Abstract
The coronavirus family has been infecting the human population for the past two decades, but the ongoing coronavirus called SARS-CoV-2 has posed an enigmatic challenge to global public health security. Since last year, the mutagenic quality of this virus is causing changes to its genetic material. To prevent those situations, the FDA approved some emergency vaccines but there is no assurance that these will function properly in the complex human body system. In point of view, a short but efficient effort has made in this study to develop an immune epitope-based therapy for the rapid exploitation of SARS-CoV-2 by applying in silico structural biology and advancing immune information strategies. The antigenic epitopes were screened from the Surface, Membrane, Envelope proteins of SARS-CoV-2 and passed through several immunological filters to determine the best possible one. According to this, 7CD4+, 10CD8+ and 5 B-cell epitopes were found to be prominent, antigenic, immunogenic, and most importantly, highly conserved among 128 Bangladeshi and 110 other infected countries SARS-CoV-2 variants. After that, the selected epitopes and adjuvant were linked to finalize the multi-epitope vaccine by appropriate linkers. The immune simulation disclosed that the engineered vaccine could activate both humoral and innate immune responses. For the prediction of an effective binding, molecular docking was carried out between the vaccine and immunological receptors (TLRs). Strong binding affinity and good docking scores clarified the stringency of the vaccines. Furthermore, MD simulation was performed within the highest binding affinity complex to observe the stability. Codon optimization and other physicochemical properties revealed that the vaccine would be suitable for a higher expression at cloning level. So, monitoring the overall in silico assessment, we anticipated that our engineered vaccine would be a plausible prevention against COVID-19.
Keywords: Molecular docking, Epitopes, In-silico cloning, Codon optimization, Molecular dynamics simulations, SARS-CoV-2
1. Introduction
The human population is rising, and its mobility has led to urbanization with the environment and ecological shifts responsible for the proliferation of numerous infectious diseases [1,2]. The human race has witnessed many of infectious diseases, which are inconsequential or even cause global disruption [1]. The new flu-like virus was supposed to emerge from Wuhan City on December 19, 2019 in China and was initially known as the novel coronavirus-2019 [[3], [4], [5]]. On February 11, 2020, the International Committee on Virus Taxonomy (ICTV) announced this virus as the severe acute respiratory syndrome-2 (SARS-2). After one month on March 11, 2020, COVID-19 was proclaimed by the World Health Organization as a pandemic condition due to 118326 confirmed cases and 4292 worldwide deaths. Up to September 25, 2021, the virus has spread rapidly to 235 countries, regions, and territories with 229,858,719 confirmed cases and 4,713,543 deaths [https://covid19.who.int]. The first case of COVID-19 in Bangladesh was detected on 8th March. Since then, 1,548,320 confirmed cases and 27,337 deaths had been registered on 25th September [https://covid19.who.int]. Coronavirus typically induces moderate respiratory tract infections such as a common cold. Still, newly developed coronavirus has many clinical symptoms such as low-grade fever, dry cough, dyspnea, exhaustion, gastrointestinal issues, diarrhea [[6], [7], [8], [9], [10], [11]]. Inflammation of the upper and lower respiratory tract has also been found to cause acute respiratory tract infection [3]. The median incubation time for COVID-19 is 3 days (range between 0 and 24 days) and the median time between first signs to death is 14 days (range between 6 and 41 days) [12,13]. Many patients with COVID-19 may experience shortness of breath and, in some cases; patients may develop septic shock, difficult to correct metabolic acidosis, or coagulation disorders [14]. It may also affect many organs; among them, kidneys and heart are very common [15,16]. Symptoms may vary from person to person; for example, some people may experience very mild symptoms without any fever and recover within 1–4 weeks, while others may develop infection and some may die [17,18]. However, the aged person with physical complications is at a high risk of fatality than the young aged and children [19]. Like other coronaviruses, COVID-19 is an enveloped single-stranded positive-sense (+SSRNA) virus [[20], [21], [22]]. The estimated genome length of SARS-CoV-2 is 30 kilo-bases (between 26000 and 32000 bases) [23,24]. Several studies reported that the full-length genome sequence of SARS-CoV-2 has a great similarity with bat coronaviruses, 45–90% to SARS-CoV and a smaller similarity of about 20–60% to MARS-CoV. For that reason, it was assumed that bat might be the original host of SARS-CoV-2, but the intermediate host is still now unknown to all [11]. The genome sequence of SARS-CoV-2 encoded sixteen non-structural and four major structural proteins [15]. These four structural proteins (Surface glycoproteins, Envelope protein, Membrane protein, and Nucleocapsid) are essential components of the viral assembly [25]. With these proteins, SARS-COV-2 paves the way for an invasion into the host [15,22,23]. Of all these structural proteins, the viral entry of SARS-CoV-2 is mediated by surface (S) glycoprotein [26]. The Membrane (M) protein consists of three structural domains, and each domain is responsible for the curvature of the membrane, the shape of the virion by binding to the nucleocapsid (N). Envelope (E) protein is involved in viral pathogenesis, the assembly and release of virion [15]. The fourth structural protein called nucleocapsid contains two different domains; both bind with the viral RNA genome, but their binding mechanism is different from each other [11]. In the case of non-structural protein, some study has reported that they are essential for the replication of SARS-CoV-2 [11].
Because of the high degree of contagiousness and its rapidly spreading to every territory, the World Health Organization declared COVID-19 a Public Health Emergency of International Concern (PHEIC) and called for the immediate implementation of adequate and effective care. As a result, scientists around the world are racing to develop a suitable vaccine candidate and some of them have already given FDA approval for emergency use. However, with the progression of the pandemic, SARS-CoV-2 continually mutated several times and have the possibility to change its pathogenicity [27]. This RNA virus is transmitted faster than any other virus in the last century [28,29]. So, it is essential to produce more and more new vaccine candidates that will stand against all of its mutational strain by giving long-lasting immunity. That is only possible by computational approaches within a short period and low cost. We thus made an effort to develop an in silico multi-epitope based vaccine by applying immunoinformatic tools. It is a subset of bioinformatics that deals with many immunological data using computational analysis. This is also a rapid computational method for the design of vaccines. Hypothetically both structural and non-structural proteins are potential for vaccine targets [30], but we only focused on the structural protein (S, M, E). Several experiments have been performed based on targeting the surface glycoprotein receptor-binding domain (RBD) domain. But, a rapid mutation on the RBD and high genomic variation of RNA viruses have a chance to escape the new strain from neutralization by currently RBD targeting antibodies [[31], [32], [33], [34], [35]]. So, considering the fact full-length surface, envelop and membrane protein have been adopted for multi-epitope vaccine constructs except for nucleocapsid. The reason for deselecting this nucleocapsid was their initial unavailability into the host cell's outside surface during infection. So, in the current study multiple immunoinformatic servers and tools have been applied to predict Cytotoxic T lymphocytes (CTL), Helper T Lymphocytes (HTL), Linear B-lymphocyte (LBL) epitopes that were highly conserved among the entire strain (In total, 238 genome sequences have been chosen to see the conservancy by prioritizing 128 Bangladeshi sequences and 110 from individually affected nations) of SARS-CoV-2 (Supplementary File-S, M, E). With these predictions, we tried to develop a suitable multi-epitope vaccine candidate by combining every selected epitope with corresponding linkers, where an adjuvant has also been associated at the first position to boost up the immunogenicity of the construct. In this way, we hoped that our designed vaccine will generate a proper immune response and could be used against SARS-CoV-2. The overall study processes have been illustrated as a graphical abstract in Fig. 1 .
Fig. 1.
Flow chart designed for the multi-epitope vaccine against SARS-COV-2. The numbers in red circle depicted the sequence of the whole process. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2. Methods and materials
2.1. Sequence retrieval and phylogenetic tree construction
The β-family coronavirus (HCOV-OC43, HCOV-HKU1, SARS, MARS, HCOV-NL63, HCOV-229E & SARS-CoV-2) surface, membrane, and envelop proteins amino acid sequences were extracted from NCBI database. The assigned accession numbers are HCoV-HKU1 (YP173238.1 = S, YP173241.1 = M, YP173240.1 = E), MERS-CoV (YP009047204.1 = S, YP009047210.1 = M, YP009047209.1 = E), HCoVNL63 (YP003767.1 = S, YP003770.1 = M, YP003769.1 = E), HCoV-229E (NP073551.1 = S, NP073555.1 = M, NP073554.1 = E), SARS-CoV-2(QJU11812.1 = S,QJU11815.1 = M, QJU11814.1 = E), HCoV-OC43 (YP009555241.1 = S, YP009555244.1 = M, YP009555243.1 = E), SARS-CoV (NP828851.1 = S, NP828855.1 = M, NP828854.1 = E). The first sequenced genome of SARS-CoV-2 in Bangladesh was chosen as a reference sequence from Child health research foundation, Dhaka [36]. To investigate their evolutionary connection, phylogenetic tree was constructed using MEGA X (neighbor-joining algorithm) using the default setting and bootstrap for 1000 replicates [37].
2.2. T-cell epitopes prediction for vaccine construct
2.2.1. CTL epitope prediction
The IEDB MHC I processing tool (http://tools.iedb.org/processing/) was applied to identify CTL (Cytotoxic T-lymphocytes) epitopes in accordance with the IEDB recommended method. To evaluate the binding interaction between peptides and MHC class I alleles, we adjusted the threshold at 50 nM (IC50 ≤ 50 strong binders) [38]. The epitopes were predicted by concentrating on three essential components: MHC-I binding, proteasomal processing and transport efficiency, and TAP transport [39]. Following prediction, the IEDB MHC I binding tool (http://tools.iedb.org/mhci/) was used to find the epitopes binding ability with 27 reference alleles using the Stabilized Matrix Method (SMM).
2.2.2. HTL epitope prediction
IEDB MHC-II Binding Predictions tool(http://tools.iedb.org/mhcii/) using SMM method was used to predict HTL epitopes against a set of 27 human reference HLAs. Epitopes were prioritized based on their IC50 value [40]. The IC50 value ≤ 50 nM demonstrated the highest binding affinity towards MHC-I & II, ≤500 nM is in the midrange, and ≤5000 nM corresponds to the lowest binding affinity [41].
2.3. B-lymphocyte epitope prediction
B-cell epitopes may be potential antigens capable of interfering with B cells [42]. As a result, the Linear B cell epitopes were predicted using the Kolaskar and Tangaoker Antigenicity and BepiPred 2.0 tools from the Immune Epitope Database Analysis Resource (IEDB-AR) server [43,44].
2.4. Antigenicity and allergen prediction of the CTL, HTL, LBL epitopes
The antigenic potential of the specified epitopes (CTL, HTL, LBL) were manually assessed using the VaxiJen2.0 server [45]. For the prediction of antigenicity, a threshold value of 0.4 was used. Non-antigenic epitopes with VaxiJen scores less than 0.4 were avoided, whereas antigenic epitopes with VaxiJen scores more than 0.4 were selected for future investigation. To minimize allergic responses to the selected epitopes, the allergenicity status of all CTL, HTL, and LBL epitopes was validated using the AllerTOP v.2.0 server [46].
2.5. Immunogenicity prediction of the CTL epitopes
The MHC class-I immunogenicity tool from the IEDB server was used to quantify the immunogenicity of cytotoxic T cell epitopes (CTL) [47]. This tool was developed to indicate the immunogenicity of peptides based on their amino-acid position and characteristics.
2.6. Toxicity prediction
The ToxinPred server (http://crdd.osdd.net/raghava/toxinpred/) was used to estimate the toxicity of the epitopes (CTL, HTL, LBL) to assure their non-toxic nature [48]. This server displays the toxicity and non-toxicity of epitopes depending on their physicochemical properties [39].
2.7. Prediction of cytokine-inducing HTL epitopes
The predicted HTL epitopes were examined further for their capacity to induce various cytokines [44]. The IFN epitope (http://crdd.osdd.net/raghava/ifnepitope/) [49], IL4pred (http://crdd.osdd.net/raghava/il4pred/) [50], and IL-10pred (http://crdd.osdd.net/raghava/IL-10pred/) servers [51,52] did the prediction using default settings.
2.8. Population coverage and conservancy analysis of the epitopes
Variants of the HLA allele were found in varying degrees of frequency all across the world [53]. As a consequence, the IEDB population analysis tool (https://tools.iedb.org/population/) was used to confirm the population coverage of our selected CTL and HTL epitopes [54]. Furthermore, a chord diagram was created through R programming languages 4.0 version to show the relationship between the country, continent, and ethnic groupings. The epitope's conservation is critical since it implies wider protection against diverse strains of SARS-COV-2. By filtering GSAID [55], a total of 238 genome sequences (128 from Bangladesh and 110 from other afflicted countries) were obtained. We attempted to cover the majority of the impacted nations' genome sequences that were accessible in the GSAID database (Supplementary File-S, M, E). Upon retrieval, multiple sequence alignment using Bioedit software was used to separate S, M, and E from these 238 whole genome sequences [56]. The recovered S, M, and E sequences were then converted to protein form using the Expasy translator program (https://web.expasy.org/translate/) [57]. To appraise the conservancy of our preferred epitopes, we used the IEDB conservancy tool [58].
2.9. Docking performance between T cell epitopes and MHC alleles
The binding affinity of CTL and HTL epitopes with their accompanying MHC alleles were compared by molecular docking approaches [3]. Predicted top-ranked epitopes (CTL, HTL) and their respective HLA allele binders (HLA-A-68; 01, HLA-A-02; 06, & HLA-DRB1-01; 01) were submitted to the Galaxy Pep-Dock server (http://galaxy.seoklab.org/cgi-bin/submit.cgi?type=PEPDOCK) [59]. The Protein Data Bank (PDB) (https://www.rcsb.org/) was used to retrieve the crystal structures of the HLA-A-02; 06 (PDB ID-3OXR), HLA-A-68; 01 (PDB ID-6PBH), and HLA-DRB1-01; 01 (PDB ID-5V4N) [60], which were then processed with BIOVIA Discovery Studio 2020 to eliminate superfluous ligands [53]. The complexes were modified with the Galaxy Refine server after docking(http://galaxy.seoklab.org/cgi-bin/submit.cgi?type=REFINE). The PRODIGY web-server (https://wenmr.science.uu.nl/prodigy/) was used to test their binding affinity [61].
