Abstract
Sister chromatid cohesion is essential for cell viability. We have isolated a novel temperature-sensitive lethal mutant named eso1-H17 that displays spindle assembly checkpoint-dependent mitotic delay and abnormal chromosome segregation. At the permissive temperature, the eso1-H17 mutant shows mild sensitivity to UV irradiation and DNA-damaging chemicals. At the nonpermissive temperature, the mutant is arrested in M phase with a viability loss due to a failure to establish sister chromatid cohesion during S phase. The lethal M-phase arrest phenotype, however, is suppressed by inactivation of a spindle checkpoint. The eso1+ gene is not essential for the onset and progression of DNA replication but has remarkable genetic interactions with those genes regulating the G1-S transition and DNA replication. The N-terminal two-thirds of Eso1p is highly homologous to DNA polymerase η of budding yeast and humans, and the C-terminal one-third is homologous to budding yeast Eco1p (also called Ctf7p), which is required for the establishment of sister chromatid cohesion. Deletion analysis and determination of the mutation site reveal that the function of the Eco1p/Ctf7p-homologous domain is necessary and sufficient for sister chromatid cohesion. On the other hand, deletion of the DNA polymerase η domain in Eso1p increases sensitivity to UV irradiation. These results indicate that Eso1p plays a dual role during DNA replication. The C-terminal region acts to establish sister chromatid cohesion, and the N-terminal region presumably catalyzes translesion DNA synthesis when template DNA contains lesions that block regular DNA replication.
Virtually all eukaryotic cells propagate through a process called the cell cycle that consists of four distinct phases, G1, S, G2, and M, whose principal role is to carry out duplication of the chromosomes and subsequent faithful distribution into daughter cells. Commitment to the initiation of the cell cycle is made at a point in late G1 phase called start or restriction point. In the fission yeast, Schizosaccharomyces pombe, passage through start requires the execution of at least two regulatory systems, Res-Cdc10-Rep (Res1p-Cdc10p and Res2p-Cdc10p-Rep2p) transcriptional factor complexes and Cdc2p-Cig2p/Cyc17p cyclin-dependent kinase complex (reviewed in references 37 and 54). Res-Cdc10-Rep complexes activate cell cycle start-specific transcription genes, which contain a cis regulatory element called the MluI cell cycle box. One of those target genes is cdc18+, whose product is a key component of the preinitiation form of origin replication complex and plays a crucial role in loading origins with the replication machinery including DNA polymerases in cooperation with minichromosome maintenance proteins (reviewed in reference 35). Cdc2p-Cig2p activity is also required for origin firing, but its critical target(s) has not been identified yet. These initiation factors and replication factors are highly conserved throughout eukaryotes.
In order to ensure faithful transmission of the duplicated chromosomes to daughter cells, the duplicated sister chromatids are linked together along their entire region during S phase until mid-M phase (13). In fact, the disorder of this linkage, called sister chromatid cohesion, can result in viability loss (10, 14, 29, 42, 50). Sister chromatid cohesion is formed by a set of proteins that are highly conserved through evolution. In the budding yeast Saccharomyces cerevisiae, there is a multiprotein complex called cohesin which contains at least four subunits: Scc1p/Mcd1p (homologous to fission yeast Rad21p), Scc3p, Smc1p, and Smc3p (reviewed in reference 34). Similarly, cohesin of Xenopus laevis egg consists of five subunits, three of which are homologues of Smc1p, Smc3p, and Scc1p/Mcd1p/Rad21p (23). From late G1 until M phase, Scc1p/Mcd1p stays bound to chromosomes but is removed in anaphase by Esp1p (homologous to fission yeast Cut1p)-dependent cleavage and then degraded in G1 by the anaphase-promoting complex–cyclosome (29, 51). Fission yeast Mis4p, which is homologous to budding yeast Scc2p and also required for sister chromatid cohesion, is not a cohesin subunit and belongs to a distinct class of proteins called adherins, which are not degraded in G1 (10). The role of adherin seems to be to load cohesin complex onto chromosomes in G1 (50). On the other hand, budding yeast Eco1p/Ctf7p, the third class of factors essential for sister chromatid cohesion, appears to be required only for the establishment of cohesion during S phase and not for the loading of adherin or cohesin onto chromosomes or for the maintenance of cohesion after S phase (42, 50).
In this report, we describe the fission yeast eso1+ gene, initially identified as the one responsible for a novel temperature-sensitive lethal cdc-like mutant that displays cell elongation and abnormal chromosome segregation. The eso1+ gene encodes an essential protein that contains two domains highly homologous to the budding yeast Rad30p or DNA polymerase η (18, 27, 40; reviewed in reference 53) and Eco1p/Ctf7p, respectively; the Eco1p/Ctf7p domain is essential for establishing sister chromatid cohesion, whereas the DNA polymerase η domain is involved in DNA repair as expected.
MATERIALS AND METHODS
Fission yeast strains, media, and genetic methods.
The S. pombe mutant strains Δmad2, mis4-242, and rad21-K1 and the strain used for visualization of Cen1-green fluorescent protein (GFP) were described previously (11, 21, 47, 49). Strains were cultured in the complete medium YE or the minimal medium MM (EMM2/PM) (2). A nitrogen (ammonium chloride)-free derivative of MM (referred to as MM-N) was used to synchronize cells in G1. When necessary, minimal medium was supplemented with leucine (250 μg/ml for MM and 50 μg/ml for MM-N) and adenine sulfate (100 μg/ml). Transformations were performed using the lithium acetate procedure as described previously (38), and cells were spread on MMA plates (15). Double mutant strains were obtained by crossing single mutants followed by tetrad analysis. 4′,6-Diamidino-2-phenylindole (DAPI) staining was done as described previously (1). Flow cytometry was performed as described previously (48). UV irradiation was performed with a UV cross-linker (XL-1500; Spectronics Co. Ltd.). Cell numbers were determined with a particle counter (Z1; Beckman Coulter, Inc.). Other general genetic manipulations of S. pombe have been described previously (2, 31).
Libraries and vectors.
The S. pombe genomic libraries were constructed by inserting restriction enzyme-digested wild-type (L972) genomic DNA into the pALSK+ vectors. The S. pombe cDNA library has been described previously (39, 45). The pALSK+ vector used for genomic DNA expression was constructed by inserting an autonomously replicating sequence (ars1) and a leucine auxotrophic selection marker gene (LEU2 of S. cerevisiae) into the pBluescript II (SK(+) vector (Stratagene) whose plasmid replication origin was replaced with that of pBR322. The pcL expression vector contains the LEU2 gene, ars1, and the simian virus 40 promoter to drive the expression of the insert. The pREP81-rad21+ plasmid was described previously (49).
Isolation of eso1+ gene.
The eso1+ gene was isolated by complementation of the temperature-sensitive eso1-H17 mutant. The h− eso1-H17 leu1-32 cells were transformed with S. pombe genomic libraries and selected on MMA plates first at 23°C for 24 h and then at 33°C for 4 days. Plasmid DNA clones were recovered in Escherichia coli from candidates and analyzed by Southern hybridization. The eso1+ cDNA was isolated from the cDNA library by colony hybridization and a 5′-RACE (rapid amplification of cDNA ends) PCR method.
Gene disruption.
Gene disruption was performed by one-step gene replacement. The 1.2-kb EcoRI-PstI fragment of the eso1+ gene was replaced with a ura4+ cassette. The linear SpeI-XhoI fragment carrying the replaced gene was transformed into the h−/h+ ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D-18 diploid strain, and stable Ura+ transformants were selected. The proper replacement of the one wild-type allele with the disrupted eso1 construct was confirmed by Southern blot analysis.
Deletion analysis and multicopy suppression analysis.
