Abstract
Up to 10% of patients with pancreatic ductal adenocarcinoma (PDAC) carry underlying germline pathogenic variants in cancer susceptibility genes. The GENetic Education Risk Assessment and TEsting (GENERATE) study aimed to evaluate novel methods of genetic education and testing in relatives of patients with PDAC. Eligible individuals had a family history of PDAC and a relative with a germline pathogenic variant in APC, ATM, BRCA1, BRCA2, CDKN2A, EPCAM, MLH1, MSH2, MSH6, PALB2, PMS2, STK11, or TP53 genes. Participants were recruited at six academic cancer centers and through social media campaigns and patient advocacy efforts. Enrollment occurred via the study website (https://GENERATEstudy.org) and all participation, including collecting a saliva sample for genetic testing, could be done from home. Participants were randomized to one of two remote methods that delivered genetic education about the risks of inherited PDAC and strategies for surveillance. The primary outcome of the study was uptake of genetic testing. From 5/8/2019–5/6/2020, 49 participants were randomized to each of the intervention arms. Overall, 90/98 (92%) of randomized participants completed genetic testing. The most frequently detected pathogenic variants included those in BRCA2 (N=15, 17%), ATM (N=11, 12%), and CDKN2A (N=4, 4%). Participation in the study remained steady throughout the onset of the Coronavirus disease (COVID-19) pandemic. Preliminary data from the GENERATE study indicate success of remote alternatives to traditional cascade testing, with genetic testing rates over 90% and a high rate of identification of germline pathogenic variant carriers who would be ideal candidates for PDAC interception approaches.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is associated with high mortality, with 90% of diagnosed individuals not surviving past 5 years (https://seer.cancer.gov) and a median survival of 11 months post-diagnosis for treated metastatic disease [1]. The aggressiveness of this disease results in a short time window for critical therapies and other intervention measures. However, when PDAC is diagnosed at an early stage, the 5-year survival increases to an estimated 40–80% (https://seer.cancer.gov) [2], a figure that has improved significantly in recent years, conceivably due to improvements in early stage detection [2]. Recent studies of the hereditary factors that increase pancreatic cancer risk have found that approximately 5–10% of cases carry germline pathogenic variants identified in multigene panel analysis [3–5] which have implications for targeted therapies in patients [6–9] and surveillance in at-risk relatives. PDAC surveillance for family members of germline pathogenic variant carriers is recommended by clinical guidelines [10, 11] and recent data have shown potential success of surveillance, including the use of endoscopic ultrasound and/or MRI, in pathogenic variant carriers, leading to the detection of PDAC at earlier stages [12–14].
Recognizing the implications of hereditary factors, clinical guidelines now recommend universal genetic testing for all patients with PDAC [10, 15]. Despite opportunities for targeted treatment and surveillance, a recent study done at a major cancer institute found that even with automated referral, fewer than 40% of cases with PDAC pursued genetic evaluation [16]. Improved communication between patients with PDAC and their relatives is crucial given the short survival associated with the disease [17]. Offering genetic testing to family members of pathogenic variant carriers that relies on the communication of results from the index (pathogenic variant-positive) patient to at-risk family members is an approach known as cascade testing. The at-risk family member typically must then arrange his/her own genetic counseling, which traditionally has required an in-person visit. Cascade testing leads to the identification of shared pathogenic variants and medical management options for the pathogenic variant carrier, such as surveillance. Studies have shown, however, that no more than 50% of cancer patients who carry pathogenic variants pursue genetic counseling and testing [18, 19], in large part due to barriers such as lack of communication among family members [20], poor understanding of the genetic condition (https://migrc.org), cost [21], and lack of access to genetic specialists who can provide genetic counseling and testing, especially to relatives in a different geographical location [22].
In addition to these barriers, the COVID-19 pandemic has led to infection control measures that have resulted in many individuals not seeking cancer preventive care [23]. The pandemic has encouraged individuals to receive health services at home, if possible, to reduce risks associated with in-person clinic visits.
Novel approaches that make genetic counseling and testing for hereditary cancer predisposition more accessible to individuals through remote means have shown promise [24–27]. To date, these approaches, including the use of telephone and/or online initiatives to educate relatives of known carriers of pathogenic variants in diverse cancer susceptibility genes, have demonstrated an increased uptake of genetic testing among invited relatives; 48–58% of invited relatives pursued genetic testing in two studies [25, 26] and participants reported low levels of distress and high satisfaction with the process [25]. To our knowledge, there has been no study on the use of remote methods for genetic education and testing that has been conducted specifically in relatives of patients with PDAC.
We sought to develop novel methods to conduct genetic education and testing for patients with increased risk for hereditary pancreatic cancer. Our goal was to design and evaluate new methods of remote genetic education and at-home saliva-based testing that address the changing landscape of genetic testing and address some of the barriers associated with traditional genetic testing. Our hypothesis was that remote methods that make both genetic education and testing more accessible by allowing participants to engage in these health services at home, without the need for in-person visits, would increase the uptake of testing relative to traditional cascade testing methods that rely on in-person visits. Moreover, we hypothesized that the addition of a genetic counselor to remote education would further increase the uptake of genetic testing and lead to greater understanding of genetic testing results, satisfaction with the genetic testing process, and downstream engagement in cancer surveillance.
Here, we report preliminary findings from the first year of a randomized study of two novel methods of remote genetic education and testing. These findings are particularly relevant in light of the COVID-19 pandemic and its challenges to in-person health care delivery [28, 29].
Materials and Methods
Stand Up To Cancer (SU2C) Multi-Site Collaboration
The SU2C-Lustgarten Foundation Pancreatic Cancer Interception Dream Team was created to formulate the clinical framework to enable cancer interception in individuals at high risk for PDAC [30]. The team’s investigators at Dana-Farber Cancer Institute, Mayo Clinic, UC San Diego, Johns Hopkins University, Weill Cornell Medical Center and MD Anderson Cancer Center designed the GENetic Education Risk Assessment and TEsting (GENERATE) study to increase the identification of individuals at high risk for PDAC through novel means of genetic education and testing. The multidisciplinary team (including genetic counselors, nurses, physicians, research scientists, patient advocates, and research coordinators) met to develop the study protocols, modify workflows, curate and/or design study questionnaires, implement recruitment procedures, and ensure easy access to information and the inclusion of support resources for participants’ downstream surveillance needs. The Dana-Farber Cancer Institute was designated as the recruitment and data coordination center. The GENERATE study protocol (Dana-Farber Protocol # 18-222) and written informed consent were approved by Dana-Farber’s Institutional Review Board (IRB) in accordance with the U.S. Common Rule, and the study was performed after receiving IRB approval.
