Abstract
SINEs (short interspersed elements) are an abundant class of transposable elements found in a wide variety of eukaryotes. Using the genomic sequencing technique, we observed that plant S1 SINE retroposons mainly integrate in hypomethylated DNA regions and are targeted by methylases. Methylation can then spread from the SINE into flanking genomic sequences, creating distal epigenetic modifications. This methylation spreading is vectorially directed upstream or downstream of the S1 element, suggesting that it could be facilitated when a potentially good methylatable sequence is single stranded during DNA replication, particularly when located on the lagging strand. Replication of a short methylated DNA region could thus lead to the de novo methylation of upstream or downstream adjacent sequences.
Most eukaryotic interspersed repeats are the result of an amplification process that depends upon the reverse transcription of an RNA intermediate. These repeats are either short or long interspersed elements (SINEs or LINEs, respectively) or retrovirus-like elements (retrotransposons and endogenous retroviruses) (18). The fraction of the genome occupied by these elements varies from 35% in humans (45) to probably more than 60% in certain plant species, such as maize (39). This level of amplification is a serious threat to the host genome, since integration to new sites can result in deleterious mutations. To limit these effects, the hosts have combined several strategies based on the direct repression of one or more steps of the mobility process or on the targeting of mobile elements away from genes (39, 51).
Transcription of retroelements is usually strongly repressed and limited either to a very small number of elements, to certain tissues, or to particular physiological conditions (25, 26, 44), suggesting that transcription represents an important control point of the mobilization process. For SINEs, despite the presence of a potentially active internal polymerase III (Pol III) promoter in a large number of elements, their specific (Pol III-dependent) transcription was shown to be weak and limited to a very small subset of elements (9, 26). In mammals and plants, transcriptional control of SINEs could be related to their high level of methylation directly blocking the initiation of transcription or contributing to the formation of transcriptionally inactive chromatin domain (10, 11, 19, 27). Thus, methylation may be part of a genome defense system which inactivates the transcription of parasitic mobile elements (54). The few transcriptionally active elements could escape methylation and/or be fortuitously associated with a transcriptional enhancer (5).
The mechanism by which a large number of SINE retroposons are methylated is for the moment unknown. Since methylation is often determined by sequence context (2), methylation of SINEs is generally thought to depend on the methylation status of their integration sites. SINEs would thus be highly methylated, because they integrate mainly in methylated regions of the genome, while the few hypomethylated (and transcriptionally active) elements would reside in hypomethylated DNA regions (7, 26, 42). Recently, the methylation status of SINEs from the sea squirt (Ciona intestinalis) genome was shown to conform to the methylation status of the surrounding DNA sequences, supporting this hypothesis (43). Alternatively, SINEs could be direct targets for DNA methylation. This targeting could be related to the repetitive nature of SINEs. Methylation of DNA repeats has been described in fungi as part of RIP and MIP phenomena (37, 41), in relation to paramutation and gene silencing in plants (29), and in the programmed methylation of the maize Mu (mutator), Ac (activator), and Spm (suppressor-mutator) DNA transposons (1, 3, 40). In these cases, methylation of repeats probably depends on homologous DNA-DNA or RNA-DNA interactions (reviewed in reference 29).
SINEs are also suspected to play a role in inducing de novo methylation. In several cases, genomic fragments containing SINEs were found to be capable of inducing de novo methylation of adjacent genomic sequences. In one case, two DNA fragments from the rat α-fetoprotein control region containing rodent SINEs were shown to promote the de novo methylation of an adjacent reporter gene (14). A second example is the methylation of the mouse aprt (adenine phosphoribosyl transferase) promoter after disruption of Sp1 elements, which may have originated from a methylation center comprising SINEs (31, 32, 53). Finally, human Alu elements were proposed as potential de novo methylation centers implicated in tumor supressor gene silencing in neoplasia (13) and in the methylation of an exon of the TP53 gene (28). Based on these reports, it has been speculated that SINEs could be good elicitors of methylation spreading (50).
The S1 element is a small (180-bp) plant SINE that was first described and studied in Brassica napus and is widely distributed among Cruciferae (8, 22). Transcription of S1 elements in B. napus is severely repressed and controled in a tissue-specific manner (9). S1 elements were recently shown to be highly methylated at symmetrical and asymmetrical positions (11). We show here that S1 elements generally insert in hypomethylated DNA regions and are de novo methylated, suggesting that they do not simply adopt the methylation status of surrounding regions, but are directly targeted by methylases. We also show that the integration of S1 element can induce directional de novo methylation of genomic flanking regions.
MATERIALS AND METHODS
Plant materials and seed germination.
B. napus seeds from the Westar cultivar and for 18 different breeding lines were obtained as a gift from DNA Landmark, Inc. Seeds were grown on solid MS media (Sigma) containing 8 g of agar per liter supplemented with Gamborg's vitamins (thiamine-HCl, 1 mg/liter; pyridoxine-HCl, 0.5 mg/liter; nicotinic acid, 0.5 mg/liter; and myoinositol, 100 mg/liter), d(+)-saccharose (30 g/liter), and MES (morpholineethanesulfonic acid [0.5 g/liter]) adjusted to pH 5.7. Plants were grown for 10 to 15 days at 23°C. Leaf material was collected, frozen in liquid nitrogen, and stored at −80°C until DNA extraction.
DNA extractions.
B. napus DNA was isolated from leaves has described previously (11) with the addition of a CsCl purification step or by using the DNeasy Plant isolation system (Qiagen).
Genomic sequencing method.
