Abstract
In animal cells, replication-dependent histone mRNAs end with a highly conserved stem-loop structure followed by a 4- to 5-nucleotide single-stranded tail. This unique 3′ end distinguishes replication-dependent histone mRNAs from all other eukaryotic mRNAs, which end with a poly(A) tail produced by the canonical 3′-end processing mechanism of cleavage and polyadenylation. The pioneering studies of Max Birnstiel’s group demonstrated nearly 40 years ago that the unique 3′ end of animal replication-dependent histone mRNAs is generated by a distinct processing mechanism, whereby histone mRNA precursors are cleaved downstream of the stem-loop, but this cleavage is not followed by polyadenylation. The key role is played by the U7 snRNP, a complex of a ~60 nucleotide U7 snRNA and many proteins. Some of these proteins, including the enzymatic component CPSF73, are shared with the canonical cleavage and polyadenylation machinery, justifying the view that the two metazoan pre-mRNA 3′-end processing mechanisms have a common evolutionary origin. The studies on U7 snRNP culminated in the recent breakthrough of reconstituting an entirely recombinant human machinery that is capable of accurately cleaving histone pre-mRNAs, and determining its structure in complex with a pre-mRNA substrate (with 13 proteins and two RNAs) that is poised for the cleavage reaction. The structure uncovered an unanticipated network of interactions within the U7 snRNP and a remarkable mechanism of activating catalytically dormant CPSF73 for the cleavage. This work provides a conceptual framework for understanding other eukaryotic 3′-end processing machineries.
Keywords: 3′-end processing, U7 snRNP, cleavage and polyadenylation, CPSF73, endonuclease, exonuclease
Introduction
In eukaryotes, protein-coding genes are transcribed by RNA polymerase II (Pol II), giving rise to primary transcripts referred to as mRNA precursors (pre-mRNAs). A set of co-transcriptional processing/maturation steps is required to convert these nascent transcripts into mature and functional mRNAs, which are subsequently exported to the cytoplasm for their translation into proteins. Three major maturation steps are involved: modification of the 5′ end to generate a 7-methylguanosine (m7G) cap, splicing to remove introns and join exons, and 3′-end processing to form a mature 3′ end [1].
In animal cells, two independent mechanisms are used for 3′-end processing [2]. The canonical mechanism of cleavage coupled to polyadenylation operates on most pre-mRNAs, requiring a large machinery with many protein factors and yielding mRNAs containing a poly(A) tail at the 3′ end (Fig. 1A) [3–5]. Replication-dependent histone pre-mRNAs are the exception to this rule as their 3′ end is generated by the U7 snRNP, which cleaves histone pre-mRNAs but does not add a poly(A) tail [6, 7]. Each processing reaction depends on the presence of two sequence elements in its pre-mRNA substrates, with those for cleavage and polyadenylation consisting of a highly conserved upstream AAUAAA hexanucleotide or its variant and a loosely defined downstream G/U-rich sequence, with the cleavage/polyadenylation site located between the two elements, 10-15 nucleotides 3′ of the hexanucleotide. In replication-dependent histone pre-mRNAs, the upstream sequence element folds into a 6-base pair stem and 4-nucleotide loop (Fig. 1B). The histone downstream element (HDE) is more variable but contains a purine-rich core with the AAAGAA consensus. Cleavage occurs after the 4th (invertebrates) or 5th (vertebrates) nucleotide 3′ of the stem, producing mature histone mRNAs with the stem-loop followed by a single-stranded tail having the ACCA or ACCCA consensus sequence at the 3′ end.
Figure 1. Sequence elements in histone pre-mRNA required for 3′-end processing.
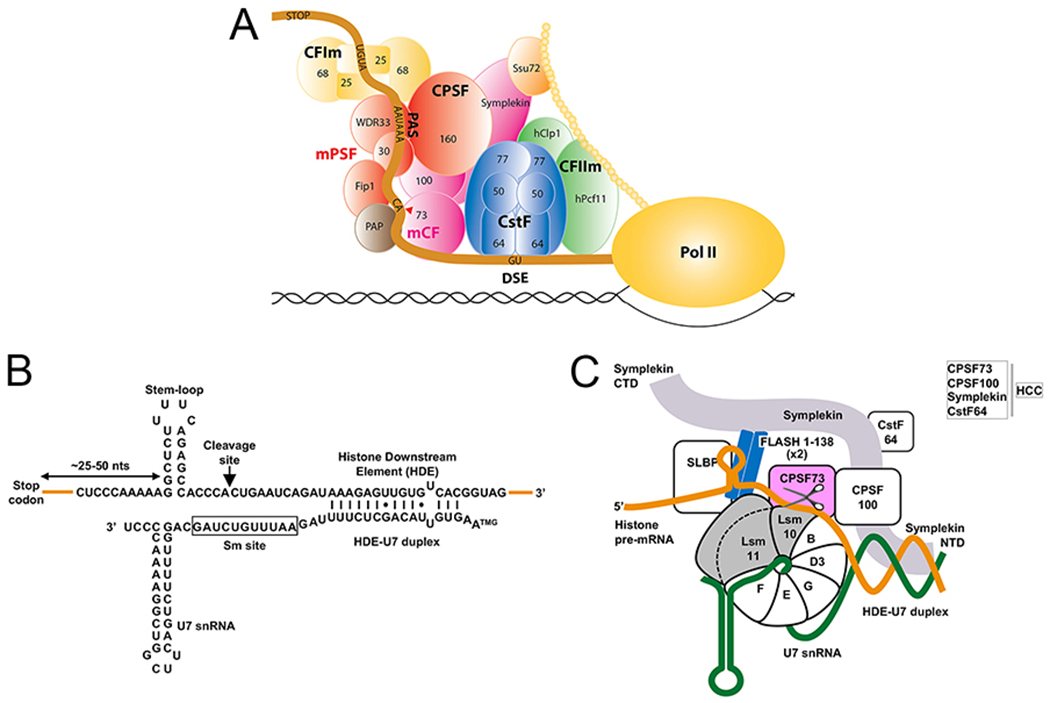
A. Schematic drawing of the mammalian canonical cleavage and polyadenylation machinery. Modified from [5]. B. Nucleotide sequence of mouse U7 snRNA and the 3′-end region of a mouse H2a histone pre-mRNA. Base pairing between the 5′ region of U7 snRNA and the HDE in histone pre-mRNA forms a duplex with twelve Watson-Crick base pairs (vertical lines), two G-U base pairs (dots) and one mismatch. TMG, trimethylguanosine cap. The unusual sequence of the Sm binding site in U7 snRNA is boxed. C. Schematic drawing of human U7 snRNP. U7 snRNA (thick green line) and the seven subunits of the Sm ring form the U7 Sm core. The U7-specific subunits of the ring are emphasized in gray. A coiled-coil dimer of the N-terminal region of FLASH interacts with the N-terminal region of Lsm11. CPSF73, the catalytic component of the histone pre-mRNA cleavage complex (HCC), is emphasized in light green. The histone pre-mRNA is in orange.
