Abstract
Background:
The gut microbiome is an important determinant of health and disease in preterm infants.
Objectives:
The objective of this paper is to share our current protocol for other neonatal intensive care units to potentially expand their existing protocols aiming to characterize the relationship between the intestinal microbiome and health outcomes in preterm infants.
Methods:
This prospective, longitudinal study planned to recruit 160 preterm infants born < 32 weeks’ gestational age or weighing < 1,500 grams and admitted to one of two-level III/IV neonatal intensive care units. During the neonatal intensive care unit period, the primary measures included events of early life pain/stress, gut microbiome, host genetic variations, and neurobehavioral assessment. During follow-up visits, gut microbiome, pain sensitivity, and medical, growth, and developmental outcomes at 4-, 8–12-, and 18–24-month corrected age (CA) were measured
Discussion:
As of February 14, 2020, 214 preterm infants have been recruited. We hypothesize that infants who experience greater levels of pain/stress will have altered gut microbiome, including potential adverse outcomes such as necrotizing enterocolitis (NEC) and host genetic variations, feeding intolerance, and/or neurodevelopmental impairments. These will differ from the intestinal microbiome of preterm infants that do not develop these adverse outcomes. To test this hypothesis, we will determine how alterations in the intestinal microbiome affect the risk of developing NEC, feeding intolerance, and neurodevelopmental impairments in preterm infants. In addition, we will examine the interaction between the intestinal microbiome and host genetics in the regulation of intestinal health and neurodevelopmental outcomes.
Keywords: gut microbiome, neurodevelopment, pain/stress, preterm infant
The bacterial flora of the intestinal tract, referred to as the gut microbiome, is increasingly being recognized as an important determinant of health and disease at all stages of life (Indrio et al., 2017). Intestinal bacteria outnumber endogenous cells by a ratio of 10:1 and benefit the host by aiding in digestion of nutrients and preventing growth of potentially pathogenic organisms (Schippa & Conte, 2014). Conversely, increased levels of harmful bacteria or microbial “dysbiosis” can lead to defects in the development and control of the immune system, which may contribute to conditions such as inflammatory bowel diseases, asthma, and even autism (Clarke et al., 2014). Each year, an estimated 15 million babies are born preterm (< 37 weeks’ gestational age [GA] globally; World Health Organization [WHO], 2018). In the preterm infant, the gut microbiome demonstrates an increased level of instability influenced by factors such as early and prolonged exposure to antibiotics and formula feeding, combined with abnormal colonizers found within the ICU environment (Pärnänen et al., 2018; Rahman et al., 2018; Sprunt et al., 1978). The heightened instability of the preterm infant’s microbiome, coupled with their unique health status, makes this population particularly susceptible to the potential effects of colonizing bacteria and/or their products.
The objective of this study was to expand existing protocols within neonatal intensive care units (NICU) that aim to characterize the relationship between the intestinal microbiome and specified health outcomes in preterm infants. Thus, we hypothesized the following: The intestinal microbiome of preterm infants who develop adverse outcomes such as necrotizing enterocolitis (NEC), feeding intolerance, cognition, language, psychosocial, emotional, and adaptive behavior delays or impairments (as identified by the NICU Network Neurobehavioral Scale [NNNS] used to assess while the infant is inpatient, the Bayley Scales of Infant and Toddler Development and the Brief Infant–Toddler Social and Emotional Assessment (BITSEA) used during outpatient visits in the NICU follow-up clinic) will differ from the intestinal microbiome of preterm infants who do not develop adverse gut outcomes. Specific Aim 1: Determine how alterations in the intestinal microbiome may affect the risk of developing NEC, feeding intolerance, cognition, language, psychosocial, emotional, and adaptive behavior delays or impairments in preterm infants cared for in the NICUs. Specific Aim 2: Examine the interaction between the intestinal microbiome and host genetics in the regulation of intestinal health and cognition, language, psychosocial, emotional, and adaptive behavior delays or impairments.
Methods
Study Design
This prospective, longitudinal study recruited preterm infants < 32 weeks’ GA or weighing < 1,500 grams at birth, with follow-up for 3 weeks in the NICU. We collected follow-up data, including the NICU Neurobehavioral Stressor Scale at 35–37 weeks’ postmenstrual age (PMA) prior to NICU discharge, and follow-up clinic visits at 4, 8–12, and 18–24 months corrected age. This study has received institutional review board (IRB) approval from the clinical site locations.