The genomic sequencing method was based on that described by Clark et al. (6). Briefly, digested DNA (1 to 5 μg) was denatured for 20 min at 37°C in 70 μl of 0.3 M NaOH. Denatured DNA was mixed with 400 μl of freshly prepared 2 M sodium metabisulfite–0.6 mM hydroquinone (pH 5) (Merck) (1.7 M–0.5 mM final concentration). The reaction mixture was incubated in a Hot Top thermal cycler (Appligene) for 18 h at 55°C with a 30-s denaturation step at 94°C every 3 h. DNA was then purified by a desalting column step (Promega Wizard DNA Clean-Up System), and eluted DNA was incubated in 0.3 M NaOH for 15 min at 37°C. After neutralization with ammonium acetate (at a 3 M final concentration), the DNA was precipitated in ethanol and resuspended in 100 μl of water.
PCRs were performed with 100 ng of treated DNA in a Crocodile III apparatus (Appligene) by using Taq Bead Hot Start polymerase (Promega). The reactions were carried out in 50 μl containing 50 pmol of each primer (the sequences of primers used to amplified bisulfite-treated DNA are available on request), 0.2 mM each deoxynucleoside triphosphate (dNTP), and 1.25 U (1 bead) of Taq Bead Hot Start polymerase in the recommended buffer. Samples were processed as follows: 1 cycle at 94°C for 5 min; followed by 1 min at proper annealing temperature (corresponding to the lowest melting temperature of the primers used); 40 cycles at 72°C for 45 s, 94°C for 30 s, and the proper annealing temperature for 30 s; and 1 cycle at 72°C for 5 min. PCR products were cloned by using the pGEM-T Easy vector (Promega) and transfected into Escherichia coli DH5α by heat shock at 42°C (38). Nucleotide sequences were determined with the T7 sequencing kit (Amersham Pharmacia Biotech).
The bisulfite reaction converts nonmethylated cytosines in DNA to uracils, while leaving 5-methylcytosines unaltered (6). To demonstrate the efficiency of the bisulfite treatment on CpG-rich S1 elements, the na16 locus (S1 with flanking regions; 750 bp total) was cloned in a pGEM-T vector, and 10 ng of this linearized plasmid was mixed with 5 μg of digested genomic DNA before bisulfite treatment. To control the treatment, we amplified this control locus by using pGEM primers designed for bisulfite-treated DNA (pG1B, GGGTGAATTGGGTTTGATGT; and pG2B, CTCCCATATAATCAACCTAC). Eighteen recombinant na16 controls were analyzed, and all clones showed a 100% conversion efficiency. All cytosines present in the S1 element or in its flanking regions were converted to uracil (139 per clone) except for methylated cytosines (4 per clone) resulting from plasmid multiplication in bacteria expressing the dcm methylation pathway. Therefore, S1 elements are not resistant to the bisulfite reaction and are treated with the same efficiency as their genomic flanking sequences.
Restriction enzyme analysis.
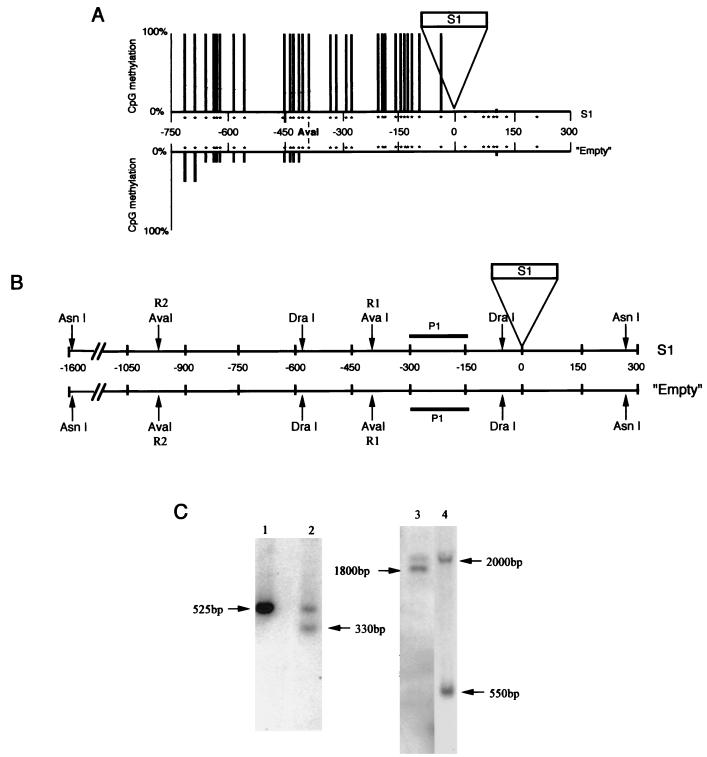
For the genomic blotting experiments, aliquots of 5 to 10 μg of genomic DNA from heterozygous plants (for the na32 locus) were digested with different combinations of restriction enzymes (DraI, TTTAAA; DraI-AvaI, CYCGRG; AsnI, ATTAAT; AsnI-AvaI), separated by electrophoresis on 1% agarose gel, transferred to nylon membranes under alkaline conditions (38), and hybridized with 32P-labelled probe P1 (see Fig. 3).
FIG. 3.