Since the two machineries recognize different pre-mRNA elements and only one of them contains the U7 snRNP, they were initially thought to lack shared components. However, similarities in the chemistry of the two cleavage reactions, including preferential cleavage after a CA dinucleotide, generation of a hydroxyl group at the 3′ end of the upstream cleavage product and the resistance of the activity to EDTA suggested otherwise, raising the possibility that the two machineries may use the same endonuclease [8, 9]. Indeed, at least three subunits of the cleavage and polyadenylation machinery, CPSF73, CPSF100 and symplekin, were shown to participate in 3′-end processing of histone pre-mRNAs (Fig. 1C). Among them, CPSF73 with its well-organized catalytic center formed at the interface of the metallo-β-lactamase and β-CASP domains [10, 11] was the primary candidate for the 3′ endonuclease in both machineries. However, the ultimate evidence for this function of CPSF73 was missing. It also remained unclear how CPSF73 finds its way to histone pre-mRNAs, which lack any sequence elements shared with the canonical mRNA precursors. Moreover, in the crystal structure of human CPSF73 alone, its catalytic center is in a closed conformation, lacking access for a single-stranded RNA substrate [11]. A similar observation was made in a study on the yeast ortholog of CPSF73, Ysh1 [12]. Thus, in both lower and higher eukaryotes CPSF73 would have to undergo a conformational rearrangement to become an active endonuclease.
In this review we will describe the spectacular recent breakthroughs in deciphering structural and functional aspects of the U7-dependent processing machinery and discuss how these findings advance our understanding of 3′-end processing of mRNA precursors in general.
Components of the U7-dependent processing machinery
The two sequence elements in histone pre-mRNAs are recognized by two different processing factors. The upstream stem-loop tightly interacts with the stem-loop binding protein (SLBP) (Fig. 1C) [13, 14], also known as the hairpin binding protein. Its centrally-located RNA binding domain (Fig. 2A) is highly conserved among evolutionarily distant organisms and is unique to SLBP orthologs. The HDE base pairs with the 5′ end of U7 snRNA (HDE-U7 duplex, Fig. 1B), a ~60-nucleotide RNA moiety of the U7 snRNP [15, 16]. Because the HDE sequence is not conserved, varying significantly among histone genes even within the same species, many histone pre-mRNAs can only form relatively short duplexes with the single functional U7 snRNA expressed in most organisms. SLBP functions in processing by facilitating the recruitment of the U7 snRNP to the pre-mRNA [17, 18]. SLBP is dispensable for in vitro processing of histone pre-mRNAs with extensive complementarity to the U7 snRNA, suggesting that it plays an auxiliary role and is not essential for the cleavage reaction. Following cleavage, SLBP remains associated with the mature 3′ end where it is accompanied by 3′hExo, a 3′-5′ exonuclease [19]. The two proteins form an intimate complex with the mature 3′ end [13] and likely cooperate in controlling translation and degradation of histone mRNAs during the cell cycle [20, 21].
Figure 2. Overall structure of an active human histone pre-mRNA 3′-end processing machinery.
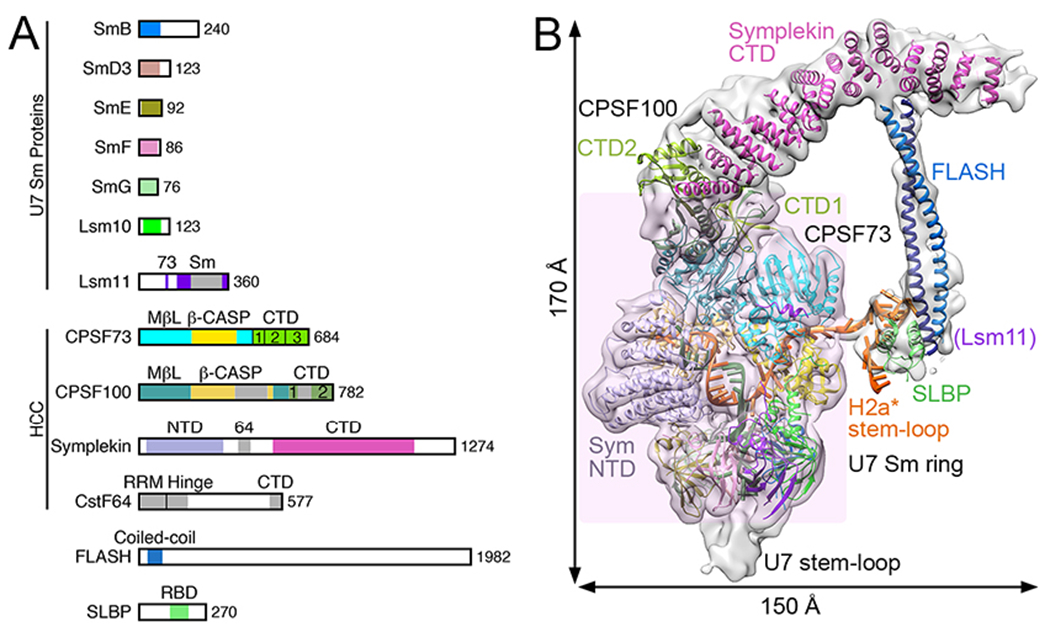
A. Domain organization of proteins in human U7 snRNP and SLBP. The domains are given different colors and named. FLASH is not drawn to scale. B. Overall structure of the reconstituted, active human U7 snRNP. The core of the U7 machinery is highlighted by a transparent pink background, while the other components are in the periphery. The overall dimensions of the machinery are also indicated. The proteins are colored as in panel A.
Near its central region, U7 snRNA contains a specific Sm binding site that differs in a few positions from the spliceosomal consensus and is also longer, encompassing 11 rather than 9 nucleotides (Fig. 1B) [22–24]. This site promotes the assembly of an unusual Sm ring around the U7 snRNA in which SmB, SmD3, SmE, SmF and SmG are shared with the spliceososmal snRNPs. The two remaining subunits, Lsm10 and Lsm11, are specific for the U7 snRNP (Fig. 1C), replacing spliceosomal SmD1 and SmD2 [25–28]. Of the two unique subunits of the ring, Lsm11 contains an unusually long N-terminal region (Fig. 2A) that mediates an essential function in processing [29–31].
U7 snRNP is limiting in cells, existing at a concentration ~100-fold lower than that of U1 snRNP, making its purification from biological sources challenging [25]. Indeed, initial biochemical approaches to isolate endogenous U7 snRNP from tissue culture cells were unsuccessful, failing to identify additional components, likely due to their loose association with the particle and/or sensitivity to proteolytic degradation [25–28]. As a result, U7 snRNA stably bound to the Sm ring (referred to as U7 Sm core here) remained for many years the only known components of U7 snRNP. Clearly, this simple composition was insufficient to explain the role of U7 snRNP in cleaving histone pre-mRNAs.