Study Setting
The study participants were recruited from two Level III/IV NICUs in Connecticut with IRB approval. Infants were identified by NICU staff or research study personnel after birth and/or admission. Parents were contacted after birth by research personnel to assess interest. Informed consent was obtained from parents before enrollment for the study. If informed consent was not obtained prior to infant’s first stool, the stool sample is collected and stored until permission is acquired. If parents do not consent, the stool sample is discarded.
Sample Size Justification
The sample size was calculated using Power Analysis & Sample Size Software ([PASS], NCSS Statistical Software, 2021) based on the method of Cohen (1988). The sample size objective is to obtain completed data from 160 preterm infants, with the estimation of 20% attrition of the initial 200 enrollment. The outcome variable was a neurobehavioral response (NNNS–Stress subscale) and predictors including controlled variables, i.e., infant sex, birth GA, delivery type, severity of illness, and body weight, and tested independent variables, i.e., daily acute and chronic pain/stress mean scores, direct breastfeeding, and skin-to-skin contact, and mean α-diversity of gut microbiome (Cong et al., 2016; Sun et al., 2020). Multiple linear regression was fit to the data, and the R-square of 0.39 was obtained by fitting NNNS–Stress. After adjusting for the controlled variables, the tested independent variables contributed a partial R-square of 0.14 in fitting NNNS–Stress. Using the design values obtained from the pilot study, a sample size of 48 achieves 80% power to detect an R-squared of 0.14 attributed to the tested independent variables using an F–test with a 5% significance level, adjusting for the controlled variables with an R-squared of 0.39. While rules for calculating power and sample size in exome-sequencing and host-microbiome interaction studies is an area of ongoing research, we are evaluating critical parameters from the sequence data to assist in developing new advanced approaches, including examining the background rate of variation, sample distribution, allele frequencies, variant annotation, and missing data, as well as employing subsequent filters on the single nucleotide polymorphism (SNP) calls (Auer et al., 2016; Blekhman et al., 2015). We anticipate that a sample size of 160 infants with completed data will have adequate power to answer the hypotheses and aims of the study.
Procedures