The eso1+ cDNA clones variously truncated at the 5′ coding region were isolated from the cDNA library and used to construct a series of amino-terminal deletion mutants. A series of carboxyl-terminally truncated mutants were constructed by gradual deletion of a full-length eso1+ cDNA with exonuclease III. All deletion mutants constructed were confirmed for their structure by DNA sequencing and inserted into the pcL vector. The full-length eso1+ gene and its deletion mutants were transformed into h− eso1-H17 leu1-32 cells and plated on MMA. Plates were incubated at 25°C for 24 h and then at 32.5 or 36°C for 4 days or at 25°C for 6 days to determine the numbers of both complemented and stably transfected cells. The complementation (suppression) activities were calculated by dividing the number of colonies formed at the restrictive temperature by the number of colonies formed at 25°C. The ability of various cell cycle control genes to rescue the eso1-H17 mutant was determined similarly.
Nucleotide sequence accession number.
The DDBJ-EMBL-GenBank accession no. for eso1+ is AB039861.
RESULTS
Isolation of the eso1-H17 mutant.
To search for new genes regulating the cell cycle in fission yeast, we generated several temperature-sensitive cell division cycle (cdc) mutants (32, 46). One mutant names eso1-H17 (essential for S-chromatin organization; see below) was chosen for further study.
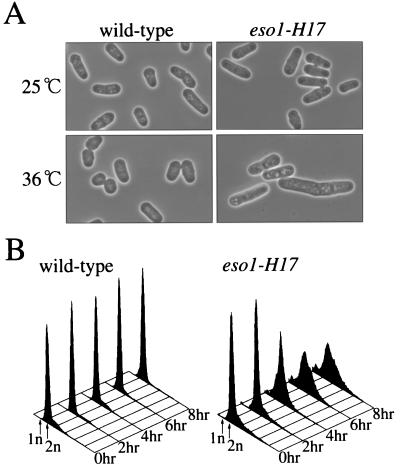
A diploid cell heterozygous for eso1+/eso1-H17 was not temperature sensitive and was indistinguishable in growth properties from wild-type diploid cells, indicating that the eso1-H17 allele is recessive. At the restrictive temperature, eso1-H17 mutant cells ceased proliferation with some cells elongated and others rounded (Fig. 1A), showing morphological heterogeneity. Upon a shift to the nonpermissive temperature, the eso1-H17 mutant cells were arrested with a broad peak of G2 DNA content as shown by flow cytometric analysis (Fig. 1B). Moreover, they quickly lost viability with the generation of aberrant nuclear morphology (see Fig. 2). Similar phenotypes are often associated with S-phase mutants (such as cut5/rad4, swi7/polα, orp1, etc.) (7, 12, 32, 41) that are defective in both DNA replication and S-phase checkpoint. To determine whether eso1-H17 is a DNA replication mutant or not, eso1-H17 cells were arrested by culturing them for 6 h at the nonpermissive temperature and analyzed for the state of chromosomes by pulsed-field gel electrophoresis, in which only completely replicated chromosomes enter the gel (17). Just like those of wild-type and G2-arrested cdc mutant cells, the chromosomes of eso1-H17 cells entered the gel (data not shown). Thus, we concluded that the eso1-H17 mutant is arrested after the completion of chromosome replication.
FIG. 1.
Phenotypes of the eso1-H17 mutant. (A) The eso1-H17 mutant shows an elongated cell morphology. Wild-type (h− leu1-32) and eso1-H17 (h− eso1-H17 leu1-32) cells were inoculated on YEA plates, incubated overnight at the indicated temperatures, and observed under the microscope. (B) The eso1-H17 mutant is arrested with a broad peak of G2 DNA content. Cells were grown to mid-log phase at 23°C in MM medium and shifted up to 34°C. Cells were sampled at 2, 4, 6, and 8 h after the temperature shift and analyzed by flow cytometry.
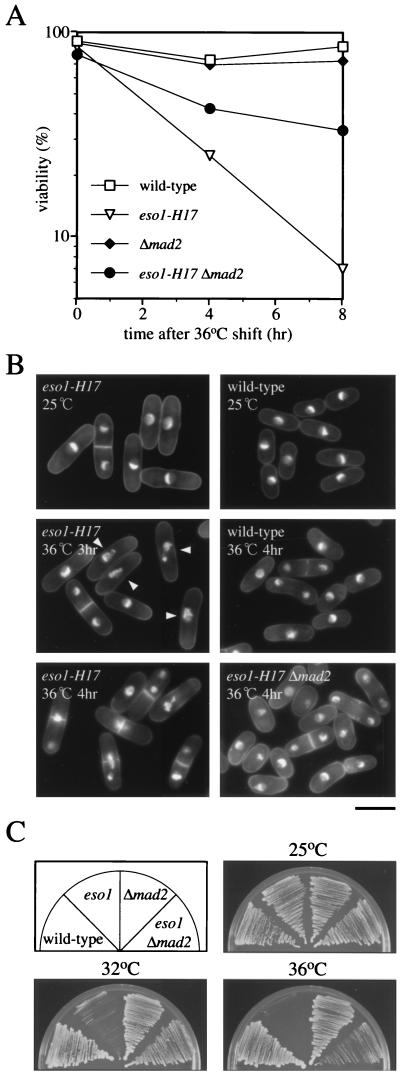
FIG. 2.
Loss of Eso1p function leads to spindle assembly checkpoint-dependent mitotic delay. (A) eso1-H17 cells quickly lost viability upon a shift to the nonpermissive temperature. Wild-type (h− leu1-32), eso1-H17 (h− eso1-H17 leu1-32), Δmad2 (h− mad2::ura4+ ura4-D18 leu1-32), and eso1-H17 Δmad2 (h− eso1-H17 mad2::ura4+ ura4-D18 leu1-32) cells were grown to mid-log phase at 25°C in YEL medium and shifted to 36°C. Cell aliquots were taken at 4 and 8 h after the temperature shift and plated on YEA at 25°C. (B) eso1-H17 cells are defective in chromosome segregation. Cells were grown to mid-log phase at 25°C in YEL medium and shifted to 36°C. Cell aliquots were taken at indicated times, fixed with glutaraldehyde, and stained with DAPI. Arrowheads indicate the cells showing a mitotic delay. Bar, 10 μm. (C) Temperature sensitivity of eso1-H17 cells is partially suppressed by deletion of the mad2+ gene. The indicated cells were inoculated on YEA plates and incubated for 3 days at the indicated temperatures.
eso1-H17 is defective in chromosome segregation.
The most remarkable phenotype of eso1-H17 cells at the restrictive temperature was rapid loss of viability (Fig. 2A) and abnormal nuclear morphology (Fig. 2B). We, therefore, examined this phenotype in depth. At 25°C, wild-type and mutant cells exhibited similar nuclear morphology. But after a shift to 36°C, the eso1-H17 mutant gradually lost interphase cells and reciprocally accumulated cells with abnormal chromosome structures. After 3 h of incubation at 36°C, a number of cells had condensed chromosomes (Fig. 2B, arrowheads within the eso1-H17 36°C 3-h panel), implying that loss of Eso1p function leads to a mitotic delay. Thereafter, many cells showed cut (septation in the absence of nuclear division) and missegregation of chromosomes (Fig. 2B, eso1-H17 36°C 4-h panel).
Arrest of eso1-H17 cells is suppressed by inactivation of a spindle assembly checkpoint.