Study Population
To be eligible, individuals must have had either 1) a first or second-degree relative with a diagnosis of PDAC AND a known germline pathogenic variant in one of 13 PDAC-predisposing genes (APC, ATM, BRCA1, BRCA2, CDKN2A, EPCAM, MLH1, MSH2, MSH6, PALB2, PMS2,STK11,TP53) OR 2) an unaffected first- or second-degree relative carrying one of these pathogenic variants and related by first- or second-degree to someone with PDAC. For example, an individual could have a blood relative with a diagnosis of PDAC that also carried a pathogenic variant in one of the 13 PDAC-predisposing genes. Alternatively, an individual could have an unaffected relative who carried one of the 13 PDAC-predisposing pathogenic variants and a different relative who had a diagnosis of PDAC. In the second case, the relative with PDAC would have to be a first- or second- degree relative of the unaffected, pathogenic variant carrier. Participants were required to provide the genetic test report of the family member with the pathogenic variant for study team review. Participants were ineligible if they were known carriers of a cancer-predisposing pathogenic variant or had had genetic counseling for cancer risk within 3 years of consent date. Willingness to share genetic test results with an identified healthcare provider and with the study team was also required (see Table 1 for complete eligibility/ineligibility criteria).
Table 1.
Eligibility and Ineligibility Criteria
| Eligibility Criteria |
|---|
| Individual who is 18 years or older |
| Individual who has signed informed consent |
| Individual who has been informed about: |
| A first or second-degree relative with a diagnosis of pancreatic ductal adenocarcinoma (PDAC) and a germline pathogenic variant in APC, ATM, BRCA1, BRCA2, CDKN2A, EPCAM, MLH1, MSH2, MSH6, PALB2, PMS2, STK11 or TP53 |
| OR |
| An unaffected first or second-degree relative with a germline pathogenic variant in one of these genes who has a first or second-degree relative with PDAC |
| The germline pathogenic variant and history of PDAC must both be on the maternal or paternal side of the family |
| Individual with a valid mailing address |
| Ineligibility Criteria |
| Individual with a known cancer pathogenic variant |
| Individual who has received prior genetic counseling for cancer risk |
| Individual who has received a bone marrow transplant, who has had a blood transfusion within the last 7 days, or who has an active malignancy (i.e. leukemia or lymphoma)* |
| Individual who is unable to sign the informed consent because of mental incompetency or psychiatric illness |
| Individual who is unwilling to complete baseline and follow-up questionnaires |
| Individual who has a life expectancy of less than 1 year |
| Individual with only an APC I1307K pathogenic variant within their family |
| Individual with only a PMS2 exons 12–15 deletion pathogenic variant within their family |
Since the study was done in relatives of PDAC cases, we also excluded any individual who had a diagnosis of PDAC
Remote Participant Consent, Enrollment and Randomization
Potential participants for the study were identified through a variety of means, including social media and targeted outreach to patients with PDAC at collaborating sites who carried a pathogenic variant. Interested patients with PDAC and their family members were provided with a brochure detailing the study requirements and providing contact information for the study team (Figure 1). To facilitate recruitment, patients with PDAC were provided with invitation letters they could send to their relatives.
Figure 1.

GENERATE Study Brochure
Interested individuals were directed to the GENERATE study website (https://GENERATEstudy.org) where they completed the electronic eligibility form to indicate their interest in joining the study and their willingness to be contacted by the study team. Ineligible individuals received an automated response that indicated they were unable to join the study and provided links to resources about cancer genetics and genetic testing. The study team contacted eligible individuals within one to two business days of receiving their completed eligibility form to review the study and address any questions. If the individual indicated their willingness to proceed, they could complete the electronic informed consent form, providing written consent, and enroll as a study participant. A paper version of the informed consent form was also available and could be provided upon request.
Study data were collected and managed using Research Electronic Data Capture (REDCap™) tools hosted at Partners Healthcare. REDCap™ is a secure, HIPAA compliant, web-based application designed to support data capture for research studies [31]. Participants uploaded a copy of the known family genetic mutation report where it was subsequently reviewed for accuracy. Incorrect family mutation reports (such as somatic test results or non-PDAC pathogenic variant reports) triggered study team follow-up with the participant. The pathogenic variant was considered to be a “non-PDAC pathogenic variant” if it was not one of the 13 PDAC-predisposing genes as listed in the eligibility criteria. Participants were randomized by family (representative of cluster randomization [32]) into one of two study arms (described more below) via an auto randomization algorithm using REDCap™.
Study Design
The GENERATE study randomized participants by family to one of two study arms: Arm 1 included remote genetic education and testing through a video-based telemedicine platform and physician-mediated testing through Color Genomics. Arm 2 included remote genetic education and testing solely through Color Genomics.
Subjects in Arm 1 underwent pre-test genetic education with a short 7-minute pre-recorded video introduced briefly by a study genetic counselor. The video provided an overview of inherited risk for pancreatic cancer and potential risks and benefits of genetic testing. Following the pre-recorded video, a genetic counselor conducted an interactive session through Doxy.me, a HIPAA compliant web-based telemedicine platform. This 15–30-minute interactive session was dedicated to answering questions and addressing any concerns. Topics that were addressed with subjects included more information about the specific genetic pathogenic variant carried by the participant’s family member, additional cancer risks and cancer risk management strategies associated with that gene. Other topics included genetic discrimination issues such as insurance coverage and other psychosocial concerns. Participants with significant distress were referred for tailored follow-up. Subjects had the opportunity to receive the live interactive session individually or with a group of family members. The pre-recorded video used in the Doxy.me session is detailed in the Storyboard which includes the script and associated images shown to participants (Supplement 1). Upon completing the educational session, participants received an e-mail with the Color Genomics GENERATE study link and could elect to proceed or not with germline testing.
Participants in Arm 2 were directed to the same Color Genomics GENERATE study link (embedded with a test code that covered the cost of testing for all participants) in which they could read about inherited cancer and testing options and elect to proceed or not with germline testing. Genetic education given on the Color Genomics website is included in Supplement 2. While the same education on the website was available to participants in both arms, those in Arm 2 did not have pre-test education other than the content provided on the Color Genomics website, as this arm was meant to mimic real-world testing through the physician-mediated model. There was no time limit given to participants in either arm for ordering testing.
