Abstract
Objective
Variants of concern (VOCs) associated with relatively high transmissibility appear to be rapidly spreading in Gabon. Therefore, it is imperative to understand the distribution of several VOCs in the population, which could have implications for transmissibility and vaccine efficacy.
Methods
Between February and May 2021, SARS-CoV-2 genomes were sequenced using the Oxford nanopore MinION method and the respective genome diversity was elucidated. Phylogenetic analysis was performed and genomes were classified using pangolin lineages.
Results
The results highlighted an increase (46%) in the alpha VOC (B.1.1.7) in the Gabonese population over the study period. In addition, an increase (31%) in the B.1.1.318 lineage, which is associated with high transmission and impaired vaccine efficacy (D614G+E484K+Y144del), was detected.
Conclusion
With the second wave ongoing, these findings highlight the need for surveillance of the SARS-CoV-2 genome in the Republic of Gabon and should provide useful guidance to policymakers in selecting an appropriate vaccine for this population.
Keywords: SARS-CoV-2, B.1.1.318, Variant of concern, B.1.1.7, Transmission, Gabon, Central Africa, Oxford Nanopore, MinION
Introduction
SARS-CoV-2 variants of concern (VOC) appear to spread more easily. Other emerging variants are also gaining attention, either known as a "variants of interest" (VOI) or "variants under investigation" (VUI), which increase transmission, warranting further studies. Since the onset of the COVID-19 pandemic, the SARS-CoV-2 genomes have accumulated genetic diversity, leading to increased transmission with altered viral properties (Kraemer et al. 2021).
Of the several new SARS-CoV-2 variants recently reported, alpha VOC B.1.1.7 (associated with the N501Y mutation) (Rambaut et al. 2020), beta VOC B.1.351 (associated with the E484K mutation) (Tegally et al. 2021), and gamma VOC P.1 (B.1.1.28.1, mutations with likely epidemiological significance E484K, K417T, and N501Y) (Graf et al. 2021), and the delta VOC B.1.617.2 (mutations with likely epidemiological significance P681R, K417N, and L452R) are of great interest. B.1.1.318 has recently been reported as a VOI, carrying D614G, D796H, E484K, P681H, T95I, Y144del, among which D614G has been implicated in increased transmissibility, and E484K with decreased vaccine efficacy.
The Republic of Gabon has evidence of a circulating alpha VOC B.1.1.7 (Zoa-Assoumou et al. 2021) and has thus far recorded 25,819 confirmed cases with 165 deaths until 30 August 2021 (WHO 2021). This may be due to factors such as age, demographics, and the constant exposure and burden of co-infections (e.g. with helminths) (Velavan et al. 2021). The current second wave, which has been ongoing since February 2021, has led to increased SARS-Cov2 genomic surveillance. This study aimed to reveal the actual distribution of SARS-CoV-2 variants circulating in the country.
Materials and Methods
The Centre de Recherches Médicales de Lambaréné undertook COVID-19 surveillance between February and May 2021. The study participants were residents living in the Lambaréné region.
NGS sequencing and phylogenetic analysis
Samples with confirmed Ct values ≤30 were subsequently sequenced using the MinION sequencing platform (Oxford Nanopore Technologies, Oxford, UK). Libraries were prepared according to the nCoV-2019 sequencing protocol (RAPID barcoding, 1200 bp amplicon) V.3 (Freed et al. 2021). Viral genome assembly was performed using the ARTIC pipeline (https://github.com/artic-network/fieldbioinformatics).
SARS-CoV-2 genomes (n=74) circulating between February and May 2021 were retrieved from the global database for influenza gene sequences (GISAID) for African (n=15) and European (n=14) lineages. Only complete viral genomes were included in the analysis, and different previously represented lineages (n=27) from 29 countries were included. All 74 genomes in the present study were aligned with Wuhan-Hu-1 strain (NC_045512.2) using the Multiple Alignment using Fast Fourier Transform (MAFTT) algorithm (Katoh et al. 2002). The phylogenetic tree was reconstructed with the maximum likelihood method with 1000 bootstrap iteration using the general time-reversible (GTR) model with rate heterogeneity (GTR+G) in the IQ-TREE server (Trifinopoulos et al. 2016). SARS-CoV-2 genomes were classified into lineages using Phylogenetic Assignment of Named Global Outbreak LINeages (pangolin) (O'Toole et al. 2021) and viral clades by Nextclade Beta (https://clades.nextstrain.org/). The final dataset was displayed with FigTree v1.4.4.