Evaluation of the upstream methylation spreading for the na32 locus. (A) The methylation status of the empty and S1 allele upstream regions was analyzed by genomic sequencing (upper strand). Only the CpG sites are indicated (by stars). The position of the R1-AvaI site is indicated. The proportion of CpG methylation is represented by bars of different lengths. Nine and seven clones were sequenced, respectively, for the empty and S1 alleles. (B) Restriction map of both alleles. Positions of the probe (P1) and of the two AvaI restriction sites (R1 and R2) are indicated. DraI and AsnI are not sensible for DNA methylation, while AvaI is inhibited by methylation. (C) Southern analysis of the methylation status of the na32 locus. DNA from heterozygous plants were digested with DraI (lane 1), DraI-AvaI (lane 2), AsnI (lane 3), or AsnI-AvaI (lane 4) and probed with the P1 fragment. The sizes of the hybridized fragments (in base pairs) are shown. The size difference between the two fragments revealed after the AsnI digestion (lane 3) can be explained by the presence or absence of the S1 element and confirms the heterozygous state of the plant studied. The R1 site is methylated for the S1 allele but unmethylated for the empty allele. The R2 sites appear to be methylated for the S1 allele.
RESULTS
S1 integrates in hypomethylated target sites.
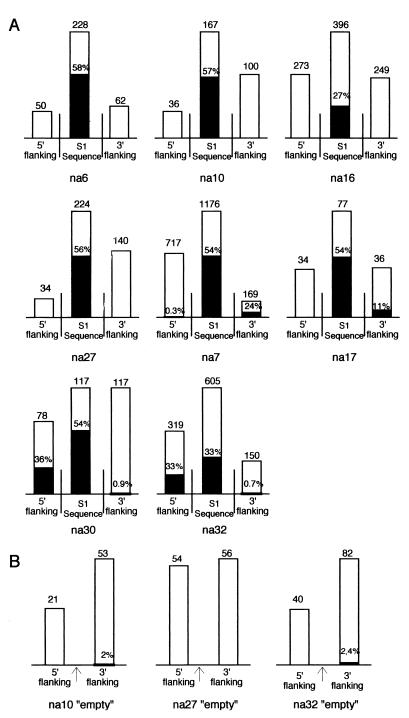
The methylation status of eight S1-containing loci (S1 plus ∼200 bp of upstream and downstream flanking sequences) was analyzed by the genomic sequencing method (see reference 6 and Materials and Methods). These loci were chosen randomly from our collection of lambda clones. All sequences flanking S1 at these loci were searched against databases, and no significant homology was detected. The results of this analysis are summarized in Fig. 1, and two examples (na16 and na32) are presented in detail in Fig. 2. In four cases (na6, na10, na16, and na27), methylated cytosine was not found in DNA sequences flanking the S1 element, suggesting that these SINEs integrated in hypomethylated target sites. Since several S1 insertions are not fixed in Brassica populations (48), we searched for heterozygous plants containing an “empty” allele (i.e., the site before S1 integration) as well as the S1-containing allele. To confirm the hypomethylated nature of S1 target sites, we analyzed the methylation status of empty alleles. Genome duplication is a common phenomenon in Brassica and was viewed as a potential problem in this approach. Genome duplication leads to the presence of several homeologous sites in the same genome. Despite the use of stringent PCR conditions that target orthologous (allelic) sites, we were concerned about the possibility of amplifying empty homeologous sites as well. We have tested this possibility by amplifying the eight genomic S1 loci by using DNA extracted from single plants (not shown; see reference 48 for examples). Empty sites were not detected for all plants (as expected if we were also targeting empty homeologous sites), and a typical allelic pattern was observed in all cases. Also, the sequencing of several PCR products for each sites did not reveal sequence divergence, as expected if duplicated sequences were also amplified. We therefore conclude that our PCR amplifications only target allelic (orthologous) sites and that the duplicate nature of the Brassica genome does not interfere with our approach.
FIG. 1.
Summary of the methylation status obtained by genomic sequencing of eight S1-containing loci (A) and three corresponding empty loci obtained from an heterozygous plant (B). For each locus (name of the locus indicated below), the height of bars is proportional to the number of cytosines analyzed (indicated above). The proportion of methylated cytosines found in S1 sequences or in upstream or downstream flanking sequences is symbolized by dark portions, and the corresponding percentages (when not 0) are indicated. The arrows for the empty loci indicate the position of insertion of the S1 element in the corresponding S1-containing site.
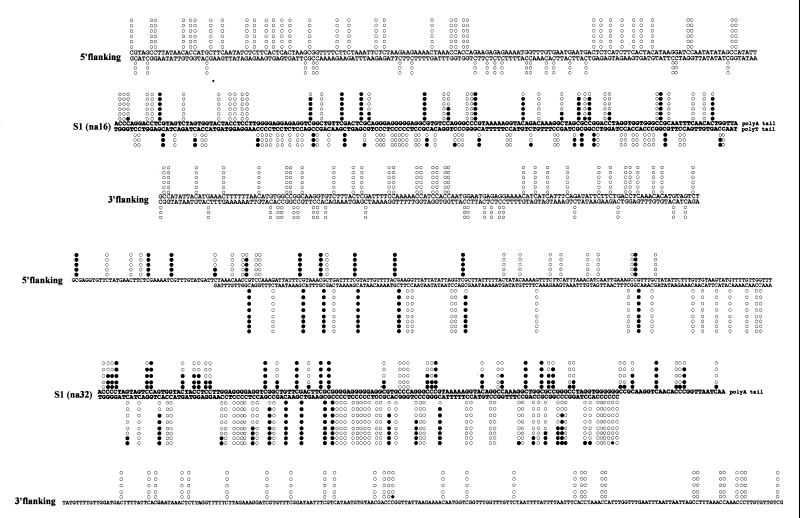
FIG. 2.