All participants had the option to speak with a genetic counselor at Color Genomics to address questions related to genetic testing. Regardless of the known pathogenic variant in the index relative, genetic testing of participants utilized the standard high-throughput Color Genomics 30-gene hereditary cancer panel which tests for pathogenic variants in the following genes: BRCA1, BRCA2, MLH1, MSH2, MSH6, PMS2, EPCAM, APC, MUTYH, MITF, BAP1, CDKN2A, CDK4, TP53, PTEN, STK11, CDH1, BMPR1A, SMAD4, GREM1, POLD1, POLE, PALB2, CHEK2, ATM, NBN, BARD1, BRIP1, RAD51C, RAD51D. Testing was performed on DNA extracted from a self-collected saliva sample. Results were classified as positive if one or more pathogenic or likely pathogenic variants was detected and negative if no variant, only benign variants, likely benign variants, or only variants of uncertain significance were detected, as previously described [33]. If a participant had a pathogenic variant, s/he was required to schedule a post-test telephone genetic counseling session with a Color Genomics genetic counselor. Otherwise, participants with negative results could log into the study’s portal to access their test results once they were notified that their results were available via email. Participants could request to speak with a study genetic counselor at any point in the process.
To identify any potential issues with the post-results disclosure, participants in Arm 1 received a post-test video-based genetic education session with a GENERATE study genetic counselor, while those in Arm 2 received an electronic post-test check-in survey from the study team who looked for potential distress and offered additional support. The number of participants who contacted study genetic counselors after the post-test check-in survey was tracked for Arm 2. In both arms, participants were provided with contact information for local cancer genetic counseling services if needed.
Study Instruments and Evaluation of Study Endpoints
Baseline questionnaires included the Demographic and Pancreatic Cancer Experience questionnaire which was developed for the GENERATE study to describe the participant population and their experience with having a relative diagnosed with PDAC and/or other cancers. This questionnaire also included metrics previously found to be associated with an individual’s experience in engaging in genetic services related to inherited PDAC susceptibility [34]. It was administered following consent but prior to randomization via the study REDCap™ electronic database (paper versions of the questionnaires were also available). Participants were not permitted to move on to the intervention without having completed the majority of the baseline questionnaires, further described below.
Additional baseline questionnaires included the Adapted Lerman Breast Cancer Worry Scale [35] and Hospital Anxiety and Depression Scale (HADS) [36], previously developed instruments that measure participants’ cancer-specific worry and general level of anxiety, respectively, as well as the Health Behaviors and Screening Questionnaire which measures participation in risk-reducing cancer screenings. The responses from these questionnaires, which were administered at baseline and additional specified time points, will be evaluated in future analyses that include other secondary post-intervention outcomes.
Street addresses, which were collected in the eligibility form from all potential participants, were used to estimate rural vs. urban location and socioeconomic status based on rural-urban commuting area (RUCA) codes [37] and area deprivation index (ADI) [38], previously validated measures. Lower RUCA codes indicate urban setting, while higher RUCA codes indicate an isolated, rural community. The ADI allows for rankings of neighborhoods using the estimated socioeconomic disadvantage of the specific region, with lower ADI scores representing the least disadvantaged groups.
Formal analyses will be conducted in future comparisons to measure the effect of the intervention on the primary outcome (uptake of genetic testing) and secondary outcomes (participant distress, cancer genetic knowledge gained [measured by the KnowGene Scale [39]], decision making, degree of family communication and uptake of surveillance procedures) in participants in the two study arms. All participants who pursued genetic testing were administered questionnaires that assess potential distress including the HADS and Multi-Dimensional Impact of Cancer Risk Assessment (MICRA). Socioeconomic status will also be assessed in a follow-up questionnaire (Supplement 3).
The projected sample size for the whole study is 500 participants, including 250 randomized to each arm. Genetic testing results will be analyzed individually for each arm, but the confounding effect of family will be assessed. Participants from the same family are randomized to the same arm. With this sample size, the study will be able to detect a 10% difference (83% vs. 93%) in the uptake of genetic testing with 80% power at 0.05 two-sided alpha level, assuming an intraclass (within family) correlation of 0.4.
In this report, which summarizes data from the first year of study enrollment, descriptive statistics summarize patient characteristics and demographics, as well as overall rates of genetic testing and pathogenic variants detected among all randomized study participants.
Results
Study Participation
In the first year of the GENERATE study, conducted from 5/8/2019–5/6/2020, 477 individuals completed the eligibility questionnaire, 131 of whom were eligible. A total of 346 individuals were ineligible due to the following: 194 did not have a relative with a known pathogenic variant, 62 carried a cancer-predisposing pathogenic variant, 54 had received genetic counseling for cancer risk, 40 did not have a healthcare provider, and 33 gave “other” as a reason. 111 eligible individuals consented; of these 107 individuals uploaded their known family mutation reports and ultimately 101 completed baseline questionnaires. Of individuals initially found to be eligible, 7 later failed screening for the following reasons: 5 uploaded somatic testing results, one had received prior cancer genetic counseling and testing and one carried a pathogenic variant in a gene not included in the 13 PDAC cancer susceptibility genes. 49 participants were randomized to Arm 1 and 49 participants were randomized to Arm 2 (Figure 2).
Figure 2. Consort Diagram of the GENERATE Study.
