Abstract
Protein kinase inhibitor (PKI) is a potent endogenous inhibitor of the cyclic AMP (cAMP)-dependent protein kinase (PKA). It functions by binding the free catalytic (C) subunit with a high affinity and is also known to export nuclear C subunit to the cytoplasm. The significance of these actions with respect to PKI's physiological role is not well understood. To address this, we have generated by homologous recombination mutant mice that are deficient in PKIα, one of the three isoforms of PKI. The mice completely lack PKI activity in skeletal muscle and, surprisingly, show decreased basal and isoproterenol-induced gene expression in muscle. Further examination revealed reduced levels of the phosphorylated (active) form of the transcription factor CREB (cAMP response element binding protein) in the knockouts. This phenomenon stems, at least in part, from lower basal PKA activity levels in the mutants, arising from a compensatory increase in the level of the RIα subunit of PKA. The deficit in gene induction, however, is not easily explained by current models of PKI function and suggests that PKI may play an as yet undescribed role in PKA signaling.
A variety of hormones, neurotransmitters, and other molecules exert their actions on target cells by means of the cyclic AMP (cAMP)-mediated signaling cascade. The generation of intracellular cAMP by stimulating G protein-coupled receptors linked to adenylyl cyclase leads to the activation of the cAMP-dependent protein kinase (PKA). This signaling cascade, one of the most versatile and multifunctional systems studied to date, is responsible for the modulation of numerous processes, such as secretion, enzyme activation, and transcription. It is also an extraordinarily well-conserved mechanism of signal transduction, since it is seen in a wide variety of organisms.
PKA is a holoenzyme, consisting of two regulatory (R) subunits and two catalytic (C) subunits. Molecules of cAMP generated within the cell bind to the R subunits, decreasing their affinity for the C subunits. This releases the C subunits to diffuse throughout the cell and phosphorylate target molecules. Apart from the R subunits, another endogenous modulator of the C subunit is also present in most tissues: protein kinase inhibitor (PKI). The PKIs are heat-stable proteins, 70 to 75 amino acids in length, that are high-affinity, specific inhibitors of PKA (24). The N-terminal region of PKI contains the sequence RRNAI, which acts as a pseudosubstrate site for PKA and is required for PKI's inhibitory activity. In addition, other amino acid residues in the N-terminal region of the protein also contribute to the interaction between PKI and the C subunit (2, 18, 19). The synthetic peptide encompassing amino acids 5 to 24 retains PKI's inhibitory activity and has been used extensively as a biochemical tool to probe the PKI signaling pathway (23). Immunocytochemical localization studies have demonstrated that the C subunit and PKI have access to both the cytoplasm and the nucleus (11, 32). By injecting synthetic C subunit and PKI, Wen et al. (32) have shown that PKI acts as a chaperone for nuclear export of the C subunit by means of a distinct leucine-rich motif within PKI. By enhancing the rate of export of the C subunit from the nucleus, PKI is thought to affect the kinetics and/or extent of PKA activity in the nucleus.
In all, there are three known isoforms of PKI, α, β, and γ, encoded by distinct genes (3, 10, 31). Each of these isoforms has a unique tissue expression pattern while sharing a nanomolar affinity for the C subunit (10, 29). PKIα mRNA is most abundant in skeletal muscle, with modest expression in the heart and brain. In contrast, PKIβ is expressed very highly in the testis, with little to no expression elsewhere. PKIγ mRNA is found at low levels in most tissues, with somewhat higher levels in the testis and heart. In some tissues that express multiple isoforms, for example, the brain or the testis, the pattern of expression is quite cell specific (25, 28).
Transfection studies in cell culture have led to the speculation that PKI, by virtue of its localization and affinity for the C subunit, serves to reset the basal activity of PKA once it is activated, in preparation for the next round of activation (33). Clearly, transfection of excess PKI relative to the C subunit reduces the transcriptional activity of PKA-regulated genes (14). Despite these extensive in vitro studies, there are no clear indications as to the physiological role of PKI. The presence of three isoforms of PKI suggests that each may serve an important role in the modulation of the cAMP-PKA signaling cascade. Their distinct patterns of tissue expression may indicate specific roles in different tissues. We addressed this question by generating targeted deletions of the PKI genes in mice. This paper describes the generation and phenotype of mouse mutants deficient in PKIα, the PKA inhibitor abundant in skeletal muscle.
MATERIALS AND METHODS
Construction of the PKIα targeting vector and generation of mutant mice.
A PKIα genomic clone was isolated from a 129SV/J mouse genomic library (21). A 7.5-kb genomic fragment containing both exons of the PKIα gene was used to construct a targeting vector, PKIα-Rec 1. An approximately 2.5-kb EcoRI-HindIII fragment of the gene encompassing exon 1 was replaced with a neomycin phosphotransferase cassette to facilitate positive selection. This strategy deleted the N-terminal two-thirds of PKIα, including the inhibitory and nuclear export domains.
Gene targeting in embryonic stem (ES) cells was performed essentially as described previously (7). PKIα-Rec 1 DNA was linearized with BamHI and then electroporated into REK3 ES cells derived from 129SV/J mice (5). Recombinant cells were selected with 200 μg of active G418 (Gibco)/ml. Resistant colonies were picked, isolated, and screened by genomic Southern blot analysis. Five clones were identified as having undergone specific homologous recombination and were microinjected into 3.5-day C57BL/6 blastocysts. These blastocysts were subsequently transferred to pseudopregnant foster mothers to yield six chimeric male mice. The chimeras were bred to C57BL/6 females, and several PKI heterozygous offspring were obtained. All experiments comparing wild-type and mutant mice used age- and sex-matched animals on the C57BL/6 × 129SV/J hybrid background.