2.10. Construction of the final vaccine structure
By combining all of the possible CTL, HTL, and LBL epitopes with suitable adjuvant and flexible linkers, a multi-epitope vaccine was constructed [62,63]. Based on the immunological filters mentioned above, the best potential epitopes were screened and combined to produce a single peptide chain [64]. Because peptides are not immunogenic on their own, an adjuvant is necessary to stimulate the immune response [65,66]. The vaccine's sequential position commenced with an adjuvant (β-defensins = GIINTLQKYYCRVRGGRCAVLSCLPKEEQIGKCSTRGRKCCRRKK) and was followed by the top CTL, HTL, and LBL epitopes [38]. The linkers used in this study were EAAAK, AYY, GPGPG, and KK, which aid in the connection of adjuvant and epitopes. The AYY linkers contribute in the formation of a favorable location for binding to the TAP transporter and enhancing epitope presentation. The GPGPG linker being utilized to promote HTL responses and to preserve the immunogenicity of helper and antibody epitopes that are conformationally dependent. The KK linkers combine B cell epitopes, and adjuvant was added to the vaccine's N-terminus with EAAAK linker, resulting in more effective separation and less contact with other vaccine domains [67].
2.11. Allergenicity prediction of the vaccine
We utilized several methods to estimate the allergenic score for the designed vaccine sequence with high accuracy [39]. The non-allergic behavior of the vaccine was validated using the AllerTop v.2.0 (https://www.ddg-pharmfac.net/AllerTOP/) [43] and AllergenFP v.1.0 (http://ddg-pharmfac.net/AllergenFP/) servers [68].
2.12. Antigenicity prediction of the vaccine
To anticipate the antigenic characteristics of the proposed vaccine construct, Vaxijen v2.0 and ANTIGENpro were both used. At 0.4 thresholds, Vaxijen v2.0 provides antigenicity with excellent accuracy (http://www.ddgpharmfac.net/vaxijen/VaxiJen/VaxiJen.html) [40,69]. ANTIGENpro (http://scratch.proteomics.ics.uci.edu/), a server that predicted total protein antigenicity based on machine learning algorithms utilizing experimentally verified microarray analysis reactivity data [30,70].
2.13. Prediction of protein solubility and transmembrane helices
The vaccine construct's solubility was determined using the SOLpro (http://scratch.proteomics.ics.uci.edu/) [71] and Protein-Sol (https://protein-sol.manchester.ac.uk/) servers [72]. SignalP-5.0 (http://www.cbs.dtu.dk/services/SignalP/data.php) [73] and TMHMM Server v2.0 (http://www.cbs.dtu.dk/services/TMHMM/) [74] were used to check for the existence of any signal peptides and transmembrane helices.
2.14. Physio-chemistry of the vaccine construct
The ExPASy ProtParam server (https://web.expasy.org/protparam/) was used to estimate the vaccine's physiochemical characteristics [75]. The vaccine candidate was fed into the server, which calculated characteristics such as molecular weight, protein half-life, instability index, theoretical pI, amino acid composition, aliphatic index, and GRAVY [76].
2.15. Cross-reactivity analysis with human proteomes
The BLASTP (https://blast.ncbi.nlm.nih.gov/Blast.cgi) search engine in the NCBI database was used to assess the sequential similarity of our epitope-based vaccine inside human proteomes. None of the antigenic sequences indicated more than a 10% similarity to the human proteome [77].
2.16. Secondary (2D) structure prediction
PSIPRED v4.0 (http://bioinf.cs.ucl.ac.uk/psipred/) server was used by two feed-forward neural networks that processed the PSI-BLAST output to predict the vaccine's secondary structure [78].
2.17. Tertiary (3D) structure prediction
RaptorX (http://raptorx.uchicago.edu/ContactMap/), a publicly available web server, was used to infer the vaccine sequence's 3D structure. The server predicts tertiary structure using a deep learning technique that integrates evolutionary coupling (EC) and sequence conservation information via an ultra-deep neural network built by two deep residual neural networks [79].
2.18. Refinement and energy minimization of the tertiary structure
To improve the optimal 3D structure of the construct, a three-step approach was explored; initially, the structure was refined by two independent servers: ModRefiner (https://zhanglab.ccmb.med.umich.edu/ModRefiner/) [80] and Galaxy Refine (http://galaxy.seoklab.org/cgi-bin/submit.cgi?type=REFINE) [81]. The energy was then minimized using Swiss-Pdb Viewer [82] and Chiron online web server (https://dokhlab.med.psu.edu/chiron/login.php) [83].
2.19. Tertiary structure validation
The ProSA-web (Protein Structure Analysis), PROCHECK (Ramachandran Plot Assessment), and ERRAT servers were deployed to validate the improved 3D structures and compare the models [84]. ProSA (https://prosa.services.came.sbg.ac.at/prosa.php) assigned to compute an overall quality score for the specified input structure. A 3D molecule view was included in the ProSA-web, facilitating the detection of the problematic part indicated by the given score [85]. The ERRAT (http://services.mbi.ucla.edu/ERRAT/) uses an empirical atom-based method to verify protein structures. This program compares the query sequence's non-bonded atomic interactions to a database of credible high-resolution crystallographic structures [86]. Finally, the Ramachandran plot, which specifies the quality of the modeled structure, was obtained using the PROCHECK server (https://servicesn.mbi.ucla.edu/PROCHECK/) [87].
2.20. Disulfide engineering of the vaccine construct
To identify the capacity of establish a disulfide bond between pairs of vaccine residues with correct geometry, the Disulfide by Design 2 server (http://cptweb.cpt.wayne.edu/DbD2/) was utilized [88].
2.21. Prediction of conformational B-cell epitopes in vaccine construct
B-lymphocytes' primary features are antigen presentation and cytokine production; in addition to generating antibodies, they provide humoral immunity in the human body [89]. ElliPro, a web server (http://tools.iedb.org/ellipro/), was used to determine conformational B-cell epitopes. Each predicted epitope is provided a PI (protrusion index) value by ElliPro. Finally, to see the epitopes, a Jmol viewer was used [90].
2.22. Molecular docking between vaccine and the receptors
Molecular docking is a computational approach that is crucial for studying protein-protein interaction patterns based on molecule binding affinity [91]. For that purpose, we docked our final vaccine construct with immune therapeutically relevant TLR2 (PDB ID-6nig), TLR3 (PDB ID-5gs0), TLR4 (PDB ID-4g8a), TLR7 (PDB ID-5gmg), and TLR8 (PDB ID-4qc0) proteins, since these interactions are critical for eliciting an adequate immune response. Because we used the TLRs structure from the protein data bank, we prepared the receptors by removing the associated ligand and water molecules using Discovery Studio 2020 software [92]. The High Ambiguity Driven Protein Docking (HADDOCK) server (http://milou.science.uu.nl/services/HADDOCK2.2/haddockserver-easy.html) was implemented for fast and accurate docking. This server used information from both biochemical bioinformatics and biophysical approaches to enhance docking sampling and scoring. The server requires active and passive residues from the vaccine and receptors as input. The CPROT server [93] predicted these interaction residues (active and passive). The clusters were polished by the HADDOCK Refinement server [94] after docking. Finally, using PDBsum (https://www.ebi.ac.uk/thorntonsrv/databases/pdbsum/Generate.html), the interaction residues between vaccine construct and TLRs were mapped [95].
2.23. Molecular dynamics (MD) simulations of the best docking complexes
GROMACS simulation package (GROMACS 2020.4) was used to perform molecular dynamics simulations for our top docking complex. MD simulation of protein-vaccine complex was carried out for 100 ns in water using CHARMM36 forcefield; trajectory and energy files were written in every 10ps [96]. The system was solvated in a truncated octahedral box, containing TIP3P water molecules. However, the simulation was performed in 0.15 M KCl by adding 122 Potassium ions and 163 Chloride ions [97]. To remove any steric incompatibilities, minimization was undertaken for 5000 steps using the Steepest Descent Method, and convergence was achieved within the maximum force 1000 (KJ mol-1 nm-1). The system was equilibrated at NVT and NPT ensembles for 100ps (50,000 steps) and 1000ps (1,000,000 steps), respectively, using time steps 0.2 and 0.1 fs. Production run for simulation was carried out at a constant temperature of 300 K and a pressure of 1 atm or bar (NPT) using weak coupling velocity-rescaling (modified Berendsen thermostat) and Parrinello-Rahman algorithms. Relaxation times were set to τ T = 0.1 ps and τ P = 2.0 ps. All bond lengths involving hydrogen atom were kept rigid at ideal bond lengths using the Linear Constraint Solver (lincs) algorithm, allowing for a time step of 2 fs. Verlet scheme was used for the calculation of non-bonded interactions. Periodic Boundary Conditions (PBC) were utilized in all x, y, z directions. Interactions within a short-range cutoff of 1.2 nm were calculated in each time step. Particle Mesh Ewald (PME) was used to calculate the electrostatic interactions and forces to account for a homogeneous medium outside the long-range cutoff. The production was run for 100ns for the complex. Visual Molecular Dynamics (VMD) software was involved to combine output trajectories from each run [98]. Finally, stability of the protein, vaccine and complex like the root means square deviation (RMSD), root mean square fluctuation (RMSF), Molecular Mechanics Poisson-Boltzmann Surface Area (MMPBSA) and Principal component analysis (PCA) were determined from the trajectories generated in the simulation [99]. The radius of gyration, temperature, pressure, density, potential was also calculated to see the compactness of the protein, vaccine and complex at different thermodynamic conditions. Bio3D package of R was used to draw all the plots [100] and MMPBSA graph was performed using MMGBSA method [101]. After plotting Xmgrace graph plotting software was used to modify all the plot [102].
2.24. mRNA structure prediction
RNAfold (http://rna.tbi.univie.ac.at/cgibin/RNAWebSuite/RNAfold.cgi) [103] and the mfold (http://unafold.rna.albany.edu/?q=mfold) web-servers [104] were used to verify the stability of the mRNA structure of the vaccine construct.
2.25. Immune simulation
C‐ImmSimserver (https://kraken.iac.rm.cnr.it/C-IMMSIM/index.php?page=0) was applied for the immunological simulation of the vaccine construct using the position‐specific scoring matrix (PSSM) [4,38]. For the majority of vaccines, the minimum recommended duration between doses 1 and 2 was 4 weeks. Our simulation took 1050-time steps (a time step of around 8 h), nearly 12 months. As a consequence, three peptide injections at time steps 1,84,170 were given four weeks apart [105].
2.26. Codon optimization and in silico cloning
The Java Codon Adaptation Tool (Jcat) (https://www.jcat.de/) was utilized for the codon optimization approach to enhance the expression of recombinant protein. Jcat developed a cDNA sequence of the vaccine to do codon optimization and reverse translation at the same time [106]. To ensure optimal protein expression, GenScript a Rare Codon Analysis Tool (https://www.genscript.com/tools/rarecodonanalysis) was used to improve the product by analyzing discrete parameters such as G-C concentration and Codon Adaptation Index (CAI) of the sequence [107]. Finally, cloning procedure was carried out using Snap Gene v4.2 software (https://snapgene.com/) by introducing the modified nucleotide sequence into the pET28a (+) expression vector [108].
3. Results
3.1. Retrieval of sequences and phylogenetic tree construction
The seven human coronavirus S, M, E protein sequences were retrieved from NCBI for the generation of phylogenetic relationship (Fig. 2 ). The analysis revealed that SARS-COV-2 had a great similarity with SARS-COV and MARS-COV (Fig. 2).
Fig. 2.
Evolutionary relationship between the family of human corona virus Surface, Membrane and Envelop protein.
3.2. Prediction of T lymphocyte epitopes
3.2.1. Prediction of CTL epitopes
Epitopes play a pivotal role in the formation of long-term immunity. We found 7 CTL epitopes using the IEDB-recommended approach described above. Several immunological filters were used to find these optimal epitopes (Table 2). All of the epitopes, as well as their positioning, antigenicity, and binding alleles, are documented in Table 1 .
Table 2.
Results of the finally selected CTL epitope segments.
| CTL Epitope | Length | Immunogenicity | Allergenicity | Toxicity | |
|---|---|---|---|---|---|
| 1. | MAYRFNGIGV | 10 | 0.3226 | Non-Allergen | Non-Toxin |
| 2. | ASFRLFARTR | 10 | 0.29647 | Non-Allergen | Non-Toxin |
| 3. | LVIGAVILR | 9 | 0.2601 | Non-Allergen | Non-Toxin |
| 4. | FASVYAWNRK | 10 | 0.25563 | Non-Allergen | Non-Toxin |
| 5. | SVLLFLAFVV | 10 | 0.24819 | Non-Allergen | Non-Toxin |
| 6. | WTAGAAAYY | 9 | 0.15259 | Non-Allergen | Non-Toxin |
| 7. | KLNDLCFTNV | 10 | 0.0961 | Non-Allergen | Non-Toxin |
Table 1.