Mitotic delay and arrest often arise by activation of a spindle assembly checkpoint. To determine whether the mitotic arrest-delay of eso1-H17 cells arose by this mechanism or not, we constructed an eso1-H17 Δmad2 double mutant strain and examined its behavior. Mad2p is not required for normal cell growth but is essential for spindle assembly checkpoint (16, 21). Unlike the eso1-H17 single mutant, the eso1-H17 Δmad2 double mutant was viable and continued to proliferate at 36°C (Fig. 2B, eso1-H17 Δmad2 36°C 4-h panel), indicating that the cell cycle arrest of the original eso1-H17 mutant was caused by the execution of a spindle assembly checkpoint. However, suppression of eso1-H17 by Δmad2 was incomplete, and its temperature sensitivity and missegregation phenotypes persisted to a certain extent in the double mutant (Fig. 2A and C). These results led us to conclude that Eso1p was essential for proper chromosome segregation but dispensable for the step of chromosome segregation per se. Since spindle assembly checkpoint was activated upon inactivation of Eso1p, this conclusion indicates that Eso1p may be required for the proper maintenance of chromosome structure.
eso1-H17 cells are sensitive to DSB.
In addition to the mitotic defect, the eso1-H17 mutant was sensitive to UV irradiation (Fig. 3A) and exposure to methyl methanesulfonate (MMS) or bleomycin (Fig. 3B) even at the permissive temperature. By contrast, the mutant was slightly resistant to base-modifying 4-nitroquinoline-1-oxide (4NQO) (Fig. 3B). Since, unlike 4NQO, UV, MMS, and bleomycin induce double-strand breaks (DSB), these data show that the eso1-H17 mutant is sensitive to DSB.
FIG. 3.
eso1-H17 cells are sensitive to DNA DSB. (A) eso1-H17 cells are sensitive to UV irradiation. Wild-type (h− leu1-32) and eso1-H17 (h− eso1-H17 leu1-32) cells were plated on YEA, irradiated with various doses of UV, and incubated for 7 days at 27°C. (B) eso1-H17 cells are sensitive to MMS and bleomycin but not to 4NQO. Approximately 104, 103, 102, and 10 cells of the wild type (upper spots) and the eso1-H17 mutant (lower spots) were spotted on normal YEA plates (referred to as control) or YEA plates containing MMS (0.004% [vol/vol]), bleomycin (0.025% [wt/vol]), or 4NQO (0.04 μM). Plates were incubated at 27°C for 3 days (control) or 4 days (others).
Unlike damage checkpoint-deficient rad mutants, the eso1-H17 cells exposed to DSB-inducing chemicals were elongated with a single nucleus and did not show any detectable checkpoint defects (data not shown). These results suggest that the eso1-H17 mutant retains proper DNA damage checkpoint but is defective in either repair of DSB or recovery from DSB-caused cell cycle arrest or both.
Eso1p is required during S phase.
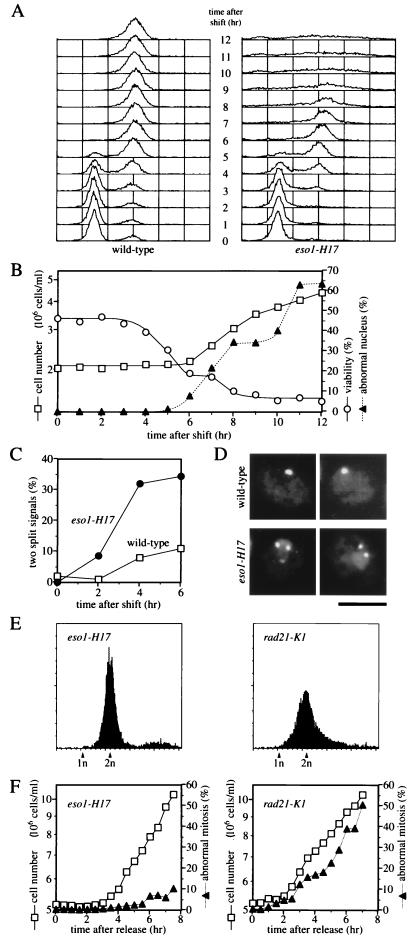
To determine in which phase of the cell cycle Eso1p is required to function, eso1-H17 cells were synchronized to G1 by nitrogen starvation and then released to start the cell cycle in nitrogen-rich growth medium at the nonpermissive temperature. Cells were harvested every hour and assayed for viability by determining the number of colonies formed at the permissive temperature. The mutant cells entered and proceeded through the phase of DNA replication without any significant delay (Fig. 4A), but their viability decreased steeply between 3 and 6 h, the period in which DNA synthesis took place (Fig. 4B). Abnormal nuclear morphologies were frequently seen between 6 and 8 h, the period of mitosis (Fig. 4B). These results indicate that the function of Eso1p needs to be executed at least during S phase for proper cell cycling.
FIG. 4.
Eso1p is required for the establishment of sister chromatid cohesion during S phase but not for its maintenance in G2 and M phases. (A) DNA replication takes place without any significant delay in eso1-H17 mutant cells. Wild-type (h− leu1-32) and eso1-H17 (h− eso1-H17 leu1-32) cells were grown to mid-log phase at 25°C and then nitrogen starved in MM-N medium for 24 h to be arrested in G1. Cells were then released by transfer into MM medium preincubated at 36°C. Cell aliquots were taken every hour and analyzed for S-phase onset and progression by flow cytometry. (B) eso1-H17 cells lose viability during S phase. To determine cell number (open squares), eso1-H17 cell aliquots were taken every hour and counted. To determine cell viability (open circles), eso1-H17 cell aliquots taken every hour were plated on YEA at 25°C. To determine percent abnormal nuclear cells (filled triangles), cells were fixed with 70% ethanol and stained with DAPI, and those with abnormal chromosome structures (overcondensed chromosomes, cut, missegregation, etc.) were counted under the fluorescence microscope. (C and D) Sister chromosomes are prematurely separated in eso1-H17 mutant cells. Wild-type (h+ Cen1-GFP) and eso1-H17 (h+ eso1-H17 Cen1-GFP) cells were arrested in G1 by nitrogen starvation at 26°C and then released in MM medium preincubated at 36°C. Live cells were observed under the fluorescence microscope. (C) Frequencies of cells showing two split Cen1-GFP signals. (D) Examples of eso1-H17 cells showing two split Cen1-GFP signals and wild-type control at 4 h. Bar, 5 μm. (E and F) Eso1p is not essential for the maintenance of cohesion in G2 and M phases. Cells of eso1-H17 (h− eso1-H17 leu1-32) and rad21-K1 (h− ura4-D18 rad21-K1-ura4+ leu1-32) mutants were grown to saturation in YEL medium at 25°C and incubated for a further 24 h to ensure growth arrest. They were then incubated at 36°C for 3 h and released in fresh YEL medium preincubated at 36°C. (E) Flow cytometric analysis of arrested cells. (F) Lack of abnormal mitosis in eso1-H17 cells released from G2 phase at the nonpermissive temperature. Cell number (open squares) was determined by counting aliquots taken every 30 min. To determine percent abnormal mitosis (filled triangles), cells were fixed with 70% ethanol and stained with DAPI, and those with abnormal mitotic chromosome structures (cut, missegregation, etc.) were counted under the fluorescence microscope.
Premature sister chromatid separation occurs in eso1-H17 cells.
The demonstration of spindle assembly checkpoint-dependent mitotic arrest, abnormal chromosome segregation, and increased sensitivity to DSB-inducing reagents as the major phenotypes of the eso1-H17 mutant led us to speculate that Eso1p might be involved in sister chromatid cohesion. To investigate this possibility, we used the Cen1-GFP system to visualize the behavior of the centromere DNA of chromosome I in live eso1-H17 cells (11, 33). Cells were synchronized; released from G1 at the nonpermissive temperature; collected at 2, 4, and 6 h after release; and immediately observed under a fluorescence microscope. DNA replication occurred at around 4 h (Fig. 4A). Unlike wild-type cells, a large number of eso1-H17 interphase cells had two split Cen1-GFP signals at 4 to 6 h, which proves defective sister chromatid cohesion (Fig. 4C). Photos of cells showing two-split Cen1-GFP dots are shown in Fig. 4D. We thus concluded that Eso1p was essential for the establishment of sister chromatid cohesion during S phase.
