Abstract
Protein ubiquitination plays an important role in the regulation of TGF-β-activated kinase 1 (TAK1)-mediated NF-κB activation. It is well established that TAK1 activation is tightly regulated with its binding partners, TAK1-binding proteins (TAB1-3). However, the tight regulation of TAK1 activation remains elusive. Here, using Trim26-knockout mice and Trim26-transgenic mice, we found that TRIM26 acts as a positive regulator of TAK1 activation by ubiquitinating its binding partner TAB1. Knockout of Trim26 inhibited TAK1 activation and downstream kinases activation, thus decreasing the induction of proinflammatory cytokines following LPS, TNF-α, and IL-1β stimulation. Mechanistically, TRIM26 catalyzes the K11-linked polyubiquitination of TAB1 at Lys294, Lys319, and Lys335 to enhance the activation of TAK1 and subsequent NF-κB and MAPK signaling. Consequently, Trim26 deficiency protects mice from LPS-induced septic shock in vivo. Moreover, Trim26 deficiency attenuates the severity of dextran sodium sulfate (DSS)-induced colitis. Thus, these finding provides a novel insight into how TAK1 activation is regulated through TRIM26-mediated ubiquitination of TAB1 and reveals the new function of TRIM26 in the regulation of the inflammatory innate immune response.
Subject terms: Acute inflammation, Microbiology, Immunological disorders
Introduction
Innate immunity provides the first line of host defense against microbial invading pathogens. Toll-like receptors (TLRs) are among the first pattern-recognition receptors to be identified as playing an essential role in the innate immune response [1–5]. TLRs initiate a signaling cascade when stimulated by ligands, like lipopolysaccharide (LPS, TLR4 ligand), polyinosinic-polycytidilic acid (Poly(I:C), TLR3 ligand), and the synthetic triacylated lipopeptide (Pam3CSK4, TLR2 ligand), which trigger the activation of nuclear factor (NF-κB) [6]. Upon stimulated with these ligands, myeloid differentiation primary response gene 88 (MyD88) and Toll/IL-1R domain-containing adapter that induces an IFN-β (TRIF) are recruited to the Toll/IL-1R domain located in the cytoplasmic tails of TLRs. MyD88/TRIF then combines with IL-1R-associated kinase 1/4 (IRAK1/4) to promote the activation of TNFR-associated factor (TRAF) 6 and transforming growth factor-activated kinase 1 (TAK1), leading to the activation of NF-κB and MAPK [4, 7, 8]. Excessive activation of TLRs signaling can cause inflammatory diseases, such as autoimmunity [9], chronic inflammation [10, 11], or allergy [12]. Thus, the TLRs-mediated inflammatory response must be tightly controlled.
TAK1 belongs to the mitogen-activated protein kinase kinase kinase family and is a core protein kinase in the TLRs-induced immune response. It can be initiated by a wide range of proinflammatory mediators, such as TLR ligands, TNF-α, and IL-1β [13, 14]. The TAK1-TABs complex consists of three regulatory subunits termed TAK1-binding protein (TAB)1, TAB2, and TAB3. TAB1 is a chaperone protein for TAK1 and is constitutively associating with TAK1, which results in the autophosphorylation and activation of TAK1 [15]. While TAB2/3, two structurally related proteins, recognize the K63-linked ubiquitin chain of TRAF molecules [16].
Several studies have demonstrated that lysine (K)63-linked and (K)48-linked polyubiquitination is critical for TAK1-TABs complex assembly and TAK1 activation. For example, Wang et al. reported that TAK1 is a ubiquitin-dependent kinase of MAPK kinase and IκB kinase (IKK), the formation of K63-linked polyubiquitin chains by TRAF6 can activate the endogenous TAK1 complex in a biochemically well-defined system [17]. The E3 ubiquitin ligase tripartite motif (TRIM)8 positively regulates TNF-α- and IL-1β-triggered NF-κB activation by catalyzing K63-linked polyubiquitination of TAK1 [18, 19]. Tripartite motif (TRIM)27 and Tripartite motif (TRIM)29 downregulates NF-kB activation by mediating lysosome-dependent degradation of TAB2 [20, 21]. The PHD domain of MEKK1 can directly promote K63-linked polyubiquitination of TAB1 and induce TGF-β-mediated TAK1 activation [22]. As polyubiquitin chains comprise several potential linkage residues, such as K6, K11, K27, K29, K33, K48, and K63 [23], whether there exist other linkage types in the regulation of TAB1 remains to be explored.
The E3 ligase TRIM26 belongs to the TRIM protein family, which is involved in various biological processes including innate immunity, cell proliferation, autophagy, and inflammatory response [24–27]. In this study, we identified TRIM26 as a positive regulator of TAK1 activation. TRIM26 interacts with TAB1 and specifically catalyzes K11-linked polyubiquitination of TAB1, which facilitates TAK1 activation and initiates downstream signaling. Our study demonstrates a novel mechanism underlying TRIM26-mediated regulation of the inflammatory response and identifies new therapeutic targets for inflammatory diseases.
Results
TRIM26 positively regulates TLRs-induced production of proinflammatory cytokines
TRIM26 is a member of the TRIM protein family, which is encoded within the locus of major histocompatibility complex class I region [28]. We speculate TRIM26 could play key roles in the regulation of immune response. Indeed, we found TRIM26 promotes nuclear IRF3 ubiquitination and degradation after virus infection to regulate type I interferon production and innate antiviral immunity [29]. However, its biological function in the inflammatory response remains unclear. To investigate the possible function of TRIM26 in inflammatory immune responses, we first examined TRIM26 expression in primary peritoneal macrophages (PMs) stimulated with various TLR ligands. The expression of Trim26 mRNA and protein was increased upon stimulation with LPS (Supplementary Fig. S1a, b). Similarly, Pam3CSK4, Poly(I:C), or R848 stimulation also enhanced Trim26 protein expression in PMs (Supplementary Fig. S1c). Notably, TNF-α and IL-1β, two proinflammatory cytokines induced upon TLR ligands stimulation, also increased Trim26 protein expression in PMs (Supplementary Fig. S1c), indicating TRIM26 may play a role in the regulation of inflammatory immune responses.
We designed a Trim26-specific small interfering (siRNA) and transfected into primary PMs to knockdown endogenous Trim26 expression. Trim26 mRNA and Trim26 protein levels were substantially reduced in Trim26-specific siRNA transfected macrophages compared to those transfected with non-targeting control siRNA (Supplementary Fig. S2a). We found knockdown of Trim26 resulted in decreased mRNA expression of Tnfα, Il6, and Il12b in PMs induced with LPS (Supplementary Fig. S2b). Further, we found Pam3CSK4-, Poly(I:C)-, and R848-induced Tnfα, Il6, and Il12b mRNA expression was much lower in Trim26 knockdown PMs (Supplementary Fig. S2c). We also designed human TRIM26-specific siRNA and transfected into THP-1 cells (human monocyte cell line). Consistent with the data obtained from mice macrophages, we found LPS-induced expression of TNF-α and IL-6 was decreased in THP-1 cells transfected with TRIM26-specific siRNA (Supplementary Fig. S2d–f). These data demonstrated that knockdown of TRIM26 suppressed TLRs-induced production of proinflammatory cytokines in both human and mouse macrophages.
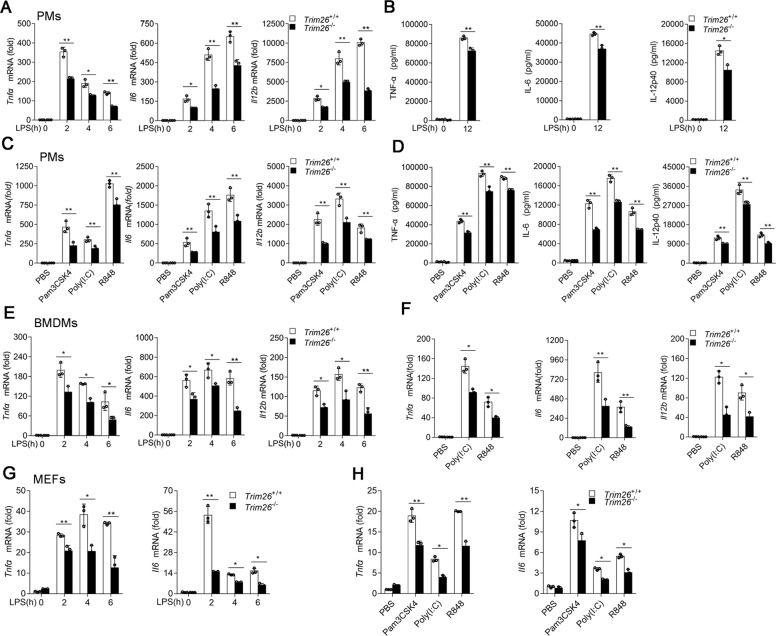
To directly evaluate the physiological role of TRIM26, we generated Trim26-knockout (Trim26−/−) mice using transcription-activation-like effector nuclease (TALEN) technology (Supplementary Fig. S3a, b). Successful deletion of Trim26 was confirmed by immunoblot analysis in PMs from Trim26−/− mice (Supplementary Fig. S3c). We prepared PMs from Trim26+/+ mice and Trim26−/− mice, followed stimulation with various TLR ligands. qPCR and ELISA analysis unveiled that induction of TNF-α, IL-6, and IL-12p40 were decreased in Trim26−/− compared with Trim26+/+ cells following LPS stimulation (Fig. 1A, B). Consistently, Trim26 deficiency also attenuated the production of proinflammatory cytokines induced by Pam3CSK4, Poly(I:C), or R848 in PMs (Fig. 1C, D). Similar results were observed in bone marrow-derived macrophages (BMDMs) and mouse embryonic fibroblasts (MEFs) prepared from Trim26+/+ and Trim26−/− mice stimulated with LPS, Pam3CSK4, Poly(I:C), or R848 (Fig. 1E–H). We also prepared PMs from Trim26-overexpressing transgenic mice (Trim26Tg) followed stimulation with LPS [29]. We found overexpression of Trim26 increased LPS-induced expression of Tnfα and Il6 mRNA (Supplementary Fig. S4a, b). Similarly, Trim26 overexpression potentiated transcription of Tnfα and Il6 mRNA following Pam3CSK4, Poly(I:C), or R848 stimulation (Supplementary Fig. S4c). Collectively, these data demonstrated that TRIM26 is a positive regulator of TLRs-induced expression of proinflammatory cytokines.
