Abstract
The B.1.1.7 (Alpha) variant has been detected in Mumbai, India during February 2021. Subsequently, we retrieved 43 sequences from specimens of 51 COVID-19 cases from Mumbai. The sequence analysis revealed that the cases were mainly affected with Alpha variant which suggests its role in community transmission of SARS-CoV-2 in Mumbai, India.
Keywords: SARS-CoV-2, Variant of concern, B.1.1.7, Community transmission, Mumbai
Over the course of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) pandemic, multiple variants have emerged across the globe. During late December 2020, a new variant of concern (VOC) B.1.1.7 (Alpha) was identified in the United Kingdom which has spread to more than 135 countries in the world including Europe, Asia, USA.1 This variant has about 17 mutations, including N501Y, P681H, HV 69–70 deletion in the spike protein and various other mutations. It has been designated as variant of concern due to its high transmissibility, with estimates ranging from 40 to 80% higher transmissibility.2 , 3 During the first wave of pandemic in India, SARS-CoV-2 cases showed downward trend since September 2020; however it again started rising in March 2021 with the number of cases being double of last year. Government of India started the genomic surveillance of newly emerging SARS-CoV-2 variants under ‘The Indian SARS-CoV-2 Consortium on Genomics (INSACOG)’. The clinical specimens of all the United Kingdom (UK) travelers were screened using next-generation sequencing (NGS) since January 2021. Genomic surveillance has detected VOC Alpha, Beta (B.1.351), Gamma (B.1.1.28.1) variants. The Alpha was found to be the second most common variant in India among the detected variants till March 2021.4
During the first week of January 2021, the new SARS-CoV-2 variant, Alpha was first detected among the UK travelers arriving in Mumbai, Maharashtra, India. Despite the strict mitigation policies, two distinct areas in Mumbai had a spurt in SARS-CoV-2 cases mainly due to dense population, informal settlements and overcrowding. With the ongoing screening of SARS-CoV-2 in government and private laboratories in Mumbai, two private laboratories in Mumbai using TaqPath kits for Real time RT-PCR detected Spike gene target failure (SGTF) among clinical specimens collected from the community during January–February 2021. SGTF has been already identified and assumed to be a surrogate marker for the detection of alpha variant.3 SGTF clinical samples of SARS-CoV-2 cases from 2 private laboratories received at Reference molecular laboratory at Kasturba Hospital for Infectious Diseases, Mumbai were sent to ICMR-National Institute of Virology (ICMR-NIV), Pune for whole genome sequencing.
Genomic characterization of the referred clinical specimens was carried out with the quantified RNA using next-generation sequencing (NGS). Briefly, the ribosomal RNA depletion was performed using NebnextrRNA depletion kit (Human/mouse/rat) followed by cDNA synthesis using the first strand and second synthesis kit. The RNA libraries were prepared using TruSeq Stranded Total RNA library preparation kit. The amplified RNA libraries were quantified and loaded on the Nexseq Illumina sequencing platform after normalization.5
Reference-based mapping was performed to retrieve the genomic sequence of the SARS-CoV-2 from clinical samples using the CLC genome workbench v20.0.4 and submitted to the public repository i.e., GISAID. A phylogenetic tree was generated using the MEGA software version.7
Out of 51 SARS-CoV-2 cases, the clinical details of 37 cases (18 male, 19 female) were available. The cases were distributed amongst all the age groups including <14 years (n = 3); 15–45 years (n = 14), 46–60 years (n = 8), and >60 years (n = 17). Out of 37 cases, 70% were asymptomatic while 27.03% had cough, 21.62% had fever, 18.92% had mild breathlessness and 13.51% had runny nose. The cases were followed till 14 days to check for the development of any kind of symptoms. The cases didn't develop disease severity and recovered completely. The COVID-19 vaccination drive has started in India from 16 January 2021. However, only health care and frontline workers were vaccinated during the initial phase of vaccination. All the study participants were not vaccinated against SARS-CoV-2 during the study period.