Eso1p function is not required for the maintenance of sister chromatid cohesion in G2-M phase.
Sister chromatid cohesion is established during DNA replication and maintained until anaphase. To investigate whether Eso1p function is also required for cohesion maintenance, eso1-H17 cells were grown to saturation in YEL medium. Under these conditions, most of the cells arrested growth with 2n DNA contents (Fig. 4E) with a low septation index (eso1 mutant, 5.6%; rad21 mutant, 9.4%), indicating that more than 80% of cells were arrested in G2 phase. The cells were exposed to 36°C, released 3 h later to resume cell cycling in fresh medium at 36°C, harvested every 30 min, and observed for mitosis. As expected, the majority of the rad21-K1 cells that have a defective cohesin subunit (49) displayed abnormal mitosis as they proceeded through cell cycling, showing that cohesin complexes are also required for the maintenance of sister chromatid cohesion (14) (Fig. 4F, right graph). In contrast, the eso1-H17 mutant cells progressed through M phase without significant abnormalities (Fig. 4F, left graph). Thus, Eso1p is not required for the maintenance of sister chromatid cohesion once it is established in S phase.
The eso1+ gene encodes a fusion between DNA polymerase η and Eco1p/Ctf7p homologues.
To clone the defective gene in eso1-H17 cells, S. pombe genomic libraries were screened for those that rescued the temperature-sensitive lethality. Two nonoverlapping clones, S1 and Bg1, could rescue the mutant up to 36°C, and further analysis showed that S1 contained the eso1+ gene (see below). Characterization of the multicopy suppressor gene on Bg1 will be described elsewhere.
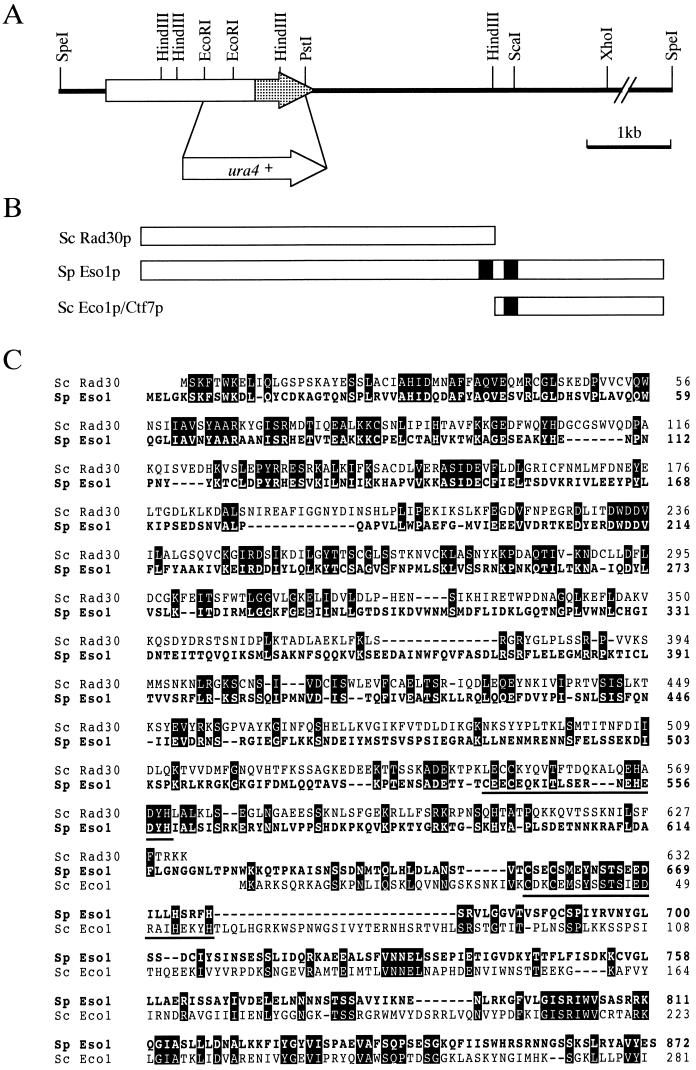
The S1 clone contained a 9.4-kb SpeI fragment, the internal 6.7-kb SpeI-XhoI fragment of which was found to be active (Fig. 5A). The unique PstI site within this fragment was essential for the activity. Nucleotide sequencing revealed the presence of a single open reading frame across the PstI site. The predicted polypeptide was an 872-amino-acid protein with an estimated molecular mass of 99 kDa. The nucleotide and predicted amino acid sequences were confirmed by sequencing a corresponding eso1+ cDNA that was isolated subsequently.
FIG. 5.
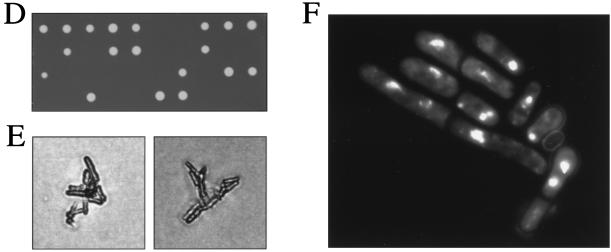
Isolation and characterization of the eso1+ gene. (A) Restriction map of the eso1+ gene. The eso1+ open reading frame is shown by an arrow, and the Eco1p/Ctf7p-homologous domain is shaded. The EcoRI-PstI region of eso1+ was replaced with a ura4+ gene cassette for generating an eso1 null mutant. (B) Schematic illustration of Eso1p of S. pombe and Rad30p and Eco1p/Ctf7p of S. cerevisiae. The zinc finger motifs are filled. (C) Amino acid homologies between Eso1p, Rad30p, and Eco1p/Ctf7p. The predicted amino acid sequence of Eso1p is shown in a single-letter code and aligned with Rad30p and Eco1p/Ctf7p. Identical amino acids are boxed. The zinc finger motifs are underlined. (D) Cells with deletions of eso1+ are lethal. Spores from eso1+/eso1::ura4+ diploid cells were tetrad dissected on YEA plates and incubated at 30°C for 4 days. (E) Terminal phenotype of Δeso1 cells. Cells that germinated from two independent Δeso1 spores on a YEA plate were photographed. (F) DAPI staining of germinating Δeso1 cells. Δeso1 spores (Ura+) derived from eso1+/eso1::ura4+ diploid cells were preferentially germinated in MM lacking uracil. Germinating cells were fixed with 70% ethanol, stained with DAPI, and photographed under the microscope.
The central region of the predicted Eso1 protein possesses two C2H2 zinc finger motifs (Fig. 5B and C). A database search revealed that the amino-terminal two-thirds of Eso1p is strongly related to Rad30p of S. cerevisiae. Rad30 protein, recently identified as DNA polymerase η, is a member of a damage-bypass replication protein family, which includes the UmuC and DinB proteins in E. coli and the Xeroderma pigmentosum variant gene product in Homo sapiens (19, 22, 26, 27, 40, 43; reviewed in reference 53). On the other hand, the carboxyl-terminal one-third of Eso1p shares significant homology with another protein family conserved among eukaryotes, which includes Eco1p/Ctf7p in S. cerevisiae and putative proteins from Arabidopsis thaliana, Mus musculus, and H. sapiens (42, 50). The genes encoding Rad30p and Eco1p/Ctf7p are not contiguous and present on chromosomes IV and VI, respectively, in budding yeast, and there is no Eco1p/Ctf7p-homologous region in the Xeroderma pigmentosum variant gene product in H. sapiens.