Two detailed examples of genomic sequencing results. (A) Plus and minus strands of the na16 locus. (B) Plus and minus strands of the na32 locus (only the plus strand for the downstream region). Cytosine methylation is indicated by solid circles, while nonmethylated sites are represented by open circles. The S1 sequence is printed in boldface.
The methylation status of the na10 and na27 empty allele was determined, and it confirmed that these S1 target sites were hypomethylated before S1 integration and remained hypomethylated following the integration event (Fig. 1). In four other S1 loci, we found a significant amount of methylated cytosine only upstream (na30 and na32) or downstream (na7 and na17) of the S1 element. These results suggest that, either these S1 elements integrated at a precise transition between methylated and hypomethylated DNA regions, or they integrated in a hypomethylated DNA region which was subsequently methylated as a consequence of the S1 integration event. As for na10 and na27, we cloned the na32 empty allele and determine its methylation status. The na32 empty allele was found to be hypomethylated (Fig. 1B), suggesting that the upstream methylation we have observed for the S1-containing alleles (33% in Fig. 1A and 2) is a direct consequence of the S1 integration event. The downstream regions of na32 empty and S1-containing alleles are both hypomethylated (2.4 and 0.7%, respectively). In this case, a single CpG site is concerned, and this preexisting methylation is not affected by the integration event. Therefore, the very low levels of methylation observed in the 5′ flanking region of na7 (0.3%) and the 3′ flanking region of na30 (0.9%) probably also preexist in the empty site and do not result from the S1 integration event. We conclude that, for the eight sites analyzed, S1 integration events took place at hypomethylated target sites and that flanking sequences could be methylated after S1 integration.
S1 elements are preferential targets for de novo methylation.
The eight S1 elements studied were found to be highly methylated (Fig. 1 and 2). The precise sequence context of S1 methylation is presented in Table 1 and is similar to the situation presented in a previous report (11). The levels of 5-methylcytosine in the eight different S1 elements range from 26% to 58%. Cytosines from the S1 element in CpG and CpNpG are preferred targets for methylation and are methylated at levels of 92 and 55%, respectively. Asymmetrical positions were also found methylated in S1 sequences with confirmation of Cp(A/T)pA as a preferred asymmetrical target site (Table 1) (11). These results suggest that S1 elements can be specific targets for methylation during or after integration in hypomethylated genomic target sites.
TABLE 1.
Proportion of 5-methylcytosine in different sequence contexts for S1
| Sequence context | % of methylated cytosinesa
|
||
|---|---|---|---|
| S1-5′FL | S1 | S1-3′FL | |
| CpG | 97 (93/96) | 92 (611/667) | 72 (23/32) |
| CpNpG | 14 (4/19) | 55 (366/671) | 0 (0/6) |
| Cp(A/T)pA | 18 (25/137) | 41 (157/382) | 11 (10/93) |
| Other | 3 (4/119) | 20 (252/1,261) | 16 (12/74) |
The number of methylated cytosines relative to the total number of cytosines analyzed for each context is presented in parentheses. S1-5′FL and S1-3′FL represent the 5′ and 3′ S1 genomic flanking regions. Only loci with DNA methylation in flanking regions were included (na30 and na32 for S1-5′FL and na7 and na17 for S1-3′FL).
Spreading of DNA methylation from S1 elements.
As discussed above, four of the eight S1 loci analyzed have a significant level of methylation in 5′ or 3′ S1 flanking sequences (Fig. 1 and 2). Since all “empty” integration sites studied were shown to be hypomethylated, this methylation of flanking sequences is probably a direct consequence of the SINE integration. We suggest that following the integration and methylation of the S1 element, methylation has spread in a directional manner from the S1 element to upstream or downstream flanking genomic regions. The methylation observed in flanking regions mainly concerns CpG sites with a reduction of 5-methylcytosine in CpNpG and asymmetrical sites compared to S1 methylation (Table 1).
The extent of the methylation spreading was tested for locus na32. DNA samples from individual heterozygous plants (i.e., presenting the empty and the S1-containing alleles at the na32 locus) were used. Using two PCR products (positions +80 to −410 and −250 to −750 with an overlapping region of 160 bp), the methylation status of the upstream regions of both alleles (on the upper strand) up to position −750 was analyzed by genomic sequencing (Fig. 3A). For the S1 allele, we observed up to position −750 a high level of methylation with the same proportion of 5-methylcytosine in different contexts, as presented in Table 1 (S1-5′FL; i.e., mainly concentrated on CpG). For the empty allele, the region is completely unmethylated up to position −417. From this position, one of the nine empty clones analyzed showed a methylation pattern similar to that of the S1 allele, while the eight other clones still maintained an unmethylated pattern up to position −720. At this position, another clone adopted an S1 (methylated) profile, while the seven others still maintained their unmethylated status up to position −750. Therefore, the transition from a methylated to an unmethylated DNA region on the empty allele is not a linear process but appears to be a stepwise process, possibly implicating frontier elements. These results suggest that, following the S1 integration event, methylation spread from the SINE to the 5′ flanking region of the na32 locus over at least 750 bp until it reached an already methylated DNA region.