Fig. 1. TRIM26 positively regulates TLRs-induced production of proinflammatory cytokines.
A qPCR analysis Tnfα, Il6, and Il12b mRNA expression in PMs prepared from Trim26+/+ and Trim26–/– mice stimulated with LPS (200 ng/ml) for the indicated times. B ELISA of TNF-α, IL-6, and IL-12p40 protein in supernatant of PMs prepared from Trim26+/+ and Trim26–/– mice stimulated with LPS for 12 h. C qPCR analysis of Tnfα, Il6, and Il12b mRNA, respectively, in PMs prepared from Trim26+/+ and Trim26–/– mice, stimulated with Pam3CSK4 (1 μg/ml), Poly(I:C) (20 μg/ml), and R848 (10 μg/ml) for 6 h. D ELISA analysis of TNF-α, IL-6, and IL-12P40 protein, respectively, in PMs prepared from Trim26+/+ and Trim26–/– mice, stimulated with Pam3CSK4 (1 μg/ml), Poly(I:C) (20 μg/ml), and R848 (10 μg/ml) for 12 h. E qPCR analysis of Tnfα, Il6, and Il12b mRNA expression in BMDMs prepared from Trim26+/+ and Trim26–/– mice, stimulated with LPS (200 ng/ml) for the indicated times. F qPCR analysis of Tnfα, Il6, and Il12b mRNA in BMDMs prepared from Trim26+/+ and Trim26–/– mice, stimulated with Poly(I:C) (20 μg/ml) and R848 (10 μg/ml) for 6 h. G qPCR analysis of Tnfα and Il6 mRNA expression in MEFs prepared from Trim26+/+ and Trim26–/– embryos (day 13.5), stimulated with LPS (200 ng/ml) for the indicated times. H qPCR analysis of Tnfα and Il6 mRNA, respectively, in MEFs prepared from Trim26+/+ and Trim26–/– embryos (day 13.5), stimulated with Pam3CSK4 (1 μg/ml), Poly(I:C) (20 μg/ml), and R848 (10 μg/ml) for 6 h. Data are shown as mean ± SD of triplicates from one representative experiment in A–H. *p < 0.05, **p < 0.01; Student’s t test. Similar results were obtained in three independent experiments.
TRIM26 enhances TLRs-induced NF-κB and MAPK signaling
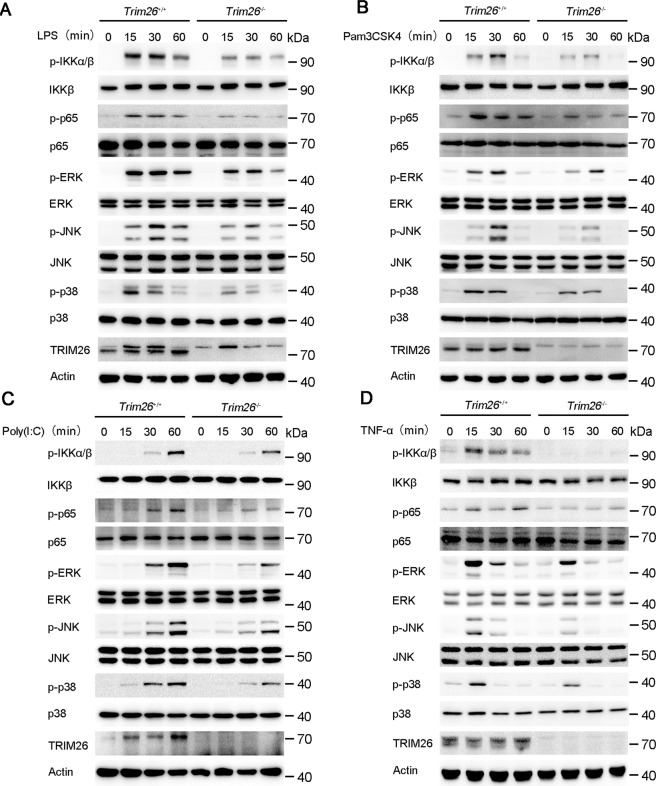
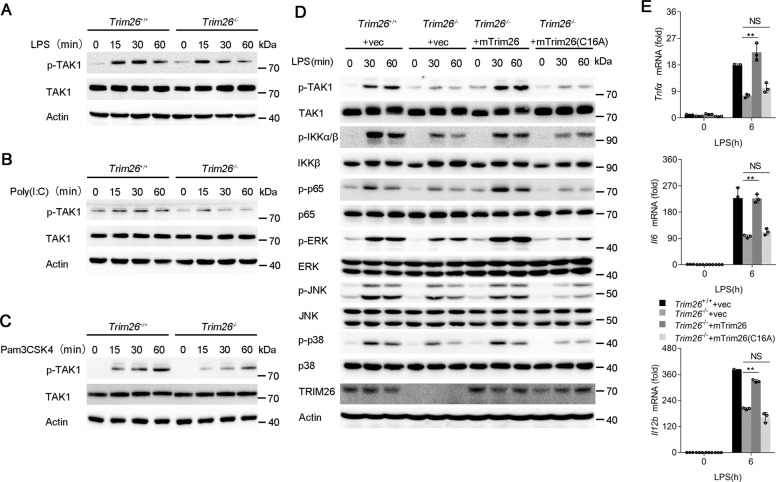
Because transcription of the proinflammatory cytokines such as Tnfα, Il6, and Il12b is mainly dependent on NF-κB and MAPK activation. Thus, we investigated the effect of TRIM26 on NF-κB and MAPK signaling. Knockout of Trim26 in PMs decreased LPS-induced phosphorylation of IKKα/β and p65, which are hallmarks of NF-κB activation (Fig. 2A). The phosphorylation of JNK, p38, and ERK in the MAPK signaling pathway was also attenuated in Trim26−/− PMs (Fig. 2A). Similarly, Poly(I:C)- or Pam3CSK4-mediated NF-κB and MAPK signaling was also decreased in PMs from Trim26−/− mice (Fig. 2B, C).
Fig. 2. TRIM26 enhances TLRs-induced NF-κB and MAPK signaling.
A Immunoblot analysis of phosphorylated and total IKKα/β, p65, ERK, JNK, and p38 in peritoneal macrophages from Trim26+/+ and Trim26–/– mice stimulated with LPS for indicated times. B–D Immunoblot analysis of phosphorylated and total IKKα/β, p65, ERK, JNK, and p38 in peritoneal macrophages from Trim26+/+ and Trim26–/– mice stimulated with Pam3CSK4(1 µg/ml), Poly(I:C) (20 µg/ml), or TNF-α (20 ng/ml) for indicated times. Similar results were obtained in three independent experiments.
TLRs-induced production of proinflammatory cytokines such as TNF-α and IL-1β would promote further production of proinflammatory cytokines using a similar signaling pathway as to TLR signaling. Consistently, the mRNA levels of Tnfα and Il6 were substantially decreased in Trim26−/− PMs after TNF-α and IL-1β stimulation compared to that in Trim26+/+ PMs (Supplementary Fig. S5a). While increased expression of Tnfα and Il6 mRNA was observed in PMs from Trim26Tg mice upon TNF-α induction (Supplementary Fig. S5b). Consistent with the above data, we found the activation of NF-κB and MAPK signaling pathway was also inhibited in PMs from Trim26−/− mice upon TNF-α stimulation (Fig. 2D). These data confirmed that TRIM26 enhances TLRs-induced NF-κB and MAPK signaling.
TRIM26 interacts with and mediates polyubiquitination of TAB1
TRIM26 regulates various biological processes by polyubiquitinating target proteins through its E3 ligase activity [24, 25, 27, 29]. In order to investigate the regulatory mechanism of TRIM26 on TLRs-induced production of proinflammatory cytokines and activation of NF-κB and MAPK signaling, we aimed to identify the molecules in the TLR signaling pathway that were targeted by TRIM26. We first transfected Myc-MyD88, Flag-IRAK1, Flag-IRAK4, or Flag-TRAF6, which are upstream molecules involved in NF-κB and MAPK activation in the TLR pathway, together with HA-ubiquitin and TRIM26 expression plasmids into HEK293T cells. Co-immunoprecipitation (Co-IP) and immunoblot analysis showed that the polyubiquitination of MyD88, IRAK1, IRAK4, and TRAF6 was not affected in the presence of TRIM26 (Supplementary Fig. S6a–d).