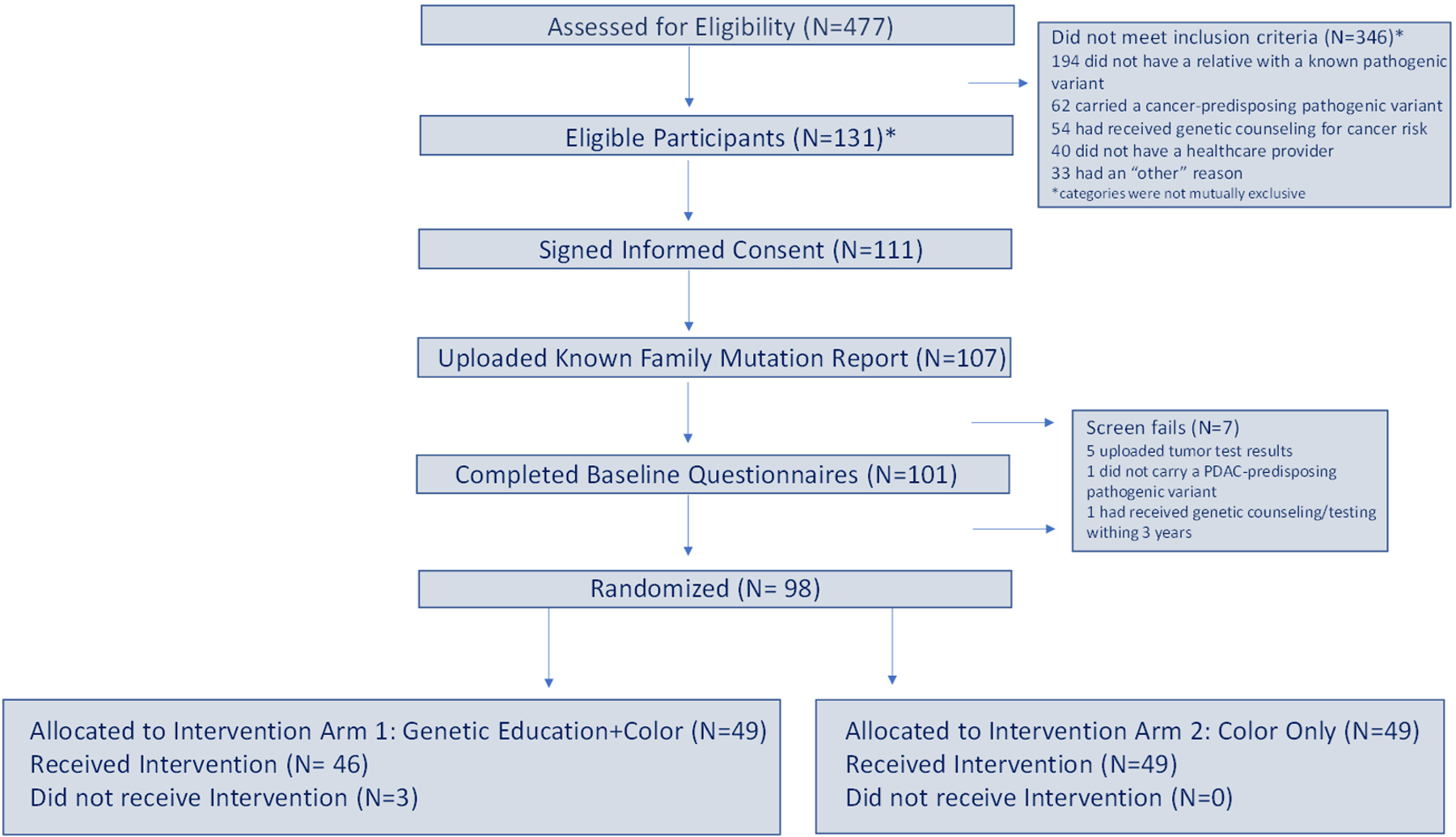
This figure details the number of individuals who fulfilled each of the necessary eligibility steps before randomization. *Of 131 “Eligible” participants, 7 were later identified as screen fails. The true number of eligible participants was therefore 124.
Demographic Characteristics of Study Participants
The mean age among randomized participants was 46. The majority of participants enrolled were Caucasian (N=95 [97%]) and of Non-Hispanic ethnicity (N=96 [98%]). Randomized participants were enrolled from throughout the US; 41 (42%) were from the Northeast, 21 (21%) were from the Midwest, 20 (20%) were from the South, and 16 (16%) were from the West (Table 2). With regard to rural/urban geographical location or socioeconomic status, study participants did not significantly differ from ineligible individuals or individuals who were eligible but did not consent (Supplement 4).
Table 2.
Demographics of Randomized Study Participants
| Total N=98 | |
|---|---|
| N (%) | |
| Age (Years) | |
| Mean +/− Standard Deviation | 46 +/− 17 |
| Range | 18–90 |
| Sex | |
| Male | 43 (44) |
| Female | 55 (56) |
| Racial Background | |
| White/Caucasian | 95 (97) |
| Black/African American | 0 |
| American Indian/Alaskan Native | 0 |
| Asian/Asian-American | 0 |
| Native Hawaiian/Other Pacific Islander | 0 |
| Two or more races | 2(2) |
| Unknown | 1(1) |
| Ethnicity | |
| Hispanic or Latino | 2 (2) |
| Non-Hispanic or Non-Latino | 96 (98) |
| Unknown | 0 |
| Geographic Location* | |
| Northeast | 41 (42) |
| Midwest | 21 (21) |
| South | 20 (20) |
| West | 16 (16) |
| Referring Institute | |
| Dana-Farber Cancer Institute | 31 (32) |
| Johns Hopkins University | 10 (10) |
| Mayo Clinic | 11 (11) |
| MD Anderson Cancer Center | 7 (7) |
| University of California, San Diego | 1 (1) |
| Weill Cornell | 0 |
| None of these institutions | 35 (36) |
| Another institution | 3 (3) |
| How did you hear about the study (can choose more than 1)? | |
| From a healthcare provider | 20 (20) |
| From a family member | 82 (84) |
| Other** | 7 (7) |
Northeast=Maine, New Hampshire, Vermont, Massachusetts, Connecticut, Rhode Island, New York, New Jersey, Pennsylvania; Midwest=North Dakota, South Dakota, Nebraska, Kansas, Minnesota, Illinois, Missouri, Iowa, Wisconsin, Michigan, Ohio, Indiana; South= Tennessee, Kentucky, Texas, Oklahoma, Arkansas, Louisiana, Mississippi, Alabama, Georgia, Florida, South Carolina, North Carolina, West Virginia, Virginia, District of Columbia, Maryland, Delaware; West= Washington, Oregon, California, Nevada, Arizona, New Mexico, Colorado, Utah, Idaho, Wyoming, Montana, Hawaii, Alaska
Other includes patient outreach through advocacy groups, internet campaigns, and social media
GENERATE Study Recruitment
A major source of participant referral was a participating study site; among randomized study participants, 60 (61%) heard about the study from one of the 6 participating study sites. However, a significant number of participants (38 [39%]) heard about the study from another institution or from a separate referral source. Participants learned about the study from healthcare providers, family members, and patient outreach efforts that included advocacy groups, the internet and social media, with the majority (N=82 [84%]) learning about the study from a family member (Table 2). Participants could have answered “yes” to more than one of these categories. The 61% who heard about the study from a participating institution could have been linked to one of the study sites because their family member was treated for PDAC at that institution (and therefore they received the brochure from the PDAC family member) or they were part of a family registry at one of the institutions. The 84% of participants who heard about the study from a family member could have heard from an unaffected relative who was also a study participant or could have heard from a relative being treated at one of the institutions. We were unable to tease out the reason for these categories. The 98 participants were enrolled from 57 different families.