To obtain definitive evidence for the authenticity of the isolated gene and to confirm the function of eso1+, cells lacking the eso1+ gene were constructed by one-step gene replacement. The sequence in eso1+ corresponding to the carboxyl-terminal half of Eso1p was replaced with the S. pombe ura4+ gene (Fig. 5A) and transfected in a diploid strain. Diploid cells disrupted for one eso1+ allele were identified and confirmed by genomic Southern hybridization. Tetrad analysis of the sporulated diploid cells revealed that only two spores were viable and that all viable spores were uracil auxotrophic (Fig. 5D). Microscopic observation showed that the Δeso1 spores germinated but arrested cell cycling after two to three divisions (Fig. 5E). Spore germination analysis showed that the Δeso1 cells resembled eso1-H17 cells and displayed abnormal chromosomes (Fig. 5F). Furthermore, Δeso1/eso1-H17 diploid cells were still temperature sensitive for growth and failed to yield any haploid spores that could grow at 36°C (data not shown). Based on these results, we concluded that the cloned gene was indeed eso1+ itself.
The C-terminal region of Eso1p is necessary and sufficient for sister chromatid cohesion.
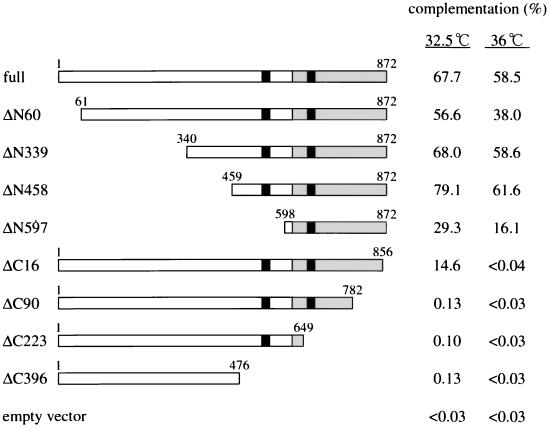
To locate the functional domain of Eso1p, a series of amino- and carboxyl-terminal deletion mutants were constructed, and their ability to rescue the eso1-H17 mutant was determined. As shown in Fig. 6, the Rad30p-homologous region was totally dispensable, and the gene truncated in the entire amino-terminal region (ΔN597) retained nearly all its ability to suppress the mutant. On the other hand, the carboxyl-terminal region homologous to Eco1p/Ctf7p was absolutely essential for function. Even a deletion of only 16 amino acids from the C terminus (ΔC16) largely abrogated the activity. Similarly, the C-terminal Eco1p/Ctf7p domain (ΔN597), but not ΔC16, suppressed the UV sensitivity of eso1-H17 cells (data not shown), indicating that the UV sensitivity of eso1-H17 cells results from poor establishment of sister chromatid cohesion. Furthermore, the eso1-H17 allele contained a point mutation that resulted in a change from glycine at position 799 to aspartic acid. These results indicate that loss of the function of the Eco1p/Ctf7p-homologous domain in Eso1p caused the eso1-H17 phenotypes.
FIG. 6.
The carboxyl-terminal Eco1p/Ctf7p-homologous domain of Eso1p is essential to rescue the eso1-H17 mutant. N-terminally and C-terminally truncated eso1+ genes were constructed and assayed for the ability to rescue the eso1-H17 mutant. The intact Eso1p is shown at the top (referred to as full). The Eco1p/Ctf7p-homologous domain is grey, and the zinc finger motifs are black. The amino acid numbers are shown at each mutant. The ability of the deletion mutants to complement eso1-H17 cells is shown in the right columns. For complementation activity, see Materials and Methods.
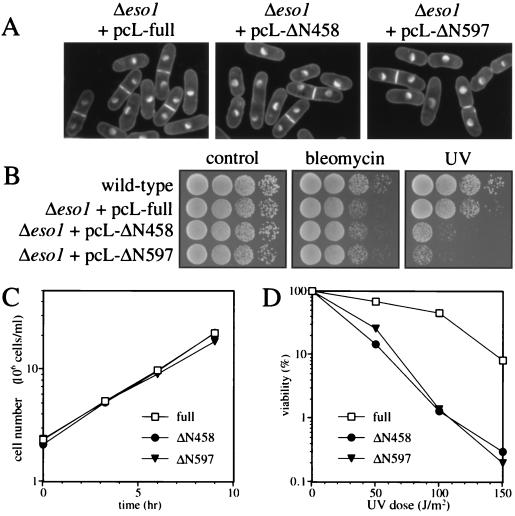
To confirm the sufficiency of the Eco1p/Ctf7p-homologous domain for the establishment of sister chromatid cohesion, we tested whether the domain could rescue the eso1 null cell. Diploid cells in which one copy of eso1+ was deleted were transformed with the eso1+ gene deletion mutants described in Fig. 6. These transformants were then sporulated, and spores were allowed to germinate on appropriately supplemented minimal medium plates. Δeso1 cells harboring not only full-length eso1+ but also the amino-terminal deletion mutants grew and formed colonies. Nuclear morphology (Fig. 7A), bleomycin sensitivity (Fig. 7B), and growth rate (Fig. 7C) of the Δeso1 cells rescued by ΔN458 or ΔN597 were indistinguishable from those of the Δeso1 cells rescued by full-length eso1+. These results indicate that the C-terminal region of Eso1p is sufficient for sister chromatid cohesion and is fully functional without the N-terminal Rad30p-homologous region.
FIG. 7.
Phenotypes of eso1 null mutants suppressed by amino-terminally truncated eso1+. (A) Nuclear morphologies of Δeso1 cells (h− ade6-M210 eso1::ura4+ ura4-D18 leu1-32) suppressed by full-length eso1+ (pcL-full) or its amino-terminal truncation mutants (pcL-ΔN458 and pcL-ΔN597). Cells were grown to mid-log phase at 30°C in YEL medium, fixed with glutaraldehyde, and stained with DAPI. (B) Bleomycin and UV sensitivities. Approximately 5 × 104, 5 × 103, 5 × 102, and 50 cells of the wild type (h− ade6-M210) and the Δeso1 mutant (h− ade6-M210 eso1::ura4+ ura4-D18 leu1-32) suppressed by pcL-full, pcL-ΔN458, and pcL-ΔN597 were spotted on normal YEA plates (referred to as control), YEA plates containing bleomycin (0.02% [wt/vol]), or normal YEA plates but with irradiation with UV (100 J/m2). Plates were incubated at 30°C for 3 days. (C) Growth curves. Δeso1 cells (h− ade6-M210 eso1::ura4+ ura4-D18 leu1-32) suppressed by pcL-full, pcL-ΔN458, or pcL-ΔN597 were grown at 30°C in MM plus adenine. (D) UV sensitivity. Δeso1 cells (h− ade6-M210 eso1::ura4+ ura4-D18 leu1-32) suppressed by pcL-full, pcL-ΔN458, or pcL-ΔN597 were plated on YEA, irradiated with various doses of UV, and incubated for 5 days at 30°C.
Deletion of the DNA polymerase η domain in Eso1p elevates UV sensitivity.
As mentioned above, expression of the C-terminal Eco1p/Ctf7p-homologous region completely suppressed the defect of sister chromatid cohesion of cells lacking eso1+. However, unlike those rescued by full-length eso1+, the Δeso1 cells rescued by ΔN458 or ΔN597 were still sensitive to UV irradiation (Fig. 7B and D). Because, in the Δeso1 cells, not only the Eco1p/Ctf7p-homologous region but also about 30% of the Rad30p-homologous region was deleted, some of this region is likely to be required for the function of the DNA polymerase η domain (Fig. 5A). We thus concluded that loss of the DNA polymerase η domain elevates UV sensitivity and tentatively identified this domain as DNA polymerase η itself.
eso1+ genetically interacts with adherin and cohesin.