PKA kinase and PKI inhibitor assays.
For kinase assays, hind leg skeletal muscle samples were homogenized in buffer (20 mM Tris, 0.1 mM EDTA, 0.5 mM EGTA, 5 mM magnesium acetate, 250 mM sucrose, 1% Triton X-100, 10 mM dithiothreitol, 0.1 mM ATP, pH 7.5) with protease inhibitors, followed by sonication and centrifugation for 10 min at 12,000 × g at 4°C. Protease inhibitors included 1 μg of pepstatin/ml, 2 μg of aprotinin/ml, 2 μg of leupeptin/ml, 125 μg of 4-(2-aminoethyl)-benzenesulfonyl fluoride/ml, and 78.5 μg of benzamidine/ml. Supernatant proteins (0.4 mg/ml) were assayed as described previously using kemptide as a substrate in the presence (total kinase) or absence (basal kinase) of 5 μM cAMP (9). Six to eight mice of each genotype were assayed separately in triplicate to obtain individual values for each mouse, which were then averaged for each genotype.
For inhibitor assays, hind leg skeletal muscle homogenates were made and assayed essentially as described previously (10). The homogenates were heated for 10 min at 95°C to inactivate endogenous kinases and then centrifuged as described above. Increasing amounts of supernatant proteins were added to a kinase assay mixture containing 1 nM purified bovine heart C subunit (Sigma). Each concentration was assayed in triplicate for two mice of each genotype, and the results are reported as percent of control C subunit activity in the absence of any added tissue extract. The experiment was repeated with similar results on a separate group of mice.
Northern blots.
For the fasting experiments, food was withdrawn from the mice in the evening, 3 h prior to lights out. They had access to water ad libitum. The mice fasted overnight for a period of 16 h before tissues were collected. For refeeding, the fasting mice were given access to mouse chow ad libitum for 6 h before tissues were collected. Isoproterenol treatment (0.5 mg/kg of body weight in 10 mM ascorbic acid-saline) was administered intraperitoneally, and tissues were harvested after 6 h. Total RNA was isolated from hind leg skeletal muscle, and Northern blots were run with 10 μg of RNA per lane as described previously (6) and subjected to phosphorimager analysis.
Skeletal-muscle cultures.
Mice were euthanized by CO2 administration, and intact gastrocnemius muscles were isolated from both legs. The muscles were rinsed in phosphate-buffered saline and placed individually in separate wells of a 6-well tissue culture plate along with 5 ml of Ham's F-10 medium. The plate was then incubated at 37°C in a tissue culture incubator for 30 min. One muscle of each pair was treated with 50 μM forskolin (diluted in dimethyl sulfoxide; Sigma), and the contralateral muscle was treated with the vehicle alone. The muscles were then returned to the incubator for a defined period of time, after which they were rinsed in phosphate-buffered saline and homogenized in 1.5 ml of boiling sodium dodecyl sulfate lysis buffer (100 mM Tris [pH 6.8], 2% sodium dodecyl sulfate, 10% glycerol). The homogenates were boiled for 10 min before being aliquoted and frozen at −80°C. Protein concentrations were determined by the bicinchoninic acid protein assay (Pierce), and the protein samples were supplemented to 10% β-mercaptoethanol and 0.1% bromophenol blue before being subjected to polyacrylamide gel electrophoresis analysis and Western blotting for total and phosphorylated (phospho)-CREB (cAMP response element binding protein).
Western blots.
For PKA subunit analysis, hind leg skeletal muscle was isolated from wild-type and knockout mice, immediately frozen on dry ice, and stored at −80°C. Samples were thawed directly in kinase homogenization buffer (10 ml/g of tissue), homogenized, sonicated, and centrifuged for 10 min at 12,000 × g at 4°C. The supernatants were aliquoted and frozen at −80°C for future use. Samples were then assayed for protein concentration by the Bradford method (Bio-Rad). Thirty-five micrograms of total protein from each animal was run in individual lanes of 10% polyacrylamide gels and transferred to a nitrocellulose membrane. The blots were then blocked for a minimum of 2 h in blocking buffer (10 mM Tris HCl [pH 8], 150 mM NaCl, 5% nonfat powdered milk, 0.05% Tween 20) and probed with anti-RIα monoclonal antibody (Signal Transduction) or anti-RIIα or anti-Cα (a kind gift from S. S. Taylor, University of California—San Diego) polyclonal antibody in blocking buffer. The blots were then washed and incubated with horseradish peroxidase-conjugated secondary antibody and visualized using the Amersham ECL system. Autoradiograms were scanned in a scanning densitometer and analyzed with ImageQuant software (Molecular Dynamics).
For the CREB and phospho-CREB Western blotting analyses, 50 μg of total protein from each muscle was run in individual lanes of a 10% polyacrylamide gel and transferred to nitrocellulose. The blots were blocked in a solution of 20 mM Tris (pH 7.6), 140 mM NaCl, 5% nonfat powdered milk, and 0.1% Tween 20 for 1 h and then probed overnight with either anti-CREB or anti-phospho-CREB antibody (New England Biolabs) in a solution of 20 mM Tris HCl (pH 7.6), 140 mM NaCl, 5% bovine serum albumin, 0.1% Tween 20. Enhanced chemiluminescence detection and autoradiogram analysis were performed as described above. Phospho-CREB levels were quantified and normalized to total CREB levels.
RESULTS
Generation of PKIα homozygous mutant mice.