Experience with Pancreatic and Other Cancers
The majority of randomized participants had a first-degree relative who carried a PDAC-predisposing pathogenic variant with 41 [42%] having a sibling, 5 [5%] a child, and 48 [49%] having a parent. Of those participants who had a second-degree relative with a PDAC-predisposing pathogenic variant, 15 [15%] had an aunt, 13 [13%] had an uncle, 9 [9%] had a grandparent, 1 [1%] had a grandchild, and 8 [8%] had a niece or nephew. Roughly one third (N=29 [30%]) of participants had more than one relative in whom genetic testing had previously identified a PDAC-predisposing pathogenic variant (Table 3).
Table 3.
Experience of Randomized Study Participants with Pancreatic and Other Cancers
| Total N=98 | |
|---|---|
| N (%) | |
| 1. Have you lost a family member to pancreatic cancer? | Yes=54 (55)/No=44 (45) |
| a. If yes, how long ago was your most recent experience losing someone to pancreatic cancer?* | |
| < 1 year | 11 (20) |
| 1–2 years | 9 (17) |
| 3–5 years | 7 (13) |
| 6–10 years | 6 (11) |
| >10 years | 21 (39) |
| b. If yes, were you a caregiver to this person?* | Yes=6 (11)/No=48 (89) |
| c. If yes, how long did you provide this care?** | |
| < 1 year | 2 (33) |
| 1–2 years | 2 (33) |
| 3–5 years | 1 (17) |
| 6–10 years | 0 |
| >10 years | 0 |
| Still providing this care | 1 (17) |
| 2. Have you lost a family member to another type of cancer (besides pancreatic)? | Yes=75 (77)/No=23 (23) |
| a. If yes, were you a caregiver to this person?* | Yes=7 (9)/No=68 (91) |
| 3. When was cancer genetics testing done in your family? | |
| Within the last 6 months | 34 (35) |
| 6 months – 1 year ago | 14 (14) |
| 1–2 years ago | 14 (14) |
| >2 years ago | 33 (34) |
| Not available | 3 (3) |
| 4. In what relative(s) was an inherited alteration in a gene found? | |
| First-degree relatives | |
| sibling | 41 (42) |
| child | 5 (5) |
| parent | 48 (49) |
| Second-degree relatives | |
| aunt | 15 (15) |
| uncle | 13 (13) |
| grandparent | 9 (9) |
| grandchild | 1 (1) |
| niece/nephew | 8 (8) |
| More than one relative | 29 (30) |
Among those who lost a family member
Among those who were caregivers
Among randomized participants, approximately half of individuals had a relative who had died from pancreatic cancer (N=54 [55%). The majority had lost a family member within the last 10 years (N=33 [61%]), with 20 (37%) losing a family member within the last 2 years. The majority of study participants had also had a relative die from another type of cancer besides pancreatic (N=75 [77%]). A minority of participants had acted as a caregiver to their family member who was lost to pancreatic cancer (N=6 [11%]), or as a caregiver to their family member lost to another form of cancer (N=7 [9%]). Among participants’ family members, approximately half (N=48 [49%]) had genetic testing done within a year of the participant consenting (Table 3).
Overall Uptake of Genetic Testing in Study Arms
Among randomized study participants, 90 (92%) completed genetic testing (Table 4). Among participants with a first-degree relative who carried a PDAC-predisposing pathogenic variant, 77 (95%) ordered genetic testing, and among those whose second-degree relative was the pathogenic variant carrier, 32 (89%) ordered genetic testing. Among participants who had both a first-degree and second-degree relative with a PDAC-predisposing pathogenic variant, 19 (95%) ordered genetic testing. The overall prevalence of PDAC-associated pathogenic variants was 51% (N=39) among participants with a first-degree relative with a PDAC-predisposing pathogenic variant, 31% (N=10) among participants with a second-degree relative with a PDAC-predisposing pathogenic variant and 42% (N=8) among those with both a first and second-degree relative with a PDAC-predisposing pathogenic variant. Pathogenic variants detected among randomized participants included BRCA2 (N=15 [17% of participants]), ATM (N=11 [12%]), CDKN2A (N=4 [4%]), BRCA1 (N=3 [3%]), MLH1, MSH2, PALB2 and PMS2 (all N=2 [2%]). 4 participants (4%) carried a pathogenic variant in an “other” gene not included in the 13 PDAC-predisposing gene pathogenic variants. These “other” genes were not counted towards the number of pathogenic variants detected (Table 4).
Table 4.
Uptake of Genetic Testing and Pathogenic Variants Detected Among 98 Randomized Participants±
| Randomized Study Participants N (%) | |
|---|---|
| Completed genetic testing | 90 (92) |
| Did not complete genetic testing | 8 (8) |
| Overall uptake of testing | |
| In first-degree relative of PV carrier (N=81) | 77 (95) |
| In second-degree relative of PV carrier (N=36) | 32 (89) |
| In both (N=20)* | 19 (95) |
| Overall prevalence of PDAC-associated pathogenic variants** | |
| In first-degree relative of PV carrier (N=77) | 39 (51) |
| In second-degree relative of PV carrier (N=32) | 10 (31) |
| In both (N=19)* | 8 (42) |
| Pathogenic variants detected (N=90) | |
| APC | 0 |
| ATM | 11 (12) |
| BRCA1 | 3 (3) |
| BRCA2 | 15 (17) |
| CDKN2A | 4 (4) |
| EPCAM | 0 |
| MLH1 | 2 (2) |
| MSH2 | 2 (2) |
| MSH6 | 0 |
| PALB2 | 2 (2) |
| PMS2 | 2 (2) |
| STK11 | 0 |
| TP53 | 0 |
| Other *** | 4 (4) |
Uptake of genetic testing as of 9/11/20
Participant has an FDR and SDR with PDAC-associated pathogenic variant
Percent in those who received results; includes pathogenic variants in APC, ATM, BRCA1, BRCA2, CDKN2A, EPCAM, MLH1, MSH2, MSH6, PALB2, PMS2, STK11, TP53
Includes pathogenic variants in CHEK2 (2) and MITF (2). 1 participant carried pathogenic variants in BRCA1/CHEK2, 1 carried BRCA2/MITF, 1 carried ATM/CHEK2, 1 carried MITF; double pathogenic variants are counted in the “other” category as well as the PDAC-predisposing pathogenic variants listed above. Pathogenic variants in CHEK2 and MITF are not believed to be associated with PDAC susceptibility.
Participation During the COVID-19 Pandemic
In the study period from 3/23/20 to 5/6/20 which included the last 44 days of the first year of enrollment (or 12% of the study period from 5/8/19 to 5/6/20), 95 participants completed the eligibility questionnaire (20% of the total during the study period from 5/8/19 to 5/6/20), 10 consented (9%), and 11 (11%) were randomized, comparable with participation rates pre-pandemic.