MHC-I epitopes with its binding allele (IC50 < 500 nM).
| CTL Epitope | Position | Allele | IC50 | Antigenicity (vaxijen v2.0) | |
|---|---|---|---|---|---|
| 1. | MAYRFNGIGV | S (902–911) | HLA-A*68:02, | 22.87 | 1.3523 |
| HLA-A*02:06, | 39.53 | ||||
| HLA-A*02:03, | 57.05 | ||||
| HLA-A*30:01, | 75.33 | ||||
| HLA-A*02:01 | 108.76 | ||||
| 2. | FASVYAWNRK | S (347–355) | HLA-A*68:01, | 15.61 | 0.5868 |
| HLA-A*33:01, | 34.68 | ||||
| HLA-A*68:01, | 76.73 | ||||
| HLA-A*30:01, | 105.43 | ||||
| HLA-A*31:01, | 139.79 | ||||
| HLA-A*11:01, | 176.32 | ||||
| HLA-A*11:01 | 218.70 | ||||
| 3. | WTAGAAAYY | S (258–266) | HLA-A*30:02, | 58.81 | 0.6306 |
| HLA-A*26:01, | 78.45 | ||||
| HLA-B*35:01, | 83.10 | ||||
| HLA-A*68:01, | 141.41 | ||||
| HLA-B*15:01, | 244.91 | ||||
| HLA-A*01:01, | 248.63 | ||||
| HLA-B*58:01, | 370.94 | ||||
| HLA-A*68:02 | 394.09 | ||||
| 4. | KLNDLCFTNV | S (386–395) | HLA-A*02:03, | 23.78 | 2.6927 |
| HLA-A*02:01, | 50.06 | ||||
| HLA-A*32:01, | 62.36 | ||||
| HLA-A*02:06, | 108.78 | ||||
| HLA-A*02:06, | 121.03 | ||||
| HLA-A*02:03 | 202.28 | ||||
| 5. | ASFRLFARTR | M (98–107) | HLA-A*31:01, | 26.33 | 0.6237 |
| HLA-A*31:01, | 99.88 | ||||
| HLA-A*68:01, | 192.30 | ||||
| HLA-A*33:01, | 200.01 | ||||
| HLA-A*30:01, | 256.43 | ||||
| HLA-A*33:01 | 496.02 | ||||
| 6. | LVIGAVILR | M (138–146) | HLA-A*68:01, | 16.35 | 0.5004 |
| HLA-A*31:01, | 100.11 | ||||
| HLA-A*11:01, | 160.07 | ||||
| HLA-A*33:01 | 403.69 | ||||
| 7. | SVLLFLAFVV | E (16-25) | HLA-A*02:01, | 9.98 | |
| HLA-A*02:06, | 14.54 | ||||
| HLA-A*02:06, | 20.92 | ||||
| HLA-A*02:06, | 29.71 | ||||
| HLA-A*02:01, | 48.43 | ||||
| HLA-A*02:01, | 63.31 | ||||
| HLA-A*02:03, | 99.07 | ||||
| HLA-A*02:03, | 107.14 | ||||
| HLA-A*68:02, | 117.92 | ||||
| HLA-A*02:03, | 344.55 | ||||
| HLA-A*30:01 | 381.92 |
3.2.2. Prediction of HTL epitopes
Utilizing SMM methods, 10 HTL epitopes were sorted (Table 3 ). The epitopes were then pass through several immunological filters and checked for their inducing capability (Table 4 ). Within each epitope, it showed the inducing ability of at least two cytokines (Table 4).
Table- 3.
MHC-II epitopes with its binding allele (IC50 < 500 nM).
| HTL Epitope | Position | Allele | IC50 | Antigenicity (vaxijen v2.0) | |
|---|---|---|---|---|---|
| 1. | MFVFLVLLPLVSSQC | S (1–15) | HLA-DRB1*01:01, | 5 | 0.5741 |
| HLA-DRB1*11:01, | 18 | ||||
| HLA-DPA1*03:01/DPB1*04:02, | 23 | ||||
| HLA-DRB1*04:05, | 41 | ||||
| HLA-DRB5*01:01, | 41 | ||||
| HLA-DPA1*01:03/DPB1*02:01, | 46 | ||||
| HLA-DPA1*02:01/DPB1*01:01, | 51 | ||||
| HLA-DRB1*15:01, | 96 | ||||
| HLA-DRB1*04:01, | 114 | ||||
| HLA-DRB1*07:01, | 118 | ||||
| HLA-DPA1*01/DPB1*04:01, | 132 | ||||
| HLA-DRB4*01:01, | 162 | ||||
| HLA-DRB1*12:01, | 232 | ||||
| HLA-DRB1*09:01, | 248 | ||||
| HLA-DRB1*08:02, | 421 | ||||
| HLA-DPA1*02:01/DPB1*05:01 | 422 | ||||
| 2. | RVVVLSFELLHAPAT | S (509–523) | HLA-DRB1*01:01, | 5 | 0.7485 |
| HLA-DPA1*02:01/DPB1*01:01, | 34 | ||||
| HLA-DPA1*03:01/DPB1*04:02, | 47 | ||||
| HLA-DPA1*01:03/DPB1*02:01, | 58 | ||||
| HLA-DRB1*03:01, | 111 | ||||
| HLA-DRB1*09:01, | 128 | ||||
| HLA-DRB1*11:01, | 147 | ||||
| HLA-DRB1*04:01, | 194 | ||||
| HLA-DRB5*01:01, | 199 | ||||
| HLA-DRB1*15:01, | 207 | ||||
| HLA-DPA1*01/DPB1*04:01, | 221 | ||||
| HLA-DRB4*01:01, | 311 | ||||
| HLA-DRB1*07:01, | 347 | ||||
| HLA-DRB1*04:05, | 366 | ||||
| HLA-DQA1*01:01/DQB1*05:01 | 372 | ||||
| 3. | LPIGINITRFQTLLA | S (229–243) | HLA-DPA1*01:03/DPB1*02:01, | 24 | 0.8156 |
| HLA-DRB4*01:01, | 42 | ||||
| HLA-DPA1*03:01/DPB1*04:02, | 52 | ||||
| HLA-DPA1*01/DPB1*04:01, | 75 | ||||
| HLA-DPA1*02:01/DPB1*01:01, | 125 | ||||
| HLA-DRB1*01:01, | 135 | ||||
| HLA-DRB1*15:01, | 168 | ||||
| HLA-DRB1*04:01, | 175 | ||||
| HLA-DRB1*09:01, | 208 | ||||
| HLA-DRB1*07:01, | 210 | ||||
| HLA-DRB1*08:02, | 223 | ||||
| HLA-DRB1*11:01, | 244 | ||||
| HLA-DRB1*04:05, | 300 | ||||
| HLA-DQA1*01:02/DQB1*06:02 | 471 | ||||
| 4. | AYYVGYLQPRTFLLK | S (264–278) | HLA-DRB1*01:01, | 26 | 0.4269 |
| HLA-DPA1*02:01/DPB1*01:01, | 70 | ||||
| HLA-DPA1*01:03/DPB1*02:01, | 97 | ||||
| HLA-DPA1*01/DPB1*04:01, | 133 | ||||
| HLA-DRB1*07:01, | 133 | ||||
| HLA-DRB1*04:05, | 151 | ||||
| HLA-DRB1*15:01, | 175 | ||||
| HLA-DPA1*03:01/DPB1*04:02, | 179 | ||||
| HLA-DRB5*01:01, | 202 | ||||
| HLA-DRB1*11:01, | 253 | ||||
| HLA-DRB1*04:01, | 263 | ||||
| HLA-DRB1*09:01 | 468 | ||||
| 5. | NIDGYFKIYSKHTPI | S (196–210) | HLA-DRB1*11:01, | 28 | 0.4465 |
| HLA-DRB1*01:01, | 41 | ||||
| HLA-DRB1*15:01, | 80 | ||||
| HLA-DRB1*07:01, | 93 | ||||
| HLA-DRB5*01:01, | 119 | ||||
| HLA-DRB1*04:01, | 297 | ||||
| HLA-DRB1*09:01, | 379 | ||||
| HLA-DRB1*04:05 | 486 | ||||
| 6. | RAAEIRASANLAATK | S (1014–1028) | HLA-DQA1*05:01/DQB1*03:1, | 0.5709 | |
| HLA-DRB1*01:01, | |||||
| HLA-DRB1*09:01, | |||||
| HLA-DRB1*13:02, | |||||
| HLA-DQA1*01:02/DQB1*06:02, | |||||
| HLA-DRB1*04:01, | |||||
| HLA-DRB1*08:02, | |||||
| HLA-DRB1*07:01, | |||||
| HLA-DRB1*11:01 | |||||
| 7. | SNLLLQYGSFCTQLN | S (750–764) | HLA-DRB1*15:01, | 46 | 0.8305 |
| HLA-DRB1*04:05, | 54 | ||||
| HLA-DPA1*01:03/DPB1*02:01, | 90 | ||||
| HLA-DRB1*01:01, | 96 | ||||
| HLA-DRB1*04:01, | 134 | ||||
| HLA-DPA1*02:01/DPB1*01:01, | 350 | ||||
| HLA-DPA1*01/DPB1*04:01 | 361 | ||||
| 8. | RTLSYYKLGASQRVA | M (174–187) | HLA-DRB1*01:01, | 8 | 0.5644 |
| HLA-DRB1*09:01, | 19 | ||||
| HLA-DRB1*07:01, | 23 | ||||
| HLA-DRB5*01:01, | 38 | ||||
| HLA-DQA1*05:01/DQB1*03:01, | 124 | ||||
| HLA-DRB1*04:01, | 251 | ||||
| HLA-DRB1*15:01, | 304 | ||||
| HLA-DRB1*11:01, | 309 | ||||
| HLA-DRB1*13:02, | 317 | ||||
| HLA-DRB1*04:05 | 444 | ||||
| 9. | PKEITVATSRTLSYY | M (165–179) | HLA-DRB1*07:01, | 33 | 0.7003 |
| HLA-DRB1*01:01, | 108 | ||||
| HLA-DRB1*09:01, | 160 | ||||
| HLA-DRB1*03:01, | 215 | ||||
| HLA-DRB5*01:01, | 281 | ||||
| HLA-DRB1*04:01, | 325 | ||||
| HLA-DRB1*11:01 | |||||
| 10. | FYVYSRVKNLNSSRV | E (56-70) | HLA-DRB1*01:01, | 16 | 0.6103 |
| HLA-DRB1*07:01, | 43 | ||||
| HLA-DRB1*11:01, | 58 | ||||
| HLA-DRB1*04:01, | 65 | ||||
| HLA-DRB1*04:05, | 68 | ||||
| HLA-DRB1*09:01, | 97 | ||||
| HLA-DRB1*13:02, | 134 | ||||
| HLA-DRB1*15:01, | 183 | ||||
| HLA-DRB5*01:01, | 21 | ||||
| HLA-DRB1*08:02, | 289 | ||||
| HLA-DPA1*03:01/DPB1*04:02 | 375 |
Table 4.
Results of the finally selected HTL epitope segments.
| HTL Epitope | Allergenicity | Toxicity | IFN server | IL4 server | IL10 server | |
|---|---|---|---|---|---|---|
| 1. | MFVFLVLLPLVSSQC | Non-Allergen | Non-Toxin | POSITIVE | Non-inducer | inducer |
| 2. | RVVVLSFELLHAPAT | Non-Allergen | Non-Toxin | POSITIVE | inducer | Non-inducer |
| 3. | LPIGINITRFQTLLA | Non-Allergen | Non-Toxin | NEGATIVE | inducer | inducer |
| 4. | AYYVGYLQPRTFLLK | Non-Allergen | Non-Toxin | POSITIVE | Non-inducer | inducer |
| 5. | NIDGYFKIYSKHTPI | Non-Allergen | Non-Toxin | POSITIVE | inducer | inducer |
| 6. | RAAEIRASANLAATK | Non-Allergen | Non-Toxin | POSITIVE | inducer | Non-inducer |
| 7. | SNLLLQYGSFCTQLN | Non-Allergen | Non-Toxin | POSITIVE | inducer | inducer |
| 8. | RTLSYYKLGASQRVA | Non-Allergen | Non-Toxin | POSITIVE | inducer | Non-inducer |
| 9. | PKEITVATSRTLSYY | Non-Allergen | Non-Toxin | POSITIVE | inducer | Non-inducer |
| 10. | FYVYSRVKNLNSSRV | Non-Allergen | Non-Toxin | NEGATIVE | inducer | inducer |
3.3. Identification of linear B cell epitopes
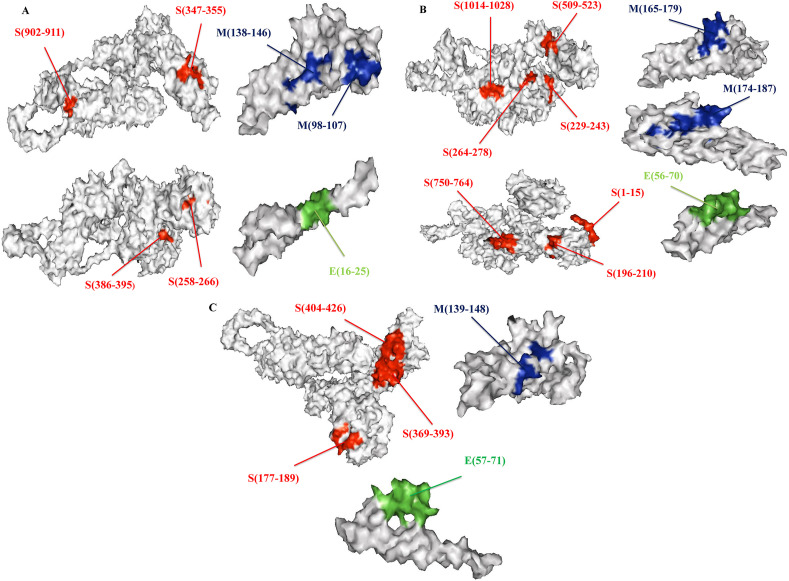
Promiscuous epitopes have been chosen based on antigenicity for the identification of linear B cell epitopes (Threshold 0.4). The epitopes were further checked for their allergenicity and toxicity (Table 5 ). We found five LBL highly antigenic epitopes ranged between 10 and 26 amino acid sequences (Table 5). Finally, the position of LBL, HTL, and CTL epitopes were visualized into the 3D structure of the surface, membrane, envelope protein by pymol software (Fig. 3 A, B, 3C).
Table 5.
B cell epitopes selected by IEDB server.
| Epitopes | Position | Antigenicity (vaxijen v2.0) | Allergenicity | Toxicity | |
|---|---|---|---|---|---|
| 1. | MDLEGKQGNFKNL | S (177–189) | 1.2592 | Non-Allergen | Non-Toxin |
| 2. | YNSASFSTFKCYGVSPTKLNDLCFT | S (369–393) | 1.4031 | Non-Allergen | Non-Toxin |
| 3. | GDEVRQIAPGQTGKIADYNYKLP | S (404–426) | 1.1017 | Non-Allergen | Non-Toxin |
| 4. | VIGAVILRGH | M (139–148) | 0.5844 | Non-Allergen | Non-Toxin |
| 5. | YVYSRVKNLNSSRVP | E (57-71) | 0.4492 | Non-Allergen | Non-Toxin |
Fig. 3.