To confirm these results and to validate the difference in methylation observed in the genomic sequencing experiment, DNA from individual heterozygous plants was digested with a restriction enzyme sensible to DNA methylation (AvaI) followed by DNA hybridization (Fig. 3B and C). We choose to analyze the methylation status of a restriction site (R1-AvaI, Fig. 3) located at position −385 on both alleles that presents a clear difference in methylation pattern in the genomic sequencing analysis (Fig. 3A). Using the P1 probe and a DraI-AvaI digest, we found two bands of similar intensities (Fig. 3C, lane 2). The first band of 525 bp is expected if the R1-AvaI site is methylated and not cleaved by the AvaI restriction enzyme. The band at 330 bp is expected if the R1-AvaI site is unmethylated and cleaved by the restriction enzyme. These results suggest that the two restriction fragments observed were generated from the two alleles (S1 and empty) that differ in their R1-AvaI site methylation status (Fig. 3A). We confirmed these results by using probe P1 and an AsnI-AvaI digest. The band near 2,000 bp has a size predicted for the S1 allele if both (R1 and R2) AvaI sites are methylated and not cleaved by the restriction enzyme, while the band at 550 bp is expected from the empty allele if the first AvaI site (R1) is unmethylated and cleaved (Fig. 3C, lane 4). These results confirm the difference in methylation status observed in the genomic sequencing between the na32 empty and S1-containing alleles at the R1-AvaI site.
DISCUSSION
S1 integrates in hypomethylated target sites.
We have shown in this study that S1 elements insert in hypomethylated regions of the B. napus genome. B. napus, like most higher plants, has a relatively high level of 5-methylcytosine. We estimated previously that almost 15% of cytosine is transformed to 5-methylcytosine in the nuclear DNA of this species (11) (compared to 2 to 7% for animal DNA [36]). It is therefore unlikely that the systematic association of S1 elements with hypomethylated portions of the B. napus genome happened by chance. We have shown recently that S1 integration (like mammalian SINE integration) occurs at nonrandom staggered breaks, probably resulting from the action of a LINE-encoded endonuclease (47). The general A/T richness of a given DNA region and the presence of short runs of pyrimidines followed by short runs of purines were shown to be a favorable context for S1 integration (47). We show here that the methylation status of the integration site is also a factor, and we suggest that the endonuclease implicated in S1 integration could be inhibited by DNA methylation (or by a chromatin structure induced by DNA methylation), thereby selecting hypomethylated DNA regions. It remains to be shown whether the methylation status also influences the target site selection of mammalian retroposons, but the similarity of target sites between plant and mammalian retroposons (47) and the observation that Alu integration sites are strongly depleted in methylatable CpG sites (15) suggest that this might be the case.
Possible mechanisms for the de novo methylation induced by S1 integration.
Our results indicate that S1 does not simply adopt the methylation level of its integration site, but is directly targeted by methylases. Recently, Pélissier et al. suggested that heavy de novo methylation at symmetrical and asymmetrical sites, like that we observed on the S1 element, is a hallmark of RNA-directed DNA methylation (34). The transposition cycle of SINEs includes the formation of an RNA-DNA intermediate following first-strand DNA synthesis (16). This RNA-DNA transposition intermediate may be recognized by methylases leading to specific de novo methylation of the element. The production of aberrant (sense and antisense [9]) S1 RNA resulting from cotranscriptional events where an S1 element is transcribed due to its presence in a Pol II transcriptional unit, could also activate a RNA-directed DNA methylation system (30). Finally, methylation of S1 elements could also happen by DNA-DNA interactions, but since S1 elements are very short repeats (less than 200 bp) they may be unable to generate a direct ectopic homologous DNA-DNA interaction (12, 37).
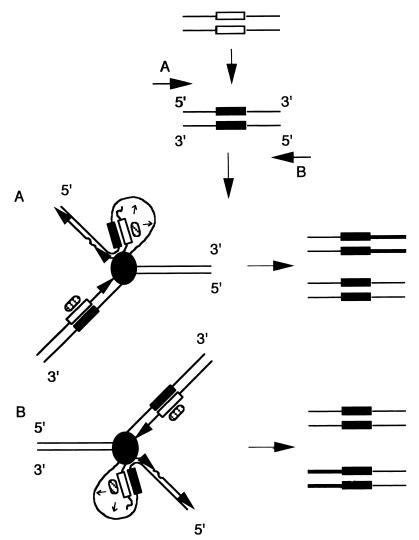
Following SINE integration, we report a de novo methylation of either its upstream or downstream flanking genomic region. We suggest that this methylation results from a vectorial spreading from the SINE into the flanking region. The restriction in the orientation of the methylation spreading is probably related to the mechanism used and deserves attention. According to in vitro studies, the recognition of a methylated cytosine in single- or double-strand oligonucleotides is sufficient to initiate methylation spreading (4, 24, 46, 49). However, methylation spreading is greatly enhanced on hemimethylated duplexes, when the methylatable cytosines are present on single-stranded DNA and when a high concentration of methylatable cytosines is available (4, 24, 46, 49). In vivo, DNA replication of a short methylated region will lead to the formation of hemimethylated DNA adjacent to single-stranded DNA only on the lagging strand and not on the leading strand (Fig. 4) (24). DNA replication could thus stimulate an oriented methylation spreading (from the hemimethylated region to the single-stranded region of the lagging strand) like the one observed in this work (Fig. 4). Methylation spreading would depend on the presence on the lagging strand of a potentially good methylatable region and the position of the replication origin in relation to the methylated locus (the SINE S1 in our case) would determine which strand (plus or minus) is the lagging strand (Fig. 4) and thus which of the flanking regions (upstream or downstream) would be de novo methylated. Although the spreading would at first only concern a small DNA region on the lagging strand, methylation can further spread after each round of replication and can expand for at least several hundred bases as observed. The oriented methylation spreading we observed in this work could thus reflect the general property of DNA replication to promote de novo methylation spreading from short methylated DNA regions.
FIG. 4.