Further progress in determining the composition of U7 snRNP was facilitated by using mammalian and Drosophila cell lines and nuclear extracts, and a combination of various biochemical and genetic approaches, including in vitro processing [32, 33], UV crosslinking [34], complementation assays [35], yeast two-hybrid cloning [36], RNAi screens [37] and single-step affinity purification schemes [38, 39]. Five proteins were added to the list of components of U7 snRNP in both vertebrates and invertebrates (Fig. 1C), with FLASH, a protein of ~2,000 amino acid residues (Fig. 2A), being the only component uniquely associated with 3′-end processing of histone pre-mRNAs. The four remaining proteins, CPSF73, CPSF100, symplekin and CstF64, were previously identified as components of the canonical cleavage and polyadenylation machinery, supporting the notion that the two 3′-end processing machineries for mRNA precursors in metazoans are evolutionarily linked. The four subunits are stably associated with each other, forming the histone pre-mRNA cleavage complex (HCC, Fig. 1C) [40]. The HCC is equivalent to the mammalian cleavage factor (mCF) in the canonical machinery (Fig. 1A) [41, 42], although mCF lacks CstF64. CPSF73 was detected by UV crosslinking in the vicinity of the scissile bond in histone pre-mRNA [34], consistent with its role as the catalytic component in U7-dependent processing.
The N-terminal ~150 amino acid residues of FLASH form a coiled-coil dimer (Fig. 2A) [31] and interact with the essential N-terminal region of Lsm11, creating a platform that recruits the HCC to the U7 snRNP, thereby converting U7 Sm core to a catalytically active processing factor (Fig. 1C). The same region of FLASH is also required for the SLBP-mediated recruitment of U7 snRNP to histone pre-mRNA [43]. It is not known whether FLASH and SLBP directly interact, or SLBP makes a contact with the FLASH-Lsm11 complex rather than FLASH alone. Previously, the recruitment of U7 snRNP by SLBP was attributed to a presumptive U7 snRNP component, ZFP100 [44–46]. However, this protein of 100 kDa and containing 18 C2H2 zinc fingers was never detected in active preparations of endogenous U7 snRNP and it may instead be involved in transcription of histone genes and only transiently bind to U7 snRNP [39]. Beside the N-terminal region, no other part of FLASH plays a direct role in processing, although its extreme C-terminus interacts with NPAT [47, 48], a universal transcriptional regulator of histone gene expression [49, 50]. Altogether, these observations demonstrate that FLASH is critical for the assembly of an active U7 snRNP, its recruitment to histone pre-mRNA through SLBP, and the proper coordination of histone gene transcription and 3′-end processing of the resultant histone pre-mRNAs.
As a result of these new findings, the U7 snRNP emerged as an RNA-guided multi-subunit endonuclease in which U7 snRNA through its 5′ end identifies the pre-mRNA substrate; FLASH and Lsm11 recruit the HCC; and CPSF73 cleaves the substrate (Fig. 1C) [51]. What remained unknown was whether the identified subunits are sufficient to support 3′-end processing activity in vitro (or perhaps some essential component(s) of the machinery were overlooked), how they function and interact with each other in the machinery, and finally how CPSF73 carries out its task given that its catalytic site is normally inaccessible to RNA [11]. Remarkably, many of these critical questions were answered by recent structural and functional studies with an active histone pre-mRNA processing complex reconstituted from 13 recombinant proteins and 2 synthetic RNAs [52, 53].
An active recombinant U7 snRNP
The reconstitution of U7 Sm core was achieved recently [52], by following the protocols developed for the spliceosomal snRNP cores [54–56]. In the presence of N-terminal segment of FLASH [31], recombinant U7 Sm core interacted with endogenous HCC from nuclear extracts, yielding semi-recombinant U7 snRNP that was capable of accurately cleaving histone pre-mRNAs [52]. The reconstitution of a fully recombinant machinery was enabled by the production of HCC or mCF, originally for studies on the canonical machinery [57]. Importantly, a mixture containing reconstituted human U7 Sm core, human HCC and human FLASH (coiled-coil region) accurately and efficiently cleaved a model H2a pre-mRNA substrate (H2a*) in the presence SLBP, demonstrating that the reconstituted human U7 snRNP is active and no other critical components are missing from the machinery [53]. A mutation in the active site of CPSF73 abolished the cleavage, convincingly showing that it is the endonuclease for the reaction. The relative ease of making various changes within the HCC and the U7 Sm core provided a previously unavailable opportunity of testing individual components and regions for their importance in processing. Using this approach, CstF64 was shown to be dispensable for the activity of recombinant U7 snRNP [53], although it may play a role in processing in vivo [58]. CstF64 is a subunit of the cleavage stimulation factor (CstF) in the canonical machinery and plays an essential role in this machinery by recognizing the G/U-rich downstream sequence element (DSE) (Fig. 1A).
A surprising discovery from these studies is that the N-terminal domain (NTD) of symplekin is required for the cleavage activity [53]. Even more surprisingly, the NTD supported processing when added to the reaction in trans, as a separate protein that is unable to stably bind to the rest of the machinery [53, 59]. The NTD is known to form a stable complex with Ssu72 and stimulate its Pol II CTD phosphatase activity [60]. It was found that Ssu72 is an inhibitor of histone pre-mRNA 3′-end processing in vitro, especially when the symplekin NTD is provided in trans [53]. The negative regulation of U7-dependent processing by Ssu72 has also been shown in vivo [61], suggesting that Ssu72 is a bona fide regulator of histone pre-mRNA 3′-end processing.
CPSF73 is both an endonuclease and a 5′-3′ exonuclease
Endonucleolytic cleavage of histone pre-mRNA by CPSF73 generates two fragments: the upstream product ending with the stem-loop that corresponds to the mature histone mRNA and the downstream cleavage product (DCP) containing the HDE. The DCP is degraded in vitro by a 5′-3′ exonuclease, likely liberating U7 snRNP from the HDE for a subsequent round of processing [62–64]. UV crosslinking identified this exonuclease as CPSF73 [64], suggesting that the single catalytic center of CPSF73 supports both endonucleolytic and 5′-3′ exonucleolytic activities, as previously demonstrated for other nucleases of the β-CASP family [8].
That the 5′-3′ exonuclease activity of the U7 snRNP is indeed provided by CPSF73 was directly demonstrated using recombinant U7 snRNP and a set of specifically designed substrates corresponding to the DCP [53, 59]. As in the case of the endonucleolytic cleavage, the 5′-3′ degradation of the DCP by the recombinant U7 snRNP was abolished by mutating two key amino acid residues of the catalytic center of CPSF73 and depended on both the presence of symplekin NTD and the ability of U7 snRNA to base pair with the substrate. The 5′-3′ exonuclease activity of CPSF73 is likely to also degrade the DCP of histone pre-mRNAs in vivo, ultimately resulting in transcription termination on histone genes, as proposed in the torpedo model, substituting for Xrn2 that plays the equivalent functions in cleavage and polyadenylation [64–68].