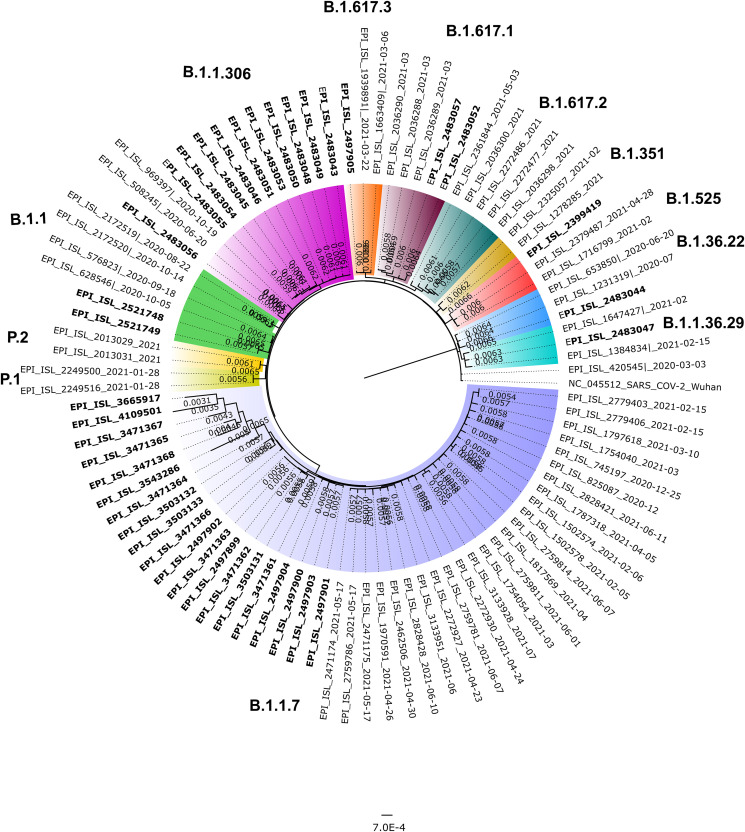
SARS-CoV-2 sequences (n = 43) were retrieved with more than 98% genome length from the clinical specimens of the COVID-19 cases. The details of the total read mapped, percent relevant reads and genome length is given in Table 1 . The lineage of the retrieved sequences was identified using the pangolin (https://pangolin.cog-uk.io/). Majority of the sequences belong to B.1.1.7 lineage (n = 23) followed by B.1.1.306 lineage (n = 11). Sequences for variant of interest (VUI): Eta [B.1.525] (n = 1), B.1.617.3 (n = 1) and Kappa [B.1.617.1] (n = 2) and other common circulating variants (n = 5) were also retrieved. Fig. 1 depicts the neighbor-joining tree for the retrieved and the reference SARS-CoV-2 sequences. The other sequences retrieved were B.1.1 (n = 2), B.1.36.22 (n = 1), B.1.1.36.29 (n = 1) and B.1.409 (n = 1)common circulating lineages. The percent nucleotide similarity (PNS) of the retrieved SARS-CoV-2 sequences in comparison to Wuhan Hu-1 isolate is given in supplementary table 1.
Table 1.
Details of the clinical samples of the Community transmission of SARS-CoV-2 cases from Mumbai.