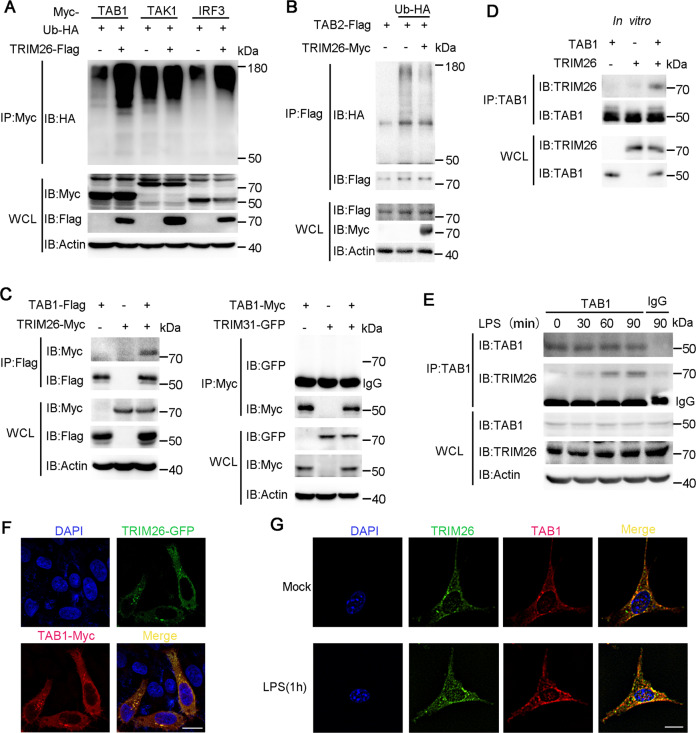
Downstream of MyD88, IRAK complex, and TRAF6 are the TAK1-TABs complex, which is recruited to TRAF6 after activation of the TLR signaling pathway [30]. We then transfected TRIM26 and HA-Ub together with Myc-TAB1, Myc-TAK1, Flag-TAB2 into HEK293T cells. We found polyubiquitination of TAB1 but not TAK1 or TAB2 was increased in the presence of TRIM26 (Fig. 3A, B). As a control, TRIM26-mediated polyubiquitination of IRF3 was easily detected (Fig. 3A), as previously reported [29]. The IKK complex consists of two catalytic subunits (IKKα and IKKβ) and a regulatory subunit IKKγ (also known as NEMO) [31], which are kinases downstream of the TAK1-TABs complex and involved in NF-κB activation. We transfected Flag-TRIM26 and HA-Ub together with Myc-IKKα, IKKβ, and IKKγ into HEK293T cells. Co-IP and immunoblot analysis showed that none of these IKK complex subunits were polyubiquitinated by TRIM26 (Supplementary Fig. S6e). These data indicated that TRIM26 may target TAB1 for polyubiquitination.
Fig. 3. TRIM26 mediates polyubiquitination of TAB1 and interacts with TAB1.
A Lysates from HEK293T cells transiently co-transfected with plasmids of Myc-TAB1, Myc-TAK1, or Myc-IRF3, Flag-TRIM26, and HA-Ub were subjected to immunoprecipitation with anti-Myc Ab, immunoblot analysis was followed with anti-HA Ab. B Lysates from HEK293T cells transiently co-transfected with plasmids of Flag-TAB2, Myc-TRIM26, and HA-Ub were subjected to immunoprecipitation with anti-Flag Ab, immunoblot analysis was followed with anti-HA Ab. C Immunoprecipitation analysis of the interaction of TRIM26 or TRIM31 with TAB1, using Myc-TRIM26, Flag-TAB1, GFP-TRIM31, or Myc-TAB1 co-transfected into HEK293T cells. D Recombinant TRIM26 and TAB1 protein were prepared in an in vitro transcription and translation system, immunoprecipitation analysis was performed using anti-TAB1 Ab, immunoblot analysis was followed with anti-TRIM26. E Lysates from peritoneal macrophages stimulated with LPS (200 ng/ml) for indicated times were subjected to immunoprecipitation analysis with anti-TAB1 Ab, followed by immunoblotting analysis with anti- TRIM26 and anti-TAB1 Ab, respectively. F Confocal microscopy of Hela cells transfected for 24 h with plasmids expressing GFP-TRIM26 and Myc-TAB1, followed by labeling of TAB1 with a Myc-specific primary antibody and an Alexa-Fluor-568-conjugated goat anti-rabbit-IgG secondary antibody (red). Scale bars, 20 μm. G Colocalization between TAB1 and Trim26 in MEFs was examined by confocal microscopy stimulated with LPS (200 ng/ml). Scale bars, 20 μm. Similar results were obtained in three independent experiments.
To further confirm TRIM26 targets TAB1, we investigated the interaction between TRIM26 and TAB1. Flag-TAB1 and Myc-TRIM26 expression plasmids were co-transfected into HEK293T cells, we found TAB1 indeed could associate with TRIM26 (Fig. 3C, left panel). While an interaction between TRIM31 and TAB1 under the same conditions was unable to be detected (Fig. 3C, right panel). To verify that TRIM26 associated with TAB1 directly, Myc-TRIM26 and His-TAB1 recombinant proteins were prepared and in vitro pull-down assay was performed. This experiment revealed that TRIM26 could form a complex with TAB1 directly (Fig. 3D). We also detected the interaction between endogenous Trim26 and TAB1 in PMs (Fig. 3E). Notably, the interaction was increased upon stimulation with LPS (Fig. 3E). Confocal microscope imaging demonstrated that TRIM26 colocalized with TAB1 (Fig. 3F). Considering that overexpressed proteins might affect their natural distributions in the cells, we also used MEFs to explore the colocalization. Consistently, confocal analysis showed the colocalization between TAB1 and Trim26 in MEFs was enhanced upon LPS stimulation (Fig. 3G). Taken together, these data suggested that TRIM26 interacts with TAB1 to regulate TLRs-induced production of proinflammatory cytokines and activation of NF-κB and MAPK signaling.
TRIM26 promotes K11-linked polyubiquitination of TAB1
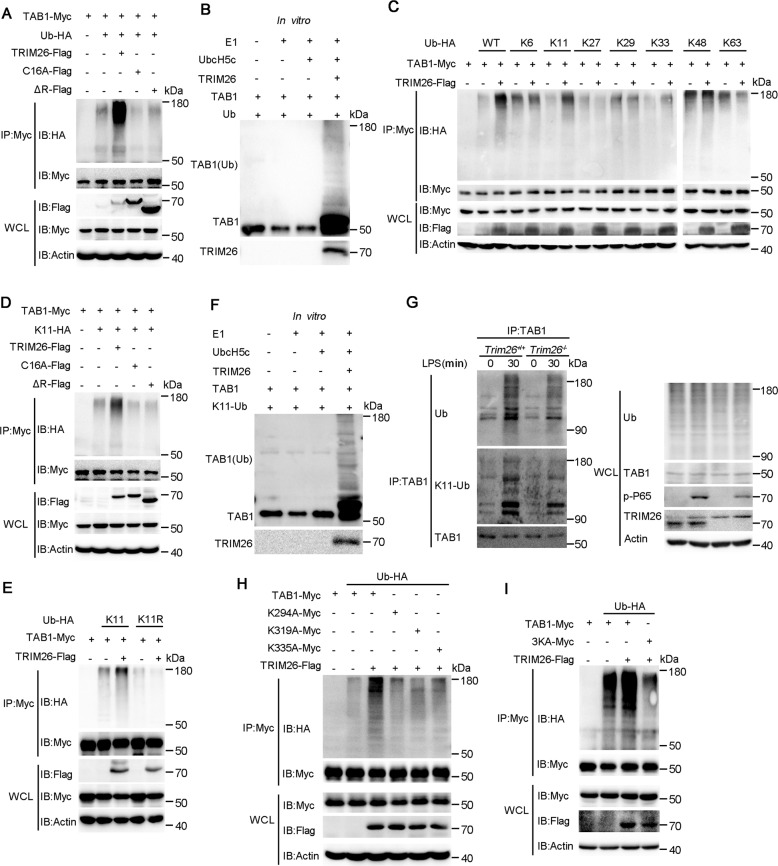
To explore whether enhanced TAB1 ubiquitination depends on the enzymatic activity of TRIM26, we transfected WT TRIM26 and RING domain mutant TRIM26 into HEK293T cells together with Myc-TAB1 and HA-Ub. TAB1 ubiquitination was easily detected in the presence of WT TRIM26 (Fig. 4A). While the substitution of the cysteine residue at position 16 of the RING domain with alanine (C16A) or deletion of the RING domain (ΔR) in TRIM26 abrogated TAB1 polyubiquitination (Fig. 4A), indicating the E3 ligase activity is required for TRIM26-mediated TAB1 ubiquitination. In vitro ubiquitination assay with recombinant TRIM26 and TAB1 protein revealed that TRIM26 can directly catalyze TAB1 ubiquitination (Fig. 4B). As different forms of ubiquitin chains play distinctive roles in the regulation of protein function [23], we investigated the type of TRIM26-induced TAB1 polyubiquitin chain. Expression plasmids for Myc-TAB1 and Flag-TRIM26 were transfected into HEK293T cells together with plasmids encoding WT ubiquitin or ubiquitin mutants containing only one single lysine residue. We found TRIM26-mediated TAB1 ubiquitination was observed in the presence of WT and K11 ubiquitin plasmid (Fig. 4C). Similarly, TRIM26-mediated K11-linked polyubiquitination of TAB1 was attenuated in TRIM26 mutants C16A and ΔR transfected HEK293T cells (Fig. 4D), indicating the E3 ligase activity of TRIM26 is indispensable required for K11-linked TAB1 ubiquitination. To directly confirm TRIM26-mediated K11-linked TAB1 polyubiquitination, we also used a ubiquitin mutant K11R, which contains all lysine residues except the lysine residue at position 11 was mutated. TAB1 was not polyubiquitinated when ubiquitin K11R was transfected into HEK293T cells in the presence or absence of TRIM26 (Fig. 4E). Considering that overexpressed proteins might affect their functions in the cells, in vitro ubiquitination assay was performed. We observed TAB1 polyubiquitination in the presence of TRIM26 and K11-linkage ubiquitin (Fig. 4F). To exclude any artificial effect caused by overexpression and determine whether TRIM26-mediated TAB1 ubiquitination occurs in physiological conditions, PMs from Trim26+/+ and Trim26−/− mice were prepared followed stimulation with LPS. Co-IP experiments showed that the level of endogenous TAB1 ubiquitination was decreased in Trim26−/− PMs compared with that in Trim26+/+ PMs upon LPS stimulation (Fig. 4G). In addition, Trim26 deficiency also inhibited K11-linked polyubiquitination of TAB1 upon LPS stimulation (Fig. 4G). Taken together, these results indicated that TRIM26 mainly mediated K11-linked polyubiquitin on TAB1 dependent on its E3 ligase activity.