In addition to acting on RNA substrates, recombinant U7 snRNP through the 5′-3′ exonuclease of CPSF73 is capable of degrading short stretches of deoxynucleotides inserted into RNA substrates, although with lower efficiency. Finally, recombinant U7 snRNP also acts as a site-specific endonuclease on single-stranded DNA substrates. Further studies are required to determine whether these DNA-specific activities of CPSF73 play any role under in vivo conditions.
Structure of an active U7 snRNP poised for the cleavage reaction
The structure of the active reconstituted U7 snRNP together with SLBP and the H2a* substrate is the long-awaited breakthrough in the field, revealing for the first time the overall architecture of an active pre-mRNA 3′-end processing machinery [53]. Moreover, the H2a* substrate is captured in the active site of CPSF73, poised for the cleavage, and therefore the structure also provides detailed molecular insights into the endonuclease reaction.
The Sm ring and most of HCC form the core of the U7 machinery, with a stable structure (Fig. 2B). In comparison, the C-terminal domain (CTD) of symplekin, FLASH, SLBP, and the stem-loop are located in the periphery of the machinery. They are more flexible and their structure cannot be defined in as much detail. The coiled-coil dimer of FLASH interacts with the CTD of symplekin, revealing a major mechanism for how HCC is recruited to the U7 Sm core. FLASH additionally interacts with SLBP and possibly Lsm11 N-terminal extension, although SLBP is not required for cleavage of the H2a* substrate whose HDE has been modified to form 15 consecutive Watson-Crick base pairs with the U7 snRNA [53]. Residues 107-118 in the N-terminal extension of Lsm11 contact the metallo-β-lactamase domain of CPSF73 (Fig. 2B), showing another interaction for this extension of Lsm11. CstF64 is disordered, and it is not required for the cleavage reaction in vitro either [53].
HCC contacts one face of the Sm ring, while the other face accommodates the 3′-end stem-loop of the U7 snRNA (Figs. 1B, 2B). This stem-loop is also flexible in structure in this machinery, and it is not required for the cleavage reaction [52]. The overall structure of the Sm ring is similar to those of spliceosomal snRNPs, with the U7 snRNA Sm site threaded through the center of the ring. However, the Sm site assumes a different conformation, including a base pair between its C8 and G11 bases, which may play an important role in incorporating Lsm10 and Lsm11 into the unique U7 snRNP.
CPSF73 is in an open conformation, with the H2a* substrate captured in its active site (Fig. 3A), providing for the first time direct insights into the catalysis by this enzyme. The scissile phosphate directly coordinates both zinc ions in the active site, and is in the correct position for nucleophilic attack by the bridging water/hydroxide ligand of the zinc ions to initiate the cleavage (Fig. 3B). Therefore, the machinery is poised for the cleavage reaction in the structure, and the reaction did not proceed in the sample used for structural studies because it was kept at 4 °C during preparation.
Figure 3. Substrate binding and catalytic mechanism of CPSF73.
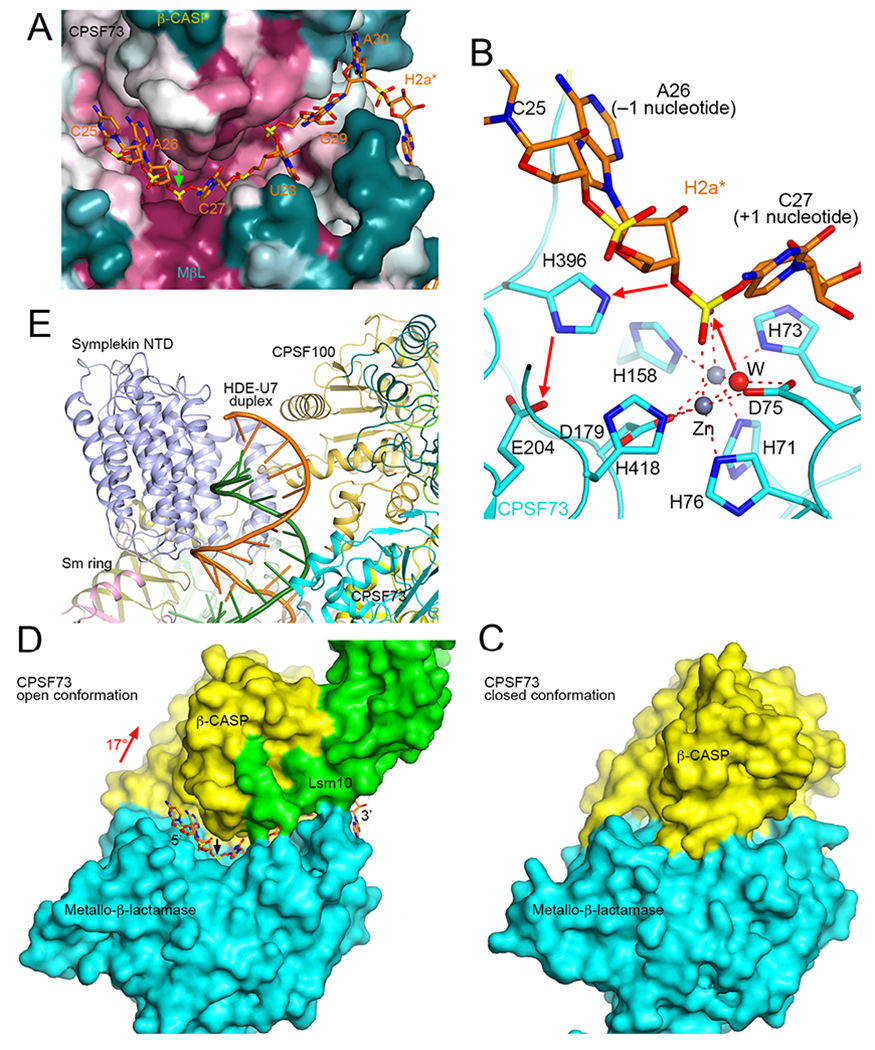
A. A canyon in the active site region of CPSF73 for binding the single-stranded pre-mRNA substrate (orange sticks). The molecular surface of CPSF73 is colored based on sequence conservation, with purple being highly conserved and dark cyan being poorly conserved [76]. B. Substrate binding mode and catalytic mechanism of CPSF73. The bridging water/hydroxide initiates the nucleophilic attack, and the 3′ oxyanion leaving group is protonated by His396, which is activated by Glu204 (red arrows). After the reaction, the upstream product carries a 3′ hydroxyl group, while the downstream cleavage product has a 5′ phosphate. C. Molecular surface of CPSF73 in a closed conformation. D. Molecular surface of CPSF73 in an open conformation. Lsm10 is also shown (green). The rotation in the β-CASP domain to achieve the open conformation is indicated. E. The HDE-U7 duplex is surrounded by symplekin NTD, CPSF100 and CPSF73.