The glycoproteins (Surface, Membrane,Envelop) 3D structure showed the location of epitopes. A represented CTL epitope, B represented HTL epitopes & C represented LBL epitopes with their appropriate positions.
3.4. Multi-epitope vaccine construction
We combined 7CTL,10HTL, and 5LBL epitopes with the help of appropriate (AAY, GPGPG, and KK) linkers. Finally, we have completed our vaccine construction through the ligation of an adjuvant by the EAAAK linker. The graphical map of the linear multi-epitope vaccine was given in (Fig. 4 ).
Fig. 4.
Graphical view of the generated vaccine construct. CTL, HTL & LBL epitopes were shown by light blue, green and violate color. Adjuvant and all the linkers were in orange and red colors. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.5. Antigenicity, allergenicity and solubility prediction of the final vaccine
The antigenicity of the vaccine was determined by the VaxiJen 2.0 and ANTIGENpro servers to be 0.5466 and 0.860981, respectively (Table S1). Furthermore, it was projected to be non-allergenic by both the AllergenFP and AllerTOP v. 2.0 servers (Table S1). The construct was also shown to be soluble in the SOLpro and protein-sol servers (Fig. S1, Table S1). The state of the engineered vaccine's transmembrane helices was anticipated in vaccine development (Fig. S2). Moreover, the lack of signal peptides also means preventing protein localization (Fig. S3).
3.6. Physiochemical properties identification
The vaccine constructions' molecular weight was calculated to be 47.145 kDa (kDa). The theoretical pI 10.01 corroborated the basic nature (Isoelectric point). The vaccine's stability was represented by the Instability Index of 28.26 (a score of less than 40 indicates a stable protein). The approximate amount of positively and negatively charged vaccine residues was 14 and 60, respectively. The half-life of mammalian reticulocytes (in vitro) was estimated to be 30 h, yeast (in vivo) to be > 20 h, and Escherichia coli to be more than 10 h (in vivo). The aliphatic index of 80.62 ensured thermostability, while GRAVY rating −0.134 confirmed the vaccine's hydrophilicity as well as its capacity to interact with diverse solvent molecules (Table S1).
3.7. Population coverage and conservation across antigen
The distribution of HLA alleles differs between nations, regions, and ethnic groups across the world. As a result, in addition to designing an effective vaccine, population coverage should be taken into account. Alleles (MHC-I, MHC-II) relevant to our selected CTL, HTL epitopes were gathered separately and combinedly for the population research. The strongest continental coverage was found in Europe, North America, East Asia, South Asia, and North Africa, while Central America had some of the weakest (Data S1, Fig. 5 C). European countries have topped the list in terms of country-specific coverage. However, in combination it covered 97.37% of the world's population. Following the advent of a pandemic, the countries most affected were China, Italy, Spain, France, Iran, and the United States. So, we were very interested to clarify how our vaccine coverage fared into these countries. The study revealed that our vaccines represent 93.29%, 98.62%, 92.85%, 98.95%, 96.15%, and 98.46% of Chinese, Italian, Spanish, French, Iranian, and United States populations respectively. Of the studied ethnic groups, the European Caucasoid ethnic population showed the highest population coverage for combined (99%), while the Asian and American ethnic populations, had more than 90% coverage (Data S1, Fig. 5B). We verified the conservation of our CTL and HTL epitopes among 238 SARS-COV-2 sequences whose genomic data was obtained from the GSAID database, as previously stated. To assess the conservancy, we noticed that all of the chosen epitopes almost spanned 100% conservancy among the sequences.
Fig. 5.
T cell epitopes population coverage with their alleles. Circular plot represented the top 49 country, 60 etnicity and 13 continent that covered high population coverage of the CTL and HTL epitopes along with their respective MHC-I, MHC-II alleles when obtained individually (MHC-I or MHC-II) and in combination (MHC-I and MHC-II). (A) Population coverage of the 49 country out of 70. (B) Population coverage of the 60 etnicity out of 83. (C) Population coverage of the 13 continent out of 20.
3.8. Secondary and tertiary structure identification of the final vaccine
From secondary structure prediction, we observed 137(44%), 102(23%), and 193(31%) amino acid sequences involved in the generation of α-helix, β-sheet, and random coil (Fig. S4, Fig. S5). The calculated RMSD value for our modeled tertiary structure was 11.391, which was the lowest among the available models (lower the RMSD value higher the quality of the structure) (see Fig. 6 ).
Fig. 6.
Representation of the tertiary structure (α-helix-red, β-sheet-blue, random coil-light green) and two terminals - (N & C). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.9. Tertiary structure refinement and validation of the final vaccine
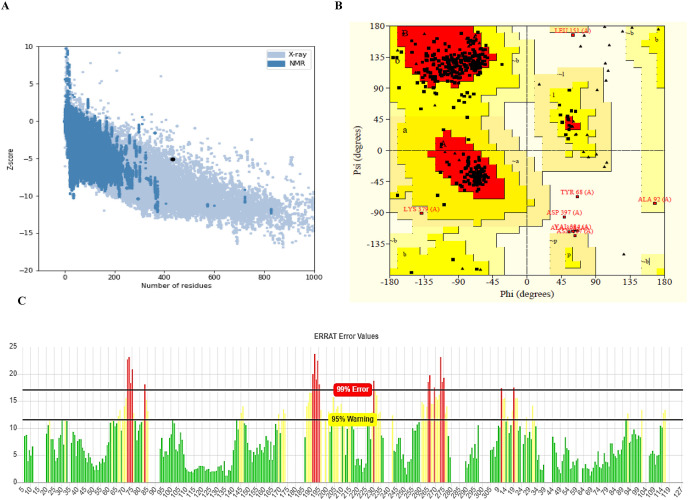
Following refinement, the best 3D model of the vaccine construct was chosen based on the RMSD (0.250), GDT-HA (0.9948), and MolProbity parameters (2.009). The structure was then subjected to energy minimization in order to keep the molecules in stable form. Ramachandran plot analysis of the refined and energy minimized model disclosed that 86.9% of residues lied in the most favorable areas, 10.8% in allowed areas and only 1.4% were disallowed areas (Fig. 7 B). The selected best model quality in ERRAT was 75.924% (Fig. 7C), while ProSA gave a Z-score −5.12, indicating that the model was in the range of native protein conformations (Fig. 7A).
Fig. 7.
(A) PROSA validated the structure with a Z score of −5.12 (B) Ramachandran plot analysis with PROCHECK server displayed a favored, allowed and disallowed area of 86.9%, 10.6% and 1.4%. (C) ERRAT validated the vaccine structure with a score of 75.924%.
3.10. Disulfide engineering of the final vaccine
In total, 39 pairs of residues that may be used in disulfide engineering have been identified (Data S2). However, after the energy value assessment, only six pairs of residues were finalized, as their value falls below the permitted range (energy should be below 2.2 kcal/mol). The position of six pairs mutations showed in Fig. S6 and their energy scores were listed in data S2.
3.11. Conformational B-cell epitope identification of final vaccine
Overall, 5 conformational B-cell epitopes were identified based on 0.5 threshold value and the maximum distance 6 (Fig. S7, Table S2). Their residues size was ranged from 3 to 79 (Table S2).
3.12. Molecular docking between T lymphocyte epitopes and MHC alleles
In general, epitopes with a preference for different alleles are the better candidates for vaccine construction. We docked our chosen epitopes (7 CTL & 10 HTL) with the alleles that were common among them. HLA-A*68:01 allele was found to be prevalent in 5 CTL epitopes and HLA-A*02:06 allele in 2 CTL epitopes (Table 1). 10 HTL epitopes, on the other hand, shared the HLA-DRB1*01:01 allele (Table 3). Such docking complexes negative binding energy were presented in (Fig. 8 ).
Fig. 8.
Performing molecular docking between alleles and CTL, HTL epitopes. A to G represented the binding between CTL epitopes with the most common MHC I alleles and H to Q represented the binding between HTL epitopes with commonly occurring MHC II alleles. HLA alleles and epitopes were represented by ribbon and stick structures. The light color indicated the templates upon which the alleles and epitopes were formed. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.13. Performing molecular docking between the vaccine and the receptors
3.13.1. Docking of the vaccine with TLR8
Representing 26% of the water refined models, HADDOCK generated 53 structures in 10 clusters. Among all other clusters, the most reliable cluster depends on the best HADDOCK score which was 558.9±19.3 (Table 6 ). The top structure of the leading cluster was then refined using the HADDOCK refining server. Where 20 structures were grouped into one cluster, that results in 100% of the water refining models. Following refinement, the top cluster's negative HADDOCK value (−227.0± 3.3) indicated a high binding affinity between the vaccine and TLR8 (Table 7 ). The refined model dynamic visualization was presented in (Fig. 9 ). Where we saw 12-hydrogen bonds have been formed and they were His172-Asn240, Arg247-Ser422, Tyr286-Asn421, Gly239-Arg547, Ser153-His699, Tyr88-Gly785, Phe145-Phe425, Thr234-Gln426, Lys73-Arg788, Arg123-Asp436, Ser127-Glu438, and Pro174-Lys328.
Table 6.
Statistics of the interaction between vaccine and TLRs.
| TLR 8 | TLR 2 | TLR 3 | TLR 4 | TLR 7 | |
|---|---|---|---|---|---|
| HADDOCK score (kcal/mol) | 558.9±19.3 | 141.6±16.3 | 89.7±27.3 | 170.0±31.2 | 173.7±40.7 |
| Cluster size | 6 | 14 | 19 | 4 | 7 |
| RMSD from the overall lowest-energy structure (Å) | 3.8±0.5 | 34.2±0.3 | 0.9±0.5 | 2.2±0.8 | 33.8±0.1 |
| Van der Waals energy (kcal/mol) | −96.9±14.2 | −135.2±14.6 | −164.3±11.6 | −98.9±15.7 | −126.4±10.2 |
| Electrostatic energy (kcal/mol) | −198.5±21.8 | −234.2±59.0 | −305.2±15.4 | −405.0±48.9 | −315.2±49.2 |
| Desolvation energy (kcal/mol) | −40.8±11.3 | −39.5±3.8 | −49.9±1.6 | −43.1±0.8 | −43.4±12.7 |
| Restraint's violation energy (kcal/mol) | 2983.0 ± 196.2 | 3630.7 ± 264.5 | 3649.5± 161.0 | 3930.1± 305.0 | 4065.4±304.1 |
| Buried Surface Area (A2) | 7363.7 ± 315.9 | 4825.7 ± 226.8 | 5209.3 ±152.0 | 3728.3 ±163.9 | 4442.0±181.3 |
| Z-Score | −2.0 | −1.8 | −2.5 | −2.5 | −1.9 |
Table 7.
HADDOCK score of the Vaccine-Receptor complex after refinement.
| Vaccine-TLRs Complex | HADDOCK score |
|---|---|
| Vaccine-TLR8 | −227.0± 3.3(kcal/mol) |
| Vaccine-TLR2 | −244.8±7.5(kcal/mol) |
| Vaccine-TLR3 | −337.6±3.3(kcal/mol) |
| Vaccine-TLR4 | −250.2±1.7(kcal/mol) |
| Vaccine-TLR7 | −256.9±4.3(kcal/mol) |
Fig. 9.
The interaction pattern between V_COV19_SN-TLR8 complex. Almost twelve hydrogen bonds have been formed between vaccine and TLR8 complex (The red color sticks denoted receptor and blue color were vaccine protein). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.13.2. Docking of the vaccine with TLR2
Representing 33% of the water refined models, HADDOCK generated 66 structures in 11 clusters. Among all other clusters, the most reliable cluster depends on the best HADDOCK score which was 141.6±16.3 (Table 6). The top structure of the leading cluster was then refined using the HADDOCK refining server. Where, 20 structures were grouped into one cluster that results 100% of the water refining models. Following refinement, the top cluster's negative HADDOCK value (−244.8± 7.5) indicated a high binding affinity between the vaccine and TLR2 (Table 7). After the interaction analysis it was seen that, 18-hydrogen bonds (ARG-SER, ARG-GLU, ASN-ARG, ARG-TYR, ARG-GLU, ALA-ARG, MET-HIS, PHE-TYR, LEU-ASP, GLY-ASP, TYR-LYS, SER-ASN, SER-SER, HIS-ASP, ILE-TYR, GLU-ARG, GLU-ARG, PRO-LYS) and 4 salt bridges (ARG-GLU, ARG-GLU, HIS-ASP, GLU-ARG) were formed between the vaccine and TLR2. Structural analysis of the docked complex with the prominent hydrogen bonds and salt bridges were depicted in (Fig. S8).
3.13.3. Docking of the vaccine with TLR3
Representing 32% of the water refined models, HADDOCK generated 65 structures in 10 clusters. Among all other clusters, the most reliable cluster depends on the best HADDOCK score which was 89.7± 27.3 (Table 6). The top structure of the leading cluster was then refined using the HADDOCK refining server. Where 20 structures were grouped into one cluster, that results in 100% of the water refining models. Following refinement, the top cluster's negative HADDOCK value (−337.6± 3.3) indicated a high binding affinity between the vaccine and TLR3 (Table 7). After the interaction analysis it was seen that, 15 hydrogen bonds (TYR-ASN, TYR-SER, TRP-SER, TYR-THR, TYR-GLN, TYR-GLN, ARG-HIS, ARG-THR, ARG-GLU, GLY-LYS, PRO-LYS, MET-LYS, VAL-TYR, SER-GLU, GLN-SER) and 2 salt bridges ARG-GLU, ASP-HIS were formed between the vaccine and TLR3. Structural analysis of the docked complex with the prominent hydrogen bonds and salt bridges were depicted in (Fig. S9).
