Fig. 2.
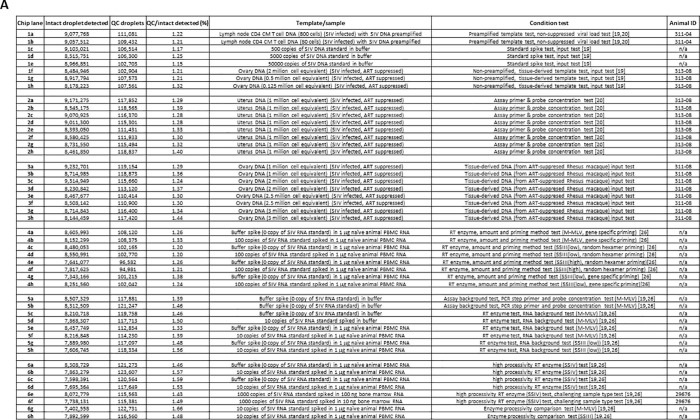
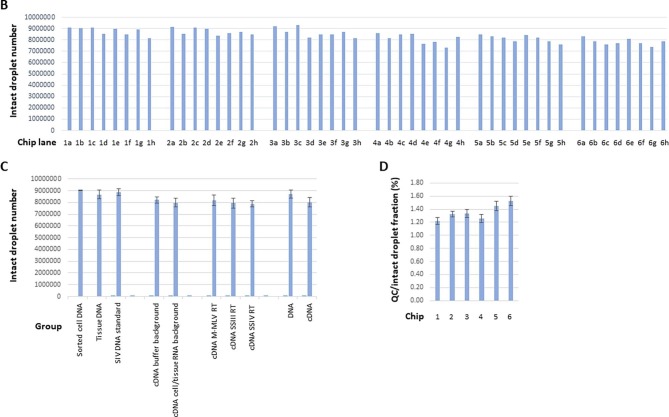
Summary of the numbers of droplets that were successfully imaged on the sense chips under various test conditions and with a variety of sample types. (A) A detailed description of the numbers of intact droplets detected during fluorescence imaging on the sense instrument, the corresponding numbers of quality control (QC) droplets (representing a fraction of the total droplets generated for each lane) during dropletization on the source instrument, the QC/intact droplet fraction, samples and test conditions, and animal IDs (if applicable). Chip 1–3 results were based on DNA templates, and chip 4–6 results were based on cDNA templates. (Chip 1) Samples tested included preamplified cell DNA from an animal infected with SIV (no ART suppression) (a and b), SIV DNA standard spike (c-e), and non-preamplified tissue DNA from an SIV-infected animal subject to ART suppression (f-g) [20], [21]. CM T cell: central memory T cell. ART: antiretroviral therapy. (Chip 2) Assay primer and probe concentration tests on tissue-derived DNA template from an SIV-infected animal subject to ART suppression [21]. Primer and probe concentration (in nM) variations tested (in the order of SGag forward, SGag reverse, SGag ddPCR probe, RCCR5 forward, RCCR5 reverse, RCCR5 ddPCR probe) were: (a) 600, 600, 200, 400, 400, 200; (b) 600, 600, 200, 600, 600, 200; (c) 600, 600, 200, 200, 200, 200 (the standard condition as indicated in section 3.4 ddPCR recipe); (d) 600, 600, 200, 200, 200, 100; (e) 600, 600, 100, 400, 400, 200; (f) 600, 600, 100, 600, 600, 200; (g) 600, 600, 100, 200, 200, 200; (h) 600, 600, 100, 200, 200, 100. (Chip 3) Tissue-derived DNA input tests. 1 million to 3 million cell equivalent DNA derived from the ovary tissue from an SIV-infected, ART suppressed Rhesus macaque were subject to ddPCR analysis. (Chip 4) Reverse transcriptase enzyme, amount and priming strategy tests. PBMC: peripheral blood mononuclear cell. M−MLV, 200 units (per manufacturer definition) of the M−MLV reverse transcriptase in each reverse transcription reaction. SSIII(low), 20 units (per manufacturer definition) of the SuperScript III reverse transcriptase in each reverse transcription reaction. SSIII(high), 200 units of the SuperScript III reverse transcriptase in each reverse transcription reaction. (Chip 5) Assay background and reverse transcriptase tests. Assay background (no template control) tests were performed both in buffer background (a-c) and in the background of RNA extracted from PBMCs from a naïve animal (e and g). M−MLV, 200 units (per manufacturer definition) of the M−MLV reverse transcriptase in each reverse transcription reaction. SSIII(low), 20 units (per manufacturer definition) of the SuperScript III reverse transcriptase in each reverse transcription reaction. (Chip 6) Enzyme processivity tests and bone marrow test. Performances of 3 reverse transcriptases with different processivity at low RNA template input were compared (a-d, g and h). In addition, the performance of the SSIV reverse transcriptase in the background of RNA derived from a high fat content tissue (i.e. bone marrow) was evaluated. The number of intact droplets detected at the sense step on chips 1–6 on average was 99.6 ± 1.2% of the number of total droplets detected at the sense instrument step. (B) Intact droplet number for each individual lane as plotted based on (A). (C) Group comparison of intact droplet numbers based on (A) and (B). The numeric values are: Sorted cell DNA group (1a, 1b), n = 2, average = 9,067,640, range = 9,057,512–9,077,768; tissue DNA group (1f-1h, 2a-2h, 3a-3h), n = 19, 8,677,637 ± 356,302; SIV DNA standard group (1c-1e), n = 3, 8,861,874 ± 307,387. cDNA buffer background group (5a-5d), n = 4, 8,224,716 ± 267,582; cDNA cell/tissue RNA background group (4a-4h, 5e-5h, 6a-6h), n = 20, 7,979,296 ± 384,328. cDNA M−MLV reverse transcriptase (RT) group (4a, 4b, 5e, 5f, 6g), n = 5, 8,167,089 ± 464,845; cDNA SSIII RT group (4c-4h, 5g, 5h, 6h), n = 9, 7,941,533 ± 409,018; cDNA SSIV RT group (6a-6f), n = 6, 7,879,446 ± 266,372. DNA group (1a-1h, 2a-2h, 3a-3h), n = 24, 8,733,167 ± 349,305; cDNA group (4a-4h, 5a-5h, 6a-6h), n = 24, 8,020,199 ± 374,283. Although not statistically significant, there was a trend which suggested that lanes that contained DNA templates as the input on average had greater intact droplet numbers than lanes that contained cDNA templates as the input. (D) Comparison of the QC droplets/intact droplets factions (in %) among the 6 chips based on (A). The numeric values are: chip 1, n = 8, 1.22 ± 0.05; chip 2, n = 8, 1.32 ± 0.05; chip 3, n = 8, 1.33 ± 0.06; chip 4, n = 8, 1.26 ± 0.06; chip 5, n = 8, 1.45 ± 0.07; chip 6, 1.53 ± 0.08.