Compared to the structure of CPSF73 alone in a closed conformation, there is a 17° rotation for its β-CASP domain, which creates a canyon between the metallo-β-lactamase and β-CASP domains that can accommodate the single-stranded pre-mRNA substrate (Figs. 3A, 3C). This canyon does not exist in the closed conformation (Fig. 3D). This large conformational change that is required for CPSF73 activation is mediated by contacts with the N- and C-terminal extensions of Lsm10 (Fig. 3C). These extensions are much shorter than the N-terminal extension of Lsm11, but their removal greatly reduced processing activity, and in fact led to cleavage at a different site in the substrate [53]. These observations suggest that the interaction between Lsm10 and CPSF73 is not only required for the catalytic activation of CPSF73 but also for its proper positioning relative to the pre-mRNA substrate and other components of U7 snRNP.
The primary trigger for the activation of CPSF73 is the recognition of the HDE-U7 duplex, which is surrounded by symplekin NTD, CPSF100, and CPSF73 of the HCC (Fig. 3E). This also explains why symplekin NTD is required for the cleavage. The recognition of the duplex by the NTD leads to a conformational rearrangement in the HCC, as compared to the structure of HCC/mCF alone [57], which in turn triggers the activation of CPSF73. Ssu72 inhibits the cleavage reaction in vitro as its binding site in the symplekin NTD directly overlaps with that of the duplex.
Conclusions
The structure of the reconstituted, active U7 snRNP captures it in a state poised for the cleavage reaction. The structural and functional observations also provide insights into how the U7 snRNP could be assembled from its components, U7 Sm core, FLASH and HCC, and how recognition of the histone pre-mRNA triggers conformational rearrangements in the HCC and the activation of CPSF73 for the cleavage reaction (Fig. 4). After the cleavage, the DCP is degraded through the 5′-3′ exonuclease activity of CPSF73. Overall, the machinery is highly dynamic structurally, and the recognition of the substrate allows the machinery to become stable (possibly transiently) and achieve the active state.
Figure 4. Function of human U7-dependent processing machinery.
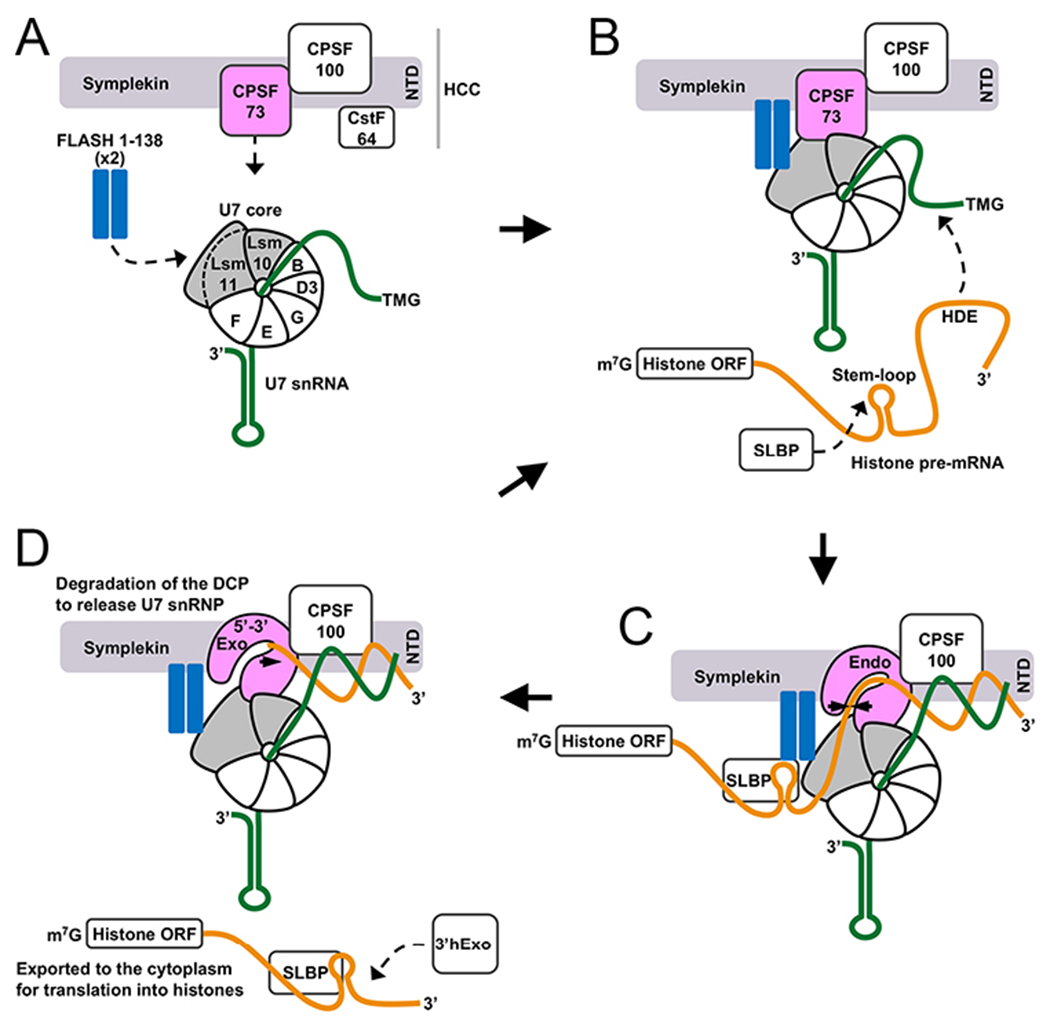
A. Assembly of human U7 snRNP. Arrows indicate binding sites for the FLASH dimer and HCC on the U7 Sm core. B. Formation of a complex between FLASH dimer and Lsm11 results in the recruitment of the HCC, with FLASH contacting symplekin, and CPSF73 contacting Lsm10 and Lsm11. C. The duplex formed between the HDE of histone pre-mRNA and U7 snRNA is recognized by the symplekin NTD, which transmits a structural signal to other components of the machinery, ultimately resulting in opening of the substrate canyon in CPSF73 and pre-mRNA binding and cleavage. D. The mature 3′ end in histone mRNA remains associated with SLBP and additionally interacts with 3′hExo. The mRNP complex is exported to the cytoplasm for translation. The downstream cleavage product (DCP) is degraded by the 5′-3′ exonuclease activity of CPSF73 to free U7 snRNP for another round of processing.