3.13.4. Docking of the vaccine with TLR4
Representing 25% of the water refined models, HADDOCK generated 50 structures in 10 clusters. Among all other clusters, the most reliable cluster depends on the best HADDOCK score which was 170.0± 31.2 (Table 6). The top structure of the leading cluster was then refined using the HADDOCK refining server. Where 20 structures were grouped into one cluster, that results in 100% of the water refining models. Following refinement, the top cluster's negative HADDOCK value (−250.2± 1.7) indicated a high binding affinity between the vaccine and TLR4 (Table 7). After the interaction analysis it was seen that, 19 hydrogen bonds (TYR-ARG, LYS-GLU, ARG-VAL, ALA-GLU, TYR-GLU, TYR-ASN, VAL-SER, TYR-PHE, ARG-ASP, ARG-ASP, ARG-ASN, SER-LYS, TYR-SER, ASN-GLN, SER-HIS, LYS-GLU, ARG-ASP, ARG-HIS, ARG-GLU) and 5 salt bridges (LYS-GLU, ARG-ASP, LYS-GLU, ARG-ASP, ARG-GLU) were formed between the vaccine and TLR4. Structural analysis of the docked complex with the prominent hydrogen bonds and salt bridges were depicted in (Fig. S10).
3.13.5. Docking of the vaccine with TLR7
Representing 35% of the water refined models, HADDOCK generated 71 structures in 9 clusters. Among all other clusters, the most reliable cluster depends on the best HADDOCK score which was 173.7± 40.7 (Table 6). The top structure of the leading cluster was then refined using the HADDOCK refining server. Where 20 structures were grouped into one cluster, that results in 100% of the water refining models. Following refinement, the top cluster's negative HADDOCK value (−256.9±4.3) indicated a high binding affinity between the vaccine and TLR7 (Table 7). After the interaction analysis it was seen that, 12 hydrogen bonds (ARG-HIS, ASN-TYR, ARG-TRP, ARG-THR, TYR-ALA, TYR-THR, SER-HIS, VAL-THR, ARG-GLU, ARG-GLU, ARG-ARG, ARG-GLU) and 2 salt bridges (ARG-GLU, ARG-GLU) were formed between the vaccine and TLR7. Structural analysis of the docked complex with the prominent hydrogen bonds and salt bridges were depicted in (Fig. S11).
3.14. Minimization of energy and MD simulations of the complex
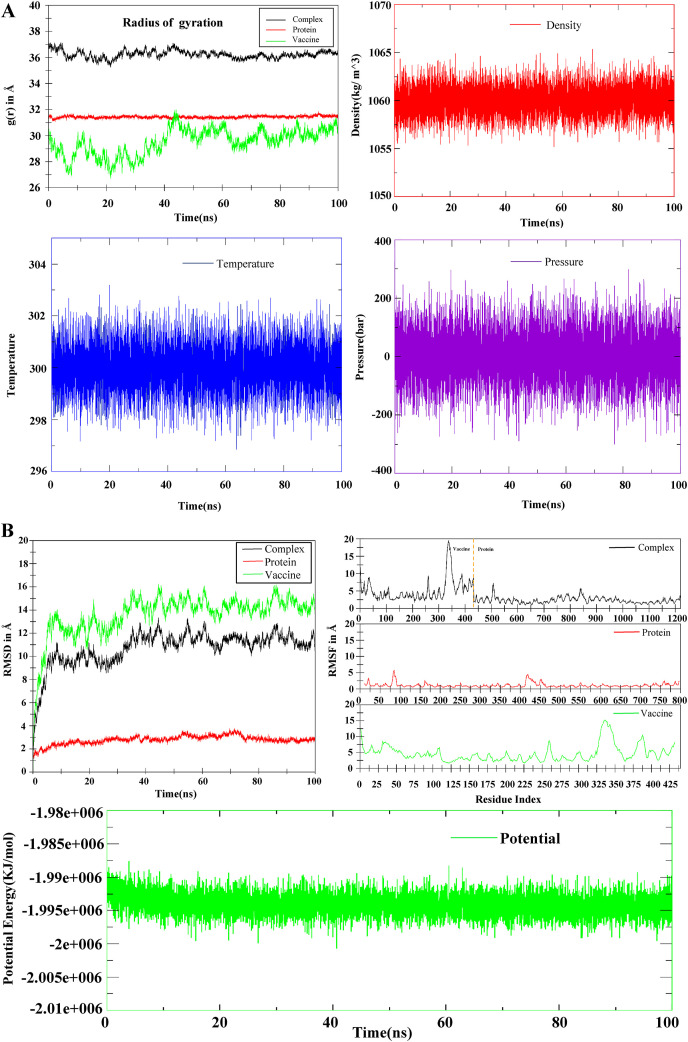
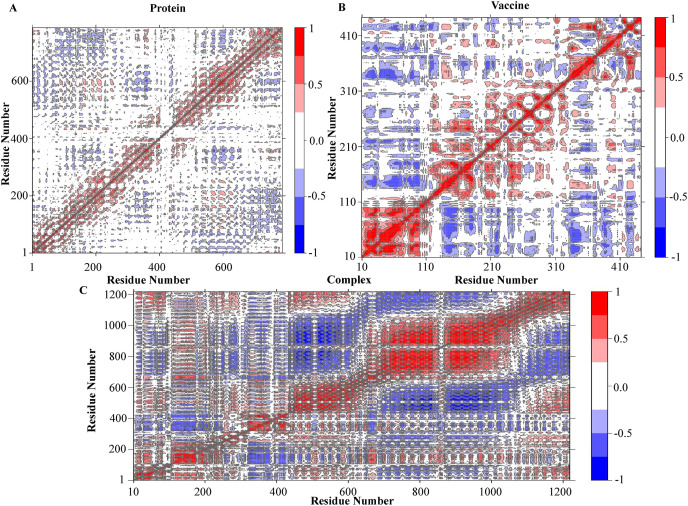
The simulation of molecular dynamics (MDS) was performed to investigate the physical movement and stability of our best docking complexes (Vaccine-TLR8) at different thermobaric conditions (Table 8 ). The simulation results have been described through the energy potential, radius of gyration, density, temperature, pressure, RMSD, RMSF, H-bond, MMGBSA and PCA analysis (Fig. 10 A,B, 11(A,B), 12). The complex is known to be energy minimized when its force reaches <1000 kJ/mol. After running the energy was found in between −1.99e+006 to −1.995e+006 which indicated the system was minimized and the complex was stable (Fig. 10B). The temperature plot revealed that the range of fluctuation was very small and the values varied between 298 and 302 K (Fig. 10A). A graph will be considered stable when the value reaches 300 K and stabilizes there. So according to this parameter, our complex was also retained its stability. The pressure value fluctuated widely throughout the 100-ns equilibration phase, but this behavior was not unexpected. Throughout the equilibration, the average fluctuation of the pressure was −200 to 200 bar (Fig. 10A). The system's average density measured for 100 ns was 1060 ± 3 kg/m^3, which also defined the complex stability (Fig. 10A). The RMSD plot showed the docked complex's structural stability and versatility. The mean RMSD value for the complex, protein and vaccine were 10.70 ± 1.49 Å, 2.77 ± 0.37 Å, 13.58 ± 1.78 Å. For vaccine, a higher RMSD indicated a change in conformation within 100ns of simulation and later it achieved a plateau. A higher RMSD of complex was due to the conformational changes in vaccine (Fig. 10B). The RMSF was calculated to determine the fluctuations of amino acid into the docked complex. RMSF showed significant rigidity of the protein structure except for residues ranging from 75 to 85 and 415–440. While RMSF of vaccine manifested slightly higher fluctuations among residues forming N-terminals, and residues ranging from 30 to 40, 255–265, 325–360, and 375–390 (Fig. 10B). The radius of the gyration plot revealed the protein's compactness along its axes (Fig. 10A). The mean RoG values were: complex = 36.23 ± 0.29 Å, protein = 31.43 ± 0.08 Å and vaccine = 29.51 ± 1.00 Å. A prominent fluctuation was observed in vaccine indicating opening and closing of structure. The protein RoG remained highly stable. The fluctuation in complex was mostly associated with the fluctuation in the vaccine molecule (Fig. 10A). The protein and vaccine binding free energy during 100ns was calculated by MMGBSA method (Fig. 11 A). The mean MMGBSA binding energy was found to be −46.53 kcal/mol (Fig. 11A). The result showed that the vaccine binds well with the protein along with few fluctuations during whole simulation time. The bindings become more stronger after 50ns of simulation time (Fig. 11A). Various energy contributions in MMGBSA binding energy (kcal/mol) for protein-vaccine complex was given in Table 8. Total number of hydrogen bonds formed within complex, protein and vaccine during 100ns of simulation time were very vital for the rigidity. Consistent fluctuating H-bonds indicated that protein and vaccine stayed bound throughout the simulation. The plot in blue color showed the number of hydrogen bonds formed between protein and vaccine. It could be said that over the course of simulation time, the interaction between protein and vaccine became more stronger, as the number of H-bonds increased. The Principal Component Analysis of the complex, protein and vaccine were calculated, where all three PCs captured 73.9%, 47.3% and 69.2% of structural variance in complex, protein and vaccine, respectively (Fig. S12). RMSF (PCA) plot represented the contribution of each residue to the first three principal components for complex, protein and vaccine. PC1 mostly captured structural variance for residues 75–90, 420–440 in protein and 5–23, 26–50, 325–355, 375–390 in vaccine (Fig. S13). Fig. 12 represented the compactness of the protein and vaccine after 100ns MD run. In this view, atoms were colored on a scale from blue to red, where blue correspond to atoms showing large motion amplitudes, and red were more rigid atoms. It could be seen that some domains of vaccine were pretty flexible and rest part of it remained in stable form. To see the correlated motion, Dynamic Cross Correlation Matrix Analysis (DCCM) was performed and visualized in Fig. 13 . In that figure colors varying from red to white to blue indicated intensity of correlated motion, where blue colors exhibited negative correlation, white showed no correlation and red color illustrated positive correlated motions between residues. Snapshots of protein-vaccine complex were taken at 0ns, 50ns and 100ns during the course of simulation time representing changing in conformations of protein and vaccine (Fig. 14 ). Both top views and side views were given for comparison (Fig. 14, Fig. S14).
Table 8.
The binding free energy (kcal/mol) for vaccine complex-protein (TLR8).
| Protein with vaccine | ΔEVDW (Van der Waal's energy) | ΔEelec (Coulombic energy) | ΔGGB (Generalized-Born Polar solvation energy) | ΔESASA (Non-Polar solvation energy) | ΔGMMPBSA (Protein-Ligand Binding energy) |
|---|---|---|---|---|---|
| Complex | −207.49 ± 25.35 (kcal/mol) | −823.29 ± 118.35 (kcal/mol) | 1008.68 ± 131.47 (kcal/mol) | −24.43 ± 2.41 (kcal/mol) | −46.53 ± 19.26 (kcal/mol) |
Fig. 10.
MD simulation between V_COV19_SN and TLR8. (A) Represented radius of gyration, Density, Temperature and Pressure during simulation of the protein, vaccine and complex. (B) Represented the RMSD, RMSF and potentials of the protein, vaccine construct and complex at the time of simulation.
Fig. 11.
(A) Binding free energy using MMGBSA method and (B) Formation of hydrogen bond during the course of 100ns MD run.
Fig. 12.
Interpolated structures of protein and vaccine along with PC1. The blue color represented large motion of amplitudes, while red was more in rigid form. This indicated some parts of it were flexible and the other parts retained its stability. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 13.
Protein residue dynamic cross correlated motions for the protein, vaccine and complex. The residues that belonged to blue color showed negative correlation, white showed no correlation and red color indicated positive correlated motions among residues. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 14.
The stability and convergence of the complex during 0ns, 50ns and 100ns MD run. The red color dictated vaccine and blue one was the protein. To see the structural changes, the complex represented here both in top view and side view. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.15. Immune simulation
Immune simulations of our final vaccine design demonstrated that it was capable of eliciting an effective immune response. In compared to IgG, high amounts of Immunoglobulin IgM were generally detected. Immunoglobulin levels (IgG1+IgG2, IgM, and IgG + IgM antibodies) were frequently enhanced in secondary and tertiary responses, with concurrent antigen decrease (Fig. 15A1). This indicated the development of immune memory as a result increased antigen clearance during subsequent exposures (Fig. 15A5). Furthermore, increased numbers of activated memory B cells revealed the vaccine's ability to elicit effective and long-lasting protection (Fig. 15 A (3,4)). Similarly, for memory formation, the TH, TR, and TC cell populations showed a strong response (Fig. 15A (5,6,7), B (1,2). A high degree of macrophage activity has been linked to dendritic and NK cell behavior (Fig. 15B (3,4,5)). Large levels of IFN- γ as well as IL-2 production were already observed. Furthermore, the innate immune system components were also involved (for example epithelial cell) (Fig. 15B6). Moreover, a lower simpson index (D) showed a variety of immune response possibilities (Fig. 15A2).
Fig. 15.
In silico Immune simulation of the V_COV19_SN construct by C-ImmSim server. (A1) Antigen and immunoglobulins, (A2) Production of cytokine and interleukins. (A3) B-cell population, (A4) B-cell population per state, (A5) TH cell population, (A6) TH cell population per state, (A7) TR (regulatory) cell population per state. (B1) TC cell population, (B2) TC-cell population per state, (B3) NK cell population, (B4) MA cell population per state, (B5) DC cell population per state, (B6) EP cell population per state, (B7) PLB cell population.
3.16. mRNA structure prediction and in silico cloning
In order to demonstrate the expression efficiency, in silico cloning was conducted into the expression vector. At first, we reverse transcribed the peptide sequence using the java codon adaption tools for optimization. The length of the nucleotide sequence following codon optimization was 1296. The optimized nucleotide sequence had an average GC content of 51.2% (Jcat) and 52.03% (Genscript) (Ideal range of GC content is between 30% and 70%) (Fig. S15A). The obtained CAI values 0.97(Jcat) and 0.85(GenScript) were deemed ideal for expression in the intended organism. Secondary structure of the mRNA on the basis of minimum free energy was also determined (Fig. S15C). The predicted structure had ΔG = −428.80 kcal/mol. There was no hairpin or pseudoknot been observed at the first nucleotide of the 5′ end (Fig. S15D). The free energy associated with their structural element was given in (Table S3). Finally, the recombinant plasmid was constructed into a pET-28a (+) vector by restriction cloning (Fig. 16 & Fig. S16).