| Sr. No | MCL No | Age | Sex | Symptomatic/asymptomatic | Address | Date of onset/testing | SARS-CoV-2 result with CT Value for E gene and ORF1 | Percentage genome retrieved | Circulating clade | Updated Pangolin Lineage | GISAID number |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | MCL-21-H-1007 | 63 | M | COUGH, BREATHLESSNESS, FEVER | Sion | 16.01.2021 | Positive | 100 | GR | B.1.1.306 | EPI_ISL_2483050 |
| 2 | MCL-21-H-970 | 65 | M | Asymptomatic | Dahisar | 28.01.2021 | Positive | 100 | GR | B.1.1.306 | EPI_ISL_2483043 |
| 3 | MCL-21-H-802 | 82 | M | Asymptomatic | Boriwali | Feb-21 | 22.06 | 97.85 | GR | B.1.1.7 | EPI_ISL_3665916 |
| 4 | MCL-21-H-808 | Not provided | F | Not provided | Not provided | Feb-21 | Not provided | 97.85 | _ | – | – |
| 5 | MCL-21-H-810 | Not provided | M | Not provided | Not provided | Feb-21 | Not provided | 99.22 | GR | B.1.1.7 | EPI_ISL_3471364 |
| 6 | MCL-21-H-812 | Not provided | F | Not provided | Not provided | Feb-21 | 18 | 99.68 | GR | B.1.1.7 | EPI_ISL_3471365 |
| 7 | MCL-21-H-813 | Not provided | M | Not provided | Not provided | Feb-21 | 21 | 99.15 | GR | B.1.1.7 | EPI_ISL_3665917 |
| 8 | MCL-21-H-814 | Not provided | M | Not provided | Not provided | Feb-21 | 15 | 98.5 | GR | B.1.1.7 | EPI_ISL_3503132 |
| 9 | MCL-21-H-815 | Not provided | M | Not provided | Not provided | Feb-21 | 15 | 98.44 | GR | B.1.1.7 | EPI_ISL_3503133 |
| 10 | MCL-21-H-816 | Not provided | F | Not provided | Not provided | Feb-21 | 19 | 99.48 | GR | B.1.1.7 | EPI_ISL_3471366 |
| 11 | MCL-21-H-817 | Not provided | F | Not provided | Not provided | Feb-21 | 18 | 96.87 | _ | – | – |
| 12 | MCL-21-H-819 | Not provided | M | Not provided | Not provided | Feb-21 | 18 | 93.92 | _ | – | – |
| 13 | MCL-21-H-820 | Not provided | M | Not provided | Not provided | Feb-21 | 26 | 98.15 | GR | B.1.1.7 | EPI_ISL_4109501 |
| 14 | MCL-21-H-821 | Not provided | M | Not provided | Not provided | Feb-21 | 21 | 99.41 | GR | B.1.1.7 | EPI_ISL_3471367 |
| 15 | MCL-21-H-822 | Not provided | F | Not provided | Not provided | Feb-21 | 15 | 99.25 | GR | B.1.1.7 | EPI_ISL_3471368 |
| 16 | MCL-21-H-823 | Not provided | M | Not provided | Not provided | Feb-21 | 28 | 99.3 | GR | B.1.1.7 | EPI_ISL_3543285 |
| 17 | MCL-21-H-824 | Not provided | M | Not provided | Not provided | Feb-21 | 19 | 99.2 | GR | B.1.1.7 | EPI_ISL_3543286 |
| 18 | MCL-21-H-1024 | 38 | M | Asymptomatic | Boriwali | 03.02.2021 | Positive | 99.82 | GR | B.1.1 | EPI_ISL_2521748 |
| 19 | MCL-21-H-1025 | 37 | F | Asymptomatic | Borilwali | 03.02.2021 | Positive | 100.02 | GR | B.1.1 | EPI_ISL_2521749 |
| 20 | MCL-21-H-1029 | 31 | F | Asymptomatic | Boriwali | 03.02.2021 | Positive | 100.02 | GR | B.1.1.306 | EPI_ISL_2483053 |
| 21 | MCL-21-H-1030 | 56 | M | COUGH FEVER | Boriwali | 03.02.2021 | Positive | 100 | GR | B.1.1.306 | EPI_ISL_2483054 |
| 22 | MCL-21-H-1031 | 32 | M | Asymptomatic | Boriwali | 03.02.2021 | Positive | 100.02 | GR | B.1.1.306 | EPI_ISL_2483055 |