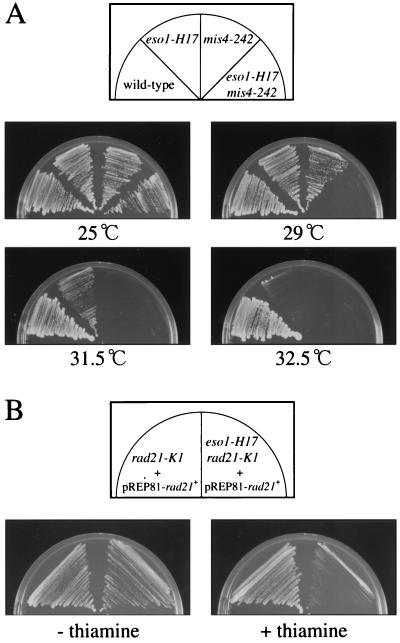
The structural and functional similarity of Eso1p to Eco1p/Ctf7p led us to test the possible genetic interactions between eso1+ and the genes encoding adherin and cohesin subunits. The eso1-H17 mutant combined with mis4-242, a temperature-sensitive mutation of adherin (10), became more thermosensitive. Unlike eso1-H17 and mis4-242 single mutants, which grow at temperatures of 32.5 and 31.5°C, respectively, the double mutant failed to grow at 29°C (Fig. 8A) and barely grew at 27°C. Moreover, the eso1-H17 mutant was synthetically lethal with rad21-K1 mutation. Tetrad dissection of spores from the cross between eso1-H17 and rad21-K1 yielded no viable double mutant cells. Moreover, as shown in Fig. 8B, the eso1-H17 rad21-K1 double mutant rescued by the pREP81-rad21+ plasmid did not grow when rad21+ in the plasmid was repressed.
FIG. 8.
eso1+ genetically interacts with the genes encoding adherin and cohesin subunits. (A) Wild-type (h− leu1-32), eso1-H17 (h− eso1-H17 leu1-32), mis4-242 (h− mis4-242 leu1-32), and eso1-H17 mis4-242 (h− eso1-H17 mis4-242 leu1-32) cells were incubated on YEA plates for 3 to 4 days at the indicated temperatures. (B) rad21-K1 (h+ ade6-M216 ura4-D18 rad21-K1-ura4+ leu1-32) and eso1-H17 rad21-K1 (h− ade6-M216 ura4-D18 eso1-H17 rad21-K1-ura4+ leu1-32) cells that were transfected with pREP81-rad21+ were inoculated on an MM-adenine plate (− thiamine) and an MM-adenine plate containing 10 μM thiamine (+ thiamine) and incubated for 6 days at 23°C.
eso1+ genetically interacts with G1-S regulators also.
The execution of eso1+ function during S phase suggests that eso1+ function might be controlled by cell cycle start factors. We, therefore, examined the ability of various cell cycle start genes to rescue the eso1-H17 mutant. As shown in Table 1, the res1+ and rep2+ genes could partially suppress the temperature sensitivity. The cdc10+, res2+, and rep1+ genes also exhibited weak suppression activities (data not shown). These genes encode a subunit of transcription factor complexes that regulate the cell cycle start-specific transcription of genes (reviewed in references 37 and 54). A major target gene of Res-Cdc10-Rep transcription factor complexes is cdc18+, which encodes a component of the prereplicative complex. However, cdc18+ itself had no detectable activity.
TABLE 1.
Various cell cycle control genes rescue the eso1-H17 mutant
| Plasmid | Suppression activity at 32.5°C (%)a |
|---|---|
| pcL-Xb | <0.03 |
| pcL-eso1+ | 63.3 |
| pcL-res1+ | 24.9 |
| pcL-rep2+ | 14.4 |
| pcL-cdc18+ | 0.06 |
| pcL-cig2+ | 18.3 |
| pcL-cdc13+ | <0.03 |
| pcL-cig1+ | <0.03 |
| pcL-pcn1+ | 5.2 |
| pcL-cdc20+ | 17.8 |
For suppression activity, see Materials and Methods.
pcL-X is a pcL vector without insert used as a negative control.
The cig2+ gene encodes a B-type cyclin that associates with Cdc2 kinase. Cig2p is thought to play an important role in the cell cycle start (24, 30), but this function is not specific to Cig2p and is shared by the other B-type cyclins, Cdc13p and Cig1p (9). However, only cig2+ suppressed the mutant. These cell cycle start genes also rescued the rad21-K1 mutant (data not shown).
In addition to the cell cycle start regulators, pcn1+ and cdc20+, which encode proliferating cell nuclear antigen (PCNA) and a catalytic subunit of DNA polymerase ɛ (8, 44, 52), respectively, components of the DNA replication machinery, displayed partial suppression. The cdc20+ cDNA having the activity contained only the carboxy-terminal one-third of the coding region devoid of the catalytic domain.
DISCUSSION
All the data presented show that eso1+, which we identified as a novel fission yeast cell cycle regulator, is a functional homologue of budding yeast ECO1/CTF7, which is required for the establishment of sister chromatid cohesion during S phase (42, 50). Like cohesin and adherin, Eso1p/Eco1p/Ctf7p seems to be evolutionarily conserved at least with respect to structure. cDNA sequence databases from higher eukaryotes contain proteins with significant amino acid homology to this family, though their function is presently unknown.
However, there is a striking difference between Eso1p and other Eco1p/Ctf7p family members. Eso1p contains a sequence highly homologous to DNA polymerase η at its N-terminal side. DNA polymerase η performs DNA synthesis on a damaged template, a critical step in postreplication repair, and in fact synthesizes a DNA strand with correct bases on cis-syn thymine-thymine dimer-containing DNA templates (18, 25, 26; reviewed in reference 53). We found that deletion of the DNA polymerase η domain of Eso1p did not affect sister chromatid cohesion but elevated sensitivity to UV damage. This result is consistent with the possibility that the N-terminal region of Eso1p indeed has a DNA polymerase η activity.
The elevated DNA damage sensitivity, particularly to DSB, of the eso1 mutant due to defective sister chromatid cohesion is highly consistent with the fact that cohesin and adherin mutants are also sensitive to UV irradiation at the permissive temperature (10, 49). The DSB sensitivity of these mutants suggests that the recombinatorial repair system may require sister chromatid cohesion for its efficient execution. This is quite reasonable, because the distance between the two homologous sequences must influence the efficiency of recombinatorial repair. In fact, the rad21 mutant was originally isolated as a DSB repair-deficient mutant (3).