Fig. 16.
In silico cloning of the V_COV19_SN. In the pET-28a (+) expression vector, V_COV19_SN was inserted where the red portion denoted the gene code for the vaccine and the vector backbone was represented in the black circle. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
From December 19, 2019, COVID-19 reportedly created life threatening situation all around the world. The virus, SARS-CoV-2 transmits from person to person via droplet has already spread to every territory worldwide. Hence, the world health organization announced this virus as a global public health emergency [97]. This inexorable viral outbreak and resulting deaths have risen day by day; on the contrary, scientists are unable to find adequate medicine to mitigate the loss of this viral disease. Since the scientific community worldwide is looking to find a viable vaccine against this expeditiously developed SARS-CoV-2 and many of them are in the clinical trial, some have been approved by the FDA, which is now available on the market [[24], [109], [110]]. But neither of these vaccinations is 100% competent to the war against SARS-2. However, it was stated that various side effects between persons and some people died after vaccination. This issue influences us a lot to design a multi-epitope vaccine through immunoinformatics approaches.
The advancement of immunoinformatics has now revolutionized the field of vaccine production. Based on multiple computational methods, we have planned to develop an antigenic multi-epitope (immunodominant) vaccine called V_COV19_SN (432aa) by targeting spike (S), membrane (M) and envelop (E) glycoprotein through a detailed genomic and proteomic study.
Due to the following features, these multi-epitope based vaccines have some benefits compared to conventional or single-epitope based vaccines: (i) It is very safe and time-saving (ii) The T cell receptors can easily identify multi classes of MHC-I and MHC-II epitopes (iii) Adjuvant, which may be associated with the vaccine, would improve the long term immune response and increase immunogenicity (iv) By this way we can also avoid the difficulty of culturing pathogens and in vitro antigen expression complication [[111], [112], [113], [114], [115], [116], [117], [118], [119]].
So, in our approach, we first identified the CTL, HTL, and LBL epitopes from the targeted S, M, E glycoproteins by the IEDB server. Then we only selected those epitopes that were antigenic, immunogenic, and non-toxic via monitoring with particular servers mentioned in method section. Finally, we connected the CTL, HTL, LBL epitopes with proper (AYY, GPGPG, KK) linkers to complete the final part of vaccine development. An adjuvant β-defensin that helps to induce the immunogenicity, longevity, and most importantly, the vaccine construct's stability has also been improved by attaching the adjuvant to the epitopes (CTL, HTL, LBL) via EAAK linkers [120,121]. Thus, the constructed multi-epitope vaccine can generate specific cellular immunity and highly potent humoral immunity against infection [[122], [123], [124]]. After the construction, we verified our vaccine's non-allergenic profile by AllerTop v.2.0 and Allergen FP v.1.0 servers. Furthermore, the Vaxijen and ANTIGENpro server's up to scratch antigenicity scores proved that the vaccine was highly antigenic.
The vaccine's physicochemical properties were also investigated using the Expasy Protparam server, where we have found the molecular weight was 47.15 kDa and the instability index was 28.26 which defines the constructed vaccine would be stable (The instability index lesser than 40 means the stability) (Table S1). The vaccine's theoretical pI was calculated to be 10.01, indicating that it was basic in nature and the aliphatic index of 80.62 confirmed its thermostability (Table S1). The vaccine's GRAVY (Grand average of hydropathicity) index −0.134 (lower the GRAVY index, greater its solubility) reflected its polar nature in water (Table S1). This solubility index was also checked by another server (Protein sol), where it again proved its effective interaction with water (Table S1). Because of its solvability and expected half-life >10 h in vivo, the constructed vaccine assured that it could be easily synthesized inside the host cell of E. coli (Table S1).
The Ramachandran map was used to verify the vaccine's structural forms (3D structure), where we found that 87.1% of the sequence was in the preferred area, 10.6% was in the additional permitted region, 0.9% was generously allowed regions, and 1.4% was in the disallowed region (Fig. 7B). ERRAT and ProSA web server (Z score server) were further used to verify the vaccine's quality, where it scored 75.924% and −5.12, indicating the vaccine's proficiency (Fig. 7C, A).
To observe the cell density behavior and optimal parameter, immune simulations were performed, where the repeated exposure have given the best immunological responses over antigen (Fig. 15). High levels of B cells and T cells population indicated humoral immunity, and the cytokines showed cell-mediated immunity (Fig. 15A(1–7), 15B(1,2)), Because of B cell proliferation and Ig isotype switching, the IFN-γ induced humoral immune responses as well [125,126]. The other (NK, MA, DC, EP and PLB) cell populations were also appreciable, suggesting a good development of immune memory against SARS-COV-2 (Fig. 15B(3–7)). Finally, a molecular docking analysis was performed between V_COV19_SN and TLRs (TLR2, TLR3, TLR4, TLR7, TLR8). Where, a stable protein-protein interaction was found with all the interactors (Fig. 9, Figs. S8–11). The highest binding affinity indicated that the interaction is more stable. Thus, in this case, we found the highest binding affinity (based on HADDOCK score) among the vaccine and TLR8, which means that the interaction between the TLR8-vaccine complex exceedingly stable compared to other complexes (Table 6). This interaction was very important for the activation of APC cells and the following CD4+, CD8+ T cells via MHC-I and MHC-II [63,127]. We also simulated molecular dynamics for 100ns because it is essential to check the vaccine's stability under in vivo conditions. As we determined the highest binding affinity between the vaccine and the TLR8, we performed MD simulation for this complex only to check its stability. By MD run it was confirmed that the interaction of our vaccine and TLR8 were stable (Fig. 10, Fig. 11). To ensure the effectiveness and efficient expression within the E. coli host, the vaccine was enhanced by codon adaptation tools and reversed translated into specific mRNA (Fig. S15). The favorable GC content, robust mRNA structure, and codon adaptability index showed its high-level capacity for effective expression in E. coli (K12) hosts (Fig. S15). Moreover, for successful cloning, two viral restriction enzymes EcoRI and BamHI were used to cut specific vaccine restriction sites. The cutting sites were then incorporated into a suitable vector pET28a (+) for in silico cloning (Fig. 16). Foroutan et al. have also adopted a similar technique to optimize codon before its expression in vitro [128]. After cloning, using IPTG (Isopropyl beta-d-1-thiogalactopyranoside) induction the recombinant vector can be propagated into E. coli cells and cultivated at 28 °C [129]. This strategy has also been applied for Pseudomonas aeruginosa, Hendra virus, Klebsiella pneumonia, Dengue, Malaria, Nipah virus and cancerous antigens [[130], [131], [132], [133], [134], [135], [136]]. Though the designed vaccine has no major limitations, still the thought was that, our vaccine construct covered almost 100% conservancy among the 128 genome sequences from Bangladesh and 110 from other affected countries. Especially it will provide an advantage to Bangladeshi population and Indian ethnic group. Also, the population coverage showed that it will be a good candidate for people all around the world. So, considering all the parameters we are very confident that by activating the immune cells and another complex signaling of the human body, our designed vaccine has the potential to produce long-lasting immunity against this deadly pathogen.
5. Conclusion
According to the current demand, we need to develop an effective vaccine or drug in an urgent basis for the management of SARS-COV-2 infections. But, it is a challenging task to produce an effective vaccine over a short period, and the possibility diminishes if the genetic material of the pathogen is RNA. Few vaccines have recently gained their FDA approval due to their promising results. However, considering the complex human body systems, there is no assurance that the vaccine will support long-lasting immunity around the globe. That's why more and more vaccine research projects are required through the immunoinformatics approaches. Using a set of immunoinformatics methods, we developed a multi-epitope dependent vaccine by combining CTL, HTL, and LBL epitopes, which were both antigenic and immunogenic. Besides, with high population coverage, our vaccine can cause robust immune responses as well. Therefore, we are very much hopeful that the vaccine built from our immunoinformatics research would be unequivocally suitable for combating this current scenario. Nevertheless, more experimental authentication is required to verify our developed vaccine as an appropriate prophylactic against this etiological agent of the COVID-19 outbreak.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
we acknowledge the contribution of different research groups around the world who generated the primary sequence data which were included in the present analysis. Specially the Research groups who sequenced the isolates in Bangladesh include the ICDDR, B, Child Health Research Foundation, National Institute of Laboratory Medicine and Referral Center, National Institute of Biotechnology, DNA Solution Ltd., COVID-19 Laboratory Center for Advanced Research in Sciences (CARS), Genomic Research Lab-BCSIR and JUST.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.imu.2021.100781.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Naz A., Shahid F., Butt T.T., Awan F.M., Ali A., Malik A. Designing multi-epitope vaccines to combat emerging coronavirus disease 2019 (COVID-19) by employing immuno-informatics approach. Front Immunol. 2020 Jul 10;11:1663. doi: 10.3389/fimmu.2020.01663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmed S., Dávila J.D., Allen A., Haklay M., Tacoli C., Fèvre E.M. Does urbanization make emergence of zoonosis more likely? Evidence, myths and gaps. Environ Urbanization. 2019 Oct;31(2):443–460. doi: 10.1177/0956247819866124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahammad I., Lira S.S. Designing a novel mRNA vaccine against SARS-CoV-2: an immunoinformatics approach. Int J Biol Macromol. 2020 Nov 1;162:820–837. doi: 10.1016/j.ijbiomac.2020.06.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kar T., Narsaria U., Basak S., Deb D., Castiglione F., Mueller D.M., Srivastava A.P. A candidate multi-epitope vaccine against SARS-CoV-2. Sci Rep. 2020 Jul 2;10(1):1–24. doi: 10.1038/s41598-020-67749-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hui D.S., Azhar E.I., Madani T.A., Ntoumi F., Kock R., Dar O., Ippolito G., Mchugh T.D., Memish Z.A., Drosten C., Zumla A. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—the latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis. 2020 Feb 1;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., et al. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N Engl J Med. 2020 Jan 29;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J.J., Dong X., Cao Y.Y., Yuan Y.D., Yang Y.B., Yan Y.Q., Akdis C.A., Gao Y.D. Clinical characteristics of 140 patients infected with SARS‐CoV‐2 in Wuhan, China. Allergy. 2020 Jul;75(7):1730–1741. doi: 10.1111/all.14238. [DOI] [PubMed] [Google Scholar]
- 8.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020 Feb 15;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehmann K.Z., Drosten C., Wendtner C., Zange M.D., Vollmar P., Rosina Ehmann D.V., Zwirglmaier K., Guggemos M.D., Seilmaier M., Niemeyer D., Kelly T.C. Virological assessment of hospitalized cases of coronavirus disease 2019. Nature. 2020;581:465–469. [Google Scholar]
- 10.Yang Y., Lu Q., Liu M., Wang Y., Zhang A., Jalali N., et al. Epidemiological and clinical features of the 2019 novel coronavirus outbreak in China. MedRxiv. 2020 Jan 1 doi: 10.1101/2020.02.10.20021675. [DOI] [Google Scholar]
- 11.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from patients with pneumonia in China. N Engl J Med. 2019;382:727–733. doi: 10.1056/NEJMoa2001017. 2020 Jan 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., et al. Clinical characteristics of 2019 novel coronavirus infection in China. N Engl J Med. 2020 Apr 1;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang W., Tang J., Wei F. Updated understanding of the outbreak of 2019 novel coronavirus (2019‐nCoV) in Wuhan, China. J Med Virol. 2020 Apr;92(4):441–447. doi: 10.1002/jmv.25689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harcourt J., Tamin A., Lu X., Kamili S., Sakthivel S.K., Murray J., Queen K., Tao Y., Paden C.R., Zhang J., Li Y. Severe acute respiratory syndrome coronavirus 2 from patient with coronavirus disease, United States. Emerg Infect Dis. 2020 Jun;26(6):1266. doi: 10.3201/eid2606.200516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong R., Chu Z., Yu F., Zha Y. Contriving multi-epitope subunit of vaccine for COVID-19: immunoinformatics approaches. Front Immunol. 2020 Jul 28;11:1784. doi: 10.3389/fimmu.2020.01784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang X., Yu Y., Xu J., Shu H., Liu H., Wu Y., Zhang L., Yu Z., Fang M., Yu T., Wang Y. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. The Lancet Respiratory Medicine. 2020 May 1;8(5):475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu R., Han H., Liu F., Lv Z., Wu K., Liu Y., Feng Y., Zhu C. Positive rate of RT-PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clin Chim Acta. 2020 Jun 1;505:172–175. doi: 10.1016/j.cca.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li L.Q., Huang T., Wang Y.Q., Wang Z.P., Liang Y., Huang T.B., Zhang H.Y., Sun W., Wang Y. COVID‐19 patients' clinical characteristics, discharge rate, and fatality rate of meta‐analysis. J Med Virol. 2020 Jun;92(6):577–583. doi: 10.1002/jmv.25757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong Y., Mo X., Hu Y., Qi X., Jiang F., Jiang Z., Tong S. Epidemiology of COVID-19 among children in China. Pediatrics. 2020 Jun 1;145(6) doi: 10.1542/peds.2020-0702. [DOI] [PubMed] [Google Scholar]
- 20.Lai C.C., Shih T.P., Ko W.C., Tang H.J., Hsueh P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents. 2020 Mar 1;55(3):105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomes C. Report of the WHO-China joint mission on coronavirus disease 2019 (COVID-19) Brazilian Journal of Implantology and Health Sciences. 2020;2(3) [Google Scholar]
