [DOI] [PubMed] [Google Scholar]
- 257.Hu Y., Ma C., Szeto T., Hurst B., Tarbet B., Wang J. Boceprevir, Calpain Inhibitors II and XII, and GC-376 Have Broad-Spectrum Antiviral Activity Against Coronaviruses. ACS Infect. Dis. 2021;7:586–597. doi: 10.1021/acsinfecdis.0c00761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Dampalla C.S., Zheng J., Perera K.D., Wong L.-Y.R., Meyerholz D.K., Nguyen H.N., Kashipathy M.M., Battaile K.P., Lovell S., Kim Y., Perlman S., Groutas W.C., Chang K.-O. Postinfection Treatment With a Protease Inhibitor Increases Survival of Mice With a Fatal SARS-CoV-2 Infection. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2101555118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Hoffman R.L., Kania R.S., Brothers M.A., Davies J.F., Ferre R.A., Gajiwala K.S., He M., Hogan R.J., Kozminski K., Li L.Y., Lockner J.W., Lou J., Marra M.T., Mitchell L.J., Murray B.W., Nieman J.A., Noell S., Planken S.P., Rowe T., Ryan K., Smith G.J., Solowiej J.E., Steppan C.M., Taggart B. Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19. J. Med. Chem. 2020;63:12725–12747. doi: 10.1021/acs.jmedchem.0c01063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., Zhu C., Hu T., Hua T., Zhang B., Yang X., Li J., Yang H., Liu Z., Xu W., Guddat L.W., Wang Q., Lou Z., Rao Z. Structure of the RNA-Dependent RNA Polymerase From COVID-19 Virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Malin J.J., Suárez I., Priesner V., Fätkenheuer G., Rybniker J. Remdesivir Against COVID-19 and Other Viral Diseases. Clin. Microbiol. Rev. 2020;34 doi: 10.1128/CMR.00162-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Qi F., Bao L., Du L., Liu S., Qin C., Sun F., Shi Z., Zhu Y., Jiang S., Lu L. Inhibition of SARS-CoV-2 (Previously 2019-nCoV) Infection by a Highly Potent Pan-Coronavirus Fusion Inhibitor Targeting its Spike Protein That Harbors a High Capacity to Mediate Membrane Fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Lee J.Y., Shin Y.S., Jeon S., Lee S.I., Noh S., Cho J.-E., Jang M.S., Kim S., Song J.H., Kim H.R., Park C.M. Design, Synthesis and Biological Evaluation of 2-Aminoquinazolin-4(3H)-one Derivatives as Potential SARS-CoV-2 and MERS-CoV Treatments. Bioorg. Med. Chem. Lett. 2021;39:127885. doi: 10.1016/j.bmcl.2021.127885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Xu J., Shi P.-Y., Li H., Zhou J. Broad Spectrum Antiviral Agent Niclosamide and Its Therapeutic Potential. ACS Infect. Dis. 2020;6:909–915. doi: 10.1021/acsinfecdis.0c00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Pillaiyar T., Laufer S. Kinases as Potential Therapeutic Targets for Anti-coronaviral Therapy. J. Med. Chem. 2021 doi: 10.1021/acs.jmedchem.1c00335. [DOI] [PubMed] [Google Scholar]
- 266.Mellott D.M., Tseng C.-T., Drelich A., Fajtová P., Chenna B.C., Kostomiris D.H., Hsu J., Zhu J., Taylor Z.W., Kocurek K.I., Tat V., Katzfuss A., Li L., Giardini M.A., Skinner D., Hirata K., Yoon M.C., Beck S., Carlin A.F., Clark A.E., Beretta L., Maneval D., Hook V., Frueh F., Hurst B.L., Wang H., Raushel F.M., O’Donoghue A.J., de Siqueira-Neto J.L., Meek T.D., McKerrow J.H. A Clinical-Stage Cysteine Protease Inhibitor Blocks SARS-CoV-2 Infection of Human and Monkey Cells. ACS Chem. Biol. 2021;16:642–650. doi: 10.1021/acschembio.0c00875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Firpo M.R., Mastrodomenico V., Hawkins G.M., Prot M., Levillayer L., Gallagher T., Simon-Loriere E., Mounce B.C. Targeting Polyamines Inhibits Coronavirus Infection by Reducing Cellular Attachment and Entry. ACS Infect. Dis. 2021;7:1423–1432. doi: 10.1021/acsinfecdis.0c00491. [DOI] [PMC free article] [PubMed] [Google Scholar]