An unanticipated finding from the structural and functional studies of the active recombinant machinery is that the catalytic activation of CPSF73 critically depends on the recognition of the HDE-U7 duplex by the symplekin NTD. The formation of a duplex between the two RNAs is a clear indication that a genuine substrate has been recruited and symplekin acts as a sensor of this event, transmitting the signal to the rest of processing machinery and ultimately triggering the conversion of the dormant endonuclease into an active enzyme. This mechanism can be viewed as an important quality control step, ensuring cleavage of only legitimate substrates and preventing undesired activity on random RNAs that are unable to form stable duplexes with the U7 snRNA. Following cleavage of histone pre-mRNA, CPSF73 uses its 5′-3′ exonuclease activity to degrade the DCP, including the region that base pairs with the U7 snRNA, hence removing the duplex RNA that is critical in the activation mechanism. Thus, once catalytically activated as an endonuclease, CPSF73 is likely to maintain its active state for the 5′-3′ exonuclease degradation of the DCP, consistent with the processive nature of this activity.
Since the discovery of Lsm10 as one of the two U7-specific ring proteins, its role in 3′-end processing of histone pre-mRNAs remained mysterious [26–28]. The structure of the active machinery demonstrates critical roles of this subunit in the activation of CPSF73 for processing [53], even though its N- and C-terminal extensions are much shorter than the N-terminal extension of Lsm11 (Fig. 2A).
When CPSF73 was UV crosslinked to the scissile bond in histone pre-mRNA during U7-dependent processing [34], the emerging view was that the U7 and canonical machineries use the same endonuclease for cleavage but differ in the mechanism of its recruitment to the substrate [69, 70]. The demonstration that symplekin NTD is required for histone pre-mRNA 3′-end processing and that Ssu72 inhibits this processing suggests a more complex picture. In the yeast canonical machinery, the roles of the two proteins are reversed, with the NTD of Pta1 (the yeast ortholog of symplekin) acting as an inhibitor of the cleavage reaction, and Ssu72 relieving the inhibitory effect of the NTD [71]. In addition, downregulation of Ssu72 in chicken cells reduces cellular concentration of polyadenylated mRNAs [61], suggesting that this aspect of cleavage and polyadenylation is conserved between lower and higher eukaryotes. It will be important to determine how higher eukaryotes control the activity of CPSF73 in the canonical machinery and determine exact roles of symplekin NTD and Ssu72 in this process.
Perspectives.
3′-end processing is a crucial step for the maturation of mRNAs. In metazoans, the canonical and U7 machineries share the same endonuclease, CPSF73, for the cleavage step of the processing. IntS11, a closely related homolog of CPSF73 and a component of Integrator, is the endonuclease that cleaves snRNA precursors and some nascent mRNAs [72–75], highlighting the evolutionary conservation of RNA 3′-end processing mechanisms that operate on RNA Pol II transcripts.
The structure of the active histone pre-mRNA 3′-end processing machinery has provided the first insights into how an active machinery is organized, how CPSF73 is activated for cleavage, and how this endonuclease binds the pre-mRNA and catalyzes the cleavage reaction.
It will be important to determine the structures of the canonical machinery and Integrator in the active state, showing how they are activated and recognize and cleave their RNA substrates. Another focus of future studies should be on determining how the symplekin NTD and Ssu72 act in opposition to each other to control the activity of the canonical and U7 machineries during co-transcriptional 3′-end processing.
Acknowledgments.
We thank Y. Sun, Y. Zhang, W.S. Aik, X.-C. Yang, W.F. Marzluff and T. Walz for their contributions to the studies on the U7 machinery. We also thank our other colleagues and collaborators for their continuing help and input. This research is supported by NIH grants R35GM118093 (to LT) and R01GM029832 (to ZD and W.F. Marzluff).
Footnotes
Conflict of Interests. The authors declare that there are no conflicts of interest.
References
- 1.Bentley DL. Coupling mRNA processing with transcription in time and space. Nat Rev Genet. 2014;15(3):163–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu H, Moore CL. On the Cutting Edge: Regulation and Therapeutic Potential of the mRNA 3′ End Nuclease. Trends Biochem Sci. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mandel CR, Bai Y, Tong L. Protein factors in pre-mRNA 3′-end processing. Cell Mol Life Sci. 2008;65:1099–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi Y, Manley JL. The end of the message: multiple protein-RNA interactions define the mRNA polyadenylation site. Genes Develop. 2015;29:889–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun Y, Hamilton K, Tong L. Recent molecular insights into canonical pre-mRNA 3′-end processing. Transcription. 2020;11:83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dominski Z, Marzluff WF. Formation of the 3′ end of histone mRNA. Gene. 1999;239(1):1–14. [DOI] [PubMed] [Google Scholar]
- 7.Dominski Z, Marzluff WF. Formation of the 3′ end of histone mRNA: getting closer to the end. Gene. 2007;396:373–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dominski Z. Nucleases of the metallo-b-lactamase family and their role in DNA and RNA metabolism. Crit Rev Biochem Mol Biol. 2007;42:67–93. [DOI] [PubMed] [Google Scholar]
- 9.Dominski Z. The hunt for the 3′ endonuclease. Wiley Interdiscip Rev RNA. 2010;1(2):325–40. [DOI] [PubMed] [Google Scholar]
- 10.Callebaut I, Moshous D, Mornon J-P, de Villartay J-P. Metallo-b-lactamase fold within nucleic acids processing enzymes: the b-CASP family. Nucl Acid Res. 2002;30:3592–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, et al. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature. 2006;444:953–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hill CH, Boreikaite V, Kumar A, Casanal A, Kubik P, Degliesposti G, et al. Activation of the endonuclease that defines mRNA 3′ ends requires incorporation into an 8-subunit core cleavage and polyadenylation factor complex. Mol Cell. 2019;73:1217–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan D, Marzluff WF, Dominski Z, Tong L. Structure of histone mRNA stem-loop, human stem-loop binding protein, and 3′hExo ternary complex. Science. 2013;339:318–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Tan D, DeRose EF, Perera L, Dominski Z, Marzluff WF, et al. Molecular mechanisms for the regulation of histone mRNA stem-loop-binding protein by phosphorylation. Proc Natl Acad Sci USA. 2014; 111:E2937–E46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schaufele F, Gilmartin GM, Bannwarth W, Birnstiel ML. Compensatory mutations suggest that base-pairing with a small nuclear RNA is required to form the 3′ end of H3 messenger RNA. Nature. 1986;323:777–81. [DOI] [PubMed] [Google Scholar]