| 23 | MCL-21-H-1033 | 24 | F | cold, cough, fever | Dahisar | 03.02.2021 | Positive | 100.01 | GR | B.1.1.306 | EPI_ISL_2483056 |
| 24 | MCL-21-H-1034 | 77 | M | fever, bodyache, cough, | Dahisar | 03.02.2021 | Positive | 100 | G | B.1.617.1 | EPI_ISL_2483057 |
| 25 | MCL-21-H-972 | 71 | M | cough breathlessness | Not provided | 06.02.2021 | Positive | 100 | GH | B.1.36.22 | EPI_ISL_2483044 |
| 26 | MCL-21-H-615 | 41 | F | Asymptomatic | Dombivali | 11.02.2021 | 26.28 | 73.1 | _ | – | – |
| 27 | MCL-21-H-616 | 8 | F | Asymptomatic | Mankhurd | 11.02.2021 | 25.05 | 41.9 | _ | – | – |
| 28 | MCL-21-H-976 | 68 | M | Breathlessness | Sion | 16.02.2021 | Positive | 100 | GR | B.1.1.306 | EPI_ISL_2483045 |
| 29 | MCL-21-H-608 | 57 | M | Asymptomatic | Bandra | 17.02.2021 | 16 | 99.68 | GR | B.1.1.7 | EPI_ISL_2497900 |
| 30 | MCL-21-H-609 | 53 | M | Asymptomatic | Mulund | 17.02.2021 | 14 | 99.73 | G | B.1.525 | EPI_ISL_2399419 |
| 31 | MCL-21-H-613 | 50 | M | Asymptomatic | Powai | 17.02.2021 | 19 | 98.91 | G | B.1.1.7 | EPI_ISL_3471363 |
| 32 | MCL-21-H-977 | 65 | M | Breathlessness | Badlapur | 17.02.2021 | Positive | 100 | GR | B.1.1.306 | EPI_ISL_2483046 |
| 33 | MCL-21-H-602 | 48 | F | Asymptomatic | Kalbadevi | 18.02.2021 | 32 | 50.05 | _ | – | – |
| 34 | MCL-21-H-603 | 25 | F | Asymptomatic | Mulund | 18.02.2021 | 21 | 98.22 | GR | B.1.1.7 | EPI_ISL_3503131 |
| 35 | MCL-21-H-604 | 37 | M | Asymptomatic | Mulund | 18.02.2021 | 17 | 99.68 | GR | B.1.1.7 | EPI_ISL_2497899 |
| 36 | MCL-21-H-605 | 64 | F | Asymptomatic | Mulund | 18.02.2021 | 22 | 98.17 | GR | B.1.1.7 | EPI_ISL_3471361 |
| 37 | MCL-21-H-606 | 54 | F | Asymptomatic | Mulund | 18.02.2021 | 25 | 97.58 | GR | B.1.1.7 | EPI_ISL_3665914 |
| 38 | MCL-21-H-607 | 71 | F | Asymptomatic | Mulund | 18.02.2021 | 22 | 99.41 | GR | B.1.1.7 | EPI_ISL_3471362 |
| 39 | MCL-21-H-610 | 66 | F | Asymptomatic | Mulund | 18.02.2021 | 15 | 99.58 | GR | B.1.1.7 | EPI_ISL_2497901 |
| 40 | MCL-21-H-611 | 12 | F | Asymptomatic | Thane | 18.02.2021 | 15 | 99.78 | GR | B.1.1.7 | EPI_ISL_2497902 |
| 41 | MCL-21-H-612 | 44 | M | Asymptomatic | Mulund | 18.02.2021 | 20 | 99.67 | GR | B.1.1.7 | EPI_ISL_2497903 |
| 42 | MCL-21-H-614 | 30 | F | Asymptomatic | Mulund | 18.02.2021 | 18 | 99.78 | GR | B.1.1.7 | EPI_ISL_2497904 |
| 43 | MCL-21-H-978 | 45 | M | Asymptomatic | Digha Navi Mumbai | 18.02.2021 | Positive | 100 | GH | B.1.36.29 | EPI_ISL_2483047 |
| 44 | MCL-21-H-801 | 2 | F | Asymptomatic | Dharavi | 23.02.2021 | Positive | 97.39 | GR | B.1.409 | EPI_ISL_3665915 |
| 45 | MCL-21-H-1020 | 21 | M | COUGH, BREATHLESSNESS, FEVER | Sion | 27.02.2021 | Positive | 99.88 | G | B.1.617.3 | EPI_ISL_2497905 |
| 46 | MCL-21-H-988 | 53 | F | COUGH, FEVER | Kandivali | 28.02.2021 | Positive | 100.01 | GR | B.1.1.306 | EPI_ISL_2483048 |
| 47 | MCL-21-H-996 | 66 | F | Asymptomatic | Kandivali | 28.02.2021 | Positive | 100 | GR | B.1.1.306 | EPI_ISL_2483049 |
| 48 | MCL-21-H-1022 | 68 | F | COUGH, BREATHLESSNESS, FEVER | Dharavi | 28.02.2021 | Positive | 100 | GR | B.1.1.306 | EPI_ISL_2483051 |