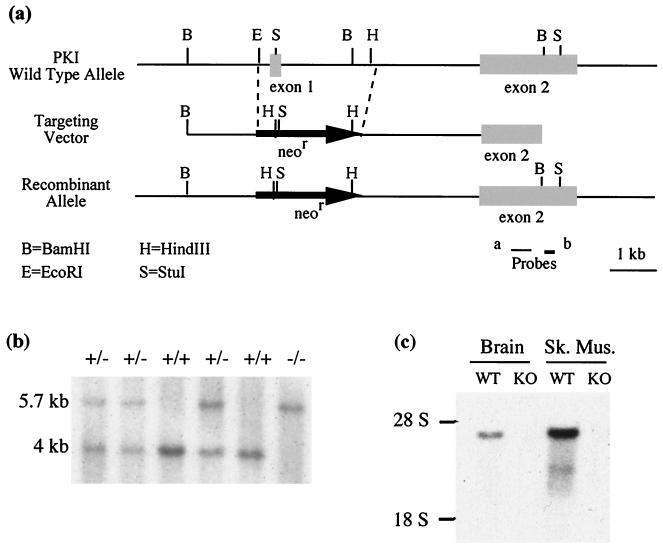
The targeting vector used to target the PKIα gene in embryonic stem cells is shown in Fig. 1a. Homologous recombination replaced the first exon of PKIα, which encodes most of the protein, with the neomycin resistance cassette. The mutant cells were used to generate heterozygous mice, which when bred gave rise to homozygous mice at the expected Mendelian ratio of approximately 25%. Genotyping of the offspring was performed by Southern blotting (Fig. 1b), and the deletion of the PKIα gene was confirmed by Northern blotting of RNA from various tissues from the mice. The homozygous (knockout) mice showed a complete absence of the PKIα transcript that is readily apparent in the wild-type mice (Fig. 1c).
FIG. 1.
Generation of PKIα knockout mice. (a) Targeting strategy at the PKIα locus. Exon 1 is replaced by the neomycin resistance cassette (neor) in a recombinant allele generated by homologous recombination. Probe b was used to identify homologous recombinant ES cells on genomic Southern blots. (b) Southern blot of tail genomic DNA from a litter derived from a heterozygote cross. A restriction digest with BamHI and StuI when hybridized with probe a shows two definitive bands; 5.7 kb indicates the recombinant allele present both in the heterozygote (+/−) and in the knockout (−/−); 4 kb represents the wild-type allele present in the wild-type mice (+/+) and the heterozygotes. (c) Northern blot of total RNA from brain and skeletal muscle from wild-type (WT) and knockout (KO) animals. Probe b was used. Note the absence of the 4.3-kb PKIα transcript in the knockout brain and skeletal muscle (Sk. Mus.).
Knockout mice were outwardly indistinguishable from the wild-type mice, exhibiting the same size and weight profiles throughout their growth (data not shown). The presence of the PKIα transcript in the Sertoli cells (but not in the germ cells) of the testis led to the hypothesis that this protein may modulate the action of follicle-stimulating hormone (FSH), which acts via the cAMP-PKA pathway to regulate spermatogenesis (28). However, the knockout mice showed normal litter sizes relative to their wild-type counterparts (7.9 ± 0.6 versus 7.3 ± 0.8, respectively; n = 13). Extensive characterization of testicular function revealed no defects in testis weights, sperm number, or sperm motility (data not shown). This suggests that PKIα does not have a significant role in FSH signaling in the testis.
PKI activity in mutant mice.
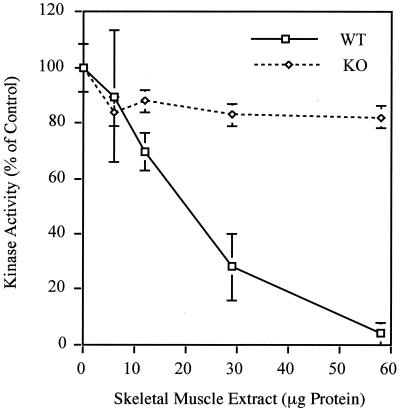
Loss of PKIα would be expected to most drastically affect skeletal muscle, the tissue that contains the greatest abundance of this protein. In addition, this tissue shows negligible levels of PKIβ and PKIγ (10, 29). We undertook a measurement of total PKI activity in this tissue from wild-type and knockout animals. The results of the assay, depicted in Fig. 2, show that addition of increasing amounts of heat-inactivated extract from wild-type muscle to an assay mix containing exogenous C subunit causes kinase activity to drop nearly to zero. The addition of a similar amount of PKIα knockout muscle extract, however, does not cause a significant decline of kinase activity. This indicates the functional absence of all PKI activity in skeletal muscle, further validating the success of the targeting strategy shown in Fig. 1. It also demonstrates the lack of functional compensation by the other two isoforms of PKI in skeletal muscle. This was further confirmed by probing a Northern blot containing skeletal muscle RNA from four wild-type and four knockout mice. PKIβ was nearly undetectable in both genotypes, while PKIγ was expressed at low levels that were unchanged in the knockouts (data not shown).
FIG. 2.
Absence of PKI activity in PKIα knockout skeletal muscle. Heat-inactivated tissue extracts from wild-type (WT) and PKIα knockout (KO) skeletal muscle were added to an assay measuring phosphorylation of the PKA substrate, kemptide, by exogenous PKA (C subunit). Kinase activity is expressed as a percentage of the control, where 100% represents kinase activity in the absence of any added tissue extract. The PKI present in wild-type muscle extract almost completely inactivates PKA. Extracts from PKIα knockouts have no significant effect even at the highest concentration of protein, demonstrating a complete absence of PKI activity in PKIα-null skeletal muscle. The error bars represent standard errors of the mean.