- 16.Bond UM, Yario TA, Steitz JA. Multiple processing-defective mutations in a mammalian histone pre-mRNA are suppressed by compensatory changes in U7 RNA both in vivo and in vitro. Genes Develop. 1991;5:1709–22. [DOI] [PubMed] [Google Scholar]
- 17.Streit A, Koning TW, Soldati D, Melin L, Schumperli D. Variable effects of the conserved RNA hairpin element upon 3′ end processing of histone pre-mRNA in vitro. Nucl Acids Res. 1993;21:1569–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dominski Z, Zheng LX, Sanchez R, Marzluff WF. Stem-loop binding protein facilitates 3′-end formation by stabilizing U7 snRNP binding to histone pre-mRNA. Mol Cell Biol. 1999;19:3561–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dominski Z, Yang X-C, Kaygun H, Dadlez M, Marzluff WF. A 3′ exonuclease that specifically interacts with the 3′ end of histone mRNA. Mol Cell. 2003;12:295–305. [DOI] [PubMed] [Google Scholar]
- 20.Sànchez R, Marzluff WF. The stem-loop binding protein is required for efficient translation of histone mRNA in vivo and in vitro. Mol Cell Biol. 2002;22(20):7093–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoefig KP, Rath N, Heinz GA, Wolf C, Dameris J, Schepers A, et al. Eril degrades the stem-loop of oligouridylated histone mRNAs to induce replication-dependent decay. Nat Struct Mol Biol. 2013;20(1):73–81. [DOI] [PubMed] [Google Scholar]
- 22.Gilmartin GM, Schaufele F, Schaffner G, Birnstiel ML. Functional analysis of the sea urchin U7 small nuclear RNA. Mol Cell Biol. 1988;8:1076–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grimm C, Stefanovic B, Schumperli D. The low abundance of U7 snRNA is partly determined by its Sm binding site. EMBO J. 1993;12:1229–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stefanovic B, Wittop Koning TH, Schümperli D. A synthetic histone pre-mRNA-U7 small nuclear RNA chimera undergoing cis cleavage in the cytoplasm of Xenopus oocytes. Nucleic Acids Res. 1995;23(16):3152–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith HO, Tabiti K, Schaffner G, Soldati D, Albrecht U, Birnstiel ML. Two-step affinity purification of U7 small nuclear ribonucleoprotein particles using complementary biotinylated 2′-O-methyl oligoribonucleotides. Proc Natl Acad Sci U S A. 1991;88(21):9784–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pillai RS, Will CL, Luhrmann R, Schumperli D, Muller B. Purified U7 snRNAs lack the Sm proteins D1 and D2 but contain Lsm10, a new 14 kDa Sm D1-like protein. EMBO J. 2001;20:5470–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pillai RS, Grimmler M, Meister G, Will CL, Luhrmann R, Fischer U, et al. Unique Sm core structure of U7 snRNPs: assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes Develop. 2003;17:2321–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schumperli D, Pillai RS. The special Sm core structure of the U7 snRNP: far-reaching significance of a small nuclear ribonucleoprotein. Cell Mol Life Sci. 2004;61:2560–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burch BD, Godfrey AC, Gasdaska PY, Salzler HR, Duronio RJ, Marzluff WF, et al. Interaction between FLASH and Lsm11 is essential for histone pre-mRNA processing in vivo in Drosophila. RNA. 2011;17:1132–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang X-C, Xu B, Sabath I, Kunduru L, Burch BD, Marzluff WF, et al. FLASH is required for the endonucleolytic cleavage of histone pre-mRNAs but is dispensable for the 5′ exonucleolytic degradation of the downstream cleavage product. Mol Cell Biol. 2011;31:1492–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aik WS, Lin MH, Tan D, Tripathy A, Marzluff WF, Dominski Z, et al. The N-terminal domains of FLASH and Lsm11 form a 2:1 heterotrimer for histone pre-mRNA 3′-end processing. PLoS One. 2017;12:e0186034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gick O, Kramer A, Keller W, Birnstiel ML. Generation of histone mRNA 3′ ends by endonucleolytic cleavage of the pre-mRNA in a snRNP-dependent in vitro reaction. EMBO J. 1986;5:1319–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mowry KL, Steitz JA. Identification of the human U7 snRNP as one of several factors involved in the 3′ end maturation of histone premessenger RNA’s. Science. 1987;238:1682–7. [DOI] [PubMed] [Google Scholar]
- 34.Dominski Z, Yang X-C, Marzluff WF. The polyadenylation factor CPSF-73 is involved in histone-pre-mRNA processing. Cell. 2005;123:37–48. [DOI] [PubMed] [Google Scholar]
- 35.Kolev NG, Steitz JA. Symplekin and multiple other polyadenylation factors participate in 3′-end maturation of histone mRNAs. Genes Develop. 2005;19:2583–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang X-C, Burch BD, Yan Y, Marzluff WF, Dominski Z. FLASH, a proapoptotic protein involved in activation of caspase-8, is essential for 3′ end processing of histone pre-mRNAs. Mol Cell. 2009;36:267–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wagner EJ, Burch BD, Godfrey AC, Salzler HR, Duronio RJ, Marzluff WF. A genome-wide RNA interference screen reveals that variant histones are necessary for replication-dependent histone pre-mRNA processing. Mol Cell. 2007;28(4):692–9. [DOI] [PubMed] [Google Scholar]
- 38.Sabath I, Skrajna A, Yang X-C, Dadlez M, Marzluff WF, Dominski Z. 3′-end processing of histone pre-mRNAs in Drosophila: U7 snRNP is associated with FLASH and polyadenylation factors. RNA. 2013;19:1726–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skrajna A, Yang X-C, Dadlez M, Marzluff WF, Dominski Z. Protein composition of catalytically active U7-dependent processing complexes assembled on histone pre-mRNA containing biotin and a photo-cleavable linker. Nucl Acids Res. 2018;46:4752–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang X-C, Sabath I, Debski J, Kaus-Drobek M, Dadlez M, Marzluff WF, et al. A complex containing the CPSF73 endonuclease and other poyadenylation factors associates with U7 snRNP and is recruited to histone pre-mRNA for 3′-end processing. Mol Cell Biol. 2013;33:28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan SL, Huppertz I, Yao C, Weng L, Moresco JJ, Yates III JR, et al. CPSF30 and Wdr33 directly bind to AAUAAA in mammalian mRNA 3′ processing. Genes Develop. 2014;28:2370–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schonemann L, Kuhn U, Martin G, Schafer P, Gruber AR, Keller W, et al. Reconstitution of CPSF active in polyadenylation: recognition of the polyadenylation signal by WDR33. Genes Develop. 2014;28:2381–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skrajna A, Yang X-C, Bucholc K, Zhang J, Hall TMT, Dadlez M, et al. U7 snRNP is recruited to histone pre-mRNA in a FLASH-dependent manner by two separate regions of the stem-loop binding protein. RNA. 2017;23:938–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dominski Z, Erkmann JA, Yang X-C, Sanchez R, Marzluff WF. A novel zinc finger protein is associated with U7 snRNP and interacts with the stem-loop binding protein in the histone pre-mRNP to stimulate 3′-end processing. Genes Develop. 