- 22.Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020 Apr;92(4):418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoque M.N., Chaudhury A., Akanda M.A., Hossain M.A., Islam M.T. Genomic diversity and evolution, diagnosis, prevention, and therapeutics of the pandemic COVID-19 disease. PeerJ. 2020 Sep 1;8:e9689. doi: 10.7717/peerj.9689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abdelmageed M.I., Abdelmoneim A.H., Mustafa M.I., Elfadol N.M., Murshed N.S., Shantier S.W., Makhawi A.M. Design of a multiepitope-based peptide vaccine against the E protein of human COVID-19: an immunoinformatics approach. BioMed Res Int. 2020 May 11:2020. doi: 10.1155/2020/2683286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Padhi A.K., Shukla R., Saudagar P., Tripathi T. High-throughput rational design of the remdesivir binding site in the RdRp of SARS-CoV-2: implications for potential resistance. Iscience. 2021 Jan 22;24(1):101992. doi: 10.1016/j.isci.2020.101992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ranga V., Niemelä E., Tamirat M.Z., Eriksson J.E., Airenne T.T., Johnson M.S. Immunogenic SARS-CoV-2 epitopes: in silico study towards better understanding of COVID-19 disease—paving the way for vaccine development. Vaccines. 2020 Sep;8(3):408. doi: 10.3390/vaccines8030408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Padhi A.K., Tripathi T. Can SARS-CoV-2 accumulate mutations in the S-protein to increase pathogenicity? ACS Pharmacology & Translational Science. 2020 Sep 8;3(5):1023–1026. doi: 10.1021/acsptsci.0c00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rabi F.A., Al Zoubi M.S., Kasasbeh G.A., Salameh D.M., Al-Nasser A.D. SARS-CoV-2 and coronavirus disease 2019: what we know so far. Pathogens. 2020 Mar;9(3):231. doi: 10.3390/pathogens9030231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Padhi A.K., Tripathi T. Targeted design of drug binding sites in the main protease of SARS-CoV-2 reveals potential signatures of adaptation. Biochem Biophys Res Commun. 2021 May 28;555:147–153. doi: 10.1016/j.bbrc.2021.03.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ojha R., Gupta N., Naik B., Singh S., Verma V.K., Prusty D., Prajapati V.K. High throughput and comprehensive approach to develop multiepitope vaccine against minacious COVID-19. Eur J Pharmaceut Sci. 2020 Aug 1;151:105375. doi: 10.1016/j.ejps.2020.105375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pallesen J., Wang N., Corbett K.S., Wrapp D., Kirchdoerfer R.N., Turner H.L., Cottrell C.A., Becker M.M., Wang L., Shi W., Kong W.P. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc Natl Acad Sci Unit States Am. 2017 Aug 29;114(35):E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song W., Gui M., Wang X., Xiang Y. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018 Aug 13;14(8) doi: 10.1371/journal.ppat.1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shang W., Yang Y., Rao Y., Rao X. The outbreak of SARS-CoV-2 pneumonia calls for viral vaccines. npj Vaccines. 2020 Mar 6;5(1):1–3. doi: 10.1038/s41541-020-0170-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou H., Chen Y., Zhang S., Niu P., Qin K., Jia W., Huang B., Zhang S., Lan J., Zhang L., Tan W. Structural definition of a neutralization epitope on the N-terminal domain of MERS-CoV spike glycoprotein. Nat Commun. 2019 Jul 11;10(1):1–3. doi: 10.1038/s41467-019-10897-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang N., Rosen O., Wang L., Turner H.L., Stevens L.J., Corbett K.S., Bowman C.A., Pallesen J., Shi W., Zhang Y., Leung K. Structural definition of a neutralization-sensitive epitope on the MERS-CoV S1-NTD. Cell Rep. 2019 Sep 24;28(13):3395–3405. doi: 10.1016/j.celrep.2019.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saha S., Malaker R., Sajib M.S., Hasanuzzaman M., Rahman H., Ahmed Z.B., Islam M.S., Islam M., Hooda Y., Ahyong V., Vanaerschot M. Complete genome sequence of a novel coronavirus (SARS-CoV-2) isolate from Bangladesh. Microbiology resource announcements. 2020 Jun 11;9(24) doi: 10.1128/MRA.00568-20. e00568-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. Mega X: molecular evolutionary genetics. [DOI] [PMC free article] [PubMed]
- 38.Rahman M.S., Hoque M.N., Islam M.R., Akter S., Alam A.R., Siddique M.A., Saha O., Rahaman M.M., Sultana M., Crandall K.A., Hossain M.A. Epitope-based chimeric peptide vaccine design against S, M and E proteins of SARS-CoV-2, the etiologic agent of COVID-19 pandemic: an in silico approach. PeerJ. 2020 Jul 27;8 doi: 10.7717/peerj.9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khan S., Khan A., Rehman A.U., Ahmad I., Ullah S., Khan A.A., Ali S.S., Afridi S.G., Wei D.Q. Immunoinformatics and structural vaccinology driven prediction of multi-epitope vaccine against Mayaro virus and validation through in-silico expression. Infect Genet Evol. 2019 Sep 1;73:390–400. doi: 10.1016/j.meegid.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 40.Rahman N., Ali F., Basharat Z., Shehroz M., Khan M.K., Jeandet P., Nepovimova E., Kuca K., Khan H. Vaccine design from the ensemble of surface glycoprotein epitopes of SARS-CoV-2: an immunoinformatics approach. Vaccines. 2020 Sep;8(3):423. doi: 10.3390/vaccines8030423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vita R., Mahajan S., Overton J.A., Dhanda S.K., Martini S., Cantrell J.R., Wheeler D.K., Sette A., Peters B. The immune epitope database (IEDB): 2018 update. Nucleic Acids Res. 2019 Jan 8;47(D1):D339–D343. doi: 10.1093/nar/gky1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hasan M., Azim K.F., Imran M.A., Chowdhury I.M., Urme S.R., Parvez M.S., Uddin M.B., Ahmed S.S. Comprehensive genome based analysis of Vibrio parahaemolyticus for identifying novel drug and vaccine molecules: subtractive proteomics and vaccinomics approach. PLoS One. 2020 Aug 19;15(8) doi: 10.1371/journal.pone.0237181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dhanda S.K., Mahajan S., Paul S., Yan Z., Kim H., Jespersen M.C., Jurtz V., Andreatta M., Greenbaum J.A., Marcatili P., Sette A. IEDB-AR: immune epitope database—analysis resource in 2019. Nucleic Acids Res. 2019 Jul 2;47(W1):W502–W506. doi: 10.1093/nar/gkz452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nain Z., Abdulla F., Rahman M.M., Karim M.M., Khan M.S., Sayed S.B., Mahmud S., Rahman S.R., Sheam M.M., Haque Z., Adhikari U.K. Proteome-wide screening for designing a multi-epitope vaccine against emerging pathogen Elizabethkingia anophelis using immunoinformatic approaches. J Biomol Struct Dyn. 2020 Nov 1;38(16):4850–4867. doi: 10.1080/07391102.2019.1692072. [DOI] [PubMed] [Google Scholar]
- 45.Doytchinova I.A., Flower D.R. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinf. 2007 Dec;8(1):1–7. doi: 10.1186/1471-2105-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dimitrov I., Flower D.R., Doytchinova I. AllerTOP-a server for in silico prediction of allergens. InBMC bioinformatics. 2013 Apr;14(6):1–9. doi: 10.1186/1471-2105-14-S6-S4. BioMed Central. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vita R., Overton J.A., Greenbaum J.A., Ponomarenko J., Clark J.D., Cantrell J.R., Wheeler D.K., Gabbard J.L., Hix D., Sette A., Peters B. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 2015 Jan 28;43(D1):D405–D412. doi: 10.1093/nar/gku938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gupta S., Kapoor P., Chaudhary K., Gautam A., Kumar R., Consortium O.S.D.D., Raghava G.P.S. Silico approach for predicting toxicity of peptides and proteins. PLoS One. 2013;8 doi: 10.1371/journal.pone.0073957. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dhanda S.K., Vir P., Raghava G.P. Designing of interferon-gamma inducing MHC class-II binders. Biol Direct. 2013 Dec;8(1):1–5. doi: 10.1186/1745-6150-8-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dhanda S.K., Gupta S., Vir P., Raghava G.P. Prediction of IL4 inducing peptides. Clin Dev Immunol. 2013;2013 doi: 10.1155/2013/263952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nagpal G., Usmani S.S., Dhanda S.K., Kaur H., Singh S., Sharma M., Raghava G.P. Computer-aided designing of immunosuppressive peptides based on IL-10 inducing potential. Sci Rep. 2017 Feb 17;7(1) doi: 10.1038/srep42851. 1-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Samad A., Ahammad F., Nain Z., Alam R., Imon R.R., Hasan M., Rahman M.S. Designing a multi-epitope vaccine against SARS-CoV-2: an immunoinformatics approach. J Biomol Struct Dyn. 2020 Jul 15:1–7. doi: 10.1080/07391102.2020.1792347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adhikari U.K., Rahman M.M. Overlapping CD8+ and CD4+ T-cell epitopes identification for the progression of epitope-based peptide vaccine from nucleocapsid and glycoprotein of emerging Rift Valley fever virus using immunoinformatics approach. Infect Genet Evol. 2017 Dec 1;56:75–91. doi: 10.1016/j.meegid.2017.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bui H.H., Sidney J., Dinh K., Southwood S., Newman M.J., Sette A. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinf. 2006 Dec;7(1):1–5. doi: 10.1186/1471-2105-7-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Elbe S., Buckland‐Merrett G. Data, disease and diplomacy: GISAID's innovative contribution to global health. Global challenges. 2017 Jan;1(1):33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hall T. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. InNucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 57.Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R.D., Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003 Jul 1;31(13):3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bhatnager R., Bhasin M., Arora J., Dang A.S. Epitope based peptide vaccine against SARS-COV2: an immune-informatics approach. J Biomol Struct Dyn. 2020 Jul 2:1–6. doi: 10.1080/07391102.2020.1787227. [DOI] [PubMed] [Google Scholar]
- 59.Lee H., Heo L., Lee M.S., Seok C. GalaxyPepDock: a protein-peptide docking tool based on interaction similarity. Nucleic Acids Res. 2015;43(W1):W431–W435. doi: 10.1093/nar/gkv495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ostertag E.M., Prak Luning, et al. DeBerardinis R.J., Moran J.V., Kazazian H.H., Jr. Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res. 2000 Mar 15;28(6):1418–1423. doi: 10.1093/nar/28.6.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xue L., Rodrigues J., Kastritis P., Bonvin A.M.J.J., Vangone A. PRODIGY: a web-server for predicting the binding affinity in protein-protein complexes. Bioinformatics. 2016 doi: 10.1093/bioinformatics/btw514. [DOI] [PubMed] [Google Scholar]
- 62.Nezafat N., Ghasemi Y., Javadi G., Khoshnoud M.J., Omidinia E. A novel multi‐epitope peptide vaccine against cancer: an in silico approach. J Theor Biol. 2014;21:349. doi: 10.1016/j.jtbi.2014.01.018. 134. [DOI] [PubMed] [Google Scholar]
- 63.Shey R.A., Ghogomu S.M., Esoh K.K., Nebangwa N.D., Shintouo C.M., Nongley N.F., Asa B.F., Ngale F.N., Vanhamme L., Souopgui J. In-silico design of a multi-epitope vaccine candidate against onchocerciasis and related filarial diseases. Sci Rep. 2019 Mar 13;9(1):1–8. doi: 10.1038/s41598-019-40833-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chauhan V., Rungta T., Rawat M., Goyal K., Gupta Y., Singh M.P. Excavating SARS‐coronavirus 2 genome for epitope‐based subunit vaccine synthesis using immunoinformatics approach. J Cell Physiol. 2021 Feb;236(2):1131–1147. doi: 10.1002/jcp.29923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li W., Joshi M.D., Singhania S., Ramsey K.H., Murthy A.K. Peptide vaccine: progress and challenges. Vaccines. 2014 Sep;2(3):515–536. doi: 10.3390/vaccines2030515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sayed S.B., Nain Z., Khan M.S., Abdulla F., Tasmin R., Adhikari U.K. Exploring lassa virus proteome to design a multi-epitope vaccine through immunoinformatics and immune simulation analyses. Int J Pept Res Therapeut. 2020 Dec;26(4):2089–2107. doi: 10.1007/s10989-019-10003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dong R., Chu Z., Yu F., Zha Y. Contriving multi-epitope subunit of vaccine for COVID-19: immunoinformatics approaches. Front Immunol. 2020 Jul 28;11:1784. doi: 10.3389/fimmu.2020.01784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dimitrov I., Naneva L., Doytchinova I., Bangov I. AllergenFP: allergenicity prediction by descriptor fingerprints. Bioinformatics. 2014 Mar 15;30(6):846–851. doi: 10.1093/bioinformatics/btt619. [DOI] [PubMed] [Google Scholar]
- 69.Solanki V., Tiwari V. Subtractive proteomics to identify novel drug targets and reverse vaccinology for the development of chimeric vaccine against Acinetobacter baumannii. Sci Rep. 2018 Jun 13;8(1):1–9. doi: 10.1038/s41598-018-26689-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Magnan Christophe N., Zeller Michael, Kayala Matthew A., Adam Vigil, Randall Arlo, Felgner Philip L., Baldi Pierre. High-throughput prediction of protein antigenicity using protein microarray data. Bioinformatics. 2010;26(December 2010):2936–2943. doi: 10.1093/bioinformatics/btq551. 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Magnan C.N., Randall A., Baldi P. SOLpro: Accurate sequence-based prediction of protein solubility. Bioinformatics. 2009;25:17. doi: 10.1093/bioinformatics/btp386. [DOI] [PubMed] [Google Scholar]
- 72.Hebditch M., Carballo-Amador M.A., Charonis S., Curtis R. Warwicker J Protein-Sol: a web tool for predicting protein solubility from sequence. Bioinformatics. 2017;33(19):3098–3100. doi: 10.1093/bioinformatics/btx345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nielsen H. Humana Press; New York, NY: 2017. Predicting secretory proteins with SignalP. InProtein function prediction; pp. 59–73. [DOI] [PubMed] [Google Scholar]
- 74.Krogh A., Larsson B., Von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001 Jan 19;305(3):567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 75.Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R.D., Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Walker J.M., editor. The proteomics protocols handbook. Humana press; 2005 Mar 9. [Google Scholar]
- 77.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 78.Buchan D.W., Minneci F., Nugent T.C., Bryson K., Jones D.T. Scalable web services for the PSIPRED protein analysis workbench. Nucleic Acids Res. 2013 Jul 1;41(W1):W349–W357. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang S., Sun S., Li Z., Zhang R., Xu J. Accurate de novo prediction of protein contact map by ultra-deep learning model. PLoS Comput Biol. 2017 Jan 5;13(1) doi: 10.1371/journal.pcbi.1005324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dong Xu, Zhang Yang. Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophys J. 2011;101:2525–2534. doi: 10.1016/j.bpj.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Heo L., Park H., Seok C. GalaxyRefine: protein structure refinement driven by side-chain repacking. Nucleic Acids Res. 2013 Jul 1;41(W1):W384–W388. doi: 10.1093/nar/gkt458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 83.Ramachandran S., Kota P., Ding F., Dokholyan N.V. Proteins: Structure, Function and Bioinformatics. 2011;79:261–270. doi: 10.1002/prot.22879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Khatoon N., Pandey R.K., Prajapati V.K. Exploring Leishmania secretory proteins to design B and T cell multi-epitope subunit vaccine using immunoinformatics approach. Sci Rep. 2017 Aug 15;7(1):1–2. doi: 10.1038/s41598-017-08842-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wiederstein M., Sippl M.J. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007 Jul 1;35(suppl_2):W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Colovos C., Yeates T.O. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 1993 Sep;2(9):1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. Procheck - a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 88.Craig D.B., Dombkowski A.A. Disulfide by Design 2.0: a web-based tool for disulfide engineering in proteins. BMC Bioinf. 2013 Dec;14(1):1–7. doi: 10.1186/1471-2105-14-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lund F.E. Cytokine-producing B lymphocytes—key regulators of immunity. Curr Opin Immunol. 2008 Jun 1;20(3):332–338. doi: 10.1016/j.coi.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ponomarenko J., Bui H.H., Li W., Fusseder N., Bourne P.E., Sette A., Peters B. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinf. 2008 Dec;9(1):1–8. doi: 10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lengauer T., Rarey M. Computational methods for biomolecular docking. Curr Opin Struct Biol. 1996 Jun 1;6(3):402–406. doi: 10.1016/s0959-440x(96)80061-3. [DOI] [PubMed] [Google Scholar]
- 92.BIOVIA, Dassault Systèmes, [Software product name], [Software version], San Diego: Dassault Systèmes.