A research coordinator was employed to facilitate the recruitment and follow-up by engaging parents during the infants’ NICU stay and scheduling follow-up visits. The primary variables measured during the NICU period include early life pain/stress events, gut microbiome, host genetic variations, and neurobehavioral assessment. During follow-up visits, gut microbiome, pain sensitivity, cognition, language, psychosocial, emotional, and adaptive behavior delays or impairments, growth parameters, and other health status and factors (described below) were measured at 4, 8–12, and 18–24-month GA (see Figures 1 and 2.). All samples were assigned a unique ID number, and research nurses systematically entered data into the Research Electronic Data Capture (REDCap) data collection system. The investigators crosschecked REDCap system and data against the clinical record in every five participants (Patridge & Bardyn, 2018).
Figure 1. Theoretical Framework and Study Aims.
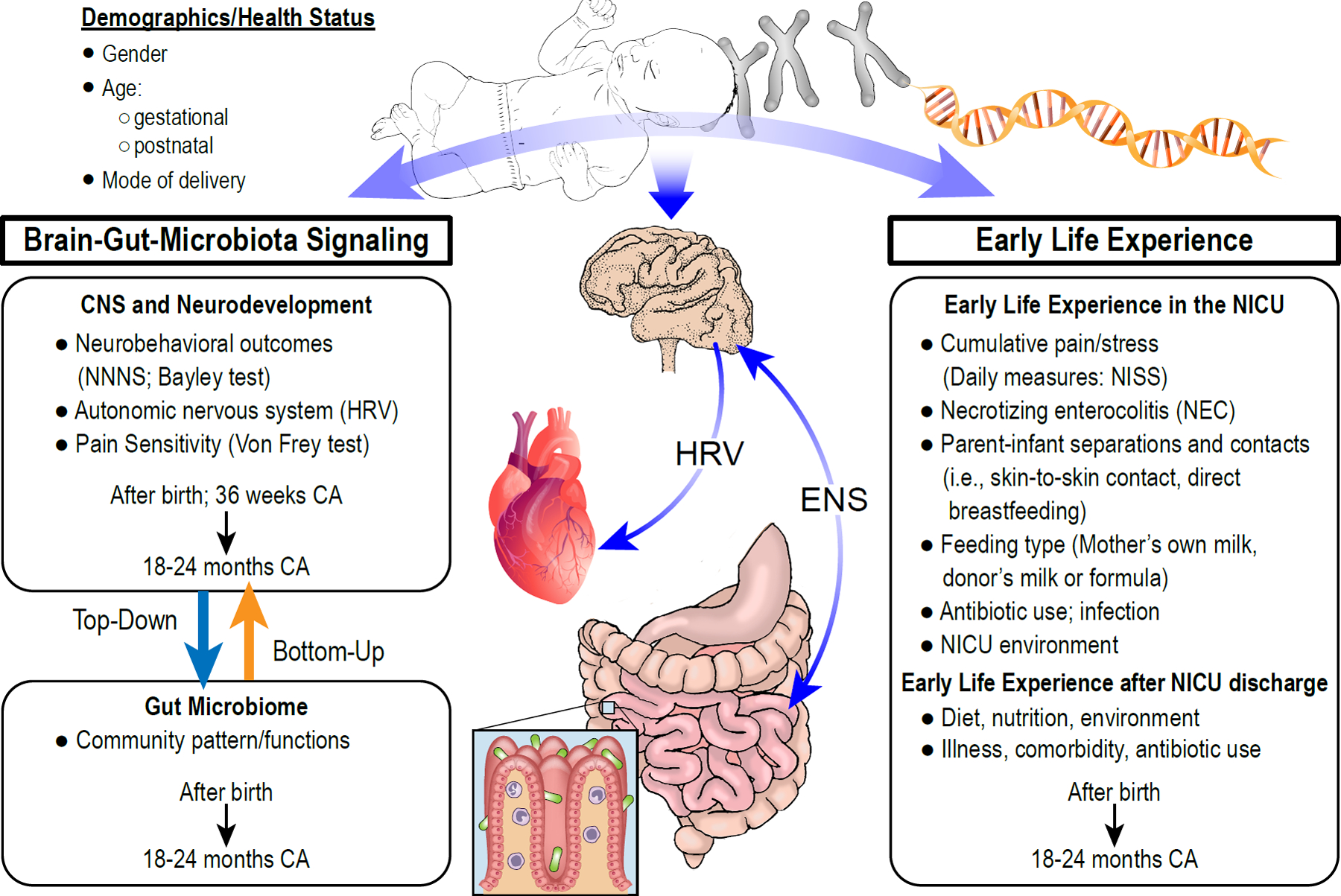
The framework incorporates the demographic, health status, and environmental factors that influence the interaction among early life experience and brain-gut-microbiota signaling. While host genotype-microbiota interactions are known to modulate the gut microbiome, the precise mechanisms leading to neurodevelopmental deficits will be thoroughly investigated in the study. Specific aims are to determine how alterations in the intestinal microbiome may impact infant health and neurodevelopmental outcomes and to examine the interaction between the intestinal microbiome and host genetics in the regulation of intestinal health and neurodevelopmental outcomes. NNNS=NICU Network Neurobehavioral Scale; HRV = heart rate variability; ENS = enteric nervous system; CA = corrected age; NISS = Neonatal Infant Stressor Scale; NEC = Necrotizing Enterocolitis.
Figure 2.
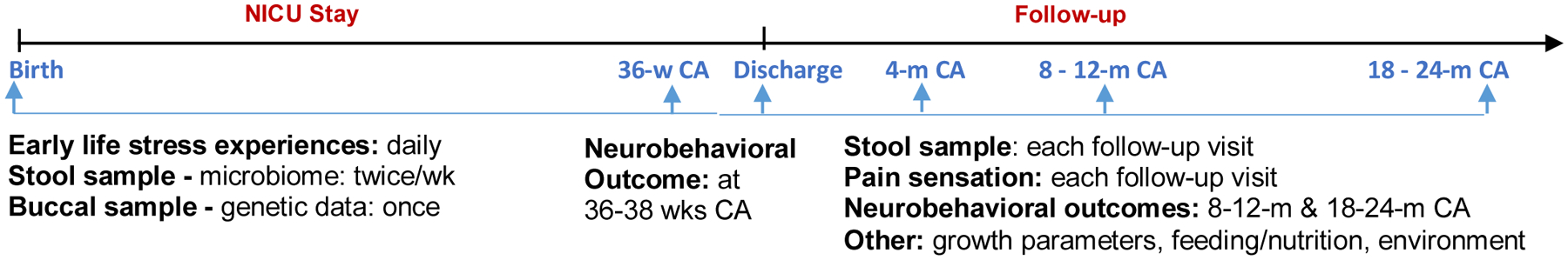
Data Collection Procedures. wk = week; m = month; CA = corrected age
As of February 14, 2020, among the 251 parents approached by the research coordinator, 214 signed the consent form, and 86 have completed all data collection time points of the study (inpatient and outpatient). Completion of the project will also help construct and maintain a neonatal specimen repository and clinical database(s), allowing specimen sharing, laboratory results, and clinical information to be available to other investigators interested in pursuing intestinal health research, thus, promoting collaboration related to microbiome and health outcomes of preterm infants.