Discussion
A major finding from the first year of study recruitment of the GENERATE study was the overall high uptake of genetic testing, with over 90% of all randomized participants completing genetic testing. We observed especially high rates of testing in those with first-degree relatives with a PDAC-predisposing pathogenic variant and those with both a first-degree relative (FDR) and a second-degree relative (SDR) with a PDAC-predisposing pathogenic variant, all of whom also had a family history of PDAC, with 95% of participants completing genetic testing.
Between 2012 and 2018, the consumer genetic testing market increased by greater than 12-fold (https://blogs.cdc.gov). While there is growing interest among the public in pursuing their own genetic testing, the rise of direct to consumer (DTC) genetic testing, where results are communicated without the guidance of a physician/counselor, presents challenges across the healthcare system (https://www.concertgenetics.com). Traditional genetic counseling/testing requires a pre-test consultation with a health care provider and results can be used to direct medical care as opposed to DTC testing where consumers can order a test directly from a commercial genetic testing company and no health care provider is required in the ordering or reporting of the test. Some commercial genetic testing laboratories have begun to offer “physician-mediated” testing [40], an intermediate approach, where the ordering physician is from a provider network affiliated with the laboratory, rather than the patient’s own provider. Challenges in accessing genetic specialists [41], coupled with the public’s interest in pursuing their own genetic testing support non-traditional methods such as physician-mediated testing for conducting effective genetic education and testing. The need to develop and evaluate these methods prompted our study.
While studies of cascade testing have shown the success of remote technologies, including the use of physician-mediated testing, these studies have either been performed in a heterogeneous mix of at-risk relatives of previously identified pathogenic variant carriers [25, 26] or limited to women at risk of hereditary breast ovarian cancer (HBOC) syndrome [27]. To our knowledge, the use of remote technologies in genetic education and testing has not been studied specifically in relatives of patients with PDAC.
The rates reflected in prior studies demonstrate the potential success of remote technologies over traditional cascade approaches with reported uptake rates of 48–58% [25, 26]. Our study far surpasses rates demonstrated with other remote technologies conducted to date. The higher rate in first-degree relatives compared to second-degree relatives is consistent with other studies of cascade testing where rates decline; one explanation may be that communication declines as distance in relationship increases [42].
The high rate of testing demonstrated among randomized participants in this study may reflect the public’s increasing receptiveness to online genetic education and the success of providing genetic testing through physician-mediated testing with a genetic testing company. This high uptake may be attributed to the ease of the methodology which included the ability to participate in genetic testing through at-home sample collection using saliva rather than blood. It may also reflect concern specific to PDAC risk which may have motivated participation in the study due to the limited survival associated with the disease. The ability to provide remote genetic education addresses many of the challenges associated with current genetic testing where family members of patients with PDAC are often challenged by physical location, access to in-person visits and cost. By providing free genetic testing, we eliminated the cost barrier. We successfully enrolled participants from diverse regions across the US by providing remote genetic education as a component of our intervention rather than genetic counseling which allowed us to cross state lines. The remote nature of the GENERATE study also allowed enrollment to keep pace with the rate before the onset of the COVID19 pandemic. An additional strength of both methodologies described in this study is their exportability to other cancers with a hereditary component. The education and genetic counseling provided via Color Genomics covers material applicable to multiple hereditary cancers. This platform has been implemented by the MAGENTA study to enroll women at increased risk for ovarian cancer [27], and could be used for studies in participants with increased risk of colorectal, stomach, and breast, among other cancer types. The interactive online education provided via the Doxy.me platform could similarly be adapted for educating those with increased risk for a diverse range of cancer types. A strength of the collaboration that was established to conduct this study is the expandable framework to study other hereditary cancers through a network of major national cancer centers to increase both study populations and scope of study goals.
Consistent with autosomal dominant inheritance, approximately half (51%) of those with an FDR with a pathogenic variant also carried a pathogenic variant and roughly one quarter of those with an SDR (31%) carried a pathogenic variant. While our eligibility criteria required participants to have a relative with one of 13 known PDAC-predisposing pathogenic variants, among those who carried a variant, the most frequently detected pathogenic variants included those in BRCA2, followed by the ATM and CDKN2A genes, reflecting what was detected in the family member and consistent with studies of germline pathogenic variants associated with PDAC [43]. Implications of identifying these high-risk individuals include strategies for early detection of PDAC lesions and prevention through novel screening approaches, which are recommended in current guidelines [11] and in the future when cutting-edge technologies are developed [30]. In addition to PDAC, carriers of PDAC-predisposing pathogenic variants have increased risk for other cancers; carriers of ATM pathogenic variants are recommended to receive risk-reducing surveillance for breast cancer [10], carriers of BRCA1/2 are recommended to undergo both ovarian and breast cancer surveillance [10], male BRCA1/2 carriers are recommended to undergo earlier screening for prostate cancer [44], and CDKN2A carriers should undergo annual screening for melanoma [10].
While we were able to achieve high rates of genetic testing in randomized participants, the initial phase of our study faced clear challenges. Despite the clinical recommendation for universal testing in PDAC [10], in practice, many patients with PDAC do not undergo genetic testing [16]. This limited the number of family members we were able to reach because at many of the study sites, patients with PDAC did not pursue genetic testing that was offered by their provider. An additional limitation of the first year of the study was the low enrollment of racial and ethnic minority participants. We are currently working with advocacy groups serving these communities and have launched a new study, the Racial/Ethnic Equity in GENetic Education and Risk Assessment and TEsting (REGENERATE) study, which aims to overcome barriers to genetic and cancer prevention education among Black/Latinx individuals at risk for PDAC. We recognize that patients with PDAC received their genetic testing at various laboratories while participants received testing solely at Color Genomics. However, the sensitivity of Color Genomics testing is greater than 99% (https://static.getcolor.com/pdfs/validationWhitePaper.pdf). For every participant with a positive result, we verified that the pathogenic variant detected matched the pathogenic variant that was detected in the positive family member. While it is possible that we had false negatives, the rates of positives reflected what we would have expected for autosomally dominant inherited traits. The only way to control for variability between labs would have been to match the testing lab where the relative was tested for each study participant, which wasn’t realistic in our study design. Thus, this is an inherent, but very minor limitation of our study.