Possible involvement of DNA replication in de novo methylation spreading. Following integration, S1 elements would be targeted for de novo methylation by an unknown mechanism (see text). After replicating the methylated SINE (black boxes), two hemimethylated regions will be formed (black and white boxes), one on the leading strand and one on the lagging strand. DNA methyltransferase (hatched oval) associated with the replication apparatus (grey circle) can recognize these hemimethylated regions and can methylate the corresponding neosynthesized strands (23) (the organization of the replication fork is presented as by Kornberg and Baker [20]). In the 3′ part of the SINE on the lagging strand, the methyltransferase is adjacent to single-stranded DNA. Methylation spreading from the SINE to flanking sequences would depend on the presence on the lagging strand of a potentially good methylatable region, and the polarity of the methylation spreading would depend on the position of replication origin and the direction of the replication fork (A or B). The initial methylation spreading (thicker line) can only concern the immediate S1 flanking region, but since de novo methylation is possible at each round of DNA replication, methylation can potentially spread by this mechanism to sequences thousand of base pairs from the SINEs. Also, we should expect that the methylated version of the locus will rapidly take over the unmethylated one following successive rounds of DNA replication.
Alternatively, we cannot exclude that methylation has spread, not from the SINE, but from a methylated region located several hundred base pairs upstream or downstream of the integration site. In that case, importation of new methylation sites by the insertion (and methylation) of the SINE would provide a magnet for de novo methylation, the SINE acting as a barrier to this spreading. However, it seems unlikely that a small methylated island like a SINE could trigger methylation at distance (∼400 bp for na32). Also, in one case in which mammalian SINEs were suspected to generate de novo methylation (32), CpG methylation was less important when located at distance from the methylation center, suggesting that methylation was originating from this center and was not spreading from external methylated sequences.
It is also interesting to note that the methylation profile of flanking sequences is different from the methylation profile of S1 sequences (Table 1). A strong reduction in the 5-methylcytosine content of CpNpG and asymmetrical sites was observed while CpG sites were equally methylated. This observation suggests that methylation of flanking regions implies a change of methylase specificity or the action of a different methylase compare to S1 elements. Interestingly, similar results were observed in tobacco plants with chromosomal inserts that were subjected to RNA-directed DNA methylation (34, 52). In these plants, viroid RNA-DNA interactions trigger specific and dense de novo methylation of viroid transgene sequences. Methylation occurred potentially at all C positions in symmetrical and nonsymmetrical sites and was essentially restricted to the viroid sequences (34). However, for one of the loci analyzed, methylation was found to spread up to 500 bp downstream from the viroid-specific sequences. (T. Pélissier and M. Wassenegger, unpublished results). In contrast to the heavy methylation pattern detected within the viroid sequences, the distribution of methylated cytosines within this area was significantly biased in favor of symmetrical sites, a pattern reminiscent of that we observed in our study.
Possible consequences of the modification of methylation patterns upon SINE integration.
In a few well-characterized cases, the insertion of a SINE in or near a gene has been shown to result in a genetic defect. In these cases, the deleterious effects result from the disruption of an open reading frame, the modification of splicing patterns causing exon skipping, or the creation of genomic instability leading to deletions and gene rearrangements (21). We show here that SINE insertion can also lead to epigenetic modifications. DNA methylation can affect chromatin organization and has been implicated in a number of important epigenetic modifications, including transcriptional regulation and gene silencing, genomic imprinting, and X-chromosome inactivation (35). The methylation spreading that we observed after S1 insertion could therefore affect gene expression. This seems likely for two reasons. First, promoters of active genes are generally undermethylated (33), and since S1 elements are targeted to hypomethylated regions, we expect the S1 element to be enriched in gene-containing regions. Second, we have shown that the de novo methylation spreading can directly affect genomic DNA at least several hundred base pairs from the insertion sites. Since inactive chromatin structure can spread from methylated region several kilobase pairs into unmethylated flanking regions (17), we estimate that a gene remote from the integration site could still be affected by an S1 integration event. Based on these observations, the local spreading of methylation observed following S1 integration could influence genes at remote genomic sites.
ACKNOWLEDGMENTS
We thank Christophe Tatout, Charles White, Zoya Auramova, and Damian Labuda for critical review of the manuscript. We also thank Benoit Landry for providing us with B. napus seeds and Alain Lenoir for technical help.
This work was supported by the CNRS (UMR 6547), by the Université Blaise Pascal, and by a European Community grant (FPIV, Molecular Tools for Biodiversity) from the BIOTECH program.