- 93.de Vries S.J., Bonvin A.M. CPORT: a consensus interface predictor and its performance in prediction-driven docking with HADDOCK. PLoS One. 2011 Mar 25;6(3) doi: 10.1371/journal.pone.0017695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Van Zundert G.C., Rodrigues J.P., Trellet M., Schmitz C., Kastritis P.L., Karaca E., Melquiond A.S., van Dijk M., De Vries S.J., Bonvin A.M. The HADDOCK2. 2 web server: user-friendly integrative modeling of biomolecular complexes. J Mol Biol. 2016 Feb 22;428(4):720–725. doi: 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 95.Laskowski R.A., Jabłońska J., Pravda L., Vařeková R.S., Thornton J.M. PDBsum: structural summaries of PDB entries. Protein Sci. 2018 Jan;27(1):129–134. doi: 10.1002/pro.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chander S., Pandey R.K., Penta A., Choudhary B.S., Sharma M., Malik R., Prajapati V.K., Murugesan S. Molecular docking and molecular dynamics simulation based approach to explore the dual inhibitor against HIV-1 reverse transcriptase and Integrase. Comb Chem High Throughput Screen. 2017 Sep 1;20(8):734–746. doi: 10.2174/1386207320666170615104703. [DOI] [PubMed] [Google Scholar]
- 97.Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. Software. 2015 Sep 1;1:19–25. [Google Scholar]
- 98.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996 Feb 1;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 99.David C.C., Jacobs D.J. Principal component analysis: a method for determining the essential dynamics of proteins. InProtein dynamics. 2014:193–226. doi: 10.1007/978-1-62703-658-0_11. [Humana Press, Totowa, NJ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Grant B.J., Skjaerven L., Yao X.Q. The Bio3D packages for structural bioinformatics. Protein Sci. 2021 Jan;30(1):20–30. doi: 10.1002/pro.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Genheden S., Kuhn O., Mikulskis P., Hoffmann D., Ryde U. The normal-mode entropy in the MM/GBSA method: effect of system truncation, buffer region, and dielectric constant. J Chem Inf Model. 2012 Aug 27;52(8):2079–2088. doi: 10.1021/ci3001919. [DOI] [PubMed] [Google Scholar]
- 102.Turner P.J. Center for Coastal and Land-Margin Research, Oregon Graduate Institute of Science and Technology; Beaverton, OR: 2005. XMGRACE, version 5.1. 19. [Google Scholar]
- 103.Lorenz R., Bernhart S.H., Zu Siederdissen C.H., Tafer H., Flamm C., Stadler P.F., Hofacker I.L. ViennaRNA package 2.0. Algorithm Mol Biol. 2011 Dec;6(1):1–4. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003 Jul 1;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Castiglione F., Mantile F., De Berardinis P., Prisco A. How the interval between prime and boost injection affects the immune response in a computational model of the immune system. Computational and mathematical methods in medicine. 2012;2012 doi: 10.1155/2012/842329. 2012 Oct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Grote A., Hiller K., Scheer M., Münch R., Nörtemann B., Hempel D.C., Jahn D. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005 Jul 1;33(suppl_2):W526–W531. doi: 10.1093/nar/gki376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Stothard P. The Sequence Manipulation Suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques. 2000;28:1102–1104. doi: 10.2144/00286ir01. [DOI] [PubMed] [Google Scholar]
- 108.Goldberg M.F., Roeske E.K., Ward L.N., Pengo T., Dileepan T., Kotov D.I., Jenkins M.K. Salmonella persist in activated macrophages in T cell-sparse granulomas but are contained by surrounding CXCR3 ligand-positioned Th1 cells. Immunity. 2018 Dec 18;49(6):1090–1102. doi: 10.1016/j.immuni.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Morse J.S., Lalonde T., Xu S., Liu W.R. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019‐nCoV. Chembiochem. 2020 Mar 2;21(5):730–738. doi: 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lane R. Sarah Gilbert: carving a path towards a COVID-19 vaccine. Lancet. 2020 Apr 18;395(10232):1247. doi: 10.1016/S0140-6736(20)30796-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lu C., Meng S., Jin Y., Zhang W., Li Z., Wang F., Wang‐Johanning F., Wei Y., Liu H., Tu H., Su D. A novel multi‐epitope vaccine from MMSA‐1 and DKK 1 for multiple myeloma immunotherapy. Br J Haematol. 2017 Aug;178(3):413–426. doi: 10.1111/bjh.14686. [DOI] [PubMed] [Google Scholar]
- 112.He R., Yang X., Liu C., Chen X., Wang L., Xiao M., Ye J., Wu Y., Ye L. Efficient control of chronic LCMV infection by a CD4 T cell epitope-based heterologous prime-boost vaccination in a murine model. Cell Mol Immunol. 2018 Sep;15(9):815–826. doi: 10.1038/cmi.2017.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Saadi M., Karkhah A., Nouri H.R. Development of a multi-epitope peptide vaccine inducing robust T cell responses against brucellosis using immunoinformatics based approaches. Infect Genet Evol. 2017 Jul 1;51:227–234. doi: 10.1016/j.meegid.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 114.Lu I.N., Farinelle S., Sausy A., Muller C.P. Identification of a CD4 T-cell epitope in the hemagglutinin stalk domain of pandemic H1N1 influenza virus and its antigen-driven TCR usage signature in BALB/c mice. Cell Mol Immunol. 2017 Jun;14(6):511–520. doi: 10.1038/cmi.2016.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jiang P., Cai Y., Chen J., Ye X., Mao S., Zhu S., Xue X., Chen S., Zhang L. Evaluation of tandem Chlamydia trachomatis MOMP multi-epitopes vaccine in BALB/c mice model. Vaccine. 2017 May 25;35(23):3096–3103. doi: 10.1016/j.vaccine.2017.04.031. [DOI] [PubMed] [Google Scholar]
- 116.Lennerz V., Gross S., Gallerani E., Sessa C., Mach N., Boehm S., Hess D., Von Boehmer L., Knuth A., Ochsenbein A.F., Gnad-Vogt U. Immunologic response to the survivin-derived multi-epitope vaccine EMD640744 in patients with advanced solid tumors. Cancer Immunol Immunother. 2014 Apr 1;63(4):381–394. doi: 10.1007/s00262-013-1516-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhu S., Feng Y., Rao P., Xue X., Chen S., Li W., Zhu G., Zhang L. Hepatitis B virus surface antigen as delivery vector can enhance Chlamydia trachomatis MOMP multi-epitope immune response in mice. Appl Microbiol Biotechnol. 2014 May;98(9):4107–4117. doi: 10.1007/s00253-014-5517-x. [DOI] [PubMed] [Google Scholar]
- 118.Davies M.N., Flower D.R. Harnessing bioinformatics to discover new vaccines. Drug Discov Today. 2007 May 1;12(9–10):389–395. doi: 10.1016/j.drudis.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 119.Kalita P., Padhi A.K., Zhang K.Y., Tripathi T. Design of a peptide-based subunit vaccine against novel coronavirus SARS-CoV-2. Microb Pathog. 2020 Aug 1;145:104236. doi: 10.1016/j.micpath.2020.104236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bonam S.R., Partidos C.D., Halmuthur S.K., Muller S. An overview of novel adjuvants designed for improving vaccine efficacy. Trends Pharmacol Sci. 2017 Sep 1;38(9):771–793. doi: 10.1016/j.tips.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 121.Lee S., Nguyen M.T. Recent advances of vaccine adjuvants for infectious diseases. Immune network. 2015 Apr;15(2):51. doi: 10.4110/in.2015.15.2.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dawood R.M., Moustafa R.I., Abdelhafez T.H., El-Shenawy R., El-Abd Y., El Din N.G., Dubuisson J., El Awady M.K. A multiepitope peptide vaccine against HCV stimulates neutralizing humoral and persistent cellular responses in mice. BMC Infect Dis. 2019 Dec;19(1) doi: 10.1186/s12879-019-4571-5. 1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gralinski L.E., Menachery V.D. Return of the coronavirus: 2019-nCoV. Viruses. 2020 Feb;12(2):135. doi: 10.3390/v12020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yong S.E., Anderson D.E., Wei W.E., Pang J., Chia W.N., Tan C.W., Teoh Y.L., Rajendram P., Toh M.P., Poh C., Koh V.T. Connecting clusters of COVID-19: an epidemiological and serological investigation. Lancet Infect Dis. 2020 Jul 1;20(7):809–815. doi: 10.1016/S1473-3099(20)30273-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fang D., Cui K., Mao K., Hu G., Li R., Zheng M., Riteau N., Reiner S.L., Sher A., Zhao K., Zhu J. Transient T-bet expression functionally specifies a distinct T follicular helper subset. J Exp Med. 2018 Nov 5;215(11):2705–2714. doi: 10.1084/jem.20180927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Snapper C.M., Paul W.E. Interferon-gamma and B cell stimulatory factor-1 reciprocally regulate Ig isotype production. Science. 1987 May 22;236(4804):944–947. doi: 10.1126/science.3107127. [DOI] [PubMed] [Google Scholar]
- 127.Shi J., Zhang J., Li S., Sun J., Teng Y., Wu M., Li J., Li Y., Hu N., Wang H., Hu Y. Epitope-based vaccine target screening against highly pathogenic MERS-CoV: an in silico approach applied to emerging infectious diseases. PLoS One. 2015 Dec 7;10(12) doi: 10.1371/journal.pone.0144475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Foroutan M., Ghaffarifar F., Sharifi Z., Dalimi A. Vaccination with a novel multi-epitope ROP8 DNA vaccine against acute Toxoplasma gondii infection induces strong B and T cell responses in mice. Comp Immunol Microbiol Infect Dis. 2020 Apr 1;69:101413. doi: 10.1016/j.cimid.2020.101413. [DOI] [PubMed] [Google Scholar]
- 129.Biswal J.K., Bisht P., Mohapatra J.K., Ranjan R., Sanyal A., Pattnaik B. Application of a recombinant capsid polyprotein (P1) expressed in a prokaryotic system to detect antibodies against foot-and-mouth disease virus serotype O. J Virol Methods. 2015 Apr 1;215:45–51. doi: 10.1016/j.jviromet.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 130.Ali M., Pandey R.K., Khatoon N., Narula A., Mishra A., Prajapati V.K. Exploring dengue genome to construct a multi-epitope based subunit vaccine by utilizing immunoinformatics approach to battle against dengue infection. Sci Rep. 2017 Aug 23;7(1):1–3. doi: 10.1038/s41598-017-09199-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ojha R., Pareek A., Pandey R.K., Prusty D., Prajapati V.K. Strategic development of a next-generation multi-epitope vaccine to prevent nipah virus zoonotic infection. ACS Omega. 2019 Aug 7;4(8):13069–13079. doi: 10.1021/acsomega.9b00944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kamthania M., Srivastava S., Desai M., Jain A., Shrivastav A., Sharma D.K. Immunoinformatics Approach to design T-cell epitope-based vaccine against hendra virus. Int J Pept Res Therapeut. 2019 Dec;25(4):1627–1637. [Google Scholar]
- 133.Pandey R.K., Bhatt T.K., Prajapati V.K. Novel immunoinformatics approaches to design multi-epitope subunit vaccine for malaria by investigating anopheles salivary protein. Sci Rep. 2018 Jan 18;8(1) doi: 10.1038/s41598-018-19456-1. 1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Dar H.A., Zaheer T., Shehroz M., Ullah N., Naz K., Muhammad S.A., Zhang T., Ali A. Immunoinformatics-aided design and evaluation of a potential multi-epitope vaccine against Klebsiella pneumoniae. Vaccines. 2019 Sep;7(3):88. doi: 10.3390/vaccines7030088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chauhan V., Rungta T., Goyal K., Singh M.P. Designing a multi-epitope based vaccine to combat Kaposi Sarcoma utilizing immunoinformatics approach. Sci Rep. 2019 Feb 21;9(1):1–5. doi: 10.1038/s41598-019-39299-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mishra S., Sinha S. Immunoinformatics and modeling perspective of T cell epitope-based cancer immunotherapy: a holistic picture. J Biomol Struct Dyn. 2009 Dec 1;27(3):293–305. doi: 10.1080/07391102.2009.10507317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.