Fig. 4. TRIM26 targets Lys294, 319, and 335 of TAB1 for K11-linked polyubiquitination.
A Immunoprecipitation analysis of the TAB1 ubiquitination in HEK293T cells transfected with plasmids encoding Myc-TAB1, HA-ubiquitin (WT) together with a control vector or plasmids encoding Flag-TRIM26(WT) or Flag-TRIM26 (C16A) or Flag-TRIM26(ΔR). B Recombinant TRIM26 and TAB1 proteins were prepared in an in vitro transcription and translation system. In vitro ubiquitination assay was performed in the presence of Ub, E1, UbcH5c, TRIM26, and TAB1. The ubiquitination of TAB1 was examined by immunoblot analysis with anti-TAB1 Ab. C The HEK293T cells were transfected with plasmids of Flag-TRIM26 and Myc-TAB1 together with HA-Ub or its mutants for 24 h and then followed by co-immunoprecipitation with anti-Myc Ab and immunoblotting analysis with anti-HA Ab. D Co-immunoprecipitation analysis of the ubiquitination of TAB1 in HEK293T cells transfected with plasmids encoding Myc-TAB1, HA-ubiquitin(K11) mutant, or HA-ubiquitin(K11R) mutant together with a control vector or plasmids encoding Flag-TRIM26(WT) or Flag-TRIM26 (C16A) or Flag-TRIM26(ΔR). E Co-immunoprecipitation analysis of the ubiquitination of TAB1 in HEK293T cells transfected with plasmids encoding Myc-TAB1, HA-ubiquitin(K11) mutant, or together with a control vector or plasmids encoding Flag-TRIM26. F In vitro ubiquitination assay was performed in the presence of K11-Ub, E1, UbcH5c, TRIM26, and TAB1. The ubiquitination of TAB1 was examined by immunoblot analysis with anti-TAB1 Ab. G Lysates from Trim26+/+ and Trim26–/– mice peritoneal macrophages stimulated with LPS (200 ng/ml) for the indicated times were subjected to immunoprecipitation with anti-TAB1 Ab followed by immunoblot analysis with anti-ubiquitin Ab. H Co-immunoprecipitation analysis of the polyubiquitination of TAB1 WT and its mutants, in which the lysine residues at position 294, 319, 335 were replaced with alanine individually, in HEK293T cells transfected with plasmids encoding Myc-TAB1 WT or its mutants, plus Flag-TRIM26 and HA-ubiquitin. I Co-immunoprecipitation analysis of the polyubiquitination of TAB1 WT and its mutant 3KA, in which all the three lysine residues at position 294, 319, 335 were replaced with alanine, in HEK293T cells transfected with plasmids encoding Myc-TAB1 WT or its mutants, plus Flag-TRIM26 and HA-ubiquitin. Similar results were obtained in three independent experiments.
MEKK1 has been characterized as an E3 ligase potentiating TAK1 activation to activate MAPKs through the ubiquitination of TAB1 [22]. Four lysine (Lys) residues of mouse TAB1, Lys294, Lys319, Lys335, and Lys350, have been identified as potential sites for K63-linked polyubiquitination mediated by MEKK1. Compared to mouse TAB1, human TAB1 does not have Lys350. Therefore, we constructed three TAB1 mutants TAB1(K294A), TAB1(K319A), and TAB1(K335A), in which the lysine residues at positions 294, 319, and 335 were replaced with alanine. Co-IP experiments showed that TRIM26 could not promote the polyubiquitination of TAB1(K294A), TAB1(K319A), and TAB1(K335A) (Fig. 4H). We further constructed a TAB1 mutant TAB1(3KA), in which all three lysine residues were replaced with alanine residue. We found that the K11-linked polyubiquitination of TAB1(3KA) was abolished in the presence of TRIM26 (Fig. 4I). Collectively, these data described that the three lysines (K294, 319, and 335) of TAB1 are the major ubiquitination sites mediated by TRIM26. Altogether, these results suggested that TRIM26 conjugates the K11-linked polyubiquitin chain at Lys294, Lys319, and Lys335 of TAB1, which leads to the activation of downstream signaling pathway.
Other reported E3 ligases for TAB1 is not involved in TRIM26-mediated K11-linked TAB1 polyubiquitination
Three E3 ligases RNF114, ITCH, and MEKK1 have been reported to promote TAB1 ubiquitination through various polyubiquitin chains [22, 32, 33]. ITCH and MEKK1 catalyzes the K48-linked and K63-linked polyubiquitination of TAB1, respectively. RNF114 forms K11-, K27-, and K48-linked polyubiquitination chains on TAB1 during the maternal-to-zygotic transition. To investigate whether TRIM26 induces K11-linked polyubiquitination exclusively, we first examined the level of K11-linked polyubiquitination on TAB1 in the presence of ITCH and MEKK1 expression plasmids. We found that both MEKK1 and ITCH unable to promote K11-linked ubiquitination of TAB1(Fig. S7a, c), indicating ITCH and MEKK1 could not catalyze K11-linked polyubiquitination of TAB1. Because RNF114 was reported to promote K11-linked TAB1 ubiquitination, we next investigated whether TRIM26 and RNF114 could work together to mediated K11-linked ubiquitination. As expected, we found overexpression of RNF114 could increase K11-linked polyubiquitination of TAB1 in HEK293T cells (Fig. S7e). We further used siRNA to knockdown the expression of endogenous TRIM26 in HEK293T cells, we found the level of K11-linked TAB1 polyubiquitination in the presence of RNF114 expression plasmid was slightly decreased in TRIM26 siRNA transfected cells (Fig. S7e), indicating both TRIM26 and RNF114 could promote K11-linked polyubiquitination of TAB1 separately.
Next, we examined whether TRIM26-mediated K11-linked polyubiquitination on TAB1 could impact the TAB1 polyubiquitination mediated by RNF114, ITCH, and MEKK1. We found overexpressed TRIM26 had no influence on MEKK1-mediated K63-linked ubiquitination, ITCH-mediated K48-linked ubiquitination, and RNF114-mediated K48-linked ubiquitination (Fig. S7b, d, f). These data suggested that RNF114, ITCH, and MEKK1 are not involved in TRIM26-mediated K11-linked TAB1 polyubiquitination.
TRIM26 enhances TAK1 phosphorylation and subsequent NF-κB and MAPK signaling
As an important component of the TAK1-TABs complex, TAB1 is essential for TAK1 kinase activity. Since we demonstrated that TRIM26 promotes K11-linked polyubiquitination of TAB1 and positively regulates TLRs-induced production of proinflammatory cytokines and activation of NF-κB and MAPK signaling, we hypothesized TRIM26 is essential for TAK1 activation. Indeed, LPS-, Poly(I:C)-, and Pam3CSK4-induced TAK1 phosphorylation was lower in PMs from Trim26−/− mice than in those from Trim26+/+ mice (Fig. 5A–C). In addition, TNF-α- or IL-1β-induced TAK1 phosphorylation was attenuated by Trim26 deficiency (Supplementary Fig. S8a, b).
Fig. 5. TRIM26 enhances phosphorylation and complex assembly of TAK1.