REFERENCES
- 1.Bennetzen J L. The mutator transposable element system of maize. Curr Top Microbiol Immunol. 1996;24:195–229. doi: 10.1007/978-3-642-79795-8_9. [DOI] [PubMed] [Google Scholar]
- 2.Bird A. The essential of DNA methylation. Cell. 1992;70:5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- 3.Brutnell T P, Dellaporta S L. Somatic inactivation and reactivation of AC associated with changes in cytosine methylation and transposase expression. Genetics. 1994;138:213–225. doi: 10.1093/genetics/138.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carotti D, Funiciello S, Palitti F, Strom R. Influence of pre-existing methylation on the de novo activity of eukaryotic DNA methyltransferase. Biochemistry. 1998;37:1101–1108. doi: 10.1021/bi971031i. [DOI] [PubMed] [Google Scholar]
- 5.Chesnokov I, Schmid C W. Flanking sequences of an Alu source stimulate transcription in vitro by interacting with sequence-specific transcription factors. J Mol Evol. 1996;42:30–36. doi: 10.1007/BF00163208. [DOI] [PubMed] [Google Scholar]
- 6.Clark S J, Harrison J, Paul C L, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deininger P L, Batzer M A, Hutchison III C A, Edgell M H. Master genes in mammalian repetitive DNA amplification. Trends Genet. 1992;8:307–311. doi: 10.1016/0168-9525(92)90262-3. [DOI] [PubMed] [Google Scholar]
- 8.Deragon J M, Landry B S, Pélissier T, Tutois S, Tourmente S, Picard G. An analysis of retroposition in plants based on a family of SINEs from Brassica napus. J Mol Evol. 1994;39:378–386. doi: 10.1007/BF00160270. [DOI] [PubMed] [Google Scholar]
- 9.Deragon J M, Gilbert N, Rouquet L, Lenoir A, Arnaud P, Picard G. A transcriptional analysis of the S1Bn (Brassica napus) family of SINE retroposons. Plant Mol Biol. 1996;32:869–878. doi: 10.1007/BF00020484. [DOI] [PubMed] [Google Scholar]
- 10.Englander E W, Wolffe A P, Howard B H. Nucleosome interactions with a human Alu element. Transcription repression and effects on template methylation. J Biol Chem. 1993;268:19565–19573. [PubMed] [Google Scholar]
- 11.Goubely C, Arnaud P, Tatout C, Heslop-Harrison J S, Deragon J M. S1 SINE retroposons are methylated at symmetrical and non-symmetrical position in Brassica napus: identification of a new methylation site in plants. Plant Mol Biol. 1999;39:243–255. doi: 10.1023/a:1006108325504. [DOI] [PubMed] [Google Scholar]
- 12.Goyon C, Rossignol J L, Faugeron G. Native DNA repeats and methylation in Ascobolus. Nucleic Acids Res. 1996;24:3348–3356. doi: 10.1093/nar/24.17.3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graff J R, Herman J G, Myohanen S, Baylin S B, Vertino P M. Mapping patterns of CpG island methylation in normal and neoplastic cells implicates both upstream and downstream regions in de novo methylation. J Biol Chem. 1997;272:22322–22329. doi: 10.1074/jbc.272.35.22322. [DOI] [PubMed] [Google Scholar]
- 14.Hass A, Schulz W. Enhancement of reporter gene de novo methylation by DNA fragments from the α-fetoprotein control region. J Biol Chem. 1994;269:1821–1826. [PubMed] [Google Scholar]
- 15.Jurka J, Klonowski P, Trifonov E N. Mammalian retroposons integrate at kinkable DNA sites. J Biomol Struct Dyn. 1998;15:717–721. doi: 10.1080/07391102.1998.10508987. [DOI] [PubMed] [Google Scholar]
- 16.Jurka J. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc Natl Acad Sci USA. 1997;94:1872–1877. doi: 10.1073/pnas.94.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kass S U, Goddard J P, Adams R L P. Inactive chromatin spreads from a focus of methylation. Mol Cell Biol. 1993;13:7372–7379. doi: 10.1128/mcb.13.12.7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kazazian H H. Mobile elements and disease. Curr Opin Genet Dev. 1998;8:343–350. doi: 10.1016/s0959-437x(98)80092-0. [DOI] [PubMed] [Google Scholar]
- 19.Kochanek S, Renz D, Doerfler W. DNA methylation in the Alu sequences of diploid and haploid primary human cells. EMBO J. 1993;12:1141–1151. doi: 10.1002/j.1460-2075.1993.tb05755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kornberg A, Baker T A. DNA replication. 2nd ed. New York, N.Y: W. H. Freeman & Co.; 1991. [Google Scholar]
- 21.Labuda D, Zietkiewick E, Mitchell G A. Alu elements as a source of genomic variation: deleterious effects and evolutionary novelties. In: Maraia R J, editor. The impact of short interspersed elements (SINEs) on the host genome. Austin, Tex: R. G. Landes Co., Springer; 1995. pp. 1–24. [Google Scholar]
- 22.Lenoir A, Cournoyer B, Warwick S I, Picard G, Deragon J M. Evolution of SINE S1 retroposons in Cruciferae plant species. Mol Biol Evol. 1997;14:934–941. doi: 10.1093/oxfordjournals.molbev.a025836. [DOI] [PubMed] [Google Scholar]
- 23.Leonhardt H, Page A W, Weier H U, Bestor T A. Targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–873. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- 24.Lindsay H, Adams R L P. Spreading of methylation along DNA. Biochem J. 1996;320:473–478. doi: 10.1042/bj3200473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu W-M, Chu W-M, Choudary P V, Schmid C W. Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Res. 1995;23:1758–1765. doi: 10.1093/nar/23.10.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu W-M, Maraia R J, Rubin C M, Schmid C W. Alu transcripts: cytoplasmic localisation and regulation by DNA methylation. Nucleic Acids Res. 1994;22:1087–1095. doi: 10.1093/nar/22.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu W-M, Schmid C W. Proposed roles for DNA methylation in Alu transcriptional repression and mutation inactivation. Nucleic Acids Res. 1993;21:1351–1359. doi: 10.1093/nar/21.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Magewu A N, Jones P A. Ubiquitous and tenacious methylation of the CpG site in codon 248 of the p53 gene may explain its frequent appearance as a mutational hot spot in human cancer. Mol Cell Biol. 1994;14:4225–4232. doi: 10.1128/mcb.14.6.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matzke M A, Matzke A J M, Eggleston W B. Paramutation and transgene silencing: a common response to invasive DNA. Trends Plant Sci. 1996;1:382–388. [Google Scholar]