Loss of an inhibitor is conventionally thought to functionally result in an increase in activity of a dynamic system. We therefore hypothesized that the effect of the PKIα deletion would be manifested by an increase in transcription of PKA-regulated genes in skeletal muscle. This tissue was chosen for analysis owing to the high level of PKI activity in the wild types and the complete absence of PKI activity in the knockouts.
Gene expression in skeletal muscle.
Few genes are known to be transcriptionally regulated by PKA in skeletal muscle. Phosphoenolpyruvate carboxykinase (PEPCK) is an enzyme involved in the gluconeogenesis pathway (17, 22) that is primarily expressed in liver but is also expressed at lower levels in skeletal muscle (16, 34). The PEPCK gene is inducible by the cAMP-PKA pathway, which can be stimulated either by fasting or by treatment with β-adrenergic agonists like isoproterenol (4, 17, 26).
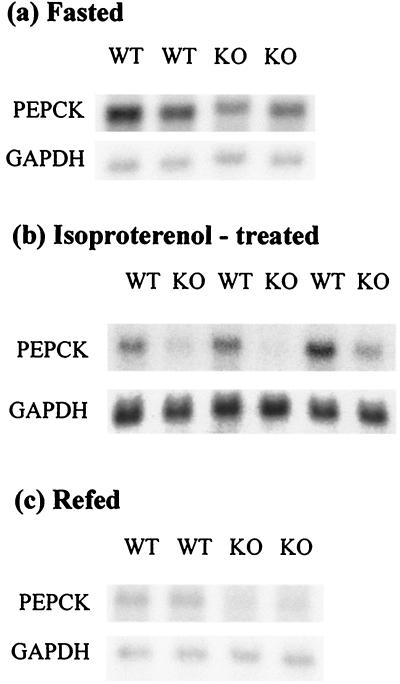
Expression of PEPCK under basal and stimulated conditions was examined in skeletal muscle by Northern blot analysis (Fig. 3). Induced expression was analyzed in two groups of mice. The first group fasted for 16 h (Fig. 3a). The second, ad libitum-fed group was treated with isoproterenol (Fig. 3b). Basal expression was examined in a third group of mice that had been refed for 6 h after fasting (Fig. 3c). Surprisingly, basal expression of PEPCK was decreased in the knockouts relative to the wild types. There was an even greater discrepancy between genotypes in the induced levels of PEPCK. In the case of isoproterenol, there was almost no induction in the knockouts at all. As might be expected, untreated mice exhibited wide variation in expression among individuals, presumably because of short-term metabolic effects, but overall the knockouts showed lower PEPCK levels than the wild types (data not shown).
FIG. 3.
Diminished expression of a PKA-responsive gene in skeletal muscle. Northern blot analysis of PEPCK mRNA levels. A riboprobe made with a PEPCK cDNA fragment as a template was used to probe the Northern blots. Each lane represents RNA from an individual wild-type (WT) or knockout (KO) mouse. The blots were reprobed for GAPDH as a control for RNA loading. (a) PEPCK levels induced by fasting are lower in the knockouts than the wild types. (b) PEPCK expression induced by treating the mice with isoproterenol is decreased in the knockouts relative to the wild-type mice. Note that the GAPDH exposure was longer than for panels a and c. (c) Basal levels of PEPCK examined after refeeding are also lower in the knockouts.
These observations countered our predictions about the effect of loss of PKI on gene transcription and required further analysis. The lower basal and induced expression cannot be explained by overcompensation by other PKI isoforms, since there is no detectable PKI activity in skeletal muscle. It could conceivably arise from a decrease in the activity of upstream effectors of gene expression, such as the transcription factor CREB or PKA itself.
CREB phosphorylation.
The 43-kDa nuclear protein CREB was originally identified as a factor that binds the conserved cAMP response element and is a target for phosphorylation by PKA (20). Phosphorylation on Ser-133 enhances the transcriptional activity of CREB and can be detected by antibodies raised specifically against phospho-CREB (12, 13). Mutagenesis of the single Ser residue renders the protein inactive as a transcription factor, suggesting that phosphorylation at this site is critical for PKA-dependent gene induction (13).
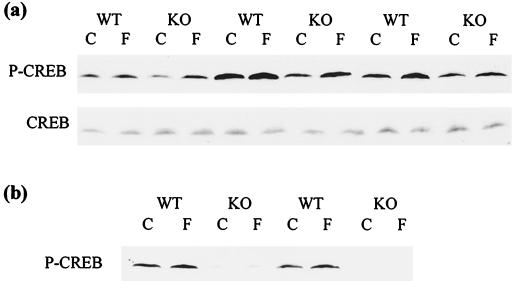
To evaluate whether the decreased gene expression was a consequence of lower levels of CREB activation, we examined phospho-CREB levels in skeletal muscle stimulated with a PKA activator. Since the time course of CREB phosphorylation and dephosphorylation is extremely short (under 1 h) (15), this experiment was better suited to in vitro skeletal muscle cultures. Organ cultures of gastrocnemius muscle were treated with forskolin, and preliminary experiments revealed that the highest expression of phospho-CREB occurred at about 8 min after stimulation, dropping substantially by 18 min. As shown in Fig. 4, phospho-CREB was present at lower levels in unstimulated (control) knockout muscle than in wild-type muscle at both time points, 8 (Fig. 4a) and 18 (Fig. 4b) min. Densitometric analysis, normalized for total CREB levels (Fig. 4a, bottom), revealed that knockouts contained about half as much phospho-CREB as wild types at the 8-min time point. Upon activation by forskolin, phospho-CREB levels rose 50% in the wild-type muscle (Fig. 4a). The fold increase in phospho-CREB levels was similar in the knockouts, resulting in stimulated phospho-CREB levels about half those of the wild types. The variability among mice in phospho-CREB levels mirrors the discrepancy of PEPCK expression among animals fed ad libitum and reflects true differences in phosphorylation, as there were equal levels of total CREB in all the lanes (Fig. 4a). Fig. 4b demonstrates an even greater difference in phosphorylation between wild types and knockouts at 18 min poststimulation, when induction by forskolin is no longer obvious. Again, levels of total CREB were unchanged (data not shown). These lower levels of phospho-CREB in the knockouts under both basal and induced conditions match the profile of gene activation that is seen in skeletal muscle (Fig. 3).