Our study was designed to assess whether the addition of a healthcare team would lead to better outcomes and higher satisfaction compared to education delivered solely through physician-mediated testing with a genetic testing company. We had hypothesized that education conducted with a genetic counselor would lead to higher uptake of testing. While we are not yet quantitatively comparing the two arms in this interim report, based on our initial data, we qualitatively observed that both arms had very high uptake of genetic testing, suggesting the effectiveness of remote education that is conducted both with the guidance of a genetic counselor and solely through the physician-mediated model. We also recognize that selection bias in our participant population may have contributed to the high rates of testing; this population represents a highly motivated group who sought a genetic education and testing study and who were able to provide genetic testing results from a family member. Recent findings from the MAGENTA study showed higher rates of genetic testing when the physician-mediated model was used compared to testing that followed telephone counseling with a genetic counselor in patients at risk of HBOC [45], contrary to what we hypothesized in the GENERATE study. This finding lends support to the use of genetic testing companies who use physician-mediated testing for genetic education and testing. Our preliminary data showed high rates of testing among all study participants, but increased participant numbers may ultimately shed light on differences between the two arms and if similar to the MAGENTA study, may endorse the use of genetics testing companies in providing pre-test genetic education.
This report presents methodology and data from the first year of study recruitment. Because we are not presenting all the data in this report, we did not present formal analysis and instead present data for all randomized participants combined rather than separated by arm. The difference between the 2 study arms was the genetic education provided; participants in Arm 1 received both Color Genomics’ education and that provided by the Doxy.me session with the genetic counselor, while those in Arm 2 only received genetic education provided by Color Genomics. In future comparisons, we will analyze results from the two study arms to look for significant differences in uptake of genetic testing and patient-related measures. We will perform a robust statistical analysis once our accrual goal has been met to determine whether there are significant differences in uptake of genetic testing, demographics, experiences of the participants, and/or differences in numbers of participants who contacted GENERATE study genetic counselors. Trends based on the first 98 participants did not indicate differences in demographics or participants’ cancer experiences between the two study arms. Future analyses that evaluate data from the administered instruments that measure participant satisfaction, distress, family communication, knowledge gained and engagement in cancer surveillance may demonstrate behaviors that changed as a result of the intervention and may or may not support the addition of a genetic counselor in the education and testing process.
In conclusion, the first year of the GENERATE study resulted in over 90% of randomized participants pursuing genetic testing for PDAC, demonstrating remarkable success of remote education and at-home participation in genetic testing in intervening against PDAC, a disease which to date, has shown limited reason for hope. The high-risk relatives identified through genetic evaluation comprise an ideal population for future cutting-edge surveillance and for implementation of cancer interception strategies with the goal of PDAC prevention.
Supplementary Material
Financial Support
This work was supported by Stand Up To Cancer-Lustgarten Foundation Pancreatic Cancer Interception Translational Cancer Research Grant (Grant Number: SU2C-AACR-DT25-17; All Authors). Stand Up To Cancer is a division of the Entertainment Industry Foundation. The indicated research grants is administered by the American Association for Cancer Research, the scientific partner of SU2C.
This work was also supported by the Mayo Clinic SPORE in Pancreatic Cancer (Grant Number: P50CA102701; G. Petersen), the Khalifa Bin Zayed Foundation, The Bowen-Chapman Family Research Fund, and by NIH grants T32 DK007533-35 (N. Rodriguez) and CA210170 (M. Goggins).
The authors disclose the following Conflicts of Interests: Dr. Matthew Yurgelun received a one-time consulting/scientific advisory board fee from Janssen Pharmaceuticals, and research funding from Janssen Pharmaceuticals; Dr. Klein receives consulting from Merck; Dr. Garber receives consulting fees from Helix Genetics; Dr. Syngal has received consultant fees from Myriad Genetics and DC Health Technologies and has rights to an inventor portion of the licensing revenue from the PREMM5 model; Dr. Maitra receives royalties from Cosmos Wisdom Biotech for a license related to a pancreatic cancer early detection test. He is also listed as an inventor on a patent licensed to Thrive Earlier Detection Ltd; Dr. Zhou, Sydney Okumura and Sherman Law are full-time employees of Color Genomics; Dr. Ocean receives consulting fees from Tyme Technologies, speaker’s bureau for Daiichi Sanyko for Injectafer.
References
- 1.Conroy T, et al. , FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med, 2011. 364(19): p. 1817–25. [DOI] [PubMed] [Google Scholar]
- 2.Blackford AL, et al. , Recent trends in the incidence and survival of Stage 1A Pancreatic Cancer: A Surveillance, Epidemiology, and End Results analysis. J Natl Cancer Inst, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yurgelun MB, et al. , Germline cancer susceptibility gene variants, somatic second hits, and survival outcomes in patients with resected pancreatic cancer. Genet Med, 2019. 21(1): p. 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shindo K, et al. , Deleterious Germline Mutations in Patients With Apparently Sporadic Pancreatic Adenocarcinoma. J Clin Oncol, 2017. 35(30): p. 3382–3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu C, et al. , Association Between Inherited Germline Mutations in Cancer Predisposition Genes and Risk of Pancreatic Cancer. Jama, 2018. 319(23): p. 2401–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Golan T and Javle M, DNA Repair Dysfunction in Pancreatic Cancer: A Clinically Relevant Subtype for Drug Development. J Natl Compr Canc Netw, 2017. 15(8): p. 1063–1069. [DOI] [PubMed] [Google Scholar]
- 7.Chiaravalli M, Reni M, and O’Reilly EM, Pancreatic ductal adenocarcinoma: State-of-the-art 2017 and new therapeutic strategies. Cancer Treat Rev, 2017. 60: p. 32–43. [DOI] [PubMed] [Google Scholar]
- 8.Kaufman B, et al. , Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J Clin Oncol, 2015. 33(3): p. 244–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Reilly EM, et al. , Randomized, Multicenter, Phase II Trial of Gemcitabine and Cisplatin With or Without Veliparib in Patients With Pancreas Adenocarcinoma and a Germline BRCA/PALB2 Mutation. J Clin Oncol, 2020. 38(13): p. 1378–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.NCCN Clinical Practice Guidelines in Oncology; Genetic/Familial High-Risk Assessment: Breast, Ovarian and Pancreatic; Version 2.2021. [DOI] [PubMed]