- 30.Mette M N, van der Winden J, Matzke M A, Matzke A J M. Production of aberrant promoter transcripts contributes to methylation and silencing of unliked homologous promoters in trans. EMBO J. 1999;18:241–248. doi: 10.1093/emboj/18.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mummaneni P, Bishop P L, Turker M S. A cis-acting element accounts for a conserved methylation pattern upstream of the mouse adenine phosphoribosyltransferase gene. J Biol Chem. 1993;268:552–558. [PubMed] [Google Scholar]
- 32.Mummaneni P, Walker K A, Bishop P L, Turker M S. Epigenetic gene inactivation induced by a cis-acting methylation center. J Biol Chem. 1995;270:788–792. doi: 10.1074/jbc.270.2.788. [DOI] [PubMed] [Google Scholar]
- 33.Naveh-Many T, Cedar H. Active gene sequences are undermethylated. Proc Natl Acad Sci USA. 1981;78:4246–4250. doi: 10.1073/pnas.78.7.4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pélissier T, Thalmeir S, Kempe D, Sanger H L, Wassenegger M. Heavy de novo methylation at symmetrical and non-symmetrical sites is a hallmark of RNA-directed DNA methylation. Nucleic Acids Res. 1999;27:1625–1634. doi: 10.1093/nar/27.7.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Razin A. CpG methylation, chromatin structure and gene silencing—a three-way connection. EMBO J. 1998;17:4905–4908. doi: 10.1093/emboj/17.17.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Razin A, Riggs A. DNA methylation and gene function. Science. 1980;210:604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- 37.Rossignol J-L, Faugeron G. Gene inactivation triggered by recognition between DNA repeats. Experientia. 1994;50:307–317. doi: 10.1007/BF01924014. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.SanMiguel P, Tikonov A, Jin Y-K, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springler P S, Edwards K J, Lee M, Avramova Z, Bennetzen J L. Nested retrotransposons in the intergenic regions of the maize genome. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 40.Schlappi M, Raina R, Fedoroff N. Epigenetic regulation of the maize Spm transposable element: novel activation of a methylated promoter by TnpA. Cell. 1994;77:427–437. doi: 10.1016/0092-8674(94)90157-0. [DOI] [PubMed] [Google Scholar]
- 41.Selker E U. Premeiotic instability of repeated sequences in Neurospora crassa. Annu Rev Genet. 1990;24:579–613. doi: 10.1146/annurev.ge.24.120190.003051. [DOI] [PubMed] [Google Scholar]
- 42.Shen M R, Batzer M A, Deininger P L. Evolution of the master Alu gene(s) J Mol Evol. 1991;33:311–320. doi: 10.1007/BF02102862. [DOI] [PubMed] [Google Scholar]
- 43.Simmen M W, Leitgeb S, Charlton J, Jones S J M, Harris B R, Clark V H, Bird A. Nonmethylated transposable elements and methylated genes in a chordate genome. Science. 1999;283:1164–1167. doi: 10.1126/science.283.5405.1164. [DOI] [PubMed] [Google Scholar]
- 44.Skowronski J, Singer M F. Expression of a cytoplasmic LINE-1 transcript is regulated in a human teratocarcinoma cell line. Proc Natl Acad Sci USA. 1985;82:6050–6054. doi: 10.1073/pnas.82.18.6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smit A F A. The origin of interspersed repeats in the human genome. Curr Opin Genet Dev. 1996;6:743–748. doi: 10.1016/s0959-437x(96)80030-x. [DOI] [PubMed] [Google Scholar]
- 46.Smith S S. Stalling of DNA methyltransferase in chromosome stability and chromosome remodelling (review) Int J Mol Med. 1998;1:147–156. [PubMed] [Google Scholar]
- 47.Tatout C, Lavie L, Deragon J M. Similar target site selection occurs in integration of plant and mammalian retroposons. J Mol Evol. 1998;47:463–470. doi: 10.1007/pl00006403. [DOI] [PubMed] [Google Scholar]
- 48.Tatout C, Warwick S, Lenoir A, Deragon J M. SINE insertion as clade markers for wild crucifer species. Mol Biol Evol. 1999;16:1614–1621. [Google Scholar]
- 49.Tollefsbol T O, Hutchison C A., III Control of methylation spreading in synthetic DNA sequences by the murine DNA methyltransferase. J Mol Biol. 1997;269:494–504. doi: 10.1006/jmbi.1997.1064. [DOI] [PubMed] [Google Scholar]
- 50.Turker M S, Bestor T H. Formation of methylation patterns in the mammalian genome. Mutat Res. 1997;386:119–130. doi: 10.1016/s1383-5742(96)00048-8. [DOI] [PubMed] [Google Scholar]
- 51.Voytas D F. Retroelements in genome organization. Science. 1996;274:737–738. doi: 10.1126/science.274.5288.737. [DOI] [PubMed] [Google Scholar]
- 52.Wassenegger M, Heimes S, Riedel L, Sanger H L. RNA-directed de novo methylation of genomic sequences in plants. Cell. 1994;76:567–576. doi: 10.1016/0092-8674(94)90119-8. [DOI] [PubMed] [Google Scholar]
- 53.Yates P A, Burman R W, Mummaneni P, Krussell S, Turker M S. Tandem B1 element located in a mouse methylation center provides a target for de novo DNA methylation. J Biol Chem. 1999;274:36357–36361. doi: 10.1074/jbc.274.51.36357. [DOI] [PubMed] [Google Scholar]
- 54.Yoder J A, Walsh C P, Bestor T H. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]