FIG. 4.
Phosphorylation of CREB is diminished in PKIα knockout skeletal muscle. Western blot analysis of phospho-CREB levels. Vehicle-treated (C) and forskolin-treated (F) skeletal muscle organ cultures were analyzed at the specified time points for both wild-type (WT) and PKIα knockout (KO) mice. (a) Eight minutes after treatment. Basal levels of phospho-CREB shown in control (C) lanes are lower in knockout than in wild-type muscles. In addition, levels of phospho-CREB in the induced (F) muscles are also lower in the knockout than in the wild type. Below is a Western blot for total CREB with the same samples. Note that total CREB content is constant in all lanes. (b) Eighteen minutes after treatment. The levels of phospho-CREB induced by forskolin are almost back to basal levels, and the difference in phospho-CREB levels between wild-type and knockout muscle is magnified.
PKA activity.
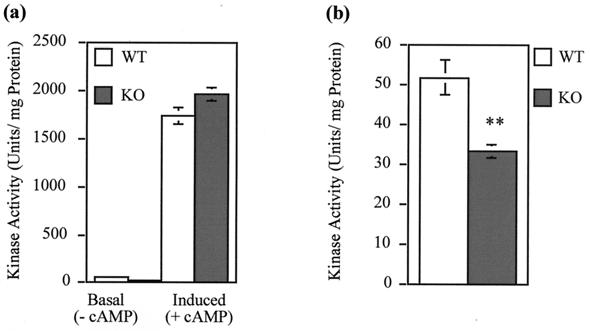
To determine whether changes in PKA activity may underlie the changes in CREB phosphorylation, a study of PKA activity was conducted with tissue extracts from wild-type and knockout skeletal muscle under two conditions. The absence of exogenous cAMP in the assay mix represents conditions of basal PKA activation, while the presence of exogenous cAMP represents total (inducible) levels of PKA. In the former instance, the knockout tissue was seen to demonstrate a significantly lower kinase activity than the wild type (Fig. 5), a result entirely consistent with the lower basal levels of gene expression and CREB phosphorylation seen in this tissue. Basal PKA activity is shown on the left in Fig. 5a and with an expanded axis in Fig. 5b. Upon addition of exogenous cAMP, total induced kinase activity levels (Fig. 5a, right) increased approximately 30-fold over basal levels. Interestingly, wild-type and knockout tissues showed similar total induced kinase activities. Note that the small increase in the knockout is not statistically significant (n = 6 knockouts and 8 wild types). Since the levels of C subunit are the same in wild-type and mutant skeletal muscle (see below), these results indicate that endogenous PKI, which is present in the wild types but not the mutants, has little or no effect in this in vitro assay of total PKA activity. Perhaps under these assay conditions, endogenous PKI, which may be only 20% as abundant as the C subunit (30), is not sufficiently concentrated to remain associated with the C subunit, despite our efforts to stabilize PKI-C subunit interactions by preparing tissue extracts in the presence of Mg and ATP. We have also determined that the cAMP concentrations that give half-maximal PKA activation in wild types and knockouts are identical (data not shown).
FIG. 5.
Basal PKA activity is decreased in PKIα knockout skeletal muscle. (a) PKA activity was assayed in skeletal muscle extracts in the absence (− cAMP) and presence (+ cAMP) of 5 μM cAMP to determine basal and total PKA activity, respectively. There is no significant difference in the total kinase activity between wild-type (WT) and knockout (KO) mice. (b) Basal activity, in the absence of exogenous cAMP, is shown with an expanded axis. The knockout extract has significantly lower basal activity than the wild type (∗∗, P = 0.003; t test). The error bars represent standard errors of the mean.
The lower basal PKA activity in the absence of PKIα raises a mechanistic question as to how this might occur. Since PKIα interacts directly with the C subunit of PKA, one possibility is that this might occur via a downregulation of C subunit in a compensatory mechanism. Alternatively, there might be compensatory changes in the R subunits.
PKA subunit levels.
Western blots for the Cα subunit of PKA in skeletal muscle showed no difference in Cα subunit between the wild-type and the knockout mice (Fig. 6), nor was there a difference in the RIIα regulatory subunit isoform. However, the regulatory subunit RIα showed an upregulation in the knockouts. The level of RIα in the knockouts was determined to be 1.6-fold that of the wild type by scanning densitometry. The compensation by only one of the two regulatory subunits present in this tissue indicates that the compensation is specific. Compensatory changes in RIα have been described earlier in RIβ and RIIβ knockout mice and have been shown to result from an increase in the stability of RIα (1). It is likely that a similar mechanism is at play here, as there is no change in RIα mRNA levels in the knockout mice (data not shown). The increase in RIα serves to bring more of the existing C subunits under regulatory control, lowering the basal kinase activity in the knockouts.
FIG. 6.
Compensatory increase in RIα regulatory subunit. Muscle extracts from wild-type (WT) and knockout (KO) mice were subjected to Western blotting and probed for the RIα, RIIα, and Cα subunits of PKA. Each lane corresponds to a tissue homogenate from a separate animal. Specific up-regulation of the RIα regulatory subunit (1.6-fold) is seen in the knockouts, with no change in the level of either the RIIα subunit or the C subunit.