Variables and Data Collection
Early Life Pain/Stress Experiences in the NICU
We collect data from infant medical records specifically focusing on cumulative pain/stress, parent–infant separation and contacts, comorbidities such as chronic lung disease, formula feeding, and antibiotic use.
Cumulative Pain/Stress Experiences:
We measure these experiences daily using the NICU Infant Stressor Scale (NISS; Newnham et al., 2009). The modified NISS designed by an expert panel consists of 68 acute and chronic procedures and interventions, ranging from diaper changes to intubation. We also collect these data from infant medical records.
Parent–Newborn Separation and Contacts
Our nursing staff record contact routinely in the infants’ medical records. They collect hr/min of breastfeeding, skin-to-skin contact with parents, or other types of contact such as touching and holding. This time record allows for quantification of stress which may result from parent–infant separation. Cong et al.’s (2017) pilot study developed a checklist to collect contact data, which has been shown to be valid and reliable (interrater reliability, r = 0.80–0.90).
Feedings
We measure feedings by recording the frequency and amount of mother’s own breastmilk (MOM), human donor milk (HDM), and formula feeding at each shift/daily through medical record review.
Antibiotics Use
We record antibiotic usage daily through medical record reviews. Type of antibiotics, dosage, and total days of use are documented. For analysis, we are calculating total days of antibiotic use and dosage.
Stool Sample Collection
Stool samples from each infant are collected twice weekly using sterile, disposable spatulas during diaper changes and then placed into a sterile specimen container (5 ml). Samples are immediately frozen upon collection at −80°C in the NICU, then transferred to a shared neonatal specimen biorepository and maintained at −80 °C until processing. After discharge from the NICU, parents collect stool samples at home using the researcher-provided microbiome collection kit, OMNIgene®●GUT (DNA Genotek, Ontario, Canada, cat. OMR-200). Parents are asked to bring the sample(s) to follow-up clinic appointments or mail in a research-provided prepaid envelope. The stool is then stored in a −80-degree freezer until analysis.
Samples and DNA Isolation Procedures
Bacterial DNA is isolated from stool samples by a protocol tested in preliminary experiments by Bomar et al. (2011) and Cong et al. (2016). According to the manufacturer’s instructions, total DNA is then isolated from ~0.25 g of stool using the MagAttract PowerSoil DNA EP Kit (Qiagen inc. ID: 27100–4-EP) on the Eppendorf epMotion liquid handling robot (epMotion 5075). Extracted DNA is quantified fluorometrically using a Synergy™ HT (BioTek, 2008) with the Quant-iT™ PicoGreen™ dsDNA Assay Kit (Molecular Probes, ThermoFisher Scientific, Eugene, OR, cat. P11495).
16S rRNA Gene-Based Analysis of Gut Microbiota
16S rRNA gene sequencing is the primary method of taking a microbial community census using the Illumina platform (Caporaso, et al, 2012). The V4 regions of bacteria within our extracted DNA are amplified with 515F and 806R primers containing Illumina adapters and Golay indices (Kozich, et al., 2013) and amplified in triplicate 15ul reactions using GoTaq (Promega, Madison, WI) with the addition of 10μg BSA (New England BioLabs, Ipswich, MA). Following amplification, PCR products are quantified and visualized using QIAxcel DNA Fast Analysis Kit (Qiagen, Hilden, Germany, cat. 929008), normalized based on the concentration of DNA in the 350–400 bp region, and pooled using the QIAgility liquid handling robot (Qiagen, 2018) at the university’s Microbial Analysis, Resources, and Services facility. The PCR products are purified using GeneRead Size Selection kit (Qiagen, Hilden, Germany, cat. Cat. ID: 180514) and paired-end sequencing (2×250) is performed on an Illumina MiSeq system. Using Mothur 1.42.3 pipeline (Schloss, et. al., 2009), operational taxonomic units (OTUs) are determined by clustering reads to the Greengenes reference 16S reference dataset (January 2018 release) at a 97% identity, and then performing de novo OTU clustering on reads that failed to cluster to a reference. Taxonomic annotation is also determined by the Greengenes reference 16S reference dataset. OTU tables, α-diversity, and β-diversity index calculated from Mothur process are then imported into R 3.6.2 for statistical analysis. The Shannon index and species of observation (SOBS) are calculated to measure the α-diversity index. The Bray-Curtis, Jaccard, and Theta YC dissimilarities index are calculated to present β-diversity of the bacterial community (Cong, et. al., 2016, 2017).