2002;16:58–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Azzouz TN, Gruber A, Schumperli D. U7 snRNP-specific Lsm11 protein: dual binding contacts with the 100 kDa zinc finger processing factor (ZFP100) and a ZFP100-independent function in histone RNA 3′ end processing. Nucl Acid Res. 2005;33:2106–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wagner EJ, Marzluff WF. ZFP100, a component of the active U7 snRNP limiting for histone pre-mRNA processing, is required for entry into S phase. Mol Cell Biol. 2006;26:6702–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang X-C, Sabath I, Kunduru L, van Wijnen AJ, Marzluff WF, Dominski Z. A conserved interaction that is essential for the biogenesis of histone locus bodies. J Biol Chem. 2014;289:33767–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bucholc K, Skrajna A, Adamska K, Yang XC, Krajewski K, Poznański J, et al. Structural Analysis of the SANT/Myb Domain of FLASH and YARP Proteins and Their Complex with the C-Terminal Fragment of NPAT by NMR Spectroscopy and Computer Simulations. Int J Mol Sci. 2020;21(15). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma T, Van Tine BA, Wei Y, Garrett MD, Nelson D, Adams PD, et al. Cell cycle-regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 2000;14(18):2298–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao J, Kennedy BK, Lawrence BD, Barbie DA, Matera AG, Fletcher JA, et al. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 2000;14(18):2283–97. [PMC free article] [PubMed] [Google Scholar]
- 51.Dominski Z, Carpousis AJ, Clouet-d’Orval B. Emergence of the b-CASP ribonucleases: Highly conserved and ubiquitous metallo-enzymes involved in messenger RNA maturation and degradation. Biochim Biophys Acta. 2013;1829:532–51. [DOI] [PubMed] [Google Scholar]
- 52.Bucholc K, Aik WS, Yang X-C, Wang K, Zhou ZH, Dadlez M, et al. Composition and processing activity of a semi-recombinant holo U7 snRNP. Nucl Acids Res. 2020;48(1508–1530). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun Y, Zhang Y, Aik WS, Yang XC, Marzluff WF, Walz T, et al. Structure of an active human histone pre-mRNA 3′-end processing machinery. Science. 2020;367(6478):700–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kambach C, Walke S, Young R, Avis JM, de la Fortelle E, Raker VA, et al. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell. 1999;96(3):375–87. [DOI] [PubMed] [Google Scholar]
- 55.Leung AKW, Kambach C, Kondo Y, Kampmann M, Jinek M, Nagai K. Use of RNA tertiary interaction modules for the crystallisation of the spliceosomal snRNP core domain. J Mol Biol. 2010;402:154–64. [DOI] [PubMed] [Google Scholar]
- 56.Leung AKW, Nagai K, Li J. Structure of the spliceosomal U4 snRNP core domain and its implication for snRNP biogenesis. Nature. 2011;473:536–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y, Sun Y, Shi Y, Walz T, Tong L. Structural insights into the human pre-mRNA 3’-end processing machinery. Mol Cell. 2020;77:800–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ruepp MD, Schweingruber C, Kleinschmidt N, Schumperli D. Interactions of CstF-64, CstF-77, and symplekin: implications on localisation and function. Mol Biol Cell. 2011;22:91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang XC, Sun Y, Aik WS, Marzluff WF, Tong L, Dominski Z. Studies with recombinant U7 snRNP demonstrate that CPSF73 is both an endonuclease and a 5′-3′ exonuclease. RNA. 2020;26(10):1345–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xiang K, Nagaike T, Xiang S, Kilic T, Beh MM, Manley JL, et al. Crystal structure of the human symplekin-Ssu72-CTD phosphopeptide complex. Nature. 2010;467:729–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wani S, Yuda M, Fujiwara Y, Yamamoto M, Harada F, Ohkuma Y, et al. Vertebrate Ssu72 regulates and coordinates 3′-end formation of RNAs transcribed by RNA polymerase II. PLoS One. 2014;9:e106040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Walther TN, Wittop Koning TH, Schümperli D, Müller B. A 5′-3′ exonuclease activity involved in forming the 3′ products of histone pre-mRNA processing in vitro. Rna. 1998;4(9):1034–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dominski Z, Yang XC, Purdy M, Marzluff WF. Differences and similarities between Drosophila and mammalian 3′ end processing of histone pre-mRNAs. Rna. 2005;11(12):1835–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang X-C, Sullivan KD, Marzluff WF, Dominski Z. Studies of the 5′ exonuclease and endonuclease activities of CPSF-73 in histone pre-mRNA processing. Mol Cell Biol. 2009;29:31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.West S, Gromak N, Proudfoot NJ. Human 5′->3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432:522–5. [DOI] [PubMed] [Google Scholar]
- 66.Kim M, Krogan NJ, Vasiljeva L, Rando OJ, Nedea E, Greenblatt JF, et al. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–22. [DOI] [PubMed] [Google Scholar]
- 67.Proudfoot NJ. Transcriptional termination in mammals: Stopping the RNA polymerase II juggernaut. Science. 2016;352(6291):aad9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eaton JD, Davidson L, Bauer DLV, Natsume T, Kanemaki MT, West S. Xrn2 accelerates termination by RNA polymerase II, which is underpinned by CPSF73 activity. Genes Develop. 2018;32:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gilmartin GM. Eukaryotic mRNA 3′ processing: a common means to different ends. Genes Develop. 2005;19:2517–21. [DOI] [PubMed] [Google Scholar]
- 70.Weiner AM. E Pluribus Unum: 3′ end formation of polyadenylated mRNAs, histone mRNAs, and U snRNAs. Mol Cell. 2005;20(2):168–70. [DOI] [PubMed] [Google Scholar]
- 71.Ghazy MA, He X, Singh BN, Hampsey M, Moore C. The essential N terminus of the Pta1 scaffold protein is required for snoRNA transcription termination and Ssu72 function but is dispensible for pre-mRNA 3′-end processing. Mol Cell Biol. 2009;29:2296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Baillat D, Hakimi M-A, Naar AM, Shilatifard A, Cooch N, Shiekhattar R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123:265–76. [DOI] [PubMed] [Google Scholar]
- 73.Baillat D, Wagner EJ. Integrator: surprisingly diverse functions in gene expression. Trends Biochem Sci. 2015;40:257–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mendoza-Figueroa MS, Tatomer DC, Wilusz JE. The Integrator Complex in Transcription and Development. Trends Biochem Sci. 2020;45(11):923–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kirstein N, Gomes Dos Santos H, Blumenthal E, Shiekhattar R. The Integrator complex at the crossroad of coding and noncoding RNA. Curr Opin Cell Biol. 2020;70:37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Armon A, Graur D, Ben-Tal N. ConSurf: an algorithmic tool for the identification of functional regions in proteins by surface mapping of phylogenetic information. J Mol Biol. 2001;307:447–63. [DOI] [PubMed] [Google Scholar]