A–C The Trim26+/+ and Trim26–/– peritoneal macrophages were stimulated with LPS (200 ng/ml), Poly(I:C) (20 μg/ml), or Pam3CSK4 (1 μg/ml) for the indicated times, and immunoblotting analysis were performed using anti-p-TAK1 or anti-TAK1 Abs. D Immunoblotting analysis of phosphorylated (p) and total TAK1, IKKα/β, p65, ERK, JNK, and p38 in Trim26+/+ and Trim26–/– peritoneal macrophages. Trim26–/– peritoneal macrophages reconstituted with vectors for mTrim26 or mTrim26(C16A) stimulated with LPS (200 ng/ml) for the indicated times. E qPCR analysis of Tnfα, Il6, and Il12b mRNA expression in Trim26+/+and Trim26–/– peritoneal macrophages. Trim26–/– peritoneal macrophages reconstituted with vectors for mouse mTrim26 or mTrim26(C16A) stimulated with LPS (200 ng/ml) for 6 h. Data are shown as mean ± SD of triplicates from one representative experiment in E. *p < 0.05, **p < 0.01 (one-way analysis of variance, ANOVA). Similar results were obtained in three independent experiments.
To directly confirm TRIM26 was required for TAK1 activation, we rescued Trim26 expression in Trim26−/− PMs through infection with lentiviral mouse Trim26 (mTrim26) overexpression vector. Overexpression of mTrim26 but not the enzymatic mutant C16A could restore LPS-induced phosphorylation of TAK1, IKKα/β, p65, ERK, JNK, and p38 (Fig. 5D). Consistently, mTrim26 instead of mTrim26(C16A) rescued the expression of Tnfα, Il6, and Il12b mRNA upon LPS stimulation (Fig. 5E).
As an E3 ligase, TRAF6 could catalyzes K63-linked polyubiquitination of TAK1, which is essential for TAK1 phosphorylation and activation of downstream kinases [34]. Co-IP and immunoblot analysis showed that TRIM26 coprecipitated with TRAF6 (Supplementary Fig. S9a). However, the interaction between TRAF6 and TAK1 was not affected in the presence of TRIM26 (Supplementary Fig. S9b). Importantly, TRAF6-mediated K63-linked polyubiquitination of TAK1 was also not impaired in the presence of TRIM26 (Fig. S9c), suggesting that TRIM26 regulates TAK1 activation independent of TRAF6. Together, these data demonstrated that TRIM26 regulates TAK1 activation and downstream NF-κB and MAPK signaling through the K11-linked ubiquitination of TAB1.
TRIM26 deficiency protects mice from LPS-induced septic shock
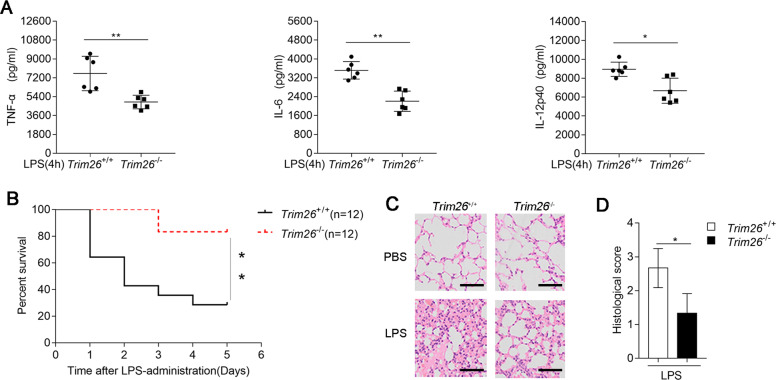
To gain insight into the functions of TRIM26 in the regulation of TLR-mediated inflammation in vivo, we monitored inflammatory cytokines expression as well as septicemia-induced death in Trim26+/+ and Trim26−/− mice after intraperitoneal (i.p.) administration of LPS. As shown in Fig. 6A, LPS-triggered serum levels of TNF-α, IL-6, and IL-12p40 were decreased in Trim26−/− mice than in WT mice. Consistent herewith, Trim26−/− mice experienced later death onset and exhibited a lower death rate (Fig. 6B) and they showed less lung inflammation and lower pathological assessment of lung severity scores (Fig. 6C, D) than their WT counterparts. Collectively, these data suggested that TRIM26 positively regulates LPS-mediated inflammatory innate immune response in vivo.
Fig. 6. TRIM26 deficiency protects mice from LPS-induced septic shock.
A Sex- and age-matched Trim26+/+ and Trim26–/– mice (n = 6) were injected intraperitoneally with LPS (40 μg/g) for 4 h. ELISA analysis of TNF-α, IL-6, and IL-12p40 in the serum of Trim26+/+ and Trim26–/– mice. B Trim26+/+ and Trim26–/– littermates (n = 12) were stimulated with LPS (40 μg/g) as in A. The survival of mice was monitored over the following 5 days. C Trim26+/+ and Trim26–/– mice were injected intraperitoneally with LPS (40 μg/g) for 4 h, H&E experiments were performed on the lung injury of the mice. D Histological analysis of colon tissues described in C. Scale bars, 100 μm. Data are shown as mean ± SD in A; n = 6 biologically independent animals. *p < 0.05, **p < 0.01; A Student’s t test; B log-rank t-est. Similar results were obtained in three independent experiments.
TRIM26 deficiency attenuates dextran sodium sulfate (DSS)-induced colitis
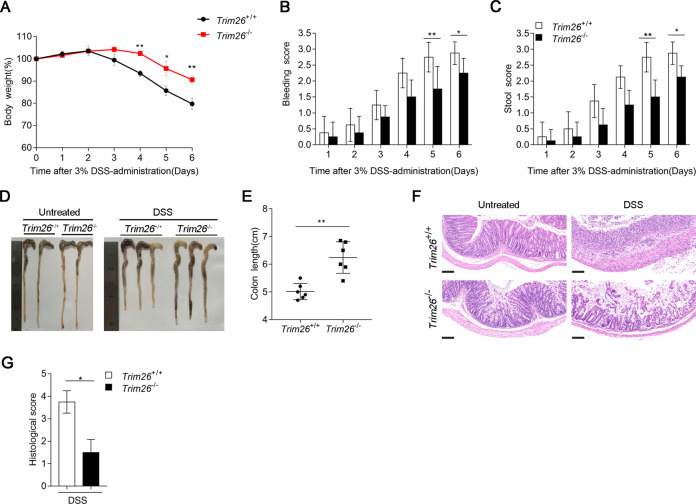
The DSS model is among the most extensively used models of inflammatory bowel disease (IBD) [35, 36]. Because NF-κB signaling plays important roles in IBD [36], thus, we examined the effects of Trim26 deficiency on DSS-induced acute colitis. First, we assessed the clinical features of age- and sex-matched WT and Trim26-deficient mice after treated with 3% DSS in the drinking water over a 6-day period. Compared with Trim26+/+ mice, Trim26−/− mice displayed attenuated colitis, as indicated by less weight loss, rectal bleeding scores, and stool consistency scores (Fig. 7A–C). To further assess the severity of colitis, colon length was measured on day 6. Colons of Trim26−/− mice were longer than those of Trim26+/+ mice administered DSS (Fig. 7D, E). Histopathological analysis revealed that the colonic mucosa of Trim26−/− mice was more intact, without apparent loss of crypt structures and mucosal ulceration, and Trim26−/− mice showed less inflammatory cell infiltration in colonic tissues than their WT littermates after DSS treatment (Fig. 7F). The pathological assessment of colitis severity scores reflected the same phenomenon (Fig. 7G). All together, these results suggested that Trim26 deficiency attenuates the severity of DSS-induced colitis.
Fig. 7. TRIM26 deficiency attenuates dextran sodium sulfate (DSS)-induced colitis.
A Sex- and age-matched Trim26+/+ and Trim26–/– mice (n = 6) were given 3% DSS in their drinking water for 6 days, body weight were measured daily. B, C Rectal bleeding score and stool consistency of Trim26+/+ and Trim26–/– mice were scored daily. D, E Macroscopic appearances and colon lengths of Trim26+/+ and Trim26–/– mice were measured on day 6. F Histopathological changes in colon tissue were examined by H&E staining. G Histological analysis of colon tissues described in f. Scale bars, 500 um. Data are shown as mean ± SD in B, C, E; n = 6 biologically independent animals. *p < 0.05, **p < 0.01; A one-way analysis of variance, ANOVA; B, C, E Student’s t test. Similar results were obtained in three independent experiments.
Discussion
TAK1 is a key regulator of NF-κB activation and proinflammatory cytokines production in response to stimulation with TLRs and cytokines. The activation of TAK1 depends on the aggregation of the TAK1-TABs complex that induces auto- or para-phosphorylation of TAK1 [17]. Over the past decade, a large number of studies have shown that TAK1 activity is extensively regulated by phosphorylation, ubiquitination, and binding to the regulatory binding partners TAB1-3 [19, 37–40]. As a critical adapter in NF-κB and MAPK signaling, TAB1 has been reported to regulate through protein ubiquitination. MEKK1 has been characterized as an essential E3 catalyzing K63-linked TAB1 polyubiquitination to activate MAPKs in response to TGF-β [22]. ITCH catalyzes the K48-linked polyubiquitination of TAB1 and promotes TAB1 degradation in TNF-α-induced inflammation [33]. However, whether TAB1 could be regulated through nonclassical protein ubiquitination remained unknown.