- 11.Goggins M, et al. , Management of patients with increased risk for familial pancreatic cancer: updated recommendations from the International Cancer of the Pancreas Screening (CAPS) Consortium. Gut, 2020. 69(1): p. 7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vasen H, et al. , Benefit of Surveillance for Pancreatic Cancer in High-Risk Individuals: Outcome of Long-Term Prospective Follow-Up Studies From Three European Expert Centers. J Clin Oncol, 2016. 34(17): p. 2010–9. [DOI] [PubMed] [Google Scholar]
- 13.Konings I, et al. , Surveillance for pancreatic cancer in high-risk individuals. BJS Open, 2019. 3(5): p. 656–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Canto MI, et al. , Risk of Neoplastic Progression in Individuals at High Risk for Pancreatic Cancer Undergoing Long-term Surveillance. Gastroenterology, 2018. 155(3): p. 740–751.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stoffel EM, et al. , Evaluating Susceptibility to Pancreatic Cancer: ASCO Provisional Clinical Opinion. J Clin Oncol, 2019. 37(2): p. 153–164. [DOI] [PubMed] [Google Scholar]
- 16.Chittenden A, Haraldsdottir S, Ukaegbu C, et al. , Implementing Systemic Genetic Counseling and Multigene Germline Testing for Individuals with Pancreatic Cancer. JCO Oncology Pract, 2020. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peters MLB, et al. , Family communication and patient distress after germline genetic testing in individuals with pancreatic ductal adenocarcinoma. Cancer, 2019. 125(14): p. 2488–2496. [DOI] [PubMed] [Google Scholar]
- 18.Cheung EL, et al. , Communication of BRCA results and family testing in 1,103 high-risk women. Cancer Epidemiol Biomarkers Prev, 2010. 19(9): p. 2211–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharaf RN, et al. , Uptake of genetic testing by relatives of lynch syndrome probands: a systematic review. Clin Gastroenterol Hepatol, 2013. 11(9): p. 1093–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forrest K, et al. , To tell or not to tell: barriers and facilitators in family communication about genetic risk. Clin Genet, 2003. 64(4): p. 317–26. [DOI] [PubMed] [Google Scholar]
- 21.Courtney E, et al. , Impact of free cancer predisposition cascade genetic testing on uptake in Singapore. NPJ Genom Med, 2019. 4: p. 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.George R, Kovak K, and Cox SL, Aligning policy to promote cascade genetic screening for prevention and early diagnosis of heritable diseases. J Genet Couns, 2015. 24(3): p. 388–99. [DOI] [PubMed] [Google Scholar]
- 23.London JW, et al. , Effects of the COVID-19 Pandemic on Cancer-Related Patient Encounters. JCO Clin Cancer Inform, 2020. 4: p. 657–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCuaig JM, et al. , Next-Generation Service Delivery: A Scoping Review of Patient Outcomes Associated with Alternative Models of Genetic Counseling and Genetic Testing for Hereditary Cancer. Cancers (Basel), 2018. 10(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frey MK, et al. , Prospective Feasibility Trial of a Novel Strategy of Facilitated Cascade Genetic Testing Using Telephone Counseling. J Clin Oncol, 2020. 38(13): p. 1389–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caswell-Jin JL, et al. , Cascade Genetic Testing of Relatives for Hereditary Cancer Risk: Results of an Online Initiative. J Natl Cancer Inst, 2019. 111(1): p. 95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rayes N, et al. , MAGENTA (Making Genetic testing accessible): a prospective randomized controlled trial comparing online genetic education and telephone genetic counseling for hereditary cancer genetic testing. BMC Cancer, 2019. 19(1): p. 648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mann DM, et al. , COVID-19 transforms health care through telemedicine: Evidence from the field. J Am Med Inform Assoc, 2020. 27(7): p. 1132–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Contreras CM, et al. , Telemedicine: Patient-Provider Clinical Engagement During the COVID-19 Pandemic and Beyond. J Gastrointest Surg, 2020. 24(7): p. 1692–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goggins MG, et al. , Intercepting Pancreatic Cancer: Our Dream Team’s Resolve to Stop Pancreatic Cancer. Pancreas, 2018. 47(10): p. 1175–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harris PA, et al. , Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform, 2009. 42(2): p. 377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Donner A and Klar N, Methods for comparing event rates in intervention studies when the unit of allocation is a cluster. Am J Epidemiol, 1994. 140(3): p. 279–89; discussion 300–1. [DOI] [PubMed] [Google Scholar]
- 33.Neben CL, et al. , Multi-Gene Panel Testing of 23,179 Individuals for Hereditary Cancer Risk Identifies Pathogenic Variant Carriers Missed by Current Genetic Testing Guidelines. J Mol Diagn, 2019. 21(4): p. 646–657. [DOI] [PubMed] [Google Scholar]
- 34.Underhill M, et al. , Relationship between individual and family characteristics and psychosocial factors in persons with familial pancreatic cancer. Psychooncology, 2018. 27(7): p. 1711–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Konings IC, et al. , Factors associated with cancer worries in individuals participating in annual pancreatic cancer surveillance. Fam Cancer, 2017. 16(1): p. 143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zigmond AS and Snaith RP, The hospital anxiety and depression scale. Acta Psychiatr Scand, 1983. 67(6): p. 361–70. [DOI] [PubMed] [Google Scholar]
- 37.Morrill R, C.J., Hart G, Meropolitan, urban, and rural commuting areas: Toward a better depiction of the United States settlement system. Urban Geography, 1999; 20:727–748. [Google Scholar]
- 38.Kind AJH and Buckingham WR, Making Neighborhood-Disadvantage Metrics Accessible - The Neighborhood Atlas. N Engl J Med, 2018. 378(26): p. 2456–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Underhill-Blazey M, et al. , Development and testing of the KnowGene scale to assess general cancer genetic knowledge related to multigene panel testing. Patient Educ Couns, 2019. 102(8): p. 1558–1564. [DOI] [PubMed] [Google Scholar]
- 40.Kilbride MK, Domchek SM, and Bradbury AR, Ethical Implications of Direct-to-Consumer Hereditary Cancer Tests. JAMA Oncol, 2018. 4(10): p. 1327–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoskovec JM, et al. , Projecting the Supply and Demand for Certified Genetic Counselors: a Workforce Study. J Genet Couns, 2018. 27(1): p. 16–20. [DOI] [PubMed] [Google Scholar]
- 42.Stoffel EM, et al. , Sharing genetic test results in Lynch syndrome: communication with close and distant relatives. Clin Gastroenterol Hepatol, 2008. 6(3): p. 333–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chaffee KG, et al. , Prevalence of germ-line mutations in cancer genes among pancreatic cancer patients with a positive family history. Genet Med, 2018. 20(1): p. 119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.NCCN Practical Guidelines in Oncology; Prostate Cancer Early Detection. Version 2.2020 2020.
- 45.Swisher EM, et al. , Results from MAGENTA: A national randomized four-arm noninferiority trial evaluating pre- and post-test genetic counseling during online testing for breast and ovarian cancer genetic risk. Journal of Clinical Oncology, 2020. 38(15_suppl): p. 1506–1506. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