Results
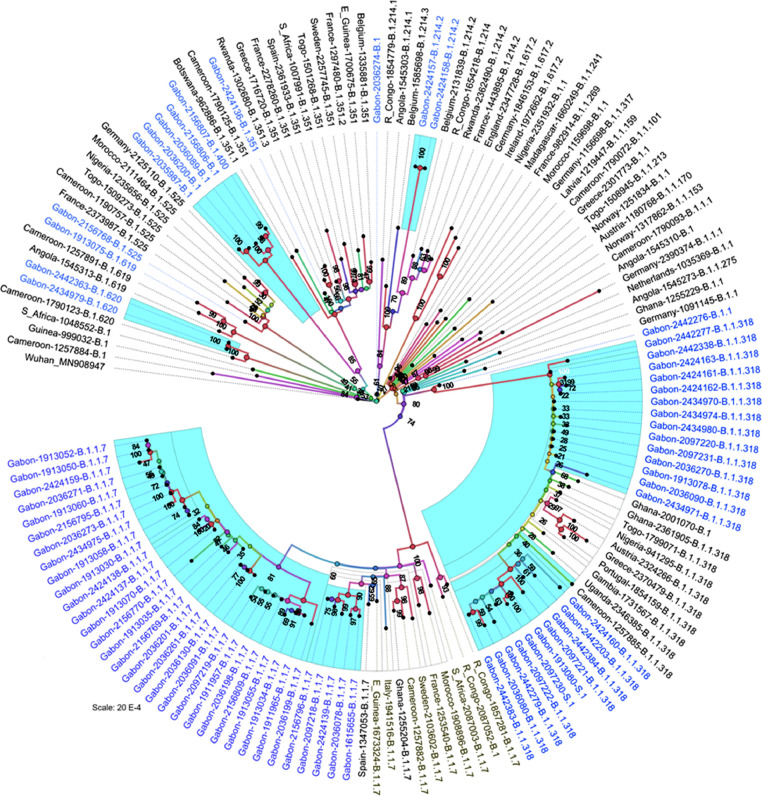
The sequenced SARS-COV-2 genomes and their respective genome diversity with distinct viral lineages are summarized in Table 1 , and the sequences were submitted to the GISAID database. Of the 71 SARS-COV-2 genomes, the alpha VOC B.1.1.7 lineage (46%) was predominant, followed by B.1.1.318 (31%), B.1 (7%), S.1, B.1.214.2 and B.1.620 (3%, respectively), and beta VOC B.1.351 (1%), B.1.400, B.1.525, B.1.619 and B.1.1 (1%, respectively). The identified alpha VOCs clustered into two GISAID clades GR (n=7/33) and GRY (n=26/33), whereas others clustered to either of the GISAID clades G, GH and GR. The proportion of B.1.1.318 increased systematically from 11% (3/27) in March, followed by 37% (7/19) in April to 75% (12/16) in May, and the B.1.1.318 genomes were representative of GISAID clades GR. Phylogenetic analysis showed that 71 SARS-CoV-2 genomes from Gabon sequestered into 10 groups, each representing a distinct SARS-CoV-2 lineage (Figure 1 ). The B.1.1.318 lineage was polyphyletic, as the cluster had sequences from Europe and parts of Africa. The B.1.1.7 lineage was paraphyletic, as the majority of the sequences from Gabon were grouped into a distinct sister clade. Among the other sequences, B.1.214.2, B.1, B.1.620, B.1.619 and B.1.525 formed monophyletic groups. Of the rest, two solitary sequences represented by B.1, B.1.351 did not cluster, and the solitary B.1.1 sequence was paraphyletic to B.1.1.318 lineage (Figure 1).
Table 1.