Here, we identified TRIM26 as a novel E3 ligase to induce a nonclassical polyubiquitination, K11-linked polyubiquitination of TAB1, which led to TAK1 phosphorylation and the subsequent transcription of proinflammatory cytokines. First, we generated Trim26−/− mice and demonstrated that Trim26 deficiency attenuated the production of proinflammatory cytokines in response to TLR ligands, TNF-α, and IL-1β stimulation. In addition, the challenge of Trim26−/− mice with LPS resulted in inflammatory responses that were less lethal than those in wild-type mice. Second, using in vitro and in vivo Co-IP assays, we showed that TRIM26 directly interacted with TAB1. We also found that TRIM26 catalyzed K11-linked polyubiquitination of TAB1 at Lys294, Lys319, and Lys335, but not other adapters like MyD88, TRAF6, TAB2. Third, we discovered that TLRs-, TNF-α-, and IL-1β-induced phosphorylation of TAK1 was markedly decreased in Trim26-deficient mice.
Yang et al. reported that RNF114 forms K11-, K27-, and K48-linked polyubiquitination chains on TAB1 during the maternal-to-zygotic transition, which leads to the degradation of TAB1 [32]. While TRIM26 only conjugates K11-linked, but not K48-linked or K27-linked polyubiquitin chain on TAB1, thus TRIM26 has no effect on the regulation of TAB1 stability. Although RNF114 and TRIM26 can both conjugates K11-linked polyubiquitination on TAB1, our experiments demonstrated that their effects are independent on each other. RNF114 could promote the polyubiquitinate of TAB1 in TRIM26 siRNA knockdown HEK293T cells (Supplementary Fig. S7e). In addition, MEKK1 and ITCH, the enzymes that have been reported to modify TAB1 polyubiquitination, unable to promote K11-linked ubiquitination of TAB1 (Fig. S7a, c). Thus, our study revealed a new mechanism underlying phosphorylation and activation of TAK1 mediated by TRIM26 catalyzing K11-linked polyubiquitination of TAB1 in the TLRs-mediated inflammatory response.
Different from K48- or K63-linked polyubiquitination chains, the function of K11-linked polyubiquitination remains unclear. In most research, like K48-linked polyubiquitination, K11 ubiquitin chains could also increase protein degradation [41, 42]. However, K11, K63, and linear ubiquitin linkages contribute to the activation of the NF-κB and MAPK signaling [43]. RNF26 conjugates K11-linked polyubiquitination of STING, thus protecting STING from K48-linked polyubiquitination and degradation [44]. How K11-linked polyubiquitination of TAB1 regulates TAK1 activation is not clear. It has been reported that ubiquitination of TAK1-TABs complex can regulate the assembly of this complex and downstream signaling [22, 38, 39]. We therefore hypothesized that K11-linked polyubiquitination of TAB1 enhancing the assembly of TAK1-TABs complex. Indeed, Co-IP experiments with TAB1 antibody showed that TAB1-TAK1 and TAB1-TAB2 associations were attenuated in Trim26-deficient PMs compared to that in WT PMs upon LPS stimulation (data not shown). Another possible explanation is that K11-linked polyubiquitination might cause a conformational change in the substrate that renders it more accessible for the binding of proteins as reported for the formation of Rv0222-SHP1-TRAF6 complex [45]. Overall, the detailed mechanism by which K11-linked polyubiquitination of TAB1 regulates the activation of TAK1 deserves further investigation.
In conclusion, using Trim26-knockout and Trim26-transgenic mice, we demonstrated that TRIM26 is a TLRs-inducible protein that acts as a positive regulator of TLRs-, TNF-α-, and IL-1β-induced inflammatory responses by promoting TAB1 K11-linked polyubiquitination and TAK1 phosphorylation. Our findings identify TRIM26 as a previously unrecognized component of the inflammatory immune response. This study reveals that through catalyzing TAB1 ubiquitination and TAK1 activation, TRIM26 plays an important role in the regulation of the inflammatory innate immune response. TRIM26 may serve as a potential target for new therapeutic interventions against inflammation-associated diseases.
Materials and methods
Mice and cells
Trim26−/− mice on a C57BL/6 background were generated by Cyagen Biosciences Inc. (Guangzhou, China) using TALEN technology. The Trim26-deficient mice were genotyped by sequencing of a PCR fragment (250 bp) of the TALEN-targeting region amplified from genomic DNA isolated from tail tips using the following primers: forward 5′-GCTTGAGCAATCCACAGGGGTATC-3′ and reverse 5′-GCCACACGGGTCGGATGTTCT-3′. Trim26Tg mice were described previously [29]. Human HEK293T, THP-1, and Hela cells were obtained from the American Type Culture Collection (Manassas, VA). Mouse primary PMs and BMDMs were prepared as described [36]. MEFs were isolated from Trim26+/+ and Trim26−/− embryos (day 13.5). The cells were cultured in DMEM supplemented with 10% FCS (Invitrogen-Gibco), 100 U/ml penicillin, and 100 μg/ml streptomycin at 37 °C in the presence of 5% CO2.
Reagents and Abs
LPS (Escherichia coli, 055:B5) was purchased from Sigma (St. Louis, MO). Poly(I:C) (tlrl-picw), R848 (tlrl-r848), and Pam3CSK4 (tlrl-pms) were purchased from Invivogen. TNF-α (410-MT-010), IL-1β (401-ML-0050) recombinant mice protein were purchased from R&D Systems. Poly(I:C), LPS, R848, Pam3CSK4, IL-1β, and TNF-α were used at a final concentration of 20 μg/ml, 200 ng/ml, 10 μg/ml, 1 μg/ml, 20 ng/ml, and 20 ng/ml, respectively. PMA (tlrl-pma) was purchased from InvivoGen. The following Abs were used at a dilution of 1:1000 for western blot analysis, unless otherwise stated: Abs targeting p65 (8242), p-p65 (Ser536) (3033), IKKβ (8943), p-IKKα(Ser176)/IKKβ(Ser177) (2078), p38 MAPK (8690), p-p38 MAPK (Thr180/Tyr182) (9215), JNK (9252), phospho-JNK (Thr183/Tyr185) (4671), p44/42 MAPK (Erk1/2) (4695), p-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (4377), p-TAK1 (Thr187) (4536), TAK1 (5206), and ubiquitin (3936) were from Cell Signaling Technology (Beverly, MA); Abs against K11-linkage ubiquitin (A18197) was from ABclonal. Abs against TRIM26 (sc-393832), TAB1 (sc-166138), and β-actin (Santa Cruz, sc-47778) were from Santa Cruz Biotechnology (Santa Cruz, CA); Ab against HA (26183) was from Thermo Fisher Scientific; Ab against Myc(A190-105A) was from Bethl Laboratories; Protein A/G PLUS-Agarose (sc-2003) used for immunoprecipitation was from Santa Cruz Biotechnology (Santa Cruz, CA); FLAG (M2) (F1804) Abs were from Sigma-Aldrich; HRP-conjugated goat anti-mice or rabbit IgG Abs used for Western blotting were from Calbiochem.
Plasmid constructs
The Flag-TRIM26 expression plasmid was described previously [29]. Plasmids encoding TRIM26 mutants, including C16A and ΔR, were generated using the KOD-Plus-Mutagenesis kit (Toyobo, Osaka, Japan). Myc-TRIM26 was generated by subcloning the TRIM26-coding sequence into the pCDNA3.1 vector. HA-ubiquitin and other plasmids were described previously [46]. All constructs were confirmed by DNA sequencing. An expression vector for Flag-TAB2 was obtained from Dr. Hui Xiao (Institut Pasteur of Shanghai, CAS, Shanghai, China). Expression vectors for HA-K11R was provided by Dr. Chen Wang (School of Life Science and Technology, China Pharmaceutical University, China). Expression vectors for TRAF6, IKKβ, and TAK1 were provided by Dr. Michael Karin (University of California at San Diego, San Diego, CA). Expression vector for ITCH-Flag was provided by Dr. Zhengfan Jiang (Peking University). Expression vector for RNF114-Flag was provided by Dr. Ran Huo (Nanjing Medical University). Expression vector for MEKK1-Flag was provided by Dr. Chao Xu (Shandong Provincial Hospital Affiliated to Shandong University). Other plasmids used in this study were described previously [29].
siRNA transfection
The siRNA target sequences for transient silencing were as follows: 5′-CCAAGGACUUCGCCAACAA-3′ (si-mTRIM26), 5′-GAGUCACAGGAACUCAUCU-3′ (si-hTRIM26), and 5′-UUCUCCGAACGUGUCACGU-3′ (si-Ctrl).
Quantitative real-time PCR
Total RNA was isolated from cells using the RNA fast 200 kit (Fastagen). Complementary DNA synthesis was performed using reverse transcriptase (Takara), followed by qPCR using A LightCycler (Roche) and QuantiTect SYBR Green PCR Kit (Roche) according to the manufacturers.
Detection of cytokine production
Mouse sera or cell supernatants were used to examine the level of cytokine secretion by ELISA. The concentrations of TNF-α, IL-6, and IL-12p40 were measured using commercial ELISA kits (Dakewe Biotech, Shenzhen, China).