| 49 | MCL-21-H-1023 | 60 | M | COUGH, BREATHLESSNESS, FEVER | Goavndi | 01.03.2021 | Positive | 99.99 | G | B.1.617.1 | EPI_ISL_2483052 |
| 50 | MCL-21-H-1316 | 40 | M | Asymptomatic | Not provided | 08.03.2021 | Positive | 77.6 | _ | – | – |
| 51 | MCL-21-H-799 | 26 | M | Asymptomatic | Santacruz | 18.03.2021 | Positive | 97.3 | _ | – | – |
Figure 1.
Phylogenetic tree of the SARS-CoV-2 sequences retrieved from Mumbai, Maharashtra: A neighbor joining tree was generated using a Tamura 3-paramter model with gamma distribution and a bootstrap replication of 1000 cycles. The sequences retrieved in this study are marked in boldfaced. The variants of concern, and the variants of interest are marked in different colors on branches; B.1.1.7 (light blue), B.1.525 (Crimson red), B.1.617.3 (orange), B.1.617.1 (brown), B.1.1.306 (pink color).
Mumbai is the India's most populous city and parts of Mumbai have impoverished tenements and large slum areas which are amongst the most densely packed human habitats in the world.6 Additionally overcrowding in public transport modes like local trains, buses are common in Mumbai. The combination of intense contact between commuters and close inhabitation favors SARS-CoV-2 transmission. This has contributed to rapid community transmission of SARS-CoV-2 among the population in Mumbai City.
The use of SGTF as surrogate marker will not be advisable as many of the SARS-CoV-2 testing laboratories do not use S-gene target for SARS-CoV-2 PCR testing. This could lead to many missed Alpha variant cases, and therefore genome sequencing is warranted.
This genome sequencing study led to identification of a mix of different variants including VOC Alpha variant and few cases of Kappa and B.1.617.3 variants in the local population of Mumbai, Maharashtra. The higher numbers of Alpha variant indicates a local transmission of the virus within the area during the month of February 2021. This could have contributed to rise in cases in this city. Further it is observed that the N501Y mutation present on the spike gene is also not present in the most of the sequences that fall in Alpha lineage. The N501Y mutation influences the binding affinity of the virus to the human ACE2 receptors7, indicating a reduced ACE2 affinity of the Alpha variant in current samples. The SARS-CoV-2 sequences retrieved from the B.1.1.306 lineage have L18F, A27S, E484K, D641G, Q675H mutations in the spike protein. E484K mutation is reported to be an escape mutation, and the presence of this mutation is a cause of concern. The presence of B.1.617.3 and Kappa variant in the samples collected during the February, 2021 indicates early circulation of these variant in Mumbai. During the second wave 2, 89, 11,744 cases and 3, 63,079 deaths have been recorded in India with a crude case fatality rate of 1.24%. As on 11th June 2021, Maharashtra reported cumulative 56, 08,753 cases during the second wave.