DISCUSSION
When PKI was initially discovered, it was thought to act as a substoichiometric inhibitor of PKA whose level might be regulated to control both basal and cAMP-stimulated PKA activity (14, 31). However, more recent studies have uncovered a potentially more dynamic role for PKI in the cAMP-regulated phosphorylation of nuclear proteins. A leucine-rich nuclear export signal was identified on PKI that allows it to act as a chaperone for the C subunit of PKA, facilitating export of the C-PKI complex from the nucleus (32). The activities of PKI as both a direct inhibitor of the C subunit and a regulator of C subunit nuclear localization suggested that it might play an important role in cAMP-mediated gene regulation. In order to examine this potential physiological role, we disrupted the gene for PKIα in mice. Knockout mice are healthy and fertile, with normal weight gain and motor behavior.
Potential compensatory mechanisms were investigated to determine whether changes in other PKI or PKA signaling components might have occurred in the PKIα knockout mice. For example, since mice have three genes encoding distinct PKI isoforms, a compensatory up-regulation in expression of PKIβ or PKIγ could partially substitute for the loss of PKIα. However, this clearly did not occur, as there was no detectable PKI activity in knockout skeletal muscle. Compensatory changes have been observed within the PKA system in mice carrying specific PKA subunit knockouts. Increases in the RIα regulatory subunit occur when there is a loss of another R isoform, e.g., in RIIα, RIβ, and RIIβ knockout mice, and the compensation results from an increased association of RIα with the C subunit to replace the missing R isoform, with a consequent increase in RIα stability (1, 8). In the PKIα knockout skeletal muscle, we observed a significant up-regulation of RIα protein, and as there was no change in the RIα mRNA level, we suggest that the same RIα protein stabilization mechanism is responsible. No change in the amount of C subunit was observed in the knockout mice, as assessed both by Western blot analysis and by an assay of total (cAMP-stimulated) kinase. We conclude that the C subunit that would normally be associated with PKIα in the wild-type mice was instead associated with the up-regulated RIα. We suggest that this change underlies the observed decrease in basal PKA activity in the knockouts. Because the interaction with RIα has a higher affinity than the interaction with PKIα (17), the C subunit associated with RIα is “locked” in a more inactive configuration.
A significant distinguishing feature of PKI and the R subunit is their subcellular localization. The preponderance of data demonstrate that R subunits are restricted to the cytoplasm, while PKI can clearly enter the nucleus. In fact, recent literature has suggested that PKI is predominantly nuclear until it binds the C subunit and chaperones it out of the nucleus (11, 33). Previous studies, however, demonstrated a substantial cytoplasmic pool of PKI associated with microtubules (27). The observed compensatory increase in RIα in the PKIα knockouts suggests that PKIα is somewhat interchangeable with R and that a significant fraction of the cytoplasmic C subunit is normally associated with PKIα. We conclude, therefore, that there is a significant cytoplasmic pool of PKIα that is replaced by RIα when PKIα is lost by targeted deletion.
The most striking defects in the PKIα mutant skeletal muscle are in gene expression and transcription factor phosphorylation, and these would not have been predicted to occur based on the known properties of PKI. The loss of PKI activity in skeletal muscle would be expected to lead to enhanced activity of the C subunit and loss of the rapid nuclear export of C subunit that is thought to help terminate the PKA signal. The expected result would be an increase in both basal and induced CREB phosphorylation and an increase in PKA-regulated gene expression. However, under basal conditions in fed animals, the phosphorylation of CREB and the level of PEPCK mRNA are significantly reduced in PKIα knockout mice. This result might appear consistent with the lower levels of basal PKA activity measured in the in vitro kinase assay, but this defect cannot be overcome by conditions that elevate cAMP and activate PKA. Fasting mice normally show an induction of PEPCK mRNA, and this response can be mimicked by administration of a nonspecific β-adrenergic receptor agonist like isoproterenol. However, in PKIα knockout mice, neither fasting nor isoproterenol is able to achieve full activation of PEPCK gene expression despite the presence of equivalent levels of total (cAMP-stimulated) PKA activity in knockout and wild-type skeletal muscle. CREB phosphorylation is also not stimulated to as high a level in PKI-deficient mice as in wild-type mice, suggesting that the defect in gene expression is in a step prior to phosphorylation of transcription factors.
One interpretation is that the cell operates at the low end of the cAMP concentration curve and that the cAMP-stimulated kinase activity levels attained in vivo are much lower than the maximal levels measured in vitro. In this scenario, the knockouts never reach the same levels of PKA activation in vivo as do the wild types but are still able to regulate their phospho-CREB and PEPCK levels, albeit around a lower set point. However, it is notable that cytoplasmic PKA signaling appears to be unaffected, since no difference was observed in skeletal muscle glycogen levels between wild types and knockouts (data not shown). An alternate interpretation of our data is that PKI actually facilitates phosphorylation of transcription factors and subsequent gene transcription in an unknown manner. Perhaps C-PKI represents a mobile pool of C subunit that could be very important, since much of the PKA holoenzyme is not only restricted to the cytoplasm but is bound to subcellular organelles and signaling complexes by association with AKAPs (A kinase anchoring proteins). It is possible that PKI regulates nuclear entry of the C subunit (rather than exclusively its export) or mediates interaction with transcription factors or other nuclear proteins. In these scenarios, a PKIα knockout would have reductions in CREB phosphorylation and PKA-regulated gene expression in the absence of any significant change in total kinase activity, precisely the phenotype we observed. However, neither of the characterized PKI activities (inhibition and nuclear export of the C subunit) currently lends support to these ideas. Further studies of these knockouts and other PKI isoform knockouts are clearly needed to explore whether the changes in CREB phosphorylation and gene expression derive from an as yet undiscovered PKI function.