High-Throughput Shotgun Sequencing
Although bacteria are the main components of the human microbiome, eukaryotic microbes and viruses are also present in the human gastrointestinal tract. After DNA is isolated using the Nextera XT library prep kit (Illumina, FC-131–1096), the libraries are sequenced on an Illumina NovaSeq 6000® as it provides longer read lengths than the HiSeq®.
Buccal Swab Sample Collection and DNA Extraction
Infant buccal epithelial tissues are collected using a soft-tipped sterile swab on the interior of each cheek at least 30 min after a feeding or oral procedure. The swab is then placed into a cryo-vial for storage and stored in a −80°C freezer at Neonatal Specimen Repository at UConn Health Center until DNA can be extracted. Total genomic DNA (gDNA) is removed from buccal cells using the Gentra® Puregene® kit (Qiagen, 2014).
Whole Exome Sequencing (WES)
Genomic DNA is prepared for WES using the Illumina TruSeq Exome Capture (45Mb) library preparation kit following manufacturer’s protocol. Each library is multiplexed and sequenced with 100bp PE on the Illumina HiSeq® 4000 sequencer (Illumina, 2015) following recommendations for sequencing coverage. The resulting short reads will be subjected to quality control (Sickle), alignment to the genome (Burrow-Wheeler Aligner [BWA]), and subsequent SNP/Indel identification (Freebayes and GenomeAnalysisToolkit [GATK]).
Genetic Information Databases and Bioinformatic Tools for Screening
The SNP detection and insertion deletion-calling approaches are implemented on a high-performance computing resource. Available resources are used to filter the data. Nonsynonymous/splice acceptor and donor site/insertions or deletions in the Database of Single Nucleotide Polymorphisms ([dbSNP], https://www.ncbi.nlm.nih.gov/snp/), HapMap samples, and 1,000 Genome (https://www.internationalgenome.org) with a frequency higher than 0.5% are removed. Synonymous changes and noncoding changes are filtered using SIFT software (http://provean.jcvi.org/index.php). Sanger sequencing in the regions of interest is used to confirm the mutation from all the samples collected in the target genes.
Neurobehavioral Outcomes
These are measured using the NNNS (Lester & Tronick, 2004), a tool designed to assess infant neurobehavioral outcomes at 36–37 weeks postmenstrual age (PMA). The NNNS allows assessment of neuro-typical or at-risk preterm infant’s neurological integrity and behavioral function between 34- and 48-weeks PMA (Sullivan et al., 2012). The three main sections of NNNS are neurological function: tone and reflexes; behavior state, sensory, and interactive processes; and signs of stress/abstinence.
Follow-Up, Cognition, Language, Psychosocial, Emotional, and Adaptive Behavior Delays or Impairment Measures
Our trained assessors use the Bayley Scale of Infant Developmental test Edition III (Bayley, 2006) and the most recent test Edition IV (Bayley & Aylward, 2019) to assess children’s cognitive, language, and motor skills between ages 1 and 42 months. This is the most widely used, valid, and reliable assessment tool for developmental assessment of preterm infants (George et al., 2015; Velikos et al., 2015). BITSEA was used to screen study participants’ social emotion and behavioral status at 18–24 months of age (Briggs-Gowan et al., 2006). The BITSEA assesses social–emotional and behavioral problems and social–emotional competencies in children 1–3 years. Higher problem scores and lower competency scores than expected for age and gender indicate risk for behavioral concerns.
Pain Sensitivity.
Pain sensitivity is tested using the noninvasive measure of flexion withdrawal reflex (WR), which assesses spinal excitability, cutaneous sensitivity, and nociceptive response in infants. The von Frey filaments (vFF) (Touch- Test™ Sensory Evaluators, Stoelting, Wood Dale, IL, cat# 58011) are used to measure infants’ WR response in the study. The lowest response to the vFF is defined as the threshold of the reflex. A trained research assistant blinded to infant history conducts the test during follow-up visits.
Demographic Characteristics, Severity of Illness Score for Neonatal Acute Physiology– Perinatal Extension–II
After birth as well as occurrence of comorbidities, such as chronic lung disease, intraventricular hemorrhage, periventricular leukomalacia, and necrotizing enterocolitis, are collected to analyze for confounding variables. Severity of illness may affect the infants’ ability to respond to stress (Richardson et al., 2001). The data are obtained from medical records.
Clinical Data
Clinical data, including post-NICU discharge medical-surgical history, medications, and hospital readmissions are collected. Detailed nutrition history including breast milk, formula, and baby and table food intake information is collected; growth parameters including weight, length and head circumferences, and clinical diagnoses are collected.