Co-immunoprecipitation and immunoblot analysis
For immunoprecipitation, whole-cell extracts were collected 24 h after transfection and were lysed in immunoprecipitation buffer containing 50 mM EDTA, 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1.0% (v/v) Nonidet P40, and protease inhibitor cocktail (Sigma). The lysates were centrifuged at 14,000 × g at 4 °C for 10 min. The supernatants were collected and incubated with protein G Plus-Agarose immunoprecipitation reagent together with 1 μg of the relevant antibodies. After incubation at 4 °C for 12 h, the beads were washed with 1 ml of immunoprecipitation buffer four times. Immunoprecipitation was eluted by boiling with a 1% (w/v) SDS sample buffer. For western blot analysis, immunoprecipitation or whole-cell lysates were subjected to SDS-PAGE, and the proteins were transferred onto nitrocellulose membranes, followed by immunoblotting with the indicated Abs, as previously described [29].
Ubiquitination assays
To evaluate the ubiquitination of TAB1 in HEK293T cells, HEK293T cells were transfected with Myc-TAB1, HA-Ub (WT), HA-Ub (K11), HA-Ub (K11R), and Flag-tagged WT or mutant TRIM26, and whole-cell extracts were immunoprecipitated with anti-Myc and analyzed by immunoblotting with an anti-HA antibody. To evaluate the ubiquitination of endogenous TAB1, macrophages were stimulated with LPS (200 ng/ml), and whole-cell lysates were immunoprecipitated with anti-TAB1 and analyzed by immunoblotting with the anti-Ub antibody.
In vitro binding and ubiquitination assays
TAB1 and TRIM26 recombinant proteins were prepared using a TNT Quick Coupled Transcription/Translation System (Promega). For the binding assay, TAB1 and TRIM26 were mixed together, followed by immunoprecipitation with the TAB1 antibody and western blotting with the TRIM26 antibody. The ubiquitination assay was conducted as described previously [29].
mTrim26 overexpression lentivirus
pLVX-IRES-Puro-mTrim26 was generated by subcloning the mTrim26-coding sequence into the pLVX-IRES-Puro vector, and the empty pLVX-IRES-Puro was used as a control. Plasmids encoding mTrim26 mutant C16A was generated using the KOD-Plus-Mutagenesis kit (Toyobo, Osaka, Japan). The lentivirus was produced by transient transfection of the pLVX-IRES-Puro-mTrim26 construct or control vector into HEK293T cells using Lipofectamine 2000 (Thermo Fisher Scientific) with pLVX-IRES-Puro, psPAX2, and pMD2.G.
In vivo LPS challenge
For endotoxicity studies, age-matched female Trim26+/+ and Trim26−/− mice (8 weeks old) were challenged with LPS (40 μg/g, administered i.p.). Mouse survival was monitored every 12 h. Female Trim26+/+ and Trim26−/− mice (8 weeks old) were i.p. injected with LPS (40 μg/g) or PBS. After 4 h, the mice were killed, blood was collected, and serum levels of TNF-α, IL-6, and IL-12p40 were measured by ELISA. Lungs from control or LPS-stimulated mice were dissected, fixed in 10% phosphate-buffered formalin, embedded in paraffin, sectioned, stained with hematoxylin and eosin (H&E) solution, and examined by light microscopy for histological changes.
DSS-induced colitis
For the colitis model establishment, age-matched female Trim26+/+ and Trim26−/− mice (6 weeks old) were provided with 3% (w/v) DSS in the drinking water for 6 days. Body weight, stool consistency, and the presence of occult blood were monitored every 24 h. Stool consistency and occult blood were scored as described previously [35, 36]. On day 6, the entire colon was excised and its length was measured. Then, 0–0.5 cm control or DSS-induced mouse colon tissues close to the rectum were collected. Colitis was assessed by H&E staining and severity scores were calculated as described previously [35, 36].
Statistical analysis
Statistical significance was determined using two-tailed Student’s t tests or two-way ANOVA analysis (where more than two groups of data were compared). Statistical significance in examining survival among Trim26+/+ and Trim26−/− mice was performed via the Kaplan–Meier survival by GraphPad Prism6.0. Values of p < 0.05 were considered to be statistically significant.
Supplementary information
Acknowledgements
We thank Dr. Hui Xiao (Institut Pasteur of Shanghai, CAS, Shanghai, China) for the Flag-TAB2 expression plasmid; Dr. Chen Wang (School of Life Science and Technology, China Pharmaceutical University, China) for the HA-K11R expression plasmid; and Dr. Michael Karin (University of California at San Diego, San Diego, CA) for the expression plasmids of TRAF6, IKKβ, and TAK1; Dr. Zhengfan Jiang (Peking University) for the Flag-ITCH expression plasmid; Dr. Ran Huo (Nanjing Medical University) for the Flag-RNF114 expression plasmid; Dr. Chao Xu (Shandong Provincial Hospital Affiliated to Shandong University) for the Flag-MEKK1 expression plasmid.
Author contributions
CG conceived and designed the study. JZ and BC performed most of the experiments with help from ZS. LZ, YZ, CM, and FY contributed to the discussion and provided reagents. CG and BL supervised the study. JZ and BL analyzed the data. CG, BL, and JZ wrote the paper.
Funding
This work was supported by the Natural Science Foundation of China (81930039, 31730026, 81525012 to CG and 31900680 to BL). This work was also supported by the Postdoctoral Science Foundation of China (BX201700146 to BL) and Shandong Provincial Natural Science Foundation (ZR2018BC021 to BL).
Data availability
The data within the article and its Supplementary Information files that support this study are available from the authors upon request.
Compliance with ethical standards
Conflict of interest
The authors declare no competing interests.
Ethical approval
All mouse experiments were carried out according to the Guidelines of the China Animal Welfare Legislation, as approved by the Ethics Committee of Scientific Research of Shandong University Qilu Hospital, Jinan, Shandong Province, China (Permit number: KYLL-2017(KS)-361).
Footnotes
Edited by: M. Piacentini
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jian Zhao, Baoshan Cai
Contributor Information
Bingyu Liu, Email: liubingyu@sdu.edu.cn.
Chengjiang Gao, Email: cgao@sdu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-021-00803-1.
References
- 1.Majer O, Liu B, Barton GM. Nucleic acid-sensing TLRs: trafficking and regulation. Curr Opin Immunol. 2017;44:26–33. doi: 10.1016/j.coi.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on toll-like receptors. Nat Immunol. 2010;11:373–84. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 3.Bonham Kevin S, Orzalli Megan H, Hayashi K, Wolf Amaya I, Glanemann C, Weninger W, et al. A promiscuous lipid-binding protein diversifies the subcellular sites of toll-like receptor signal transduction. Cell. 2014;156:705–16. doi: 10.1016/j.cell.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barton GM, Medzhitov R. Toll-like receptor signaling pathways. Science. 2003;300:1524–5. doi: 10.1126/science.1085536. [DOI] [PubMed] [Google Scholar]
- 5.Fitzgerald KA, Kagan JC. Toll-like receptors and the control of immunity. Cell. 2020;180:1044–66. doi: 10.1016/j.cell.2020.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin S-C, Lo Y-C, Wu H. Helical assembly in the MyD88–IRAK4–IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–90. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu J, Chen ZJ. Innate immune sensing and signaling of cytosolic nucleic acids. Annu Rev Immunol. 2014;32:461–88. doi: 10.1146/annurev-immunol-032713-120156. [DOI] [PubMed] [Google Scholar]
- 8.Gay NJ, Symmons MF, Gangloff M, Bryant CE. Assembly and localization of Toll-like receptor signalling complexes. Nat Rev Immunol. 2014;14:546–58. doi: 10.1038/nri3713. [DOI] [PubMed] [Google Scholar]
- 9.Elshabrawy HA, Essani AE, Szekanecz Z, Fox DA, Shahrara S. TLRs, future potential therapeutic targets for RA. Autoimmun Rev. 2017;16:103–13. doi: 10.1016/j.autrev.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burgueno JF, Abreu MT. Epithelial Toll-like receptors and their role in gut homeostasis and disease. Nat Rev Gastroenterol Hepatol. 2020;17:263–78. doi: 10.1038/s41575-019-0261-4. [DOI] [PubMed] [Google Scholar]
- 11.Joosten LA, Abdollahi-Roodsaz S, Dinarello CA, O’Neill L, Netea MG. Toll-like receptors and chronic inflammation in rheumatic diseases: new developments. Nat Rev Rheumatol. 2016;12:344–57. doi: 10.1038/nrrheum.2016.61. [DOI] [PubMed] [Google Scholar]