ACKNOWLEDGMENTS
This work was funded by grants from the National Institutes of Health (HD 33057 to R.L.I. and GM32875 to G.S.M.). E.A.G. was supported by the Andrew Mellon Foundation and NIH Training Grant T32 DK07247.
We thank Thong Su and R. Scott Frayo for valuable technical assistance. We also thank D. Granner for the PEPCK cDNA and S. Taylor for the Cα antibody.
REFERENCES
- 1.Amieux P S, Cummings D E, Motamed K, Brandon E P, Wailes L A, Le K, Idzerda R L, McKnight G S. Compensatory regulation of RIalpha protein levels in protein kinase A mutant mice. J Biol Chem. 1997;272:3993–3998. doi: 10.1074/jbc.272.7.3993. [DOI] [PubMed] [Google Scholar]
- 2.Baude E J, Dignam S S, Reimann E M, Uhler M D. Evidence for the importance of hydrophobic residues in the interactions between the cAMP-dependent protein kinase catalytic subunit and the protein kinase inhibitors. J Biol Chem. 1994;269:18128–18133. [PubMed] [Google Scholar]
- 3.Beale E G, Dedman J R, Means A R. Isolation and characterization of a protein from rat testis which inhibits cyclic AMP-dependent protein kinase and phosdiesterase. J Biol Chem. 1977;252:6322–6327. [PubMed] [Google Scholar]
- 4.Beebe S J, Koch S R, Chu D T, Corbin J D, Granner D K. Regulation of phosphoenolpyruvate carboxykinase gene transcription in H4IIE hepatoma cells: evidence for a primary role of the catalytic subunit of 3′,5′-cyclic adenosine monophosphate-dependent protein kinase. Mol Endocrinol. 1987;1:639–647. doi: 10.1210/mend-1-9-639. [DOI] [PubMed] [Google Scholar]
- 5.Brandon E P, Gerhold K A, Qi M, McKnight G S, Idzerda R L. Derivation of novel embryonic stem cell lines and targeting of cyclic AMP-dependent protein kinase genes. Recent Prog Horm Res. 1995;50:403–408. doi: 10.1016/b978-0-12-571150-0.50028-7. [DOI] [PubMed] [Google Scholar]
- 6.Brandon E P, Logue S F, Adams M R, Qi M, Sullivan S P, Matsumoto A M, Dorsa D M, Wehner J M, McKnight G S, Idzerda R L. Defective motor behavior and neural gene expression in RIIbeta-protein kinase A mutant mice. J Neurosci. 1998;18:3639–3649. doi: 10.1523/JNEUROSCI.18-10-03639.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brandon E P, Zhuo M, Huang Y Y, Qi M, Gerhold K A, Burton K A, Kandel E R, McKnight G S, Idzerda R L. Hippocampal long-term depression and depotentiation are defective in mice carrying a targeted disruption of the gene encoding the RI beta subunit of cAMP-dependent protein kinase. Proc Natl Acad Sci USA. 1995;92:8851–8855. doi: 10.1073/pnas.92.19.8851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burton K A, Johnson B D, Hausken Z E, Westenbroek R E, Idzerda R L, Scheuer T, Scott J D, Catterall W A, McKnight G S. Type II regulatory subunits are not required for the anchoring-dependent modulation of Ca2+ channel activity by cAMP-dependent protein kinase. Proc Natl Acad Sci USA. 1997;94:11067–11072. doi: 10.1073/pnas.94.20.11067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clegg C H, Correll L A, Cadd G G, McKnight G S. Inhibition of intracellular cAMP-dependent protein kinase using mutant genes of the regulatory type I subunit. J Biol Chem. 1987;262:13111–13119. [PubMed] [Google Scholar]
- 10.Collins S P, Uhler M D. Characterization of PKIgamma, a novel isoform of the protein kinase inhibitor of cAMP-dependent protein kinase. J Biol Chem. 1997;272:18169–18178. doi: 10.1074/jbc.272.29.18169. [DOI] [PubMed] [Google Scholar]
- 11.Fantozzi D A, Harootunian A T, Wen W, Taylor S S, Feramisco J R, Tsien R Y, Meinkoth J L. Thermostable inhibitor of cAMP-dependent protein kinase enhances the rate of export of the kinase catalytic subunit from the nucleus. J Biol Chem. 1994;269:2676–2686. [PubMed] [Google Scholar]
- 12.Ginty D D, Kornhauser J M, Thompson M A, Bading H, Mayo K E, Takahashi J S, Greenberg M E. Regulation of CREB phosphorylation in the suprachiasmatic nucleus by light and a circadian clock. Science. 1993;260:238–241. doi: 10.1126/science.8097062. [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez G A, Montminy M R. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- 14.Grove J R, Avruch J. Probing cAMP gene regulation with a recombinant protein kinase inhibitor. In: Cohen P, Foulkes J G, editors. The hormonal control of gene transcription. New York, N.Y: Elsevier Science Publishers; 1991. pp. 173–196. [Google Scholar]
- 15.Hagiwara M, Brindle P, Harootunian A, Armstrong R, Rivier J, Vale W, Tsien R, Montminy M R. Coupling of hormonal stimulation and transcription via the cyclic AMP-responsive factor CREB is rate limited by nuclear entry of protein kinase A. Mol Cell Biol. 1993;13:4852–4859. doi: 10.1128/mcb.13.8.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanson R W, Garber A J. Phosphoenolpyruvate carboxykinase. I. Its role in gluconeogenesis. Am J Clin Nutr. 1972;25:1010–1021. doi: 10.1093/ajcn/25.10.1010. [DOI] [PubMed] [Google Scholar]