Extensive mitigation policies like effective contact tracing of the local and international travelers, strict isolation and quarantine protocols, imposing a ban on public gatherings, community awareness and engagement, stringent lockdown proved to be appropriate strategies to break the chain of virus transmission. Besides this, robust and proactive genomic surveillance of emerging SARS-CoV-2 variants is imperative for the timely identification and control of their transmission in the community. Rapidly increasing vaccine coverage in India would be the most effective way to inhibit further deadly waves of COVID-19.
Ethical approval
The study was approved by the Institutional Biosafety Committee and Institutional Human Ethics Committee of ICMR-NIV, Pune, India under project ‘Molecular epidemiological analysis of SARS-COV-2 circulating in different regions of India’ (20-3-18N).
Financial support & sponsorship
Financial support was provided by the Department of Health Research, Ministry of Health & Family Welfare, New Delhi, at ICMR-National Institute of Virology, Pune under project ‘Molecular epidemiological analysis of SARS-COV-2 circulating in different regions of India’ (20-3-18N).
Author contributions
JS, PDY contributed to study design, data collection, data analysis, interpretation and writing and critical review. SA, AS, SP, DAN, MG, RRS, DYP, MD, NK contributed to data collection, data analysis data interpretation, writing and critical review.
Data availability statement
Data set of this study is available within article and any other information will be available from the corresponding author upon reasonable request.
Declaration of competing interest
No conflict of interest exists among authors.
Acknowledgement
Authors gratefully acknowledge the encouragement and support extended by Prof. (Dr.) Balram Bhargava, Secretary to the Government of India Department of Health Research, Ministry of Health & Family Welfare & Director-General, ICMR New Delhi; Prof. (Dr.) Priya Abraham, Director, ICMR-NIV, Pune and Dr. Nivedita Gupta, Head, Virology unit, Epidemiology and Communicable Diseases, ICMR, New Delhi. Authors are also thankful to the team members of Maximum Containment Facility, ICMR-NIV, Pune including Dr. Abhinendra Kumar, Mr. Yash Joshi, Mrs. Savita Patil, Mrs. Triparna Majumdar, Ms. Pranita Gawande, Ms. Jyoti Yemul, Mrs. Ashwini Waghmare and Mrs. Kaumudi Kalele for providing excellent technical support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmii.2021.10.004.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.PANGO lineages. 2021, June 8. https://cov-lineages.org/lineage_list.html Available from.
- 2.Volz E., Mishra S., Chand M., Barrett J.C., Johnson R., Geidelberg L., et al. Transmission of SARS-CoV-2 lineage B. 1.1. 7 in England: insights from linking epidemiological and genetic data. medRxiv. 2021 doi: 10.1101/2020.12.30.20249034. 2020-12. [DOI] [Google Scholar]
- 3.Davies N.G., Abbott S., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J.D., et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B. 1.1. 7 in England. Science. 2021;(6538):372. doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Indian SARS-CoV-2 genome sequencing consortia (INSACOG) 2021, June 8. https://dbtindia.gov.in/insacog Available from.
- 5.Yadav P.D., Nyayanit D.A., Shete A.M., Jain S., Majumdar T.P., Chaubal G.Y., et al. Complete genome sequencing of Kaisodi virus isolated from ticks in India belonging to Phlebovirus genus, family Phenuiviridae. Ticks tick-borne dis. 2019;10(1):23–33. doi: 10.1016/j.ttbdis.2018.08.012. [DOI] [PubMed] [Google Scholar]
- 6.Golechha M. COVID-19 containment in asia's largest urban slum dharavi-Mumbai, India: lessons for policymakers globally. J Urban Health. 2020;97(6):796–801. doi: 10.1007/s11524-020-00474-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laffeber Charlie, de Koning K., Kanaar Roland, Joyce H., Lebbink G. Experimental evidence for enhanced receptor binding by rapidly spreading SARS-CoV-2 variants. J Mol Biol. 2021 Jul 23;433(15):167058. doi: 10.1016/j.jmb.2021.167058. Epub 2021 May 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data set of this study is available within article and any other information will be available from the corresponding author upon reasonable request.