Proportion of Gabonese SARS-CoV-2 viral lineages.
| PANGO lineage | Frequency (%) | GISAID ID |
|---|---|---|
| B.1.1.7 (N=33) | 46 | EPI_ISL_1615655; EPI_ISL_1911957; EPI_ISL_1911965; EPI_ISL_1913030; EPI_ISL_1913034; EPI_ISL_1913035; EPI_ISL_1913050; EPI_ISL_1913052; EPI_ISL_1913055; EPI_ISL_1913058; EPI_ISL_1913060;EPI_ISL_1913070; EPI_ISL_2036078; EPI_ISL_2036091; EPI_ISL_2036130; EPI_ISL_2036198; EPI_ISL_2036199; EPI_ISL_2036201; EPI_ISL_2036261; EPI_ISL_2036271; EPI_ISL_2036273; EPI_ISL_2097218; EPI_ISL_2156769; EPI_ISL_2156770;EPI_ISL_2156795; EPI_ISL_2156809; EPI_ISL_2424137; EPI_ISL_2424138; EPI_ISL_2424139; EPI_ISL_2424159; EPI_ISL_2434975 |
| B.1.1.318 (N=22) | 31 | EPI_ISL_1913078; EPI_ISL_2036080; EPI_ISL_2036090; EPI_ISL_2036270; EPI_ISL_2097220- EPI_ISL_2097222; EPI_ISL_2097231; EPI_ISL_2424160- EPI_ISL_2424163; EPI_ISL_2434970; EPI_ISL_2434971; EPI_ISL_2434974; EPI_ISL_2434980; EPI_ISL_2442203; EPI_ISL_2442277; EPI_ISL_2442279; EPI_ISL_2442338; EPI_ISL_2442383; EPI_ISL_2442384 |
| B.1 (N=5) | 7 | EPI_ISL_2035987; EPI_ISL_2036089; EPI_ISL_2036200; EPI_ISL_2036274; EPI_ISL_2156806 |
| S.1 (N=2) | 3 | EPI_ISL_1913080; EPI_ISL_2097230 |
| B.1.214.2 (N=2) | 3 | EPI_ISL_2424157; EPI_ISL_2424158 |
| B.1.620 (N=2) | 3 | EPI_ISL_2434979; EPI_ISL_2442363 |
| B.1.400 (N=1) | 1 | EPI_ISL_2156807 |
| B.1.525 (N=1) | 1 | EPI_ISL_2156768 |
| B.1.619 (N=1) | 1 | EPI_ISL_1913075 |
| B.1.1 (N=1) | 1 | EPI_ISL_2442276 |
| B.1.351 (N=1) | 1 | EPI_ISL_2424136 |
Figure 1.
Maximum likelihood phylogenetic tree of currently available SARS-CoV-2 genomes from the Republic of Gabon (nodes and branches highlighted in cyan; names in blue). Numbers above branches represent percentages of bootstrap values (1000 replicates). All the full-length genomes retrieved from the GISAID (global database for influenza gene sequences) labelled as country of origin, GISAID ID., and PANGOLIN lineage. Branch lengths are drawn according to the number of nucleotide substitutions per site.
Discussion
The observation of an increase in incidence of B.1.1.318 from 11% to 75% between March and May 2021 is of interest. Variant B.1.1.318 was discovered in January 2021. Phylogenetic analysis (Figure 1) revealed that all 33 isolates in the B.1.1.7 clade formed a distinct cluster. On the other hand, 22 isolates fell into two relatively cohesive phylogenetic clusters of B.1.1.318 genomes, with the first ancestral to the second, suggesting two distinct genetic divergences, the first due to early introduction by imported cases and subsequently evolving within Africa. Similarly, the SARS-CoV-2 genomes from clades B.1.214.2, B.1.351, B.1.525, B.1.19 and B.1.620 suggest that they could be recent introductions to the Gabonese and are therefore less abundant.
Taken together, this study highlights circulating new variants of potential epidemiological importance.
Conflicts of interest
The authors declare no conflict of interest in the submitted work.
Acknowledgments
Acknowledgments
We thank all researchers and staff of CERMEL who are working for COVID-19 diagnosis. The authors gratefully acknowledge the encouragement of the Gabonese Government. We also thank the COPIL committee and the Ministry of Health. The authors TPV and AAA thank the support of PANDORA-ID NET. AAA and MGP are members of CANTAM (EDCTP-RegNet2015- 1045)
Author's contribution
AAA and BL designed the study procedures. GPM performed all experimental procedures. MNM, RB, SOOB, GNO, JYH, SZA, EGR, and RMN were involved in sampling procedures and collating the meta data. SRP and SR supported experimental procedures and performed phylogenetic analysis. JBLD, JFDS, SB, BL, and PGK contributed to the study design and patient recruitment. AAA, GPM and TPV analysed the data and wrote the manuscript.