- 17.Hanson R W, Reshef L. Regulation of phosphoenolpyruvate carboxykinase (GTP) gene expression. Annu Rev Biochem. 1997;66:581–611. doi: 10.1146/annurev.biochem.66.1.581. [DOI] [PubMed] [Google Scholar]
- 18.Hauer J A, Taylor S S, Johnson D A. Binding-dependent disorder-order transition in PKI alpha: a fluorescence anisotropy study. Biochemistry. 1999;38:6774–6780. doi: 10.1021/bi983074k. [DOI] [PubMed] [Google Scholar]
- 19.Knighton D R, Zheng J H, Ten Eyck L F, Xuong N H, Taylor S S, Sowadski J M. Structure of a peptide inhibitor bound to the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253:414–420. doi: 10.1126/science.1862343. [DOI] [PubMed] [Google Scholar]
- 20.Montminy M R, Bilezikjian L M. Binding of a nuclear protein to the cyclic-AMP response element of the somatostatin gene. Nature. 1987;328:175–178. doi: 10.1038/328175a0. [DOI] [PubMed] [Google Scholar]
- 21.Olsen S R, Uhler M D. Isolation and characterization of cDNA clones for an inhibitor protein of cAMP-dependent protein kinase. J Biol Chem. 1991;266:11158–11162. [PubMed] [Google Scholar]
- 22.Pilkis S J, Granner D K. Molecular physiology of the regulation of hepatic gluconeogenesis and glycolysis. Annu Rev Physiol. 1992;54:885–909. doi: 10.1146/annurev.ph.54.030192.004321. [DOI] [PubMed] [Google Scholar]
- 23.Scott J D, Fischer E H, Demaille J G, Krebs E G. Identification of an inhibitory region of the heat-stable protein inhibitor of the cAMP-dependent protein kinase. Proc Natl Acad Sci USA. 1985;82:4379–4383. doi: 10.1073/pnas.82.13.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scott J D, Fischer E H, Takio K, Demaille J G, Krebs E G. Amino acid sequence of the heat-stable inhibitor of the cAMP-dependent protein kinase from rabbit skeletal muscle. Proc Natl Acad Sci USA. 1985;82:5732–5736. doi: 10.1073/pnas.82.17.5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seasholtz A F, Gamm D M, Ballestero R P, Scarpetta M A, Uhler M D. Differential expression of mRNAs for protein kinase inhibitor isoforms in mouse brain. Proc Natl Acad Sci USA. 1995;92:1734–1738. doi: 10.1073/pnas.92.5.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snell K, Duff D A. Muscle phosphoenolpyruvate carboxykinase activity and alanine release in progressively starved rats. Int J Biochem. 1979;10:423–426. doi: 10.1016/0020-711x(79)90066-1. [DOI] [PubMed] [Google Scholar]
- 27.Tash J S, Welsh M J, Means A R. Protein inhibitor of cAMP-dependent protein kinase: production and characterization of antibodies and intracellular localization. Cell. 1980;21:57–65. doi: 10.1016/0092-8674(80)90114-2. [DOI] [PubMed] [Google Scholar]
- 28.Van Patten S M, Donaldson L F, McGuinness M P, Kumar P, Alizadeh A, Griswold M D, Walsh D A. Specific testicular cellular localization and hormonal regulation of the PKIalpha and PKIbeta isoforms of the inhibitor protein of the cAMP-dependent protein kinase. J Biol Chem. 1997;272:20021–20029. doi: 10.1074/jbc.272.32.20021. [DOI] [PubMed] [Google Scholar]
- 29.Van Patten S M, Howard P, Walsh D A, Maurer R A. The alpha- and beta-isoforms of the inhibitor protein of the 3′,5′-cyclic adenosine monophosphate-dependent protein kinase: characteristics and tissue- and developmental-specific expression. Mol Endocrinol. 1992;6:2114–2122. doi: 10.1210/mend.6.12.1491692. [DOI] [PubMed] [Google Scholar]
- 30.Walsh D A, Ashby C D. Protein kinases: aspects of their regulation and diversity. Recent Prog Horm Res. 1973;29:329–359. doi: 10.1016/b978-0-12-571129-6.50012-9. [DOI] [PubMed] [Google Scholar]
- 31.Walsh D A, Ashby C D, Gonzalez C, Calkins D, Fischer E H, Krebs E G. Purification and characterization of a protein inhibitor of adenosine 3′,5′-monophosphate-dependent protein kinases. J Biol Chem. 1971;246:1977–1985. [PubMed] [Google Scholar]
- 32.Wen W, Harootunian A T, Adams S R, Feramisco J, Tsien R Y, Meinkoth J L, Taylor S S. Heat-stable inhibitors of cAMP-dependent protein kinase carry a nuclear export signal. J Biol Chem. 1994;269:32214–32220. [PubMed] [Google Scholar]
- 33.Wiley J C, Wailes L A, Idzerda R L, McKnight G S. Role of regulatory subunits and protein kinase inhibitor (PKI) in determining nuclear localization and activity of the catalytic subunit of protein kinase A. J Biol Chem. 1999;274:6381–6387. doi: 10.1074/jbc.274.10.6381. [DOI] [PubMed] [Google Scholar]
- 34.Zimmer D B, Magnuson M A. Immunohistochemical localization of phosphoenolpyruvate carboxykinase in adult and developing mouse tissues. J Histochem Cytochem. 1990;38:171–178. doi: 10.1177/38.2.1688895. [DOI] [PubMed] [Google Scholar]