- 12.Stewart CR, Stuart LM, Wilkinson K, van Gils JM, Deng J, Halle A, et al. CD36 ligands promote sterile inflammation through assembly of a Toll-like receptor 4 and 6 heterodimer. Nat Immunol. 2010;11:155–61. doi: 10.1038/ni.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oeckinghaus A, Hayden MS, Ghosh S. Crosstalk in NF-kappaB signaling pathways. Nat Immunol. 2011;12:695–708. doi: 10.1038/ni.2065. [DOI] [PubMed] [Google Scholar]
- 14.Ajibade AA, Wang HY, Wang RF. Cell type-specific function of TAK1 in innate immune signaling. Trends Immunol. 2013;34:307–16. doi: 10.1016/j.it.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 15.Shibuya H, Yamaguchi K, Shirakabe K, Tonegawa A, Gotoh Y, Ueno N, et al. TABi: An Activator Of the TAK1 MAPKKK in TGF-β signal transduction. Science. 1996;272:1179–82. doi: 10.1126/science.272.5265.1179. [DOI] [PubMed] [Google Scholar]
- 16.Kanayama A, Seth RB, Sun L, Ea C-K, Hong M, Shaito A, et al. TAB2 and TAB3 activate the NF-κB pathway through binding to polyubiquitin chains. Mol Cell. 2004;15:535–48. doi: 10.1016/j.molcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 17.Wang C, Deng L, Hong M, Akkaraju GR, Inoue J-I, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–51. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 18.Li Q, Yan J, Mao AP, Li C, Ran Y, Shu HB, et al. Tripartite motif 8 (TRIM8) modulates TNF- and IL-1-triggered NF-B activation by targeting TAK1 for K63-linked polyubiquitination. Proc Natl Acad Sci USA. 2011;108:19341–6. doi: 10.1073/pnas.1110946108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan Y, Yu Y, Shi Y, Sun W, Xie M, Ge N, et al. Lysine 63-linked polyubiquitination of TAK1 at lysine 158 is required for tumor necrosis factor α- and Interleukin-1β-induced IKK/NF-κB and JNK/AP-1 activation. J Biol Chem. 2010;285:5347–60. doi: 10.1074/jbc.M109.076976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dou Y, Xing J, Kong G, Wang G, Lou X, Xiao X, et al. Identification of the E3 ligase TRIM29 as a critical checkpoint regulator of NK cell functions. J Immunol. 2019;203:873–80. doi: 10.4049/jimmunol.1900171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen SY, Zhang HP, Li J, Shi JH, Tang HW, Zhang Y, et al. Tripartite motif‐containing 27 attenuates liver ischemia/reperfusion injury by suppressing TAK1 via TAB2/3 degradation. Hepatology. 2020;73:738–58. [DOI] [PMC free article] [PubMed]
- 22.Charlaftis N, Suddason T, Wu X, Anwar S, Karin M, Gallagher E. The MEKK1 PHD ubiquitinates TAB1 to activate MAPKs in response to cytokines. EMBO J. 2014;33:2581–96. doi: 10.15252/embj.201488351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Huizen M, Kikkert M. The role of atypical ubiquitin chains in the regulation of the antiviral innate immune response. Front. Cell Dev. Biol. 2020;7:1–8. [DOI] [PMC free article] [PubMed]
- 24.Ran Y, Zhang J, Liu LL, Pan ZY, Nie Y, Zhang HY, et al. Autoubiquitination of TRIM26 links TBK1 to NEMO in RLR-mediated innate antiviral immune response. J Mol Cell Biol. 2016;8:31–43. doi: 10.1093/jmcb/mjv068. [DOI] [PubMed] [Google Scholar]
- 25.Edmonds MJ, Carter RJ, Nickson CM, Williams SC, Parsons JL. Ubiquitylation-dependent regulation of NEIL1 by Mule and TRIM26 is required for the cellular DNA damage response. Nucleic Acids Res. 2017;45:726–38. doi: 10.1093/nar/gkw959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hatakeyama S. TRIM family proteins: roles in autophagy, immunity, and carcinogenesis. Trends Biochemical Sci. 2017;42:297–311. doi: 10.1016/j.tibs.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 27.Tao J, Luo M, Sun H, Zhao H-M, Sun Q-S, Huang Z-M. Overexpression of tripartite motif containing 26 inhibits non-small cell lung cancer cell growth by suppressing PI3K/AKT signaling. Kaohsiung J Med Sci. 2020;36:417–22. doi: 10.1002/kjm2.12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer M, Gaudieri S, Rhodes DA, Trowsdale J. Cluster of TRIM genes in the humanMHC class I region sharing the B30.2 domain. Tissue Antigens. 2002;61:63–71. doi: 10.1034/j.1399-0039.2003.610105.x. [DOI] [PubMed] [Google Scholar]
- 29.Wang P, Zhao W, Zhao K, Zhang L, Gao C. TRIM26 negatively regulates interferon-beta production and antiviral response through polyubiquitination and degradation of nuclear IRF3. PLoS Pathog. 2015;11:e1004726. doi: 10.1371/journal.ppat.1004726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo L, Lucas RM, Liu L, Stow JL. Signalling, sorting and scaffolding adaptors for Toll-like receptors. J Cell Sci. 2019;133:1–10. doi: 10.1242/jcs.239194. [DOI] [PubMed] [Google Scholar]
- 31.Hinz M, Scheidereit C. The IkappaB kinase complex in NF-kappaB regulation and beyond. EMBO Rep. 2014;15:46–61. doi: 10.1002/embr.201337983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Y, Zhou C, Wang Y, Liu W, Liu C, Wang L, et al. The E3 ubiquitin ligase RNF 114 and TAB1 degradation are required for maternal-to-zygotic transition. EMBO Rep. 2017;18:205–16. doi: 10.15252/embr.201642573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Theivanthiran B, Kathania M, Zeng M, Anguiano E, Basrur V, Vandergriff T, et al. The E3 ubiquitin ligase Itch inhibits p38α signaling and skin inflammation through the ubiquitylation of Tab1. Sci Signal. 2015;8:ra22. doi: 10.1126/scisignal.2005903. [DOI] [PubMed] [Google Scholar]
- 34.Chen ZJ. Ubiquitination in signaling to and activation of IKK. Immunological Rev. 2012;246:95–106. doi: 10.1111/j.1600-065X.2012.01108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song H, Liu B, Huai W, Yu Z, Wang W, Zhao J, et al. The E3 ubiquitin ligase TRIM31 attenuates NLRP3 inflammasome activation by promoting proteasomal degradation of NLRP3. Nat Commun. 2016;7:1–11. doi: 10.1038/ncomms13727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu H, Liu H, Zhao X, Zheng Y, Liu B, Zhang L, et al. IKIP Negatively Regulates NF-kappaB Activation and Inflammation through Inhibition of IKKalpha/beta Phosphorylation. J Immunol. 2020;204:418–27. doi: 10.4049/jimmunol.1900626. [DOI] [PubMed] [Google Scholar]
- 37.Hirata Y, Takahashi M, Morishita T, Noguchi T, Matsuzawa A. Post-translational modifications of the TAK1-TAB complex. Int J Mol Sci. 2017;18:1–17. [DOI] [PMC free article] [PubMed]
- 38.Lei CQ, Wu X, Zhong X, Jiang L, Zhong B, Shu HB. USP19 inhibits TNF-alpha- and IL-1beta-triggered NF-kappaB activation by deubiquitinating TAK1. J Immunol. 2019;203:259–68. doi: 10.4049/jimmunol.1900083. [DOI] [PubMed] [Google Scholar]
- 39.Zhou Q, Cheng C, Wei Y, Yang J, Zhou W, Song Q, et al. USP15 potentiates NF-kappaB activation by differentially stabilizing TAB2 and TAB3. FEBS J. 2020;287:3165–83. doi: 10.1111/febs.15202. [DOI] [PubMed] [Google Scholar]
- 40.Hu MM, Yang Q, Zhang J, Liu SM, Zhang Y, Lin H, et al. TRIM38 inhibits TNF- and IL-1-triggered NF- B activation by mediating lysosome-dependent degradation of TAB2/3. Proc Natl Acad Sci USA. 2014;111:1509–14. doi: 10.1073/pnas.1318227111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dynek JN, Goncharov T, Dueber EC, Fedorova AV, Izrael-Tomasevic A, Phu L, et al. c-IAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. EMBO J. 2010;29:4198–209. doi: 10.1038/emboj.2010.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng Y, Liu Q, Wu Y, Ma L, Zhang Z, Liu T, et al. Zika virus elicits inflammation to evade antiviral response by cleaving cGAS via NS1-caspase-1 axis. EMBO J. 2018;37:1–18. [DOI] [PMC free article] [PubMed]
- 43.Kist M, Komuves LG, Goncharov T, Dugger DL, Yu C, Roose-Girma M, et al. Impaired RIPK1 ubiquitination sensitizes mice to TNF toxicity and inflammatory cell death. Cell Death Differ. 2021;28:985–1000. doi: 10.1038/s41418-020-00629-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qin Y, Zhou MT, Hu MM, Hu YH, Zhang J, Guo L, et al. RNF26 temporally regulates virus-triggered type I interferon induction by two distinct mechanisms. PLoS Pathog. 2014;10:e1004358. doi: 10.1371/journal.ppat.1004358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L, Wu J, Li J, Yang H, Tang T, Liang H, et al. Host-mediated ubiquitination of a mycobacterial protein suppresses immunity. Nature. 2020;577:682–8. doi: 10.1038/s41586-019-1915-7. [DOI] [PubMed] [Google Scholar]
- 46.Song G, Liu B, Li Z, Wu H, Wang P, Zhao K, et al. E3 ubiquitin ligase RNF128 promotes innate antiviral immunity through K63-linked ubiquitination of TBK1. Nat Immunol. 2016;17:1342–51. doi: 10.1038/ni.3588. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data within the article and its Supplementary Information files that support this study are available from the authors upon request.