Quality of the Child-Care Environment
This is measured using The Index of Child-Care Environment (ICCE), a screening questionnaire completed by the caregiver examines: (a) human stimulation, (b) avoidance of restriction, (c) social stimulation, and (d) social support to measure the environment in which the child is being reared (Sugisawa et al., 2010).
Data Analysis
Given the longitudinal, repeat measure nature of this study, both will be conducted within and between neonate comparisons. To address the first specific aim, differences in microbial species and diversities will be compared between groups (e.g., feeding intolerance vs. no feeding intolerance) and to determine changes in microbiota before and after an outcome of interest (e.g., NEC) using appropriate paired and unpaired tests. Principal component analysis (or similar dispersion analytic techniques) will be used to evaluate the dispersion of microbial populations between groups. Once optimal thresholds are identified, multivariable regression may be performed. To identify factors that will be retained in the final models, we will use parsimonious model-building techniques. Similar analyses will be conducted to determine optimal proportions of specific bacteria related to an outcome of interest, such as neurobehavioral development. To explore the second specific aim, host genetic variations will be associated with specific gut microbiome patterns and functions in the NICU and at follow-up visits by using functional linear regression model and adjusting potential covariates. The interaction between early life pain/stress, host genotypic profiles, and gut microbiome in the regulation of neurodevelopmental outcomes will be analyzed by joint time trajectory models with adjusting potential covariates. The potential covariates include but not limited to sex, feeding types, antibiotics use, and sample collection location.
Discussion
Preterm infants are subjected to numerous stressors, including cumulative painful and stressful events, maternal–infant separation, feeding intolerance, hypoxia, and infection, resulting in a high risk of neurodevelopmental deficits and chronic mental and metabolic illness later in life. It is increasingly accepted that gut microbiota plays a crucial role in human health; however, it is still a missing piece in unraveling the short- and long-term effect of early life stress on early programming of neuroimmune systems and neurobehavioral outcomes. Meanwhile, the interplay between the host genetics and microbial genomes has recently emerged, and as such, host-microbial genomics holds equivalent significance for infant development. Yet, these complex mechanisms remain largely unknown. Understanding how early life stress, host genetics, and gut microbiome shape brain and behavior is an extremely worthwhile new frontier in life and nursing science. By identifying the precise mechanisms that contribute to neurodevelopmental deficits, we anticipate that the knowledge gained from our study can lead to the development of novel biologically based assessment measures to identify infants at risk and targeted intervention strategies in this vulnerable population to further improve health in infancy and later life.
Conclusion
This protocol is aiding us in determining how alterations in the intestinal microbiome affect the risk of developing NEC, feeding intolerance, cognition, language, psychosocial, emotional, and adaptive behavior delays or impairments in preterm infants cared for in the NICUs. In addition, examining the interaction between the intestinal microbiome and host genetics in the regulation of intestinal health and cognition, language, psychosocial, emotional, and adaptive behavior delays or impairments might aid in developing interventions to mitigate negative sequelae.
Table 1:
Data Collection Time Points and Measurements
| Measurements | Time | Data / Variables |
|---|---|---|
| During NICU Hospitalization | ||
| Infant demographics and other characteristics | At enrollment | Chart: mixed data |
| Early life experience: Cumulative Pain/Stressor: NICU Infant Stressor Scale (NISS) | Chart review, daily | NISS scores: continuous data, does not include infant state as preterm infant states do not always align with term infant states (i.e. crying or not crying) |
| Early life experience: Feeding type: mother’s own milk, human donor milk, formula | Chart review, daily | Feeding type: categorical data |
| Early life experience: Antibiotic use | Chart review, daily | Days of antibiotic use: continuous data |
| Gut Microbiome: Stool samples: 16S rRNA gene; metagenomics and metabolomics | Twice / week | Microbial abundance, diversity, indicator species, functions: mixed data |
| Host genetic traits: buccal swab: whole exome sequencing | Once | Genetic variations, SNPs: mixed data |
| Neurodevelopment: NICU Network Neurobehavioral Scale (NNNS) | 36–37 wks CA | 13 subscales scores: continuous data, infant state taken into consideration |
| Follow-up Visits at 4, 8–12, and 18–24 month corrected age (CA) | ||
| Gut Microbiome: Stool samples: 16S rRNA gene; metagenomics and metabolomics | Each follow-up visit | Microbial abundance, diversity, indicator species: mixed data |
| Cognition, Language, Psycho-Social, Emotional and Adaptive Behavior: Bayley Scale of Infant Developmental III Test and BITSEA scale | 8–12 m & 18–24 m CA follow-up visits | 3 Bayley composite scores and 2 BITSEA composite scores: continuous data |
| Neurodevelopment: Pain sensitivity test – Von Frey test | Each follow-up visit | Pain threshold scores: continuous data, infant state taken into consideration |
| Other: Growth parameters, feeding and nutrition, parent interview / questionnaire. | Each follow-up visit | Mixed data |
| Other: Index of the Child Care Environment (ICCE), caregiver interview/questionnaire | 18–24 m CA follow-up visit | Mixed data |
Acknowledgements
Research reported in this publication was supported by the National Institute of Nursing Research of the National Institutes of Health under Award Number 5R01NR016928-02 (Cong, X.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
The Connecticut Children’s Medical Center Institutional Review Board provided consent, Approval No. 16–001. Written informed consent was obtained from parents of participants of the study. All subjects have been de-identified and given a subject ID number to ensure confidentiality.