It is of considerable importance to elucidate how Eso1p interacts with adherin and cohesin and how Eso1p established sister chromatid cohesion during S phase. Up to now, little was known about the molecular basis of the establishment of sister chromatid cohesion during DNA synthesis. Adherin and cohesin are essential, but not in themselves sufficient, for sister chromatid cohesion. Sister chromatids are separated precociously in the eco1-1 mutant though cohesin complexes stay bound to chromosomes (50). Similarly, we found no difference in the localization of Mis4GFPp and Rad21GFPp between wild-type and eso1-H17 mutant cells (K. Tanaka, unpublished data). Thus, the Eso1/Eco1/Ctf7 protein family is perhaps not required for the loading of adherin and cohesin onto chromosomes but has a role in connecting nascent sister chromatids that have been duplicated in S phase. Eso1 protein is present throughout the cell cycle at a constant level (K. Tanaka, unpublished data), indicating that, if it is, the activity is regulated at the posttranslational level. As shown in Fig. 6A, the truncation mutant ΔC16, in which G799, the residue mutated in eso1-H17, is intact, is partially active at 32.5°C but totally inactive at 36°C. This result suggests that the C-terminal region of Eso1p may physically interact with another protein(s). We found that eso1+ genetically interacts with pcn1+ (encoding PCNA) and cdc20+ (encoding a catalytic subunit of DNA polymerase ɛ). Similarly, POL30 (encoding PCNA) was isolated as a high-copy suppressor of the ctf7-203 mutant in budding yeast (42). Interestingly, the Cdc20 protein truncated at the DNA polymerase domain retained eso1-H17 suppression activity. The carboxyl-terminal region of Cdc20p is essential for cell viability (8), and the DNA polymerase domain of polymerase ɛ is dispensable for growth in budding yeast (20). Although the molecular aspect of interactions of Eso1p with DNA polymerase ɛ and PCNA is totally unknown, Eso1p might colocalize with the replication machinery and thereby be promoted to interact with the nascent DNA loaded with adherin and cohesin. Polymerase ɛ and PCNA might have a role in colocalization and/or activation of Eso1p. The presence of the DNA polymerase η domain in Eso1p is also consistent with this possibility.
One of our remarkable findings is that the eso1-H17 mutant is partially suppressed by the cell cycle start genes, such as res1+, rep2+, and cig2+. The effect seems to be indirect because the expression of eso1+ is not regulated by the Res-Cdc10-Rep transcriptional activator complexes (T. Yonekawa, unpublished data). Res-Cdc10-Rep may act via the cig2+ gene, since induction of its mRNA at the G1-S boundary fully depends on these transcriptional activators (28, 30, 36; K. Tanaka, unpublished data), but seemingly not via the rad21+ gene, because the expression of rad21+ gene is not Cdc10p dependent though it is cell cycle regulated with a peak during G1-S transition (4). Interestingly, only cig2+, not the cdc13+ or the cig1+ gene, was able to suppress the eso1 mutant, though the cell cycle start function is not specific to Cig2p and is shared by other B-type cyclins (9). cig2+ also rescues the temperature sensitivity of the rad21-K1 mutant. These results raise the possibility that the Cig2 cyclin has a novel function as an activator of sister chromatid cohesion, though it is inessential since Δcig2 cells are viable (5, 6, 36). Eso1p possesses a single Cdc2 kinase phosphorylation consensus sequence (SPKR; from position 505 to 508), but mutations that change the serine residue to alanine or aspartic acid did not significantly influence Eso1p activity, suggesting that Eso1p is unlikely to be a direct functional substrate of Cig2p-Cdc2p kinase (T. Yonekawa, unpublished data).
ACKNOWLEDGMENTS
We thank Tomohiro Matsumoto, Kazuo Tatebayashi, and Koei Okazaki for providing strains, plasmids, and libraries.
This work was supported by grants from the Ministry of Education, Science and Culture of Japan and a CREST (Core Research for Evolutional Science and Technology) Research Project from JST (Japan Science and Technology Corporation).
REFERENCES
- 1.Adachi Y, Yanagida M. Higher order chromosome structure is affected by cold-sensitive mutations in a Schizosaccharomyces pombe gene crm1+ which encodes a 115-kD protein preferentially localized in the nucleus and its periphery. J Cell Biol. 1989;108:1195–1207. doi: 10.1083/jcb.108.4.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfa C, Fantes P, Hyams J, McLeod M, Warbrick E. Experiments with fission yeast. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- 3.Birkenbihl R P, Subramani S. Cloning and characterization of rad21, an essential gene of Schizosaccharomyces pombe involved in DNA double-strand-break repair. Nucleic Acids Res. 1992;20:6605–6611. doi: 10.1093/nar/20.24.6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birkenbihl R P, Subramani S. The rad21 gene product of Schizosaccharomyces pombe is a nuclear, cell cycle-regulated phosphoprotein. J Biol Chem. 1995;270:7703–7711. doi: 10.1074/jbc.270.13.7703. [DOI] [PubMed] [Google Scholar]
- 5.Bueno A, Russell P. Two fission yeast B-type cyclins, Cig2 and Cdc13, have different functions in mitosis. Mol Cell Biol. 1993;13:2286–2297. doi: 10.1128/mcb.13.4.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connolly T, Beach D. Interaction between the Cig1 and Cig2 B-type cyclins in the fission yeast cell cycle. Mol Cell Biol. 1994;14:768–776. doi: 10.1128/mcb.14.1.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D'Urso G, Grallert B, Nurse P. DNA polymerase alpha, a component of the replication initiation complex, is essential for the checkpoint coupling S phase to mitosis in fission yeast. J Cell Sci. 1995;108:3109–3118. doi: 10.1242/jcs.108.9.3109. [DOI] [PubMed] [Google Scholar]
- 8.D'Urso G, Nurse P. Schizosaccharomyces pombe cdc20+ encodes DNA polymerase epsilon and is required for chromosomal replication but not for the S phase checkpoint. Proc Natl Acad Sci USA. 1997;94:12491–12496. doi: 10.1073/pnas.94.23.12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fisher D L, Nurse P. A single fission yeast mitotic cyclin B p34cdc2 kinase promotes both S-phase and mitosis in the absence of G1 cyclins. EMBO J. 1996;15:850–860. [PMC free article] [PubMed] [Google Scholar]
- 10.Furuya K, Takahashi K, Yanagida M. Faithful anaphase is ensured by Mis4, a sister chromatid cohesion molecule required in S phase and not destroyed in G1 phase. Genes Dev. 1998;12:3408–3418. doi: 10.1101/gad.12.21.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goshima G, Saitoh S, Yanagida M. Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes Dev. 1999;13:1664–1677. doi: 10.1101/gad.13.13.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grallert B, Nurse P. The ORC1 homolog orp1 in fission yeast plays a key role in regulating onset of S phase. Genes Dev. 1996;10:2644–2654. doi: 10.1101/gad.10.20.2644. [DOI] [PubMed] [Google Scholar]
- 13.Guacci V, Hogan E, Koshland D. Chromosome condensation and sister chromatid pairing in budding yeast. J Cell Biol. 1994;125:517–530. doi: 10.1083/jcb.125.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gutz H, Heslot H, Leupold U, Loprieno N. Schizosaccharomyces pombe. In: King R C, editor. Handbook of genetics. Vol. 1. New York, N.Y: Plenum Press; 1974. pp. 395–446. [Google Scholar]
- 16.He X, Patterson T E, Sazer S. The Schizosaccharomyces pombe spindle checkpoint protein mad2p blocks anaphase and genetically interacts with the anaphase-promoting complex. Proc Natl Acad Sci USA. 1997;94:7965–7970. doi: 10.1073/pnas.94.15.7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hennessy K M, Lee A, Chen E, Botstein D. A group of interacting yeast DNA replication genes. Genes Dev. 1991;5:958–969. doi: 10.1101/gad.5.6.958. [DOI] [PubMed] [Google Scholar]