Data availability statement
The data supporting reported results are available on request.
Funding
AIDCO: mobilization of research funds for Covid-19 as a public health emergency EDCTP RIA 2020 EF-2961.
Ethical Statement
Informed consent was obtained from all subjects involved in the study.
References
- Freed Nikki E., Vlková Markéta, Faisal Muhammad B., Silander Olin K. Rapid and Inexpensive Whole-Genome Sequencing of SARS-CoV-2 Using 1200 Bp Tiled Amplicons and Oxford Nanopore Rapid Barcoding. Biology Methods and Protocols. 2021;5(1):1–7. doi: 10.1093/biomethods/bpaa014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf, Tiago, Gonzalo Bello, Taina Moreira Martins Venas, Elisa Cavalcante Pereira, Anna Carolina Dias Paixao, Luciana Reis Appolinario, Renata Serrano Lopes, et al. 2021. “<p>Identification of SARS-CoV-2 P.1-Related Lineages in Brazil Provides New Insights about the Mechanisms of Emergence of Variants of Concern</P>.” https://doi.org/10.21203/rs.3.rs-580195/v1.
- Katoh Kazutaka, Misawa Kazuharu, Kuma Kei Ichi, Miyata Takashi. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Research. 2002;30(14):3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer Moritz U.G., Hill Verity, Ruis Christopher, Dellicour Simon, Bajaj Sumali, McCrone John T., Baele Guy, et al. Spatiotemporal Invasion Dynamics of SARS-CoV-2 Lineage B.1.1.7 Emergence. Science. 2021;373(6557):889–895. doi: 10.1126/science.abj0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole Áine, Scher Emily, Underwood Anthony, Jackson Ben, Hill Verity, McCrone John T, Colquhoun Rachel, et al. Assignment of Epidemiological Lineages in an Emerging Pandemic Using the Pangolin Tool. Virus Evolution. 2021;7(2):1–9. doi: 10.1093/ve/veab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut Andrew, Loman Nick, Pybus Oliver, Barclay Wendy, Barrett Jeff, Carabelli Alesandro, Connor Tom, Peacock Tom, Robertson David, Volz Erik. Preliminary Genomic Characterisation of an Emergent SARS-CoV-2 Lineage in the UK Defined by a Novel Set of Spike Mutations - SARS-CoV-2 Coronavirus /NCoV-2019 Genomic Epidemiology - Virological. Virological.Org. 2020:1–9. https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563 [Google Scholar]
- Tegally Houriiyah, Wilkinson Eduan, Giovanetti Marta, Iranzadeh Arash, Fonseca Vagner, Giandhari Jennifer, Doolabh Deelan, et al. Detection of a SARS-CoV-2 Variant of Concern in South Africa. Nature. 2021;592(7854):438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- Trifinopoulos Jana, Nguyen Lam Tung, Haeseler Arndt von, Minh Bui Quang. W-IQ-TREE: A Fast Online Phylogenetic Tool for Maximum Likelihood Analysis. Nucleic Acids Research. 2016;44(W1):W232–W235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velavan Thirumalaisamy P., Meyer Christian G., Esen Meral, Kremsner Peter G., Ntoumi Francine. COVID-19 and Syndemic Challenges in ‘Battling the Big Three’: HIV, TB and Malaria. International Journal of Infectious Diseases. 2021;106(April 2020):29–32. doi: 10.1016/j.ijid.2021.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. 2021. “WHO Coronavirus (COVID-19) Dashboard; Accessed on 25 August 2021.” https://covid19.who.int/region/afro/country/ga.
- Zoa-Assoumou Samira, Ndeboko Bénédicte, Manouana Gédéon Prince, Houechenou Rotimi Myrabelle Avome, Bikangui Rodrigue, Mveang-Nzoghe Amandine, Ondo Georgelin Nguema, et al. SARS-CoV-2 Emerging Variants in Africa: View from Gabon. The Lancet Microbe. 2021;2(8):e349. doi: 10.1016/S2666-5247(21)00125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data supporting reported results are available on request.