The authors have no conflicts of interest to report.
Contributor Information
Ming-Hui Chen, University of Connecticut, Department of Statistics, Storrs, CT.
Angela Starkweather, University of Connecticut, School of Nursing, Storrs, CT.
Kendra Maas, University of Connecticut, Microbial Analysis, Resources, and Services, 181 Auditorium Road, Unit 3032, Storrs, CT.
Xiaomei S. Cong, University of Connecticut, School of Nursing, Storrs, CT.
References
- Auer PL, Reiner AP, Wang G, Kang HM, Abecasis GR, Altshuler D, Bamshad MJ, Nickerson DA, Tracy RP, Rich SS, NHLBI GO Exome Sequencing Project, & Leal SM (2016). Guidelines for large-scale sequence-based complex trait association studies: Lessons learned from the NHLBI exome sequencing project. American Journal of Human Genetics, 99, 791–801. 10.1016/j.ajhg.2016.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayley N (2006) Bayley Scales of Infant and Toddler Development - Third Edition. San Antonio, TX: Harcourt Assessment. [Google Scholar]
- Bayley N & Aylward GP (2019). Bayley Scales of Infant and Toddler Development – Fourth Edition. Bloomington, MN: Pearson NCS, Inc. [Google Scholar]
- Blekhman R, Goodrich JK, Huang K, Sun Q, Bukowski R, Bell JT, Spector TD, Keinan A, Ley RE, Gevers D. and Clark AG. (2015). Host genetic variation impacts microbiome composition across human body sites. Genome Biology, 16, 191. 10.1186/s13059-015-0759-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomar L, Maltz M, Colston S, & Graf J (2011). Directed culturing of microorganisms using metatranscriptomics. MBio, 2, e00012–11. 10.1128/mBio.00012-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs-Gowan MJ, & Carter AS (2006). BITSEA: Brief infant–toddler social and emotional assessment. Examiner’s manual. Harcourt Assessment. [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, & Knight R (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. The ISME journal, 6(8), 1621–1624. 10.1038/ismej.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke G, O’Mahony SM, Dinan TG, & Cryan JF (2014). Priming for health: Gut microbiota acquired in early life regulates physiology, brain and behavior. Acta Paediatrica, 103, 812–819. 10.1111/apa.12674 [DOI] [PubMed] [Google Scholar]
- Cohen J (1988). Statistical power analysis for the behavioral sciences (2nd ed.). Lawrence Erlbaum Associates. [Google Scholar]
- Cong X, Xu W, Janton S, Henderson WA, Matson A, McGrath JM, Maas K, & Graf J (2016). Gut microbiome developmental patterns in early life of preterm infants: Impacts of feeding and gender. PLoS ONE, 11, e0152751. 10.1371/journal.pone.0152751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong X, Wu J, Vittner D, Xu W, Hussain N, Galvin S, Fitzsimons M, McGrath JM, & Henderson WA (2017). The impact of cumulative pain/stress on neurobehavioral development of preterm infants in the NICU. Early Human Development, 108, 9–16. 10.1016/j.earlhumdev.2017.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George JM, Boyd RN, Colditz PB, Rose SE, Pannek K, Fripp J, Lingwood BE, Lai MM, Kong AHT, Ware RS, Coulthard A, Finn CM, & Bandaranayake SE (2015). PPREMO: A prospective cohort study of preterm infant brain structure and function to predict neurodevelopmental outcome. BMC Pediatrics, 15, 123. 10.1186/s12887-015-0439-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indrio F, Martini S, Francavilla R, Corvaglia L, Cristofori F, Mastrolia SA, Neu J, Rautava S, Russo Spena G, Raimondi F, & Loverro G (2017). Epigenetic matters: The link between early nutrition, microbiome, and long-term health development. Frontiers in Pediatrics, 5, 178. 10.3389/fped.2017.00178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozich JJ, Westcott SL, Baxter NT, Highlander SK, & Schloss PD (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Applied and Environmental Microbiology, 79(17), 5112–5120. 10.1128/AEM.01043-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester BM, & Tronick EZ (2004). The neonatal intensive care unit network neurobehavioral scale procedures. Pediatrics, 113, 641–667. [PubMed] [Google Scholar]