- 18.Johnson R E, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Poleta. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 19.Johnson R E, Kondratick C M, Prakash S, Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 20.Kesti T, Flick K, Keranen S, Syvaoja J E, Wittenberg C. DNA polymerase epsilon catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol Cell. 1999;3:679–685. doi: 10.1016/s1097-2765(00)80361-5. [DOI] [PubMed] [Google Scholar]
- 21.Kim S H, Lin D P, Matsumoto S, Kitazono A, Matsumoto T. Fission yeast Slp1: an effector of the Mad2-dependent spindle checkpoint. Science. 1998;279:1045–1047. doi: 10.1126/science.279.5353.1045. [DOI] [PubMed] [Google Scholar]
- 22.Kim S R, Maenhaut-Michel G, Yamada M, Yamamoto Y, Matsui K, Sofuni T, Nohmi T, Ohmori H. Multiple pathways for SOS-induced mutagenesis in Escherichia coli: an overexpression of dinB/dinP results in strongly enhancing mutagenesis in the absence of any exogenous treatment to damage DNA. Proc Natl Acad Sci USA. 1997;94:13792–13797. doi: 10.1073/pnas.94.25.13792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Losada A, Hirano M, Hirano T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev. 1998;12:1986–1997. doi: 10.1101/gad.12.13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin-Castellanos C, Labib K, Moreno S. B-type cyclins regulate G1 progression in fission yeast in opposition to the p25rum1 cdk inhibitor. EMBO J. 1996;15:839–849. [PMC free article] [PubMed] [Google Scholar]
- 25.Masutani C, Araki M, Yamada A, Kusumoto R, Nogimori T, Maekawa T, Iwai S, Hanaoka F. Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J. 1999;18:3491–3501. doi: 10.1093/emboj/18.12.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 27.McDonald J P, Levine A S, Woodgate R. The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics. 1997;147:1557–1568. doi: 10.1093/genetics/147.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McInerny C, Kersey P J, Creanor J, Fantes P A. Positive and negative roles for cdc10 in cell cycle gene expression. Nucleic Acids Res. 1995;23:4761–4768. doi: 10.1093/nar/23.23.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- 30.Mondesert O, McGowan C H, Russell P. Cig2, a B-type cyclin, promotes the onset of S in Schizosaccharomyces pombe. Mol Cell Biol. 1996;16:1527–1533. doi: 10.1128/mcb.16.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 32.Murakami H, Okayama H. A kinase from fission yeast responsible for blocking mitosis in S phase. Nature. 1995;374:817–819. doi: 10.1038/374817a0. [DOI] [PubMed] [Google Scholar]
- 33.Nabeshima K, Nakagawa T, Straight A F, Murray A, Chikashige Y, Yamashita Y M, Hiraoka Y, Yanagida M. Dynamics of centromeres during metaphase-anaphase transition in fission yeast: Dis1 is implicated in force balance in metaphase bipolar spindle. Mol Biol Cell. 1998;9:3211–3225. doi: 10.1091/mbc.9.11.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nasmyth K. Separating sister chromatids. Trends Biochem Sci. 1999;24:98–104. doi: 10.1016/s0968-0004(99)01358-4. [DOI] [PubMed] [Google Scholar]
- 35.Nishitani H, Nurse P. The cdc18 protein initiates DNA replication in fission yeast. Prog Cell Cycle Res. 1997;3:135–142. doi: 10.1007/978-1-4615-5371-7_11. [DOI] [PubMed] [Google Scholar]
- 36.Obara-Ishihara T, Okayama H. A B-type cyclin regulates conjugation via interacting with cell cycle ‘start’ genes in fission yeast. EMBO J. 1994;13:1863–1872. doi: 10.1002/j.1460-2075.1994.tb06455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okayama H, Nagata A, Jinno S, Murakami H, Tanaka K, Nakashima N. Cell cycle control in fission yeast and mammals: identification of new regulatory mechanism. Adv Cancer Res. 1996;69:17–62. doi: 10.1016/s0065-230x(08)60859-3. [DOI] [PubMed] [Google Scholar]
- 38.Okazaki K, Okazaki N, Kume K, Jinno S, Tanaka K, Okayama H. High-frequency transformation method and library transducing vectors for cloning mammalian cDNAs by trans-complementation of Schizosaccharomyces pombe. Nucleic Acids Res. 1990;18:6485–6489. doi: 10.1093/nar/18.22.6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okazaki N, Okazaki K, Tanaka K, Okayama H. The ste4+ gene, essential for sexual differentiation of Schizosaccharomyces pombe, encodes a protein with a leucine zipper motif. Nucleic Acids Res. 1991;19:7043–7047. doi: 10.1093/nar/19.25.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roush A A, Suarez M, Friedberg E C, Radman M, Siede W. Deletion of the Saccharomyces cerevisiae gene RAD30 encoding an Escherichia coli DinB homolog confers UV radiation sensitivity and altered mutability. Mol Gen Genet. 1998;257:686–692. doi: 10.1007/s004380050698. [DOI] [PubMed] [Google Scholar]
- 41.Saka Y, Yanagida M. Fission yeast cut5+, required for S phase onset and M phase restraint, is identical to the radiation-damage repair gene rad4+ Cell. 1993;74:383–393. doi: 10.1016/0092-8674(93)90428-s. [DOI] [PubMed] [Google Scholar]
- 42.Skibbens R V, Corson L B, Koshland D, Hieter P. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 1999;13:307–319. doi: 10.1101/gad.13.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith B T, Walker G C. Mutagenesis and more: umuDC and the Escherichia coli SOS response. Genetics. 1998;148:1599–1610. doi: 10.1093/genetics/148.4.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sugino A, Ohara T, Sebastian J, Nakashima N, Araki H. DNA polymerase epsilon encoded by cdc20+ is required for chromosomal DNA replication in the fission yeast Schizosaccharomyces pombe. Genes Cells. 1998;3:99–110. doi: 10.1046/j.1365-2443.1998.00169.x. [DOI] [PubMed] [Google Scholar]
- 45.Sugiyama A, Tanaka K, Okazaki K, Nojima H, Okayama H. A zinc finger protein controls the onset of premeiotic DNA synthesis of fission yeast in a Mei2-independent cascade. EMBO J. 1994;13:1881–1887. doi: 10.1002/j.1460-2075.1994.tb06457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suto K, Nagata A, Murakami H, Okayama H. A double-strand break repair component is essential for S phase completion in fission yeast cell cycling. Mol Biol Cell. 1999;10:3331–3343. doi: 10.1091/mbc.10.10.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takahashi K, Yamada H, Yanagida M. Fission yeast minichromosome loss mutants mis cause lethal aneuploidy and replication abnormality. Mol Biol Cell. 1994;5:1145–1158. doi: 10.1091/mbc.5.10.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanaka K, Okazaki K, Okazaki N, Ueda T, Sugiyama A, Nojima H, Okayama H. A new cdc gene required for S phase entry of Schizosaccharomyces pombe encodes a protein similar to the cdc10+and SWI4 gene products. EMBO J. 1992;11:4923–4932. doi: 10.1002/j.1460-2075.1992.tb05599.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tatebayashi K, Kato J, Ikeda H. Isolation of a Schizosaccharomyces pombe rad21ts mutant that is aberrant in chromosome segregation, microtubule function, DNA repair and sensitive to hydroxyurea: possible involvement of Rad21 in ubiquitin-mediated proteolysis. Genetics. 1998;148:49–57. doi: 10.1093/genetics/148.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Toth A, Ciosk R, Uhlmann F, Galova M, Schleiffer A, Nasmyth K. Yeast cohesin complex requires a conserved protein, Eco1p (Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 1999;13:320–333. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- 52.Waseem N H, Labib K, Nurse P, Lane D P. Isolation and analysis of the fission yeast gene encoding polymerase delta accessory protein PCNA. EMBO J. 1992;11:5111–5120. doi: 10.1002/j.1460-2075.1992.tb05618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woodgate R. A plethora of lesion-replicating DNA polymerases. Genes Dev. 1999;13:2191–2195. doi: 10.1101/gad.13.17.2191. [DOI] [PubMed] [Google Scholar]
- 54.Woollard A, Nurse P. G1 regulation and checkpoints operating around START in fission yeast. Bioessays. 1995;17:481–490. doi: 10.1002/bies.950170604. [DOI] [PubMed] [Google Scholar]