- NCSS Statistical Software. (2021). Power Analysis & Sample Size (PASS). https://www.ncss.com/software/pass/ [Google Scholar]
- Nelson MC, Morrison HG, Benjamino J, Grim SL, & Graf J (2014). Analysis, optimization and verification of Illumina-generated 16S rRNA gene amplicon surveys. PLoS ONE, 9, e94249. 10.1371/journal.pone.0094249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newnham CA, Inder TE, & Milgrom J (2009). Measuring preterm cumulative stressors within the NICU: The Neonatal Infant Stressor Scale. Early Human Development, 85, 549–555. 10.1016/j.earlhumdev.2009.05.002 [DOI] [PubMed] [Google Scholar]
- Pärnänen K, Karkman A, Hultman J, Lyra C, Bengtsson-Palme J, Larsson DGJ, Rautava S, Isolauri E, Salminen S, Kumar H, Satokari R, & Virta M (2018). Maternal gut and breast milk microbiota affect infant gut antibiotic resistome and mobile genetic elements. Nature Communication, 9, 3891. 10.1038/s41467-018-06393-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patridge EF, & Tania P Bardyn TP. (2018). Research Electronic Data Capture (REDCAP). Journal of the Medical Library Association, 106, 142–144. 10.5195/jmla.2018.319 [DOI] [Google Scholar]
- Rahman SF, Olm MR, Morowitz MJ, & Banfield JF (2018). Machine learning leveraging genomes from metagenomes identifies influential antibiotic resistance genes in the infant gut microbiome. mSystems, 3, e00123–17. 10.1128/mSystems.00123-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson DK, Corcoran JD, Escobar GJ, Lee SK (2001). SNAP-II and SNAPPE-II: Simplified newborn illness severity and mortality risk scores. Journal of Pediatrics, 138, 92–100. 10.1067/mpd.2001.109608 [DOI] [PubMed] [Google Scholar]
- Schippa S, & Conte MP (2014). Dysbiotic events in gut microbiota: Impact on human health. Nutrients, 6, 5786–5805. 10.3390/nu6125786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, & Weber CF (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied and Environmental Microbiology, 75(23), 7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprunt K, Leidy G, & Redman W (1978). Abnormal colonization of neonates in an intensive care unit: Means of identifying neonates at risk of infection. Pediatric Research, 12, 998–1002. 10.1203/00006450-197810000-00010 [DOI] [PubMed] [Google Scholar]
- Sugisawa Y, Shinohara R, Tong L, Tanaka E, Watanabe T, Onda Y, Kawashima Y, Hirano M, Tomisaki E, Mochizuki Y, Morita K, Gan-Yadam A, Yato Y, Yamakawa N, Anme T, & Japan Children’s Study Group. (2010). The trajectory patterns of parenting and the social competence of toddlers: A longitudinal perspective. Journal of Epidemiology, 20, S459–S465. 10.2188/jea.je20090172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan MC, Miller RJ, Fontaine LA, & Lester B (2012). Refining neurobehavioral assessment of the high-risk infant using the NICU Network Neurobehavioral Scale. Journal of Obstetric, Gynecologic & Neonatal Nursing, 41, 17–23. 10.1111/j.1552-6909.2011.01322.x [DOI] [PubMed] [Google Scholar]
- Sun Z, Xu W, Cong X, Li G, & Chen K (2020). Log-contrast regression with functional compositional predictors: Linking preterm infants’ gut microbiome trajectories to neurobehavioral outcome. Annals of Applied Statistics, 14, 1535–1556. 10.1214/20-AOAS1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velikos K, Soubasi V, Michalettou I, Sarafidis K, Nakas C, Papadopoulou V, Zafeiriou D, & Drossou V (2015). Bayley–III scales at 12 months of corrected age in preterm infants: Patterns of developmental performance and correlations to environmental and biological influences. Research in Developmental Disabilities, 45, 110–119. 10.1016/j.ridd.2015.07.014 [DOI] [PubMed] [Google Scholar]
- World Health Organization. (2018). Preterm Birth. https://www.who.int/news-room/fact-sheets/detail/preterm-birth.


